Abstract
Background
Liver fibrosis risk is a heritable trait, the outcome of which is the net deposition of extracellular matrix by hepatic stellate cell-derived myofibroblasts. Whereas nucleotide sequence variations have been extensively studied in liver fibrosis, the role of copy number variations (CNV) in which genes exist in abnormal numbers of copies (mostly due to duplication or deletion) has had limited exploration.
Methods
The impact of the XPO4 CNV on histological liver damage was examined in a cohort comprised 646 Caucasian patients with biopsy-proven MAFLD and 170 healthy controls. XPO4 expression was modulated and function was examined in human and animal models.
Findings
Here we demonstrate in a cohort of 816 subjects, 646 with biopsy-proven metabolic associated liver disease (MAFLD) and 170 controls, that duplication in the exportin 4 (XPO4) CNV is associated with the severity of liver fibrosis. Functionally, this occurs via reduced expression of hepatic XPO4 that maintains sustained activation of SMAD3/SMAD4 and promotes TGF-β1-mediated HSC activation and fibrosis. This effect was mediated through termination of nuclear SMAD3 signalling. XPO4 demonstrated preferential binding to SMAD3 compared to other SMADs and led to reduced SMAD3-mediated responses as shown by attenuation of TGFβ1 induced SMAD transcriptional activity, reductions in the recruitment of SMAD3 to target gene promoters following TGF-β1, as well as attenuation of SMAD3 phosphorylation and disturbed SMAD3/SMAD4 complex formation.
Interpretation
We conclude that a CNV in XPO4 is a critical mediator of fibrosis severity and can be exploited as a therapeutic target for liver fibrosis.
Funding
ME and JG are supported by the Robert W. Storr Bequest to the Sydney Medical Foundation, University of Sydney; a National Health and Medical Research Council of Australia (NHMRC) Program Grant (APP1053206) and Project and ideas grants (APP2001692, APP1107178 and APP1108422). AB is supported by an Australian Government Research Training Program (RTP) scholarship. EB is supported by Horizon 2020 under grant 634413 for the project EPoS.
Keywords: MAFLD, XPO4, Fibrosis, TGFβ
Research in context.
Evidence before this study
Fibrosis is a highly heritable and polygenic trait; if persistent, it leads to tissue scarring, organ failure, and eventually, death. The role CNVs, a more recently appreciated class of large-scale germline variants on gene function and liver fibrosis remains under-characterized.
Added value of this study
We examined the relationship between CNV in exportin 4 (XPO4) (13q12.11) with liver damage in MAFLD in a large cohort of patients of Caucasian ethnicity and delineated its effector mechanism. We demonstrated that duplication in the XPO4 CNV is associated with the severity of liver fibrosis. Functionally, this occurs via reduced expression of hepatic XPO4 that maintains sustained activation of SMAD3/SMAD4 and promotes TGF-β1-mediated hepatic stellate cell activation and fibrosis. This effect was mediated through termination of nuclear SMAD3 signalling and disturbed SMAD3/SMAD4 complex formation.
Implications of all the available evidence
Our findings suggest that the CNV in XPO4 gene is a genetic risk marker for the severity of liver fibrosis in MAFLD and may be a biomarker for risk. Moreover, XPO4 has anti-fibrotic activity via regulation of TGF-β/SMAD3 signalling suggesting that targeting this gene has therapeutic potential.
Alt-text: Unlabelled box
1. Introduction
Fibrosis is a highly heritable and polygenic trait characterized by the net deposition of extracellular matrix (ECM) in and around injured tissues; if persistent, fibrosis leads to tissue scarring, organ failure, and eventually, death [1,2]. Fibrotic diseases including chronic liver lung and kidney disease and heart failure contribute to ∼45% of mortality in affluent countries [3,4] and thus is a substantial burden on health systems. Limited therapeutic options are currently available that are directed specifically at the fibrosis process. Thus, a better understanding of the molecular basis controlling the fibrosis response is required to facilitate the development of novel therapies.
The advent of genomics generated much interest as a pathway that could lead to novel drug discovery [5,6]. Several analyses of pharmaceutical pipelines have suggested that drugs for targets in which a genetic link to disease is evident has nearly double the likelihood of successful clinical development with improved cost-effectiveness, compared to those not having such a link [7,8].
Out of all the classes of genetic variation that include single nucleotide polymorphisms (SNPs), insertions-deletions (inDels) and copy number variation (CNV), SNPs have occupied a preeminent position among studied human genetic variations [9]. Genome-wide association studies (GWAS) and large candidate gene studies have identified several SNPs that contribute to an increased likelihood of fibrosis risk and progression [10], [11], [12], [13], [14]. However, these SNPs only explain a proportion of the known disease heritability, reflecting the existence of ‘missing heritability’ [10,15]. Thus, other types of genetic variation likely exist to explain this missing heritability.
The role CNVs, a more recently appreciated class of large-scale germline variants on gene function and liver fibrosis remains under-characterized. CNVs due to duplication or deletion of a sequence is a form of structural variation defined as changes in the number of DNA copies (in comparison to the reference) of >1 kb; CNVs cover as much as ∼ 5%-25% of the human genome. Functionally, CNVs comprise ∼17.7% of the detected variations in gene expression [16] and they do so by altering gene dosage or disrupting the sequences of genes within partially overlapped regions [17,18] so can exert a profound effect on the risk for disease [19], [20], [21].
A genome-wide CNV association study in a small cohort of Malaysian subjects identified a CNV in exportin 4 (XPO4) to be correlated with the progression of metabolic (dysfunction) associated fatty liver disease (MAFLD) [22]. XPO4 maps to chromosome 13q and belongs to the importin β family which acts as a bidirectional nuclear transport receptor. Decreased XPO4 expression has been shown to associate with hepatocellular carcinoma [23,24]. Not surprisingly, similar to SNPs, CNVs exhibit substantial variability in both frequency and effect size across ethnicities [25,26]. Thus, it is crucial to investigate whether the effect of this CNV can be replicated to other populations and if so, to understand the underlying functional mechanism of action.
In this work, we investigated the role of the CNV in XPO4 (13q12.11) with liver damage in MAFLD in a large cohort of patients of Caucasian ethnicity and delineated its effector mechanism.
2. Methods
2.1. Patient cohort
The study comprised 646 Caucasian patients with biopsy-proven MAFLD who had blood samples available for genetic analyses and representative of their wider populations. In addition, 170 healthy controls were recruited. Subjects with evidence of secondary causes of steatosis or alternative diagnoses were excluded including alcohol abuse (men, >30 g/day; women, >20 g/day), total parenteral nutrition, chronic viral hepatitis (hepatitis B and hepatitis C), autoimmune liver diseases, hereditary hemochromatosis, α1-antitrypsin deficiency, Wilson's disease and drug-induced liver injury. The healthy control group was enrolled from Westmead hospital, Sydney. They reported no history of chronic liver disease.
2.2. Ethics
Ethics approvals from all participating sites were obtained from their corresponding Human Research Ethics Committees: The Sydney West Area Health Service, the University of Sydney, Human Research Ethics Committee (HREC/17/WMEAD/433), Australia; the Sir Charles Gairdner Hospital Human Research Ethics Committee, WA, Australia; Ethical Committee of the Città della Salute e della Scienza University Hospital, Torino, Italy; the Ethics committee of the Catholic University of the Sacred Heart, Rome, Italy (Fondazione Policlinico Gemelli IRCCS: ID 2576); Ethics Committee of Valme University Hospital, Seville; the Clinical Research Ethics Committee of the La Princesa University Hospital (PI-228/07); Ethics Committee name of the CEIC-Cantabria, Santander, Spain; and the Ethics Committee of the Hospital Clinico Universitario de Valladolid, Spain (F0-P07-10). Written informed consent for genetic testing was obtained from all participants.
All animal procedures were approved by the Western Sydney Local Health District Animal Ethics Committee and were conducted in accordance with the Animal Experimentation guidelines of the National Health and Medical Research Council (NHMRC) of Australia.
2.3. Clinical and laboratory assessment
Demographic and clinical data were obtained including age, gender, ethnicity, height and weight. Body mass index (BMI) was calculated as weight divided by the square of the height (kg/m2). Diabetes was defined as fasting blood glucose ≥ 7.0 mmol/L, previous diagnosis of diabetes or the use of antidiabetic drugs. The homeostasis model assessment (HOMA-IR) was calculated as: (fasting serum insulin [μU/mL] × fasting serum glucose [mmol/L])/22.5 [27]. At the time of liver biopsy, a fasting blood sample was obtained and routine biochemical tests were performed. Additional blood samples were drawn and frozen at −80°C for future research.
2.4. Determination of XPO4 CNV
The XPO4 13q12.11 CNV was genotyped using TaqMan CNV real-time, quantitative PCR assays (Assay Hs03857719_cn). Reactions were run at least as duplicates with PCR cycling conditions as follows: 1 PCR cycle at 95°C for 10 min, followed by 40 cycles at 95°C for 15 sec and 60°C for 1 min. Negative controls were introduced for every run to ensure genotyping quality. CN was assigned from the raw Cq values using CopyCaller™ software (version 2.0; Applied Biosystems). This software provides extensive diagnostics for the validity of the results and employs a clustering algorithm and assigns the cluster with the most samples as CN= 2.
