Abstract
Mammal forelimbs are highly diverse, ranging from the elongated wing of a bat to the stout limb of the mole. The mammal forelimb has been a long-standing system for the study of early developmental patterning, proportional variation, shape change, and the reduction of elements. However, most of this work has been performed in mice, which neglects the wide variation present across mammal forelimbs. This review emphasizes the critical role of non-model systems in limb evo-devo and highlights new emerging models and their potential. We discuss the role of gene networks in limb evolution, and touch on functional analyses that lay the groundwork for further developmental studies. Mammal limb evo-devo is a rich field, and here we aim to synthesize the findings of key recent works and the questions to which they lead.
Keywords: Mammal, Forelimb, Evo-devo, evolution, development, Non-model
Mammals have adapted to a diverse array of habitats including but not limited to aerial, aquatic, arboreal, and subterranean environments. Key to mammals’ success is the form and function of their forelimbs, which enables them to explore diverse environments. Like all tetrapods, the mammalian limb is divided into three segments: the stylopod (humerus), zeugopod (radius/ulna), and autopod (carpals, metacarpals, and digits). Though its basic patterning is a relative constant, the mammalian forelimb is highly adaptable within this standard framework. The stylopod, zeugopod, and autopod are present in all mammals, but their forms have diversified to allow species to fly, swim, climb, dig, and more. By reconciling development with evolutionary theory, evo-devo has the potential to reveal the mechanisms by which this diversification occurs, and thus has brought incredible advancements to our understanding of how mammals have evolved into disparate and successful forms[1–3].
To date, our knowledge of the mechanisms underpinning mammalian limb development have largely relied on mice due to their substantial benefits as a genetically tractable model organism. However, relying on a single generalist species in this way neglects much of the morphological and functional diversity that characterizes mammals’ adaptation to a wide array of environments[4].
In recent years, this lack of diversity has begun to be remedied by further inclusion of non-model organisms [Box 1]. This increase in the number of viable organisms for evo-devo studies vastly expands the scope of our understanding of mammalian limb evolution. In this review, we highlight the contributions and potential of non-model organisms to improve our understanding of mammalian limb evo-devo. We note major recent findings across several mammal groups and suggest future directions for the field based on this increase in diversity and potential of study systems.
Box 1:
Here we refer to non-model organisms as any organism outside of the typically studied species for a clade. Some groups have been more frequently studied in mammals (e.g., bats, opossums, pigs), but relative to mice, they lack the vast majority of tools needed to perform deep developmental studies, so we refer to them as non-model organisms as well.
Working with Non-Model Organisms:
A key challenge to the use of non-model organisms is the procurement of specimens. Bats, for example, are not easily maintained in lab conditions, and harvesting embryos from any lab-raised colonies can be highly inefficient. Ungulates are also logistically difficult to maintain as a lab animal. Other groups, like the monotremes and xenarthrans, are far too rare to generate large quantities of embryos. There are several ways this is overcome. Developmental studies of bats typically rely on field-caught specimens, as many species are substantially abundant that embryos can be sustainably collected. Field collection is also the source of pregnant moles used in developmental studies[39,40,42]. While ungulates are not easily maintained in a lab, their embryos can be sourced from several places, including farms, breeders, and museums [30,62]. The few developmental studies of monotremes rely on these types of sources as well.
Benefits of Museum Specimens:
Museum specimens can offer enormous amounts of data on species that are hard to access. One stellar example of this is a recent study that coordinated with museums around the world to CT scan a developmental series of the extinct thylacine, showing that even the rarest samples can be studied non-destructively[63]. Recent work on echidnas capitalized on historical thin sections supplemented by a breeding colony to reveal how the middle ear and jaw may have functioned during the transition from the ancestral state, demonstrating the power of studying unique mammals[64,65]. Similarly, studies of xenarthrans can and have relied on museum specimens for developmental analyses[66].
Non-Model Organisms in Evo-Devo: Precedent
The evo-devo perspective seeks to resolve the drivers of evolution by uniting the variation of developmental processes studied in the lab with the millions of years of evolution observed in nature and the fossil record[5–8]. This unification of biological scales is aided by the inclusion of a diverse array of model systems, which provide a fuller picture of how developmental processes differ and generate the variation necessary for natural selection. This is not a new goal; the importance of non-model organisms to evo-devo has long been recognized[8–12]. Excitingly, in the past decade, improved access to imaging and next-generation sequencing has finally allowed new models to begin to achieve their potential in evo-devo. In this section, we will explore how new models have been assimilated into evo-devo studies, offering an example of how the difficulties of developing new study systems can be overcome.
