Abstract
The evolution the novel corona virus disease (COVID-19) as a pandemic has inflicted several thousand deaths per day endangering the lives of millions of people across the globe. In addition to thermal scanning mechanisms, chest imaging examinations provide valuable insights to the detection of this virus, diagnosis and prognosis of the infections. Though Chest CT and Chest X-ray imaging are common in the clinical protocols of COVID-19 management, the latter is highly preferred, attributed to its simple image acquisition procedure and mobility of the imaging mechanism. However, Chest X-ray images are found to be less sensitive compared to Chest CT images in detecting infections in the early stages. In this paper, we propose a deep learning based framework to enhance the diagnostic values of these images for improved clinical outcomes. It is realized as a variant of the conventional SqueezeNet classifier with segmentation capabilities, which is trained with deep features extracted from the Chest X-ray images of a standard dataset for binary and multi class classification. The binary classifier achieves an accuracy of 99.53% in the discrimination of COVID-19 and Non COVID-19 images. Similarly, the multi class classifier performs classification of COVID-19, Viral Pneumonia and Normal cases with an accuracy of 99.79%. This model called the COVID-19 Super pixel SqueezNet (COVID-SSNet) performs super pixel segmentation of the activation maps to extract the regions of interest which carry perceptual image features and constructs an overlay of the Chest X-ray images with these regions. The proposed classifier model adds significant value to the Chest X-rays for an integral examination of the image features and the image regions influencing the classifier decisions to expedite the COVID-19 treatment regimen.
Keywords: COVID-19, Chest X-Ray, Super pixel, GMM, SqueezeNet
1. Introduction
The novel corona virus disease (COVID-19) has emerged as a pandemic threat and public health concern over the world. Existing institutional arrangements and prevailing healthcare priorities in COVID-19 management are focused on a person-centered, cure-centric system rather than being socially sustainable. Health systems need to be revamped, be socially and economically sustainable, and address systemic drivers that currently limit accessibility, equity and affordability of care. COVID-19 pandemic has re-emphasized the urgent need to redesign health systems to prioritize the broader social determinants of health. This redesign will need to be approached in multiple ways to ensure a long-term healthcare model that acknowledges the current constraints on the existing systems.
Recent researches on COVID-19 management advocate the need for building a sustainable and health environment employing artificial intelligence and touch-less approaches (Megahed & Ghoneim, 2020). In line with this, an extensive survey on deep learning approaches for COVID-19 detection and containment in smart cities is presented in Bhattacharya et al. (2021) and Murugappan, Bourisly, Krishnan, Maruthapillai, and Muthusamy (2021). The authors review several deep learning paradigms for medical image analysis in COVID-19 outbreak prediction, infection tracking, diagnosis, treatment, and drug research. This paper provides deep insights on various deep learning approaches in combatting COVID-19 and advocates the need to design COVID-19 detection systems with optimum accuracy. The mortalities due to COVID-19 pandemic can be considerably reduced by early detection of the infections, isolation of the subjects and administration of anti-viral drugs. At present, the Reverse Transcription Polymerase Chain Reaction (RT-PCR) (Wang, Xu, et al., 2020) test on clinical specimens, is the most widely employed screening protocol which is time consuming, highly sensitive to infinitesimal DNA contamination and less accurate.
Along with examinations of symptoms and pathogenic testing, imaging examinations are found indispensable in the diagnosis in the screening, detection, diagnosis and prognosis of COVID-19 (Aradhya et al., 2021, Bhapkar et al., 2021, Dey et al., 2020, Kaiser et al., 2021, Murugappan et al., 2021, Singh et al., 2021). In a study on imaging modalities in the diagnosis of COVID-19, Yang et al. (2020) have shown that Computed Tomography (CT) images are very effective in capturing Ground Glass Opacity (GGO), consolidations and patchy areas in the peripherals of the lungs, in the early and advanced stages of infections. This investigation also reveals that Chest X Ray (CXR) images are less sensitive to these characteristics in the early stages whereas progressive opacities and consolidations are captured well in the advanced stages. However, CXR is recommended as an initial screening tool due the difficulties encountered in establishing CT scanning facilities in low resource settings and shifting patients to the CT scanning suites.
With the infiltration of COVID-19, time consumption in CT scanning and susceptibility of infections at the CT scanning sites, there is a growing need for the detection of valuable diagnostic features from CXR images. Significant research has been conducted in this context and deep learning models with high sensitivity towards COVID 19 features in CXR images have been proposed. A deep Convolutional Neural Network (CNN) called the COVID-Net (Wang, Lin, & Wong, 2020) based on residual Projection Expansion Projection Extension (PEPX) demonstrates an accuracy of 93.3% and 91% sensitivity on an exclusive dataset.
In addition to COVID-19 detection, segmentation of infections can provide great insights to accelerate clinical decisions. Generally, UNet (Ronneberger, Fischer, & Brox, 2015) an improved version of the Convolutional Network (CNN) architecture, is widely employed in the semantic segmentation of biomedical images. It follows a symmetric encoder–decoder structure with several upsampling and downsampling layers with skip connections between every level to propagate gradients from low resolution images to higher resolution ones. Several variants of UNet have been introduced in the recent years to harness the potential of the symmetric UNet architecture.
The Residual Dilated Attention Gate-UNet (RDA-UNet) (Zhuang, Li, Joseph Raj, Mahesh, & Qiu, 2019) is built with residual units and attention gates in each layer. In addition, dilated kernels are adopted to improve the network performance by expanding the receptive field. This model employed in lesion detection from breast ultrasound images was enhanced with a Generative Adversarial Network (GAN) to eliminate false positives and precise segmentation of boundaries. This model called the Residual Dilated Attention Gate UNet Wasserstein GAN (RDA UNET WGAN) (Negi, Raj, Nersisson, Zhuang, & Murugappan, 2020) trains faster and is highly stable compared to the RDA-UNet. Generally, a GAN comprises a generator and discriminator which compete with each other. The generator produces new samples with the same probability distribution as the training samples and the discriminator attempts to determine whether the samples are genuine or fake. Once the generator can give a set of samples that have high similarity to the training samples, it can generate incrementally higher-quality samples. WGAN is a GAN variant implemented with two separate neural networks and a gradient reversal layer, for computing exact gradients of the 1-Wasserstein distance, contributing to improved stability.
The Attention Gate-Dense Network-Improved Dilation Convolution-UNET (ADID (Raj et al., 2021), for lung lesion segmentation from chest CT images for COVID-19 detection is implemented with attention gate mechanism, dense networks and dilation convolution mechanisms. The attention module focuses on the target lesions of arbitrary shapes and sizes for precise segmentations. The dense connections between the dilation convolution layers and the skip connection reduce the gradients and enhance the localization ability. The Eff-UNet (Baheti, Innani, Gajre, & Talbar, 2020) is built with a compound scaled EfficientNet as encoder and a UNet decoder for segmentation in unstructured environments. This model incorporates low level spatial information and high-level features for precise segmentation of objects from road scenes. The Multiscale Statistical U-Net (MSU-Net) (Wang et al., 2019) for cardiac MRI segmentation employs a Statistical Convolutional Neural Network (SCNN) which models the inputs as multi scale canonical distributions to speed up the segmentation process, also exploiting spatio-temporal relationships between the samples. Further, the UNet is realized as a parallel architecture to statistically process the inputs. A detailed analysis of the architectures of UNet and its variants reveal that the performance of these networks considerably increases with the addition attention gate, densenet and dilation convolution modules.
Evincing the need for an integral model for COVID-19 detection and infection segmentation, we propose a light weight deep learning classifier based on SqueezeNet (Iandola et al., 2016) for COVID-19 detection from CXR images. This model is also coupled with a segmentation module to semantically separate the Region of Interest (ROI) using super pixels for a thorough examination of the images. We call our proposed framework as the COVID-19 Super pixel SqueezNet (COVID-SSNet) model. The contributions of this research are as below.
-
1.
A lightweight model for COVID-19 detection and segmentation which more than 99% accuracy for binary and three class classifications is proposed.
-
2.
It is established that a classification-segmentation pipeline is ideal for COVID-19 detection and infection segmentation.
-
3.
Gradient based class activations coupled with super pixel segmentation provide best classification and segmentation abilities of the SqueezeNet based model.
Performance evaluation of our model with a standard dataset and, interpretations of visual and quantitative results signify its superiority compared to the state-of-the-art classification and segmentation models.
This paper is organized as below. In Section 2, we present a review of the relevant work in the context of our research. The dataset, methods employed in realizing the framework and details of implementation are described in Section 3. In Section 4, we present the architecture of the proposed framework with schematic diagrams. In Section 5, we describe our experimental results, performance analyses and comparisons. The paper is concluded in Section 6.
2. State-of-the-art
2.1. Deep CNN models for COVID-19 detection from CXR images
In this section we give a comprehensive review on the existing deep learning models for COVID-19 detection from CXR images. Radiological studies (Salehi, Abedi, Balakrishnan, & Gholamrezanezhad, 2020) show that GGOs, peripheral distribution and bilateral involvement are widely observed in CXR images. A deep CNN model for COVID-19 detection must be capable of learning these features from the CXR images.
