Abstract
Background
Circular RNAs (circRNAs) had been identified as a non‐coding RNA associated with many types of cancer in recent years. However, the involvement of hsa_circ_0008274 in lung adenocarcinoma (LUAD) has not been explored. The aim of our research is to explore the biological mechanism and function of hsa_circ_0008274 in LUAD.
Methods
The hsa_circ_0008274, miR‐578, and high mobility group AT‐Hook 2 (HMGA2) mRNA expression levels were detected via qRT‐PCR. Cell Counting Kit‐8 (CCK‐8) Transwell assay and wound healing assay were performed to measure the cell proliferation, invasion, and migration ability. Luciferase reporter and Western blotting experiments were performed to characterize the competing endogenous RNA (ceRNA) mechanism of hsa_circ_0008274.
Results
Our findings determined that the expression of hsa_circ_0008274 in LUAD was significantly decreased. Cell experiments showed that overexpressed hsa_circ_0008274 could reduce the proliferation and invasion ability of LUAD cells. Moreover, miRNA‐578 could identify as a miRNA sponge of hsa_circ_0008274. Overexpressed hsa_circ_0008274 reduced the proliferation and invasion of LUAD cells caused by miR‐578 mimics. Increasing the expression of miR‐578 can aggravate the proliferation and invasion of LUAD cells and block the inhibition of proliferation and invasion of LUAD cells mediated by overexpressed hsa_circ_0008274. Subsequent data indicate that HMGA2 of the tumor‐promoting gene is the target gene of miR‐578. The upregulation of HMGA2 partially reversed the tumor inhibitory effect of LUAD cells induced by overexpressed hsa_circ_0008274 or miR‐578 mimics.
Conclusions
In summary, our data show that the overexpression of hsa_circ_0008274 repressed the proliferation and invasion of LUAD through downregulating miR‐578 and activating HMGA2.
Keywords: HMGA2, hsa_circ_0008274, lung adenocarcinoma, miR‐578
Overexpression of hsa_circ_0008274 repressed the proliferation and invasion of LUAD through downregulating miR‐578 and activating HMGA2.
INTRODUCTION
Non‐small cell lung cancer (NSCLC) accounts for 85% of all lung cancer incidence, and their 5‐year survival rate is <20%, of which lung adenocarcinoma (LUAD) has leapt to the top of the NSCLC.1, 2 Although recent studies have explored various molecular mechanisms of the pathogenesis of LUAD, our understanding of the emergence and development of LUAD is still far from complete. Therefore, the study of the biological mechanism of LUAD is of great significance for the development of effective and promising potential therapeutic targets for LUAD.
Circular RNA (circRNAs) is a novel type of non‐coding RNA, which has attracted more and more attention because of its ability of regulation and management in cancers.3, 4 At present, circRNA have been considered as an important regulator of gene expression.5, 6 The sponge of microRNA (miRNA) is one of the important mechanisms of circRNAs.7 MiRNA is an earlier subtype of non‐coding RNA, which regulates gene expression after transcription by interacting with 3′‐UTR of mRNAs.8 MiRNA binds to messenger RNAs (mRNAs) with complementary sequences, resulting in mRNA degradation and translation limitations.8 Hence, through competitive binding with miRNAs, circRNAs exerts the role of competing endogenous RNA and releases the inhibitory function of miRNAs on mRNA. CircRNA/miRNA/mRNA axis is an important mechanism of gene expression regulation.
It has been reported that many kinds of circRNAs were abnormally high or low expression in LUAD, which played a key role in the occurrence and development of LUAD through different mechanisms.9, 10, 11 However, little is known about the biological role of hsa_circ_0008274 in LUAD. The purpose of this study is to detect the expression and function of hsa_circ_0008274 in LUAD and to explore its potential regulatory mechanism.
MATERIALS AND METHODS
Clinical samples
From May to September of 2020, 57 specimens of lung cancer tissues from patients with LUAD and matched adjacent normal tissues were collected from the Affiliated Hospital of Qingdao University. All participants provided a signed informed consent form, and this study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the Ethics Committee of The Affiliated Hospital of Qingdao University (QYFYKY 2020‐09‐03‐2).