2.5. Liver Histopathology
Liver biopsies were scored by an expert liver pathologist in each participating centre unaware of clinical or genetic data. Histological scoring was based on the system proposed by Kleiner et al. [28] Steatosis was graded from 0 to 3, lobular inflammation from 0 to 3 and hepatocellular ballooning from 0 to 2. Fibrosis was staged from 0 to 4 with 4 representing cirrhosis. The activity score (NAS) was calculated to quantify disease activity [28]. All biopsies were of appropriate size and included enough portal tracts for pathological grading and staging of histological features. The inter-observer agreement between pathologists was studied previously and was excellent for steatosis (κ = 0.85) and good for fibrosis (κ =0.78) [29].
2.6. Animal experimentation protocols
Three different models of murine liver fibrosis were employed. Bile duct ligation: C57BL/6 male mice (6–8 weeks) were subject to bile duct ligation (BDL) or Sham for 2 weeks to induce fibrosis as described previously [30]. Carbon tetrachloride (CCl4)-induced fibrosis: male C57BL/6 mice were injected intraperitoneally with CCl4 (1 ml kg−1 body weight) twice a week for 4 weeks to induce liver fibrosis. Control mice received an equivalent volume of olive oil. Methionine and choline-deficient (MCD) diet: experimental steatohepatitis was induced by administration of a methionine and choline-deficient (MCD) diet [31]. Male C57BL/6 mice were fed either the MCD diet or a control diet supplemented with methionine and choline as previously described [32]. Mice had ad libitum access to food and water. These experiments were conducted in 6-8 mice per group to detect a difference of 1.5 s.d. in the steatohepatitis Clinical Research Network (CRN) score. Finally, on completing each experiment, mice were sacrificed. They were weighed and euthanised with ketamine (100 mg/kg body weight) and xylazine (20 mg/kg body weight) in normal saline through an intraperitoneal injection. The mice were assessed for adequacy of anaesthetic by loss of pain reflexes.
2.7. Cell culture
LX‐2 cells and JS-1, immortalized human and mouse HSC lines respectively were cultured with in modified Eagle's medium (Life Technologies) supplemented with 10% non–heat‐inactivated fetal bovine serum and 1% penicillin/streptomycin solution (EuroClone). Mycoplasma testing and STR profiling of cell lines are done routinely. Primary human HSCs (ScienCell) were seeded on uncoated plastic dishes and cultivated with Iscove's modified medium (Dulbecco's medium; EuroClone, Italy) supplemented with 20% fetal bovine serum, 1% glutamine 200 mM, sodium pyruvate 100 mM, nonessential amino acid solution 100 ×, and an antibiotic antimycotic solution 100 × (Gibco Life Technologies, Carlsbad, CA). Primary HSCs between passages 1 and 8 were used for this study.
2.8. XPO4 silencing
Transfection was performed with Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions using 50 - 75 nM of specific Silencer siRNA (Supplementary table 1) or Silencer Select negative Control for 24 hours, then treated with or without TGF-β1 (2.5 ug/ml) and harvested after 48 hours. The efficiency of transfection was evaluated by real‐time PCR and western blot.
2.9. XPO4 overexpression
Transfection was performed with FuGENE HD (Promega) according to the manufacturer's instructions using Human XPO4 (NM_022459.4) DYK tagged ORF clone (Genscript CAT# OHU03871D) and Xpo4 (NM_020506) Mouse Myc-DDK-tagged ORF Clone (Origene CAT#: MR220257) or suitable empty vector for 24 hours and then treated with or without TGF-β1 (2.5–5.0 ug/ml) and harvested after 48hours later. The efficiency of transfection was evaluated by real‐time PCR and western blot.
2.10. Real‐time PCR analysis
RNA was isolated from primary HSCs, LX‐2 cells, JS-1 cells, and from liver tissues from mice and human liver biopsies by RNeasy (Qiagen) according to the manufacturer's instructions. 1000 ng of total RNA was reverse-transcribed into cDNA using qScript cDNA SuperMix (Quantabio, cat no 95048-500). Three microliters of diluted complementary DNA were loaded together with SYBR Select Master Mix on a 96‐well plate for real‐time PCR analysis (Applied Biosystems, Thermo Fisher Scientific). Taqman Probes and Forward and reverse primers for each gene of interest were specifically designed and used at 10 pmol/μL; GAPDH was used as the house-keeping gene. Primer sequences are provided in (supplementary table 2).
2.11. Western Blot
Whole cell extracts were fractionated by SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane using a transfer apparatus according to the manufacturer's protocol (Bio-Rad). Following transfer, membranes were blocked with 5% non-fat milk in TBST (50 mM Tris, pH 7.4, 150 mM NaCl, 0.05% Tween -20) for 1 hour at room temperature. Membranes were washed twice with TBST and incubated with primary antibodies (Supplementary table 3) diluted in 5% skim milk/TBST, overnight at 4°C. Following incubation, membranes were washed three times for 10 min with TBST and incubated with secondary antibodies, diluted in 5% skim milk/TBST, for 1 hour in dark. Blots were washed with TBST three times and developed with SuperSignal West Pico PLUS Chemiluminescent and SuperSignal West Femto Maximum Sensitivity Substrates (Life Technologies, Australia) according to the manufacturer's protocol. Membranes were scanned using ChemiDoc Touch Imaging system (Bio-Rad).
2.12. Immunohistochemistry
Slides of human and mouse liver were stained using the BOND RX Fully Automated Research Stainer (Leica Biosystems) using the Bond Polymer Refine Detection method (DS9800 Leica).
2.13. Immunofluorescence analysis
Cells were rinsed twice with PBS, fixed with 4% buffered paraformaldehyde and permeabilized with 0.5% Triton X-100 for 15 minutes. Cells were then incubated with the primary antibody overnight at 4°C and then with the secondary antibody conjugated to rhodamine or FITC (Molecular Probes) for 1 hour at room temperature. Cells were examined using a deconvolutional microscope (Zeiss).
2.14. Cell proliferation
Using the Biotrak cell proliferation ELISA system (Amersham, Little Chalfont, UK), DNA synthesis was measured by incorporation of the pyrimidine analogue bromodeoxyuridine (BrdU) into the DNA of proliferating cells. LX2 cells were cultured in 96 well plates for 24 hours, then further transfected with XPO4 plasmid or siRNA or their appropriate control in the presence or absence of TGF-β1 for 24 hours. Cells were then labelled with 10 μM BrdU for 24 hours. After removing the culture medium, cells were fixed and the incorporated BrdU detected by the subsequent substrate reaction according to the manufacturer's protocol.
2.15. Proximity ligation assay
LX-2 cells cultured in Nunc Lab-Tek II chamber slides (Thermo Fisher Scientific) were treated for 2h with or without 2.5ng/ml TGFβ. Cells were washed 3 times with PBS, fixed with 4% PFA for 20 min on ice, permeabilized with 0.1% triton, blocked for 1h at 37 degrees (humidity chamber) using Duolink Blocking Solution (Sigma-Aldrich), and incubated with both SMAD3 (Thermo Fisher Scientific, MA5-15663, mouse monoclonal antibody, diluted 1:500) and XP04 (Abcam, ab222384, rabbit polyclonal antibody, 1μg/ml) specific antibodies overnight at 4°C. Both antibodies were diluted using the Duolink Antibody Diluent (Sigma-Aldrich). The Duolink proximity ligation assay (Sigma-Aldrich) was performed to identify SMAD3 and XPO$4 interactions. PLA probes preparation, ligation, amplification, final wash step and preparation for imaging was done according to the manufacturer's instructions. Cells were visualized using the Olympus FV1000 confocal microscope and signals were identified and counted. For each treatment ± TGFβ, ten representative pictures/condition were analysed.
2.16. Surface Plasmon Resonance Assay
Various concentrations of proteins dissolved in water were manually printed onto the bare gold-coated (thickness 47 nm) PlexArray Nanocapture Sensor Chip (Plexera Bioscience, Seattle, WA, US) at 40% humidity. Each concentration was printed in replicate and each spot contained 0.2 uL of sample solution. The chip was incubated at 80% humidity at 4°C overnight, then rinsed with 10 x PBST for 10 min, 1 x PBST for 10 min, and deionized water twice for 10 min. The chip was then blocked with 5% (w/v) non-fat milk in water overnight, washed with 10 x PBST for 10 min, 1 x PBST for 10 min, and deionized water twice for 10 min before being dried under a stream of nitrogen prior to use. SPRi measurements were performed with PlexAray HT (Plexera Bioscience, Seattle, WA, US). Collimated light (660 nm) passes through the coupling prism, reflects off the SPR-active gold surface, and is received by the CCD camera. Buffers and samples were injected by a non-pulsatile piston pump into the 30 uL flowcell that was mounted on the coupling prism. Each measurement cycle contained four steps: washing with PBST running buffer at a constant rate of 2 uL/s to obtain a stable baseline, sample injection at 5 uL/s for binding, surface washing with PBST at 2 uL/s for 300 s, and regeneration with 0.5% (v/v) H3PO4 at 2 uL/s for 300 s. All measurements were performed at 25°C. The signal changes after binding and washing (in AU) are recorded as the assay value. Selected protein-grafted regions in the SPR images were analyzed, and the average reflectivity variations of the chosen areas were plotted as a function of time. Real-time binding signals were recorded and analyzed by the Data Analysis Module (DAM, Plexera Bioscience, Seattle, WA, US). Kinetic analysis was performed using BIAevaluation 4.1 software (Biacore, Inc.) via profacgen (NY, USA).
