Summary
Hand1 and Hand2 are transcriptional factors, and knockout mice of these genes show left and right ventricular hypoplasia, respectively. However, their function and expression in human cardiogenesis are not well studied. To delineate their expressions and assess their functions in human cardiomyocytes (CMs) in vitro, we established two triple-reporter human induced pluripotent stem cell lines that express HAND1mCherry, HAND2EGFP and either MYH6-driven iRFP670 or tagBFP constitutively and investigated their expression dynamics during cardiac differentiation. On day 5 of the differentiation, HAND1 expression marked cardiac progenitor cells. We profiled the CM subpopulations on day 20 with RNA sequencing and found that mCherry+ CMs showed higher proliferative ability than mCherry− CMs and identified a gene network of LEF1, HAND1, and HAND2 to regulate proliferation in CMs. Finally, we identified CD105 as a surface marker of highly proliferative CMs.
Keywords: HAND1, HAND2, cardiomyocyte, development, pluripotent stem cell
Graphical abstract
Highlights
-
•
Expression of HAND1 marks cardiovascular progenitor cells
-
•
LEF1 is a key regulator of proliferating cardiomyocytes
-
•
CD105 expression marks highly proliferative cardiomyocytes
HAND1 and HAND2 are important cardiac transcriptional factors, but their functions in human cardiogenesis are difficult to analyze. Okubo et al. established HAND1mCherry and HAND2EGFP reporter induced pluripotent stem cells and showed that HAND1 marks cardiovascular progenitors and regulates the gene network of cardiomyocyte proliferation. They further showed that CD105 can be used as a surface marker for proliferative cardiomyocytes.
Introduction
The heart structure is initiated when lateral plate mesoderm (LPM) differentiates into cardiovascular progenitor cells (CPCs) and cardiomyocytes (CMs) (Garry and Olson, 2006; Wu et al., 2008). CPCs originate from two populations in the cardiac crescent, the first and second heart field (FHF and SHF, respectively), which mainly contribute to the left ventricle (LV) and the right ventricle (RV), atria, and outflow tract (OFT), respectively (Cai et al., 2003; Meilhac and Buckingham, 2018). Nevertheless, little is known about the mechanism that forms the heart structure during cardiogenesis.
Hand1 and Hand2 are related basic-helix-loop-helix transcriptional factors (TFs) and required for the morphological development of the heart in mice (Cserjesi et al., 1995; George and Firulli, 2019; Srivastava et al., 1995). Consistently, knockout of these genes causes severe hypoplasia of the heart dose dependently (McFadden et al., 2005). Hand1 and Hand2 are first expressed in LPM and cardiac crescent in mice (Cserjesi et al., 1995; de Soysa et al., 2019; Srivastava et al., 1997). From the heart tube stage, the expression of Hand1 is restricted to the LV and OFT regions in heart development (Barnes et al., 2010; de Soysa et al., 2019; Firulli et al., 1998; McFadden et al., 2005; Meilhac and Buckingham, 2018). Hand1 knockout is embryonic lethal in mice due to extraembryonic defects (Firulli et al., 1998; Riley et al., 1998), and mice with conditional deletions of Hand1 demonstrated defects in looping, poorly organized ventricular septa, and LV hypoplasia and died within 3 days after birth (McFadden et al., 2005). On the other hand, mice with overexpressed Hand1 showed disrupted heart morphogenesis with an elevated proliferation of cells and failed expansion of the LV in vivo (Risebro et al., 2006; Togi et al., 2004). By contrast, Hand2 knockout mice showed hypoplasia of the RV, a thinner myocardium in the ventricle, and embryonic lethality, suggesting that Hand2 is essential for SHF cells (Srivastava et al., 1997; Tsuchihashi et al., 2011). Recent single-cell RNA sequencing (RNA-seq) analysis showed that Hand2-deficient mice could give rise to RV cells but not OFT cells (de Soysa et al., 2019). These studies have suggested that Hand1 and Hand2 play critical roles from LPM to heart organogenesis, but the regulations and functions are hidden by the spatiotemporal complexity and heterogeneity of the heart. Additionally, the expression dynamics and functions of HAND1 and HAND2 in human cardiogenesis have hardly been investigated.
In humans, mutant HAND1 causes hypoplastic left heart syndrome, suggesting human HAND1 has a similar role to Hand1 (Reamon-Buettner et al., 2008, 2009). A single-cell RNA-seq study of human embryonic heart reported that the expression of HAND1 was enriched in ventricular CMs, especially in the LV at the early stage (5 weeks of gestation) (Cui et al., 2019). The same study also found that the expression of HAND2 was widely spread but higher in atrial CMs than in ventricular CMs. Although understanding the mechanism of human heart development is beneficial for the future development of cardiac regenerative medicine, detailed analysis of human cardiogenesis has been difficult due to the limited availability of human cell sources. Human induced pluripotent stem cells (hiPSCs) recapitulate many features of cardiac lineage specification, making them an attractive model in vitro to study human developmental mechanisms and different CM subpopulations (Burridge et al., 2012; Protze et al., 2019; Randolph and Lian, 2019).
In the present study, to investigate the expression dynamics and molecular functions of HAND1 and HAND2 in human in vitro CM differentiation, we established HAND1/HAND2 double-reporter hiPSCs to observe the expression dynamics and identify subpopulations during the cardiac differentiation process. By combining a third reporter fluorescent protein to mark MYH6-positive cardiac cells, we characterized HAND1+ and HAND1− subpopulations in the early stage and identified a new surface marker and gene regulatory network of proliferative CMs in the later stage.
Results
Establishing the HAND1mCherry, HAND2EGFP, and MYH6-iRFP670 triple-reporter hiPSC line
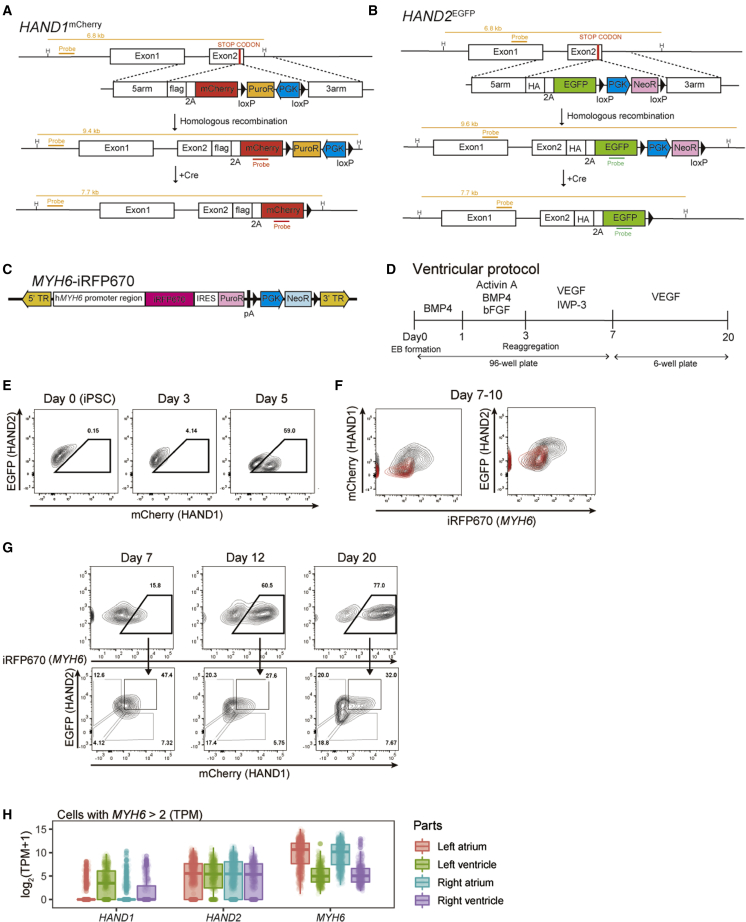
To delineate the expression of HAND1 and HAND2 noninvasively and to assess their functions in differentiating human CMs in vitro, we established HAND1mCherry, HAND2EGFP, and MYH6-iRFP670 triple-reporter hiPSCs. We targeted an allele of the HAND1 gene on chromosome 5 using the CRISPR-Cas9 system to insert mCherry and 2A self-cleaving peptide in front of the stop codon. Simultaneously, we introduced a single copy of the EGFP gene into the HAND2 gene on chromosome 4 to generate double-reporter HAND1mCherry, HAND2EGFP hiPSCs (Figures 1A and 1B and S1A–S1E) (Mali et al., 2013). In addition, we used the piggyBac vector system to induce a near-infrared fluorescent protein, iRFP670, under the control of the MYH6 promoter activity to monitor CMs to this double-reporter hiPSC line (Figures 1C, S1F, and S1G) (Funakoshi et al., 2016; Woltjen et al., 2009). This triple-reporter line was differentiated into CMs using the modified embryonic body (EB)-based protocol (Funakoshi et al., 2016; Yang et al., 2008) (Figure 1D). We observed the expression dynamics of these fluorescent proteins during differentiation by fluorescence-activated cell sorting (FACS) from day 1 of the differentiation to day 20 (Figures S2A–S2C). HAND1mCherry was first detected from day 3, and mCherry+ and mCherry− populations were observed from day 5 (Figures 1E and S2D). Later, EGFP expression began, and iRFP670+ CMs were induced from the EGFP and mCherry high population on day 7 (Figures 1, S2D, and S2E). On day 20, four subpopulations (mCherry− EGFP−, mCherry− EGFP+, mCherry+ EGFP−, and mCherry+ EGFP+) were observed in iRFP670+ CMs, although the signal for mCherry+ EGFP− CMs was low (Figures 1G and S2E) compared with widely expressed EGFP, but mCherry was localized (Figure S2E). An analysis of single-cell RNA-seq data of human developmental heart revealed that cells highly expressing HAND1 were enriched in the LV, but most cells expressed HAND2 (Figure 1H) (Cui et al., 2019). These findings suggested that the different expressions of HAND1 and HAND2 in our in vitro model reflect their expressions in the developing heart.