Cell culture and transfection
The LUAD cell lines A549, CALU‐3, SPC‐A‐1, and GLC‐82, as well as normal human bronchial epithelial (HBE) cell line were purchased from the Cell Bank of Chinese Academy of Sciences (Shanghai, China), which were cultured in 37°C and 5% CO2. Small interfering RNAs (siRNAs) were used to knock down hsa_circ_0008274 expression and si‐normal control (si‐NC) was used as a control, which were purchased from Sangon Biotech (Shanghai, China). The overexpression transcript of (UDP‐Glucose Glycoprotein Glucosyltransferase 2) UGGT2 was inserted into pLVX vector to construct the overexpressed hsa_circ_0021087 vector. The miR‐578 mimics and inhibitors were synthesized by GenePharma Company (Shanghai, China). The CDS region of HMGA2 encoding full‐length protein was subcloned into pcDNA3.1 vector to construct HMGA2 expression vector. The cells were transfected with liposome 3000 transfection reagent (Invitgen, Thermo Fisher Science).
Verification of circular structure
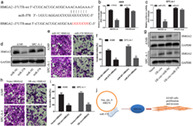
The circular structure of hsa_circ_0008274 was verified by convergent primers and primers, and the linear transcripts and circular transcripts of UGGT2 were amplified. In addition, the circular junction was sequenced to confirm the hsa_circ_0008274 sequence. At the same time, ribonuclease R (RNase R) was used to verify the expression of linear and circular UGGT2 in LUAD cells.
RNA extraction and RT‐qPCR assay
TRIzol reagent (Invitgen) was applied to extract total RNA and reversing transcribed into cDNA by RR047A kit (Beijing, Takara). Glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) was used as the internal control of hsa_circ_0008274 and UGGT2, and U6 was used as the internal reference of miR‐578
Cell counting kit‐8 assay
A549 and SPC‐A‐1 were inoculated in 96‐well plates and cultured overnight. LUAD cells were transfected and cultured for 2 days, treated with cell counting kit‐8 (CCK‐8) solution of 10 mL/well (Dojindo, Tokyo, Japan), and incubated for 2 hours. In the end, the absorbance was measured at 450 nm.
Colony formation assay
LUAD cells were inoculated in 6‐well plates and cultured for ~14 days. The colonies were fixed, stained, and counted by inverted microscope.
Cell invasion assay
The invasive potential of LUAD was detected by 24‐well Transwell chamber. LUAD cells were inoculated into the upper cavity. A total of 10% fetal bovine serum (FBS) was contained in the lower compartment culture medium, but not in the upper compartment culture medium. After 24 hours of culture, the LUAD cells on the lower surface were fixed with 70% ethanol and stained with 0.1% crystal violet, and the cells above were measured by inverted microscope.
Wound‐healing assay
The transfected LUAD cells were inoculated into a 6‐well plate until 80% of the subfusion. After scraping and culturing for 2 days. The images were collected at 0 and 48 hours, respectively, and the relative width was measured.
Luciferase reporter assay
The hsa_circ_0008274 of cDNA fragment containing wild‐type or mutant miR‐578 binding site was amplified and cloned into pmirGLO vector. The reporter vector with NC mimics or miR‐578 mimics was co‐transfected into A549 and SPC‐A‐1 cells. After 48 hours of culture, the luciferase activity was detected.
Western blot assay
The total protein was extracted and transferred to polyvinylidene fluoride (PVDF) membrane. After sealing with 5% skim milk, we incubated the first antibody and the second antibody. The main antibodies used in this research are HMGA2 and β‐actin (Univ, Shanghai, China).
Statistical analysis
All data were performed and analyzed via SPSS software (version 23.0) and GraphPad Prism software (version 5.0), the statistically index was limited as p < 0.05. Differences in two groups were assessed as average ± SD, and χ2 test was used to assess the differences of categoric variables.
RESULTS
Identification hsa_circ_0008274 expression in LUAD cells and tissues
Liang et al. have demonstrated that hsa_circ_0008274 was downregulated in LUAD.12 Before verifying the level of hsa_circ_0008274 expression, we first verified its circular structure after treatment with RNase R. The expression of linear UGGT2 was significantly lower than that of circular (Figure 1(a)), and we have verified it in cells and tissues. The hsa_circ_0008274 expression was downregulated in multiple LUAD cell lines (i.e., A549, Calu‐3, SPC‐A‐1, and GLC‐82) compared to HBE cell (Figure 1(b)). Consistently, the expression of hsa_circ_0008274 in 57 LUAD tissues was significantly lower than that in paracancerous tissues (Figure 1(c)). Subsequently, to identify the above candidate circRNA, the prognostic value of their host genes in LUAD samples was analyzed using a Kaplan–Meier plotter (Figure 1(d)). It was confirmed that the lower the expression of UGGT2 gene in LUAD, the worse the prognosis (p = 0.017).
