Abstract
Purpose:
Mantle Cell Lymphoma (MCL) is a fatal subtype of Non-Hodgkin’s Lymphoma. SOX11 transcription factor is overexpressed in the majority of nodal MCL. We have previously reported that B-cell specific overexpression of SOX11 promotes MCL pathogenesis via critically increasing BCR signaling in vivo. SOX11 is an attractive target for MCL therapy however no small molecule inhibitor of SOX11 has been identified to date. Although transcription factors are generally considered undruggable, the ability of SOX11 to bind to the minor groove of DNA led us to hypothesize that there may exist cavities at the protein-DNA interface that are amenable to targeting by small molecules.
Experimental Design:
Using a combination of in silico predictions and experimental validations, we report here the discovery of three structurally related compounds (SOX11i) that bind SOX11, perturb its interaction with DNA and effect SOX11-specific anti-MCL cytotoxicity.
Results:
We find mechanistic validation of on-target activity of these SOX11i in the inhibition of BCR signaling and the transcriptional modulation of SOX11 target genes, specifically, in SOX11 expressing MCL cells. One of the three SOX11i, exhibits relatively superior in vitro activity and displays cytotoxic synergy with Ibrutinib in SOX11 expressing MCL cells. Importantly, this SOX11i induces cytotoxicity specifically in SOX11-positive Ibrutinib-resistant MCL patient samples and inhibits BTK phosphorylation in a xenograft mouse model derived from one of these subjects.
Conclusions:
Taken together, our results provide a foundation for therapeutically targeting SOX11 in MCL by a novel class of small molecules.
Keywords: Transcription factor, SOX11, Mantle Cell Lymphoma, Ibrutinib, BTK-phosphorylation, cytotoxicity
Introduction:
MCL is a lethal sub-type of B-cell lymphoma and represents approximately 6% of all Non-Hodgkin’s Lymphoma (NHL), the most common hematological malignancy worldwide1. Despite advances in chemotherapy and immunotherapy, MCL patients have a median survival of 7–8 years, exhibit a continuous pattern of relapse and in clinical trials their survival curves have not yet reached a plateau. Upon relapse, targeted agents such as bortezomib, lenalidomide and BTKi Ibrutinib have shown excellent clinical activity in MCL patients but are not curative2–4. Thus, advances in therapeutics for this disease are urgently needed5. The pathogenesis of MCL is characterized by cell cycle dysregulation and Cyclin D1 (CCND1) overexpression caused by the t(11;14)6,7 translocation, leading to clonal expansion of malignant B lymphocytes. A subset of CCND1 negative MCL patients have been identified with more than half of them harboring translocations that lead to overexpression of other CCND2/3 due to rearrangements with immunoglobulin genes8. However, the CCND1/2/3 overexpression gene signatures in MCL patients are indistinguishable from each other and mouse models overexpressing CCND1 do not recapitulate the MCL disease phenotype9 suggesting their overexpression alone is not sufficient to drive the disease and that additional genetics lesions are necessary for MCL pathogenesis10. Indeed, whole-exome sequencing of primary MCL patient samples has identified recurrent mutations in TP53, CDKN2A, CDKN2C, ATM, KMT2D, KMT2B, KMT2C, SMARCA4 and NOTCH isoforms among other genes in subsets of patients11–15. Despite these findings, early transformation events that precipitate these genetic events are not well understood, and mouse models with these individual mutations do not recapitulate disease phenotype. The current management of MCL involves anti-CD20 antibodies and chemotherapy. The Bruton’s tyrosine kinase inhibitor (BTKi) Ibrutinib16 and the BCL2 inhibitor (BCL2i) Venetoclax17,18 are emerging MCL treatment options that produce high response rates but modest durations of response of 17.5 and 14 months, respectively, in this patient population. However, resistance to one or both of these agents frequently develops in MCL and new treatments, especially for chemorefractory MCL resistant to BTKi or BCL2i are the most urgent unmet need in clinical practice19.
SOX11 belongs to the SOXC family of high mobility group (HMG) transcription factors, which consists of SOX4, SOX11, and SOX1220. HMG transcription factors bind to DNA and facilitate conformational changes that allow binding of other transcription factors, usually leading to activation or repression of downstream genes. In early B cells, SOX4, a closely related SOX11 family member with identical target binding motifs, can inhibit differentiation via direct inhibition of WNT signaling21. SOXC transcription factors have overlapping roles in central and peripheral nerve development, and it is likely that these factors play a role in B cell development, as SOX11-null mice do not form spleens, and SOX4 deficient mice do not form B lymphocytes22.
SOX11 protein is expressed in the majority (78–93%) of MCL patients and is specific for MCL as compared to other types of NHL23–25. SOX11 is expressed even in the 5%−10% of CCND1-negative MCL patients6. In serial biopsies obtained from MCL patients, SOX11 is found to be expressed specifically in pre-malignant lymph nodes, suggesting SOX11 upregulation is an early event in the malignant transformation of B lymphocytes in MCL26. Microarray studies performed on primary MCL samples in our lab confirm SOX11 expression to be significantly higher in CD19+ tumor cells as compared to naïve B cells from healthy controls27. Further, SOX11 expression in MCL is associated with poor prognosis23.
SOX11 depletion by RNAi in human MCL cell lines leads to reduced tumor growth in xenograft models28. Additional studies corroborate the finding that SOX11 contributes to MCL pathogenesis in part via regulation of BLIMP1 downstream targets29. Our group published a ChIP-seq analysis in the MCL cell line Granta-519 that identified direct binding targets of SOX11 within critical pathways involved in cell proliferation and cell cycle control, including the WNT, Protein Kinase A and TGF-β receptor signaling cascades27. We developed the first murine models of SOX11 overexpression (Eμ-SOX11) which consistently develops an aberrant oligoclonal CD5+CD19+CD23- B cell proliferation that is identical to clinical MCL30. In addition, RNA-sequencing of CD5+CD19+ splenocytes from SOX11 overexpressing mice show increased B cell receptor (BCR) signaling compared to wild type controls. This is particularly relevant as constitutively active BCR signaling has been recently implicated in human MCL31–33. Ibrutinib, a BCR signaling inhibitor (BTKi), shows significant therapeutic activity with a 60% overall response rate in relapsed MCL patients16. Still, the majority of patients relapse after Ibrutinib treatment, and relapses are associated with acquired mutations in the binding pocket for Ibrutinib33. Since SOX11 is upstream of BTK, identification of SOX11 targets that are key intermediates for MCL pathogenesis may reveal new therapeutic targets for patients that relapse on Ibrutinib and/or Venetoclax (BCL2i), which is emerging as a promising therapy for MCL17,18.
SOX11 presents a potential new target in MCL, but transcription factors in general are considered “undruggable” because most have interaction surfaces that are devoid of pockets and grooves for the binding of small molecules. Nonetheless, because SOX proteins bind to the minor groove of DNA and derive part of their specificity through a severe distortion of the DNA, we hypothesized that this might lend to cavities at the protein-DNA interface that are amenable to targeting by small molecules. Indeed, putative binding pockets for small molecules could be identified at the protein-DNA interface of a SOX11–DNA homology model we built using the crystal structure of the SOX4-DNA binding domain (DBD) as a template34. Using a combination of in silico screening predictions and experimental validations, we report here first-in-class small molecule inhibitors of SOX11 with on-target anti-MCL activity in BTKi -resistant cellular models in vitro and in patient-derived ex vivo models of Ibrutinib resistance.
Methods:
Surface Plasmon Resonance (SPR) Assay
10μg/ml (His)6-SOX11-DBD was captured on the gold-coated sensor chip (SCR NiHC200M) (XanTec bioanalytics) by injecting at 10 μl/min for 180 seconds. Compounds were dissolved in 25 mM Tris, 150 mM NaCl, 5 mM MgCl2, 0.005% Tween-20, 2% DMSO, pH 7.4 and injected at 25 μl/min for 425 seconds. The signals were measured at 25ºC using Reichert 2SPR (Reichert Technologies) The association and dissociation times were set at 120 and 300 seconds, respectively. Three wash steps of 180 seconds at 25 were included between each experimental run to ensure a steady baseline. The ka, kd, and Kd values were determined using Reichert’s TraceDrawer software. Average and standard deviation were calculated from two or more separate experiments.
Fluorescence Resonance Energy Transfer (FRET) studies
50 nM (His)6-mNeonGreen-SOX11-DBD was incubated with compounds at room temperature for 20 minutes in 96-well plates followed by 37.5 nM AlexaFluor-568 conjugated double-stranded oligonucleotide (prepared as FAM-ds-oligo) for 20 minutes. Plates were placed in a Safire2 (Tecan Group) spectroscope with excitation at 488 (mNeonGreen; λex/em=506/517 nm) and emission was recorded from 512 to 620 nm (AlexaFluor-568; λex/em=578/603 nm). Control wells included buffer alone, compound alone, mNeonGreen-SOX11-DBD (donor) alone and Alexa568-dsDNA oligo (acceptor) alone and were used for background subtraction. Non-interference from compound intrinsic fluorescence signals was confirmed since <2% signal in excess of buffer alone control was observed for each compound. FRET ratios were calculated according to the TechNote TNPJ100.04 from ProZyme (https://prozyme.com/pages/tech-notes).
