Abstract
The SAGA coactivator complex is essential for eukaryotic transcription and comprises four distinct modules, one of which contains the ubiquitin hydrolase USP22. In yeast, the USP22 ortholog deubiquitylates H2B, resulting in Pol II Ser2 phosphorylation and subsequent transcriptional elongation. In contrast to this H2B‐associated role in transcription, we report here that human USP22 contributes to the early stages of stimulus‐responsive transcription, where USP22 is required for pre‐initiation complex (PIC) stability. Specifically, USP22 maintains long‐range enhancer–promoter contacts and controls loading of Mediator tail and general transcription factors (GTFs) onto promoters, with Mediator core recruitment being USP22‐independent. In addition, we identify Mediator tail subunits MED16 and MED24 and the Pol II subunit RBP1 as potential non‐histone substrates of USP22. Overall, these findings define a role for human SAGA within the earliest steps of transcription.
Keywords: epigenetic, pre‐initiation complex, SAGA, transcription, USP22
Subject Categories: Post-translational Modifications, Proteolysis & Proteomics; Protein Biosynthesis & Quality Control; Transcription
Loss of human USP22 affects the stability of the pre‐initiation complex, rather than histone H2B ubiquitylation, during stimulus‐responsive transcription.

Introduction
Formation of the pre‐initiation complex (PIC) is among the central events in eukaryotic transcription. The PIC consists of ~45 proteins, including the RNA polymerase II (Pol II) enzyme complex and the general transcription factors (GTFs) TFIID, TFIIA, TFIIB, TFIIF, TFIIE, and TFIIH (Duttke, 2015; Schilbach et al, 2017). At many loci, additional multiprotein complexes participate in the early events of transcription. These include the Mediator complex which, among other functions, coordinates transmission of information between activator‐bound regulatory enhancers and the promoter‐bound PIC (Malik & Roeder, 2010; El Khattabi et al, 2019). These events and the proteins involved exhibit remarkable evolutionary conservation.
Also critical to the regulation of transcription across eukaryotes is the SAGA coactivator complex. SAGA is a 2MDa complex consisting of approximately 20 proteins structurally divided into modules with discrete biochemical functions (Grant et al, 1997; Roberts & Winston, 1997; Grant et al, 1998; Ben‐Shem et al, 2020; Cheon et al, 2020; Soffers & Workman, 2020; Grant et al, 2021; Helmlinger et al, 2021). As we and others have shown, these modules include an activator‐docking module comprising the TRRAP subunit, a core module that mediates recruitment of TATA‐binding protein (TBP) (Sharov et al, 2017; Papai et al, 2020; Wang et al, 2020), an acetyltransferase module (HAT), and a deubiquitylase module (DUB) (Brownell et al, 1996; McMahon et al, 1998; Henry et al, 2003; Cornelio‐Parra et al, 2021). The role of the acetyltransferase module has been extensively studied, and its catalytic subunit GCN5/KAT2A was the first KAT to be directly linked to transcription via acetylation of histones (Georgakopoulos & Thireos, 1992; Brownell et al, 1996; Grant et al, 1997), which leads to recruitment of the chromatin remodeler SWI/SNF (Chandy et al, 2006) and the super‐elongation complex (Gates et al, 2017). The DUB module of SAGA was discovered more recently and has been less well characterized. In yeast, the DUB module contains Ubp8p as its catalytic core, and its removal of monoubiquitin from histone H2B at residue K123 (K120 in humans) mediates transcriptional elongation (Henry et al, 2003; Wyce et al, 2007). While most studies of yeast SAGA function suggest that it is present primarily at stress‐inducible genes (Basehoar et al, 2004; Huisinga & Pugh, 2004), more recent findings suggest that SAGA and TFIID overlap in their activator function at all Pol II‐transcribed genes (Baptista et al, 2017; Warfield et al, 2017; Donczew et al, 2020). These two complexes share subunits of the TAF module and similar structural features therein (Wu et al, 2004; Bieniossek et al, 2013), yet SAGA exhibits unique capabilities via its two enzymatic modules.
Additional layers of complexity have impaired our ability to decipher the specific role of the human Ubp8p ortholog USP22 within SAGA (Zhang et al, 2008b; Zhao et al, 2008). In yeast, for example, mutation or deletion of ATXN7 ortholog Sgf73 results in increased ubiquitylation of histone H2B (Yan & Wolberger, 2015; Hsu et al, 2019). However, in higher eukaryotes such as Drosophila, the DUB module can bind chromatin independently of SAGA (Mohan et al, 2014; Li et al, 2017). Moreover, human USP22 has numerous substrates beyond H2B, including another subunit of the histone octamer, H2A (Zhang et al, 2008a), as well as several non‐histone substrates (Atanassov et al, 2009; Atanassov & Dent, 2011; Gao et al, 2014; Gennaro et al, 2018). Furthermore, most studies of the SAGA DUB module in humans have manipulated the regulatory subunit ATXN7L3 rather than USP22 directly (Lang et al, 2011; Bonnet et al, 2014). In Drosophila, biochemical defects associated with loss of ATXN7L3 were initially interpreted as a loss of USP22 function. However, a recent study from the Dent group demonstrated that ATXN7L3 forms catalytic complexes with other DUBs beyond USP22 (Atanassov et al, 2016). Since these other DUBs are not incorporated into SAGA, phenotypes associated with ATXN7L3 include both SAGA‐dependent and SAGA‐independent effects.
Given the prevalence of USP22 overexpression in aggressive cancer phenotypes (Wang & Dent, 2014), efforts are underway to target its catalytic activity. Understanding the precise contributions of USP22 to transcription in humans is key to the rational design and implementation of these strategies. Remarkably, genome‐wide studies of SAGA and USP22 in humans remain limited. As reported here, direct analysis of events in the transcription cycle that are regulated by USP22 reveals that it is indeed required for target gene transcription in response to ER stress. This requirement of USP22 persists for the biological consequences of ER stress, including apoptosis. Comprehensive analysis of loci encoding ER stress response genes unexpectedly revealed no correlation between a transcriptional dependence upon USP22 and alterations in H2B ubiquitylation levels. Instead, USP22 participates in the earliest events of PIC formation, with the Mediator core module subunit MED1 the only protein assessed whose binding to the promoter is not dependent on USP22. Consistent with this, USP22 contributes to the stability of long‐range enhancer–promoter contacts at ER stress‐activated target genes. Moreover, a proteome‐wide screen identified several PIC subunits whose ubiquitylation inversely correlates with USP22 activity. These findings demonstrate that human SAGA is indeed required for stimulus‐responsive transcription and reveal an unexpected role for USP22 in regulating PIC stability.
Results
SAGA and USP22 inducibly bind to the promoter region of ER stress response genes
Several pieces of evidence in metazoans suggest that the DUB module, which contains USP22 as its catalytic core, may have functions outside H2B deubiquitylation and transcriptional elongation. For example, in humans, USP22 has several well‐characterized substrates beyond H2B (Atanassov et al, 2009; Atanassov & Dent, 2011; Gao et al, 2014; Gennaro et al, 2018). In addition, the USP22 accessory proteins ATXN7L3 and ENY2 form alternative DUB complexes with the USP22‐related enzymes USP27X and USP51 in humans (Atanassov et al, 2016), and neither of these DUBs have apparent orthologs in yeast. These and other complexities of the metazoan system have been a barrier to understanding the role of the DUB module in human SAGA. In order to precisely define the contributions of USP22 to the mammalian transcription cycle, we utilized the ER stress response, a known SAGA‐dependent transcriptional program (Nagy et al, 2009), as a useful platform for these studies.
We first defined the genome occupancy patterns for the two catalytic SAGA modules during the human ER stress response by performing ChIP‐seq in HCT116 colorectal cancer cells for three SAGA proteins: the KAT GCN5, the DUB USP22, and the USP22 accessory factor ATXN7L3 (Fig EV1, EV2, EV3, EV4). This analysis was performed before and after treatment with Thapsigargin, whose non‐competitive inhibition of the sarco‐endoplasmic reticulum Ca2+ ATPase (SERCA) pump proteins induces the ER stress response (Lytton et al, 1991; Furuya et al, 1994); to minimize secondary transcriptional effects, we induced ER stress for 2 h only. The results of this analysis showed distinct binding profiles of the three SAGA members assessed, with little overlap between them (Fig EV1D). GCN5 and ATXN7L3 peaks were identified at over 1,500 and 4,000 loci each, respectively, while USP22 peaks numbered only in the low hundreds. We therefore prioritized peaks likely to represent SAGA complex binding by relaxing our criteria for a SAGA binding event, identifying those shared by all three proteins in at least one replicate and at least one condition. This modified peak calling yielded 311 peaks shared between all three members, 293 of which were associated with gene promoters, consistent with findings from Drosophila where SAGA binds primarily to the TSS of genes (Li et al, 2017). That GCN5 and ATXN7L3 are both members of non‐SAGA complexes (Wang et al, 2008; Atanassov et al, 2016) may explain why they occupy more sites than USP22. We analyzed the 293 SAGA‐bound promoters and found that 29 showed stress‐induced recruitment of SAGA (Figs 1A and B, and EV1, Appendix Fig S1). Normalized ChIP‐seq signals for the individual SAGA subunits demonstrated inducible binding of GCN5, USP22, and ATXN7L3 after treatment (Fig 1C). While GCN5 demonstrated slightly higher basal occupancy than the other two SAGA subunits in untreated conditions, levels of induced binding were tightly correlated between all three subunits (Fig 1D–F). These findings suggest that recruitment of a fully assembled SAGA complex occurs at a relatively restricted set of loci in humans.
Figure EV1. Related to Fig 1, SAGA and USP22 inducibly bind to the promoter region of ER stress response genes.

-
A, BChIP‐qPCR for USP22 at the CHOP and ERP70 loci before and after Thaps treatment, with and without shRNA‐mediated depletion of USP22. Three independent experiments are represented as mean ± SEM, with significance measured by Student’s t‐test. ***P < 0.005. X‐axis labels on CHOP indicate coordinates relative to the TSS.
-
CChIP‐qPCR for USP22 and ATXN7L3 at bound (GRP78, SEL1L) and unbound (FNDC3B) genes. Three independent experiments are represented as mean ± SEM. Desert serves as a negative control.
-
DVenn diagram overlap of SAGA peaks prior to filtering for ER stress responsiveness as measured by increases in SAGA subunit ChIP‐seq signal following Thaps treatment.
Figure EV2. Related to Fig 3, USP22 is required for transcription of ER stress response genes.

-
ACells were treated for 2 h with 100 nM Thapsigargin in the presence or absence of shRNAs targeting USP22. Whole‐cell lysate was subjected to immunoblotting with the indicated antibodies.
-
BHCT116 cells were treated with 100 nM Thapsigargin for the indicated times in the presence or absence of shRNA targeting USP22. RNA was isolated and subjected to qRT–PCR against the indicated transcripts. Three independent experiments are represented as mean ± SEM.
-
C, DCells were treated as in (A) and subjected to ChIP‐qPCR for ATF4 and XBP1s at the indicated gene enhancers and promoters. Three independent experiments are represented as mean ± SEM.
Source data are available online for this figure.
