Abstract
Background
Autophagy inhibits tumorigenesis by limiting inflammation. LncRNAs regulate gene expression at various levels as RNAs; thus, both autophagy and lncRNAs are closely related to the occurrence and development of tumours.
Methods
A total of 232 autophagy-related genes were used to construct a coexpression network to extract autophagy-related lncRNAs. A prognostic signature was constructed by multivariate regression analysis. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was applied to analyse enrichment in cancer-related pathways. Immune infiltration analysis was used to analyse the relationship between the prognostic signature and the tumour microenvironment.
Results
Nine autophagy-related lncRNAs were used to construct a prognostic model for non-small-cell lung cancer. The median risk score was used to discriminate the high- and low-risk groups, and the low-risk group was found to have better survival. Because KEGG pathway analysis showed that the prognostic signature was enriched in some immune pathways, further analysis of immune infiltration was conducted, and it was found that the prognostic signature did play a unique role in the immune microenvironment. Additionally, the prognostic signature was associated with clinical factors.
Conclusion
We constructed a prognostic model of autophagy-related lncRNAs that can predict the prognosis of non-small-cell lung cancer.
Keywords: Autophagy, Cancer, lncRNAs, Signature, Immune infiltration
Introduction
Lung cancer is one of the most serious malignancies that threatens health, and its incidence has increased, mainly owing to the increase in smoking, a known risk factor [1]. Autophagy is a conserved catabolic cellular process in which damaged or unusable proteins or other cytoplasmic components are transported to double-membrane vesicles (autophagosomes) and then enter lysosomes or vacuoles for degradation [2]. The level of autophagy is a key threshold for promoting cell survival or inducing cell death in response to environmental stress [3–5]. Autophagy plays a related role in tumorigenesis and anticancer treatment by modulating inflammation, hypoxia, immunosuppression and metabolism in the tumour microenvironment. In particular, impaired autophagic flux is associated with chronic inflammation, immunosuppression, matrix formation, cancer stem cell generation, angiogenesis, metastasis, and metabolic reprogramming in the tumour microenvironment. The tumour microenvironment is composed of a variety of immune cells, mesenchymal cells, extracellular matrix components and active mediators (such as cytokines, chemokines, growth factors, and humoural factors), in addition to tumour cells. The tumour microenvironment can be divided into an immune microenvironment based on immune cells and a nonimmune microenvironment based on fibroblasts. An abnormal tumour microenvironment is closely related to resistance to cell death, promotion of proliferation, avoidance of immune destruction, maintenance of inflammation or induction of angiogenesis [6]. During autophagy, cytoplasmic material is degraded in lysosomes. Because lysosomes have a distinct membrane as a safety mechanism to prevent the leakage of their degradation enzymes, autophagy involves complex membrane dynamics. There are three types of autophagy involving different modes of delivery of cargo to lysosomes: macroautophagy, microautophagy, and chaperone-mediated autophagy. Macroautophagy is the main regulatory form of autophagy occurring in response to environmental and physiological signals. Microautophagy involves direct phagocytosis of cytoplasmic contents by lysosomes, while chaperone-mediated autophagy involves the translocation of chaperone auxiliary substrate proteins (and potentially DNA and RNA molecules) across the lysosomal membrane [7]. Long noncoding RNAs (lncRNAs) have been widely reported to regulate pathophysiological processes through mechanisms such as gene imprinting, histone modification, chromatin remodelling, transcriptional interference, nuclear transport, transcriptional activation, and cell cycle regulation. LncRNAs are mainly transcribed by RNA polymerase II. LncRNAs are important elements of the mammalian transcriptome that control a variety of cellular mechanisms and regulate cellular processes, such as cell metabolism, drug resistance, growth, proliferation, invasion, metastasis, and apoptosis, ensuring homeostasis. They may be oncogenic or tumour suppressive by directly or indirectly affecting the transcription of a variety of proteins through transcriptional and post-transcriptional changes. The main regulatory mechanisms of lncRNAs are the stabilization of proteins in the nucleus and the sponging of miRNAs in the cytoplasm. They can also act as competitive endogenous RNAs (ceRNAs) by competitively binding to microRNAs (miRNAs) and thus inhibiting their function [8]. In this study, 9 autophagy-related lncRNAs with prognostic value (AC020765.2, AC254562.3, AL031666.1, LINC01426, MMP2-AS1, AC102953.2, AP000695.2, LINC00941 and NKILA) in patients were identified using multivariate Cox regression analysis; a prognostic signature was then established based on these prognostic lncRNAs, which may serve as an independent prognostic factor in lung cancer.
Materials and methods
Isolation and sorting of lncRNAs and mRNAs
Data, including transcriptome profile data and clinical information, for all included patients with lung adenocarcinoma and squamous cell carcinoma were downloaded from The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov/). The data were sorted by a Perl script (https://www.perl.org), and a total of 108 normal samples and 1037 tumour samples were obtained; at the same time, we deleted entries with missing information. Through transferring the annotations to a human genes format, we performed ID conversion and distinguished lncRNAs and mRNAs.
Autophagy gene and lncRNA screening
The autophagy gene list was obtained from the Human Autophagy Database (HADb, https://autophagy.lu/clustering/index.html). When extracting autophagy genes, we performed an averaging operation on genes that appeared multiple times; normal samples and low-expression genes (autophagy-related mRNAs or lncRNAs with expression < 0.5) were deleted. Pearson correlation analysis was applied to identify correlations between the lncRNAs and autophagy-related genes. A lncRNA with a correlation coefficient |R2| > 0.3 and P < 0.001 was considered to be an autophagy-related lncRNA.
Signature development
Univariate and multivariate Cox regression analysis was performed to evaluate the prognostic value of autophagy-related lncRNAs. To establish the risk score, lncRNAs with a P-value < 0.01 in the univariate analysis were included in the multivariate stepwise Cox regression analysis. The following formula was used to determine the risk score for each patient: β gene1 × expr gene 1 + β gene 2 × expr gene 2 + … + β gene n × expr gene n. Cox regression analysis was performed to establish a signature for predicting survival. Specifically, we assigned risk scores by calculating the linear sum of the lncRNA expression levels weighted by the corresponding regression coefficients (β). The β values were calculated by log transformation of the hazard ratio (HR) from the multivariate Cox regression analysis. The high-risk and low-risk groups were established based on the median risk score. The lncRNAs expression values were defined as the expression level of gene n (expr gene n) [9].
Construction of the lncRNA-mRNA interaction network by cox regression analysis
It was vital to match the autophagy-related lncRNAs and mRNAs according to the Cox regression analysis results; thus, the network visualized with Cytoscape (version 3.7.1) could highlight the connections and mechanisms involved in the development of lung cancer. Furthermore, as the number of lncRNAs was high, it was valuable to create a signature comprising a limited number of variables and the best Akaike information criterion (AIC).
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) is an method for analysing whole-genome expression profile chip data by comparing genes with predefined gene sets [10]. This method operates through analysis of gene sets and can thus be used to determine whether the gene set shows a statistically significant difference between the two biological states. In this study, we verified whether genes differentially expressed between the two groups are enriched during autophagy.
Analysis of immune infiltrates
TIMER is a comprehensive resource for systematic analysis of immune infiltrates across diverse cancer types (https://cistrome.shinyappes.io/timer/) and was used to evaluate potential relationships between the risk grouping and tumour-infiltrating immune cells. TIMER employs a recently published statistical method known as deconvolution to deduce the prevalence of TIIGs from gene expression profiles. To approximate the abundance of TIIGs, the TIMER database uses TCGA data for 10,897 samples across 32 types of cancer. To assess the relative variations in gene expression amongst sets in the samples [11, 12], we used a deconvolution algorithm based on gene expression called CIBERSORT (http://cibersort.stanford.edu/). With CIBERSORT, we measured the immune response of 22 TIICs to evaluate their association with the risk grouping in lung cancer and to reveal correlations amongst TIICs. We used standard annotation files to establish gene expression datasets and used the default signature matrix with 1000 permutations. Through Monte Carlo sampling, the approximate P-value for the deconvolution was used to determine the confidence levels of the outcomes. To analyse the influence of the high and low risk groupings on the immune microenvironment, we utilized 999 tumour samples that we classified into two groups. To determine the types of lymphocytes affected by the grouping, we set the P-value threshold at < 0.05 [13, 14].
Statistical analysis
Survival status was the basis for the univariate Cox regression analysis, and R software (version 3.6.2) was used to generate Kaplan-Meier curves. GSEA (http://www.broadinstitute.org/gsea/index.jsp) was used to discriminate two sets of functional annotations. Statistical significance was assumed at a threshold two-tailed P < 0.05.
Results
Collation of transcriptome data
We identified 14,142 lncRNAs that were extracted from TCGA datasets, and a total of 210 autophagy-related genes were downloaded from the Human Autophagy Database (HADb, http://autophagy.lu/clustering//index.html). We conducted coexpression analysis of the autophagy genes and lncRNAs to identify autophagy-related lncRNAs (|R2| > 0.3 and P < 0.001). According to the properties of the genes, we separated 1496 identified lncRNAs for further identification of prognostic genes. We combined the two data types futime (survival time) and fustate (survival status), which were obtained from the clinical data (downloaded from TCGA), into the lncRNA expression matrix. Patients with incomplete clinical information (futime, fustate, age, sex, grade, state or TNM) were excluded from the following analysis.
Construction of the cox prognostic model
Through univariate Cox regression analysis, 18 lncRNAs were found to have prognostic value for lung cancer (P ≤ 0.01), and these lncRNAs were subjected to multivariate Cox regression analysis. A risk score formula based on AC020765.2, AC254562.3, AL031666.1, LINC01426, MMP2-AS1, AC102953.2, AP000695.2, LINC00941 and NKILA had the lowest AIC (Akaike information criterion); among these lncRNAs, five were favourable factors (AC020765.2, AC254562.3, AL031666.1, LINC01426, MMP2-AS1) and four were considered unfavourable prognostic factors (AC102953.2, AP000695.2, LINC00941 and NKILA). The risk assessment score for the prediction of overall survival was calculated as follows: expAL031666.1 × 0.176009 + expAC020765.2 × 0.138779 + expAC102953.2 × 0.103983 + expAP000695.2 × 0.145198 + expNKILA × 0.048298 + expMMP2-AS1 × 0.187273 + expLINC01426 × 0.086274 + expAC254562.3 × 0.150477 + expLINC00941 × 0.054391 (Fig. 1) [15, 16].
Fig. 1.
Construction of prognostic signature. The red curve in the survival curve represented the high expression of lncRNA, and the blue curve represented the ground expression of lncRNA. The low expression of five lncRNAs related to autophagy had better survival, while the high expression of the other four lncRNAs related to autophagy had better survival
Visualization of co-expression network
To better present the connections among and mechanisms linking prognosis-related autophagy lncRNAs and mRNAs, we first visualized the coexpression results with Cytoscape and constructed heat maps for the lncRNAs and mRNAs in the coexpression network to show the differences in the expression data (Fig. 2A-C). The distribution of the different patients, who were separated into two groups by the median value of the risk score, was significantly different in Cox regression analysis of the autophagy-related genes, while it was not significantly different between the two groups in the Cox regression analysis of all genes (Fig. 3A-B) [17]. The risk survival curves indicated that the five-year survival rates in the low-risk (CI: 0.446–0.579) and high-risk (CI: 0.32–0.443) groups were higher than 0.5 and 0.38, respectively (P < 0.0001) (Fig. 3C). Next, we constructed a Sankey diagram to further classify the lncRNAs as protective lncRNAs (the higher the expression of the lncRNA, the lower the risk) or risky lncRNAs and more comprehensively visualize their connections (Fig. 3D).