Bats:
Bats are an excellent example of how a non-model organism can be integrated into the field of evo-devo. As a diverse, specialized, and highly successful group, they have provided insights into the developmental drivers of limb ratios, membrane formation, sensory adaptations, and cranial evolution[13–20]. In the context of limb evolution, their elongated limbs and wing membranes provide a starting point to answer many questions about how limb ratios change and unique morphologies develop to allow colonization of new niches[20–22].
Recent work on bat limbs has improved our understanding of the origins and evolution of mammalian flight and highlights the important contributions of studies of modern and fossil anatomy to evo-devo. A study of the timing of ossification (bone formation) in developing bats found that the autopod and sternum ossify very late[23]. These regions are considered to be highly adaptable in bats, perhaps, in part, due to this delay in ossification. This may represent one mechanism by which development can shape evolution; elements that ossify late during development may have a higher tendency to vary.
Fossil evidence has shown that the earliest bats likely had more limited flight capabilities than modern bats. The aerodynamic variables of a primitive fossil bat show that early chiropterans may have only relied on flight to commute, rather than hunt the way many modern bats do[22,24–26]. The evolution of flight style is thus an excellent target of further research, as the aerodynamic properties of bat wings widely vary, and this variation may be influenced by the autopod’s delayed ossification[26]. How this may relate to regulatory changes that drive limb elongation has yet to be explored[17,20]. Further investigations of developmental timing and its evolutionary consequences may benefit from a reconstruction of the ancestral ossification sequence of bats or a direct comparison of ossification sequences between flight styles.
Ungulates:
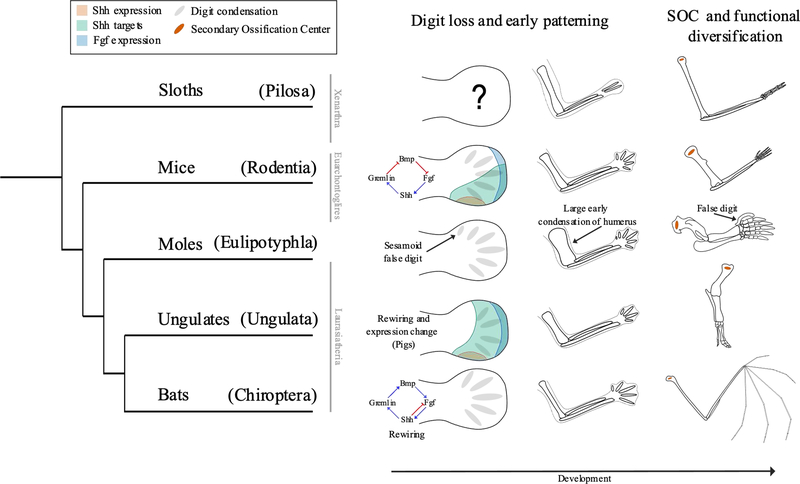
The autopod of ungulates represents varying levels of digit loss, and thereby can be used as a tool for improving our understanding of evolution by the reduction of elements. Studies of ungulates have revealed that multiple mechanisms have evolved to reduce digit number, affecting both early and late developmental processes [Fig. 1][27]. For example, in horses and camels, digits are lost after initial patterning occurs due to expanded regions of cell death. In pigs and cows, however, changes in gene regulation of Ptch1 alter early patterning, causing digit reduction by a wholly different mechanism[27,28].
Figure 1: Diversity of Mammal Limb Development -.
Schematic of limb development for sloths, mice, moles, ungulates, and bats through developmental time. Changes in developmental networks and expression patterns are indicated where known. Gray ovals in the hand plate and autopod represent early digit condensations. Orange ovals in the humerus represent secondary ossification centers. Bmp — Bone morphogenic protein; Fgf — Fibroblast growth factor; Shh — Sonic hedgehog. Expression patterns and networks from [56,17], respectively. Phylogeny based on [67–69].
Recent work has relied on embryos, fossils, and functional analyses to elucidate the extent and cause of digit reduction in horses. An analysis of embryonic material has found that horses go through a stage with five discrete digit condensations. Thus, even the most extreme case of digit reduction in ungulates shows evidence of a pentadactyl state[29]. This study also found that splint metapodials of horses are formed not only from the adjacent metapodials, but rather the fusion of both metapodials on each side[29]. Evidence for this has also been found in fossil horses, in which the splint metacarpals are clearly composed of two fused bones[30].
