Abstract
Background
The emergence of novel variants of concern of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) demands fast and reliable detection of such variants in local populations.
Methods
Here we present a cost-efficient and fast workflow combining a prescreening of SARS-CoV-2-positive samples using reverse transcription polymerase chain reaction melting curve analysis with multiplexed IP-RP-HPLC-based single nucleotide primer extensions.
Results
The entire workflow from positive SARS-CoV-2 testing to base-specific identification of variants requires about 24 hours.
Conclusions
We applied the sensitive method to monitor local variant of concern outbreaks in SARS-CoV-2-positive samples collected in a confined region of Germany.
Keywords: mutation screening, SARS-CoV-2, SIRPH, SNuPE, variants of concern
During the ongoing coronavirus disease 2019 (COVID-19) pandemic, recent reports of new severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants with changes in viral functionalities or pathogenic aspects have caused major concern. Since December 2020, several emerging variants of concern (VOCs) with increased transmission were reported from UK (B.1.1.7), South Africa (B.1.351), Brazil (P.1), and India (B.1.617). To allow for the timely adaptation of public health interventions, it is of utmost importance to monitor and precisely follow the ingression and distribution of these VOCs as quickly as possible. They are each characterized by a set of mutations that can be used for variant monitoring. B.1.1.7, B.1.351, and P.1 are characterized by the spike protein mutation N501Y. In addition, B.1.351 and P.1 also share the mutation E484K, but are discriminated by the mutation V1176F, which is only present in P.1 [1–3]. The amino acid substitution E484K in the receptor-binding domain of the spike protein was reported to be associated with immune escape from neutralizing antibodies, which is especially worrisome regarding vaccination or reinfection [4]. More recently, E484K has also been reported in some B.1.1.7 strains [5]. B.1.617, which is characterized by the mutations L452R and P681R, divides into 3 sublineages. B.1.617.1 and B.1.617.3 show the mutation E484Q, which apparently has similar functional consequences as E484K [6].
The gold standard for the identification of virus variants is sequencing of full viral genomes. Currently, reverse transcription polymerase chain reaction (RT-PCR) melting curve analysis approaches followed by confirmation through viral genome sequencing are widely used. However, the required time for next-generation sequencing (NGS) and the extra costs call for faster and more cost-effective test systems. Previously, we proposed a pooling approach for high-throughput RT-PCR testing to identify SARS-CoV-2 infection in large cohorts of asymptomatic people [7]. Here we extend this strategy to obtain a fast and accurate base-specific detection of VOCs by combining a sequential RT-PCR-based melting curve analysis as a prescreening approach of SARS-CoV-2-positive samples with a multiplexed IP-RP-HPLC-based single nucleotide primer extension approach (SIRPH) [8, 9]. This protocol allows a fast and comprehensive generation of “single base sequencing” signatures for a relevant spectrum of VOCs to assign the variant lineages within the (local) pandemic situation.
METHODS
SARS-CoV-2 (Pool) Testing and RT-PCR Melting Curve Analysis
Respiratory samples were tested individually (expected high positivity rate) or in pools (expected low positivity rate, 16 samples per pool) performed with Biomek i5 Span8 (Beckman Coulter) by automated SARS-CoV-2 dual target RT-PCR (E and ORF1) using Cobas 6800 (Roche Diagnostics). To identify VOC candidates, positive samples were extracted using RNAdvance Viral and Biomek i5 MC96 (Beckman Coulter) and subjected to N501Y RT-PCR melting curve analysis (VirSNiP SARS-CoV-2 Spike N501Y TibMolBiol). N501Y-positive cases were further tested for delH69/V70, E484K, and V1167F by additional melting curve RT-PCR (VirSNiP SARS-CoV-2 TibMolBiol). It should be noted that due to newly emerging variants we recently changed our VOC screening schedule. All SARS-CoV-2-positive samples are tested for N501Y, delH69/V70, and E484K using a multiplexed RT-PCR melting curve analysis (VirSNiP SARS del69,70 + 484K + 501Y). If positive for N501Y and E484K simultaneously, the sample is tested in addition for the presence of V1176F to differentiate between B.1.351 and P.1. Samples that test negative in the multiplexed RT-PCR melting curve analysis are followed up with an additional screening for L452R, E484Q, and P681R to identify B.1.617. All analyses were performed according to the manufacturers` instructions. The study was approved by the local ethics committee of the Saarland University Medical Center at Saarland Ärztekammer.
