Abstract
The angiotensin Converting Enzyme 2 (ACE2) receptor is a key component of the renin-angiotensin-aldesterone system (RAAS) that mediates numerous effects in the cardiovascular system. It is also the cellular point of contact for the coronavirus spike protein. Cleavage of the receptor is both important to its physiological function as well as being necessary for cell entry by the virus. Shedding of ACE2 by the metalloprotease ADAM17 releases a catalytically active soluble form of ACE2, but cleavage by the serine protease TMPRSS2 is necessary for virion internalization. Complicating the issue is the observation that circulating ACE2 can also bind to the virus effectively blocking attachment to the membrane-bound receptor. This work investigates the possibility that the inflammatory response to coronavirus infection can abrogate shedding by ADAM17, thereby favoring cleavage by TMPRSS2 and thus cell entry by the virion.
Keywords: COVID-19, ACE2, ADAM17, TMPRSS2, Heat shock response, Small Hsp
Graphical abstract
1. Introduction
Similar to the earlier SARS-CoV (Severe Acute Respiratory Syndrome CoronaVirus) SARS-CoV-2, the causative agent of COVID-19, invades the cell through attachment of the spike (S) protein to the ACE2 receptor, leading to internalization of the virion [1]. ACE2 is a type I transmembrane protein that can exist in either cellular (membrane-bound) or circulating (soluble) forms [2]. Circulating ACE2 is cleaved, or shed, from full length ACE2 on the cell membrane by ADAM17 (A Disintegrin And Metalloprotease), and it has been demonstrated that recombinant soluble ACE2 effectively blocks the association of the SARS-CoV spike protein with ACE2 [3], suggesting that high levels of shed ACE2 could inhibit COVID-19 infectivity. By contrast the serine protease TMPRSS2 (TransMembranePRotease2) augments the viral infectivity of SARS-CoV by both priming the S protein for membrane fusion as well as cleaving the ACE2 receptor [4]. Cleavage of ACE2 by ADAM17 was found to be dispensible for activation of SARS-CoV cell entry, but it remains unclear how TMPRSS2 transcends ADAM17 for cleavage of ACE2 following infection by the virus.
ADAM17 is expressed as a zymogen, where autoinhibition by a large prodomain maintains the enzyme in a closed, or inactive, state [5]. Conformational displacement during cellular routing yields an open the conformation, allowing for proteolytic degradation by intracellular proteinases to generate the active enzyme. The remaining ectodomain consists of catalytic, a disintegrin and a membrane proximal domain (MPD), as well as a CANDIS (Conserved Adam seventeeN Dynamic Interaction Sequence) adjacent to the juxtamembrane region, Fig. S1A. Phosphatidylserine (PS) is the most abundant negatively charged phospholipid in the cell membrane and its translocation to the outer leaflet of the lipid bilayer is implicated in ADAM sheddase activity [6]. The PS-binding motif is a triplet cluster of cationic residues, R_KK in ADAM10 [7] and RK_K in ADAM17 [8], that mediates the interaction of the protease with the membrane. In ADAM17 this motif is located in the MPD. Given that most ADAM17 substrates are cleaved at positions very close to the membrane of the same cell [9], and that increased shedding in vivo does not follow from overexpression of the protease [10], this leads to a scenario for PS-activation whereby interaction of the cationic motif in the MPD with the PS-exposed bilayer reorients the catalytic domain proximate to both its substrate and the cell membrane, Fig. S1B [11]. In the quiescent state the protease is assumed to orient freely at the cell surface and coordination of these spatio-temporal events is critical for PS-activation. Dysregulation of the sheddase activity of ADAM17 through disruption of this repositioning of the catalytic domain would be expected to influence the severity of COVID-19, since ADAM17 is responsible for the regulated shedding of ACE2.
