Abstract
Disease-relevant human induced pluripotent stem cells (iPSCs) are generated worldwide for research purposes; however, without robust and practical ethical, legal, and quality standards, there is a high risk that their true potential will not be realized. Best practices for tissue procurement, iPSC reprogramming, day-to-day cultivation, quality control, and data management aligned with an ethical and legal framework must be included into daily operations to ensure their promise is maximized. Here we discuss key learning experiences from 7 years of operating the European Bank for induced Pluripotent Stem Cells (EBiSC) and recommend how to incorporate solutions into a daily management framework.
Keywords: EBiSC, iPSC, quality control, quality, ethics, legal, biobank, repository, reprogramming, banking, data management, guidance
Highlights
-
•
Ethics for iPSCs must be explicit, GDPR compliant, and allow future research
-
•
iPSC use restrictions are linked to consent, reprogramming, and gene editing
-
•
Quality control must be implemented from primary tissue handling onward
-
•
Robust data management is essential to ensure privacy and enable data sharing
The European Bank for induced Pluripotent Stem Cells discusses common pitfalls during iPSC generation and recommends robust and practical solutions for ethical, legal, and quality frameworks. Through these recommendations, high-quality iPSC lines can be efficiently distributed, maximizing the use of tissue samples that have been gifted to researchers to accelerate disease research and the discovery of novel therapeutic solutions.
Main text
Introduction
Reprogramming somatic cells into induced pluripotent stem cells (iPSCs) creates the opportunity to establish more accurate, novel, powerful models for the study of human cells and tissues not otherwise readily obtainable, such as previous commonly used cell lines and donated human tissue (Takahashi et al., 2007; Takahashi and Yamanaka, 2006). This led to significant investment around the world to establish large collections of iPSCs, e.g., CIRM, WiCell (United States) and CiRA (Japan) (Guhr et al., 2018; Huang et al., 2019). Since 2014, the European Bank for induced Pluripotent Stem Cells (EBiSC) has brought European experts in reprogramming, biobanking, and the pharmaceutical industry together to create a large collection of iPSC lines for research (www.EBiSC.org). This investment provides a single access point to >900 iPSC lines from donors affected by more than 30 genetic diseases, with standardized culture conditions and quality control (QC).
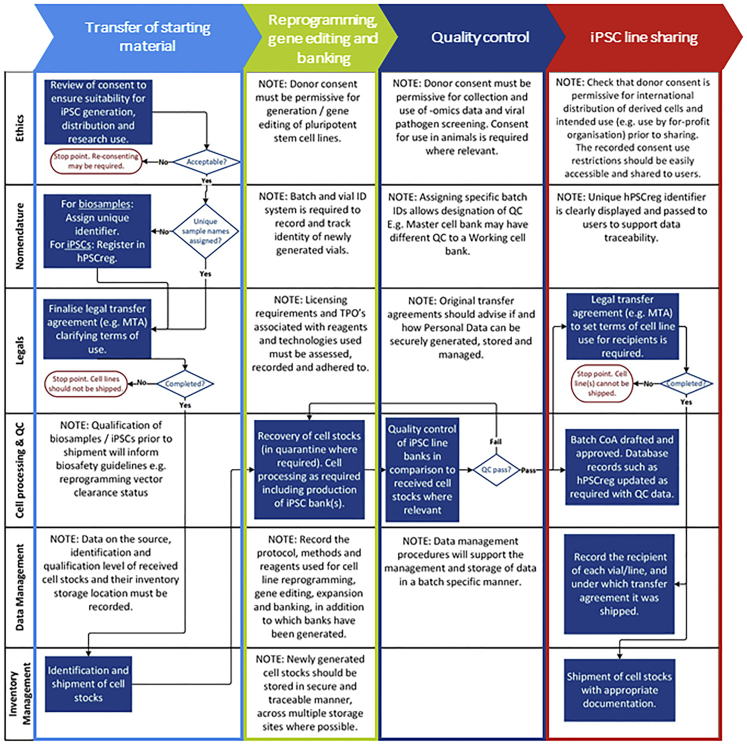
Given the long history of wasted biomedical resources through cell lines of poor quality or complex and restricted access agreements, EBiSC was created to protect a huge public investment into iPSC research across Europe (€432 million/$474 million by 2018) and elsewhere in the world, which has given rise to an estimated 10,000 iPSC lines (Guhr et al., 2018; Huang et al., 2019). EBiSC plays a crucial role in collecting iPSCs for broad, non-profit distribution with a core goal of simplifying access to high-quality, disease-relevant iPSCs for both commercial and non-profit organizations to use for research (Allsopp et al., 2019). Where demand has been identified, additional collaborative research projects continue to generate new disease-relevant cohorts of well-characterized iPSCs (e.g., IMI-ADAPTED; https://www.IMI-ADAPTED.eu/). Centralized iPSC repositories such as EBiSC help ensure that iPSC resources generated within these research projects are protected and made sustainable long term (De Sousa et al., 2017; Rao, 2013). Institutional core iPSC facilities also now commonly provide researchers with access to affordable and centralized iPSC reprogramming, gene editing, banking, and characterization (https://coredinates.org/), thereby increasing accessibility by supporting non-expert users and reducing costs. With the frequency of core facilities on the rise, there is great potential for generating high numbers of iPSC line cohorts that could progress disease research. However, establishing consistent standards and processes that are robust and realistic is vital to truly accelerate iPSC use and ultimately affect human healthcare (Allsopp et al., 2019). Best practice on the collection of human tissue samples, data management, QC, and a legal framework that is both compliant with local policy and simplifies transfer to external users must be part of, and not additional to, the core infrastructure that manages routine tasks (Figure 2). By standardizing and improving the quality of iPSCs and the tissue samples used to make them, cell lines and datasets can be easily exchanged between academia and industry, increasing outputs and efficiency of research.
Figure 2.
A cohesive and coordinated approach toward best practice and resource sustainability
Generating iPSC resources that are sustainable long term is dependent on ensuring best practice in a multi-pronged and coherent manner. Different aspects of best practices do not operate in isolation but rather interact and are dependent on each other throughout the process. An exemplar cell processing pathway is shown here based on the receipt of starting cell material, cell processing (e.g., reprogramming and iPSC banking), QC, and distribution of qualified vials to other researchers. It is shown that outputs of different activities (shown here in rows), including defined processes for ethical and legal governance, banking, QC, data management, inventory, and labeling, feed into each other and should be implemented in a coordinated manner. Implementation of a single criterion is not sufficient and increases the risk of restricting usage of valuable iPSC resources. By incorporating all aspects into central infrastructure in an aligned and cohesive manner, distribution and downstream use of iPSCs can be simplified and quality of the resource ensured.
