Abstract
Background
Chlamydia trachomatis (Ct) infection ascending to the upper genital tract can cause infertility. Direct association of genetic variants as contributors is challenging because infertility may not be diagnosed until years after infection. Investigating the intermediate trait of ascension bridges this gap.
Methods
We identified infertility genome-wide association study (GWAS) loci using deoxyribonucleic acid from Ct-seropositive cisgender women in a tubal factor infertility study and Ct-infected cisgender women from a longitudinal pelvic inflammatory disease cohort with known fertility status. Deoxyribonucleic acid and blood messenger ribonucleic acid from 2 additional female cohorts with active Ct infection and known endometrial infection status were used to investigate the impact of infertility single-nucleotide polymorphisms (SNPs) on Ct ascension. A statistical mediation test examined whether multiple infertility SNPs jointly influenced ascension risk by modulating expression of mediator genes.
Results
We identified 112 candidate infertility GWAS loci, and 31 associated with Ct ascension. The SNPs altered chlamydial ascension by modulating expression of 40 mediator genes. Mediator genes identified are involved in innate immune responses including type I interferon production, T-cell function, fibrosis, female reproductive tract health, and protein synthesis and degradation.
Conclusions
We identified Ct-related infertility loci and their potential functional effects on Ct ascension.
Keywords: chlamydia disease, genital tract ascension, GWAS, host genetics, infertility
Untreated cervical Chlamydia trachomatis (Ct) infection can ascend to the upper genital tract, eliciting inflammation that results in pelvic inflammatory disease (PID). Pelvic inflammatory disease, clinical or subclinical, can lead to tubal factor infertility (TFI). Tubal factor infertility occurs when a woman’s fallopian tube(s) are damaged, preventing fertilization or implantation. A recent Dutch cohort study, which followed over 5500 women for 8 years, found a 4.2-fold increased risk for TFI in women who experienced chlamydial infection versus Ct-negative women [1]. Although Ct is recognized as an important cause of TFI, most infected women do not develop infertility, possibly because the infection is limited to the cervix. Previous studies of women with Ct-related infertility identified host genetic variants associated with altered disease risk but focused on pathophysiological pathways implicated in animal models of chlamydial disease. Direct association of genetic variants as contributors to Ct infection-induced infertility is challenging because infertility may not be diagnosed until years after resolution of infection. To bridge this divide, we investigated the intermediate trait of Ct ascension because this is a prerequisite for fallopian tube inflammation and TFI.
Genome-wide association studies (GWAS) support broad and unbiased detection of genetic variation. However, applying GWAS to identification of single-nucleotide polymorphism(s) (SNP) associated with ascending Ct infection is not feasible, because it would require thousands of participants with endometrial Ct infection status diagnosed by an invasive procedure. Furthermore, the complex immunopathology that drives PID and subsequently TFI is likely modulated by multiple genetic variants. Most loci identified by GWAS are not mapped to protein-coding regions, so the mechanism underlying their effect can be difficult to interpret. Nevertheless, when genetic variants affect transcription factor binding sites in the promoter region of a regulated gene to influence its expression, typically within 1 megabyte (Mb) on either side of the transcriptional start site, this can assist in functional annotation [2].
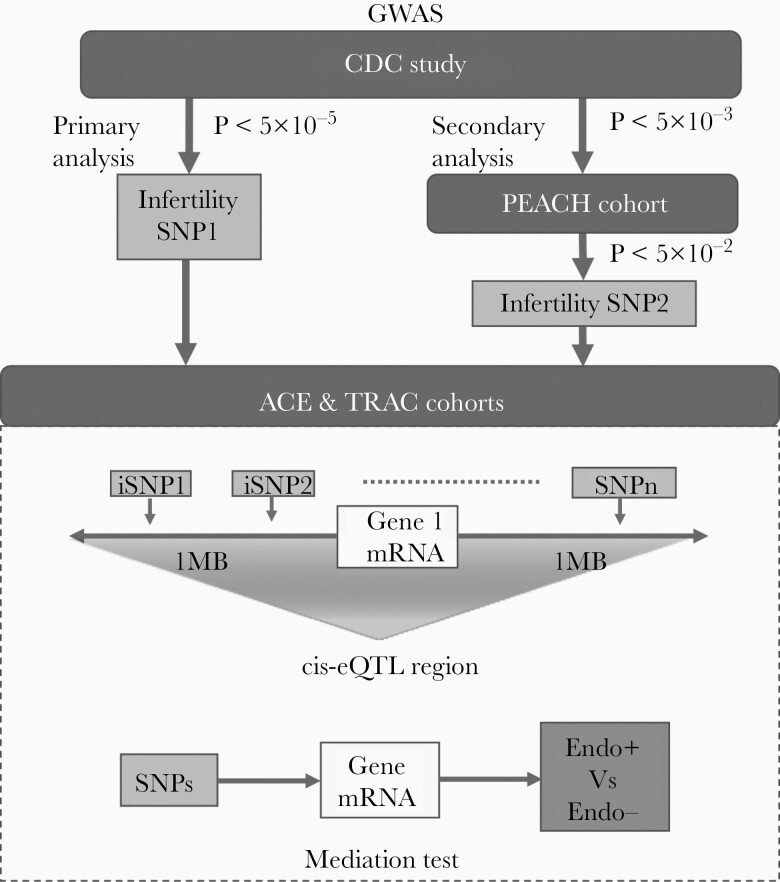
In this study, we attempted to overcome these limitations of GWAS by using multiple relevant patient cohorts (Figure 1). We identified GWAS loci of infertility from 2 studies: (1) in a case-control study, we used deoxyribonucleic acid (DNA) from Ct-seropositive cisgender women, with TFI (cases) or with nontubal-factor infertility by other causes (controls); and (2) in a longitudinal cohort study, we included cisgender women with documented PID and Ct infection who developed infertility and others who became pregnant. We next explored the functional impact of candidate infertility SNPs on Ct ascension from an independent cohort with DNA and messenger ribonucleic acid (mRNA) data from Ct-infected women with (Endo+) or without (Endo−) endometrial infection. Finally, we used robust statistical mediation analysis to examine whether multiple simultaneously expressed infertility SNPs influence the risk of Ct ascension through modulation of mediator gene expression. These analyses identified important candidates that should be further investigated for their potential roles in Ct-induced infertility.