Statistical analysis
Statistical comparisons were made using 2-tailed Student’s t-test with Prism software (GraphPad Prism, RRID:SCR_002798) to calculate p-values: P ≤ 0.05 (*), P ≤ 0.01 (**), P ≤ 0.001 (***). The average and mean standard deviation were calculated after experiments were done three times unless indicated.
Primary Patient Samples and PDX Model
All studies involving human samples were performed under Ohio State University Institutional Review Board committee–approved protocols, through which informed written consent was obtained, and deidentified samples were utilized. Primary MCL cells were obtained from the peripheral blood of four patients with acquired Ibrutinib resistant MCL. Patient characteristics are presented in Table S4. Cells were thawed and placed in RPMI-1640 medium supplemented with 10% FBS and 1 mM sodium pyruvate for 3 hours. Cells were then treated with either compound or DMSO control. All animal studies were carried out in accordance with the guidelines of the Institute for Animal Studies at OSU. The MCL patient derived xenograft (PDX) mouse model was generated by engrafting peripheral blood mononuclear cells from Patient # 2 (Pt. 2) with acquired Ibrutinib-resistant MCL intravenously into NSG mice and subsequently passaged 5 times. CD5+CD19+ MCL cells used for the described experiments were purified from the spleen of mice once removal criteria were met.
Cell Culture
MCL cell lines JeKo-1, Mino, Z-138 and JVM-2 were purchased from American Type Culture Collection (ATCC). All cell lines were authenticated by short tandem repeat DNA profiling and routinely tested for Mycoplasma infection as per standard practice (MycoAlert, Lonza Bioscience). All MCL cell lines were maintained in RPMI 1640 medium (Mod.) 1X with L-Glutamine (Corning) supplemented with 10% Fetal Bovine Serum (Gemini Bio-Products) and 1% penicillin-streptomycin 100X solution (Corning). The cells were grown at 37°C in a humidified incubator containing 5% CO2.
Cell Viability Assay
Cell viability was determined by a fluorometric resazurin reduction method (CellTiter-Blue; Promega) following the manufacturer’s instructions. 100,000 cells in 100 μl of RPMI 1640 medium were plated in 96 well flat bottom Falcon Polystyrene Microplates (Corning) and treated with compounds (8 replicates per condition). Cells were incubated for 72 hours. After incubation, 20 μl CellTiter-Blue Cell was added to each well and incubated for another 2 hours. Plates were put into a fluorescence plate reader that records fluorescence at 560/590 nm to get optical density (OD) values. The number of viable cells in each treated well was calculated, based on the linear least squares regression of the standard curve (OD vs. cell concentration). The viability of cells treated with compounds was normalized to the viability of cells treated with 0.2% Dimethyl sulfoxide (DMSO). Cell counts were confirmed with Trypan Blue Exclusion Assay on the Countess automated cell counter (Invitrogen) according to the manufacturer’s specifications. The viability of cells treated with compounds for 48 hours was normalized to the viability of cells treated with 0.2% DMSO.
Microarray Assay
Gene expression profiles of tumor samples from patients newly diagnosed with MCL before any treatment and naïve B-cells (NBCs) from healthy donor specimens were used to evaluate mRNA expression levels of SOX C transcription factors SOX4, SOX11, and SOX12 (GEO accession number GSE70910)16,56. Sample collection and laboratory studies were in compliance with institutional review board and Helsinki protocols. CD19+ cells from 26 MCL patients treated at the National Institutes of Health were purified by magnetic bead sorting from peripheral blood or lymph node products before freezing to ensure greater than 90% purity. For controls, purified normal immunoglobulin D positive (IgD+) NBCs were obtained with the use of magnetic bead sorting from specimens from 5 healthy donors undergoing routine tonsillectomy (for non-neoplastic indications) at the Children’s Hospital at Montefiore (Bronx, NY). Briefly, total RNA was extracted using the RNeasy kit (Qiagen) and profiled by Affymetrix Human Genome U133 Plus 2.0 arrays according to the manufacturer’s instructions. CEL files were processed using Affymetrix Expression Console software and normalized by the Robust Multi-Averaging (RMA) method.
Annexin V and 7-AAD Staining
Cells were treated with compounds and harvested after 24 hours. Subsequent to harvesting, cells were prepped for analysis using the eBioscience Annexin V Apoptosis Detection Kit (ThermoFisher Scientific) and following the manufacturer’s instructions. Cells were analyzed using the BD LSRFortessa flow cytometer. Data analysis was done using Cytobank15. For combination studies, isobolograms were made using the values for affected fraction of cells and combination index (CI) values were determined using CompuSyn software (http://www.biosoft.com/w/calcusyn.htm). The CI value correlates with the effect of combination treatment. A CI of <0.9 is considered synergistic, a CI of ⩾ 0.9 or ⩽ 1.1 is considered additive, and a CI of >1.1 is considered antagonistic.
Analysis by flow cytometry on primary samples
After treatment, cells were stained with 5 μl FITC-Annexin V antibody (BD Bioscience) and 5 μl propidium iodide solution (BD Biosciences) for 15 min at room temperature in the provided binding buffer before they were analyzed on a BD LSRFortessa flow cytometer.
PhosphoFlow
Cells were treated with compounds and harvested after 24 hours. Cells were washed and fixed by using the eBioscience Foxp3 / Transcription Factor Fixation/Permeabilization Concentrate and Diluent kit (ThermoFisher Scientific) and following the manufacturer’s instructions. Fixed cells were stained with Alexa Fluor 647 Mouse Anti-BTK (pY223)/Itk (pY180) (BD Bioscience) and analyzed using the BD LSRFortessa flow cytometer. Data analysis was done using Cytobank15.
Quantitative real time polymerase chain reaction (RT-PCR)
1 million cells/ml were treated with compounds and incubated for 24 hours. RNA was extracted using the RNeasy Plus Mini Kit (Qiagen) according to the manufacturer’s protocols. cDNA was prepared using SuperScript VILO Master Mix (ThermoFisher Scientific) and detected by SsoFast EvaGreen Supermix (Bio-Rad Laboratories) on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories). Gene expression was normalized to hypoxanthine phosphoribosyltransferase (HPRT) and expressed relative to cells treated with 0.2% DMSO using the 2-ΔΔCt formula. Thermal cycler conditions were: initial step of 30 sec at 95°C followed by 40 cycles of 5 sec at 95°C (denature) and 5 sec at 60°C (anneal/extend) followed by 5 min at 55 ºC to 95 ºC in increments of 0.5 ºC.
Primers used for quantitative RT-PCR
HPRT Forward Primer: 5`-AAAGGACCCCACGAAGTGTT −3`
HPRT Reverse Primer: 5`-TCAAGGGCATATCCTACAACAA-3`
SMAD3 Forward Primer: 5`-TGGACGCAGGTTCTCCAAAC-3`
SMAD3 Reverse Primer: 5`-CCGGCTCGCAGTAGGTAAC-3`
PAX5 Forward Primer: 5`-GAGCGGGTGTGTGACAATGA-3`
PAX5 Reverse Primer: 5`-GCACCGGAGACTCCTGAATAC-3`
PPP3CA Forward Primer: 5`- CCAAGTCACCGGCTTACAG −3`
PPP3CA Reverse Primer: 5`- CCTCCTTCATAAGATGCGCCTT −3`
Immunoblot Analysis
For whole cell extracts, cells were lysed in cold RIPA buffer containing cOmplete, Mini Protease Inhibitor Cocktail and phosphatase inhibitors ((Sigma-Aldrich) and cell lysates were clarified by centrifugation. Proteins were analyzed by immunoblot using standard procedures. Primary antibodies to the following proteins were used: Actin Antibody (Santa Cruz
Biotechnology Cat# sc-1615, RRID:AB_630835) and Anti-SOX11 antibody (Atlas Antibodies Cat#AMAb90502, RRID:AB_2665568). For primary cells antibodies to the following proteins were used: SOX11 (Abcam); BTK, phospho BTK (Tyr223), and GAPDH (Cell Signaling Technology). Blot patterns were analyzed using Image-J software16, providing a quantitative measure of protein expression (ImageJ, RRID:SCR_003070).