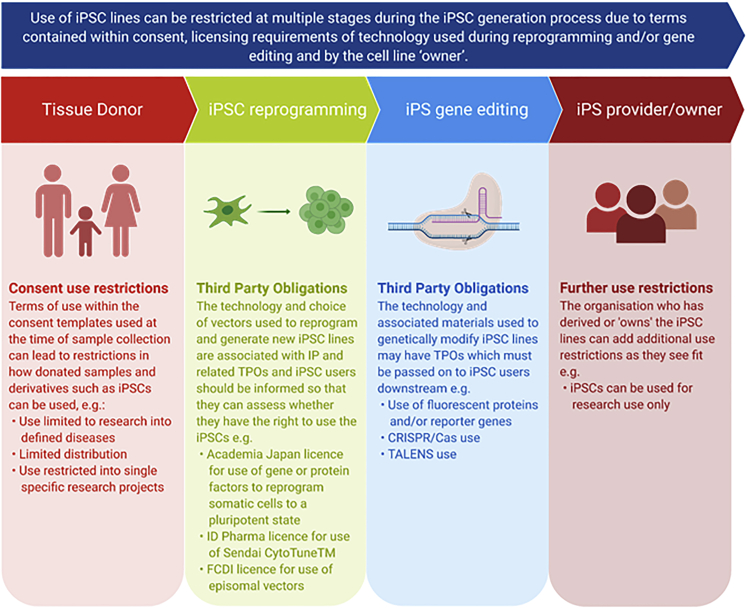
To date, EBiSC has collected >900 iPSC lines, generated from >740 primary tissue samples collected across >27 clinical sites (within >30 different research studies) and shared by >20 depositing institutions (including commercial, academic, and small/medium enterprise [SME] organizations) across Europe and the United States. With the goal of standardizing quality and reproducibility, learnings from this process have highlighted a number of aspects that are recognized by EBiSC as critical roadblocks that must be considered when establishing an iPSC repository or core facility. Here we lay out recommendations for how future iPSC research projects, facilities, and repositories (hereafter referred to as repositories) can address these frequent issues and benefit from EBiSC's experience. Restricted use of iPSC lines and third-party licensing obligations can be incurred at multiple time points, starting with donor agreements and primary tissue provision through to reprogramming, gene editing, and distribution, hence early planning and review before any tissue samples are collected is essential (Figure 1). Similarly, qualification of primary tissue and/or its early passage cell cultures can greatly ease downstream processing and should be aligned with the planned qualification of iPSCs. Cell culture best practice and a robust quality management system (QMS) can support implementing, maintaining, and improving processes for efficient iPSC line generation, storage, and distribution. Through incorporation of these recommendations, high-quality iPSCs that meet community-agreed standards can be generated and efficiently distributed, maximizing the use of tissue samples that have been gifted to researchers to accelerate disease research and the discovery of novel therapeutic solutions, and enhancing scientific reproducibility across the life-science research landscape.
Figure 1.
IPSC use and licensing restrictions can be introduced at multiple stages during the iPSC generation process
Consent use restrictions can originate from terms of use contained within consent templates used to collect the original donated biosample. Intellectual property associated with technology and reagents used during reprogramming and gene editing may carry licensing requirements and third-party obligations that need to be respected and passed on to users. The iPSC line provider or owner can then include any use restrictions as they see fit: for example, that distributed iPSCs cannot be used for direct exploitation. All restrictions for use need to be clearly assessed, respected, and recorded.
Collection of consent for iPSC research
Without study participants being willing to donate tissue samples for iPSC generation, this field of research would be severely hampered. Hence, the use of participant information sheets (PIS) and informed consent forms (ICF) that are clear and explicit on how donated tissue and data (hereafter referred to as donations) will be used is essential to maintain trust with study participants. From the researcher's perspective, avoiding unintended restrictions on downstream use due to poorly worded or unclear consent templates is essential to maximize donations and avoid volunteers who have donated tissue having to go through re-consenting for use (note: exact guidance on the local requirements for submission, review, and approval of consent templates, health and safety requirements for tissue collection, and the management of storing and distributing donated samples and associated data should be followed at all times, but are not outlined here due to global variances; Andrews et al., 2015).
Out of >740 samples used for EBiSC iPSC generation between 2014 and 2019, only 27 tissue samples were directly collected for EBiSC purposes; hence, a detailed and robust review to ensure that third-party consent documentation does not preclude iPSC generation, characterization, and distribution has consistently been an essential first step when assessing the vast majority of incoming iPSCs. EBiSC has developed specific review criteria incorporating Lomax et al.’s DISCUSS principles (Lomax et al., 2013) (key for the use of tissue donated prior to the publication and wide adoption of iPSC generation), hPSCreg (Human Pluripotent Stem Cell Registry; www.hPSCreg.eu) data validation standards, and guidance from ethical advisors (Table 1). This review framework aims to enable the use of donations gifted for disease research, balanced with respecting the intended use that was consented for. These criteria are compliant with the European General Data Protection Regulations (GDPR), which came into European Union (EU) law in 2018 to ensure adequate protection of volunteer demographic data while at the same time permitting sharing of “personal” datasets (such as whole-genome sequencing) to support research and maximize use of associated iPSCs. Note that similar local data protection laws exist or are forthcoming internationally and must be similarly adhered to where applicable. EBiSC review of local consent templates is primarily based on (1) the template PIS; (2) the template ICF; (3) local ethical review; and (4) confirmation of full ICF completion by volunteers and positive responses to optional clauses (note that, if collecting donations from individuals under the age of consent or from adults who lack mental capacity to understand the terms of consent, the donor's legal representative will generally consent on the individual's behalf, requiring specific templates for the guardian. Where appropriate, these may be accompanied by capacity-appropriate PIS using images and/or simpler language and assent templates to record willingness to donate; these should also be reviewed to ensure consistency of message). Review criteria essentially strive to ensure that consent has been voluntarily given, that the volunteer has been fully informed on how the donation will be used both immediately and in the future (including using iPSCs in animal studies), that donation will not affect the donor’s medical care, and that their privacy will be protected at all times. If review criteria could not be met, depositors were asked to assess in conjunction with the relevant clinical teams if study participants could be re-consented with consent documentation that did meet explicit terms of use. For a limited number of patients, re-consenting was not possible, hence the use of limited consent templates resulted in iPSCs that were unable to be widely shared and used, restricting the prospective beneficial impact of these gifted samples for research discovery.
Table 1.