Figure EV3. Related to Fig 4, USP22 is required for efficient recruitment of Pol II to activator‐induced target genes independent of Ub‐H2B.

-
AFlowchart depicting ChIP‐seq subsetting.
-
BMetagene profiles for Pol II (top) and Ub‐H2B (bottom) ChIP‐seq for all genes not directly bound by USP22 and insensitive to USP22 knockdown, termed “Insensitive” genes.
-
C, DChIP‐qPCR for ATXN7L3 and GCN5 at the indicated loci, before and after ER stress, in the presence and absence of shRNA targeting USP22, represented from three independent experiments as mean ± SEM.
Figure EV4. Related to Fig 6, deubiquitylation of Pol II is controlled by USP22.
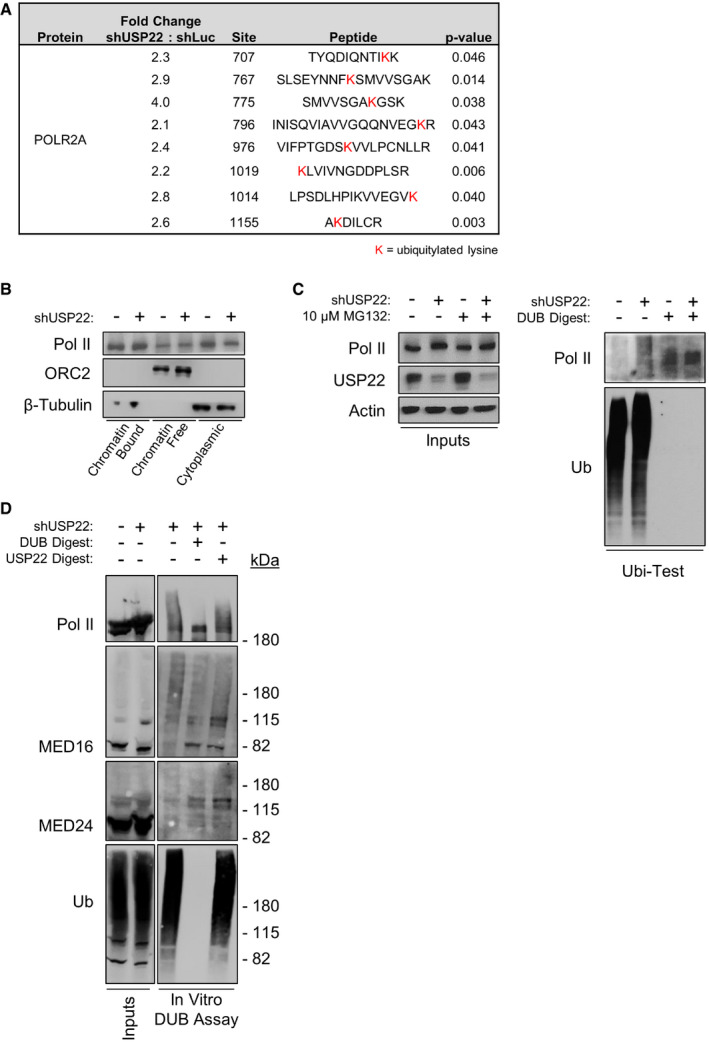
- High‐throughput proteomic analysis of Pol II following depletion of USP22, with significance measured by Student’s t‐test. Specific ubiquitylated lysines detected are highlighted in red text.
- HCT116 cells were treated for 2 h with 100 nM Thapsigargin in the presence or absence of shRNA targeting USP22. Cells were harvested and fractionated, and fractions were subjected to immunoblotting with the indicated antibodies. Loading controls ORC2 and β‐tubulin are duplicate images from Fig 6.
- In vitro deubiquitylation UbiTest of endogenously ubiquitylated Pol II. HCT116 cells were treated with MG132 in the presence or absence of shRNA targeting USP22. Lysates were generated using buffer containing protease cocktail inhibitor, pan‐DUB inhibitor PR619, and the JAMM protease inhibitor o‐phenanthroline. Ubiquitylated proteins were purified on ubiquitin‐binding resin, and eluates were either undigested (lanes 1 and 2) or digested with USP2 to strip polyubiquitin (lanes 3 and 4) and reduce target proteins to unit length. Digestion reactions were subjected to immunoblotting with the indicated antibodies. shRNA control USP22 and loading controls actin and Ub are duplicate images from Fig 6.
- In vitro deubiquitylation of endogenously ubiquitylated proteins. HCT116 cells were treated as in (C). Ubiquitylated proteins were purified on ubiquitin‐binding resin, and eluates were either undigested (lane 3) or digested with non‐specific DUB (“USP2”, lane 4) or human DUB module (“USP22”, lane 5) to reduce target protein ubiquitylation. Digestion reactions were subjected to immunoblotting with the indicated antibodies.
Source data are available online for this figure.
Figure 1. SAGA and USP22 inducibly bind to the promoter region of ER stress response genes.
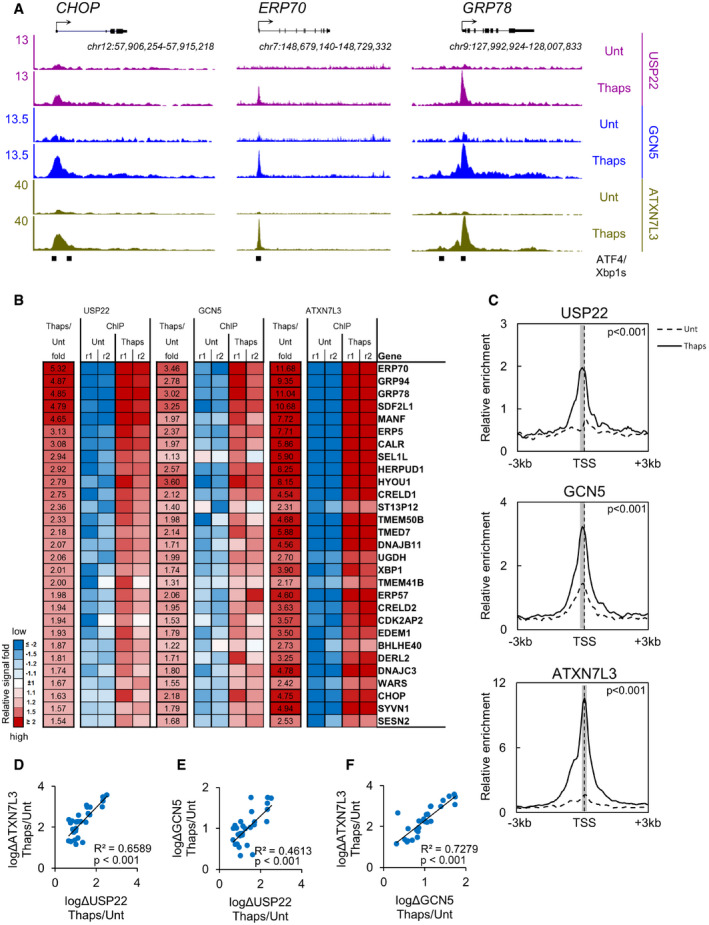
- A
-
BHeat map of USP22, GCN5, and ATXN7L3 association at the TSS for 29 genes whose promoters undergo significant recruitment of USP22 following induction of ER stress.
-
CMean ChIP‐seq signal for USP22, GCN5, and ATXN7L3 surrounding gene promoters from (B). Nonparametric Mann–Whitney analysis of 7 bins (gray shade) surrounding maximal peak in Thaps‐treated conditions is reported.
-
D–FLinear regression analysis of SAGA member recruitment to target gene promoters following induction of ER stress.
USP22 is required for the biological consequences of ER stress
Having observed direct recruitment of USP22 to stress‐induced genes, we assessed the functional requirement for this recruitment. Thapsigargin‐mediated inhibition of SERCA is irreversible and inevitably results in apoptosis (Lytton et al, 1991; Furuya et al, 1994). Depletion studies demonstrated that full induction of this apoptotic response requires USP22 (Fig 2A and B, Appendix Fig S3). Consistent with these results from Cas3/7 and Annexin V staining, PARP and caspase 3 cleavage also were impaired following depletion of USP22 (Fig 2C). These data suggest that recruitment of USP22 to ER stress response gene promoters is required for full functionality of the ER stress response.
Figure 2. USP22 is required for the biological consequences of ER stress.

- HCT116 cells were treated with 100 nM Thapsigargin for 24 h. Cells were stained with dye recognizing cleaved caspases 3 and 7 and imaged every 4 h. Scale bar = 400 µm.
- Quantification of apoptotic populations as indicated by Cas3/7 fluorescence from (A). Three independent experiments are represented as mean ± SEM, with significance calculated by two‐way ANOVA between conditions over time (f(3) = 50.530, P < 0.001); significant pairwise comparisons by Tukey’s HSD post hoc assessment are indicated by *.
- Whole‐cell lysates from the indicated time points were subjected to immunoblotting with the indicated antibodies.
Source data are available online for this figure.
USP22 regulates transcription of ER stress response genes
Given the direct recruitment of USP22 to ER stress response genes and its regulation of the biological response to ER stress, we interrogated the requirement of USP22 for the transcription of ER stress response genes. In the presence of an ER stress stimulus, unfolded proteins accumulate in the ER, leading to sequestration of GRP78, a chaperone protein, from transmembrane proteins ATF6, PERK, and IRE1, the latter two of which self‐dimerize, autophosphorylate (Sidrauski & Walter, 1997; Harding et al, 1999), and activate translation of ER stress transcription factors ATF4 and Xbp1s, respectively. ATF6, itself a transcription factor, is cleaved from the ER membrane and transits through the Golgi to the nucleus (Haze et al, 1999), where in concert with ATF4 and Xbp1s, it activates ER stress response genes such as GRP78 and additional ER stress response transcription factors such as CHOP (Yamamoto et al, 2007; Bergmann & Molinari, 2018). Ruling out an effect on upstream signaling events, depletion of USP22 did not directly affect activation of these transcription factors (Fig EV2A). Transcript levels of the ATF6 gene itself decreased without USP22, but its initial activation as a transcription factor occurs post‐transcriptionally when the ATF6 protein is cleaved from ER membrane and translocates to the nucleus via the Golgi (Haze et al, 1999). Although our antibody to ATF6 detected no cleaved isoform following induction of ER stress with Thapsigargin, we also observed no decrease in uncleaved ATF6 protein post‐induction, suggesting that in our system, this cleavage event does not occur in response to Thapsigargin and therefore does not contribute to the transcriptional defect observed upon loss of USP22 (Fig EV2B). Moreover, binding of activators to target gene promoters and enhancers, whose binding sites were determined by publicly available genome‐wide binding profiles of ATF4 and Xbp1s (Chen et al, 2014; Gowen et al, 2015), was also unaffected by loss of USP22 (Fig EV2C and D). However, upregulation of ER stress response genes CHOP and GRP78 was defective without USP22 (Fig 3A and Appendix Fig S4A). Kinetic analysis revealed that defects in the transcription of ER stress response genes following USP22 depletion appeared permanent rather than simply delayed (Fig 3B and Appendix Fig S4B). Moreover, immunoblotting for the GRP78 and CHOP proteins reflects the patterns observed at the transcript level, where the shUSP22 condition remains perpetually impaired in its upregulation of these ER stress targets. Assessment of histone H2B in whole‐cell lysates showed reduced Ub‐H2B levels, in line with previous observations where alternate DUB complexes overcompensate for loss of USP22 (Atanassov et al, 2016).