Narin, Kaya, and Pamuk (2020) have employed three pre-trained deep learning models for COVID-19 detection from CXR images. The authors have tested the convolutional ResNet50, InceptionV3 and Inception ResNet V2 networks on a small dataset with 50 COVID-19 and 50 normal CXR images and report the highest classification accuracy of 98% with ResNet50. Similarly, the MobileNetv2 and VGG19 models demonstrate high accuracy and specificity compared to the Inception, Xception and Inception ResNet V2 models, tested with two relatively larger datasets comprising CRX images of COVID-19, pneumonia and normal cases in Apostolopoulos and Mpesiana (2020). Though two class and three class accuracies are best for the VGG19, specificity is reportedly high with the MobileNetv2 which signifies low falsified negative predictions of individual disease classes.
In Apostolopoulos, Aznaouridis, and Tzani (2020), the MobileNetv2 is extended to the detection of six pulmonary diseases including edema, effusion, fibrosis, emphys, pneumonia and Covid19, achieving 99.18% accuracy in COVID-19 detection. Inspired by the high classification accuracy of this model, the authors advocated further investigation of the extracted features to standardize them as potential biomarkers. Farooq and Hafeez (2020) have introduced Covid-ResNet in the detection of COVID-19, viral and bacterial infections and normal cases from the COVIDx dataset, achieving an overall accuracy of 96.23%. This classifier, a variant of the pre-trained ResNet50 model is adaptively fine-tuned in three stages with decaying learning rates, ensuring the consistency of the weights in the layers, contributing to the stability of the classifier. Covidx-net (Hemdan, Shouman, & Karar, 2020) is a unified framework of binary classifiers for COVID-19 detection from a small subset of CXR images from the public COVID-19 dataset. This framework comprising seven deep CNNs, all trained with unique learning rates demonstrates 90% accuracy with VGG19 and Densenet models. Capsule (Hinton, Sabour, & Frosst, 2018) networks based on Expectation Maximization (EM) are demonstrated to exhibit better accuracy on smaller datasets compared to CNNs. The COVID-CAPS (Afshar et al., 2020) is a binary COVID-19 classifier built on a capsule network, shown to achieve an accuracy of 95.7% and 98.3% without and with pre-training respectively on a small CXR dataset. The performance of this model matches the one proposed in Sethy, Behera, Ratha, and Biswas (2020) which employs ResNet50 for deep feature extraction and Support Vector Machine (SVM) for classification.
In addition to classification, CNN models are also employed in the segmentation of lungs from CXR images in the COVID-19 diagnostic pipeline. Lung segmentation from CXR images is essential in the detection of lung nodules, quantification and staging of infections. The adversarial U-Net (Gaál, Maga, & Lukács, 2020) architecture for lung segmentation is found to generalize well with arbitrary CXR datasets with a Dice Score of 97.5%. Similarly, COVID_MTNet (Alom, Rahman, Nasrin, Taha, & Asari, 2020) is an integral framework for COVID-19 detection and segmentation of infected regions from both CT and CXR images. It is implemented with two distinct networks, an Inception Recurrent Residual Convolutional Neural Network (IRRCNN) model for COVID-19 detection and a NABLA-N (Alom, Aspiras, Taha, & Asari, 2019) network for segmentation of infected regions.
A deep learning framework for COVID-19 detection presented in Sharifrazi et al. (2021) follows a fusion approach with sobel filtering, CNN and SVM. Initially, the edges of the CXR images are extracted with the sobel filters and fed as input to the CNN. The CNN outputs are further classified by a SVM and a Sigmoid function separately. Experimental results show that combination of Sobel, CNN and SVM provides the best classification accuracy of 99.02%.
A deep CNN called the (DeTraC) (Abbas, Abdelsamea, & Gaber, 2021) performs decomposition, transfer learning and composition operations in sequence, to eliminate irregularities in the test data, extracting principal components of the CXR images. This process reduces the size of the CXR feature space and the original classes are decomposed into a subset of classes. An ImageNet (Deng et al., 2009) pretrained CNN is then trained with these components and the class labels corresponding to the decomposed classes are assigned. VGGNet (Alam, Ahsan, Based, Haider, & Kowalski, 2021) is a CNN classifier model built by training a VGG-19 model with fused features extracted from CXR images with Histogram Oriented Gradient (HOG) descriptors and a customized convolutional neural network (CNN). In this model Anisotropic diffusion is applied to filter the noise from the test images before classification. Following COVID-19 identification from a test image, Watershed segmentation is used to segment the infected regions.
A customized CNN model called the CoroDet (Hussain et al., 2021) performs COVID-19 detection, binary, 3-class and 4-class classification of CXR and CT images. It is built with convolution, pooling and dense layers incrementally evaluating combinations of the layers and activation functions. The 22 layer model achieves classification accuracies of 91.2%, 94.2% and 99.1% for 4-class, 3-class and binary classifications of COVID-19, Viral Pneumonia, bacterial pneumonia and normal cases. A comprehensive review (Nayak, Nayak, Sinha, Arora, & Pachori, 2021) on the application of deep learning approaches for COVID-19 detection from CXR images evaluates the binary classification performance of eight pre-trained models. Experimental analyses with pre-trained AlexNet, VGG-16, GoogleNet, MobileNet-V2, SqueezeNet, ResNet-34, ResNet-50 and Inception-V3 models on normal and COVID-19 images show that the best classification accuracy of 98.33% is achieved by ResNet-34. Though the structure of ResNet-50 is deeper than ResNet-34, it achieves an accuracy of 97.50 for a batch size of 32. This result signifies that better results can be achieved by comparatively less deeper networks trained at optimal learning rates.
2.2. Interpretation of outputs in CNN virus detection models
Though the CNN models are demonstrated to exhibit high accuracy in the discrimination of COVID-19 and other bacterial and viral pneumonia from CT and CXR image, their behavior is not understandable due to their intrinsic black box nature. According to Shi et al. (2020), analyses of CXR images reveal frequent occurrences of GGOs in peripheral, posterior, medial and basal areas, air space consolidation, traction bronchiectasis, traction bronchiectasis and septal thickening in COVID-19 patients. Rather than completely relying on the quantitative performance metrics such as accuracy and precision, examination of the Region of Interest (ROI) activating the classifier models can improve the clinical decisions. In this context, several research works have been performed for visualization and interpretation of the CNN models by localization of the ROIs relevant to the input classified. In Wang et al. (2017), the authors have shown that a disease detection and localization framework can be constructed with a multi class classifier and gradient based algorithm to detect and localize pneumonia in CXR images. Similarly, discrimination of pneumonia versus normal cases and bacterial versus viral pneumonia from CXR images, performed with an Inception V3 model was followed by an occlusion test to identify the significant ROI contributing to the decision of the network in Kermany et al. (2018).
Similarly, the Chexnet (Rajpurkar et al., 2017) a 121 layered dense CNN trained with CXR images determines the probability of 14 pathologies and localizes pneumonia with activation maps. A two branch Attention Guided CNN (AG-CNN) (Guan et al., 2018) based on ResNet, fuses global features extracted from the CXR images with those from salient regions of these images, segmented with a mask derived from the heatmaps. It is shown that best accuracy is achieved with combined features rather than classifications with global and local features. Class Activation Mapping (CAM) (Zhou, Khosla, Lapedriza, Oliva, & Torralba) which generates heatmaps from feature maps, highlighting salient regions of the images is vital in the visual interpretation of the behaviors of the CNN models. A generalized Gradient-weighted Class Activation Mapping (Grad-CAM) (Selvaraju et al., 2017), constructs class discerning localization maps from the gradients of the target class and the image feature maps for diverse CNN classifier models. In the detection of pneumonia from pediatric (Rajaraman, Candemir, Kim, Thoma, & Antani, 2018) CXRs, Grad-CAM is used to visualize the outputs of a customized VGG16 to localize the ROI. Similarly, the DarkCovidNet (Ozturk et al., 2020) model for binary and multi class COVID-19 classification with CXR images, also presents the heat maps to the radiologists to analyze the behavior of the model. The radiologists find that these maps are effective in the discrimination of normal COVID-19 cases, manifesting high intensity concentrations in the affected regions. Chowdhury, Rahman, et al. (2020) report a classification accuracy of 98.3% for both binary and multi class COVID-19 classifications with a SqueezeNet model. The authors show that the discriminating features of COVID-19, viral pneumonia and normal images are detected by the 14th layer of the SqueezeNet model.
