Abstract
A complex network of transcription factor interactions propagates across the larval eye disc to establish columns of evenly-spaced R8 precursor cells, the founding cells of Drosophila ommatidia. After the recruitment of additional photoreceptors to each ommatidium, the surrounding cells are organized into their stereotypical pattern during pupal development. These support cells – comprised of pigment and cone cells – generate the hexagonal honeycomb lattice on top of which lenses are secreted. Since the proteins and processes essential for correct eye patterning are conserved, elucidating how these function and change during Drosophila eye patterning can substantially advance our understanding of transcription factor and signaling networks, cytoskeletal structures, adhesion complexes, and the biophysical properties of complex tissues during their morphogenesis. Our understanding of many of these aspects of Drosophila eye patterning is largely descriptive. Many important questions, especially relating to the regulation and integration of cellular events, remain.
Graphical Abstract
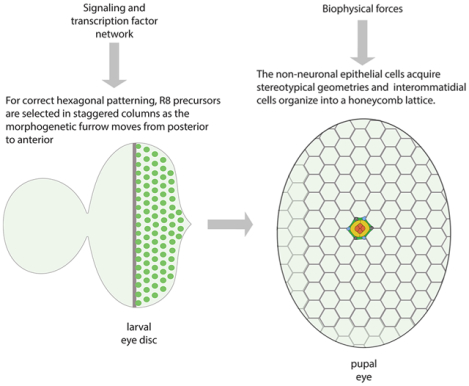
Introduction
The Drosophila pupal eye is a beautiful and engaging model for exploring processes that drive tissue patterning. The eye is composed of about 800 ommatidia (unit eyes) arranged in staggered columns and each ommatidium contains a cluster of eight photoreceptors surrounded by defined accessory cells. These adopt distinctive shapes and organize into a simple repeating pattern that gives the pupal eye its ordered honeycomb-lattice appearance (Figure 1A) (Cagan and Ready, 1989a; Ready et al., 1976; Wolff and Ready, 1993). Each ommatidium provides a template over which the lenses are generated and in the adult eye these have the appearance of abutting bubbles (Figure 1B). Organization of the lenses is dependent on correct patterning of the underlying eye neuroepithelium. This patterning occurs several days before generation of the lenses even begins, and requires a) the precise staggered placement of R8 photoreceptor precursors and, consequently, photoreceptor clusters across the eye field and subsequently b) correct organization of accessory cells. This review discusses these two intricate aspects of Drosophila eye development.
Figure 1: The Drosophila compound eye is highly ordered.
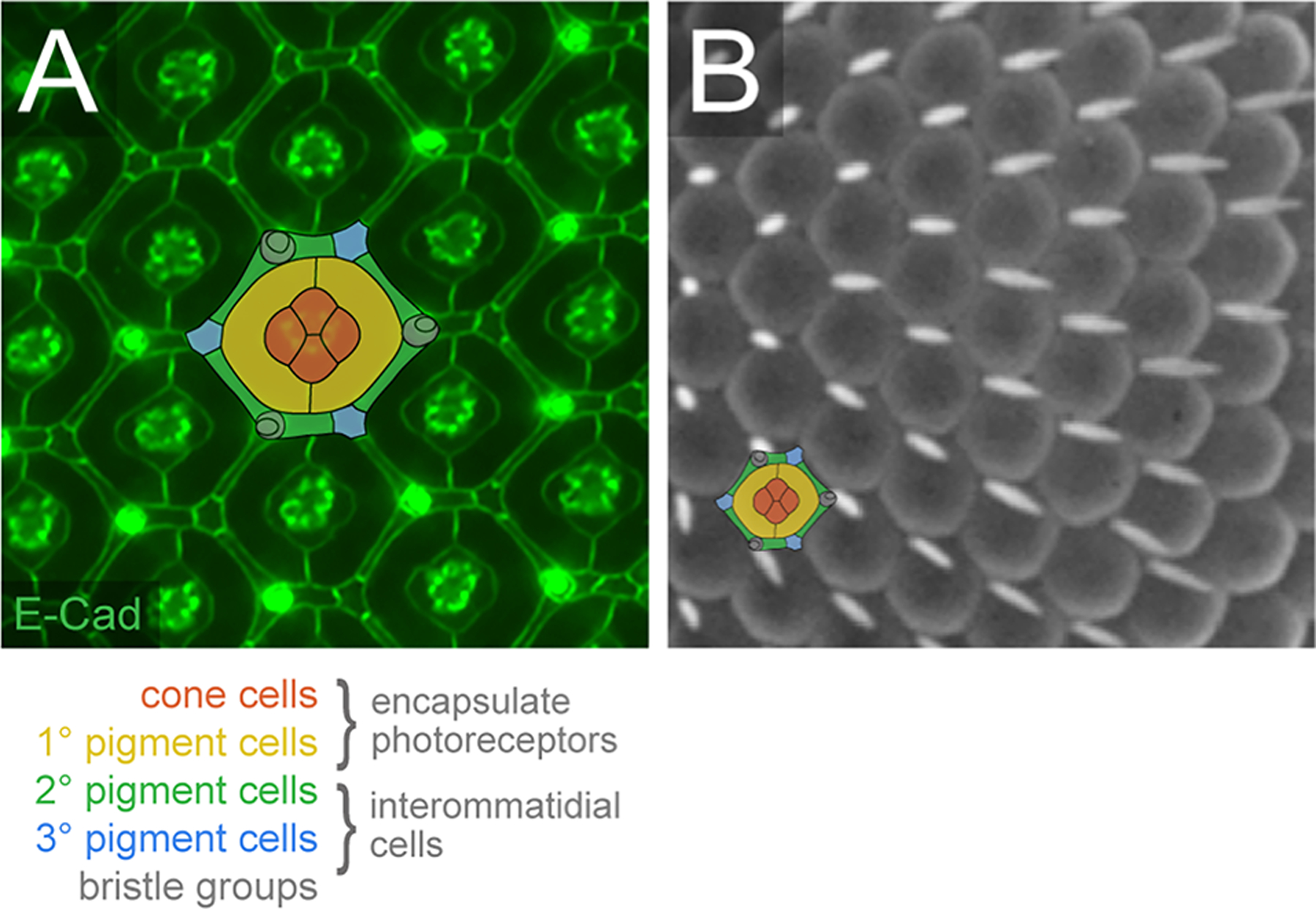
(A) Small region of the pupal eye at 40 h APF. AJs have been detected with antibodies to E-Cadherin. The epithelial support cells are color-coded, as indicated. (B) Small region of a scanning electron micrograph of the adult eye. A single ommatidium is illustrated to emphasize the cells that lie below the rounded lenses, although in the adult eye the IC lattice is more compressed than illustrated. (Images: RIJ)
Selecting R8 photoreceptor precursors
Ommatidial development begins with the selection of the R8 photoreceptor precursors, which are required for subsequent recruitment of seven more photoreceptor precursors per ommatidium (Frankfort et al., 2001; Jarman et al., 1994; Ready et al., 1976; Tomlinson and Ready, 1987). Hence, arrangement of ommatidia in staggered rows across the eye field relies on the orderly selection of correctly-spaced R8 precursors. This occurs during the final day of Drosophila larval development, after the eye primordium (the eye imaginal disc) has already been specified as such by a network of retinal determination genes, grown to reach its appropriate size, and patterned with respect to its dorsal-ventral and anterior-posterior axes (Kumar, 2012; Treisman, 2013).
The R8s are established as the morphogenetic furrow (MF) travels as a wave across the eye disc, beginning at the posterior (Figure 2A and B) (Kumar, 2020). The MF is a moving indentation in the tissue, generated by synchronized constriction of the apical domains and apical-basal axis of eye disc cells, driven by coordinated actin-myosin and microtubule activities in response to a moving front of abutting Hedgehog (Hh) and Decapentaplegic (Dpp) signaling activities (Benlali et al., 2000; Corrigall et al., 2007; Escudero et al., 2007; Fernandes et al., 2014; Heberlein et al., 1993). As the MF flows across the eye disc, successive rows of R8 precursors (and consequently ommatidia) are born at a rate of ~70–120 mins per row (Basler and Hafen, 1989; Campos-Ortega and Hofbauer, 1977; Ready et al., 1976; Wolff and Ready, 1993). Setting up the correct spacing of these ommatidia depends on regulation of the proneural basic type II helix-loop-helix (bHLH) transcription factor Atonal (Ato) which specifies the R8 precursor fate (Dokucu et al., 1996; Jarman et al., 1994; Jarman et al., 1995).
Figure 2: Patterning begins in the larval eye with selection of the R8-precursors.
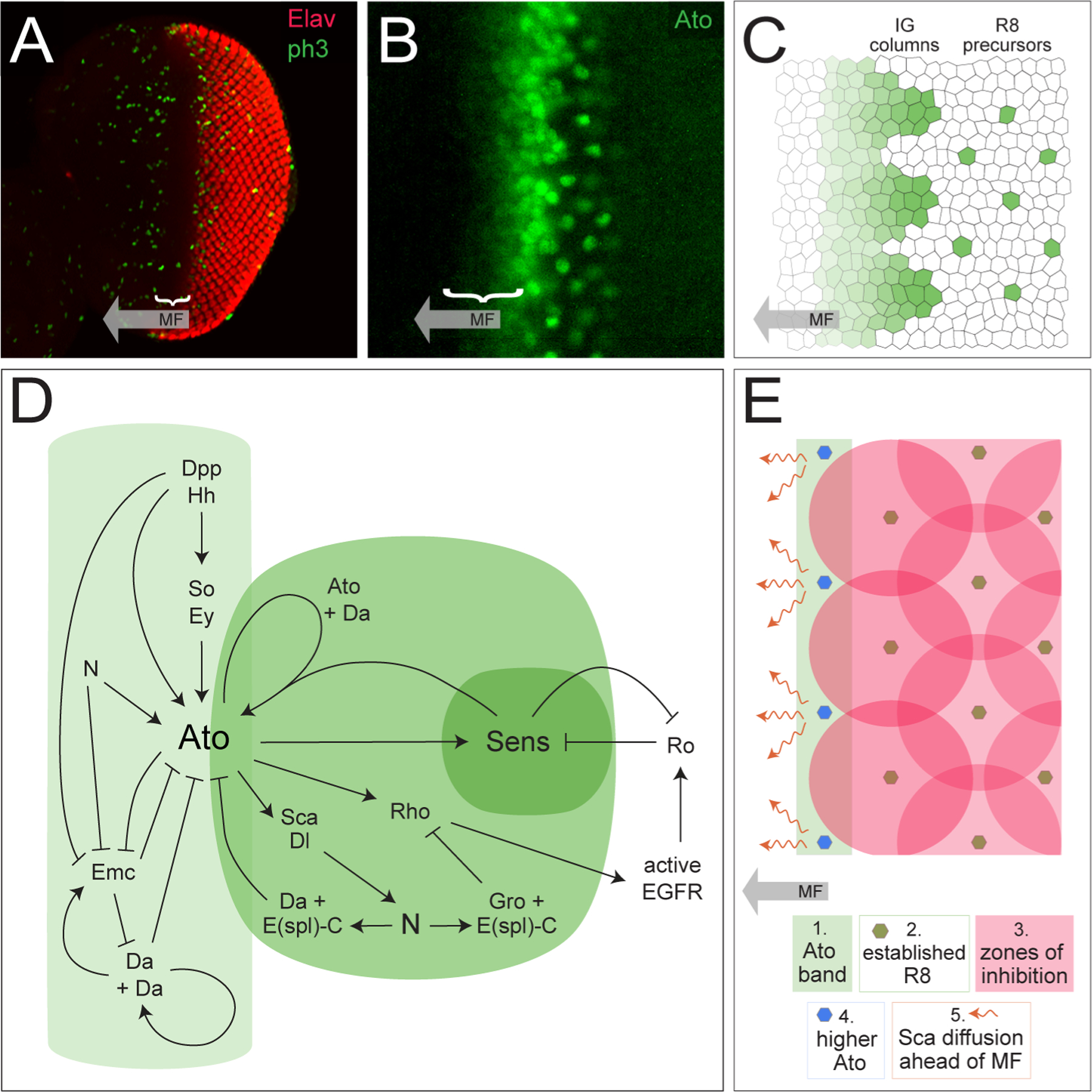
(A) A third instar larval eye disc, with photoreceptors detected with anti-Elav (red) and dividing cells detected with anti-phospho-histone 3B. The MF is indicated with a bracket as it proceeds from posterior (right) to anterior (left). (Image: RIJ) (B) Small region of the eye disc, at higher magnification than the image in A., with Ato detection (Image: Susan Spencer) (C) Cartoon of ato expression. Cell outlines do not accurately reflect the shapes of cells in the larval disc. (D) The network of major signals and interactions that regulate ato expression. Light green indicates interactions that regulate the ato band; medium and dark green represents Ato in the IGs and R8 precursors. (E) Graphical summary of R8 selection as the MF moves from posterior, as per models/computer simulations. (Drawings: RIJ, inspired by (Courcoubetis et al., 2019; Lubensky et al., 2011)).