Fever is a complex cytokine-mediated physiological response to infection, and the initial presentation of fever in COVID-19 is likely a manifestation of the body's immune response to viral replication. However a persistent high fever (>39 °C) is considered to be an indicator of severe infection [12]. The heat shock response (HSR) is a highly conserved evolutionary response not just to thermal stress but is also critical to the resolution of the inflammatory response to infection. Whereas the major risk factors for fatal outcomes in COVID-19 patients share the characteristic of being inflammatory diseases linked to a defective HSR, the small heat shock proteins (sHsp) Hsp20 (or HspB6) and Hsp27 (or HspB1) exhibit widely recognized anti-inflammatory capabilities linked to cardiprotection [13], protection against free radicals and environmental toxins [14] and against the consequences of viral infection [15]. The sHsp subunit has a mass of 14–40 kDa and is characterized by a highly conserved α-crystallin domain (ACD) flanked by an N-terminal domain (NTD) and a C-terminal extension (CTE). sHsps form large polydisperse assemblies of varying subunit size through the extension of the CTE from one homodimer into the α-crystallin domain of the neighboring one, a motif called “patching” [16]. A temperature-regulated subunit exchange releases the chaperone-active dimer from the oligomeric assembly, exposing substrate binding sites that allow dynamic and reversible interaction with a wide array of targets (over 450 in the case of Hsp27), providing stress tolerance through the maintenance of cellular proteostasis [17]. In the case of tubulin and F-actin this interaction serves to stabilize the cytoskeleton by affecting the spatio-temporal organization of the components and preventing reorganization [18]. We have previously demonstrated how the M. tuberculosis Hsp Acr can exploit the host innate response by dysregulating shedding of the inflammatory cytokine CXCL16 by ADAM10 [19]. Among the over 80 cell-bound substrates of ADAM17 at least 9 are cytokines or cytokine receptors that have been reported as triggering inflammation when dysregulated [[20], [21], [22]].
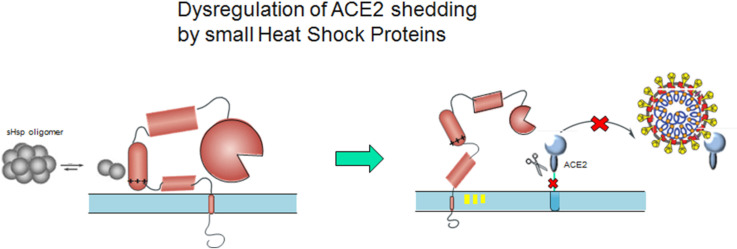
This work investigates whether the HSR in COVID-19 can also serve to dysregulate the cleavage of ACE2 by ADAM17. By mapping substrate binding sites in the ACD of human Hsp20 to non-canonical patching sequences in ADAM17 we postulate that temporal accessibility of these sites allows for dynamic formation of an ADAM17-Hsp20 complex. Modeling of the full ectodomain of ADAM17 explains how this association can disrupt the repositioning of the ADAM17 catalytic domain, thus preventing ACE2 cleavage from the membrane, Fig. S1C.
2. Methods
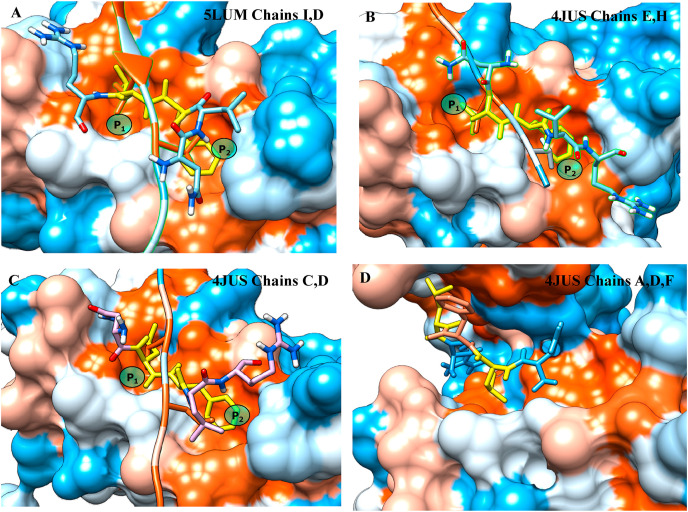
Crystal structures for a variety of human Hsp20 tetramers stabilized by interaction of the ACD with patching sequences contained within the NTD are available from the RCSB (www.rcsb.org) as Protein Data Bank (pdb) code entries 5LUM [23] and 4JUS [24]. After adding hydrogens the structures were subjected to a short energy minimization using the CHARMm force field [25] as implemented in the Discovery Studio program suite. Protein-peptide docking using the DINC 2.0 (Docking INCrementally) protocol was used to characterize the strength of the protein-protein interactions (PPI) responsible for the putative complexation of sHsps with ADAM17. DINC is an incremental, meta-docking method optimized for the docking of longer peptides [26]. The approach was validated by confirming that for the experimental assemblies the lowest energy pose obtained by docking differed from the crystal structure by a heavy-atom root mean square difference (RMSD) of less than 1.0 Å, Fig. 1 .