Figure 1.
Data analysis strategy and selection criteria used in this study. CDC, Centers for Disease Control and Prevention; cis-eQTL, cis-expression quantitative trait loci; GWAS, genome-wide association study; MB, megabyte; mRNA, messenger ribonucleic acid; PEACH, Pelvic Inflammatory Disease Evaluation and Clinical Health cohort; SNP, single-nucleotide polymorphism; TRAC, T cell Response Against Chlamydia study.
METHODS
All study participants provided written informed consent before initiation of study procedures. Institutional Review Boards (IRB) for Human Subjects Research at the University of North Carolina, the University of Pittsburgh, and the University of Alabama at Birmingham approved the study. The Centers for Disease Control and Prevention (CDC) human subjects review determined that CDC investigators were not engaged in human subjects research for this study and that CDC IRB approval was not required.
Study Population and Study Design
Whole blood DNA from participants recruited into a CDC-funded case-control study was used for primary analysis to identify potential GWAS loci of Ct-related TFI [3]. This discovery dataset included all Ct-seropositive cisgender women, tested by anti-immunoglobulin (Ig)G for Ct major outer membrane protein (OmpA), anti-IgG1 and IgG3 to Ct elementary bodies, or anti-Ct HSP60, participating in the CDC-funded study. All participants in the CDC-funded study were infertile, with cases having evidence of tubal blockage as determined by hysterosalpingography or laparoscopy, whereas controls showed tubal patency. A total of 382 females with Ct-seropositivity and infertility were genotyped, including 113 with TFI (cases) and 269 with non-TFI infertility (controls).
Cisgender women diagnosed with Ct-induced PID with DNA available for analysis from the Pelvic Inflammatory Disease Evaluation and Clinical Health (PEACH) study were included in a secondary analysis to identify additional infertility loci [4]. A total of 61 women with Ct-induced PID, including 15 infertile cases and 46 fertile controls, were genotyped.
THe DNA and RNA data from Ct-infected women [5] participating in a randomized clinical trial of enhanced anaerobic treatment for women with PID (The Anaerobes and Clearance of Endometritis, [ACE] trial) [6] or the T cell Response Against Chlamydia (TRAC) study [7] were used to investigate whether infertility genetic loci were also associated with differential risk of endometrial Ct infection through alteration of mediator gene expression levels (Figure 1). Blood mRNA and DNA from 143 Ct-infected women, 71 who were Ct positive only at the cervix, and 72 who were also Ct positive on endometrial biopsy, was profiled and genotyped.
Details of these study cohorts are described in Supplementary Methods.
Genotyping, Imputation, and Data Quality Control in the Centers for Disease Control and Prevention-Funded Study, Pelvic Inflammatory Disease Evaluation and Clinical Health, ACE, and T Cell Response Against Chlamydia Cohorts
Genotyping was performed with Illumina BeadChip arrays, as described in Supplementary Methods.
Genetic Association Analysis for Chlamydia trachomatis-Related Infertility Loci in the Centers for Disease Control and Prevention-Funded Study and Pelvic Inflammatory Disease Evaluation and Clinical Health Cohort
Candidate infertility loci determined by GWAS were defined as (1) loci with P < 5 × 10E-5 in the CDC-funded study (primary analysis) or (2) by requiring the GWAS-determined loci in the CDC-funded study to meet a less stringent statistical cutoff of P < 5 × 10E-3 along with a genetic association at P < .05 in the PEACH cohort (secondary analysis). Genetic association analyses were conducted by logistic regression in PLINK (version 1.9). Population stratification was adjusted using the first 15 principal components, which explained over 80% of the total variance.
Genetic Association of Infertility Loci With Chlamydia trachomatis Ascension in ACE and T Cell Response Against Chlamydia Cohorts
The association of genotypes of candidate infertility loci with Ct ascension in ACE and TRAC cohorts was tested in PLINK by logistic regression. Population stratification was adjusted using the first 15 principal components, which explained over 80% of the total variance.
Messenger Ribonucleic Acid Array Data Acquisition and Processing in ACE and T Cell Response Against Chlamydia Cohorts
Total RNA was isolated from whole blood of ACE and TRAC participants and profiled by BeadChip microarray [5].