Reverse Transcription
Isolated RNA was reverse transcribed using random hexamer primers (NEB) and Maxima Reverse Transcriptase H- (ThermoFisher) following the manufacturer’s recommendations. cDNA from the same preparation was submitted to PCRs for single nucleotide primer extension (SNuPE) and NGS Library generation.
SNuPE and IP-RP-HPLC (SIRPH)
Two microliters of cDNA was used as the template in a 30-μL reaction in the presence of 3 mM of Tris–HCl (pH 8.8), 0.7 mM of (NH4)2SO4, 50 mM of KCl, 2.5 mM of MgCl2, 0.06 mM of each dNTP, 3 U HotFire DNA polymerase (Solis BioDyne), and 167 nM of primers (Supplementary Table 1). PCRs were performed at 95°C for 15 minutes followed by 35 cycles at 95°C/60 seconds, 54°C (58°C for A570D + D614G)/30 seconds, 72°C/30 seconds, and a final extension 72°C/5 minutes. Five μL of PCR products was treated with 1U of ExoCIAP (mixture of Exonuclease I [Jena Bioscience] and Calf Intestine Alkaline Phosphatase [Calbiochem]) for 30 minutes at 37°C. To inactivate the ExoCIAP enzymes, the reaction was incubated for 15 minutes at 80°C. Afterwards, 14 μL of primer extension mastermix (50 mM of Tris–HCl, pH 9.5, 2.5 mM of MgCl2, 0.05 mM of ddNTPs, 1.6 μM of each SNuPE primer, and 2.5 U of Termipol DNA polymerase [Solis BioDyne]) were added. Primer extension reactions were performed at 96°C for 2 minutes, followed by 50 cycles at 96°C/30 seconds, 50°C/30 seconds, and 60°C/20 seconds. Separation of products was conducted on an XBridge BEH C18 2.5 µm 4.6 mm × 50 mm column (Waters) at 0.9 mL/min at 50°C by continuously mixing buffer B (0.1 M TEAA, 25% acetonitril) with buffer A (0.1 M TEAA) (Supplementary Table 2). HPLC runs for the entire set of potentially mutated sites (PMS) took ~60 minutes per sample. Mutations were determined by relative retention times in comparison with the nonmutated reference.
Sanger Sequencing
Sanger sequencing of exemplary PCR products generated for SIRPH analysis was performed at Macrogen (Amsterdam) using 40 ng of purified PCR product and 10 pmol of corresponding PCR primers.
Viral Genome Sequencing
NGS Libraries for viral genome sequencing were generated using the primer design from the ARTIC, version 0.3, protocol [10] modified for Illumina sequencing. We added Illumina-compatible adapter sequences directly to the ARTIC primers (Supplementary Table 3). To obtain a more equal coverage, we introduced a third multiplex pool with 5 primer pairs covering difficult genomic regions (Supplementary Table 3). Multiplex PCRs were carried out as described in the ARTIC protocol. After purification, PCR products were submitted to an indexing PCR with TruSeq primers (Illumina) for 8 cycles. After final purification and normalization, samples were sequenced on a MiSeq (Illumina).
NGS Data Processing
Viral genome data were processed using CoVpipe (https://gitlab.com/RKIBioinformaticsPipelines/ncov_minipipe). After trimming of adapter/primer sequences and low-quality reads (<Q30), reads were aligned to the SARS-CoV-2 reference NC_045512.2. Reads mapping to the human genome were excluded. Variant calling was performed with default parameters, and consensus sequences were masked for lowly covered sites (<20×). Visualization was achieved with Auspice and Nextstrain [11].