Fig. 1.
The lowest energy poses obtained by docking of the experimental chain (shown as ribbon structure with the patching residue in redstick) and the ADAM17 621NLFLR625 sequence (shown as stick representation with the patching residues shown in yellow for Hsp20 A: from reference 23; B,C: from reference 24; and D: the unique patching sequence only predicted for 621NLFLR625 (ACR surface colored using the Eisenberg hydrophobicity scale where red is the most hydrophobic and blue is the least). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
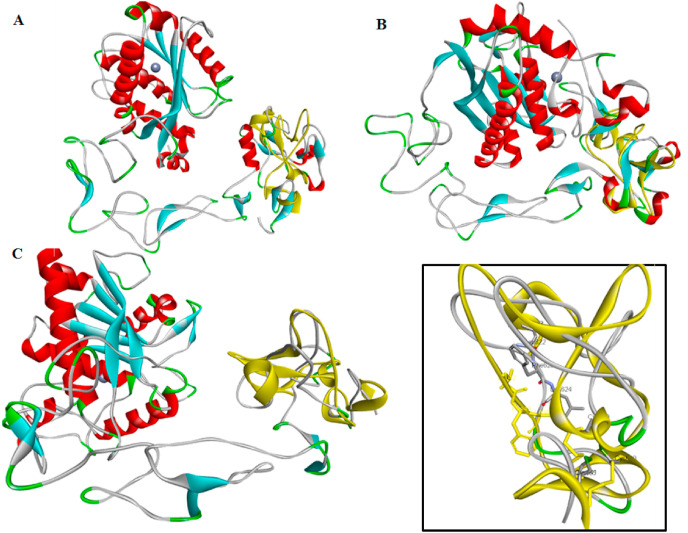
Homology modeling was used to generate a complete structure of the ADAM17 ectodomain. Two models were generated, one using a recently characterized x-ray structure of the ADAM10 ectodomain [27], available as Protein Data Bank (pdb) code entry 6BE6, and the other using vascular apoptosis-inducing protein-1 (VAP1) [28], a snake venom homolog of mammalian ADAM (pdb id 2ERO) as templates. Whereas the VAP1 template, Fig. 2 A, displays the unique C-shaped structure implicated in ADAM17 regulation [11], the ADAM10 ectodomain, Fig. 2B, exhibits a more compact arrangement where part of the MPD occludes key elements of the catalytic domain. After insertion and refinement of missing residues using the MODELLER loop refinement algorithm [29] the query sequence of hADAM17 (Uniprot ID P78536) was aligned to the template, and the alignment used to create a set of 20 homology models. The best model is selected as the structure with the smallest value of the normalized discrete optimized molecule energy (DOPE) [30]. Finally the catalytic domain was replaced by grafting the x-ray crystal structure of the domain (1BKC) [31] in place of the homology model, hydrogens were added and the structures subjected to energy minimization using the CHARMm force field.
Fig. 2.
