Abstract
Mitogen-activated protein kinase (MAPK) signaling is required for plant cell death responses to invading microbial pathogens. Iron- and reactive oxygen species (ROS)-dependent ferroptotic cell death occurs in rice (Oryza sativa) during an incompatible rice–Magnaporthe oryzae interaction. Here, we show that rice MAP kinase (OsMEK2 and OsMPK1) signaling cascades are involved in iron- and ROS-dependent ferroptotic cell death responses of rice to M. oryzae infection using OsMEK2 knock-out mutant and OsMEK2 and OsMPK1 overexpression rice plants. The OsMPK1:GFP and OsWRKY90:GFP transcription factor were localized to the nuclei, suggesting that OsMPK1 in the cytoplasm moves into the nuclei to interact with the WRKY90. M. oryzae infection in ΔOsmek2 knock-out plants did not trigger iron and ROS accumulation and lipid peroxidation, and also downregulated OsMPK1, OsWRKY90, OsRbohB, and OsPR-1b expression. However, 35S:OsMEK2 overexpression induced ROS- and iron-dependent cell death in rice. The downstream MAP kinase (OsMPK1) overexpression induced ROS- and iron-dependent ferroptotic cell death response to virulent M. oryzae infection. The small-molecule ferroptosis inhibitor ferrostatin-1 suppressed iron- and ROS-dependent ferroptotic cell death in 35S:OsMPK1 overexpression plants. However, the small-molecule inducer erastin triggered iron- and lipid ROS-dependent, but OsMEK2-independent, ferroptotic cell death during M. oryzae infection. Disease (susceptibility)-related cell death was lipid ROS-dependent, but iron-independent in the ΔOsmek2 knock-out mutant during the late M. oryzae infection stage. These combined results suggest that OsMEK2 and OsMPK1 expression positively regulates iron- and ROS-dependent ferroptotic cell death, and blast disease (susceptibility)-related cell death was ROS-dependent but iron-independent in rice–M. oryzae interactions.
Keywords: cell death, ferroptosis, iron, Magnaporthe oryzae, mitogen-activated protein kinase (MAPK) signaling, reactive oxygen species (ROS), rice
Introduction
Plants have evolved effective innate immune system responses to avert the invasion of microbial pathogens in their natural habitat (Dodds and Rathjen, 2010; Schwessinger and Ronald, 2012; Fu and Dong, 2013). Plant immune system responses are mediated by pattern-triggered immunity (PTI) and effector-triggered immunity (ETI), which are effectively upregulated inside plant cells in response to pathogen infection (Jones and Dangl, 2006). PTI is activated by plant perception of conserved microbial structures, called pathogen-associated molecular patterns (PAMPs), via the transmembrane pattern recognition receptors (PRRs) (Zipfel, 2008). ETI is activated by plant recognition of specific pathogen effector molecules via intracellular nucleotide-binding leucine-rich repeat (NLR) receptors, called resistance (R) proteins (Jones and Dangl, 2006; Block and Alfano, 2011; Oh and Martin, 2011). The two immune systems trigger a series of molecular signaling events that lead to diverse cellular responses including transcriptional reprogramming, synthesis of defense-related proteins, reactive oxygen species (ROS) burst, and iron- and ROS-dependent ferroptotic cell death (Boller and He, 2009; Dangol et al., 2019).
Mitogen-activated protein (MAP) kinase (MAPK) signaling pathways have pivotal roles in plant defense, immunity, and hypersensitive cell death responses to pathogen attack (Ishihama et al., 2011; Meng and Zhang, 2013; Thulasi Devendrakumar et al., 2018). However, the downstream signaling networks activated by defense-related MAPKs have not been completely defined in plants. Plant MAPK cascades proceed through three central kinases: MAPK kinase kinase (MAPKKK); MAPK kinase (MAPKK), also known as MAPK and ERK (extracellular signal-regulated kinase) kinase (MEK); and MAP kinase (MAPK or MPK) (Meng and Zhang, 2013). These kinases are sequentially phosphorylated as MAPKKKs activate downstream MAPKKs (MEKs), which subsequently activate MAPKs (Rodriguez et al., 2010). Phosphorylation of MAPKs may promote their nuclear translocation to target other kinases, proteins, or transcription factors in the nucleus (Khokhlatchev et al., 1998; Rodriguez et al., 2010). MAPKs can activate transcription factors such as WRKYs. The Arabidopsis genome encodes 60 MAPKKKs, 10 MAPKKs, and 20 MAPKs (Ichimura et al., 2002). A previous study showed that Arabidopsis innate immune responses are mediated by a MAP kinase signaling cascade (MEKK1, MKK4/MKK5, and MPK3/MPK6) and WRKY22/WRKY29 transcription factors (Asai et al., 2002). Arabidopsis MPK3 and MPK6 are involved in ETI (Tsuda et al., 2009; Meng and Zhang, 2013; Su et al., 2018). Pathogen-responsive MAPK cascades (MEKK1-MKK4/MKK5-MPK3/MPK6 and MEKK1-MKK1/2-MPK4) have pivotal roles in defense signaling against pathogen attack in Arabidopsis thaliana (Pitzschke et al., 2009; Rasmussen et al., 2012; Meng and Zhang, 2013). Nearly two decades ago, Yang et al. (2001) identified a tobacco MAPKK (NtMEK2) upstream of both salicylic acid-induced protein kinase (SIPK) and wounding-induced protein kinase (WIPK). Expression of a constitutively active NtMEK2 mutant induced hypersensitive response (HR)-like cell death and defense responses in tobacco. Many kinases in MAPK cascades, including MAPKKK, MEK, SIPK/WIPK, and MAPK, are involved in N gene-mediated resistance to tobacco mosaic virus in tobacco (Jin et al., 2002, 2003; Liu et al., 2004). Tobacco WRKY/MYB transcription factors downstream of MAPK cascades have crucial roles in regulating N-mediated resistance to TMV (Liu et al., 2004). Silencing of MEK2 (SlMKK2), SlMPK2, and SlMKK4 in tomato disrupted the resistance to infection by Xanthomonas campestris pv. vesicatoria (Xcv) and Botrytis cinerea (Melech-Bonfil and Sessa, 2011; Li et al., 2014).
The rice genome contains 74 MAPKKK, 8 MAPKK, and 17 MAPK genes (Hamel et al., 2006; Reyna and Yang, 2006; Rao et al., 2010; Yang et al., 2015). We previously identified 74 non-redundant interactors with rice MAPKs and performed high-resolution mapping of the MAPK interactome network, which controls different signaling pathways underlying the cellular and physiological responses in rice (Singh et al., 2012). Rice MAP kinase kinase1 (OsMEK1) physically interacts with rice MAP kinase1 (OsMPK1), OsMPK6, and OsMPK5. OsMEK2 interacts with OsMPK1 and OsMPK6. OsMEK6 interacts with OsMPK1 and OsMPK5 (Singh et al., 2012). Rice MAP kinase kinases (OsMAP2Ks or OsMEKs) may regulate multiple signaling pathways affecting many biological processes by associating with different sets of rice MAPK interactomes (Singh and Jwa, 2013). However, few kinase components in rice MAPK cascades are involved in immunity and defense responses in rice–pathogen interactions (Singh and Jwa, 2013; Yang et al., 2015). OsMKK10-2–mediated activation of OsMPK6 via specific phosphorylation subsequently induced WRKY45 expression and blast (Magnaporthe oryzae) resistance in rice plants (Ueno et al., 2015). OsMPKK10-2 is involved in disease resistance and drought tolerance (Ma et al., 2017), physically interacts with OsMPK6 and OsMPK3, and phosphorylates the two OsMPKs, leading to X. oryzae pv. oryzicola (Xoo) resistance and drought tolerance (Ma et al., 2017). The MAP kinase module OsMKK3-OsMPK7-OsWRKY30 is involved in induced resistance to Xanthomonas oryzae pv. oryzicola (Xoo) infection in rice (Jalmi and Sinha, 2016).
Cell death is a fundamental biological process that occurs during development, senescence, immunity, and stress resistance in multicellular organisms. ROS bursts are involved in basal immune responses, NLR-mediated hypersensitive cell death, and disease-associated cell death in plants (Greenberg and Yao, 2004; Choi et al., 2012, 2013; Jwa and Hwang, 2017). Ferroptosis differs from apoptosis, necrosis, and autophagy, and was first discovered in mammalian cells as a form of non-apoptotic iron-dependent cell death (Dixon et al., 2012). Ferroptotic cell death requires the accumulation of ROS, iron, and lipid peroxides (Stockwell et al., 2017). Iron homeostasis and ROS burst have important roles in activating defense responses against plant pathogens (Liu et al., 2007; Aznar et al., 2015). We recently reported that iron- and ROS-dependent ferroptosis occurs in rice–Magnaporthe oryzae interactions (Dangol et al., 2019). This is the first plant pathosystem in which ferroptotic cell death was discovered (Caseys, 2019). Avirulent M. oryzae infection triggers iron and ROS (H2O2) accumulation at the cell death sites in rice tissues (Dangol et al., 2019). Iron is required for lipid peroxide accumulation. Iron and ROS accumulation and lipid peroxidation are blocked by the iron chelator deferoxamine, the lipophilic antioxidant ferrostatin-1, the actin polymerization inhibitor cytochalasin E, and the NADPH-oxidase inhibitor diphenyleneiodonium (DPI), thereby restricting HR cell death in rice (Dangol et al., 2019). By contrast, the RAS-selective lethal small molecule inducer erastin triggered iron-dependent ROS accumulation and glutathione depletion, which ultimately promoted M. oryzae-induced ferroptotic cell death. Rice NADP-malic enzyme (NADP-ME) and NADPH-oxidase (Rboh) are ROS sources that have been proposed to be involved in iron- and ROS-dependent ferroptotic cell death (Singh et al., 2016; Dangol et al., 2019).
Our previous study identified rice mitogen-activated protein (MAP) kinase kinase 2 (OsMEK2) as a rice MAP interactor (Singh et al., 2012). Rice MAP kinase (OsMPK1) is an interactor of OsMEK2 and actively involved in M. oryzae infection (Singh et al., 2012; Ueno et al., 2015). Here, we used OsMEK2 and OsMPK1 to investigate whether rice MAPKs are involved in the signaling network that mediates ferroptotic cell death in rice–M. oryzae interactions. OsMEK2 knock-out via T-DNA insertion in rice cultivar Dongjin (DJ) suppressed iron- and ROS-dependent ferroptotic cell death, which ultimately induced susceptible responses to avirulent M. oryzae 007 infection. However, OsMEK2 overexpression in rice DJ induced iron- and ROS-dependent ferroptotic cell death against M. oryzae 007 infection. Treatment of the OsMEK2-knockout mutants with erastin induced the ROS burst and iron accumulation, which caused ferroptotic cell death in ΔOsmek2 knock-out plants in response to M. oryzae infection. Disease (susceptibility)-related cell death at the late stage of M. oryzae infection in ΔOsmek2 knock-out plants is ROS-dependent and iron-independent. During M. oryzae infection, OsMEK2 knock-out and overexpression differentially regulated the expression of OsMPK1, OsMPK6, and the OsWRKY90 transcription factor in the rice MAPK signaling pathways. OsMPK1 overexpression in susceptible rice cultivar Nipponbarre (NB) induced iron- and ROS-mediated ferroptotic cell death against M. oryzae PO6-6 infection. Treatment with ferrostatin-1 suppressed iron- and ROS-dependent ferroptotic cell death in 35S:OsMPK1 overexpression leaf sheaths during infection. These combined results indicate that OsMEK2 and OsMPK1 expression via MAPK signaling pathway positively regulates iron- and ROS-dependent ferroptotic cell death and plant immunity to M. oryzae infection.
Materials and Methods
Plant Materials and Growth Conditions
The WT rice cultivars Dongjin (DJ) and Nipponbarre (NB) and the ΔOsmek2 knock-out, 35S:OsMEK2 and 35S:OsMPK1 overexpression lines were used in this study. ΔOsmek2 T-DNA insertion knock-out mutant seeds were provided by the Rice Functional Genomic Express Database (RiceGE) managed by the Salk Institute Genomic Analysis Laboratory1 (Jeon et al., 2000). DJ and NB rice seeds were obtained from the National Institute of Crop Science, South Korea2. Rice seeds were germinated in water for 5 days and then planted in plastic pots containing Baroker soil (Seoul Bio, South Korea). Rice plants were grown in growth chambers at 28°C under white fluorescent light (150 μmol photons m–2 s–1) with a 16 h photoperiod and 60% relative humidity, as described previously (Dangol et al., 2019).
Fungal Cultures and Growth Conditions
The rice blast fungal strains Magnaporthe oryzae 007 and PO6-6 were provided by the Center for Fungal Genetic Resources, Seoul National University, Seoul, South Korea3. M. oryzae 007 was avirulent (incompatible) and M. oryzae PO6-6 was virulent (compatible) to the rice cultivar DJ. The rice cultivar NB was susceptible to M. oryzae PO6-6 infection. The fungal cultures were stored at –20°C and cultured on rice bran agar media (20 g rice bran, 20 g sucrose, and 20 g agar in 1 L Milli-Q water). M. oryzae strains were grown at 25°C in the dark for 2 weeks. M. oryzae sporulation was induced by removing aerial mycelia from the fungal culture plates, followed by their incubation under a continuous fluorescent light (80 μmol photons m–2 s–1) for 2–3 days at 25°C.
Fungal Inoculation of Rice Tissues and Infection Evaluation
Conidial suspensions of M. oryzae strains were inoculated on rice leaves and leaf sheaths as described previously (Singh et al., 2016; Dangol et al., 2019). M. oryzae conidia were harvested from the sporulated culture plates using a 0.025% Tween 20 (Sigma-Aldrich) solution. The conidial concentration was adjusted to 4 × 105 conidia mL–1. The conidial suspension was spray-inoculated over the surface of 2-week-old rice seedlings. The inoculated seedlings were incubated at 25–28°C for 24 h under dark and moist conditions, and then were moved to normal conditions (16 h light/8 h dark). Disease phenotypes were observed at 5 days after inoculation and classified with respect to susceptible (large, elliptical, grayish, and expanded lesions) and resistant (slightly elongated, necrotic brownish spots) reactions.