Probabilistic Estimation of Expression Residuals Factor Analysis for Confounding Factors in ACE and T Cell Response Against Chlamydia Cohorts
We applied the probabilistic estimation of expression residuals (PEER) method [8] to infer and manage confounding factors affecting gene expression levels, as described in Supplementary Methods.
Mediation Test in ACE and T Cell Response Against Chlamydia Cohorts
After examining the association of genotypes at infertility loci with Ct ascension in ACE and TRAC cohorts at the individual SNP level, we estimated the joint effect of multiple SNPs on ascension and investigated whether they acted by modulating expression of mediator gene(s). Candidate mediator genes were tested if the infertility locus lay within 1 Mb on either side of the gene’s transcriptional start site. To assess the gene mediation effects of our infertility loci on ascension, we used Generalized Multi-SNP Mediation Intersection-Union Test ([GSMUT] [9]) (Supplementary Methods). To determine the directionality of effects of mediator gene expression levels on the risk of ascension, we compared expression levels of mediator genes between Endo+ and Endo− women in the ACE and TRAC cohorts by logistic regression. For multiple testing correction, the false-discovery rate (FDR) was estimated by using the fdrtool package in R. P < .1, corresponding to 19.8% FDR, were considered suggestively significant.
RESULTS
Population Characteristics
Participant characteristics from the CDC-funded infertility study and the PEACH cohort are summarized in Supplementary Tables 1 and 2, respectively. In both studies, age, education, insurance, substance use, self-reported infection history, coinfections at enrollment, and contraceptive method use had no statistically significant differences between cases and controls. Potential population stratification in cases and controls was controlled by adjusting the major principal components of genetic ancestry in the statistical model. Characteristics of ACE and TRAC participants used in GSMUT analysis of association with Ct ascension are summarized in Supplementary Table 3. Oral contraceptive pill use and Neisseria gonorrhoeae coinfection were significantly increased in Endo+ women [7] and adjusted as covariates.
Identification of Infertility Loci in Centers for Disease Control and Prevention-Funded and Pelvic Inflammatory Disease Evaluation and Clinical Health Studies and Their Association With Chlamydia trachomatis Ascension in ACE and T Cell Response Against Chlamydia
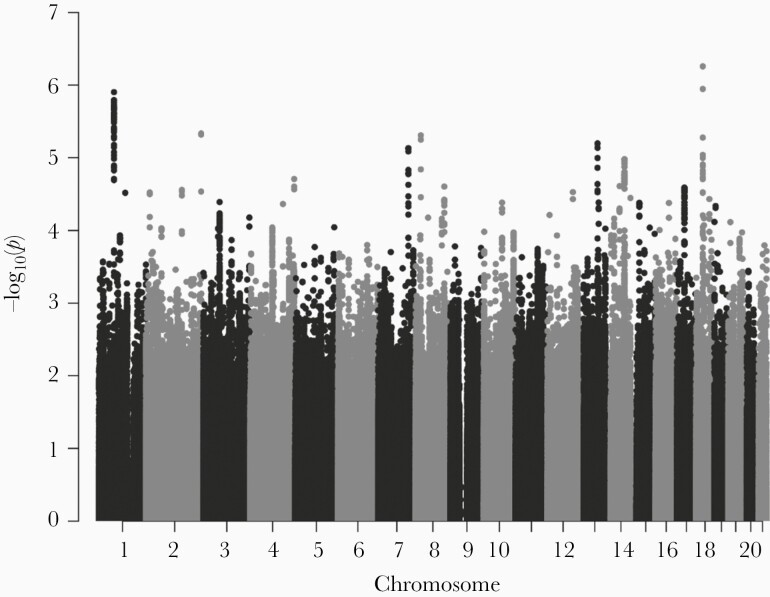
The Manhattan plot of GWAS results from CDC-funded study participants is shown in Figure 2. The GWAS infertility loci determined from primary analysis of the CDC-funded study, predominantly white women without a history of gonorrhea, are detailed in Supplementary Table 4. Twenty-six loci were found, mapping to 15 distinct chromosomes. Infertility loci determined from the secondary analysis incorporating the PEACH cohort, primarily black women with a high rate of gonococcal coinfection, are detailed in Supplementary Table 5. Eighty-six loci on 21 chromosomes were identified. We then determined whether these infertility GWAS loci were also associated with Ct ascension in ACE and TRAC; findings are summarized in Supplementary Tables 4 and 5. At the individual SNP level, 48 SNPs from the primary analysis and 65 SNPs from the secondary analysis were modestly associated with ascending infection (P < .20). None were statistically significant after multiple-testing correction. Therefore, we formally tested the combined effect of multiple SNPs on the risk of ascending infection through altered expression of a mediator gene by the causal mediation test, GSMUT.
Figure 2.
Manhattan plot summarizing the infertility genome-wide association study (GWAS) results of the Centers for Disease Control and Prevention-funded study. Each dot represents 1 single-nucleotide polymorphism (SNP). The x-axis indicates the physical base-pair position of each tested SNP; y-axis indicates the −log(10) P values from GWAS.
