Abstract
Cell growth is orchestrated by a number of interlinking cellular processes. Components of the TOR pathway have been proposed as potential regulators of cell growth, but little is known about their immediate effects on protein synthesis in response to TOR‐dependent growth inhibition. Here, we present a resource providing an in‐depth characterisation of Schizosaccharomyces pombe phosphoproteome in relation to changes observed in global cellular protein synthesis upon TOR inhibition. We find that after TOR inhibition, the rate of protein synthesis is rapidly reduced and that notable phosphorylation changes are observed in proteins involved in a range of cellular processes. We show that this reduction in protein synthesis rates upon TOR inhibition is not dependent on S6K activity, but is partially dependent on the S. pombe homologue of eIF4G, Tif471. Our study demonstrates the impact of TOR‐dependent phospho‐regulation on the rate of protein synthesis and establishes a foundational resource for further investigation of additional TOR‐regulated targets both in fission yeast and other eukaryotes.
Keywords: phosphoproteomics, protein synthesis, TOR regulation
Subject Categories: Metabolism; Post-translational Modifications, Proteolysis & Proteomics; Signal Transduction
Phosphoproteomic analysis of Schizosaccharomyces pombe reveals a TOR‐dependent network of phosphoproteins involved in the regulation of protein synthesis rates.
Introduction
Cell growth, the accumulation of biomass and cell reproduction, underpin the overall growth of all living organisms. Biomass is made up of cellular components and macromolecules, a major constituent of which is protein. Protein biosynthesis is considered the largest energy‐consuming process in actively growing cells, accounting for ˜30% in mammalian cells and ˜50% in bacterial cells of total energy consumption (Buttgereit & Brand, 1995; Russell & Cook, 1995). The control of the global cellular rate of protein synthesis is therefore central to understanding cellular growth control. In eukaryotic cells, the target of rapamycin (TOR) signalling pathway is thought to play a key role in regulating cell growth. TOR is a serine/threonine protein kinase that is a member of the phosphatidylinositol kinase‐related kinase (PIKK) family. It was discovered in budding yeast through characterising mutants resistant to the growth inhibitor rapamycin (Heitman et al, 1991) and is conserved across eukaryotes (Brown et al, 1994). The TOR kinases form two protein complexes, TOR complex 1 (TORC1) and TOR complex 2 (TORC2). Studies in both yeasts and mammalian cells have suggested TORC1 as the major complex involved in cellular growth control (Loewith et al, 2002; Wullschleger et al, 2006; Weisman et al, 2007; Yang et al, 2013). Despite the large body of work that has been undertaken to characterise TOR‐regulated processes, surprisingly little is known about how components downstream of TOR signalling are involved in altering the rates of protein synthesis. Therefore, to understand eukaryotic growth control, it is critical to elucidate how TOR‐regulated pathways modulate growth processes, particularly the rate of global cellular protein synthesis.
Previous studies have focussed on the cellular effects of inhibiting the TOR kinases. Rapamycin treatment leads to a progressive decrease in the rate of protein synthesis in budding yeast (Barbet et al, 1996) and to a lesser extent in mammalian cells (Beretta et al, 1996). More potent and selective inhibitors of TOR, such as Torin1, specifically target the catalytic site of TOR (Thoreen et al, 2009), inhibiting cellular growth in both mammalian (Thoreen et al, 2012) and fission yeast (Atkin et al, 2014) cells. Torin1 inhibits the function of both TOR kinases in fission yeast, which are found in two distinct protein complexes, TORC1 (contains the Tor2 kinase) and TORC2 (contains the Tor1 kinase) (Weisman et al, 2007). Inhibition of growth with Torin1 is also reversible and does not induce cell death even after 24 h of incubation (Atkin et al, 2014) and thus provides a useful means to perturb TOR signalling and enable physiological studies upon inhibition of TOR activity.
As TOR and other components within the TOR‐signalling pathway are protein kinases, changes in protein phosphorylation have been investigated to identify potential targets involved in regulating cellular growth processes after TOR inhibition. The S6 kinases and translation initiation‐related factors, including the 4E‐binding protein (4E‐BP) and eIF4F complex, are among the most widely discussed proteins as potential downstream targets of TOR in having an impact on protein synthesis upon inhibition (Schmelzle & Hall, 2000; Averous & Proud, 2006; De Virgilio & Loewith, 2006; Ma & Blenis, 2009). Early studies in budding yeast suggested that Sch9 (a S6K orthologue) is a major target of TORC1 and is thought to regulate translation initiation through transcriptional regulation of RNA pol III and pol I‐dependent genes, many of which are ribosome biogenesis (ribi)‐related genes (Urban et al, 2007); (Huber et al, 2009). However, studies in mice have shown that a complete knockout of S6K1 did not affect global translational activity (Garelick et al, 2013), raising questions about the role of the S6 kinases and their downstream phosphorylation targets in regulating translational activity.
Phosphoproteomic studies undertaken in both budding yeast and mammalian cells have uncovered interactions between TOR and various pathways that regulate growth, including the PKA (Soulard et al, 2010) and insulin signalling pathways (Hsu et al, 2011; Yu et al, 2011), as well as pyrimidine and amino acid biosynthesis (Robitaille et al, 2013; Oliveira et al, 2015). However, it still remains unclear how these observed phosphoproteomic changes act to influence cellular growth or protein synthesis. Most of these phosphorylation studies have lacked sufficient temporal resolution to correlate phosphosphorylation changes with alterations in growth processes. Their phosphosite analyses were also limited by the mass spectrometry techniques employed. Recent advancements have increased the number of samples that can be processed in parallel, as well as detection sensitivity and reliability, enhancing the molecular resolution and improving the quantitative phosphoproteomic analyses considerably.
Using these improved techniques, we have carried out an in‐depth phosphoproteomics study in fission yeast to characterise the changes at fine temporal resolution after inhibition of TOR kinase activity with Torin1. Our study has generated a database of proteins and phosphosite targets potentially involved in modulating protein synthesis and other growth‐related processes. Functional categorisation of these proteins, based on cellular processes, provides a more comprehensive overview of the essential functions that may be involved in coordinating growth control. Many of the target proteins identified are conserved in both budding yeast and humans, extending the relevance of the database. By monitoring protein synthesis rates in parallel with TOR inhibition, we are able to correlate the phosphorylation changes with the kinetics of protein synthesis inhibition. This an improved approach to identify potential targets that may have roles in the regulation of protein synthesis, and are dependent on the TOR‐signalling pathway.
Results
TOR activity is a major regulator of cellular protein synthesis rates
The rate of global cellular protein synthesis was monitored by measuring the rate of incorporation of the methionine analogue L‐homoproparylglycine (HPG) (Knutsen et al, 2015; Green & Pelkmans, 2016), after inhibition of TOR with Torin1 in fission yeast. The methionine analogue was tagged with a fluorescent label using click chemistry, with the signal detected at a single‐cell level using flow cytometry. Fluorescent signal measured in a population of exponentially growing fission yeast cells was normally distributed on a logscale (Fig 1A), and the median rate of analogue incorporation, after an initial lag of 5 min, was linear over 60 min (Fig 1B). Therefore to ensure that the incorporation of the analogue was within the linear range, a pulse of 15 min or longer was used to calculate the rate of protein synthesis (detailed in Materials and Methods).
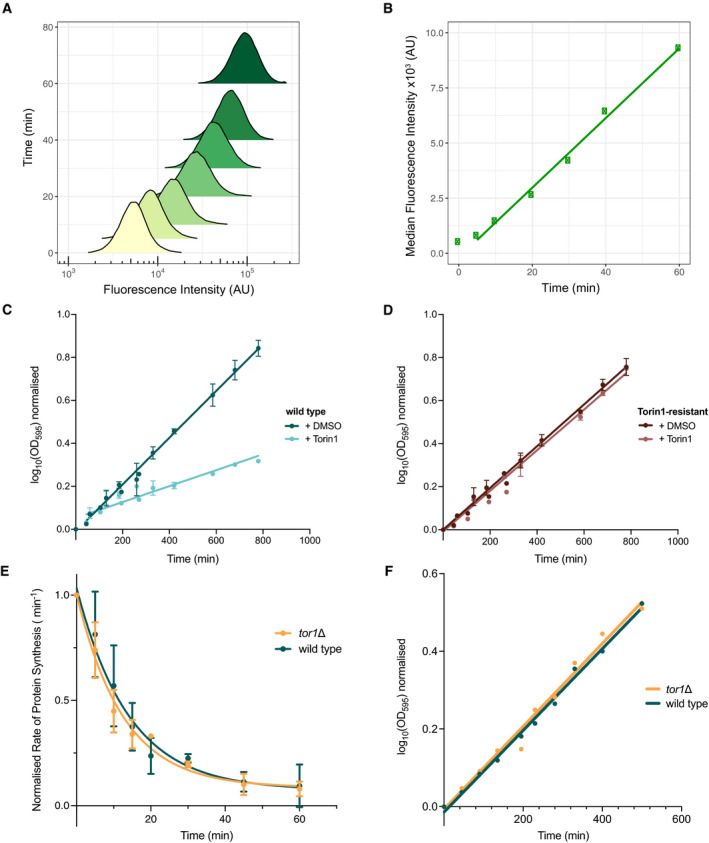
Figure 1. TOR activity is a major regulator of cellular protein synthesis rates.
-
ADensity plot showing the measured fluorescence intensity distribution of individual cells for each time point after HPG addition. Time is represented on the y‐axis, with the bottom of the distribution plot aligning to the corresponding time point.
-
BMedian fluorescence intensity of the distribution plots in A plotted against the indicated time after HPG addition. Linear regression line fitted from 5 min onwards.
-
C, DGrowth rates as measured by the change in optical density (OD595) over time for wild type (C) and Torin1‐resistant (D) cells upon 5 µM of Torin1 treatment (at T = 0 min). Respective strains were treated with the DMSO vehicle alone to control for potential effects of the solvent on cell growth. Logarithmic values of the OD595 increase are plotted against time in minutes. Data aggregated from three independent experiments with error bars representing the standard deviations (SD). Linear regressions were fitted using the curve‐fitting function on Prism9. Ratios in the rate of mass doubling (as calculated from gradient of regression lines detailed in Materials and Methods) of Torin1‐treated cells to DMSO control were 0.34 in wild type (C) and 0.97 for Torin1‐resistant (D) cells.
-
ERate of protein synthesis as measured by the amount of HPG incorporation after 20 min of differing time intervals (0–60 min) of 5 µM Torin1 for wild type (green) and tor1∆ (yellow). Values are normalised to T = 0 min. Exponential one‐phase decay non‐linear regressions were fitted using the curve‐fitting function on Prism9. Data aggregated from 3 independent experiments with error bars representing the standard deviations (SD).
-
FGrowth rates for wild type and tor1∆, as measured by the change in optical density (OD595) over time. Linear regressions fitted with Prism9.
The selective ATP‐competitive inhibitor Torin1 completely inhibits colony formation at 5 µM (Lie et al, 2018). We demonstrated that when it was added to exponentially growing fission yeast cells in liquid culture, growth as measured by increase in optical density (OD595) began to reduce by 80–90 min, with the rate of mass doubling reducing to a third compared to cells treated with the DMSO solvent alone (Fig 1C). This experiment was repeated using the Torin1‐resistant, tor2‐G2040D mutant (Atkin et al, 2014), and the rate of mass doubling was found to be unchanged after Torin1 addition (Fig 1D). This established that the mutant was resistant to inhibitory growth effects of the drug in terms of cellular mass increase and was subsequently used to control for off‐target effects of the Torin1 inhibitor.
To investigate the effects of TOR inhibition on protein synthesis, the rate of synthesis was monitored in cells treated with Torin1 at 5 µM for differing lengths of time (Fig 1E). In wild type, the synthesis rate fell to 50% of the starting level within 10 min of Torin1 addition, 75% by 20 min and 90% by 40 min. This shows that the majority of protein synthesis is dependent upon TOR activity, and therefore, we would expect that the activity of potential TOR regulatory targets would also be altered within 10–20 min. We next assayed the protein synthesis rate in a tor1∆ mutant strain, which eliminates the non‐essential TORC2. There was no measurable difference in the growth rates between wild type and tor1∆ cells (Fig 1F), and the changes in rates of protein synthesis upon TOR inhibition were identical (Fig 1E). These observations are consistent with the idea that the essential TORC1 is the major complex responsible for the rapid inhibition of protein synthesis and that changes in phosphorylation levels relevant to regulating protein synthesis should be detectable within 20 min and significant by 40 min of TOR inhibition.
