Abstract
Purpose:
TP53 aberration (TP53 mutation and/or 17p deletion) is the most important predictive marker in chronic lymphocytic leukemia (CLL). While each TP53 aberration is considered an equal prognosticator, the prognostic value of carrying isolated (single-hit) or multiple (multi-hit) TP53 aberrations remains unclear, particularly in the context of targeted agents.
Experimental Design:
We performed deep sequencing of TP53 using baseline samples collected from 51 TP53 aberrant patients treated with ibrutinib in a phase II study (NCT01500733).
Results:
We identified TP53 mutations in 43 patients (84%) and del(17p) in 47 (92%); 9 and 42 patients carried single-hit and multi-hit TP53, respectively. The multi-hit TP53 subgroup was enriched with younger patients who had prior treatments and unmutated immunoglobulin heavy-chain variable region gene. We observed significantly shorter overall survival, progression-free survival (PFS), and time-to-progression (TTP) in patients with multi-hit TP53 compared with those with single-hit TP53. Clinical outcomes were similar in patient subgroups stratified by 2 or >2 TP53 aberrations. In multivariable analyses, multi-hit TP53 CLL was independently associated with inferior PFS and TTP. In sensitivity analyses, excluding mutations below 1% VAF demonstrated similar outcome. Results were validated in an independent population-based cohort of 112 CLL patients treated with ibrutinib.
Conclusions:
In this study, single-hit TP53 defines a distinct subgroup of patients with an excellent long-term response to single-agent ibrutinib, while multi-hit TP53 is independently associated with shorter PFS. These results warrant further investigations on prognostication and management of multi-hit TP53 CLL.
Keywords: Prognostication, targeted therapy, TP53 mutation, deep sequencing, NGS
INTRODUCTION
Mutations in the TP53 gene and deletion of the short arm of chromosome 17 [del(17p)] are associated with genomic instability and an adverse outcome in cancer (1, 2). In chronic lymphocytic leukemia (CLL), TP53 aberrations [del(17p) and/or TP53 mutation] predict unfavorable prognosis with shorter time to treatment initiation, higher likelihood of developing refractoriness to or early relapse after chemoimmunotherapy (CIT), and inferior overall survival (OS) (3–5). The prevalence of TP53 aberrations differ by treatment settings; del(17p) affecting 4–8% of newly diagnosed CLL patients, and up to 40% at relapse (6–8). International guidelines in CLL recommend universal testing for TP53 aberrations before any line of therapy using fluorescence in situ hybridization (FISH) to detect del(17p) and Sanger or next-generation sequencing (NGS) to detect TP53 mutations down to 5–10% variant allele frequency (VAF) (9–11).
Although del(17p) and TP53 mutations frequently co-occur, a monoallelic TP53 aberration can be found in 30%−50% of TP53 aberrant CLL patients, which has been associated with various poor outcome in patients treated with CIT (12–17). A recent study in myelodysplastic syndrome (MDS) demonstrated inferior outcome for patients with more than one TP53 aberration (multi-hit) as compared to patients with only one TP53 aberration (single-hit); the latter group demonstrated outcome similar to patients with TP53 wild-type MDS (18). Similar data has been published for patients with acute myeloid leukemia (AML) receiving venetoclax-based therapies (19). Findings in CLL have been mixed. In a cohort of patients with newly diagnosed CLL, patients with concurrent del(17p) and TP53 mutation had worse survival than those with a single TP53 aberration only, suggesting the gene-dosage effect of TP53 (15). Likewise, previous studies similarly support that monoallelic TP53 do not impair the p53 protein completely in the era of chemotherapy (13, 20–22). Others, however, showed that an isolated TP53 mutation was linked to negative outcomes comparable to those with concurrent aberrations in newly diagnosed patients and upon chemotherapy (14, 23). Further, there is an ongoing debate for the clinically meaningful allelic burden of TP53 mutation. TP53 mutations detected at low VAF (Sanger negative) are associated with poor outcomes once CLL-directed CIT begins (17, 24). For del(17p) by FISH, patients with a higher proportion of cells affected (<25% vs. 25%−75% vs. >75%) have shorter time to first treatment (25).
Despite significant advances in targeted therapy and long-term outcomes in CLL, patients with TP53 aberration have a shorter duration of response to ibrutinib compared to those without TP53 aberrations (26–29). In patients treated with the BTK inhibitor ibrutinib, both clonal selection due to previous therapy and genomic instability in TP53 aberrated clones have been suggested as predisposing factors for the acquisition of mutations leading to treatment resistance (30, 31).
Traditionally, TP53 aberrations are considered as a dichotomized prognostic marker (aberrated or not). Even so, advances in novel treatment and sequencing call for better delineation of differences in TP53 aberrations. We thus applied sensitive NGS with a limit of detection (LOD) at 0.2% to investigate clinical outcomes in 51 patients treated with ibrutinib carrying either a single or multiple TP53 hits. Whether single-hit or multi-hit TP53 impacts clinical outcome on novel therapies as recently demonstrated in MDS and AML, remains unknown in CLL (18, 19).
MATERIALS AND METHODS
Patients with TP53 aberrant CLL in a phase II study of single-agent ibrutinib were included (clinicaltrials.gov: NCT01500733) (32). TP53 aberrations were defined by detection of 1) del(17p) based on FISH, 2) TP53 mutations based on targeted NGS, or 3) tissue expression of p53 based on immunohistochemistry (26). Immunohistochemistry was only performed in two patients as supplemental evidence of TP53 alteration. FISH cutoff for del(17p) was 8% or more interphase cells counted according to the local laboratory cutoff. All patients received ibrutinib 420 mg/day until progressive disease (PD) or intolerable side effects occurred. The study was approved by the Institutional Review Board and conducted according to the Declaration of Helsinki. Written informed consent was obtained from all patient.
We analyzed baseline samples collected prior to the initiation of ibrutinib for mutations in the TP53 gene (exons 2–10 +2 base-pair intronic overlap) by deep NGS using methods previously described (22). In brief, the target region was amplified by PCR using 100 ng genomic DNA extracted from peripheral blood mononuclear cells. Library preparation was performed according to the manufacturer’s protocol (Roche Nimblegen, Madison, WI, USA) and sequenced as paired-end on a NextSeq and MiSeq (2×150 and 2×125 base PE, respectively; Illumina, San Diego, CA, USA). To obtain comparable sequencing depth among samples, NextSeq FASTQ files were subsampled to 20%. We used a bioinformatics pipeline developed in CLC Biomedical Genomics Workbench 3.0 (Qiagen, Hilden, Germany) mapping to the hg19 reference genome. Variants were cross-referenced in the International Agency for Research on Cancer TP53 database (http://p53.iarc.fr) excluding synonymous mutations and validated single nucleotide polymorphisms. The median coverage was 43,744x (95% above 11,000x), and the LOD of the assay was 0.2% VAF as previously described (22).