2.17. Luciferase assay
For TGF-β1–induced transcriptional reporter assays, LX-2 and JS-1 cells were transfected with one of SBE-Luc reporters, SBE4-Luc, ARE-Luc or SBE2-Luc reporter plasmids (Addgene). Transfection efficiency was normalized by cotransfection with the negative control plasmid. Luciferase activity was measured using a Dual Luciferase Assay System (BPSbioscience Catalog #: 60683-1) as per the manufacturer's instructions.
2.18. ROS measurement
JS-1 cells were transfected with Xpo4 or control vectors for 24 hour and then treated with TGF-β1 or vehicle control for 6 hours. Intracellular ROS was measured using the redox-sensitive dye DCFDA/H2DCFDA- cellular ROS Assay Kit (Abcam, ab113851). Fluorescence intensities were estimated by plate reader analysis.
2.19. RNA-seq
2.19.1. RNA-seq library generation
Total RNA was isolated from the JS1 cell line transfected with Xpo4 (NM_020506) Mouse Myc DDK-tagged ORF Clone (Origene CAT#: MR220257) or empty vector treated with or without TGF-β1 (5.0 ng/ml) using the RNA mini kit (Qiagen). RNA purity and integrity were confirmed using an Agilent Bioanalyzer. Sequencing libraries were prepared from 100–500 ng of total RNA using the TrueSeq RNA sample preparation kit v2 (Illumina). Briefly, mRNA was purified, fragmented, and used for first- and second-strand cDNA synthesis followed by adenylation of 3′ _ends. Samples were ligated to unique adaptors and subjected to PCR amplification. Libraries were then validated using the 2100 BioAnalyzer (Agilent), normalized and pooled for sequencing. RNA-seq libraries prepared from three biological replicates for each condition were sequenced on the Illumina HiSeq 2000 using barcoded multiplexing and a 100-bp read length.
2.20. High-throughput sequencing and analysis
Image analysis and base calling were done with Illumina CASAVA-1.8.2. This yielded a median of 29.9M usable reads per sample. . Read alignment and junction mapping to genome build GRCh38 was accomplished using STAR version 2.7.2b (Dobin, 2013 #573). Known splice junctions from mm10 were supplied to the aligner and de novo junction discovery was also permitted. Differential gene expression analysis and statistical testing were performed using Cuffdiff2 version 2.2.1 (Trapnell, 2013 #572), employing the Ensembl genome annotation. Transcript expression was calculated as gene-level relative abundance in fragments per kilobase of exon model per million mapped fragments and employed correction for transcript abundance bias (Trapnell, 2013 #572). RNA-seq results for genes of interest were also explored visually using the University of California Santa Cruz (UCSC) Genome Browser. The software tools gene set enrichment analysis (GSEA) and ingenuity pathway analysis (IPA) were used for pathway analyses.
2.21. Chromatin Immunoprecipitation (ChIP-qPCR)
JS1 cells were transfected with Xpo4 (NM_020506) Mouse Myc DDK-tagged ORF Clone (Origene CAT#: MR220257) or empty vector for 24 hours and then treated with or without TGF-β1 (5.0 ng/ml) for another 24 hours. Cells were then harvested for Chip assay. The experimental procedure for ChIP was described previously (Barish, 2010 #574). Briefly, after fixation, nuclei were isolated, lysed in buffer containing 1% SDS, 10 mM EDTA, 50 mM Tris-HCl pH 8.0, and protease inhibitors, and sheared with a Diagenode Bioruptor to chromatin fragment sizes of 200–1000 base pairs. Chromatin was immunoprecipitated with Smad3 antibody (Abcam cat# ab28379). ChIP-qPCR assays were performed using biological triplicates with primers listed in supplementary table 4.
2.22. Collagen Assay
JS-1 cells were transfected with Xpo4 (NM_020506) Mouse Myc DDK-tagged ORF Clone (Origene CAT#: MR220257) or empty vector for 24 hours and then treated with or without TGF-β1 (5.0 ng/ml) for another 24 hours then the collagen assay was performed using the Sircol™ Collagen Assay kit following the manufacturer's instructions.
2.23. Statistical methods
Results are expressed as mean ± SEM (standard error of the mean), median and interquartile range or number (percentage) as appropriate. The Student's t-test or Analysis of variance (ANOVA) or non-parametric, i.e. Wilcoxon-Mann-Whitney U-test or Kruskal-Wallis tests were used to compare quantitative data as appropriate. χ2 and Fisher-exact tests were used for comparison of frequency data and to evaluate the relationships between groups. All tests were two-tailed and p values <0.05 were considered significant. Shapiro-Wilk test was used to test the assumption of normality as appropriate. Multivariable regression modelling with backward elimination was undertaken to test independent associations of the XPO4 CNV with the following steatosis; ballooning, inflammation; and fibrosis using ordinal regression models for ordinal traits and multiple logistic regression models were fitted to binary traits incorporating biologically relevant covariates associated with the risk of liver disease progression (age, gender, Body Mass Index (BMI), ALT and presence of type 2 diabetes mellitus (T2DM)). For this analysis, steatosis was dichotomized as mild (S1) or moderate/severe (S2–S3); inflammation was dichotomized as absent/mild (A0–A1) or moderate/severe (METAVIR score A2–A3), and fibrosis as no fibrosis (F0) and fibrosis (F1–F4). Results are expressed as odds ratios and 95% confidence intervals (CI).
List of siRNAs and negative control used.
| SiRNA name | Cat no | company |
|---|---|---|
| SMARTpool: ON-TARGETplus Mouse Xpo4 siRNA | L-062054-01-0010 | Dharmacon |
| ON-TARGETplus Human DNMT1 (1786) siRNA - SMARTpool, | L-004605-00-0005 | Dharmacon |
| ON-TARGETplus Human DNMT3A (1788) siRNA - SMARTpool | L-006672-01-0005 | Dharmacon |
| ON-TARGETplus Human DNMT3B (1789) siRNA - SMARTpool | L-006395-00-0005 | Dharmacon |
| Human XPO4 | 4390771 | Ambion |
| ON-TARGETplus Non-targeting Pool | D-001810-10-05 | Dharmacon |
| Control#1 siRNA | 4390843 | Ambion |
Primer sequences used for RT-PCR.
| Primer Name | Forward (5’ to 3’) | Reverse (5’ to 3’) |
|---|---|---|
| Human XPO4 | GGC AAA GAC TTA TCT CCT GGT GG | CTG TAT CTG CTC CGA ACG CTG T |
| Human GAPDH | CCT GCA CCA CCA ACT GCT TA | GGC CAT CCA CAG TCT TCT GAG |
| Human αSMA | GAC AAT GGC TCT GGG CTC TGT AA | ATG CCA TGT TCT ATC GGG TAC TT |
| Human Mmp9 | GAA CCA ATC TCA CCG ACA GG | GCC ACC CGA GTG TAA CCA TA |
| Human TIMP-1 | ACTTCCACAGGTCCCACAAC | CACTGTGCATTCCTCACAGC |
| Human CTGF | AAT GCT GCG AGG AGT GGGT | CGG CTC TAA TCA TAG TTG GGT CT |
| Mouse XPO4 | GGC AGC ATC GAG TCA | CCC GAA CAT ACT TTT |
| Mouse GAPDH | CAC CAT CTC CCA GGA GCG CAG | CCT TCT CCA TGG TGG TGC AGA |
| Mouse TGF-β | GTGGGGACTTCTTGGCACT | GAGTGTCCACGACGGTGAG |
| Mouse Col1a1 | AGGAGAACCAGGTGACGAAG | CCCCAGCTTCTCCTTTCTCT |
| Mouse α-SMA | CTCTCTTCCAGCCATCTTTCAT | TATAGGTGGTTTCGTGGATGC |
| Mouse TIMP-1 | GCAAAGAGCTTTCTCAAAGAC | AGGGATAGATAAACAGGGAAACACT |
| Mouse MMP-9 | GATGCGTGGAGAGTCGAAAT | CACCAAACTGGATGACGATG |
Antibodies used for Western blot.
| Antibody name | Cat no | company |
|---|---|---|
| Anti-Xpo4 antibody | NB100-56495 | Novus Biologicals |
| Anti- α-Smooth Muscle Actin Antibody | 14968 | Cell Signaling Technology |
| Anti-beta Actin antibody | ab8227 | Abcam |
| Anti-Smad 3 antibody | 9513 | Cell Signaling Technology |
| Anti-Smad4 antibody | 38454 | Cell Signaling Technology |
| Anti-GAPDH antibody | ab8245 | Abcam |
List of primer sequences for Chromatin Immunoprecipitation (ChIPip-q PCR)
| Gene name | Primer Sequence |
|---|---|
| m Acta2_-214A | ACAACTGCTCAAATGCCCAG |
| m Acta2_-214B | TGAGGAATGTGCAAACCGTG |
| m Col1a1_-334A | TCCTTAGTTCTTCCCCAGGATG |
| m Col1a1_-334B | ACAAAGGGATCAGTGTCAGG |
| m Timp2_-154A | CTGGATTGGCACTCACACAC |
| m Timp2_-154B | CCTGAGTTCCACCCAGAGTC |
| m Ccn2/Ctgf_-32A | AGACGGAGGAATGTGGAGTG |
| m Ccn2/Ctgf_-32B | TCACATTCCTCCCCACCTTC |
2.24. Role of funding source
The funders had no role in study design, data collection, data analyses, interpretation, or writing of report.