RESULTS
Single nucleotide primer extension coupled to IP-RP-HPLC detection (SIRPH) is a powerful and fast approach for point mutation analysis of known genomic variants. This assay is characterized by primers that anneal with their 3’-end directly adjacent to PMS. Using ddNTPs, the primer can only be extended by a single nucleotide complementary to the genomic information on the annealed DNA strand. Due to the incorporation of different bases in wild-type and mutant, extended primers differ in hydrophobicity, which allows their separation by IP-RP-HPLC. We adapted this approach for screening of VOCs in the SARS-CoV-2 genome in a multiplex assay that allows simultaneous detection of several mutations.
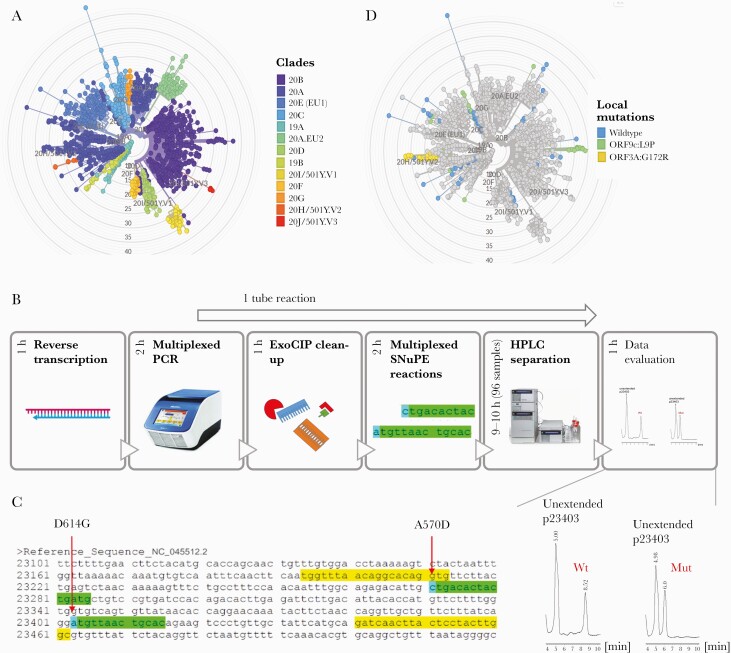
We first established the method on 80 SARS-CoV-2-positive samples collected in the southwest of Germany (Saarland) from Nov/Dec 2020 for B.1.1.7, B.1.351, and P.1 (Figure 1A). We designed primers for our SNuPE assay (Figure 1B, C; Supplementary Table 1) to determine the nucleotide at the PMS D614G (nucleotide exchange: A→G in all clades except 19A), N501Y present in all 3 VOCs (A→T), and the discriminatory PMS A570D (C→A in B.1.1.7), P681H (C→A in B.1.1.7), T716I (C→T in B.1.1.7), E484K (G→A in B.1.351 and P.1), and V1176F (G→T in P.1). We also analyzed 2 local mutations, L9P (T→C) and G172R (G→C), which we observed in a high frequency in NGS data of earlier samples (Sep/Oct 2020) (Figure 1D) from Saarland to follow their further ingression and distribution.
Figure 1.
Multiplexed IP-RP-HPLC-based SIRPH. A, Phylogenetic tree of SARS-CoV-2 sequences derived from GISAID references (accessed on 01/29/2021) and sequencing data from Saarland (March–October 2020, available on GISAID using the identifiers listed in Supplementary Table 7) colored by clade. Variants of concern are found in clades 20I/501Y.V1 (B.1.1.7), 20H/501Y.V2 (B.1.351), and 20J/501Y.V3 (P.1). B, SIRPH Workflow: Isolated RNA is reverse-transcribed, followed by a multiplexed PCR of regions of interest. After ExoCIP clean-up of residual dNTPs and primers, multiplexed SNuPE reactions are performed that are separated by IP-RP-HPLC. The whole workflow can be performed within 1 day. C, Exemplary SNuPE primer design (green) for the PMS (blue) A570D and D614G within a single PCR product (yellow). D, Distribution of the local mutations ORF9c:L9P (green) and ORF3A:G172R (yellow) in samples from Saarland (March–October 2020). E, Exemplary HPLC separation of SNuPE reactions at the PMS D614G showing a profile for wild-type (A) and mutation (G). Abbreviations: Mut, mutation; PCR, polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SIRPH, single nucleotide primer extension approach; SNuPE, single nucleotide primer extension; Wt, wild-type.