2.3. Super pixel based knowledge infusion in deep learning models
Super pixels are groups of semantically similar pixels, carrying high-level information which provide a compact representation of images. While conventional deep learning models are trained to learn features from raw images, recently computer vision models are constructed as CNNs infused with domain knowledge captured using super pixels. A hybrid model employing a CNN to capture spatial information and Graph Neural Network (GNN) to capture information from super pixels is proposed in Chhablani, Sharma, Pandey, and Dash (2021). This model Evaluated with standard and domain specific image datasets demonstrates that superior prediction performances can be achieved by incorporating domain knowledge with CNN classifiers. In line with this, the SP-CNN (Xie, Gao, Jin, & Zhao, 2021) based on super pixel pooling, encoder–decoder architecture and transfer learning is proposed for classification of Hyper Spectral Images (HSI). Experimental results with benchmark datasets show that best classification accuracies are achieved by fusing spatial structures captured by the super pixels, with spectral features.
Similarly, a deep learning framework for skin lesion classification is proposed in Afza, Sharif, Mittal, Khan, and Jude Hemanth (2021). Initially, the dermoscopy images are subjected to super pixel segmentation to separate the lesions. Then a pretrained ResNet-50 model is trained with the lesion images for feature extraction by transfer learning. The features extracted are further optimized with a bio inspired algorithm and then classified with a naïve Bayesian classifier. In Tello-Mijares and Woo (2021), a two-level classifier employing Simple Linear Iterative Clustering (SLIC) (Achanta et al., 2012) super pixel clustering approach for automated detection of GGOs and pulmonary infiltrates is presented for follow-up assessment in COVID-19 patients. From a detailed review of literature on deep learning models for COVID-19 detection, interpretations of the classifier outputs and significance of super pixel driven classifier models, it is evident that unified approaches incorporating super pixel segmentation and deep learning approaches are getting wide research attention recently. Efficient deep learning classifier models leveraging super pixel segmentation for COVID-19 detection are yet to be benchmarked. In this research, we strive to bridge this gap with a novel classifier for COVID-19 detection from chest X-ray images, which can be generalized to other modalities.
3. Materials and methods
In this section, we describe our dataset and the methods employed in the construction of the proposed classification-segmentation framework.
3.1. Model assumptions
The proposed model is realized with a classification-segmentation pipeline with the following assumptions.
-
1.
Due to variable nature of COVID-19 symptoms, COVID-19 detection from segmented ROI may lead to ambiguous feature learning of the model. Hence, the proposed model is realized as a cascaded classification-segmentation framework which facilitates learning biomarkers by visualizing classifier outputs.
-
2.
Gradients of a target class flowing to the final learnable layer of a classifier highlight the discriminating features influencing the classifier decisions. Hence color based GMM super pixel segmentation is employed to localize the homogeneous regions of the Grad-CAM which guide the classifier in assigning a target label. This capability helps to identify infections of various degrees ranging from mild to intensive manifestations.
3.2. Dataset and experimental setup
In this research, we have constructed training and testing datasets from the award winning Kaggle CXR public database (Chowdhury et al., 2020), comprising 219 COVID-19 positive images, 1345 viral pneumonia images and 1341 normal images. Originally these images are of dimension 1024 × 1024 in png format. The database is divided into two distinct training and testing subsets, with images resized to 227 × 227. Then the training dataset is augmented with images generated by applying the translation, rotation and scaling operations on the training images. The description of the dataset is given in Table 1. The proposed framework is implemented with Matlab 2021b software in an i7-7700K processor with 32GB DDR4 RAM equipped with a NVIDIA GeForce GTX1060 3GB Graphics card.
Table 1.
Description of training and testing dataset.
| Infection | No. of images in database |
No. of training images |
No. of training images after image augmentation |
No. of testing images |
|
|---|---|---|---|---|---|
| Binary classification |
Multiclass classification |
||||
| COVID-19 | 219 | 119 | 476 | 100 | 100 |
| Viral Pneumonia | 1345 | 645 | 2580 | 1400 | 700 |
| Normal | 1341 | 641 | 2564 | 700 | |
3.3. SqueezeNet architecture
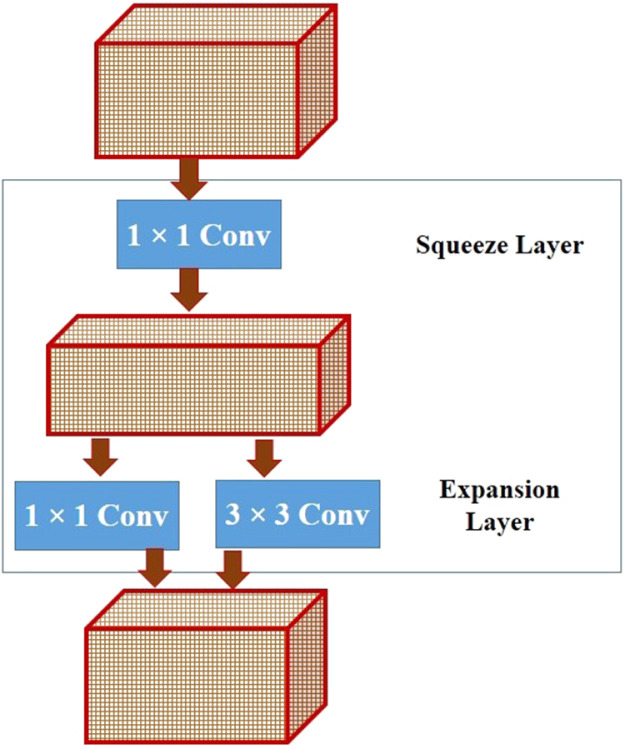
As stated earlier, we have employed a variant of the SqueezNet model for extraction of deep features from the candidate images for classification in this research. This model characterized by fewer parameters is considerably smaller and highly accurate compared to the conventional CNNs. It consists of a stack of fire modules shown in Fig. 1, comprising a squeeze layer of 1 × 1 filters and an expansion layer comprising 1 × 1 and 3 × 3 filters. While the squeeze layer downsamples the input channels, the expansion layer performs cross channel pooling with the 1 × 1 filters and captures the spatial structures with the 3 × 3 filters, resulting in expansion.
Fig. 1.
Fire module in SqueezeNet.
The conventional SqueezNet classifier architecture proposed in Alom et al. (2019) is shown in Fig. 2 which comprises the convolutional layers, fire modules, max pooling layers and soft max layers.
Fig. 2.
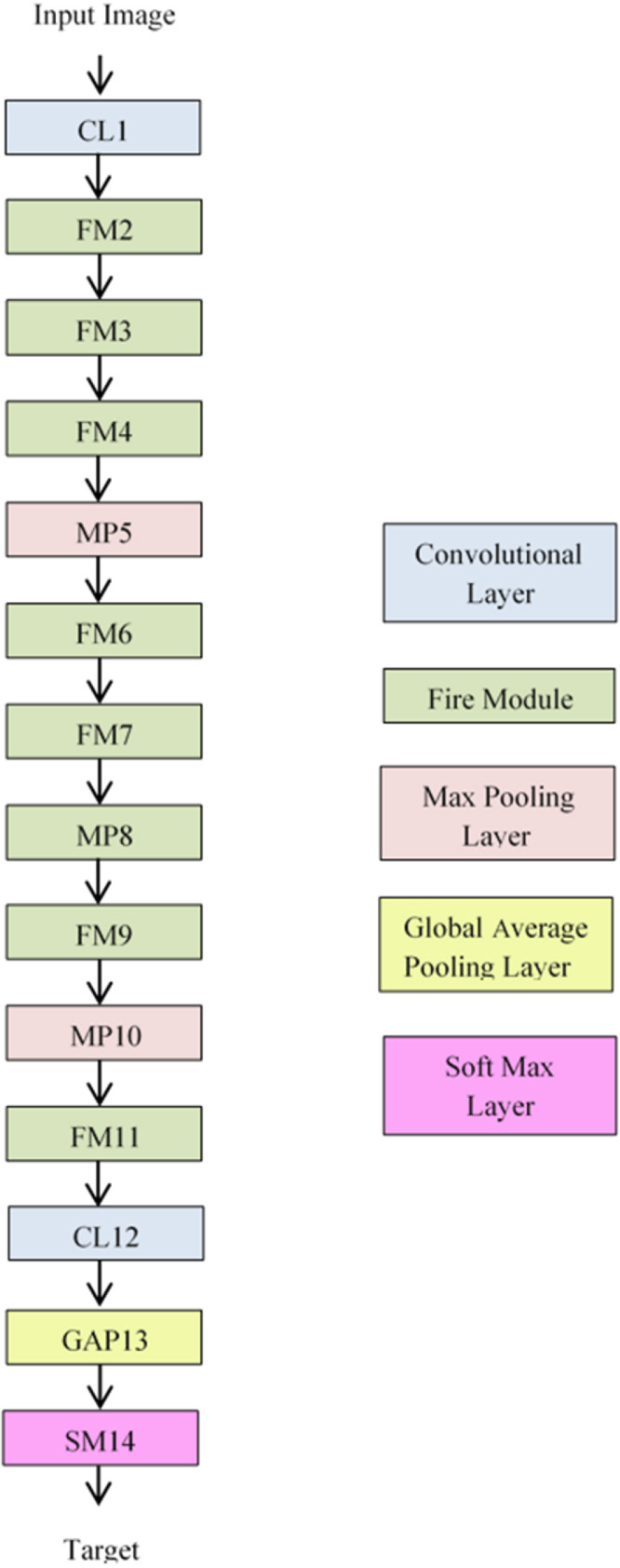
SqueezeNet architecture.