3. Results
3.1. Duplication of the XPO4 CNV increases the severity of liver fibrosis in patients with MAFLD
The baseline clinical features of MAFLD patients are depicted in Table 1. In this cohort, XPO4 CNVs ranged from 0 to 8 copies per individual. 267 (41.3%) were copy number neutral, 52 (8.1%) had deletions, while 327 (50.6%) had duplications. In contrast, in the control cohort, 162 (95.3%) were copy number neutral, 5 (2.9%) had deletions and 3 (1.8%) had duplications. The very low prevalence of duplications and deletions in the general population suggested to us a potential pathogenic role for XPO4 in MAFLD.
Table 1.
Characteristics of the patient cohort with MAFLD.
| Variables | MAFLD cohort (n=646) |
|---|---|
| Age (years) | 48.4 (38-57) |
| Male (%) | 351 (54.3) |
| BMI (Kg/m2) | 33.3 (29-42) |
| ALT (IU/L) | 55 (34-84) |
| AST (IU/L) | 36 (26-52) |
| GGT (IU/L) | 57 (32-114) |
| Platelets (x109/L) | 247 (199-298) |
| Diabetics (%) | 167 (25.9) |
| Cholesterol (mmol/L) | 4.84 (4.1-5.7) |
| Triglycerides (mmol/L) | 1.6 (1.12-2.26) |
| HDL-C (mmol/L) | 1.2 (1-1.51) |
| LDL-C (mmol/L) | 2.8 (2.21-3.51) |
| Blood glucose (mmol/L) | 5.5 (4.95-6.58) |
| HOMA-IR | 3.46 (2.22-5.61) |
|
Steatosis grade 2-3 (%) |
287 (44.4) |
| Inflammation 2-3 (%) | 188 (29.2) |
| Fibrosis Stage 2-4 (%) | 242 (37.5) |
Values are median with interquartile range or frequency and percentage.
Therefore, we investigated the association of XPO4 CNVs with the spectrum of liver damage related to MAFLD. CNV duplication was associated with more advanced fibrosis, hepatocellular ballooning, severe inflammation and with reduced hepatic steatosis (p<0.009, for all) compared to combined non-duplications (neutral and deletions combined) (Fig. 1). In addition, carriers of the CNV duplication had higher mean fibrosis (1.44 vs. 1.28, p=0.01), inflammation (1.26 vs. 0.89, p=0.0001) and hepatocellular ballooning (1.2 vs. 0.76, p=0.0001) but lower steatosis (1.57 vs. 1.71, p=0.01) compared to combined non-duplications (neutral and deletions combined) using the Student's t-test in all comaprisons. Similar findings were observed when we compared CNV duplications to both CNV deletions and neutral status (Supplementary Fig. 1). In multivariable logistic regression analysis adjusted for age, sex, BMI, ALT and T2DM, CNV duplication conferred an increased risk for the presence of fibrosis (OR=1.98; 95% CI: 1.36-2.89, p=0.0001) (Supplementary Table 1). The results were virtually similar in three other models, one including HOMA-IR with the above mentioned variables, a second including AST instead of ALT and a third including AST/ALT ratio instead of ALT.
Fig. 1.
In MAFLD, the XPO4 CNV is associated with the severity of liver damage in patients with MAFLD. CNV gain (duplication) compared to non-gain (CNV neutral or deletion) in XPO4 in a cohort of MAFLD (n=646). p values represent the significance of a trend in the prevalence of more severe degrees of histological damage according to different copy number status.
3.2. XPO4 correlates with liver fibrosis in humans and murine models
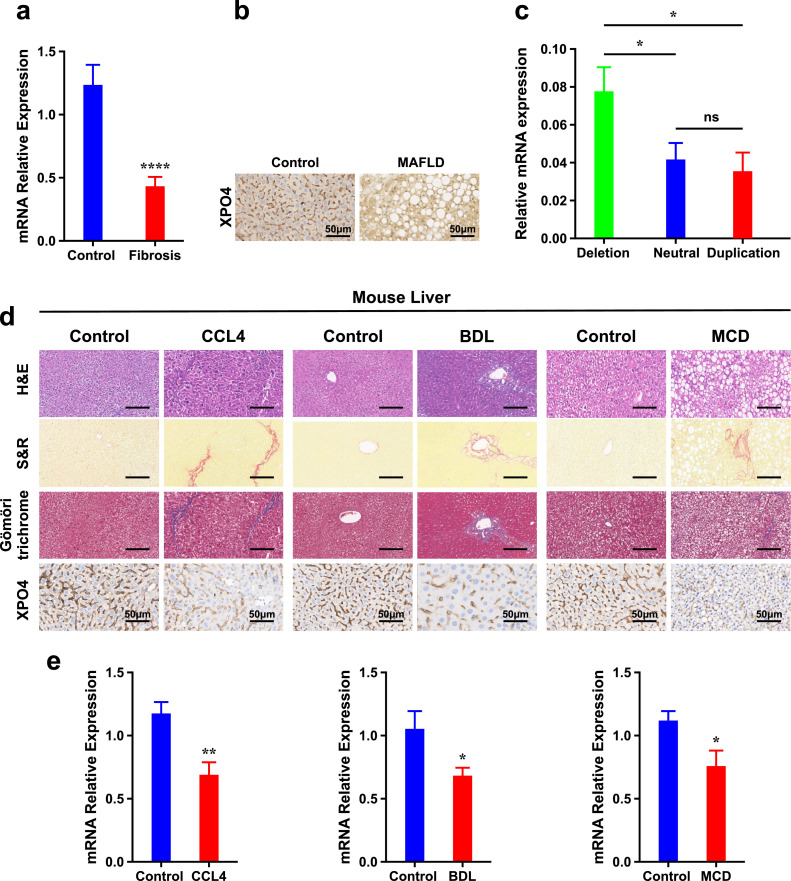
To investigate the involvement of XPO4 to fibrosis, we examined its hepatic expression in control subjects (n=25) and patients with MAFLD related fibrosis (≥F1) (n=38). XPO4 mRNA levels were considerably decreased in the livers of patients with fibrosis compared to that in controls (p < 0.0001, using the Student's t-test) (Fig. 2a). Consistently, immunohistochemistry revealed that XPO4 was decreased in the liver of patients with MAFLD related fibrosis compared to that from controls (Fig. 2b). In addition, we observed that copy number amplification of XPO4 was negatively correlated with its mRNA expression; expression levels of XPO4 decreased when comparing those with deletions to those without (P<0.05, using the ANOVA test) (Fig. 2c), indicating that copy number gain contributes to down-regulation of XPO4.
Fig. 2.
XPO4 expression is decreased with fibrosis in humans and in multiple fibrosis models in mice. a) Relative hepatic mRNA levels of XPO4 measured by RT-PCR and normalized to GAPDH as internal control in MAFLD patients with fibrosis (n=38) and in controls (n=28). b) Immunohistochemistry (IHC) staining for XPO4 in liver sections from representative MAFLD patients with fibrosis and in controls with normal liver histology. c) Relative hepatic mRNA levels of XPO4 by XPO4 CNV status (duplication, neutral or deletion). d) Hematoxylin–eosin (H&E), Sirius red and Gömöri trichrome staining and IHC for Xpo4 in liver sections from representative mice treated with vehicle or carbon tetrachloride (CCl4) (1 ml kg−1 body weight, i.p., twice a week, for 4 weeks), bile duct ligation (BDL) or Sham operation, or maintained on a methionine and choline–deficient (MCD) or control diet (CD) for 6 weeks (as a model of metabolic (dysfunction) associated steatohepatitis related fibrosis. e) Relative hepatic mRNA levels of Xpo4 measured by RT-PCR and normalized to Gapdh as internal control in mice from the CCl4, BDL or MCD models. Results are expressed as mean ± SEM; statistical significance of the differences between groups was calculated using the unpaired two-sample Student's t-test or ANOVA test. *P < 0.05, ** P<0.01, *** P<0.001.
We proceeded to determine whether xpo4 is similarly downregulated in experimental liver injury models. Compared with controls, liver sections of mice treated with carbon tetrachloride (CCL4) for 4 weeks showed increased collagen deposition indicated by hematoxylin and eosin (H&E), Sirius red and trichrome staining (Fig. 2d). Consistent with the human data, xpo4 mRNA levels by real time quantitative PCR (RT-qPCR) and intensity of staining by IHC in liver sections was less than that in control mice (Fig. 2d,e). To ensure that the changes were not model-specific, the observations were replicated in two other models of liver injury, namely, bile duct ligation (BDL) for 2 weeks and in mice fed a methionine and choline–deficient (MCD) diet for 6 weeks. The latter model is more representative of fibrosis associated with steatohepatitis (Fig. 2d,e).