Recommendations for ethical and legal requirements
| # | Guidance 1. Recommendations for ethical and legal requirements | |
|---|---|---|
| Consent: EBiSC recommends that iPSC research projects ensure the PIS and ICF selected for use explicitly cover: | ||
| 1.1 | that tissue samples and demographic data have been freely and voluntarily donated for research use and that lack of donation will not affect their medical care in any way | |
| 1.2 | that samples will be used to generate derivative cell lines, such as iPSC lines, that can be retained indefinitely in culture | |
| 1.3 | that collected samples and derived cell lines may be characterized, including genomic analysis | |
| 1.4 | that derived cell lines and data (including genomic data) may be shared with researchers worldwide, including for-profit and not-for-profit organizations | |
| 1.5 | the rights of the donor regarding data associated with them, their donated sample(s), and any associated dataset(s); e.g., that genomic data may be stored in secure databases and shared to researchers as managed access data. This includes the inherent risks of data sharing, how these risks will be minimized, and how these data may be stored and shared | |
| 1.6 | how the study participant's identity will be protected at all times | |
| 1.7 | that the study participant will get no financial or legal benefits from donating a sample, including lack of financial benefit from any products or services that may be derived from the sample or derivatives | |
| 1.8 | whether derived cell lines may be used in research using animal models | |
| 1.9 | that derived cell lines may be used in as-yet undefined research activities, for the purpose of biological research purposes, avoiding any unintentional research use restrictions such as limiting research use to specific disease areas | |
| 1.10 | that the donor has the right to withdraw consent for use of the donated sample, but that only the original tissue sample will be destroyed and, if iPSC lines have already been generated, the lines and associated dataset(s) cannot be destroyed. However, any “pseudonymized” link between the donor and the donated sample can be completely severed. See section “nomenclature and data management” for further information on data management | |
| LEGAL: EBiSC recommends that iPSC research projects ensure the following legal aspects are assessed at project start: | ||
| 1.11 | investigate and understand licensing implications of using iPSC derivation reagents and methods and ensure these are understood and recorded before committing any resource to technical procedures such as reception of donor materials, iPSC generation, and gene editing | |
| 1.12 | clarify and agree ownership of iPSC lines and which party/parties may administer responsibility for deposition or distribution | |
| 1.13 | ensure transfer of any and all cell material is explicitly recorded in a locally acceptable legal agreement (such as a MTA or other) that is signed by all involved parties. This includes transfer of cell material between collaborative partners and for distribution of cell lines to users | |
| 1.14 | where feasible, operate an intellectual property horizon scanning activity focused on general iPSC resource operation by utilizing industry links and liaison with relevant patent office information | |
To further simplify the consent collection and review process, a template PIS and ICF are available for researchers to download via the EBiSC website. These templates must be adapted to include study-specific details and submitted for local ethical review in line with local guidelines. Through use of these templates, EBiSC aims to ensure that individuals making donations are fully aware of the intended downstream use and that samples and derivatives may be used in as-yet unknown scientific methodologies, but that appropriate measures will be implemented to assure that their data, privacy, and confidentiality will be protected at all times. It is critical that adoption of these templates should be done as a very first step during project or facility development due to the lengthy time likely required for local ethical review and approval, prior to actual use with study participants. It is also recognized that commercial companies occupy a significant proportion of the iPSC research landscape; hence, use and exploitation of derivatives by commercial companies is recommended for inclusion in consent forms to maximize possibilities for iPSC usage in the future, even if not intended at the time of donation. Obtaining consent for commercial use at the start greatly opens up options downstream, without the need for participant re-contacting/re-consenting, which can be impossible years after cells have been used in developmental research. Best practice is to explicitly inform donors how their personal data (as defined by GDPR), including genetic data, will be collected, used and shared and their rights to withdraw consent over both use of their donated samples and associated data (Morrison et al., 2017). Note that local regulatory bodies will likely have region-specific guidance, which should be consulted. As genomic data are unique to each individual, it will always carry a theoretical risk of identification, however small that risk may be. Hence participants should always be made aware of this and be advised as to how their privacy will be protected (Gymrek et al., 2013; Isasi et al., 2014). Further guidance on how personal data can be protected and anonymized is provided in section “nomenclature and data management.”
Templates of the ethically reviewed and approved PIS and ICF should be securely stored and linked to donated samples, including any details of local ethical review, such as the name of the ethical review board and an associated reference number. The use of iPSC-specific databases such as hPSCreg is recommended, whereby detailed information on the consent templates used, the terms of consent contained within, and details on the local ethical review performed can be recorded in a cell line-specific manner to enable simple communication of the terms of consent to end users. It is recommended to avoid inclusion of disease-associated restrictions (for example, “Your donated sample will only be used for research into Parkinson disease and other movement disorders”), which may limit downstream use of iPSCs, particularly as our understanding of the interconnection between multiple pathologies deepens (Figure 1). Indeed, more than 50% of lines currently deposited in EBiSC are derived from disease-restricted consent. Considering these aspects as a first step when planning to collect donated samples for iPSC generation can help to ensure that the effort and resource that go into sample collection are truly maximized.
iPSC line ownership, licensing, and third-party restrictions
The legal landscape that regulates and supports the iPSC landscape is complex and, if not properly assessed and managed, can lead to delays in distribution and use of generated iPSCs, even where lines are only being exchanged between collaborators (see Table 1 for guidance). It is strongly recommended to ensure that transfer of all cell samples between different organizations, including the transfer of primary material from the clinical source to the reprogramming center, is explicitly recorded in documentation signed by all involved parties. Whether through use of a material transfer agreement (MTA) or other, documentation should clearly state (1) the source and “ownership” of cells, including a list of relevant anonymized and unique donor or primary tissue identifiers (see section “nomenclature and data management”); (2) the purposes for which cells can be used, including permitted and/or prohibited use in a commercial context and any ethical limitations (e.g., preventing primary material from being used for gamete generation); (3) who will own any derivatives, such as iPSCs or differentiated cell populations (and thus take responsibility for ensuring adequate licensing provisions for cell line generation); (4) how research outputs such as iPSCs, genomic datasets, or publications should be managed, accessed, and shared; (5) warranties, liabilities, and indemnification across organizations; and (6) conditions for agreement termination (Volarevic et al., 2018).
Many patents have been filed that are associated with various stages of the iPSC reprogramming process, including IPS Academia Japan's (IPS-AJ) patent for use of the reprogramming factors POU5F1, KLF4, MYC, and SOX2 to reprogram somatic cell types back into a pluripotent state, specific reprogramming vectors (e.g., Sendai CytoTune), gene editing technologies (e.g., CRISPR-Cas9), and other reagents (e.g., fluorescent markers for generation of reporter lines), and others (Morita et al., 2019; Tessensohn and Yamamoto, 2009). iPSC research organizations may require various licenses dependent on the organizational type (i.e., non-profit or for profit) and the scope of use (e.g., basic research versus use within compound screening assays). Note that licenses may be required for both generation and use of iPSCs and/or their derivatives (such as differentiated cell populations), and requirements apply at the point of reagent purchase, not at the point of use or final product distribution. To add complexity, applicability of licensing varies internationally. With the registration of iPSC-related patents continuing to rapidly rise across the world, keeping on top of all patents is a costly and extremely time-consuming task; however, due diligence is critically important. Hence, EBiSC recommends early mapping of planned iPSC research activities and use of any associated protocols and reagents against licensing requirements and any associated licensing requirements investigated and recorded. This is particularly important if investing research toward direct exploitation. For example, specific reprogramming vectors for the generation of iPSCs generally carry with them clear licensing requirements that must be abided by and third-party obligations that need to be passed on to users (Morita et al., 2019). It is recommended to seek early advice from local legal and technical transfer teams so that licensing requirements in the appropriate geographical and organizational context are centrally understood, recorded, and built into the local framework for distribution of cells. It is critical that this takes place before iPSC reprogramming and/or gene editing takes place to avoid wasted time, effort, and money. For example, a collaborative iPSC research program involving commercial organizations may require partners to carry the appropriate IPS-AJ license depending on the material being shared and its intended downstream use. It is preferable to establish this early rather than later on when cell shipment and project progress may be delayed while licenses are obtained.
Licensing restrictions aside, deposition into EBiSC requires agreement with the “owner”/depositor of the line(s) that EBiSC can store and distribute the named iPSCs on the depositor’s behalf. Note that the depositor is generally defined as the organization who either generated the iPSCs or collected the original tissue samples, with joint deposition recognizing that a cohort of lines may have been heavily dependent on contributions from multiple parties. Early discussion and agreement as to which institution is the primary owner of the iPSCs, and hence has the legal right to register and distribute the line(s), is recommended, with prior experience showing that failure to do so can result in extreme delays and the amendment of whole consortium project agreements or, in the worst cases, inability to deposit with a central supplier at all. Furthermore, the user may waste significant resources on generation of data that cannot be published or generate intellectual property associated with a new discovery that may then belong to another party.
Reprogramming, banking, and QC
iPSC researchers internationally will be familiar with the hefty investment of time and resources often required for iPSC research. Use of poor-quality iPSCs is not only a waste of research resources and public funds but also puts publications at risk of retraction, blocks the utility of patient samples gifted for this purpose, and impedes the development of clinical and therapeutic outcomes from iPSC research, which is surely the goal for all iPSC researchers. It can be argued here that publicly funded stem cell researchers have a social responsibility in this regard, as they are responsible for dissemination of iPSC resources for use within other research activities.