Middle-aged leaf sheaths (5–7 cm lengths) of 4- or 5-week-old rice plants were inoculated with M. oryzae conidial suspensions (4 × 105 conidia mL–1). Inoculated leaf sheaths were incubated in a moistened box with 100% relative humidity at 25°C under dark conditions. After incubation for different times, epidermal layers were excised from the leaf sheaths, cut into 1.5 cm lengths, and fixed on glass microscope slides. The infected epidermal cells from each of three epidermal sheaths were observed under the microscope and divided into two infection phenotypes: cells with viable IH and cells with HR cell death. For the quantification of invasive hyphae (IH) and HR cell death, approximately 500 infected cells in each of the leaf sheaths were counted at least three times from one representative of three independent experiments.
Identification of T-DNA Insertion in ΔOsmek2 Mutants
ΔOsmek2 T-DNA insertion mutant seeds from RiceGE (Jeon et al., 2000) were screened by PCR using the left gene-specific primer (LP), the right gene-specific primer (RP), and the T-DNA right border primer (RB). The gene-specific primers are listed in Supplementary Table 1. To verify homozygosity in the knock-out mutant plants, PCR was performed with LP and RP primers of the gene, and homozygous plants were identified by the lack of specific PCR products. The LP and RB primers were used for PCR analysis to confirm the presence of the T-DNA insertion in ΔOsmek2 knock-out mutant plants. Quantitative real-time RT-PCR and immunoblotting analyses were performed to verify whether transcriptional and translational expression of OsMEK2 were blocked in ΔOsmek2 knock-out mutants.
Rice Transformation
The full length cDNAs of OsMEK2 and OsMPK1 were amplified from rice cDNA library and inserted into plant expression vector pCAMLA under the control of CaMV 35S promoter, followed by selection of hygromycin gene (hph). The constructed CaMV 35S:OsMEK2 and CaMV 35S:OsMPK1 were introduced in the rice cultivars DJ and NB, respectively, by Agrobacterium tumefaciens-mediated transformation, as described previously with slight modification (Hiei et al., 1994).
Briefly, 35S:OsMEK2 and 35S:OsMPK1 were delivered into rice calli using A. tumefaciens strain LBA4404 (Hoekema et al., 1983; Lee et al., 2005). The transformed calli were selected on the selection media containing gradually increasing concentrations of hygromycin (30 mg/L and 60 mg/L). After rooting and shooting, rice seedlings were transferred in water for 4 days. After adaption in water, rice seedlings were raised in soil in a growth chamber. The positive transformants from T0 generation were selected by PCR using hygromycin primers. Next, seeds of T1 generation were analyzed on the hygromycin-containing media and T2 generation seeds were used for homozygote selection. The functional analysis was performed from T3 generation. The levels of gene expression were determined by immunoblot analysis and qRT-PCR. The primers used for the experiments are listed in Supplementary Table 1.
Real-Time RT-PCR Analyses
Total RNA was isolated from rice tissue using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. The gene expression levels were analyzed by reverse-transcription PCR (RT-PCR) and real-time quantitative PCR (qRT-PCR). First-strand cDNA was synthesized from 2 μg total RNA in 20 μL reaction mixture using a cDNA synthesis kit (Invitrogen) according to the manufacturer’s instructions. Prepared cDNA (1 μL) was used as a template for both RT-PCR and qRT-PCR. The qRT-PCR was performed using TOPrealTM qPCR 2 × PreMIX (SYBR Green with low ROX; Enzynomics, Daejeon, South Korea) according to the manufacturer’s instructions. Relative gene expression levels were determined using rice 18S ribosomal RNA or rice ubiquitin as an internal standard gene. Gene-specific primers used for the real-time RT-PCR analysis are listed in Supplementary Table 1.
Protein Isolation and Immunoblot Assay
Rice proteins were extracted using trichloroacetic acid (TCA)/acetone extraction buffer [TCAAEB; 10% (w/v) trichloroacetic acid and 0.07% β-mercaptoethanol in 100 mL acetone] as described previously (Cho et al., 2006). Rice leaves were ground to a fine powder using liquid nitrogen in a mortar and pestle. Then, proteins were precipitated with TCAAEB and washed three times with wash buffer [0.07% β-mercaptoethanol, 2 mM ethylenediaminetetraacetic acid (EDTA), and EDTA-free protease inhibitor cocktail tablet (Roche) in a final volume of 100 mL acetone]. Protein precipitates were air-dried at room temperature, stored at –80°C for at least 24 h, and solubilized in lysis buffer containing thiourea and Tris (LB-TT) {7 M urea, 2 M thiourea, 4% (w/v) 3-[(3-cholamidopropyl) dimethylammonio]-l-propanesulfonate (CHAPS), 18 mM Tris-HCI (pH 8.0), 14 mM Trizma base, two EDTA-free protease inhibitor cocktail tablets (Roche), 0.2% (v/v) Triton X-100, and 50 mM dithiothreitol (DTT) in a final volume of 100 mL}. After centrifuging at 15,000 × g for 15 min at 4°C, the supernatants were precipitated using pre-chilled acetone and solubilized in LB-TT buffer.
The OsMEK2 protein expression levels in DJ and ΔOsmek2 mutant plants were determined by 10% SDS-polyacrylamide gel electrophoresis (PAGE) followed by immunoblot analysis using rabbit polyclonal anti-MEK2 antibody (EnoGene® E580135-A-SE). Immuno-reactive target bands were detected by Odyssey® CLx Imaging System (LI-COR Biosciences). Equal gel loading was checked by Ponceau S staining.
Erastin and Ferrostatin-1 Treatment
The small molecule cell death inducer erastin was used to investigate whether erastin treatment triggered ferroptotic cell death in rice leaf sheath cells as described previously (Dangol et al., 2019). M. oryzae conidia (4 × 105 conidia mL–1) were mixed with 10 μM erastin (Sigma-Aldrich, St. Louis, MO, United States) and inoculated on leaf sheaths. The erastin-treated and M. oryzae-inoculated leaf sheath tissues were incubated in the dark at 25°C. The ferroptosis inhibitor, ferrostatin-1 (Fer-1, Sigma-Aldrich), was treated as described previously (Dangol et al., 2019). Rice epidermal layers were excised from the M. oryzae-infected leaf sheaths and then vacuum-infiltrated in 10 μM Fer-1 solution for 10 min, followed by their incubation for 24 h.
CM-H2DCFDA Assay and DAB Staining
Cellular ROS (H2O2) localization in rice leaf sheath cells was visualized using 5- (and 6-) chloromethyl-2′,7′-dichlorofluorescin diacetate acetyl ester (CM-H2DCFDA) and 3,3′-diaminobenzidine (DAB) staining methods as described previously (Shin et al., 2005; Dangol et al., 2019). Briefly, thin epidermal layers of rice leaf sheaths were excised and cut into equal pieces, followed by incubation in 1 mL water for 5 min to remove wound-induced ROS. Epidermal sheath samples were incubated in 2 μM CM-H2DCFDA (Molecular Probes Life Technologies, Eugene, OH, United States) in 1 × phosphate-buffered saline (PBS) buffer in the dark for 30 min on a horizontal shaker. The incubated sheath samples were washed twice with 1 × PBS buffer for 5 min in the dark. ROS localization inside the epidermal sheath cells was observed immediately under a fluorescence microscope.
For DAB staining, epidermal layers of rice leaf sheaths were vacuum-infiltrated with 1 mg mL–1 DAB (Sigma-Aldrich, St. Louis, MO, United States) solution for 5 min, followed by overnight destaining with ethanol:acetic acid:glycerol (3:1:1, v/v/v). ROS localization in the DAB-stained epidermal cells was observed under a microscope. The DAB-stained cells were categorized into two phenotypes: Type I, infected cells that display no or weak DAB staining; and Type II, infected cells that display strong DAB staining. DAB-stained 500 cells with different phenotypes were counted from each of infected sheaths. The counted cell numbers were then converted into the percentages of DAB-stained cells.
Chemiluminescence Assay for ROS Measurement
The chemiluminescence assay was used to measure ROS production in rice leaf sheaths as described previously (Singh et al., 2016; Dangol et al., 2019) with minor modifications. Epidermal layers of treated and M. oryzae-inoculated rice leaf sheaths were cut into small pieces (0.5 cm length) and incubated in 1 mL of sterilized Milli-Q water for 5 min to remove wound-induced ROS. Then, a piece of epidermal layer was added into a mixture of 30 μL luminol (Bio-Rad, Hercules, CA, United States), 1 μL horseradish peroxidase (Jackson Immunoresearch, West Grove, PA, United States), and 69 μL Milli-Q water in each well of a 96-well plate. Chemiluminescence (RLU, relative luminescent units) was detected from the ROS signals after 5 min incubation using a GloMax® 96 Microplate Luminometer (Promega, Madison, WI, United States).
Malondialdehyde (MDA) Assay
The malondialdehyde (MDA) assay was performed to determine lipid peroxidation in rice leaf sheath tissues as described previously (Zhang et al., 2009; Dangol et al., 2019). Lipid peroxidation is the degradation of lipids due to oxidative damage in plant cells. Briefly, the ground tissue powder of rice leaf sheath was mixed with the reaction solution [0.5% (w/v) thiobarbituric acid, 20% (v/v) trichloroacetic acid (TCA), and 0.25 mL 175 mM NaCl in 2 mL of 50 mM Tris-Cl, pH 8.0]. The mixed reaction was then incubated in boiling water for 5 min, cooled on ice for 5 min, and centrifuged at 14,000 × g for 5 min. The MDA concentration (C) in the resulting supernatant was determined by measuring supernatant absorbances (OD, optical density) at 450, 532, and 600 nm, and then calculating MDA concentration according to the following equation: C = 6.45 × (OD532–OD600) – (0.56 × OD450).
Ferric Ion (Fe3+) Detection by Prussian Blue Staining
Prussian blue staining was performed to detect ferric ion (Fe3+) in rice leaf sheaths as described previously (Liu et al., 2007; Dangol et al., 2019). Briefly, epidermal layers of rice leaf sheaths were excised and incubated in equal volumes (1:1, v/v) of 7% potassium ferrocyanide and 2% hydrochloric acid (HCl) for 15 h at room temperature. Prussian blue (ferric ferrocyanides, which combine with Fe3+ inside leaf sheath epidermal cells) was observed as a bright blue color under a fluorescence microscope. Prussian blue-stained cells were categorized into two phenotypes: Type I, cells that contain IH but are weakly or not Prussian blue-stained; and Type II, strongly Prussian blue-stained cells with only a few poor hyphae. Prussian blue-stained 500 cells with different phenotypes were counted from each of infected sheaths. The counted cell numbers were then converted into the percentages of Prussian blue-stained cells.
Subcellular Localization of OsMEK2, OsMPK1, and OsWRKY90 in N. benthamiana Leaves
The full length cDNA of OsMEK2, OsMPK1, and OsWRKY90 were amplified from the rice cDNA library with the gene-specific primers containing attB1 and attB2 sites, as described in Supplementary Table 1. The amplified PCR products were used as a template for the second PCR using attB1 and attB2 primers. The second PCR products were sub-cloned into the pDONRTM201 entry vector using BP clonase (Invitrogen) to create entry clones. The entry clones were recombined into the Gateway binary vector pGWB552 tagged with G3 green fluorescent protein (G3GFP) using LR clonase (Invitrogen) (Nakagawa et al., 2007; Dangol et al., 2017).
The binary plasmids containing OsMEK2, OsMPK1, and OsWRKY90 were transformed into A. tumefaciens GV3101. Recombinant agrobacteria were prepared for infiltration using a protocol as described previously with slight modification (Sainsbury and Lomonossoff, 2008). Briefly, single colonies of recombinant agrobacteria were incubated into the liquid LB media (10 g/L tryptone, 5 g/L yeast extract; 10 g/L NaCl, pH 7) containing spectinomycin (100 μg/L) for overnight at 28°C with continuous shaking. Harvested recombinant agrobacteria were resuspended to an OD600 = 0.2 in MMA (10 mM MES pH 5.6, 10 mM MgCl2, 150 μM acetosyringone). The agrobacterial suspension was incubated for 2 h at room temperature, and infiltrated into the abaxial leaves of 6-week-old Nicotiana benthamiana plants using a blunt tipped plastic syringe. Subcellular localization of 00:GFP, OsMEK2:GFP, OsMPK1:GFP, and OsWRKY90:GFP in N. benthamiana epidermal cells 36 h after agroinfiltration were microscopically observed following 4′,6-diamidino-2-phenylindole (DAPI, 5 μg/ml) staining for 10 min. Nuclear localization of the proteins was visualized by counterstaining the nuclei of the cells with DAPI.
Microscopy
Images were captured using a fluorescence microscope (Zeiss equipped with Axioplan 2; Campbell, CA, United States) with 40× oil-immersion objective lens. CM-H2DCFDA-specific fluorescence was visualized under the fluorescence microscope using a combination of excitation (450–490 nm) and emission (515–565 nm) green fluorescence (GF) filters. Subcellular images were also taken using a fluorescence microscope (Olympus, Japan) using bright field, GF (Ex/Em: 488/498–548 nm), and DAPI (Ex/Em: 405/421–523 nm) filters.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: OsMEK1 (Os01g32660), OsMEK2 (Os06g05520), OsMEK3 (Os03g12390), OsMEK4 (Os02g46760), OsMEK5 (Os06g09190), OsMEK6 (Os02g54600), OsMEK7 (Os06g09180), OsMEK8 (Os06g27890), OsMPK1 (Os06g06090), OsMPK6 (Os10g38950), OsWNK1 (Os07g38530), OsWRKY90 (Os09g30400), OsNADP-ME (Os01g52500), OsRbohB (Os01g25820), OsPR-1b (Os01g28450), OsPAL1 (Os04g43760), OsAPX1 (Os0.g17690), OsAPX2 (Os07g49400), OsUbiqutin (Os06g46770), 18S rRNA (XR_003238819.1), AtMKK1 (At4g26070), AtMKK2 (At4g29810), AtMEK3 (NP_198860), AtMEK4 (At1g51660), AtMEK5 (At3g21220), AtMKK6 (At5g56580), AtMKK9 (At1g73500), AtWNK9 (At3g04910), NtNPK2 (BAA06731), SlMKK1 (NP_001234744), SlMKK2 (NP_001234588), NbMEK2 (LOC107818847), and NtMEK2 (AF325168).