Data Sharing Statement
For original data, please contact samir.parekh@mssm.edu
Results:
Specific overexpression of SOX11 among SOXC family members in MCL
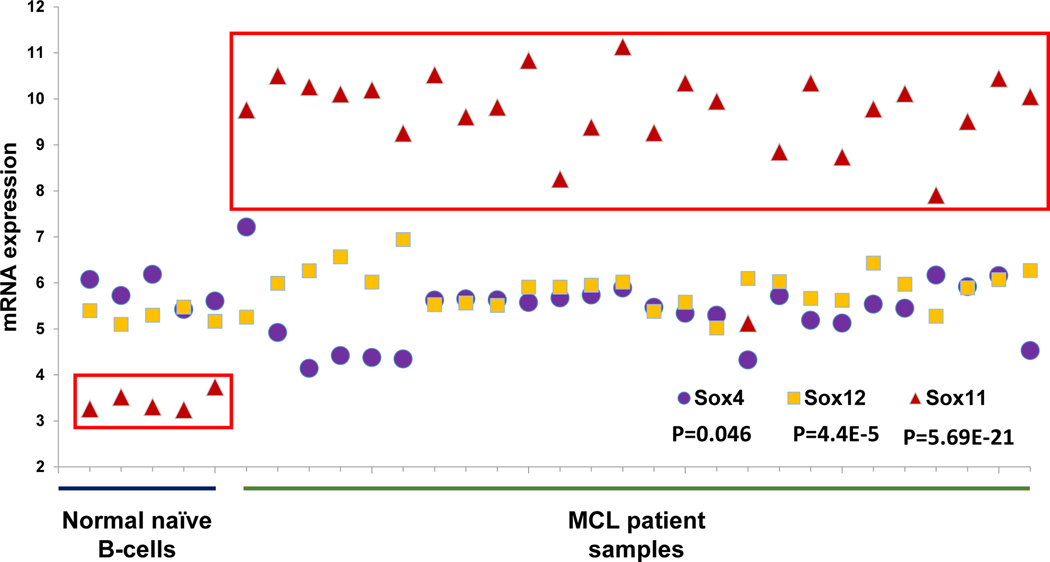
The SOXC family consists of three members, SOX4, SOX11 and SOX12, with high sequence and structural homology. To validate SOX11 as a target for development of small molecule inhibitors, we used microarray analysis to examine the expression levels of SOX4, SOX11 and SOX12 in naïve B-cells from five healthy donors and 26 MCL patients (Figure 1). While we observed similar baseline expression levels of SOX4 and SOX12 in both healthy and MCL derived B-cells, the mRNA expression of SOX11 was specifically increased approximately 6-fold in MCL patient samples. These findings are consistent with previous immuno-histochemical studies in MCL patients23,35–37.
Figure 1. SOX11 is overexpressed in MCL patient samples.
Microarray analysis on 5 normal naïve B-cell patients and 26 MCL patients. SOX C transcription factors (SOX4, SOX11 and SOX12) were probed for mRNA expression levels in each patient. Student’s t-test was used to calculate p-values.
Prediction of small molecule binders of SOX11-DBD
Previous ChIP-Seq experiments suggest that SOX11 mediates the overexpression of several oncogenes by binding directly to their DNA regulatory elements27. SOX proteins, including SOX11 recognize and bind to the cognate DNA sequence (~4 [TTGT])34 via a small positively charged DNA binding domain (DBD). Amino acids from the SOX-DBD insert into the minor groove of the DNA recognition sequence, resulting in sharp bending of the DNA, while the conformation of the protein remains largely unchanged38. We reasoned that small molecules that bind to the DNA interacting surface of SOX11 could have the potential to preclude DNA binding.
To enable in silico screening of small molecules that bind to the SOX11-DBD, we derived a homology model based on the closest transcription factor (TF) with an available crystal structure, i.e., the SOX4-DBD34 (see details in the Methods section). There are only four different residues between SOX4 and SOX11 in this domain: Q71, L99, D104, and Q110 in murine SOX4 correspond to K61, M89, E94, and R100 in human SOX11, respectively.
The first in silico screening at this SOX11-DBD homology model using the NCGC pharmaceutical collection (NPC; http://tripod.nih.gov/npc/) dataset39 prioritized 67 compounds for experimental testing (see Methods section for details). The fluorescence anisotropy (FA) assay used to assess inhibition of SOX11-DNA interaction in the presence of 20 μM of each compound identified Flavitan as a potential inhibitor (Supplementary Figure S1A) because of its > 50% inhibition of DNA binding (Supplementary Figure S1B). However, this molecule was not studied further because of its high polarity, high molecular weight, and inactivity in MCL cell lines. Instead, its binding pose within the SOX11-DBD was used as a reference to prioritize additional hits from virtual screening of approximately 10 million ready-to-dock molecules from the ZINC version 12 Drugs Now subset (see virtual screening workflow in Supplementary Figure S2; refer to Methods for details). Specifically, results of a structural interaction fingerprint (SIFt)40 assessment of the binding pose of Flavitan were compared to SIFt results obtained for the identified 112 top-scoring ligands from the aforementioned virtual screening using SIFt Tanimoto similarity. A total of 31 ligands with a SIFt Tanimoto similarity ≥ 0.4 with the SIFt assessment of Flavitan were selected for experimental analysis, but only five of them (Cpd A-E) were purchased.
Validation of small-molecule binders of SOX11-DBD
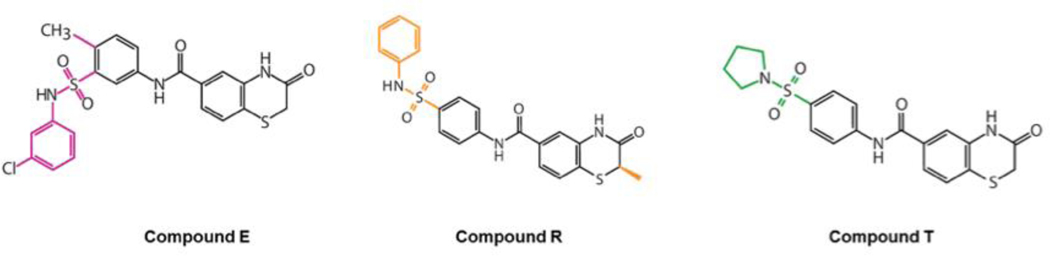
Next, we tested the effect of compounds A-E on the viability of SOX11 expressing Z-138 MCL cell line versus the SOX11-negative JVM-2 MCL cell line (Supplementary Figure S3). Of the five compounds assayed, compound E displayed selective growth inhibition of Z-138 indicating that compound E may be exerting its cellular activity by binding to SOX11. Based on these results, we identified structural analogs with at least 80% similarity to compound E by querying the ZINC database. Of these, we procured two compounds that were available for purchase which we refer to as compound R and compound T (Figure 2).
Figure 2. Structure of compounds predicted to bind SOX11-DBD.
The 2-D chemical structures of compounds E, R and T.