Ato is at first expressed in a dorso-ventral band of cells straddling the anterior bank of the MF (Figure 2B and C). As the MF passes, ato expression is reduced to evenly-spaced groups of 10–15 cells termed intermediate groups (IGs) and eventually to just one cell per IG that becomes the R8 precursor (Baker et al., 1996; Dokucu et al., 1996; Jarman et al., 1994; Jarman et al., 1995). Ato functions as a homodimer or, more commonly, in complex with the type I bHLH protein Daughterless (Da) (Jarman et al., 1993) and activity of this heterodimer is augmented by interaction of an additional proneural transcription regulator, Senseless (Sens) (Acar et al., 2006; Powell et al., 2008). Restriction of ato-expression requires multiple layers of positive and negative regulation and is the focus of the discussion below. Most of these regulatory interactions are illustrated in Figure 2D. However, discussion of R8 selection must include brief consideration of Sens, which is required for R8 differentiation. Expression of sens is promoted by Ato within a subset of IG cells before Sens is restricted, like Ato, to single R8s (Frankfort et al., 2001; Pepple et al., 2008). However, once individual R8 precursors are selected, ato expression rapidly fades from these cells and any remaining Ato is phosphorylated to repress its function (Quan et al., 2016). In contrast, expression of sens persists.
Broad ato expression
Initial ato expression in a continuous strip of cells is promoted by the RDGs Eyeless (Ey), Sine Oculis (So) and Dacshund (Dac) that regulate a 3’ cis-regulatory ato enhancer (Sun et al., 1998; Tanaka-Matakatsu and Du, 2008; Zhang et al., 2006). Expression of these RDGs is promoted by Dpp activity in the MF which in turn is regulated by Hh, although Hh signaling also stimulates ato independently of Dpp (Borod and Heberlein, 1998; Dominguez, 1999; Firth and Baker, 2009; Heberlein et al., 1995). Expression of ato rapidly increases in groups of cells that emerge as IGs, in part due to autoregulation mediated by a 5’ ato enhancer recognized by Ato/Da heterodimers whose function is enhanced by Sens (Acar et al., 2006; Brown et al., 1996; Melicharek et al., 2008; Powell et al., 2008; Sun et al., 1998). At the same time, Da/Da homodimers are proposed to dampen ato expression at the 3’ ato enhancer (Lim et al., 2008; Melicharek et al., 2008). Several chromatin remodeling factors have also been found necessary for correct utilization of the 3’ and 5’ ato enhancers (Melicharek et al., 2008).
Notch (N) signaling also contributes to robust ato expression in IGs. The N-responsive transcription factor Suppressor-of-Hairless (Su(H)) represses ato when N is inactive, a block that is lifted by N activity within the ato stripe (Li and Baker, 2001). In addition, transcription of extramacrochaete (emc) is inhibited by Notch activity within the morphogenetic furrow (Baonza and Freeman, 2001). Dpp and Hh activity also repress emc expression (Li and Baker, 2019). Emc is an HLH protein that lacks a DNA-binding domain and, if not reduced, would bind Ato or Da in the Atonal-strip and IGs to generate non-functional dimers, disrupting Ato and Da autoregulation and compromising expression of other targets (Baker and Yu, 1997; Bhattacharya and Baker, 2011; Brown et al., 1995; Li and Baker, 2001). The relationships between Ato and Da, and Emc are complex. Da promotes emc transcription and stabilizes Emc proteins when complexed with them, whilst Ato somehow renders Emc unstable, possibly via disrupting Emc/Da complexes (Bhattacharya and Baker, 2011; Li and Baker, 2018, 2019). Such disruption would potentiate Ato and Da function. A second HLH protein, Hairy, was originally also implicated in repressing ato and da expression but this was not supported by subsequent investigations. (Bhattacharya and Baker, 2012).
N-mediated lateral inhibition of ato
Whilst N signaling promotes ato, this relationship rapidly changes as N mediates lateral inhibition that restricts ato expression to IGs and then R8 precursors. Proneural transcription factors including Ato elevate expression Dl, thus stimulating N activity in adjacent cells and consequently expression of N/Su(H) targets including Enhancer of Split Complex (E(spl)-C) genes (Bailey and Posakony, 1995; Heitzler et al., 1996; Hinz et al., 1994; Jennings et al., 1994; Kunisch et al., 1994; Ligoxygakis et al., 1998; Powell et al., 2001). In addition, Ato drives expression of Scabrous (Sca) which is secreted, interacts with N, and enhances N activity in nearby cells (Baker et al., 1990; Baker and Zitron, 1995; Lee et al., 1996; Lee et al., 2000; Mlodzik et al., 1990). The E(spl)-C proteins are bHLH transcriptional repressors that dimerize with Groucho (Gro) or Da (which is also upregulated by N activity) to repress ato expression in IG cells (Chanut et al., 2000; Delidakis and Artavanis-Tsakonas, 1992; Knust et al., 1992; Lim et al., 2008; Zhang and Du, 2015). Hence Ato, and subsequently also Dl and sca, is dampened in cells where E(spl)-C proteins are active. To prevent this happening too soon – that is, to permit the initial neurogenic role of N – it seems that activity of E(spl)-C proteins requires their phosphorylation and this is momentarily delayed (Bandyopadhyay et al., 2016; Bose et al., 2014; Trott et al., 2001). Ultimately, Ato and N activities rapidly become complementary via this system of lateral inhibition which transforms the ato band into a row of well-spaced R8 precursors (Baker et al., 1996; Baker and Zitron, 1995).
The role of EGFR signaling
Like N signaling, Epidermal Growth Factor Receptor (EGFR) signaling is utilized at multiple points in early eye development so that it has been tricky to clearly delineate roles in ommatidial spacing from, for example, EGFR’s requirement for photoreceptor recruitment, cell survival, and cell proliferation. Multiple studies have described manipulations to EGFR signaling that disrupt ommatidial spacing (eg.(Baonza et al., 2001; Brown et al., 2006; Brown et al., 2007; Chen and Chien, 1999; Dominguez et al., 1998; Spencer et al., 1998; Yang and Baker, 2001) whilst, conversely, activating a temperature-sensitive EGFR allele in eye development was reported to cause no defects in ommatidial spacing (Kumar et al., 1998; Rodrigues et al., 2005). However, that EGFR signaling contributes to limiting the R8 precursor fate to just one cell per ommatidium is clear. Ato/Da complexes drive expression of rhomboid (rho) (Baonza et al., 2001; Chen and Chien, 1999) which processes the EGFR ligand Spitz (Spi) so that it is secreted from Ato+ cells (Lee et al., 2001; Urban et al., 2001). Subsequently EGFR is activated in surrounding cells, which is essential for recruitment of a further seven photoreceptor precursors per cluster (Freeman, 1996, 1997). In addition, EGFR activity drives expression of rough (ro) in a subset of photoreceptor precursors as they are recruited to each cluster (Dokucu et al., 1996; Kimmel et al., 1990). Rough is a homeodomain transcription factor that represses sens, and therefore ato, hence restricting the R8 fate to just one cell per IG and within each of these R8 precursors Sens, in turn, represses ro to maintain the R8 fate (Frankfort et al., 2001; Pepple et al., 2008). To limit the potency of this Rho-EGFR-Ro network in restricting the R8 fate, rho is subject to additional layers of regulation. For example, as N becomes activated in IG cells, E(spl)-C proteins team up with Groucho (Gro) to repress rho expression, restricting Rho to the R8 precursor (the R2/R5 precursors later express rho, once recruited to the cluster) (Zhang and Du, 2015).
Computer simulations of R8 specification
Of course, the MF is a moving wave of morphogenesis and the momentary engagement of signaling and transcriptional interactions is coordinated with its movement. That R8 precursors are evenly spaced in staggered columns suggests mechanisms ensure this particular and robust R8 placement. Several computational models have been developed to account for dynamic propagation of R8 columns when paired with computational simulations (Courcoubetis et al., 2019; Gavish and Barkai, 2016; Gavish et al., 2016; Lubensky et al., 2011; Pennington and Lubensky, 2010; Zhu et al., 2016). For these models, velocity of MF propagation, and hence initiation of the band of ato expression, is taken to be constant. With the exception of the model generated by Zhu et al. (2016), the network of transcription and signaling interactions associated with activation, inhibition and autoregulation of Ato is generally simplified in order to minimize the number of parameters and equations integrated into each simulation. Together the, the parameters used mainly capture rates of signal production, diffusion, and range of signaling (local, short-distance or cell autonomous), rates of signal decay, threshold levels required for activating or inhibiting responses, and the propagating response of Ato. Models also rely on the correct patterning of an initial column of R8s and hence do not model R8 patterning at the very posterior part of the eye disc. Together these simulations emphasize the importance of Scabrous as a key short-range diffusing signal that is key in ato repression (Courcoubetis et al., 2019; Gavish et al., 2016; Lubensky et al., 2011). Only cells just outside of Sca’s range can generate Ato at levels sufficient for those same cells to then generate and secrete Sca, so that patterning is propagated (Figure 2E). Simulations also suggest that Sca is generated and secreted rapidly and can diffuse ahead of the MF. Hence the band of ato-expressing cells that is observed in eye discs at the anterior front of the MF overlays a pre-pattern of Sca that is observed only later as IGs. Modelling also emphasized that R8 patterning can withstand some fluctuation in the rate of MF progression and Ato production: introducing a small amount of noise into their modeling parameters (more or less akin to fluctuations in signaling and gene expression expected in the eye) generated minor variations in the number and distribution of Ato-positive cells that could subsequently be cleaned up by Dl-N signaling (Courcoubetis et al., 2019; Gavish et al., 2016)). Importantly, introducing noise in the absence of Sca severely disrupted R8 patterning in simulations, suggesting that Sca enables robust R8 spacing in the eye despite stochastic gene expression and signaling activities (Courcoubetis et al., 2019).