Figure 3. USP22 regulates transcription of ER stress response genes.

- HCT116 cells were treated with 100 nM Thapsigargin for the indicated times in the presence or absence of shRNA targeting USP22. Cells were harvested, and whole‐cell lysate was subjected to immunoblotting with the indicated antibodies.
- Cells were treated as in (A), but over an extended series of time points. RNA was isolated and subjected to qRT–PCR against the indicated transcripts. Three independent experiments are represented as mean ± SEM.
- A Tetracycline (Tet)‐inducible, shRNA‐resistant USP22 transgene was stably introduced into HCT116 cells. Cells were treated as in (A), with and without expression of the transgene via Tet treatment. Whole‐cell lysates were subjected to immunoblotting with the indicated antibodies.
- RNA was isolated from conditions described in (C) subjected to qRT–PCR analysis against the CHOP mRNA. Three independent experiments are represented as mean ± SEM, with significance measured by Student’s t‐test. ***P < 0.005.
Source data are available online for this figure.
To ensure that the effects observed in the RNAi studies were direct consequences of the absence of USP22, we rescued USP22 expression with a conditional, shRNA‐resistant allele and largely restored ER stress‐induced upregulation of CHOP protein (Fig 3C) and mRNA (Fig 3D). Of note, autophosphorylation of PERK and PERK‐mediated phosphorylation of translation factor EIF2a (p‐)EIF2α represents initial activation events of the ER stress response that precede transcription of ER stress target genes and were probed here as controls for successful stimulation (Fig 3C and Appendix Fig S4A). Neither phosphorylation event, as observed by the shift in PERK migration or the increase in p‐EIF2α band intensity, was affected by USP22 depletion, illustrating that the pre‐transcriptional response to stress is not impaired by loss of USP22. Furthermore, depletion of USP22 with several distinct shRNA constructs elicited similar defects in the transcription of ER stress response genes (Appendix Fig S4C and D). Collectively, these data confirm that transcription of the ER stress response requires USP22.
USP22 is required for recruitment of Pol II to ER stress response genes independent of H2B deubiquitylation
Ubiquitylation of H2B has long been associated with elongating Pol II (Xiao et al, 2005; Tanny et al, 2007). In yeast, H2B ubiquitylation at actively transcribed genes occurs following loading and phosphorylation of Pol II at Ser5 of the carboxy‐terminal domain (CTD), and subsequent deubiquitylation by the USP22 ortholog Ubp8p is required for optimal gene activation (Henry et al, 2003; Wyce et al, 2007). To assess whether H2B ubiquitylation status is the rate‐limiting factor controlled by USP22 as part of its role in transcription in metazoans, we performed ChIP‐seq for Pol II and Ub‐H2B in untreated and Thapsigargin‐treated cells, with and without depletion of USP22. By using changes in Pol II occupancy and Ub‐H2B levels in the gene body as a surrogate for transcriptional activity, our analysis is sensitive to early transcription of target genes, where these changes may precede detection of accumulated mRNA. To this point, many genes that exhibit significant increases in Pol II binding after stress induction were defined by previous studies as members of the second wave of ER stress response transcription (Dombroski et al, 2010; Bergmann et al, 2018), yet our ChIP‐seq analyses identified them as target genes at just 2 h after stress induction. This approach revealed 1,447 Thapsigargin‐responsive genes as true targets of the ER stress response transcriptional program based on increased Pol II occupancy at the promoters and coding regions (Figs 4A and EV3A). Depletion of USP22 impaired stress‐induced accumulation of Pol II at many ER stress response genes, both at the promoters and throughout the coding regions (Fig 4A and Appendix Fig S2). Interestingly, ER stress‐induced recruitment of Pol II was sensitive to loss of USP22 at 283 genes that were either directly bound by SAGA (“Bound” group, 28 genes, Fig 4B, upper panel) or not bound by SAGA (“Unbound” group, 255 genes, Fig 4C, upper panel); with the exception of XBP1, which was insensitive to depletion of USP22, the 28 genes in the Bound group are the same genes identified in Fig 1B. An additional 1,164 ER stress‐induced genes were insensitive to USP22 depletion (Fig EV3B). Mean Pol II occupancy was markedly higher across the bound group than the unbound group. It is noteworthy that ER stress‐induced recruitment of ATXN7L3 and GCN5 was unaffected by depletion of USP22 (Fig EV3C and D). These data suggest that USP22 is dispensable for stable recruitment of the SAGA complex to gene promoters. However, the presence of USP22 is essential for maximal transcription of these genes.
Figure 4. USP22 is required for recruitment of Pol II to ER stress response genes independent of H2B deubiquitylation.

- Metagene profiles for Pol II (upper panel) and Ub‐H2B (middle panel) ChIP‐seq for USP22‐dependent, USP22‐bound genes. Nonparametric Mann–Whitney analyses of 7 bins (gray shade) surrounding TSS (TSS) and the midpoint of the profiles (Body) in Thaps‐treated conditions are reported. mRNA analyses of select bound gene transcripts are presented (lower panel). Three independent experiments are represented as mean ± SEM, with significance measured by Student’s t‐test. *P < 0.05, **P < 0.01, and ***P < 0.005.
- Metagene profiles for Pol II (upper panel) and Ub‐H2B (middle panel) ChIP‐seq for USP22‐dependent, USP22‐unbound genes. Nonparametric Mann–Whitney analyses of 7 bins (gray shade) surrounding TSS (TSS) and the midpoint of the profiles (Body) in Thaps‐treated conditions are reported. mRNA analysis of select unbound gene transcripts are presented (lower panel). Three independent experiments are represented as mean ± SEM, with significance measured by Student’s t‐test. **P < 0.01 and ***P < 0.005.
Depletion of USP22 impaired recruitment of Pol II to both USP22‐bound and USP22‐unbound ER stress response genes. Given that in yeast, deubiquitylation of histone H2B by the SAGA DUB module is a necessary step in inducible transcription (Henry et al, 2003; Wyce et al, 2007), depletion of USP22 should result in aberrant increases in H2B ubiquitylation at USP22‐sensitive genes. However, we observed no such increases at any USP22‐sensitive, stress‐induced genes, whether bound or unbound. Depletion of USP22 had no effect on either basal or stress‐induced levels of Ub‐H2B at bound genes (Fig 4B, middle panel) and instead reduced stress‐induced Ub‐H2B levels at unbound genes (Fig 4C, middle panel). While levels of Ub‐H2B did not dramatically change upon USP22 depletion, stress‐induced recruitment of Pol II at both gene groups was impaired. This implies a broad requirement for USP22 in stimulus‐responsive transcription, even without its direct binding to all affected loci. While the defect in Pol II occupancy in USP22‐depleted cells is presumed to reflect a decrease level of transcription at these loci, direct examination of individual mRNA levels for both bound and unbound genes provided a direct confirmation (Fig 4B and C, lower panels). Although these results do not exclude deubiquitylation of H2B as a USP22‐regulated event in metazoans, they suggest it is not the rate‐limiting step controlled by human USP22 in transcription, as it is for Ubp8p in yeast (Wyce et al, 2007). Rather, USP22 participates prior to or in conjunction with recruitment of Pol II.
USP22 mediates PIC stability at ER stress response genes
Yeast Ubp8p removes monoubiquitin from H2B following phosphorylation of Ser5 of the Pol II CTD, allowing Ser2‐mediated phosphorylation by Ctk1, which ultimately facilitates elongation (Wyce et al, 2007). Since our genome‐wide analysis shows a USP22‐dependent effect on Pol II recruitment to stress‐induced genes, we assessed the requirement for USP22 in the phosphorylation of the Pol II CTD during transcription. Using a set of tiling primer pairs that span the CHOP locus (Fig 5A), we quantified changes in total Pol II as well as the pSer5 and pSer2 isoforms of Pol II. In addition to impairing stress‐induced recruitment of total Pol II across the gene, depletion of USP22 caused defects in both pSer5 Pol II and pSer2 Pol II (Fig 5B). This is in contrast to prior studies in yeast and humans where only pSer2 Pol II is affected by loss of Ubp8p or USP22, respectively (Wyce et al, 2007; Chipumuro & Henriksen, 2012). Stress‐induced phosphorylation of Ser5 and Ser2 was also defective at ERP70 and GRP78 in the absence of USP22 (Fig 5C). These data suggest that within the ER stress transcriptional response, phosphorylation of both initiation‐ and elongation‐linked sites in the Pol II CTD is dependent on USP22.
Figure 5. USP22 mediates PIC stability at ER stress response genes.

-
ASchematic of the CHOP locus. Black boxes indicate amplicons for subsequent ChIP‐qPCR analysis.
-
BHCT116 cells were treated for 2 h with 100 nM Thapsigargin in the presence or absence of shRNA targeting USP22. Cells were cross‐linked, harvested, and subjected to ChIP‐qPCR for total RNA Pol II, pSer5 Pol II, and pSer2 Pol II at the loci across CHOP (indicated in (A)). Three independent experiments are represented as mean ± SEM.
-
CChIP‐qPCR for total RNA Pol II, pSer5 Pol II, and pSer2 Pol II at ERP70, GRP78, and a gene desert (Prom = gene promoter and Coding = downstream coding region—see Appendix Table S1 for primer sequences). Three independent experiments are represented as mean ± SEM. with significance measured by Student’s t‐test. *P < 0.05, **P < 0.01, and ***P < 0.005.
-
D–GCells were treated as in (B) and subjected to ChIP‐qPCR for TBP, TFIIB, TFIIF, and TAF7 at the indicated gene promoters. Three independent experiments are represented as mean ± SEM, with significance measured by Student’s t‐test. *P < 0.05 and ***P < 0.005.
-
HAn aliquot of cells from (D‐G) were harvested for whole‐cell lysate and subjected to immunoblotting with the indicated antibodies.
Source data are available online for this figure.
In vitro, assembly of the PIC comprises a series of concerted steps, the first of which is TFIID‐ or SAGA‐mediated recruitment of TATA‐binding protein (TBP) (Weinzierl et al, 1993; Grant et al, 1998; Lee et al, 2000). This is followed by binding of Mediator and TFIIB, Pol II and TFIIF, and ultimately TFIIH, whose CDK7 subunit catalyzes phosphorylation of Pol II CTD at Ser5. In yeast, the DUB module of SAGA acts post‐PIC assembly to deubiquitylate H2B (Wyce et al, 2007). However, in the human cells examined here, USP22 aids the stable association of TBP, TFIIB, and TFIIF to stress‐induced gene promoters (Fig 5D–F). Additionally, recruitment of the TFIID‐specific subunit TAF7 subunit suggests that ER stress response transcription factors simultaneously recruit SAGA and TFIID for efficient transcription, at least at this early time point (Fig 5G), which is consistent with previous studies (Donczew et al, 2020). Total protein levels of these general transcription factors were unaffected by USP22 depletion, confirming that their decreased occupancy at ER stress response genes was due specifically to impaired recruitment (Fig 5H). These findings implicate metazoan USP22 in unexpectedly early steps in transcriptional activation.