Figure 1.
HAND1mCherry, HAND2EGFP, and MYH6-iRFP670 triple-reporter hiPSC line
(A and B) Scheme of the establishment of the HAND1 and HAND2 double-reporter line with mCherry and EGFP, respectively, using the CRISPR-Cas9 system and removal of the selection cassettes using the Cre/loxP system. 5arm, 5′ homologous arm; 3arm, 3′ homologous arm; 2A, 2A peptide; HA, HA-tag; flag, FLAG tag; PGK, promoter sequence of phosphoglycerate-kinase 1; PuroR, puromycin resistance gene; NeoR, neomycin resistance gene. HindⅢ (H) was used to digest genomic DNA for Southern blotting. Orange lines indicate external and internal probes for Southern blotting with expected band sizes.
(C) Construction of the MYH6-iRFP670 reporter with the piggyBac transposon system.
(D) Scheme of the ventricular CM differentiation protocol.
(E) Representative FACS plots of the expression dynamics of mCherry and EGFP on days 0, 3, and 5.
(F) Representative FACS plots of the expressions of mCherry and EGFP with iRFP670 of the triple reporter (black) and parental 409B2 (red) on day 7 of the differentiation.
(G) Representative FACS plots of subpopulations based on the expressions of mCherry and EGFP in iRFP670+ CMs on days 7, 12, and 20.
(H) Box plots of the expressions of HAND1, HAND2, and MYH6 in LV, RV, left atrium, right atrium of the developing heart. Cells that showed MYH6 > 2 were used for the single-cell RNA-seq data in GSE106118.
HAND1 expression marks CPCs in the early stage in vitro
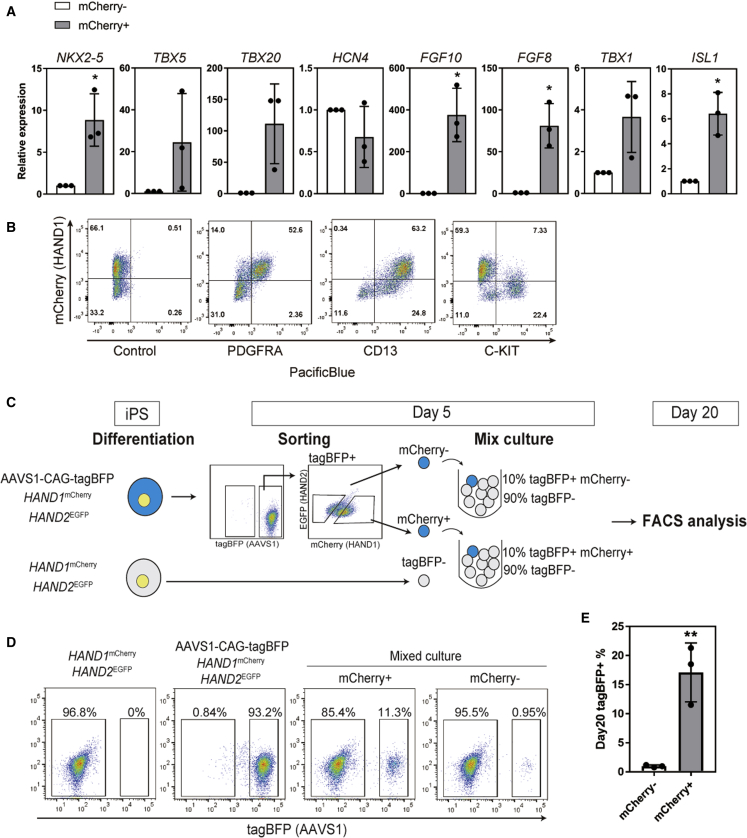
We investigated the mCherry+ and mCherry− populations on day 5 separately. mCherry+ cells showed higher expression levels of cardiac genes for FHF (NKX2-5, TBX5, TBX20, and HCN4) and SHF (TBX1, ISL1, FGF10, and FGF8) than mCherry− cells with one exception, HCN4 (Figure 2A) (Andersen et al., 2018; Bruneau et al., 1999; Cai et al., 2003; Meilhac and Buckingham, 2018; Singh et al., 2005). In addition, the expressions of two CPC markers, PDGFRA and CD13, were consistent with the expression of mCherry. C-KIT, a surface marker of the earliest hematopoietic and vascular progenitors, was expressed in mCherry− cells (Figure 2B) (Kattman et al., 2011; Skelton et al., 2016; Yang et al., 2008). These results suggest that mCherry+ cells on day 5 are CPCs.
Figure 2.
HAND1+ cells on day 5 contribute to the CPC population
(A) Gene expressions of the cardiac TFs NKX2-5, TBX5, TBX20, HCN4, FGF10, FGF8, TBX1, and ISL1 in mCherry+ cells relative to mCherry− cells isolated on day 5 (n = 3 independent experiments). ∗p < 0.05 by Welch's t test.
(B) Representative FACS plots of mCherry expression with antibodies for PDGFRA, CD13, and C-KIT on day 5.
(C) Scheme of the mixed coculture system for tracing mCherry+ and mCherry− cells in EBs. To label the cells, CAG promoter-driven tagBFP was knocked into the AAVS1 locus of HAND1mCherryHAND2EGFP double-reporter hiPSCs (AAVS1-CAG-tagBFP). The tagBFP-labeled (tagBFP+) mCherry− and mCherry+ cells were isolated and mixed with non-labeled parental double-reporter cells (tagBFP−) on day 5. On day 20, the mixed cultures were analyzed by FACS.
(D) Representative FACS plots of the tagBFP expression on day 20.
(E) Percentages of tagBFP+ cells on day 20 in EBs mixed with tagBFP+ mCherry+ cells and tagBFP− cells or with tagBFP+ mCherry− cells and tagBFP− cells on day 5 (n = 3 independent experiments). ∗∗p < 0.01 by unpaired t test. Data represent means ± SD.
To determine if mCherry− cells had cardiac lineage fate potential, we tried to culture day 5 mCherry+ and mCherry− cell populations; however, isolated mCherry− cells did not reaggregate or reproduce EBs. Therefore, we established a mixed coculture system to trace mCherry− cells on day 5 by labeling the HAND1mCherry and HAND2EGFP double-reporter cell line with constitutively expressing tagBFP (Figures 2C and S3). tagBFP-labeled and non-labeled double-reporter iPSCs were differentiated into CMs simultaneously (Figure 2C). On day 5, mCherry+ and mCherry− cells were isolated from the tagBFP+ population. We then mixed the isolated tagBFP-labeled mCherry+ and mCherry− cells separately with the non-labeled parental double-reporter cells (tagBFP−) at a 1:9 ratio and continued the differentiation process until day 20. On day 20, the percentage of tagBFP+ cells in the mixed cocultures was analyzed by FACS. The percentage of tagBFP+ mCherry+ cells in the mixed culture was over 10%, but the percentage of tagBFP+ mCherry− cells was less than 2% (Figures 2D and 2E). These results indicate that mCherry− cells did not differentiate into cardiac cells after day 5. Taken together, the expression of HAND1 at the early stage marks CPCs derived from hiPSCs.
HAND1 is upregulated by BMP4 at the early and late stages
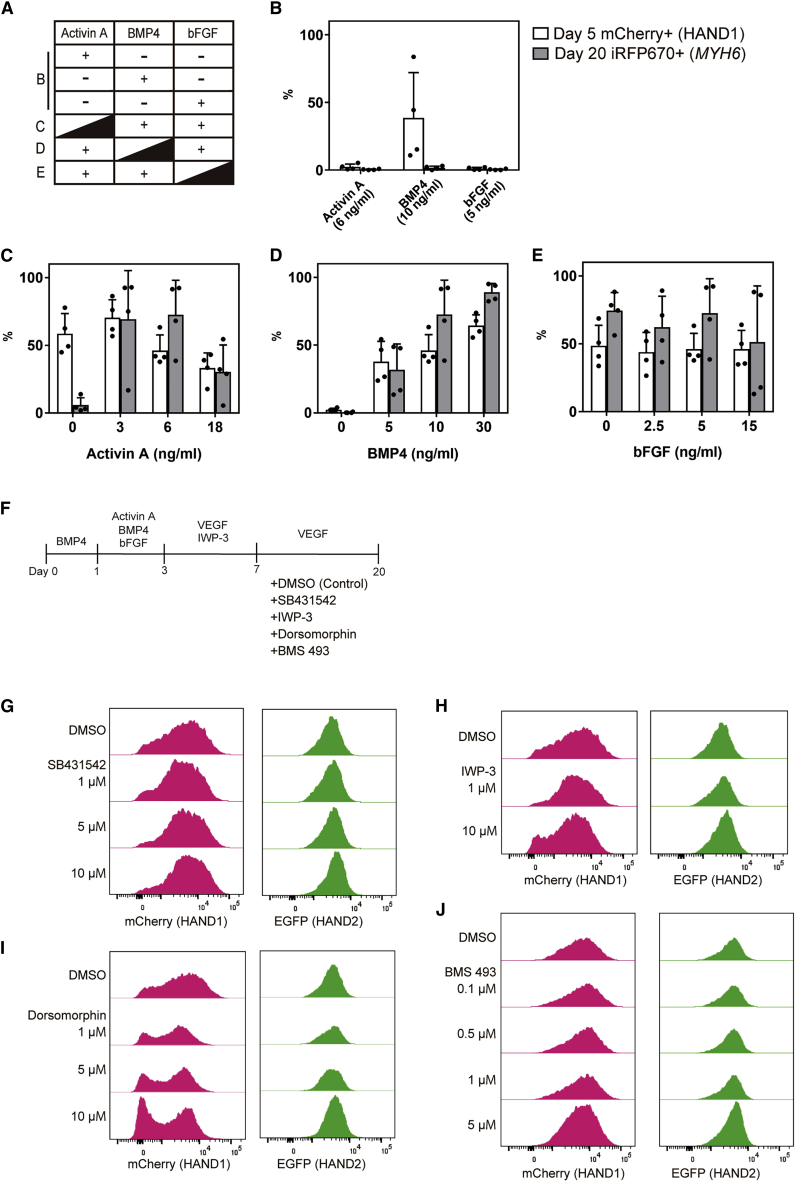
To examine the regulation of HAND1 and HAND2 expressions, we manipulated the concentrations of cytokines in the differentiation protocol. For the early stage, we manipulated Activin A, bone morphogenetic protein 4 (BMP4), and basic fibroblast growth factor (bFGF) in day 0 and day 1 media and analyzed the percentages of mCherry+ cells on day 5 and iRFP670+ CMs on day 20 (Figures 3A, S4A, and S4B). BMP4 induced mCherry+ cells, but neither Activin A nor bFGF did (Figure 3B). Increasing the Activin A concentration led to fewer mCherry+ and iRFP670+ cells, but 0 ng/mL Activin A generated mCherry+ cells and no iRFP670+ CMs (Figure 3C). In contrast, increasing the BMP4 concentration promoted the number of mCherry+ and iRFP670+ cells (Figure 3D). Finally, increasing the bFGF concentration had no significant effect on the number of mCherry+ or iRFP670+ cells (Figure 3E). In these conditions, we did not observe differences in the distributions of HAND1 and HAND2 of day 20 CMs. These results indicate that the expression of HAND1 at the early stage is upregulated by BMP4 and downregulated by Activin A. In addition, they suggest that the expression of HAND1 is necessary but not sufficient to specify hiPSCs into CPCs.