Models also highlight aspects of R8 patterning that are not well understood or challenge common assumptions. For example, in the computer simulations of Courcoubetis et al. (2019), R8 spacing was accurate for the first few columns but began to deteriorate further away from the first R8 column. Accordingly, Courcoubetis et al (2019) comment that larger Drosophila eyes might be more susceptible to errors in R8 placement. The implication here is that to maintain accurate R8 spacing, the size of the eye field must be limited, implying that growth and patterning mechanisms may be integrated. This suggestion remains to be tested and may be at odds with observations of Drosophila strains and other Diptera species that have large eyes containing large numbers of ommatidial columns that appear to be correctly patterned (see Casares and McGregor, 2021) for a discussion on fly eye size). Alternate suggestions are that the breakdown in R8 patterning observed in simulations reflects a minor deficit in modeling parameters or that additional factors operate toward the anterior of the eye field to maintain continued uniform R8 placement.
Pupal eye patterning
After R8 specification, seven additional photoreceptor precursors are recruited to form each photoreceptor cluster and each ommatidium becomes encapsulated by four cone cells and two primary (1°) pigment cells (Kumar, 2012; Treisman, 2013). Many undifferentiated interommatidial cells (ICs) remain between each ommatidium and from ~17 hours-after-puparium-formation (h APF) these rearrange into two rows and then intercalate to bring them into single file (Figure 3A) (Hellerman et al., 2015; Johnson et al., 2011; Larson et al., 2008). Mechanosensory bristles emerge, their position broadly determined by selection of bristle cell precursors during larval development (Meserve and Duronio, 2017). The bristles become positioned to occupy three of six vertices of the hexagon that is gradually shaped about each ommatidium. Apoptosis eliminates superfluous ICs and all remaining cells adopt distinctive geometries to give rise to the highly ordered array of secondary (2°) and tertiary (3°) pigment cells observed by ~40 h APF (for development at 25°C, Figure 1A). Because of the beautiful cell shapes and near-perfect patterning of the pupal eye, it is an excellent model for studies of morphogenesis and the principles that regulate pupal eye patterning are emerging. Biophysical, adhesion and cytoskeletal properties of the fully-patterned 40 h APF pupal eye have mainly been considered, whilst the patterning events of ~10–22 h earlier have received less attention.
Figure 3: Local cell movements, growth and shape changes characterize the early pupal eye.
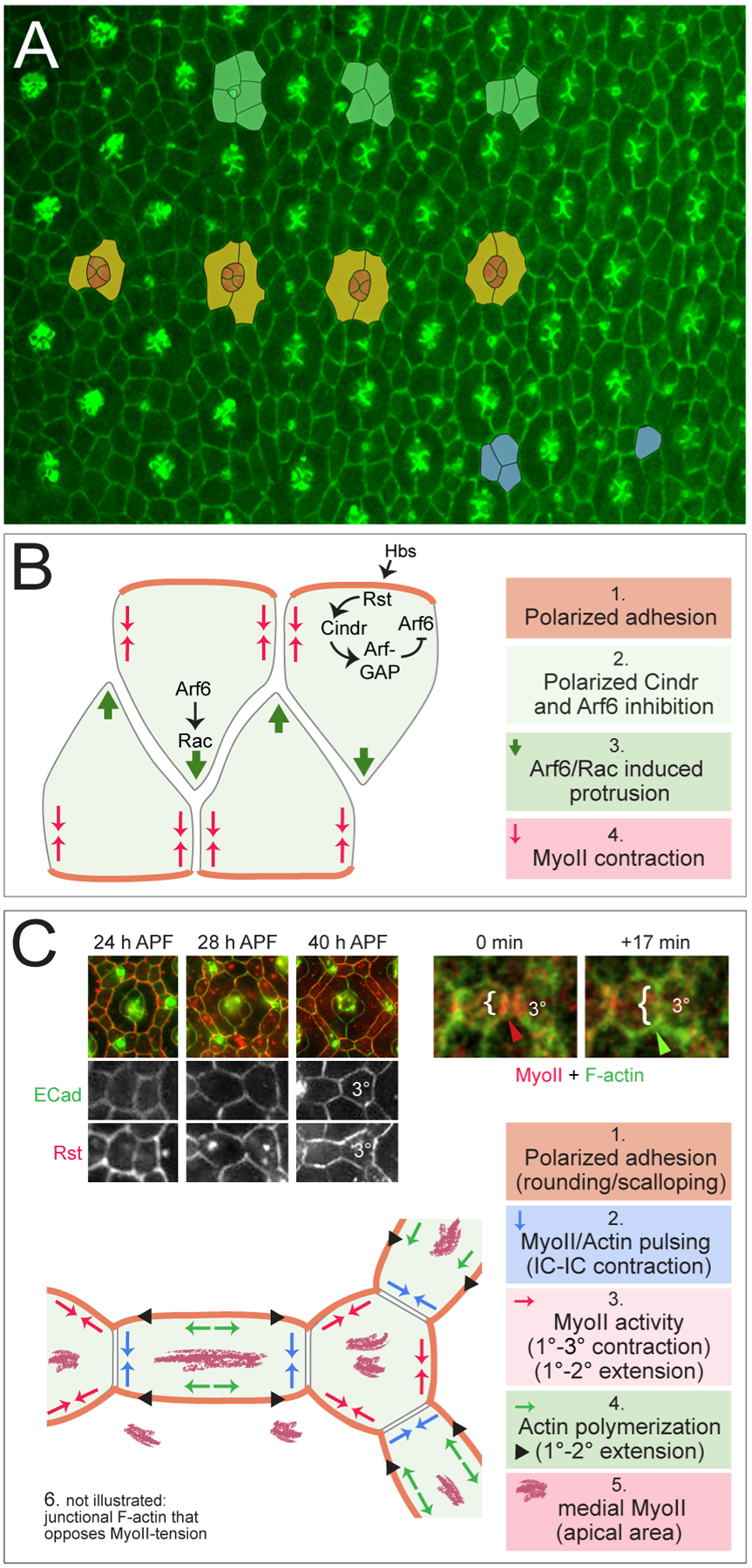
(A) Central region of the pupal eye at 22 h APF, which is marked by a gradient of development evident for another ~8 h. Examples of intercalating ICs are marked in green, cone cells in orange and encircling 1° cells in yellow. Three ICs that will compete for a single 3° niche are in blue. Posterior is to the right. (Image adapted from (Hellerman et al., 2015). (B) Model of the molecular regulation of intercalation. See text for details and note that model requires experimental confirmation. (C) At right, Rst (red) and E-cad (green) in ommatidia and, in panels below, at higher magnification in ICs. Rst becomes excluded from IC-IC boundaries and concentrated at 1°-IC where it complexes with Hbs (not shown) (Image adapted from (Johnson et al., 2012). At left, live-imaging of MyoII (red) and actin (green) in ICs. Bracket indicates contraction and expansion of the 2°−3° junction; arrowheads reflect associated MyoII and F-actin accumulation. (Image adapted from (Del Signore et al., 2018). Below, model of dynamic cytoskeletal and junction events that drive reshaping of the ICs. See text for details.
IC intercalation
ICs positioned along the horizontal sides of ommatidia intercalate to bring them into single file, with cells adherent to ventral ommatidia projecting dorsally, and vice versa (Figure 3A, B)(Hellerman et al., 2015). Similar cell movements establish the 3° cell position, which is fought over by two or three cells: each is bound to two ommatidia and pushes toward a third to secure the 3° position (Figure 3A)(Hellerman et al., 2015).
To intercalate, ICs extend large projections that protrude between IC neighbors. Regulation of adhesion is important during this process and analyses of core AJ components (eg. E-Cadherin) indicated that more AJ complexes accumulate at IC-1° boundaries than at IC-IC boundaries, suggesting differences in the stability of these AJs (DeAngelis et al., 2020). These differences are amplified by polarized distribution of members of the immunoglobulin cell adhesion molecule (IgCAM) family including Roughest (Rst) and Hibris (Hbs). These generate heterodimeric adhesion complexes, essential for correct pupal eye morphogenesis, (Araujo et al., 2003; Bao and Cagan, 2005; Bao et al., 2010; Grzeschik and Knust, 2005; Reiter et al., 1996; Wolff and Ready, 1991). Rst and Hbs have complementary expression patterns: rst becomes restricted to ICs whilst hbs expression is driven by N activity in neighboring 1°s and the extracellular domains of these IgCAMs interact, driving isolation of Rst/Hbs complexes to AJs between ICs and 1° cells (Bao et al., 2010). Reducing the function of Rst or Hbs disrupts IC intercalation, and live-imaging of rst mutant eyes suggests this is because ICs do not remain stably adherent to adjacent 1°s whilst stretching toward target 1°s of opposite ommatidia (Larson et al., 2008). A second pair of IgCAMs, Sticks-and-stones (Sns) and Kin of Irre-C (Kirre), are similarly expressed in complementary patterns in the eye but their contribution to patterning and IC intercalation seems less important (Bao, 2014).
With IC-1° adhesion secured, the ICs are able to extend projections to intercalate. These projections rely on the GTPase Arf6 which has been shown to promote Rac1 and Arp2/3 activity to stimulate F-actin remodeling in vitro, and is required for successful IC intercalation (Hu et al., 2009; Humphreys et al., 2013; Johnson et al., 2011; Koo et al., 2007). One model proposes that Arf6 activity is restricted to where the projections will form in ICs because the negative Arf6 regulators - ArfGAPs including ArfGAP3 and ASAP - are localized elsewhere (Figure 3B) (Johnson et al., 2011). Specifically, the prediction is that since the ArfGAPs interact with the adaptor protein Cindr, they are recruited to IC-1° cell boundaries because Cindr in turn binds the Rst/Hbs complexes localized to these AJs (Bao and Cagan, 2005; Johnson et al., 2012; Johnson et al., 2011). This model requires testing. For example, it’s not been clearly shown that the ArfGAPs and Cindr accumulate at higher levels at IC-1° cell boundaries in comparison to AJs between neighboring ICs, although disrupting their activities does modify intercalation (Johnson et al., 2011; Johnson et al., 2008). In addition, direct spatial analyses of Arf6 and Rac activation to confirm their activity within IC projections have not been done. Finally, Cindr and its vertebrate orthologs contribute to cytoskeletal stability because they have intrinsic F-actin capping function and can recruit other actin capping proteins and actin regulators (eg. Cortactin) (Bruck et al., 2006; Edwards et al., 2014; Johnson et al., 2008; Tang and Brieher, 2012; Zhao et al., 2013). Hence, the prediction is that in ICs Cindr and other actin-stabilizing proteins are sequestered to IC-1° AJs, so that cytoskeleton remodeling is permitted elsewhere to promote intercalation.