3.4. Gaussian mixture model superpixels
Super pixels are group of pixels with common characteristics such as pixel intensity, color and energy. Images are segmented into super pixels of arbitrary shape and size, for object detection in computer vision problems such as semantic labeling, semantic segmentation, object tracking etc. The SLIC algorithm constructs super pixels which are approximately equal in size by clustering the pixels based on their proximity and similarity in color, guided by a weight factor. However, these pixels are not accurate as the SLIC segmentation results in several isolated regions, influenced by the number of the clusters assumed by the kmeans segmentation algorithm.
| (1) |
| (2) |
The Gaussian Mixture Model (GMM) (Smith, Kindermans, Ying, & Le, 2018) super pixels exhibit better segmentation accuracies than the SLIC algorithm, as each super pixel is modeled as a Gaussian representation, by randomly choosing a distribution initially from a group of Gaussian distributions. A Gaussian distribution is associated with a covariance , mean and the mixing probability, such that the condition in Eq. (1) is satisfied. Maximum Likelihood (ML) estimation is performed to find if the data points are fitted within each Gaussian.
For an arbitrary data point x, the probability that it belongs to a Gaussian is given in Eq. (2) where the latent variable takes the value 1 when belongs to the cluster n and 0 otherwise.
| (3) |
| (4) |
| (5) |
For a Gaussian , the mixing coefficient is given in Eq. (3) which is the probability that a data point belongs to . The set of all latent variables is represented as and the probability with which all the data points belong to the Gaussians is given in Eq. (4). For a given data point , the probability that it belongs to a Gaussian is given in Eq. (5).
| (6) |
The probability of a latent variable such that a data point exists in a Gaussian n is given in Eq. (6). The parameters of this model determined by EM are collectively expressed as .
| (7) |
Given a 2D image I of dimension MK, the total number of pixels labeled , can be assigned to a super pixel which is analogous to the clustering P data points into Gaussians. For a pixel , the superpixel label is computed as in Eq. (7)).
It can be seen that these labels are computed from the posterior probability after evaluation of for a cluster n, which facilitates precise grouping of pixels compared to the SLIC algorithm. In this paper, we employ superpixel segmentation to segmentation the ROI from the heat maps.
4. Proposed work
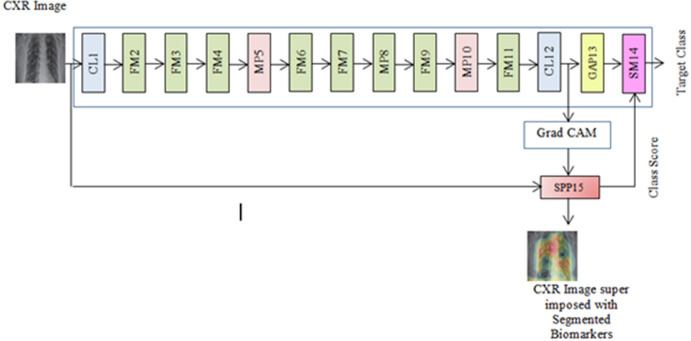
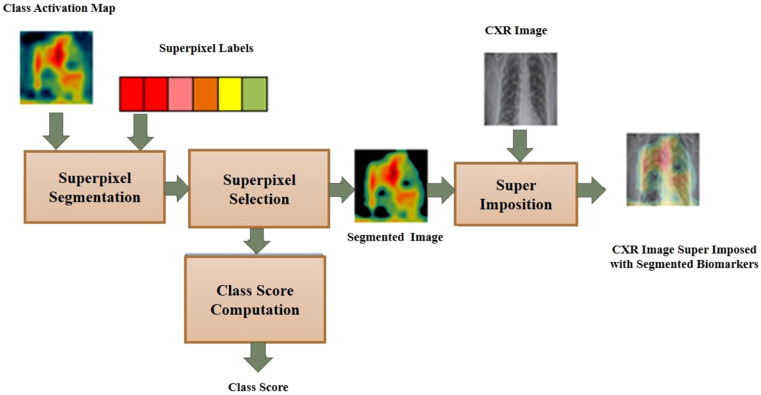
The proposed COVID-SSNet model realized as an integral framework merging the classification and segmentation processes is illustrated in Fig. 3. This is built by extending the standard SqueezeNet classification model with Grad-CAM and super pixel pooling. In this framework, we apply the Grad-CAM on the image feature maps from the final convolutional layer CL12 to construct the activation map. This heat map is given as input to the Super Pixel Pooling Layer (SPP) for segmentation and a normalized class score in the range [0 1] is computed from the active super pixels. The schematic of this layer depicting the super pixel segmentation operations is shown in Fig. 4.
Fig. 3.
SqueezeNet architecture.
Fig. 4.
Superpixel pooling.
Followed by classification, we segment the activation maps to separate the ROI attributing to the classification decision. In an activation map, red regions heavily influence the classification decisions while the blue regions are less significant. From an empirical analysis on the super pixel labeling based on mean RGB components, we perceive that the super pixels are assigned one of the labels from a set of colors Red, Orange, Red-Orange, Maroon, Yellow, Olive, Gray, Blue, Black . The potential regions focused by the network contain super pixels labeled Red, Orange, Red-Orange, Maroon, Yellow and Olive. The super pixels of the activation maps are selectively merged based on their color label using Algorithm (shown in Fig. 5) to construct the segmented activations maps which are then overlaid on the test images for localization of the infected regions.
Fig. 5.
Pseudocode for activation map segmentation.
The computation of the class score for the segmented heat map is described below. For each input image , let be its heat map and be the set of superpixels of . For each superpixel, , an activation score is assigned corresponding to the mean color of the super pixel as 6-Red, 5- Orange, 4-Red-Orange, 3-Maroon, 2-Yellow, 1-Olive, 0- Gray, Blue, Black. The class score is computed as the summation of the activation scores in Eq. (8) and normalized in the range [0 1] by min–max normalization.
| (8) |
The softmax layer SM14 assigns the target class label to the input images based on the class score of each image and the input from the Global Average Pooling layer GAP13.
5. Experimental results and discussion
In this paper, we follow deep transfer learning employing the pre-trained SqueezeNet model which is trained on the ImageNet dataset. We further fine-tune this network training it with the training dataset constructed from Singh et al. (2021). The SqueezeNet model is initially trained on the training dataset for binary and three class classifications and tested with the respective test datasets. As shown in Table 1, the dataset is separated into training and testing subsets. For binary classification, the images ascribing to the viral pneumonia and the normal cases are merged for training and testing.
The hyperparameters of the proposed framework are given in Table 2. The values of these parameters are finalized after several empirical evaluations of the stability of the proposed system. The Stochastic Gradient Descent with Momentum (SGDM) optimizer employs the momentum, a moving average of the gradients. SGDM iteratively updates the weight of the network with a linear combination of the gradients and the previous updates, while training the model. Here, the contribution of the previous updates is maximized by assuming 0.9 for the momentum. The framework was initially trained for 50 epochs with a batch size of 32 and a constant learning rate of 0.001. With the model weights optimized by SGDM and varying the batch size, best accuracy was obtained at 100 epochs for a batch size of 128.
Table 2.
Hyper parameters of COVID-SSNet.
| Parameter | Value |
|---|---|
| Maximum Epochs | 100 |
| MiniBatchSize | 32,64,128 |
| Momentum | 0.9 |
| Learning Rate | 0.001 |
| L2 Regularization | 0.0001 |
| Optimization | SGDM |
For fine-tuning the pre-trained SqueezeNet for binary and three class classifications for COVID-19 detection, the last learnable convolutional layer, was modified with 2 and 3 filters separately to discern the target class of the input image. Further, to prevent the model from over-fitting, L2 regularization is applied to keep the weights and bias values smaller. The regularization factor is assumed to be 0.0001 to reduce the network weights slowly.