Gene Mediation Effects of Infertility Loci on Ascension Identified by Generalized Multi-SNP Mediation Intersection-Union Test
Two loci, LINC01750 and SORBS2, were identified from the primary GWAS analysis of infertility that influenced the risk of ascension in women from the ACE and TRAC cohorts through 5 mediator genes. Secondary analysis using data from both the CDC-funded and PEACH cohorts identified 29 loci that affected the risk of ascension through modulating expression of 36 mediator genes (Supplementary Table 6). All of these mediations were suggestively significant after adjustment for multiple comparisons (FDR <0.2). The association of infertility loci with Ct ascension via mediator genes provides potential functional annotation. Because information on directionality is compromised with GSMUT, we directly compared mediator gene expression levels in Endo+ and Endo− women in ACE and TRAC to determine whether mediator genes were up- or down-regulated with ascension. Table 1 provides general descriptions of mediator gene functionality, as well as directionality of mediator gene expression, in Endo+ versus Endo− women.
Table 1.
Mediator Gene Functions and Directionality
| Overall Function | Gene Name | Specific Function | Expression in Endo+ Versus Endo− Women | Citation |
|---|---|---|---|---|
| Innate Immune Response | ||||
| ABR | Regulation of inflammation | ↑ | [26] | |
| ASB13 | Innate immune response regulator | ↑ | [27] | |
| TLR3a | Type I interferon induction | ↑ | [28] | |
| NPR2 | C-type natriuretic peptide receptor which inhibits IFN-γ mediated gene expression | ↑ | [20] | |
| PSG3 | IFN-γ responsiveness | ↑ | [29] | |
| PVR (CD155) | NK cell effector function | ↑ | [30] | |
| Mir620 | Targets CCL2 (MCP-1) | ↑ | [31] | |
| COG6 | Regulation of inflammation | ↓ | [32] | |
| MEIS2 | Myeloid cell differentiation | ↓ | [33] | |
| PSG2 | IDO and TGF-β activity | ↓ | [34] | |
| SLC5A1 | Glucose transport; innate immune cell function | ↓ | [35] | |
| Adaptive Immune Response | ||||
| PFKP | Glycolysis; T-cell function | ↑ | [23] | |
| TLR3a | T-cell activation | ↑ | [36] | |
| LARGE | T-cell development | ↓ | [24] | |
| LGMN | T-cell activation; Class II antigen presentation | ↓ | [25] | |
| Fibrosis | ||||
| PLOD2 | Fibrotic enzyme | ↑ | [37] | |
| TLR3a | Wound healing, fibrosis | ↑ | [38] | |
| Proliferation and Apoptosis | ||||
| CSE1L | Cell proliferation | ↓ | [39] | |
| HERPUD1 | Apoptosis regulation | ↓ | [40] | |
| TMEM192 | Apoptosis regulation | ↓ | [41] | |
| XRN1 | Mitochondrial function | ↓ | [42] | |
| Protein Synthesis and Degradation | ||||
| ASB13 | Protein degradation | ↑ | [43] | |
| HERPUD1 | Destruction of misfolded proteins | ↓ | [44] | |
| MRM1 | Ribosomal RNA transferase | ↑ | [45] | |
| MTRF1 | Mitochondrial translation | ↑ | [46] | |
| UROS | Methionine synthesis | ↑ | [47] | |
| WDR33 | Protein synthesis; cell differentiation | ↑ | [48] | |
| Gene Regulation | ||||
| HELTb | Transcriptional repressor | ↓ | [40] | |
| ZNF512 | Transcription regulation | ↑ | [13] | |
| Cell Membrane Formation and Function | ||||
| PCDH17 | Cell adhesion | ↑ | [49] | |
| PIGO | Cell membrane phospholipid synthesis | ↑ | ||
| SLC26A5 | Membrane anion transporter | ↑ | [50] | |
| CADPS | Vesicle exocytosis | ↓ | [s1] | |
| CEPT1b | Cell membrane phospholipid synthesis | ↓ | [s2] | |
| LR2BPb | Ligand endocytosis | ↓ | [s3] | |
| Intracellular Energy Transfer | ||||
| ABR | GTPase | ↑ | [s4] | |
| NDUFB4 | NADH dehydrogenase | ↑ | [s5] | |
| Female Reproductive Tract Health | ||||
| OVGP1b | Fertilization, implantation, embryonic development | ↑ | [10] | |
| NPR2 | Female reproductive tract development | ↑ | [s6] | |
| GAS2L2 | Regulates ciliary orientation | ↑ | [s7] | |
| Miscellaneous | EMR4 | Human pseudogene | ↑ | [s8] |
| RPP38-DT | Unknown function | ↑ | ||
| FAM162B | Unknown function | ↓ | ||
| HEATR9 | Unknown function | ↓ | ||
| KRTAP3-1 | Keratin-associated protein | ↓ | [s9] |
Abbreviations: IDO, indoleamine dioxygenase; IFN, interferon; MCP-1, monocyte chemoattractant protein-1; NK, natural killer; RNA, ribonucleic acid; TGF, transforming growth factor.
aTLR3 (P < .01) overlapped from primary and secondary analyses coincident with loci for chlamydial ascension.
bMediator genes (P < .01) from primary analysis of Centers for Disease Control and Prevention (CDC) cohort (P < 5 × 10E-05) coincident with loci for chlamydial ascension.