The very tip of an intercalating IC’s protrusion is marked by a tricellular AJ (tAJ), characterized by localization of another IgCAM protein Sidekick (Sdk) (Letizia et al., 2019). The tension at tAJs is expected to be high, and Sdk responds to and regulates this tension via recruitment of Polychaetoid (Pyd, Drosophila ZO-1) and Canoe (Cno, Drosophila Afadin), which are proposed to tether Sdk to the actin cytoskeleton (Letizia et al., 2019). Live-imaging of sdk mutants or retinas expressing pydRNAi revealed that both proteins are required for the formation of IC protrusions and IC movement (Letizia et al., 2019; Seppa et al., 2008), but how these tAJ proteins contribute to extension of an IC protrusion has not been resolved. It should be noted though that in response to pydRNAi expression, increased density of AJ components has been observed across the entire pupal eye, and this too would restrict IC movement during intercalation (Seppa et al., 2008).
Finally, data presented by Letizia et al. (2019) provide clues of a mechanism that remodels the AJs between two ICs being separated by a third protruding IC. Specifically, live-imaging of actin and myosin at this IC interface revealed that Myosin-II (MyoII) accumulates at this shrinking IC-IC junction (Letizia et al., 2019). This suggests that myosin-induced contraction promotes AJ destruction to permit intercalation (Figure 3B). A similar requirement for MyoII activity in junction remodeling has been described in other tissues during cell rearrangements (Harris, 2018; Heer and Martin, 2017; Pinheiro and Bellaiche, 2018). The pupal eye provides the opportunity to now explore mechanisms that coordinate myosin activity and junction separation with the protrusive activities of neighboring ICs.
Refining the IC lattice
Once ICs are organized into single file around ommatidia, two important events sculpt the lattice into its final pattern. First, apoptosis removes numerous excess ICs to leave just nine about an ommatidium (six 2°s and three 3°s, Figure 1A). Apoptosis is promoted by N, Wingless (Wg) and Jun N-terminal kinase (JNK) signaling activities and opposed by EGFR and Yorkie (Yrk) (Bushnell et al., 2018; Cagan and Ready, 1989b; Cordero et al., 2004; DeAngelis et al., 2020; Freeman, 1996; Miller and Cagan, 1998; Querenet et al., 2015; Sawamoto et al., 1994; Sawamoto et al., 1998; Wolff and Ready, 1991; Yu et al., 2002). Except for Wg, these signals are similarly associated with cell death or survival in other tissues. Precisely how these signals are spatially integrated in the retina to balance activation of the apoptotic pathway with cell survival remains an open question. However, that apoptosis is positionally regulated is clear: ICs closer to bristles are more vulnerable to apoptosis (Monserrate and Brachmann, 2007). In addition, expression of the chromatin-remodeling protein Trithorax-like (Trl) in cone cells is crucial for the removal of excess ICs positioned on the oblique sides of the IC hexagon but is less important for pruning ICs located along horizontal sides of ommatidia (Dos-Santos et al., 2008). This suggests that Trl regulates gene expression in cone cells to promote an apoptosis-inducing signal or inhibit a cell-survival signal that emanates from the cone cells and orients toward ICs positioned obliquely.
Elimination of the appropriate number of excess ICs ensures the remaining cells can adopt simple 2° and 3° shapes. Dramatic increases in IC number, observed for example when Hippo activity is altered to enhance proliferation in the larval eye disc, causes ommatidia to be surrounded by numerous rows of small ICs. However, when activity of the core apoptotic machinery is impaired, as long as the number of grouped ICs at an intercalating cluster is below ~8 (rather than the usual ~5), these can still intercalate and pack to generate a hexagonal lattice (RIJ, unpublished)(DeAngelis et al., 2020; Larson et al., 2010)). Conversely, when intercalation fails, elimination of at least some misplaced ICs can generate a simpler lattice (RIJ, unpublished)(Letizia et al., 2019). Hence apoptosis provides robustness for lattice patterning.
Second, biophysical factors mould the 2° and 3° cells but they travel a bumpy road before adopting their final shapes. Initially ICs transition through a ~10–12 h period of being more rounded and live-imaging revealed that their apical areas fluctuate periodically during this stage (Blackie et al., 2020). These fluctuations are associated with pulsing MyoII activity, discussed below. The 2°s will eventually elongate to become rectangular, whilst the 3°s become hexagonal, with reduced apical areas (Figure 3C). These changes correlate with growth of the ommatidium, observed as a ~30% increase in the apical area of the 1° and cone cell cores (Larson et al., 2010). Ommatidial growth probably promotes narrowing of the ICs and facilitates 2° cell anisotropy, but cytoskeletal and adhesion dynamics are likely more dominant factors in this regard.
Rounding of ICs is coupled with scalloping of the 1° cells and ‘pinching’ of IC-IC boundaries (Figure 3C). This transitional phenotype correlates with preferential accumulation of the IgCAM complexes Rst/Hbs and Kirre/Sns (discussed above) at 1°-IC junctions, driving expansion of this cell interface to promote scalloping (Bao and Cagan, 2005; Bao et al., 2010). Accordingly, when IgCAMs fail to accumulate at 1°-IC boundaries and instead distribute evenly about IC junctions, ICs fail to round (scalloping fails) and IC-IC boundaries in addition remain extended (Johnson et al., 2012). This IgCAM-centric view of rounding/scalloping is not at odds with a second perspective in which contraction of IC-IC boundaries tugs on 1°s via tAJs to induce scalloping.
Indeed, live-imaging of IC-IC boundaries revealed their repeated contraction and expansion until ICs acquire their final shapes (Del Signore et al., 2018). As in other tissues, contraction correlated with periodic accumulation of MyoII and the myosin-activators Rho1 and Rok at junctions, whilst expansion correlated with actin polymerization associated with Arp2/3 (Figure 3C). Live-imaging and clonal analyses revealed that within 3° cells, repeated MyoII contraction is essential to appropriately reduce the length of 1°−3° cell boundaries over time but MyoII contributes less to narrowing the 2°−3° junction length. Instead, repeated MyoII contraction within 2° cells constricts the 2°−3° interface to narrow 2°s into slim rectangles. At the same time, 2°s are elongated because they are ‘pulled’ by contraction by neighboring 3°s (mediated by MyoII activity along the 1°−3° cell boundary) and because Arp2/3-mediated actin polymerization along 1°−2° cell contacts supports expansion of this cell interface (Figure 3C) (Del Signore et al., 2018). In a separate study, cyclical assembly of an apical-medial meshwork of MyoII was observed in retinal cells that correlated with pulsed constriction of the apical cell area and was suggested to also enable ICs to achieve their correct shapes and sizes (Blackie et al., 2020). Data presented from this work also hints that contraction of apical-medial MyoII in 1°s amplifies their scalloping (Blackie et al., 2020).
The analyses of Del Signore and colleagues beautifully demonstrate that cytoskeletal dynamics in emerging 2° and 3° cells differ. This is the first molecular difference between 2° and 3° cells to be described. In addition, they demonstrate that there are different requirements for actin and MyoII activities at different IC junctions. How the Rho GTPases or other signals that regulate actin and myosin are organized to implement polarized cytoskeletal activities in ICs is not known. Also unclear is how the cyclical activity of actin and myosin eventually ceases at IC junctions so that they can be stabilized.
A gradual increase in the density of AJ complexes is observed as eye-patterning proceeds, suggesting that cells become locked in place once achieving their appropriate shapes (DeAngelis et al., 2020). Several proteins and pathways have been identified for their role in promoting stable AJs in ICs (and in 1° cells) and impairing their function or expression leads to punctate distribution or decreased density of AJs and lattice mis-patterning. These regulators include the GTPase Rho1 that represses endocytosis of E-Cadherin (Warner and Longmore, 2009; Yashiro et al., 2014); the kinase Csk (and it’s regulators ASPP and RASSF8 that are recruited to AJs by MAGI) which targets Src to impede AJ turnover (Langton et al., 2009; Vidal et al., 2006; Zaessinger et al., 2015); the guanine exchange factor Vav that is activated by the EGFR (Martin-Bermudo, 2015); Decapentaplegic (Dpp, a Drosophila TGF-β) signaling (Cordero et al., 2007); and the adaptor protein Cindr (discussed above, (Johnson et al., 2008). AJs are also compromised when the function of Yorkie (Yki) or its cofactor Mask is impaired and expression of a large variety of adhesion and cytoskeleton-related genes is regulated downstream of their activities, indicating a broad requirement for appropriate Hippo activity to achieve correct adhesion during Drosophila eye morphogenesis (DeAngelis et al., 2020).
Patterning the core: 1° and cone cells
Patterning of the cone and 1° cells occurs alongside that of the IC lattice. Their beautiful stereotypical geometries acquired by ~40 h APF (Figure 1A) have attracted considerable analyses as examples of cell configurations that conform to biophysical properties (Gemp et al., 2011; Hilgenfeldt et al., 2008; Kafer et al., 2007). But how do these cells come to be arranged in this way?
The four cone cells (CCs) are recruited to each ommatidium during larval development after ommatidia have acquired their full set of photoreceptor precursors (Wolff and Ready, 1993). A variety of signals originating from photoreceptors, including EGFR, N, JNK and Wg activities, and transcription factors including Prospero (Pros), Pax2 (also known as Shaven and Sparkling) and Cut (Ct) contribute to recruitment and development of the cone cells (Charlton-Perkins et al., 2011a). These are recruited in pairs, beginning with the anterior and posterior CCs that by ~17 h APF are easily observed in their position above photoreceptor clusters and in direct contact with each other, whilst the dorsal and ventral cone cell pair are apically excluded (Figures 3A and 4A). Unlike other epithelial cells of the pupal eye, the cone cells express N-Cadherin (N-Cad), possibly in response to Pax2 which in mammals regulates expression of N-Cad (Christophorou et al., 2010; Hayashi and Carthew, 2004). The anterior/posterior CCs initially have higher levels of Pax2 than dorsal/ventral CCs (Charlton-Perkins et al., 2011b), which, together with their earlier selection, may drive the anterior/posterior CCs to express N-Cad first or at higher levels, to mediate their initial attachment. However, this configuration rapidly changes and the CC group undergoes a classic T1-T2-T3 junction exchange to bring the dorval/ventral cones into contact (Figure 4A and B).
Figure 4: Junction and cytoskeletal factors drive shaping of cone and 1° cells.