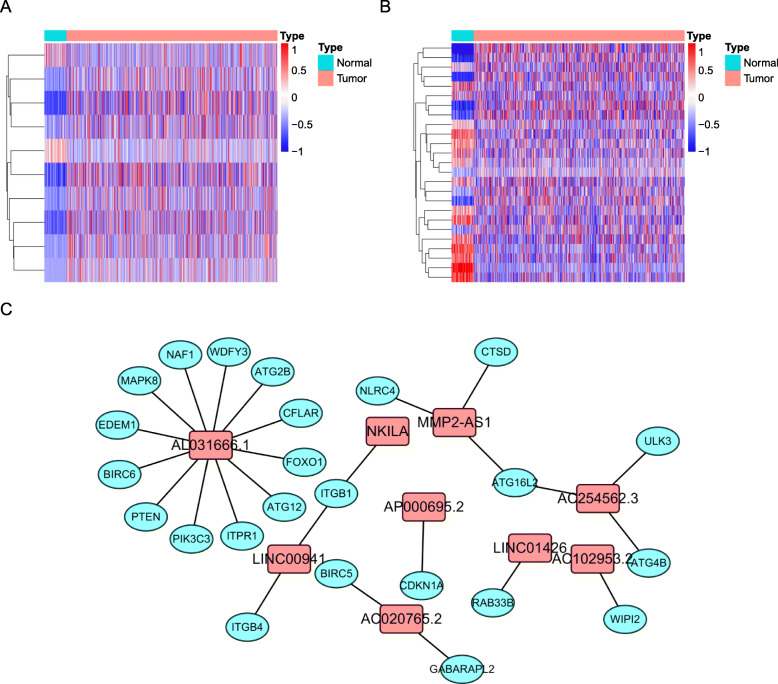
Fig. 2.
Co-expression network construction. (A) Expression of lncRNA related to autophagy in the prognostic signature of heat map between normal and tumor samples. (B) Expression of mRNA related to autophagy in the co-expression network in heat map between normal and tumor samples. Red represented high expression, blue represented ground expression, and the depth of color represented the level of expression. (C) The co-expression network showed the link between lncRNA and autophagy-related mRNA
Fig. 3.
Presentation mode of the prognostic signature. (A) Principal component analysis diagram (PCA) showed the distribution of all genes. (B) Principal component analysis chart (PCA) showed the distribution of prognostic signature. Green represented low risk, and red represented high risk. (C) Kaplan-meier curve reflected the survival significance of the prognostic model. (D) The alluvial map was used to further show the relationship between autophagy-related mRNA and autophagy-related lncRNA, and their risk types
Validation of the cox prognostic model
The risk curve had three parts, and the patients’ risk increased sequentially from left to right. The first part implied that we divided these patients into the high-risk and low-risk groups on the basis of the median value of the risk score. The survival status plot indicated that as the risk value increased, the patients’ survival time decreased. The expression heat map indicated that risk increased with increased expression of some lncRNAs (LINC00941, AP000695.2, NKILA and AC102953.2) and with decreased expression of other lncRNAs (AC020765.2, AC254562.3, AL031666.1, LINC01426 and MMP2-AS1) (Fig. 4A). To evaluate whether the constructed model was independent of other clinical traits as a predictive factor, we performed an independent prognostic analysis. It was found that clinical stage (P < 0.001), T stage (P < 0.001), M stage (P = 0.007), N stage (P < 0.001) and risk score (P < 0.001) were directly related to the prognosis of patients (Fig. 4B), while multivariate Cox regression analysis showed that only T stage (P = 0.023), N stage (P = 0.029) and risk score (P < 0.001) were statistically independent predictive factors (Fig. 4C) [18]. The prediction efficiency of the model was evaluated by ROC curve analysis, and it can be seen that the area under the curve was 0.685 for one-year survival, 0.648 for two-year survival, and 0.638 for three-year survival. This finding indicates that the predictive efficiency of our model is good (Fig. 4D). The area under the red curve in the ROC curve with multiple indicators was 0.673, suggesting that our model had promising power for predicting the clinical outcome of patients. Furthermore, among the clinical traits, risk score had the largest area under the curve value; thus, our model was also superior to other clinical traits for predicting the survival of patients (Fig. 5A) [19].
Fig. 4.
Validation of prognostic signature. (A) The risk plot showed that as the risk score increases, the proportion of patient deaths increases, which in turn indicateed the reliability of the model. The risk heat map further showed the difference in the expression of prognostic lncRNA between the high and low risk groups. (B) Univariate Cox regression analysis showed that stage, T, N, M and riskscore had prognostic significance for non-small cell lung cancer. (C) Multivariate Cox regression analysis displayed that age, T, N and riskscore were independent prognostic factors
Fig. 5.
Immune Infiltration Analysis. (A) The area under the ROC curve was used to predict the predictive power of the prognostic model. The largest area under the prognostic model curve indicated the best prediction effect. (B) KEGG enrichment analysis showed that the prognostic model is mainly enriched in the cancer pathway. (C) The scatter plot showed that the prognostic model is significantly correlated with B cells, CD4+ T cells and macrophages. (D) The violin chart was used to show the difference between the 21 groups of immune cells in the high and low risk groups. Red represents high expression and blue represents low expression
Gene set enrichment analysis
Further functional annotation was conducted through GSEA, and the results revealed that the differentially expressed genes between the two groups were enriched in metabolism-related and tumour-related pathways. GSEA showed that a total of 45 gene sets were significantly enriched with a nominal P value < 0.05. Among the identified pathways, several pathways were well established in cancers, including small cell lung cancer, pathways in cancer, thyroid cancer, P53 signaling pathway, and WNT signaling pathway; all of these pathways promote the development of tumours, indicating that these pathways were active in high-risk patients, while other pathways were silent in high-risk patients. The PAR signalling pathway participates in lipid metabolism and induces anticancer effects in human tumours. Butanoate metabolism, which significantly increases the metabolic stress on tumour cell mitochondria, promotes specific apoptosis of lung cancer cells and inhibits tumour growth. Fatty acid metabolism, α-linolenic acid metabolism and arachidonic acid metabolism inhibit the initiation and metastasis of cancer (Fig. 5B) [20, 21].
Differences in immune cells in the high- and low-risk groups
Independent tumour-infiltrating lymphocytes contribute to the prediction of overall survival and the status of sentinel lymph nodes. Hence, TIMER was applied to analyse the possible relationships between risk grouping and immune infiltration in lung cancer. As shown in Fig. 5C, a negative correlation existed between risk classification and the numbers of B cells (P-value = 7.192 × 10–6), macrophages (P-value = 0.022) and CD4+ T cells (P-value = 0.027) [22, 23]. To estimate whether there is a difference between the tumour immune microenvironment in the two groups of patients, 999 tumour patients were divided into a low-risk group and a high-risk group, which contained 500 and 499 patients, respectively. The comprehensive CIBERSORT algorithm was employed to characterize the infiltration of 21 different immune cells based on gene expression profiles, with patients separated into two groups based on the median risk score. Markers of activated memory CD4+ T cells and M0 macrophages showed low expression in the low-risk group, while markers of resting mast cells showed high expression in the low-risk group (Fig. 5D). The correlation heat map obtained with the 22 types of immune cells revealed that CD8+ T cells correlated positively with activated memory CD4+ T cells but showed a negative relationship with resting memory CD4+ T cells (Fig. 6A). The immune score and matrix score of the patients can help to determine the degree of immune cell infiltration in the tumour microenvironment and the tumour purity. Analysis of the matrix microenvironment showed that there was no significant difference among the patients when they were divided into two groups based on the median value of the matrix score, while when the patients were divided into two groups based on the median value of the immune score, significant differences were revealed in the immune microenvironment (Fig. 6B-C) [24–27].
Fig. 6.
Immune microenvironment analysis. (A) Correlation heat map showed the correlation between 22 immune cells and prognostic models. (B) We predicted the content of stromal cells through matrix scores, and then analyzed the differences in stromal cells between high and low risk groups. (C) We used the immune score to predict the content of immune cells, and then analyzed the differences in immune cells in the high and low risk groups
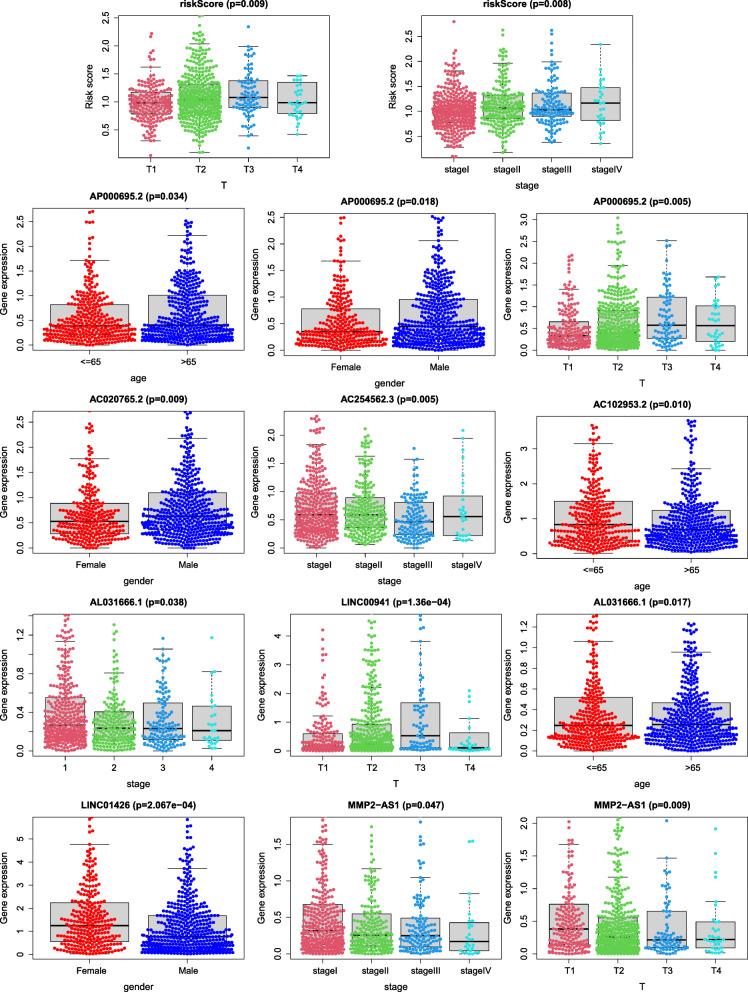
Correlation analysis of clinical parameters
We conducted correlation analysis of clinical parameters to assess whether the risk score and the lncRNAs in the Cox prognostic model are related to clinical traits. We found that the risk model was significantly related to clinical stage and T stage. In addition, AP00695.2 in the model was significantly related to age, sex and T stage, while AC020765.2 was related to sex, AC254562.3 was related to clinical stage, AC102953.2 was significantly different between ages, and LINC00941 was related to T stage. The differences were significant: AL031666.1 was significantly different between ages and stages, LINC01426 was significantly different between sexes, and MMP2-AS1 had an obvious difference between clinical stages and T stages (Fig. 7).