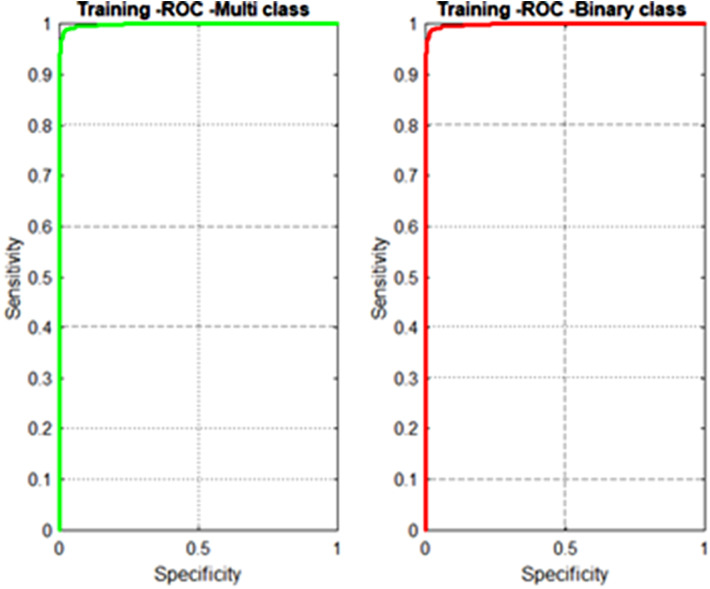
By transfer learning, we train the SqueezeNet model with 476 COVID 19 images, 2580 Viral Pneumonia images and 2564 normal images for multi class classification. Similarly, for binary classification the model is trained with 476 COVID 19 images and 5144 Non-COVID-19 images. The training accuracies of the proposed model for binary and multi class classification are shown as Receiver Operator Characteristics (ROC) curves in Fig. 6 signifying a high training accuracy.
Fig. 6.
ROC curves for training binary and multiclass classifiers.
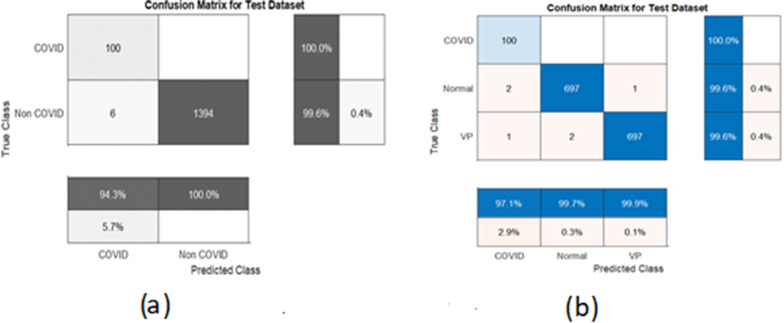
We exercise these trained models with the test dataset and show the confusion matrices in Fig. 7. We see that the COVID-19 cases are classified with an accuracy of 100% for both classifications with a few misclassifications with viral pneumonia and normal cases.
Fig. 7.
(a) Confusion matrix for binary classification (b) Confusion matrix for multi class classification.
5.1. Explainable artificial intelligence analysis
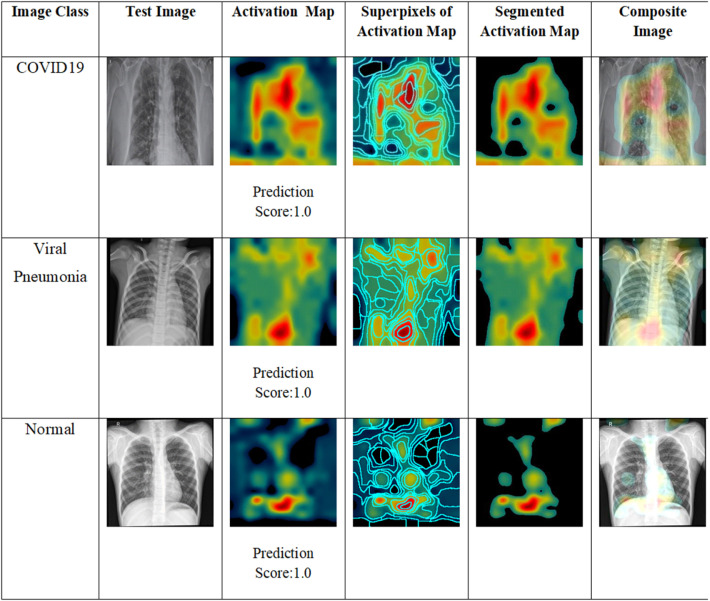
Though deep learning models demonstrate superior data representation and learning abilities across multiple domains, they appear to be a black box to the end-user. XIA studies are performed to understand the behavior of the models with respect to inputs. Particularly, in classifier models, attribution is a kind of XIA approach to determine the contribution of an input feature which strongly attributes the decision of the classifier. This process generates an attribution map which visualizes the significant input features influencing the classifier decision. In this research, XIA is an implicit part of the proposed framework as the activation maps are generated by Grad-CAM and segmented in the super pixel pooling layer. The behavior of the proposed framework is described in this section with illustrations and numeric prediction scores for with correctly and erroneously classified samples as below.
The test images, activation maps, segmented activation maps and composite images are shown in Fig. 8 for the correctly classified images. It is seen that the discriminant features of the three classes are clearly visible in the activation maps. Instead of construction of overlays of the test images and activation maps, we perform super pixel segmentation on these maps to separate the semantically identical regions. It is evident that the segmented activation maps carry the most representative discriminating features which are combined with the images to construct the composite images. It is also observed that the fine traces of hazy patches, consolidations and GGOs are captured and segmented by the proposed model. For the selected test image, COVID-19 is detected with score 1.0 by the proposed model focusing on the upper and left lower region of the image and the hazy patches. Similarly, viral pneumonia is detected with a score 1.0 with strong activation in the lower middle and upper left regions of the image and strong hazy patches in the upper right, upper left and lower middle regions of the image. The normal case is predicted with strong activations in the lower middle and lower right regions of the images with an accuracy of 1.0.
Fig. 8.
Network activation for correct classifications.
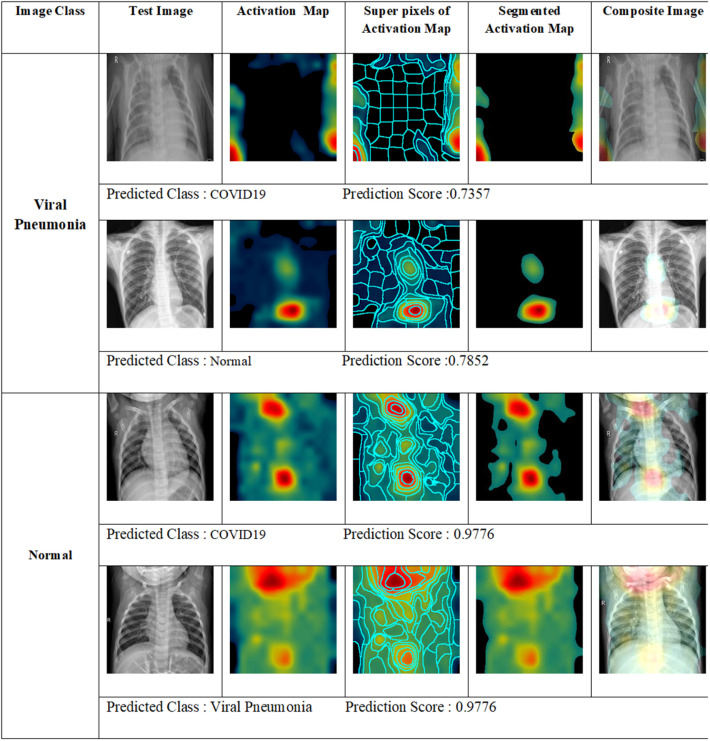
From the confusion matrices generated for the test dataset, we see that the COVID-19 images are classified with 100% accuracy with no misclassifications. We present a visual analysis of the misclassified images with the corresponding activation maps as shown in Fig. 9. We see that the activation maps of the viral pneumonia images are misclassified into COVID-19 and normal with prediction scores 0.7357 and 0.7852 respectively.
Fig. 9.
Network activation for misclassifications.
The misclassifications are attributed to the overlapping and non-specific nature of the COVID-19 symptoms. A detailed analysis of the misclassified samples with the class scores and visualizations can evaluate the degree of overlap between the symptoms. Further investigation in this line can provide deep insights on the relationship between manifestation of the symptoms and disease progression.
5.2. Comparative analyses
We present a comparison of our proposed model with state-of-the-art classifiers based on diverse CNN based classifiers modeled for COVID-19 detection from CXR images in Table 3. It is seen that the highest classification accuracy is achieved by our model.
Table 3.