A ribbon structure of VAP1 ectodomain, available from RCSB as id 2ERO, aligned with the closed conformation of the MPD of ADAM17, shown in yellow; B structure of ADAM10 ectodomain, available from RCSB as id 6BE6, aligned with the closed conformation of the MPD of ADAM17, shown in yellow; C homology model of ADAM17 ectodomain structure generated using the VAP1 template, inset homology-modeled MPD aligned with the closed conformation of the MPD of ADAM17, shown in yellow, with the 622LFL624 patching sequences shown in stick form. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
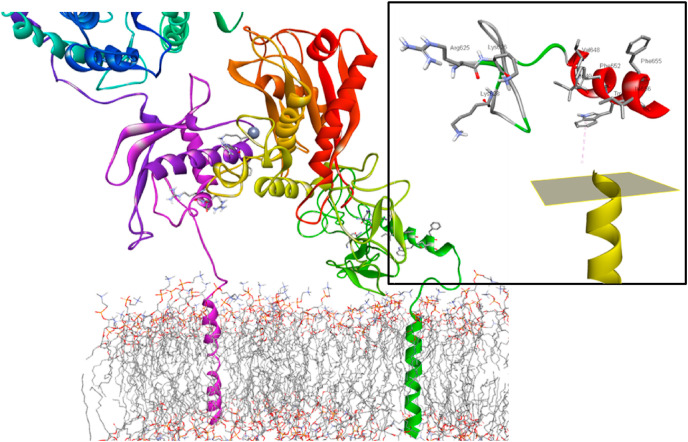
The ClusPro docking server [32] was employed to generate the ACE2/ADAM17 complex, using the homology models for the ADAM17 ectodomain and the complete ACE2 receptor, available as pdb id 6M1D [33]. This direct, rigid-body protocol involves Fast Fourier Transform (FFT)-based global sampling of the rotational/translational space, followed by clustering of the one thousand lowest-energy structures, finishing with a CHARMM minimization to remove steric clashes. Ranking is based on cluster population, using the cluster center as the model complex. The structure generated in Fig. 3 represents the center of the most populated cluster when scored using either electrostatic-favored, hydrophobic-favored, Van der Waals-electrostatic-favored, or balanced weight coefficients. The ACE2 attractor set included residues Arg652, Lys657, Arg659, Arg708, Ser709 and Arg710. The attractor set for the ligand was chosen as the His405, Glu406, His409, Gly412 and His415 residues that incorporate the zinc-binding consensus motif for ADAM17.
Fig. 3.
The homology-modeled ADAM17 ectodomain, generated using the snake venom homolog VAP1 as template and with CANDIS and juxtamembrane regions added, complexed to the ACE2, available from RCSB as id 6M1D, and embedded in a POPC bilayer as described in methods; inset orientation of the RK_K side chains and 652FWDF655 CANDIS segment relative to the plane of the membrane.
The orientation of the complex relative to the cell membrane was determined by embedding the ACE2 transmembrane (TM) region of the complex in an explicit membrane configuration containing a 1-palmitoyl-2-oleoylphosphatidylcholine (POPC) bilayer. The optimal orientation was defined as the tilt angle of the TM relative to the membrane normal that corresponds to the minimum solvation energy [34]. After embedding and orienting the ADAM17 TM, the C-terminus of ADAM17 was extended to add the α-helical CANDIS domain [35]. Finally the CANDIS and TM domains of ADAM17 were connected using the MODELLER loop refinement algorithm.
3. Results and discussion
The quaternary structure of sHsps derives from the extension of the CTE of one homodimer into the ACD of the neighboring subunit, patching a hydrophobic groove in the ACD with a conserved IXI/V motif. This hydrophobic groove is a cleft defined by the β4 and β8 sheets running anti-parallel. Because of two short 310 helices the CTE must run at an angle to the β4/β8 strands covering one pocket, P 1, defined in Hsp20 by the side chains of Vβ4/Vβ4/Vβ8/Sβ8, and a second pocket, P 2 defined by Iβ4/Vβ4/Sβ8/Lβ8. In addition to this canonical patch mediated by the CTE crystal structures have revealed ACD tetrameric assemblies for a variety of human sHsps that arise from extensive inter-dimer patching of the hydrophobic groove by tripeptide motifs found in the NTD directly adjacent to the ACD, that resemble but are distinct from the canonical IXI sequence. Table 1 . Because of the angle of the patch to the β4/β8 fold some of these sequences only cover a single pocket. Similar sequences (shown in bold) are to be found in both ADAM10 (651DGPLAR_KK659) and ADAM17 (621NLFLRK_K628) proximate to the cationic sequence identified as responsible for PS-activation. By facilitating a protein-protein interaction (PPI) between the ADAM10 and the pathogenic sHsp Mtb Acr we have previously shown how these residues provide a mechanism for the heat shock activation of ADAM10, providing an immunomodulatory role for the mycobacterium in the formation of the tubercular granuloma [19]. The ΔΔGbinding, i.e. free energy of binding for the comparable ADAM17 sequence relative to that calculated for the experimental patch, Table 1, confirms the feasibility of forming such a dynamic sHsp/ADAM17 complex. However even with leucine residues isomeric to the canonical patching isoleucine the flexibility of the Phe632 methylene group allows the aryl ring to occupy one of the pockets in preference to the leucine, Fig. 1A–C. This same flexibility also allows for a unique double patching scheme where the leucine side chains cover the P1 pockets of two different ACD subunits, Fig. 1D, a motif not mirrored in the crystal structures. Homology models of the ectodomain provide insight as to how these potential interactions might affect the proteolytic activity of ADAM17.