Fig. 7.
Clinical correlation analysis. The prognostic model was significantly correlated with T and stage, which implied the accuracy and reliability of the model. While Part of the lncRNAs in the model were also significantly related to clinical factors respectively
Discussion
Lung cancer, which is one of the most fatal malignancies, is currently a major health issue worldwide. It is of great urgency to find a way to predict the overall survival rate of patients with lung cancer. Epigenetic modifications of genes, especially lncRNAs, have shown close links to lung cancer [28, 29]. LncRNAs, as supplemental genes/miRNAs, are promising predictors of the risk of lung cancer recurrence. Due to technological limitations, problems still exist in functional research lncRNAs in comparison with coding RNAs. Therefore, it is vital to establish a risk model to better predict the prognosis of lung cancer.
In this research, we identified 9 prognostic autophagy-related lncRNAs and divided patients into high-risk and low-risk groups based on the median risk score. Through univariate and multivariate Cox regression analysis, we concluded that the risk model is an independent prognostic factor.
Autophagy originates with the formation of membrane structures called phagocytic cells or membranes. After phagocytic cell formation begins, the double membrane grows to surround the cellular contents during a process called the autophagic extension phase. Autophagy can promote the survival of tumour cells but can also lead to cell death. It can be enhanced or inhibited by anticancer agents. Upregulation of autophagy during cancer treatment can promote either the survival or death of tumour cells. Although little is known about the role of autophagy in cancer therapy to date, recent studies have suggested that therapeutic autophagy will become a new approach for lung cancer treatment [30–32]. First, autophagy can have a tumour-suppressive function. Autophagy is a valuable mechanism used by cells to maintain cell integrity and genome stability. The absence of autophagy genes naturally interferes with this homeostasis; thus, it may initiate cell tumour development. Furthermore, a variety of autophagy mechanisms contribute to tumour suppression. Under stress, autophagy is activated to remove damaged proteins and organelles, including mitochondria. Inhibition or lack of autophagy leads to an increase in the reactive oxygen species level, resulting in accumulation of DNA damage, which is manifested as gene amplification, increased numbers of double-strand breaks and polyploid nuclei. This increase in DNA damage may lead to higher susceptibility to the onset and development of cancer.
In addition, autophagy is a carcinogenic process, and both mechanical tissue and genetic research support this hypothesis. When the intracellular and extracellular environments are deficient and cells are under metabolic stress, autophagy is activated as an adaptation mechanism. In the early stages of tumour formation, cancer cells often experience hypoxia and an environment in which nutrients are limited due to tumour growth because of the lack of an effective blood supply. These conditions cause metabolic stress and lead to reduced mitochondrial oxidative phosphorylation. Subsequently, cancer cell proliferation is suppressed, and cells can enter a dormant state. During dormancy, tumour cells rely on autophagy as a survival strategy, thereby repurposing nutrients to promote cell survival. When the environmental stress is ameliorated, cancer cells can resume proliferation. In fact, defective autophagy causes lung tumours to halt progression and become benign eosinophilic tumours, which are characterized by an abundant cytoplasm and high mitochondrial quality [33]. Recent studies have shown that LINC01426 can act as a predictive gene of SQCLC and GC. LINC00941 was defined as an optimal diagnostic lncRNA biomarker for HNSCC, GC and LUAD. AP000695.2 was used as one of the indicators for constructing a prognostic model of gastric adenocarcinoma [34]. NKILA was found to promote tumour immune evasion by sensitizing T cells to activation-induced cell death [35]. NKILA was also found to suppress nasopharyngeal carcinoma carcinogenesis and metastasis via NF-kappaB pathway inhibition [36]. NKILA was shown to suppress TGF-beta-induced epithelial-mesenchymal transition by blocking NF-kappaB signalling in breast cancer [37].
Equally important, our study used the TIMER database to reveal connections between the risk signature and immune infiltration levels in lung cancer. We found that the associations of the risk signature with B cells, CD4+ T cells and macrophages were the strongest. Moreover, our CIBERSORT analysis revealed that the expression of markers of activated mast cells, M0 macrophages and activated memory CD4+ T cells was increased in the high-risk group, whereas the expression of markers of naive B cells, T follicular helper cells, resting dendritic cells and resting mast cells was decreased. Our results could indicate a possible mechanism by which the lncRNAs in the risk signature regulate the functions of mast cells in tumours. Mast cells are multifunctional cells, and related studies have confirmed that they are related to the pathological process of neoplastic diseases. For example, mast cells can promote tumour angiogenesis by releasing heparin or dissolve surrounding connective tissue by releasing proteolytic enzymes, which is beneficial for tumour growth and metastasis. In contrast, other studies have shown that mast cells surrounding the tumour have the role of tumour defence and host protection. Combining previous findings with our research, we can propose a corresponding explanation that activated mast cells promote tumour growth and resting mast cells inhibit tumour growth.
In conclusion, we demonstrated that the lncRNAs investigated using this model can serve as therapeutic targets for precision treatment of lung cancer. Practical research will be conducted to further verify their biological functions and explore the underlying molecular mechanisms.
Acknowledgments
Not applicable.
Authors’ contributions
Xinyang Zhang participated in the design of the article’s ideas and the implementation of the specific steps, while Yu Cao provided assistance in the compilation of the data for this study, and Li Chen participated in the revision and approval of the article. The author(s) read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
All datas were from TCGA databases. Which are publicly available (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors have no conflicts of interest to declare.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yu Cao joint first authors
References
- 1.Tanner NT, Thomas NA, Ward R, Rojewski A, Gebregziabher M, Toll BA, Silvestri GA. Association of Cigarette Type and Nicotine Dependence in patients presenting for lung Cancer screening. Chest. 2020;158(5):2184–2191. doi: 10.1016/j.chest.2020.05.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou Z, Zhang Y, Gao J, Hao X, Shan C, Li J, Liu C, Wang Y, Li P. Circular RNAs act as regulators of autophagy in cancer. Mol Ther Oncolytics. 2021;21:242–254. doi: 10.1016/j.omto.