For external validation, we included a population-based cohort of 205 CLL patients treated with ibrutinib (33). Data were collected retrospectively from the Danish CLL registry and patient records (6). TP53 mutations detected by either NGS or Sanger in this cohort had a LOD of 5%−20% (6).
Single-hit and multi-hit TP53 were defined as having only one TP53 aberration [del(17p) or a single TP53 mutation] and more than one TP53 aberrations [del(17p) and TP53 mutation(s); or multiple TP53 mutations], respectively. Primary outcomes were OS and progression-free survival (PFS) from the initiation of ibrutinib therapy. Death was censored for time-to-progression (TTP), whereas it was considered a competing risk for the cumulative incidence of relapse (CIR). A pairwise log-rank test was applied for OS, PFS, and TTP; Gray’s test was applied for CIR. The study was adequately powered to detect a 30% survival difference using a two-sided significance level of 0.05. Discrimination capabilities were assessed by Harrell’s C-index. We included all baseline characteristics in multivariable Cox regression analysis applying Firth’s penalized likelihood in cases of zero recorded events (34). Statistical analyses were performed with R version 4.0.3 using survival, survminer, coxphf, and Publish (35).
RESULTS
Patient characteristics
Baseline characteristics of 51 CLL patients with TP53 aberration treated with ibrutinib are summarized in Table 1. Efficacy and safety data of the study were previously reported (26). Due to eligibility criteria of the study, most patients had high-risk features including advanced Rai stage, relapsed/refractory (R/R) disease, elevated β2-microglobulin (B2M), immunoglobulin heavy-chain variable region gene (IGHV) unmutated status (U-CLL), and del(17p).
Table 1.
Baseline patient characteristics stratified on TP53 aberrational status (single-hit vs. multi-hit TP53)
| Total (n = 51) | Single-hit TP53 (n = 9) |
Multi-hit TP53 (n = 42) |
|||
|---|---|---|---|---|---|
| Characteristic | No. (%) | No. (%) | No. (%) | ||
| Age, years | |||||
| Median (IQR) | 62 (59–69) | 69 (66–76) | 62 (59–67.8) | ||
| Sex | |||||
| Female | 20 (39.2) | 4 (44.4) | 16 (38.1) | ||
| Male | 31 (60.8) | 5 (55.6) | 26 (61.9) | ||
| Rai stage | |||||
| 1 | 13 (25.5) | 1 (11.1) | 12 (28.6) | ||
| 2 | 5 (9.8) | 1 (11.1) | 4 (9.5) | ||
| 3 | 8 (15.7) | 2 (22.2) | 6 (14.3) | ||
| 4 | 25 (49.0) | 5 (55.6) | 20 (47.6) | ||
| B2M, mg/L | |||||
| Median (IQR) | 3.9 (2.9–5.8) | 3.3 (2.5–3.7) | 4.0 (3.0–6.1) | ||
| IGHV status | |||||
| M-CLL | 17 (33.3) | 7 (77.8) | 10 (23.8) | ||
| U-CLL | 34 (66.7) | 2 (22.2) | 32 (76.2) | ||
| del(17p) | |||||
| No | 4 (7.8) | 1 (11.1) | 3 (7.1) | ||
| Yes | 47 (92.2) | 8 (88.9) | 39 (92.9) | ||
| del(17p) positivity by FISH, % | |||||
| Median (IQR) | 56% (18.8–85.8) | 16.5% (14.5–53.8) | 62% (30.8–87.5) | ||
| Treatment status | |||||
| TN | 34 (66.7) | 8 (88.9) | 26 (61.9) | ||
| RR | 17 (33.3) | 1 (11.1) | 16 (38.1) |
Abbreviations: ab, aberration; B2M, β2-microglobulin; IGHV, immunoglobulin heavy-chain variable region; M-CLL, mutated IGHV status; U-CLL, unmutated IGHV status; del(17p), deletion of chromosome 17p; TN, treatment-naïve; RR, relapsed-refractory, IQR, interquartile range.
One patient had del(17p) detected by microarray.
TP53 mutations identified
At baseline, we identified 220 TP53 mutations in 43 (84%) patients with a median VAF of 0.6% [interquartile range (IQR), 0.3%−1.6%]. The vast majority of the mutations were low burden with VAFs below 10% (177 mutations or 80%) including 146 minor TP53 mutations (VAF < 1%). The remaining 43 TP53 mutations had high burden VAFs (> 10%; Fig. 1A). At the individual patient level, 39 (76%) patients had at least one high burden mutation and four (8%) patients carried only low burden mutations. Mutations are characterized in detail in Fig. 1B and Supplementary Table S1 (16).
Figure 1.