Figure 3.
Effects of cytokines on HAND1 and HAND2 expression in ventricular and atrial differentiation
(A) Scheme of the cytokines in day 0–1 differentiation medium used for the experiments in (B)–(E).
(B) Percentages of mCherry+ cells on day 5 and iRFP670+ cells on day 20 with only Activin A, BMP4 or bFGF in day 0–1 medium (n = 4 independent experiments).
(C–E) Percentages of mCherry+ cells on day 5 and iRFP670+ CMs on day 20 cultured with various concentrations of Activin A (C), BMP4 (D), and bFGF (E) in day 0–1 medium (n = 4 independent experiments). All other cytokine concentrations followed the ventricular protocol. Data represent means ± SD.
(F) Scheme of the differentiation protocol for the experiments in (G)–(J).
(G–J) Representative histogram of mCherry and EGFP expressions on day 20 in iRFP670+ cells cultured with SB431542 (G), IWP-3 (H), dorsomorphin (I), and BMS 493 (J).
We also manipulated the concentrations of vascular endothelial growth factor (VEGF) and IWP-3, a WNT signal inhibitor, in day 3 medium and VEGF and bFGF in maintenance medium after day 7 (Figures S4F and S4G). No effects were found on the mCherry expression on day 5 or the subpopulation distribution on day 20.
To further investigate the role of the signals induced by cytokines, we used several chemical inhibitors of cytokine signals, including SB431542 (TGFβ signal inhibitor), IWP-3, dorsomorphin (BMP signal inhibitor), and BMS 493 (retinoic acid [RA] signal inhibitor), from day 7 until day 20 in the cardiac differentiation (Figure 3F). The expression of mCherry decreased with the administration of IWP-3 and especially with dorsomorphin (Figures 3G–3J and S4H–S4K). Therefore, HAND1 is regulated by the BMP signal at the early stage and late stage. In contrast, we did not find a regulator for HAND2 expression.
Expressions of HAND1 and HAND2 are down- and upregulated in the atrial induction protocol, respectively
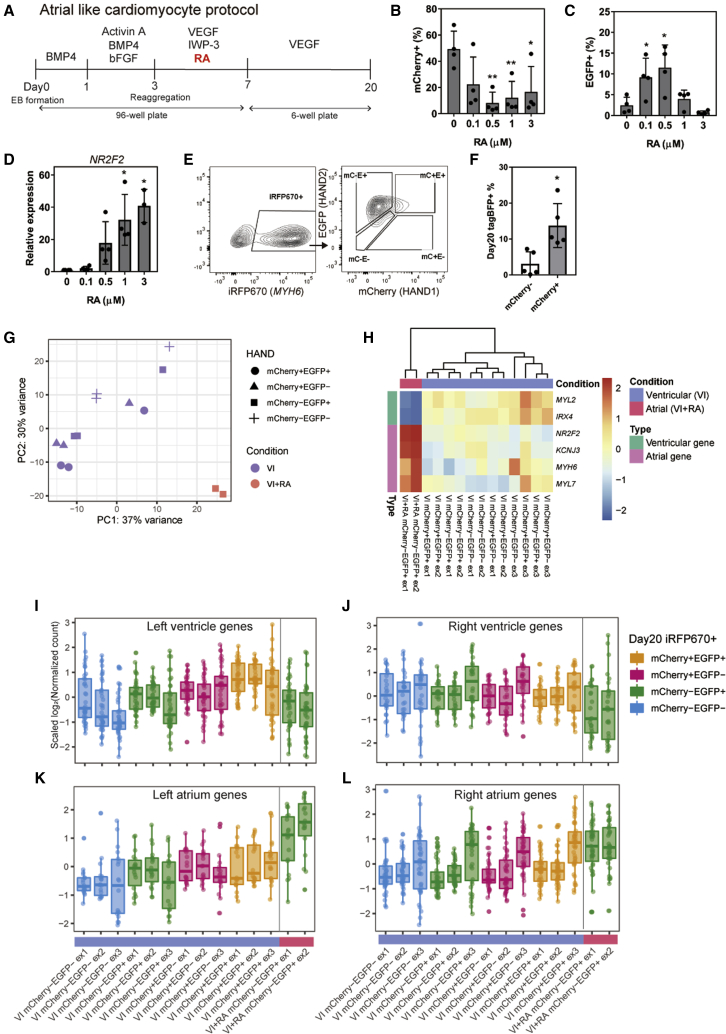
HAND2 is expressed widely in the developing heart of both species, but it is especially high in human atrial CMs (Cui et al., 2019). Mouse atrial CMs are generated from the SHF by RA signaling in vivo (de Soysa et al., 2019; Hochgreb et al., 2003; Rochais et al., 2009). Some studies have reported that human atrial CMs are generated from human pluripotent stem cells by RA in vitro (Devalla et al., 2015; Lee et al., 2017). Since CMs induced with our protocol are mainly ventricular CMs (Takaki et al., 2020), to promote atrial CM differentiation, we administered RA to the triple-reporter hiPSCs on day 3 (Figure 4A). In this differentiation condition, the relationship between HAND2 and SHF during human in vitro cardiogenesis was investigated. The addition of 0.5 μM RA resulted in more EGFP+ cells and fewer mCherry+ cells in association with the upregulation of NR2F2, an atrial marker gene, on day 7 (Figures 4B–4D). On day 20, the majority of iRFP670+ CMs induced by RA addition was mCherry− EGFP+ (Figures 4E, S5A, and S5B). The results showed that HAND1 and HAND2 are down- and upregulated by RA, respectively, in atrial CM differentiation. We also confirmed the expression of HAND1 on day 5 of the atrial protocol marks CPCs using the coculture tracing system shown in Figure 2C: mCherry+ cells were maintained in the atrial protocol, but mCherry− cells were not, consistent with the ventricular protocol, suggesting that the expression of HAND1 on day 5 marks CPCs in both conditions (Figure 4F).
Figure 4.
Retinoic acid in the atrial protocol upregulated HAND2 and downregulated HAND1
(A) Scheme of the RA-modified atrial CM differentiation protocol.
(B and C) Percentages of mCherry+ cells (B) and EGFP+ cells (C) on day 7 at different RA concentrations (n = 4 independent experiments). ∗p < 0.05, ∗∗p < 0.01 by one-way ANOVA with Dunnett's test comparing with 0 μM.
(D) Gene expression level of NR2F2, an atrial marker gene, on day 7 at different RA concentrations. (n = 3–4 independent experiments). ∗p < 0.05, ∗∗p < 0.01 by one-way ANOVA with Dunnett's test comparing with 0 μM.
(E) Representative FACS plots of iRFP670, mCherry (mC), and EGFP (E) expressions on day 20 (n = 5 independent experiments).
(F) Percentages of tagBFP+ cells on day 20. EBs were mixed with tagBFP+ mCherry+ cells and tagBFP− cells or with tagBFP+ mCherry− cells and tagBFP− cells on day 5 (n = 3 independent experiments). ∗p < 0.05 by unpaired t test. Data represent means ± SD.
(G) Principal component analysis plot of ventricular CMs (VEGF + IWP-3 on day 3; VI) and atrial CMs (VEGF + IWP-3 + RA on day 3; VI + RA) (n = 3 independent experiments).
(H) Heatmap of scaled expression levels of MYL7, MYH6, KCNJ3, and NR2F2 (atrial marker genes) and MYL2 and IRX4 (ventricular marker genes) from the RNA-seq data of day 20 CMs.
(I–L) Box plots of the scaled log2 normalized counts of chamber-specific genes in each subpopulation. The upper and lower quartiles are indicated by the boxes, and the median by the lines within each box.