Results
Identification of the Rice MAPK Interactor OsMEK2
In our previous study, we isolated rice mitogen-activated protein kinase kinases (MAPKKs or MEKs) that interacted with OsMAPKs using yeast two-hybrid analysis (Singh et al., 2012). Amino acid sequences of the isolated rice MEKs were aligned with those of Arabidopsis AtMAPKKs, and subsequently categorized into Groups A–D (Ichimura et al., 2002) (Supplementary Figure 1). The plant MAPKKs contained 11 conserved subdomains (Supplementary Figure 2). All of the aligned MAPKKs contained the active site domain [D(I/L/V)KP] and the conserved motif (S/T-X5-S/T, where X represents any amino acid residue) (Supplementary Figure 1A). The rice MAPKKs OsMEK1 and OsMEK2 belonged to Group A serine (S)/threonine (T) kinases. OsMEK1 amino acid sequence shared 54% and 53% homology with AtMKK1 and AtMKK2, respectively, whereas OsMEK2 had 62% sequence homology with both AtMKK1 and AtMKK2. A phylogenetic tree was generated to compare OsMEKs with Arabidopsis MAPKKs (Kumar et al., 2016) (Supplementary Figure 1B). OsMEK2 shares 65–66% homology with N. benthamiana NbMEK2 and tomato SlMKK1 (Supplementary Figure 2). OsMEK2 was phylogenetically close to NbMEK2, SlMKK1, AtMKK1 and AtMKK2 (Supplementary Figure 3). Based on the sequence alignment data of rice MAPKKs, OsMEK2 was selected to investigate whether rice MAPKKs are required for ferroptotic cell death signaling in this study.
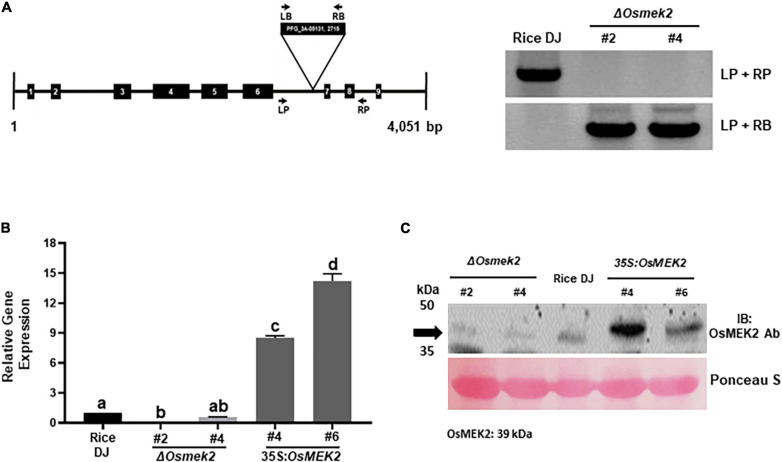
OsMEK2 was knocked out in rice cultivar DJ by T-DNA insertion mutagenesis (Jeon et al., 2000). The OsMEK2 genomic DNA sequence contains nine exons and eight introns (Supplementary Figure 4). The T-DNA insertion mutant, ΔOsmek2, was identified in the intronic region between the sixth and seventh exons (Figure 1A). The genotypes of ΔOsmek2 (M5) progeny were analyzed with primer sets LP + RP and LP + LB to detect transgene and homo/hetero selection, respectively (Figure 1A). The ΔOsmek2 #2 andΔOsmek2 #4 mutants (M5) were identified as T-DNA insertion homozygous plants that lacked the specific PCR products.
FIGURE 1.
Genotyping, transcriptional, and immunoblotting analyses of ΔOsmek2 knock-out and 35S:OsMEK2 overexpression lines. (A) Genotyping of ΔOsmek2 plants. The schematic diagram shows the T-DNA insertion site in the OsMEK2 gene. Exons and introns are depicted by solid boxes and lines, respectively. The T-DNA insertion ΔOsmek2 knock-out plants (M5) were detected using the gene primers (LP + RP) and the vector primers (LP + RB). LP, left primer; RP, right primer; LB, left border; RB, right border. (B) Transcriptional analysis of OsMEK2 expression in wild-type (WT) rice (cultivar DJ), ΔOsmek2 #2 and #4, and 35S:OsMEK2 #4 and #6 plants using quantitative real-time RT-PCR. (C) SDS-PAGE and immunoblotting assays of OsMEK2 expression in wild-type (WT) rice (cultivar DJ), ΔOsmek2 #2 and #4 and 35S:OsMEK2 #4 and #6 plants using OsMEK2 Ab (EnoGene® E580135-A-SE) (∼39 kDa). Ab, antibody; IB, immunoblot; PAGE, polyacrylamide gel electrophoresis.
OsMEK2 expression in ΔOsmek2 knock-out and 35S:OsMEK2 overexpression lines was examined by quantitative real-time RT-PCR and immunoblotting (Figures 1B,C). The qRT-PCR and immunoblot analyses indicated that OsMEK2 was not expressed in ΔOsmek2 #2 and #4 knock-out lines, but distinctly expressed in 35S:OsMEK2 #4 and #6 overexpression lines, compared to the wild-type rice DJ. These combined data indicate that the OsMEK2 gene is knocked out in the selected ΔOsmek2 #2 and #4 lines, but overexpressed in 35S:OsMEK2 #4 and #6 lines. Avirulent M. oryzae 007 infection caused susceptible response in ΔOsmek2 #2 and #4 lines, but resistant response in 35S:OsMEK2 #4 and #6 overexpression lines (Supplementary Figure 5). M. oryzae 007 grew well and produced invasive hyphae (IH) in the leaf sheath cells of ΔOsmek2 #2 and #4 knock-out plants, but induced hypersensitive cell death in wild-type (WT) rice DJ and 35S:OsMEK2 #4 and #6 overexpression plants (Supplementary Figures 5A,B). The ΔOsmek2 #2 and 35S:OsMEK2 #4 lines were selected to use in this study, because ΔOsmek2 #2 and #4 lines and 35S:OsMEK2 #4 and #6 lines exhibited the same susceptible and resistant responses to M. oryzae 007 infection, respectively.
The OsMEK2 Gene Is Required for Cell Death and Resistant Responses to M. oryzae Infection
Quantitative real-time RT-PCR analyses showed that avirulent M. oryzae 007 infection triggered the induction of OsMEK2 expression at early infection times (1–12 hpi). During M. oryzae 007 infection, OsMEK2 expression was up-regulated from 1 to 12 hpi and then back to the background level at 24 hpi (Figure 2). In contrast, OsMEK2 expression by virulent M. oryzae PO6-6 infection remained unchanged at 1, 6, and 12 hpi, but up-regulated at 24 hpi. Notably, infection with avirulent M. oryzae 007 significantly reduced OsMEK2 expression at late infection stages (72–96 hpi). These results suggest that early induction of OsMEK2 expression is involved in rice defense signaling in the incompatible rice–M. oryzae interaction (Figure 2).
FIGURE 2.
Quantitative real-time RT-PCR analysis of time-course expression of OsMEK2 in rice leaf sheaths in the compatible and incompatible interactions of rice with Magnaporthe oryzae. Rice leaf sheaths were sampled at different time points after inoculation with virulent and avirulent M. oryzae PO6-6 and 007, respectively. OsMEK2 expression was analyzed by quantitative RT-PCR. Relative gene expression of OsMEK2 at each time point was calculated by normalizing with respect to expression of the internal control OsUbiquitin gene. Data represent the mean ± SD from three independent experiments. Asterisks above the columns indicate significant differences as analyzed by Student’s t-test (P < 0.05). hpi, hours post-inoculation.
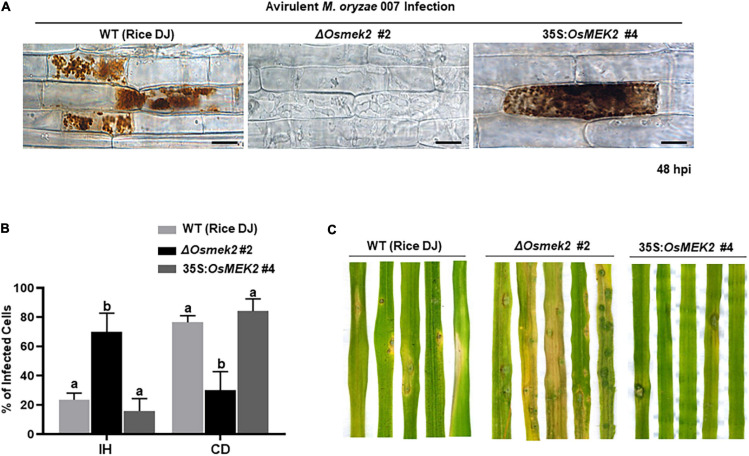
We investigated whether OsMEK2 is required for cell death and resistant responses to M. oryzae 007 infection using ΔOsmek2 #2 and 35S:OsMEK2 #4 rice plants (Figure 3). M. oryzae 007 grew poorly and caused cell death responses in leaf sheath epidermal cells of rice DJ and 35S:OsMEK2 #4 overexpression plants at 48 hpi (Figures 3A,B). By contrast, the blast fungus grew well with plentiful invasive hyphae (IH) in the invaded ΔOsmek2 #2 leaf sheath cells. Avirulent M. oryzae 007 infection induced significantly more hypersensitive cell death in rice DJ and 35S:OsMEK2 #4 leaf sheaths than in ΔOsmek2 #2 leaf sheaths (Figure 3B). Whole-leaf disease phenotypes were observed at 5 days after inoculation with M. oryzae 007 (Figure 3C). Rice DJ and 35S:OsMEK2 #4 leaves displayed a typical resistant reaction with small necrotic, and brownish restricted lesions. By contrast, ΔOsmek2 #2 mutant leaves displayed a typical susceptible reaction with large grayish lesions (Figure 3C). These combined results indicate that OsMEK2 knock-out in rice DJ rendered resistance ineffective and induced susceptibility (disease) in response to avirulent M. oryzae infection. However, OsMEK2 overexpression in rice DJ enhanced the cell death and resistance responses to rice blast disease.
FIGURE 3.
Avirulent Magnaporthe oryzae 007 infection causes susceptible responses in the ΔOsmek2 #2 knock-out plants, but resistant responses in the wild-type rice and 35S:OsMEK2 #4 overexpression plants. (A) Images of rice sheath epidermal cells infected with M. oryzae 007 (48 hpi). Rice leaf sheaths were inoculated with a conidial suspension (4 × 105 conidia mL–1). M. oryzae 007 grew well and produced invasive hyphae in the OsMEK2 knock-out (ΔOsmek2 #2) rice, but induced hypersensitive cell death in wild-type (WT) rice cultivar DJ and OsMEK2-overexpressed (35S:OsMEK2 #4) plants. Images were captured using a fluorescence microscope. hpi, hours post-inoculation. Scale bars = 20 μm. (B) Quantification of cell death and invasive hyphae in rice sheath cells infected with M. oryzae 007 (48 hpi). Results are presented as mean values ± SD; n = 4 leaf sheaths from different plants. Different letters above the bars indicate significantly different means (P < 0.05), as analyzed by Fisher’s protected least significant difference (LSD) test. IH, invasive hyphae; CD, cell death. (C) Disease types of rice leaves in wild-type rice (DJ), ΔOsmek2 #2 and 35:OsMEK2 #4 plants. Two-week-old rice seedlings were spray-inoculated with a conidial suspension (4 × 105 conidia mL–1) of M. oryzae 007. Diseased leaves were photographed at 5 days after inoculation. Disease types indicate a resistant-type lesion (necrotic brownish spots) and a susceptible-type lesion (large grayish, and expanded lesions). Experiments were repeated three times with similar results.
OsMEK2 Knock-Out and Overexpression Differentially Regulates MAPKs, WRKY and Defense-Related Gene Expression in Rice During M. oryzae Infection
A previous study reported that rice OsMAP2K2 (OsMEK2) interacted with and phosphorylated OsMAPKs, such as OsMPK1 and OsMPK6 (Singh et al., 2012). We analyzed OsMPK1, OsMPK6, and OsWRKY90 expression in leaf sheaths of rice DJ, ΔOsmek2 #2 knock-out and 35S:OsMEK2 #4 overexpression plants during avirulent M. oryzae 007 infection (Figure 4). OsMEK2 knock-out in rice DJ plants distinctly downregulated OsMPK1 expression throughout the course of M. oryzae infection. However, OsMEK2 overexpression did not upregulate expression of OsMPK1 and OsMPK6 in rice DJ plants. By contrast, OsMPK6 downregulation in ΔOsmek2 #2 leaf sheath cells was observed at early infection stages 3–12 hpi. OsMPK6 (or OsMPK1) activation by OsMKK10-2 is required for the induction of OsWRKY45 expression and blast resistance in rice (Ueno et al., 2015). OsMPK1 is the pathogen-responsive MAPK that is involved in disease resistance (Singh et al., 2012; Ueno et al., 2015). Bimolecular fluorescence complementation (BiFC) analysis in rice leaf sheath indicates that OsMPK1 physically interacts with the OsWRKY80 transcription factor (Singh et al., 2012) and subsequently OsWRKY90 (Shen et al., 2012) as its downstream target. In plant disease resistance networks, WRKY transcription factors can associate with MAPK cascades and regulate downstream defense-related genes in the nucleus (Pandey and Somssich, 2009; Ishihama et al., 2011; Jalmi and Sinha, 2016). Avirulent M. oryzae 007 infection significantly upregulated OsWRKY90 expression in rice DJ and 35S:OsMEK2 #4 leaf sheaths, but did not affect OsWRKY90 expression in ΔOsmek2 #2 leaf sheaths at all tested time points after inoculation (Figure 4). This indicates that OsMEK2, the rice MAP2K, targets the OsWRKY90 transcription factor to function as a positive regulator of resistance to M. oryzae infection. Rice plant resistance to M. oryzae infection is markedly enhanced by overexpression of OsWRKY45, OsWRKY53, and OsWRKY89 (Chujo et al., 2007; Shimono et al., 2007; Wang et al., 2007).
FIGURE 4.
Quantitative real-time RT-PCR analysis of time-course expression of the OsMEK2 interactors OsMPK1, OsMPK6, and OsWRKY90 in leaf sheaths of wild-type (WT) rice (cultivar DJ), ΔOsmek2 #2 and 35S:OsMEK2 #4 plants infected with avirulent Magnaporthe oryzae 007. Leaf sheaths of wild-type (cultivar DJ), ΔOsmek2 #2 and 35S:OsMEK2 #4 plants were sampled at different time points after inoculation, followed by total RNA extraction. Relative gene expression of OsMPK1, OsMPK6, and OsWRKY90 (Os09g30400) at each time point was obtained by normalizing with respect to the expression of the internal control OsUbiquitin (Os06g46770) gene. Data represent the means ± SD from three independent experiments. Different letters above the bars indicate significantly different means (P < 0.05), as analyzed by Fisher’s protected least significant difference (LSD) test.