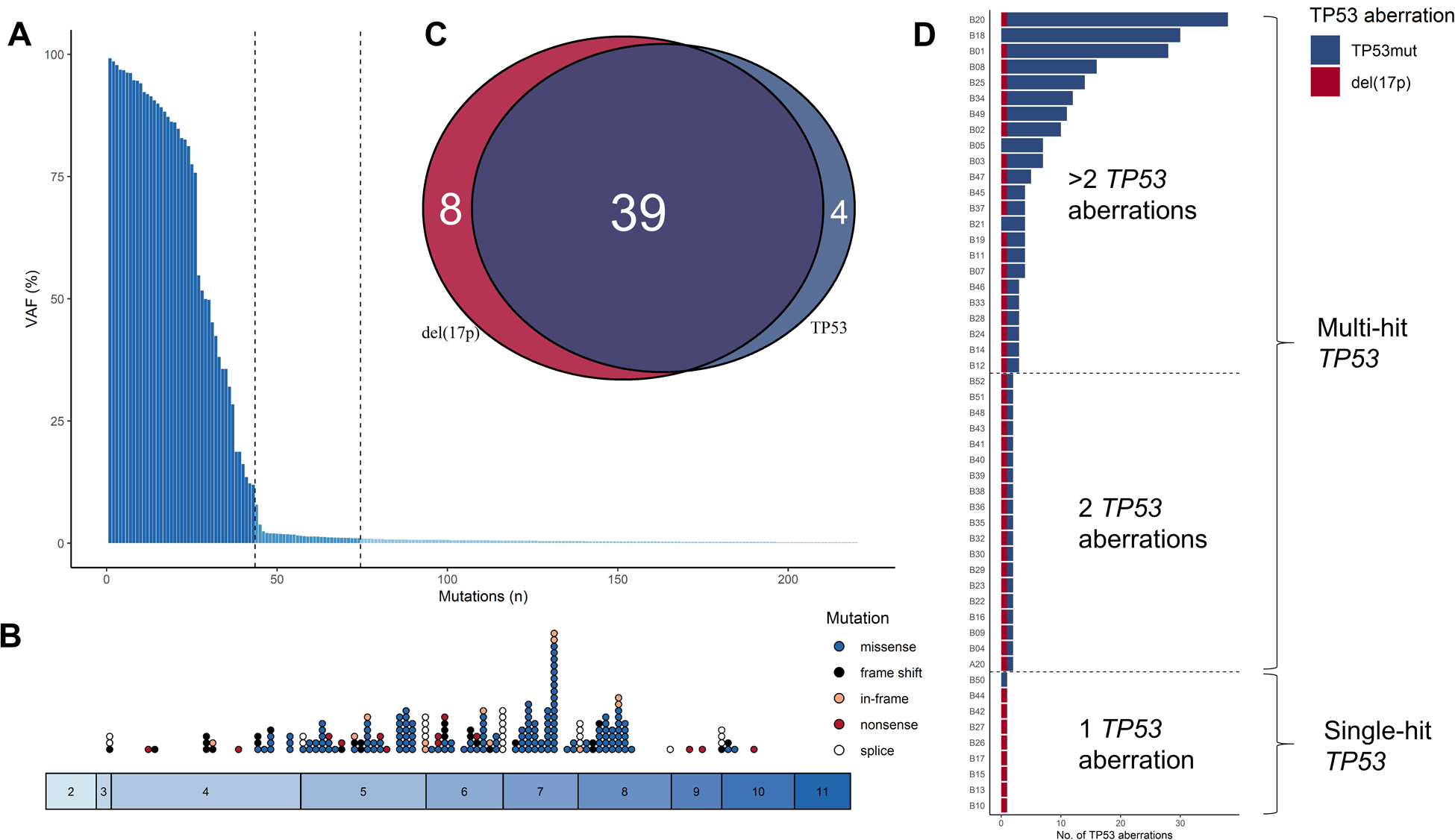
Characterization of TP53 mutations. (A) Overall, 220 mutations were identified: 43 high burden [variant allele frequency (VAF) >10%] and 177 low burden mutations (VAF ≤10%) including 146 minor mutations (VAF <1%). (B) Mutations were mainly identified in hot spot exons 5–9. (C) In all 51 patients, 17p deletion [del(17p); red] and TP53 mutations (blue) mainly cooccurred (purple). (D) Patients were grouped into single-hit and multi-hit TP53 aberrations.
Among 51 patients analyzed, 47 (92%) patients carried del(17p), and 39 (76%) had concurrent del(17p) and TP53 mutations (Fig. 1C). The median number of mutations per patient was one (IQR, 1–3 mutations), and mutations were enriched among patients with R/R disease as compared to treatment-naïve patients [median 1 (IQR, 1–2) vs. 3 (IQR, 1–13) mutations, respectively. P = 0.006; Wilcoxon signed-rank test]. Nine patients had single-hit TP53 and 42 had multi-hit TP53 including 19 patients with only 2 TP53 aberrations. Among the 42 patients with multi-hit TP53, three carried TP53 mutations only, 39 patients had del(17p) with one (n=19) or more (n=20) concomitant TP53 mutations (Fig. 1D). All four patients carrying only TP53 mutations had at least one high burden mutation (VAF >10%); one patient carried only a single TP53:c.847C>T encoding functional p53. Patients with multi-hit TP53 were younger and more frequently U-CLL compared to patients with single-hit TP53 (Table 1).
Correlation of the number and allele burden of TP53 aberrations with outcomes on ibrutinib
With a median follow-up time of 6.3 years (IQR, 6.1–7.2 years), 16 patients had died and 23 had PD; four patients died of causes unrelated to disease progression (26). OS and PFS were significantly longer for patients with single-hit TP53 compared to those with multi-hit TP53 (P ≤ 0.042; Fig. 2A–B). For patients with multi-hit TP53, the median OS was not reached, while median PFS was 4.6 years (95% CI, 3.7 years to not reached). For patients with single-hit TP53, 5-year OS and PFS were 100% compared with 69% and 43%, respectively, for patients with multi-hit TP53. We observed no difference in OS and PFS in multi-hit patients with 2 and >2 TP53 aberrations (P ≥ 0.32; Fig. 2C–D) or between patients with TP53 mutations only and those with del(17p) regardless of TP53 mutational status (Supplementary Fig. S1). TTP and CIR were significantly superior in patients with single-hit TP53 compared to patients with multi-hit TP53, whereas similar TTP and CIR were demonstrated for multi-hit TP53 patients carrying 2 and >2 TP53 aberrations (Supplementary Fig. S2).
Figure 2.