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klionsky DJ, Abdel-Aziz AK, Abdelfatah S, Abdellatif M, Abdoli A, Abel S, Abeliovich H, Abildgaard MH, Abudu YP, Acevedo-Arozena A, Adamopoulos IE, Adeli K, Adolph TE, Adornetto A, Aflaki E, Agam G, Agarwal A, Aggarwal BB, Agnello M, Agostinis P, Agrewala JN, Agrotis A, Aguilar PV, Ahmad ST, Ahmed ZM, Ahumada-Castro U, Aits S, Aizawa S, Akkoc Y, Akoumianaki T, Akpinar HA, al-Abd AM, al-Akra L, al-Gharaibeh A, Alaoui-Jamali MA, Alberti S, Alcocer-Gómez E, Alessandri C, Ali M, Alim al-Bari MA, Aliwaini S, Alizadeh J, Almacellas E, Almasan A, Alonso A, Alonso GD, Altan-Bonnet N, Altieri DC, Álvarez ÉMC, Alves S, Alves da Costa C, Alzaharna MM, Amadio M, Amantini C, Amaral C, Ambrosio S, Amer AO, Ammanathan V, An Z, Andersen SU, Andrabi SA, Andrade-Silva M, Andres AM, Angelini S, Ann D, Anozie UC, Ansari MY, Antas P, Antebi A, Antón Z, Anwar T, Apetoh L, Apostolova N, Araki T, Araki Y, Arasaki K, Araújo WL, Araya J, Arden C, Arévalo MA, Arguelles S, Arias E, Arikkath J, Arimoto H, Ariosa AR, Armstrong-James D, Arnauné-Pelloquin L, Aroca A, Arroyo DS, Arsov I, Artero R, Asaro DML, Aschner M, Ashrafizadeh M, Ashur-Fabian O, Atanasov AG, Au AK, Auberger P, Auner HW, Aurelian L, Autelli R, Avagliano L, Ávalos Y, Aveic S, Aveleira CA, Avin-Wittenberg T, Aydin Y, Ayton S, Ayyadevara S, Azzopardi M, Baba M, Backer JM, Backues SK, Bae DH, Bae ON, Bae SH, Baehrecke EH, Baek A, Baek SH, Baek SH, Bagetta G, Bagniewska-Zadworna A, Bai H, Bai J, Bai X, Bai Y, Bairagi N, Baksi S, Balbi T, Baldari CT, Balduini W, Ballabio A, Ballester M, Balazadeh S, Balzan R, Bandopadhyay R, Banerjee S, Banerjee S, Bánréti Á, Bao Y, Baptista MS, Baracca A, Barbati C, Bargiela A, Barilà D, Barlow PG, Barmada SJ, Barreiro E, Barreto GE, Bartek J, Bartel B, Bartolome A, Barve GR, Basagoudanavar SH, Bassham DC, Bast RC, Jr, Basu A, Batoko H, Batten I, Baulieu EE, Baumgarner BL, Bayry J, Beale R, Beau I, Beaumatin F, Bechara LRG, Beck GR, Jr, Beers MF, Begun J, Behrends C, Behrens GMN, Bei R, Bejarano E, Bel S, Behl C, Belaid A, Belgareh-Touzé N, Bellarosa C, Belleudi F, Belló Pérez M, Bello-Morales R, Beltran JSO, Beltran S, Benbrook DM, Bendorius M, Benitez BA, Benito-Cuesta I, Bensalem J, Berchtold MW, Berezowska S, Bergamaschi D, Bergami M, Bergmann A, Berliocchi L, Berlioz-Torrent C, Bernard A, Berthoux L, Besirli CG, Besteiro S, Betin VM, Beyaert R, Bezbradica JS, Bhaskar K, Bhatia-Kissova I, Bhattacharya R, Bhattacharya S, Bhattacharyya S, Bhuiyan MS, Bhutia SK, Bi L, Bi X, Biden TJ, Bijian K, Billes VA, Binart N, Bincoletto C, Birgisdottir AB, Bjorkoy G, Blanco G, Blas-Garcia A, Blasiak J, Blomgran R, Blomgren K, Blum JS, Boada-Romero E, Boban M, Boesze-Battaglia K, Boeuf P, Boland B, Bomont P, Bonaldo P, Bonam SR, Bonfili L, Bonifacino JS, Boone BA, Bootman MD, Bordi M, Borner C, Bornhauser BC, Borthakur G, Bosch J, Bose S, Botana LM, Botas J, Boulanger CM, Boulton ME, Bourdenx M, Bourgeois B, Bourke NM, Bousquet G, Boya P, Bozhkov PV, Bozi LHM, Bozkurt TO, Brackney DE, Brandts CH, Braun RJ, Braus GH, Bravo-Sagua R, Bravo-San Pedro JM, Brest P, Bringer MA, Briones-Herrera A, Broaddus VC, Brodersen P, Brodsky JL, Brody SL, Bronson PG, Bronstein JM, Brown CN, Brown RE, Brum PC, Brumell JH, Brunetti-Pierri N, Bruno D, Bryson-Richardson RJ, Bucci C, Buchrieser C, Bueno M, Buitrago-Molina LE, Buraschi S, Buch S, Buchan JR, Buckingham EM, Budak H, Budini M, Bultynck G, Burada F, Burgoyne JR, Burón MI, Bustos V, Büttner S, Butturini E, Byrd A, Cabas I, Cabrera-Benitez S, Cadwell K, Cai J, Cai L, Cai Q, Cairó M, Calbet JA, Caldwell GA, Caldwell KA, Call JA, Calvani R, Calvo AC, Calvo-Rubio Barrera M, Camara NOS, Camonis JH, Camougrand N, Campanella M, Campbell EM, Campbell-Valois FX, Campello S, Campesi I, Campos JC, Camuzard O, Cancino J, Candido de Almeida D, Canesi L, Caniggia I, Canonico B, Cantí C, Cao B, Caraglia M, Caramés B, Carchman EH, Cardenal-Muñoz E, Cardenas C, Cardenas L, Cardoso SM, Carew JS, Carle GF, Carleton G, Carloni S, Carmona-Gutierrez D, Carneiro LA, Carnevali O, Carosi JM, Carra S, Carrier A, Carrier L, Carroll B, Carter AB, Carvalho AN, Casanova M, Casas C, Casas J, Cassioli C, Castillo EF, Castillo K, Castillo-Lluva S, Castoldi F, Castori M, Castro AF, Castro-Caldas M, Castro-Hernandez J, Castro-Obregon S, Catz SD, Cavadas C, Cavaliere F, Cavallini G, Cavinato M, Cayuela ML, Cebollada Rica P, Cecarini V, Cecconi F, Cechowska-Pasko M, Cenci S, Ceperuelo-Mallafré V, Cerqueira JJ, Cerutti JM, Cervia D, Cetintas VB, Cetrullo S, Chae HJ, Chagin AS, Chai CY, Chakrabarti G, Chakrabarti O, Chakraborty T, Chakraborty T, Chami M, Chamilos G, Chan DW, Chan EYW, Chan ED, Chan HYE, Chan HH, Chan H, Chan MTV, Chan YS, Chandra PK, Chang CP, Chang C, Chang HC, Chang K, Chao J, Chapman T, Charlet-Berguerand N, Chatterjee S, Chaube SK, Chaudhary A, Chauhan S, Chaum E, Checler F, Cheetham ME, Chen CS, Chen GC, Chen JF, Chen LL, Chen L, Chen L, Chen M, Chen MK, Chen N, Chen Q, Chen RH, Chen S, Chen W, Chen W, Chen XM, Chen XW, Chen X, Chen Y, Chen YG, Chen Y, Chen Y, Chen YJ, Chen YQ, Chen ZS, Chen Z, Chen ZH, Chen ZJ, Chen Z, Cheng H, Cheng J, Cheng SY, Cheng W, Cheng X, Cheng XT, Cheng Y, Cheng Z, Chen Z, Cheong H, Cheong JK, Chernyak BV, Cherry S, Cheung CFR, Cheung CHA, Cheung KH, Chevet E, Chi RJ, Chiang AKS, Chiaradonna F, Chiarelli R, Chiariello M, Chica N, Chiocca S, Chiong M, Chiou SH, Chiramel AI, Chiurchiù V, Cho DH, Choe SK, Choi AMK, Choi ME, Choudhury KR, Chow NS, Chu CT, Chua JP, Chua JJE, Chung H, Chung KP, Chung S, Chung SH, Chung YL, Cianfanelli V, Ciechomska IA, Cifuentes M, Cinque L, Cirak S, Cirone M, Clague MJ, Clarke R, Clementi E, Coccia EM, Codogno P, Cohen E, Cohen MM, Colasanti T, Colasuonno F, Colbert RA, Colell A, Čolić M, Coll NS, Collins MO, Colombo MI, Colón-Ramos DA, Combaret L, Comincini S, Cominetti MR, Consiglio A, Conte A, Conti F, Contu VR, Cookson MR, Coombs KM, Coppens I, Corasaniti MT, Corkery DP, Cordes N, Cortese K, Costa MC, Costantino S, Costelli P, Coto-Montes A, Crack PJ, Crespo JL, Criollo A, Crippa V, Cristofani R, Csizmadia T, Cuadrado A, Cui B, Cui J, Cui Y, Cui Y, Culetto E, Cumino AC, Cybulsky AV, Czaja MJ, Czuczwar SJ, D’Adamo S, D’Amelio M, D’Arcangelo D, D’Lugos AC, D’Orazi G, da Silva JA, Dafsari HS, Dagda RK, Dagdas Y, Daglia M, Dai X, Dai Y, Dai Y, Dal Col J, Dalhaimer P, Dalla Valle L, Dallenga T, Dalmasso G, Damme M, Dando I, Dantuma NP, Darling AL, Das H, Dasarathy S, Dasari SK, Dash S, Daumke O, Dauphinee AN, Davies JS, Dávila VA, Davis RJ, Davis T, Dayalan Naidu S, de Amicis F, de Bosscher K, de Felice F, de Franceschi L, de Leonibus C, de Mattos Barbosa MG, de Meyer GRY, de Milito A, de Nunzio C, de Palma C, de Santi M, de Virgilio C, de Zio D, Debnath J, DeBosch BJ, Decuypere JP, Deehan MA, Deflorian G, DeGregori J, Dehay B, del Rio G, Delaney JR, Delbridge LMD, Delorme-Axford E, Delpino MV, Demarchi F, Dembitz V, Demers ND, Deng H, Deng Z, Dengjel J, Dent P, Denton D, DePamphilis ML, der CJ, Deretic V, Descoteaux A, Devis L, Devkota S, Devuyst O, Dewson G, Dharmasivam M, Dhiman R, di Bernardo D, di Cristina M, di Domenico F, di Fazio P, di Fonzo A, di Guardo G, di Guglielmo GM, di Leo L, di Malta C, di Nardo A, di Rienzo M, di Sano F, Diallinas G, Diao J, Diaz-Araya G, Díaz-Laviada I, Dickinson JM, Diederich M, Dieudé M, Dikic I, Ding S, Ding WX, Dini L, Dinić J, Dinic M, Dinkova-Kostova AT, Dionne MS, Distler JHW, Diwan A, Dixon IMC, Djavaheri-Mergny M, Dobrinski I, Dobrovinskaya O, Dobrowolski R, Dobson RCJ, Đokić J, Dokmeci Emre S, Donadelli M, Dong B, Dong X, Dong Z, Dorn GW, 2nd, Dotsch V, Dou H, Dou J, Dowaidar M, Dridi S, Drucker L, du A, du C, du G, du HN, du LL, du Toit A, Duan SB, Duan X, Duarte SP, Dubrovska A, Dunlop EA, Dupont N, Durán RV, Dwarakanath BS, Dyshlovoy SA, Ebrahimi-Fakhari D, Eckhart L, Edelstein CL, Efferth T, Eftekharpour E, Eichinger L, Eid N, Eisenberg T, Eissa NT, Eissa S, Ejarque M, el Andaloussi A, el-Hage N, el-Naggar S, Eleuteri AM, el-Shafey ES, Elgendy M, Eliopoulos AG, Elizalde MM, Elks PM, Elsasser HP, Elsherbiny ES, Emerling BM, Emre NCT, Eng CH, Engedal N, Engelbrecht AM, Engelsen AST, Enserink JM, Escalante R, Esclatine A, Escobar-Henriques M, Eskelinen EL, Espert L, Eusebio MO, Fabrias G, Fabrizi C, Facchiano A, Facchiano F, Fadeel B, Fader C, Faesen AC, Fairlie WD, Falcó A, Falkenburger BH, Fan D, Fan J, Fan Y, Fang EF, Fang Y, Fang Y, Fanto M, Farfel-Becker T, Faure M, Fazeli G, Fedele AO, Feldman AM, Feng D, Feng J, Feng L, Feng Y, Feng Y, Feng W, Fenz Araujo T, Ferguson TA, Fernández ÁF, Fernandez-Checa JC, Fernández-Veledo S, Fernie AR, Ferrante AW, Jr, Ferraresi A, Ferrari MF, Ferreira JCB, Ferro-Novick S, Figueras A, Filadi R, Filigheddu N, Filippi-Chiela E, Filomeni G, Fimia GM, Fineschi V, Finetti F, Finkbeiner S, Fisher EA, Fisher PB, Flamigni F, Fliesler SJ, Flo TH, Florance I, Florey O, Florio T, Fodor E, Follo C, Fon EA, Forlino A, Fornai F, Fortini P, Fracassi A, Fraldi A, Franco B, Franco R, Franconi F, Frankel LB, Friedman SL, Fröhlich LF, Frühbeck G, Fuentes JM, Fujiki Y, Fujita N, Fujiwara Y, Fukuda M, Fulda S, Furic L, Furuya N, Fusco C, Gack MU, Gaffke L, Galadari S, Galasso A, Galindo MF, Gallolu Kankanamalage