Analyses of physical stress and internal geometry performed on fossil horse limbs reveal a potential functional driver of digit loss. As body mass increases, the side digits of fossil horses fail to accommodate for increasing loads, and instead the middle digit must expand in size and alter its internal structure. Thus, increasing body size may have facilitated the loss of digits, as a single digit can functionally outperform multiple digits with the same total bone mass[31].
These new findings lead us to ask if all digit reduction mechanisms constitute a similar response to changes in body mass and show similar levels of incomplete digit loss, or if the functional pressures and developmental mechanisms of reduction are unrelated.
As exemplified by bats and ungulates, non-model organisms provide depth and clarity to evo-devo. Further diversification of study species thus can provide an increasingly holistic understanding of limb evolution.
Non-Model Organisms in Evo-Devo: New Systems
In the past few years, a wide array of species has been more thoroughly investigated, providing the foundation from which we can better understand the mechanisms underlying the diversification and specialization of mammal limbs.
Monotremes:
As an outgroup to placental and marsupial mammals, monotremes may provide insights into the ancestral mammalian limb condition. A pair of recent descriptive studies of echidna musculature and skeletal anatomy have revealed that the architectural diversity and range of motion in echidna limbs are highly reduced, potentially representing a conservative phenotype[32,33]. This conflicts with our understanding of monotreme limb development based on ossification sequences, which is highly derived in that the stylopod ossifies later than the zeugopod, a trait shared only by some moles[34]. Thus, monotremes offer a unique system to study mammalian limb evolution as they represent both an outgroup to most mammals and an instance of highly derived limb development. Further investigations of monotremes and comparisons with further outgroups may reveal how the elaboration of early developmental mechanisms has generated the diversity present today.
Moles:
Moles are of particular interest due to their extremely unique forelimb anatomy linked to their fossorial lifestyle[35,36]. Recent work has delved into the function of mole forelimbs, revealing how their unique stylopod and autopod function while walking and digging[37,38]. X-ray imaging has revealed that, while walking, moles rely heavily on their sesamoid false digit, allowing most true digits to be reserved for digging. [38]. This improved functional understanding informs studies of the autopod’s development. The development of the sesamoid false digit has been attributed to a heterochronic shift in Sox9 expression, but the extent to which this influences the rest of the autopod or other limb elements remains to be explored[39]. How, for example, might the development of the false digit and the functional consequences of reserving other digits for digging contribute to the robust nature of other autopod bones?
When burrowing, movement varies only in the distal portions of the limb when different behaviors are employed[37]. This may suggest that the stylopod is more conserved while the zeugopod is more variable. A developmental study of the mole stylopod noted that the abnormal morphology of the humerus is established when the cartilaginous scaffold is just forming. Later in development the humerus is twisted, contributing to the moles’ unique cranially angled limb posture[38,40]. These findings suggest that, contrary to the expectations stated above, early limb development can be highly modified, but the mechanisms underlying this have not been investigated. Further investigations of the development and variation of these structures and the range of would help clarify our understanding of how the limb evolves.
Thus far, a limited amount of work has been done on mole development, but with newfound understanding of limb function in this group, there is greater incentive to investigate the development of their false digits, joint morphology, and limb shape[39–43].
Xenarthra:
Xenarthrans exhibit great extant and extinct ecomorphological diversity, making them particularly interesting to the study of evolution. An analysis of fossil and modern xenarthran humeri found that phylogenetically distant arboreal species converged on the same limb morphology. This was true for fossorial species as well. Furthermore, the evolutionary rates of fossorial humeri were much lower than those of arboreal species, suggesting that some underlying processes may bias the evolution of ecologically linked morphologies[44] This, coupled with a recent analysis of cortical compactness in slow arboreal mammals (including xenarthrans) and work studying bone microstructure, may lend insight into how skeletal architecture affects and is affected by development and evolution[44,45]. For example, an analysis of long bone structure via CT scanning of embryonic xenarthrans could reveal the timing of developmental processes that may underlie their unique limb morphologies.
By more thoroughly investigating non-model systems, we can begin to reveal the processes by which limb elements are elongated, reduced, deformed, and restructured [Fig. 1]. These mechanisms generate the variation that natural selection acts upon, allowing us to better understand the process of evolution.