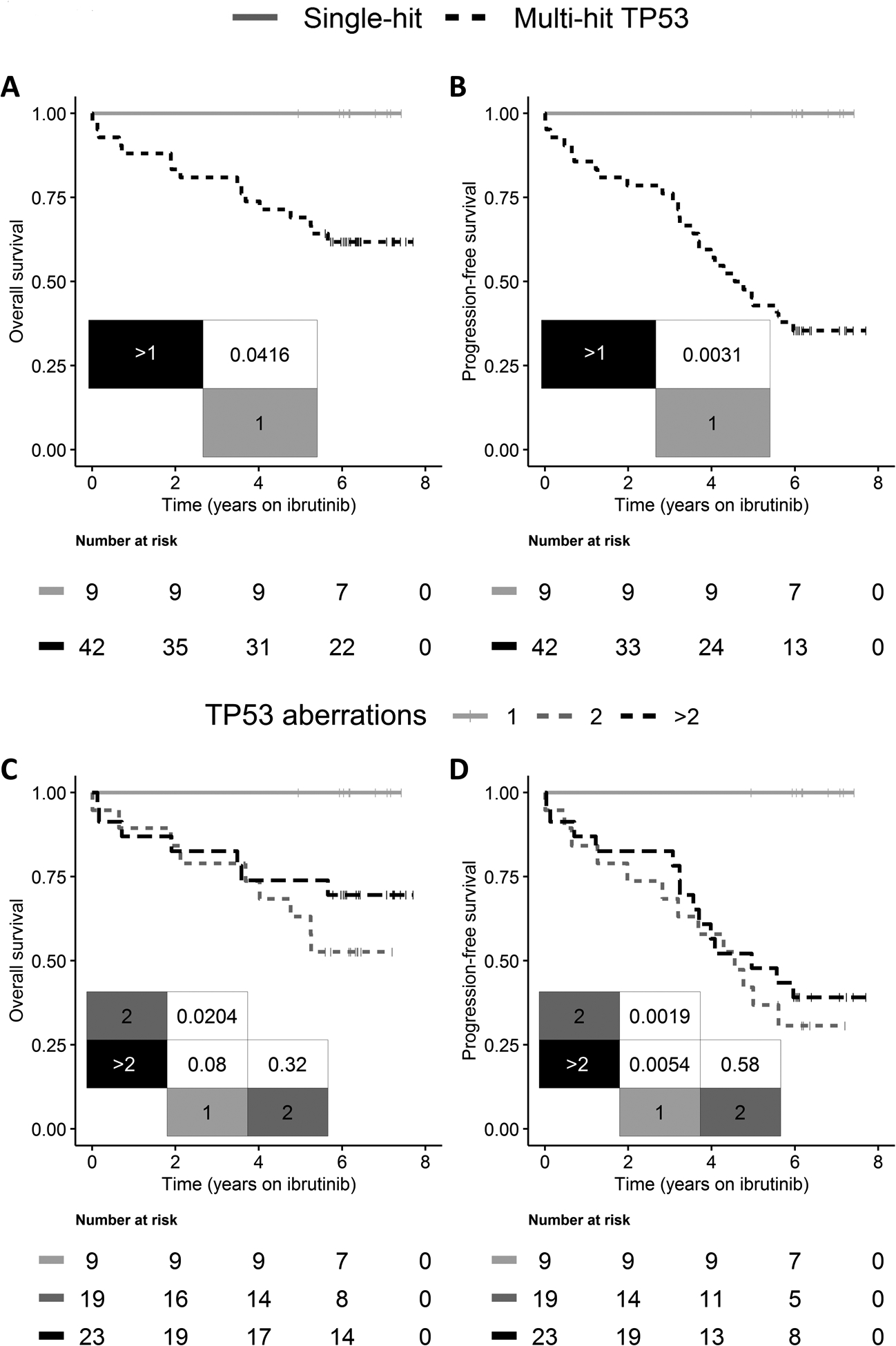
(A, C) Overall survival and (B, D) progression-free survival following initiation of single-agent ibrutinib stratified on (A-B) single-hit (gray) vs. multi-hit (black striped) TP53 and in (C-D) patients carrying 1 (gray) vs. 2 (dark gray striped) vs. > 2 (black striped) TP53 aberrations. Overall survival and progression-free survival were significantly shorter in patients with multi-hit TP53. However, stratifying multi-hit TP53 patients further into carrying 2 and > 2 TP53 aberrations demonstrated similar outcome.
To determine the optimal VAF cutoff that allows the selection of a clinically relevant burden of TP53 mutations, we tested cutoffs ranging from 0.2%−10% VAF (Supplementary Fig. S3). The discrimination capability for OS increased with higher VAF cutoff as more patients were considered to have single-hit TP53, whereas the discrimination capability for PFS and TTP decreased with increasing VAF cutoff. By using a VAF cutoff of 1%, we observed similar outcome and discrimination capabilities as compared to using a VAF cutoff of 0.2%, while a VAF cutoff of 2% resulted in a less favorable PFS and TTP for patients with single-hit TP53 aberration as well as lower discrimination capabilities (Supplementary Fig. S3).
Without any events among patients with single-hit TP53, we used Cox regression with Firth’s penalized likelihood (34). Multivariable analyses confirmed that multi-hit TP53 was independently associated with adverse PFS and TTP, while the analysis was unable to demonstrate any independent prognostic markers for OS (Fig. 3 and Supplementary Fig. S4). Due to the observed enrichment of multi-hit TP53 in younger patients with R/R disease and U-CLL (Table 1), we performed subgroup analyses by log-rank for these three baseline characteristics. In line with results from the multivariable analyses, patients with multi-hit TP53 had a consistently inferior outcome in all analyses compared to those with single-hit TP53 (Supplementary Fig. S5). Within the multi-hit TP53 subset, only R/R CLL was an inferior prognostic marker leading to further stratification of the subset in all outcomes.
Figure 3.
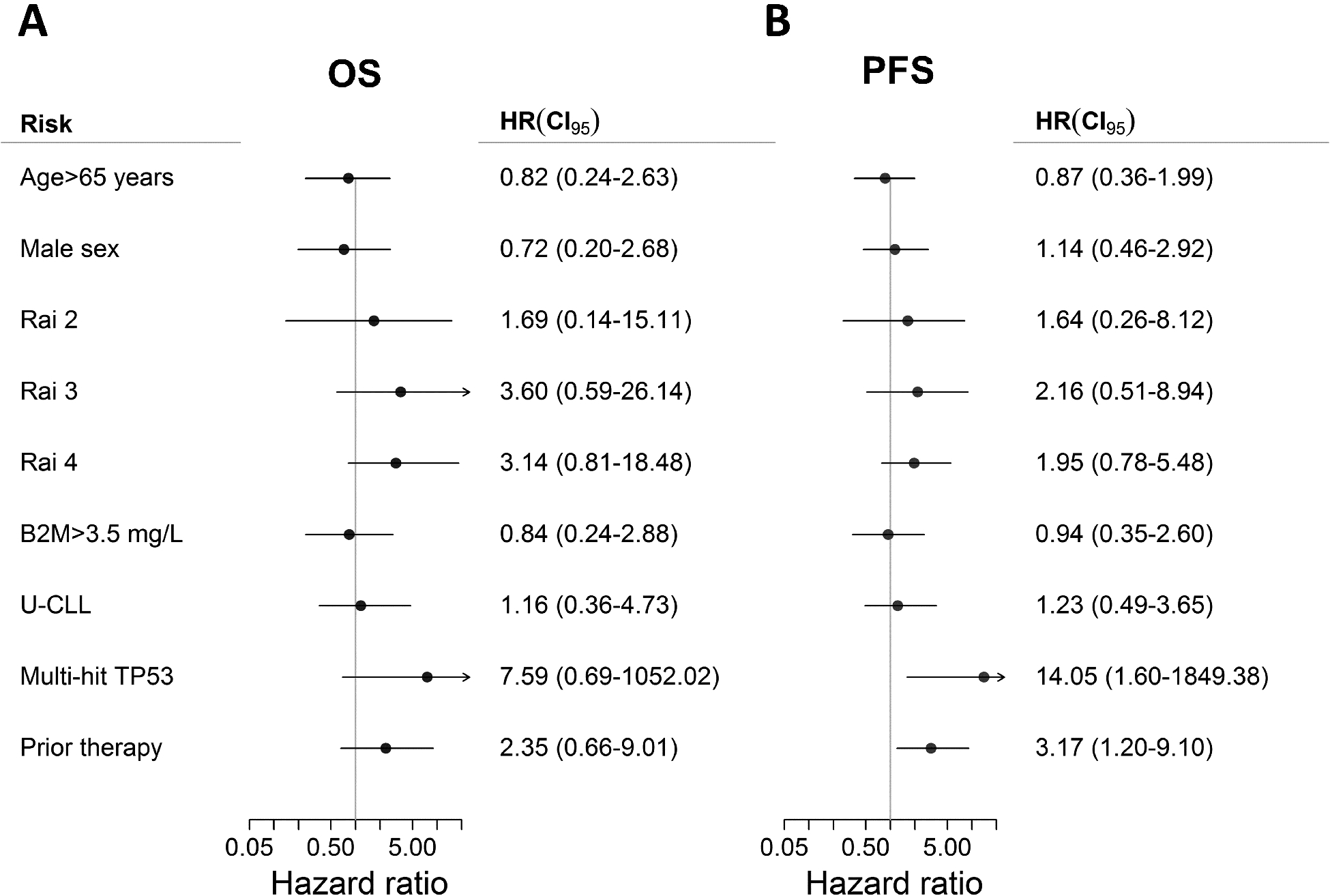
Multivariable analyses with Firth’s penalized likelihood. (A) No baseline risk factors demonstrated impact on overall survival (OS), whereas (B) multi-hit TP53 and prior therapy were independently associated with shorter progression-free survival (PFS).