Notably, mutations in close vicinity of each other that could be covered within the same PCR product (Figure 1C; Supplementary Figure 1) were assessed simultaneously in a multiplexed SNuPE reaction. After RNA extraction and reverse transcription, cDNA was used as a template for 4 different PCR reactions yielding PCR products harboring the PMS (Figure 1B). In a following 1-tube reaction, PCR products were treated with exonuclease I and calf intestinal phosphatase (ExoCIP) to remove primers and nucleotides, heat-inactivated, and subjected to SNuPE followed by IP-RP-HPLC separation (Figure 1B) in 96 sample formats. In these SNuPE reactions, we either assessed 2 PMS at once (E484K, N501Y, P681H, T716I, L9P, G172R) or 1 PMS (A570D, D614G) per reaction (Supplementary Figure 1), resulting in a total of 5 HPLC runs per sample. In addition, different point mutations that occur at the same genomic position such as E484K and E484Q can be discriminated within the same SNuPE reaction.
All 80 samples of our initial cohort showed the mutation D614G, which emerged early in the pandemic and is now predominant in samples from across the globe [12]. The PMS characteristic of the 3 analyzed VOCs (E484K, N501Y, P681H, T716I, A570D) were in concordance with the original SARS-CoV-2 sequence (Supplementary Table 4). Our local mutations revealed interesting patterns over time (Supplementary Table 4). The spread of the ORF9c mutation L9P dropped from 35.3% in Sep/Oct 2020 (n = 51) to 6.25% in Nov/Dec 2020 (n = 80), while the ORF3A mutation G172R remained more abundant (33.3% to 27.5%).
For validation, we performed full viral genome sequencing on all 80 samples (Ct values 12–26) (Supplementary Table 4). For variant calling, we only considered sites with coverage of at least 20 (Supplementary Table 4). Especially in the region harboring our local L9P variant, we observed high coverage-related dropout rates in the sequencing data. Thus, we performed additional validation of exemplary samples by Sanger sequencing (Supplementary Figure 2). All SNuPE results were in full concordance with viral genome sequencing and Sanger sequencing–based variant calling (Supplementary Table 4, Supplementary Figure 2), demonstrating the reliability and robustness of our SNuPE approach.
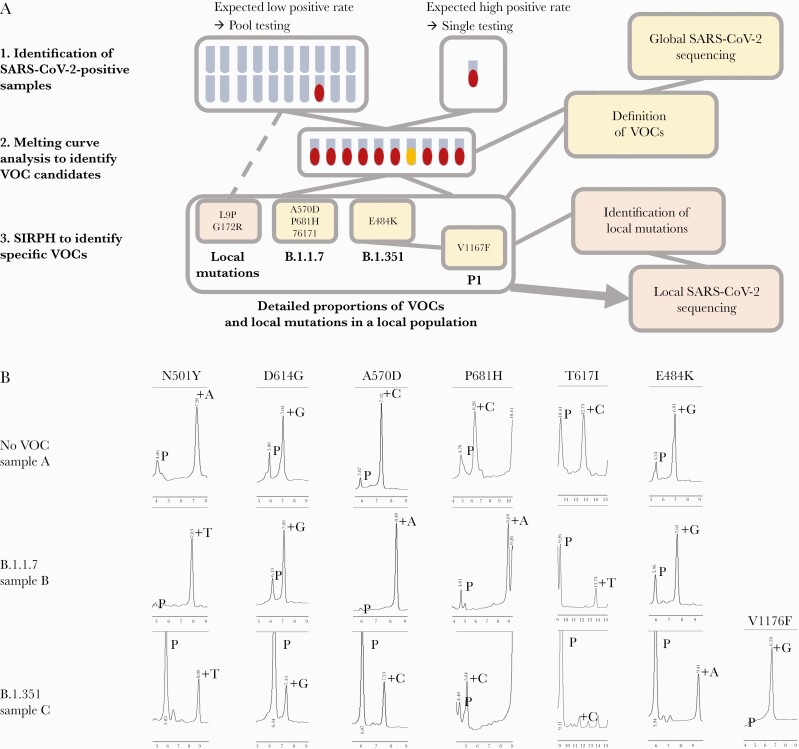
Inspired by these results, we developed a rapid screening strategy for prospective high-throughput SARS-CoV-2 and subsequent VOC testing (Figure 2A). Asymptomatic individuals, that is, health care workers of the local university hospital, were screened by SARS-CoV-2 pool testing in pools of 16, while samples from symptomatic people or public health services (contact tracing) with expected positivity rates above 2% were investigated by individual SARS-CoV-2 RT-PCR. Positive nucleic acid extracts were immediately subjected to N501Y mutation-specific RT-PCR melting curve analysis to identify candidates of currently circulating VOCs. Subsequent SNuPE analysis was performed to clearly identify and assign the respective VOCs.