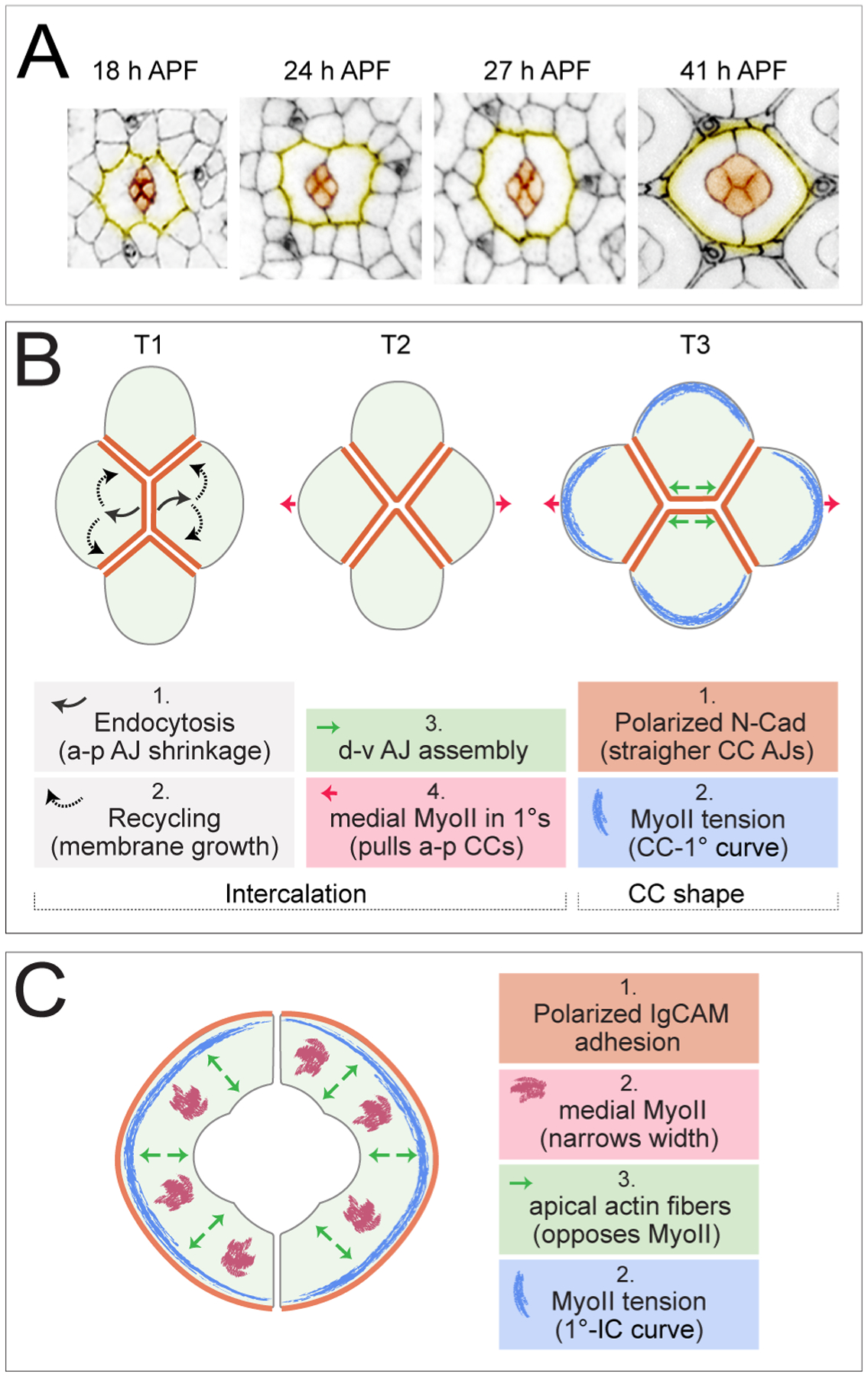
(A) Single ommatidia, with cone cells in orange and the outlines of 1° cells in yellow. Images are presented to scale. (Adapted from (Johnson, 2020)). (B) Illustration of the T1-T2-T3 transition of cone cells with factors that drive intercalation and the acquisition of the final CC shapes and (C) illustration of the factors that influence the final 1° shapes. See text for details. Note experimental confirmation of the models presented is incomplete.
Analyses of T1-T2 junction exchanges in other tissues have revealed a requirement for MyoII to shorten the junction analogous to the anterior/posterior CC interface, and a requirement for apical-medial MyoII in these same cells to simultaneously ‘pull’ on the junction to enhance its instability (Figure 4B)(Blankenship et al., 2006; Kasza et al., 2014; Levayer and Lecuit, 2013; Rauzi et al., 2010; Simoes Sde et al., 2014; Tamada et al., 2012; Warrington et al., 2013). Transition from the T2-T3 conformation is then promoted by continued apical-medial MyoII contraction in anterior/posterior cells to prize these apart whilst AJs are assembled between dorsal and ventral cells (Bardet et al., 2013; Collinet et al., 2015; Simoes Sde et al., 2010; Yu and Fernandez-Gonzalez, 2016). However, detailed analyses of MyoII in the pupal eye showed little accumulation at CC-CC boundaries and little apical-medial activity (Blackie et al., 2021). Further, genetic perturbation of MyoII did not disrupt CC junction exchange (Blackie et al., 2021). Instead, it was endocytosis of membrane from the anterior/posterior CC interface that was found to reduce this junction and it’s plausible that transition to the T2 stage is mediated by recycling of these vesicles to adjacent CC membranes to promote their extension (Figure 4B) (Blackie et al., 2021). Then, subsequent elongation of the dorsal/ventral CC boundary may be promoted by mechanical coupling of the anterior and posterior CCs to the 1°s which, unlike CCs, do have an active apical-medial myosin meshwork (discussed below): in data presented by Blackie et al. (2020), the apical-medial myosin network in 1° cells was ablated and in the images they present of these experiments, the dorsal/ventral CC junction appears shorter. Finally, two systems have been implicated in stabilizing the T3 conformation of CCs: N signaling activity in the dorsal and ventral CCs, and the Ig-CAMs Rst (also required in the dorsal/ventral pair) and Hbs (required in all cone cells) (Blackie et al., 2021; Grillo-Hill and Wolff, 2009). Precisely how these systems maintain the dorsal/ventral CC junction is unclear.
The four cone cells eventually acquire spectacular energetically-favorable shapes, governed by differences in adhesion at different AJs and polarized cytoskeletal activity (Figure 4A and B). Expression of N-Cad only in CCs ensures that adhesion between them is stronger than between CCs and 1°s, segregating these cell types (Hayashi and Carthew, 2004). N-Cad AJs accumulate between CCs to straighten and elongate this interface whilst limiting expansion of the remaining CC membranes which are shaped to minimize their surface areas (Hayashi and Carthew, 2004; Hilgenfeldt et al., 2008; Kafer et al., 2007). Curiously, accumulation of MyoII is inhibited downstream of N-Cad, excluding it from CC-CC boundaries, presumably to prevent contraction of these AJs (Chan et al., 2017). Surface/cortical tension is also lower between CCs than at the rounded CC-1° membrane, as predicted by the differential adhesion hypothesis and mathematical models, and these differences in tension also contribute to CC shape (Foty and Steinberg, 2005; Kafer et al., 2007; Kong et al., 2019; Steinberg, 1963). Finally, MyoII accumulates in cone cells along the CC-1° boundaries, adding another factor that shapes this border (Aigouy and Le Bivic, 2016; Chan et al., 2017).
The 1° cell fate is established in the pupa from ~14 h APF by activity of the transcription factor Lozenge (Lz) and N, which responds to increased Dl in neighboring CCs (Cagan and Ready, 1989b; Nagaraj and Banerjee, 2007). Each 1° pair grows rapidly to surround the CCs and photoreceptors of an ommatidium, and they form secure AJs where they meet (Figure 3A, 4A). As discussed earlier, 1°s transition through a period of scalloping. During this time their apical areas pulse, driven by a network of apical-medial myosin but, unlike in other cell types where pulsing correlates with gradual shrinking of cell area, the 1° cells grow (Blackie et al., 2020). In fact, apical-medial myosin is crucial in limiting the final 1° size (disruption of this network results in enlarged 1°s), implying that mechanisms oppose apical-medial MyoII contractility (Blackie et al., 2020; Deng et al., 2020). We speculate that it is the actin cytoskeleton that opposes MyoII. Careful preservation of the cytoskeleton reveals apical F-actin strands oriented perpendicularly to the 1°-IC boundary and traversing the width of 1°s (DeAngelis et al., 2020; Johnson et al., 2008). We predict that these function like apical stress fibers to oppose contractile forces and maintain 1° cell shape (RIJ, unpublished). In addition, Spectrin was recently shown to link the cortical cytoskeleton to the apical plasma membrane in 1°s (Deng et al., 2020). This linkage is crucial to restrict the size of 1°s, oppose contractile tension, and transmit cytoskeletal tension to the cortical membrane. Interestingly, Deng and colleagues found that Spectrin interacted with Arp2/3-generated actin (Deng et al., 2020), which may lie above the apical stress fibers that we observe.
Once they acquire their final smooth shape, accumulated MyoII is observed in 1°s along the concave surface of 1°-IC boundaries suggesting that MyoII-is crucial for generating or maintaining the rounded shape of this surface (Figure 4B) (Aigouy and Le Bivic, 2016; DeAngelis et al., 2020). Some experimental data support this idea: in published images of single 1° cells with higher MyoII activity, one can observe that the 1°-IC boundaries are more concave than those of their wild type neighbors (Blackie et al., 2020; Deng et al., 2020). Conversely, single 1°s with reduced myosin activity have less concave 1°-IC boundaries (Warner and Longmore, 2009). Additionally, if MyoII is severely reduced in adjoining IC and 1°s (Warner and Longmore, 2009), or across the eye as it is when the Yki cofactor Mask is impaired, the 1°-IC border is straighter, emphasizing different requirements for myosin across this cell interface (DeAngelis et al., 2020). Accumulation of MyoII at 1°-IC boundaries is reminiscent of MyoII at the CC-1° interface. Indeed, higher mechanical tension at these cell interfaces is reflected in accumulation of tension-sensing proteins including those that go on to modify Hippo signaling (Deng et al., 2020). In fact, it turns out that Yki, and Mask, are required for multiple aspects of pupal eye patterning (DeAngelis et al., 2020). Hence, whilst for many years the Drosophila eye has been the tissue of choice for screens to identify components of the Hippo pathway, the pupal eye – which is post-mitotic – now provides a model to study Hippo’s contribution to tissue morphogenesis.
Closing notes
Those wanting to work with the larval or pupal eye as a model for tissue patterning are in luck: there are numerous open questions, several of which have been raised in this review. In addition, helpful guides on how to work with these tissues are available (Baker et al., 2014; DeAngelis and Johnson, 2019; Hsiao et al., 2012; Tea et al., 2014; Wolff, 2007). It’s worth mentioning here that it is helpful to align pupal eye images with respect to the tissue’s dorsal-ventral axes so that the 1° pairs are oriented as presented in the figures of this review, as is conventional (although anterior-posterior orientation differs amongst research groups).
Highlights.
The Drosophila eye is a highly-organized neuroepithelium.
A transcription factor network establishes columns of evenly-spaced R8 precursors.
Computational models describe propagation of R8 placement across the eye.
Accessory cells adopt stereotypical positions and shapes essential for correct eye patterning.
Morphogenesis of accessory cells requires conserved processes.