S, Galluzzi L, Galy V, Gammoh N, Gan B, Ganley IG, Gao F, Gao H, Gao M, Gao P, Gao SJ, Gao W, Gao X, Garcera A, Garcia MN, Garcia VE, García-del Portillo F, Garcia-Escudero V, Garcia-Garcia A, Garcia-Macia M, García-Moreno D, Garcia-Ruiz C, García-Sanz P, Garg AD, Gargini R, Garofalo T, Garry RF, Gassen NC, Gatica D, Ge L, Ge W, Geiss-Friedlander R, Gelfi C, Genschik P, Gentle IE, Gerbino V, Gerhardt C, Germain K, Germain M, Gewirtz DA, Ghasemipour Afshar E, Ghavami S, Ghigo A, Ghosh M, Giamas G, Giampietri C, Giatromanolaki A, Gibson GE, Gibson SB, Ginet V, Giniger E, Giorgi C, Girao H, Girardin SE, Giridharan M, Giuliano S, Giulivi C, Giuriato S, Giustiniani J, Gluschko A, Goder V, Goginashvili A, Golab J, Goldstone DC, Golebiewska A, Gomes LR, Gomez R, Gómez-Sánchez R, Gomez-Puerto MC, Gomez-Sintes R, Gong Q, Goni FM, González-Gallego J, Gonzalez-Hernandez T, Gonzalez-Polo RA, Gonzalez-Reyes JA, González-Rodríguez P, Goping IS, Gorbatyuk MS, Gorbunov NV, Görgülü K, Gorojod RM, Gorski SM, Goruppi S, Gotor C, Gottlieb RA, Gozes I, Gozuacik D, Graef M, Gräler MH, Granatiero V, Grasso D, Gray JP, Green DR, Greenhough A, Gregory SL, Griffin EF, Grinstaff MW, Gros F, Grose C, Gross AS, Gruber F, Grumati P, Grune T, Gu X, Guan JL, Guardia CM, Guda K, Guerra F, Guerri C, Guha P, Guillén C, Gujar S, Gukovskaya A, Gukovsky I, Gunst J, Günther A, Guntur AR, Guo C, Guo C, Guo H, Guo LW, Guo M, Gupta P, Gupta SK, Gupta S, Gupta VB, Gupta V, Gustafsson AB, Gutterman DD, H.B. R, Haapasalo A, Haber JE, Hać A, Hadano S, Hafrén AJ, Haidar M, Hall BS, Halldén G, Hamacher-Brady A, Hamann A, Hamasaki M, Han W, Hansen M, Hanson PI, Hao Z, Harada M, Harhaji-Trajkovic L, Hariharan N, Haroon N, Harris J, Hasegawa T, Hasima Nagoor N, Haspel JA, Haucke V, Hawkins WD, Hay BA, Haynes CM, Hayrabedyan SB, Hays TS, He C, He Q, He RR, He YW, He YY, Heakal Y, Heberle AM, Hejtmancik JF, Helgason GV, Henkel V, Herb M, Hergovich A, Herman-Antosiewicz A, Hernández A, Hernandez C, Hernandez-Diaz S, Hernandez-Gea V, Herpin A, Herreros J, Hervás JH, Hesselson D, Hetz C, Heussler VT, Higuchi Y, Hilfiker S, Hill JA, Hlavacek WS, Ho EA, Ho IHT, Ho PWL, Ho SL, Ho WY, Hobbs GA, Hochstrasser M, Hoet PHM, Hofius D, Hofman P, Höhn A, Holmberg CI, Hombrebueno JR, Yi-Ren Hong CWH, Hooper LV, Hoppe T, Horos R, Hoshida Y, Hsin IL, Hsu HY, Hu B, Hu D, Hu LF, Hu MC, Hu R, Hu W, Hu YC, Hu ZW, Hua F, Hua J, Hua Y, Huan C, Huang C, Huang C, Huang C, Huang C, Huang H, Huang K, Huang MLH, Huang R, Huang S, Huang T, Huang X, Huang YJ, Huber TB, Hubert V, Hubner CA, Hughes SM, Hughes WE, Humbert M, Hummer G, Hurley JH, Hussain S, Hussain S, Hussey PJ, Hutabarat M, Hwang HY, Hwang S, Ieni A, Ikeda F, Imagawa Y, Imai Y, Imbriano C, Imoto M, Inman DM, Inoki K, Iovanna J, Iozzo RV, Ippolito G, Irazoqui JE, Iribarren P, Ishaq M, Ishikawa M, Ishimwe N, Isidoro C, Ismail N, Issazadeh-Navikas S, Itakura E, Ito D, Ivankovic D, Ivanova S, Iyer AKV, Izquierdo JM, Izumi M, Jäättelä M, Jabir MS, Jackson WT, Jacobo-Herrera N, Jacomin AC, Jacquin E, Jadiya P, Jaeschke H, Jagannath C, Jakobi AJ, Jakobsson J, Janji B, Jansen-Dürr P, Jansson PJ, Jantsch J, Januszewski S, Jassey A, Jean S, Jeltsch-David H, Jendelova P, Jenny A, Jensen TE, Jessen N, Jewell JL, Ji J, Jia L, Jia R, Jiang L, Jiang Q, Jiang R, Jiang T, Jiang X, Jiang Y, Jimenez-Sanchez M, Jin EJ, Jin F, Jin H, Jin L, Jin L, Jin M, Jin S, Jo EK, Joffre C, Johansen T, Johnson GVW, Johnston SA, Jokitalo E, Jolly MK, Joosten LAB, Jordan J, Joseph B, Ju D, Ju JS, Ju J, Juárez E, Judith D, Juhász G, Jun Y, Jung CH, Jung SC, Jung YK, Jungbluth H, Jungverdorben J, Just S, Kaarniranta K, Kaasik A, Kabuta T, Kaganovich D, Kahana A, Kain R, Kajimura S, Kalamvoki M, Kalia M, Kalinowski DS, Kaludercic N, Kalvari I, Kaminska J, Kaminskyy VO, Kanamori H, Kanasaki K, Kang C, Kang R, Kang SS, Kaniyappan S, Kanki T, Kanneganti TD, Kanthasamy AG, Kanthasamy A, Kantorow M, Kapuy O, Karamouzis MV, Karim MR, Karmakar P, Katare RG, Kato M, Kaufmann SHE, Kauppinen A, Kaushal GP, Kaushik S, Kawasaki K, Kazan K, Ke PY, Keating DJ, Keber U, Kehrl JH, Keller KE, Keller CW, Kemper JK, Kenific CM, Kepp O, Kermorgant S, Kern A, Ketteler R, Keulers TG, Khalfin B, Khalil H, Khambu B, Khan SY, Khandelwal VKM, Khandia R, Kho W, Khobrekar NV, Khuansuwan S, Khundadze M, Killackey SA, Kim D, Kim DR, Kim DH, Kim DE, Kim EY, Kim EK, Kim HR, Kim HS, Hyung-Ryong Kim, Kim JH, Kim JK, Kim JH, Kim J, Kim JH, Kim KI, Kim PK, Kim SJ, Kimball SR, Kimchi A, Kimmelman AC, Kimura T, King MA, Kinghorn KJ, Kinsey CG, Kirkin V, Kirshenbaum LA, Kiselev SL, Kishi S, Kitamoto K, Kitaoka Y, Kitazato K, Kitsis RN, Kittler JT, Kjaerulff O, Klein PS, Klopstock T, Klucken J, Knævelsrud H, Knorr RL, Ko BCB, Ko F, Ko JL, Kobayashi H, Kobayashi S, Koch I, Koch JC, Koenig U, Kögel D, Koh YH, Koike M, Kohlwein SD, Kocaturk NM, Komatsu M, König J, Kono T, Kopp BT, Korcsmaros T, Korkmaz G, Korolchuk VI, Korsnes MS, Koskela A, Kota J, Kotake Y, Kotler ML, Kou Y, Koukourakis MI, Koustas E, Kovacs AL, Kovács T, Koya D, Kozako T, Kraft C, Krainc D, Krämer H, Krasnodembskaya AD, Kretz-Remy C, Kroemer G, Ktistakis NT, Kuchitsu K, Kuenen S, Kuerschner L, Kukar T, Kumar A, Kumar A, Kumar D, Kumar D, Kumar S, Kume S, Kumsta C, Kundu CN, Kundu M, Kunnumakkara AB, Kurgan L, Kutateladze TG, Kutlu O, Kwak SA, Kwon HJ, Kwon TK, Kwon YT, Kyrmizi I, la Spada A, Labonté P, Ladoire S, Laface I, Lafont F, Lagace DC, Lahiri V, Lai Z, Laird AS, Lakkaraju A, Lamark T, Lan SH, Landajuela A, Lane DJR, Lane JD, Lang CH, Lange C, Langel Ü, Langer R, Lapaquette P, Laporte J, LaRusso NF, Lastres-Becker I, Lau WCY, Laurie GW, Lavandero S, Law BYK, Law HKW, Layfield R, le W, le Stunff H, Leary AY, Lebrun JJ, Leck LYW, Leduc-Gaudet JP, Lee C, Lee CP, Lee DH, Lee EB, Lee EF, Lee GM, Lee HJ, Lee HK, Lee JM, Lee JS, Lee JA, Lee JY, Lee JH, Lee M, Lee MG, Lee MJ, Lee MS, Lee SY, Lee SJ, Lee SY, Lee SB, Lee WH, Lee YR, Lee YH, Lee Y, Lefebvre C, Legouis R, Lei YL, Lei Y, Leikin S, Leitinger G, Lemus L, Leng S, Lenoir O, Lenz G, Lenz HJ, Lenzi P, León Y, Leopoldino AM, Leschczyk C, Leskelä S, Letellier E, Leung CT, Leung PS, Leventhal JS, Levine B, Lewis PA, Ley K, Li B, Li DQ, Li J, Li J, Li J, Li K, Li L, Li M, Li M, Li M, Li M, Li M, Li PL, Li MQ, Li Q, Li S, Li T, Li W, Li W, Li X, Li YP, Li Y, Li Z, Li Z, Li Z, Lian J, Liang C, Liang Q, Liang W, Liang Y, Liang YT, Liao G, Liao L, Liao M, Liao YF, Librizzi M, Lie PPY, Lilly MA, Lim HJ, Lima TRR, Limana F, Lin C, Lin CW, Lin DS, Lin FC, Lin JD, Lin KM, Lin KH, Lin LT, Lin PH, Lin Q, Lin S, Lin SJ, Lin W, Lin X, Lin YX, Lin YS, Linden R, Lindner P, Ling SC, Lingor P, Linnemann AK, Liou YC, Lipinski MM, Lipovšek S, Lira VA, Lisiak N, Liton PB, Liu C, Liu CH, Liu CF, Liu CH, Liu F, Liu H, Liu HS, Liu HF, Liu H, Liu J, Liu J, Liu J, Liu L, Liu L, Liu M, Liu Q, Liu W, Liu W, Liu XH, Liu X, Liu X, Liu X, Liu X, Liu Y, Liu Y, Liu Y, Liu Y, Liu Y, Livingston JA, Lizard G, Lizcano JM, Ljubojevic-Holzer S, LLeonart ME, Llobet-Navàs D, Llorente A, Lo CH, Lobato-Márquez D, Long Q, Long YC, Loos B, Loos JA, López MG, López-Doménech G, López-Guerrero JA, López-Jiménez AT, López-Pérez Ó, López-Valero I, Lorenowicz MJ, Lorente M, Lorincz P, Lossi L, Lotersztajn S, Lovat PE, Lovell JF, Lovy A, Lőw P, Lu G, Lu H, Lu JH, Lu JJ, Lu M, Lu S, Luciani A, Lucocq JM, Ludovico P, Luftig MA, Luhr M, Luis-Ravelo D, Lum JJ, Luna-Dulcey L, Lund AH, Lund VK, Lünemann JD, Lüningschrör P, Luo H, Luo R, Luo S, Luo Z, Luparello C, Lüscher B, Luu L, Lyakhovich A, Lyamzaev KG, Lystad AH, Lytvynchuk L, Ma AC, Ma C, Ma M, Ma NF, Ma QH, Ma X, Ma Y, Ma Z, MacDougald OA, Macian F, MacIntosh GC, MacKeigan JP, Macleod KF, Maday S, Madeo F, Madesh M, Madl T, Madrigal-Matute J, Maeda A, Maejima Y, Magarinos M, Mahavadi P, Maiani E, Maiese K, Maiti P, Maiuri MC, Majello B, Major MB, Makareeva E, Malik F, Mallilankaraman K, Malorni W, Maloyan A, Mammadova N, Man GCW, Manai F, Mancias JD, Mandelkow EM, Mandell MA, Manfredi AA, Manjili MH, Manjithaya R, Manque P, Manshian BB, Manzano R, Manzoni C, Mao K, Marchese C, Marchetti S, Marconi AM, Marcucci F, Mardente S, Mareninova OA, Margeta M, Mari M, Marinelli S, Marinelli O, Mariño G, Mariotto S, Marshall RS, Marten MR, Martens S, Martin APJ, Martin