We next investigated the expression patterns of some defense-related genes that are induced in response to M. oryzae 007 infection in rice, such as pathogenesis-related protein 1b (OsPR-1b), phenylalanine ammonia lyase1 (OsPAL1), ascorbate peroxidase1 (OsAPX1), and OsAPX2 (Nakashita et al., 2001; Agrawal et al., 2003; Xie et al., 2011). OsPR-1b expression was induced in rice DJ and 35S:OsMEK2 #4 at all tested times, whereas it was only induced in ΔOsmek2 #2 at 96 hpi (Supplementary Figure 6). OsPAL1 was distinctly induced in 35S:OsMEK2 #4 leaf sheaths during infection. OsPAL1 expression patterns did not significantly differ in rice DJ and ΔOsmek2 #2 leaf sheath cells at 12–96 hpi. OsAPX1 and OsAPX2 expression was gradually upregulated in ΔOsmek2 #2 leaf sheath cells at 12–72 hpi (Supplementary Figure 6). These results indicate that OsMEK2 expression positively regulates OsPR-1b and OsPAL1 expression in rice during M. oryzae infection.
OsMEK2 Is Required for ROS and Ferric Ion Accumulation and Lipid Peroxidation in Rice–M. oryzae Interactions
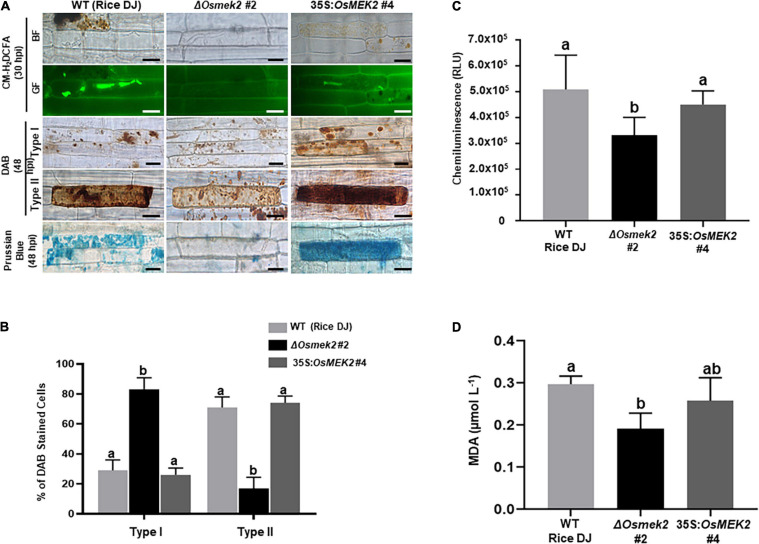
We analyzed ROS and ferric ion (Fe3+) accumulation and lipid [malondialdehyde (MDA)] peroxidation in leaf sheath cells of rice DJ, 35S:OsMEK2 #4 and ΔOsmek2 #2 plants during avirulent M. oryzae 007 infection to determine whether OsMEK2 is involved in iron- and ROS-dependent ferroptotic cell death (Figure 5). CM-H2DCFDA (green fluorescence) and DAB (dark brown) staining revealed that ROS (H2O2) strongly accumulated inside and around invasive hyphae (IH) in rice DJ and 35S:OsMEK2 #4 epidermal cells at 30–48 hpi (Figure 5A). By contrast, ROS did not accumulate around invasive hyphae (IH) in ΔOsmek2 #2 epidermal cells after avirulent M. oryzae 007 infection. The ROS-sensitive CM-H2DCFDA dye is an indicator that can be used to monitor ROS localization in living plant cells (Kristiansen et al., 2009). CM-H2DCFDA-specific ROS-localized fluorescence was clearly visible around invasive hyphae (IH) and cellular membranes in rice DJ and 35S:OsMEK2 #4 cells, whereas ROS-localized fluorescence was absent or weakly visible around invasive hyphae (IH) in ΔOsmek2 #2 cells at 30 hpi (Figure 5A). DAB is oxidized by H2O2 in the presence of peroxidase to generate a dark brown precipitate, which indicates the presence and distribution of H2O2 in plant cells (Fryer et al., 2002; Kristiansen et al., 2009). We classified DAB-stained cells into two phenotypes: Type I infected cells display no or weak DAB staining, and Type II infected cells display strong DAB staining (Figure 5B). Most of the infected cells displayed strong brown staining (Type II phenotype) in rice DJ and 35S:OsMEK2 #4 cells. By contrast, significantly fewer ΔOsmek2 #2 cells displayed DAB staining at 48 hpi. A chemiluminescent assay with a luminometer revealed that ROS levels were significantly lower in ΔOsmek2 #2 cells than in DJ and 35S:OsMEK2 #4 cells at 48 hpi (Figures 5A–C).
FIGURE 5.
OsMEK2 knock-out and overexpression in wild-type rice DJ differentially regulates ROS and ferric Ion (Fe3+) accumulation and lipid peroxidation in leaf sheaths infected with avirulent Magnaporthe oryzae 007. (A) CM-H2DCFDA (green fluorescence), DAB, and Prussian blue (blue color) staining shows accumulation of ROS (H2O2) and ferric ion (Fe3+) in rice leaf sheath epidermal cells of the wild-type (WT) rice cultivar DJ, OsMEK2 knock-out (ΔOsmek2 #2) and OsMEK2-overexpressed (35:OsMEK2 #4) plants during M. oryzae infection. Scale bar = 20 μm. (B) Quantification of DAB-stained cells at 48 h after inoculation. The DAB-stained cells were categorized into two phenotypes: Type I, infected cells that display no or weak DAB staining; and Type II, infected cells that display strong DAB staining. Results are presented as mean values ± SD; n = 4 leaf sheaths from different plants. (C) Quantification of ROS production in rice leaf sheaths at 48 h after inoculation. ROS production was quantified by a luminol-based assay using a GloMax® 96 Microplate Luminometer (Promega). Values are means ± SD of total relative luminescent units (RLU) (n = 10). (D) Lipid (MDA) peroxidation determination in rice leaf sheaths at 48 h after inoculation. Results are presented as mean values ± SD; n = 4 leaf sheaths from different plants. Images were captured using a fluorescence microscope (Zeiss equipped with Axioplan 2) with bright field and a combination of excitation (450–490 nm) and emission (515–565 nm) GF filters. Experiments were repeated three times with similar results. Different letters above the bars indicate significantly different means (P < 0.05), as analyzed by Fisher’s protected least significant difference (LSD) test. BF, bright field; GF, green fluorescence; hpi, hours post-inoculation; MDA, malondialdehyde.
Ferric ion (Fe3+) accumulation and localization in rice cells was detected by Prussian blue (blue color) staining of rice leaf sheath cells at 48 hpi with avirulent M. oryzae 007 (Figure 5A). Rice DJ and 35S:OsMEK2 #4 epidermal cells displayed strong blue staining, whereas ΔOsmek2 #2 epidermal cells did not display blue stain. Next, we analyzed oxidative damage and lipid (MDA) peroxidation in rice leaf sheath cells at 48 hpi with M. oryzae 007 (Figure 5D) by performing the MDA assay as described previously (Zhang et al., 2009; Dangol et al., 2019). Lipid peroxidation levels were significantly lower in ΔOsmek2 #2 cells than in rice DJ. The MDA level in 35S:OsMEK2 #4 was similar to that in rice DJ. These combined results indicate that OsMEK2 has crucial roles in ROS and Fe3+ accumulation and lipid peroxidation during the ferroptotic cell death response in rice.
Erastin Triggers Iron- and ROS-Dependent Ferroptotic Cell Death in ΔOsmek2 Knock-Out Mutant Plants During M. oryzae Infection
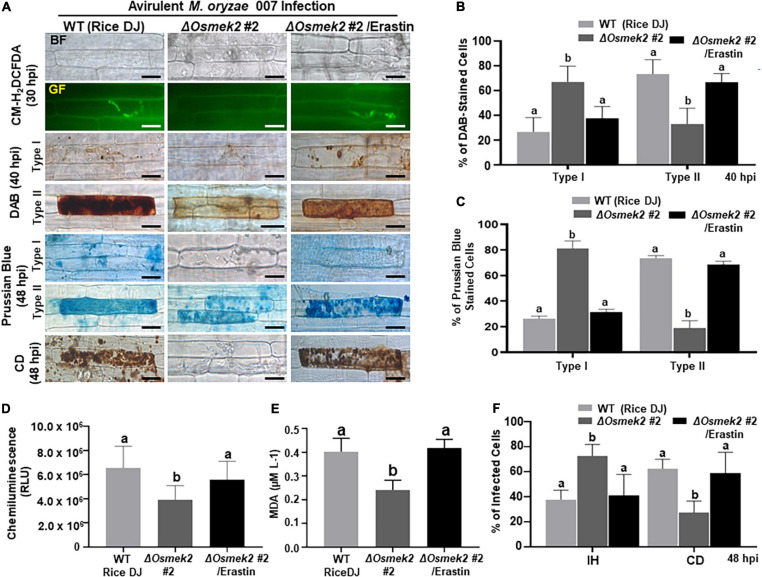
Erastin is a small molecule inducer that triggers ferroptotic cell death in mammals and plants (Dixon et al., 2012; Dangol et al., 2019). Treatment with 10 μM erastin triggered ROS (H2O2) and Fe3+ accumulation and cell death response in ΔOsmek2 #2 leaf sheaths during avirulent M. oryzae 007 infection (Figure 6). However, mock (water) or 10 μM erastin treatment did not trigger ROS (H2O2) and Fe3+ accumulation in healthy rice DJ leaf sheaths (Supplementary Figure 7). CM-H2DCFDA and DAB staining detected H2O2 accumulation in ΔOsmek2 #2 leaf sheath cells at 30∼48 hpi with M. oryzae conidial suspension containing 10 μM erastin (Figures 6A,B). Erastin treatment during M. oryzae 007 infection induced H2O2 accumulation in ΔOsmek2 #2 cells at 48 hpi as detected with a luminometer (Figure 6D). Erastin induced Fe3+ accumulation and increased the number of Prussian blue-stained cells in ΔOsmek2 #2 leaf sheaths at 48 hpi (Figures 6A,C). Iron-dependent MDA peroxidation was upregulated at 48 hpi in ΔOsmek2 #2 leaf sheath cells by treating with erastin (Figure 6E). Erastin treatment significantly enhanced the cell death response in ΔOsmek2 #2 cells during M. oryzae infection (Figures 6A,F). These combined results indicate that erastin triggers iron- and lipid ROS-dependent, but OsMEK2-independent, ferroptotic cell death in rice during M. oryzae infection.
FIGURE 6.
The small molecule inducer erastin triggers iron- and ROS-dependent ferroptotic cell death in the compatible ΔOsmek2–Magnaporthe oryzae interaction. Leaf sheaths of WT rice DJ and ΔOsmek2 #2 knock-out plants were inoculated with conidial suspensions (4 × 105 conidia/mL) of M. oryzae 007 containing 10 μM erastin. (A) Erastin treatment recovered ROS and ferric ion (Fe3+) accumulation and cell death in leaf sheaths of ΔOsmek2 #2 plants during avirulent M. oryzae 007 infection. ROS accumulation in leaf sheath epidermal cells was detected by CM-H2DCFDA (green fluorescence) and DAB (dark brown color) staining. Prussian blue (blue color) staining shows ferric ion accumulation in rice cells. The images are representative of different leaf sheath samples from three independent experiments. Scale bars = 20 μm. (B) DAB-stained cell phenotypes. DAB-stained cells were divided into two phenotypes: Type I, cells that contain invasive hyphae (IH) but are weakly or not DAB-stained; and Type II, strongly DAB-stained cells with only a few poor hyphae. (C) Quantification of Prussian blue-stained cells. Prussian blue-stained cells were divided into two phenotypes: Type I, cells that contain invasive hyphae (IH) but are weakly or not Prussian blue-stained; and Type II, strongly Prussian blue-stained with only a few poor hyphae. (D) Quantification of ROS accumulation. ROS accumulation was monitored using a GloMax® 96 Microplate Luminometer (Promega). Values are means ± SD of total relative luminescent units (RLU) (n = 10). (E) Determination of lipid (MDA) peroxidation in leaf sheaths at 48 h after inoculation. Results are presented as means ± SD; n = 4 leaf sheaths from different plants. (F) Quantification of infected cell phenotypes in rice leaf sheaths. Images were taken using a fluorescence microscope (Zeiss equipped with Axioplan 2) with bright field and green fluorescence (GF) filters. Experiments were repeated three times with similar results. Results are presented as means ± SD; n = 4 leaf sheaths. Different letters above the bars indicate significantly different means (P < 0.05), as analyzed by Fisher’s protected least significant difference (LSD) test. IH, invasive hyphae; CD, cell death; BF, bright field; GF, green fluorescence; hpi, hours post-inoculation.