Non-Model Organisms in Evo-Devo: Comparative Studies and Generating Variation
While recent work has improved the diversity of our study systems, there have been several advancements in our understanding of the generation of diversity through comparative development. This work has relied on traditional models but is steadily expanding to include other groups as well.
An analysis of transcriptomic data from opossum, mouse, pig, and bat limbs has shown that variation in gene expression levels increases between species as development proceeds[46]. This trend leads to further questions about how variation arises in development and what mechanisms generate it. The study of gene regulation offers a deeper look into these processes and has been applied across several systems[47–49].
Gene regulation and the networks of interactions that underlie it are important factors in developmental evolution[50–52]. Previous work in non-model systems has found that changes in regulatory networks and elements facilitate the elongation of limbs in bats and the retention of the interdigital membrane [Fig. 1][17,53,54]. Additionally, regulatory changes are thought to facilitate the reduction of digits in some ungulates, as the evolution of expression patterns of key genes that are conserved elsewhere seems to cause digit loss during autopod development[27,28,55]. In a comparison of mouse and pig limbs, significant rewiring of regulatory networks in the pig autopod were found, leading to differences in the expression of genes important to the development of the limb bud’s anterior-posterior axis[56]. Thus, network rewiring may be one mechanism driving digit reduction in artiodactyls. Further comparisons of regulatory networks within different species will advance our understanding of how regulatory changes facilitate the development and evolution of the limb.
Other work in mice has revealed how changes to development can overcome physical constraints on diversity. The growth plate, which facilitates bone elongation, has been the focus of many recent studies, as the proportions of long bones vary widely across mammals[28,57–60]. The secondary ossification center (SOC), located at the epiphyses of long bones of mammals, is closely associated with the growth plate, yet only recently has it been considered in an evo-devo context. A thorough study combining CT scanning, immuno-histochemistry, and structural measurements of tissues identified the potential evolutionary significance of this structure. The SOC shields the growth plate from physical stress during development, preventing unnecessary cell death and allowing mammal limbs to develop in otherwise intolerable conditions[61]. While this work was only done in mice, it implies that the SOC permits the development of the diverse array of limb morphologies present in mammals. Non-model systems may thus reveal the significance of this structure to the evolution of mammal limbs.
Non-Model Organisms in Evo-Devo: Future Questions
Recent work in non-model organisms and novel techniques tested in mice provide new ways to address various old and emerging questions of mammal limb evo-devo.
Regarding gene regulation, we now have the tools to ask how changes to the wiring of regulatory networks generate differences across a diverse array of species. This has begun to be addressed in bat and pig limbs but can be applied to the variety of models discussed here to understand the regulatory changes that may facilitate the formation of the mole humerus and other unique limb morphologies. Following the methods of Tissieres et al. (2020), it is now feasible to compare gene network evolution across different species and confirm the phenotypic effects of network rewiring in mice. Xenarthra and moles, for example, have much to offer to our understanding of regulatory evolution in the mammal autopod, as they offer additional and largely uninvestigated cases of variation by digit loss and sesamoid expansion, respectively [Fig. 1].
Recent advancements in our understanding of the function of the mole forelimb and the potential evolutionary consequences of the SOC can guide investigations of the evolutionary role of physical constraints. For example, we may now ask how the unique function and morphology of the mole forelimb influence its development and the development of adjacent structures. Hypotheses on the SOC’s evolutionary influence from Xie et al. (2019) would greatly benefit from tests in other species. The presence, development, and morphology of the SOC should be studied in groups that represent diverse long bone phenotypes, like the moles, xenarthrans, ungulates, and monotremes. How, for example, are the unique functional constraints of fossorial or arboreal animals influenced by the SOC, which has only been studied in a general terrestrial species [Fig. 1]? After better understanding the SOC across mammalian taxa, we can return to the mouse and attempt experimental manipulations to confirm its role as a mediator of physical constraints.
The potential of non-model organisms is vast. The difficulties of working with non-model groups are not negligible, but they can, in many cases, be overcome. More importantly, for a field focused on developmental generation of variation, any inclusion of diversity is a net benefit. In the past two years, the field of evo-devo has expanded the breadth of available models and produced new findings on how developmental processes affect evolution. The unification of these advancements has much to offer to evo-devo, and there is no doubt that these new tools, models, and theories will together advance our conceptualization of the evolution of life.
Funding
AOH is funded by a National Science Foundation Graduate Research Fellowship (DGE-1650604). The Sears lab is supported by the Division of Integrative Organismal Systems of the National Science Foundation (1257873 and 2017803) and the National Institute of Health (HD050042–01).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare that they have no conflicts of interest.