EBiSC has previously reported significant challenges when collecting older iPSCs from a range of academic and SME depositors, improved by disseminating a standardized QC regime across partners, supporting users with an advice and training framework, and collecting cohorts of lines generated and deposited by other large iPSC research projects using established quality frameworks (De Sousa et al., 2017; O'Shea et al., 2020). By incorporating QC testing into donation and early iPSC processing, lines can be generated that have a robust historical dataset to support ongoing QC and any subsequent investigations into unusual cell line behavior (Table 2). Once cell lines are transferred from a supplier such as EBiSC, users should implement routine QC testing into day-to-day processes to ensure iPSC quality (Table 2). If performing cell reprogramming directly, at an absolute minimum, collected primary samples and/or derived cell populations should be screened for human viral pathogens and the presence of contaminants such as bacteria, yeast, or fungi, and cell line identity should be recorded (Table 2). By including extra qualification on primary samples, such as confirming the presence of disease-associated genotype and recording genomic health (e.g., karyotype, SNPa), users can fully interpret and understand these data when performed on the iPSC(s) and distinguish between genomic aberrations that were present in the individual and those that have been introduced during the reprogramming process.
Table 2.
Recommendations for iPSC quality control from primary tissue to reprogramming, banking, routine QC, and gene editing.
| # |
Guidance 2. iPSC QC recommendations |
||||
|---|---|---|---|---|---|
| Assay | Primary tissue | Early reprogrammed clone(s) | iPSC line(s): When generating master or working banks | iPSC line(s) monitoring during routine culture | |
| 2.1 | Cell line identity (STR allele profile recorded) | Required | Required | Required | Every 6–8 weeks or 10–12 passages on lines in culture (competence of users and use of facility should be taken into account here. Novice users and/or heavily used laboratory spaces can increase risk of cell line identity switches/contamination, so routine screening should be implemented on a more frequent basis). When cell lines have been accessed from an external source, always ask for the STR profile to use as a reference point. |
| 2.2 | Genetic stability (such as G banding, SNP, or aCGH) | Preferable | Required | Every 6 weeks or 10 passages if extended culture is required, after any significant selection event such as single cell cloning, or if morphology or growth rate alters in culture | |
| 2.3 | Negative result for bacteria, yeast, and fungi screening using TSB and FTM inoculation | Required | Required | Required | Visual, daily. a more sensitive screen, such as TSB and FTM inoculation, should be performed prior to sharing any cultures to other researchers and when receiving cultures from external sources |
| 2.4 | Negative result for Mycoplasma screening using high-sensitivity method | Required | Required | Required | Every 3–4 weeks or 5–6 passages if extended culture is required. Perform if morphology or growth cycle alters in culture. From a practical perspective, this could be a monthly screen of every in vitro cell line in culture within a single laboratory. This should also be performed prior to sharing any cultures with other researchers and when receiving cultures from external sources |
| 2.5 | Negative result for human viral pathogens (HIV1, HIV2, HBV, and HCV) screening | Required (if not performed on the sample donor) | Only if not done on primary tissue/donor | ||
| 2.6 | Morphology | Required | Required | Required | Visual, ideally daily or whenever cultures are checked |
| 2.7 | Viability and recovery post thaw should be assessed and specific recovery requirements, such as high-density seeding or the temporary use of rho-kinase inhibitors, recorded | Required | Required | ||
| 2.8 | Clearance or silencing of reprogramming vector | Required | Only if not done on earlier clones | ||
| 2.9 | Expression of markers associated with undifferentiated hPSCs, assessed using flow cytometry. Recommended to include both transcriptional regulators (e.g., POU5F1) and surface markers (e.g., SSEA-1, SSEA-4) | Required | Recommended to be performed prior to initiation of differentiation experiments. Perform if morphology or growth cycle alters in culture |
||
| 2.10 | Pluripotency: assessment of differentiation potential through in vitro trilineage differentiation and germ layer marker expression | Required (scientific consensus agrees that a functional differentiation assay is the most robust way to assess pluripotency. Solely assessing expression of markers such as POU5F1 and TRA-1-60 does not take into account mutations and or accurately indicate functional pluripotent efficacy; Andrews and Stacey, 2015; Stacey et al., 2019) | Perform on early master stocks, long-term cultures established for experimental purposes, if morphology or growth cycle alters in culture or if issues with established differentiation protocols or similar are observed | ||
| 2.11 | Confirmation of genetic lesion for disease-relevant lines | Preferable | Preferable (mandatory for gene-edited lines, see Guidance 3) | Confirmation of genetic lesion should be included as a routine cell line identity check, if using multiple gene-edited lines from the same donor | |
| Guidance 3. Additional qualification for iPSC lines that have been genetically edited | |||
|---|---|---|---|
| # | Assay | Parental iPSC line(s) | Genetically modified iPSC line(s) |
| 3.1 | Genomic array such as G banding, SNP, or aCGH | Required | Required |
| 3.2 | Sequence of target locus | Required | Required |
| 3.3 | Impact on protein expression where appropriate (e.g., gene knockouts, insertion of reporter or inducible gene expression systems) | Required | |
| 3.4 | Assessment of off-target effects (assess potential off-target effects using database prediction tools and sequence most likely affected loci; e.g., NGS or WGS) | Required | |
| 3.5 | Assessment of OnTEs (e.g., quantitative-genotyping PCR) | Required | |
aCGH, array comparative genomic hybridization FTM, fluid thioglycolate medium; hPSCs, human pluripotent stem cells; NGS, next-generation sequencing; TSB, tryptone soya broth; WGS, whole-genome sequencing.
Best practice for generating iPSC line banks
Once a stable iPSC line is established, maintaining distribution stocks by continuous passaging exposes cells to risk of incurring genetic changes over time or total loss of the cell line due to cross-contamination, microbial contamination, etc. Thus, it is common best practice during iPSC generation to periodically cryopreserve cells to ensure backup stocks are available from key points during the process (e.g., early passage clones) and create a stable resource that can be referred back to long term. The latter production of master stocks (well-characterized batches or lots made up of cryopreserved vials in secure long-term storage for replenishment of working stocks) and working stocks (larger qualified batches of vials that can be used for day-to-day experimental use) ensure that suitable, well-qualified vials are always available, while making certain that additional stocks are safeguarded for future batch production (Geraghty et al., 2014; Stacey et al., 2013). QC can be implemented at different levels across master and working batches according to researcher needs and should be performed directly on banked vials. Batch identifiers to segregate individual master and working banks and implementation of a process to track batch-specific details such as passage number and culture methods are recommended (Table 3) (Andrews et al., 2015; Stacey et al., 2013). Low-temperature-suitable vial labels should be printed with indelible ink (i.e., alcohol resistant), generated using a label printer (and not handwritten), be resistant to loss of adhesive qualities in liquid nitrogen storage, and use unique cell line and batch nomenclature (Table 4). Appropriate systems should be in place to easily link physical vials to batch-specific records to ensure that all information is readily available (see sections “quality assurance framework” and “nomenclature and data management”).
Table 3.
Recommendations for a practical quality assurance framework
| # | Guidance 4. Recommendations for a practical quality assurance framework |
|---|---|
| 4.1 | Ensure that SOPs are written for all core processes, such as routine iPSC culture, generating batches, managing cryo-storage storage, and guidance on health and safety procedures. These SOPs should be centrally available and version controlled |
| 4.2 | Implement a system to investigate issues, find root failures, and implement corrective actions across all staff members to avoid repeat occurrences. This can be done in a practical approach, which does not need to be cumbersome |
| 4.3 | Have a documented system of staff training with a training manual in key laboratory procedures and principles of best practices |
| 4.4 | Introduce a system for assigning batch (or lot) identifiers and including batch-specific information, such as culture conditions and passage numbers, on vial labels |
| 4.5 | An inventory of materials stored within fridges and freezers is recommended with a yearly check included to keep records up to date with responsibility assigned to specific staff member(s) |
| 4.6 | Ensure that, prior to distribution of any cell stocks, critical QC such as screening for Mycoplasma and checking cell line identity is completed and the data available for cell line recipients |
| 4.7 | Keep records of all incoming and outgoing cell material, including source, recipient, sample type, date of transfer, and pseudonymized/anonymized sample identifiers |
| 4.8 | Routinely monitor performance of key equipment, such as fridges, freezers, and biosafety cabinets, including regular maintenance where required |
Table 4.
Recommendations for secure iPSC data management
| # |
Guidance 5. Recommendations for iPSC data management |
|
|---|---|---|
| Data type | Solutions | |
| 5.1 | Donor sample nomenclature | Implement robust procedures for assigning pseudonymized/anonymized identifiers for donor samples. Properties associated with assigned samples such as anonymized donor ID, age at sample collection, sex, and disease diagnosis should be carefully recorded within a secure internal database. Most importantly, the paper trail of consent to the sample donation must be kept intact. As long as accurate records are thus maintained, the sample ID can be a simple institutional identifier with increasing numerical digits; e.g., XY1, XY2 |
| 5.2 | IPSC line nomenclature | Implement robust procedures for assigning pseudonymized/anonymized identifiers for early reprogrammed clones and iPSC lines, using established tools such as hPSCreg wherever possible |
| 5.3 | Batch identifiers | Implementation of a simple identifier system for cell line batches (or lots) |
| 5.4 | Vial labeling | Use of unique printed labels that are suited to low temperatures, long-term liquid nitrogen storage, printed using indelible ink, with machine-readable barcodes and human-readable identifiers |
| 5.5 | Donor demographics and basic clinical data | Submission of cell line data to hPSCreg |
| 5.6 | Detailed clinical datasets and genomic datasets | Submission of data to a local secure managed access data repository, many of which are available internationally, such as the European Genome-Phenome Archive or the NIH-NCBI Database of Genotypes and Phenotypes |
| 5.7 | Day-to-day iPSC handling | Using standard forms for day-to-day data collection increases standardization of what and how data are captured when being performed by multiple people, minimizing variabilities. Formal laboratory notebooks can take this role; however, it is key that the data points recorded are standardized. Key reagents (media, matrix, dissociation agents, etc.) should be logged, including lot number and expiry date. Morphology, confluency, split ratio/seeding density, and passage number should also be recorded. Summary details (such as passaging method) are recorded in hPSCreg |
Cell harvesting and cryopreservation are critical steps during iPSC expansion and banking, with inappropriate procedures and reagents increasing the risk of issues post thaw such as poor recovery, appearance of genetic variants, and even complete loss of viability. Prior to cryopreservation, iPSCs should be scaled up to required cell number and harvested (normally as either single cells or clumps) when at appropriate confluency: generally ∼70%–80% but this will vary across different iPSC lines. The goal of batch production is that each vial within a batch should have uniform characteristics and will perform comparably when thawed. Hence, after harvesting iPSCs into suspension, cells from multiple cell culture plates/flasks must be pooled into a single vessel (i.e., batch) for addition of cryopreservant and gentle mixing before aliquoting into individual vials. If vials are numbered, start aliquoting at vial 1 and fill vials in ascending order, allowing consistency of performance to be demonstrated.
iPSC researchers commonly rely on slow-rate freezing using dimethyl sulfoxide (DMSO)-based cryoprotectants (Andrews and Stacey, 2015; Capes-Davis and Freshney, 2021). It is worth noting that procedures that are functional at lower volumes (e.g., manual closing of 20 vials during cryopreservation) can become cumbersome and impractical when handling multiple iPSC lines in parallel and/or performing cryopreservation of high cell numbers (e.g., manually closing 100 vials during cryopreservation). Prior to scaling up the banking volume of iPSCs, it is recommended to assess the facility, equipment, cell stage, and downstream use and consider (1) whether iPSCs should be cryopreserved in suspension as single cells or clumps; (2) the use of technological solutions, such as controlled rate coolers; (3) the selected cryopreservation format and its functionality within the current storage facilities and processes; and lastly (4) selection of a recovery protocol that is simple, robust, and can be easily adopted by cell line recipients. EBiSC currently cryopreserves all iPSCs as dissociated clumps in 1 mL of cryopreservation medium containing 10% (v/v) DMSO, in 2 mL of internally screw-threaded cryovials with silicone O rings (to prevent contamination of the thread in handling and storage) at a concentration of 1–2 × 106 cells, allowing end users to use a single recovery protocol that is robust and commonly recognizable by most cell culture researchers. Detailed EBiSC protocols for iPSC harvesting and cryopreservation are available in Capes-Davis and Freshney (2021).
Post cryopreservation, stocks should be split between different −150°C freezers and/or liquid nitrogen dewars, and, if possible, at different storage sites to secure against unforeseen infrastructure disruptions. It is strongly recommended to only store iPSCs at −80°C transiently post cryopreservation. It is critical that up-to-date content records are kept of all storage systems, particularly for those where regular inventory checks are more difficult because they maintain temperatures at below −50°C. EBiSC can attest to external research groups reporting that issues such as incorrect cell line identity, poor viability, karyological abnormalities, and even loss of all iPSC line stocks were traced back to fluxes in storage temperature, poor inventory of vials, and inappropriate vial labeling in their facilities. It should be noted that specific local regulations may carry requirements that affect sample storage and labeling, and best practice is to check local guidance (Van Den Heuvel et al., 2020)
QC recommendations
Across the iPSC research landscape, a core group of assays are defined as being required for adequate characterization of iPSCs, as shown in Table 2 (Andrews and Stacey, 2015; O'Shea et al., 2020; Sullivan et al., 2018). After batch generation, vial(s) should be thawed according to local procedures and given time to recover prior to performing batch QC. For sterility/Mycoplasma screening, samples should be collected from confluent cultures. More details are available in O'Shea et al. (2020).
During routine iPSC culture, specific frequent-failure quality criteria, such as cell authenticity and Mycoplasma, should be monitored routinely to ensure iPSC integrity and avoid expenditure of resources on incorrect or poor-quality iPSCs (Table 2). Incorrect cell line identity remains one of the biggest issues across all in vitro cell line research areas, as EBiSC has previously reported (De Sousa et al., 2017). If thawing iPSCs from external sources, it is recommended to check whether cell material has been screened for human viral pathogens (HIV1, HIV2, hepatitis B virus [HBV], hepatitis C virus [HCV]) prior to handling and to collect a sample for short tandem repeat (STR) testing as soon as possible post thaw for immediate analysis. Users should generate their own working iPSC banks, characterize them as per Table 2, and culture each vial for experimental purposes for a maximum of 6 weeks (8–10 passages). If extended periods of culture are required for experimental purposes, QC should be performed routinely on each in-use cell line to ensure early detection of cell line contaminations or genetic drift (see Table 2 for more details on screening frequencies). Good cell culture practice can help to prevent cell line identity issues from occurring at all; different cell lines should never be handled in a biosafety cabinet together at the same time, and biosafety cabinets should always be cleaned between different lines using an appropriate disinfectant, such as 70% isopropanol (Geraghty et al., 2014; Pamies et al., 2020). However, even with appropriate cell culture safeguards in place, mistakes can happen in a busy laboratory, and it is essential to be on guard and have the appropriate cell line identity screening processes in place. Many commercial kits are available that amplify the defined European set of STR standard markers, including 15 allele markers plus AMEL to determine the donor's biological sex (Welch et al., 2012).
Gene editing and QC
Genetically modified iPSCs have become a powerful tool for basic research and disease modeling (Czerwińska et al., 2019). Genetic modifications can be performed using multiple technologies, including zinc finger nucleases, transcription activator-like effector nucleases, and the CRISPR-CAS systems (Czerwińska et al., 2019). The latter, CRISPR-CAS, is very effective, straightforward to use, and hence is the most highly adopted system. However, any instance whereby nucleases are used to cut DNA always leaves the possibility of incorrect repair. Therefore it is critical to confirm the resulting sequence at each allele around the intended locus (Table 2). Importantly, as the nuclease is directed to cut the DNA via the guide RNA of the CRISPR-CAS complex, which recognizes the DNA over a stretch of 20 base pairs followed by the Protospacer adjacent motif (PAM) sequence (NGG/NRG), there is the risk that areas with homology to this sequence are also cut, followed by non-homologous end joining with the potential to create insertion/deletions and on-target effects (Weisheit et al., 2020). Use of database prediction tools when designing guide RNAs will predict the efficiency of guides and the likelihood of any off-target effects, which can be followed up by DNA sequence analysis post editing (Liu et al., 2020). Particular care should be taken regarding recent reports of on-target effects (OnTEs). Most of these OnTEs are small deletions or insertions of a few bases at the CRISPR cutting site on one or both alleles. A PCR carried out with primers flanking these OnTEs will amplify and reveal the deletions or insertions by Sanger sequencing. However, OnTEs can also be larger deletions or insertions at the CRISPR cutting site of several hundred or thousand bases. If they are only present on one allele, larger OnTEs can remain undetected by standard PCR and Sanger sequencing. Large deletions on one allele can delete the primer binding sites, meaning that the affected allele is then not amplified. Thus, sequencing analyses can appear homozygous as desired but only represent the sequence of the unaffected allele. Since it is unpredictable how large these deletions are, there is no guarantee for a primer design. Large insertions on one allele, even if they are within the primer region, can prevent allele amplification since the PCR product becomes too big. Again, this leads to the illusion of a homozygous sequence. New techniques such as quantitative-genotyping PCR can detect OnTEs and should be incorporated into routine iPSC screening post modification, as, if undetected, these OnTEs can have distinct downstream effects on disease modeling (Schmid et al., 2019; Weisheit et al., 2020). Recommended QC is outlined in Table 2 and should be implemented with consideration to the specific modification that has been performed, in addition to general iPSC QC guidance.
Quality assurance framework
A quality assurance framework for iPSC biobanking, containing standardized operating procedures (SOPs) for common processes, should be used regardless of organizational type (Table 3). This should be implemented based on actual requirements; for example, an academic research team may only need a flexible system whereby core SOPs are centrally stored; cryovial inventory is recorded, maintained, and checked annually; and large-scale issues affecting laboratory operations (e.g., a Mycoplasma contamination) are recorded, the investigation logged, and corrective/preventive actions followed up on. However, if operating as a core facility generating iPSC lines and derivatives en masse, more stringent controls are likely necessary.
It is essential to have a central record of the source and background of all cell material. Optimally, incoming cell material will have been screened (either at the donor or sample level) prior to transfer. If data are not available, the QMS must guide users to appropriate personal protective equipment and use of correct biosafety level according to local guidelines, with the assumption that cell material is infectious until screening shows otherwise. The use of unscreened cell material not only risks contravening ethical and legal governance and the health of staff but also risks using a line not of the expected origin and spreading microbiological contaminants to other cell lines in culture. Each laboratory sharing cell material has a responsibility to keep records of what has been shared, when it was transferred, under which legal agreement (e.g., MTA), and whom it was transferred to/from.
Correspondingly managed under the QMS umbrella should be defined working areas within the laboratory space. It is strongly recommended to use separate zones for (1) incoming cell lines from external sources; (2) the culture of primary cell types, such as fibroblast lines; and (3) “clean” cultures that have been screened and tested negative for microbiological agents. Separate laboratory spaces should be employed for these zones wherever possible; however, if space is limited, separate incubators and a daily rota whereby clean cells are strictly cultured before unscreened lines are handled can be used, including, of course, a rigorous cleaning regime between cultures to ensure that any adventitious agents are not transferred (Stacey, 2011). Routine maintenance and monitoring of key equipment is required to ensure staff safety and integrity of ongoing cell cultures.
Nomenclature and data management
iPSC database tools such as hPSCreg are critical for (1) providing procedures to help maintain data privacy; (2) standardizing disease phenotyping data; and (3) making data FAIR (findable, accessible, interoperable, and reusable; Wilkinson et al., 2016). HPSCreg allows the creation of cell line-specific records to store information regarding the source, characteristics, and ethical provenance of iPSC lines (Seltmann et al., 2016). Certification of cell line records in hPSCreg provides the registrant with a certificate allowing the usage of the cell line in European Commission (EC)-funded research. Hence, hPSCreg sits at the core of EBiSC's data management approach. Cell line depositors register their lines into hPSCreg and enter anonymized data on the donor and associated iPSCs. Data points are then standardized and displayed on the EBiSC public cell line catalog, giving users a comprehensive overview of each iPSC line.
Cell line nomenclature
Unique cell line names are essential to ensure full traceability and allow discrimination between iPSC lines from different donors and cohorts, particularly when sharing iPSCs across collaborating institutions (Luong et al., 2011). The hPSCreg database currently has >2,720 iPSC lines registered and provides a standardized and unique naming format to allow visual traceability between parental iPSC lines and associated sub-clones (Kurtz et al., 2018). Unique cell line identifiers are key to enable long-term traceability of a specific iPSC back to primary tissue samples, data archives, and associated publications. The use of unique hPSCreg nomenclature protects the privacy and confidentiality of tissue sample donors in compliance with applicable local regulations. HPSCreg identifications (IDs) also provide a solution that takes into account repeated reprogramming of the same donor material, as cells from different reprogramming procedures could yield lines with different properties due to unique genetic changes. This standard cell line nomenclature used by hPSCreg and endorsed by members of the stem cell community serves to alleviate ambiguity in stem cell line naming by providing a rule-based nomenclature.
Systems will also be required for unambiguous coding and tracking of donor samples, particularly when samples may be shared with other external researchers. To ensure a sample donor's privacy, any information relating to the donor should not be used for cell line naming, including donor initials, birth date, or genotype. Despite the ease of numbering, cell line identifiers should not be a series of commonly used abbreviations, such as hFB1, hFB2, etc., as such practices are prone to repetition by multiple research groups. For example, iPSCs from the same donor have already been deposited into EBiSC that were reprogrammed by multiple different organizations, and their common fibroblast origin was not always clearly recorded. In the absence of a single, widely adopted nomenclature system for non-clinical research, samples should be named consistently within a single organization as recommended by the International Society for Biological and Environmental Repositories (Campbell et al., 2018), with additional guidance available in Table 4. The critical factor is ensuring that internal records are unambiguous, accurate, maintained, and secure.