FIGURE 1.
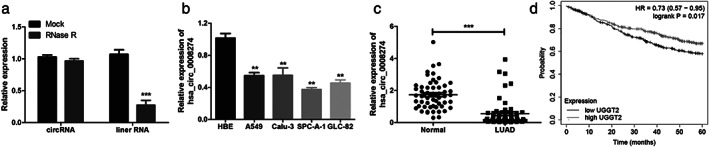
Hsa_circ_0008274 expression analysis in LUAD tissues and cell lines. (a) The expression levels of circular and linear UGGT2 in A549 and SPC‐A‐1 cells treated with RNase R. (b) qRT‐PCR analysis of hsa_circ_0008274 expression in A549, CALU3, SPC‐A‐1, GLC‐82, and human bronchial epithelial cells (HBE). (c) Hsa_circ_0008274 expression analysis of LUAD and paired normal lung tissues. (d) The OS rates of LUAD patients with high and low expression of mRNA UGGT2 was evaluated by Kaplan–Meier method. **p < 0.01, ***p < 0.001
Overexpression of hsa_circ_0008274 inhibited the proliferation and invasion of LUAD cells
In view of the low expression of hsa_circ_0008274 in LUAD, we studied the overexpression of hsa_circ_0008274 on the proliferation, colony formation, and invasion of LUAD cells. The overexpression vector hsa_circ_0008274 was transfected into A549 and SPC‐A‐1 cells to upregulate hsa_circ_0008274 expression (Figure 2(a)). The results of CCK‐8 assay showed that overexpressed hsa_circ_0008274 could significantly suppress the proliferation of A549 and SPC‐A‐1 cells (Figure 2(b)). Colony formation assay further confirmed the inhibitory effect of overexpression of hsa_circ_0008274 on cell proliferation, that is, fewer clones appeared when hsa_circ_0008274 was overexpressed (Figure 2(c)). The overexpression of hsa_circ_0008274 inhibited the invasive potential of A549 and SPC‐A‐1 cells (Figure 2(d)). Similarly, it also suppressed the migration ability of LUAD (Figure 2(e)). Therefore, these data show that the overexpression of hsa_circ_0008274 has an anti‐tumor effect in LUAD cells.
FIGURE 2.
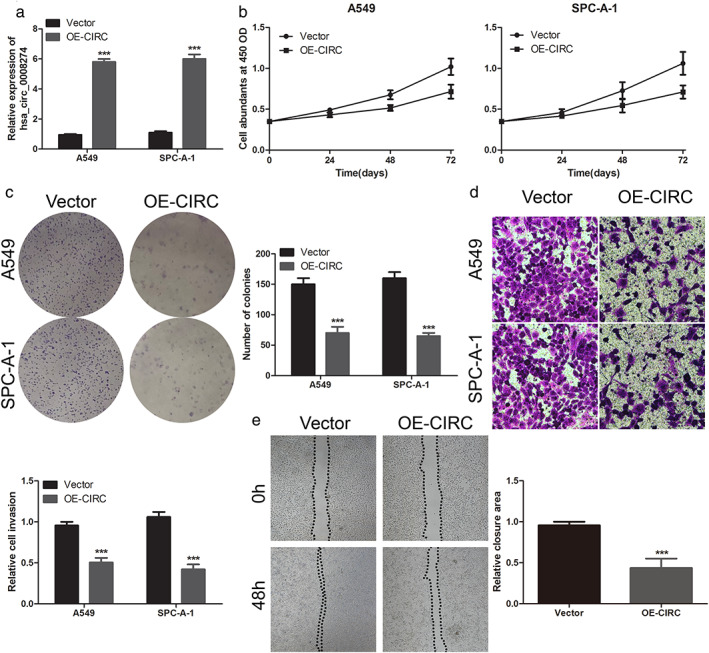
Overexpressed hsa_circ_0008274 inhibited the proliferation and invasion of LUAD cells. (a) After overexpressing, relative hsa_circ_0008274 expression was determined by RT‐qPCR. (b) CCK‐8 assay was assessed to detect cell proliferation. (c) Colony‐forming ability was applied by colony formation assay. (d),(e) Evaluation of invasion and metastasis potential were measured by Transwell invasion assay and wounding assay. **p < 0.01 and ***p < 0.001
MiR‐578 promoted the proliferation and invasion of LUAD cells
As shown in Figure 3(a), Liang et al., as we predicted by websites, had identified miR‐578 and miR‐633 could become possible targets for hsa_circ_0008274,12 the binding sites were shown in Additional Figure. Next, the miR‐578 and miR‐633 expression was determined in LUAD cell lines and HBE cell. The data showed that the expression of miR‐578 in LUAD cells was significantly higher than that in normal cells, but miR‐633 was not significant (Figure 3(b)). In addition, upregulation of miR‐578 significantly promoted the proliferation and invasion of LUAD cells, whereas inhibition of miR‐578 inhibited the proliferation (Figure 3(c)) and invasion (Figure 3(d)) of LUAD cells. Overexpressed miR‐578 and hsa_circ_0008274 could promote LUAD cell progression (Figure 3(e)–(g)). Collectively, these results demonstrated that miR‐578 acted as a tumor active miRNA in LUAD.