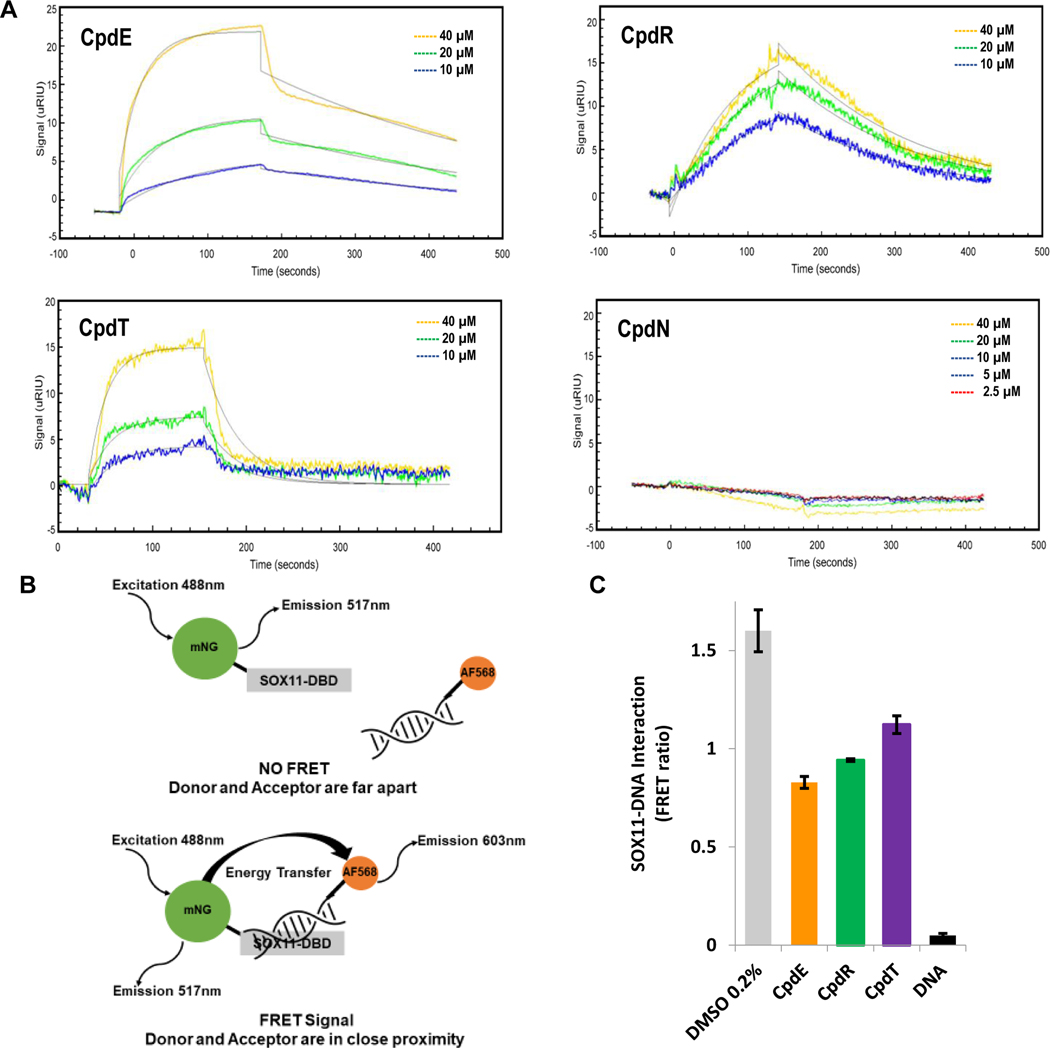
We used surface plasmon resonance to determine if compound E, compound R and compound T (referred to as SOX11i) bind directly to (His)6-SOX11-DBD immobilized on a Ni-NTA gold chip. As can be seen in the sensograms in Figure 3A, all three compounds were found to bind SOX11-DBD with micromolar affinities (Table S1). Compound N, an inactive compound from our screen, was also assayed and did not associate with SOX11-DBD while Sm4, a pan-SOXi small molecule44 and a duplex DNA containing the SOX11 binding motif displayed expected affinities (Table S1). To evaluate whether the binding of these compounds to SOX11-DBD deterred the binding of DNA, we sought to employ the FA assay (described above and Methods) which measures fluorescence anisotropy at fluorescein excitation and emission. Unexpectedly, the intrinsic fluorescence of the compounds complicated data analysis. These compounds have spectral properties over a broad emission range and hence were unsuitable to be evaluated by the FA assay. As an alternative, we developed a FRET based assay in which a duplex DNA carrying the SOX11 binding sequence is labeled with the fluorophore Alexa Fluor 568 on the 5’-end of one strand and assessed against the SOX11-DBD fused to an N-terminal (His)6-mNeonGreen (mNG) fluorescent protein. Binding of SOX11-DBD to the DNA brings the fluorophores closer due to DNA bending, leading to a concomitant increase in FRET (Figure 3B). Small molecules that disrupt SOX11-DBD and DNA binding are expected to reduce FRET efficiency. We observed biochemical inhibition of the SOX11-DNA interaction by compounds E, R and T (Figure 3C). Compound E at 20 μM caused the most inhibition of the SOX11-DBD:DNA interaction with unlabeled DNA used as a competitive positive control. To confirm the inhibition of SOX11-DBD:DNA interaction by SOX11i we employed an orthogonal electrophoretic mobility shift assay (EMSA). For all three SOX11i, we observed a dose-dependent reduction in the mobility shift of DNA mediated by incubation with SOX11-DBD (Supplementary Figure S4) with Compound E showing the lowest IC50 value.
Figure 3. Compounds bind directly to SOX11-DBD and interfere with DNA binding.
(A) Surface plasmon resonance shows that compounds E, R and T bind to SOX11 and have KD values in the μM range calculated by using ka and kd values. Compound N, an inactive compound from our screen, did not show an association and its kinetics constants were not calculable. Each experiment was done two or more times and representative sensograms show binding signal (μRIU) of compounds over time (seconds) to SOX11-DBD. Table S1 shows averages with mean standard deviations for each kinetic constant. (B) Diagram illustrating how the FRET assay works. (C) FRET bar graph shows that compounds E, R and T inhibit SOX11-DNA interaction with a decrease in FRET ratio. DNA with no fluorophore attached was used as a competitive positive control. The average and mean standard deviation were calculated after experiments were done three times.
In order to ensure the cellular growth inhibition and biochemical activity we observed was not the result of SOX11i binding directly to DNA we employed a Topoisomerase-based DNA unwinding assay that detects small molecule intercalators of DNA. Ibrutinib was used a negative control and the established DNA intercalator, ethidium bromide as a positive control. We were able to confirm that all three SOX11i and Ibrutinib do not directly bind DNA at 20 μM; that is, they did not interfere with the DNA unwinding activity of topoisomerase I whereas ethidium bromide completely inhibited its ability to relax supercoiled plasmid DNA (Supplementary Figure S5).
SOX11i are cytotoxic in MCL cells in a SOX11 dependent manner
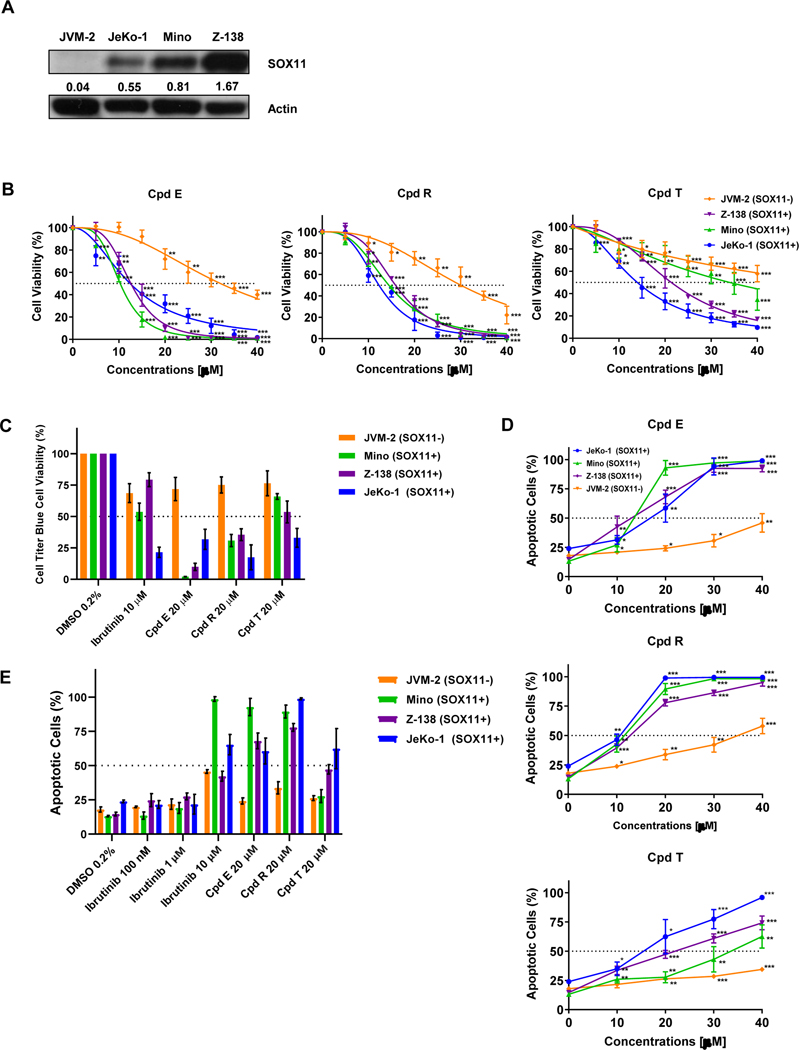
We next expanded our single dose observation of SOX11-specific growth inhibition of Z-138 cells by Compound E by conducting two different dose-response assays (0–40 μM) in three SOX11-positive MCL cell lines (JeKo-1, Mino and Z-138) versus the SOX11-negative JVM-2 MCL cell line (See Figure 4A for SOX11 status).