Data management
iPSCs for disease modeling and drug discovery have such potential due to the associated datasets describing the donor and iPSC line's background (Table 4). Both basic data (such as donor disease background or iPSC reprograming method) and high-level clinical data (e.g., age of disease onset) can be stored in hPSCreg and are then available to support other researchers. Special provision must be given to sensitive personal data, which can be used to identify an individual. In such cases, additional datasets, such as family medical history, detailed clinical information, medications, and genomics, should be shared via a secure managed-access data repository, such as the European Genome-Phenome Archive or the NIH-NCBI database of genotypes and phenotypes, to secure longevity of datasets collected and allow researcher access to these sensitive datasets by application to a data access committee (DAC) (Lappalainen et al., 2017). If access is granted by the DAC, the requesting researcher must honor the terms of data access and usage by signing a data access agreement (DAA). It is worth noting that any iPSC-linked consent use restrictions or other third-party obligations still apply. Terms of use may be jurisdiction-specific and should be clearly laid out in the DAA. For example, the GDPR has implications for data management and sharing in the EU (Morrison et al., 2017) and also between EU regions and other countries. Despite the stringent control of personal data, mechanisms must be available to allow researchers to use STR data for the purpose of cell line identity authentication. Such measures could be to share STR data through a DAA, or, alternatively, to enable cell line identity checking via a web-based tool that compares query STR profiles with a database of known STR profiles, without revealing reference STR profiles to unauthorized persons (Robin et al., 2020).
Conclusion
The importance of a research initiative planning for sustainability right from the start is hopefully clear from the above guidance. Ensuring that the ethical, legal, and qualification framework is appropriately established from the outset is critical to avoiding wasting resources on cell lines that ultimately cannot be shared. From EBiSC's experience not only in establishing EBiSC as a sustainable entity but also through extensive collaboration with other large and small iPSC research groups across Europe, a lack of planning from the outset can lead to extensive delays in sharing tools and data. Even if not practically intended immediately, it is recommended to establish processes with the mindset that all iPSCs must be broadly shareable, as the associated ethical, legal, and QC requirements that go along with that will ensure high-quality and robust resource outputs and reproducibility of scientific data, even if never distributed externally.
For fixed-term projects, such as a research initiative that is generating new iPSCs for use in disease modeling, three key questions should be addressed at project start: (1) where will iPSCs be stored after the end of the funding period; (2) who will own and maintain stocks and distribute them if other researches want access; and (3) how will genomic, phenotypic, and clinical datasets be stored so that they can be readily linked to the iPSCs in a research context? Making the most of publicly available tools such as hPSCreg will not only support keeping track of cell line details but also ensure that research outputs from publicly funded projects are findable and accessible after project end of life. Accurate and up-to-date cell line records ensure that, when iPSCs are shared with other users, the associated data needed (such as donor sex, disease diagnosis, and STR profile) are secure, readily available, and easily retrievable even in the event of staff changes. iPSC repositories such as EBiSC (with others available internationally) provide a secure home for iPSC lines and data and will control access to both through defined access agreements. These agreements may limit downstream use by including explicit prohibitions on attempts to identify sample donors and use in a commercial context. EBiSC can also support researchers in sustainability planning with advice available online (www.EBiSC.org) and through direct correspondence. Key is that standards outlined here are implemented in a coherent and multi-pronged manner, as each aspect is inherently linked with the others throughout the iPSC biobanking process. Figure 2 shows an exemplar cell processing pathway and highlights how each of the criteria discussed here (rows) affect each procedural step (columns). For example, robust data management procedures are required to record ethical and legal provenance to the starting material, which QC data are available (and thus under which biosafety level cells should be handled), and the location of stored vials. Through broad adherence to standards outlined here, we as a community can raise the quality and efficiency of iPSC-based research and thus accelerate mutual goals toward development of new therapeutics and improved understanding of healthy and diseased states.
Author contributions
Conceptualization, R.S. and S.C.M.; writing – original draft, R.S., S.C.M., N.M., B.H., and G.S.; writing – review & editing, R.S., S.C.M., N.M., B.H., B.S., A.C.S., G.S., P.D.S., and A.C.; supervision, H.Z.; project administration, R.S., S.C.M., and H.Z.; funding acquisition, H.Z. and S.C.M.
Declaration of interests
The authors declare no competing interests.
Acknowledgments
Many thanks to Dr. Timothy E. Allsopp, Dr. Andreas Ebneth, and Kevin Bruce for their broad input into development of EBiSC processes. Thank you to Dr. Ralf Kettenhofen for support with formalizing Figures 1 and 2. Figures were created with BioRender.com. EBiSC2 has received funding from the Innovative Medicines Initiative 2 Joint Undertaking (JU) under grant agreement no. 821362. The JU receives support from the EU's Horizon 2020 research and innovation program and EFPIA.
References
- Allsopp T.E., Ebneth A., Cabrera-Socorro A. Deploying human pluripotent stem cells to treat central nervous system disorders : facts, challenges and realising the potential. Stem Cell Res. 2019;41:101581. doi: 10.1016/j.scr.2019.101581. [DOI] [PubMed] [Google Scholar]
- Andrews P., Stacey G. International stem cell banking initiative (ISCBI) Regen. Med. 2015;10:1–44. doi: 10.2217/rme.14.93. [DOI] [Google Scholar]
- Andrews P.W., Baker D., Benvinisty N., Miranda B., Bruce K., Brüstle O., Choi M., Choi Y.M., Crook J.M., De Sousa P.A. Points to consider in the development of seed stocks of pluripotent stem cells for clinical applications: International Stem Cell Banking Initiative (ISCBI) Regen. Med. 2015 doi: 10.2217/rme.14.93. [DOI] [PubMed] [Google Scholar]
- Campbell L.D., Astrin J.J., Desouza Y., Giri J., Patel A.A., Rawley-Payne M., Rush A., Sieffert N. The 2018 revision of the ISBER best practices: summary of changes and the editorial. Biopreserv. Biobank. 2018;16:3–6. doi: 10.1089/bio.2018.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capes-Davis A., Freshney R.I. Wiley-Blackwell; 2021. Freshney’s Culture of Animal Cells: A Manual of Basic Technique and Specialized Applications, 8th Ed, Chapter 23. [Google Scholar]
- Czerwińska P., Mazurek S., Kołodziejczak I., Wiznerowicz M. Gene delivery methods and genome editing of human pluripotent stem cells. Rep. Pract. Oncol. Radiother. 2019;24:180–187. doi: 10.1016/j.rpor.2019.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sousa P.A., Steeg R., Wachter E., Bruce K., King J., Hoeve M., Khadun S., McConnachie G., Holder J., Kurtz A. Rapid establishment of the European Bank for induced Pluripotent Stem Cells (EBiSC) - the Hot Start experience. Stem Cell Res. 2017;20:105–114. doi: 10.1016/j.scr.2017.03.002. [DOI] [PubMed] [Google Scholar]
- Geraghty R.J., Capes-Davis A., Davis J.M., Downward J., Freshney R.I., Knezevic I., Lovell-Badge R., Masters J.R., Meredith J., Stacey G.N. Guidelines for the use of cell lines in biomedical research. Br. J. Cancer. 2014;111:1021–1046. doi: 10.1038/bjc.2014.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guhr A., Kobold S., Seltmann S., Seiler Wulczyn A.E.M., Kurtz A., Löser P. Recent trends in research with human pluripotent stem cells: impact of research and use of cell lines in experimental research and clinical trials. Stem Cell Rep. 2018;11:485–496. doi: 10.1016/j.stemcr.2018.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gymrek M., McGuire A., Golan D., Halperin E., Erlich Y. Identifying personal genomes by surname inference. Science. 2013;339:321–324. doi: 10.1126/science.1229566. [DOI] [PubMed] [Google Scholar]
- Huang C.Y., Liu C.L., Ting C.Y., Chiu Y.T., Cheng Y.C., Nicholson M.W., Hsieh P.C.H. Human iPSC banking: barriers and opportunities. J. Biomed. Sci. 2019;26:1–14. doi: 10.1186/s12929-019-0578-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isasi R., Andrews P.W., Baltz J.M., Bredenoord A.L., Burton P., Chiu I.-M., Hull S.C., Jung J.-W., Kurtz A., Lomax G.P. Identifiability and privacy in pluripotent stem cell research. Cell Stem Cell. 2014;14:427–430. doi: 10.1016/j.stem.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz A., Seltmann S., Bairoch A., Bittner M.S., Bruce K., Capes-Davis A., Clarke L., Crook J.M., Daheron L., Dewender J. A standard nomenclature for referencing and authentication of pluripotent stem cells. Stem Cell Rep. 2018;10:1–6. doi: 10.1016/j.stemcr.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen I., Almeida-king J., Kumanduri V., Senf A., Dylan J., Saunders G., Kandasamy J., Caccamo M., Barker J., Jokinen P. The European Genome-phenome Archive of human data consented for biomedical research. Nat. Genet. 2017;47:692–695. doi: 10.1038/ng.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Zhang Y., Zhang T. Computational approaches for effective CRISPR guide RNA design and evaluation. Comput. Struct. Biotechnol. J. 2020;18:35–44. doi: 10.1016/j.csbj.2019.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomax G.P., Hull S.C., Lowenthal J., Rao M., Isasi R. The DISCUSS Project: induced pluripotent stem cell lines from previously collected research biospecimens and informed consent: points to consider. Stem Cells Transl. Med. 2013:727–730. doi: 10.5966/sctm.2013-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong M.X., Auerbach J., Crook J.M., Daheron L., Hei D., Lomax G., Loring J.F., Ludwig T., Schlaeger T.M., Smith K.P. A call for standardized naming and reporting of human ESC and iPSC lines. Cell Stem Cell. 2011;8:357–359. doi: 10.1016/j.stem.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Morita Y., Okura H., Matsuyama A. Patent application trends of induced pluripotent stem cell technologies in the United States, Japanese, and European applications. Biores. Open Access. 2019;8:45–58. doi: 10.1089/biores.2018.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison M., Bell J., George C., Harmon S., Munsie M., Kaye J. The European general data protection regulation: challenges and considerations for iPSC researchers and biobanks. Regen. Med. 2017;12:693–703. doi: 10.2217/rme-2017-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea O., Steeg R., Chapman C., MacKintosh P., Stacey G.N. Development and implementation of large-scale quality control for the European Bank for induced Pluripotent Stem Cells. Stem Cell Res. 2020;45:101773. doi: 10.1016/j.scr.2020.101773. [DOI] [PubMed] [Google Scholar]
- Pamies D., Leist M., Coecke S., Bowe G., Allen D., Gstraunthaler G., Bal-Price A., Pistollato F., DeVries R., Hartung T. Good cell and tissue culture practice 2.0 (GCCP 2.0) - Draft for stakeholder discussion and call for action. ALTEX. 2020;37:490–492. doi: 10.14573/altex.2007091. [DOI] [PubMed] [Google Scholar]
- Rao M. IPSC crowdsourcing: a model for obtaining large panels of stem cell lines for screening. Cell Stem Cell. 2013;13:389–391. doi: 10.1016/j.stem.2013.09.005. [DOI] [PubMed] [Google Scholar]
- Robin T., Capes-Davis A., Bairoch A. CLASTR: the Cellosaurus STR similarity search tool - a precious help for cell line authentication. Int. J. Cancer. 2020;146:1299–1306. doi: 10.1002/ijc.32639. [DOI] [PubMed] [Google Scholar]
- Schmid B., Prehn K.R., Nimsanor N., Garcia B.I.A., Poulsen U., Jørring I., Rasmussen M.A., Clausen C., Mau-Holzmann U.A., Ramakrishna S. Generation of a set of isogenic, gene-edited iPSC lines homozygous for all main APOE variants and an APOE knock-out line. Stem Cell Res. 2019;34 doi: 10.1016/j.scr.2018.11.010. [DOI] [PubMed] [Google Scholar]
- Seltmann S., Lekschas F., Robert M., Stachelscheid H., Bittner M., Zhang W., Kidane L., Seriola A., Veiga A., Stacey G., Kurtz A. hPSCreg –– the Human Pluripotent Stem Cell Registry. Nucleic Acids Res. 2016;44:757–763. doi: 10.1093/nar/gkv963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey G.N. vol. 731. Springer Protocols; 2011. Chapter 7. Cell culture contamination; pp. 79–91. (Cancer Cell Culture Methods and Protocols). [DOI] [Google Scholar]
- Stacey G.N., Andrews P.W., Barbaric I., Boiers C., Chandra A., Cossu G., Csontos L., Frith T.J., Halliwell J.A., Hewitt Z. Stem cell culture conditions and stability: a joint workshop of the PluriMes consortium and pluripotent stem cell platform. Regen. Med. 2019;14:243–255. doi: 10.2217/rme-2019-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey G.N., Crook J.M., Hei D., Ludwig T. Banking human induced pluripotent stem cells: lessons learned from embryonic stem cells? Cell Stem Cell. 2013;13:385–388. doi: 10.1016/j.stem.2013.09.007. [DOI] [PubMed] [Google Scholar]
- Sullivan S., Stacey G.N., Akazawa C., Aoyama N., Baptista R., Bedford P., Bennaceur Griscelli A., Chandra A., Elwood N., Girard M. Quality control guidelines for clinical-grade human induced pluripotent stem cell lines. Regen. Med. 2018;13:859–866. doi: 10.2217/rme-2018-0095. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tessensohn J.A., Yamamoto S. Accelerated patent examination procedures spur Japanese university innovation. Nat. Biotechnol. 2009;27:815–818. doi: 10.1038/nbt0909-815. [DOI] [PubMed] [Google Scholar]
- Van Den Heuvel R., Den Hond E., Colles A., Nelen V., Van Campenhout K., Schoeters G. Biobank@VITO: biobanking the general population in Flanders. Front. Med. 2020;7:1–9. doi: 10.3389/fmed.2020.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volarevic V., Markovic B.S., Gazdic M., Volarevic A., Jovicic N. Ethical and safety issues of stem cell-based therapy. Int. J. Med. Sci. 2018;15:36–45. doi: 10.7150/ijms.21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisheit I., Kroeger J.A., Malik R., Klimmt J., Crusius D., Dannert A., Dichgans M., Paquet D. Detection of deleterious on-target effects after HDR-mediated CRISPR editing. Cell Rep. 2020;31:107689. doi: 10.1016/j.celrep.2020.107689. [DOI] [PubMed] [Google Scholar]
- Welch L.A., Gill P., Phillips C., Ansell R., Morling N., Parson W., Palo J.U., Bastisch I. European Network of Forensic science Institutes (ENFSI): evaluation of new commercial STR multiplexes that include the European Standard Set (ESS) of markers. Forensic Sci. Int. Genet. 2012;6:819–826. doi: 10.1016/j.fsigen.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Wilkinson M.D., Dumontier M., Aalbersberg Ij.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.W., da Silva Santos L.B., Bourne P.E. Comment: the FAIR Guiding Principles for scientific data management and stewardship. Sci. Data. 2016;3:1–9. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]