3.3. XPO4 is highly expressed in hepatic stellate cells
Next, to dissect the function of XPO4 during liver fibrosis, we analysed its expression in different cell types of the liver, namely hepatocytes, hepatic stellate cells (HSC), and hepatic sinusoidal endothelial cells (HSEC). The highest expression level of XPO4 was found in HSCs followed by HSEC; expression levels were low in hepatocytes (Supplementary Fig. 2a).
HSCs activate and differentiate into myofibroblasts that produce extracellular matrix during the process of fibrosis in vivo, and spontaneously activate under culture conditions [33]. Consistent with a role in fibrosis, XPO4 mRNA decreased from the first to the fourth passage (P < 0.01), opposite to the increasing levels of the profibrogenic cytokine connective tissue growth factor (CTGF) (P < 0.05) (Supplementary Fig. 2b,c). We tested whether transforming growth factor beta (TGF-β), a major HSC activator and a potent fibrosis promoting stimulus might regulate the expression of XPO4. In line with the downregulation of XPO4 in HSCs during hepatic fibrosis, stimulation of a human HSC cell line (LX-2), a mouse HSC cell line (JS-1), and primary HSCs with recombinant TGFβ1 resulted in a significant decrease in the expression of XPO4 mRNA and protein levels that correlating with an increase in α-Smooth muscle actin (α-SMA) and CTGF expression in these cells (Supplementary Fig. 2d-f).
In total, these data indicate that XPO4 has high expression in HSCs, is downregulated during liver fibrosis and during culture activation of hepatic stellate cells, raising the possibility that it could be involved in the process of HSC activation in fibrosis.
3.4. XPO4 regulates the phenotype of hepatic stellate cells
To further explore the role of XPO4 in fibrosis and HSC activation in a systematic unbiased manner, we overexpressed Xpo4 in JS-1 cells and undertook RNA-Seq analysis. As shown in Supplementary Fig.s 3a and b, transfection of an XPO4 plasmid into JS-1 cells for 48 hours led to increased XPO4 mRNA and protein expression compared with the negative control. Ingenuity pathways analysis (IPA, Ingenuity systems, Inc., Redwood City, CA) analysis revealed that hepatic fibrosis/hepatic stellate cell activation was one of the top significant overlapping pathways to be altered in XPO4 overexpressed cells Fig. 2a. GSEA analysis similarly found significant enrichment in gene sets associated with extracellular matrix and collagen-related pathways in the control cells compared to XPO4 overexpressed cells Fig. 3b. Further, XPO4 overexpression comprehensively suppressed profibrotic gene expression, both in the absence and presence of TGFβ1 as illustrated in Fig. 3c.
Fig. 3.
Inhibition of HSC activation and the modulation of TGFβ1 effects by XPO4. a) Ingenuity pathways analysis (IPA, Ingenuity systems, Inc., Redwood City, CA) revealed that hepatic fibrosis/hepatic stellate cell activation was one of the top significant overlapping pathway altered in XPO4 overexpressed cells compared to control. b) GSEA analysis similarly found significant enrichment in gene sets associated with extracellular matrix and collagen-related pathways in control cells compared to XPO4 overexpressed cells c) Heat map of fold change of genes involved in fibrosis in murine JS-1 HSCs in Xpo4 overexpressed cells compared to control for 24 hours followed by TGFβ1 (2.5 ng/ml) treatment or vehicle control for 24 h using log2-transformed mRNA-Seq expression data, n = 4 per group. d) Acta2, Col1a1, Ctgf, Tgfb1, Mmp9 and Timp1 qRT-PCR analysis in murine HSCs transfected with control or Xpo4 vector for 24 hours followed by TGFβ1 (2.5 ng/ml) or vehicle control treatment for 24 h. e) RT-PCR analysis for the same panel of genes in murine HSCs transfected with control or Xpo4 siRNA for 24 hours followed by TGFβ1 (2.5 ng/ml) or vehicle control treatment for 24 h. f) COL1A1 level as measured by Sircol™ Collagen Assay, g) Acta2 expression as assessed by immunofluorescence and h) measured fluorescence intensity and i) ROS generation as measured by fluorescence intensity was significantly reduced in Xpo4-vector-transfected JS-1 cells treated with or without TGFβ1 (2.5 ng/mL for 24 h) compared to control. Results are expressed as mean ± SEM of at least three independent experiments performed in triplicate; statistical significance of the differences between groups was calculated using the unpaired two-sample Student's t-test or ANOVA test. *P < 0.05, ** P<0.01, *** P<0.001.
To validate the RNA sequencing data, RT-PCR analysis was used to quantify changes in mRNA expression of a subset of fibrosis markers. Xpo4 overexpression led to a dramatic decrease in the expression of Acta2, Col1a1 and Ctgf in cells treated with or without TGFβ1 (Fig. 3d). Conversely, knockdown using Xpo4 siRNA decreased Xpo4 mRNA (Supplementary Fig. 3c) and led to increased mRNA expression of the same genes (Fig. 3e). Furthermore, increased Xpo4 expression led to decreased collagen levels measured by a collagen quantitative assay (Fig. 3f), the α-SMA protein level by immunofluorescence (Fig. 3,h) and reactive oxygen species (ROS) level measured by fluorescence intensity (Fig. 3i). Similar findings were obtained in human LX-2 cells (Supplementary Fig. 4) suggesting that the effect of XPO4 is conserved across species.
During fibrosis resolution, increased activity of extracellular matrix (ECM)-degrading enzymes such as matrix metalloproteinases (MMPs) play a pivotal role [34]. XPO4 overexpression increased MMP-9, while its inhibition decreased levels in both LX-2 and JS-1 cells (Fig. 3 and Supplementary Fig. 4). Modulation of XPO4 expression on HSCs did not affect cell proliferation at basal levels and in response to TGFβ1 in both cell lines (Supplementary Fig. 5).
3.5. XPO4 has a preferential direct interaction with SMAD3
To understand how XPO4 affects TGFβ1 signalling during HSC activation we proceeded to identify XPO4 interaction with different SMADs, the major mediators of TGF-β signalling [35]. It has been reported that XPO4 interacts with SMAD3 [36], but whether this effect applies to all cell types responding to TGF-β, including HSCs is unknown. Further, it is not known whether XPO4 is a non-specific exporter for all SMADs or whether it interacts selectively with SMAD3. To this end, we measured the binding affinity of XPO4 to SMADs 1-6 using surface plasmon resonance (SPR) spectroscopy where XPO4 is immobilized on the sensor chip. A higher binding profile was observed with SMAD3, with a binding constant (KD) of 4.88 × 10−9 M, with medium binding demonstrated for SMAD1, SMAD2 and SMAD4 and low binding for SMAD5 and SMAD6 (Fig. 4a-f).
Fig. 4.
XPO4 interaction with SMADs. a-f) Surface plasmon resonance (SPR) sensorgrams for the interaction between XPO4 and SMAD proteins (SMAD1, SMAD2, SMAD3, SMAD4, SMAD5 and SMAD6) shown as dissociation constant (KD) values. g) The XPO4 interaction with SMAD3 was confirmed using a proximity ligation assay in the presence or absence of TGF-β1. Confocal microscopy demonstrated enhancement in fluorescent signals (red fluorescent spots), indicative of an XPO4:SMAD3 interaction (within 40 nm) with and without TGF-β1. No signal was observed in the negative control assay containing the 2 antibodies with one probe only. h) Proximity ligation signals were quantified and are presented as signal per nuclei. Data are representative of two independent experiments. i) Schematic illustration of SMAD3 deletion mutants. The numbers indicate the amino acid positions. j) Surface plasmon resonance (SPR) sensorgrams of the interaction between XPO4 and SMAD3 deletion mutants shown as dissociation constant (KD) values. Results are expressed as mean ± SEM; the p-value was calculated using the unpaired two-sample Student's t-test. *P < 0.05, ** P<0.01, *** P<0.001. KD value: >10−6: low;10−8-10−6: medium; 10−9-10−8: high binding.
To confirm the XPO4/SMAD3 interaction in HSCs, we performed a Duolink proximity ligation assay (PLA) with XPO4 and SMAD3 specific antibodies. PLA generates fluorescence when 2 proteins are within 40 nm [37]. As shown by the interaction signals per cell, this confirmed strong binding of XPO4 with SMAD3 at baseline that was reduced after TGFβ1 stimulation, (Fig. 4g,h). Negative control PLA experiments omitting either XPO4 or SMAD3 probe demonstrated no signal amplification, confirming the validity of the assay. Moreover, using the UCSC genome browser, SMAD3 binds to the XPO4 gene principally at intronic sequences in the 5’ region (Supplementary Fig. 6).