Deubiquitylation of PIC components MED16, MED24, and RPB1 is controlled by USP22
As deubiquitylation of Ub‐H2B was not defective in USP22‐depleted cells, we employed an unbiased approach to identify additional substrates of USP22. Using affinity purification and mass spectrometry, we quantified USP22‐dependent changes in the ubiquitin‐modified proteome (Kim et al, 2011). We identified a number of high‐confidence, candidate non‐histone proteins whose ubiquitylation status was altered by loss of USP22, many of which are directly involved in transcription. Primary among these were the Mediator tail subunits MED16 and MED24 and the major Pol II subunit RPB1 (Figs 6A and EV4A). H2B was also identified in this screen (Table EV1; see Dataset EV1 for complete dataset), but levels decreased rather than increased in response to USP22 depletion, consistent with previous observations (Atanassov et al, 2016). Depletion of USP22 did not alter steady‐state levels of MED16, MED24, or Pol II (Figs 6B and EV4B), suggesting that their ubiquitylation status is not linked to proteasome‐mediated degradation. To confirm that changes in ubiquitylation of these candidates are USP22‐dependent, we utilized an independent method that relies on purification of ubiquitylated proteins using tandem ubiquitin‐binding entities (Hjerpe et al, 2009). This analysis validated both the Mediator tail MED24 and the Pol II subunit RPB1 as hyperubiquitylated in the absence of USP22 (Figs 6C and EV4C). Furthermore, recombinantly expressed and purified human DUB module was capable of directly deubiquitylating MED16, MED24, and Pol II in an in vitro digestion reaction, as indicated by reduction of visible streaking with MED16 and Pol II as well as increases in unit‐length protein (Fig EV4D). The appearance of higher molecular weight bands in the DUB‐digested conditions suggests the persistence of ubiquitylated residues or other post‐translational modifications insensitive to DUB or USP22 activity.
Figure 6. Deubiquitylation of PIC components MED16 and MED24 is controlled by USP22.
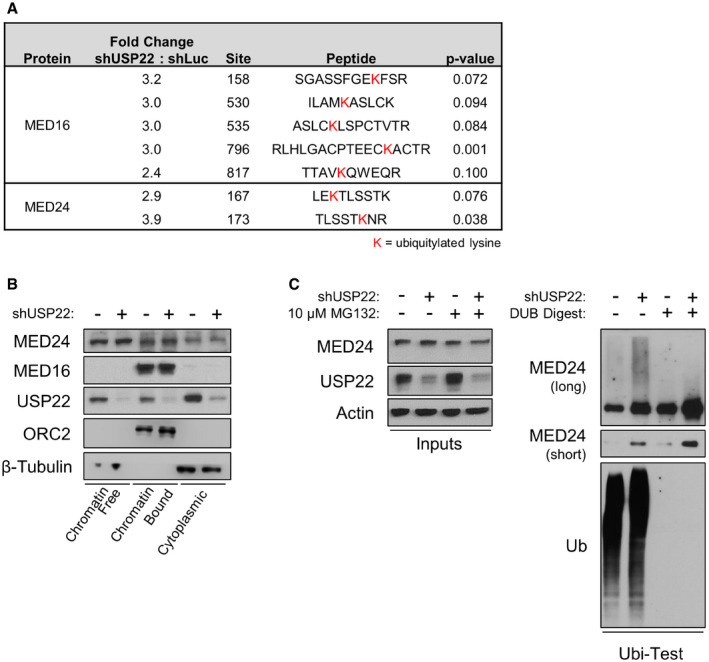
- High‐throughput proteomic analysis of Mediator tail subunits MED16 and MED24 following depletion of USP22, with significance measured by Student’s t‐test. Specific ubiquitylated lysines detected are highlighted in red text.
- HCT116 cells were treated for 2 h with 100 nM Thapsigargin in the presence or absence of shRNA targeting USP22. Cells were harvested and fractionated, and fractions were subjected to immunoblotting with the indicated antibodies.
- In vitro deubiquitylation UbiTest of endogenously ubiquitylated MED24. HCT116 cells were treated with MG132 in the presence or absence of shRNA targeting USP22. Lysates were generated using buffer containing protease cocktail inhibitor, pan‐DUB inhibitor PR619, and the JAMM protease inhibitor o‐phenanthroline. Ubiquitylated proteins were purified on ubiquitin‐binding resin, and eluates were either undigested (lanes 1 and 2) or digested with USP2 to strip polyubiquitin (lanes 3 and 4) and reduce target proteins to unit length. Digestion reactions were subjected to immunoblotting with the indicated antibodies.
Source data are available online for this figure.
USP22 is required for efficient recruitment of the Mediator tail module to ER stress response gene promoters and enhancers
Mediator comprises 25–30 proteins in eukaryotes to form a 1.8 MDa complex that bridges UAS/enhancer elements to the promoter‐bound PIC (Davis et al, 2002). This Mediator‐regulated looping of enhancer–promoter regions is essential for transcription in many contexts (Park et al, 2005; Wang et al, 2005; Kagey et al, 2010). Mediator contains four modules: head, middle, tail, and kinase, most of which have been characterized in detail structurally and biochemically (Tsai et al, 2014; Robinson et al, 2016; Nozawa et al, 2017; Schilbach et al, 2017; Tsai et al, 2017; Zhao et al, 2021). In vitro transcription studies with reconstituted Mediator complex show that the middle and head modules form the essential core Mediator required for transcription, while the tail module is dispensable (Jeronimo et al, 2016). However, the tail module is the primary contact between sequence‐specific transcription factors bound at enhancers and the promoter of the cognate target gene (Park et al, 2000; Stevens et al, 2002; Zhang et al, 2004; Thakur et al, 2009; Brzovic et al, 2011; El Khattabi et al, 2019). Evidence from yeast suggests that the Mediator tail is required for efficient transcription of SAGA‐dependent genes, but not for transcription of TFIID‐dependent genes (Ansari et al, 2012; Paul et al, 2015).
To assess the functional relationship between the Mediator tail module and USP22 during transcription, we performed ChIP‐seq for both middle (MED1) and tail (MED16) modules of Mediator in USP22‐competent and USP22‐depleted cells before and after induction of ER stress. Depletion of USP22 impaired MED16 binding at stress‐induced gene promoters and enhancers, yet elicited no discernible effect on either basal or stress‐induced binding of MED1 (Fig 7A). This is consistent with previous observations in yeast where the middle and head modules can associate with the PIC at the core promoter in the absence of the tail module (Jeronimo et al, 2016; Petrenko et al, 2016).
Figure 7. USP22 is required for efficient recruitment of the Mediator tail module to ER stress response gene promoters and enhancers.
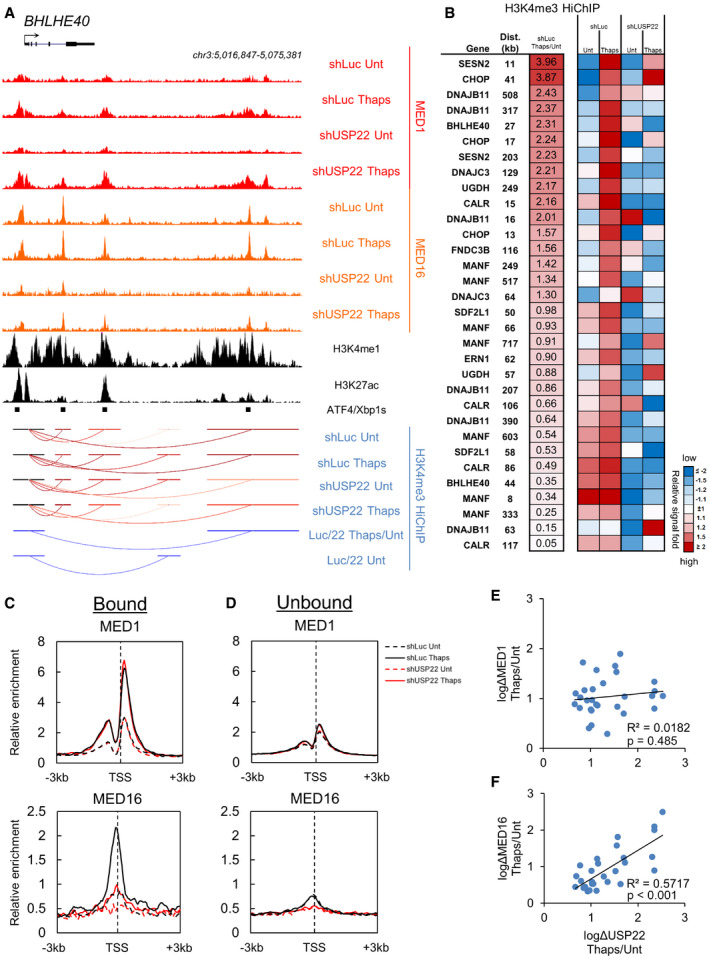
-
AGenome browser image of ChIP‐seq for middle and tail Mediator subunits and H3K4me3 HiChIP tracks before and after induction of ER stress at ER stress response gene BHLHE40. H3K4me1 and H3K27ac tracks are from the ENCODE Consortium (GEO accessions GSM945858 and GSM945853). ATF4/Xbp1s binding sites are from publicly available datasets (GEO accessions GSE69304 and GSE49952).
-
BH3K4me3 HiChIP signal (log2) at promoter–enhancer loops before and after induction of ER stress or USP22 depletion at indicated bound group genes.
-
C, DMean ChIP‐seq signal MED1 and MED16 surrounding gene promoters from gene groups defined in Fig 4.
-
E, FLinear regression analysis of Mediator and USP22 recruitment to USP22‐bound target gene promoters following induction of ER stress.
Given the established role of the Mediator tail in bridging activator‐bound enhancers with target gene promoters, we assessed long‐range DNA interactions to gain insight into the potential mechanism of USP22‐mediated control of tail Mediator binding to ER stress response loci. We performed Hi‐C followed by ChIP‐seq (HiChIP) for H3K4me3, a histone modification known to be found at active gene promoters. HiChIP confers practical and experimental advantages over Hi‐C because it allows for targeted assessment of long‐range interactions by isolating only Hi‐C products containing the epitope to the ChIP antibody of choice, thus reducing the overall complexity of the dataset and reducing the number of sequencing reads required to achieve sufficient coverage across the interactome. Several long‐range interactions can be observed at the BHLHE40 promoter, a USP22‐sensitive, SAGA‐bound ER stress response gene (Fig 7A). Two long‐range interactions with the BHLHE40 promoter change in response to ER stress, the more distal of which overlaps MED16 ChIP signal, contains an XBP1s binding site, and is deficient in forming a promoter–enhancer loop in the absence of shUSP22 (Thaps/Luc shLuc/shUSP22). To support this observation in a broader context, a heat map of ER stress promoter–enhancer loops depicts bound group gene promoters whose interaction frequencies increase following ER stress and are defectively regulated following loss of USP22 (Fig 7B and Table EV2). These data suggest that USP22 participates in the stabilization of long‐range promoter–enhancer loops and that disruption of these interactions via depletion of USP22 correlates with deficient upregulation of target gene transcription.