To profile each subpopulation, we analyzed the RNA-seq data of day 20 iRFP670+ CMs in the four subpopulations from the ventricular induction protocol and the mCherry− EGFP+ population from the RA-modified (atrial) induction protocol (Figure 4G). The CMs generated from the atrial protocol showed a higher expression of atrial genes and lower expression of ventricular genes compared with all subpopulations from the ventricular protocol (Figure 4H), indicating mCherry− EGFP+ CMs prepared from the atrial protocol were atrial CMs. Because HAND1 is widely expressed in the ventricle, although its expression is higher in the LV than RV (Figure 1H), we investigated if HAND1− (mCherry−) CMs of the ventricular protocol are RV-like CMs. No clear RV-specific expression patterns were observed in mCherry− CMs from the ventricular protocol (Figures 4I–4L). On the other hand, we found mCherry− EGFP+ atrial CMs had higher expressions of atrial genes, and mCherry+ EGFP+ CMs tended to have higher expressions of LV-specific genes.
Next we investigated the expression profiles of CMs induced in the monolayer culture. When iRFP670+ CMs were differentiated from day 5, the percentage of the mCherry− EGFP+ population was higher in EB-based CMs (Figures S5C–S5E), suggesting that monolayer CMs were a heterogeneous population of atrial and LV/RV CMs.
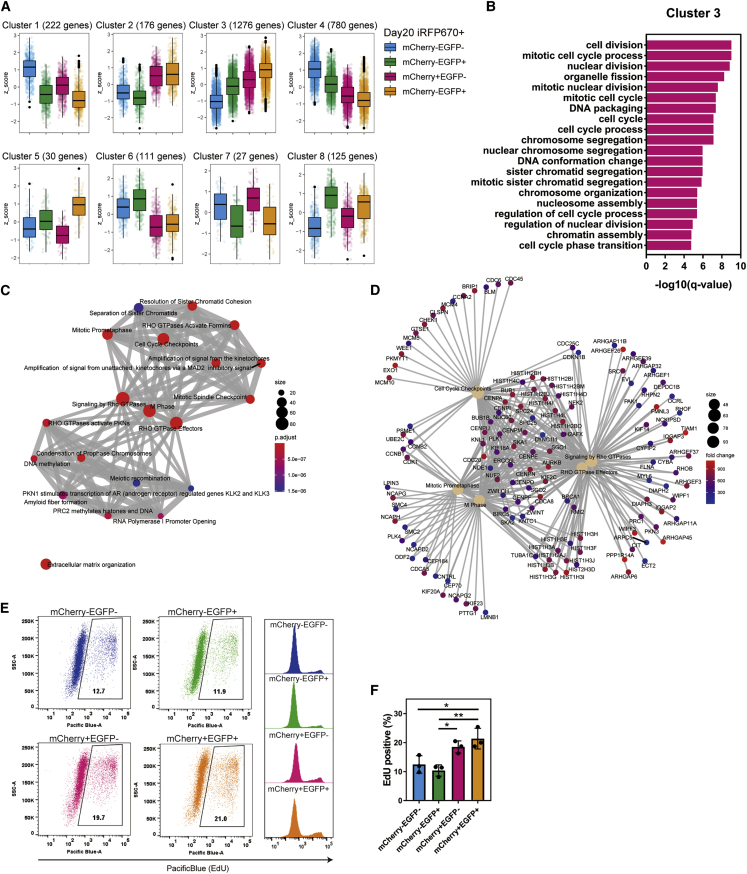
Profiling the four subpopulations of ventricular CMs
To characterize each ventricular subpopulation, we extracted differentially expressed genes and clustered them (Table S1). We found two major clusters, cluster 3 (1,276 genes) and cluster 4 (780 genes), which were highly expressed in mCherry+ CMs and mCherry− CMs, respectively (Figure 5A). A gene ontology (GO) analysis showed enriched GO terms for the cell cycle in cluster 3 (Figure 5B). Consistently, an enrichment analysis of Reactome pathways found cell cycle-related pathways were highly enriched in cluster 3 (Figures 5C and 5D and Table S2). Thus, we examined the 5-ethynil-2'-deoxyuridine (EdU) positive ratio of each subpopulation isolated from iRFP670+ CMs on day 20, finding that mCherry+ CMs showed significantly higher percentages of EdU+ cells than mCherry− CMs (Figures 5E and 5F). Previous studies indicated that CMs proliferate less in the mature heart during cardiogenesis in vivo and in vitro (Funakoshi et al., 2016; Takeuchi, 2014). Therefore, we investigated the maturation levels of the CM subpopulations by staining for α-actinin to observe the sarcomere structure and compared the expression levels of two immaturity-related genes, MYH6 and TNNI1, and two maturation marker genes, MYH7 and TNNI3, but found no clear difference between the four cell populations (Figures S5F and S5G) (Cui et al., 2019; Friedman et al., 2018). These results suggest that the expression of HAND1 is correlated with the proliferation ability of CMs but not with the maturation level.
Figure 5.
Characterization of subpopulations isolated from iRFP670+ CMs on day 20
(A) Genes that showed a significant expression difference between the four subpopulations based on the likelihood ratio test (adjustment p value <0.05) were clustered into eight clusters. Box plots of the scaled values (Z score) show the upper and lower quartiles by boxes and the median as a line within each box.
(B) Top 20 enriched GO terms in biological process for the genes in cluster 3.
(C) Enrichment map of the Reactome pathway from the gene set enrichment analysis using the genes in cluster 3.
(D) Network plot of the most significantly enriched terms with the genes in cluster 3.
(E) Representative FACS plot of EdU assays.
(F) Percentages of EdU+ cells in each subpopulation isolated on day 20 (n = 3 independent experiments). ∗p < 0.05, ∗∗p < 0.01 by one-way ANOVA with Tukey's multiple comparisons test. Data represent means ± SD.
Regulatory network of CM proliferation
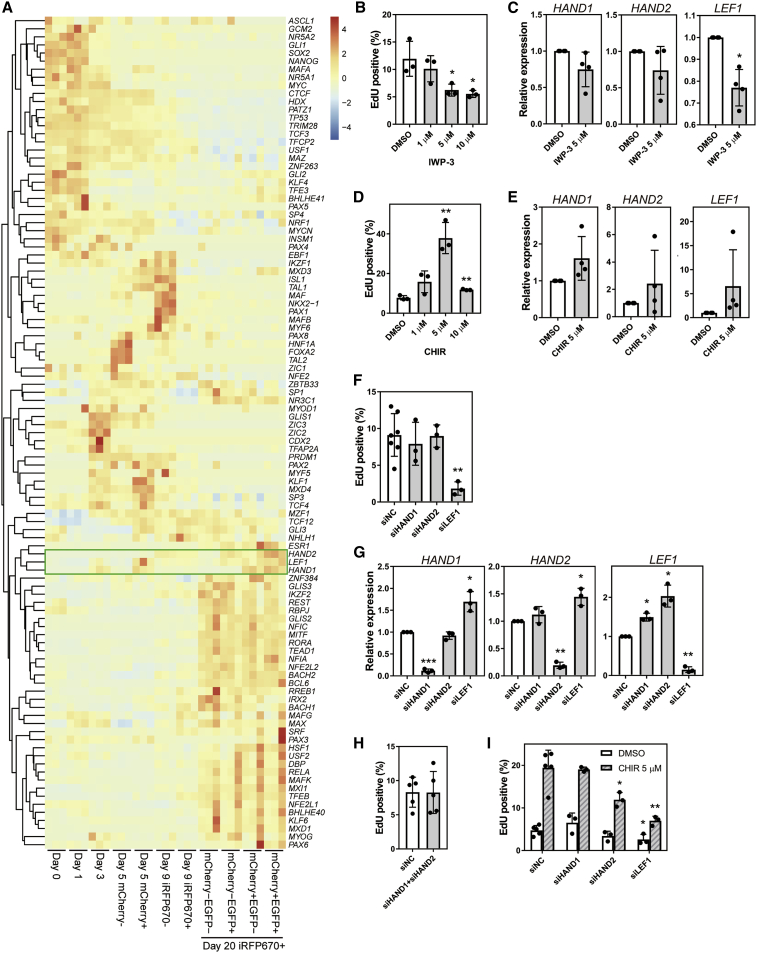
To understand the molecular mechanism determining highly and lowly proliferative CMs, we performed an upstream analysis of cluster three genes, which represent highly expressed genes in the mCherry+ population, using the geneXplain platform (Kel et al., 2006; Koschmann et al., 2015). As a result, a total of 103 TFs were predicted as upstream factors. Within these 103 TFs, LEF1, whose expression was significantly higher in mCherry+ EGFP+ CMs than mCherry− CMs, belongs to the same cluster as HAND1, indicating it has the most similar expression to HAND1 during differentiation (day 0 to day 20) (Figures 6A and S6A). LEF1 is a factor in the WNT signaling pathway and activates its target genes by cooperating with β-catenin (Clevers, 2006). Also in the enrichment analysis using the Reactome pathway, the WNT-related pathways “TCF dependent signaling in response to WNT” and “Signaling by WNT” were significantly enriched (Table S2) (Jassal et al., 2020). An analysis of RNA-seq data found that WNT signaling genes showed stage-specific expressions, including LEF1 expression, which increased from day 5 (Figure S6B). Taken together, the upstream analysis of differentially expressed genes between HAND1+/− populations highlight the importance of WNT signaling in CM proliferation.
Figure 6.