We next determined the minimal region crucial for the XPO4-SMAD3 interaction. A series of N-terminal– and C-terminal–truncated SMAD3 mutants were generated (Fig. 4i) and surface plasmon resonance was undertaken to map the domain of SMAD3 that associated with XPO4 with high affinity and specificity. This revealed interaction between SMAD3 and XPO4 through cooperative binding to both the MH1 and MH2 domains (Fig. 4j).
3.6. XPO4 regulates TGF-β/SMAD3-induced transcriptional activity
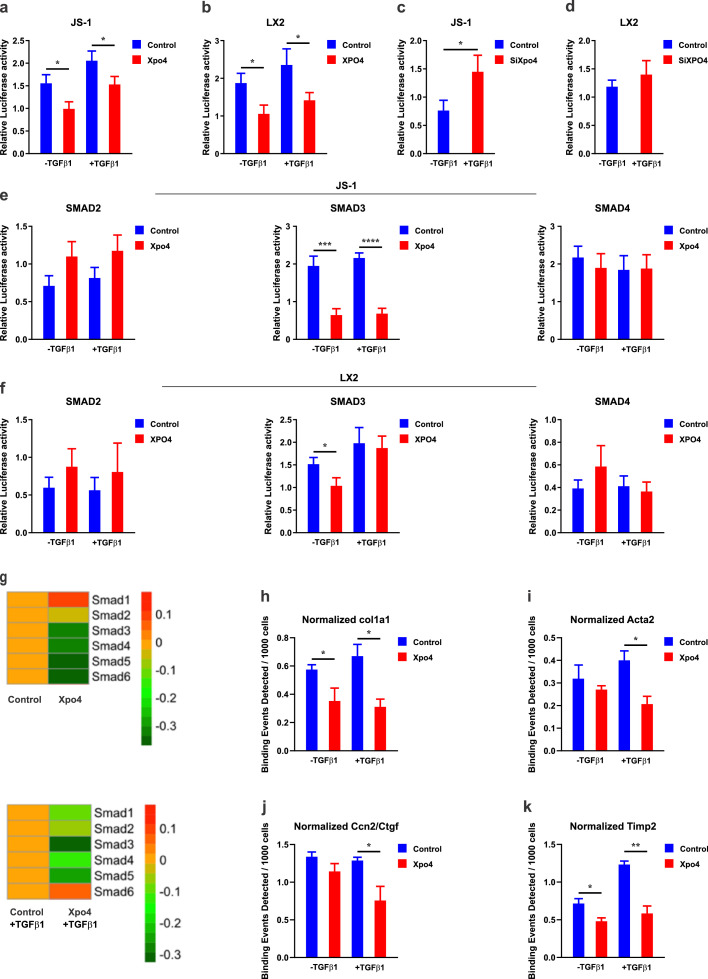
We next investigated how XPO4 modulates TGF-β1-SMAD signalling in HSC using various SMAD-dependent gene reporters. As expected, increased expression of XPO4 caused a decrease in basal and TGFβ1-dependent transcription from the SMAD-binding element (SBE)-Luc reporter in both JS-1 and LX-2 cells (Fig. 5a,b), with an opposite effect using siRNA (Fig. 5c,d). To specify the effect on different SMADs, we tested the effect of XPO4 on SBE4-Luc, a SMAD3-specific luciferase reporter construct, ARE-Luc, a TGF-β1–responsive but SMAD2-specific luciferase reporter and SBE2-Luc, a SMAD4-specific luciferase reporter. Increased XPO4 in LX-2 and JS-1 cells resulted in a decrease in the activity of SBE4-Luc, but not the activity of ARE-Luc or SBE2-Luc (Fig. 5e,f). In addition, using our RNA-Seq on JS-1 cells, we found that Xpo4 overexpression decreased Smad3 expression, but not other Smads from 1-6 (Fig. 5g).
Fig. 5.
XPO4 regulates TGF-β/SMAD3-induced transcriptional activity in HSCs. JS-1 (mouse) and LX-2 (human) HSCs were transfected with a control or XPO4 vector for 24 hours followed by TGFβ1 (2.5 ng/ml) or vehicle treatment for 24 h. The luciferase activity was measured by a dual luciferase assay for the SMAD-binding element (SBE)-Luc reporter in a) JS-1 cells and b) LX-2-cells. JS-1 and LX-2 cells were transfected with a control or XPO4 siRNA for 24 hours followed by TGFβ1 (2.5 ng/ml) or vehicle treatment for 24 h. The luciferase activity was measured by a dual luciferase assay for the SMAD-binding element (SBE)-Luc reporter in c) JS-1 cells and d) LX-2-cells. JS-1 and LX-2 cells were transfected with a control or XPO4 vector for 24 hours followed by TGFβ1 (2.5 ng/ml) or vehicle treatment for 24 h. The luciferase activity was measured by a dual luciferase assay for the SBE4-Luc, a SMAD3-specific luciferase reporter construct, ARE-Luc, a TGF-β1–responsive but SMAD2-specific luciferase reporter and SBE2-Luc, a SMAD4-specific luciferase reporter in e)JS-1 and f) LX-2 cells. g) Heat map of the SMAD 1-6 genes expression in XPO4 overexpression cells compared to control for 24 hours followed by TGFβ1 (2.5 ng/ml) or vehicle treatment for 24 h. Only Smad3 showed significant alteration wit Xpo4 overexpression at both conditions. JS-1 cells were transfected with a control or XPO4 vector for 24 hours followed by TGFβ1 (2.5 ng/ml) or vehicle treatment for 24 h and ChIP-qPCR was undertaken. Cells were immunoprecipitated with Smad3 antibody and analysed by qPCR using primers flanking h) Col1a1, i) Acta2, j) Ccn2/Ctgf and k) Timp2. XPO4 overexpression attenuated binding of SMADs to the promoters of these pro-fibrotic genes. Values are mean ±SEM for triplicate samples. Statistical significance was determined by the unpaired two-sample Student's t-test. P < 0.05, ** P<0.01, *** P<0.001.
Finally, chromatin immunoprecipitation (ChIP) studies demonstrated that Smad3 binding to the promoters of pro-fibrotic genes such as Col1a1, Acta2, Ctgf and Timp2 in response to TGFβ1 is significantly reduced by Xpo4 overexpression (Fig. 5-k), further confirming a direct role of XPO4 in modulating profibrotic gene expression via a preferential impact on TGFβ1/SMAD3 signalling.
3.7. XPO4 decreases SMAD3 phosphorylation level and nuclear retention
Phosphorylation of SMADs by the activated TGFβ1 receptor complex is an essential event in the initiation of TGFβ1 signal transduction [35]. Thus, we monitored the status of SMAD3 phosphorylation as well as of SMAD4 in response to TGFβ1 stimulation of HSCs. As shown in Fig. 6a, while TGFβ1 induced robust SMAD3 phosphorylation in control cells, increased XPO4 reduced the phosphorylation level of SMAD3 in response to TGFβ1, with no effect on SMAD4.
Fig. 6.
XPO4 decreases SMAD3 phosphorylation and nuclear retention and disturbs the SMAD3/SMAD4 complex in HSCs. a) Immunoblot for SMAD3, p-SMAD3 and SMAD4. XPO4 was overexpressed in LX-2 cells for 48 hours and continuously treated with 2.5 ng ml−1 TGFβ1 for 240 min. b) LX-2 cells were transfected with a control or XPO4 vector for 48 hours followed by TGFβ1 (2.5 ng/ml) or vehicle treatment for 1 hour. The cells were stained with an anti-SMAD3 and then by a FITC-conjugated anti-rabbit secondary antibody (green, for SMAD3); nuclei were visualized with DAPI (blue). Scale bars: 20 μm. c) Intensity of nuclear SMAD3 in cells was quantified using ImageJ software. The percentages of nuclear SMAD3 shown at the right represents the mean of two independent experiments; error bars indicate the SEM. Scale bars: 20 μm. d) LX-2 cells were transfected with a control or XPO4 vector for 48 hours followed by TGFβ1 (2.5 ng/ml) or vehicle treatment for 1 hour. The cells were stained with an anti-SMAD3 or anti-SMAD4 antibody and then by a rhodamine-conjugated anti-mouse secondary antibody (red, for SMAD4) or a FITC-conjugated anti-rabbit secondary antibody (green, for SMAD3); the nuclei were visualized with DAPI (blue). Scale bar: 20 μm. Values are mean ±SEM for triplicate samples. Statistical significance was determined by the unpaired two-sample Student's t-test. P < 0.05, ** P<0.01, *** P<0.001. e) Proposed scheme illustrating the effect of XPO4 on TGF-β1-mediated SMAD3 activation in HSCs. XPO4 has preferential interaction with SMAD3 and inhibits TGFβ1-mediated SMAD3 phosphorylation. XPO4 also decreases the nuclear retention of SMAD3 and disturbs the SMAD3/SMAD4 complex in HSCs at baseline and in response to TGFβ1. The inhibition of HSC response to TGFβ1 may contribute to the anti-fibrotic effect of XPO4.