Using the groups of USP22‐sensitive ER stress response genes defined earlier (Fig 4B and C), binding patterns revealed robust recruitment of both MED1 and MED16 to USP22‐Bound genes (Fig 7C). By contrast, we observed only modest recruitment of MED1 after ER stress and minimal changes in MED16 after depletion of USP22 at unbound genes (Fig 7D).
Our ChIP‐seq analyses show that core Mediator binds ER stress response gene promoters together with USP22 and the rest of SAGA. In fact, MED1 ChIP signal is found at all SAGA‐Bound gene promoters defined in Fig 4. However, assessment of ER stress‐induced changes in MED1 and USP22 binding shows poor correlation (Fig 7E). This is often due to the fact that MED1 is already bound to these promoters prior to ER stress. By contrast, both USP22 and MED16 are recruited to SAGA‐bound promoters in a stress‐dependent manner and to a similar extent across these loci (Fig 7F). Additionally, regions proximal to gene promoters in the USP22‐bound group contained binding sites for transcription factors Xbp1s or ATF4 as well as enhancer marks H3K4me1 and H3K27ac (Appendix Fig S2). Depletion of USP22 resulted in a marked reduction of MED16 binding in unstressed conditions in both bound and unbound gene groups (Fig 7C and D), indicating that the tail module and SAGA need not co‐occupy a specific locus for USP22 to exert its effects on long‐range promoter–enhancer interactions. Collectively, these findings suggest that regulation of MED16 and MED24 deubiquitylation by USP22 may stabilize tail module binding at gene enhancers to promote PIC stability and subsequent transcription.
Discussion
The precise biochemical events in the transcription cycle that are controlled by individual SAGA subunits have been difficult to define, particularly in metazoans. This is based in part on subunit promiscuity (Helmlinger & Tora, 2017). For example, the TRRAP subunit of SAGA is also a component of a distinct acetyltransferase complex, NuA4/TIP60. TAFs 5, 6, 9, 10, and 12 exist in both SAGA and TFIID. hGCN5 can be replaced within SAGA by its paralog acetyltransferase PCAF/KAT2B, and both of these enzymes exist outside SAGA as subunits of the related ATAC (Wang et al, 2008), ADA (Eberharter et al, 1999), and other complexes (Brand et al, 1999; Martinez et al, 2001; Soffers et al, 2019; Torres‐Zelada et al, 2019). Most relevant to the findings presented here, the SAGA DUB subunits ATXN7L3 and ENY2 are essential for USP22 activity as well as ubiquitin hydrolases USP27X and USP51; prior to the recent discovery of ATXN7L3’s multiplicity of DUB partners, transcription‐related events identified as ATXN7L3‐dependent were attributed solely to USP22 activity as part of SAGA. This final distinction is evident even in the current stimulus‐responsive transcriptional program, where depletion of the common subunit ATXN7L3 increases global (Appendix Fig S5A) and local (Appendix Fig S5B) Ub‐H2B levels in unstressed cells, while depletion of USP22 or GCN5 did not have this effect.
The present study focuses on defining the precise role in transcription played by the human SAGA DUB module and its catalytic core USP22. The central underpinning for this analysis comes from comprehensive yeast studies of the USP22 ortholog Ubp8p, which deubiquitylates K123 of H2B at the 5′ end of ORFs to facilitate Pol II CTD phosphorylation and transcriptional elongation (Henry et al, 2003; Wyce et al, 2007). Our genome‐wide occupancy studies in humans included USP22, GCN5, ATXN7L3, Pol II, and Ub‐H2B and revealed a set of findings that uncover stark contrasts to the paradigm established in yeast. Most striking is the finding that the human USP22 DUB module of SAGA affects PIC stability itself. This result contrasts with a previous in vitro study which designates SAGA function primarily after PIC formation (Chen et al, 2012), reflecting previously reported disparities regarding contributions to PIC stability in vitro versus in vivo (Petrenko et al, 2019). Our genome‐wide analysis of USP22 in activated transcription reveals a highly restricted set of genes directly occupied by USP22 and a broader set of stimulus‐responsive genes that indirectly require USP22 activity for their transcription. In further contrast to yeast SAGA, loss of metazoan USP22 has no impact on H2B ubiquitylation at its directly bound target genes and results in a paradoxical decrease in H2B ubiquitylation at induced genes that require USP22 indirectly. Instead, genome occupancy studies implicate recruitment of Mediator tail subunits as among the primary transcriptional events for which USP22 is essential.
Our long‐range interaction analysis at ER stress‐responsive genes demonstrates a requirement for the SAGA DUB module to regulate enhancer–promoter contacts. These observations, coupled with the identification of Mediator tail subunits MED16 and MED24 as direct substrates of USP22, suggest a model where the ubiquitylation status of these subunits controls promoter–enhancer interactions. The tail module of Mediator typically bridges enhancer‐bound activators and promoter‐bound Mediator middle and head modules to form enhancer–promoter loops essential for activated transcription (Kagey et al, 2010; Phillips‐Cremins et al, 2013; Jeronimo et al, 2016; Robinson et al, 2016), although it should be noted that this physical “bridge” model has recently been challenged, instead proposing that Mediator acts as a functional transmission hub from TF‐bound enhancers to PIC‐bound promoters (El Khattabi et al, 2019). Our genome‐wide studies implicate USP22 as a central regulator of these Mediator tail functions, based on the finding that USP22 is required for induction of Mediator tail binding to promoters, while core Mediator binding does not require USP22. Even where basal levels of Mediator tail binding are decreased in USP22‐depleted cells, core Mediator binding appears unaffected. Analysis of a selected subset of enhancers in this pathway reveals a correlation between USP22 and Mediator tail binding. This link between SAGA and Mediator tail echoes previous studies showing that loss of tail affects expression of only a subset of genes and that this subset is enriched for SAGA‐dependent targets (Ansari et al, 2012; Paul et al, 2015). In line with our observations, Arabidopsis MED16 is required for recruitment of Mediator and Pol II during the induction of the cold on‐regulated (COR) transcriptional program (Hemsley et al, 2014).
It is also noteworthy that we observe little evidence for any of the three SAGA subunits occupying enhancers, yet significant evidence of promoter‐bound SAGA. This is consistent with at least one previous study in Drosophila, in which Ada2b, Spt3, and Sgf11 were interrogated as surrogates for SAGA and were shown to localize over TSSs genome‐wide (Li et al, 2017). Additionally, a limited number of genes showed low levels of SAGA binding across their coding regions after stress induction, consistent with previous studies (Govind et al, 2007; Wyce et al, 2007; Weake et al, 2011). Finally, findings reported here present strong evidence for a more modular view of SAGA than previously held, with many loci occupied by the hGCN5/KAT2A acetyltransferase module of SAGA independently of the USP22 subunit. The DUB module subunit ATXN7L3 is present at a greater number of hGCN5‐bound loci than is USP22, likely reflecting the role ATXN7L3 plays as an essential partner of SAGA‐independent DUBs USP27X and USP51 (Atanassov et al, 2016). Additionally, the discovery that USP22 controls the ubiquitylation status of multiple Mediator/PIC subunits suggests a potential role for SAGA in transcriptional regulation, though additional experiments are necessary to further elucidate the mechanism behind these interactions. The apparently metazoan‐specific activity of SAGA on Mediator function reported here is consistent with observations that human Mediator itself has a set of functions and even individual subunits that do not appear in yeast (Sato et al, 2004; Ishikawa et al, 2006; Bourbon, 2008; Malovannaya et al, 2011; Huang et al, 2012). Numerous studies have shown that USP22 activity is essential across many forms of cancer (Yang et al, 2018). In light of successful efforts to therapeutically target similar epigenetic regulators, an increased understanding of USP22’s mechanism of action may have near‐term clinical implications.
Materials and Methods
ChIP and ChIP‐seq
ChIP‐seq was performed as described previously (Schmidt et al, 2009) with minor modifications. Antibodies were conjugated to either Dynabeads Protein A (rabbit) or Dynabeads Protein G (mouse) (Thermo Fisher). Cells were fixed in 1% formaldehyde for 10 min at room temperature, followed by quenching with 0.18 M glycine for 5 min. Cells were harvested and chromatin was extracted as described. Chromatin was sheared to 100–300 bp fragments using the Q800R2 Sonicator with circulating chiller (QSonica), for 25 min at 50% amplitude, 20 s on/20 s off. Sheared chromatin was incubated with antibodies overnight at 4°C against indicated proteins. Wash steps were performed as described with the exception of ChIP samples for USP22, pSer5 Pol II BioLegend #: 920304), and pSer2 Pol II (BioLegend #: 920204), which were washed in buffers containing 0.025% SDS instead of 0.1% SDS. pSer5 and pSer2 Pol antibodies were bound to beads as previously described (Morris et al, 2005). Immunoprecipitated chromatin underwent elution, reverse cross‐linking, RNAse A treatment, proteinase K digestion, and phenol:chloroform extraction. For ChIP‐seq, 10 ng of DNA from inputs and IPs was used to prepare libraries using Accel‐NGS 2S Plus DNA Library Kit (Swift Biosciences, MI) following the manufacturer’s protocol with an additional wash at the end using SPRIselect beads (Beckman Coulter, IN) to remove any larger products that could interfere with clustering on the instrument. Final libraries at a concentration of 1 nM were sequenced on NextSeq 500 using 75 bp single‐end chemistry. Each IP condition was performed in biological replicate. For ChIP and ChIP‐seq, antibodies used were USP22 (Novus Biologicals #: NBP1‐49644), GCN5 (Santa Cruz #: sc‐20698), ATXN7L3 (Bethyl Laboratories #: A302‐800A), Pol II (Santa Cruz #: sc‐899), Ub‐H2B (Millipore #: 05‐1312), MED1 (Bethyl Laboratories #: A300‐793A), MED16 (Abcam #: ab130996), TBP (Abcam #: ab818), TFIIB (Santa Cruz #: sc‐225)), TFIIF (Abcam #: ab28179), TAF7 (Santa Cruz #: sc‐101167), Ub‐H2B (Millipore #: 05‐1312), ATF4 (Cell Signaling #: 11815), and XBP1s (Cell Signaling #: 12782). For ChIP‐qPCR, primer sequences are available in Appendix Table S1.
ChIP‐seq analysis
ChIP‐seq data were aligned using bowtie (Langmead et al, 2009) against the hg19 version of the human genome, and HOMER (Heinz et al, 2010) was used to generate bigwig files and call significant peaks vs input of samples using ‐style factor option (‐style histone option for ubH2B) with duplicate reads ignored. Peaks that passed FDR< 5% threshold were considered significant. ChIP‐seq signals for any regions were derived from bigwig files using bigWigAverageOverBed tool from UCSC toolbox (Kent et al, 2010) with mean0 option and normalized within each binding factor using upper quartile normalization to account for ChIP pulldown efficiency differences between experiments. Fold differences between samples were calculated with average input signal 0.35 used as a floor for the minimum allowed signal. Ensemble GRCh37.p7 transcriptome information was used to identify the main gene transcript and its transcription start site (TSS) as a transcript with a significant Pol2 peak and the best Pol2 signal vs input within 500 bp from the start site. 16,932 genes with identified main transcripts were considered. Regions with 500 bp around TSS, 500 bp around 3 kb downstream from TSS, and gene body (TSS to end) were used to calculate normalized enrichment. Estimation of significance of a region’s differential binding between groups pairs was done using DESeq2 (Love et al, 2014) and FDR< 5% results were considered significant. ChIP‐seq profiles showing distribution of signal 3 kb around TSS, upstream from TSS, and downstream from gene end were done using 50 bp bins. Distribution of the ChIP‐seq signal across the gene body was done using 2% gene length bins. Average signal for multiple genes was calculated using geometric mean.