Bibliography
*Of importance **Of outstanding importance
- 1.Mariani FV, Martin GR: Deciphering skeletal patterning: clues from the limb. Nature 2003, 423:319–325. [DOI] [PubMed] [Google Scholar]
- 2.Zeller R, López-Ríos J, Zuniga A: Vertebrate limb bud development: moving towards integrative analysis of organogenesis. Nature Reviews Genetics 2009, 10:845–858. [DOI] [PubMed] [Google Scholar]
- 3.Young JJ, Tabin CJ: Saunders’s framework for understanding limb development as a platform for investigating limb evolution. Developmental Biology 2017, 429:401–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polly PD: Limbs in Mammalian Evolution. InFins into Limbs: Evolution, Development, and Transformation 2007, 245–268. [Google Scholar]
- 5.Wagner GP, Larsson HCE: What is the promise of developmental evolution? III. The crucible of developmental evolution. Journal of Experimental Zoology 2003, 300B:1–4. [DOI] [PubMed] [Google Scholar]
- 6.Müller GB, Newman SA: The innovation triad: an EvoDevo agenda. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution 2005, 304B:487–503. [DOI] [PubMed] [Google Scholar]
- 7.Raff RA: Written in stone: fossils, genes and evo—devo. Nat Rev Genet 2007, 8:911–920. [DOI] [PubMed] [Google Scholar]
- 8.Müller GB: Evo—devo: extending the evolutionary synthesis. Nature Reviews Genetics 2007, 8:943–949. [DOI] [PubMed] [Google Scholar]
- 9.Milinkovitch MC, Tzika A: Escaping the mouse trap: the selection of new Evo-Devo model species. J Exp Zool 2007, 308B:337–346. [DOI] [PubMed] [Google Scholar]
- 10.Minelli A, Baedke J: Model organisms in evo-devo: promises and pitfalls of the comparative approach. History and Philosophy of the Life Sciences 2014, 36:42–59. [DOI] [PubMed] [Google Scholar]
- 11.Franz- Odendaal TA, Hockman D: Non- model organisms and unique approaches are needed for the future of evo- devo. Dev Dyn 2019, 248:618–619. [DOI] [PubMed] [Google Scholar]
- 12.Wei F: A new era for evolutionary developmental biology in non-model organisms. Sci China Life Sci 2020, 63:1251–1253. [DOI] [PubMed] [Google Scholar]
- 13.Cretekos CJ, Rasweiler IV JJ, Behringer RR: Comparative studies on limb morphogenesis in mice and bats: a functional genetic approach towards a molecular understanding of diversity in organ formation. Reprod Fertil Dev 2001, 13:691. [DOI] [PubMed] [Google Scholar]
- 14.Weatherbee SD, Behringer RR, Rasweiler JJ, Niswander LA: Interdigital webbing retention in bat wings illustrates genetic changes underlying amniote limb diversification. Proceedings of the National Academy of Sciences 2006, 103:15103–15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams RA: Morphogenesis in Bat Wings: Linking Development, Evolution and Ecology. Cells Tissues Organs 2008, 187:13–23. [DOI] [PubMed] [Google Scholar]
- 16.Santana SE, Grosse IR, Dumont ER: Dietary Hardness, Loading Behavior, and the Evolution of Skull Form in Bats: Evolution of skull form in Bats. Evolution 2012, 66:2587–2598. [DOI] [PubMed] [Google Scholar]
- 17.Cooper LN, Sears KE: How to Grow a Bat Wing. In Bat Evolution, Ecology, and Conservation. Edited by Adams RA, Pedersen SC. Springer; New York; 2013:3–20. [Google Scholar]
- 18.Dumont ER, Samadevam K, Grosse I, Warsi OM, Baird B, Davalos LM: Selection for Mechanical Advantage Underlies Multiple Cranial Optima in New World Leaf-Nosed Bats: Cranial optima in New World Leaf-Nosed Bats. Evolution 2014, 68:1436–1449. [DOI] [PubMed] [Google Scholar]
- 19.Sadier A, Davies KT, Yohe LR, Yun K, Donat P, Hedrick BP, Dumont ER, Dávalos LM, Rossiter SJ, Sears KE: Multifactorial processes underlie parallel opsin loss in neotropical bats. eLife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sears KE, Behringer RR, Rasweiler JJ, Niswander LA: Development of bat flight: Morphologic and molecular evolution of bat wing digits. Proceedings of the National Academy of Sciences 2006, 103:6581–6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eckalbar WL, Schlebusch SA, Mason MK, Gill Z, Parker AV, Booker BM, Nishizaki S, Muswamba-Nday C, Terhune E, Nevonen KA, et al. : Transcriptomic and epigenomic characterization of the developing bat wing. Nature Genetics 2016, 48:528–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Speakman JR: The evolution of flight and echolocation in bats: another leap in the dark. Mammal Review 2001, 31:111–130. [Google Scholar]