TOR‐dependent phosphoproteomic changes identify conserved proteins with potential roles in modulating rates of protein synthesis
Based on these results, we carried out a proteomics and phosphoproteomics time course study to identify the molecular changes responsible for the reduction in rates of biomass accumulation and protein synthesis after TOR inhibition. Exponentially growing wild type and Torin1‐resistant cells were treated with Torin1 (5 µM) and sampled at intervals from 5 to 85 min. The samples were labelled with isobaric tandem mass tag labels (TMT10plex) for relative quantification of both the protein and individual phospho‐peptide levels (detailed in Materials and Methods).
A total of 3,467 proteins were identified across all samples, accounting for ˜70% of the fission yeast proteome (Table EV1). After controlling for off‐target effects of DMSO (the Torin1 solvent; see Materials and Methods), none of the proteins showed more than a 2‐fold change in protein levels relative to the wild type starting levels (T = 0 min), in either wild type or Torin1‐resistant cells, after 40 min of Torin1 treatment (Fig 2A), when the rate of protein synthesis was reduced by around 90% (Fig 1E). A closer look at the frequency distribution of the changes in wild type cells showed that 98% of the proteins (3,323/3,393) were within a ± 1.3‐fold range for their changes in protein levels (Fig 2B). And for those that showed changes between 1.3 to 2‐fold, corresponding changes were also observed in Torin1‐resistant cells, as indicated by the positively correlated values on Fig 2A (y = x in light grey, plotted as guide), suggesting that these are likely due to non‐specific and off‐target effects of DMSO or Torin1 treatment. The combined observations in Fig 2A and B therefore indicate that the changes in protein levels are unlikely to play a role in regulating the rate of protein synthesis after TOR inhibition.
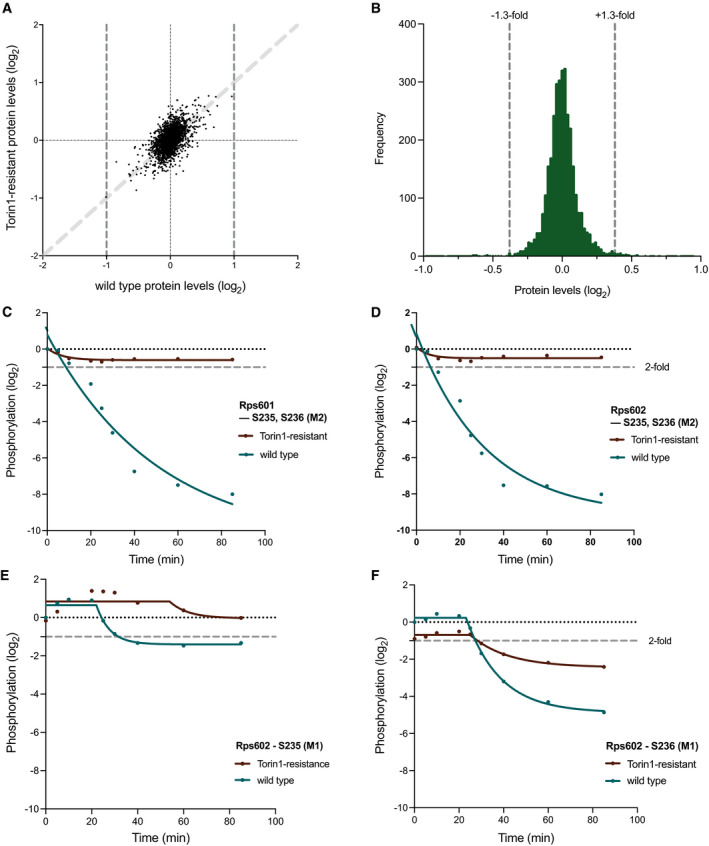
Figure 2. TOR‐dependent phosphoproteomic changes identifies conserved proteins with potential roles in modulating rates of protein synthesis.
-
AScatter plot showing the protein levels in wild type cells after 40 min of Torin1 (5 µM) addition plotted against the corresponding values for the same protein in Torin1‐resistant cells after 40 min. Dark grey dashed lines at x = ±1 indicate the 2‐fold threshold, of which no proteins exceeded in the study. The light grey dashed line represents y = x, plotted as guide to indicate positive correlation in protein levels between wild type and Torin1‐resistant cells.
-
BFrequency distribution of the proteins levels after 40 min of Torin1 treatment in wild type cells (n = 3,380), with 98% of all proteins (3,314/3,380) within a 1.3‐fold boundary, as indicated by the grey dashed lines at ± 0.38.
-
C–FPhosphorylation changes of the conserved phosphosites, S235 and S236, on Rps601(C) and Rps602 (D–F), over 85 min of Torin1 (5 µM) treatment wild type (green) and Torin1‐resistant (brown) cells. M‐values (in brackets) represent the different multiplicities of the same phosphosites. Lines were fitted with the curve‐fitting function on Prism9 using the non‐linear regressions exponential one‐phase decay (C, D) or plateau followed by one‐phase decay (E, F) functions, respectively. Grey dashed lines indicate 2‐fold threshold.
We next analysed the phosphoproteome following TOR inhibition. To confirm the effectiveness of Torin1 inhibition, we first analysed the phosphorylation behaviour of the 40S ribosomal protein S6 (RPS6), a well‐established biochemical read‐out of TORC1 and TORC2 activity (Nakashima et al, 2010; Du et al, 2012; Atkin et al, 2014). In fission yeast, there are two copies of the gene encoding for the Rps6 protein, rps601 and rps602, each containing two well‐conserved phosphosites at positions S235 and S236, both of which were identified in our study. In both proteins, the two phosphosites exhibited rapid dephosphorylation kinetics in the respective proteins, exceeding close to 4‐fold within 20 min of Torin1 treatment in wild type cells (Fig 2C and D). For the Torin1‐resistant mutant, the phosphorylation levels remained constant throughout the time course, demonstrating that the dephosphorylation observed in Rps6 phosphosites in wild type cells is from the inhibition of TOR kinases activity, and not due to off‐target effects of Torin1 treatment. Further information can be inferred from the multiplicity (denoted by the M‐number in brackets) of the two phosphosites on each of the proteins. In this case, M2 indicates that two phosphorylation events were detected within the same fragmented peptide at the same time phosphorylation was detected on the specified phosphosite. We also detected the M1 versions of the S235 and S236 phosphosites in this experiment, but the phosphorylation changes were much less immediate than the M2 phosphosites (Fig 2E and F). This suggests that their phosphorylation behaviours are likely dependent on each other, possibly cooperative.
This phosphorylation study identified a total of 20,072 phosphosites on 2,271 phosphoproteins, over 3 times the number of phosphosites identified from previous TOR phosphoproteomics studies in both budding yeast and mammalian cells. We controlled for non‐specific and off‐target phosphorylation changes caused by DMSO or Torin1 treatment by taking into account the phosphorylation values of the Torin1‐resistant strain at each time point (detailed in Materials and Methods, Table EV2). A total of 1,043 phosphosites were identified to show a more than 2‐fold increase or decrease in phosphorylation at the end of the 85‐min time course, accounting for 5% of the overall detected phosphosites. Of these, 650 phosphosites on 264 phosphoproteins exhibited a more than 2‐fold phosphorylation change relative to starting levels within 40 min of Torin1 addition (Table EV3). Among the 264 phosphoproteins, 160 have homologues in both budding yeast and human cells, forming a database of proteins under immediate phosphorylation control upon TOR kinase inhibition (conservation indicated on Table EV3). An accompanying data visualisation tool (Shiny app) has been made available online for visualising the individual phosphorylation kinetics (https://tmak.shinyapps.io/TM_App‐Table_EV3/), and an illustrated example of the workflow is provided in Fig EV1 and detailed in Materials and Methods.
Figure EV1. Illustrated example for data visualisation tool.
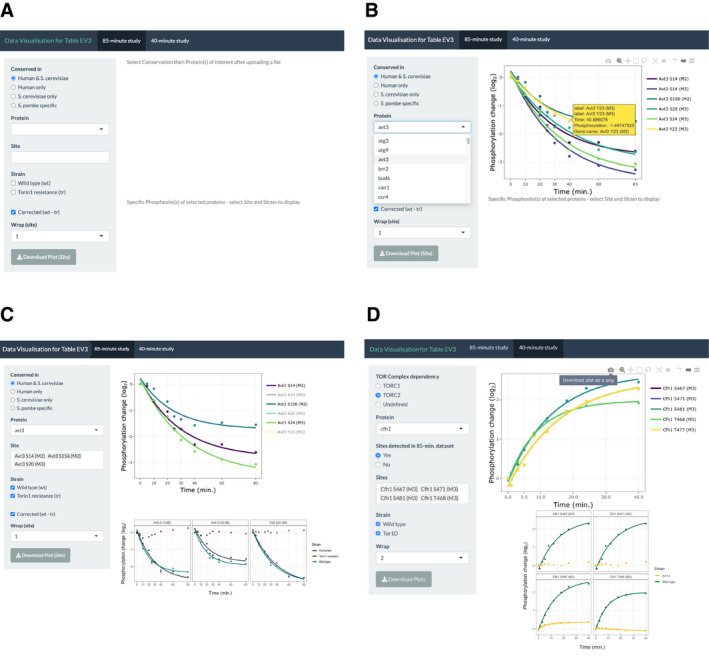
- Initial interface of the first “85‐min study” tab upon opening the application from the given link.
- Demonstration of initial filtering of protein list based on conservation and subsequent display of graph with all phosphosites in top panel.
- Illustration of individual phosphosite and specification of strain dataset to display in lower facet panels.
- Interface of the second “40‐min study tab” interface with additional filtering criteria on sidebar panel and demonstration of the “Wrap” and “Download” functions.
Cellular coordination of growth control
To gain an overview of the proteins involved in the coordination of cellular growth control, we looked to see what key functional processes are represented among the proteins that underwent rapid phosphorylation changes upon TOR kinase inhibition. Since the TOR‐signalling pathway and its interacting network are highly conserved across eukaryotes, we focussed on the 160 conserved proteins containing homologues in both human cells and budding yeast. We classified the proteins based on their Gene Ontology (GO) Biological Process annotations (from PomBase Lock et al, 2019) and found that 90% of the proteins (143/160) were represented by 17 major categories (Table 1). As it is likely that the core regulatory proteins needed to regulate growth are also required for cellular survival, we looked first to see which of the proteins are encoded by essential genes. Of the conserved proteins, 23% (36/160) were found to be essential for cell viability (Table EV3), a third of which (12/36) have roles in cytoplasmic translation (GO:0002181) and ribosome biogenesis (GO:0042254). This supports the view that translation is a major point of regulation for TOR‐dependent growth control.
Table 1.