Table 1.
Non-canonical patching of the β4/β8 groove in the α-crystallin domain of Hsp20.
| Structure | ΔΔGbinding (kcals mol−1) | Patching residues (bold) |
||||
|---|---|---|---|---|---|---|
| Pocket P1 | Pocket P2 | |||||
| hADAM17 peptide NLFLR+5LUM (1) | Arg625 | Leu624 | – | Phe623 | Leu622 | |
| hADAM17 peptide NLFLR+4JUS (2) | Asn621 | Leu622 | – | Phe623 | Leu624 | |
| Human Hsp20 ACD complexa (5LUM)(3) | 1–3 = +0.2 | Pro4 | Val5 | Pro6 | Val7 | Gln8 |
| Human Hsp20b chains C + D (4JUS) (4) | 2–4 = +0.2 | Ala65 | Val64 | Pro63 | Leu62 | Ala61 |
| Human Hsp20b chain, E + H (4JUS) (5) | 2–5 = −1.2 | Gln66 | – | Val67 | Pro68 | Thr69 |
Patching as reported in Ref. 23.
Patching between different ΔN56 oligomers, Ref. 24.
The VAP1 and ADAM10 ectodomain structures provide markedly different templates for homology modeling of ADAM17. Comparing the MPDs of both templates with the ADAM17 MPD structure previously characterized using heteronuclear NMR spectroscopy [36] it can be seen that there is dramatically better alignment for the more compact ADAM10 structure, Fig. 2B, than for the C-shaped VAP1 structure, Fig. 2A. The MPD is known to play the role of a molecular switch for the shedding activity of ADAM17, with the NMR-based structure representing the inactive, or closed, conformation. This closed conformation differs from the open, or active, conformation of the MPD most critically with respect to two disulfide linkages, present as C600–C635 and C630–C641 in the closed conformation but as C600–C630 and C635–C641 in the open state. Interestingly protein disulfide isomerase (PDI), a stress protein essential in maintaining homeostasis [37], induces a conformational change in the MPD from the active to inactive state, but not the reverse. While the MPD of ADAM10 has disulfide linkages (C594–C639 and C632–C645) aligned to those found in the inactive state of ADAM17, two of the disulfide linkages in the crystal structure of VAP1, C556–C598 and C592–C603 show no such alignment, suggesting a closed state in the former and an open, or active, conformation for the latter. Congruent with this analysis is the result that whereas no functional complex was generated for the docking of the ADAM10-generated homology model to ACE2, the model generated using the snake venom homologVAP1 successfully docked to the ACE2 neck region that encompasses residues of the cleavage motif, Fig. 3.
The interface contacts for the ACE2/ADAM17 complex are summarized in Table S1, and can be characterized as representing three distinct regions centering on the ACE2 residues Lys657, Arg652 and Arg710 respectively, Fig. S2. While sharing many of the same contacts found in an earlier model of the complex, composed of the ACE2 receptor and just the catalytic domain of ADAM17 [38], the model in Fig. 3 displays more contacts involving a greater variety of intermolecular forces. This difference is most likely the result of our use of only 11 attractor residues, versus 22 in the earlier model, allowing for a less restrictive sampling of the conformational space. Comparative analysis indicates that these model complexes are essentially interconvertible through a simple translation the length of one turn of the a-helical interface. While both models confirm extensive contacts with the ACE2 residues Arg652, Lys657 and Arg710, previously identified as essential for binding recognition, neither predicts any participation for residues Arg708 and Ser709, the proposed site for cleavage of ACE2 by ADAM17 [39].