Abbreviations: B2M, β2-microglobulin; U-CLL, unmutated immunoglobulin heavy-chain variable region gene (IGHV) status.
External validation of the number of TP53 aberrations
We utilized an independent group of 205 CLL patients in a Danish nationwide cohort treated with ibrutinib outside clinical trials (33). We excluded the 92 patients without TP53 aberrations and one patient receiving combination targeted therapy from further analyses. Among 112 patients analyzed, the median age was 73 years (IQR, 66–77) and 77 (69%) had R/R disease. FISH was performed in all 112 patients, while TP53 was sequenced in 61 (54%) of patients using NGS and Sanger with a LOD of 5% and 20% VAF, respectively. As a result, 94 patients carried del(17p) with no or unknown TP53 mutation including 43 patients who were tested for the mutation and had wild-type TP53 (single-hit TP53); 18 carried both del(17p) and at least 1 TP53 mutation (multi-hit TP53). Except for del(17p) enriched in patients with multi-hit TP53, all other baseline characteristics were similar in the two groups (Table S2).
At a median follow-up of 2.3 years (IQR 1.5–3.3 years), patients with multi-hit TP53 demonstrated inferior OS and PFS than those with single-hit TP53 or no known aberration (P = 0.002; Fig. 4). Restricting the analyses to include only patients with known TP53 mutational status based on Sanger or NGS, we observed longer, but insignificant OS and PFS in patients with single-hit TP53 compared to multi-hit TP53 (P = 0.11 and P = 0.08, respectively; Supplementary Fig. S6).
Figure 4.
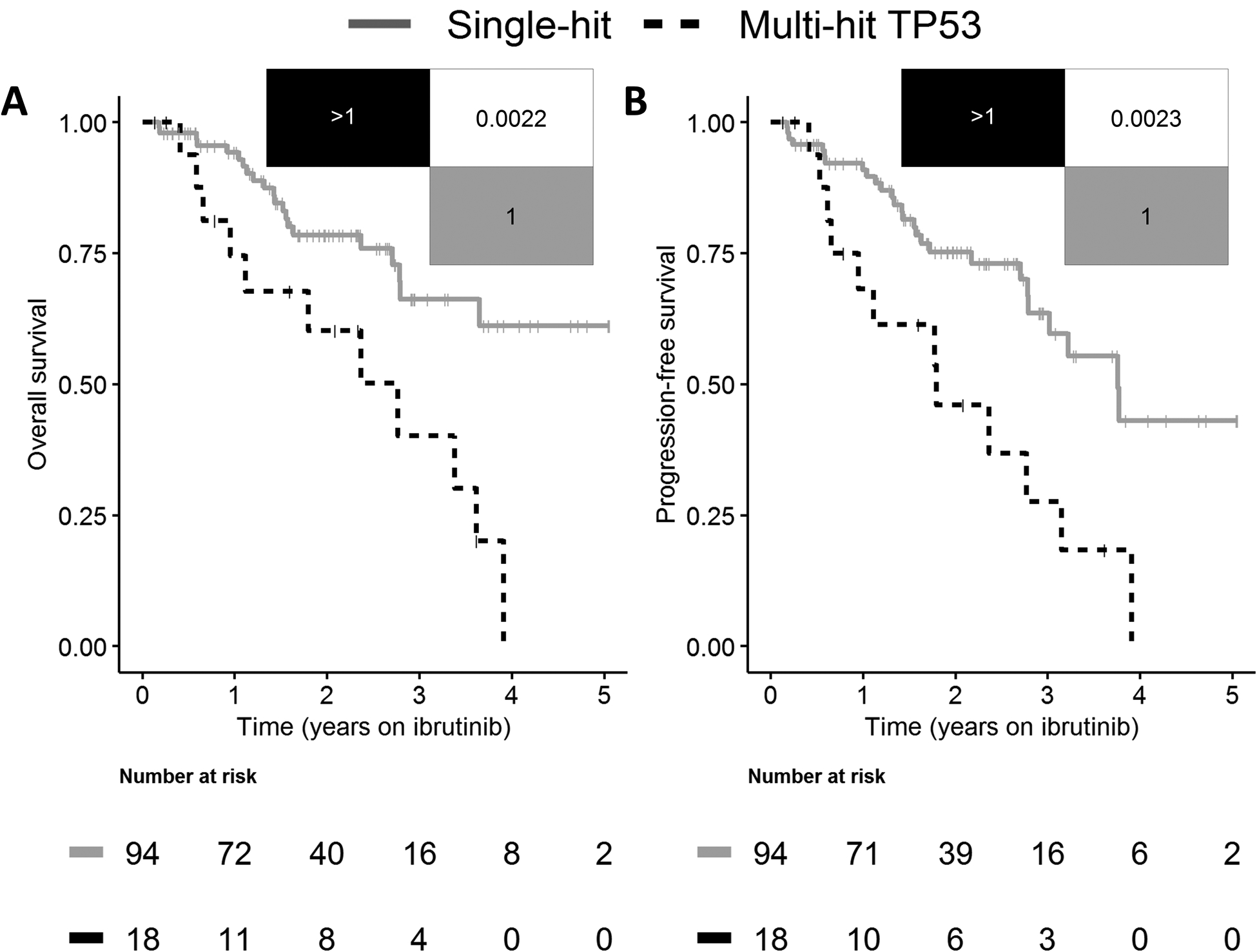
Validation of multi-hit TP53 in 112 TP53 aberrant patients with chronic lymphocytic leukemia following initiation of single-agent ibrutinib. (A) Overall survival and (B) progression-free survival in patients with single-hit (gray) and multi-hit (black striped) TP53. Overall survival and progression-free survival were significantly shorter in patients with multi-hit TP53.