17 major GO Biological Process categories for the 160 conserved proteins with phosphosites showing more than 2‐fold change within 40 min of Torin1 addition.
| GO term | Frequency | Gene names (Essential genes in bold) |
|---|---|---|
| Signalling (GO:0023052) | 37 (5) | amk2, atg1, bzz1, cek1, far11, gtr2, igo1, iml1, inp53, kcs1, mor2, orb6, pab1, par1, pik1, pom1, pom2, ppk19, ppk25, ppk30, ras1, rga3, rgf1, rnc1, rng2, sat1, sck1, sck2, sea2, sea3, sec73, SPAC824.09c, SPBC16E9.02c, stm1, syt22, win1, wis1 |
| Vesicle‐Mediated Transport (GO:0016192) | 33 (10) | bzz1, cdc15, ede1, gga21, glo3, gyp1, irs4, mpd2, mvp1, nxt3, orb6, osh2, pan1, pep12, pob1, ppk19, sat1, sec16, sec2, sec27, sec31, sec5, sec73, sec9, SPAC824.09c, SPCC622.14, sst4, syb1, syt22, trs130, vps17, vti1, wdr44 |
| Cytoplasmic Translation (GO:0002181) | 15 (10) | erf1, hrs1, rpl8,rps101, rps601, rps602, slh1, SPAC29E6.06c , sum2, sum3, sup35, tif212, tif225, tif303, tif471 |
| Ribosome Biogenesis (GO:0042254) | 15 (4) | enp1, noc12, nup124, nup146, nup40, pxr1, rei1, rpl8, rps101, rps601, rps602, SPBC16H5.08c, SPBC4.02c, tif471, tif6 |
| Actin Cytoskeleton Organization (GO:0030036) | 13 (2) | bud6, bzz1, cdc15, ede1, imp2, myp2, orb6, pan1, pom1, ppk30, pxl1, rng2, tea4 |
| Autophagy (GO:0006914) | 13 (1) | atg1, atg11, atg13, atg3, atg9, gtr2, iml1, irs4, nxt3, osh2, ppk19, sec16, ubx3 |
| Establishment or Maintenance of Cell Polarity (GO:0007163) | 13 (2) | alp14, bud6, cdc15, mor2, mto1, orb6, osh2, pom1, ras1, syt22, tea1, tea3, tea4 |
| Chromatin Organization (GO:0006325) | 12 (2) | bdf2, eaf6, eaf7, hta2, nap1, nap2, pst3, set2, sgf29, shf1, SPBC4.02c, taf12 |
| Membrane Organization (GO:0061024) | 12 (6) | cdc15, glo3, mvp1, ned1, pep12, sec16, sec31, sec9, syb1, ubx3, vps17, vti1 |
| Protein‐Containing Complex Assembly (GO:0065003) | 12 (6) | alp14, brr2, bud6, mto1, nap1, nap2, rng2, sec16, taf12, tif212, tif303, tif6 |
| Transcription, DNA‐Templated (GO:0006351) | 12 (6) | eaf7, hsf1, maf1, nup40, pst3, rpc37, set2, sfp1, sgf29, taf12, tfa1, tfg1 |
| Nucleocytoplasmic Transport (GO:0006913) | 11 (2) | kap123, ned1, npp106, nup124, nup146, nup40, nup61, ras1, rei1, SPBC16H5.08c, tif6 |
| mRNA Metabolic Process (GO:0016071) | 9 (1) | brr2, ccr4, cip1, cip2, not2, not3, rnc1, sts5, vts1 |
| DNA Repair (GO:0006281) | 8 (2) | eaf6, eaf7, hta1, hta2, pms1, pst3, rad21, ung1 |
| Mitotic Cell Cycle Phase Transition (GO:0044772) | 8 (0) | cek1, igo1, pab1, par1, pom1, rad25, sck2, wis1 |
| Mitotic Cytokinesis (GO:0000281) | 8 (1) | cdc15, igo1, imp2, myp2, pom1, pom2, pxl1, rng2 |
| Transmembrane Transport (GO:0055085) | 8 (1) | abc2, avt3, can1, pmc1, sbh1, sec62, SPCC1672.11c, stm1 |
| Uncategorised | 17 (0) | ada1, dml1, gde1, gpd1, izh3, knk1, nrp1, ntp1, nvj2, pcc1, pcy1, scw1, SPAC22A12.14c, SPBC30D10.05c, tcb2, tmf1, ubp7 |
Next, we performed a GO enrichment analysis on the 160 conserved proteins to identify other cellular processes that are potentially involved in TOR‐dependent growth control (Table EV4). We found that proteins responsible for establishing and maintaining cell polarity (GO:0061246) were among the most highly enriched categories (14.5‐fold). Other structural organisation‐related categories were also seen to be enriched, with proteins involved in cortical actin cytoskeleton organisation (GO:0030865) and organelle localisation (GO:0051640), exhibiting a 4.2‐ and 2.9‐fold enrichment, respectively (Table EV4). This highlights that the structural organisation of a cell is heavily regulated upon TOR inhibition.
As TOR is a major signalling kinase within the cell, it is unsurprising that many signalling‐related proteins are heavily regulated upon TOR inhibition. Overall, signalling‐related proteins (GO: 0023052) account for the largest group of proteins (37/160) that exhibit rapid phosphorylation changes upon TOR inhibition. In particular, enrichment analysis identified a 5.6‐fold enrichment for those involved in the negative regulation of intracellular signal transduction (GO:1902532; Table EV4). Other notable categories include proteins responsible for retrograde transport from the endosome to Golgi (GO:0042147; 7.6‐fold), which are partly represented by vesicle‐mediated transport proteins (GO:0016192; 2.5‐fold), as well as those involved in autophagy‐related processes (G0:0000422; 9.9‐fold and GO:0000045; 9.1‐fold) (Table EV4). These observations provide evidence that TOR‐dependent growth regulation is highly coordinated and involves multiple major cellular processes.
TORC1‐dependent phosphorylation regulates the rate of protein synthesis
Given that the S. pombe TORC2 (containing Tor1) is not involved in the rapid inhibition of protein synthesis (Fig 1E), we focussed on TORC1, the complex containing the essential Tor2 kinase. Using similar proteomic and phosphoproteomic approaches, we investigated the relative phosphorylation changes in wild type and tor1∆ cells to focus on potential targets that could play a critical role in modulating the rates of protein synthesis following TOR inhibition. We performed a shorter 40‐min time course experiment, by which time the rate of protein synthesis is much reduced, and sampled at higher frequency to gain a better temporal resolution of the protein and phosphorylation changes occurring at early time points following Torin1 addition (see Materials and Methods). This second time course largely replicates the critical part of the first time course, and examination of the same phosphosites from the two experiments indicates that the data are reproducible.
At the protein level, 3707 proteins were detected in both the wild type and tor1∆ cells across all time points (Table EV1). None of the detected proteins showed more than a 2‐fold change in expression levels in either of the two strains after 40 min of Torin1 treatment (Fig EV2). We investigated whether there were inherent differences in the protein levels between cells containing both functional TOR complexes (wild type) or only the essential TOR complex 1 (tor1∆). The corresponding levels of individual proteins in tor1∆ cells were compared to the levels measured in wild type cells before background subtraction and before Torin1 addition. 26 proteins showed over a 1.5‐fold difference in expression in tor1∆ compared to wild type cells before Torin1 treatment (Protein IDs in red in Table EV1 and red in Fig EV2), indicating that the expression of these proteins is affected by the absence of TORC2 during steady‐state growth. Inhibition of TOR activity, however, did not cause a change in relative expression of these proteins when compared to their background expression levels in the respective strains, suggesting that the absence of the non‐essential Tor1 kinase does not have an effect on protein expression upon inhibition of TOR activity (highlighted in red in Fig EV2). The observation of unchanged protein expression upon TOR inhibition in either of the two strains supports our earlier conclusion that changes in the rates of protein synthesis upon TOR inhibition are unlikely to involve changes in protein levels.
Figure EV2. Relative protein expression levels in wild type and tor1∆ after 40 min of Torin1 treatment.
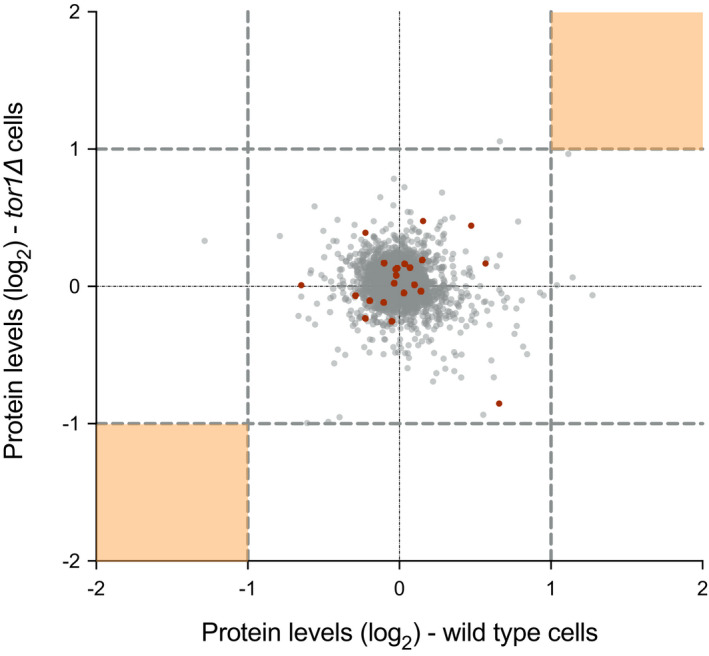
Scatter plot of the relative expression levels of individual protein in wild type and tor1∆ cells from the 40‐min proteomic time course experiment. Twofold thresholds are represented by grey dashed lines for either direction in the respective strains. Orange shaded areas indicate the expected regions with more than 2‐fold changes in relative protein expression levels in both wild type and tor1∆ cells, of which none were detected in the study. Points highlighted in red indicate the 23 proteins showing over 1.5‐fold difference in expression levels between wild type and tor1∆ before Torin1 treatment. But the relative expression levels of these proteins remain within a 2‐fold range upon inhibition of TOR activity inhibition.
A total of 7,162 phosphosites were identified across all 20 samples in this experiment, representing a total of 1,532 phosphoproteins, 89% of which (6,340/7,162) were also in common with the 85‐min time course (Table EV2). 372 phosphosites on 177 proteins showed a more than 2‐fold change within 40 min of Torin1 treatment in wild type cells (marked on Table EV3). To focus on the changes that are dependent upon TORC1, we first excluded phosphosites that were dependent on TORC2. 43 of the 374 sites showed a more than 2‐fold change in phosphorylation in wild type cells, but remained below a 1.3‐fold difference in phosphorylation in tor1∆ cells within 40 min of Torin1 addition (Fig 3A and B; marked as “TORC2” on Table EV3 and visualisable on Shiny App, see Materials and Methods). This indicates that the phosphorylation changes detected in wild type cells are dependent upon the presence of the non‐essential TORC2 complex and hence were excluded from further analysis.
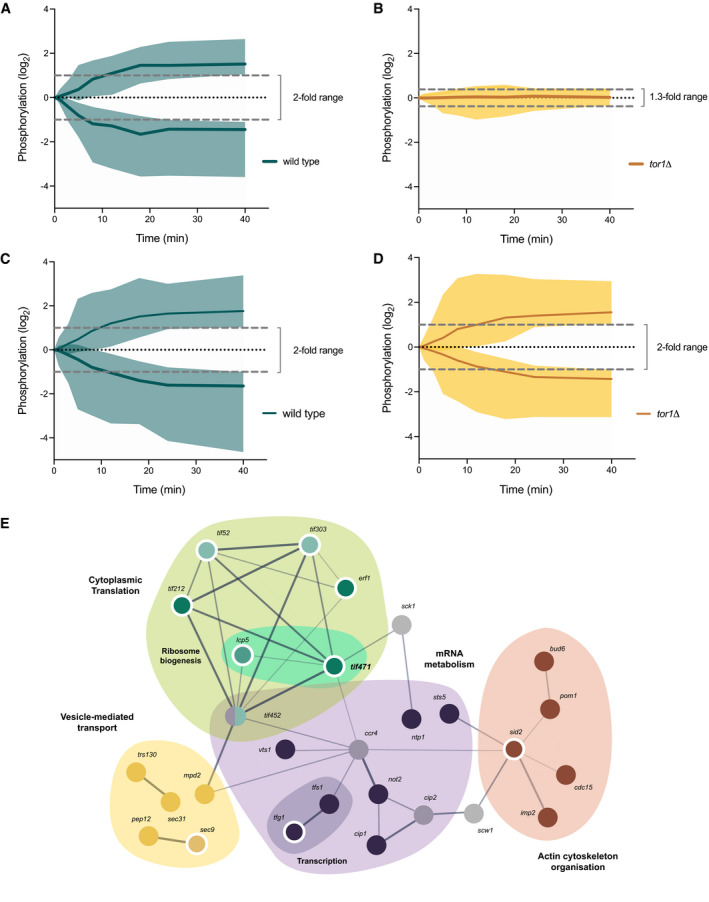
Figure 3. Identifying a core TORC1‐dependent protein interaction network in regulating rates of protein synthesis.