- 23.López-Aguirre C, Hand SJ, Koyabu D, Son NT, Wilson LAB: Postcranial heterochrony, modularity, integration and disparity in the prenatal ossification in bats (Chiroptera). BMC Evolutionary Biology 2019, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Amador LI, Simmons NB, Giannini NP: Aerodynamic reconstruction of the primitive fossil bat Onychonycteris finneyi (Mammalia: Chiroptera). Biol Lett 2019, 15:20180857. * Examines aerodynamic properties of two of the oldest known bat fossils. Find low aerodynamic efficiency relative to modern bats, and similar performance to gliding mammals when the handwing is removed. Hypothesizes that the handwing evolved rapidly to facilitate the transition between gliding and powered flight. Excellent recent example of the power of fossil material to guide evolutionary developmental studies.
- 25.Simmons NB, Seymour KL, Habersetzer J, Gunnell GF: Primitive Early Eocene bat from Wyoming and the evolution of flight and echolocation. Nature 2008, 451:818–821. [DOI] [PubMed] [Google Scholar]
- 26.Amador LI, Almeida FC, Giannini NP: Evolution of Traditional Aerodynamic Variables in Bats (Mammalia: Chiroptera) within a Comprehensive Phylogenetic Framework. J Mammal Evol 2020, 27:549–561. [Google Scholar]
- 27.Cooper KL, Sears KE, Uygur A, Maier J, Baczkowski K-S, Brosnahan M, Antczak D, Skidmore JA, Tabin CJ: Patterning and post-patterning modes of evolutionary digit loss in mammals. Nature 2014, 511:41–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saxena A, Towers M, Cooper KL: The origins, scaling and loss of tetrapod digits. Phil Trans R Soc B 2017, 372:20150482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kavanagh KD, Bailey CS, Sears KE: Evidence of five digits in embryonic horses and developmental stabilization of tetrapod digit number. Proc R Soc B 2020, 287:20192756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Solounias N, Danowitz M, Stachtiaris E, Khurana A, Araim M, Sayegh M, Natale J: The evolution and anatomy of the horse manus with an emphasis on digit reduction. R Soc open sci 2018, 5:171782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McHorse BK, Biewener AA, Pierce SE: Mechanics of evolutionary digit reduction in fossil horses (Equidae). Proc R Soc B 2017, 284:20171174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Regnault S, Pierce SE: Pectoral girdle and forelimb musculoskeletal function in the echidna ( Tachyglossus aculeatus ): insights into mammalian locomotor evolution. R Soc open sci 2018, 5:181400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Regnault S, Fahn-Lai P, Norris RM, Pierce SE: Shoulder Muscle Architecture in the Echidna (Monotremata: Tachyglossus aculeatus) Indicates Conserved Functional Properties. J Mammal Evol 2020, 10.1007/s10914-020-09498-6. [DOI] [Google Scholar]
- 34.Weisbecker V: Monotreme Ossification Sequences and the Riddle of Mammalian Skeletal Development: Monotreme ossification sequence. Evolution 2011, 65:1323–1335. [DOI] [PubMed] [Google Scholar]
- 35.Sánchez-Villagra MR, Menke PR, Geisler JH: Patterns of evolutionary transformation in the humerus of moles (Talpidae, Mammalia): a character analysis. Mammal Study 2004, 29:163–170. [Google Scholar]
- 36.Sánchez-Villagra MR, Menke PR: The mole’s thumb — evolution of the hand skeleton in talpids (Mammalia). Zoology 2005, 108:3–12. [DOI] [PubMed] [Google Scholar]
- 37.Lin Y-F, Konow N, Dumont ER: How moles destroy your lawn: the forelimb kinematics of eastern moles in loose and compact substrates. J Exp Biol 2019, 222:jeb182436. [DOI] [PubMed] [Google Scholar]
- 38. Lin Y-F, Konow N, Dumont ER: How moles walk; it’s all thumbs. Biol Lett 2019, 15:20190503. * Uses X-ray reconstruction of moving morphology to understand the kinematics of how moles walk. Finds that the sesamoid false digit in the mole autopod bears much of the weight while walking, revealing the functional role of this abnormality. Find that moles have a secondarily derived sprawling posture, highlighting them as a group with high amounts of limb specialization.