H3K4me3 HiChIP and HiChIP analysis
HiChIP was performed as described previously (Mumbach et al, 2016). Cells were fixed in 1% formaldehyde for 10 min at room temperature, followed by quenching with 0.125 M glycine for 5 min. Cells were harvested and cross‐linked chromatin was extracted, MboI‐digested, biotin‐labeled, and ligated as described. Chromatin was sheared to 100–300 bp fragments using the Q800R2 Sonicator with circulating chiller (QSonica), for 25 min at 50% amplitude, 20 s on/20 s off. Sheared chromatin was incubated with anti‐H3K4me3 (Sigma #: 07‐473) for 2 h at 4°C against indicated proteins. Wash, elution, reverse cross‐linking, biotin purification, and Tn5 tagmentation steps were performed as described. Libraries were constructed using Nextera XT Index Kit (Illumina) and size selected using AMPure XP beads (Beckman Coulter) as described.
Raw HiChIP data were aligned using bowtie2 (v2.2.4) algorithm (Li & Durbin, 2009) against hg19 and processed using HiC‐Pro (v2.11.3) software (Servant et al, 2015) with default parameters. Loops across all samples were identified based on HiC‐Pro output using hichipper (v0.7.7) R package (Lareau & Aryee, 2018), and significantly identified loops that passed FDR< 1% threshold were taken for further analysis. Paired comparisons between conditions were performed using diffloop (v1.16.0) R package (Lareau & Aryee, 2018), and results that passed FDR< 5% were considered significant.
Cell culture, viral infection, transfection, and treatment
The human cell line HCT116 was obtained from American Type Culture Collection (ATCC). The ectopic USP22‐inducible HCT116 tetracycline hydrochloride (Tet)‐operable cell line was generated by cloning pcDNA3.1 FLAG‐USP22 into the pLenti6.3/V5/TO‐DEST vector using the Virapower™ T‐Rex™ system (Thermo Fisher). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Mediatech) supplemented with 10% fetal calf serum (FBS, Gemini Bio‐Products) and 2.5 μg/ml Plasmocin (Invivogen) at 37°C in 5% CO2.
As indicated, cells were infected with lentiviral shRNA plasmids corresponding to USP22 (XM_042698.6‐914s1c1, NM_015276.1‐545s21c1, NM_015276.1‐1366s21c1, XM_042698.6‐1921s1c1), ATXN7L3 (NM_001098833.1‐1008s21c1), GCN5 (NM_021078.1‐2484s1c1), PCAF (NM_003884.3‐987s1c1), and control luciferase shRNA (SHC007) that were obtained from the TRC collection (Sigma‐Aldrich) as previously published. Cells were selected with 5 μg/ml puromycin (Sigma‐Aldrich) 24 h after infection.
For indicated experiments, cells were treated with 100 nM Thapsigargin (Sigma‐Aldrich), 10 μM MG132 (SelleckChem), 8 ng/ml tetracycline hydrochloride (Sigma‐Aldrich), 50 μM PR619 (Life Sensors), and 5 μM o‐phenanthroline (LifeSensors).
Apoptosis assays
Cells were treated with 100 nM Thapsigargin for the indicated time points. For Cas3/7 visualization, cells were plated in triplicate in 6‐well plates along with cleaved caspase 3/7 dye and images were captured every 4 h on the Incucyte Live Cell Analysis System (Sartorius). For flow cytometry, cells were harvested and labeled with Annexin V and propidium iodide. Fluorescence was measured on the BD FACSCalibur (BD Biosciences).
Immunoblotting, cellular fractionation, and mRNA analysis
Cells were harvested and lysed in a Nonidet P‐40‐based whole‐cell lysis buffer supplemented with protease inhibitor cocktail (Sigma‐Aldrich). Lysate concentration was determined using the bicinchoninic acid (BCA) assay and analyzed by SDS–PAGE using antibodies against USP22 (Novus Biologicals #: NBP1‐49644), PARP (Cell Signaling #: 9532), caspase 3 (Cell Signaling #: 9662), actin (Santa Cruz #: sc‐8432), GAPDH (Cell Signaling #: 2118), CHOP (Cell Signaling #: 2895), GRP78 (Cell Signaling #: 3177), Ub‐H2B (Millipore #: 05‐1312), histone H2B (Santa Cruz #: sc‐10108), PERK (Cell Signaling # 5683), TBP (Abcam #: ab818), TFIIF (Abcam #: ab28179), Pol II (Santa Cruz #: sc‐9001), MED24 (Bethyl Laboratories #: A301‐472A), MED16 (Abcam #: ab130996), ORC2 (BD Pharmingen #: 559266) and β‐tubulin (Sigma‐Aldrich #: T4026), ubiquitin (Santa Cruz #: sc‐9133), ATF4 (Cell Signaling #: 11815), ATF6 (Abcam #: ab11909), Xbp1s (Cell Signaling #: 12782), GCN5 (Santa Cruz #: sc‐20698), ATXN7L3 (Bethyl Laboratories #: A302‐800A), MED16 (Abcam #: ab130996), and pan‐14‐3‐3 (Santa Cruz #: sc‐1657). Cellular fractionations were performed as described previously (Carey et al, 2009).
Total RNA was extracted using TRIzolTM (Thermo Fisher) and reverse‐transcribed using the High‐Capacity cDNA Reverse Transcription Kit (Thermo Fisher) according to the manufacturer’s instructions. qRT–PCR was performed using Fast SYBRTM Green (Thermo Fisher) as described (Zhang et al, 2005). mRNA levels between samples were normalized to 18S transcript levels. Primer sequences are available in Appendix Table S1.
UbiScan®
Samples were analyzed using the PTMScan® method as previously described (Rush et al, 2005; Stokes et al, 2012; Stokes et al, 2015). Briefly, HCT116 cells were infected with shRNA against either control (shLuc) or USP22 (shUSP22). Cellular extracts were prepared in 8 M urea buffer, sonicated, centrifuged, reduced with DTT, and alkylated with iodoacetamide. Total protein (15mg) for each sample was digested with trypsin and purified over C18 columns for enrichment with the Ubiquitin K‐GG Remnant Motif antibody (#5562). Enriched peptides were purified over C18 STAGE tips(Rappsilber et al, 2003) and analyzed by mass spectrometry as described previously(Gennaro et al, 2018). A 2.5‐fold cutoff was used to denote changes between samples and analytical % CV values were calculated for each peptide to determine reproducibility across runs.
UbiTest®
HCT116 cells were harvested after 6 days of knockdown and 4 h after proteasome inhibition with MG132. Cells were lysed in RIPA buffer supplemented with protease cocktail inhibitor, PR619, and o‐phenanthroline. Protein lysate was incubated with α‐Ub TUBE1 agarose resin, eluted, and digested with global DUB USP2 for 2 h to remove polyubiquitylation, as recommended by the manufacturer (LifeSensors). Undigested and digested samples were analyzed by SDS–PAGE and immunoblotting.
For the in vitro USP22 digestion reactions, protein lysate was incubated with α‐Ub TUBE1 agarose resin, washed, and eluted. Elutions were then incubated with either buffer, recombinant USP2 (a non‐specific DUB), or hDUB module recombinantly expressed from baculoviral constructs and purified from Sf9 cells. Undigested and digested samples were analyzed by SDS–PAGE and immunoblotting.
Statistical analysis
Data collected from at least three independent experiments are presented as mean ± SEM. For ChIP‐seq profiles, statistical analysis was performed using non‐parametric Mann–Whitney tests between values from all replicates of one comparison group versus another; significance is explicitly indicated. Linear regression analysis was performed in Microsoft Excel; R 2 values and significance are explicitly indicated. For the apoptosis time course, two‐way ANOVA followed by Tukey’s HSD as post hoc assessment of pairwise comparisons was performed in Microsoft Excel; significant pairwise comparisons are indicated with *. For comparison between two groups, statistical testing was performed using SPSS with differences between two groups determined by Student’s t‐test; significance is denoted as *P < 0.05; **P < 0.01; and ***P < 0.005.
Author contributions
TJS, SMS, and SBM designed research. TJS, VJG, MAT, DD, KLP, SB, and FW performed experiments. TJS, DD, AVK, and CM analyzed data. KEK and TB contributed new reagents and analytic tools. TJS and SBM wrote the manuscript. All authors discussed the results and commented on the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Review Process File
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Dataset EV1
Source Data for Expanded View and Appendix
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 5
Source Data for Figure 6
Acknowledgements
These studies were supported in part via NIH grants R01CA182569 (to KEK and SBM) and F31CA165475 (to TJS). We gratefully acknowledge B.F. Pugh (Penn State University) for providing advice and Jonathan Ruffin (IQVIA) for providing bioinformatics analysis. FW and TB are employed by Progenra Inc., a biotech company engaged in targeting DUBs for therapeutic benefit. The National Cancer Institute‐supported SKCC Flow Cytometry and Cancer Genomics facilities and the Wistar Bioinformatics facility were used in these studies.
The EMBO Journal (2021) 40: e102509.