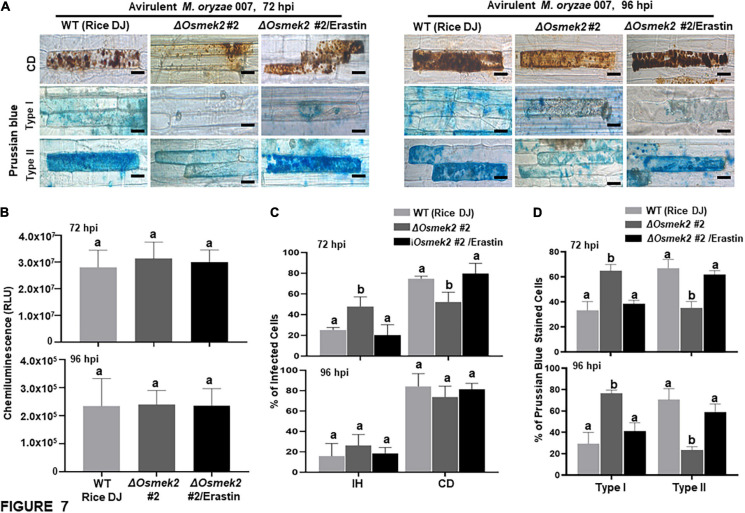
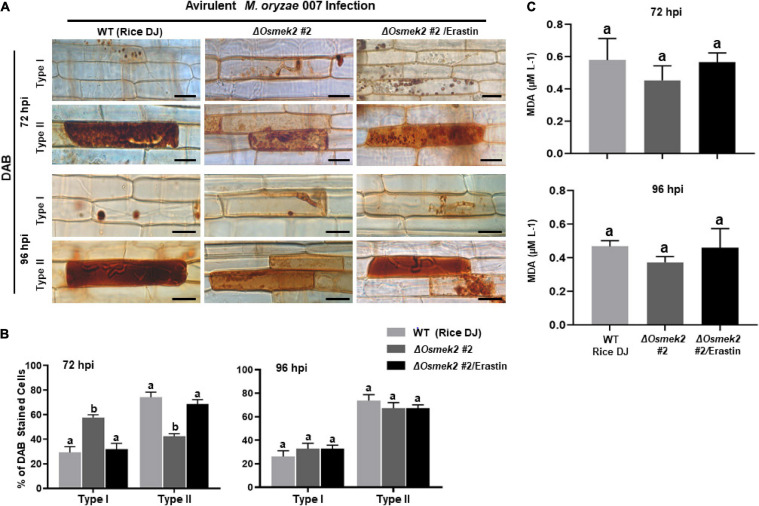
Disease-Related Cell Death Is ROS-Dependent but Iron-Independent in ΔOsmek2 Knock-Out Mutant Plants During the Late Stage of M. oryzae Infection
Reactive oxygen species (H2O2) and Fe3+ did not accumulate in healthy rice DJ leaf sheaths at 72 and 92 h after treatment with 10 μM erastin (Supplementary Figure 7). Erastin treatment strongly induced HR cell death, ROS and Fe3+ accumulation, and lipid peroxidation in ΔOsmek2 #2 leaf sheaths at 72 and 96 hpi with avirulent M. oryzae 007, similar to that observed in rice DJ (Figures 7, 8). By contrast, M. oryzae 007 infection induced disease-related cell death but not Fe3+ accumulation in erastin-untreated leaf sheaths of the susceptible ΔOsmek2 #2 cells at 72 and 96 hpi (Figures 7A,C,D). However, the chemiluminescent assay indicated that the high ROS levels observed in the ΔOsmek2 #2 cells were similar to those observed in rice DJ and erastin-treated ΔOsmek2 #2 leaf sheaths at 72 and 96 hpi (Figure 7B). ROS accumulation, MDA peroxidation, and cell death phenotypes were distinctly enhanced in the ΔOsmek2 #2 cells (Figures 7A,C, 8A–C); however, increased Fe3+ accumulation was not observed at 72 and 96 hpi (Figures 7A,D). Avirulent M. oryzae 007 infection did not increase the number of Prussian blue-stained cells in ΔOsmek2 #2 leaf sheaths at 72 and 96 hpi (Figure 7D). These combined results indicate that disease-related cell death is ROS-dependent but iron-independent in the compatible rice–M. oryzae interaction. Increased ROS production and lipid peroxidation in M. oryzae-infected tissues may induce susceptibility-related cell death that facilitates subsequent fungal invasion and infection. However, intracellular iron accumulation may not be required for disease-related cell death in compatible rice–M. oryzae interactions.
FIGURE 7.
Disease-related cell death is ROS-dependent and iron-independent in the compatible ΔOsmek2–Magnaporthe oryzae interaction. ΔOsmek2 #2 leaf sheaths were inoculated with conidial suspensions (4 × 105 conidia mL–1) of M. oryzae 007 containing 10 μM erastin. M. oryzae 007 infection induced disease-related cell death, but ΔOsmek2 #2 leaf sheath cells did not accumulate ferric ions (Fe3+) at 96 h after inoculation. (A) Erastin treatment induces cell death and ferric ion (Fe3+) accumulation in ΔOsmek2 #2 leaf sheath cells during avirulent M. oryzae 007 infection. Prussian blue (blue color) staining shows ferric ion accumulation in rice cells. The images are representative of different leaf sheath samples from three independent experiments. Scale bars = 20 μm. (B) Quantification of ROS accumulation in leaf sheath cells. ROS quantities were monitored using a GloMax® 96 Microplate Luminometer (Promega). Values are means ± SD of total relative luminescent units (RLU) (n = 10). (C) Quantification of infected cell phenotypes in rice leaf sheaths. Results are presented as mean values ± SD; n = 4 leaf sheaths from different plants. (D) Quantification of Prussian blue-stained cells. Prussian blue-stained cells were categorized into two phenotypes: Type I, cells that contain invasive hyphae (IH) but are weakly or not Prussian blue-stained; and Type II, strongly Prussian blue-stained with only a few poor hyphae. Images were taken using a fluorescence microscope (Zeiss equipped with Axioplan 2). Experiments were repeated three times with similar results. Results are presented as means ± SD; n = 4 leaf sheaths. Different letters above the bars indicate significantly different means (P < 0.05), as analyzed by Fisher’s protected LSD test. IH, invasive hyphae; CD, cell death; BF, bright field; GF, green fluorescence; hpi, hours post-inoculation.
FIGURE 8.
ROS accumulation and lipid peroxidation in leaf sheaths of wild-type (WT) rice (cultivar DJ) and ΔOsmek2 #2 plants at 72 and 96 h after inoculation with avirulent Magnaporthe oryzae 007 with 10 μM erastin. Rice ΔOsmek2 #2 leaf sheaths were inoculated with conidial suspensions (4 × 105 conidia mL–1) of avirulent M. oryzae 007 with and without 10 μM erastin. Avirulent M. oryzae 007 infection induced ROS production and lipid peroxidation in leaf sheaths of the susceptible ΔOsmek2 plants at 96 hpi, which was similar to those in erastin-treated leaf sheaths of the susceptible ΔOsmek2 #2 plants. (A) DAB-stained cell phenotypes at 72 and 96 h after inoculation. DAB-stained cells were categorized into two phenotypes: Type I, cells that contain invasive hyphae (IH) but are weakly or not DAB-stained; and Type II, strongly DAB-stained cells with only a few poor hyphae. Scale bars = 20 μm. (B) Quantification of DAB-stained cells at 72 and 96 h after inoculation. DAB-stained cells were categorized into two phenotypes: Type I, infected cells that display no or weak DAB staining; Type II, infected cells that display strong DAB staining. (C) Determination of lipid (MDA) peroxidation in rice leaf sheaths at 72 and 96 h after inoculation. Results are presented as mean values ± SD; n = 4 leaf sheaths from different plants. Images were captured using a fluorescence microscope (Zeiss equipped with Axioplan 2). Results are presented as mean values ± SD; n = 4 leaf sheaths from different plants. Different letters above the bars indicate significantly different means (P < 0.05) as analyzed by Fisher’s protected LSD test. Experiments were repeated three times with similar results. hpi, hours post-inoculation.
OsMPK1 Overexpression Induces Iron- and ROS-Dependent Ferroptotic Cell Death in Rice During M. oryzae Infection
Rice MAP kinase (OsMPK1) is an interactor of OsMEK2 and actively involved in M. oryzae infection (Singh et al., 2012; Ueno et al., 2015). Genomic DNA sequence of OsMPK1 contains six exons and five introns (Supplementary Figure 8). Amino acid sequence alignments of OsMPK1 with other plant MPKs indicated that OsMPK1 shares 67.95–96.20% homology with MAPKs of rice, Arabidopsis, tomato, and maize (Supplementary Figure 9). In particular, OsMPK1 has high levels of identity with OsMPK6 (96.20%) and AtMPK6 (83.76%) (Supplementary Figure 9). OsMPK1 was also phylogenetically close to OsMPK6 and AtMPK6 (Supplementary Figure 10). We overexpressed OsMPK1 in the susceptible rice cultivar Nipponbarre (NB) under the control of CaMV 35S promoter. OsMPK1 was distinctly overexpressed in leaf sheath cells of 35S:OsMPK1-transformed plants (Supplementary Figure 11). 35S:OsMPK1 overexpression induced a hypersensitive cell death with poorly grown invasive hyphae (IH) in leaf epidermal cells at 48 hpi (Figures 9A,B). DAB and CM-H2DCFDA staining showed accumulation of ROS around the IH in 35S:OsMPK1 leaf sheath cells at 36–48 hpi during virulent M. oryzae PO6-6 infection (Figures 9A,D). Chemiluminescence assay with a luminometer revealed that ROS levels distinctly increased in 35S:OsMPK1 overexpression cells at 48 hpi (Figure 9E). Prussian blue staining of Fe3+ showed strong accumulation of ferric ion in 35S:OsMPK1 cells at 48 hpi (Figures 9A,C). Lipid (MDA) peroxidation levels were significantly higher in 35S:OsMPK1 overexpression cells than in rice NB cells at 48 hpi (Figure 9F). By contrast, ferostatin-1(Fer-1) treatments distinctly inhibited iron- and ROS-dependent ferroptotic cell death during infection, which ultimately led to the restored normal hyphal growth in 35S:OsMPK1 overexpression cells (Figure 9). ROS and ferric ion accumulation and lipid peroxidation nearly disappeared in Fer-1-treated 35S:OsMPK1 leaf sheaths during infection (Figures 9A,B,D–F). The combined results indicate that OsMPK1 is involved in ROS and Fe3+ accumulation and lipid peroxidation leading to the ferroptotic cell death during M. oryzae infection.
FIGURE 9.
OsMPK1 overexpression induces ROS and ferric Ion (Fe3+) accumulation, lipid peroxidation and cell death in rice leaf sheaths during virulent Magnaporthe oryzae PO6-6 infection. Leaf sheaths of the susceptible wild-type (WT) cultivar NB and OsMPK1-overexpressed (35S:OsMPK1) plants were inoculated with the conidial suspension (4 × 105 conidia/mL) of virulent M. oryzae PO6-6 and then treated with 10 μM Fer-1. (A) Microscopic images of cell death and ROS and ferric Ion (Fe3+) accumulation in rice sheath cells untreated or treated with 10 μM Fer-1 at 48 hpi. ROS accumulation in the infected leaf sheath epidermal cells was detected by CM-H2DCFDA (green fluorescence) and DAB (dark brown color) staining. Prussian blue (blue color) staining shows ferric ion accumulation in rice cells. The images are representatives of different leaf sheath samples from three independent experiments. Scale bars = 20 μm. (B) Quantification of cell death (CD) and invasive hyphae (IH) in rice sheath cells at 48 hpi. (C) Quantification of Prussian blue-stained cells. Prussian blue-stained cells were divided into two phenotypes: Type I, cells that contain invasive hyphae (IH) but are weakly or not Prussian blue-stained; and Type II, strongly Prussian blue-stained with only a few poor hyphae. (D) DAB-stained cell phenotypes. DAB-stained cells were divided into two phenotypes: Type I, cells that contain invasive hyphae (IH) but are weakly or not DAB-stained; and Type II, strongly DAB-stained cells with only a few poor hyphae. (E) Quantification of ROS accumulation. ROS accumulation was monitored using a GloMax® 96 Microplate Luminometer (Promega). Values are means ± SD of total relative luminescent units (RLU) (n = 10). (F) Determination of lipid peroxidation by MDA (malondialdehyde) assay. Images were taken using a fluorescence microscope (Zeiss equipped with Axioplan 2) with bright field and green fluorescence (GF) filters. Experiments were repeated three times with similar results. Results are presented as mean values ± SD; n = 4 leaf sheaths from different plants. Different letters above the bars indicate significantly different means (P < 0.05) as analyzed by Fisher’s protected LSD test. Fer-1, ferrostatin-1, hpi, hours post-inoculation.
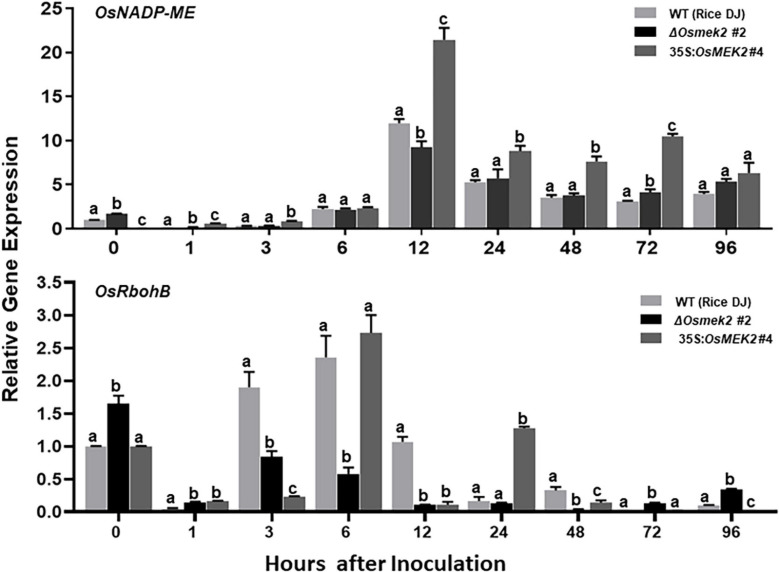
OsMEK2 Expression Positively Regulates OsNADP-ME and OsRbohB Expression in Rice During M. oryzae 007 Infection
We recently reported that rice NADP-malic enzyme (OsNADP-ME) and respiratory burst oxidase homolog (OsRboh, NADPH-oxidase) are involved in Fe3+ and ROS accumulation during cell death and defense responses in rice (Dangol et al., 2019). Interaction of N. benthamiana WRKY8 with MAPKs induce the downstream target genes NADP-ME and Rboh, resulting in the ROS burst (Yoshioka et al., 2003; Ishihama et al., 2011). RbohB activation via MAPK cascades is required for the pathogen-responsive ROS burst (Adachi and Yoshioka, 2015). Here, we analyzed the expression of OsNADP-ME2-3 (Singh et al., 2016) and OsRbohB (Wong et al., 2007) in wild-type rice DJ, ΔOsmek2 #2 knock-out and 35S:OsMEK2 #4 overexpression plants during avirulent M. oryzae 007 infection (Figure 10). OsNADP-ME2-3 expression patterns did not differ in rice DJ and ΔOsmek2 #2 plants, except for a reduction in ΔOsmek2 #2 plants at 12 hpi. However, OsNADP-ME2-3 was distinctly expressed in 35S:OsMEK2 #4 plants at 12 and 72 hpi. OsRbohB expression was significantly downregulated in ΔOsmek2 #2 plants, but distinctly upregulated at 96 hpi, compared to that in rice DJ (Figure 10). These combined results indicate that OsMEK2 expression positively regulates OsNADP-ME and OsRbohB expression during avirulent M. oryzae infection.
FIGURE 10.