Nuclear localization of SMAD3 and its direct binding to target gene promoters is required for downstream TGFβ1 signalling [38]. To characterise the molecular mechanism of XPO4 regulation of SMAD3 activity in HSCs, we determined whether XPO4 might modify the nuclear translocation of SMAD3. To this end, the location of SMAD3 in the nuclear and cytoplasmic fractions of TGFβ1 or vehicle treated HSCs were determined using immunofluorescence staining. As expected, TGFβ1 treatment robustly increased the nuclear localization of SMAD3, while XPO4 overexpression decreased the level of nuclear SMAD3 and concomitantly was associated with increased levels of cytoplasmic SMAD3, compared to control cells (Fig. 6b, c).
3.8. XPO4 disturbs the SMAD3/SMAD4 complex
Lastly, we tested whether XPO4 limits the binding of SMAD3 to SMAD4 since the formation of the SMAD3/SMAD4 complex is necessary for the nuclear retention of SMAD3 [38]. As visualized by immunofluorescence, SMAD3 and SMAD4 colocalized in the nucleus when induced by TGFβ1 in LX-2 cells, whereas overexpression of XPO4 reduced this nuclear colocalization (Fig. 6d). This confirmed that XPO4 plays an essential in limiting the sustained activation of the SMAD3/SMAD4 complex (Fig. 6e).
4. Discussion
In this work, we found that duplication of CNV 13q12.11 in XPO4 is associated with an increased severity of liver fibrosis in patients with MAFLD. Mechanistically, this resulted in decreased expression of hepatic XPO4, which maintains sustained SMAD3/SMAD4 activation and promotes TGFβ1 mediated HSC activation and fibrosis following liver injury.
The modest size of genetic effects detected so far confirms the multifactorial aetiology of fibrosis and suggests that as with other complex diseases, additional genetic influences exist that are yet to be identified [9]. CNVs are a common form of genetic variation playing an important role in the generation of genetic diversity often influencing gene expression and transcriptional regulation to a greater extent than the more commonly described single nucleotide polymorphism (SNPSs) [39], [40], [41]. The clinical consequences of a CNV depend on many factors including ethnic background or environmental factors [42]. Here, in a large cohort of Caucasian patients, we demonstrated that duplication of a rare and modest size CNV (18 Kb) in the XPO4 gene conveys an increased risk for severe fibrosis in patients with MAFLD. Approximately 80% of common CNVs (minor allele frequency > 5%) have been well-tagged by SNPs [43]. However, most low-frequency CNVs and multi-allelic CNVs are poorly tagged by SNP-based GWAS but they could be informative in terms of the discovery of missing heritability [43]. In our control cohort, ∼ 95% of individuals had two copies (copy number neutral), while < 2% has a duplication; in marked contrast the prevalence of duplication in adults with MAFLD was >50%, supporting a likely functional role. Our findings are also consistent with the original CNV GWAS in a small Malaysian cohort (49 patients with MAFLD (39 with steatohepatitis and 10 with simple steatosis) and 49 matched controls) that found this CNV exclusively in patients with but not in controls [22]. In a subsequent study from the same group in a cohort of 249 patients with MAFLD, the CNV duplication was observed in 34% of the steatohepatitis cohort but in 19% of controls [44]. The reasons of this discordance is not clear.
TGF-β is the principal cytokine mediating fibrosis and though therapies that neutralize TGFβ show anti-fibrotic activity, the benefits are compromised by unwanted systemic effects and the differential contribution of each R-SMAD [45,46]. SMADs are a family of structurally related intracellular proteins and possess intrinsic nucleocytoplasmic shuttling capacity which enables them to transmit TGF-β signals from the cell membrane to the nucleus serving as signalling effectors for TGF-β [47]. A major mechanisms that governs the duration and strength of signalling is the continued presence of these effectors/mediators in the nucleus [47]. In contrast to SMAD3 that promotes tissue fibrosis, SMAD2 protects against TGFβ-mediated fibrosis [48]. Thus, identifying potential specific regulators of TGFβ signaling might be a more promising approach. XPO4 expression we demonstrate was decreased with advanced liver fibrosis in humans and in murine models of fibrosis. From the cohort data, increasing fibrosis also associated with increasing copies of the XPO4 CNV. Our studies confirmed the inhibitory effects of XPO4 on HSC activation through antagonistic effects on TGF-β signalling. This effect was mediated through termination of nuclear SMAD3 signalling. XPO4 demonstrated preferential binding to SMAD3 compared to other SMADs and led to reduced SMAD3-mediated responses as shown by attenuation of TGFβ1 induced SMAD transcriptional activity, reductions in the recruitment of SMAD3 to target gene promoters following TGF-β1, as well as attenuation of SMAD3 phosphorylation and disturb SMAD3/SMAD4 complex formation [38]. Decreased retention of SMAD3 in the nucleus leads to attenuation of TGF-β/SMAD3–mediated transcriptional activity and target gene expression (Fig. 6e), while it has no impact on other SMAD family members. Collectively, these findings suggest that XPO4 might be a novel target to treat liver fibrosis with potentially less side effects. As TGF-β/SMAD3 is implicated in the progression of fibrosis in virtually all liver diseases, it would be interesting to explore the role of XPO4 CNVs in other liver disease in future studies.
The study has some limitations. The detailed characteristics of subjects in the control cohort were not available. Therefore, their matching to subjects with MAFLD was not feasible. However, the main focus of this study was to explore the impact of XPO4 CNV on the histological severity of MAFLD and to delineate the functional mechanisms. Second, details of medications for patients with MAFLD were not available for a large proportion of patients and so we could not undertake further analyses for their correlation with the XPO4 CNV.
In conclusion, our findings suggest that the CNV in XPO4 gene is a risk genetic variation for the severity of liver fibrosis in MAFLD and may be a biomarker for risk. Moreover, XPO4 has anti-fibrotic activity via regulation of TGF-β/SMAD3 signalling, suggesting the therapeutic potential of this molecule.
Contributors
M.E., M.M, A.B conceived the research; M.M, A.B, A.A, R.E, O.L, A.K, S.H, S.A, C.L, functional studies, CNV analysis and bioinformatics; patient enrolment, clinical phenotype data collation, sample acquisition/DNA preparation and intellectual contribution was performed by all authors; the manuscript was written and revised by M.E., M.M, A.B and J.G; A.B and M.M verified the underlying data. All authors critically reviewed the manuscript and approved the final submitted manuscript.
Declaration of Competing Interest
TB currently acts as an advisor to Abbvie, Alexion, Bayer, BMS, Gilead, Intercept, Janssen, MSD/Merck, Merz, Novartis, Sequana Medical, and Spring Bank. He has received speaking honoraria from Abbvie, Alexion, Bayer, BMS, Eisai, Gilead, Intercept, Ipsen, Janssen, MSD/Merck, Merz, Novartis, Sirtex and Sequana Medical in the past 2 years. He has received grant support from Abbvie, BMS, Gilead, Humedics, Intercept, Janssen, MSD/Merck, Merz, Novartis, and Sequana Medical. EB reports grants from GILEAD, personal fees from NOVO NORDISK, personal fees from PFIZER, outside the submitted work All other authors declare no competing financial interests.
Acknowledgments
Acknowledgements
We would like to thank all the patients for their participation in this study. We would like to thank “The Tumour Bank of The Children's Hospital at Westmead” for use of the Tumour Bank's automated IHC stainer. Confocal microscopy was performed at the Westmead Scientific Platforms, which are supported by the Westmead Research Hub, the Cancer Institute New South Wales, the National Health and Medical Research Council and the Ian Potter Foundation.
Data sharing statement
RNA-seq source data have been deposited at NCBI GEO dataset and are publicly available under the accession number: GSE180010. The rest of the data are available within the main text and supplementary file. The study did not generate new materials or codes.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103521.
Contributor Information
Jacob George, Email: jacob.george@sydney.edu.au.
Mohammed Eslam, Email: mohammed.eslam@sydney.edu.au.