DISCUSSION
We here demonstrate excellent OS and PFS in CLL patients with single-hit TP53 treated with ibrutinib monotherapy. Multi-hit TP53 was an independent risk factor associated with adverse PFS, with results validated in a population-based external cohort of CLL patients treated with ibrutinib.
CLL patients have heterogeneous outcomes during treatment with targeted agents. Several studies, including subgroup analyses of individual studies and pooled analyses of multiple trials, have identified clinical variables prognostic for ibrutinib treated patients (28, 32). Predictors of unfavorable outcomes on ibrutinib include TP53 aberration, complex karyotype, history of previous treatments, and elevated biochemical factors such as B2M and lactate dehydrogenase (36–38). However, previous prognostic studies in CLL patients treated with ibrutinib have several limitations including various treatment regimens used in the cohort (37), high rates of missing data for TP53 mutation (37), and a relatively small number of patients with TP53 aberration (28). Unlike previous studies, all patients in the present study were TP53 aberrant, systematically analyzed by deep NGS and uniformly treated with single-agent ibrutinib.
While previous studies have demonstrated an additive effect of TP53 mutation and del(17p) on clinical outcome following CIT (13, 15, 20), we here show for the first time an additive effect of TP53 aberrations in patients treated with single-agent ibrutinib. By including all patients with TP53 aberrant CLL from a prospective study, we sought to limit selection bias. Results were externally validated, although outcomes for single-hit TP53 patients were not as impressive as for patients in the phase II study (2-year PFS of 75% vs. 100%). This is likely a result of an older patient population (median 73 years vs. 62 years) with a higher proportion of R/R status (69% vs. 33%) in the validation cohort as multi-hit TP53 aberration and prior treatment demonstrated independent prognostic impact in multivariable analysis. Limiting the comparison of results in the two cohorts, we note the markedly shorter follow-up time with broader IQR in the validation cohort. Further, we could not perform a multivariate analysis of the validation cohort due to high rates of missing data in variables.
Interestingly, no differences in outcomes were detectable between multi-hit patients carrying 2 or >2 TP53 aberrations. In agreement with our findings, a study recently reported poor prognosis of multi-hit TP53 for patients with MDS and AML, whereas patients harboring TP53 wild-type and single-hit demonstrated favorable outcomes (18, 19). The study on MDS indicates that biallelic targeting of the TP53 locus is present in multi-hit TP53 cases including patients carrying multiple mutations (18). Although our study does not provide data on allele-specific aberrations, the fact that two or more TP53 aberrations correlate with poor outcomes on ibrutinib is consistent with the two-hit hypothesis (12), in which tumor suppressor genes require biallelic disruption to cause a phenotypic change (i.e. loss of p53 function) (39). However, with up to 37 distinct mutations in one patient sample in the present study, we speculate that multiple TP53 aberrations could also represent intratumoral heterogeneity and differences in the burden of TP53 aberrations suggest interclonal selection (2, 30, 31, 40). Such early clonal shifts associated with TP53 aberrations have previously been linked to an increased risk of progression on ibrutinib (41).
The number of patients with TP53 aberrations in our study is similar to other studies reporting TP53 aberrations in CLL (14, 15, 17, 20, 22, 24, 25, 28, 42). However, as the proportion patients with del(17p) was high (92%), only few patients had isolated TP53 mutations, and the proportion of patients with single-hit was lower than expected (13–15, 17, 20, 24). This probably reflects that most TP53 aberrant patients in the original phase II study were included based on del(17p). Moreover, in the population-based validation cohort, a different cutoff for TP53 mutation VAF was used. With the LOD for TP53 mutations in the population-based cohort being considerably higher (5–20%) compared to that applied to the phase II study (0.2% VAF), a different distribution of patients with single- and multi-hit TP53 is seen. In addition, patients in population-based cohorts are routinely screened for del(17p) by FISH, while testing for TP53 mutations is inconsistent, despite clear clinical recommendations (9–11), which may have led to selection bias in the validation cohort. Data for the validation cohort were collected retrospectively, and more TP53 aberrations could have been identified if all pretreatment samples had been available for deep sequencing. The threshold for del(17p) by FISH is markedly higher (10% of 200 cells analyzed) than that of TP53 mutations by deep NGS (0.2% VAF), thus no data on outcome for patients with minor del(17p) subclones by FISH are available. Multiple del(17p) and biallelic del(17p) are rare, thus FISH may still be considered sufficient for detection of del(17p) with a cutoff above 10% affected cells (20, 43). By deep NGS, we could identify numerous minor TP53 mutations below 1% VAF (146 or 66%) enriched in patients previously treated with CIT, consistent with previous studies (22, 42). As DNA was extracted from peripheral blood mononuclear cells, we are unable to discern whether such minor mutations exist in CLL or non-CLL cells. Like our study, others have not found TP53 mutations enriched in elderly patients with CLL (15).
Current clinical guidelines for TP53 mutational analysis in CLL recommend reporting of TP53 mutations down to 5% VAF with a caveat that the significance of mutations below 10% VAF is unknown (10). To investigate a clinically meaningful VAF cutoff, we gradually excluded low burden TP53 mutations for the current study. By excluding minor mutations below VAF of 1%, a similar predictive performance for OS, PFS and TTP was demonstrated, while excluding low burden mutations below 2% VAF resulted in an inferior performance to predict PFS and TTP. Collectively, VAF cutoff of 1% for TP53 mutations was a clinically meaningful threshold for patients treated with ibrutinib, similar to findings from previous studies on CIT (17, 22, 24, 42). Ongoing efforts to improve inter-laboratory reproducibility of high-sensitivity DNA assays for TP53 are needed before a lower LOD can be implemented in clinical practice (44).
In summary, the number of TP53 aberrations assessed by the combination of deep NGS at 1% LOD and FISH correlates with OS and is an independent prognostic factor for PFS and TTP in patients with CLL on single-agent ibrutinib. Patients with multi-hit TP53 aberrations may thus be prioritized for clinical trials. Whether novel therapies including combination targeted therapy or chimeric antigen receptor T-cell therapy benefit this patient population remains unknown (45–49). Patients with single-hit TP53, who mainly had isolated del(17p), can achieve durable responses to ibrutinib monotherapy. An assessment of TP53 aberrations by using both FISH and deep NGS should be performed in all CLL patients considered for treatment with ibrutinib.