Quantitative real-time RT-PCR analysis of time-course expression of OsNADP-ME and OsRbohB in rice leaf sheaths infected with avirulent Magnaporthe oryzae 007. Leaf sheaths of the wild-type (WT) rice cultivar DJ, OsMEK2 knock-out (ΔOsmek2 #2) and OsMEK2-overexpressed (35:OsMEK2 #4) plants were sampled at different time points after inoculation, followed by total RNA extraction. Relative gene expression levels of OsNADP-ME (Os01g52500) and OsRbohB (Os01g25820) at each time point were calculated by normalizing with respect to the expression of the internal control 18S rRNA (XR_003238819.1) gene. Data represent the means ± SD from three independent experiments. Different letters above the bars indicate significantly different means (P < 0.05), as analyzed by Fisher’s protected LSD test.
Subcellular Localization of OsMEK2, OsMPK1, and OsWRKY90
The subcellular localization study of MAP kinase signaling proteins is important for understanding their biological functions in plant cells. In this study, we investigated subcellular localization of green fluorescent protein (GFP)-tagged 35S:OsMEK2 (OsMEK2:GFP), OsMPK1:GFP, and OsWRKY90:GFP in N. benthamiana leaves using A. tumefaciens-mediated transient expression (Figure 11). The nuclei inside cells were counterstained with DAPI to help verify nuclear localization of GFP-tagged proteins. The control GFP construct (00:GFP) was ubiquitously detected in the cytoplasm of N. benthamiana cells. OsMEK2:GFP was localized mainly to the cytoplasm, but also to some nuclei in N. benthamiana cells. OsMPK1:GFP was localized to both the cytoplasm and nuclei. However, the OsWRKY90:GFP transcription factor was located inside the nuclei, but not in the cytoplasm. These results indicate that OsMEK2 interacts with OsMPK1 in the cytoplasm, and OsMPK1 moves into the nuclei to interact with the OsWRK90 transcription factor.
FIGURE 11.
Subcellular localization of OsMEK2, OsMPK1, and OsWRKY90 at 36 h after agroinfiltration into Nicotiana benthamiana leaves. 4′,6-diamidino-2-phenylindole (DAPI) staining was used to visualize nuclei in N. benthamiana epidermal cells. Images of subcellular localization of 00:GFP, OsMEK2:GFP, OsMPK1:GFP, and OsWRKY90:GFP were taken with a fluorescence microscope using bright field, GF (green fluorescence) and DAPI filters. GFP, green fluorescent protein. Scale bars = 50 μm.
Discussion
Plant mitogen-activated protein kinase (MAPK) cascades are involved in signaling multiple defense responses, the HR, and cell death responses during pathogen invasion and infection (Meng and Zhang, 2013; Thulasi Devendrakumar et al., 2018). We recently reported a ferroptotic cell death response in rice (Oryza sativa) during Magnaporthe oryzae infection (Dangol et al., 2019). Ferroptosis is a form of non-apoptotic iron-dependent cell death that was first discovered in oncogenic mammalian cells (Dixon et al., 2012). OsMEK2 interacts with OsMPK1 (Singh et al., 2012). Here, we demonstrated that rice MAP kinase (OsMEK2 and OsMPK1) signaling was required for iron- and ROS-dependent ferroptotic cell death in rice–M. oryzae interactions, and blast disease (susceptibility)-related cell death was ROS-dependent but iron-independent in the susceptible ΔOsmek2 mutant plants.
We previously reported that OsMEK2 physically interacts with and phosphorylates downstream OsMPK1 and OsMPK6 (Singh et al., 2012). MAPK kinase (MEK)–MAPK interactions may have functional roles in HR cell death responses and MAPK signaling networks during M. oryzae infection in rice plants. In the present study, OsMEK2 knock-out in rice DJ plants induced a susceptible (disease) response to M. oryzae infection in ΔOsmek2 knock-out plants; however, OsMEK2 overexpression in 35S:OsMEK2 plants redeemed hypersensitive cell death response against M. oryzae infection. These results suggest the HR-mediated resistance signaling of OsMEK2 during M. oryzae infection. OsMEK2 knock-out and overexpression differentially regulated OsMPK1, OsMPK6, and OsWRKY90 expression in ΔOsmek2 and 35S:OsMEK2 plants, especially during early stages of M. oryzae infection. These results indicated that OsMEK2 expression distinctly induced the downstream OsMPK1 and OsMPK6 signaling responses to M. oryzae infection. OsMPK1 physically interacts with the OsWRKY80 (Singh et al., 2012) and OsWRKY90 (Shen et al., 2012) transcription factors. In plant disease resistance networks, WRKY transcription factors can associate with MAP kinases in the nuclei and regulate downstream defense-related gene expression (Pandey and Somssich, 2009; Ishihama et al., 2011; Jalmi and Sinha, 2016). OsMEK2 expression distinctly induced pathogenesis-related protein 1b (OsPR-1b) but not ascorbate peroxidase1/2 (OsAPX1/2) during M. oryzae infection. PR-1 proteins are markers of defense responses to pathogen infection in rice (Mitsuhara et al., 2008). Thus, OsMEK2 signaling may trigger the MAP kinase cascade pathways leading to HR-mediated resistance to M. oryzae infection.
Mitogen-activated protein kinase signaling cascades are highly conserved in diverse plant species and involved in plant defense responses (Tanaka et al., 2009; Melech-Bonfil and Sessa, 2011; Oh et al., 2013; Ma et al., 2017). Activation of the MAPKK-MAPK cascades is associated with programmed cell death (PCD) in plants (Thulasi Devendrakumar et al., 2018). ROS mediate cellular defense responses against pathogen invasion in plants (Apel and Hirt, 2004; Mittler et al., 2004). Ferric ion (Fe3+) is essential in plants for HR cell death and defense- and disease-related iron homeostasis (Liu et al., 2007; Dangol et al., 2019). Iron is required for intracellular lipid peroxide accumulation (Stockwell et al., 2017; Dangol et al., 2019). Pathogen-responsive MAPKs may trigger the early ROS burst during plant defense and cell death responses (Meng and Zhang, 2013). Our study suggests that OsMEK2 activation is one of the earliest signaling events involved in iron- and ROS-dependent ferroptotic cell death in rice. Iron and ROS accumulation was not induced in ΔOsmek2 knock-out leaf sheaths during early M. oryzae infection. The ROS burst in rice may originate from the plasma membrane NADPH-oxidase (OsRbohB), which is activated during early M. oryzae infection. MAPKs could phosphorylate WRKY transcription factors to subsequently activate NADPH oxidases (Rbohs), which are essential for potent and prolonged ROS burst (Adachi et al., 2015; Jwa and Hwang, 2017). Early MAPK kinase (OsMEK2) signaling seems likely to activate OsMPK1, OsWRKY90, and NADPH-oxidase (OsRbohB), ultimately leading to the iron- and ROS-dependent ferroptotic cell death response. In our study, overexpression of 35S:OsMPK1 significantly induced iron and ROS accumulation during infection. However, the ferroptosis inhibitor, ferrostatin-1 (Fer-1), suppressed iron- and ROS-dependent ferroptotic cell death, which ultimately led to the restored normal invasive hyphal growth in 35S:OsMPK1 overexpression plants. Ma et al. (2017) demonstrated that overexpression of rice MPK6 (OsMPK1 in current study) reduced susceptibility in rice cultivar Zhonghua 11 against Xanthomonas oryzae pv. oryzae infection. Our results suggest that the OsMEK2-OsMPK1-OsWRKY90 cascades positively regulate ferroptotic cell death in rice against M. oryzae infection. Rice MPKK10.2 and MPK6 cascades induced resistance against X. oryzae pv. oryzae infection (Ma et al., 2017). Plant MAP kinases have been demonstrated to differentially regulate WRKY transcription factors in defense-related signaling pathways (Eulgem and Somssich, 2007; Pandey and Somssich, 2009; Ishihama and Yoshioka, 2012). Jalmi and Sinha (2016) reported positive involvement of the OsMKK3-OsMPK7-OsWRKY30 module in inducing rice resistance against X. oryzae infection. Rice OsMPK7 interacts with and phosphorylates OsWRKY30 to mediate resistance against X. oryzae infection (Jalmi and Sinha, 2016). Erastin is an oncogenic RAS-selective lethal (RSL) small molecule that effectively damages human cancer cells but does not affect isogenic normal cells (Dolma et al., 2003). Dixon et al. (2012) first discovered that erastin induces cellular iron-dependent lipid ROS accumulation in mammalian cells, leading to the unique iron-dependent non-apoptotic cell death (ferroptosis). The ferroptosis inducer erastin inhibits glutathione peroxidase 4 (GPX4) activity to elevate cytoplasmic lipid ROS levels (Yang and Stockwell, 2016; Stockwell et al., 2017). GPX4 is an inhibitor of lipid peroxidation (Ursini et al., 1982), and reduces membrane phospholipid hydroperoxides to suppress ferroptosis (Stockwell et al., 2017). In this study, we showed that erastin treatment of ΔOsmek2 knock-out mutant rice triggered iron and ROS accumulation and lipid peroxidation, leading to iron- and lipid ROS-dependent ferroptotic cell death during M. oryzae infection. The erastin-induced ferroptotic cell death in rice is iron- and lipid ROS-dependent, but is independent of the rice MAPK kinase OsMEK2. In our earlier study, we validated these results by showing that erastin treatment triggered OsMADP-ME2–independent ferroptotic cell death in rice (Dangol et al., 2019). NADP-ME provides the cytoplasmic electron donor NADPH for ROS production (Singh et al., 2016; Jwa and Hwang, 2017). Thus, erastin-mediated induction of ferroptotic cell death in rice may not require specific cell death-related plant genes, such as OsMEK2 and OsNADP-ME2. Plant and mammalian genes that are specifically regulated by erastin to trigger ferroptotic cell death have not yet been identified (Stockwell et al., 2017; Hirschhorn and Stockwell, 2019). Erastin-induced ferroptotic cell death in rice does not appear to be genetically controlled, but may occur non-specifically.
Disease-related cell death occurs during compatible (susceptible) interactions between plants and pathogens (Greenberg, 1997; Richberg et al., 1998). In this study, we showed that M. oryzae infection induced disease-related cell death that was not dependent on iron accumulation in rice cells at the late infection stage. ROS accumulation, lipid peroxidation, and cell death phenotypes distinctly increased in the ΔOsmek2 knock-out at the late M. oryzae infection stages (72 and 96 hpi); however, significant iron accumulation did not occur. Iron-independent and ROS-dependent cell death at late infection stages in the compatible rice–M. oryzae interaction is distinct from ferroptotic HR cell death, but similar to necrosis-like cell death. Necrotic cell death caused by compatible plant interactions with necrotrophic pathogens is dependent on ROS accumulation (Mengiste, 2012). A toxin or secreted virulence factor from the microbial pathogen may directly kill plant cells or trigger an endogenous cell death program (Greenberg, 1997). Cell death in compatible interactions may derive from pathogen-mediated necrosis rather than host-induced PCD (Morel and Dangl, 1997; Gilchrist, 1998). ROS accumulation and lipid peroxidation during M. oryzae infection at the late stages are involved in disease (susceptibility)-related cell death in rice, as suggested previously (Govrin and Levine, 2000; Greenberg and Yao, 2004; Torres et al., 2006; Choi et al., 2013; Jwa and Hwang, 2017). However, intracellular iron accumulation may not be required for disease-related cell death in compatible rice–M. oryzae interactions. By contrast, iron accumulation is likely essential for the induction of ferroptotic cell death to restrict avirulent M. oryzae invasion into rice cells.
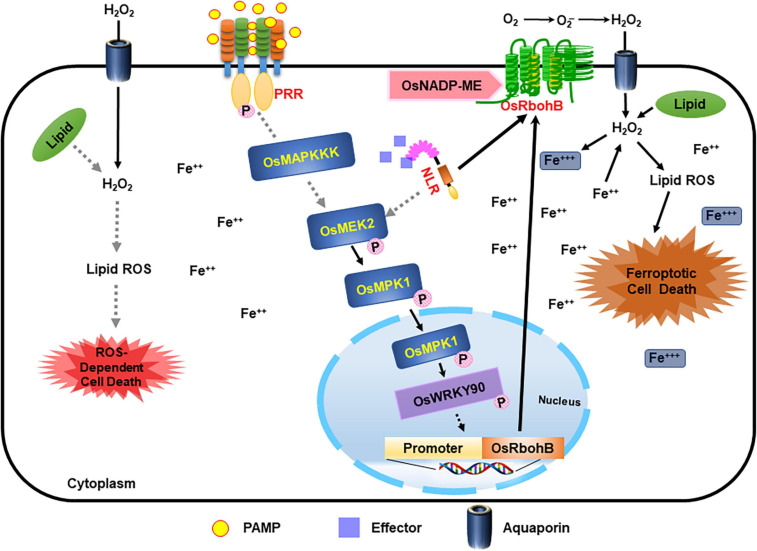
Here, we combine our cumulative data to propose the following working model: OsMEK2-OsMPK1-OsWRKY90 signaling positively regulates iron- and ROS-dependent ferroptotic HR cell death in rice–M. oryzae interactions (Figure 12). The invasion of avirulent M. oryzae 007 into rice cells activates rice MAP kinases (OsMEK2 and OsMPK1) via different hypothesized MAPK signaling pathways. The superoxide (O2–) produced from apoplastic oxygen (O2) can be converted to hydrogen peroxide (H2O2) by superoxide dismutase (SOD) (Figure 12; Marino et al., 2012; Kadota et al., 2015). Apoplastic ROS (H2O2) produced by plasma membrane-bound Rbohs during ETI migrates across the plasma membrane using aquaporin channels and into the cell (Bienert and Chaumont, 2014; Jwa and Hwang, 2017). The increased accumulation of iron (Fe3+) and lipid ROS triggers lipid peroxidation and subsequent ferroptotic cell death (Figure 12; Dixon et al., 2012; Stockwell et al., 2017; Dangol et al., 2019). The invasion of avirulent M. oryzae into rice cells activates rice MAP kinases (OsMEK2 and OsMPK1) via different hypothesized MAPK signaling pathways. The perception of PAMPs or pathogen effectors via membrane-bound PRRs or NLRs, respectively, activates OsMAP kinase cascades in rice cells, as proposed previously (Jones and Dangl, 2006; Zipfel, 2008). MAP kinase kinases (MEKs) activate MAP kinases, which migrate from the cytoplasm to the nucleus and regulate transcriptional reprogramming (Morris, 2001; Ahlfors et al., 2004). OsMEK2 interacts with and phosphorylates downstream rice MAP kinase 1 (OsMPK1) and MAP kinase 6 (OsMPK6) in the cytoplasm (Singh et al., 2012). OsMPK1 moves from the cytoplasm into the nucleus to interact with the OsWRKY90 transcription factor (Figure 11; Singh et al., 2012). OsMEK2 expression triggers OsMPK1-OsWRKY90 signaling pathways in the nucleus (Figure 11), which may lead to the upregulation of OsNADP-malic enzyme and rice NADPH-oxidase B (OsRbohB) (Figure 12). The MAPK-WRKY pathway activates Rbohs, leading to a prolonged and robust ROS burst (Adachi et al., 2015). The de novo synthesis of OsRbohB and its trafficking to the plasma membrane is involved in iron- and ROS-dependent ferroptotic death in rice cells. Rice MAP kinase 1 (OsMPK1) also may target OsWRKY90 to bind to specific sequences of some defense-related genes such as OsPR-1b. Disease (susceptibility)-related cell death is lipid ROS-dependent, but iron-independent, in the compatible rice–M. oryzae interaction (Figure 12). Iron accumulation may not mediate disease-related cell death in rice. Iron-independent disease-related cell death is likely a necrosis-type cell death that is distinct from ferroptotic cell death in incompatible rice–M. oryzae interactions. However, iron- and ROS-dependent ferroptotic cell death in rice is a generally regulated form of cell death that is common in incompatible rice–M. oryzae interactions (Dangol et al., 2019). Iron accumulation in ferroptotic cells may be harmful to the invaded hyphae of avirulent M. oryzae. Further research on the functions of this ROS-dependent disease-related cell death is required to determine how virulent M. oryzae infection suppresses ferroptotic cell death and induces disease-related cell death, which may be beneficial for pathogen growth in planta.