FIGURE 3.
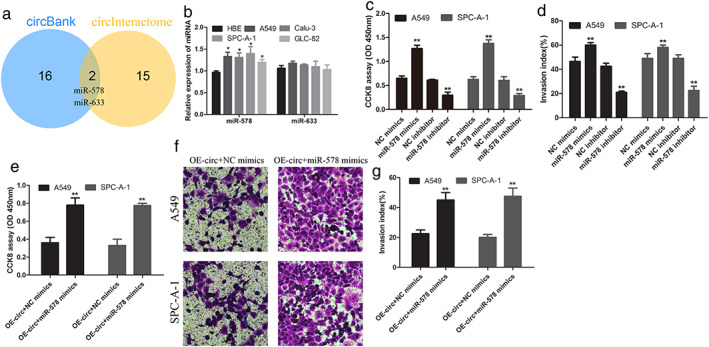
miR‐578 restrained the proliferation and invasion of LUAD cells. (a) MiR‐578 and miR‐633 are the intersection of circBank and circinteractome databases. (b) The expression of miR‐578 and miR‐633 in lung adenocarcinoma cell lines compared to normal lung cell, only miR‐578 expression was increased. After miR‐578 mimics and inhibitor, (c,e) cell proliferation was measured by the CCK‐8 assay, and (d) Transwell invasion assay was applied to examine invasive potential. (f),(g) After overexpressed circRNA with or without miR‐578, the invasion of cell was detected. *p < 0.05, **p < 0.01
Additional Figure: The binding sites between miR‐578 and hsa_circ_0008274 were shown. The red sequences represent its binding position.
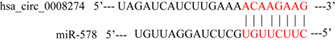 The downregulated hsa_circ_0008274 released miR‐578, and silences HMGA2 expression, which in turn affected the proliferation and invasion of LUAD
The downregulated hsa_circ_0008274 released miR‐578, and silences HMGA2 expression, which in turn affected the proliferation and invasion of LUAD
HMGA2 expression was decreased by miR‐578 in LUAD cells
To reveal the potential mechanism by which miR‐578 regulates the progression of LUAD cell lines, we tried to identify the target gene of miR‐578. HMGA2, as we predicted via bioinformatic software, had been identified as an oncogene in NSCLC13, 14 and became our research target. The 3′‐UTR of HMGA2 contains the possible binding site of miR‐578, and the HMGA2‐mut vector is constructed for follow‐up experiments (Figure 4(a)). Luciferase reporter assay displayed that miR‐578 mimics significantly decreased the luciferase activity of wild‐type HMGA2 but had no significant effect on mutant HMGA2 (Figure 4(b),(c)).
FIGURE 4.
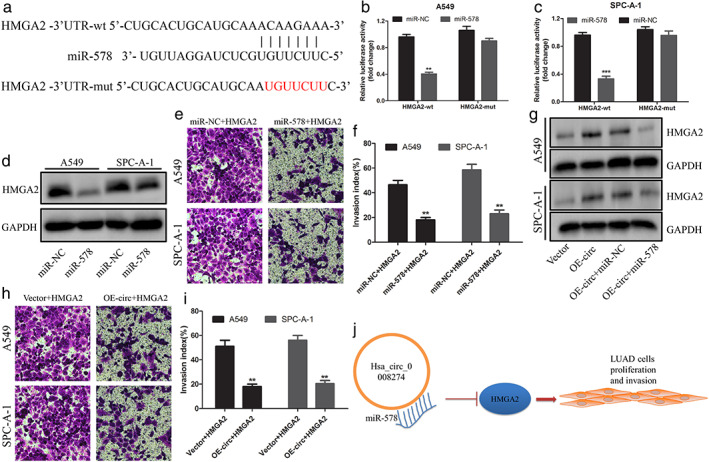
HMGA2 overexpression reversed hsa_circ_0008274 upregulated‐ or miR‐578 mimics‐mediated antitumor function. (a) The binding sequences between miR‐578 and HMGA2‐3′UTR were shown. The red sequences represent its mutant. (b),(c) The luciferase activity of HMGA2‐wt and HMGA2‐mut in A549 and SPC‐A‐1 cells was detected by dual‐luciferase reporter assay after transfection of miR‐578 or miR‐NC. (d) HMGA2 expression was determined by Western blot with or without miR‐578 mimics. (e),(f) Cell invasion was evaluated by the Transwell invasion. (g) The HMGA2 protein expression was measured after overexpressed circRNA cotransfected with or without miR‐578 mimics. (h),(i) Cell invasion was evaluated by the Transwell invasion. (j) The schematic diagram revealed that the downregulated hsa_circ_0008274 released miR‐578 and silences HMGA2 expression, which, in turn, affected the proliferation and invasion of LUAD
HMGA2 was regulated by overexpressed hsa_circ_0008274 or miR‐578 mimics‐mediated antitumor effect
To determine that HMGA2 is the target gene of hsa_circ_0008274/miR‐578 axis, we carried out the rescue experiment of HMGA2. We found that the expression of HMGA2 decreased significantly in LUAD cells transfected with miR‐578 mimics (Figure 4(d)), and the invasive ability of LUAD cells decreased when miR‐578 and HMGA2 were transfected into LUAD cells at the same time (Figure 4(e),(f)). As expected, overexpressed of hsa_circ_0008274 markedly enhanced the level of HMGA2 expression, but the expression level of HMGA2 was reversed when hsa_circ_0008274 overexpression and miR‐578 mimics were transfected into LUAD cells (Figure 4(g)). Similarly, overexpressed of HMGA2 remarkably reversed the hsa_circ_0008274 overexpression‐mediated inhibitory effect on LUAD cells (Figure 4(h),(i)). Overall, the above data confirmed that HMGA2 was the biological target of hsa_circ_0008274‐miR‐578 axis in regulating LUAD cells progression.
DISCUSSION
With the development of modern medical technology, the diagnosis rate of early lung cancer is getting higher and higher, especially LUAD, but it still needs to be screened by early biomarkers.15 The high stability of circRNA in tissues and plasma have attracted wide attention in recent years and have been used as a biological target for early diagnosis and treatment.16 However, there are still a large number of exact roles of circRNA in LUAD that have not been discovered. In‐depth revelation of the biological mechanism of circRNA is of great significance for the discovery of promising biomarkers in patients with LUAD. As previous studies have confirmed, the expression of hsa_circ_0008274 in LUAD shows a decreasing trend.17, 18, 19 Our study aimed to discover that hsa_circ_0008274 expression is downregulated in LUAD tissues, and cell lines compared with normal, in vitro experiments show that overexpression of hsa_circ_0008274 inhibits the proliferation and invasion of LUAD. Hence, our findings confirmed an oncogenic function of downregulated hsa_circ_0008274 in LUAD.
MiRNA sponging is also suitable for hsa_circ_0008274.7, 12 In our research, we identified miR‐578 as the miRNA target of hsa_circ_0008274. It had been reported that miR‐578 was a cancer‐related miRNA in hepatocellular carcinoma, breast cancer, and osteosarcoma.20, 21, 22 Our study reported that the expression of miR‐578 is upregulated in LUAD cells, and the miR‐578 mimics promoted the proliferation and invasion of LUAD, indicating that miR‐578 played a role in promoting cancer in LUAD. Interestingly, the overexpression of hsa_circ_0008274 can inhibit the proliferation and invasion of LUAD cells caused by the miR‐578 mimics. Therefore, our study confirmed that miR‐578 was the biological target of hsa_circ_0008274 in LUAD. The low expression of hsa_circ_0008274 in LUAD tumor tissue may be one of the reasons for the high expression of miR‐578, suggesting that hsa_circ_0008274‐miR‐578 axis plays a crucial role in the progression of LUAD.
More and more evidence shows that HMGA2 plays a valuable role in tumor development.14, 23, 24, 25 Silencing HMGA2 produced an effect on gastric cell proliferation and migration.26 These studies showed that the tumor‐promoting effect of HMGA2 has been widely reported. Additionally, the carcinogenic effect of HMGA2 had also been confirmed in NSCLC.27, 28 However, there are still unsolved problems about the imbalance of HMGA2 in LUAD. In our research, we identified HMGA2 as the hypothetical target gene of miR‐578. Our data showed that miR‐578 negatively regulated the expression of HMGA2 in LUAD cells. In addition, the expression of miR‐578 was increased in LUAD, and the inhibition of miR‐578 played an antitumor function. It is worth noting that the miR‐578 mimics significantly reversed the tumorigenicity of LUAD cells mediated by HMGA2, which confirmed that HMGA2 is the functional target of miR‐578 in LUAD.
In summary, this study demonstrated that the overexpression of hsa_circ_0008274 weakened the expression of miR‐578, which affected HMGA2, and, therefore, inhibited the proliferation and invasion of LUAD (Figure 4(j)). These results suggest that hsa_circ_0008274 played a significant role in the progression of LUAD. Moreover, our research showed that hsa_circ_0008274‐miR‐578‐HMGA2 was a novel axis in LUAD, which provided a new idea for understanding the molecular pathogenesis and therapeutic targets of LUAD.
CONFLICT OF INTEREST
The authors declare no conflict of interests.
ACKNOWLEDGMENTS
This study is supported by the Natural Science Foundation of Shandong Province (ZR2020MH234), Science and Technology Benefiting People in the west coast of Qingdao New District (2020‐57), and Postdoctoral Applied Research Project in Qingdao.
Wang M, Ma M, Yang Y, et al. Overexpression of hsa_circ_0008274 inhibited the progression of lung adenocarcinoma by regulating HMGA2 via sponging miR‐578. Thorac Cancer. 2021;12:2258–2264. 10.1111/1759-7714.14059
Maolong Wang and Minge Ma contributed equally to this work.
Contributor Information
Yong Sun, Email: sunyong@qdu.edu.cn.
Wenjie Jiao, Email: jiaowj@qduhospital.cn.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. [DOI] [PubMed] [Google Scholar]
- 2.Petrelli NJ, Winer EP, Brahmer J, Dubey S, Smith S, Thomas C, et al. Clinical cancer advances 2009: major research advances in cancer treatment, prevention, and screening–a report from the American Society of Clinical Oncology. J Clin Oncol. 2009;27:6052–69. [DOI] [PubMed] [Google Scholar]
- 3.Harper K, Mcdonnell E, Whitehouse A. CircRNAs: From anonymity to novel regulators of gene expression in cancer (Review). Int J Oncol. 2019. 10.3892/ijo.2019.4904. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Lu T, Wang Q, Liu J, Jiao W. Circular RNAs: crucial regulators in the human body (review). Oncol Rep. 2018;40:3119–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeng X, Lin W, Guo M, Zou Q. A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput Biol. 2017;13:e1005420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205–11. [DOI] [PubMed] [Google Scholar]
- 7.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8. [DOI] [PubMed] [Google Scholar]
- 8.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA‐binding sites in the 5' UTR as in the 3' UTR. Proc Natl Acad Sci U S A. 2007;104:9667–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei S, Zheng Y, Jiang Y, Li X, Geng J, Shen Y, et al. The circRNA circPTPRA suppresses epithelial‐mesenchymal transitioning and metastasis of NSCLC cells by sponging miR‐96‐5p. EBioMedicine. 2019;44:182–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu H, Han X, Ren J, Ren K, Li Z, Sun Z. Circular RNA HIPK3 induces cell proliferation and inhibits apoptosis in non‐small cell lung cancer through sponging miR‐149. Cancer Biol Ther. 2020;21:113–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Xu R, Zhang D, Lu T, Yu W, Wo Y, et al. Circ‐ZKSCAN1 regulates FAM83A expression and inactivates MAPK signaling by targeting miR‐330‐5p to promote non‐small cell lung cancer progression. Transl Lung Cancer Res. 2019;8:862–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang L, Zhang L, Zhang J, Bai S, Fu H. Identification of circRNA‐miRNA‐mRNA networks for exploring the fundamental mechanism in lung adenocarcinoma. Onco Targets Ther. 2020;13:2945–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arora S, Singh P, Rahmani AH, Almatroodi SA, Dohare R, Syed MA. Unravelling the Role of miR‐20b‐5p, CCNB1, HMGA2 and E2F7 in Development and Progression of Non‐Small Cell Lung Cancer (NSCLC). Biology. 2020;9(8):201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu L, Ma Y, Zhang H, Lu QJ, Yang L, Jiang GN, et al. HMGA2 regulates circular RNA ASPH to promote tumor growth in lung adenocarcinoma. Cell Death Dis. 2020;11:593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Millares L, Barreiro E, Cortes R, Martinez‐Romero A, Balcells C, Cascante M, et al. Tumor‐associated metabolic and inflammatory responses in early stage non‐small cell lung cancer: local patterns and prognostic significance. Lung Cancer. 2018;122:124–30. [DOI] [PubMed] [Google Scholar]
- 16.Yin Y, Long J, He Q, Li Y, Liao Y, He P, et al. Emerging roles of circRNA in formation and progression of cancer. J Cancer. 2019;10:5015–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen T, Yang Z, Liu C, Wang L, Yang J, Chen L, et al. Circ_0078767 suppresses non‐small‐cell lung cancer by protecting RASSF1A expression via sponging miR‐330‐3p. Cell Prolif. 2019;52:e12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao J, Li L, Wang Q, Han H, Zhan Q, Xu M. CircRNA expression profile in early‐stage lung adenocarcinoma patients. Cell Physiol Biochem. 2017;44:2138–46. [DOI] [PubMed] [Google Scholar]
- 19.Ding X, Zhang S, Li X, Feng C, Huang Q, Wang S, et al. Profiling expression of coding genes, long noncoding RNA, and circular RNA in lung adenocarcinoma by ribosomal RNA‐depleted RNA sequencing. FEBS Open Bio. 2018;8:544–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu A, Li Y, Kong M, Zhu B, Liu R, Bao F, et al. Upregulated hsa_circ_0005785 facilitates cell growth and metastasis of hepatocellular carcinoma through the miR‐578/APRIL axis. Front Oncol. 2020;10:1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Z, Wang F, Xiong Y, Wang N, Gu Y, Qiu X. CircZFR functions as a sponge of miR‐578 to promote breast cancer progression by regulating HIF1A expression. Cancer Cell Int. 2020;20:400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji X, Shan L, Shen P, He M. Circular RNA circ_001621 promotes osteosarcoma cells proliferation and migration by sponging miR‐578 and regulating VEGF expression. Cell Death Dis. 2020;11:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Wang J, Wu J. Emerging roles for HMGA2 in colorectal cancer. Transl Oncol. 2020;14:100894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ouchi K, Miyachi M, Yagyu S, Kikuchi K, Kuwahara Y, Tsuchiya K, et al. Oncogenic role of HMGA2 in fusion‐negative rhabdomyosarcoma cells. Cancer Cell Int. 2020;20:192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S, Mo Q, Wang X. Oncological role of HMGA2 (review). Int J Oncol. 2019;55:775–88. [DOI] [PubMed] [Google Scholar]
- 26.Liu Z, Lu Y, Jiang Q, Yang Y, Dang C, Sun R. miR‐491 inhibits BGC‐823 cell migration via targeting HMGA2. Int J Biol Markers. 2019;34:364–72. [DOI] [PubMed] [Google Scholar]
- 27.Wang B, Pan LY, Kang N, Shen XY. PP4R1 interacts with HMGA2 to promote non‐small‐cell lung cancer migration and metastasis via activating MAPK/ERK‐induced epithelial‐mesenchymal transition. Mol Carcinog. 2020;59:467–77. [DOI] [PubMed] [Google Scholar]
- 28.Zeng Z, Zhao G, Rao C, Hua G, Yang M, Miao X. Knockdown of lncRNA ZFAS1‐suppressed non–small cell lung cancer progression via targeting the miR‐150‐5p/HMGA2 signaling. J Cell Biochem. 2020;121(8‐9):3814–3824. 10.1002/jcb.29542. [DOI] [PubMed] [Google Scholar]