Acknowledgements
Our understanding of Drosophila eye development is due to the efforts of numerous researchers who have worked with this tissue over the past ~110 years. My thanks to them, and apologies to those who have not been cited in this review. Grants from the National Institutes of Health (R15GM114729) and National Science Foundation (DBI-1828327) provided support for this review.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acar M, Jafar-Nejad H, Giagtzoglou N, Yallampalli S, David G, He Y, Delidakis C, Bellen HJ, 2006. Senseless physically interacts with proneural proteins and functions as a transcriptional co-activator. Development 133, 1979–1989. [DOI] [PubMed] [Google Scholar]
- Aigouy B, Le Bivic A, 2016. The PCP pathway regulates Baz planar distribution in epithelial cells. Sci Rep 6, 33420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo H, Machado LC, Octacilio-Silva S, Mizutani CM, Silva MJ, Ramos RG, 2003. Requirement of the roughest gene for differentiation and time of death of interommatidial cells during pupal stages of Drosophila compound eye development. Mech Dev 120, 537–547. [DOI] [PubMed] [Google Scholar]
- Bailey AM, Posakony JW, 1995. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev 9, 2609–2622. [DOI] [PubMed] [Google Scholar]
- Baker NE, Li K, Quiquand M, Ruggiero R, Wang LH, 2014. Eye development. Methods 68, 252–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NE, Mlodzik M, Rubin GM, 1990. Spacing differentiation in the developing Drosophila eye: a fibrinogen-related lateral inhibitor encoded by scabrous. Science 250, 1370–1377. [DOI] [PubMed] [Google Scholar]
- Baker NE, Yu S, Han D, 1996. Evolution of proneural atonal expression during distinct regulatory phases in the developing Drosophila eye. Curr Biol 6, 1290–1301. [DOI] [PubMed] [Google Scholar]
- Baker NE, Yu SY, 1997. Proneural function of neurogenic genes in the developing Drosophila eye. Curr Biol 7, 122–132. [DOI] [PubMed] [Google Scholar]
- Baker NE, Zitron AE, 1995. Drosophila eye development: Notch and Delta amplify a neurogenic pattern conferred on the morphogenetic furrow by scabrous. Mech Dev 49, 173–189. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay M, Bishop CP, Bidwai AP, 2016. The Conserved MAPK Site in E(spl)-M8, an Effector of Drosophila Notch Signaling, Controls Repressor Activity during Eye Development. PLoS One 11, e0159508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, 2014. Notch controls cell adhesion in the Drosophila eye. PLoS Genet 10, e1004087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Cagan R, 2005. Preferential adhesion mediated by Hibris and Roughest regulates morphogenesis and patterning in the Drosophila eye. Dev Cell 8, 925–935. [DOI] [PubMed] [Google Scholar]
- Bao S, Fischbach KF, Corbin V, Cagan RL, 2010. Preferential adhesion maintains separation of ommatidia in the Drosophila eye. Dev Biol 344, 948–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baonza A, Casci T, Freeman M, 2001. A primary role for the epidermal growth factor receptor in ommatidial spacing in the Drosophila eye. Curr Biol 11, 396–404. [DOI] [PubMed] [Google Scholar]
- Baonza A, Freeman M, 2001. Notch signalling and the initiation of neural development in the Drosophila eye. Development 128, 3889–3898. [DOI] [PubMed] [Google Scholar]
- Bardet PL, Guirao B, Paoletti C, Serman F, Leopold V, Bosveld F, Goya Y, Mirouse V, Graner F, Bellaiche Y, 2013. PTEN controls junction lengthening and stability during cell rearrangement in epithelial tissue. Dev Cell 25, 534–546. [DOI] [PubMed] [Google Scholar]
- Basler K, Hafen E, 1989. Dynamics of Drosophila eye development and temporal requirements of sevenless expression. Development 107, 723–731. [DOI] [PubMed] [Google Scholar]
- Benlali A, Draskovic I, Hazelett DJ, Treisman JE, 2000. act up controls actin polymerization to alter cell shape and restrict Hedgehog signaling in the Drosophila eye disc. Cell 101, 271–281. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Baker NE, 2011. A network of broadly expressed HLH genes regulates tissue-specific cell fates. Cell 147, 881–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Baker NE, 2012. The role of the bHLH protein hairy in morphogenetic furrow progression in the developing Drosophila eye. PLoS One 7, e47503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackie L, Tozluoglu M, Trylinski M, Walther RF, Schweisguth F, Mao Y, Pichaud F, 2021. A combination of Notch signaling, preferential adhesion and endocytosis induces a slow mode of cell intercalation in the Drosophila retina. Development 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackie L, Walther RF, Staddon MF, Banerjee S, Pichaud F, 2020. Cell-type-specific mechanical response and myosin dynamics during retinal lens development in Drosophila. Mol Biol Cell 31, 1355–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship JT, Backovic ST, Sanny JS, Weitz O, Zallen JA, 2006. Multicellular rosette formation links planar cell polarity to tissue morphogenesis. Dev Cell 11, 459–470. [DOI] [PubMed] [Google Scholar]
- Borod ER, Heberlein U, 1998. Mutual regulation of decapentaplegic and hedgehog during the initiation of differentiation in the Drosophila retina. Dev Biol 197, 187–197. [DOI] [PubMed] [Google Scholar]
- Bose A, Majot AT, Bidwai AP, 2014. The Ser/Thr phosphatase PP2A regulatory subunit widerborst inhibits notch signaling. PLoS One 9, e101884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KE, Baonza A, Freeman M, 2006. Epithelial cell adhesion in the developing Drosophila retina is regulated by Atonal and the EGF receptor pathway. Dev Biol 300, 710–721. [DOI] [PubMed] [Google Scholar]
- Brown KE, Kerr M, Freeman M, 2007. The EGFR ligands Spitz and Keren act cooperatively in the Drosophila eye. Dev Biol 307, 105–113. [DOI] [PubMed] [Google Scholar]
- Brown NL, Paddock SW, Sattler CA, Cronmiller C, Thomas BJ, Carroll SB, 1996. daughterless is required for Drosophila photoreceptor cell determination, eye morphogenesis, and cell cycle progression. Dev Biol 179, 65–78. [DOI] [PubMed] [Google Scholar]
- Brown NL, Sattler CA, Paddock SW, Carroll SB, 1995. Hairy and emc negatively regulate morphogenetic furrow progression in the Drosophila eye. Cell 80, 879–887. [DOI] [PubMed] [Google Scholar]
- Bruck S, Huber TB, Ingham RJ, Kim K, Niederstrasser H, Allen PM, Pawson T, Cooper JA, Shaw AS, 2006. Identification of a novel inhibitory actin-capping protein binding motif in CD2-associated protein. J Biol Chem 281, 19196–19203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell HL, Feiler CE, Ketosugbo KF, Hellerman MB, Nazzaro VL, Johnson RI, 2018. JNK is antagonized to ensure the correct number of interommatidial cells pattern the Drosophila retina. Dev Biol 433, 94–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagan RL, Ready DF, 1989a. The emergence of order in the Drosophila pupal retina. Dev Biol 136, 346–362. [DOI] [PubMed] [Google Scholar]
- Cagan RL, Ready DF, 1989b. Notch is required for successive cell decisions in the developing Drosophila retina. Genes Dev 3, 1099–1112. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega JA, Hofbauer A, 1977. Cell clones and pattern formation: On the lineage of photoreceptor cells in the compound eye ofDrosophila. Wilehm Roux Arch Dev Biol 181, 227–245. [DOI] [PubMed] [Google Scholar]
- Casares F, McGregor AP, 2021. The evolution and development of eye size in flies. Wiley Interdiscip Rev Dev Biol 10, e380. [DOI] [PubMed] [Google Scholar]
- Chan EH, Chavadimane Shivakumar P, Clement R, Laugier E, Lenne PF, 2017. Patterned cortical tension mediated by N-cadherin controls cell geometric order in the Drosophila eye. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanut F, Luk A, Heberlein U, 2000. A screen for dominant modifiers of ro(Dom), a mutation that disrupts morphogenetic furrow progression in Drosophila, identifies groucho and hairless as regulators of atonal expression. Genetics 156, 1203–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton-Perkins M, Brown NL, Cook TA, 2011a. The lens in focus: a comparison of lens development in Drosophila and vertebrates. Mol Genet Genomics 286, 189–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton-Perkins M, Whitaker SL, Fei Y, Xie B, Li-Kroeger D, Gebelein B, Cook T, 2011b. Prospero and Pax2 combinatorially control neural cell fate decisions by modulating Ras- and Notch-dependent signaling. Neural Dev 6, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CK, Chien CT, 1999. Negative regulation of atonal in proneural cluster formation of Drosophila R8 photoreceptors. Proc Natl Acad Sci U S A 96, 5055–5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophorou NA, Mende M, Lleras-Forero L, Grocott T, Streit A, 2010. Pax2 coordinates epithelial morphogenesis and cell fate in the inner ear. Dev Biol 345, 180–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinet C, Rauzi M, Lenne PF, Lecuit T, 2015. Local and tissue-scale forces drive oriented junction growth during tissue extension. Nat Cell Biol 17, 1247–1258. [DOI] [PubMed] [Google Scholar]
- Cordero J, Jassim O, Bao S, Cagan R, 2004. A role for wingless in an early pupal cell death event that contributes to patterning the Drosophila eye. Mech Dev 121, 1523–1530. [DOI] [PubMed] [Google Scholar]
- Cordero JB, Larson DE, Craig CR, Hays R, Cagan R, 2007. Dynamic Decapentaplegic signaling regulates patterning and adhesion in the Drosophila pupal retina. Development 134, 1861–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigall D, Walther RF, Rodriguez L, Fichelson P, Pichaud F, 2007. Hedgehog signaling is a principal inducer of Myosin-II-driven cell ingression in Drosophila epithelia. Dev Cell 13, 730–742. [DOI] [PubMed] [Google Scholar]
- Courcoubetis G, Ali S, Nuzhdin SV, Marjoram P, Haas S, 2019. Threshold response to stochasticity in morphogenesis. PLoS One 14, e0210088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis MW, Johnson RI, 2019. Dissection of the Drosophila Pupal Retina for Immunohistochemistry, Western Analysis, and RNA Isolation. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis MW, McGhie EW, Coolon JD, Johnson RI, 2020. Mask, a component of the Hippo pathway, is required for Drosophila eye morphogenesis. Dev Biol 464, 53–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Signore SJ, Cilla R, Hatini V, 2018. The WAVE Regulatory Complex and Branched F-Actin Counterbalance Contractile Force to Control Cell Shape and Packing in the Drosophila Eye. Dev Cell 44, 471–483 e474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delidakis C, Artavanis-Tsakonas S, 1992. The Enhancer of split [E(spl)] locus of Drosophila encodes seven independent helix-loop-helix proteins. Proc Natl Acad Sci U S A 89, 8731–8735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Yang L, Wen P, Lei H, Blount P, Pan D, 2020. Spectrin couples cell shape, cortical tension, and Hippo signaling in retinal epithelial morphogenesis. J Cell Biol 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokucu ME, Zipursky SL, Cagan RL, 1996. Atonal, rough and the resolution of proneural clusters in the developing Drosophila retina. Development 122, 4139–4147. [DOI] [PubMed] [Google Scholar]