KR, Martin S, Martin S, Martín-Segura A, Martín-Acebes MA, Martin-Burriel I, Martin-Rincon M, Martin-Sanz P, Martina JA, Martinet W, Martinez A, Martinez A, Martinez J, Martinez Velazquez M, Martinez-Lopez N, Martinez-Vicente M, Martins DO, Martins JO, Martins WK, Martins-Marques T, Marzetti E, Masaldan S, Masclaux-Daubresse C, Mashek DG, Massa V, Massieu L, Masson GR, Masuelli L, Masyuk AI, Masyuk TV, Matarrese P, Matheu A, Matoba S, Matsuzaki S, Mattar P, Matte A, Mattoscio D, Mauriz JL, Mauthe M, Mauvezin C, Maverakis E, Maycotte P, Mayer J, Mazzoccoli G, Mazzoni C, Mazzulli JR, McCarty N, McDonald C, McGill MR, McKenna SL, McLaughlin BA, McLoughlin F, McNiven MA, McWilliams TG, Mechta-Grigoriou F, Medeiros TC, Medina DL, Megeney LA, Megyeri K, Mehrpour M, Mehta JL, Meijer AJ, Meijer AH, Mejlvang J, Meléndez A, Melk A, Memisoglu G, Mendes AF, Meng D, Meng F, Meng T, Menna-Barreto R, Menon MB, Mercer C, Mercier AE, Mergny JL, Merighi A, Merkley SD, Merla G, Meske V, Mestre AC, Metur SP, Meyer C, Meyer H, Mi W, Mialet-Perez J, Miao J, Micale L, Miki Y, Milan E, Milczarek M, Miller DL, Miller SI, Miller S, Millward SW, Milosevic I, Minina EA, Mirzaei H, Mirzaei HR, Mirzaei M, Mishra A, Mishra N, Mishra PK, Misirkic Marjanovic M, Misasi R, Misra A, Misso G, Mitchell C, Mitou G, Miura T, Miyamoto S, Miyazaki M, Miyazaki M, Miyazaki T, Miyazawa K, Mizushima N, Mogensen TH, Mograbi B, Mohammadinejad R, Mohamud Y, Mohanty A, Mohapatra S, Möhlmann T, Mohmmed A, Moles A, Moley KH, Molinari M, Mollace V, Møller AB, Mollereau B, Mollinedo F, Montagna C, Monteiro MJ, Montella A, Montes LR, Montico B, Mony VK, Monzio Compagnoni G, Moore MN, Moosavi MA, Mora AL, Mora M, Morales-Alamo D, Moratalla R, Moreira PI, Morelli E, Moreno S, Moreno-Blas D, Moresi V, Morga B, Morgan AH, Morin F, Morishita H, Moritz OL, Moriyama M, Moriyasu Y, Morleo M, Morselli E, Moruno-Manchon JF, Moscat J, Mostowy S, Motori E, Moura AF, Moustaid-Moussa N, Mrakovcic M, Muciño-Hernández G, Mukherjee A, Mukhopadhyay S, Mulcahy Levy JM, Mulero V, Muller S, Münch C, Munjal A, Munoz-Canoves P, Muñoz-Galdeano T, Münz C, Murakawa T, Muratori C, Murphy BM, Murphy JP, Murthy A, Myöhänen TT, Mysorekar IU, Mytych J, Nabavi SM, Nabissi M, Nagy P, Nah J, Nahimana A, Nakagawa I, Nakamura K, Nakatogawa H, Nandi SS, Nanjundan M, Nanni M, Napolitano G, Nardacci R, Narita M, Nassif M, Nathan I, Natsumeda M, Naude RJ, Naumann C, Naveiras O, Navid F, Nawrocki ST, Nazarko TY, Nazio F, Negoita F, Neill T, Neisch AL, Neri LM, Netea MG, Neubert P, Neufeld TP, Neumann D, Neutzner A, Newton PT, Ney PA, Nezis IP, Ng CCW, Ng TB, Nguyen HTT, Nguyen LT, Ni HM, Ní Cheallaigh C, Ni Z, Nicolao MC, Nicoli F, Nieto-Diaz M, Nilsson P, Ning S, Niranjan R, Nishimune H, Niso-Santano M, Nixon RA, Nobili A, Nobrega C, Noda T, Nogueira-Recalde U, Nolan TM, Nombela I, Novak I, Novoa B, Nozawa T, Nukina N, Nussbaum-Krammer C, Nylandsted J, O’Donovan TR, O’Leary SM, O’Rourke EJ, O’Sullivan MP, O’Sullivan TE, Oddo S, Oehme I, Ogawa M, Ogier-Denis E, Ogmundsdottir MH, Ogretmen B, Oh GT, Oh SH, Oh YJ, Ohama T, Ohashi Y, Ohmuraya M, Oikonomou V, Ojha R, Okamoto K, Okazawa H, Oku M, Oliván S, Oliveira JMA, Ollmann M, Olzmann JA, Omari S, Omary MB, Önal G, Ondrej M, Ong SB, Ong SG, Onnis A, Orellana JA, Orellana-Muñoz S, Ortega-Villaizan MDM, Ortiz-Gonzalez XR, Ortona E, Osiewacz HD, Osman AHK, Osta R, Otegui MS, Otsu K, Ott C, Ottobrini L, Ou JHJ, Outeiro TF, Oynebraten I, Ozturk M, Pagès G, Pahari S, Pajares M, Pajvani UB, Pal R, Paladino S, Pallet N, Palmieri M, Palmisano G, Palumbo C, Pampaloni F, Pan L, Pan Q, Pan W, Pan X, Panasyuk G, Pandey R, Pandey UB, Pandya V, Paneni F, Pang SY, Panzarini E, Papademetrio DL, Papaleo E, Papinski D, Papp D, Park EC, Park HT, Park JM, Park JI, Park JT, Park J, Park SC, Park SY, Parola AH, Parys JB, Pasquier A, Pasquier B, Passos JF, Pastore N, Patel HH, Patschan D, Pattingre S, Pedraza-Alva G, Pedraza-Chaverri J, Pedrozo Z, Pei G, Pei J, Peled-Zehavi H, Pellegrini JM, Pelletier J, Peñalva MA, Peng D, Peng Y, Penna F, Pennuto M, Pentimalli F, Pereira CMF, Pereira GJS, Pereira LC, Pereira de Almeida L, Perera ND, Pérez-Lara Á, Perez-Oliva AB, Pérez-Pérez ME, Periyasamy P, Perl A, Perrotta C, Perrotta I, Pestell RG, Petersen M, Petrache I, Petrovski G, Pfirrmann T, Pfister AS, Philips JA, Pi H, Picca A, Pickrell AM, Picot S, Pierantoni GM, Pierdominici M, Pierre P, Pierrefite-Carle V, Pierzynowska K, Pietrocola F, Pietruczuk M, Pignata C, Pimentel-Muiños FX, Pinar M, Pinheiro RO, Pinkas-Kramarski R, Pinton P, Pircs K, Piya S, Pizzo P, Plantinga TS, Platta HW, Plaza-Zabala A, Plomann M, Plotnikov EY, Plun-Favreau H, Pluta R, Pocock R, Pöggeler S, Pohl C, Poirot M, Poletti A, Ponpuak M, Popelka H, Popova B, Porta H, Porte Alcon S, Portilla-Fernandez E, Post M, Potts MB, Poulton J, Powers T, Prahlad V, Prajsnar TK, Praticò D, Prencipe R, Priault M, Proikas-Cezanne T, Promponas VJ, Proud CG, Puertollano R, Puglielli L, Pulinilkunnil T, Puri D, Puri R, Puyal J, Qi X, Qi Y, Qian W, Qiang L, Qiu Y, Quadrilatero J, Quarleri J, Raben N, Rabinowich H, Ragona D, Ragusa MJ, Rahimi N, Rahmati M, Raia V, Raimundo N, Rajasekaran NS, Ramachandra Rao S, Rami A, Ramírez-Pardo I, Ramsden DB, Randow F, Rangarajan PN, Ranieri D, Rao H, Rao L, Rao R, Rathore S, Ratnayaka JA, Ratovitski EA, Ravanan P, Ravegnini G, Ray SK, Razani B, Rebecca V, Reggiori F, Régnier-Vigouroux A, Reichert AS, Reigada D, Reiling JH, Rein T, Reipert S, Rekha RS, Ren H, Ren J, Ren W, Renault T, Renga G, Reue K, Rewitz K, Ribeiro de Andrade Ramos B, Riazuddin SA, Ribeiro-Rodrigues TM, Ricci JE, Ricci R, Riccio V, Richardson DR, Rikihisa Y, Risbud MV, Risueño RM, Ritis K, Rizza S, Rizzuto R, Roberts HC, Roberts LD, Robinson KJ, Roccheri MC, Rocchi S, Rodney GG, Rodrigues T, Rodrigues Silva VR, Rodriguez A, Rodriguez-Barrueco R, Rodriguez-Henche N, Rodriguez-Rocha H, Roelofs J, Rogers RS, Rogov VV, Rojo AI, Rolka K, Romanello V, Romani L, Romano A, Romano PS, Romeo-Guitart D, Romero LC, Romero M, Roney JC, Rongo C, Roperto S, Rosenfeldt MT, Rosenstiel P, Rosenwald AG, Roth KA, Roth L, Roth S, Rouschop KMA, Roussel BD, Roux S, Rovere-Querini P, Roy A, Rozieres A, Ruano D, Rubinsztein DC, Rubtsova MP, Ruckdeschel K, Ruckenstuhl C, Rudolf E, Rudolf R, Ruggieri A, Ruparelia AA, Rusmini P, Russell RR, Russo GL, Russo M, Russo R, Ryabaya OO, Ryan KM, Ryu KY, Sabater-Arcis M, Sachdev U, Sacher M, Sachse C, Sadhu A, Sadoshima J, Safren N, Saftig P, Sagona AP, Sahay G, Sahebkar A, Sahin M, Sahin O, Sahni S, Saito N, Saito S, Saito T, Sakai R, Sakai Y, Sakamaki JI, Saksela K, Salazar G, Salazar-Degracia A, Salekdeh GH, Saluja AK, Sampaio-Marques B, Sanchez MC, Sanchez-Alcazar JA, Sanchez-Vera V, Sancho-Shimizu V, Sanderson JT, Sandri M, Santaguida S, Santambrogio L, Santana MM, Santoni G, Sanz A, Sanz P, Saran S, Sardiello M, Sargeant TJ, Sarin A, Sarkar C, Sarkar S, Sarrias MR, Sarkar S, Sarmah DT, Sarparanta J, Sathyanarayan A, Sathyanarayanan R, Scaglione KM, Scatozza F, Schaefer L, Schafer ZT, Schaible UE, Schapira AHV, Scharl M, Schatzl HM, Schein CH, Scheper W, Scheuring D, Schiaffino MV, Schiappacassi M, Schindl R, Schlattner U, Schmidt O, Schmitt R, Schmidt SD, Schmitz I, Schmukler E, Schneider A, Schneider BE, Schober R, Schoijet AC, Schott MB, Schramm M, Schröder B, Schuh K, Schüller C, Schulze RJ, Schürmanns L, Schwamborn JC, Schwarten M, Scialo F, Sciarretta S, Scott MJ, Scotto KW, Scovassi AI, Scrima A, Scrivo A, Sebastian D, Sebti S, Sedej S, Segatori L, Segev N, Seglen PO, Seiliez I, Seki E, Selleck SB, Sellke FW, Selsby JT, Sendtner M, Senturk S, Seranova E, Sergi C, Serra-Moreno R, Sesaki H, Settembre C, Setty SRG, Sgarbi G, Sha O, Shacka JJ, Shah JA, Shang D, Shao C, Shao F, Sharbati S, Sharkey LM, Sharma D, Sharma G, Sharma K, Sharma P, Sharma S, Shen HM, Shen H, Shen J, Shen M, Shen W, Shen Z, Sheng R, Sheng Z, Sheng ZH, Shi J, Shi X, Shi YH, Shiba-Fukushima K, Shieh JJ, Shimada Y, Shimizu S, Shimozawa M, Shintani T, Shoemaker CJ, Shojaei S, Shoji I, Shravage BV, Shridhar V, Shu CW, Shu HB, Shui K, Shukla AK, Shutt TE, Sica V, Siddiqui A, Sierra A, Sierra-Torre V, Signorelli S, Sil P, Silva BJA, Silva JD, Silva-Pavez E, Silvente-Poirot S, Simmonds RE, Simon AK, Simon