Appendix. Supplementary materials
References
- 1.Vilar-Gomez E, Calzadilla-Bertot L, Wong VWS, Castellanos M, Aller-de la Fuente R, Metwally M, Eslam M. Fibrosis severity as a determinant of cause-specific mortality in patients with advanced nonalcoholic fatty liver disease: a multi-national cohort study. Gastroenterology. 2018;155:443. doi: 10.1053/j.gastro.2018.04.034. -+ [DOI] [PubMed] [Google Scholar]
- 2.Loomba R, Schork N, Chen CH, Bettencourt R, Bhatt A, Ang B, Nguyen P. Heritability of hepatic fibrosis and steatosis based on a prospective twin study. Gastroenterology. 2015;149:1784–1793. doi: 10.1053/j.gastro.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wynn TA. Cellular and molecular mechanisms of fibrosis. J Pathol. 2008;214:199–210. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eslam M, George J. Genetic contributions to NAFLD: leveraging shared genetics to uncover systems biology. Nat Rev Gastroenterol Hepatol. 2020;17(1):40–52. doi: 10.1038/s41575-019-0212-0. [DOI] [PubMed] [Google Scholar]
- 5.Plenge RM, Scolnick EM, Altshuler D. Validating therapeutic targets through human genetics. Nat Rev Drug Discovery. 2013;12:581–594. doi: 10.1038/nrd4051. [DOI] [PubMed] [Google Scholar]
- 6.Eslam M, George J. Genetic insights for drug development in NAFLD. Trends Pharmacol Sci. 2019;40:506–516. doi: 10.1016/j.tips.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Cook D, Brown D, Alexander R, March R, Morgan P, Satterthwaite G, Pangalos MN. Lessons learned from the fate of AstraZeneca's drug pipeline: a five-dimensional framework. Nat Rev Drug Discovery. 2014;13:419–431. doi: 10.1038/nrd4309. [DOI] [PubMed] [Google Scholar]
- 8.Nelson MR, Tipney H, Painter JL, Shen JD, Nicoletti P, Shen YF, Floratos A. The support of human genetic evidence for approved drug indications. Nat Genet. 2015;47:856. doi: 10.1038/ng.3314. -+ [DOI] [PubMed] [Google Scholar]
- 9.Eslam M, Valenti L, Romeo S. Genetics and epigenetics of NAFLD and NASH: clinical impact. J Hepatol. 2018;68:268–279. doi: 10.1016/j.jhep.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 10.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozlitina J, Smagris E, Stender S, Nordestgaard BG, Zhou HH, Tybjaerg-Hansen A, Vogt TF. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014;46:352. doi: 10.1038/ng.2901. -+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Speliotes EK, Yerges-Armstrong LM, Wu J, Hernaez R, Kim LJ, Palmer CD, Gudnason V. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLos Genet. 2011;7 doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Metwally M, Bayoumi A, Romero-Gomez M, Thabet K, John M, Adams LA, Huo X. A polymorphism in the Irisin-encoding gene (FNDC5) associates with hepatic steatosis by differential miRNA binding to the 3′UTR. J Hepatol. 2019;70(3):494–500. doi: 10.1016/j.jhep.2018.10.021. [DOI] [PubMed] [Google Scholar]
- 14.Zheng KI, Fan JG, Shi JP, Wong VW, Eslam M, George J, Zheng MH. From NAFLD to MAFLD: a "redefining" moment for fatty liver disease. Chin Med J (Engl) 2020;133:2271–2273. doi: 10.1097/CM9.0000000000000981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozlitina J, Smagris E, Stender S, Nordestgaard BG, Zhou HH, Tybjaerg-Hansen A, Vogt TF. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014;46:352–356. doi: 10.1038/ng.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gamazon ER, Stranger BE. The impact of human copy number variation on gene expression. Brief Funct Genom. 2015;14:352–357. doi: 10.1093/bfgp/elv017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou J, Lemos B, Dopman EB, Hartl DL. Copy-number variation: the balance between gene dosage and expression in drosophila melanogaster. Genome Biol Evol. 2011;3:1014–1024. doi: 10.1093/gbe/evr023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stankiewicz P, Lupski JR. Structural Variation in the Human Genome and its Role in Disease. Annu Rev Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 21.Zhang F, Gu WL, Hurles ME, Lupski JR. Copy Number Variation in Human Health, Disease, and Evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zain SM, Mohamed R, Cooper DN, Razali R, Rampal S, Mahadeva S, Chan WK. Genome-wide analysis of copy number variation identifies candidate gene loci associated with the progression of non-alcoholic fatty liver disease. PLoS One. 2014;9 doi: 10.1371/journal.pone.0095604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zender L, Xue W, Zuber J, Semighini CP, Krasnitz A, Ma B, Zender P. An oncogenomics-based in vivo rnai screen identifies tumor suppressors in liver cancer. Cell. 2008;135:852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang F, Fan YC, Mu NN, Zhao J, Sun FK, Zhao ZH, Gao S. Exportin 4 gene expression and DNA promoter methylation status in chronic hepatitis B virus infection. J Viral Hepat. 2014;21:241–250. doi: 10.1111/jvh.12136. [DOI] [PubMed] [Google Scholar]
- 25.Huang RS, Chen PX, Wisel S, Duan SW, Zhang W, Cook EH, Das S. Population-specific GSTM1 copy number variation. Hum Mol Genet. 2009;18:366–372. doi: 10.1093/hmg/ddn345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ulloa AE, Chen JY, Vergara VM, Calhoun V, Liu JY. Association between copy number variation losses and alcohol dependence across african american and european american ethnic groups. Alcoholism-Clin Experiment Res. 2014;38:1266–1274. doi: 10.1111/acer.12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chitturi S, Farrell G, Frost L, Kriketos A, Lin R, Liddle C, Samarasinghe D. Serum leptin in NASH correlates with hepatic steatosis but not fibrosis: A manifestation of lipotoxicity? Hepatology. 2002;36:403–409. doi: 10.1053/jhep.2002.34738. [DOI] [PubMed] [Google Scholar]
- 28.Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, Ferrell LD. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 29.Kazankov K, Barrera F, Moller HJ, Rosso C, Bugianesi E, David E, Ibrahim Kamal Jouness R. The macrophage activation marker sCD163 is associated with morphological disease stages in patients with non-alcoholic fatty liver disease. Liver Int. 2016;36(10):1549–1557. doi: 10.1111/liv.13150. [DOI] [PubMed] [Google Scholar]
- 30.Dong ZX, Su L, Brymora J, Bird C, Xie Q, George J, Wang JH. Resistin mediates the hepatic stellate cell phenotype. World J Gastroenterol. 2013;19:4475–4485. doi: 10.3748/wjg.v19.i28.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.George J, Pera N, Phung N, Leclercq I, Hou JY, Farrell G. Lipid peroxidation, stellate cell activation and hepatic fibrogenesis in a rat model of chronic steatohepatitis. J Hepatol. 2003;39:756–764. doi: 10.1016/s0168-8278(03)00376-3. [DOI] [PubMed] [Google Scholar]
- 32.Abdelaal M, le Roux CW, Docherty NG. Morbidity and mortality associated with obesity. Ann Transl Med. 2017;5:161. doi: 10.21037/atm.2017.03.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedman SL. Molecular regulation of hepatic fibrosis, an integrated cellular response to tissue injury. J Biol Chem. 2000;275:2247–2250. doi: 10.1074/jbc.275.4.2247. [DOI] [PubMed] [Google Scholar]
- 34.Duarte S, Saber J, Fujii T, Coito AJ. Matrix metalloproteinases in liver injury, repair and fibrosis. Matrix Biol. 2015;44-46:147–156. doi: 10.1016/j.matbio.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hata A, Chen YG. TGF-beta Signaling from Receptors to Smads. Cold Spring Harb Perspect Biol. 2016;8 doi: 10.1101/cshperspect.a022061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurisaki A, Kurisaki K, Kowanetz M, Sugino H, Yoneda Y, Heldin CH, Moustakas A. The mechanism of nuclear export of Smad3 involves exportin 4 and Ran. Mol Cell Biol. 2006;26:1318–1332. doi: 10.1128/MCB.26.4.1318-1332.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Awan Z, Tay ES, Eyre NS, Wu LE, Beard MR, Boo I, Drummer HE. Calsyntenin-1 mediates hepatitis C virus replication. J Gen Virol. 2016;97:1877–1887. doi: 10.1099/jgv.0.000511. [DOI] [PubMed] [Google Scholar]
- 38.Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, Redon R, Werner J. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 2007;39:1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henrichsen CN, Chaignat E, Reymond A. Copy number variants, diseases and gene expression. Hum Mol Genet. 2009;18:R1–R8. doi: 10.1093/hmg/ddp011. [DOI] [PubMed] [Google Scholar]
- 41.Schlattl A, Anders S, Waszak SM, Huber W, Korbel JO. Relating CNVs to transcriptome data at fine resolution: Assessment of the effect of variant size, type, and overlap with functional regions. Genome Res. 2011;21:2004–2013. doi: 10.1101/gr.122614.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zarrei M, MacDonald JR, Merico D, Scherer SW. A copy number variation map of the human genome. Nat Rev Genet. 2015;16:172–183. doi: 10.1038/nrg3871. [DOI] [PubMed] [Google Scholar]
- 43.McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, Shapero MH. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 44.Zain SM, Mohamed Z, Pirmohamed M, Tan HL, Alshawsh MA, Mahadeva S, Chan WK. Copy number variation in exportin-4 (XPO4) gene and its association with histological severity of non-alcoholic fatty liver disease. Sci Rep. 2015;5 doi: 10.1038/srep13306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu HH, Chen DQ, Wang YN, Feng YL, Cao G, Vaziri ND, Zhao YY. New insights into TGF-beta/Smad signaling in tissue fibrosis. Chem Biol Interact. 2018;292:76–83. doi: 10.1016/j.cbi.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 46.Schuppan D, Kim YO. Evolving therapies for liver fibrosis. J Clin Invest. 2013;123:1887–1901. doi: 10.1172/JCI66028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dai F, Duan X, Liang YY, Lin X, Feng XH. Coupling of dephosphorylation and nuclear export of Smads in TGF-beta signaling. Methods Mol Biol. 2010;647:125–137. doi: 10.1007/978-1-60761-738-9_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meng XM, Huang XR, Chung AC, Qin W, Shao X, Igarashi P, Ju W. Smad2 protects against TGF-β/Smad3-mediated renal fibrosis. J Am Soc Nephrol. 2010;21:1477–1487. doi: 10.1681/ASN.2009121244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.