- 39.Bickelmann C, Mitgutsch C, Richardson MK, Jiménez R, de Bakker MA, Sánchez-Villagra MR: Transcriptional heterochrony in talpid mole autopods. EvoDevo 2012, 3:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bickelmann C, Jiménez R, Richardson MK, Sánchez-Villagra MR: Humerus development in moles (Talpidae, Mammalia). Acta Zoologica 2014, 95:283–289. [Google Scholar]
- 41.Mitgutsch C, Richardson MK, Jiménez R, Martin JE, Kondrashov P, de Bakker MAG, Sánchez-Villagra MR: Circumventing the polydactyly ‘constraint’: the mole’s ‘thumb.’ Biol Lett 2012, 8:74–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bickelmann C, van der Vos W, de Bakker M A G, Jiménez R, Maas S, Sánchez- Villagra MR: Hox gene expression in the specialized limbs of the Iberian mole (Talpa occidentalis). Evolution & Development 2017, 19:3–8. [DOI] [PubMed] [Google Scholar]
- 43.Sansalone G, Colangelo P, Loy A, Raia P, Wroe S, Piras P: Impact of transition to a subterranean lifestyle on morphological disparity and integration in talpid moles (Mammalia, Talpidae). BMC Evol Biol 2019, 19:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Serio C, Raia P, Meloro C: Locomotory Adaptations in 3D Humerus Geometry of Xenarthra: Testing for Convergence. Front Ecol Evol 2020, 8:139. [Google Scholar]
- 45.Alfieri F, Nyakatura JA, Amson E: Evolution of bone cortical compactness in slow arboreal mammals. Evolutionary Biology; 2020. [DOI] [PubMed] [Google Scholar]
- 46.Maier JA, Rivas-Astroza M, Deng J, Dowling A, Oboikovitz P, Cao X, Behringer RR, Cretekos CJ, Rasweiler JJ, Zhong S, et al. : Transcriptomic insights into the genetic basis of mammalian limb diversity. BMC Evol Biol 2017, 17:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sears KE: Quantifying the impact of development on phenotypic variation and evolution: Development and phenotypic variation. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution 2014, 322:643–653. [DOI] [PubMed] [Google Scholar]
- 48.Sears KE, Maier JA, Rivas-Astroza M, Poe R, Zhong S, Kosog K, Marcot JD, Behringer RR, Cretekos CJ, Rasweiler JJ, et al. : The Relationship between Gene Network Structure and Expression Variation among Individuals and Species. PLoS Genet 2015, 11:e1005398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petit F, Sears KE, Ahituv N: Limb development: a paradigm of gene regulation. Nature Reviews Genetics 2017, 18:245–258. [DOI] [PubMed] [Google Scholar]
- 50.Levine M, Davidson EH: Gene regulatory networks for development. 2005, [DOI] [PMC free article] [PubMed]
- 51.Davidson EH, Erwin DH: Gene Regulatory Networks and the Evolution of Animal Body Plans. Science 2006, 311:796–800. [DOI] [PubMed] [Google Scholar]
- 52. Sadier A: How Do Gene Networks Promote Morphological Evolution. In Old Questions and Young Approaches to Animal Evolution. Edited by Martín-Durán JM, Vellutini BC. Springer International Publishing; 2019:209–234. * Summarizes the importance of gene regulatory networks to morphological evolution. Highlights how networks affect various levels of development and how they may result in changes to morphology. Concludes with a summary of current methods used to further understand GRNs and their role in evolution.
- 53.Sears K, Maier JA, Sadier A, Sorensen D, Urban DJ: Timing the developmental origins of mammalian limb diversity. genesis 2018, 56:e23079. [DOI] [PubMed] [Google Scholar]
- 54.Cretekos CJ, Wang Y, Green ED, Martin JF, Rasweiler JJ, Behringer RR: Regulatory divergence modifies limb length between mammals. Genes Dev 2008, 22:141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zuniga A: Next generation limb development and evolution: old questions, new perspectives. Development 2015, 142:3810–3820. [DOI] [PubMed] [Google Scholar]
- 56. Tissières V, Geier F, Kessler B, Wolf E, Zeller R, Lopez-Rios J: Gene Regulatory and Expression Differences between Mouse and Pig Limb Buds Provide Insights into the Evolutionary Emergence of Artiodactyl Traits. Cell Reports 2020, 31:107490. ** Reveals regulatory divergence in artiodactyl limbs based on ATAC-seq data. Finds loss of asymmetry due to Ptch1 expression restriction and widespread regulatory differences between mouse and pig limbs. Provides methodology for identifying regulatory changes associated with differentially expressed genes.