The orientation of the 622LFL624 sequence capable of patching ADAM17 to a sHsp is shown in Fig. 2 inset for both the closed state of the MPD, as characterized by NMR, and in the MPD of the homology-modeled ADAM17 ectodoamin capable of docking to ACE2. To better understand the effect of sHsp interaction on ADAM17 proteolysis the orientation of the complex relative to the cell membrane was modeled by extending the C-terminus of ADAM17 to add the α-helical CANDIS domain and connected to the ADAM17 TM embedded an in a POPC bilayer. As well as being involved in substrate recognition the CANDIS segment also plays a role in regulating sheddase activity, with the hydrophobic side of the CANDIS helix being essential for ADAM17 activity [40]. This effect is primarily hydrophobic, with the 652FWDF655 segment having a significant impact on membrane binding and ADAM17 activity. In our membrane-bound model of the ACE2/ADAM17 complex these hydrophobic residues are more or less parallel to the plane of the membrane, as are the RK_K side chains critical for PS-activation, Fig. 3 inset. A 90° rotation coupled with a membrane-directional translation would bring both segments into direct contact with the bilayer. Under a rigid-body approximation such a movement would also translate the zinc-binding motif away from Arg652 and Lys657 and towards Arg708 and Ser709, the proposed site of ACE2 cleavage by ADAM17.
The model in Fig. 3 provides a mechanism for the dysregulation of PS-mediated ACE2 shedding by ADAM17. Release of chaperone-active dimers from oligomeric SHsp assemblies exposes a hydrophobic cleft within the ACD. Patching of this cleft by a sequence in the open conformation of the ADAM17 MPD prevents the re-orientation of both the RK_K cationic sequence and the CANDIS helix necessary for proteolysis. In this scenario proper positioning of the ADAM17 active site proximate to the ACE2 cleavage site is inhibited even as substrate recognition remains unaffected. Other scenarios are also feasible, such as the possibility that interaction of the closed conformation of the MPD with the sHsp prevents conversion of ADAM17 to the active form by a so-far undiscovered isomerase. Whatever the underlying reality the analysis presented here provides a nexus for understanding how ACE2, ADAM17, PS and the HSR can cumulatively facilitate SARS-CoV-2 infectivity as well as exacerbate the inflammatory effects of COVID-19.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors wish to acknowledge the Welch Foundation (Grant# BH-0018) for its continuing support of the Chemistry Department at St. Edward's University.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrc.2021.08.040.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U.S.A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gross L.Z.F., Sacerdoti M., Piiper A., Zeuzem S., Leroux A.E., Biondi ACE2 R.M. The receptor that enables infection by SARS-CoV-2: biochemistry, structure, allostery and evaluation of the potential development of ACE2 modulators ChemMedChem. 2020;15:1682–1690. doi: 10.1002/cmdc.202000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heurich A., Hofmann-Winkler H., Gierer S., Liepold T., Jahn O., Pöhlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2014;88:1293–1307. doi: 10.1128/JVI.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leonard J.D., Lin F., Milla M.E. Chaperone-like properties of the prodomain of TNFalpha-converting enzyme (TACE) and the functional role of its cysteine switch. Biochem. J. 2005;387:797–805. doi: 10.1042/BJ20041727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sommer A., Kordowski F., Büch J., Maretzky T., Evers A., Andrä J., Düsterhöft S., Michalek M., Lorenzen I., Somasundaram P., Tholey A. Phosphatidylserine exposure is required for ADAM17 sheddase function. Nat. Commun. 2016;7:1–14. doi: 10.1038/ncomms11523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bleibaum F., Sommer A., Veit M., Rabe B., Andrä J., Kunzelmann K., Bhakdi S. ADAM10 sheddase activation is controlled by cell membrane asymmetry. J. Mol. Cell Biol. 2019;11:979–993. doi: 10.1093/jmcb/mjz008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sommer A., Bhakdi S., Reiss K. How membrane asymmetry regulates ADAM17 sheddase function. Cell Cycle. 2016;15:2995. doi: 10.1080/15384101.2016.1211449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horiuchi K. A brief history of tumor necrosis factor alpha--converting enzyme: an overview of ectodomain shedding. Keio J. Med. 2013;62:29–36. doi: 10.2302/kjm.2012-0003-re. [DOI] [PubMed] [Google Scholar]