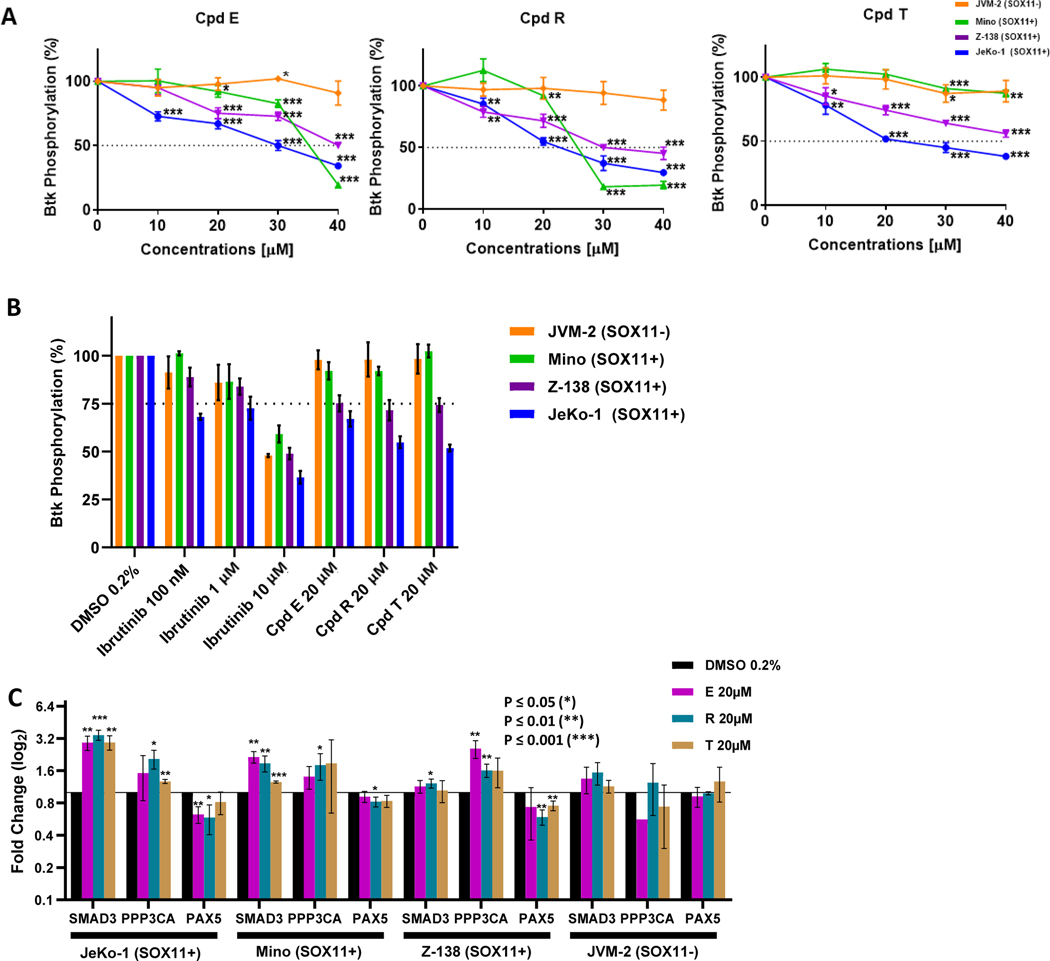
Figure 4. SOX11i are cytotoxic in SOX11 expressing MCL cell lines.
(A) Western blot showing the expression of SOX11 in JVM-2, JeKo-1, Mino and Z-138 Mantle Cell Lymphoma cell lines. The numbers represent SOX11 to Actin ratio. (B) Line graph showing cell titer blue cell viability percentage in JVM-2 (orange), Z-138 (purple), Mino (green) and JeKo-1 (blue) after 72 hours of treatment with compound E, compound R and compound T. Cells were treated with 0 to 40 μM of each compound. The average and mean standard deviation were calculated after experiments were done three times. Student-T Test was used to calculate p-values: P ≤ 0.05 (*), P ≤ 0.01 (**), P ≤ 0.001 (***). (C) Bar graph showing Cell Titer Blue cell viability percentage of cells treated with 0.2% of DMSO, 10μM Ibrutinib or 20 μM of each SOX11i. (D) Line graph showing apoptotic percentage using Annexin V and 7-AAD staining and flow cytometry in JVM-2 (orange), Z-138 (purple), Mino (green) and JeKo-1 (blue) after 72 hours of treatment with compound E, compound R and compound T. Cells were treated with 0 to 40 μM of each compound. The average and mean standard deviation were calculated after experiments were done three times. Student’s t-test was used to calculate p-values: P ≤ 0.05 (*), P ≤ 0.01 (**), P ≤ 0.001 (***). (E) Bar graph showing percent apoptotic cells by FACS analysis of Annexin V and 7-AAD staining after 72 hours of treatment with 0.2% of DMSO, Ibrutinib (100nm, 1μM, 10 μM) or 20 μM of each SOX11i.
We used the metabolic Cell Titer Blue assay to determine viability of these MCL cell lines upon treatment with SOX11i for 72 hours. Both Compound E and Compound R had robust growth inhibitory activity in SOX11+ cells (IC50: 12–16 μM) as compared to SOX11- JVM-2 cells (IC50: 30–32 μM) with a 2.5 to 2.7-fold selectivity. Compound T had less effect in SOX11+ cells (IC50: 14–34 μM); however, it was still selective as IC50 was not reached in JVM-2 cells (Figure 4B–C; Table S2).
We further tested the SOX11-dependent growth inhibitory activity of SOX11i by si-RNA mediated knock down of SOX11 in SOX11+ JeKo-1 cells. An increase in survival of si-SOX11 cells compared to control cells treated with 5μM Compound R confirmed its SOX11-dependency and on-target cytotoxicity (Supplementary Figure S6). Additionally, we treated PBMCs from two healthy donors with Compound R for 24, 48 and 72 hours and found viability profiles matching those for SOX11- JVM-2 cells (Supplementary Figure S7).
The SOX11-specific growth inhibition exerted by these SOX11i could be the result of cytotoxic cell death. We performed flow cytometric analysis by staining SOX11i treated MCL cells with Annexin V and 7-AAD to directly assay and quantify early and late apoptotic cell death. As can be seen in Figure 4D–E and Table S3, Compound E had an apoptotic IC50 value of 15 μM in JeKo-1, 12 μM in Z-138, and 12 μM in Mino cells whereas in JVM-2 IC50 was not reached. Compound T had an IC50 of 14 μM in JeKo-1, 19 μM in Z-138 and 33 μM in Mino cells while IC50 was not reached in JVM-2 cells. Compound R was relatively more active with IC50 values of 10 μM in JeKo-1, 12 μM in Z-138 and 11 μM in Mino cells. The IC50 value in JVM-2 was 35 μM, showing SOX11 selectivity of similar magnitude to growth inhibitory assays.
SOX11i inhibit key SOX11-dependent intracellular pathways and validated target genes
One of the characteristics of MCL is increased BCR signaling. Eμ-SOX11 transgenic mice overexpressing SOX11 specifically in B cells have increased activating tyrosine phosphorylation of Bruton’s tyrosine kinase (pBTK-Y223) and Phospholipase C γ (pPLCγ-Y759)30. Upon observing SOX11 specific anti-MCL cytotoxicity we tested whether the SOX11i affect pBTK-Y223 levels in SOX11-expressing MCL cell lines, JeKo-1, Z-138 and Mino, using JVM-2 cells as a comparator. Cells were treated with compounds E, R and T (10 – 40 μM range) for 24 hours and flow cytometry was used to measure pBTK -Y223 levels. Compound E inhibited pBTK levels in all three SOX11+ cell lines with an IC50 of 28–43 μM (Figure 5A). Compound R was relatively more active with IC50 ranging from 23–34 μM. Compound T was the least active, reaching IC50 for only one of the three cell lines (JeKo-1). Reassuringly, pBTK level in SOX11-negative JVM-2 cells was unaffected by all three SOX11i upto 40 μM, further confirming a SOX11-specific mechanism of action. Positive control BTKi Ibrutinib on the other hand, reduced pBTK level in all four cell lines irrespective of SOX11 expression status (Figure 5B).
Figure 5. SOX11i inhibit SOX11-dependent signaling and gene expression.
(A) Line graph showing percentage of BTK phosphorylation in JVM-2 (orange), Mino (green), Z-138 (purple) and JeKo-1 (blue) after 24 hours of treatment with compound E, compound R and compound T. Cells were treated with 10 μM, 20 μM, 30 μM and 40 μM of each compound. The average and mean standard deviation were calculated after experiments were done three times. Student’s t-test was used to calculate p-values: P ≤ 0.05 (*), P ≤ 0.01 (**), P ≤ 0.001 (***). (B) Bar graph showing percentage of BTK phosphorylation in JVM-2 (orange), Mino (green), Z-138 (purple) and JeKo-1 (blue) after 24 hours of treatment with 0.2% of DMSO, 20 μM of compound E, 20 μM of compound R, 20 μM of compound T, 1 μM of Ibrutinib and 10 μM of Ibrutinib. (C) mRNA expression of SMAD3, PPP3CA and PAX5 after JeKo-1, Mino, and Z-138 were treated with 0.2% of DMSO (black), 20 μM of compound E (pink), 20 μM of compound R (teal), and 20 μM of compound T (gold) for 24 hours. The average and mean standard deviation were calculated after experiments were done three times. Student’s t-test was used to calculate p-values: P ≤ 0.05 (*), P ≤ 0.01 (**), P ≤ 0.001 (***).
For further mechanistic validation of the inhibition of the transcriptional activity of SOX11 by SOX11i, we investigated the expression levels of SOX11 target genes. Mothers against decapentaplegic homolog 3 (SMAD3) and Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform encoded by the PPP3CA gene, are MCL specific target genes of SOX11 and mediators for the transforming growth factor-β (TGF-β) pathway. Both these genes are repressed by SOX11 overexpression in MCL cell lines Granta-519 and Z-138 and their modulation by SOX11 has been validated by ChIP-qPCR and reporter assays27. Conversely, Paired Box 5 (PAX5), a regulator of terminal B cell differentiation is a validated MCL-specific gene activated by SOX11 that promotes a more aggressive disease phenotype28.
Treatment of three SOX11+ cell lines JeKo-1, Mino and Z138 with 20 μM SOX11i E, R and T for 24 hours caused a significant de-repression of both SMAD3 and PPP3CA target genes and downregulation of PAX5. These transcriptional effects were virtually absent in SOX11-negative JVM-2 cells (Figure 5C). These results provide further evidence that these SOX11i interfere with SOX11-DNA interaction and thus inhibit both activating and repressing functions of SOX11.