WNT signaling and proliferation in CMs
(A) Heatmap and clustering of scaled expression levels of 103 TFs predicted as upstream factors from the RNA-seq data of days 0, 3, 5 (isolated mCherry− and mCherry+ populations), day 9 (isolated iRFP670− and iRFP670+ populations), and day 20 subpopulations in iRFP670+ CMs (n = 3 independent experiments). Green box highlights HAND1, LEF1, and HAND2.
(B) Percentage of EdU+ cells on day 20 among EGFP+ CMs isolated on day 15 following WNT inhibitor (IWP-3) administration on days 16–18 (n = 3 independent experiments). ∗p < 0.05 by one-way ANOVA with Dunnett's test comparing with DMSO.
(C) Real-time qPCR results for HAND1, HAND2, and LEF1 expression under IWP-3 treatment (n = 4 independent experiments). ∗p < 0.05, by Welch's t test.
(D) Percentage of EdU+ cells on day 20 among EGFP+ CMs isolated on day 15 following GSK3 inhibitor CHIR99021 (CHIR) administration on days 16–18 (n = 3 independent experiments). ∗∗∗p < 0.001 by one-way ANOVA with Dunnett's test comparing with DMSO.
(E) Real-time qPCR results of HAND1, HAND2, and LEF1 expression under CHIR treatment (n = 4 independent experiments).
(F) Percentage of EdU+ cells after treatment with siRNAs for negative control (siNC), HAND1 (siHAND1), HAND2 (siHAND2), and LEF1 (siLEF1) (n = 3–7 independent experiments). ∗∗p < 0.01 by one-way ANOVA with Dunnett's test comparing with siNC.
(G) Real-time qPCR results for HAND1, HAND2, and LEF1 expression after siRNA treatment on day 20. EGFP+ CMs isolated on day 15 (n = 3 independent experiments). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 by Welch's t test.
(H) Percentage of EdU+ cells with simultaneous knockdown of HAND1 and HAND2 (n = 5 independent experiments).
(I) Percentage of EdU+ cells after CHIR and siRNA combination treatment (n = 3–5 independent experiments). ∗p < 0.05, ∗∗∗p < 0.001 by one-way ANOVA with Dunnett's test comparing with siNC. Data represent means ± SD.
We therefore hypothesized that HAND1, HAND2, and LEF1 control the cell cycle in CMs via WNT signaling. To test this hypothesis, a single CM reporter hiPSC line, 201B7 MYH6-EGFP, was differentiated and isolated on day 15 (Funakoshi et al., 2016). The CMs were then seeded and cultured in a dish until day 20 to investigate the direct effects of IWP-3 and CHIR99021 (CHIR), a GSK-3 inhibitor that induces lower β-catenin phosphorylation levels and enhances WNT signaling, from days 16 to 18. The administration of IWP-3 lowered the EdU+ ratio as well as the expression levels of LEF1 (Figures 6B and 6C). Conversely, CHIR administration upregulated the EdU+ ratio and tended to increase the expression levels of HAND1, HAND2, and LEF1 (Figures 6D and 6E).
To test the direct effects of HAND1, HAND2, and LEF1 on the cell cycle in CMs, we used small interfering RNAs (siRNAs) to knock down their expressions. MYH6-EGFP+ CMs were purified by FACS and transfected with siRNAs on day 15. The EdU+ ratio and expressions of HAND1, HAND2, and LEF1 were measured on day 20 by real-time quantitative PCR (qPCR). The knockdown of LEF1 caused a significant reduction in the EdU+ ratio, but the knockdown experiments of HAND1 or HAND2 did not (Figures 6F, 6G, S6C and S6D). We also performed an RNA-seq analysis of the three knockdowns to elucidate the effects of WNT signaling genes (Figure S6E). The knockdown of HAND1 or HAND2 did not change the expressions of WNT signaling genes. On the other hand, CTNNB1, DKK3, and GSK3B were downregulated by LEF1 knockdown, as was LEF1 itself (Figure S6E). Thus, only the knockdown of LEF1 affected the expression levels of WNT signaling factors.
It was reported that the hypoplasia in Hand1- and Hand2-deficient mice is dose dependent (McFadden et al., 2005). Therefore, we next suppressed the expressions of HAND1 and HAND2 simultaneously but again found no change in the EdU+ ratio (Figure 6H). Finally, we found that the knockdown of HAND2 caused a significant reduction in the EdU+ ratio if WNT signaling was activated by CHIR, suggesting a proliferation role of HAND2 (Figure 6I). However, the knockdown of HAND1 did not show this effect. This difference may be attributable to the higher frequency of HAND2+ CMs compared with HAND1+ CMs.
Interestingly, the HAND1 and HAND2 knockdown experiments significantly enhanced the expression of LEF1, while conversely the LEF1 knockdown significantly increased the expressions of HAND1 and HAND2 (Figure 6G). These observations suggested that HAND1 and HAND2 regulate LEF1 expression and that LEF1 regulates HAND1 and HAND2 expression. We checked these direct regulations using published chromatin immunoprecipitation sequencing (ChIP-seq) data (Table S3) (Boeva et al., 2017; Consortium, 2012; Durbin et al., 2018; Hemming et al., 2018; Tsankov et al., 2015). We found HAND1 and HAND2 bind upstream of the LEF1 locus without active histone mark (H3K27Ac), especially in human cell lines, including GIST-T1, BE2C, CLB-Ga, K562, and HEK293, and human embryonic stem cell (ESC)-derived mesodermal cells on day 5, suggesting HAND1 and HAND2 directly downregulate LEF1 expression by binding to the upstream region (Figure S6F). The ChIP-seq data also indicated that HAND1, HAND2, and LEF1 directly bind to the loci of CCND1 and CCND2, two cell cycle regulators. The overexpression of CCND1 induces human CMs to proliferate (Mohamed et al., 2018). Furthermore, the data showed that LEF1 binds to the upstream loci of HAND1 and HAND2. Overall, these data suggest that a gene regulatory network for the proliferation of CMs involves the binding of HAND1, HAND2, and LEF1 to each other's loci.
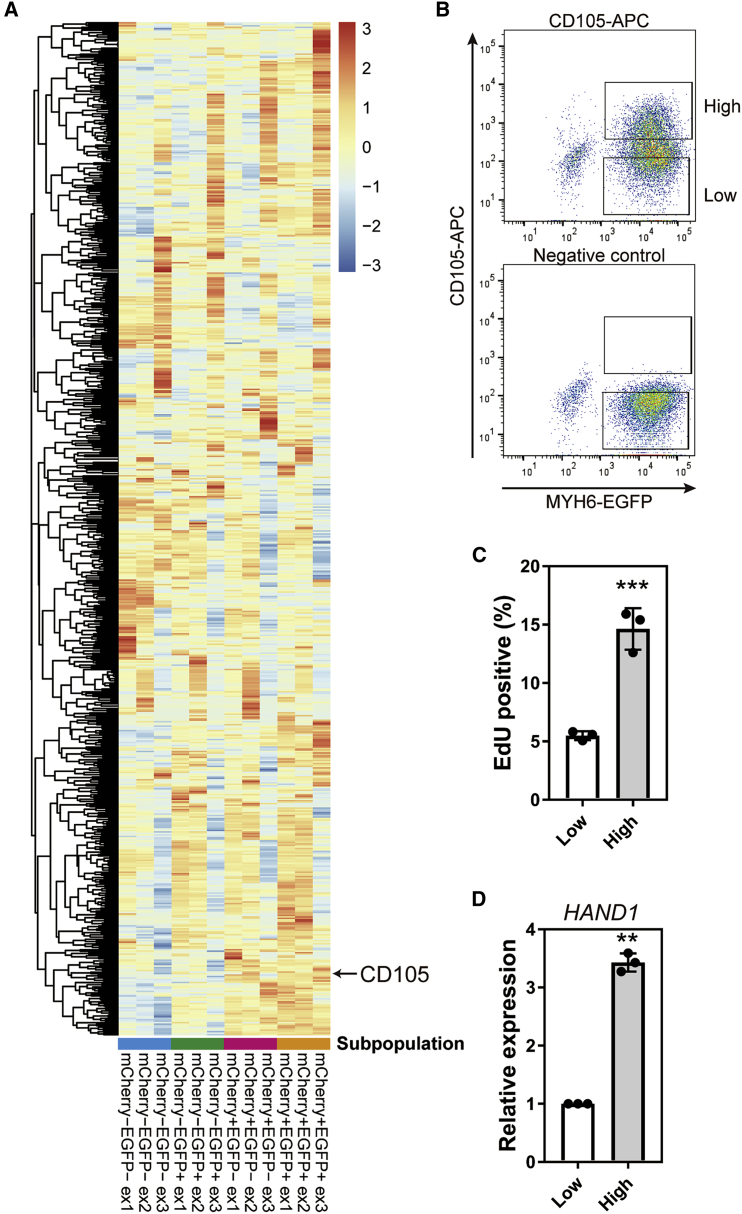
CD105 is a marker of highly proliferative CMs
To further investigate the relationship between CM proliferation and cardiac TFs in CMs, we focused on marker proteins for proliferative CMs. We picked up cell surface genes among the differentially expressed genes from our RNA-seq data in the CM subpopulations. CD105 (also known as endoglin, encoded by ENG) showed a higher expression in mCherry+ CMs (Figure 7A). We sorted CD105-high (top 30%) and CD105-low (bottom 30%) populations from EGFP+ CMs on day 20 (Figure 7B). The EdU+ ratio and HAND1 expression level of CD105-high CMs were higher than those of CD105-low CMs (Figures 7C and 7D). We also tested the utility of CD105 to sort lineage (CD31, CD49a, CD140b, CD90)-negative and signal regulatory protein alpha (SIRPA)-positive CMs derived from 692D2 (on-feeder hiPSC line) and 1390D4 (feeder-free hiPSC line). We found a high EdU ratio in the CD105+ population, which also showed a high expression level of HAND1 (Figures S7A–S7D). CD105 is a TGFβ receptor (Arthur et al., 2000). Therefore, we investigated whether TGFβ signaling is associated with cell cycle activity by adding SB431542 on days 16–18 to the isolated CMs. The inhibitor caused no change in the EdU+ ratio (Figures S7E and S7F), suggesting that TGFβ signaling is not involved in cell cycle regulation.