-
A–DSummary profile plots illustrating the minimum and maximum range of phosphorylation for the respective phosphorylation sites in wild type (in green) and tor1∆ (in yellow) cells, with lines indicating the median values of the phosphorylation profiles and the shaded areas representing the range of values. Values are normalised to their respective values at T = 0 min in each strain. Top two panels represent the 43 TORC2‐dependent phosphosites showing more than 2‐fold change in wild type (A) but within 1.3‐fold difference in tor1∆ cells (B). Bottom two panels illustrate the 158 phosphosites showing relative phosphorylation changes of more than 2‐fold in both wild type (C) and tor1∆ (D) cells.
-
EInteraction network generated from the interactome analysis of the 49 potential TORC1‐dependent proteins conserved among fission yeast, budding yeast and human, based on collated annotations from the STRING database. Interactions between proteins were scored based on both functional and physical interactions from experimental, text mining and database sources only. Connections were drawn upon reaching a minimum interaction score of 0.4, and the lines were weighted to indicate the relative strength of the annotated interactions. The resultant network of the 28 connected proteins were functionally categorised based on their GO biological processes as annotated on PomBase and represented by the following colours, respectively: cytoplasmic translation (GO:0002181) and ribosome biogenesis (GO:0042254) in green, mRNA metabolism (GO:0016071) and transcription (GO:0006351) in purple, vesicle‐mediated transport (GO:0016192) in yellow and actin cytoskeleton organisation (GO:0030036) in brown. Grey nodes indicate proteins that were not assigned to the highlighted functional categories. Proteins that were identified as common in both phosphoproteomics studies are denoted by the darker hues of the respective colour categories, and the nodes of essential proteins are outlined in white (9/28).
Following exclusion of the TORC2‐dependent sites, we analysed the remaining phosphosites and identified 158 phosphosites on 83 proteins that showed a more than 2‐fold change for both wild type and tor1∆ within 40 min (Fig 3C and D; marked as “TORC1” on Table EV3). Among these potential TORC1‐dependent proteins, many were found to be conserved across eukaryotes, with 59% (49/83) sharing homologues in both budding yeast and humans, and a further 28% (23/83) with homologues identified in either one. Using experimental evidence annotated on the STRING database (Szklarczyk et al, 2019), we subsequently performed an interactome analysis on the 49 conserved proteins (see Materials and Methods) and found that 28 of these proteins were functionally or physically interacting. Within this group of interacting proteins, 4 main clusters emerged based on GO biological process categorisation: cytoplasmic translation, mRNA metabolism, vesicle‐mediated transport and actin cytoskeletal organisation. This forms a core network of phospho‐regulated proteins that may have roles in controlling cellular growth (Fig 3E; Table 2).
Table 2.
All GO biological process categories for the 28 conserved interacting proteins.
| GO term | Frequency | Gene names |
|---|---|---|
| mRNA Metabolic Process (GO:0016071) | 8 | ccr4, cip1, cip2, not2, sts5, tfs1, tif452, vts1 |
| Cytoplasmic Translation (GO:0002181) | 6 | erf1, tif212, tif303, tif452, tif471, tif52 |
| Vesicle‐Mediated Transport (GO:0016192) | 6 | cdc15, mpd2, pep12, sec31, sec9, trs130 |
| Actin Cytoskeleton Organization (GO:0030036) | 5 | bud6, cdc15, imp2, pom1, sid2 |
| Establishment/ Maintenance of Cell Polarity (GO:0007163) | 4 | bud6, cdc15, pom1, sid2 |
| Membrane Organization (GO:0061024) | 4 | cdc15, pep12, sec31, sec9 |
| Mitotic Cytokinesis (GO:0000281) | 4 | cdc15, imp2, pom1, sid2 |
| Protein‐Containing Complex Assembly (GO:0065003) | 4 | bud6, tfs1, tif212, tif303 |
| Signaling (GO:0023052) | 3 | pom1, sck1, sid2 |
| Mitotic Cell Cycle Phase Transition (GO:0044772) | 2 | pom1, sid2 |
| Ribosome Biogenesis (GO:0042254) | 2 | lcp5, tif471 |
| Transcription, DNA‐Templated (GO:0006351) | 2 | tfg1, tfs1 |
| Cell wall organization or biogenesis (GO:0071554) | 1 | scw1 |
The role of the S6 proteins kinases and translation initiation factor eIF4G on the control of protein synthesis rates
This study has identified a range of putative targets that potentially regulate the rate of protein synthesis in response to TORC1. Of particular interest are the S6 proteins kinase (S6K) family, which are among the most widely discussed targets, hence we decided to further investigate and understand their potential role in TOR regulation of protein synthesis. There are 3 functional homologues of the S6Ks in fission yeast, Psk1, Sck1 and Sck2 (Nakashima 2012). After 40 min of Torin1 treatment, we found that the relative protein expression of all 3 proteins remained unchanged in both wild type and tor1∆ cells (Fig 4A). Sck1 was identified in our interactome analysis as a potentially conserved TORC1‐dependent target (Fig 3E), and one phosphosite in particular (T632) showed a greater than 2‐fold phosphorylation decrease within 18 min of Torin1 addition (Fig 4B). Furthermore, in the earlier phosphoproteomics experiment, we also identified one Sck2 phosphosite that showed a more than 2‐fold phosphorylation decrease in wild type cells within 30 min of Torin1 addition (Fig 4C). Previously identified downstream substrates for these protein kinases include the Rps601 and 602 proteins, which showed over 8‐fold reductions within 40 min of Torin1 addition (Fig 2C and D). The changes in phosphorylation kinetics of the S6 protein kinases and their downstream targets therefore suggest that they could be potential modulators of protein synthesis downstream of the TOR kinase, as implicated in other studies (reviewed in (Saxton & Sabatini, 2017)).
Figure 4. The effects of TOR inhibition on S6K phosphorylation and rates of protein synthesis.
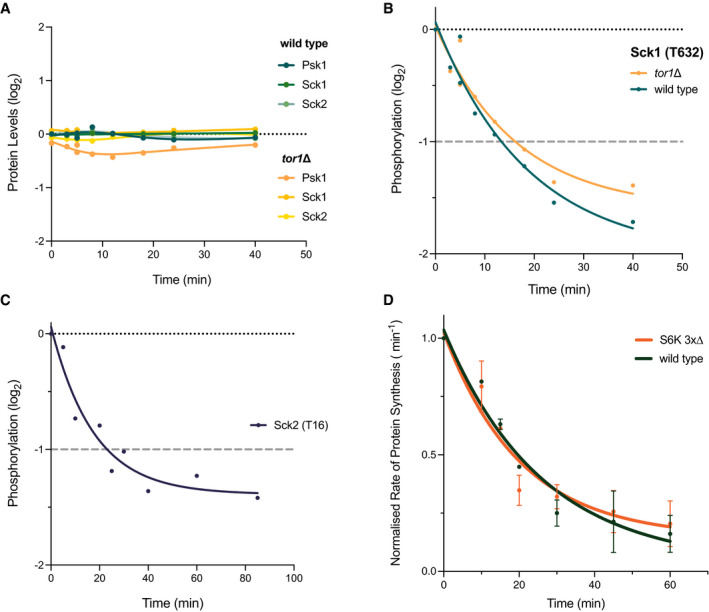
- Relative protein expression levels of the 3 S6 kinase homologues, Psk1, Sck1 and Sck2, plotted against time of Torin1 addition. Respective proteins are represented by the different coloured lines as detailed in the legend.
- Relative phosphorylation level of the T632 phosphosite identified on Sck1 in the 40‐min time course study. Phosphorylation levels of both wild type (green) and tor1∆ (yellow) are presented relative to the value for wild type cells at T = 0 min. Grey dashed line indicates 2‐fold threshold.
- Relative phosphorylation of the T16 phosphosite identified on Sck2 in the 85‐min time course study (background adjusted detailed in Materials and Methods). Grey dashed line indicates 2‐fold threshold.
- Changes in the rates of protein synthesis upon Torin1 treatment (5 µM) measured for wild type and the S6 kinase deletion mutant (S6K 3x∆) plotted over time. Non‐linear regression exponential decay lines were fitted using the curve‐fitting function on Prism9. Error bars represent the standard deviation (SD) of data aggregated from 3 independent experiments.
To investigate this possibility, we undertook a genetic analysis of the S6K homologues. Previous genetic analyses have suggested that deletion of any one or combinations of the three S6K homologues does not affect cell viability in fission yeast (Nakashima et al, 2012). We therefore wanted to investigate whether the S6Ks have a role in modulating the rates of protein synthesis in response to growth inhibitory signals from the TOR pathway. We constructed a triple deletion mutant (S6K 3x∆) of the S6 kinase homologues and measured the rates of protein synthesis following Torin1 treatment. After 60 min of Torin1 addition, we found no significant difference in the rates of inhibition in protein synthesis between S6K 3x∆ and wild type (Fig 4D). This result indicates that the immediate decrease in protein synthesis rates upon TOR inhibition is not dependent upon the S6Ks or their downstream targets.
Another set of putative targets of TOR‐dependent regulation on protein synthesis are those related to cytoplasmic translation. These represent a notable proportion of the 27 proteins that are part of the conserved TORC1‐dependent core interactome network, as identified in the cytoplasmic translation cluster (Fig 3E). In particular, 9 of the 27 proteins are encoded by genes essential for cell viability (outlined in white in Fig 3E), and are considered as stronger candidates for protein synthesis regulation given the central role protein synthesis has in cellular growth. Among these 9 proteins, 4 of them are Translation Initiation Factors (TIFs), and together with Lcp5, an essential ribonucleoprotein involved in 18S rRNA maturation, these 5 proteins play crucial roles in recruiting and engaging the ribosome for initiating translation. Additionally, one other translation‐related protein Erf1 was identified in our analysis, which is shown to have functions at the end of the translational process, and is responsible for coordinating the release of newly synthesised peptides from the active ribosome.
Of these 6 essential translation‐related proteins, 3 were found to have common phosphorylation sites in both our phosphoproteomics studies and showed greater than 2‐fold changes within 40 min of Torin1 addition. Tif212 is the fission yeast homologue of the eukaryotic translation initiation factor eIF2 β‐subunit and is one of the 3 subunits of the heterotrimeric GTP‐binding protein required for the recruitment of the initiation methionine residue Met‐tRNAi. One Tif212 phosphorylation site was identified in the TORC1‐dependency analysis, showing rapid dephosphorylation in both wild type and tor1∆ cells within 12 min of Torin1 addition (Fig 5A). Rapid phosphorylation changes were also observed for the translation release factor Erf1, with levels changing more than 2‐fold in both wild type and tor1∆ cells within 10 min of Torin1 addition (Fig 5B).
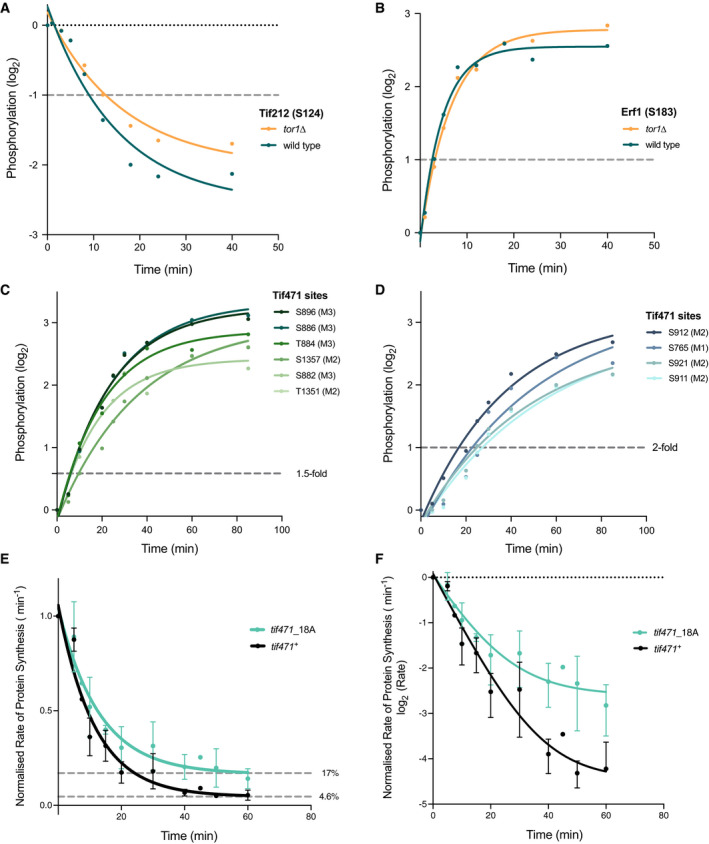
Figure 5. Examining potential essential translation‐related targets and the functional importance of phosphorylation on rates of protein synthesis upon TOR kinase inhibition.