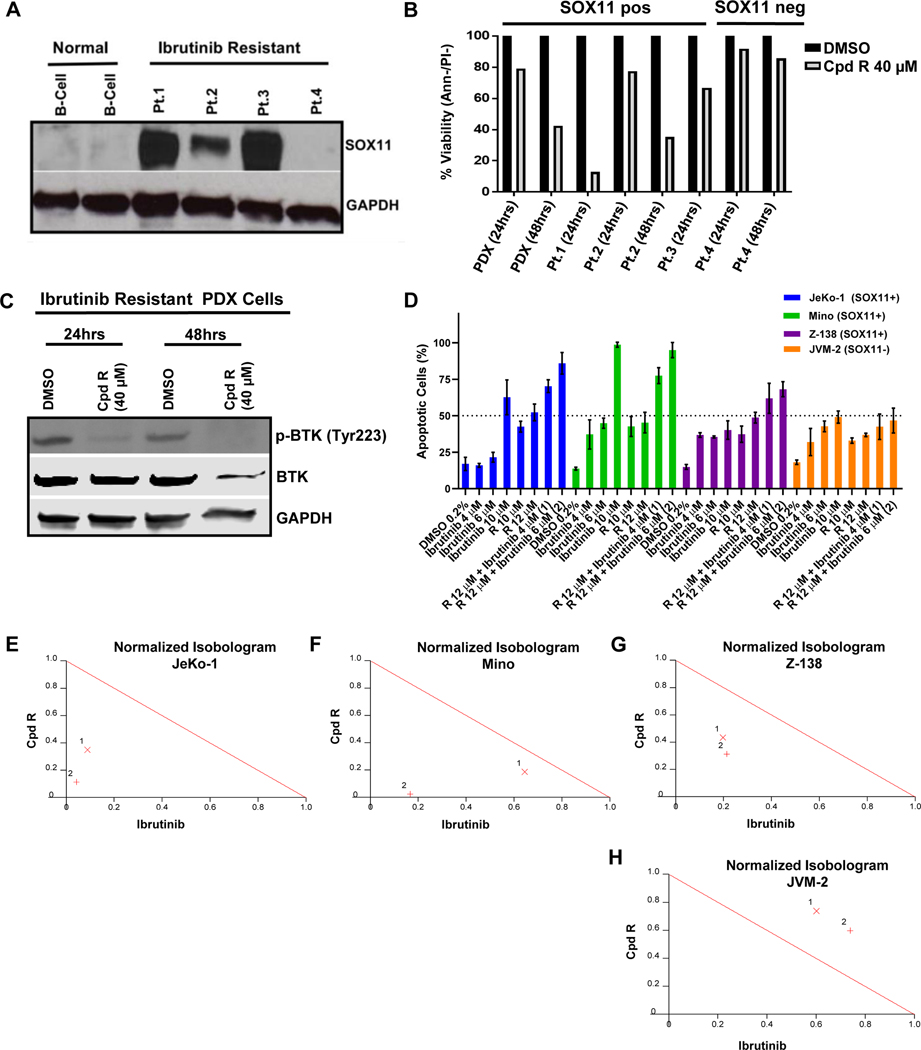
Compound R is cytotoxic in Ibrutinib-resistant primary MCL and synergizes with Ibrutinib in MCL cell lines
Since compound R showed the strongest in vitro SOX11-specific anti-MCL activity and was efficacious in the majority of other assays, it became our SOX11i of choice for testing in Ibrutinib resistant primary MCL. SOX11 expression was compared in primary samples from four Ibrutinib-resistant MCL patients (see Table S4 for patient characteristics) and two healthy volunteers. As expected, the two healthy volunteers were SOX11-negative, however, three of the four Ibrutinib resistant MCL patients were positive for SOX11 expression (Figure 6A). Treatment with compound R induced cytotoxicity specifically in the three SOX11-positive Ibrutinib-resistant (Pt. 1, 2, 3) but not in the SOX11-negative Ibrutinib-resistant (Pt. 4) MCL patient samples (Figure 6B). In a patient derived xenograft (PDX) mouse model of one of these SOX11-positive subjects (Pt. 2), Compound R treatment led to complete loss of BTK phosphorylation confirming its on-target mechanism of action (Figure 6C). We also tested Compound R in combination with Ibrutinib in SOX11+ JeKo-1, Z-138 and Mino cells and found strong synergy between the two agents (Figure 6D) as depicted in the normalized isobolograms (Figure 6E–G). A similar experiment with SOX11-negative JVM-2 cells showed antagonism between Compound R and Ibrutinib (Figure 6H) further confirming the on-target activity of Compound R.
Figure 6. Compound R induces cell death in Ibrutinib-resistant primary MCL and is synergistic with Ibrutinib in SOX11+ MCL cell lines.
(A) Immunoblot analysis showing SOX11 expression in whole cell lysates from 4 primary MCL patient samples with acquired resistance to Ibrutinib compared to normal B lymphocytes from 2 healthy donors. GAPDH was used as a loading control. (B) The same 4 Ibrutinib resistant MCL primary patient samples in addition to cells obtained from an MCL PDX mouse model (derived from Pt. 2) were treated with 40 μM of compound R for 24 and 48 hours. Viability of the cells was obtained by Annexin V−/propidium iodide−staining and flow cytometry at the indicated time points. Graph shows normalized percentage of live cells relative to the DMSO treated control from 1 experiment. (C) Immunoblot analysis showing the expression levels of total BTK and phosphorylated BTK (p-BTKTyr223) in whole cell lysates from the Ibrutinib resistant PDX cells after treatment with 40 μM of compound R at the indicated time points. Decrease in p-BTK level was noted upon treatment with compound R as of 24 hours compared to the DMSO control. GAPDH used as loading control. (D) Bar graph showing the viability of JeKo-1, Z-138, Mino and JVM-2 after 72 hours of treatment with compound R alone and in combination with Ibrutinib at different concentrations. Viability was obtained by using Annexin V and 7-AAD staining and flow cytometry. The average and mean standard deviation were calculated after experiments were done three times. (E-H) Normalized isobolograms derived from combination index values obtained for JeKo-1 (E), Z-138 (F), Mino (G) and JVM-2 (H) cells treated with combinations of Compound R and Ibrutinib two different sets of concentrations show synergy between the two agents in all three SOX11-positive cells lines but antagonism in the SOX11-negative cell line. The theoretical oblique red line is an isobole with a CI value of 1.0 denoting an additive effect. As seen here, CI values of <0.9 which fall well below the red line indicate strong synergism (http://www.biosoft.com/w/calcusyn.htm).
Discussion:
TF SOX11 is expressed in the majority of MCL patients and is an important contributor to MCL pathogenesis. It is developmentally regulated, and is not expressed in adult tissues. As such, SOX11 represents a potential new drug target in MCL, but as a TF, it would be considered “undruggable”. Although, substantial success has been achieved in targeting protein-protein interactions mediated by TFs41 or by targeting a ligand-binding domain outside of the DNA binding domain (DBD)42, targeting protein-DNA interactions still constitutes a major challenge. The overall convexity of DBDs (to bind the DNA major groove, for example) and the lack of pockets usually found in other targets such as kinases and GPCRs make it difficult to target DBDs with small molecules. Still, a small molecule (FDI-6) targeting the TF FOXM1 DBD43 has been developed, as well as another one (Sm4) targeting the SOX18 DBD, which also inhibits SOX11 DBD albeit at sub-millimolar concentrations44. Our in silico screening using a SOX11-DBD homology model led to the successful identification of three structurally related compounds (SOX11i; E, R, and T) that bind the SOX11 DBD and perturb its interaction with DNA. Specifically, our binding data from cell free assays (SPR) confirm that SOX11i (E, R and T) physically bind to SOX11 DBD with micromolar affinities and FRET and EMSA assays show that the compounds interfere with SOX11:DNA binding in vitro. During the preparation of our manuscript, Dodonova et al.45 published the X-ray crystal structure of the SOX11-DBD in complex with DNA (PDB code 6T78). Importantly, our 3-D homology model of SOX11-DBD utilized for virtual screening is confirmed by their experimentally determined structure with an RMSD of 1Å (Supplementary Figure S8).
TP53 mutated MCL is associated with the worst prognosis12,46,47. Our study utilized four different MCL cell lines. Z-138 and JVM-2 have wild-type TP53, whereas MINO (missense V147G mutant), JeKo-1 (deleted) have defective TP53. Our data demonstrate that sensitivity to SOX11i (e.g. Compound R) is SOX11-specific and independent of TP53 status since Z-138, MINO and JeKo-1 are all sensitive but JVM-2 is not. Hence, SOX11i could provide a much-needed avenue for treatment of TP53 mutated MCL.