FIGURE 12.
Proposed model of rice MAP kinase kinase 2 (OsMEK2) signaling pathways leading to iron- and ROS-dependent ferroptotic cell death in incompatible rice–Magnaporthe oryzae interactions. M. oryzae invasion in rice cells activates rice MAP kinase kinase 2 (OsMEK2) via different hypothesized signaling pathways (gray dotted lines). The perception of PAMPs or pathogen effectors via membrane-bound PRRs or NLRs, respectively, activates MAP kinases in plant cells. Active OsMEK2 triggers OsMPK1-OsWRKY90 pathways in the nucleus, which may lead to upregulation of NADP-malic enzyme (ME) and NADPH-oxidase (OsRbohB). The de novo synthesis of OsRbohB and its trafficking to the plasma membrane contributes to iron- and ROS-dependent ferroptotic death in rice cells. Disease (susceptibility)-related cell death is ROS-dependent and iron-independent in the compatible ΔOsmek2–M. oryzae interaction. MEK, mitogen-activated protein kinase kinase; MPK, mitogen-activated protein kinase; NADP-ME, NADP-malic enzyme; NLR, nucleotide-binding leucine-rich repeat; PAMP, pathogen-associated molecular pattern; PRR, pattern recognition receptor; WRKY, tryptophan (W), arginine (R), lysine (K), tyrosine (Y) transcription factor.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
N-SJ designed the research. SD, NKN, RS, YC, JW, and H-GL carried out all the experiments. SD, NKN, BKH, and N-SJ analyzed the data and wrote the manuscript. BKH and N-SJ reviewed and edited the final manuscript. All authors read and agreed to the published version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Funding. This work was performed with the support of the Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ015966012021), Rural Development Administration, Republic of Korea. This research was also supported by the National Research Foundation (NRF) of Korea, the Korea Government (Grant No. 2019R1F1A1059830).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.710794/full#supplementary-material
Amino acid sequence alignment and phylogenetic tree of rice MAPKKs (OsMEKs) with Arabidopsis MAPKKs. (A) Amino acid sequence alignment of OsMEKs with Arabidopsis MAPKKs. Rice MAPKKs are aligned with Arabidopsis MAPKKs, which are categorized into four groups (Group A–D) using Clustal Omega (EMBL-EBI). The MAPKK active site [D(I/L/V)K] and conserved domain [S/T-X-S/T] are located between kinase subdomains VII and VIII. (B) Phylogenetic tree of OsMEKs with Arabidopsis MAPKKs was constructed using the neighbor-joining method based on Molecular Evolutionary Genetics Analysis Version 7.0 (MEGA7) (Kumar et al., 2016). Accession numbers of the plant MAPKKs are OsMEK1 (Os01g32660), OsMEK2 (Os06g05520), OsMEK3 (Os03g12390), OsMEK4 (Os02g46760), OsMEK5 (Os06g09190), OsMEK6 (Os02g54600), OsMEK7 (Os06g09180), OsMEK8 (Os06g27890), OsWNK1 (Os07g38530), AtMKK1 (At4g26070), AtMKK2 (At4g29810), AtMEK3 (NP_198860), AtMEK4 (At1g51660), AtMEK5 (At3g21220), AtMKK6 (At5g56580), AtMKK9 (At1g73500), and AtWNK9 (At3g04910).
Amino acid sequence alignment of rice MAPKKs (OsMEKs) with Arabidopsis, tomato, and tobacco MEKs. Amino acid sequence of OsMEK2 was aligned with MAP kinase kinases of Arabidopsis, tomato, and tobacco using CLUSTAL OMEGA (EMBL-EBI). Accession numbers of the aligned plant MAPKKs are OsMEK1 (Os01g32660), OsMEK2 (Os06g05520), OsMEK3 (Os03g12390), OsMEK6 (Os02g54600), OsMEK8 (Os06g27890), AtMKK1 (At4g26070), AtMKK2 (At4g29810), AtMEK3 (NP_198860), AtMEK4 (At1g51660), AtMEK5 (At3g21220), SlMKK2 (NP_001234588), NtMEK2 (AF325168), and NbMEK2 (LOC107818847). Asterisks and dots at the bottom of sequences indicate identical and similar amino acids, respectively. Domain numbers (I∼XI) on the top of sequences indicate conserved subdomains. The conserved consensus motif (GXGXXG) in conserved subdomain I is boxed. The active motif [D(I/L/V)K] and the conserved consensus motif [S/TXXXXXS/T, serine(S)/threonine (T) residues] between conserved subdomains VII and VIII are shown in yellow.
Circular phylogenetic tree of rice MAPKKs (OsMEKs) with Arabidopsis, tomato, and tobacco MAPKs. Phylogenetic tree was constructed using the neighbor-joining method based on Molecular Evolutionary Genetics Analysis Version 7.0 (MEGA7) (Kumar et al., 2016). Accession numbers of the plant MAPKKs are OsMEK1 (Os01g32660), OsMEK2 (Os06g05520), OsMEK3 (Os03g12390), OsMPK8a (Os06g27890), AtMEK3 (NP_198860), AtMEK4 (At1g51660), AtMEK5 (At3g21220), AtMKK1 (At4g26070), AtMKK2 (At4g29810), SlMKK1 (NP_001234744), SlMKK2 (NP_001234588), NtMEK2 (AF325168), NtNPK2 (BAA06731), and NbMEK2 (LOC107818847).
Nucleotide sequences and deduced amino acid sequences of rice MAP kinase kinase 2 (OsMEK2) genomic DNA. Small letters represent nucleotide sequences of exons and introns. Capital letters represent deduced amino acid sequences of exons. Initiation and termination codons of the OsMEK2 coding region are represented by asterisks. Exon-intron splice junctions (gt/ag) are represented by bold letters. Numbers at the right refer to nucleotide and amino acid (in parentheses) residue positions in the respective sequence.
Avirulent Magnaporthe oryzae 007 infection causes susceptible responses in ΔOsmek2 knock-out lines, but resistant responses in the wild-type rice DJ and 35S:OsMEK2 overexpression lines. Rice leaf sheaths were inoculated with a conidial suspension (4 × 105 conidia mL–1). (A) Images of rice sheath epidermal cells infected with M. oryzae 007 (48 hpi). M. oryzae 007 grew well and produced invasive hyphae in the ΔOsmek2 #2 and #4 knock-out plants, but induced hypersensitive cell death in wild-type (WT) rice DJ and 35S:OsMEK2 #4 and #6 overexpression plants. Images were captured using a fluorescence microscope. hpi, hours post-inoculation. Scale bars = 20 μm. (B) Quantification of invasive hyphae (IH) and cell death (CD) in rice sheath cells infected with M. oryzae 007 (48 hpi). Results are presented as mean values ± SD; n = 4 leaf sheaths from different plants. Different letters above the bars indicate significantly different means (P < 0.05), as analyzed by Fisher’s protected least significant difference (LSD) test. IH, invasive hyphae; CD, cell death.
Quantitative real-time RT-PCR analysis of time-course expression of defense-response genes OsPR-1b, OsPAL1, OsAPX1, and OsAPX2 in wild-type (WT) rice (cultivar DJ), ΔOsmek2 #2, and 35S:OsMEK2 #4 plants during Magnaporthe oryzae 007 infection. Leaf sheaths of wild-type (cultivar DJ), ΔOsmek2 #2, and 35S:OsMEK2 #4 rice plants were sampled at different time points after inoculation, followed by total RNA extraction. Relative gene expression levels of defense-responsive genes OsPR-1b (Os01g28450), OsPAL1 (Os04g43760), OsAPX1 (Os0.g17690), and OsAPX2 (Os07g49400) at each time point were obtained by normalizing the gene expression with respect to expression of the internal control 18S rRNA gene. Data represent the means ± SD from three independent experiments. Different letters above the bars indicate significantly different means (P < 0.05) as analyzed by Fisher’s protected LSD test.
Images of Prussian blue-stained and DAB-stained leaf sheath cells of wild-type (WT) rice (cultivar DJ) at different time points after erastin treatment. Rice DJ leaf sheaths were treated with mock (water) and 10 μM erastin. Images were taken using a fluorescence microscope (Zeiss equipped with Axioplan 2). Prussian blue and DAB staining in erastin-treated rice leaf sheaths did not detect ferric ion and ROS accumulation, respectively, at different time points after treatment. Experiments were repeated three times with similar results. hpt, hours post-treatment. Scale bars = 20 μm.
Nucleotide sequences and deduced amino acid sequences of rice MAP kinase 1 (OsMPK1) genomic DNA. Small letters represent nucleotide sequences of exons and introns. Capital letters represent deduced amino acid sequences of exons. Initiation and termination codons of the OsMPK1 coding region are represented by asterisks. Numbers at the right refer to nucleotide and amino acid (in parentheses) residue positions in the respective sequence.
Amino acid sequence alignment of rice MAP kinase 1 (OsMPK1) with other plant MPKs. Amino acid sequence of OsMPK1 was aligned with plant MAP kinases using CLUSTAL OMEGA (EMBL-EBI). Accession numbers of the aligned plant MPKs are OsMPK1 (Os06g06090), OsMPK6 (Os10g38950), AtMPK6 (At2g43790), AtMPK3 (At3g45640), AtMPK4 (At4g01370), AtMPK11 (At1g01560), NbMKK1 (BAE95414), LeMPK1 (NP_001234011), LeMPK2 (NP_001234355), LeMPK3 (NP_001234360), and ZmMPK5 (PW05027).
Phylogenetic tree of rice MAP kinase 1 (OsMPK1) with plant MPKs. Accession numbers: MPKs are OsMPK1 (Os06g06090), OsMPK6 (Os10g38950), AtMPK6 (At2g43790), AtMPK3 (At3g45640), AtMPK4 (At4g01370), AtMPK11 (At1g01560), NbMKK1 (BAE95414), LeMPK1 (NP_001234011), LeMPK2 (NP_001234355), LeMPK3 (NP_001234360), and ZmMPK5 (PW05027).
Transcriptional analysis of OsMPK1 expression in leaf sheath epidermal cells of wild-type (WT) rice (cultivar NB) and 35S:OsMPK1 plants using qRT-PCR. Relative gene expression of OsMPK1 was obtained by normalizing with respect to the expression of the internal control OsUbiquitin gene. Data represent the means ± SD from three independent experiments. Asterisks indicate statistically significant differences (Student’s t-test, P < 0.01).
Primers used in this study.