- Dominguez M, 1999. Dual role for Hedgehog in the regulation of the proneural gene atonal during ommatidia development. Development 126, 2345–2353. [DOI] [PubMed] [Google Scholar]
- Dominguez M, Wasserman JD, Freeman M, 1998. Multiple functions of the EGF receptor in Drosophila eye development. Curr Biol 8, 1039–1048. [DOI] [PubMed] [Google Scholar]
- Dos-Santos N, Rubin T, Chalvet F, Gandille P, Cremazy F, Leroy J, Boissonneau E, Theodore L, 2008. Drosophila retinal pigment cell death is regulated in a position-dependent manner by a cell memory gene. Int J Dev Biol 52, 21–31. [DOI] [PubMed] [Google Scholar]
- Edwards M, Zwolak A, Schafer DA, Sept D, Dominguez R, Cooper JA, 2014. Capping protein regulators fine-tune actin assembly dynamics. Nat Rev Mol Cell Biol 15, 677–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero LM, Bischoff M, Freeman M, 2007. Myosin II regulates complex cellular arrangement and epithelial architecture in Drosophila. Dev Cell 13, 717–729. [DOI] [PubMed] [Google Scholar]
- Fernandes VM, McCormack K, Lewellyn L, Verheyen EM, 2014. Integrins regulate apical constriction via microtubule stabilization in the Drosophila eye disc epithelium. Cell Rep 9, 2043–2055. [DOI] [PubMed] [Google Scholar]
- Firth LC, Baker NE, 2009. Retinal determination genes as targets and possible effectors of extracellular signals. Dev Biol 327, 366–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foty RA, Steinberg MS, 2005. The differential adhesion hypothesis: a direct evaluation. Dev Biol 278, 255–263. [DOI] [PubMed] [Google Scholar]
- Frankfort BJ, Nolo R, Zhang Z, Bellen H, Mardon G, 2001. senseless repression of rough is required for R8 photoreceptor differentiation in the developing Drosophila eye. Neuron 32, 403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M, 1996. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell 87, 651–660. [DOI] [PubMed] [Google Scholar]
- Freeman M, 1997. Cell determination strategies in the Drosophila eye. Development 124, 261–270. [DOI] [PubMed] [Google Scholar]
- Gavish A, Barkai N, 2016. A two-step patterning process increases the robustness of periodic patterning in the fly eye. J Biol Phys 42, 317–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavish A, Shwartz A, Weizman A, Schejter E, Shilo BZ, Barkai N, 2016. Periodic patterning of the Drosophila eye is stabilized by the diffusible activator Scabrous. Nat Commun 7, 10461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemp IM, Carthew RW, Hilgenfeldt S, 2011. Cadherin-dependent cell morphology in an epithelium: constructing a quantitative dynamical model. PLoS Comput Biol 7, e1002115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillo-Hill BK, Wolff T, 2009. Dynamic cell shapes and contacts in the developing Drosophila retina are regulated by the Ig cell adhesion protein hibris. Dev Dyn 238, 2223–2234. [DOI] [PubMed] [Google Scholar]
- Grzeschik NA, Knust E, 2005. IrreC/rst-mediated cell sorting during Drosophila pupal eye development depends on proper localisation of DE-cadherin. Development 132, 2035–2045. [DOI] [PubMed] [Google Scholar]
- Harris TJC, 2018. Sculpting epithelia with planar polarized actomyosin networks: Principles from Drosophila. Semin Cell Dev Biol 81, 54–61. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Carthew RW, 2004. Surface mechanics mediate pattern formation in the developing retina. Nature 431, 647–652. [DOI] [PubMed] [Google Scholar]
- Heberlein U, Singh CM, Luk AY, Donohoe TJ, 1995. Growth and differentiation in the Drosophila eye coordinated by hedgehog. Nature 373, 709–711. [DOI] [PubMed] [Google Scholar]
- Heberlein U, Wolff T, Rubin GM, 1993. The TGF beta homolog dpp and the segment polarity gene hedgehog are required for propagation of a morphogenetic wave in the Drosophila retina. Cell 75, 913–926. [DOI] [PubMed] [Google Scholar]
- Heer NC, Martin AC, 2017. Tension, contraction and tissue morphogenesis. Development 144, 4249–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzler P, Bourouis M, Ruel L, Carteret C, Simpson P, 1996. Genes of the Enhancer of split and achaete-scute complexes are required for a regulatory loop between Notch and Delta during lateral signalling in Drosophila. Development 122, 161–171. [DOI] [PubMed] [Google Scholar]
- Hellerman MB, Choe RH, Johnson RI, 2015. Live-imaging of the Drosophila pupal eye. J Vis Exp, 52120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgenfeldt S, Erisken S, Carthew RW, 2008. Physical modeling of cell geometric order in an epithelial tissue. Proc Natl Acad Sci U S A 105, 907–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz U, Giebel B, Campos-Ortega JA, 1994. The basic-helix-loop-helix domain of Drosophila lethal of scute protein is sufficient for proneural function and activates neurogenic genes. Cell 76, 77–87. [DOI] [PubMed] [Google Scholar]
- Hsiao HY, Johnston RJ Jr., Jukam D, Vasiliauskas D, Desplan C, Rister J, 2012. Dissection and immunohistochemistry of larval, pupal and adult Drosophila retinas. J Vis Exp, 4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Shi B, Jarzynka MJ, Yiin JJ, D’Souza-Schorey C, Cheng SY, 2009. ADP-ribosylation factor 6 regulates glioma cell invasion through the IQ-domain GTPase-activating protein 1-Rac1-mediated pathway. Cancer Res 69, 794–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys D, Davidson AC, Hume PJ, Makin LE, Koronakis V, 2013. Arf6 coordinates actin assembly through the WAVE complex, a mechanism usurped by Salmonella to invade host cells. Proc Natl Acad Sci U S A 110, 16880–16885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarman AP, Grau Y, Jan LY, Jan YN, 1993. atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell 73, 1307–1321. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Grell EH, Ackerman L, Jan LY, Jan YN, 1994. Atonal is the proneural gene for Drosophila photoreceptors. Nature 369, 398–400. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Sun Y, Jan LY, Jan YN, 1995. Role of the proneural gene, atonal, in formation of Drosophila chordotonal organs and photoreceptors. Development 121, 2019–2030. [DOI] [PubMed] [Google Scholar]
- Jennings B, Preiss A, Delidakis C, Bray S, 1994. The Notch signalling pathway is required for Enhancer of split bHLH protein expression during neurogenesis in the Drosophila embryo. Development 120, 3537–3548. [DOI] [PubMed] [Google Scholar]
- Johnson RI, 2020. Adhesion and the Cytoskeleton in the Drosophila Pupal Eye, in: Singh A, Kango-Singh M (Eds.), Molecular Genetics of Axial Patterning, Growth and Disease in the Drosophila Eye. Springer, Cham, pp. 189–213. [Google Scholar]
- Johnson RI, Bao S, Cagan RL, 2012. Interactions between Drosophila IgCAM adhesion receptors and cindr, the Cd2ap/Cin85 ortholog. Dev Dyn 241, 1933–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RI, Sedgwick A, D’Souza-Schorey C, Cagan RL, 2011. Role for a Cindr-Arf6 axis in patterning emerging epithelia. Mol Biol Cell 22, 4513–4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RI, Seppa MJ, Cagan RL, 2008. The Drosophila CD2AP/CIN85 orthologue Cindr regulates junctions and cytoskeleton dynamics during tissue patterning. J Cell Biol 180, 1191–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafer J, Hayashi T, Maree AF, Carthew RW, Graner F, 2007. Cell adhesion and cortex contractility determine cell patterning in the Drosophila retina. Proc Natl Acad Sci U S A 104, 18549–18554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasza KE, Farrell DL, Zallen JA, 2014. Spatiotemporal control of epithelial remodeling by regulated myosin phosphorylation. Proc Natl Acad Sci U S A 111, 11732–11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel BE, Heberlein U, Rubin GM, 1990. The homeo domain protein rough is expressed in a subset of cells in the developing Drosophila eye where it can specify photoreceptor cell subtype. Genes Dev 4, 712–727. [DOI] [PubMed] [Google Scholar]
- Knust E, Schrons H, Grawe F, Campos-Ortega JA, 1992. Seven genes of the Enhancer of split complex of Drosophila melanogaster encode helix-loop-helix proteins. Genetics 132, 505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W, Loison O, Chavadimane Shivakumar P, Chan EH, Saadaoui M, Collinet C, Lenne PF, Clement R, 2019. Experimental validation of force inference in epithelia from cell to tissue scale. Sci Rep 9, 14647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo TH, Eipper BA, Donaldson JG, 2007. Arf6 recruits the Rac GEF Kalirin to the plasma membrane facilitating Rac activation. BMC Cell Biol 8, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP, 2012. Building an ommatidium one cell at a time. Dev Dyn 241, 136–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP, 2020. Catching the next wave: patterning of the eye by the morphogenetic furrow, in: Singh A, Kango-Singh M (Eds.), Molecular Genetics of Axial Patterning, Growth and Disease in Drosophila Eye, 2 ed. Springer Nature, Switzerland. [Google Scholar]
- Kumar JP, Tio M, Hsiung F, Akopyan S, Gabay L, Seger R, Shilo BZ, Moses K, 1998. Dissecting the roles of the Drosophila EGF receptor in eye development and MAP kinase activation. Development 125, 3875–3885. [DOI] [PubMed] [Google Scholar]
- Kunisch M, Haenlin M, Campos-Ortega JA, 1994. Lateral inhibition mediated by the Drosophila neurogenic gene delta is enhanced by proneural proteins. Proc Natl Acad Sci U S A 91, 10139–10143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langton PF, Colombani J, Chan EH, Wepf A, Gstaiger M, Tapon N, 2009. The dASPP-dRASSF8 complex regulates cell-cell adhesion during Drosophila retinal morphogenesis. Curr Biol 19, 1969–1978. [DOI] [PubMed] [Google Scholar]
- Larson DE, Johnson RI, Swat M, Cordero JB, Glazier JA, Cagan RL, 2010. Computer simulation of cellular patterning within the Drosophila pupal eye. PLoS Comput Biol 6, e1000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DE, Liberman Z, Cagan RL, 2008. Cellular behavior in the developing Drosophila pupal retina. Mech Dev 125, 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EC, Hu X, Yu SY, Baker NE, 1996. The scabrous gene encodes a secreted glycoprotein dimer and regulates proneural development in Drosophila eyes. Mol Cell Biol 16, 1179–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EC, Yu SY, Baker NE, 2000. The scabrous protein can act as an extracellular antagonist of notch signaling in the Drosophila wing. Curr Biol 10, 931–934. [DOI] [PubMed] [Google Scholar]
- Lee JR, Urban S, Garvey CF, Freeman M, 2001. Regulated intracellular ligand transport and proteolysis control EGF signal activation in Drosophila. Cell 107, 161–171. [DOI] [PubMed] [Google Scholar]
- Letizia A, He D, Astigarraga S, Colombelli J, Hatini V, Llimargas M, Treisman JE, 2019. Sidekick Is a Key Component of Tricellular Adherens Junctions that Acts to Resolve Cell Rearrangements. Dev Cell 50, 313–326 e315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levayer R, Lecuit T, 2013. Oscillation and polarity of E-cadherin asymmetries control actomyosin flow patterns during morphogenesis. Dev Cell 26, 162–175. [DOI] [PubMed] [Google Scholar]