Supplementary Material
Translational relevance.
Testing for TP53 aberrations is recommended prior to treatment of patients with chronic lymphocytic leukemia (CLL) as TP53 confers resistance to chemoimmunotherapy. While TP53 aberrant patients receive targeted treatments, TP53 aberration remains a predictor of poor outcome. Deletion of chromosome 17p [del(17p)] and TP53 mutations mostly cooccur, while each lesion may be found as the sole TP53 aberration – especially when using deep next-generation sequencing. Although currently considered equal prognostic markers, patients treated with single-agent ibrutinib carrying only a single TP53 hit [del(17p) or a TP53 mutation] demonstrate excellent progression-free and overall survival on ibrutinib compared to those with multiple TP53 hits. Thus, testing for both del(17p) by FISH and TP53 mutations by deep next-generation sequencing should be performed to improve risk stratification of and facilitate clinical trials testing novel treatment approaches in multi-hit TP53 CLL.
ACKNOWLEDGEMENTS
We thank patients included in this study. We also thank the treating physicians in the US and Denmark. Inhye Ahn acknowledges support from the American Society of Hematology Scholar Award.
Disclosure of Potential Conflicts of Interest:
AW received research funding from Pharmacyclics, Acerta Pharma, Merck, Verastem, Nurix, and Genmab. CUN received support, consultancy fees and/or travel grants from Abbvie, Gilead, Janssen, Roche, CSL Behring, Acerta, Genmab, Sunesis and Astra Zeneca outside this work. The phase II study (NCT01500733) was, in part, supported by Pharmacyclics LLC, an AbbVie company, who have read and commented on the manuscript.
Financial support:
CB was supported by the Danish Cancer Society. XT, AW, and IEA are supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute. CUN received research funding from the Danish Cancer Society and the Novo Nordisk Foundation (grant NNF16OC0019302) for this trial.
REFERENCES
- 1.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donehower LA, Soussi T, Korkut A, Liu Y, Schultz A, Cardenas M, et al. Integrated Analysis of TP53 Gene and Pathway Alterations in The Cancer Genome Atlas. Cell reports. 2019;28:3010. [DOI] [PubMed] [Google Scholar]
- 3.Stilgenbauer S, Schnaiter A, Paschka P, Zenz T, Rossi M, Dohner K, et al. Gene mutations and treatment outcome in chronic lymphocytic leukemia: results from the CLL8 trial. Blood. 2014;123:3247–54. [DOI] [PubMed] [Google Scholar]
- 4.Bahlo J, Kutsch N, Bauer K, Bergmann MA, Byrd J, Chaffee KG, et al. An international prognostic index for patients with chronic lymphocytic leukaemia (CLL-IPI): a meta-analysis of individual patient data. Lancet Oncol. 2016;17:779–90. [DOI] [PubMed] [Google Scholar]
- 5.Burger JA. Treatment of Chronic Lymphocytic Leukemia. N Engl J Med. 2020;383:460–73. [DOI] [PubMed] [Google Scholar]
- 6.da Cunha-Bang C, Geisler CH, Enggaard L, Poulsen CB, de Nully Brown P, Frederiksen H, et al. The Danish National Chronic Lymphocytic Leukemia Registry. Clin Epidemiol. 2016;8:561–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–6. [DOI] [PubMed] [Google Scholar]
- 8.Zenz T, Gribben JG, Hallek M, Dohner H, Keating MJ, Stilgenbauer S. Risk categories and refractory CLL in the era of chemoimmunotherapy. Blood. 2012;119:4101–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Dohner H, et al. iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood. 2018;131:2745–60. [DOI] [PubMed] [Google Scholar]
- 10.Malcikova J, Tausch E, Rossi D, Sutton LA, Soussi T, Zenz T, et al. ERIC recommendations for TP53 mutation analysis in chronic lymphocytic leukemia-update on methodological approaches and results interpretation. Leukemia. 2018;32:1070–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eichhorst B, Robak T, Montserrat E, Ghia P, Niemann CU, Kater AP, et al. Chronic lymphocytic leukaemia: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2020. [DOI] [PubMed] [Google Scholar]
- 12.Knudson AG Jr. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68:820–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malcikova J, Smardova J, Rocnova L, Tichy B, Kuglik P, Vranova V, et al. Monoallelic and biallelic inactivation of TP53 gene in chronic lymphocytic leukemia: selection, impact on survival, and response to DNA damage. Blood. 2009;114:5307–14. [DOI] [PubMed] [Google Scholar]
- 14.Zenz T, Eichhorst B, Busch R, Denzel T, Habe S, Winkler D, et al. TP53 Mutation and Survival in Chronic Lymphocytic Leukemia. J Clin Oncol. 2010;28:4473–9. [DOI] [PubMed] [Google Scholar]
- 15.Stengel A, Kern W, Haferlach T, Meggendorfer M, Fasan A, Haferlach C. The impact of TP53 mutations and TP53 deletions on survival varies between AML, ALL, MDS and CLL: an analysis of 3307 cases. Leukemia. 2017;31:705–11. [DOI] [PubMed] [Google Scholar]
- 16.Campo E, Cymbalista F, Ghia P, Jager U, Pospisilova S, Rosenquist R, et al. TP53 aberrations in chronic lymphocytic leukemia: an overview of the clinical implications of improved diagnostics. Haematologica. 2018;103:1956–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nadeu F, Delgado J, Royo C, Baumann T, Stankovic T, Pinyol M, et al. Clinical impact of clonal and subclonal TP53, SF3B1, BIRC3, NOTCH1, and ATM mutations in chronic lymphocytic leukemia. Blood. 2016;127:2122–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernard E, Nannya Y, Hasserjian RP, Devlin SM, Tuechler H, Medina-Martinez JS, et al. Implications of TP53 allelic state for genome stability, clinical presentation and outcomes in myelodysplastic syndromes. Nat Med. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DiNardo CD, Tiong IS, Quaglieri A, MacRaild S, Loghavi S, Brown FC, et al. Molecular patterns of response and treatment failure after frontline venetoclax combinations in older patients with AML. Blood. 2020;135:791–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu L, Kim HT, Kasar S, Benien P, Du W, Hoang K, et al. Survival of Del17p CLL Depends on Genomic Complexity and Somatic Mutation. Clin Cancer Res. 2017;23:735–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blakemore SJ, Clifford R, Parker H, Antoniou P, Stec-Dziedzic E, Larrayoz M, et al. Clinical significance of TP53, BIRC3, ATM and MAPK-ERK genes in chronic lymphocytic leukaemia: data from the randomised UK LRF CLL4 trial. Leukemia. 2020;34:1760–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brieghel C, Kinalis S, Yde CW, Schmidt AY, Jonson L, Andersen MA, et al. Deep targeted sequencing of TP53 in chronic lymphocytic leukemia: clinical impact at diagnosis and at time of treatment. Haematologica. 2019;104:789–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rossi D, Cerri M, Deambrogi C, Sozzi E, Cresta S, Rasi S, et al. The prognostic value of TP53 mutations in chronic lymphocytic leukemia is independent of Del17p13: implications for overall survival and chemorefractoriness. Clin Cancer Res. 2009;15:995–1004. [DOI] [PubMed] [Google Scholar]