NOTE: The remaining mediator genes (P < .01) are from secondary analysis of CDC cohort (P < 5 × 10E-03) and Pelvic Inflammatory Disease Evaluation and Clinical Health (PEACH) cohort (P < 5 × 10E-02) coincident with loci for chlamydial ascension.
The LINC01750 locus for infertility was also a locus for ascension mediated through altering OVGP1 and CEPT1 expression. OVGP1 encodes a protein specific to oviduct and endometrial epithelial cells [10]; its transcription was modestly increased in Endo+ women. CEPT1 encodes a dual specificity enzyme that catalyzes synthesis of 2 major cell membrane phospholipids [11] that control lipid-driven proinflammatory cascades [12]. CEPT1 expression was modestly decreased in Endo+ women. The SORBS2 locus for infertility was associated with altered risk for Ct ascension through modulation of 3 mediator genes: LRP2BP, HELT, and TLR3. LRP2BP encodes lipid receptor-related protein 2 (LRP2) binding protein. THe LRP2, or megalin, is a receptor found in the plasma membrane of many epithelial cells that promotes endocytosis. HELT encodes a basic-helix-loop-helix protein belonging to a superfamily of DNA-binding transcription factors that regulate cell cycle, apoptosis, and differentiation [13]. Expression of LRP2BP and HELT was modestly decreased in Endo+ women. TLR3 was the only mediator gene identified from both primary and secondary analyses, and it was modestly increased in Endo+ women. TLR3 is an intracellular pattern recognition receptor, detecting double-stranded RNA to induce nuclear factor-κB activation and production of type I interferons (IFNs).
Secondary analysis identified 29 loci influencing the risk of ascension in cisgender women from ACE and TRAC through 36 mediator genes. The mediator genes encode proteins implicated in innate inflammation, T-cell function, wound healing, fibrosis, cell proliferation and apoptosis, protein synthesis and degradation, gene regulation, cell membrane formation and function, cell metabolism, and overall health of the female reproductive tract (Table 1).
Discussion
We conducted a GWAS for TFI in women from a CDC-funded infertility study who were seropositive for Ct. Among loci for infertility from primary analysis, 2 (LINC01750 and SORBS2 loci) influenced Ct ascension via modulation of 5 mediator genes: OVGP1, CEPT1, LR2BP, HELT, and TLR3. Our finding that infertility loci modulate expression of an oviduct epithelium-specific gene, OVGP1, suggests that genetic polymorphisms affecting production of this protein may alter reproductive tract responses vital to fertility. Animal studies indicate that OVGP1 aids in sperm capacitation, fertilization, and early embryonic development [14]. OVGP1 transcription levels were increased in Endo+ versus Endo− women, possibly reflecting increased production in response to Ct-induced inflammation and activation of epithelial repair mechanisms. CEPT1 is responsible for synthesis of phosphatidylcholine, a constituent of Ct membranes that must be acquired from its mammalian host to support chlamydial proliferation. Altered expression could thus control chlamydial ascension by influencing bacterial burden. Furthermore, changes in cell membrane phospholipid composition alter cellular recognition of danger-associated molecular patterns and downstream inflammatory signaling [12], which possibly modulate tubal damage during chlamydial invasion. Changes in cellular endocytosis and trafficking of cargo to lysosomes via differential expression of LRP2BP could affect Ct ascension via altered uptake or killing of bacteria or through changes in uptake of danger molecules released from damaged infected cells. HELT could alter Ct ascension through control of reproductive tract epithelial cell cycling, apoptosis, and differentiation [13].
Because TLR2 and TLR4 play roles in recognition of Ct and are expressed in the human female genital tract, associations between polymorphisms in these TLRs and Ct sequelae were investigated in several prior candidate gene studies, which gave inconsistent findings. The relatively small sample size in these studies and/or lack of information related to prior chlamydial infection make their interpretation challenging. Although TLR3 is present in the female genital tract, we did not find previous studies testing its genetic association with human Ct disease. In this study, we identified infertility variants in or near the TLR3 gene. Activation of TLR3 and IFN regulatory factor 3 (IRF3)-induced production of type I IFNs during chlamydial infection could enhance Ct infection and ascension through inhibition of T-cell and IFN-γ responses essential for chlamydial control [15]. We previously reported that women with Ct-induced PID and endometritis display elevated type-I IFN-induced gene transcription in peripheral blood [5], and that women with endometrial Ct infection secrete higher levels of type-I IFN-induced chemokines compared with women with only cervical infection [5]. Mouse models of genital tract chlamydial infection gave conflicting results, finding either no effect of TLR3 deficiency [16] or altered shedding and pathology [17]. Detecting increased TLR3 transcripts in Endo+ women supports a deleterious role for this pathogen recognition receptor during chlamydial infection.
TLR3 was the only gene identified by an additional infertility variant using data from both the CDC-funded and PEACH cohorts. Women in PEACH were often coinfected with gonorrhea, which promotes prominent neutrophil responses and epithelial cell death. It is possible that TLR3 functions indirectly through sensing self-RNA released from damaged cells [18]. Finally, stimulation of TLR3 influences wound healing and fibrosis [19], with direct implications for development of TFI.