Figure 7.
Identification of CD105 as a surface marker of proliferative CMs
(A) Heatmap of the expression level of cell surface (GO: 0009986) genes in each subpopulation of CMs on day 20 by RNA-seq data.
(B) Representative FACS plots of MYH6-EGFP reporter hiPSCs (201B7) using CD105-APC antibody on day 20. CD105-high and CD105-low populations were isolated from the upper 30% and lower 30% of EGFP+ CMs, respectively.
(C) Percentages of EdU+ cells on day 23 in EGFP+ and CD105-APC-high or -low CMs isolated on day 20 (n = 3 independent experiments). ∗∗∗p < 0.001 by unpaired t test.
(D) Expression level of HAND1 in CD105-high and -low CMs isolated on day 20 (n = 3 independent experiments). ∗∗p < 0.01 by Welch's t test. Data represent means ± SD.
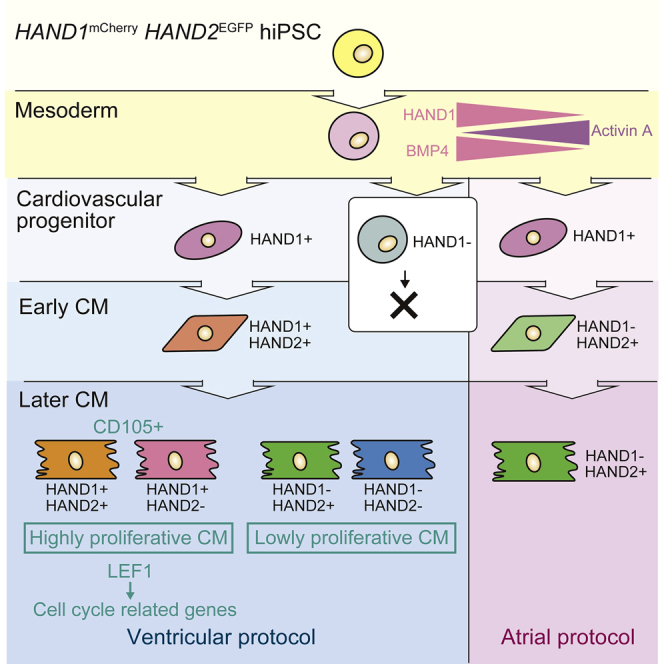
Taken together, our in vitro differentiation model of the expression dynamics of HAND1 and HAND2 (Figure S7G) highlights a cell cycle regulation system involving LEF1, HAND1, and HAND2 and revealed CD105 as a marker of proliferative CMs.
Discussion
HiPSC reporter systems are useful tools for assessing the developmental mechanisms of cardiogenesis. These systems can clarify the role of cardiac TFs by tracing and profiling cell subpopulations. Previously, studies using Tbx1/Hcn4 and TBX5/NKX2-5 double-reporter iPSC lines investigated the dynamics of these genes and subpopulations (Andersen et al., 2018; Zhang et al., 2019). However, the expression dynamics of HAND1 and HAND2 in human cardiogenesis still remain to be clarified. In this study, we established a HAND1/HAND2 double-reporter hiPSC line and investigated the expression dynamics of the two TFs during cardiac differentiation for the first time.
According to a previous study investigating gene expressions, heart cell types develop differently between human and mouse in vivo (Cui et al., 2019). In mice, the expressions of both Hand1 and Hand2 begin from LPM, and there is little spatial overlap between the two in the heart tube (Cserjesi et al., 1995; de Soysa et al., 2019; Srivastava et al., 1997). On the other hand, we observed that the expression of HAND1-mCheery in hiPSCs started from day 3 of the differentiation, which is the mesodermal stage, and the expression of HAND2-EGFP started between days 5 and 7, demonstrating the difference between human in vitro and mouse in vivo differentiation.
According to the gene expression profiles of ventricular CMs, our reporter did not show evidence of RV CM differentiation. One reason could be that the HAND1− CM subpopulation on day 20 is heterogeneous. Another possible reason is that our ventricular protocol did not produce RV CMs. Protze et al. (2019) hypothesized that RV CMs are generated from a low Activin A concentration, but experimental confirmation is still awaited. Thus, more study on the different cellular properties of LV and RV CMs is needed in order to produce RV CMs in vitro.
Previous reports have shown that CHIR via canonical WNT signaling can enhance the proliferation of CMs derived from hiPSCs (Buikema et al., 2020; Mills et al., 2019). In the present study, we found LEF1 expression is a key transcriptional regulator of the cell cycle in CMs. In mice, Lef1 is a canonical WNT signaling factor detected in embryonic heart transiently at E13.5–E17.5 and directly binds to the promoter region of CCND2 and CCND1 (Shtutman et al., 1999; Ye et al., 2019), which are required for the progression of G1 in the cell cycle. These previous reports as well as our findings support the direct regulation of LEF1 and cell cycle progression in CMs under WNT signaling.
Some previous reports have shown that Hand1 overexpression and conditional knockout cause overgrowth of the heart tube and smaller LV size, respectively, in mice, indicating Hand1 promotes proliferation (McFadden et al., 2005; Risebro et al., 2006). Recently, an LV-specific Hand1 enhancer was identified, and LV-specific Hand1-deficient mice from that study had a similar phenotype to the aforementioned conditional knockout mice (McFadden et al., 2005; Vincentz et al., 2017). However, LV-specific Hand1 and Hand2 dual-deficient mice showed an overgrowth of the myocardium, suggesting Hand1 and Hand2 suppress the proliferation of CMs (Vincentz et al., 2017). Thus, mouse studies indicate that Hand1 and Hand2 can both promote and suppress proliferation in a context-dependent manner. In the present study, we observed that the knockdown of HAND1 did not change the EdU+ ratio, but HAND2 or LEF1 knockdown reduced the proliferation. Also, we observed that the expression of LEF1 was upregulated by the knockdown of HAND1 and HAND2, indicating the expression of HAND1 and HAND2 suppresses CM proliferation. These findings suggest a gene regulatory network of the cell cycle in CMs for precise cell proliferation depends on HAND1, HAND2, and LEF1.
Finally, we found CD105 as a marker of highly proliferative CMs. CD105 is highly expressed in endothelial and mesenchymal cells and also a TGFβ signaling receptor. Its deficiency in mouse is embryonic lethal as a result of cardiovascular abnormalities (Arthur et al., 2000), but little is known about CD105 function in human CMs. In the present study, TGFβ signaling inhibition did not change the proliferation capacity of CMs, suggesting that CD105 has little functional contribution to the proliferation as a TGFβ signaling receptor even though it can act as marker of proliferative CMs.
To conclude, our results indicate that HAND1 has different roles in the early and late stages of in vitro differentiation. The first expression of HAND1, on day 3–5 (early stage), is induced by BMP4. Our tracing system showed that HAND1 marks CPCs in both our ventricular and atrial protocols, suggesting its role in the cell fate decision to CMs. In the later stage, our data indicated that HAND1 and HAND2 regulate LEF1 to affect the proliferation capacity of CMs via WNT signaling. Of note, atrial CMs showed a strong expression of HAND2 and little expression of HAND1, suggesting that regulatory mechanisms for the proliferation of atrial CMs are different from those of ventricular CMs. We also found evidence that CD105 is a marker of proliferative CMs. However, further molecular analysis is needed to reveal how HAND1 and HAND2 repress LEF1 and how the three factors orchestrate the development of the four-chambered heart morphology during cardiac organogenesis.
Experimental procedures
Establishment of triple-reporter lines
To establish the HAND1mCherry and HAND2EGFP double-reporter line, the CRISPR-Cas9 system was used to knock in a FLAG-2A-mCherry cassette (floxed PGK-puromycin resistance) and an HA-2A-EGFP cassette (floxed-PGK-neomycin resistance) at the HAND1 and HAND2 loci, respectively. To establish the triple-reporter line with constitutive tagBFP expression in addition to the double-reporter line, a CAG-tagBFP cassette for AAVS1 was knocked in using TALEN. All vectors of these knockin experiments were transfected into hiPSCs by electroporation with an NEPA 21 (NEPAGENE) following a previously described method with modifications (Li et al., 2015). MYH6-EGFP reporter hiPSCs were established as previously reported (Funakoshi et al., 2016). In this study, we modified the MYH6-EGFP piggyBac vector to change the EGFP coding sequence to an iRFP670 coding sequence. All DNA oligos/primers and vectors are listed in Table S4, and detailed protocols are described in the supplemental information.
hiPSC culture and cardiomyocyte induction
The cell culture and induction protocol for ventricular CMs were done as reported previously, with some modification (Funakoshi et al., 2016; Yang et al., 2008). Specifically, we alternatively used 96-well and 6-well plates coated with poly(2-hydroxyethyl methacrylate) (HEMA, Sigma-Aldrich). The atrial differentiation protocol was the same except 0.5 μM all-trans RA (Wako) was added to the day 3 medium. The use of hiPSCs were approved by the Ethics Committee of Kyoto University.
Flow cytometry and immunostaining
All analysis and sorting were performed with FACS AriaⅡ (Becton Dickinson) and analyzed by FlowJo (Becton Dickinson). All antibodies are listed in Table S5, and detailed protocols are described in the supplemental information.