HU, Simons M, Singh A, Singh LP, Singh R, Singh SV, Singh SK, Singh SB, Singh S, Singh SP, Sinha D, Sinha RA, Sinha S, Sirko A, Sirohi K, Sivridis EL, Skendros P, Skirycz A, Slaninová I, Smaili SS, Smertenko A, Smith MD, Soenen SJ, Sohn EJ, Sok SPM, Solaini G, Soldati T, Soleimanpour SA, Soler RM, Solovchenko A, Somarelli JA, Sonawane A, Song F, Song HK, Song JX, Song K, Song Z, Soria LR, Sorice M, Soukas AA, Soukup SF, Sousa D, Sousa N, Spagnuolo PA, Spector SA, Srinivas Bharath MM, St. Clair D, Stagni V, Staiano L, Stalnecker CA, Stankov MV, Stathopulos PB, Stefan K, Stefan SM, Stefanis L, Steffan JS, Steinkasserer A, Stenmark H, Sterneckert J, Stevens C, Stoka V, Storch S, Stork B, Strappazzon F, Strohecker AM, Stupack DG, Su H, Su LY, Su L, Suarez-Fontes AM, Subauste CS, Subbian S, Subirada PV, Sudhandiran G, Sue CM, Sui X, Summers C, Sun G, Sun J, Sun K, Sun MX, Sun Q, Sun Y, Sun Z, Sunahara KKS, Sundberg E, Susztak K, Sutovsky P, Suzuki H, Sweeney G, Symons JD, Sze SCW, Szewczyk NJ, Tabęcka-Łonczynska A, Tabolacci C, Tacke F, Taegtmeyer H, Tafani M, Tagaya M, Tai H, Tait SWG, Takahashi Y, Takats S, Talwar P, Tam C, Tam SY, Tampellini D, Tamura A, Tan CT, Tan EK, Tan YQ, Tanaka M, Tanaka M, Tang D, Tang J, Tang TS, Tanida I, Tao Z, Taouis M, Tatenhorst L, Tavernarakis N, Taylor A, Taylor GA, Taylor JM, Tchetina E, Tee AR, Tegeder I, Teis D, Teixeira N, Teixeira-Clerc F, Tekirdag KA, Tencomnao T, Tenreiro S, Tepikin AV, Testillano PS, Tettamanti G, Tharaux PL, Thedieck K, Thekkinghat AA, Thellung S, Thinwa JW, Thirumalaikumar VP, Thomas SM, Thomes PG, Thorburn A, Thukral L, Thum T, Thumm M, Tian L, Tichy A, Till A, Timmerman V, Titorenko VI, Todi SV, Todorova K, Toivonen JM, Tomaipitinca L, Tomar D, Tomas-Zapico C, Tomić S, Tong BCK, Tong C, Tong X, Tooze SA, Torgersen ML, Torii S, Torres-López L, Torriglia A, Towers CG, Towns R, Toyokuni S, Trajkovic V, Tramontano D, Tran QG, Travassos LH, Trelford CB, Tremel S, Trougakos IP, Tsao BP, Tschan MP, Tse HF, Tse TF, Tsugawa H, Tsvetkov AS, Tumbarello DA, Tumtas Y, Tuñón MJ, Turcotte S, Turk B, Turk V, Turner BJ, Tuxworth RI, Tyler JK, Tyutereva EV, Uchiyama Y, Ugun-Klusek A, Uhlig HH, Ułamek-Kozioł M, Ulasov IV, Umekawa M, Ungermann C, Unno R, Urbe S, Uribe-Carretero E, Üstün S, Uversky VN, Vaccari T, Vaccaro MI, Vahsen BF, Vakifahmetoglu-Norberg H, Valdor R, Valente MJ, Valko A, Vallee RB, Valverde AM, van den Berghe G, van der Veen S, van Kaer L, van Loosdregt J, van Wijk SJL, Vandenberghe W, Vanhorebeek I, Vannier-Santos MA, Vannini N, Vanrell MC, Vantaggiato C, Varano G, Varela-Nieto I, Varga M, Vasconcelos MH, Vats S, Vavvas DG, Vega-Naredo I, Vega-Rubin-de-Celis S, Velasco G, Velázquez AP, Vellai T, Vellenga E, Velotti F, Verdier M, Verginis P, Vergne I, Verkade P, Verma M, Verstreken P, Vervliet T, Vervoorts J, Vessoni AT, Victor VM, Vidal M, Vidoni C, Vieira OV, Vierstra RD, Viganó S, Vihinen H, Vijayan V, Vila M, Vilar M, Villalba JM, Villalobo A, Villarejo-Zori B, Villarroya F, Villarroya J, Vincent O, Vindis C, Viret C, Viscomi MT, Visnjic D, Vitale I, Vocadlo DJ, Voitsekhovskaja OV, Volonté C, Volta M, Vomero M, von Haefen C, Vooijs MA, Voos W, Vucicevic L, Wade-Martins R, Waguri S, Waite KA, Wakatsuki S, Walker DW, Walker MJ, Walker SA, Walter J, Wandosell FG, Wang B, Wang CY, Wang C, Wang C, Wang C, Wang CY, Wang D, Wang F, Wang F, Wang F, Wang G, Wang H, Wang H, Wang H, Wang HG, Wang J, Wang J, Wang J, Wang J, Wang K, Wang L, Wang L, Wang MH, Wang M, Wang N, Wang P, Wang P, Wang P, Wang P, Wang QJ, Wang Q, Wang QK, Wang QA, Wang WT, Wang W, Wang X, Wang X, Wang Y, Wang Y, Wang Y, Wang YY, Wang Y, Wang Y, Wang Y, Wang Y, Wang Z, Wang Z, Wang Z, Warnes G, Warnsmann V, Watada H, Watanabe E, Watchon M, Wawrzyńska A, Weaver TE, Wegrzyn G, Wehman AM, Wei H, Wei L, Wei T, Wei Y, Weiergräber OH, Weihl CC, Weindl G, Weiskirchen R, Wells A, Wen RH, Wen X, Werner A, Weykopf B, Wheatley SP, Whitton JL, Whitworth AJ, Wiktorska K, Wildenberg ME, Wileman T, Wilkinson S, Willbold D, Williams B, Williams RSB, Williams RL, Williamson PR, Wilson RA, Winner B, Winsor NJ, Witkin SS, Wodrich H, Woehlbier U, Wollert T, Wong E, Wong JH, Wong RW, Wong VKW, Wong WWL, Wu AG, Wu C, Wu J, Wu J, Wu KK, Wu M, Wu SY, Wu S, Wu SY, Wu S, Wu WKK, Wu X, Wu X, Wu YW, Wu Y, Xavier RJ, Xia H, Xia L, Xia Z, Xiang G, Xiang J, Xiang M, Xiang W, Xiao B, Xiao G, Xiao H, Xiao HT, Xiao J, Xiao L, Xiao S, Xiao Y, Xie B, Xie CM, Xie M, Xie Y, Xie Z, Xie Z, Xilouri M, Xu C, Xu E, Xu H, Xu J, Xu JR, Xu L, Xu WW, Xu X, Xue Y, Yakhine-Diop SMS, Yamaguchi M, Yamaguchi O, Yamamoto A, Yamashina S, Yan S, Yan SJ, Yan Z, Yanagi Y, Yang C, Yang DS, Yang H, Yang HT, Yang H, Yang JM, Yang J, Yang J, Yang L, Yang L, Yang M, Yang PM, Yang Q, Yang S, Yang S, Yang SF, Yang W, Yang WY, Yang X, Yang X, Yang Y, Yang Y, Yao H, Yao S, Yao X, Yao YG, Yao YM, Yasui T, Yazdankhah M, Yen PM, Yi C, Yin XM, Yin Y, Yin Z, Yin Z, Ying M, Ying Z, Yip CK, Yiu SPT, Yoo YH, Yoshida K, Yoshii SR, Yoshimori T, Yousefi B, Yu B, Yu H, Yu J, Yu J, Yu L, Yu ML, Yu SW, Yu VC, Yu WH, Yu Z, Yu Z, Yuan J, Yuan LQ, Yuan S, Yuan SSF, Yuan Y, Yuan Z, Yue J, Yue Z, Yun J, Yung RL, Zacks DN, Zaffagnini G, Zambelli VO, Zanella I, Zang QS, Zanivan S, Zappavigna S, Zaragoza P, Zarbalis KS, Zarebkohan A, Zarrouk A, Zeitlin SO, Zeng J, Zeng JD, Žerovnik E, Zhan L, Zhang B, Zhang DD, Zhang H, Zhang H, Zhang H, Zhang H, Zhang H, Zhang H, Zhang H, Zhang HL, Zhang J, Zhang J, Zhang JP, Zhang KYB, Zhang LW, Zhang L, Zhang L, Zhang L, Zhang L, Zhang M, Zhang P, Zhang S, Zhang W, Zhang X, Zhang XW, Zhang X, Zhang X, Zhang X, Zhang X, Zhang XD, Zhang Y, Zhang Y, Zhang Y, Zhang YD, Zhang Y, Zhang YY, Zhang Y, Zhang Z, Zhang Z, Zhang Z, Zhang Z, Zhang Z, Zhang Z, Zhao H, Zhao L, Zhao S, Zhao T, Zhao XF, Zhao Y, Zhao Y, Zhao Y, Zhao Y, Zheng G, Zheng K, Zheng L, Zheng S, Zheng XL, Zheng Y, Zheng ZG, Zhivotovsky B, Zhong Q, Zhou A, Zhou B, Zhou C, Zhou G, Zhou H, Zhou H, Zhou H, Zhou J, Zhou J, Zhou J, Zhou J, Zhou K, Zhou R, Zhou XJ, Zhou Y, Zhou Y, Zhou Y, Zhou ZY, Zhou Z, Zhu B, Zhu C, Zhu GQ, Zhu H, Zhu H, Zhu H, Zhu WG, Zhu Y, Zhu Y, Zhuang H, Zhuang X, Zientara-Rytter K, Zimmermann CM, Ziviani E, Zoladek T, Zong WX, Zorov DB, Zorzano A, Zou W, Zou Z, Zou Z, Zuryn S, Zwerschke W, Brand-Saberi B, Dong XC, Kenchappa CS, Li Z, Lin Y, Oshima S, Rong Y, Sluimer JC, Stallings CL, Tong CK. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition) Autophagy. 2021;17(1):1–382. doi: 10.1080/15548627.2020.1797280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hou HH, Pan HJ, Liao WY, Lee CH, Yu CJ. Autophagy in fibroblasts induced by cigarette smoke extract promotes invasion in lung cancer cells. Int J Cancer. 2020;147(9):2587–2596. doi: 10.1002/ijc.33127. [DOI] [PubMed] [Google Scholar]
- 5.Wang J, Liu D, Sun Z, Ye T, Li J, Zeng B, Zhao Q, Rosie Xing H. Autophagy augments the self-renewal of lung cancer stem cells by the degradation of ubiquitinated p53. Cell Death Dis. 2021;12(1):98. doi: 10.1038/s41419-021-03392-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang R, Zeh H, Lotze M, et al. The multifaceted effects of autophagy on the tumor microenvironment. Adv Exp Med Biol. 2020;1225:99–114. doi: 10.1007/978-3-030-35727-6_7. [DOI] [PubMed] [Google Scholar]