- 57.Marchini M, Rolian C: Artificial selection sheds light on developmental mechanisms of limb elongation. Evolution 2018, 72:825–837. [DOI] [PubMed] [Google Scholar]
- 58.Rubin S, Agrawal A, Stegmaier J, Svorai J, Addadi Y, Villoutreix P, Stern T, Zelzer E: 3D MAPs discovers the morphological sequence chondrocytes undergo in the growth plate and the regulatory role of GDF5 in this process. Developmental Biology; 2020. [DOI] [PMC free article] [PubMed]
- 59. Cooper KL: Developmental and Evolutionary Allometry of the Mammalian Limb Skeleton. Integrative and Comparative Biology 2019, 59:1356–1368. ** Reviews current understanding and outstanding questions of the evolution and development of skeletal proportions. Highlights the mechanisms by which limb proportions may change and the role of allometry in evolution.
- 60. Rolian C: Endochondral ossification and the evolution of limb proportions. WIREs Developmental Biology 2020, 9:e373. * Reviews mechanisms of endochondral growth and development. Discusses the role of endochondral growth and constraints in evolution and allometry. Together with Cooper, 2019, provides a well-rounded description of the development of long bone growth and its role in limb evolution.
- 61. Xie M, Gol’din P, Herdina AN, Estefa J, Medvedeva EV, Li L, Newton PT, Kotova S, Shavkuta B, Saxena A, et al. : Secondary ossification center induces and protects growth plate structure. Evolutionary Biology; 2020. ** Combines developmental and mechanical analyses of long bones to understand the function of the secondary ossification structure in mammals. Proposes that the SOC shields hypertrophic chondrocytes from the stresses of terrestrial locomotion, permitting greater diversity of limb forms.
- 62.Ross D, Marcot JD, Betteridge KJ, Nascone-Yoder N, Bailey CS, Sears KE: Constraints on Mammalian Forelimb Development: Insights from developmental disparity. Evolution 2013, 67:3645–3652. [DOI] [PubMed] [Google Scholar]
- 63.Newton AH, Spoutil F, Prochazka J, Black JR, Medlock K, Paddle RN, Knitlova M, Hipsley CA, Pask AJ: Letting the ‘cat’ out of the bag: pouch young development of the extinct Tasmanian tiger revealed by X-ray computed tomography. R Soc open sci 2018, 5:171914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anthwal N, Fenelon JC, Johnston SD, Renfree MB, Tucker AS: Transient role of the middle ear as a lower jaw support across mammals. eLife 2020, 9:e57860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anthwal N, Tucker AS: The TMJ Disc Is a Common Ancestral Feature in All Mammals, as Evidenced by the Presence of a Rudimentary Disc During Monotreme Development. Front Cell Dev Biol 2020, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hautier L, Oliver JD, Pierce SE: An Overview of Xenarthran Developmental Studies with a Focus on the Development of the Xenarthrous Vertebrae. J Mammal Evol 2018, 25:507–523. [Google Scholar]
- 67.Meredith RW, Janecka JE, Gatesy J, Ryder OA, Fisher CA, Teeling EC, Goodbla A, Eizirik E, Simao TLL, Stadler T, et al. : Impacts of the Cretaceous Terrestrial Revolution and KPg Extinction on Mammal Diversification. Science 2011, 334:521–524. [DOI] [PubMed] [Google Scholar]
- 68.Foley NM, Springer MS, Teeling EC: Mammal madness: is the mammal tree of life not yet resolved? Phil Trans R Soc B 2016, 371:20150140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jebb D, Huang Z, Pippel M, Hughes GM, Lavrichenko K, Devanna P, Winkler S, Jermiin LS, Skirmuntt EC, Katzourakis A, et al. : Six reference-quality genomes reveal evolution of bat adaptations. Nature 2020, 583:578–584. [DOI] [PMC free article] [PubMed] [Google Scholar]