RNA extraction and real-time qPCR
RNA was purified using QIAZOL reagent and the miRNeasy Micro Kit (QIAGEN), and cDNA was synthesized using ReverTra Ace (TOYOBO) with poly T primer or ReverTra Ace qPCR RT Master Mix with gDNA Remover (TOYOBO). Transcripts were amplified using the TaqMan probes (Applied Biosystems) listed in Table S6 and TaqMan Universal Master Mix II with Uracil-N-glycosylase (UNG) (Applied Biosystems). Real-time qPCR analysis was performed using StepOne (Applied Biosystems). Gene expression levels were calculated using the 2−ΔΔCT method with GAPDH or ACTB as the reference gene.
Knockdown experiments
For knockdown, the reverse transfection method was used with Lipofectamine RNAiMAX Reagent (Invitrogen) following the manufacturer's instructions. On day 15, 5 nmol of each siRNA (Silencer Select, Ambion) was transfected into isolated MYH6-EGFP+ CMs as single-cell suspensions (Table S7). The suspensions were seeded at 3–4 × 105 cells/well on a fibronectin-coated 12-well plate. On day 16, the medium was refreshed. On day 20, the cells were collected with QIAZOL and M-PER (Thermo).
RNA-seq and data analysis
Details of the experiment procedure and analysis are described in the supplemental information.
Statistics
GraphPad Prism (Version 7.05) was used to statistically analyze EdU data and gene expression data. All bar charts with error bars represent means ± standard deviation (SD).
Data and code availability
The accession number for the RNA-seq reported in this paper is GEO: GSE156394.
Author contributions
C.O. and Y.Y. conceived this project and wrote the manuscript. C.O. performed the computational and experimental analyses. A.H. supported the CRISPR experiments. S.Y. supervised the project. A.I., M. Nishikawa, and M. Narita helped with the experimental analyses. All authors contributed to the final article.
Conflicts of interests
S.Y. is a scientific advisor without salary of iPS Academia Japan, Inc. and Altos Labs, Inc. Y.Y. receives a research grant from Takeda Pharmaceutical Company Ltd. Kyoto University has filed a patent application relevant to this work. C.O and Y.Y are the investigators of record listed on the patent application.
Acknowledgments
We thank Dr. Peter Karagiannis for critical reading of the manuscript, Dr. Knut Woltjen for supporting experiments using the piggyBac transposon and TALEN, Dr. Takuya Yamamoto for providing some scripts and advice for the RNA-seq analysis, Dr. Kanae Mitsunaga for providing advice on the FACS analysis, Mr. Shunsuke Kihara for performing the confocal imaging, Dr. Hirohide Saito for providing the template oligos of tagBFP and iRFP670, and Dr. Keisuke Okita for providing the Cre expression vector. We are also grateful to Ms. Sayaka Takeshima, Ms. Yoko Uematsu, and Ms. Kaoru Shimizu for their administrative support. This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI 16J02610 to C.O. and 17H04176 to Y.Y. This work was also supported by The Leducq Foundation (18CVD05), the Research Center Network for Realization of Regenerative Medicine, Japan Agency for Medical Research and Development (AMED) (JP19bm0104001, JP19bm0204003, JP19bm0804008, and JP20bm0804022), the Research on Regulatory Science of Pharmaceuticals and Medical Devices, AMED (JP19mk0104117), the Research Project for Practical Applications of Regenerative Medicine, AMED (JP19bk0104095), the iPS Cell Research Fund, and the Secom Science and Technology Foundation.
Published: July 22, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.06.014.
Supplemental information
Significantly enriched pathways were listed (adjustment p value < 0.05)
References
- Andersen P., Tampakakis E., Jimenez D.V., Kannan S., Miyamoto M., Shin H.K., Saberi A., Murphy S., Sulistio E., Chelko S.P. Precardiac organoids form two heart fields via Bmp/Wnt signaling. Nat. Commun. 2018;9:3140. doi: 10.1038/s41467-018-05604-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur H.M., Ure J., Smith A.J., Renforth G., Wilson D.I., Torsney E., Charlton R., Parums D.V., Jowett T., Marchuk D.A. Endoglin, an ancillary TGFbeta receptor, is required for extraembryonic angiogenesis and plays a key role in heart development. Dev. Biol. 2000;217:42–53. doi: 10.1006/dbio.1999.9534. [DOI] [PubMed] [Google Scholar]
- Barnes R.M., Firulli B.A., Conway S.J., Vincentz J.W., Firulli A.B. Analysis of the Hand1 cell lineage reveals novel contributions to cardiovascular, neural crest, extra-embryonic, and lateral mesoderm derivatives. Dev. Dyn. 2010;239:3086–3097. doi: 10.1002/dvdy.22428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeva V., Louis-Brennetot C., Peltier A., Durand S., Pierre-Eugene C., Raynal V., Etchevers H.C., Thomas S., Lermine A., Daudigeos-Dubus E. Heterogeneity of neuroblastoma cell identity defined by transcriptional circuitries. Nat. Genet. 2017;49:1408–1413. doi: 10.1038/ng.3921. [DOI] [PubMed] [Google Scholar]
- Bruneau B.G., Logan M., Davis N., Levi T., Tabin C.J., Seidman J.G., Seidman C.E. Chamber-specific cardiac expression of Tbx5 and heart defects in Holt-Oram syndrome. Dev. Biol. 1999;211:100–108. doi: 10.1006/dbio.1999.9298. [DOI] [PubMed] [Google Scholar]
- Buikema J.W., Lee S., Goodyer W.R., Maas R.G., Chirikian O., Li G., Miao Y., Paige S.L., Lee D., Wu H. Wnt activation and reduced cell-cell contact synergistically induce massive expansion of functional human iPSC-derived cardiomyocytes. Cell Stem Cell. 2020;27:50–63.e55. doi: 10.1016/j.stem.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge P.W., Keller G., Gold J.D., Wu J.C. Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell. 2012;10:16–28. doi: 10.1016/j.stem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C.L., Liang X., Shi Y., Chu P.H., Pfaff S.L., Chen J., Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cserjesi P., Brown D., Lyons G.E., Olson E.N. Expression of the novel basic helix-loop-helix gene eHAND in neural crest derivatives and extraembryonic membranes during mouse development. Dev. Biol. 1995;170:664–678. doi: 10.1006/dbio.1995.1245. [DOI] [PubMed] [Google Scholar]
- Cui Y.L., Zheng Y.X., Liu X.X., Yan L.Y., Fan X.Y., Yong J., Hu Y.Q., Dong J., Li Q.Q., Wu X.L. Single-cell transcriptome analysis maps the developmental track of the human heart. Cell Rep. 2019;26:1934. doi: 10.1016/j.celrep.2019.01.079. [DOI] [PubMed] [Google Scholar]
- de Soysa T.Y., Ranade S.S., Okawa S., Ravichandran S., Huang Y., Salunga H.T., Schricker A., del Sol A., Gifford C.A., Srivastava D. Single-cell analysis of cardiogenesis reveals basis for organ-level developmental defects. Nature. 2019;572:120. doi: 10.1038/s41586-019-1414-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devalla H.D., Schwach V., Ford J.W., Milnes J.T., El-Haou S., Jackson C., Gkatzis K., Elliott D.A., Chuva de Sousa Lopes S.M., Mummery C.L. Atrial-like cardiomyocytes from human pluripotent stem cells are a robust preclinical model for assessing atrial-selective pharmacology. EMBO Mol. Med. 2015;7:394–410. doi: 10.15252/emmm.201404757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin A.D., Zimmerman M.W., Dharia N.V., Abraham B.J., Iniguez A.B., Weichert-Leahey N., He S., Krill-Burger J.M., Root D.E., Vazquez F. Selective gene dependencies in MYCN-amplified neuroblastoma include the core transcriptional regulatory circuitry. Nat. Genet. 2018;50:1240–1246. doi: 10.1038/s41588-018-0191-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli A.B., McFadden D.G., Lin Q., Srivastava D., Olson E.N. Heart and extra-embryonic mesodermal defects in mouse embryos lacking the bHLH transcription factor Hand1. Nat. Genet. 1998;18:266–270. doi: 10.1038/ng0398-266. [DOI] [PubMed] [Google Scholar]
- Friedman C.E., Nguyen Q., Lukowski S.W., Helfer A., Chiu H.S., Miklas J., Levy S., Suo S., Han J.J., Osteil P. Single-cell transcriptomic analysis of cardiac differentiation from human PSCs reveals HOPX-dependent cardiomyocyte maturation. Cell Stem Cell. 2018;23:586–598.e8. doi: 10.1016/j.stem.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi S., Miki K., Takaki T., Okubo C., Hatani T., Chonabayashi K., Nishikawa M., Takei I., Oishi A., Narita M. Enhanced engraftment, proliferation, and therapeutic potential in heart using optimized human iPSC-derived cardiomyocytes. Sci. Rep-uk. 2016;6 doi: 10.1038/srep19111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garry D.J., Olson E.N. A common progenitor at the heart of development. Cell. 2006;127:1101–1104. doi: 10.1016/j.cell.2006.11.031. [DOI] [PubMed] [Google Scholar]
- George R.M., Firulli A.B. Hand factors in cardiac development. Anat. Rec. (Hoboken) 2019;302:101–107. doi: 10.1002/ar.23910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemming M.L., Lawlor M.A., Zeid R., Lesluyes T., Fletcher J.A., Raut C.P., Sicinska E.T., Chibon F., Armstrong S.A., Demetri G.D. Gastrointestinal stromal tumor enhancers support a transcription factor network predictive of clinical outcome. Proc. Natl. Acad. Sci. U S A. 2018;115:E5746–E5755. doi: 10.1073/pnas.1802079115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochgreb T., Linhares V.L., Menezes D.C., Sampaio A.C., Yan C.Y., Cardoso W.V., Rosenthal N., Xavier-Neto J. A caudorostral wave of RALDH2 conveys anteroposterior information to the cardiac field. Development. 2003;130:5363–5374. doi: 10.1242/dev.00750. [DOI] [PubMed] [Google Scholar]