-
A, BPhosphosites identified on (A) Tif212 and (B) Erf1, respectively, showing 2‐fold phosphorylation change within 20 min of Torin1 (5 µM) addition. Both wild type (green) and tor1∆ (yellow) phosphorylation values are relative to the starting phosphorylation levels before Torin1 addition in wild type cells (T = 0 min). Grey dashed lines indicate 2‐fold threshold.
-
C, DRelative phosphorylation levels of the 10 common phosphosites of Tif471 plotted against time after Torin1 treatment. Phosphorylation profiles are grouped based on phosphorylation kinetics, where 6 sites showed more than 1.5‐fold increase by 10 min (C), and the remaining 4 exceeded 2‐fold by 30 min (D). Grey dashed lines indicate 2‐fold threshold.
-
E, FChanges in rates of protein synthesis of the tif471_18A mutant compared to the wild type control (tif471 +) upon Torin1 (5 µM) treatment. Residual rates of protein synthesis after 60 min of Torin1 treatment was 17% for the tif471_18A phosphomutant and 4.6% for the tif471 + control relative to starting levels in the respective strains. The two graphs represent the untransformed (E) and log2 transformed (F) rates respectively. Error bars represent the standard deviation (SD) of data aggregated from three independent experiments.
Thirdly, the fission yeast homologue of eIF4G, Tif471 was identified as one of the most heavily phospho‐regulated proteins, with a total of 27 phosphosites exhibiting TOR‐dependent changes across the two studies. In particular, 10 sites showed rapid TORC1‐dependent phosphorylation changes, all exceeding a 2‐fold change by 30 min, and 6 showing a greater than 1.5‐fold change within 10 min (Fig 5C and D). To determine whether the phosphosites identified on Tif471 play a regulatory role for protein synthesis, we constructed a non‐phosphorylatable mutant of Tif471 and examined the rates of protein synthesis following Torin1 treatment. A total of 18 serine or threonine residues were replaced with alanine (see Materials and Methods for mutation sites), and the resulting tif471_18A mutant strain is viable. The mutant strain also has a similar growth rate to the control, which has a tagged wild type sequence at the endogenous locus (tif471 +) (Fig EV3), as well as similar protein expression levels (unpublished observations).
Figure EV3. Relative growth rates in tif471_18A mutant is comparable to wild type cells.
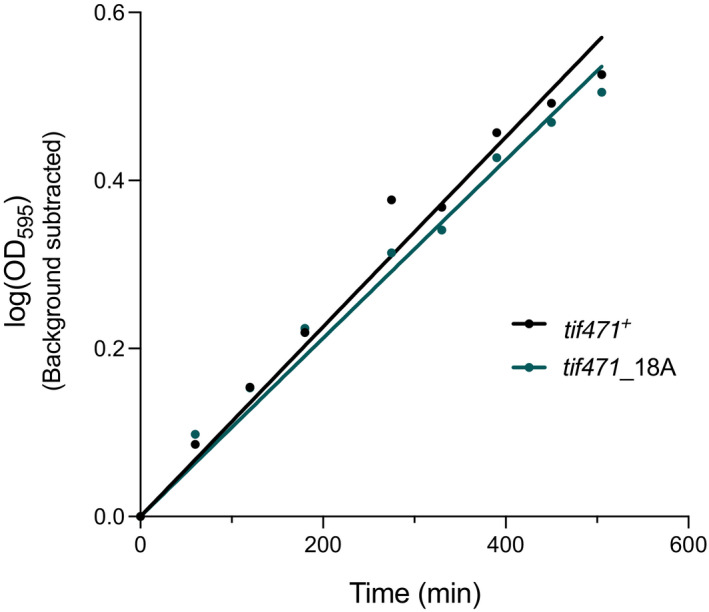
Representative plot of the relative growth rates as measured by the change in optical density (OD595) over time for tif471_18A mutant and control (tif471 +) cells. Values are background subtracted and gradients of the regression lines were 0.0011 for both strains, fitted using simple linear regression on Prism 9.
When comparing the rates of protein synthesis after 60 min of Torin1 treatment, we noted that the residual rate for tif471_18A was reduced to 17% of its starting level, over 3 times higher than the control (5%) (Fig 5E and F). This demonstrates that the changes in phosphorylation states of these Tif471 phosphosites contribute to the control of cellular rates of protein synthesis in response to TOR activity. As a reduction in protein synthesis rates still occurs in the tif471_18A mutant, other factors in addition to Tif471 will likely be involved in regulating protein synthesis in response to TOR inhibition, and are yet to be identified.
Discussion
We have investigated how the TOR‐signalling pathway regulates eukaryotic cell growth, with a focus on understanding the control of global cellular protein synthesis. Using fission yeast, we describe an in‐depth analysis of the proteomic and phosphoproteomic changes that occur after inhibition of both TOR protein kinases with Torin1 in two time course studies at fine temporal resolution. Our results establish that TORC2, which contains the non‐essential Tor1 kinase, is dispensable for regulating global cellular protein synthesis, and so we have characterised changes that are primarily dependent on the essential Tor2 kinase containing TORC1. A total of 20,894 phosphosites located on 2,306 different phosphoproteins were identified in the two studies. Of these, 908 phosphosites on 343 proteins exhibited a more than 2‐fold change in phosphorylation levels after 40 min of Torin1 treatment, by which time protein synthesis rates were reduced over 90%. Two‐thirds of these proteins (213/316) shared homologues in both budding yeast and human cells, and a total of 88% (279/316) of these had homologues in at least one of these two organisms.
Our database of phosphoproteomic changes after TOR kinase inhibition provides an in‐depth resource for investigating the TOR pathways' involvement in regulating cell growth, with many of the identified phosphoproteins conserved among eukaryotes. We investigated two possible candidates, the S6 protein kinases and the translation initiation factor, eIF4G (Tif471), examining their potential roles in cell growth control with respect to protein synthesis. The S6 kinases have widely been suggested as downstream regulators of the TOR kinase pathways and implicated in growth control via modulation of translation initiation activity (see reviews in (Wullschleger et al, 2006; Ma & Blenis, 2009; Saxton & Sabatini, 2017)). However, following deletion of all three S6 kinase fission yeast homologues (Psk1, Sck1 & Sck2), we observed no effects on the rate of rapid protein synthesis reduction upon inhibition of the TOR kinases.
One of the proteins most heavily regulated by phosphorylation identified in our study was the translation initiation factor Tif471, the fission yeast homologue of eIF4G. In a phosphomutant of Tif471 (tif471_18A), we find that in response to TOR inhibition, the residual rate of protein synthesis is reduced to 17% of the starting level, more than three times higher than in wild type. This, to our knowledge, is the first demonstration of a component regulated by phosphorylation as a consequence of TOR inhibition that has an immediate impact on reducing the global cellular rates of protein synthesis. The partial inhibition observed suggests that there are multiple points of control distributed within the TOR downstream network, and complete inhibition will require the coordination of various components.
The detailed temporal and molecular resolution of our study provides a comprehensive data resource for further evaluation of potential TOR‐dependent regulatory targets, and the accompanying data visualisation tool allows for the data to be readily accessible. Gene ontology (GO) analyses of the TOR‐dependent phosphoproteome identified a range of molecular processes with four broad categories that were particularly enriched: signalling‐related processes, cell polarity and actin cytoskeletal organisation, vesicle‐mediated transport and autophagy, and translation‐related processes.
The switching off of growth will require a coordinated response from multiple major cellular processes, so it is unsurprising that several signalling network pathways have been implicated in TOR regulatory control. The MAPK‐signalling pathway is thought to be responsible for maintaining structural integrity of a cell through actin cytoskeletal reorganisation (Samaj et al, 2004; Madrid et al, 2016). Our study identified the MAPKK homologue Wis1, and downstream signalling cascade components including the RAS protein homologue Ras1, and regulators of the Rho GTPases, Rga1 and Rgf1. In addition, components of the SIN‐MOR pathways, Sid2 and Nak1, responsible for coordinating cytoskeletal rearrangements in fission yeast (Gupta et al, 2013) were also identified. Protein homologues of these pathways are conserved in mammalian cells as part of the Hippo and NDR‐kinase signalling networks and have been implicated in growth control. These proteins are also often found to be misregulated in cancer (Halder & Johnson, 2011).
Across the eukaryotes, components of the actin cytoskeletal network and those involved in cellular spatial organisation have previously been implicated as targets of the non‐essential Tor1 containing complex TORC2 (Loewith et al, 2002; Jacinto et al, 2004; Sarbassov et al, 2004; Cybulski & Hall, 2009; Rispal et al, 2015). In particular, TORC2 has been identified as a key component of PI3K/Akt signalling (Sarbassov et al, 2004; Hresko & Mueckler, 2005), and in fission yeast, it has been suggested that it is regulated by the Rab‐family GTPase, Rhy1 and its regulators (Tatebe & Shiozaki, 2010; Tatebe et al, 2010). In this study, we identified a conserved component of the GEF complex Sat1, which is involved in Rab GTPase activation to regulate TORC2‐Akt signalling (Tatebe et al, 2010). Interestingly, phosphosites on proteins involved in actin cytoskeletal organisation including Bud6 and Imp2 also exhibited TORC1‐dependent phosphorylation changes upon addition of Torin1, suggesting a potential overlap between the TORC1 and TORC2 complexes in the remodelling aspects of cytoskeletal structures within a cell.
In line with a recent fission yeast study, where vesicle‐mediated transport proteins were altered by TOR‐dependent cellular nutritional states (Lie et al, 2018), we showed that phosphorylation of vesicle‐mediated transport and autophagy‐related proteins are dependent on TOR activity. A number of essential cellular growth functions are supported by vesicular transport processes, including Golgi trafficking, exocytosis and endocytosis, as well as proteosomal and lysosomal pathways. Therefore, coordination of anabolic and catabolic processes appears to be involved in cellular decision‐making required for cell growth, perhaps in part regulated through interplay between TOR and autophagy‐related processes (reviewed in (Dunlop & Tee, 2014)).
Consistent with previous studies suggesting translation initiation as a major point of control for a TOR‐dependent regulation of growth (reviewed in (Ma & Blenis, 2009; Loewith & Hall, 2011)), a number of the translation initiation components were identified in our study. Notably, 5 of the 7 translation‐related proteins categorised as part of the TORC1‐dependent core interactome network were translation initiation factors (Tif‐). Other components of the translation machineries were also identified, including Erf1, the translation termination and release factor, and Lcp5, the U3 snoRNP‐associated protein, shown to be involved in 18S rRNA maturation in budding yeast (Wiederkehr et al, 1998). The human orthologue of Lcp5, Neuroguidin (NGDN) has also been shown to repress translation through interactions with the cap‐binding eukaryotic initiation factor 4E (eIF4E), preventing the eIF4E‐4G interaction require for mRNA cap‐dependent translation initiation (Jung et al, 2006). This supports the potential role of eIF4G in translational control.
In addition, our study revealed a cluster of conserved proteins related to mRNA metabolism that exhibited TORC1‐dependent phosphorylation changes, and some of these proteins have been shown to interact with translation initiation machineries. These include Ccr4 and Not2, which are components of the CCR4‐Not complex, and have been shown to regulate mRNA fate and turnover (Parker & Song, 2004; Wiederhold & Passmore, 2010). Ccr4 co‐localises with P‐bodies and is essential for P‐body formation in mammalian cells (Cougot et al, 2004; Andrei et al, 2005). The P‐body exoribonuclease Sts5, which is the homologue of SSD1 in budding yeast, was also identified in our study. SSD1 is shown to colocalise with mRNAs associated with growth zones, and was implicated in P‐body and stress‐granule dependent translational repression, after exposure to cellular stress in budding yeast (Jansen et al, 2009; Kurischko et al, 2011). This suggests that TOR‐dependent translational regulation is not limited to the regulation of translation initiation machineries, and may also involve factors that influence mRNA fate and turnover.