Using combined data from the CDC-funded and PEACH cohorts, we identified additional infertility GWAS loci mediating Ct ascension. ABR, ASB13, COG6, and SLC5A1 regulate innate inflammatory responses. NPR2 is the primary receptor for C-type natriuretic peptide, which down-regulates IFN-γ-mediated inflammation and kynurenine generation by IFN-γ-induced indoleamine dioxygenase (IDO) [20]. PSG3 and PVR influence IFN-γ responses and natural killer (NK) cell effector function, respectively, and PSG2 is linked to changes in IDO activity. Detection of these genes is relevant because IFN-γ-induced IDO is a primary host defense mechanism against Ct [21] and NK cell activation drives CD4+ T cells towards IFN-γ-producing Th1 cells that are key to chlamydial control [22]. PFKP is essential for T-cell metabolism [23] and LARGE for T-cell development [24]. LGMN is an upstream activator of the Cathepsin L-Mediated Intracellular C3 Activation (CTSL-C3)-IFN-γ axis in human CD4+ T cells and an important promoter of human Th1 responses [25]. CD4+ T cells isolated from LGMN-deficient mice display a specific defect in IFN-γ secretion and Th1 responses [25], so alterations in LGMN could influence chlamydial growth and infection duration.
Infertility loci were also associated with mediator genes involved in apoptosis and cell proliferation, protein synthesis and degradation, and overall gene regulation, potentially influencing tissue damage and healing. Our identification of genes involved in the formation and function of cell membranes, endocytosis, exocytosis, and anion transporters is striking because Ct invades host epithelial cells by a panoply of mechanisms and resides within a protective vacuole, stealing lipids from the host endoplasmic reticulum-Golgi apparatus.
Our use of GSMUT to test the association of SNPs and ascension mediated by altered gene expression is a strength of our study because this method enables examination of the combined effects of multiple SNPs acting completely through mediator genes, as well as effects partially mediated through these genes. However, a limitation is its inability to determine directionality and magnitude of mediation effects.
Although control participants in the CDC-funded study had patent tubes by hysterosalpingography (HSG) or laparoscopy, they were infertile, which may increase false-negative findings. Mild scarring, undetected by HSG, could have prevented pregnancy. We attempted to overcome sample size limitations by combining results from the CDC-funded and PEACH cohorts and by analyzing tentative loci for their association with alterations in risk for Ct ascension in 2 additional patient cohorts. Our mediation test linked coincident infertility and ascension loci to differential expression of mediator genes associated with Ct ascension. The biological functions of their gene products suggest that they are plausible candidates for proteins that influence spread of Ct.
Conclusions
Our study highlights the potential for systematic genetics and mediation testing to dissect a complex disease and identify potential mediation genes involved in its pathogenesis. Data indicate that development of tubal pathology is influenced by multiple inflammatory pathways. A larger, independent cohort with documented prior Ct infection and reproductive outcomes (infertility versus fertility) would enable us to validate these findings. Nevertheless, this current study has yielded novel molecular targets that can be investigated in functional studies in vitro or in animal models to better determine their relationship to Ct infection and development of tubal disease, with the ultimate goals of determining therapeutics adjunctive to antibiotics and advancing development of biomarkers that indicate risk for ascension and aid in the diagnosis of PID.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank the women who agreed to participate in this study; Ingrid Macio, Melinda Petrina, Carol Priest, Abi Jett, and Lorna Rabe for their efforts in the clinic and the microbiology laboratory; the staff at the Allegheny County Health Department Sexually Transmitted Disease Clinic for their support; and Centers for Disease Control and Prevention staff Jesse Thomas and Allie Coor for assisting with the consultation process. We also thank Dr. Nicholas A. Cataldo for carefully reviewing this manuscript.
Disclaimer. The study represents the view of the authors and does not necessarily represent the official position of the Centers for Disease Control and Prevention.
Financial support. This work was funded by the National Institute of Allergy and Infectious Diseases at the National Institutes of Health through R01 AI119164 and U19 AI084024 (to T. D.) and U19 AI144181 (to T. D., C. M. C., and X. Z.) and Centers for Disease Control and Prevention, Prevention Research Centers through 5U48DP001915 (to W. M. G.) and 5U48DP001918 to (H. C. W. and C. L. H.).