Figure 2.
High-throughput screening strategy for variant monitoring in a local population. A, Iterative workflow from identification of SARS-CoV-2-positive samples (1) followed by variant candidate identification by RT-PCR melting curve analysis (2) and final identification of specific VOCs as defined by global sequencing efforts. Detailed proportions of VOCs or local variants of interest identified by SIRPH analysis can then be used for selection of samples in local sequencing projects. It should be noted that the panel of analyzed mutations both in the melting curve analysis and our SIRPH assay is adapted continuously to newly emerging mutations. B, HPLC separation profiles of the VOC characterizing PMS set for an exemplary non-VOC (sample A), a B.1.1.7 (sample B) and a B.1.351 (sample C) variant as identified in the workflow described in (A). Abbreviations: Mut, mutation; PMS, potentially mutated sites; RT-PCR, reverse transcription polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SIRPH, single nucleotide primer extension approach; VOC, variant of concern; Wt, wild-type.
We applied this approach for a routine test on a large cohort to identify VOCs in Saarland generating reliable and highly accurate results. From 2059 pooled and 2987 individual respiratory samples from nasopharyngeal swabs collected between January 25 and 31, 2021, 542 samples tested SARS-CoV-2 positive by RT-PCR. Within these, we identified 13 VOC candidates by RT-PCR melting curve analysis–based screening during this early phase of VOC introduction into Saarland. All candidates were submitted to SIRPH analysis for confirmation and further VOC assignment, revealing the presence of all characteristic mutations 501Y, 570D, 681H, and 716I in 12 out of 13 predefined samples, clearly classifying them as B.1.1.7, while 1 sample was identified as B.1.351 (Figure 2B; Supplementary Table 5). The whole process lasted about 24 hours from positive RT-PCR testing to PMS sequence validation.
Now, we routinely use this approach for VOC screening in Saarland. Given the dynamic situation of the pandemic with newly emerging variants, we continuously adapt our SIRPH analysis to new VOCs such as the recent lineage B.1.617. We added an SIRPH assay for L452R and extended the E484 assay to discriminate between E484, E484K, and E484Q (Supplementary Figure 1, Supplementary Table 1), allowing the identification of B.1.617 lineages (Supplementary Table 6).