We have demonstrated that protein synthesis rates are rapidly reduced upon TOR‐dependent growth inhibition and that this reduction is not dependent on S6 kinase activity. We propose that translation‐related proteins remain key potential candidates for regulating eukaryotic cell growth, and identify the eIF4G homologue in fission yeast, Tif471, as one of the regulators of protein synthesis inhibition through TOR‐dependent phosphorylation. Our list of potential TOR phosphorylation targets shows that TOR‐dependent growth control requires coordination of a range of cellular processes for timely and effective regulation, and our study provides a foundational resource for potential targets to be further investigated in both fission yeast and other eukaryotes.
Materials and Methods
Reagents and Tools table
| Reagent/Resource | Reference or source | Identifier or catalog number |
|---|---|---|
| Experimental models | ||
| S. pombe: 972 h‐ (wild type) | Nurse Lab | PN1 (TM11) |
| S. pombe: tor2_G2040D (Torin1‐resistant) | Petersen Lab (Atkin et al, 2014) | JP1669 (TM134) |
| S. pombe: tor1∆::HphMX6 h‐ | This study | TM97 |
| S. pombe: psk1∆::HphMX6 Sck1∆::NatMX6 Sck2∆::KanMX6 h‐ (S6K 3x∆) | This study | TM74 |
| S. pombe: 972/972h‐/h‐ | This study | TM21 |
| S. pombe: tif471+‐v5‐HphMX6 h‐ | This study | TM121 |
| S. pombe: tif471_18A‐v5‐HphMX6 h‐ | This study | TM156 |
| Recombinant DNA | ||
| Plasmid: pSC‐A‐Amp‐tif471(wt)‐v5‐HphMX6 | This study | TMp16 |
| Plasmid: pSC‐A‐Amp‐tif471_13A‐v5‐HphMX6 | This study | TMp20 |
| Plasmid: pSC‐A‐Amp‐tif471_18A‐v5‐HphMX6 | This study | TMp21 |
| Oligonucleotides and sequence‐based reagents | ||
| PCR primer: ATTCCTGAAGGATCTGATGGAAGC | This study | pTM101 |
| PCR primer: CATTAGACTTAAAGGAGCCGCA | This study | pTM102 |
| PCR primer: TTTGGAAAATTTTAATTTCATTC | This study | pTM111 |
| PCR primer: TTTACCCTCTTCCTCAGC | This study | pTM112 |
| PCR primer: acggaaagctgaggaagagggtaaaCGGGAAGCTGATAAAAAC | This study | pTM113 |
| PCR primer: atgaatgaaattaaaattttccaaaGCATTTCTCAATTACAATGTAG | This study | pTM114 |
| PCR primer: gaatcgatcgaggtttcgccgcttctggtgctggat | This study | pTM141 |
| PCR primer: atccagcaccagaagcggcgaaacctcgatcgattc | This study | pTM142 |
| PCR primer: ctggttttggtggtcctgcagagagaaagggcatt | This study | pTM143 |
| PCR primer: aatgccctttctctctgcaggaccaccaaaaccag | This study | pTM144 |
| PCR primer: ctctcgatccggcgcaaatgctcacgcccatgctggccct | This study | pTM145 |
| PCR primer: gggccagcatgggcgtgagcatttgcgccggatcgagag | This study | pTM146 |
| PCR primer: ACCGACTTTTCTGCTTTGGT | This study | spTM64 |
| PCR primer: AGCAATCCACCATTTAGCTCT | This study | spTM67 |
| PCR primer: TCCGTGATGACTTACACCGC | This study | spTM72 |
| PCR primer: TGAGTTAGAATCTTTTTGACCCCC | This study | spTM75 |
| Chemicals, enzymes and other reagents | ||
| Complete mini Protease Inhibitor Cocktail | Sigma Aldrich | Cat # 11836153001 |
| PhosSTOP phosphatase Inhibitor tablets | Sigma Aldrich | Cat # PHOSS‐RO |
| Propidium iodide solution | Biotium | Cat # 40017 |
| Torin1 | Torcris | Cat # 4247 |
| Pierce Trypsin Protease, MS Grade | ThermoFisher | Cat # 90058 |
| Software | ||
| FlowJo v10.3.0 | FlowJo | https://www.flowjo.com/ |
| Prism v9.0.0 | GraphPad | https://www.graphpad.com/scientific‐software/prism/ |
| Rstudio v1.3 | Rstudio | https://rstudio.com/products/rstudio/rstudio‐desktop |
| Perseus v1.4.0.2.3 | Perseus | https://maxquant.net/perseus/ |
| MaxQuant v1.5.0.13 | MaxQuant | https://www.maxquant.org/ |
| Online Databases | ||
| PomBase | PomBase | https://www.pombase.org/ |
| PANTHER (GO Enrichment) | Gene Ontology | http://geneontology.org/ |
| Clustal Omega | EMBL‐EBI | https://www.ebi.ac.uk/Tools/msa/clustalo/ |
| Deposited Data | ||
| Mass spectrometry files | This study | https://www.ebi.ac.uk/pride/archive/ PRIDE: PXD023664 |
| Data Visualisation Tool | This study | https://tmak.shinyapps.io/TM_App‐Table_EV3/ |
| Other | ||
| Ultrospec 2100 pro UV/visible spectrophotometer | Amersham Biosciences | Cat # 80‐2112‐21 (BioChrom) |
| Click‐iT® HPG Alexa Fluor® Protein Synthesis Assay Kits | ThermoFisher | Cat # C10428 |
| Dynabeads Protein A | ThermoFisher | Cat # 10002D |
| Dynabeads M‐270 Epoxy | ThermoFisher | Cat # 14302D |
| TMT 10plex Isobaric Label Reagent Set 1 × 0.8 mg | ThermoFisher | Cat # 90110 |
| High‐Select™ TiO2 Phosphopeptide Enrichment Kit | ThermoFisher | Cat # A32993 |
| High‐Select™ Fe‐NTA Phosphopeptide Enrichment Kit | ThermoFisher | Cat # A32992 |
| Pierce™ High pH Reversed‐Phase Peptide Fractionation Kit | ThermoFisher | Cat # 84868 |
| EASY‐Spray C18 column, 75 mm × 50 cm | ThermoFisher | Cat # ES803 |
| Orbitrap Fusion Lumos Tribrid Mass Spectrometer | ThermoFisher | Cat # IQLAAEGAAPFADBMBCX |
| BD LSRFortessa | BD Biosciences | Cat # 649225 |
| UltiMate 3000 HPLC System | ThermoFisher | Cat # 5041.0010 |
| QuikChange Multi Site‐Directed Mutagenesis | Agilent | Cat # 200514 |
Methods and Protocols
S. pombe genetics and cloning
Full genotype of all strains, plasmids, gDNA fragments and primers used in this study are listed in the Reagents Table. Standard S. pombe media and methods as previously detailed in (Moreno et al, 1991). Gene deletions and tagging fragments were prepared using PCR based amplifications described in (Bahler et al, 1998), with primers designed on the Bähler laboratory website (Resources). S6K 3x∆ mutant was generated through consecutive lithium acetate transformations and selection, and verified through colony PCR checking of both the gene deletion construct and absence of wild type gene copies.
The tif471_18A mutant was constructed by first tagging the tif471 gene at the endogenous locus with a v5‐HphMX tag at the 3′ end of the open reading frame (ORF) (subsequently used as the tif471 + control). The tif471 genes were amplified from the extracted by PCR with primers (pTM101 & 102) 1.06 kb either side of the ORF. The amplified gene was subcloned into the pSC‐A‐Amp vector using the StrataClone Blunt PCR Cloning Kit (TMp16). The 18 alanine mutations were introduced through 2 consecutive cloning steps, first replacing amino acid position 706 onwards until the end of the ORF with the synthetic gene fragment (gBlock) synthesised by Integrated DNA Technologies (IDT). The gBlock fragment contained 13 alanine mutations: S912A, S919A, S921A, S1293A, S1300A, S1333A, S1349A, T1351A, S1353A and S1357A. The 3′ end the endogenous gene on TMp16 was replaced by the fragment at the using the NEBuilder HiFi DNA Assembly kit and primers pTM111‐114. The final 5 alanine mutations, S882A, T884A, S886A, S896A and S896A, were added by 2 subsequent reactions using the Agilent QuikChange Multi Site‐Directed Mutagenesis Kit and primers pTM141‐146, resulting in the TMp21 plasmid containing all 18 alanine mutants on Tif471.
The entire locus containing mutated amino acid residues was then amplified by PCR with pTM101 and 102 and purified using the QIAquick PCR Purification Kit, then transformed into stable wild type diploid cells (TM21) and selected with antibiotic resistance. The resultant heterozygous diploid was subsequently sporulated into haploids, to check for a 50:50 segregation of wild type to mutant haploids through random spore analysis, and mutant haploids were isolated. Alanine mutations were checked with specifically designed primers on Primer3Plus website (spTM64, 67, 72 and 75) by DNA sequencing from extracted gDNA.
Growth rate measurement and calculations
Cells were inoculated in static liquid media overnight at corresponding temperature and subsequently diluted to within exponential growth cell density (OD ˜ 0.1 ‐ 0.8). Optical density (OD) was measured at wavelength (λ) = 595 nm using the Ultrospec 2100 pro UV/visible spectrophotometer (Amersham Biosciences). OD595 = 0.1 approximated to 1.26 × 106 cells/ml. All cell cultures were maintained in exponential growth for 48 h in the culturing conditions defined above before any experiments were conducted. Growth rate calculations were made based on the OD measurements, where the logarithm (base 10) of the OD was taken (Log(OD)) and plotted against time. Using the linear regression line fitting function on Prism9, the gradient of the fitted line was used to calculate the generation time (gt) using the formula , where m is represented by the gradient of the regression line. Error bars indicate standard deviations (SD) of three independent experiments unless stated otherwise.
Protein synthesis assay
Cells were grown in EMMG at 25°C and maintained in exponential growth (OD 0.1–0.8) for 48 h in liquid culture; cells were diluted to OD 0.4–0.5 for the start of the assay. The main culture was then split into individual cultures for each time point and allowed to recover for at least 1 h before beginning the assay. The individual cultures were treated with Torin1 (added at 1:1,000 from 5 mM stock in DMSO to final concentration 5 µM) for different lengths of time accordingly. The methionine analogue L‐homopropargylglycine (HPG) was added (from a 50 mM stock solution to a final concentration of 50 µM) to cultures 15 min before the first sampling of each time point. Two sample were taken at 10 min apart for the individual time points, and the rate of protein synthesis was calculated from the difference between the two values and then divided through by 10 min.
Sample processing
420 µl of samples were collected at the indicated time points and fixed in 980 µl of 100% ice‐cold ethanol to reach final concentration of 70% ethanol and then kept on ice for 20–30 min. Sample processing and labelling procedures were based on (Knutsen et al, 2015). Cells were washed and permeabilised for 20 min with 0.5ml of PBS and 1% Triton X‐100 in PBS, respectively, then washed and blocked in 0.5 ml of 1% BSA in PBS for 1 h. All steps were performed at room temperature and incubations were with rotation on tube rotators, protected from light.
Composition of reaction mix was modified from the Click‐iT™ HPG Alexa Fluor™ 488 Protein Synthesis Assay Kit (Invitrogen). Each sample was stained with 375 µl of reaction mix per reaction, containing 321.56 µl of 1× Click‐iT HPG reaction buffer, 15 µl Copper (II) sulphate (Cu2SO4) (100 mM), 0.94 µl 488 Alexa Fluor® azide and 37.5 µl 1× Click‐iT HPG buffer additive. Reaction mix was removed, and samples were washed with 500 µl of sodium citrate (50 mM). Cells were subsequently washed and resuspended in 500 µl of sodium citrate and treated with RNase (0.1 mg/ml) for 3 h to overnight at 37°C, protected from light. Following RNase digestion, cells were pelleted and resuspended in 500 µl of sodium citrate with propidium iodide (PI) added to a final concentration of 2 µg/ml. Samples were loaded onto 96‐well U bottom plates and ran on BD‐LSR‐Fortessa flow cytometers using the High Throughput Sampler (HTS), and 100,000 events were recorded per sample. Sample flow rate was set to 1 µl/s, with a mixing volume of 100 µl for 2 mixes at 180 µl/s mixing speed, and a wash volume of 400 µl.