Our SOX11 inhibitors represent a proof of concept that DNA-TF interaction can be targeted to abrogate aberrant gene regulation and cellular growth in cancer. Our current molecules are early tool compounds for probing downstream effects of TF inhibition in vitro. The SOX11i we have discovered have potencies in the micromolar range and they also have limited aqueous solubility which has prevented us from testing them in our transgenic and PDX mouse models. We are actively pursuing chemical modifications to make inhibitors that are more potent, selective and with desirable pharmacological properties for in vivo application.
Cellular resistance to chemotherapy is a major obstacle to cancer treatment in MCL. The most significant advancement in the treatment of this challenging disease is the introduction of molecularly targeted therapies such as BTKi and BCL2i. However, patients develop resistance to BTKi due to a number of mechanisms including C481S mutations in the BTK binding pocket preventing Ibrutinib binding and median survival is less than 4 months after BTKi resistance48. Similarly, resistance to BCL2i has been described due to upregulation of the AKT pathway49. The development of resistance to therapeutics in BTKi and BCL2i setting represents the most urgent unmet need in MCL. Our data using primary MCL cells and PDX models demonstrate that our lead SOX11i, compound R, can overcome Ibrutinib resistance. The approval of anti-CD19 CAR-T cells50 and ongoing trials of CD20xCD3 bispecific antibodies51,52 represent promising new approaches for relapsed MCL patients, but are not currently curative. Given the room for improvement in MCL therapeutics, our results lay the foundation for future optimization of the chemical probes identified thus far towards inhibitors for therapeutic application in treating naïve and relapsed/refractory patients with MCL.
These novel small molecule inhibitors will also be useful for understanding the pathogenesis of other SOX11 positive malignancies such as epithelial ovarian tumors53,54, medulloblastoma55, gliomas56 and basal like breast cancer57, and ultimately expand therapeutic options for SOX11-expressing human cancers.
Supplementary Material
Statement of Translational Relevance:
Mantle cell lymphoma (MCL) is a fatal subtype of non-Hodgkin’s lymphoma. New treatments for relapsed MCL are the most urgent unmet need in clinical practice. The transcription factor SOX11 is a bona-fide MCL oncogene overexpressed in 80–90% MCL patients and we have shown that it drives MCL tumor development by augmenting BCR signaling in vivo. Here we report the discovery of the first-in-class small molecule inhibitors of SOX11 with on-target anti-MCL activity in cell lines and in patient-derived ex vivo models of Ibrutinib resistance. Our results represent a foundation for the development of a novel class of anti-MCL agents with therapeutic application in treatment naïve as well as relapsed/refractory settings. The broader implication that our work provides is a model for rational, structure guided therapeutic discovery of inhibitors of the SOX family of transcription factors.
Acknowledgements:
We are grateful for support from Celgene Consortium Funding, NIH R01 CA244899, NIGMS-funded Integrated Pharmacological Sciences Training Program T32 GM062754 and Tisch Cancer Center Development Grant. Computations were run on resources available through the Scientific Computing Facility at the Icahn School of Medicine at Mount Sinai supported by the Office of Research Infrastructure of the National Institutes of Health under award numbers S10OD018522 and S10OD026880.
Financial support: We are grateful for support from Celgene Consortium Funding, NIH R01 CA244899, NIGMS-funded Integrated Pharmacological Sciences Training Program T32 GM062754 and Tisch Cancer Center Development Grant. Computations were run on resources available through the Scientific Computing Facility at the Icahn School of Medicine at Mount Sinai supported by the Office of Research Infrastructure of the National Institutes of Health under award numbers S10OD018522 and S10OD026880.
Conflict of interest disclosure statement:
S. Parekh is a paid consultant for Foundation Medicine; and has received research funding from Celgene and Karyopharm. M. Filizola has received research funding from Celgene. A.K. Aggarwal has received research funding from Celgene. J. Jin has received research funding from Celgene. No potential conflicts of interest were disclosed by the other authors.
Patents: S.S. Jatiani, S. Christie, R. Jain, A. Kapoor, C. Lee, H. Ü. Kaniskan, F. Meng, J. Jin, M. Filizola, A.K. Aggarwal and S. Parekh are named inventors on a provisional patent application filed by Icahn School of Medicine at Mount Sinai.
References:
- 1.Fernandez V, Hartmann E, Ott G, Campo E, Rosenwald A. Pathogenesis of mantle-cell lymphoma: all oncogenic roads lead to dysregulation of cell cycle and DNA damage response pathways. J Clin Oncol. 2005;23(26):6364–6369. [DOI] [PubMed] [Google Scholar]
- 2.Eskelund CW, Kolstad A, Jerkeman M, et al. 15-year follow-up of the Second Nordic Mantle Cell Lymphoma trial (MCL2): prolonged remissions without survival plateau. Br J Haematol. 2016November;175(3):410–418.3 [DOI] [PubMed] [Google Scholar]
- 3.Gerson JN, Handorf E, Villa D, et al. Survival Outcomes of Younger Patients With Mantle Cell Lymphoma Treated in the Rituximab Era. J Clin Oncol. 2019February20;37(6):471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vose JM. Mantle cell lymphoma: 2017 update on diagnosis, risk-stratification, and clinical management. Am J Hematol. 2017August;92(8):806–813. [DOI] [PubMed] [Google Scholar]
- 5.Schieber M, Gordon LI, Karmali R. Current overview and treatment of mantle cell lymphoma. F1000Res. 2018July25;7:F1000 Faculty Rev-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng W, Fu K, Quintanilla-Fend L, Lim M, Ondrejka S, Hsi ED. Cyclin D1-negative blastoid mantle cell lymphoma identified by SOX11 expression. Am J Surg Pathol. 2012;36(2):214–9. [DOI] [PubMed] [Google Scholar]
- 7.Parekh S, Weniger MA, Wiestner A. New molecular targets in mantle cell lymphoma. Semin Cancer Biol. 2011;21(5):335–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martín-Garcia D, Navarro A, Valdés-Mas R, et al. CCND2 and CCND3 hijack immunoglobulin light-chain enhancers in cyclin D1- mantle cell lymphoma. Blood. 2019;133(9):940–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lovec H, Grzeschiczek A, Kowalski MB, Möröy T. Cyclin D1/bcl-1 cooperates with myc genes in the generation of B-cell lymphoma in transgenic mice. EMBO J. 1994August1;13(15):3487–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bodrug SE, Warner BJ, Bath ML, Lindeman GJ, Harris AW, Adams JM. Cyclin D1 transgene impedes lymphocyte maturation and collaborates in lymphomagenesis with the myc gene. EMBO J. 1994;13:2124–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beà S, Valdés-Mas R, Navarro A, et al. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc Natl Acad Sci U S A. 2013;110(45):18250–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eskelund CW, Dahl C, Hansen JW, et al. TP53 mutations identify younger mantle cell lymphoma patients who do not benefit from intensive chemoimmunotherapy. Blood. 2017October26;130(17):1903–1910. [DOI] [PubMed] [Google Scholar]
- 13.Agarwal R, Chan YC, Tam CS, et al. Dynamic molecular monitoring reveals that SWI-SNF mutations mediate resistance to ibrutinib plus venetoclax in mantle cell lymphoma. Nat Med. 2019January;25(1):119–129. [DOI] [PubMed] [Google Scholar]
- 14.Ferrero S, Rossi D, Rinaldi A, et al. KMT2D mutations and TP53 disruptions are poor prognostic biomarkers in mantle cell lymphoma receiving high-dose therapy: a FIL study. Haematologica. 2020June;105(6):1604–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain P, Zhang S, Kanagal-Shamanna R, et al. Genomic profiles and clinical outcomes of de novo blastoid/pleomorphic MCL are distinct from those of transformed MCL. Blood Adv. 2020March24;4(6):1038–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang ML, Rule S, Martin P, et al. Targeting BTK with ibrutinib in relapsed or refractory mantle-cell lymphoma. N Engl J Med. 2013;369(6):507–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davids MS, Roberts AW, Seymour JF, et al. Phase I First-in-Human Study of Venetoclax in Patients With Relapsed or Refractory Non-Hodgkin Lymphoma. J Clin Oncol. 2017;35(8):826–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tam CS, Anderson MA, Pott C, et al. Ibrutinib plus Venetoclax for the Treatment of Mantle-Cell Lymphoma. N Engl J Med. 2018;378(13):1211–1223. [DOI] [PubMed] [Google Scholar]
- 19.Cheah CY, Chihara D, Romaguera JE, et al. Patients with mantle cell lymphoma failing ibrutinib are unlikely to respond to salvage chemotherapy and have poor outcomes. Ann Oncol. 2015;26(6):1175–9. [DOI] [PubMed] [Google Scholar]