DISCUSSION
Our presented strategy shows that a combination of prescreening by (pooled) SARS-CoV-2 and subsequent RT-PCR melting curve analysis, together with a confirmatory SIRPH analysis, generates fast and reliable results to speed up monitoring of VOCs in larger cohorts. We demonstrate how this strategy can quickly be implemented for VOC monitoring to rationalize local public health interventions by supporting the tracing of VOCs. While whole-genome sequencing will remain the gold standard for virus monitoring and the detection of new mutations, the proposed screening by PCR/SIRPH has several advantages. First of all, the method is robust, fast (24 hours), and very cost-effective compared with NGS confirmation. Notably, it robustly calls regions with coverage dropouts in sequencing data (Supplementary Figure 2). It can even be applied to samples with low viral load (Ct 28–36) (Supplementary Table 6) that typically fail or result in low-quality data during sequencing. Furthermore, the method is flexible and can be implemented on a variety of existing RP-HPLC systems. In addition, we perform SNuPE assays as “1-tube” reactions that can be multiplexed. New mutation-specific assays can be rapidly established within 2 working days to include newly emerging variants, allowing for fast and precise population surveillance. The combined pool PCR/SIRPH approach can be used to flank sequencing surveillance programs; in Germany, only up to 5% (or 10% when positive cases drop below 70 000 cases per week) of randomly chosen positive cases will be full genome–sequenced. The emergence and spreading of local variants call for fast and thorough monitoring of the entire population in a timely manner. As time is crucial for surveillance, our method offers base-specific results in 1 day, as compared with NGS-based sequencing, which usually requires up to 5 working days. Ongoing sequencing of full viral genomes in a proportion of samples will be used to screen for the development of new SARS-CoV-2 variants. In an iterative process, the information gained by sequencing can be used to easily adapt the SIRPH approach to new VOCs that may emerge in the future course of the pandemic.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
The authors wish to acknowledge the contributions of the technicians Katrin Thieser, Luisa Scheidhauer, Helga Appel, and Barbara Best.
Financial support. This work was supported by Staatskanzlei Saarland (SaarCoSeq) to Sigrun Smola and Jörn Walter and by the Ministry of Health Saarland (SaarCoMM) to Prof. Smola. Abdulrahman Salhab was supported by the German Federal Ministry of Research and Education grant for de.NBI (031L0101D).
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Patient consent. The study was approved by the local ethics committee of the Saarland University Medical Center at Saarland Ärztekammer. The study does not include factors necessitating patient consent.
Data accession. Fasta files of full viral genome sequencing can be accessed on GISAID (https://www.gisaid.org/) using the identifiers listed in Supplementary Table 7.
References
- 1.Davies NG, Barnard RC, Jarvis CI, et al. . Estimated transmissibility and severity of novel SARS-CoV-2 variant of concern 202012/01 in England. Epidemiology. In press. [Google Scholar]
- 2.Tegally H, Wilkinson E, Giovanetti M, et al. . Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. Epidemiology. In press. [Google Scholar]
- 3.Voloch CM, Francisco RdS Jr, de Almeida LGP, et al. . Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. J Virol 2021; 95:e00119–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greaney AJ, Starr TN, Gilchuk P, et al. . Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe 2021; 29:44–57.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Public Health England. Investigation of novel SARS-CoV-2 variant: variant of concern 202012/01 (technical briefing 5). 2021. Available at: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/959426/Variant_of_Concern_VOC_202012_01_Technical_Briefing_5.pdf. Accessed 14 January 2021.
- 6.Winger A, Caspari T. The spike of concern—the novel variants of SARS-CoV-2. Viruses 2021; 13:1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lohse S, Pfuhl T, Berkó-Göttel B, et al. . Pooling of samples for testing for SARS-CoV-2 in asymptomatic people. Lancet Infect Dis 2020; 20:1231–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tierling S, Sers C, Lehmann A, Walter J. A fast, cost-efficient and sensitive approach for KRAS mutation detection using multiplexed primer extension with IP/RP-HPLC separation. Int J Cancer 2012; 130:567–74. [DOI] [PubMed] [Google Scholar]
- 9.Hoogendoorn B, Owen MJ, Oefner PJ, et al. . Genotyping single nucleotide polymorphisms by primer extension and high performance liquid chromatography. Hum Genet 1999; 104:89–93. [DOI] [PubMed] [Google Scholar]
- 10.Tyson JR, James P, Stoddart D, et al. . Improvements to the ARTIC multiplex PCR method for SARS-CoV-2 genome sequencing using nanopore. Genomics. In press. [Google Scholar]
- 11.Hadfield J, Megill C, Bell SM, et al. . Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 2018; 34:4121–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Volz E, Hill V, McCrone JT, et al. . Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell 2021; 184:64–75.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.