- 24.Rossi D, Khiabanian H, Spina V, Ciardullo C, Bruscaggin A, Fama R, et al. Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia. Blood. 2014;123:2139–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tam CS, Shanafelt TD, Wierda WG, Abruzzo LV, Van Dyke DL, O’Brien S, et al. De novo deletion 17p13.1 chronic lymphocytic leukemia shows significant clinical heterogeneity: the M. D. Anderson and Mayo Clinic experience. Blood. 2009;114:957–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farooqui MZ, Valdez J, Martyr S, Aue G, Saba N, Niemann CU, et al. Ibrutinib for previously untreated and relapsed or refractory chronic lymphocytic leukaemia with TP53 aberrations: a phase 2, single-arm trial. Lancet Oncol. 2015;16:169–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Byrd JC, Hillmen P, O’Brien S, Barrientos JC, Reddy NM, Coutre S, et al. Long-term follow-up of the RESONATE phase 3 trial of ibrutinib vs ofatumumab. Blood. 2019;133:2031–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Brien S, Furman RR, Coutre S, Flinn IW, Burger JA, Blum K, et al. Single-Agent Ibrutinib in Treatment-Naive and Relapsed/Refractory Chronic Lymphocytic Leukemia: A 5-Year Experience. Blood. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahn IE, Tian X, Wiestner A. Ibrutinib for Chronic Lymphocytic Leukemia with TP53 Alterations. N Engl J Med. 2020;383:498–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahn IE, Underbayev C, Albitar A, Herman SE, Tian X, Maric I, et al. Clonal evolution leading to ibrutinib resistance in chronic lymphocytic leukemia. Blood. 2017;129:1469–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woyach JA, Furman RR, Liu TM, Ozer HG, Zapatka M, Ruppert AS, et al. Resistance mechanisms for the Bruton’s tyrosine kinase inhibitor ibrutinib. N Engl J Med. 2014;370:2286–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahn IE, Farooqui MZH, Tian X, Valdez J, Sun C, Soto S, et al. Depth and durability of response to ibrutinib in CLL: 5-year follow-up of a phase 2 study. Blood. 2018;131:2357–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aarup K, Rotbain EC, Enggaard L, Pedersen RS, Bergmann OJ, Thomsen RH, et al. Real-world outcomes for 205 patients with chronic lymphocytic leukemia treated with ibrutinib. Eur J Haematol. 2020;105:646–54. [DOI] [PubMed] [Google Scholar]
- 34.Firth D Bias Reduction of Maximum Likelihood Estimates. Biometrika. 1993;80:27–38. [Google Scholar]
- 35.R Core Team. R: A language and environment for statistical computing. 2020[cited2021 02-20]; Available from: https://www.R-project.org/
- 36.O’Brien SM, Jaglowski S, Byrd JC, Bannerji R, Blum KA, Fox CP, et al. Prognostic Factors for Complete Response to Ibrutinib in Patients With Chronic Lymphocytic Leukemia: A Pooled Analysis of 2 Clinical Trials. JAMA oncology. 2018;4:712–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soumerai JD, Ni A, Darif M, Londhe A, Xing G, Mun Y, et al. Prognostic risk score for patients with relapsed or refractory chronic lymphocytic leukaemia treated with targeted therapies or chemoimmunotherapy: a retrospective, pooled cohort study with external validations. The Lancet Haematology. 2019;6:e366–e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahn IE, Tian X, Ipe D, Cheng M, Albitar M, Tsao LC, et al. Prediction of Outcome in Patients With Chronic Lymphocytic Leukemia Treated With Ibrutinib: Development and Validation of a Four-Factor Prognostic Model. J Clin Oncol. 2020:Jco2000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landau DA, Sun C, Rosebrock D, Herman SEM, Fein J, Sivina M, et al. The evolutionary landscape of chronic lymphocytic leukemia treated with ibrutinib targeted therapy. Nat Commun. 2017;8:2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malcikova J, Stano-Kozubik K, Tichy B, Kantorova B, Pavlova S, Tom N, et al. Detailed analysis of therapy-driven clonal evolution of TP53 mutations in chronic lymphocytic leukemia. Leukemia. 2015;29:877–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leeksma AC, Baliakas P, Moysiadis T, Puiggros A, Plevova K, van der Kevie-Kersemaekers AM, et al. Genomic arrays identify high-risk chronic lymphocytic leukemia with genomic complexity: a multi-center study. Haematologica. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.European Research Initiave on CLL: Projects. 2020[cited2020-04-22]; Available from:
- 45.Jain N, Keating M, Thompson P, Ferrajoli A, Burger J, Borthakur G, et al. Ibrutinib and Venetoclax for First-Line Treatment of CLL. N Engl J Med. 2019;380:2095–103. [DOI] [PubMed] [Google Scholar]
- 46.Hillmen P, Rawstron AC, Brock K, Munoz-Vicente S, Yates FJ, Bishop R, et al. Ibrutinib Plus Venetoclax in Relapsed/Refractory Chronic Lymphocytic Leukemia: The CLARITY Study . J Clin Oncol 2019;37:2722–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turtle CJ, Hay KA, Hanafi LA, Li D, Cherian S, Chen X, et al. Durable Molecular Remissions in Chronic Lymphocytic Leukemia Treated With CD19-Specific Chimeric Antigen Receptor-Modified T Cells After Failure of Ibrutinib. J Clin Oncol. 2017;35:3010–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med. 2011;365:725–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rogers KA, Huang Y, Ruppert AS, Abruzzo LV, Andersen BL, Awan FT, et al. Phase II Study of Combination Obinutuzumab, Ibrutinib, and Venetoclax in Treatment-Naïve and Relapsed or Refractory Chronic Lymphocytic Leukemia. J Clin Oncol. 2020:Jco2000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