Data availability
The datasets produced in this study are available in the following databases:
ChIP‐seq data: Gene Expression Omnibus GSE121798 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE121798)
HiChIP data: Gene Expression Omnibus GSE158108 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE158108)
References
- Ansari SA, Ganapathi M, Benschop JJ, Holstege FC, Wade JT, Morse RH (2012) Distinct role of mediator tail module in regulation of SAGA‐dependent, TATA‐containing genes in yeast. EMBO J 31: 44–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanassov BS, Dent SY (2011) USP22 regulates cell proliferation by deubiquitinating the transcriptional regulator FBP1. EMBO Rep 12: 924–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanassov BS, Evrard YA, Multani AS, Zhang Z, Tora L, Devys D, Chang S, Dent SY (2009) Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance. Mol Cell 35: 352–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanassov BS, Mohan RD, Lan X, Kuang X, Lu Y, Lin K, McIvor E, Li W, Zhang Y, Florens Let al (2016) ATXN7L3 and ENY2 coordinate activity of multiple H2B deubiquitinases important for cellular proliferation and tumor growth. Mol Cell 62: 558–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista T, Grunberg S, Minoungou N, Koster MJE, Timmers HTM, Hahn S, Devys D, Tora L (2017) SAGA is a general cofactor for RNA polymerase II transcription. Mol Cell 68: 130–143.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basehoar AD, Zanton SJ, Pugh BF (2004) Identification and distinct regulation of yeast TATA box‐containing genes. Cell 116: 699–709 [DOI] [PubMed] [Google Scholar]
- Ben‐Shem A, Papai G, Schultz P (2020) Architecture of the multi‐functional SAGA complex and the molecular mechanism of holding TBP. FEBS J 288: 3135–3147 [DOI] [PubMed] [Google Scholar]
- Bergmann TJ, Fregno I, Fumagalli F, Rinaldi A, Bertoni F, Boersema PJ, Picotti P, Molinari M (2018) Chemical stresses fail to mimic the unfolded protein response resulting from luminal load with unfolded polypeptides. J Biol Chem 293: 5600–5612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann TJ, Molinari M (2018) Three branches to rule them all? UPR signalling in response to chemically versus misfolded proteins‐induced ER stress. Biol Cell 110: 197–204 [DOI] [PubMed] [Google Scholar]
- Bieniossek C, Papai G, Schaffitzel C, Garzoni F, Chaillet M, Scheer E, Papadopoulos P, Tora L, Schultz P, Berger I (2013) The architecture of human general transcription factor TFIID core complex. Nature 493: 699–702 [DOI] [PubMed] [Google Scholar]
- Bonnet J, Wang CY, Baptista T, Vincent SD, Hsiao WC, Stierle M, Kao CF, Tora L, Devys D (2014) The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes Dev 28: 1999–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbon HM (2008) Comparative genomics supports a deep evolutionary origin for the large, four‐module transcriptional mediator complex. Nucleic Acids Res 36: 3993–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand M, Yamamoto K, Staub A, Tora L (1999) Identification of TATA‐binding protein‐free TAFII‐containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J Biol Chem 274: 18285–18289 [DOI] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD (1996) Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84: 843–851 [DOI] [PubMed] [Google Scholar]
- Brzovic PS, Heikaus CC, Kisselev L, Vernon R, Herbig E, Pacheco D, Warfield L, Littlefield P, Baker D, Klevit REet al (2011) The acidic transcription activator Gcn4 binds the mediator subunit Gal11/Med15 using a simple protein interface forming a fuzzy complex. Mol Cell 44: 942–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey MF, Peterson CL, Smale ST (2009) Dignam and roeder nuclear extract preparation. Cold Spring Harb Protoc 2009: pdb.prot5330 [DOI] [PubMed] [Google Scholar]
- Chandy M, Gutierrez JL, Prochasson P, Workman JL (2006) SWI/SNF displaces SAGA‐acetylated nucleosomes. Eukaryot Cell 5: 1738–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Iliopoulos D, Zhang Q, Tang Q, Greenblatt MB, Hatziapostolou M, Lim E, Tam WL, Ni M, Chen Yet al (2014) XBP1 promotes triple‐negative breast cancer by controlling the HIF1alpha pathway. Nature 508: 103–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XF, Lehmann L, Lin JJ, Vashisht A, Schmidt R, Ferrari R, Huang C, McKee R, Mosley A, Plath Ket al (2012) Mediator and SAGA have distinct roles in Pol II preinitiation complex assembly and function. Cell Rep 2: 1061–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon Y, Kim H, Park K, Kim M, Lee D (2020) Dynamic modules of the coactivator SAGA in eukaryotic transcription. Exp Mol Med 52: 991–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipumuro E, Henriksen MA (2012) The ubiquitin hydrolase USP22 contributes to 3'‐end processing of JAK‐STAT‐inducible genes. FASEB J 26: 842–854 [DOI] [PubMed] [Google Scholar]
- Cornelio‐Parra DV, Goswami R, Costanzo K, Morales‐Sosa P, Mohan RD (2021) Function and regulation of the Spt‐Ada‐Gcn5‐Acetyltransferase (SAGA) deubiquitinase module. Biochim Biophys Acta 1864: 194630 [DOI] [PubMed] [Google Scholar]
- Davis JA, Takagi Y, Kornberg RD, Asturias FA (2002) Structure of the yeast RNA polymerase II holoenzyme: mediator conformation and polymerase interaction. Mol Cell 10: 409–415 [DOI] [PubMed] [Google Scholar]
- Dombroski BA, Nayak RR, Ewens KG, Ankener W, Cheung VG, Spielman RS (2010) Gene expression and genetic variation in response to endoplasmic reticulum stress in human cells. Am J Hum Genet 86: 719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donczew R, Warfield L, Pacheco D, Erijman A, Hahn S (2020) Two roles for the yeast transcription coactivator SAGA and a set of genes redundantly regulated by TFIID and SAGA. Elife 9: e50109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duttke SH (2015) Evolution and diversification of the basal transcription machinery. Trends Biochem Sci 40: 127–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A, Sterner DE, Schieltz D, Hassan A, Yates JR 3rd, Berger SL, Workman JL (1999) The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae . Mol Cell Biol 19: 6621–6631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Khattabi L, Zhao H, Kalchschmidt J, Young N, Jung S, Van Blerkom P, Kieffer‐Kwon P, Kieffer‐Kwon KR, Park S, Wang Xet al (2019) A pliable mediator acts as a functional rather than an architectural bridge between promoters and enhancers. Cell 178: 1145–1158.e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya Y, Lundmo P, Short AD, Gill DL, Isaacs JT (1994) The role of calcium, pH, and cell proliferation in the programmed (apoptotic) death of androgen‐independent prostatic cancer cells induced by thapsigargin. Can Res 54: 6167–6175 [PubMed] [Google Scholar]
- Gao Y, Lin F, Xu P, Nie J, Chen Z, Su J, Tang J, Wu Q, Li Y, Guo Zet al (2014) USP22 is a positive regulator of NFATc2 on promoting IL2 expression. FEBS Lett 588: 878–883 [DOI] [PubMed] [Google Scholar]
- Gates LA, Shi J, Rohira AD, Feng Q, Zhu B, Bedford MT, Sagum CA, Jung SY, Qin J, Tsai MJet al (2017) Acetylation on histone H3 lysine 9 mediates a switch from transcription initiation to elongation. J Biol Chem 292: 14456–14472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennaro VJ, Stanek TJ, Peck AR, Sun Y, Wang F, Qie S, Knudsen KE, Rui H, Butt T, Diehl JAet al (2018) Control of CCND1 ubiquitylation by the catalytic SAGA subunit USP22 is essential for cell cycle progression through G1 in cancer cells. Proc Natl Acad Sci USA 115: E9298–E9307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgakopoulos T, Thireos G (1992) Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J 11: 4145–4152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind CK, Zhang F, Qiu H, Hofmeyer K, Hinnebusch AG (2007) Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol Cell 25: 31–42 [DOI] [PubMed] [Google Scholar]
- Gowen BG, Chim B, Marceau CD, Greene TT, Burr P, Gonzalez JR, Hesser CR, Dietzen PA, Russell T, Iannello Aet al (2015) A forward genetic screen reveals novel independent regulators of ULBP1, an activating ligand for natural killer cells. Elife 4: e08474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen‐Hughes T, Allis CD, Winston Fet al (1997) Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev 11: 1640–1650 [DOI] [PubMed] [Google Scholar]
- Grant PA, Schieltz D, Pray‐Grant MG, Steger DJ, Reese JC, Yates JR 3rd, Workman JL (1998) A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94: 45–53 [DOI] [PubMed] [Google Scholar]
- Grant PA, Winston F, Berger SL (2021) The biochemical and genetic discovery of the SAGA complex. Biochim Biophys Acta 1864: 194669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D (1999) Protein translation and folding are coupled by an endoplasmic‐reticulum‐resident kinase. Nature 397: 271–274 [DOI] [PubMed] [Google Scholar]
- Haze K, Yoshida H, Yanagi H, Yura T, Mori K (1999) Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell 10: 3787–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK (2010) Simple combinations of lineage‐determining transcription factors prime cis‐regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmlinger D, Papai G, Devys D, Tora L (2021) What do the structures of GCN5‐containing complexes teach us about their function? Biochim Biophys Acta 1864: 194614 [DOI] [PubMed] [Google Scholar]
- Helmlinger D, Tora L (2017) Sharing the SAGA. Trends Biochem Sci 42: 850–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsley PA, Hurst CH, Kaliyadasa E, Lamb R, Knight MR, De Cothi EA, Steele JF, Knight H (2014) The Arabidopsis mediator complex subunits MED16, MED14, and MED2 regulate mediator and RNA polymerase II recruitment to CBF‐responsive cold‐regulated genes. Plant Cell 26: 465–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL (2003) Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA‐associated Ubp8. Genes Dev 17: 2648–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjerpe R, Aillet F, Lopitz‐Otsoa F, Lang V, England P, Rodriguez MS (2009) Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin‐binding entities. EMBO Rep 10: 1250–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CH, Chen YJ, Yang CN (2019) Loss of function in SAGA deubiquitinating module caused by Sgf73 H93A mutation: a molecular dynamics study. J Mol Graph Model 91: 112–118 [DOI] [PubMed] [Google Scholar]
- Huang Y, Li W, Yao X, Lin QJ, Yin JW, Liang Y, Heiner M, Tian B, Hui J, Wang G (2012) Mediator complex regulates alternative mRNA processing via the MED23 subunit. Mol Cell 45: 459–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga KL, Pugh BF (2004) A genome‐wide housekeeping role for TFIID and a highly regulated stress‐related role for SAGA in Saccharomyces cerevisiae . Mol Cell 13: 573–585 [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Tachikawa H, Miura Y, Takahashi N (2006) TRIM11 binds to and destabilizes a key component of the activator‐mediated cofactor complex (ARC105) through the ubiquitin‐proteasome system. FEBS Lett 580: 4784–4792 [DOI] [PubMed] [Google Scholar]
- Jeronimo C, Langelier MF, Bataille AR, Pascal JM, Pugh BF, Robert F (2016) Tail and kinase modules differently regulate core mediator recruitment and function in vivo . Mol Cell 64: 455–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SSet al (2010) Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D (2010) BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics 26: 2204–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJet al (2011) Systematic and quantitative assessment of the ubiquitin‐modified proteome. Mol Cell 44: 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G, Bonnet J, Umlauf D, Karmodiya K, Koffler J, Stierle M, Devys D, Tora L (2011) The tightly controlled deubiquitination activity of the human SAGA complex differentially modifies distinct gene regulatory elements. Mol Cell Biol 31: 3734–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory‐efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau CA, Aryee MJ (2018) hichipper: a preprocessing pipeline for calling DNA loops from HiChIP data. Nat Methods 15: 155–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Causton HC, Holstege FC, Shen WC, Hannett N, Jennings EG, Winston F, Green MR, Young RA (2000) Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405: 701–704 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows‐Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Seidel CW, Szerszen LT, Lange JJ, Workman JL, Abmayr SM (2017) Enzymatic modules of the SAGA chromatin‐modifying complex play distinct roles in Drosophila gene expression and development. Genes Dev 31: 1588–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol 15: 550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytton J, Westlin M, Hanley MR (1991) Thapsigargin inhibits the sarcoplasmic or endoplasmic reticulum Ca‐ATPase family of calcium pumps. The Journal of biological chemistry 266: 17067–17071 [PubMed] [Google Scholar]