- 20.Dy P, Penzo-Méndez A, Wang H, Pedraza CE, Macklin WB, Lefebvre V. The three SoxC proteins--Sox4, Sox11 and Sox12--exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 2008;36(9):3101–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mallampati S, Sun B, Lu Y, et al. Integrated genetic approaches identify the molecular mechanisms of Sox4 in early B-cell development: intricate roles for RAG1/2 and CK1ε. Blood. 2014;123(26):4064–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sock E, Rettig SD, Enderich J, Bösl MR, Tamm ER, Wegner M. Gene targeting reveals a widespread role for the high-mobility-group transcription factor Sox11 in tissue remodeling. Mol Cell Biol. 2004;24(15):6635–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernàndez V, Salamero O, Espinet B, et al. Genomic and gene expression profiling defines indolent forms of mantle cell lymphoma. Cancer Res. 2010;70(4):1408–18. [DOI] [PubMed] [Google Scholar]
- 24.Leshchenko VV, Kuo PY, Shaknovich R, et al. Genomewide DNA-methylation analysis reveals novel targets for drug development in mantle cell lymphoma. Blood. 2010;116(7):1025–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dictor M, Ek S, Sundberg M, et al. Strong lymphoid nuclear expression of SOX11 transcription factor defines lymphoblastic neoplasms, mantle cell lymphoma and Burkitt’s lymphoma. Haematologica. 2009;94(11):1563–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carvajal-Cuenca A, Sua LF, Silva NM, et al. In situ mantle cell lymphoma: clinical implications of an incidental finding with indolent clinical behavior. Haematologica. 2012;97(2):270–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuo PY, Leshchenko VV, Fazzari MJ, et al. High-resolution chromatin immunoprecipitation (ChIP) sequencing reveals novel binding targets and prognostic role for SOX11 in mantle cell lymphoma. Oncogene. 2015;34(10):1231–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vegliante MC, Palomero J, Pérez-Galán P, et al. SOX11 regulates PAX5 expression and blocks terminal B-cell differentiation in aggressive mantle cell lymphoma. Blood. 2013;121(12):2175–85. [DOI] [PubMed] [Google Scholar]
- 29.Palomero J, Vegliante MC, Eguileor A, et al. SOX11 defines two different subtypes of mantle cell lymphoma through transcriptional regulation of BCL6. Leukemia. 2016;30(7):1596–9. [DOI] [PubMed] [Google Scholar]
- 30.Kuo PY, Jatiani SS, Rahman AH, et al. SOX11 augments BCR signaling to drive MCL-like tumor development. Blood. 2018;131(20):2247–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rinaldi A, Kwee I, Taborelli M, et al. Genomic and expression profiling identifies the B-cell associated tyrosine kinase Syk as a possible therapeutic target in mantle cell lymphoma. Br J Haematol. 2006;132(3):303–16. [DOI] [PubMed] [Google Scholar]
- 32.Cinar M, Hamedani F, Mo Z, Cinar B, Amin HM, Alkan S. Bruton tyrosine kinase is commonly overexpressed in mantle cell lymphoma and its attenuation by Ibrutinib induces apoptosis. Leuk Res. 2013;37(10):1271–7. [DOI] [PubMed] [Google Scholar]
- 33.Chiron D, Di Liberto M, Martin P, et al. Cell-cycle reprogramming for PI3K inhibition overrides a relapse-specific C481S BTK mutation revealed by longitudinal functional genomics in mantle cell lymphoma. Cancer Discov. 2014;4(9):1022–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jauch R, Ng CK, Narasimhan K, Kolatkar PR. The crystal structure of the Sox4 HMG domain-DNA complex suggests a mechanism for positional interdependence in DNA recognition. Biochem J. 2012;443(1):39–47. [DOI] [PubMed] [Google Scholar]
- 35.Chen YH, Gao J, Fan G, Peterson LC. Nuclear expression of Sox11 is highly associated with mantle cell lymphoma but is independent of t(11;14)(q13;q32) in non-mantle cell B-cell neoplasms. Mod Pathol. 2010;23(1):105–112. [DOI] [PubMed] [Google Scholar]
- 36.Mozos A, Royo C, Hartmann E, et al. SOX11 expression is highly specific for mantle cell lymphoma and identifies the cyclin D1-negative subtype. Haematologica. 2009;94(11):1555–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Asplund AC, Porwit A, et al. The subcellular Sox11 distribution pattern identifies subsets of mantle cell lymphoma: correlation to overall survival. Br J Haematol. 2008;143(2):248–252. [DOI] [PubMed] [Google Scholar]
- 38.Werner MH, Huth JR, Gronenborn AM, Clore GM. Molecular basis of human 46X,Y sex reversal revealed from the three-dimensional solution structure of the human SRY-DNA complex. Cell. 1995;81(5):705–14. [DOI] [PubMed] [Google Scholar]
- 39.Huang R, Southall N, Wang Y, et al. The NCGC pharmaceutical collection: a comprehensive resource of clinically approved drugs enabling repurposing and chemical genomics. Sci Transl Med. 2011;3(80):80ps16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deng Z, Chuaqui C, Singh J. Structural interaction fingerprint (SIFt): a novel method for analyzing three-dimensional protein-ligand binding interactions. J Med Chem. 2004;47(2):337–44. [DOI] [PubMed] [Google Scholar]
- 41.Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303(5659):844–8. [DOI] [PubMed] [Google Scholar]
- 42.Burris TP, Solt LA, Wang Y, et al. Nuclear receptors and their selective pharmacologic modulators. Pharmacol Rev. 2013;65(2):710–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gormally MV, Dexheimer TS, et al. Suppression of the FOXM1 transcriptional programme via novel small molecule inhibition. Nat Commun. 2014;5:5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fontaine F, Overman J, Moustaqil M, et al. Small-Molecule Inhibitors of the SOX18 Transcription Factor. Cell Chem Biol. 2017;24(3):346–359. [DOI] [PubMed] [Google Scholar]
- 45.Dodonova SO, Zhu F, Dienemann C, et al. Nucleosome-bound SOX2 and SOX11 structures elucidate pioneer factor function. Nature 2020. 580(7805): 669–672. [DOI] [PubMed] [Google Scholar]
- 46.Jain P, Wang M. Mantle cell lymphoma: 2019 update on the diagnosis, pathogenesis, prognostication, and management. Am J Hematol. 2019June;94(6):710–725. [DOI] [PubMed] [Google Scholar]
- 47.Elhassadi E, Hennessy B, Kumar S, et al. TP53 status in mantle cell lymphoma (MCL)-a 10 year single center experience. Presented at: 2019 European Hematology Association Congress; June 13–16, 2019; Amsterdam, The Netherlands. Abstract PF493. [Google Scholar]
- 48.Hershkovitz-Rokah O, Pulver D, Lenz G, Shpilberg O. Ibrutinib resistance in mantle cell lymphoma: clinical, molecular and treatment aspects. Br J Haematol. 2018;181(3):306–319. [DOI] [PubMed] [Google Scholar]
- 49.Pham LV, Huang S, Zhang H, et al. Strategic Therapeutic Targeting to Overcome Venetoclax Resistance in Aggressive B-cell Lymphomas. Clin Cancer Res. 2018;24(16):3967–3980. [DOI] [PubMed] [Google Scholar]
- 50.Wang M, Munoz J, Goy A, et al. KTE-X19 CAR T-Cell Therapy in Relapsed or Refractory Mantle-Cell Lymphoma. N Engl J Med. 2020;382(14):1331–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bannerji R, Advani RH, Brown JR, et al. Safety and Preliminary Clinical Activity of REGN1979, an Anti-CD20 x Anti-CD3 Bispecific Antibody, in Patients with B-NHL Previously Treated with CD20-Directed Antibody Therapy. Blood. 2017;130:1550. [Google Scholar]
- 52.Hutchings M, Mous R, Clausen MR, et al. Subcutaneous Epcoritamab Induces Complete Responses with an Encouraging Safety Profile across Relapsed/Refractory B-Cell Non-Hodgkin Lymphoma Subtypes, Including Patients with Prior CAR-T Therapy: Updated Dose Escalation Data. Blood. 2020; 136 (Supplement 1): 45–46. [Google Scholar]
- 53.Brennan DJ, Ek S, Doyle E, et al. The transcription factor Sox11 is a prognostic factor for improved recurrence-free survival in epithelial ovarian cancer. Eur J Cancer. 2009;45(8):1510–7. [DOI] [PubMed] [Google Scholar]
- 54.Sernbo S, Gustavsson E, Brennan DJ, et al. The tumour suppressor SOX11 is associated with improved survival among high grade epithelial ovarian cancers and is regulated by reversible promoter methylation. BMC Cancer. 2011;11:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Bont JM, Kros JM, Passier MM, et al. Differential expression and prognostic significance of SOX genes in pediatric medulloblastoma and ependymoma identified by microarray analysis. Neuro Oncol. 2008;10(5):648–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weigle B, Ebner R, Temme A,et al. Highly specific overexpression of the transcription factor SOX11 in human malignant gliomas. Oncol Rep. 2005;13(1):139–44. [PubMed] [Google Scholar]
- 57.Shepherd JH, Uray IP, Mazumdar A, et al. The SOX11 transcription factor is a critical regulator of basal-like breast cancer growth, invasion, and basal-like gene expression. Oncotarget. 2016;7(11):13106–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.