Voltage for corresponding parameters was set based on defined criteria. Side scatter (SSC) adjusted to have ˜90% of the cells between 7500 and 15,000 (roughly one doubling). Voltage for the 530/30 blue laser channel (for detection of 488) was set to have the maximum range of intensity whilst keeping 95% of the events below the maximum 250,000 (AU). For the 610/20 yellow laser channel (PI), voltage was set to have the 2C DNA content peak centred around 50 (AU). Cells were manually gated on FlowJo (version.10.3.0) for doublet discrimination, and only cells with 2C DNA content or below were included for further analysis. For the initial experiment in establishing HPG incorporation as a measure for protein synthesis at steady state, median values were calculated for whole populations after data clean‐up and values were exported for statistical analysis on R. Linear regression lines were fitted using the linear model of the geom_smooth function. Error bars indicate standard deviations (SD) of three independent experiments unless stated otherwise.
Proteomic and phosphoproteomic studies
Two time course studies were conducted to investigate the proteomic and phosphoproteomic changes upon Torin1 (5 µM) addition, with the first time point (T = 0 min) taken just before Torin1 or DMSO addition. A DMSO sample treated for the duration of the respective time courses was used as a control to account for non‐specific phosphorylation changes. Time points for the 85‐min time course were taken at 0, 5, 10, 20, 25, 30, 40, 60 and 85 (+DMSO 85) min, and for the 40‐min time course, they were at 0, 1, 3, 5, 8, 12, 18, 24 and 40 (+DMSO 40) min.
Sample preparation for mass spectrometry
Each protein sample (300 µg) was reduced with 5 mM dithiothreitol (DTT) for 25 min at 56°C, alkylated with 10 mM iodoacetamide for 30 min at room temperature in the dark and then quenched with 7.5 M DTT. Samples were digested using SP3 on‐bead methodology (Hughes et al, 2019) with the variation that 50 mM HEPES (pH 8.5) was used in place of ammonium bicarbonate. Briefly, proteins were bound to the SP3 beads (10:1 beads:protein (w/w)) in 50% ethanol (v/v) and then washed three times in 80% ethanol, prior to resuspension in 50 mM HEPES (pH 8.5) with 1:40 (trypsin:protein (w/w)) overnight at 37°C. The digested samples were arranged in sets of ten and labelled using the TMT10plex Isobaric Label Reagent Set (Thermo Fisher), as per the manufacturer’s instructions. Following labelling and mixing, multiplexed samples were desalted using a C18 SepPak column. Phosphopeptide enrichment was performed by Sequential Enrichment from Metal Oxide Affinity Chromatography (SMOAC, Thermo Fisher) with initial enrichment using the High‐Select TiO2 Phosphopeptide Enrichment Kit followed by the High‐Select Fe‐NTA Phosphopeptide Enrichment Kit (both Thermo Scientific) for the non‐bound flow through fractions. Phosphopeptides and non‐bound flow through fractions were desalted and fractionated using the High pH Reversed‐Phase Peptide Fractionation Kit (Pierce) and analysed on an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher) coupled to an UltiMate 3000 HPLC system for online liquid chromatographic separation. Each run consisted of a 3 h gradient elution from a 75 μm × 50 cm C18 column.
MaxQuant (version 1.5.0.13) was used for all data processing. The data were searched against a PomBase (Lock et al, 2019) extracted S. pombe proteome FASTA file. Default MaxQuant parameters were used with the following adjustments with Phospho(STY) being added as a variable modification, MaxQuant output files were imported into Perseus (version 1.4.0.2).
Quantification and statistical analysis
All reporter intensities were log2 transformed and only phosphosites or proteins which were quantified in all 20 channels were retained. Reported intensities were normalised by subtracting the median value for a specific TMT channel and then by subtracting the median value of each phosphosite or protein across the time course. Both protein and phosphorylation levels were compared to the corresponding values measured in wild type cells at 0 min before Torin1 or DMSO (control) addition for the two respective experiments.
Proteomic analysis
Changes in protein levels are normalised to the starting protein levels at time T = 0 min in wild type cells for the respective experiments before Torin1 or DMSO addition (labelled “wt.0.min” and “WT 00 min” for the 85‐min and 40‐min experiments, respectively, in Table EV1). For both time course studies, proteins showing non‐specific (DMSO dependent) changes were excluded from subsequent analysis if they showed more than 1.5‐fold change (± 0.585) in the DMSO treated sample at the end of the respective time courses, either at 85 (“wt.85.DMSO”) or 40 min (“WT 40 min DMSO”). For the 85‐min time course experiment, proteins were excluded in further analysis if they showed more than 1.5‐fold difference (within the ± 0.0585 range) in starting protein levels in the Torin1‐resistance strain (“tr.0.min” in Table EV1). Frequency distribution analysis was carried out on values from the “wt.40.min” column (Table EV1) following data clean‐up in Prism9 (n = 3,380, bin width = 0.2). For the 40‐min time course study, proteins that showed more than a 1.5‐fold difference (± 0.585) between the 40‐min Torin1‐treated sample and the DMSO control in either wild type (“WT 40 min ‐ DMSO control”) or tor1∆ (“Tor1D 40min ‐ DMSO control”) cells were excluded from subsequent analysis in order to account for DMSO non‐specific changes. (Columns mentioned highlighted in green in Table EV1).
Phosphoproteomic analysis
Phosphosites were first filtered for more than 2‐fold change in phosphorylation in wild type cells (“wt.85.min” and “WT 40 min”) at the end of the respective 85‐min and 40‐min time course experiments (Tables EV2 and EV3). Non‐specific DMSO changes were accounted for by only including sites that showed more than 2‐fold difference in phosphorylation after subtracting values of the DMSO treated control from the Torin1‐treated samples, respectively, i.e. “wt 85min ‐ DMSO control” column for the 85‐min experiment, and the “WT 40min ‐ DMSO control” column for the 40‐min experiment. For the 85‐min time course experiment, sites that showed more than 1.5‐fold (± 0.585) change in phosphorylation levels at the end of the time course (T = 85 min) relative to the start (T = 0 min) in the Torin1‐resistant strain (“tr.85.min relative control”) were excluded. Phosphorylation levels were subsequently corrected for non‐specific Torin1‐related changes in the 85‐min time course by subtracting the phosphorylation values of the Torin1‐resistant strain from the values measured in wild type cells in the corresponding time points for the individual phosphosites. Values are then background subtracted to have the relative phosphorylation levels before Torin1 addition (T = 0 min) starting at zero for direct comparison among sites and subsequent fold change analyses (labelled “sub.x.min” in Tables EV2 and EV3; header in blue). Sites exhibiting more than 2‐fold change by 40 min of Torin1 treatment (“sub.40.min”) and sustaining the change until the end of 85 min (“sub.85.min”) were considered potential TOR‐dependent phosphosites.
In the 40‐min time course, potential sites were further categorised as TORC1‐ or TORC2‐dependent based on the phosphorylation changes in wild type and tor1∆ cells relative to their respective phosphorylation levels before Torin1 addition (T = 0 min). Sites showing more than 2‐fold change by 40 min in wild type cells (“WT 40 min” and “WT 40min ‐ DMSO control”) but less than 1.3‐fold in tor1∆ (“Tor1D norm 40 min” and “Tor1D 40min ‐ DMSO control”) were classified as TORC2‐dependent sites, and TORC1‐dependent sites were defined as those showing more than 2‐fold change in both wild type and tor1∆ cells (marked on Table EV3 respectively as “TORC2” and “TORC1” under “Complex dependency”; one of the filtering criteria on the online Data Visualisation Tool). The change in relative phosphorylation levels is represented in XY plots where the x‐axis indicates time after Torin1 addition and the y‐axis shows the level of phosphorylation relative to the starting level before Torin1 addition. All plots for the 85‐min time course represent the background corrected and zero normalised values (“sub.x.min”; x indicates the corresponding time points, corresponding column titles highlighted in blue in Table EV2) unless stated otherwise. Plots for the 40‐min time course represent phosphorylation values that are both normalised to the started values at wild type T = 0 min (“WT XX min” & “Tor1D norm XX min”; XX indicates the corresponding time points, column titles highlighted in blue in Table EV2). Lines represented on the graphs are drawn using the curve‐fitting function on Prism9 to fit one‐phase exponential decay non‐linear regressions unless stated otherwise.
Data visualisation tool
A data visualisation tool was developed for visualising the phosphorylation data of the 908 TOR‐dependent phosphosites from the two phosphoproteomics time courses (data based on Table EV3). An illustrated example of the application interface is provided in Fig EV1A–D, and a publicly accessible link is made available online on https://tmak.shinyapps.io/TM_App‐Table_EV3/. Tabs on the top bar provide individual access to the “85‐min study” and “40‐min study” datasets, respectively, with subsequent filtering selections specific to each dataset (Fig EV1A). Prompts on the graph place‐holders in grey specify the filtering selections required for plotting and displaying the graphs.
For the “85‐min study” tab, the “Conserved in” selection allows for sub‐setting of the protein list based on conservation, and subsequently, the list of proteins available for selection gets updated in the “Protein” field based on the selected criteria (Fig EV1B). A graph in the top panel is then displayed with all the phosphosites of the selected protein plotted, with the corrected phosphorylation values plotted over time in minutes. Further information on the individual sites including specific phosphorylation values can be accessed by hovering the mouse over each line at the specific time point of interest (Fig EV1B). Individual phosphosites can also be specifically selected or unselected by single‐clicking on the site of interest in the legend on the right (Fig EV1C). Double‐clicking on an individual site specifically selects and displays that site in isolation. The “Site” field in the sidebar panel is updated according to the selected protein, and multiple sites can be selected for visualisation of the individual phosphosites in the lower facet panels (Fig EV1C). The “Strain” field provides an option for the data from the independent strains or the corrected relative phosphorylation values (selected by default) to be displayed in isolation or overlaid onto the same graph for each individual phosphosite in the lower panel. All lines were fitted with a non‐linear regression using the “nls” method in the stat_smooth Self‐Starting Nis Asymptotic Regression Model function from the R ggplot package. Note that if a line cannot be fitted due to a “singular gradient” error, the line will be omitted and only points are plotted on the graph.
In the “40‐min study” tab, similar navigation and selection fields can be found, but the “TOR Complex dependency” filter is in place of “Conserved in” field in the other panel. A further sub‐categorisation selection field is included between the “Protein” and “Site” fields to select for sites that may or may not be in common with the 85‐min dataset (Fig EV1D). The “Strain” field allows for the phosphorylation profiles of wild type (selected by default) or tor1∆ strains to be displayed individually or overlaid on the same graph for each phosphosite. The “Wrap” field provides the option of displaying the facet panels in the specified number of rows selected from the dropdown menu and is available on both tabs. Both the displayed plots can also be downloaded in either of the tabs by using the “Download Plot (Sites)” button in the sidebar panel for downloading the faceted plots of the individual phosphosites, or the camera icon as shown in Figure EV1D for the graph in the top panel.
GO categorisation and enrichment analysis
The corresponding proteins of the identified phosphosites were categorised based on Gene Ontology (GO) biological processes (Tables 1 and 2) as annotated on PomBase (Lock et al, 2019). Note that proteins can belong to more than one corresponding category. Gene deletion viability and information on protein conservation and homologues were also based on PomBase annotations. Subsequent GO enrichment analysis was carried out for the 149 conserved proteins from the 85‐min time course that contained annotated homologues in both Saccharomyces cerevisiae and human. The analysis was performed using the enrichment analysis tool from the PANTHER classification system (Mi et al, 2013) with respect to the GO biological process in S. pombe, and the provided reference list was based on the list of identified proteins from the proteomic analysis from the same study. Table 2 is arranged and ranked according to the fold enrichment and the categories are representative of processes that showed more than a 2‐fold enrichment.
Interactome analysis
The analysis was performed on the 49 conserved proteins from the 83 proteins that were identified to contain TORC1‐dependent phosphosites. Using the STRING database (Szklarczyk et al, 2019), the potential interactions were scored based on both functional and physical interactions from experimental, text mining and database sources only. Two nodes were connected upon reaching a minimum score of 0.4, and the lines were weighted to indicate the relative strength of the annotated interactions. 28 proteins are represented on the interactome network and are marked on Table EV3.