Supplement sponsorship. This supplement is sponsored by the Centers for Disease Control and Prevention.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1.Hoenderboom BM, van Benthem BHB, van Bergen JEAM, et al. . Relation between Chlamydia trachomatis infection and pelvic inflammatory disease, ectopic pregnancy and tubal factor infertility in a Dutch cohort of women previously tested for chlamydia in a chlamydia screening trial. Sex Transm Infect 2019; 95:300–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Consortium GT. Human genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015; 348:648–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorwitz RJ, Wiesenfeld HC, Chen PL, et al. . Population-attributable fraction of tubal factor infertility associated with chlamydia. Am J Obstet Gynecol 2017; 217:336.e1–e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ness RB, Soper DE, Holley RL, et al. . Effectiveness of inpatient and outpatient treatment strategies for women with pelvic inflammatory disease: results from the Pelvic Inflammatory Disease Evaluation and Clinical Health (PEACH) randomized trial. Am J Obstet Gynecol 2002; 186:929–37. [DOI] [PubMed] [Google Scholar]
- 5.Zheng X, O’Connell CM, Zhong W, et al. . Discovery of blood transcriptional endotypes in women with pelvic inflammatory disease. J Immunol 2018; 200:2941–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiesenfeld HC, Meyn LA, Darville T, Macio IS, Hillier SL. A Randomized controlled trial of ceftriaxone and doxycycline, with or without metronidazole, for the treatment of acute pelvic inflammatory disease. Clin Infect Dis 2021; 72:1181–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Russell AN, Zheng X, O’Connell CM, et al. . Analysis of factors driving incident and ascending infection and the role of serum antibody in Chlamydia trachomatis genital tract infection. J Infect Dis 2016; 213:523–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stegle O, Parts L, Piipari M, Winn J, Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat Protoc 2012; 7:500–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong W, Darville T, Zheng X, Fine J, Li Y. Generalized multi-SNP mediation intersection-union test. Biometrics 2020; doi: 10.1111/biom.13418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laheri S, Ashary N, Bhatt P, Modi D. Oviductal glycoprotein 1 (OVGP1) is expressed by endometrial epithelium that regulates receptivity and trophoblast adhesion. J Assist Reprod Genet 2018; 35:1419–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright MM, McMaster CR. PC and PE synthesis: mixed micellar analysis of the cholinephosphotransferase and ethanolaminephosphotransferase activities of human choline/ethanolamine phosphotransferase 1 (CEPT1). Lipids 2002; 37:663–72. [DOI] [PubMed] [Google Scholar]
- 12.Ruiz M, Jové M, Schlüter A, et al. . Altered glycolipid and glycerophospholipid signaling drive inflammatory cascades in adrenomyeloneuropathy. Hum Mol Genet 2015; 24:6861–76. [DOI] [PubMed] [Google Scholar]
- 13.Davis RL, Turner DL. Vertebrate hairy and enhancer of split related proteins: transcriptional repressors regulating cellular differentiation and embryonic patterning. Oncogene 2001; 20:8342–57. [DOI] [PubMed] [Google Scholar]
- 14.Buhi WC. Characterization and biological roles of oviduct-specific, oestrogen-dependent glycoprotein. Reproduction 2002; 123:355–62. [DOI] [PubMed] [Google Scholar]
- 15.Nagarajan UM, Prantner D, Sikes JD, et al. . Type I interferon signaling exacerbates Chlamydia muridarum genital infection in a murine model. Infect Immun 2008; 76: 4642–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagarajan UM, Sikes J, Prantner D, et al. . MyD88 deficiency leads to decreased NK cell gamma interferon production and T cell recruitment during Chlamydia muridarum genital tract infection, but a predominant Th1 response and enhanced monocytic inflammation are associated with infection resolution. Infect Immun 2011; 79:486–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carrasco SE, Hu S, Imai DM, et al. . Toll-like receptor 3 (TLR3) promotes the resolution of Chlamydia muridarum genital tract infection in congenic C57BL/6N mice. PLoS One 2018; 13:e0195165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai Y, Di Nardo A, Nakatsuji T, et al. . Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat Med 2009; 15:1377–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Q, Fang D, Fang J, et al. . Impaired wound healing with defective expression of chemokines and recruitment of myeloid cells in TLR3-deficient mice. J Immunol 2011; 186:3710–7. [DOI] [PubMed] [Google Scholar]
- 20.Day A, Jameson Z, Hyde C, Simbi B, Fowkes R, Lawson C. C -type natriuretic peptide (CNP) inhibition of interferon-gamma-mediated gene expression in human endothelial cells in vitro. Biosensors (Basel) 2018; 8:86–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kane CD, Vena RM, Ouellette SP, Byrne GI. Intracellular tryptophan pool sizes may account for differences in gamma interferon-mediated inhibition and persistence of chlamydial growth in polarized and nonpolarized cells. Infect Immun 1999; 67:1666–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tseng CT, Rank RG. Role of NK cells in early host response to chlamydial genital infection. Infect Immun 1998; 66:5867–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones N, Cronin JG, Dolton G, et al. . Metabolic adaptation of human CD4+ and CD8+ T-cells to T-cell receptor-mediated stimulation. Front Immunol 2017; 8:1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liou LY, Walsh KB, Vartanian AR, et al. . Functional glycosylation of dystroglycan is crucial for thymocyte development in the mouse. PLoS One 2010; 