- 10.Yoda M., et al. Systemic overexpression of TNFalpha-converting enzyme does not lead to enhanced shedding activity in vivo. PloS One. 2013;8 doi: 10.1371/journal.pone.0054412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grötzinger J., Lorenzen I., Düsterhöft S. Molecular insights into the multilayered regulation of ADAM17: the role of the extracellular region. Biochim. Biophys. Acta Mol. Cell Res. 2017;1864:2088–2095. doi: 10.1016/j.bbamcr.2017.05.024. [DOI] [PubMed] [Google Scholar]
- 12.Deborah H.L., Ng, Yee Choy Chiaw, Chan Yi-Hao, Young Barnaby E., Fong Siew-Wai, Ng Lisa F.P., Laurent Renia, Lye David C., Chia Po Ying. National centre for infectious diseases COVID-19 outbreak research team, fever patterns, cytokine profiles, and outcomes in COVID-19. Open Forum Infectious Diseases. 2020;7:375. doi: 10.1093/ofid/ofaa375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwards H.V., Cameron R.T., Baillie G.S. The emerging role of HSP20 as a multifunctional protective agent. Cell. Signal. 2011;23:1447–1454. doi: 10.1016/j.cellsig.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Ferns G., Shams S., Shafi S. Heat shock protein 27: its potential role in vascular disease. Int. J. Exp. Pathol. 2006;87:253–274. doi: 10.1111/j.1365-2613.2006.00484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vahabpour R., Soleymani S., Roohvand F., Zabihollahi R., Bolhassani A. In vitro anti-viral effects of small heat shock proteins 20 and 27: a novel therapeutic approach. Curr. Pharmaceut. Biotechnol. 2019;20:1011–1017. doi: 10.2174/1389201020666190729104648. [DOI] [PubMed] [Google Scholar]
- 16.Healy E.F., King P.J. A mechanism of action for small heat shock proteins. Biochem. Biophys. Res. Commun. 2012;417:268–273. doi: 10.1016/j.bbrc.2011.11.098. [DOI] [PubMed] [Google Scholar]
- 17.Ecroyd H., Carver J.A. Crystallin proteins and amyloid fibrils. Cell. Mol. Life Sci. 2009;66:62–81. doi: 10.1007/s00018-008-8327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohto-Fujita Eri, Hayasaki Saaya, Atomi Aya, Fujiki Soichiro, Watanabe Toshiyuki, Boelens Wilbert C., Shimizu Miho, Atomi Yoriko. Dynamic localization of αB-crystallin at the microtubule cytoskeleton network in beating heart cells. J. Biochem. 2020;168:125–137. doi: 10.1093/jb/mvaa025. [DOI] [PubMed] [Google Scholar]
- 19.Healy E.F., Goering L.M., Hauser C.R., King P.J. An immunomodulatory role for the Mycobacterium tuberculosis Acr protein in the formation of the tuberculous granuloma. FEBS Lett. 2021;595:284–293. doi: 10.1002/1873-3468.13998. [DOI] [PubMed] [Google Scholar]
- 20.Zunke F., Rose-John S. The shedding protease ADAM17: physiology and pathophysiology. Biochim. Biophys. Acta Mol. Cell Res. 2017;1864:2059–2070. doi: 10.1016/j.bbamcr.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 21.Dreymueller D., Pruessmeyer J., Groth E., Ludwig A. The role of ADAM-mediated shedding in vascular biology. Eur. J. Cell Biol. 2012;91:472–485. doi: 10.1016/j.ejcb.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 22.Scheller J., Chalaris A., Garbers C., Rose-John S. ADAM17: a molecular switch to control inflammation and tissue regeneration. Trends Immunol. 2011;32:380–387. doi: 10.1016/j.it.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 23.Sluchanko N.N., Beelen S., Kulikova A.A., Weeks S.D., Antson A.A., Gusev N.B., Strelkov S.V. Structural basis for the interaction of a human small heat shock protein with the 14-3-3 universal signaling regulator. Structure. 2017;25:305–316. doi: 10.1016/j.str.