- Malik S, Roeder RG (2010) The metazoan Mediator co‐activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet 11: 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malovannaya A, Lanz RB, Jung SY, Bulynko Y, Le NT, Chan DW, Ding C, Shi Y, Yucer N, Krenciute Get al (2011) Analysis of the human endogenous coregulator complexome. Cell 145: 787–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, Chait BT, Roeder RG (2001) Human STAGA complex is a chromatin‐acetylating transcription coactivator that interacts with pre‐mRNA splicing and DNA damage‐binding factors in vivo . Mol Cell Biol 21: 6782–6795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD (1998) The novel ATM‐related protein TRRAP is an essential cofactor for the c‐Myc and E2F oncoproteins. Cell 94: 363–374 [DOI] [PubMed] [Google Scholar]
- Mohan RD, Dialynas G, Weake VM, Liu J, Martin‐Brown S, Florens L, Washburn MP, Workman JL, Abmayr SM (2014) Loss of Drosophila Ataxin‐7, a SAGA subunit, reduces H2B ubiquitination and leads to neural and retinal degeneration. Genes Dev 28: 259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DP, Michelotti GA, Schwinn DA (2005) Evidence that phosphorylation of the RNA polymerase II carboxyl‐terminal repeats is similar in yeast and humans. The Journal of biological chemistry 280: 31368–31377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumbach MR, Rubin AJ, Flynn RA, Dai C, Khavari PA, Greenleaf WJ, Chang HY (2016) HiChIP: efficient and sensitive analysis of protein‐directed genome architecture. Nat Methods 13: 919–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Z, Riss A, Romier C, le Guezennec X, Dongre AR, Orpinell M, Han J, Stunnenberg H, Tora L (2009) The human SPT20‐containing SAGA complex plays a direct role in the regulation of endoplasmic reticulum stress‐induced genes. Mol Cell Biol 29: 1649–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa K, Schneider TR, Cramer P (2017) Core Mediator structure at 3.4 A extends model of transcription initiation complex. Nature 545: 248–251 [DOI] [PubMed] [Google Scholar]
- Papai G, Frechard A, Kolesnikova O, Crucifix C, Schultz P, Ben‐Shem A (2020) Structure of SAGA and mechanism of TBP deposition on gene promoters. Nature 577: 711–716 [DOI] [PubMed] [Google Scholar]
- Park JM, Kim HS, Han SJ, Hwang MS, Lee YC, Kim YJ (2000) In vivo requirement of activator‐specific binding targets of mediator. Mol Cell Biol 20: 8709–8719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SW, Li G, Lin YP, Barrero MJ, Ge K, Roeder RG, Wei LN (2005) Thyroid hormone‐induced juxtaposition of regulatory elements/factors and chromatin remodeling of Crabp1 dependent on MED1/TRAP220. Mol Cell 19: 643–653 [DOI] [PubMed] [Google Scholar]
- Paul E, Zhu ZI, Landsman D, Morse RH (2015) Genome‐wide association of mediator and RNA polymerase II in wild‐type and mediator mutant yeast. Mol Cell Biol 35: 331–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrenko N, Jin Y, Wong KH, Struhl K (2016) Mediator undergoes a compositional change during transcriptional activation. Mol Cell 64: 443–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrenko N, Jin Y, Dong L, Wong KH, Struhl K (2019) Requirements for RNA polymerase II preinitiation complex formation in vivo . ELife 8: e43654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips‐Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Yet al (2013) Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153: 1281–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ishihama Y, Mann M (2003) Stop and go extraction tips for matrix‐assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 75: 663–670 [DOI] [PubMed] [Google Scholar]
- Roberts SM, Winston F (1997) Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 147: 451–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PJ, Trnka MJ, Bushnell DA, Davis RE, Mattei PJ, Burlingame AL, Kornberg RD (2016) Structure of a complete mediator‐RNA polymerase II pre‐initiation complex. Cell 166: 1411–1422.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ (2005) Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol 23: 94–101 [DOI] [PubMed] [Google Scholar]
- Sato S, Tomomori‐Sato C, Parmely TJ, Florens L, Zybailov B, Swanson SK, Banks CA, Jin J, Cai Y, Washburn MPet al (2004) A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol Cell 14: 685–691 [DOI] [PubMed] [Google Scholar]
- Schilbach S, Hantsche M, Tegunov D, Dienemann C, Wigge C, Urlaub H, Cramer P (2017) Structures of transcription pre‐initiation complex with TFIIH and mediator. Nature 551: 204–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, Odom DT (2009) ChIP‐seq: using high‐throughput sequencing to discover protein‐DNA interactions. Methods 48: 240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant N, Varoquaux N, Lajoie BR, Viara E, Chen CJ, Vert JP, Heard E, Dekker J, Barillot E (2015) HiC‐Pro: an optimized and flexible pipeline for Hi‐C data processing. Genome Biol 16: 259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov G, Voltz K, Durand A, Kolesnikova O, Papai G, Myasnikov AG, Dejaegere A, Ben Shem A, Schultz P (2017) Structure of the transcription activator target Tra1 within the chromatin modifying complex SAGA. Nat Commun 8: 1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Walter P (1997) The transmembrane kinase Ire1p is a site‐specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 90: 1031–1039 [DOI] [PubMed] [Google Scholar]
- Soffers JHM, Li X, Saraf A, Seidel CW, Florens L, Washburn MP, Abmayr SM, Workman JL (2019) Characterization of a metazoan ADA acetyltransferase complex. Nucleic Acids Res 47: 3383–3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soffers JHM, Workman JL (2020) The SAGA chromatin‐modifying complex: the sum of its parts is greater than the whole. Genes Dev 34: 1287–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens JL, Cantin GT, Wang G, Shevchenko A, Berk AJ (2002) Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science 296: 755–758 [DOI] [PubMed] [Google Scholar]
- Stokes MP, Farnsworth CL, Moritz A, Silva JC, Jia X, Lee KA, Guo A, Polakiewicz RD, Comb MJ (2012) PTMScan direct: identification and quantification of peptides from critical signaling proteins by immunoaffinity enrichment coupled with LC‐MS/MS. Mol Cell Proteomics 11: 187–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes MP, Farnsworth CL, Gu H, Jia X, Worsfold CR, Yang V, Ren JM, Lee KA, Silva JC (2015) Complementary PTM profiling of drug response in human gastric carcinoma by immunoaffinity and IMAC methods with total proteome analysis. Proteomes 3: 160–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny JC, Erdjument‐Bromage H, Tempst P, Allis CD (2007) Ubiquitylation of histone H2B controls RNA polymerase II transcription elongation independently of histone H3 methylation. Genes Dev 21: 835–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur JK, Arthanari H, Yang F, Chau KH, Wagner G, Naar AM (2009) Mediator subunit Gal11p/MED15 is required for fatty acid‐dependent gene activation by yeast transcription factor Oaf1p. J Biol Chem 284: 4422–4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres‐Zelada EF, Stephenson RE, Alpsoy A, Anderson BD, Swanson SK, Florens L, Dykhuizen EC, Washburn MP, Weake VM (2019) The Drosophila Dbf4 ortholog Chiffon forms a complex with Gcn5 that is necessary for histone acetylation and viability. J Cell Sci 132: jcs214072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai KL, Tomomori‐Sato C, Sato S, Conaway RC, Conaway JW, Asturias FJ (2014) Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell 157: 1430–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai KL, Yu X, Gopalan S, Chao TC, Zhang Y, Florens L, Washburn MP, Murakami K, Conaway RC, Conaway JWet al (2017) Mediator structure and rearrangements required for holoenzyme formation. Nature 544: 196–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Dienemann C, Stutzer A, Urlaub H, Cheung ACM, Cramer P (2020) Structure of the transcription coactivator SAGA. Nature 577: 717–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Dent SY (2014) Functions of SAGA in development and disease. Epigenomics 6: 329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Carroll JS, Brown M (2005) Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell 19: 631–642 [DOI] [PubMed] [Google Scholar]
- Wang YL, Faiola F, Xu M, Pan S, Martinez E (2008) Human ATAC is a GCN5/PCAF‐containing acetylase complex with a novel NC2‐like histone fold module that interacts with the TATA‐binding protein. J Biol Chem 283: 33808–33815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfield L, Ramachandran S, Baptista T, Devys D, Tora L, Hahn S (2017) Transcription of nearly all yeast RNA polymerase II‐transcribed genes is dependent on transcription factor TFIID. Mol Cell 68: 118–129.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Dyer JO, Seidel C, Box A, Swanson SK, Peak A, Florens L, Washburn MP, Abmayr SM, Workman JL (2011) Post‐transcription initiation function of the ubiquitous SAGA complex in tissue‐specific gene activation. Genes Dev 25: 1499–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinzierl RO, Dynlacht BD, Tjian R (1993) Largest subunit of Drosophila transcription factor IID directs assembly of a complex containing TBP and a coactivator. Nature 362: 511–517 [DOI] [PubMed] [Google Scholar]
- Wu PY, Ruhlmann C, Winston F, Schultz P (2004) Molecular architecture of the S. cerevisiae SAGA complex. Mol Cell 15: 199–208 [DOI] [PubMed] [Google Scholar]
- Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, Eick D, Hughes TR, Krogan NJ, Strahl BD, Berger SL (2007) H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA‐related complex. Mol Cell 27: 275–288 [DOI] [PubMed] [Google Scholar]
- Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD (2005) Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol Cell Biol 25: 637–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, Harada A, Mori K (2007) Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell 13: 365–376 [DOI] [PubMed] [Google Scholar]
- Yan M, Wolberger C (2015) Uncovering the role of Sgf73 in maintaining SAGA deubiquitinating module structure and activity. J Mol Biol 427: 1765–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Zang H, Luo Y, Wu J, Fang Z, Zhu W, Li Y (2018) High expression of USP22 predicts poor prognosis and advanced clinicopathological features in solid tumors: a meta‐analysis. OncoTargets Ther 11: 3035–3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Sumibcay L, Hinnebusch AG, Swanson MJ (2004) A triad of subunits from the Gal11/tail domain of Srb mediator is an in vivo target of transcriptional activator Gcn4p. Mol Cell Biol 24: 6871–6886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XY, DeSalle LM, Patel JH, Capobianco AJ, Yu D, Thomas‐Tikhonenko A, McMahon SB (2005) Metastasis‐associated protein 1 (MTA1) is an essential downstream effector of the c‐MYC oncoprotein. Proc Natl Acad Sci USA 102: 13968–13973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XY, Pfeiffer HK, Thorne AW, McMahon SB (2008a) USP22, an hSAGA subunit and potential cancer stem cell marker, reverses the polycomb‐catalyzed ubiquitylation of histone H2A. Cell Cycle 7: 1522–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB (2008b) The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell‐cycle progression. Mol Cell 29: 102–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Young N, Kalchschmidt J, Lieberman J, El Khattabi L, Casellas R, Asturias FJ (2021) Structure of mammalian mediator complex reveals Tail module architecture and interaction with a conserved core. Nat Commun 12: 1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov Aet al (2008) A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell 29: 92–101 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Review Process File
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Dataset EV1
Source Data for Expanded View and Appendix
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 5
Source Data for Figure 6
Data Availability Statement
The datasets produced in this study are available in the following databases:
ChIP‐seq data: Gene Expression Omnibus GSE121798 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE121798)
HiChIP data: Gene Expression Omnibus GSE158108 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE158108)