Performance comparison with state-of-the-art methods.
| Reference | Model | Test images | Accuracy | Observations |
|---|---|---|---|---|
| Proposed Model | COVID-SSNet | 100 COVID19 1400 Non-COVID19 100 COVID 700 Viral Pneumonia 700 Normal |
0.9953 0.9960 |
The highest classification accuracy is achieved for binary and three class classifications. |
| Sethy et al. (2020) | ResNet50 + SVM | 25 COVID-19 (+) 25 COVID-19 (-) |
95.38 | ResNet50 is employed in feature extraction only. SVM adds computational overheads. |
| Chowdhury, Rahman, et al. (2020) | SqueezeNet | 60 COVID-19 60 Viral Pneumonia 60 Normal 60 COVID-19 60 Normal |
98.3 98.3 |
SqueezeNet demonstrates superior performance compared to AlexNet, ResNet18 and DenseNet201 with few parameters. |
| Wang et al. (2020) | COVID-Net | 53 COVID-19 (+) 5526 COVID-19 (-) 8066 Normal |
92.4 | Long-range connectivity among network layers increase the computational complexity. |
| Hemdan et al. (2020) | COVIDX-Net VGG19 DenseNet201 |
25 COVID-19 (+) 25 Normal |
90.0 | Best classification results are achieved by VGG19 and DenseNet201 compared to ResNetV2, InceptionV3, InceptionResNetV2, Xception, and MobileNetV2. Best performance is attributed to the composite operation of Densenet and the highly non-linear activation function of VGG19 |
| Narin et al. (2020) | ResNet50 InceptionV3 InceptionResNetV2 |
50 COVID-19 (+) 50 COVID-19 (-) |
98.0 97.0 87.0 |
ResNet50 model provides good classification performance attributed to its depth and skip connections. |
| Sharifrazi et al. (2021) | CNN+SVM+Sobel | 77 COVID-19 (+) 256 Normal |
99.02 | The architecture of the CNN is not explicitly specified for a fair comparison. |
| Abbas et al. (2021) | Deep CNN (DeTraC) | 105 COVID-19 (+) 80 Normal 11 SARS |
93.1 | DeTraC Decompose, Transfer, and Compose operations are performed to eliminated irregularities in the CXR images before training. |
| Alam et al. (2021) | Fusion features (CNN+HOG) + VGG19 pre-train model |
1979 COVID-19 3111 normal |
99.49 | The proposed model is very complex. It employs HoG and CNN for feature extraction. After fusion, the features are gien as input to VGG19. |
| Hussain et al. (2021) | CoroDet | 500 COVID-19 800 Viral Pneumonia 800 Normal 500 COVID-19 800 Normal |
94.2 99.1 |
22-layer customized CNN is built iteratively trying different layer, activation function and optimization parameter combinations. |
| Nayak et al. (2021) | ResNet-34 | 203 COVID-19 (+) 203 Normal |
98.33 | Best classification accuracy is achieved by ResNet-34 compared to ResNet-50, GoogleNet, VGG-16, AlexNet, MobileNet-V2, Inception-V3 and SqueezeNet. Superior performance of ResNet-34 is attributed to better performance-accuracy trade-off. |
In addition to the state-of-the art models, the proposed model is compared with two SqueezeNet based COVID-19 detection models. For a fair comparison, the models proposed in Ban, Liu, and Cao (2018) and Ucar and Korkmaz (2020) are evaluated with the dataset described in Table 1. The performance metrics including balanced scores and training times are evaluated as in Table 4. Generally, the decision to use a certain batch size often is driven by some intuition regarding the computation time of the model and the effort to train the model on a specific dataset. Larger batch sizes usually lead to faster convergence and a higher accuracy. Large batch sizes are preferred to maximize the performance as the gradient updates are very large. Conversely, small batch sizes influence updates of small sizes on the gradients. The batch sizes determine the samples drawn from the dataset for training. For data spread analysis, the squeezeNet based models are evaluated with 3 batch sizes viz. 32, 64 and 128. The mean values of the accuracy, sensitivity, specificity, precision, F1-score and Mathews Correlation Coefficient (MCC) metrics with standard deviation, evaluated with Eqs. (9)–(14) are given in Table 4.
| (9) |
| (10) |
| (11) |
| (12) |
| (13) |
| (14) |
where, TP — True Positive, TN — True Negative, FP — False Positive, FN — False Negative.
Table 4.
Performance comparison with SqueezeNet.
| Mod. | Clas. | ILR | Acc. | Sens. | Spec. | Prec. | F1S | MCC | TT (ms) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2-class | 0.001 (C) | 99.53±0.003 | 99.51±0.001 | 99.62±0.009 | 99.71±0.008 | 99.65±0.001 | 99.41±0.004 | 1570 |
| 3-class | 0.001 (C) | 99.60±0.0026 | 99.71±0.015 | 99.80±0.017 | 99.81±0.003 | 99.21±0.006 | 99.25±0.007 | 1586 | |
| 2 | 2-class | 0.0003 (VWE) | 98.71±0.093 | 96.68±0.106 | 98.52±0.130 | 98.47±0.129 | 98.54±0.122 | 98.13±0.104 | 1972 |
| 3-class | 0.0003 (VWE) | 98.13±0.102 | 96.20±0.128 | 98.12±0.098 | 98.33±0.184 | 98.29±0.974 | 98.02±0.982 | 1920 | |
| 3 | 2-class | 0.001 (VWE) | 99.01±0.140 | 99.12±0.135 | 98.51±0.274 | 98.89±0.099 | 98.93±0.196 | 98.32±0.197 | 1932 |
| 3-class | 0.001 (VWE) | 98.81±0.162 | 98.74±0.150 | 98.82±0.167 | 98.71±0.154 | 98.82±0.159 | 98.52±0.165 | 1941 | |
Legend– Mod.: Model; Clas.: Classification; ILR: Initial Learning Rate; C: Constant; VWE: Varies With Epochs; Acc.: Accuracy; Sens.: Sensitivity; Spec.: Specificity; Prec.: Precision; F1S: F1Score; MCC: Mathews Correlation Coefficient; TT: Training time.
1: Proposed – COVID-SSNet; 2: Chowdhury, Rahman, et al. (2020) – SqueezeNet; 3: Ucar and Korkmaz (2020) – Bayes-SqueezeNet.
From the above table, it is seen that our COVID-SSNet demonstrates superior performance compared to the conventional SqueezeNet model for binary and multi class classifications. In this paper, in addition to the conventional performance measures, the balanced measures F1 score and MCC are employed to evaluate the classifier models under imbalanced training and testing data. The MCC in the range [ −1.0 +1.0] evaluates closer to +1.0, when the balanced ratio of the TP, FP, TN and FN measures is high. When MCC is around 1.0, the classification is perfect. F1 score is computed as the weighted average of the precision and sensitivity metrics. Precision is the ratio of the number of correct positive predictions out of the total number of positive predictions and sensitivity is the ratio of the number of correct positive predictions to the total number of positive samples. F1 in the range [0.0 1.0] evaluates to 1.0 under ideal classification.
Though the model proposed in Chowdhury, Rahman, et al. (2020) is also based on SqueezeNet and SGDM, it observed that the performance of the model is comparatively lower. This model is trained with a momentum of 0.9, and an initial learning rate of 0.0003. Though the momentum is similar to our model, low classification accuracy is ascribed to lower learning rate which results in slow convergence of this model. Further, it is seen that the performance of the Bayesian optimization based squeezeNet classifier proposed in Ucar and Korkmaz (2020) is also inferior to the proposed SGDM optimized COVID-SSNet. This degradation of the Bayesian SqueezeNet is attributed to the estimation of posteriors in optimizing the learning rate by approximation. The training times are also found to be better for the proposed model, which show that SGDM optimization can stabilize the model, while the learning rate is constant.
Additionally, analysis of the standard deviation also shows that the proposed model is highly stable irrespective of the batch size. It implies that the impact of data spread across batches has a minimal effect on the proposed model. Whereas, for the other two models, the training samples drawn in different batches affect their performance due to variable learning rates. Further, this inference also aligns with the results of Smith et al. (2018) which show that best classification accuracy can be achieved by increasing the batch size without decaying the learning rate.
Further, the segmentation capabilities of the proposed model are evaluated with the objective metrics Dice, Structure Measure (SM) (Fan, Cheng, Liu, Li, & Borji, 2017) and Enhance -Alignment Measure (EM) (Fan et al., 2018). The ground truths required for these evaluations are obtained with the Ground Truth Labeler App of Matlab 2021b under the guidance of a medical expert.
The Dice metric which is the measure of the overlap between the prediction and the ground truth is given in Eq. (15).
| (15) |
The SM which evaluates the similarity between the segmented output and the ground truth mask is given in Eq. (16), where , , , and refer to the object-aware similarity, region-aware similarity, balance factor between and , prediction and the ground truth respectively. We have evaluated with =0.5, the default specified in Fan et al. (2017).
| (16) |
The EM which is a measure of the global and local similarity between binary maps is given in Eq. (17), where and are the width and height of ground truth mask and is the enhanced alignment matrix.
| (17) |
Given a prediction , is constructed by constructing a binary mask, thresholding each pixel in the range [0 255], The alignment matrix captures the pixel-level and image-level similarity between the prediction and based on the global means. We present a comparison of the segmentation results of the proposed model for consolidations, GGOs and hazy patches in Table 5. It is seen that the segmentation metrics are better for GGOs compared to consolidations and hazy patches which shows that the structure of consolidations and hazy patches are ambiguous compared to GGOs. It is also seen that the EM which measures the global and local similarity differ with the thresholds employed in the construction of the alignment matrix . Closer observation of the metrics shows that EM values are smaller when the threshold is high for this dataset. However, it may differ with other datasets due to the inherent image features.
Table 5.