5:e9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freeley S, Cardone J, Günther SC, et al. . Asparaginyl endopeptidase (Legumain) supports human Th1 induction via cathepsin L-mediated intracellular C3 activation. Front Immunol 2018; 9:2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cunnick JM, Schmidhuber S, Chen G, et al. . Bcr and Abr cooperate in negatively regulating acute inflammatory responses. Mol Cell Biol 2009; 29:5742–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee MN, Roy M, Ong SE, et al. . Identification of regulators of the innate immune response to cytosolic DNA and retroviral infection by an integrative approach. Nat Immunol 2013; 14:179–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsumoto M, Seya T. TLR3: interferon induction by double-stranded RNA including poly(I:C). Adv Drug Deliv Rev 2008; 60:805–12. [DOI] [PubMed] [Google Scholar]
- 29.Saito H, Morita Y, Fujimoto M, Narazaki M, Naka T, Kishimoto T. IFN regulatory factor-1-mediated transcriptional activation of mouse STAT-induced STAT inhibitor-1 gene promoter by IFN-gamma. J Immunol 2000; 164:5833–43. [DOI] [PubMed] [Google Scholar]
- 30.Fuchs A, Cella M, Giurisato E, Shaw AS, Colonna M. Cutting edge: CD96 (tactile) promotes NK cell-target cell adhesion by interacting with the poliovirus receptor (CD155). J Immunol 2004; 172:3994–8. [DOI] [PubMed] [Google Scholar]
- 31.Kan AA, van Erp S, Derijck AA, et al. . Genome-wide microRNA profiling of human temporal lobe epilepsy identifies modulators of the immune response. Cell Mol Life Sci 2012; 69:3127–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson SD, Sudman M, Ramos PS, et al. . The susceptibility loci juvenile idiopathic arthritis shares with other autoimmune diseases extend to PTPN2, COG6, and ANGPT1. Arthritis Rheum 2010; 62:3265–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujino T, Yamazaki Y, Largaespada DA, et al. . Inhibition of myeloid differentiation by Hoxa9, Hoxb8, and Meis homeobox genes. Exp Hematol 2001; 29:856–63. [DOI] [PubMed] [Google Scholar]
- 34.Blois SM, Sulkowski G, Tirado-González I, et al. . Pregnancy-specific glycoprotein 1 (PSG1) activates TGF-β and prevents dextran sodium sulfate (DSS)-induced colitis in mice. Mucosal Immunol 2014; 7:348–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma P, Khairnar V, Madunić IV, et al. . SGLT1 deficiency turns listeria infection into a lethal disease in mice. Cell Physiol Biochem 2017; 42:1358–65. [DOI] [PubMed] [Google Scholar]
- 36.Tabiasco J, Devêvre E, Rufer N, et al. . Human effector CD8+ T lymphocytes express TLR3 as a functional coreceptor. J Immunol 2006; 177:8708–13. [DOI] [PubMed] [Google Scholar]
- 37.van der Slot AJ, Zuurmond AM, Bardoel AF, et al. . Identification of PLOD2 as telopeptide lysyl hydroxylase, an important enzyme in fibrosis. J Biol Chem 2003; 278:40967–72. [DOI] [PubMed] [Google Scholar]
- 38.Farina GA, York MR, Di Marzio M, et al. . Poly(I:C) drives type I IFN- and TGFβ-mediated inflammation and dermal fibrosis simulating altered gene expression in systemic sclerosis. J Invest Dermatol 2010; 130:2583–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Behrens P, Brinkmann U, Wellmann A. CSE1L/CAS: its role in proliferation and apoptosis. Apoptosis 2003; 8:39–44. [DOI] [PubMed] [Google Scholar]
- 40.Paredes F, Parra V, Torrealba N, et al. . HERPUD1 protects against oxidative stress-induced apoptosis through downregulation of the inositol 1,4,5-trisphosphate receptor. Free Radic Biol Med 2016; 90:206–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Z, Lv YJ, Song YP, et al. . Lysosomal membrane protein TMEM192 deficiency triggers crosstalk between autophagy and apoptosis in HepG2 hepatoma cells. Oncol Rep 2012; 28:985–91. [DOI] [PubMed] [Google Scholar]
- 42.Sinturel F, Bréchemier-Baey D, Kiledjian M, Condon C, Bénard L. Activation of 5’-3’ exoribonuclease Xrn1 by cofactor Dcs1 is essential for mitochondrial function in yeast. Proc Natl Acad Sci U S A 2012; 109:8264–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu P, Verhaar AP, Peppelenbosch MP. Signaling size: ankyrin and SOCS box-containing ASB E3 ligases in action. Trends Biochem Sci 2019; 44:64–74. [DOI] [PubMed] [Google Scholar]
- 44.Lee KW, Okot-Kotber C, LaComb JF, Bogenhagen DF. Mitochondrial ribosomal RNA (rRNA) methyltransferase family members are positioned to modify nascent rRNA in foci near the mitochondrial DNA nucleoid. J Biol Chem 2013; 288:31386–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soleimanpour-Lichaei HR, Kühl I, Gaisne M, et al. . mtRF1a is a human mitochondrial translation release factor decoding the major termination codons UAA and UAG. Mol Cell 2007; 27:745–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muñoz JJ, Roca C, Santos JL, Arroyo M, de Salamanca RE. Effect of zinc or S-adenosyl-l-methionine on long term administration of low doses of lead to rats. Pharmacol Toxicol 1993; 73:189–91. [DOI] [PubMed] [Google Scholar]
- 47.Schönemann L, Kühn U, Martin G, et al. . Reconstitution of CPSF active in polyadenylation: recognition of the polyadenylation signal by WDR33. Genes Dev 2014; 28:2381–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cassandri M, Smirnov A, Novelli F, et al. . Zinc-finger proteins in health and disease. Cell Death Discov 2017; 3:17071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baranova I, Kovarikova H, Laco J, et al. . Identification of a four-gene methylation biomarker panel in high-grade serous ovarian carcinoma. Clin Chem Lab Med 2020; 58:1332–40. [DOI] [PubMed] [Google Scholar]
- 50.Li J, Xia F, Reithmeier RA. N-glycosylation and topology of the human SLC26 family of anion transport membrane proteins. Am J Physiol Cell Physiol 2014; 306:C943–60. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.