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weeks S.D., Baranova E.V., Heirbaut M., Beelen S., Shkumatov A.V., Gusev N.B., Strelkov S.V. Molecular structure and dynamics of the dimeric human small heat shock protein HSPB6. J. Struct. Biol. 2014;185:342–354. doi: 10.1016/j.jsb.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 25.Brooks B.R., Bruccoleri R.E., Olafson B.D., States D.J., Swaminathan S., Karplus, Charmm M. A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 26.Antunes D.A., Moll M., Devaurs D., Jackson K.R., Lizée G., Kavraki L.E. DINC 2.0: a new protein–peptide docking webserver using an incremental approach. Canc. Res. 2017;77:e55–e57. doi: 10.1158/0008-5472.CAN-17-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seegar T.C., Killingsworth L.B., Saha N., Meyer P.A., Patra D., Zimmerman B., Kruse A.C. Structural basis for regulated proteolysis by the α-secretase ADAM10. Cell. 2017;171:1638–1648. doi: 10.1016/j.cell.2017.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takeda S., Igarashi T., Mori H., Araki S. Crystal structures of VAP1 reveal ADAMs' MDC domain architecture and its unique C-shaped scaffold. EMBO J. 2006;25:2388–2396. doi: 10.1038/sj.emboj.7601131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fiser András, Kinh Gian Do Richard, Šali Andrej. Modeling of loops in protein structures. Protein Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen M.Y., Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maskos K., Fernandez-Catalan C., Huber R., Bourenkov G.P., Bartunik H., Ellestad G.A., Reddy P., Wolfson M.F., Rauch C.T., Castner B.J., Davis R. Crystal structure of the catalytic domain of human tumor necrosis factor-α-converting enzyme. Proc. Natl. Acad. Sci. U.S.A. 1998;95:3408–3412. doi: 10.1073/pnas.95.7.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozakov D., Hall D.R., Xia B., Porter K.A., Padhorny D., Yueh C., Beglov D., Vajda S. The ClusPro web server for protein–protein docking. Nat. Protoc. 2017;12:255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spassov V.Z., Yan L., Szalma S. Introducing an implicit membrane in generalized Born/solvent accessibility continuum solvent models. J. Phys. Chem. B. 2002;106(2002):8726–8738. [Google Scholar]
- 35.Düsterhöft S., Höbel K., Oldefest M., Lokau J., Waetzig G.H., Chalaris A., Garbers C., Scheller J., Rose-John S., Lorenzen I., Grötzinger J. A disintegrin and metalloprotease 17 dynamic interaction sequence, the sweet tooth for the human interleukin 6 receptor. J. Biol. Chem. 2014;289:16336–16348. doi: 10.1074/jbc.M114.557322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Düsterhöft S., Jung S., Hung C.W., Tholey A., Sönnichsen F.D., Grötzinger J., Lorenzen I. Membrane-proximal domain of a disintegrin and metalloprotease-17 represents the putative molecular switch of its shedding activity operated by protein-disulfide isomerase. J. Am. Chem. Soc. 2013;135:5776–5781. doi: 10.1021/ja400340u. [DOI] [PubMed] [Google Scholar]
- 37.Wan Q., Song D., Li H., He M.L. Stress proteins: the biological functions in virus infection, present and challenges for target-based antiviral drug development. Signal transduction and targeted therapy. 2020;5:1–40. doi: 10.1038/s41392-020-00233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stevens B.R. BioRxiv; 2020. TMPRSS2 and ADAM17 Interactions with ACE2 Complexed with SARS-CoV-2 and B0AT1 Putatively in Intestine, Cardiomyocytes, and Kidney. [Google Scholar]
- 39.Lai Z.W., Hanchapola I., Steer D.L., Smith A.I. Angiotensin-converting enzyme 2 ectodomain shedding cleavage-site identification: determinants and constraints. Biochemistry. 2011;50:5182–5194. doi: 10.1021/bi200525y. [DOI] [PubMed] [Google Scholar]
- 40.Düsterhöft S., Michalek M., Kordowski F., Oldefest M., Sommer A., Röseler J., Reiss K., Grötzinger J., Lorenzen I. Extracellular juxtamembrane segment of ADAM17 interacts with membranes and is essential for its shedding activity. Biochemistry. 2015;54:5791–5801. doi: 10.1021/acs.biochem.5b00497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.