Segmentation metrics.
| Infection | Metrics |
|||
|---|---|---|---|---|
| Dice | EM (Threshold 64) | EM (Threshold 128) | SM | |
| GGO | 0.9206 | 0.9401 | 0.9072 | 0.8545 |
| Consolidations | 0.8542 | 0.9272 | 0.8957 | 0.8529 |
| Hazy Patches | 0.8792 | 0.9164 | 0.8484 | 0.8317 |
In addition to classification, our model presents extended capabilities to segment the biomarkers and construct composite CXR images, enhancing their diagnostic values. It is seen that the activation regions featuring GGOs, consolidations and fine patches are localized by the proposed model. From an extensive literature review and clinical case studies we understand that bilateral, basal, medial, peripheral, multifocal, subpleural and posterior GGOs manifest in the CXR images of COVID-19 patients. However, GGOs seem to be unilateral and follow a central distribution in the CXR images of viral pneumonia patients. Air space consolidations are found to coexist with the GGOs in COVID-19 and viral pneumonia cases. Studies also show that people with normal X-rays are also infected with COVID-19. From Afza et al. (2021), it is apparent that COVID-19 imaging features are variable and non-specific and correlate with that of Middle East respiratory syndrome coronavirus (MERS-COV), H7N, H7N9, Human parainfluenza and Respiratory syncytial viruses.
The proposed model simplifies the separation of potential ROIs from the images through geometric, spatial and morphological analyses of the activation regions for the differential diagnosis of COVID-19 to identify the potential biomarkers. Further, it can be trained exclusively with these biomarkers and extended to severity analysis.
There are three potential limitations of the proposed network which need to be addressed.
-
1.
Lack of benchmarking standard datasets for performance evaluation is a major limitation with COVID-19 detection systems. Though several CXR and CT image datasets are introduced in recent works on COVID-19 detection, it is difficult to select suitable datasets to train and evaluate a model for comparative performance analyses due to the diversities with respect to the size and subset classes. The proposed model is compared with several models in Table 3, based on quantitative results reported in existing literature. Further, only two SqueezeNet based models are evaluated with the dataset considered in this research for want of time.
-
2.
In this research, super pixel segmentation of the activation maps is performed with constant values of horizontal and vertical pixel window sizes which constrain the size of the super pixels. Varying these windows can facilitate capturing biomarkers of arbitrary sizes ranging from very fine nodules to large infection patterns. Evaluating the performance of the proposed model for different window sizes is computationally expensive.
-
3.
Severity analysis, a major concern in COVID-19 management is not dealt in this research. With a suitable mathematical model, the quantitative class scores obtained from the Grad-CAMs can be mapped to disease severity. The above limitations drive the enhancement of the proposed model as below.
A voluminous labeled dataset comprising images of various infections featuring diverse morphological structures can be constructed to train the proposed model for COVID-19 detection, multi class segmentation and severity analysis from images of different modalities. This dataset will serve as an image corpus for deep learning approaches at various stages in COVID-19 management. Further, GMM super pixels segmentation can be enhanced to multi scale segmentation for segmentation of infections without size constraints.
It is evident that the proposed framework follows domain adaptation kind of transfer learning which fine-tunes the ImageNet pretrained SqueezeNet model with a completely different and a relatively very smaller chest X-ray dataset. The classification ability of this framework is testified with the classification results. Further, this framework can be extended to detect COVID-19 and segment infections from images of other modalities as well including chest CT images. It can be accomplished by freezing the weights of the COVID-SSNet for feature extraction and training it with multi-modal image datasets.
6. Conclusion
In this paper, we have proposed a novel deep learning framework for COVID-19 detection and segmentation of the infected regions from CXR images. We have constructed a variant of the conventional SqueezeNet model incorporating a super pixel pooling layer for segmentation of the infected region. We have shown that this model achieves more than 99% of training and testing accuracies for binary and multi class classifications with a standard dataset signifying its reliability. The segmentation of infected regions and their localization in CXR images achieved with the SqueezeNet is highly significant compared to the state-of-the-art models. The proposed system demonstrates it classification efficacy, feature driven classification decision and localization of infections. Contrary to the observations that CXR images have less diagnostic value, this model enhances the potential clinical values of these images. With the growing severity of the pandemic day by day, many countries have introduced mobile CXR units to capture images from the susceptible and infected people. The proposed model is ideal in the present scenario for rapid diagnosis and prognosis towards reducing the morbidity and mortality of COVID-19 cases. This model also paves way for further research on investigation of the symptoms in the segmented regions and combinations of occurrences in CXR images, for standardizing them as potential biomarkers of COVID-19.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Abbas A., Abdelsamea M.M., Gaber M.M. Classification of covid-19 in chest X-ray images using detrac deep convolutional neural network. Applied Intelligence: The International Journal of Artificial Intelligence, Neural Networks, and Complex Problem-Solving Technologies. 2021;51(2):854–864. doi: 10.1007/s10489-020-01829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achanta R., Shaji A., Smith K., Lucchi A., Fua P., Süsstrunk S. Slic superpixels compared to state-of-the-art superpixel methods. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2012;34(11):2274–2282. doi: 10.1109/TPAMI.2012.120. [DOI] [PubMed] [Google Scholar]
- Afshar P., Heidarian S., Naderkhani F., Oikonomou A., Plataniotis K.N., Mohammadi A. Covid-caps: A capsule network-based framework for identification of covid-19 cases from X-ray images. Pattern Recognition Letters. 2020;138:638–643. doi: 10.1016/j.patrec.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afza F., Sharif M., Mittal M., Khan M.A., Jude Hemanth D. A hierarchical three-step superpixels and deep learning framework for skin lesion classification. Methods. 2021 doi: 10.1016/j.ymeth.2021.02.013. [DOI] [PubMed] [Google Scholar]
- Alam N.-A., Ahsan M., Based M.A., Haider J., Kowalski M. Covid-19 detection from chest X-ray images using feature fusion and deep learning. Sensors. 2021;21(4) doi: 10.3390/s21041480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alom M.Z., Aspiras T., Taha T.M., Asari V.K. 2019. Skin cancer segmentation and classification with nabla-n and inception recurrent residual convolutional networks. arXiv preprint arXiv:1904.11126, pp. 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alom M.Z., Rahman M., Nasrin M.S., Taha T.M., Asari V.K. 2020. Covid_mtnet: Covid-19 detection with multi-task deep learning approaches. arXiv preprint arXiv:2004.03747, pp. 1–11. [Google Scholar]
- Apostolopoulos I.D., Aznaouridis S.I., Tzani M.A. Extracting possibly representative covid-19 biomarkers from X-ray images with deep learning approach and image data related to pulmonary diseases. Journal of Medical and Biological Engineering. 2020;40:462–469. doi: 10.1007/s40846-020-00529-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Physical and Engineering Sciences in Medicine. 2020;43(2):635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aradhya V.M., Mahmud M., Agarwal B., Kaiser M. One shot cluster based approach for the detection of covid-19 from chest X-ray images. Cognitive Computation. 2021:1–9. doi: 10.1007/s12559-020-09774-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baheti, B., Innani, S., Gajre, S., & Talbar, S. (2020). Eff-unet: A novel architecture for semantic segmentation in unstructured environment. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition (CVPR) workshops.
- Ban Z., Liu J., Cao L. Superpixel segmentation using gaussian mixture model. IEEE Transactions on Image Processing. 2018;27(8):4105–4117. doi: 10.1109/TIP.2018.2836306. [DOI] [PubMed] [Google Scholar]
- Bhapkar H.R., Mahalle P.N., Shinde G.R., Mahmud M. In: COVID-19: Prediction, decision-making, and its impacts. Santosh K., Joshi A., editors. Springer; Singapore: 2021. Rough sets in COVID-19 to predict symptomatic cases; pp. 57–68. (Lecture notes on data engineering and communications technologies). [Google Scholar]
- Bhattacharya S., Reddy Maddikunta P.K., Pham Q.-V., Gadekallu T.R., Siva Rama Krishnan S, Chowdhary C.L., et al. Deep learning and medical image processing for coronavirus (covid-19) pandemic: A survey. Sustainable Cities and Society. 2021;65 doi: 10.1016/j.scs.2020.102589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhablani G., Sharma A., Pandey H., Dash T. 2021. Superpixel-based domain-knowledge infusion in computer vision. [Google Scholar]
- Chowdhury M.E., Rahman T., Khandakar A., Mazhar R., Kadir M.A., Mahbub Z.B., et al. Can ai help in screening viral and covid-19 pneumonia? IEEE Access. 2020;8:132665–132676. [Google Scholar]
- Chowdhury M.E., et al. 2020. Covid-19 radiography database: Covid-19 chest X-ray database. [Google Scholar]
- Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., & Fei-Fei, L. (2009). Imagenet: A large-scale hierarchical image database. In 2009 IEEE conference on computer vision and pattern recognition (pp. 248–255).