Author contributions
TM and PN conceived the experiments. TM designed and performed the experiments. AWJ performed mass spectrometry and processed raw data. TM analysed processed data. TM and PN wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Review Process File
Expanded View Figure PDF
Table EV1
Table EV2
Table EV3
Table EV4
Acknowledgements
This work was supported by the Francis Crick Institute which receives its core funding from Cancer Research UK (FC01121), the UK Medical Research Council (FC01121) and the Wellcome Trust (FC01121). In addition, this work was supported by the Wellcome Trust Grant to PN [grant number 214183 and 093917], The Lord Leonard and Lady Estelle Wolfson Foundation and the Woosnam Foundation and Breast Cancer Research Foundation (BCRF‐20‐117). For the purpose of Open Access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission. We are grateful to Scott Curran, Jessica Greenwood, Jacky Hayles, Clovis Basier, Emma Roberts and Ryoko Mandeville for critical reading of the manuscript and help with experiments. The Protein Analysis and Proteomics Platform at the Crick for mass spectrometry, Hefin Rhys for critical advice on data visualisation tool development and Janni Petersen for strains.
The EMBO Journal (2021) 40: e107911.
Data availability
The datasets produced in this study are available in the following databases: Mass spectrometry proteomics data: PRIDE PXD023664 (https://www.ebi.ac.uk/pride/archive/projects/PXD023664).
References
- Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera‐Pomar R, Luhrmann R (2005) A role for eIF4E and eIF4E‐transporter in targeting mRNPs to mammalian processing bodies. RNA 11: 717–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkin J, Halova L, Ferguson J, Hitchin JR, Lichawska‐Cieslar A, Jordan AM, Pines J, Wellbrock C, Petersen J (2014) Torin1‐mediated TOR kinase inhibition reduces Wee1 levels and advances mitotic commitment in fission yeast and HeLa cells. J Cell Sci 127: 1346–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averous J, Proud CG (2006) When translation meets transformation: the mTOR story. Oncogene 25: 6423–6435 [DOI] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A 3rd, Steever AB, Wach A, Philippsen P, Pringle JR (1998) Heterologous modules for efficient and versatile PCR‐based gene targeting in Schizosaccharomyces pombe . Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN (1996) TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell 7: 25–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beretta L, Gingras AC, Svitkin YV, Hall MN, Sonenberg N (1996) Rapamycin blocks the phosphorylation of 4E‐BP1 and inhibits cap‐dependent initiation of translation. EMBO J 15: 658–664 [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL (1994) A mammalian protein targeted by G1‐arresting rapamycin‐receptor complex. Nature 369: 756–758 [DOI] [PubMed] [Google Scholar]
- Buttgereit F, Brand MD (1995) A hierarchy of ATP‐consuming processes in mammalian cells. Biochem J 312(Pt 1): 163–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N, Babajko S, Seraphin B (2004) Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol 165: 31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybulski N, Hall MN (2009) TOR complex 2: a signaling pathway of its own. Trends Biochem Sci 34: 620–627 [DOI] [PubMed] [Google Scholar]
- De Virgilio C, Loewith R (2006) Cell growth control: little eukaryotes make big contributions. Oncogene 25: 6392–6415 [DOI] [PubMed] [Google Scholar]
- Du W, Halova L, Kirkham S, Atkin J, Petersen J (2012) TORC2 and the AGC kinase Gad8 regulate phosphorylation of the ribosomal protein S6 in fission yeast. Biol Open 1: 884–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop EA, Tee AR (2014) mTOR and autophagy: a dynamic relationship governed by nutrients and energy. Semin Cell Dev Biol 36: 121–129 [DOI] [PubMed] [Google Scholar]
- Garelick MG, Mackay VL, Yanagida A, Academia EC, Schreiber KH, Ladiges WC, Kennedy BK (2013) Chronic rapamycin treatment or lack of S6K1 does not reduce ribosome activity in vivo. Cell Cycle 12: 2493–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green VA, Pelkmans L (2016) A systems survey of progressive host‐cell reorganization during rotavirus infection. Cell Host Microbe 20: 107–120 [DOI] [PubMed] [Google Scholar]
- Gupta S, Mana‐Capelli S, McLean JR, Chen CT, Ray S, Gould KL, McCollum D (2013) Identification of SIN pathway targets reveals mechanisms of crosstalk between NDR kinase pathways. Curr Biol 23: 333–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Johnson RL (2011) Hippo signaling: growth control and beyond. Development 138: 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hall MN (1991) Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253: 905–909 [DOI] [PubMed] [Google Scholar]
- Hresko RC, Mueckler M (2005) mTOR.RICTOR is the Ser473 kinase for Akt/protein kinase B in 3T3‐L1 adipocytes. J Biol Chem 280: 40406–40416 [DOI] [PubMed] [Google Scholar]
- Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MBet al (2011) The mTOR‐regulated phosphoproteome reveals a mechanism of mTORC1‐mediated inhibition of growth factor signaling. Science 332: 1317–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber A, Bodenmiller B, Uotila A, Stahl M, Wanka S, Gerrits B, Aebersold R, Loewith R (2009) Characterization of the rapamycin‐sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev 23: 1929–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CS, Moggridge S, Muller T, Sorensen PH, Morin GB, Krijgsveld J (2019) Single‐pot, solid‐phase‐enhanced sample preparation for proteomics experiments. Nat Protoc 14: 68–85 [DOI] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN (2004) Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol 6: 1122–1128 [DOI] [PubMed] [Google Scholar]
- Jansen JM, Wanless AG, Seidel CW, Weiss EL (2009) Cbk1 regulation of the RNA‐binding protein Ssd1 integrates cell fate with translational control. Curr Biol 19: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung MY, Lorenz L, Richter JD (2006) Translational control by neuroguidin, a eukaryotic initiation factor 4E and CPEB binding protein. Mol Cell Biol 26: 4277–4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutsen JH, Rodland GE, Boe CA, Haland TW, Sunnerhagen P, Grallert B, Boye E (2015) Stress‐induced inhibition of translation independently of eIF2alpha phosphorylation. J Cell Sci 128: 4420–4427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurischko C, Kuravi VK, Herbert CJ, Luca FC (2011) Nucleocytoplasmic shuttling of Ssd1 defines the destiny of its bound mRNAs. Mol Microbiol 81: 831–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie S, Banks P, Lawless C, Lydall D, Petersen J (2018) The contribution of non‐essential Schizosaccharomyces pombe genes to fitness in response to altered nutrient supply and target of rapamycin activity. Open Biol 8: 180015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock A, Rutherford K, Harris MA, Hayles J, Oliver SG, Bahler J, Wood V (2019) PomBase 2018: user‐driven reimplementation of the fission yeast database provides rapid and intuitive access to diverse, interconnected information. Nucleic Acids Res 47: D821–D827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Hall MN (2011) Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189: 1177–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN (2002) Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell 10: 457–468 [DOI] [PubMed] [Google Scholar]
- Ma XM, Blenis J (2009) Molecular mechanisms of mTOR‐mediated translational control. Nat Rev Mol Cell Biol 10: 307–318 [DOI] [PubMed] [Google Scholar]
- Madrid M, Vazquez‐Marin B, Franco A, Soto T, Vicente‐Soler J, Gacto M, Cansado J (2016) Multiple crosstalk between TOR and the cell integrity MAPK signaling pathway in fission yeast. Sci Rep 6: 37515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Thomas PD (2013) PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res 41: D377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe . Methods Enzymol 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Nakashima A, Otsubo Y, Yamashita A, Sato T, Yamamoto M, Tamanoi F (2012) Psk1, an AGC kinase family member in fission yeast, is directly phosphorylated and controlled by TORC1 and functions as S6 kinase. J Cell Sci 125: 5840–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima A, Sato T, Tamanoi F (2010) Fission yeast TORC1 regulates phosphorylation of ribosomal S6 proteins in response to nutrients and its activity is inhibited by rapamycin. J Cell Sci 123: 777–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira AP, Ludwig C, Zampieri M, Weisser H, Aebersold R, Sauer U (2015) Dynamic phosphoproteomics reveals TORC1‐dependent regulation of yeast nucleotide and amino acid biosynthesis. Sci Signal 8: rs4 [DOI] [PubMed] [Google Scholar]
- Parker R, Song H (2004) The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol 11: 121–127 [DOI] [PubMed] [Google Scholar]
- Rispal D, Eltschinger S, Stahl M, Vaga S, Bodenmiller B, Abraham Y, Filipuzzi I, Movva NR, Aebersold R, Helliwell SBet al (2015) Target of rapamycin complex 2 regulates actin polarization and endocytosis via multiple pathways. J Biol Chem 290: 14963–14978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robitaille AM, Christen S, Shimobayashi M, Cornu M, Fava LL, Moes S, Prescianotto‐Baschong C, Sauer U, Jenoe P, Hall MN (2013) Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science 339: 1320–1323 [DOI] [PubMed] [Google Scholar]
- Russell JB, Cook GM (1995) Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiol Rev 59: 48–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaj J, Baluska F, Hirt H (2004) From signal to cell polarity: mitogen‐activated protein kinases as sensors and effectors of cytoskeleton dynamicity. J Exp Bot 55: 189–198 [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument‐Bromage H, Tempst P, Sabatini DM (2004) Rictor, a novel binding partner of mTOR, defines a rapamycin‐insensitive and raptor‐independent pathway that regulates the cytoskeleton. Curr Biol 14: 1296–1302 [DOI] [PubMed] [Google Scholar]
- Saxton RA, Sabatini DM (2017) mTOR signaling in growth, metabolism, and disease. Cell 168: 960–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzle T, Hall MN (2000) TOR, a central controller of cell growth. Cell 103: 253–262 [DOI] [PubMed] [Google Scholar]
- Soulard A, Cremonesi A, Moes S, Schutz F, Jeno P, Hall MN (2010) The rapamycin‐sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol Biol Cell 21: 3475–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta‐Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork Pet al (2019) STRING v11: protein‐protein association networks with increased coverage, supporting functional discovery in genome‐wide experimental datasets. Nucleic Acids Res 47: D607–D613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebe H, Morigasaki S, Murayama S, Zeng CT, Shiozaki K (2010) Rab‐family GTPase regulates TOR complex 2 signaling in fission yeast. Curr Biol 20: 1975–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebe H, Shiozaki K (2010) Rab small GTPase emerges as a regulator of TOR complex 2. Small GTPases 1: 180–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM (2012) A unifying model for mTORC1‐mediated regulation of mRNA translation. Nature 485: 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS (2009) An ATP‐competitive mammalian target of rapamycin inhibitor reveals rapamycin‐resistant functions of mTORC1. J Biol Chem 284: 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman Het al (2007) Sch9 is a major target of TORC1 in Saccharomyces cerevisiae . Mol Cell 26: 663–674 [DOI] [PubMed] [Google Scholar]
- Weisman R, Roitburg I, Schonbrun M, Harari R, Kupiec M (2007) Opposite effects of tor1 and tor2 on nitrogen starvation responses in fission yeast. Genetics 175: 1153–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederhold K, Passmore LA (2010) Cytoplasmic deadenylation: regulation of mRNA fate. Biochem Soc Trans 38: 1531–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederkehr T, Pretot RF, Minvielle‐Sebastia L (1998) Synthetic lethal interactions with conditional poly(A) polymerase alleles identify LCP5, a gene involved in 18S rRNA maturation. RNA 4: 1357–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN (2006) TOR signaling in growth and metabolism. Cell 124: 471–484 [DOI] [PubMed] [Google Scholar]
- Yang H, Rudge DG, Koos JD, Vaidialingam B, Yang HJ, Pavletich NP (2013) mTOR kinase structure, mechanism and regulation. Nature 497: 217–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SPet al (2011) Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 332: 1322–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Review Process File
Expanded View Figure PDF
Table EV1
Table EV2
Table EV3
Table EV4
Data Availability Statement
The datasets produced in this study are available in the following databases: Mass spectrometry proteomics data: PRIDE PXD023664 (https://www.ebi.ac.uk/pride/archive/projects/PXD023664).