- Jassal B., Matthews L., Viteri G., Gong C., Lorente P., Fabregat A., Sidiropoulos K., Cook J., Gillespie M., Haw R. The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48:D498–D503. doi: 10.1093/nar/gkz1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman S.J., Witty A.D., Gagliardi M., Dubois N.C., Niapour M., Hotta A., Ellis J., Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Kel A., Voss N., Jauregui R., Kel-Margoulis O., Wingender E. Beyond microarrays: finding key transcription factors controlling signal transduction pathways. Bmc Bioinformatics. 2006;7 doi: 10.1186/1471-2105-7-S2-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koschmann J., Bhar A., Stegmaier P., Kel A.E., Wingender E. Upstream analysis": an integrated promoter-pathway analysis approach to causal interpretation of microarray data. Microarrays (Basel) 2015;4:270–286. doi: 10.3390/microarrays4020270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Protze S.I., Laksman Z., Backx P.H., Keller G.M. Human pluripotent stem cell-derived atrial and ventricular cardiomyocytes develop from distinct mesoderm populations. Cell Stem Cell. 2017;21:179–194.e4. doi: 10.1016/j.stem.2017.07.003. [DOI] [PubMed] [Google Scholar]
- Li H.L., Fujimoto N., Sasakawa N., Shirai S., Ohkame T., Sakuma T., Tanaka M., Amano N., Watanabe A., Sakurai H. Precise correction of the dystrophin gene in Duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports. 2015;4:143–154. doi: 10.1016/j.stemcr.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden D.G., Barbosa A.C., Richardson J.A., Schneider M.D., Srivastava D., Olson E.N. The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development. 2005;132:189–201. doi: 10.1242/dev.01562. [DOI] [PubMed] [Google Scholar]
- Meilhac S.M., Buckingham M.E. The deployment of cell lineages that form the mammalian heart. Nat. Rev. Cardiol. 2018;15:705–724. doi: 10.1038/s41569-018-0086-9. [DOI] [PubMed] [Google Scholar]
- Mills R.J., Parker B.L., Quaife-Ryan G.A., Voges H.K., Needham E.J., Bornot A., Ding M., Andersson H., Polla M., Elliott D.A. Drug screening in human PSC-cardiac organoids identifies pro-proliferative compounds acting via the mevalonate pathway. Cell Stem Cell. 2019;24:895–907.e6. doi: 10.1016/j.stem.2019.03.009. [DOI] [PubMed] [Google Scholar]
- Mohamed T.M.A., Ang Y.S., Radzinsky E., Zhou P., Huang Y., Elfenbein A., Foley A., Magnitsky S., Srivastava D. Regulation of cell cycle to stimulate adult cardiomyocyte proliferation and cardiac regeneration. Cell. 2018;173:104–116.e2. doi: 10.1016/j.cell.2018.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protze S.I., Lee J.H., Keller G.M. Human pluripotent stem cell-derived cardiovascular cells: from developmental biology to therapeutic applications. Cell Stem Cell. 2019;25:311–327. doi: 10.1016/j.stem.2019.07.010. [DOI] [PubMed] [Google Scholar]
- Randolph L.N., Lian X.L. Beyond purple hearts: a colorful approach to isolate distinct heart cells from human iPSCs. Cell Stem Cell. 2019;24:675–677. doi: 10.1016/j.stem.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reamon-Buettner S.M., Ciribilli Y., Inga A., Borlak J. A loss-of-function mutation in the binding domain of HAND1 predicts hypoplasia of the human hearts. Hum. Mol. Genet. 2008;17:1397–1405. doi: 10.1093/hmg/ddn027. [DOI] [PubMed] [Google Scholar]
- Reamon-Buettner S.M., Ciribilli Y., Traverso I., Kuhls B., Inga A., Borlak J. A functional genetic study identifies HAND1 mutations in septation defects of the human heart. Hum. Mol. Genet. 2009;18:3567–3578. doi: 10.1093/hmg/ddp305. [DOI] [PubMed] [Google Scholar]
- Riley P., Anson-Cartwright L., Cross J.C. The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nat. Genet. 1998;18:271–275. doi: 10.1038/ng0398-271. [DOI] [PubMed] [Google Scholar]
- Risebro C.A., Smart N., Dupays L., Breckenridge R., Mohun T.J., Riley P.R. Hand1 regulates cardiomyocyte proliferation versus differentiation in the developing heart. Development. 2006;133:4595–4606. doi: 10.1242/dev.02625. [DOI] [PubMed] [Google Scholar]
- Rochais F., Mesbah K., Kelly R.G. Signaling pathways controlling second heart field development. Circ. Res. 2009;104:933–942. doi: 10.1161/CIRCRESAHA.109.194464. [DOI] [PubMed] [Google Scholar]
- Shtutman M., Zhurinsky J., Simcha I., Albanese C., D'Amico M., Pestell R., Ben-Ze'ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. U S A. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M.K., Christoffels V.M., Dias J.M., Trowe M.O., Petry M., Schuster-Gossler K., Burger A., Ericson J., Kispert A. Tbx20 is essential for cardiac chamber differentiation and repression of Tbx2. Development. 2005;132:2697–2707. doi: 10.1242/dev.01854. [DOI] [PubMed] [Google Scholar]
- Skelton R.J., Brady B., Khoja S., Sahoo D., Engel J., Arasaratnam D., Saleh K.K., Abilez O.J., Zhao P., Stanley E.G. CD13 and ROR2 permit isolation of highly enriched cardiac mesoderm from differentiating human embryonic stem cells. Stem Cell Rep. 2016;6:95–108. doi: 10.1016/j.stemcr.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D., Cserjesi P., Olson E.N. A subclass of bHLH proteins required for cardiac morphogenesis. Science. 1995;270:1995–1999. doi: 10.1126/science.270.5244.1995. [DOI] [PubMed] [Google Scholar]
- Srivastava D., Thomas T., Lin Q., Kirby M.L., Brown D., Olson E.N. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat. Genet. 1997;16:154–160. doi: 10.1038/ng0697-154. [DOI] [PubMed] [Google Scholar]
- Takaki T., Inagaki A., Chonabayashi K., Inoue K., Miki K., Ohno S., Makiyama T., Horie M., Yoshida Y. Optical recording of action potentials in human induced pluripotent stem cell-derived cardiac single cells and monolayers generated from long QT syndrome type 1 patients (vol 2019, 7532657, 2019) Stem Cells Int. 2020;2020 doi: 10.1155/2019/7532657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T. Regulation of cardiomyocyte proliferation during development and regeneration. Dev. Growth Differ. 2014;56:402–409. doi: 10.1111/dgd.12134. [DOI] [PubMed] [Google Scholar]
- Togi K., Kawamoto T., Yamauchi R., Yoshida Y., Kita T., Tanaka M. Role of Hand1/eHAND in the dorso-ventral patterning and interventricular septum formation in the embryonic heart. Mol. Cell Biol. 2004;24:4627–4635. doi: 10.1128/MCB.24.11.4627-4635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankov A.M., Gu H., Akopian V., Ziller M.J., Donaghey J., Amit I., Gnirke A., Meissner A. Transcription factor binding dynamics during human ES cell differentiation. Nature. 2015;518:344–349. doi: 10.1038/nature14233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchihashi T., Maeda J., Shin C.H., Ivey K.N., Black B.L., Olson E.N., Yamagishi H., Srivastava D. Hand2 function in second heart field progenitors is essential for cardiogenesis. Dev. Biol. 2011;351:62–69. doi: 10.1016/j.ydbio.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincentz J.W., Toolan K.P., Zhang W., Firulli A.B. Hand factor ablation causes defective left ventricular chamber development and compromised adult cardiac function. PLoS Genet. 2017;13:e1006922. doi: 10.1371/journal.pgen.1006922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woltjen K., Michael I.P., Mohseni P., Desai R., Mileikovsky M., Hamalainen R., Cowling R., Wang W., Liu P., Gertsenstein M. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.M., Chien K.R., Mummery C. Origins and fates of cardiovascular progenitor cells. Cell. 2008;132:537–543. doi: 10.1016/j.cell.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Soonpaa M.H., Adler E.D., Roepke T.K., Kattman S.J., Kennedy M., Henckaerts E., Bonham K., Abbott G.W., Linden R.M. Human cardiovascular progenitor cells develop from a KDR plus embryonic-stem-cell-derived population. Nature. 2008;453:524–U526. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- Ye B., Li L., Xu H., Chen Y., Li F. Opposing roles of TCF7/LEF1 and TCF7L2 in cyclin D2 and Bmp4 expression and cardiomyocyte cell cycle control during late heart development. Lab. Invest. 2019;99:807–818. doi: 10.1038/s41374-019-0204-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.Z., Termglinchan V., Shao N.Y., Itzhaki I., Liu C., Ma N., Tian L., Wang V.Y., Chang A.C.Y., Guo H.C. A human iPSC double-reporter system enables purification of cardiac lineage subpopulations with distinct function and drug response profiles. Cell Stem Cell. 2019;24:802. doi: 10.1016/j.stem.2019.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Significantly enriched pathways were listed (adjustment p value < 0.05)
Data Availability Statement
The accession number for the RNA-seq reported in this paper is GEO: GSE156394.