- Dey N., Rajinikanth V., Fong S., Kaiser M., Mahmud M. Social-group-optimization assisted kapur’s entropy and morphological segmentation for automated detection of covid-19 infection from computed tomography images. Cognitive Computation. 2020;12(5):1011–1023. doi: 10.1007/s12559-020-09751-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, D.-P., Cheng, M.-M., Liu, Y., Li, T., & Borji, A. (2017). Structure-measure: A new way to evaluate foreground maps. In Proceedings of the IEEE international conference on computer vision.
- Fan D.-P., Gong C., Cao Y., Ren B., Cheng M.-M., Borji A. 2018. Enhanced-alignment measure for binary foreground map evaluation. [Google Scholar]
- Farooq M., Hafeez A. 2020. Covid-resnet: A deep learning framework for screening of covid19 from radiographs. arXiv preprint arXiv:2003.14395, pp. 1–5. [Google Scholar]
- Gaál G., Maga B., Lukács A. 2020. Attention u-net based adversarial architectures for chest X-ray lung segmentation. arXiv preprint arXiv:2003.10304, pp. 1–7. [Google Scholar]
- Guan Q., Huang Y., Zhong Z., Zheng Z., Zheng L., Yang Y. 2018. Diagnose like a radiologist: Attention guided convolutional neural network for thorax disease classification. arXiv preprint arXiv:1801.09927, pp. 1–10. [Google Scholar]
- Hemdan E.E.-D., Shouman M.A., Karar M.E. 2020. Covidx-net: A framework of deep learning classifiers to diagnose covid-19 in X-ray images. arXiv preprint arXiv:2003.11055, pp. 1–14. [Google Scholar]
- Hinton, G. E., Sabour, S., & Frosst, N. (2018). Matrix capsules with em routing. In International conference on learning representations (pp. 1–15).
- Hussain E., Hasan M., Rahman M.A., Lee I., Tamanna T., Parvez M.Z. Corodet: A deep learning based classification for covid-19 detection using chest X-ray images. Chaos, Solitons & Fractals. 2021;142 doi: 10.1016/j.chaos.2020.110495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iandola F.N., Han S., Moskewicz M.W., Ashraf K., Dally W.J., Keutzer K. 2016. Squeezenet: Alexnet-level accuracy with 50× fewer parameters and 0.5 MB model size. arXiv preprint arXiv:1602.07360, pp. 1–13. [Google Scholar]
- Kaiser M.S., Al Mamun S., Mahmud M., Tania M.H. In: COVID-19: Prediction, decision-making, and its impacts. Santosh K., Joshi A., editors. Springer; Singapore: 2021. Healthcare robots to combat COVID-19; pp. 83–97. (Lecture notes on data engineering and communications technologies). [Google Scholar]
- Kermany D.S., Goldbaum M., Cai W., Valentim C.C., Liang H., Baxter S.L., et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122–1131. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- Megahed N.A., Ghoneim E.M. Antivirus-built environment: Lessons learned from covid-19 pandemic. Sustainable Cities and Society. 2020;61 doi: 10.1016/j.scs.2020.102350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugappan M., Bourisly A.K., Krishnan P.T., Maruthapillai V., Muthusamy H. In: Computational modelling and imaging for SARS-CoV-2 and COVID-19. Prabha S., Karthikeyan P., Kamalanand K., Selvaganesan N., editors. CRC Press; 2021. Artificial intelligence based covid-19 detection using medical imaging methods: A review; pp. 1–22. [Google Scholar]
- Narin A., Kaya C., Pamuk Z. 2020. Automatic detection of coronavirus disease (covid-19) using X-ray images and deep convolutional neural networks. arXiv preprint arXiv:2003.10849, pp. 1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak S.R., Nayak D.R., Sinha U., Arora V., Pachori R.B. Application of deep learning techniques for detection of covid-19 cases using chest X-ray images: A comprehensive study. Biomedical Signal Processing and Control. 2021;64 doi: 10.1016/j.bspc.2020.102365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi A., Raj A.N.J., Nersisson R., Zhuang Z., Murugappan M. Rda-unet-wgan: an accurate breast ultrasound lesion segmentation using wasserstein generative adversarial networks. Arabian Journal for Science and Engineering. 2020;45(8):6399–6410. [Google Scholar]
- Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of covid-19 cases using deep neural networks with X-ray images. Computers in Biology and Medicine. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A.N.J., Zhu H., Khan A., Zhuang Z., Yang Z., Mahesh V.G., et al. Adid-unet—a segmentation model for covid-19 infection from lung ct scans. PeerJ Computer Science. 2021;7 doi: 10.7717/peerj-cs.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaraman S., Candemir S., Kim I., Thoma G., Antani S. Visualization and interpretation of convolutional neural network predictions in detecting pneumonia in pediatric chest radiographs. Applied Sciences. 2018;8(10):1715. doi: 10.3390/app8101715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajpurkar P., Irvin J., Zhu K., Yang B., Mehta H., Duan T., et al. 2017. Chexnet: Radiologist-level pneumonia detection on chest X-rays with deep learning. arXiv preprint arXiv:1711.05225, pp. 1–7. [Google Scholar]
- Ronneberger O., Fischer P., Brox T. In: Medical image computing and computer-assisted intervention. Navab N., Hornegger J., Wells W.M., Frangi A.F., editors. Cham. Springer International Publishing; 2015. U-net: Convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- Salehi S., Abedi A., Balakrishnan S., Gholamrezanezhad A. Coronavirus disease 2019 (covid-19): a systematic review of imaging findings in 919 patients. American Journal of Roentgenology. 2020;215(1):87–93. doi: 10.2214/AJR.20.23034. [DOI] [PubMed] [Google Scholar]
- Selvaraju, R. R., Cogswell, M., Das, A., Vedantam, R., Parikh, D., & Batra, D. (2017). Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE international conference on computer vision (pp. 618–626).
- Sethy P.K., Behera S.K., Ratha P.K., Biswas P. 2020. Detection of coronavirus disease (covid-19) based on deep features and support vector machine. Preprints, pp. 1–10. [Google Scholar]
- Sharifrazi D., Alizadehsani R., Roshanzamir M., Joloudari J.H., Shoeibi A., Jafari M., et al. Fusion of convolution neural network, support vector machine and sobel filter for accurate detection of covid-19 patients using X-ray images. Biomedical Signal Processing and Control. 2021;68 doi: 10.1016/j.bspc.2021.102622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J., et al. Radiological findings from 81 patients with covid-19 pneumonia in wuhan, china: a descriptive study. The Lancet Infectious Diseases. 2020;20(4):425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.K., Kumar A., Mahmud M., Kaiser M.S., Kishore A. Covid-19 infection detection from chest X-ray images using hybrid social group optimization and support vector classifier. Cognitive Computation. 2021:1–13. doi: 10.1007/s12559-021-09848-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.L., Kindermans P.-J., Ying C., Le Q.V. 2018. Don’t decay the learning rate, increase the batch size. [Google Scholar]
- Tello-Mijares S., Woo L. Computed tomography image processing analysis in covid-19 patient follow-up assessment. Journal of Healthcare Engineering. 2021;2021 doi: 10.1155/2021/8869372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ucar F., Korkmaz D. Covidiagnosis-net: Deep bayes-squeezenet based diagnosis of the coronavirus disease 2019 (covid-19) from X-ray images. Medical Hypotheses. 2020;140 doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Lin Z.Q., Wong A. Covid-net: a tailored deep convolutional neural network design for detection of covid-19 cases from chest X-ray images. Scientific Reports. 2020;10(1):19549. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Peng, Y., Lu, L., Lu, Z., Bagheri, M., & Summers, R. (2017). Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In IEEE CVPR (pp. 3462–3471).
- Wang T., Xiong J., Xu X., Jiang M., Yuan H., Huang M., et al. In: Medical image computing and computer assisted intervention. Shen D., Liu T., Peters T.M., Staib L.H., Essert C., Zhou S., Yap P.-T., Khan A., editors. Cham. Springer International Publishing; 2019. Msu-net: Multiscale statistical u-net for real-time 3d cardiac mri video segmentation; pp. 614–622. [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., et al. Detection of SARS-CoV-2 in different types of clinical specimens. Journal of the American Medical Association. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F., Gao Q., Jin C., Zhao F. Hyperspectral image classification based on superpixel pooling convolutional neural network with transfer learning. Remote Sensing. 2021;13(5) [Google Scholar]
- Yang W., Sirajuddin A., Zhang X., Liu G., Teng Z., Zhao S., et al. The role of imaging in 2019 novel coronavirus pneumonia (covid-19) European Radiology. 2020;30(9):4874–4882. doi: 10.1007/s00330-020-06827-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, B., Khosla, A., Lapedriza, A., Oliva, A., & Torralba, A. (2016). Learning deep features for discriminative localization. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 2921–2929).
- Zhuang Z., Li N., Joseph Raj A.N., Mahesh V.G., Qiu S. An rdau-net model for lesion segmentation in breast ultrasound images. PLoS One. 2019;14(8) doi: 10.1371/journal.pone.0221535. [DOI] [PMC free article] [PubMed] [Google Scholar]