- 7.Russo M, Russo GL. Autophagy inducers in cancer. Biochem Pharmacol. 2018;153:51–61. doi: 10.1016/j.bcp.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Thin KZ, Liu X, Feng X, Raveendran S, Tu JC. LncRNA-DANCR: a valuable cancer related long non-coding RNA for human cancers. Pathol Res Pract. 2018;214(6):801–805. doi: 10.1016/j.prp.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Miao H, Chen D, Li R, et al. Identification of an immune-related six-long noncoding RNA signature as a novel prognosis biomarker for adenocarcinoma of lung. Biosci Rep. 2021;41:1. doi: 10.1042/BSR20202444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qi-Dong X, Yang X, Lu JL, et al. Development and validation of a nine-redox-related long noncoding RNA signature in renal clear cell carcinoma. Oxidative Med Cell Longev. 2020;2020:6634247. doi: 10.1155/2020/6634247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu F, Wu H. Identification of prognostic biomarkers and molecular targets among JAK family in breast Cancer. J Inflamm Res. 2021;14:97–114. doi: 10.2147/JIR.S284889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye GC, Liu YF, Huang L, Zhang CY, Sheng YL, Wu B, Han L, Wu CL, Dong B, Qi Y. Key microRNAs and hub genes associated with poor prognosis in lung adenocarcinoma. Aging (Albany NY) 2021;13(3):3742–3762. doi: 10.18632/aging.202337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Z, Wei X, Pan Y, Xu J, Si Y, Min Z, Yu B. A new risk factor indicator for papillary thyroid cancer based on immune infiltration. Cell Death Dis. 2021;12(1):51. doi: 10.1038/s41419-020-03294-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cong S, Guo Q, Cheng Y, He Y, Zhao X, Kong C, Ning S, Zhang G. Immune characterization of ovarian Cancer reveals new cell subtypes with different prognoses, immune risks, and molecular mechanisms. Front Cell Dev Biol. 2020;8:614139. doi: 10.3389/fcell.2020.614139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie X, Wang Y, Zhang S, Li J, Yu Z, Ding X, Ye L, Gong P, Zhu Q, Li J, Chen Z, Yao X, du Z, Zeng Q, Chen H, Yang Z, Chen G. A novel five-lncRNA signature panel improves high-risk survival prediction in patients with cholangiocarcinoma. Aging (Albany NY) 2021;13(2):2959–2981. doi: 10.18632/aging.202446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niu X, Sun J, Meng L, Fang T, Zhang T, Jiang J, Li H. A five-lncRNAs signature-derived risk score based on TCGA and CGGA for glioblastoma: potential prospects for treatment evaluation and prognostic prediction. Front Oncol. 2020;10:590352. doi: 10.3389/fonc.2020.590352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ikegami T, Taniguchi A, Okada T, et al. Functionalization using polymer or silane? A practical test method to characterize hydrophilic interaction chromatography phases in terms of their functionalization method. J Chromatogr A. 2020;1638:461850. doi: 10.1016/j.chroma.2020.461850. [DOI] [PubMed] [Google Scholar]
- 18.Huang TT, Li SH, Chen YH, Lu HI, Lo CM, Fang FM, Chou SY, Chiu YC, Chou YP, Wang YM. Definitive chemoradiotherapy for clinical T4b esophageal cancer - treatment outcomes, failure patterns, and prognostic factors. Radiother Oncol. 2021;157:56–62. doi: 10.1016/j.radonc.2021.01.007. [DOI] [PubMed] [Google Scholar]
- 19.Alshabibi AS, Suleiman ME, Tapia KA, et al. Impact of time awake and hours slept at night on Radiologists' mammogram interpretations. J Am Coll Radiol. 2021;18(5):730–738. doi: 10.1016/j.jacr.2020.12.023. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Hughes KR, Raghani RM, Ma J, Orbach S, Jeruss JS, Shea LD. Cargo-free immunomodulatory nanoparticles combined with anti-PD-1 antibody for treating metastatic breast cancer. Biomaterials. 2021;269:120666. doi: 10.1016/j.biomaterials.2021.120666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu S, Huang Z, Deng X, Zou X, Li H, Mu S, Cao B. Identification of key candidate biomarkers for severe influenza infection by integrated bioinformatical analysis and initial clinical validation. J Cell Mol Med. 2021;25(3):1725–1738. doi: 10.1111/jcmm.16275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rojas K, Baliu-Piqué M, Manzano A, Saiz-Ladera C, García-Barberán V, Cimas FJ, Pérez-Segura P, Pandiella A, Győrffy B, Ocana A. In silico transcriptomic mapping of integrins and immune activation in basal-like and HER2+ breast cancer. Cell Oncol (Dordr) 2021;44(3):569–580. doi: 10.1007/s13402-020-00583-9. [DOI] [PubMed] [Google Scholar]
- 23.Xu WX, Zhang J, Hua YT, Yang SJ, Wang DD, Tang JH. An integrative Pan-Cancer analysis revealing LCN2 as an oncogenic immune protein in tumor microenvironment. Front Oncol. 2020;10:605097. doi: 10.3389/fonc.2020.605097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pinto A, Marangon I, Méreaux J, Nicolás-Boluda A, Lavieu G, Wilhelm C, Sarda-Mantel L, Silva AKA, Pocard M, Gazeau F. Immune reprogramming precision photodynamic therapy of peritoneal metastasis by scalable stem-cell-derived extracellular vesicles. ACS Nano. 2021;15(2):3251–3263. doi: 10.1021/acsnano.0c09938. [DOI] [PubMed] [Google Scholar]
- 25.Tarancon R, Mata E, Uranga S, et al. Therapeutic efficacy of pulmonary live tuberculosis vaccines against established asthma by subverting local immune environment. EBioMedicine. 2021;103186. [DOI] [PMC free article] [PubMed]
- 26.Oshi M, Kim TH, Tokumaru Y, Yan L, Matsuyama R, Endo I, Cherkassky L, Takabe K. Enhanced DNA repair pathway is associated with cell proliferation and worse survival in hepatocellular carcinoma (HCC) Cancers (Basel) 2021;13(2):323. doi: 10.3390/cancers13020323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang KC, Diermeier SD, Yu AT, Brine LD, Russo S, Bhatia S, Alsudani H, Kostroff K, Bhuiya T, Brogi E, Pappin DJ, Bennett CF, Rigo F, Spector DL. MaTAR25 lncRNA regulates the Tensin1 gene to impact breast cancer progression. Nat Commun. 2020;11(1):6438. doi: 10.1038/s41467-020-20207-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun CC, Zhu W, Li SJ, Hu W, Zhang J, Zhuo Y, Zhang H, Wang J, Zhang Y, Huang SX, He QQ, Li DJ. FOXC1-mediated LINC00301 facilitates tumor progression and triggers an immune-suppressing microenvironment in non-small cell lung cancer by regulating the HIF1alpha pathway. Genome Med. 2020;12(1):77. doi: 10.1186/s13073-020-00773-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu N, Jiang M, Liu H, Chu Y, Wang D, Cao J, Wang Z, Xie X, Han Y, Xu B. LINC00941 promotes CRC metastasis through preventing SMAD4 protein degradation and activating the TGF-beta/SMAD2/3 signaling pathway. Cell Death Differ. 2021;28(1):219–232. doi: 10.1038/s41418-020-0596-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gremke N, Polo P, Dort A, Schneikert J, Elmshäuser S, Brehm C, Klingmüller U, Schmitt A, Reinhardt HC, Timofeev O, Wanzel M, Stiewe T. mTOR-mediated cancer drug resistance suppresses autophagy and generates a druggable metabolic vulnerability. Nat Commun. 2020;11(1):4684. doi: 10.1038/s41467-020-18504-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cochrane CR, Vaghjiani V, Szczepny A, Jayasekara WSN, Gonzalez-Rajal A, Kikuchi K, McCaughan G, Burgess A, Gough DJ, Watkins DN, Cain JE. Trp53 and Rb1 regulate autophagy and ligand-dependent hedgehog signaling. J Clin Invest. 2020;130(8):4006–4018. doi: 10.1172/JCI132513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen T, Cai LD, Liu YH, Li S, Gan WJ, Li XM, Wang JR, Guo PD, Zhou Q, Lu XX, Sun LN, Li JM. Ube2v1-mediated ubiquitination and degradation of Sirt1 promotes metastasis of colorectal cancer by epigenetically suppressing autophagy. J Hematol Oncol. 2018;11(1):95. doi: 10.1186/s13045-018-0638-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson CM, Macleod KF. Autophagy and cancer cell metabolism. Int Rev Cell Mol Biol. 2019;347:145–190. doi: 10.1016/bs.ircmb.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zha Z, Zhang P, Li D, et al. Identification and construction of a long noncoding RNA prognostic risk model for stomach adenocarcinoma patients. Dis Markers. 2021;2021:8895723. doi: 10.1155/2021/8895723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang D, Chen J, Yang L, Ouyang Q, Li J, Lao L, Zhao J, Liu J, Lu Y, Xing Y, Chen F, Su F, Yao H, Liu Q, Su S, Song E. NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation-induced cell death. Nat Immunol. 2018;19(10):1112–1125. doi: 10.1038/s41590-018-0207-y. [DOI] [PubMed] [Google Scholar]
- 36.Zhang W, Guo Q, Liu G, Zheng F, Chen J, Huang D, Ding L, Yang X, Song E, Xiang Y, Yao H. NKILA represses nasopharyngeal carcinoma carcinogenesis and metastasis by NF-κB pathway inhibition. PLoS Genet. 2019;15(8):e1008325. doi: 10.1371/journal.pgen.1008325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu W, Chen F, Cui X, Yang L, Chen J, Zhao J, Huang D, Liu J, Yang L, Zeng J, Zeng Z, Pan Y, Su F, Cai J, Ying Z, Zhao Q, Song E, Su S. LncRNA NKILA suppresses TGF-β-induced epithelial-mesenchymal transition by blocking NF-κB signaling. Int J Cancer. 2018;143(9):2213–2224. doi: 10.1002/ijc.31605. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datas were from TCGA databases. Which are publicly available (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga).