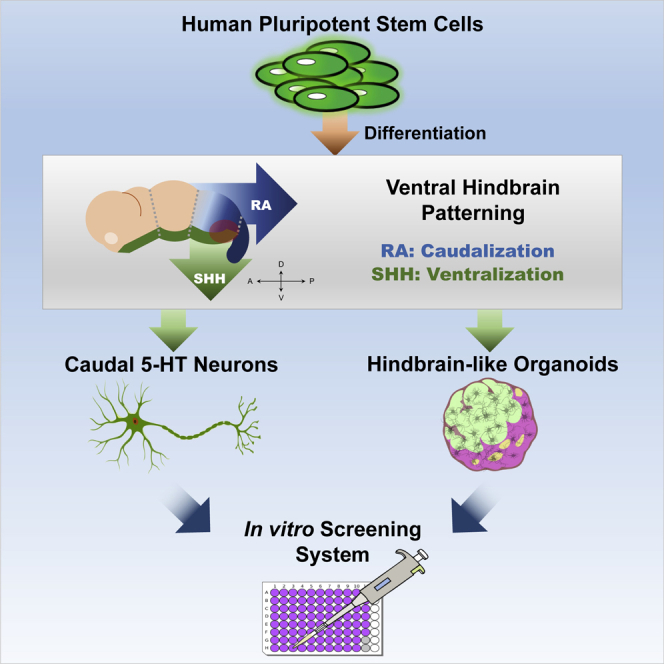
Summary
Serotonin (5-HT) neurons, the major components of the raphe nuclei, arise from ventral hindbrain progenitors. Based on anatomical location and axonal projection, 5-HT neurons are coarsely divided into rostral and caudal groups. Here, we propose a novel strategy to generate hindbrain 5-HT neurons from human pluripotent stem cells (hPSCs), which involves the formation of ventral-type neural progenitor cells and stimulation of the hindbrain 5-HT neural development. A caudalizing agent, retinoid acid, was used to direct the cells into the hindbrain cell fate. Approximately 30%–40% of hPSCs successfully developed into 5-HT-expressing neurons using our protocol, with the majority acquiring a caudal rhombomere identity (r5–8). We further modified our monolayer differentiation system to generate 5-HT neuron-enriched hindbrain-like organoids. We also suggest downstream applications of our 5-HT monolayer and organoid cultures to study neuronal response to gut microbiota. Our methodology could become a powerful tool for future studies related to 5-HT neurotransmission.
Keywords: hindbrain, human pluripotent stem cells, neuronal development, organoids, serotonin neurons
Graphical abstract
Highlights
-
•
Activation of SHH and RA signaling induces 5-HT neuronal fate from hPSCs
-
•
The generated 5-HT neurons have caudal hindbrain characteristics
-
•
Hindbrain-like organoids may form from hPSCs by activation of SHH and RA signaling
-
•
5-HT neurons in monolayer and organoid culture can be used as a screening platform
In this article, Shim and colleagues show that the generation of caudal hindbrain-type 5-HT neurons as well as 5-HT-neuron-enriched hindbrain-like organoids can be achieved through the modulation of SHH and RA signaling in differentiating hPSCs. The authors further propose that these 5-HT neurons in a monolayer and organoid culture system could be utilized as a screening platform for small molecules that trigger the release of 5-HT.
Introduction
Serotonin (5-hydroxytryptamine; 5-HT) is a neurotransmitter expressed in both the peripheral and the central nervous systems (CNS). 5-HT neurons comprise a large portion of nine raphe nuclei (B1–9) along the rostrocaudal axis of the brain stem. The rostral clusters (B5–9) project their axons mainly into the forebrain, including the cortex, striatum, hippocampus, and hypothalamus, and contribute to the regulation of mood, circadian rhythm, appetite, and cognition. Caudal clusters of raphe nuclei (B1–4) innervate the cerebellum and spinal cord, where they play important roles in homeostasis, involuntary movement, pain control, and motor activity (Alonso et al., 2013). Several psychiatric drugs, such as citalopram, fluoxetine, and paroxetine, function by inhibiting the 5-HT reuptake system, showing the importance of 5-HT in human psychology (Hiemke and Härtter, 2000). In the brain, 5-HT neurons constitute a large proportion of the brain-stem raphe nuclei that localize in the basal plate of the pons and medulla. While the cell bodies are restricted in the raphe nuclei, the axons of 5-HT neurons innervate nearly all parts of the CNS, including the forebrain and spinal cord (Alonso et al., 2013). 5-HT neurons are generated from the ventral progenitor domain, which is positive for NKX2.2 and adjacent to the floor plate. This occurs under the effect of Sonic hedgehog (SHH), a ventralizing signaling molecule released from the notochord (Pattyn et al., 2003). The 5-HT neurons are developed in several clusters called rhombomeres (r1–r3 and r5–r8) located at the caudal side of the isthmus organizer along the rostrocaudal axis (Alonso et al., 2013).
The ability of human pluripotent stem cells (hPSCs) to multiply and differentiate into all three germ layers has made them powerful tools in regenerative medicine, as well as neuroscience. Previous efforts to generate neurons from hPSCs efficiently by inhibiting SMAD signaling, using a combination of small molecules, has become a fundamental differentiation strategy in human neuroscience (Chambers et al., 2009). This protocol was further developed by Kriks et al. (2011) to add regional and subtype specificity in differentiating neurons by adding certain signaling molecules at calculated time points before inducing neuronal maturation (Kriks et al., 2011). These achievements made it possible to create a human-based platform for disease modeling and transplantation studies (Burbulla et al., 2017; Chung et al., 2016; Schweitzer et al., 2020).
As in the midbrain dopamine neuron differentiation paradigm (Kriks et al., 2011), we patterned the cells into a ventral hindbrain fate and induced their differentiation into 5-HT neurons. Both dopamine and 5-HT neurons differentiate from the ventral part of the neural tube and are separated by the isthmus organizer. Therefore, they are likely to share similar patterning molecules. Retinoic acid (RA) is an acidic derivative of vitamin A. Retinaldehyde dehydrogenase 2 (Raldh2) catalyzes RA synthesis in the mesoderm, and RA diffuses into the neuroectoderm. In the hindbrain, RA is known to induce caudalization in a concentration-dependent manner by directly regulating the expression of the HOX gene family (Okada et al., 2004). The absence of RA in Raldh2-deficient mice caused severe defects in hindbrain segmentation, whereby the area of the region expressing the r5–r8-specific genes was reduced (Vitobello et al., 2011). In this study, we selected RA as a caudalizing agent to direct ventral-type neural progenitor cells (NPCs) into a hindbrain cell fate, due to the prominent caudalizing activity of RA (Niederreither et al., 2000). It has been previously reported that inducing ventralization in hPSC-derived hindbrain NPCs resulted in the development of 5-HT neurons of the r3 subtype (Lu et al., 2016). However, in the present study, we tried a distinct approach to direct 5-HT neuron differentiation by early ventral acquisition together with caudalization. Here, we suggest a novel differentiation protocol for hPSC-derived 5-HT neurons that results in the generation of 5-HT neurons with a caudal subtype.
Stem cell differentiation studies have primarily relied on two-dimensional culture systems that enable control over the uniformity of the cell population. Recent developments in organoid culture systems complement monolayer culture by providing self-assembly properties and a higher level of cell-to-cell interaction that exist in endogenous organs (Fatehullah et al., 2016). In neuroscience, the application of organoid technology from hPSCs was pioneered through the establishment of cerebral-type organoids (Lancaster et al., 2013). Organoids with midbrain region specification have also been developed by adopting a differentiation protocol developed for monolayer midbrain neurons into an organoid culture system (Jo et al., 2016; Qian et al., 2016). Here, we propose that by modifying our two-dimensional 5-HT neuron differentiation protocol, we can generate three-dimensional hindbrain-5-HT-neuron organoids. In addition, we demonstrate the practical application of our monolayer and organoid cultures.
Results
Establishment of a 5-HT neuron-differentiation protocol from hPSCs
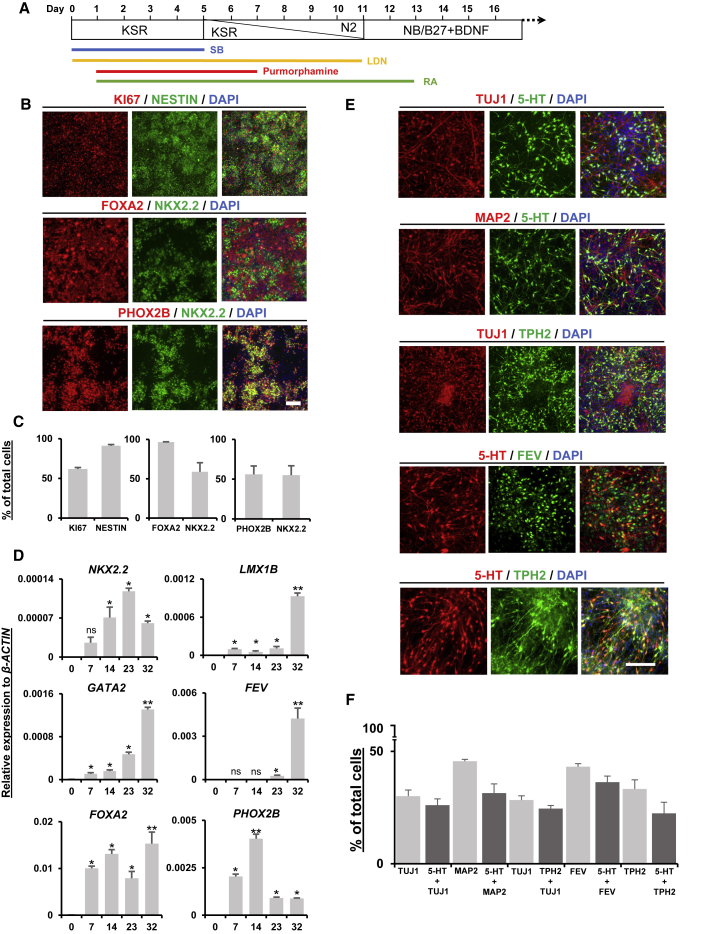
Based on the previous success in generating midbrain-type dopamine neurons, we hypothesized that the generation of hindbrain-type 5-HT neurons would involve anteroposterior patterning of the ventral precursors. In the midbrain dopamine-neuron protocol, short-time activation of SHH signaling during the early days of differentiation was sufficient to induce a ventral identity. In this study, LDN139189 and SB431542 (LSB) were used to inhibit SMAD signaling and induce neuroepithelial identity, while purmorphamine was utilized to activate smoothened in the SHH signaling pathway (Briscoe and Ericson, 1999; Chambers et al., 2009; Kriks et al., 2011). RA was used as a strong caudalizing factor to induce the hindbrain cell fate, replacing the activation of WNT signaling, which is necessary for midbrain patterning. RA treatment was administered at various concentrations, lengths, and timing of exposure to optimize the production of 5-HT neurons (Figures S1B–S1D). This step was necessary because there is only a short time window for regional determination during the early days of neural induction from hPSCs (Kriks et al., 2011; Metzis et al., 2018). The expression of 5-HT was analyzed by immunostaining 30 days after differentiation, and the percentage of 5-HT-positive (+) cells was calculated. The results showed that the highest percentage of 5-HT neurons was induced by the highest concentration (2 μM) of RA treatment administered at the early time points (day 1 or 3) with the longest exposure duration (13 days; Figures 1A and S1B–S1D). After 11 days of differentiation, the expression of the ventral regional markers, NKX2.2 and FOXA2, was upregulated together with that of the NPC marker, NESTIN, and proliferation marker, KI67, which suggests a ventral NPC stage (Figures 1B and 1C). Since 5-HT and motor neurons arise from shared progenitors, the expression of PHOX2B, a transcription factor exclusively expressed by visceral motor neurons, was assessed (Jacob et al., 2007). NKX2.2 expression was less co-labeled with FOXA2 (NKX2.2+/FOXA2+) on day 11 than with PHOX2B (NKX2.2+/PHOX2B+). However, this tendency was reversed following the day of differentiation. On differentiation day 19, the number of NKX2.2+/FOXA2+ cells increased, while that of NKX2.2+/PHOX2B+ cells decreased. After 30 days of differentiation, as the progenitors matured into neurons, NKX2.2 expression was downregulated and remained only weakly expressed in some of the neurons (Figures S1E and S1F). This pattern was supported by quantitative real-time PCR (qPCR) assay, which suggested a continuous increase in the transcription factors FOXA2, GATA2, LMX1B, and FEV, while NKX2.2 and PHOX2B expression peaked on days 23 and 14, respectively (Figure 1D). NKX2.2 has been suggested to activate GATA2 downstream of SHH signaling (Craven et al., 2004). GATA2, in turn, activates LMX1B and FEV, which are both known regulators of 5-HT neuron maturation and subtype specification (Craven et al., 2004; Krueger and Deneris, 2008). FEV is the human homolog of rat Pet-1, and it is known to be exclusively expressed in hindbrain 5-HT neurons and not in any other neuronal types (Hendricks et al., 1999). The maturation of 5-HT neurons was assessed on differentiation day 30 by the expression of the neuronal markers, MAP2 and TUJ1. We found that 30%–40% of the DAPI-stained cells were expressing MAP2, TUJ1, and FEV, together with 5-HT. Tryptophan hydroxylase 2 (TPH2), the rate-limiting enzyme of 5-HT synthesis in 5-HT neurons (Gutknecht et al., 2008), was also found to be co-expressed with 5-HT (Figures 1E and 1F). We also observed a portion of TH+ (14.8%), GABA+ (8.4%), glutamate+ (11.7%) neurons; unspecified proliferating KI67+ cells (12.1%); and E-cadherin+ non-neural flat-shaped cells (4.3%) (Figure S1G), indicating that heterogeneity still existed in our neuronal populations. However, we did not find any expression of OCT3/4, indicating the absence of undifferentiated hPSCs (data not shown). In addition, we checked the expression of cholinergic (CHAT) and motor (HB9) neuron markers, but those were absent from our populations (data not shown). To observe further maturation of the 5-HT neurons, culture was prolonged and MAP2+ massive neurite extensions were observed in the 60-day-old neurons, together with the expression of 5-HT and TPH2 (Figure S1H). To test the consistency of our protocol in other cell lines, we utilized two human induced pluripotent stem cell (hiPSC) lines, L2131 and L2122, which have been shown in previous studies to differentiate into neurons (Chung et al., 2016; Seibler et al., 2011). 5-HT neurons successfully differentiated from hPSCs with comparable efficiency using both of these cell lines (Figure S1I). Altogether, our findings suggest that our protocol can consistently induce hPSCs into hindbrain-type 5-HT neurons across multiple hPSC lines. The resulting neurons are also available for long-term culture, providing simplicity and flexibility for various downstream applications.
Figure 1.
Protocol for the differentiation of 5-HT neurons from hPSCs
(A) Schematic representation of 5-HT neuron differentiation from hPSCs. KSR, knockout serum replacement medium.
(B) Representative images of immunocytochemical analysis of proliferating NPC (KI67 and NESTIN), ventral (NKX2.2 and FOXA2), and visceral motor neuron (PHOX2B) markers on differentiation day 11.
(C) Quantification of immunostaining images in (B) as the percentage of positive cells with the indicated antibodies among the DAPI-stained cells per microscopic field.
(D) Time-dependent expression of GATA2, FEV, LMX1B, NKX2.2, FOXA2, and PHOX2B measured by qPCR. The y axis indicates the relative expression of mRNA normalized to β-ACTIN expression. Significantly different from day 0 at ∗p < 0.05 and ∗∗p < 0.0001; ns, not significant; n = 4 independent experiments.
(E) Immunocytochemistry analysis of 5-HT, TPH2, TUJ1, FEV, and MAP2 on differentiation day 30.
(F) The percentage of single-positive or double-positive cells in (E) with the indicated antibodies among the DAPI-stained cells per microscopic field. All data are presented as the mean ± SEM (C, D, and F). Scale bar, 200 μm. See also Figure S1.
Regional characterization of 5-HT neurons
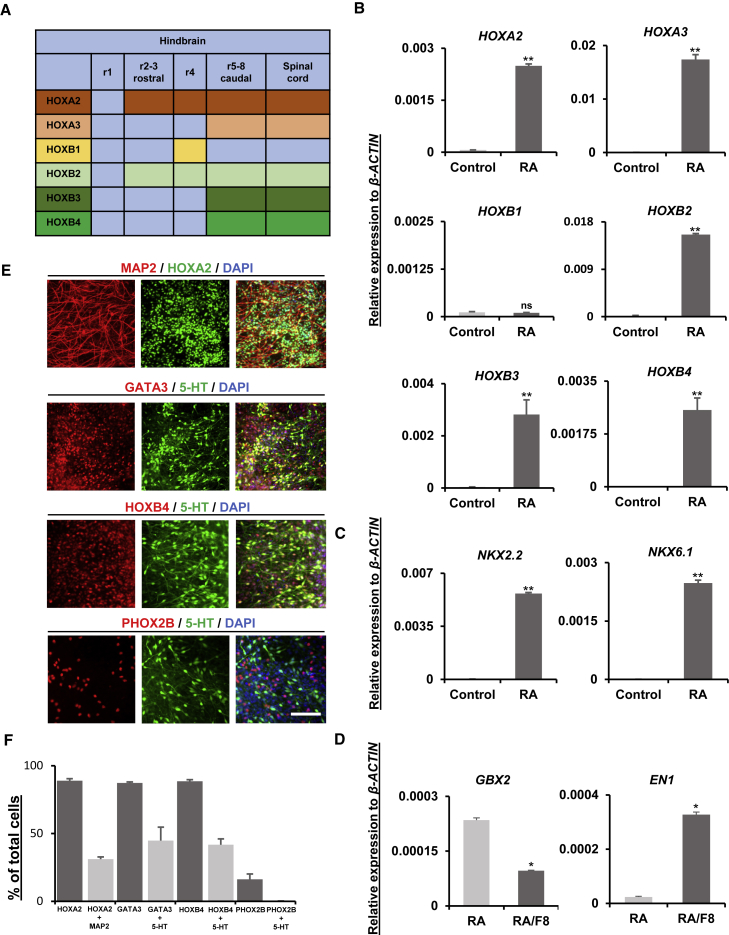
The majority of 5-HT neurons are located in the brainstem raphe nuclei; these are roughly divided into two clusters to form the rostral and caudal raphe nuclei. 5-HT neurons of the rostral raphe innervate the midbrain and various forebrain regions, including the basal ganglia, hippocampus, and hypothalamus. 5-HT neurons that reside in the caudal raphe project their axons to the caudal hindbrain and spinal cord. According to animal model developmental studies, the rostral cluster of the raphe nuclei develops from the r1–r3 regions, while the r5–r8 regions give rise to caudal raphe nuclei (Alonso et al., 2013). A schematic diagram of the expression of HOX family members in r1–r8 and the spinal cord, based on a previous study with E9.5 mouse embryos, is depicted in Figure 2A (Hunt et al., 1991). To understand which rhombomere our 5-HT neurons were derived from, mRNA samples were collected on differentiation day 30. Ventral-type neurons differentiated from human embryonic stem cells (hESCs), with LSB and purmorphamine and without any caudalizing factors, were harvested at an appropriate time point and used as controls (Kriks et al., 2011). High expression of HOXA2, HOXA3, HOXB2, HOXB3, and HOXB4 was observed by qPCR, while low expression was observed for HOXB1. This pattern closely resembled that observed in caudal rhombomeres r5–r8 (Figure 2B). The expression of the ventral markers, NKX2.2 and NKX6.1, was highly upregulated compared with non-patterned neurons differentiated with only dual SMAD signaling inhibition as the control (Figure 2C; Chambers et al., 2009). We also checked the expression of GBX2 and EN1 to confirm the fate choice of the 5-HT neurons. For comparison, an EN1-expressing population was generated with RA and FGF8 treatments (Ye et al., 1998). 5-HT neurons generated with our protocol showed significantly lower EN1 expression compared with FGF8-treated populations, further confirming that the majority of 5-HT neurons were of the caudal hindbrain type (Figure 2D). In addition, the expression of HOXA2 and HOXB4, as well as GATA3, a caudal hindbrain marker, was clarified at the individual cell level by immunostaining. The expression of PHOX2B was also observed in small portions of the cells, but they were not co-expressed with 5-HT neurons (Figures 2E and 2F).
Figure 2.
Regional characterization of 5-HT neurons
(A) Illustrative table of the regional markers along the rostrocaudal hindbrain axis.
(B) Relative mRNA expression of rhombomere-specific genes (HOXA2, HOXA3, HOXB1, HOXB2, HOXB3, and HOXB4), measured by qPCR on differentiation day 30 in neurons treated with LSB and purmorphamine, with or without 2 μM RA treatment.
(C and D) Validation of the ventral (NKX2.2 and NKX6.1) (C) and rostrocaudal hindbrain (GBX2 and EN1) (D) marker expression on differentiation day 30 with real-time PCR. The y axis indicates the relative expression of mRNA normalized to β-ACTIN expression. Significantly different from control values at ∗p < 0.05 and ∗∗p < 0.0001; ns, not significant; n = 3 independent experiments.
(E) Immunocytochemical analysis of the regional hindbrain markers (HOXA2, GATA3, HOXB4, and PHOX2B) with MAP2 and 5-HT.
(F) Percentage of single-positive or double-positive cells in (E) with the indicated antibodies among the DAPI-stained cells per microscopic field. All data are presented as the mean ± SEM (B–D and F). Scale bar, 200 μm. F8, FGF8.
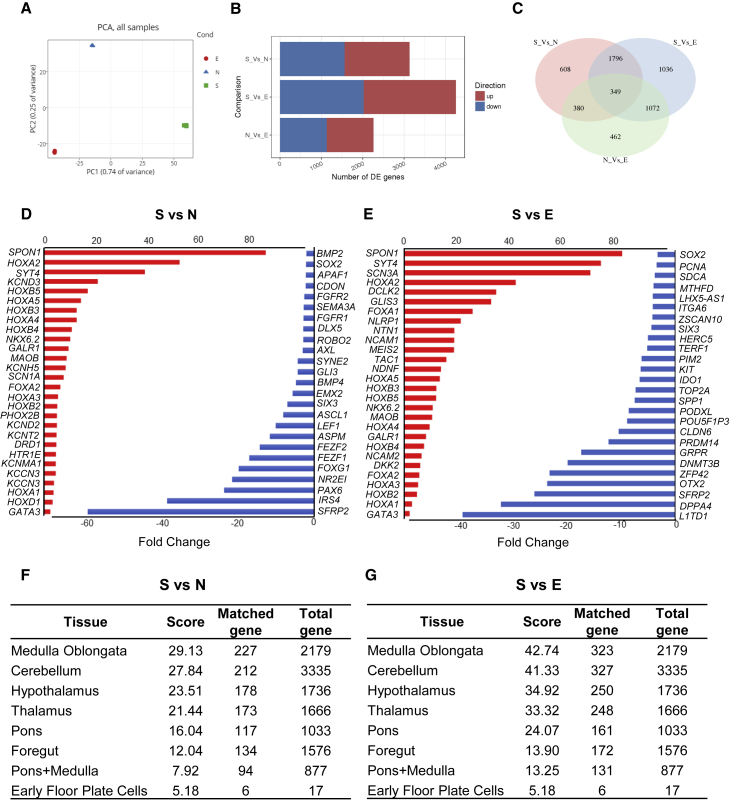
To further investigate the regional identity of the 5-HT neurons, we performed a transcriptome analysis by comparing the microarray profiles from undifferentiated hESCs (group E), non-patterned neurons (group N; differentiation day 25), and 5-HT neurons (group S; differentiation day 30; Figures 3A–3C). The microarray data showed high enrichment of the HOX gene family, GATA3, and ventral markers such as SPON1, FOXA2, and the NKX family in 5-HT neurons (Figures 3D and 3E). However, HOXA6, 7, and 10 and HOXB6, 7, 8, and 9, which are usually expressed by the spinal cord, were not detected. This suggests that the majority of hPSC-derived 5-HT neurons produced with our protocol were r5–r8-like 5-HT neurons. The enrichment pattern of the HOX gene family in this study was similar to that shown in the previous studies (Okaty et al., 2015; Zeisel et al., 2018). For a broader examination of the genes upregulated in 5-HT neuron populations, several statistical analyses were performed. The KEGG pathway analysis of genes upregulated in 5-HT neurons showed enrichment in several pathways (Figures S2A–S2D). We filtered the genes to keep the significantly differentially expressed genes in the S versus N or S versus E comparison. We applied K-means clustering to the expression data of these genes. Gene ontology enrichment of specific clusters, which included genes that were upregulated in 5-HT neurons, was performed (Figures S3A–S3D). The GeneAnalytics Tissues & Cell analysis (http://geneanalytics.genecards.org) was used to further characterize the 5-HT neurons (Fuchs et al., 2016). Genes upregulated in 5-HT neurons were compared against a database derived from various human tissues and cell types. The results revealed a relatively high matching score with the medulla oblongata and cerebellum, which confirmed the resemblance of our 5-HT neuron populations to these brain regions (Figures 3F and 3G). We also selected several representative genes from the upregulated gene list, including some cell-surface markers highly expressed in hindbrain, and checked for their expression in the Allen Human Brain Atlas (Allen Institute for Brain Science, 2010; Hawrylycz et al., 2012). We found that these genes were also highly expressed in the “raphe nuclei of the medulla,” which further supports the caudal hindbrain identification of our 5-HT neurons (Figures S3E and S3F).
Figure 3.
Transcriptome analysis of 5-HT neurons
(A) Principle-component analysis plot of nine independent samples for 5-HT neurons (S, three samples), non-patterned neurons (N, three samples), and undifferentiated hESCs (E, three samples).
(B) Bar plot summarizes the comparison of the numbers of significant differentially expressed genes between N and E with S.
(C) Venn diagram presents the overlap between the significantly differentially expressed genes.
(D and E) Selected lists of differentially expressed genes comparing S versus N (D) or S versus E (E). Red bars depict the genes upregulated in S (positive fold change), and blue bars indicate genes downregulated in S (negative fold change). All genes that are shown had an adjusted p < 0.05.
(F and G) GeneAnalytics Tissues & Cells analysis comparing upregulated gene lists of S versus N (F) or S versus E (G) with a database of genes expressed in specific human tissues and organs. DE, differentially expressed; PCA, principal-component analysis. See also Figures S2 and S3.
Generation of three-dimensional hindbrain-like 5-HT-organoids
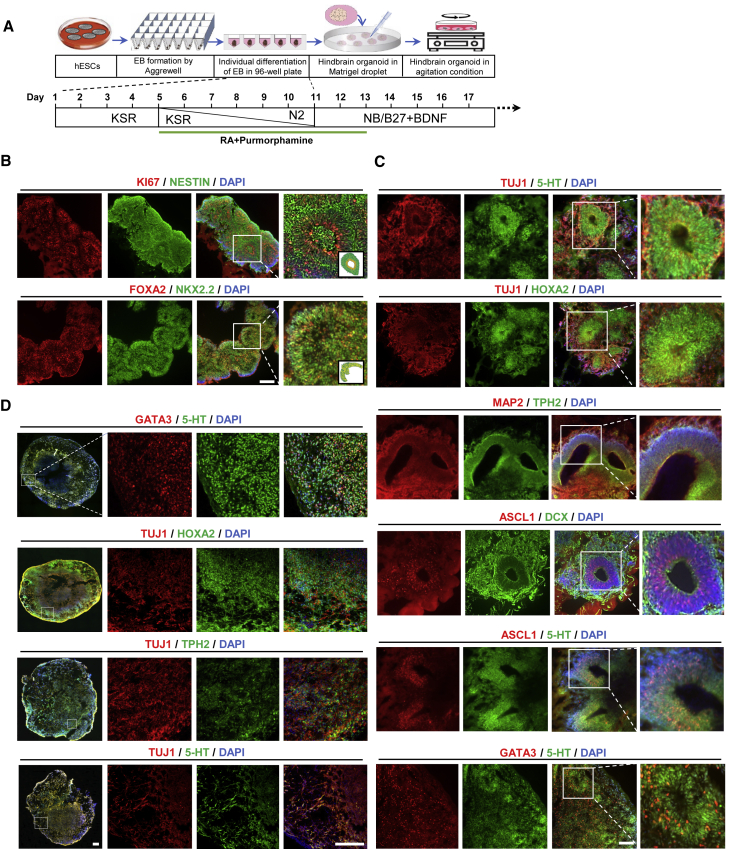
Similar to a previous region-specific-organoid study, we developed a strategy to generate hindbrain-like 5-HT organoids based on our novel monolayer protocol. Embryoid body (EB)-like structures were first induced by seeding the hPSCs into 24-well AggreWell plates (Figure 4A). As in the two-dimensional 5-HT neuron protocol, starting from day 1, differentiation was induced by growing EBs in medium supplemented with purmorphamine and RA (without LDN193189 and SB431542). The addition of RA was shown to induce the expression of HOXB4 while abolishing the anterior marker expression, OTX2, in the neural tube-like structures of mouse stem cell-derived EBs (Meinhardt et al., 2014). However, our early attempt failed to produce organoids with ideal budding and multilayered structures. Instead, a plentiful amount of neural fibers was found stretched out from the EBs, indicating a rapid neural-fate induction (Figure S4A). Among the added factors, RA has been shown to induce neural acquisition in mouse embryonic stem cells (Okada et al., 2004). Considering the rapid induction of neural fates as the reason behind the failure to produce ideal organoid shapes, we examined whether organoid formation could be optimized by modulating the timing of the RA treatment (Figure S4C). Interestingly, when the RA treatment was delayed to day 7, the EBs differentiated into nicely shaped organoids, with buds and layers, at the cost of a decreased yield of 5-HT neurons (Figures S4B and S4D–S4E). Based on this, days 5–13 were chosen as the optimal times for RA treatments that still produced proper organoid structures without substantially compensating 5-HT neuron yields (Figures S4D and S4E).
Figure 4.
Generation of three-dimensional hindbrain-like 5-HT-organoids
(A) Schematic representation of the hindbrain-like 5-HT-organoid generation process.
(B) Representative images of immunocytochemical analysis of proliferating NPC markers (KI67 and NESTIN) and floor-plate markers (NKX2.2 and FOXA2) on differentiation day 17 (three left columns). (Insets) Schematic of the structural characteristics in organoids, as shown in the enlarged image (right).
(C and D) Immunocytochemistry analysis of 5-HT, HOXA2, TPH2, TUJ1, ASCL1, DCX, GATA3, and MAP2 on differentiation day 30 (C) and of 5-HT, HOXA2, TPH2, TUJ1, and GATA3 on differentiation day 60 (D). Scale bars, 200 μm. See also Figure S4.
On day 17, EBs were cryosectioned and immunostained with ventral NPC-specific markers. The abundant presence of KI67+/NESTIN+ cells, as well as cell co-expression of NKX2.2 and FOXA2, suggested that the hPSC-derived EBs had been successfully induced into a ventral NPC stage. At this stage, the formation of a layered structure became apparent by the expression of KI67 in the inner layer, which was surrounded by radial-shaped cells positive for the NPC marker, NESTIN (Figure 4B). These data ensured that the organoid differentiation protocol followed a similar developmental paradigm with respect to our monolayer 5-HT neuron protocol. After 30 days of differentiation, when EBs had completely turned from spherical into an organoid-like shape, cryosection and immunostaining were again performed. As shown in Figure 4C, our organoid protocol generated 5-HT neurons, as indicated by co-expression of 5-HT or HOXA2 with TUJ1 in nearly all parts of the organoid. Co-expression of MAP2 with TPH2 confirmed the mature phenotype of 5-HT neurons in the organoids. At this stage, organoids showed a clear tubular structure that resembled the developing hindbrain (Gutzmann and Sive, 2010), which was positive for the 5-HT neuron progenitor marker, mammalian achaete-scute homolog-1 (ASCL1), and migrating cell markers, doublecortin (DCX) and 5-HT (Figure 4C). The expression of GATA3, a marker of the caudal hindbrain, was found only in a portion of 5-HT-expressing cells. Considering that the three-dimensional structure of organoids might prevent uniform exposure to RA, it was likely that mixed subtypes of 5-HT neurons are present in the 5-HT organoids (Figure 4C). We also checked for the expression of CHAT and HB9 in the organoids and, unlike the monolayer, a portion of CHAT+ and HB9+ cells was found (Figure S4F). However, we did not observe any TH+ cells in the organoid populations (data not shown). The expression of CHAT+ neurons was also found in the brain-stem organoids described in another study (Eura et al., 2020); however, the absence of TH+ and the abundance of 5-HT+ neurons as well as other hindbrain-specific markers in our organoid populations indicated a narrow patterning into the 5-HT-enriched hindbrain region. Another round of cryosection and immunostaining was performed on the hindbrain-like organoids after 60 days of culture. Bipolar-shaped neuron-like cells expressing 5-HT and GATA3 were observed in some of the organoids, together with TUJ1 and HOXA2. However, this pattern of expression was inconsistent across individual organoids. 5-HT neurons were enriched in some organoids, while the other organoids showed reduced expression of 5-HT neurons compared with day 30. In addition, the tubular structure could no longer be observed at this stage, suggesting a limitation in our protocol for long-term maintenance of hindbrain-like organoids without loss of 5-HT expression (Figure 4D). To show the replicability of our protocol, we performed hindbrain-like organoid differentiation with one of the hiPSC lines previously used in the monolayer culture (Figure S4G).
Although qPCR may not be ideal for the analysis of a heterogeneous population, such as an organoid, it is still necessary to examine the expression of region-specific and 5-HT neuron-specific markers in the majority of cells at the mRNA level. mRNA samples were isolated from 30-day-old organoids, and the gene expression was compared with that of organoids differentiated using a previously published protocol without any patterning (Lancaster et al., 2013). We found a significant upregulation of HOXA2 and hindbrain marker GBX2, while the forebrain-midbrain marker OTX2 was expressed at a very low level (Figure S4H). Moreover, the expression of the ventral marker NKX2.2, as well as the 5-HT neuron markers GATA2, LMX1B, and FEV, was significantly increased (Figure S4I). Altogether, this mRNA-level analysis confirmed that organoids produced by our newly established protocol showed not only the proper budding structure but also hindbrain-like marker expression and were composed mainly of 5-HT neurons.
Physiological analysis and the potential applications of 5-HT monolayer and organoid cultures
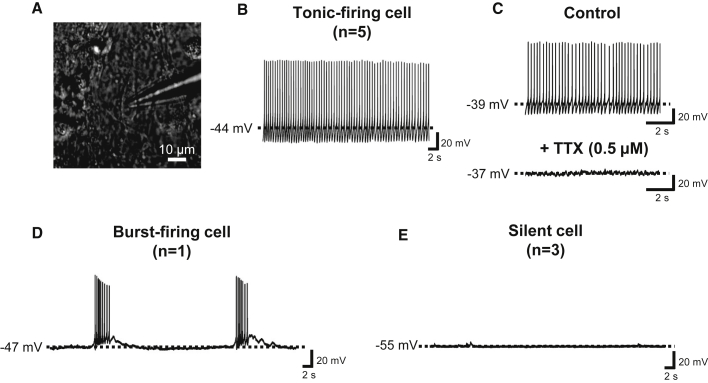
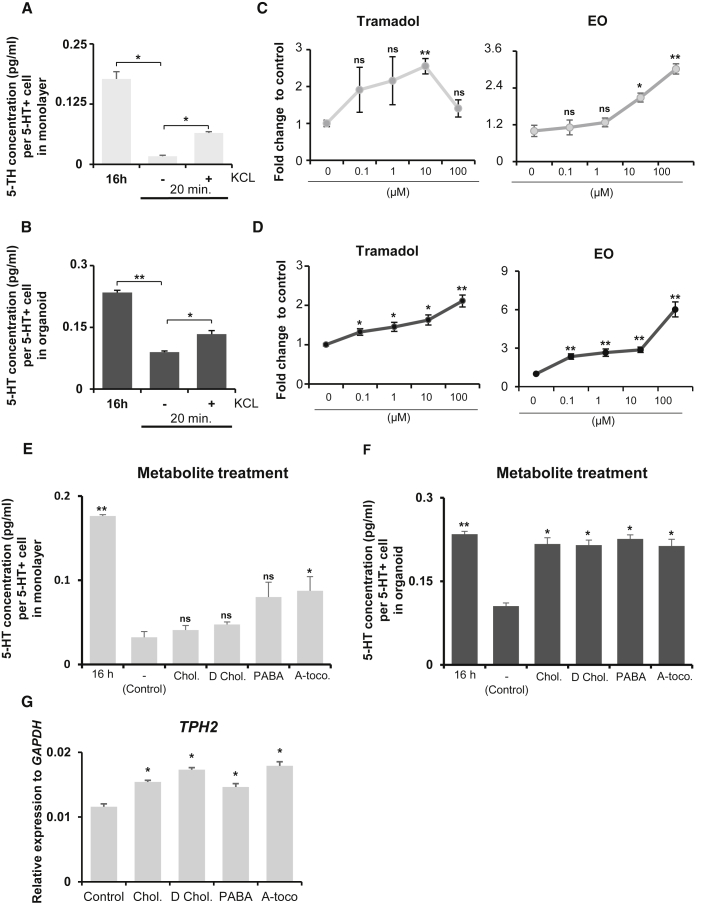
After establishing a novel method to generate caudal-type 5-HT neurons and hindbrain-like 5-HT organoids, we wanted to demonstrate the functionality of our populations. First, we performed an electrophysiological analysis on our neurons in the monolayer cultures. We targeted putative 5-HT neurons in culture (differentiation day 60) for whole-cell patch-clamp recordings (Figure 5A). We recorded nine cells in current-clamp modes, and found that five cells (55.5%) showed tonic-firing patterns with an average baseline membrane potential of −39.6 ± 1.1 mV (Figure 5B). In order to examine if Na+ is responsible for the action potential (AP) of these tonic-firing cells, we applied tetrodotoxin (TTX; 0.5 μM), a voltage-gated Na+ channel blocker. We found that the treatment of cells with TTX completely blocks the AP of tonic-firing cells (Figure 5C). These results suggest that the hPSCs have successfully differentiated to neurons that can fire APs mediated by the voltage-gated Na+ channels. We also noted that one of nine cells (11.1%) was a burst-firing cell (Figure 5D), and the remaining three cells (33.3%) did not fire APs (Figure 5E). The resting membrane potential of these silent cells was −55.0 ± 6.4 mV. Next, neurotransmitter release was triggered by KCl-induced depolarization to further clarify the functionality of our 5-HT neurons and organoids. The medium was collected on differentiation day 65 (monolayer) and day 85 (organoid) after 16 h (overnight) of incubation. In the same populations, fresh medium was incubated for 20 min, with and without 56 mM KCl treatment. 5-HT release into the medium was measured using an enzyme-linked immunosorbent assay, as presented in Figures 6A and 6B. Both the monolayer and the organoid cultures showed significant upregulation of 5-HT release upon KCl-evoked depolarization, indicating the functionality of the generated neurons and organoids. To evaluate their potential as human-derived models for drug screening as downstream pharmacological applications, we selected two drugs that worked on the 5-HT reuptake system, tramadol and escitalopram oxalate (EO), and separately tested them on both monolayer and organoid cultures. Tramadol is an analgesic opiate that regulates pain by acting on mu-opioid receptors, while EO is a selective 5-HT reuptake inhibitor. Due to its selective effect on the 5-HT system, EO is prescribed to treat major depressive disorder. Both tramadol and EO inhibit 5-HT reuptake into the presynaptic axon terminal and, thus, increase the level of extracellular 5-HT (Garnock-Jones and McCormack, 2010; Sansone and Sansone, 2009). We discovered that treatment with either tramadol or EO was able to boost 5-HT release in both the monolayer and the organoid cultures in a dose-dependent manner (Figures 6C and 6D). However, high concentrations of tramadol (100 μM) were toxic to the monolayer 5-HT neuron culture, as indicated by apparent cell death and decreased 5-HT levels.
Figure 5.
Electrophysiological analysis of 5-HT monolayer cultures
(A) Bright-field illumination of a cell targeted with a glass micropipette electrode for whole-cell patch-clamp recordings.
(B) Current-clamp recording trace of a tonic-firing cell (n = 5).
(C) Current-clamp recording traces of a tonic-firing cell before (top) and after (bottom) TTX application.
(D) Current-clamp recording trace of a burst-firing cell (n = 1).
(E) Current-clamp recording trace of a silent cell (n = 3). Dashed lines indicate baseline or resting membrane potential. n, the number of recorded cells.
Figure 6.
Applications of 5-HT monolayer and organoid cultures
(A and B) 5-HT released from 5-HT neuron monolayer (A; day 65) and organoid (B; day 85) cultures after overnight (16 h) and 20 min incubation with or without 56 mM KCl treatment.
(C and D) Fold change in 5-HT release in monolayer (C) and organoid (D) cultures after an hour treatment with tramadol or EO at various concentrations.
(E and F) 5-HT released from the monolayer (E; day 65) and organoid (F; day 85) cultures after overnight (16 h) or 1 h incubation with or without treatment with the following metabolites: cholate (78 μM), deoxycholate (25 μM), p-aminobenzoate (1 μM), or α-tocopherol (8 μM). The y axis indicates the 5-HT concentration per individual 5-HT neuron in 24 well dish (A and E) or in organoid (B and F).
(G) Relative expression of TPH2 in the 5-HT neuron monolayer culture (day 65) after 1 h incubation with or without metabolite treatments. The y axis indicates the relative expression of mRNA normalized to GAPDH expression. Significantly different from cultures after an hour incubation without KCl treatment (A and B) and corresponding control (C–G) values at ∗p < 0.05 and ∗∗p < 0.0001; ns, not significant. n = 3 independent experiments. All data are presented as the mean ± SEM. A-toco, α-tocopherol; Chol, cholate; D Chol, deoxycholate; PABA, p-aminobenzoate.
An interesting study by Yano et al. (2015) revealed that certain metabolite products of gut microbiota could trigger the upregulation of tryptophan hydroxylase 1 (Tph1) expression and the sequential 5-HT release from enterochromaffin cells in intestines (Yano et al., 2015). TPH1 works as the rate-limiting enzyme of peripheral 5-HT synthesis. Its isozyme, TPH2, shares an identical role in regulating 5-HT synthesis in the brain (Gutknecht et al., 2008). Therefore, we wondered if the same metabolites that upregulated Tph1 expression in enterochromaffin cells could also induce 5-HT release from hindbrain 5-HT neurons. We selected four metabolites that had been used in the previous study: cholate, deoxycholate, p-aminobenzoate, and α-tocopherol (Yano et al., 2015). The results showed that an hour of exposure to all tested metabolites induced the upregulation of 5-HT release in both monolayer and organoid cultures. Even though only α-tocopherol stimulation was significant in monolayer neurons (Figure 6E), all four metabolites successfully provoked significant 5-HT release in organoids (Figure 6F). Next, we investigated whether these metabolites triggered 5-HT release by increasing 5-HT synthesis. mRNA samples were collected 1 h after metabolite exposure, and the expression of TPH2 was quantified by qPCR. The results showed significant upregulation of TPH2 gene expression, supporting our hypothesis (Figure 6G). Taken together, these experiments showed that specific metabolites secreted by gut microbiota increased 5-HT synthesis by regulating TPH2 expression. At the same time, they demonstrated the immense potential of 5-HT monolayer culture and organoids generated with our newly established protocols as a human-based platform for drug screening and as a model to study human physiology.
Discussion
Considering the various subtypes of 5-HT neurons and their different functions, studies on 5-HT networks are required. In addition, most of our knowledge about 5-HT neurons comes from rodent-based studies, making the need for relevant human-based model platforms even more important.
Hindbrain 5-HT neurons are located close to the midbrain dopamine neurons, separated only by the isthmus organizer. Shifting the position of the isthmus along the anteroposterior axis increased the number of one neuronal subtype at the expense of another (Brodski et al., 2003), showing a close developmental profile between dopamine and 5-HT neurons. Here, we propose a novel strategy to generate hindbrain 5-HT neurons from hPSCs, which is mediated by the formation of ventralized NPCs. A strong caudalizing agent, RA, was used to drive the cells into a hindbrain cell fate. Interestingly, along with the differentiation, we observed the co-expression of NKX2.2 with PHOX2B, a motor neuron progenitor marker, particularly in the early days of differentiation (day 11). This was not surprising since motor and 5-HT neurons develop from shared progenitors that express Nkx2.2. As found in mouse development, motor neurons are born earlier from Phox2b+/Nkx2.2+ progenitors in the ventral hindbrain. However, as Foxa2 starts to be expressed in the ventralmost part of the Nkx2.2+ region, the expression of Phox2b is suppressed, and the Pet1-expressing 5-HT neurons are born (Jacob et al., 2007). This is in line with our data that show the downregulation of PHOX2B+/NKX2.2+ cells as the number of FOXA2+/NKX2.2+ cells increases.
In terms of efficiency, 30%–40% of the 5-HT neurons obtained from the monolayer cultures can be problematic for downstream applications, such as for clinical purposes. Therefore, it is necessary to develop a strategy to sort out the 5-HT-positive neurons created by our protocol. In a previously published midbrain study, the lineage-specific reporter line of NURR1-GFP was utilized for disease modeling studies (Riessland et al., 2019). Similarly, 5-HT neuron lineage-specific reporter lines, such as FEV-GFP, can be used in future applications. Alternatively, region- or type-specific cell surface markers can be used to sort 5-HT neurons. This strategy has also been applied to sort midbrain dopamine neurons using CORIN, a floor-plate-specific cell surface marker (Kikuchi et al., 2017). Several cell surface markers suggested by microarray data, such as SPON1, L1CAM, and NTN1, can be utilized for this kind of sorting.
In a previously published protocol, patterning was initiated by the formation of rostral hindbrain progenitors for 7 days, followed by another 2 weeks of ventralization step and, finally, neuronal differentiation that resulted in the generation of rostral-type 5-HT neurons (Lu et al., 2016). In this study, we generated 5-HT neurons based on the paradigm of ventralization and rostrocaudal axis determination that occur simultaneously in neuroepithelial cells. Moreover, the patterning steps in our protocol were executed in the first 13 days of differentiation, in line with previous studies showing that, for proper differentiation, the axial determination should be completed before neurogenesis is started (Kriks et al., 2011; Metzis et al., 2018). It is still unclear which strategy reflects the actual human brain development. Nevertheless, it is interesting that both strategies could successfully produce 5-HT neurons with distinct regional subtype identities. Here, we successfully generated 5-HT neurons with a gene expression profile that was similar to that of caudal rhombomeres (r5–r8).
Even though the protocol for cerebral and midbrain-type organoids has been established, to date, no hindbrain-type 5-HT-neuron organoids have been developed from hPSCs. Therefore, we modified our monolayer protocol into a three-dimensional culture to generate hindbrain-like 5-HT organoids. In this study, we established a protocol for the generation of 5-HT-neuron-enriched hindbrain-like organoids with developmental-specific structural characteristics. Nevertheless, there were still some limitations in our organoid culture system. Although we could observe enriched 5-HT expression overlapping with ASCL1 in neural tube-like structures, this observation may not be in line with a previous mouse study that showed that the precursors of 5-HT neurons migrate from the ASCL1+ region of the neural tube as they reach maturation (Pattyn et al., 2004). In addition, the level of variation in 5-HT expression found in individual organoids after prolonged culture (60 days) was high. One possible explanation is the cellular stress that has been suggested as a general phenomenon in organoid culture. Activation of the stress pathway in developing organoids impaired cell-subtype specifications, including those necessary for the proper migration and formation of multiple-layer structures (Bhaduri et al., 2020). We also observed a cavity in the center of 60-day-old organoids that might be caused by cell death. Another possible reason for the lack of multiple-layer structures is that, in the developing brain, patterning factors are released from specific organizers to affect nearby areas. However, in our protocol, patterning factors available in the medium affected the whole organoid area, resulting in the lack of certain cell types required for the migration and formation of multiple layers. Altogether, these observations suggest that further optimization is required to support the development and survival of 5-HT neurons in long-term organoid cultures.
Treatment with drugs that inhibit the 5-HT reuptake system resulted in higher 5-HT levels in the medium, indicating the great potential of 5-HT neurons and organoids generated with our protocol as a model for drug screening purposes. We further explored the application of 5-HT neurons to study the relationship between the brain and the gut microbiota. There is good evidence that a substance in the gut could somehow affect neurons in the CNS. Injection of α-synuclein fibrils into the mouse intestine led to the formation of endogenous α-synuclein fibrils in the brain (Kim et al., 2019). Moreover, previous studies supported that the existence of gut microbiota might affect the 5-HT turnover rate in the CNS and induce anxious behavior in mice (Diaz Heijtz et al., 2011).
Despite growing interest, our understanding of the mechanisms that link the gut-microbiota-brain axis is limited. Here, we demonstrated that four microbiota-derived metabolites that have been shown to induce 5-HT release from enterochromaffin cells (Yano et al., 2015) could also stimulate 5-HT neurons in both monolayer and organoid cultures. The level of increase was not always powerful, which makes sense, since microbiota metabolites are not psychostimulants. Moreover, powerful stimulation of 5-HT release would be toxic for the neurons themselves, and thus, a mild 5-HT release may be a model more equivalent to actual physiologic conditions. Even though the experimental setting was modest, the results were intriguing. Treatment with α-tocopherol has been shown to reduce depressive-like behaviors in an animal model (Manosso et al., 2013), and the level of α-tocopherol in the blood serum of patients with major depression was found to be lower than that in healthy controls (Maes et al., 2000). How α-tocopherol affects behavior and the involvement of 5-HT neurons in this phenomenon are not clear. We suggest that indigenous bacteria in the healthy gut may provide a continuous supply of useful metabolites for 5-HT neurons in the CNS. Metabolic products from the beneficial microbiota may pass from blood vessels into the brain, where they can affect 5-HT neurons. This is supported by the fact that the selected components used in this study have been reported to be capable of crossing the blood-brain barrier (Ferri et al., 2015).
In this study, we established a novel protocol to differentiate hPSCs into caudal-type 5-HT neurons, which we expanded further into a three-dimensional culture system for the generation of functional hindbrain-like 5-HT-organoids. These achievements will provide a great tool to improve future studies and to accelerate our understanding of human 5-HT neuron development and related diseases.
Experimental procedures
Detailed descriptions of experimental procedures can be found in the supplemental information.
Human pluripotent stem cell lines
The hPSCs used in this study were H9 hESC (WA09, XX, Wicell), L2122 (XX), and L2131 (XY) hiPSC lines. All lines were confirmed in previous studies (Chung et al., 2016; Seibler et al., 2011). hPSCs were maintained in Essential 8 medium (Gibco, Grand Island, NY) according to the manufacturer's instructions as described in detail in the supplemental experimental procedures. This study was reviewed by the SCH-IRB (protocol 201408-BM-024-01).
Summarized human 5-HT neuron differentiation
hPSCs were dissociated into single cells and seeded onto Matrigel (Corning, Corning, NY)-coated culture dishes. To initiate differentiation, the maintenance medium was replaced with knockout serum replacement medium in combination with N2 medium (Gibco). Medium was supplemented with 200 nM LDN193189 (STEMCELL Technologies, Vancouver, BC, Canada) and 10 μM SB431542 (Tocris, Ellisville, MO) and 2 μM each purmorphamine (Tocris) and RA (Sigma, St. Louis, MO). To support neuronal differentiation, the medium was switched at day 11 into NB/B27 medium (Gibco) supplemented with 20 ng/mL BDNF (Peprotech, London, UK). At day 20, the cells were passaged at a 1:2 ratio onto freshly prepared Matrigel-coated dishes.
Data and code availability
Transcriptome analysis was executed using the Affymetrix Whole Transcript expression array according to the manufacturer's protocol (GeneChip WT PLUS reagent kit; Thermo Fisher Scientific, Waltham, MA). Raw data of the microarray are accessible through GEO Series accession GEO: GSE167278.
Author contributions
P.V. and V.V. designed and performed the experiments, analyzed the data, and wrote the manuscript. L.P. performed the experiments, analyzed the data, and wrote the manuscript. Y.O., V.B.J., P.S., and J-W. Sohn conducted the experiments and analyzed the data. G.F. and D.Y. analyzed the microarray data. Y.K.L., J.K.Y., and J-w. Shim designed the concept, supervised the whole project, analyzed the data, and wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgments
This work was supported by grants NRF-2017M3A9B4062401, NRF-2017R1A2B4003018, NRF-2021R1A2C1005940, NRF-2016K1A4A3914725, and NRF-2019R1A5A8083404 funded by the National Research Foundation of Korea (NRF) of the Ministry of Science and ICT, Republic of Korea. The authors would like to thank Dr. Dimitri Krainc from Harvard Medical School, Boston, MA, USA, for providing two hiPSC lines (L2122 and L2131). G.F. is the incumbent David and Stacey Cynamon Research Fellow Chair in Genetics and Personalized Medicine.
Published: July 8, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.06.006.
Contributor Information
Yun Kyung Lee, Email: yunklee@sch.ac.kr.
Jeong Kyo Yoon, Email: jkyoon@sch.ac.kr.
Jae-won Shim, Email: shimj@sch.ac.kr.
Supplemental information
References
- Allen Institute for Brain Science . internet resource; 2010. Allen Human Brain Atlas. [Google Scholar]
- Alonso A., Merchán P., Sandoval J.E., Sánchez-Arrones L., Garcia-Cazorla A., Artuch R., Ferrán J.L., Martínez-De-La-Torre M., Puelles L. Development of the serotonergic cells in murine raphe nuclei and their relations with rhombomeric domains. Brain Struct. Funct. 2013;218:1229–1277. doi: 10.1007/s00429-012-0456-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaduri A., Andrews M.G., Mancia Leon W., Jung D., Shin D., Allen D., Jung D., Schmunk G., Haeussler M., Salma J. Cell stress in cortical organoids impairs molecular subtype specification. Nature. 2020;578:142–148. doi: 10.1038/s41586-020-1962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J., Ericson J. The specification of neuronal identity by graded sonic hedgehog signalling. Semin. Cell Dev. Biol. 1999;10:353–362. doi: 10.1006/scdb.1999.0295. [DOI] [PubMed] [Google Scholar]
- Brodski C., Weisenhorn D.M.V., Signore M., Sillaber I., Oesterheld M., Broccoli V., Acampora D., Simeone A., Wurst W. Location and size of dopaminergic and serotonergic cell populations are controlled by the position of the midbrain-hindbrain organizer. J. Neurosci. 2003;23:4199–4207. doi: 10.1523/JNEUROSCI.23-10-04199.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbulla L.F., Song P., Mazzulli J.R., Zampese E., Wong Y.C., Jeon S., Santos D.P., Blanz J., Obermaier C.D., Strojny C. Dopamine oxidation mediates mitochondrial and lysosomal dysfunction in Parkinson’s disease. Science. 2017;357:1255–1261. doi: 10.1126/science.aam9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S.Y., Kishinevsky S., Mazzulli J.R., Graziotto J., Mrejeru A., Mosharov E.V., Puspita L., Valiulahi P., Sulzer D., Milner T.A. Parkin and PINK1 patient iPSC-derived midbrain dopamine neurons exhibit mitochondrial dysfunction and α-synuclein accumulation. Stem Cell Reports. 2016;7:664–677. doi: 10.1016/j.stemcr.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven S.E., Lim K.C., Ye W., Engel J.D., de Sauvage F., Rosental A. Gata2 specifies serotonergic neurons downstream of sonic hedgehog. Development. 2004;131:1165–1173. doi: 10.1242/dev.01024. [DOI] [PubMed] [Google Scholar]
- Diaz Heijtz R., Wang S., Anuar F., Qian Y., Björkholm B., Samuelsson A., Hibberd M.L., Forssberg H., Pettersson S. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. U S A. 2011;108:3047–3052. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eura N., Matsui T.K., Luginbühl J., Matsubayashi M., Nanaura H., Shiota T., Kinugawa K., Iguchi N., Kiriyama T., Zheng C. Brainstem organoids from human pluripotent stem cells. Front. Neurosci. 2020;14:538. doi: 10.3389/fnins.2020.00538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatehullah A., Tan S.H., Barker N. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 2016;18:246–254. doi: 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- Ferri P., Angelino D., Gennari L., Benedetti S., Ambrogini P., Del Grande P., Ninfali P. Enhancement of flavonoid ability to cross the blood-brain barrier of rats by co-administration with α-tocopherol. Food Funct. 2015;6:394–400. doi: 10.1039/c4fo00817k. [DOI] [PubMed] [Google Scholar]
- Fuchs S.B.A., Lieder I., Stelzer G., Mazor Y., Buzhor E., Kaplan S., Bogoch Y., Plaschkes I., Shitrit A., Rappaport N. GeneAnalytics: an integrative gene set analysis tool for next generation sequencing, RNAseq and microarray data. OMICS. 2016;20:139–151. doi: 10.1089/omi.2015.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnock-Jones K.P., McCormack P.L. Escitalopram: a review of its use in the management of major depressive disorder in adults. CNS Drugs. 2010;24:769–796. doi: 10.2165/11204760-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Gutknecht L., Waider J., Kraft S., Kriegebaum C., Holtmann B., Reif A., Schmitt A., Lesch K.P. Deficiency of brain 5-HT synthesis but serotonergic neuron formation in Tph2 knockout mice. J. Neural Transm. 2008;115:1127–1132. doi: 10.1007/s00702-008-0096-6. [DOI] [PubMed] [Google Scholar]
- Gutzmann J.H., Sive H. Epithelial relaxation mediated by the myosin phosphatase regulator Mypt1 is required for brain ventricle lumenexpansion and hindbrain morphogenesis. Development. 2010;137:795–804. doi: 10.1242/dev.042705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz M.J., Lein E.S., Guillozet-Bongaarts A.L., Shen E.H., Ng L., Miller J.A., Van De Lagemaat L.N., Smith K.A., Ebbert A., Riley Z.L. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks T., Francis N., Fyodorov D., Deneris E.S. The ETS domain factor Pet-1 is an early and precise marker of central serotonin neurons and interacts with a conserved element in serotonergic genes. J. Neurosci. 1999;19:10348–10356. doi: 10.1523/JNEUROSCI.19-23-10348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiemke C., Härtter S. Pharmacokinetics of selective serotonin reuptake inhibitors. Pharmacol. Ther. 2000;85:11–28. doi: 10.1016/s0163-7258(99)00048-0. [DOI] [PubMed] [Google Scholar]
- Hunt P., Gulisano M., Cook M., Sham M.H., Faiella A., Wilkinson D., Boncinelli E., Krumlauf R. A distinct Hox code for the branchial region of the vertebrate head. Nature. 1991;353:861–864. doi: 10.1038/353861a0. [DOI] [PubMed] [Google Scholar]
- Jacob J., Ferri A.L., Milton C., Prin F., Pla P., Lin W., Gavalas A., Ang S.L., Briscoe J. Transcriptional repression coordinates the temporal switch from motor to serotonergic neurogenesis. Nat. Neurosci. 2007;10:1433–1439. doi: 10.1038/nn1985. [DOI] [PubMed] [Google Scholar]
- Jo J., Xiao Y., Sun A.X., Cukuroglu E., Tran H.D., Göke J., Tan Z.Y., Saw T.Y., Tan C.P., Lokman H. Midbrain-like organoids from human pluripotent stem cells contain functional dopaminergic and neuromelanin-producing neurons. Cell Stem Cell. 2016;19:248–257. doi: 10.1016/j.stem.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi T., Morizane A., Doi D., Magotani H., Onoe H., Hayashi T., Mizuma H., Takara S., Takahashi R., Inoue H. Human iPS cell-derived dopaminergic neurons function in a primate Parkinson’s disease model. Nature. 2017;548:592–596. doi: 10.1038/nature23664. [DOI] [PubMed] [Google Scholar]
- Kim S., Kwon S.H., Kam T.I., Panicker N., Karuppagounder S.S., Lee S., Lee J.H., Kim W.R., Kook M., Foss C.A. Transneuronal Propagation of Pathologic α-synuclein from the gut to the brain models Parkinson’s disease. Neuron. 2019;103:627–641.e7. doi: 10.1016/j.neuron.2019.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriks S., Shim J.W., Piao J., Ganat Y.M., Wakeman D.R., Xie Z., Carrillo-Reid L., Auyeung G., Antonacci C., Buch A. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 2011;480:547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger K.C., Deneris E.S. Serotonergic transcription of human FEV reveals direct GATA factor interactions and fate of pet-1-deficient serotonin neuron precursors. J. Neurosci. 2008;28:12748–12758. doi: 10.1523/JNEUROSCI.4349-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Renner M., Martin C.A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Zhong X., Liu H., Hao L., Huang C.T.L., Sherafat M.A., Jones J., Ayala M., Li L., Zhang S.C. Generation of serotonin neurons from human pluripotent stem cells. Nat. Biotechnol. 2016;34:89–94. doi: 10.1038/nbt.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes M., De Vos N., Pioli R., Demedts P., Wauters A., Neels H., Christophe A. Lower serum vitamin E concentrations in major depression. Another marker of lowered antioxidant defenses in that illness. J. Affect. Disord. 2000;58:241–246. doi: 10.1016/s0165-0327(99)00121-4. [DOI] [PubMed] [Google Scholar]
- Manosso L.M., Neis V.B., Moretti M., Daufenbach J.F., Freitas A.E., Colla A.R., Rodrigues A.L.S. Antidepressant-like effect of α-tocopherol in a mouse model of depressive-like behavior induced by TNF-α. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2013;46:48–57. doi: 10.1016/j.pnpbp.2013.06.012. [DOI] [PubMed] [Google Scholar]
- Meinhardt A., Eberle D., Tazaki A., Ranga A., Niesche M., Wilsch-Bräuninger M., Stec A., Schackert G., Lutolf M., Tanaka E.M. 3D reconstitution of the patterned neural tube from embryonic stem cells. Stem Cell Reports. 2014;3:987–999. doi: 10.1016/j.stemcr.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzis V., Steinhauser S., Pakanavicius E., Gouti M., Stamataki D., Ivanovitch K., Watson T., Rayon T., Mousavy Gharavy S.N., Lovell-Badge R. Nervous system regionalization entails axial allocation before neural differentiation. Cell. 2018;175:1105–1118.e17. doi: 10.1016/j.cell.2018.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederreither K., Vermot J., Schuhbaur B., Chambon P., Dollé P. Retinoic acid synthesis and hindbrain patterning in the mouse embryo. Development. 2000;127:75–85. doi: 10.1242/dev.127.1.75. [DOI] [PubMed] [Google Scholar]
- Okada Y., Shimazaki T., Sobue G., Okano H. Retinoic-acid-concentration-dependent acquisition of neural cell identity during in vitro differentiation of mouse embryonic stem cells. Dev. Biol. 2004;275:124–142. doi: 10.1016/j.ydbio.2004.07.038. [DOI] [PubMed] [Google Scholar]
- Okaty B.W., Freret M.E., Rood B.D., Brust R.D., Hennessy M.L., deBairos D., Kim J.C., Cook M.N., Dymecki S.M. Multi-scale molecular Deconstruction of the serotonin neuron system. Neuron. 2015;88:774–791. doi: 10.1016/j.neuron.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattyn A., Vallstedt A., Dias J.M., Samad O.A., Krumlauf R., Rijli F.M., Brunet J.F., Ericson J. Coordinated temporal and spatial control of motor neuron and serotonergic neuron generation from a common pool of CNS progenitors. Genes Dev. 2003;17:729–737. doi: 10.1101/gad.255803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattyn A., Simplicio N., Van Doorninck H., Goridis C., Guillemot F., Brunet J.F. Ascl1/Mash1 is required for the development of central serotonergic neurons. Nat. Neurosci. 2004;7:589–595. doi: 10.1038/nn1247. [DOI] [PubMed] [Google Scholar]
- Qian X., Nguyen H.N., Song M.M., Hadiono C., Ogden S.C., Hammack C., Yao B., Hamersky G.R., Jacob F., Zhong C. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riessland M., Kolisnyk B., Kim T.W., Cheng J., Ni J., Pearson J.A., Park E.J., Dam K., Acehan D., Ramos-Espiritu L.S. Loss of SATB1 induces p21-dependent cellular Senescence in Post-mitotic dopaminergic neurons. Cell Stem Cell. 2019;25:514–530.e8. doi: 10.1016/j.stem.2019.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansone R.A., Sansone L.A. Tramadol: seizures, serotonin syndrome, and coadministered antidepressants. Psychiatry (Edgmont). 2009;6:17. [PMC free article] [PubMed] [Google Scholar]
- Schweitzer J.S., Song B., Herrington T.M., Park T.Y., Lee N., Ko S., Jeon J., Cha Y., Kim K., Li Q. Personalized iPSC-derived dopamine progenitor cells for Parkinson’s disease. N. Engl. J. Med. 2020;382:1926–1932. doi: 10.1056/NEJMoa1915872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibler P., Graziotto J., Jeong H., Simunovic F., Klein C., Krainc D. Mitochondrial parkin recruitment is impaired in neurons derived from mutant PINK1 induced pluripotent stem cells. J. Neurosci. 2011;31:5970–5976. doi: 10.1523/JNEUROSCI.4441-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitobello A., Ferretti E., Lampe X., Vilain N., Ducret S., Ori M., Spetz J.-F., Selleri L., Rijli F.M. Hox and Pbx factors control retinoic acid synthesis during hindbrain segmentation. Dev. Cell. 2011;20:469–482. doi: 10.1016/j.devcel.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano J.M., Yu K., Donaldson G.P., Shastri G.G., Ann P., Ma L., Nagler C.R., Ismagilov R.F., Mazmanian S.K., Hsiao E.Y. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W., Shimamura K., Rubenstein J.L., Hynes M.A., Rosenthal A. FGF and Shh signals control dopaminergic and serotonergic cell fate in the anterior neural plate. Cell. 1998;93:755–766. doi: 10.1016/s0092-8674(00)81437-3. [DOI] [PubMed] [Google Scholar]
- Zeisel A., Hochgerner H., Lönnerberg P., Johnsson A., Memic F., van der Zwan J., Häring M., Braun E., Borm L.E., La Manno G. Molecular architecture of the mouse nervous system. Cell. 2018;174:999–1014.e22. doi: 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Transcriptome analysis was executed using the Affymetrix Whole Transcript expression array according to the manufacturer's protocol (GeneChip WT PLUS reagent kit; Thermo Fisher Scientific, Waltham, MA). Raw data of the microarray are accessible through GEO Series accession GEO: GSE167278.