- Li K, Baker NE, 2018. Regulation of the Drosophila ID protein Extra macrochaetae by proneural dimerization partners. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Baker NE, 2019. Transcriptional and post-transcriptional regulation of extra macrochaetae during Drosophila adult peripheral neurogenesis. Dev Biol 449, 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Baker NE, 2001. Proneural enhancement by Notch overcomes Suppressor-of-Hairless repressor function in the developing Drosophila eye. Curr Biol 11, 330–338. [DOI] [PubMed] [Google Scholar]
- Ligoxygakis P, Yu SY, Delidakis C, Baker NE, 1998. A subset of notch functions during Drosophila eye development require Su(H) and the E(spl) gene complex. Development 125, 2893–2900. [DOI] [PubMed] [Google Scholar]
- Lim J, Jafar-Nejad H, Hsu YC, Choi KW, 2008. Novel function of the class I bHLH protein Daughterless in the negative regulation of proneural gene expression in the Drosophila eye. EMBO Rep 9, 1128–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubensky DK, Pennington MW, Shraiman BI, Baker NE, 2011. A dynamical model of ommatidial crystal formation. Proc Natl Acad Sci U S A 108, 11145–11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melicharek D, Shah A, DiStefano G, Gangemi AJ, Orapallo A, Vrailas-Mortimer AD, Marenda DR, 2008. Identification of novel regulators of atonal expression in the developing Drosophila retina. Genetics 180, 2095–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meserve JH, Duronio RJ, 2017. A population of G2-arrested cells are selected as sensory organ precursors for the interommatidial bristles of the Drosophila eye. Dev Biol 430, 374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DT, Cagan RL, 1998. Local induction of patterning and programmed cell death in the developing Drosophila retina. Development 125, 2327–2335. [DOI] [PubMed] [Google Scholar]
- Mlodzik M, Baker NE, Rubin GM, 1990. Isolation and expression of scabrous, a gene regulating neurogenesis in Drosophila. Genes Dev 4, 1848–1861. [DOI] [PubMed] [Google Scholar]
- Monserrate JP, Brachmann CB, 2007. Identification of the death zone: a spatially restricted region for programmed cell death that sculpts the fly eye. Cell Death Differ 14, 209–217. [DOI] [PubMed] [Google Scholar]
- Nagaraj R, Banerjee U, 2007. Combinatorial signaling in the specification of primary pigment cells in the Drosophila eye. Development 134, 825–831. [DOI] [PubMed] [Google Scholar]
- Pennington MW, Lubensky DK, 2010. Switch and template pattern formation in a discrete reaction-diffusion system inspired by the Drosophila eye. Eur Phys J E Soft Matter 33, 129–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepple KL, Atkins M, Venken K, Wellnitz K, Harding M, Frankfort B, Mardon G, 2008. Two-step selection of a single R8 photoreceptor: a bistable loop between senseless and rough locks in R8 fate. Development 135, 4071–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro D, Bellaiche Y, 2018. Mechanical Force-Driven Adherens Junction Remodeling and Epithelial Dynamics. Dev Cell 47, 3–19. [DOI] [PubMed] [Google Scholar]
- Powell LM, Deaton AM, Wear MA, Jarman AP, 2008. Specificity of Atonal and Scute bHLH factors: analysis of cognate E box binding sites and the influence of Senseless. Genes Cells 13, 915–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell PA, Wesley C, Spencer S, Cagan RL, 2001. Scabrous complexes with Notch to mediate boundary formation. Nature 409, 626–630. [DOI] [PubMed] [Google Scholar]
- Quan XJ, Yuan L, Tiberi L, Claeys A, De Geest N, Yan J, van der Kant R, Xie WR, Klisch TJ, Shymkowitz J, Rousseau F, Bollen M, Beullens M, Zoghbi HY, Vanderhaeghen P, Hassan BA, 2016. Post-translational Control of the Temporal Dynamics of Transcription Factor Activity Regulates Neurogenesis. Cell 164, 460–475. [DOI] [PubMed] [Google Scholar]
- Querenet M, Goubard V, Chatelain G, Davoust N, Mollereau B, 2015. Spen is required for pigment cell survival during pupal development in Drosophila. Dev Biol 402, 208–215. [DOI] [PubMed] [Google Scholar]
- Rauzi M, Lenne PF, Lecuit T, 2010. Planar polarized actomyosin contractile flows control epithelial junction remodelling. Nature 468, 1110–1114. [DOI] [PubMed] [Google Scholar]
- Ready DF, Hanson TE, Benzer S, 1976. Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol 53, 217–240. [DOI] [PubMed] [Google Scholar]
- Reiter C, Schimansky T, Nie Z, Fischbach KF, 1996. Reorganization of membrane contacts prior to apoptosis in the Drosophila retina: the role of the IrreC-rst protein. Development 122, 1931–1940. [DOI] [PubMed] [Google Scholar]
- Rodrigues AB, Werner E, Moses K, 2005. Genetic and biochemical analysis of the role of Egfr in the morphogenetic furrow of the developing Drosophila eye. Development 132, 4697–4707. [DOI] [PubMed] [Google Scholar]
- Sawamoto K, Okano H, Kobayakawa Y, Hayashi S, Mikoshiba K, Tanimura T, 1994. The function of argos in regulating cell fate decisions during Drosophila eye and wing vein development. Dev Biol 164, 267–276. [DOI] [PubMed] [Google Scholar]
- Sawamoto K, Taguchi A, Hirota Y, Yamada C, Jin MH, Okano H, 1998. Argos induces programmed cell death in the developing Drosophila eye by inhibition of the Ras pathway. Cell Death Differ 5, 262–270. [DOI] [PubMed] [Google Scholar]
- Seppa MJ, Johnson RI, Bao S, Cagan RL, 2008. Polychaetoid controls patterning by modulating adhesion in the Drosophila pupal retina. Dev Biol 318, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes Sde M, Blankenship JT, Weitz O, Farrell DL, Tamada M, Fernandez-Gonzalez R, Zallen JA, 2010. Rho-kinase directs Bazooka/Par-3 planar polarity during Drosophila axis elongation. Dev Cell 19, 377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes Sde M, Mainieri A, Zallen JA, 2014. Rho GTPase and Shroom direct planar polarized actomyosin contractility during convergent extension. J Cell Biol 204, 575–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer SA, Powell PA, Miller DT, Cagan RL, 1998. Regulation of EGF receptor signaling establishes pattern across the developing Drosophila retina. Development 125, 4777–4790. [DOI] [PubMed] [Google Scholar]
- Steinberg MS, 1963. Reconstruction of tissues by dissociated cells. Some morphogenetic tissue movements and the sorting out of embryonic cells may have a common explanation. Science 141, 401–408. [DOI] [PubMed] [Google Scholar]
- Sun Y, Jan LY, Jan YN, 1998. Transcriptional regulation of atonal during development of the Drosophila peripheral nervous system. Development 125, 3731–3740. [DOI] [PubMed] [Google Scholar]
- Tamada M, Farrell DL, Zallen JA, 2012. Abl regulates planar polarized junctional dynamics through beta-catenin tyrosine phosphorylation. Dev Cell 22, 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka-Matakatsu M, Du W, 2008. Direct control of the proneural gene atonal by retinal determination factors during Drosophila eye development. Dev Biol 313, 787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang VW, Brieher WM, 2012. alpha-Actinin-4/FSGS1 is required for Arp2/3-dependent actin assembly at the adherens junction. J Cell Biol 196, 115–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tea JS, Cespedes A, Dawson D, Banerjee U, Call GB, 2014. Dissection and mounting of Drosophila pupal eye discs. J Vis Exp, e52315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson A, Ready DF, 1987. Neuronal differentiation in Drosophila ommatidium. Dev Biol 120, 366–376. [DOI] [PubMed] [Google Scholar]
- Treisman JE, 2013. Retinal differentiation in Drosophila. Wiley Interdiscip Rev Dev Biol 2, 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott RL, Kalive M, Paroush Z, Bidwai AP, 2001. Drosophila melanogaster casein kinase II interacts with and phosphorylates the basic helix-loop-helix proteins m5, m7, and m8 derived from the Enhancer of split complex. J Biol Chem 276, 2159–2167. [DOI] [PubMed] [Google Scholar]
- Urban S, Lee JR, Freeman M, 2001. Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell 107, 173–182. [DOI] [PubMed] [Google Scholar]
- Vidal M, Larson DE, Cagan RL, 2006. Csk-deficient boundary cells are eliminated from normal Drosophila epithelia by exclusion, migration, and apoptosis. Dev Cell 10, 33–44. [DOI] [PubMed] [Google Scholar]
- Warner SJ, Longmore GD, 2009. Distinct functions for Rho1 in maintaining adherens junctions and apical tension in remodeling epithelia. J Cell Biol 185, 1111–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrington SJ, Strutt H, Strutt D, 2013. The Frizzled-dependent planar polarity pathway locally promotes E-cadherin turnover via recruitment of RhoGEF2. Development 140, 1045–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T, 2007. Dissection techniques for pupal and larval Drosophila eyes. CSH Protoc 2007, pdb prot4715. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF, 1991. Cell death in normal and rough eye mutants of Drosophila. Development 113, 825–839. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF, 1993. Pattern formation in the Drosophila retina. The Development of Drosophila melanogaster, edited by Bate M and Arias AM. Cold Spring Harbor Laboratory Press, Cold Spring Harbor., 1277–1325. [Google Scholar]
- Yang L, Baker NE, 2001. Role of the EGFR/Ras/Raf pathway in specification of photoreceptor cells in the Drosophila retina. Development 128, 1183–1191. [DOI] [PubMed] [Google Scholar]
- Yashiro H, Loza AJ, Skeath JB, Longmore GD, 2014. Rho1 regulates adherens junction remodeling by promoting recycling endosome formation through activation of myosin II. Mol Biol Cell 25, 2956–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JC, Fernandez-Gonzalez R, 2016. Local mechanical forces promote polarized junctional assembly and axis elongation in Drosophila. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SY, Yoo SJ, Yang L, Zapata C, Srinivasan A, Hay BA, Baker NE, 2002. A pathway of signals regulating effector and initiator caspases in the developing Drosophila eye. Development 129, 3269–3278. [DOI] [PubMed] [Google Scholar]
- Zaessinger S, Zhou Y, Bray SJ, Tapon N, Djiane A, 2015. Drosophila MAGI interacts with RASSF8 to regulate E-Cadherin-based adherens junctions in the developing eye. Development 142, 1102–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Du W, 2015. Groucho restricts rhomboid expression and couples EGFR activation with R8 selection during Drosophila photoreceptor differentiation. Dev Biol 407, 246–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Ranade S, Cai CQ, Clouser C, Pignoni F, 2006. Direct control of neurogenesis by selector factors in the fly eye: regulation of atonal by Ey and So. Development 133, 4881–4889. [DOI] [PubMed] [Google Scholar]
- Zhao J, Bruck S, Cemerski S, Zhang L, Butler B, Dani A, Cooper JA, Shaw AS, 2013. CD2AP links cortactin and capping protein at the cell periphery to facilitate formation of lamellipodia. Mol Cell Biol 33, 38–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Owen MR, Mao Y, 2016. The spatiotemporal order of signaling events unveils the logic of development signaling. Bioinformatics 32, 2313–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]