References
- Adachi H., Nakano T., Miyagawa N., Ishihama N., Yoshioka M., Katou Y., et al. (2015). WRKY transcription factors phosphorylated by MAPK regulate a plant immune NADPH oxidase in Nicotiana benthamiana. Plant Cell 27 2645–2663. 10.1105/tpc.15.00213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi H., Yoshioka H. (2015). Kinase-mediated orchestration of NADPH oxidase in plant immunity. Brief. Funct. Genomics 14 253–259. 10.1093/bfgp/elv004 [DOI] [PubMed] [Google Scholar]
- Agrawal G. K., Jwa N. S., Iwahashi H., Rakwal R. (2003). Importance of ascorbate peroxidases OsAPX1 and OsAPX2 in the rice pathogen response pathways and growth and reproduction revealed by their transcriptional profiling. Gene 322 93–103. 10.1016/j.gene.2003.08.017 [DOI] [PubMed] [Google Scholar]
- Ahlfors R., Macioszek V., Rudd J., Brosché M., Schlichting R., Scheel D., et al. (2004). Stress hormone-independent activation and nuclear translocation of mitogen-activated protein kinases in Arabidopsis thaliana during ozone exposure. Plant J. 40 512–522. 10.1111/j.1365-313X.2004.02229 [DOI] [PubMed] [Google Scholar]
- Apel K., Hirt H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55 373–399. 10.1146/annurev.arplant.55.031903.141701 [DOI] [PubMed] [Google Scholar]
- Asai T., Tena G., Plotnikova J., Willmann M. R., Chiu W. L., Gomez-Gomez L., et al. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415 977–983. 10.1038/415977a [DOI] [PubMed] [Google Scholar]
- Aznar A., Chen N. W. G., Thomine S., Dellagi A. (2015). Immunity to plant pathogens and iron homeostasis. Plant Sci. 240 90–97. 10.1016/j.plantsci.2015.08.022 [DOI] [PubMed] [Google Scholar]
- Bienert G. P., Chaumont F. (2014). Aquaporin-facilitated transmembrane diffusion of hydrogen peroxide. Biochim. Biophys. Acta 1840 1596–1604. 10.1016/j.bbagen.2013.09.017 [DOI] [PubMed] [Google Scholar]
- Block A., Alfano J. R. (2011). Plant targets for Pseudomonas syringae type III effectors: virulence targets or guarded decoys? Curr. Opin. Microbiol. 14 39–46. 10.1016/j.mib.2010.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T., He S. Y. (2009). Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324 742–744. 10.1126/science.1171647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caseys C. (2019). Ferroptosis: a companion of ROS in fighting Magnaporthe in rice. Plant Cell 31 13–14. 10.1105/tpc.18.00970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K., Torres N. L., Subramanyam S., Deepak S. A., Sardesai N., Han O. (2006). Protein extraction/solubilization protocol for monocot and dicot plant gel-based proteomics. J. Plant Biol. 49 413–420. 10.1007/bf03031120 [DOI] [Google Scholar]
- Choi D. S., Hwang I. S., Hwang B. K. (2012). Requirement of the cytosolic interaction between pathogenesis-related protein10 and leucine-rich repeat protein1 for cell death and defense signaling in pepper. Plant Cell 24 1675–1690. 10.1105/tpc.112.095869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H. W., Kim D. S., Kim N. H., Jung H. W., Ham J. H., Hwang B. K. (2013). Xanthomonas filamentous hemagglutinin-like protein Fha1 interacts with pepper hypersensitive-induced reaction protein CaHIR1 and functions as a virulence factor in host plants. Mol. Plant Microbe Interact. 26 1441–1454. 10.1094/MPMI-07-13-0204-R [DOI] [PubMed] [Google Scholar]
- Chujo T., Takai R., Akimoto-Tomiyama C., Ando S., Minami E., Nagamura Y., et al. (2007). Involvement of the elicitor-induced gene OsWRKY53 in the expression of defense-related genes in rice. Biochim. Biophys. Acta 1769 497–505. 10.1016/j.bbaexp.2007.04.006 [DOI] [PubMed] [Google Scholar]
- Dangol S., Chen Y., Hwang B. K., Jwa N. S. (2019). Iron- and reactive oxygen species-dependent ferroptotic cell death in rice-Magnaporthe oryzae interactions. Plant Cell 31 189–209. 10.1105/tpc.18.00535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangol S., Singh R., Chen Y., Jwa N. S. (2017). Visualization of multicolored in vivo organelle markers for co-localization studies in Oryza sativa. Mol. Cells 40 828–836. 10.14348/molcells.2017.0045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon S. J., Lemberg K. M., Lamprecht M. R., Skouta R., Zaitsev E. M., Gleason C. E., et al. (2012). Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149 1060–1072. 10.1016/j.cell.2012.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P. N., Rathjen J. P. (2010). Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11 539–548. 10.1038/nrg2812 [DOI] [PubMed] [Google Scholar]
- Dolma S., Lessnick S. L., Hahn W. C., Stockwell B. R. (2003). Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell 3 285–296. 10.1016/s1535-6108(03)00050-3 [DOI] [PubMed] [Google Scholar]
- Eulgem T., Somssich I. E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10 366–371. 10.1016/j.pbi.2007.04.020 [DOI] [PubMed] [Google Scholar]
- Fryer M. J., Oxborough K., Mullineaux P. M., Baker N. R. (2002). Imaging of photo-oxidative stress responses in leaves. J. Exp. Bot. 53 1249–1254. 10.1093/jexbot/53.372.1249 [DOI] [PubMed] [Google Scholar]
- Fu Z. Q., Dong X. N. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64 839–863. 10.1146/annurev-arplant-042811-105606 [DOI] [PubMed] [Google Scholar]
- Gilchrist D. G. (1998). Programmed cell death in plant disease: the purpose and promise of cellular suicide. Annu. Rev. Phytopathol. 36 393–414. 10.1146/annurev.phyto.36.1.393 [DOI] [PubMed] [Google Scholar]
- Govrin E., Levine A. (2000). The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 10 751–757. 10.1016/s0960-9822(00)00560-1 [DOI] [PubMed] [Google Scholar]
- Greenberg J. T. (1997). Programmed cell death in plant–pathogen interactions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 525–545. 10.1146/annurev.arplant.48.1.525 [DOI] [PubMed] [Google Scholar]
- Greenberg J. T., Yao N. (2004). The role and regulation of programmed cell death in plant-pathogen interactions. Cell Microbiol. 6 201–211. 10.1111/j.1462-5822.2004.00361 [DOI] [PubMed] [Google Scholar]
- Hamel L. P., Nicole M. C., Sritubtim S., Morency M. J., Ellis M., Ehlting J., et al. (2006). Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 11 192–198. 10.1016/j.tplants.2006.02.007 [DOI] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6 192–198. [DOI] [PubMed] [Google Scholar]
- Hirschhorn T., Stockwell B. R. (2019). The development of the concept of ferroptosis. Free Radic. Biol. Med. 133 130–143. 10.1016/j.freeradbiomed.2018.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekema A., Hirsch P. R., Hooykaas P. J. J., Schilperoort R. A. (1983). A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 303 179–180. 10.1038/303179a0 [DOI] [Google Scholar]
- Ichimura K., Shinozaki K., Tena G., Sheen J., Henry Y., Champion A., et al. (2002). Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 7 301–308. 10.1016/S1360-1385(02)02302-6 [DOI] [PubMed] [Google Scholar]
- Ishihama N., Yamada R., Yoshioka M., Katou S., Yoshioka H. (2011). Phosphorylation of the Nicotiana benthamiana WRKY8 transcription factor by MAPK functions in the defense response. Plant Cell 23 1153–1170. 10.1105/tpc.110.081794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama N., Yoshioka H. (2012). Post-translational regulation of WRKY transcription factors in plant immunity. Curr. Opin. Plant Biol. 15 431–437. 10.1016/j.pbi.2012.02.003 [DOI] [PubMed] [Google Scholar]
- Jalmi S. K., Sinha A. K. (2016). Functional involvement of a mitogen activated protein kinase module, OsMKK3-OsMPK7-OsWRK30 in mediating resistance against Xanthomonas oryzae in rice. Sci. Rep. 6:37974. 10.1038/srep37974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J. S., Lee S., Jung K. H., Jun S. H., Jeong D. H., Lee J., et al. (2000). T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 22 561–570. 10.1046/j.1365-313x.2000.00767 [DOI] [PubMed] [Google Scholar]
- Jin H., Axtell M. J., Dahlbeck D., Ekwenna O., Zhang S., Staskawicz B., et al. (2002). NPK1, an MEKK1-like mitogen-activated protein kinase kinase kinase, regulates innate immunity and development in plants. Dev. Cell 3 291–297. 10.1016/S1534-5807(02)00205-8 [DOI] [PubMed] [Google Scholar]
- Jin H., Liu Y., Yang K. Y., Kim C. Y., Baker B., Zhang S. (2003). Function of a mitogen-activated protein kinase pathway in N gene-mediated resistance in tobacco. Plant J. 33 719–731. 10.1046/j.1365-313X.2003.01664.x [DOI] [PubMed] [Google Scholar]
- Jones J. D., Dangl J. L. (2006). The plant immune system. Nature 444 323–329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Jwa N. S., Hwang B. K. (2017). Convergent evolution of pathogen effectors toward reactive oxygen species signaling networks in plants. Front. Plant Sci. 8:1687. 10.3389/fpls.2017.01687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y., Shirasu K., Zipfel C. (2015). Regulation of the NADPH oxidase RBOHD d during plant immunity. Plant Cell Physiol. 56 1472–1480. 10.1093/pcp/pcv063 [DOI] [PubMed] [Google Scholar]
- Khokhlatchev A. V., Canagarajah B., Wilsbacher J., Robinson M., Atkinson M., Goldsmith E., et al. (1998). Phosphorylation of the MAP kinase ERK2 promotes its homodimerization and nuclear translocation. Cell 93 605–615. 10.1016/s0092-8674(00)81189-7 [DOI] [PubMed] [Google Scholar]
- Kristiansen K. A., Jensen P. E., Møller I. M., Schulz A. (2009). Monitoring reactive oxygen species formation and localization in living cells by use of the fluorescent probe CM-H2DCFDA and confocal laser microscopy. Physiol. Plant. 136 369–383. 10.1111/j.1399-3054.2009.01243.x [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Kim S. H., Jung Y. H., Kim J. A., Lee M. O., Choi P. G., et al. (2005). Molecular cloning and functional analysis of rice (Oryza sativa L.) OsNDR1 on defense signaling pathway. Plant Pathol. J. 21 149–157. 10.5423/ppj.2005.21.2.149 [DOI] [Google Scholar]
- Li X., Zhang Y., Huang L., Ouyang Z., Hong Y., Zhang H., et al. (2014). Tomato SlMKK2 and SlMKK4 contribute to disease resistance against Botrytis cinerea. BMC Plant Biol. 14:166. 10.1186/1471-2229-14-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Greenshields D. L., Sammynaiken R., Hirji R. N., Selvaraj G., Wei Y. (2007). Targeted alterations in iron homeostasis underlie plant defense responses. J. Cell Sci. 120 596–605. 10.1242/jcs.001362 [DOI] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Dinesh-Kumar S. P. (2004). Involvement of MEK1 MAPKK, NTF6 MAPK, WRKY/MYB transcription factors, COI1 and CTR1 in N-mediated resistance to tobacco mosaic virus. Plant J. 38 800–809. 10.1111/j.1365-313X.2004.02085.x [DOI] [PubMed] [Google Scholar]
- Ma H., Chen J., Zhang Z., Ma L., Yang Z., Zhang Q., et al. (2017). MAPK kinase 10.2 promotes disease resistance and drought tolerance by activating different MAPKs in rice. Plant J. 92 557–570. 10.1111/tpj.13674 [DOI] [PubMed] [Google Scholar]
- Marino D., Dunand C., Puppo A., Pauly N. (2012). A burst of plant NADPH oxidases. Trends Plant Sci. 17 9–15. 10.1016/j.tplants.2011.10.001 [DOI] [PubMed] [Google Scholar]
- Melech-Bonfil S., Sessa G. (2011). The SlMKK2 and SlMPK2 genes play a role in tomato disease resistance to Xanthomonas campestris pv. vesicatoria. Plant Signal. Behav. 6 154–156. 10.4161/psb.6.1.14311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Zhang S. (2013). MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51 245–266. 10.1146/annurev-phyto-082712-102314 [DOI] [PubMed] [Google Scholar]
- Mengiste T. (2012). Plant immunity to necrotrophs. Annu. Rev. Phytopathol. 50 267–294. 10.1146/annurev-phyto-081211-172955 [DOI] [PubMed] [Google Scholar]
- Mitsuhara I., Iwai T., Seo S., Yanagawa Y., Kawahigasi H., Hirose S., et al. (2008). Characteristic expression of twelve rice PR1 family genes in response to pathogen infection, wounding, and defense-related signal compounds (121/180). Mol. Genet. Genomics 279 415–427. 10.1007/s00438-008-0322-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Gollery M., Breusegem F. V. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9 490–498. 10.1016/j.tplants.2004.08.009 [DOI] [PubMed] [Google Scholar]
- Morel J. B., Dangl J. L. (1997). The hypersensitive response and the induction of cell death in plants. Cell Death Differ. 4 671–683. 10.1038/sj.cdd.4400309 [DOI] [PubMed] [Google Scholar]
- Morris P. C. (2001). MAP kinase signal transduction pathways in plants. New Phytol. 151 67–89. 10.1046/j.1469-8137.2001.00167 [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Suzuki T., Murata S., Nakamura S., Hino T., Maeo K., et al. (2007). Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71 2095–2100. 10.1271/bbb.70216 [DOI] [PubMed] [Google Scholar]
- Nakashita H., Yoshioka K., Takayama M., Kuga R., Midoh N., Usami R., et al. (2001). Characterization of PBZ1, a probenazole-inducible gene, in suspension-cultured rice cells. Biosci. Biotechnol. Biochem. 65 205–208. 10.1271/bbb.65.205 [DOI] [PubMed] [Google Scholar]
- Oh C. S., Hwang J. N., Choi M. S., Kang B. C., Martin G. B. (2013). Two leucines in the N-terminal MAPK-docking site of tomato SlMKK2 are critical for interaction with a downstream MAPK to elicit programmed cell death associated with plant immunity. FEBS Lett. 587 1460–1465. 10.1016/j.febslet.2013.03.033 [DOI] [PubMed] [Google Scholar]
- Oh C. S., Martin G. B. (2011). Effector-triggered immunity mediated by the Pto kinase. Trends Plant Sci. 16 132–140. 10.1016/j.tplants.2010.11.001 [DOI] [PubMed] [Google Scholar]
- Pandey S. P., Somssich I. E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150 1648–1655. 10.1104/pp.109.138990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke A., Djamei A., Bitton F., Hirt H. (2009). A major role of the MEKK1–MKK1/2–MPK4 pathway in ROS signalling. Mol. Plant 2 120–137. 10.1093/mp/ssn079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao K. P., Richa T., Kumar K., Raghuram B., Sinha A. K. (2010). In silico analysis reveals 75 members of mitogen-activated protein kinase kinase kinase gene family in rice. DNA Res. 17 139–153. 10.1093/dnares/dsq011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M. W., Roux M., Petersen M., Mundy J. (2012). MAP kinase cascades in Arabidopsis innate immunity. Front. Plant Sci. 3:169. 10.3389/fpls.2012.00169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyna N. S., Yang Y. (2006). Molecular analysis of the rice MAP kinase gene family in relation to Magnaporthe grisea infection. Mol. Plant Microbe Interact. 19 530–540. 10.1094/MPMI-19-0530 [DOI] [PubMed] [Google Scholar]
- Richberg M. H., Aviv D. H., Dangl J. L. (1998). Dead cells do tell tales. Curr. Opin. Plant Biol. 1 480–485. 10.1016/s1369-5266(98)80039-3 [DOI] [PubMed] [Google Scholar]
- Rodriguez M. C. S., Petersen M., Mundy J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61 621–649. 10.1146/annurev-arplant-042809-112252 [DOI] [PubMed] [Google Scholar]
- Sainsbury F., Lomonossoff G. P. (2008). Extremely high-level and rapid transient protein production in plants without the use of viral replication. Plant Physiol. 148 1212–1218. 10.1104/pp.108.126284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger B., Ronald P. C. (2012). Plant innate immunity: perception of conserved microbial signatures. Annu. Rev. Plant Biol. 63 451–482. 10.1146/annurev-arplant-042811-105518 [DOI] [PubMed] [Google Scholar]
- Shen H., Liu C., Zhang Y., Meng X., Zhou X., Chu C., et al. (2012). OsWRKY30 is activated by MAP kinases to confer drought tolerance in rice. Plant Mol. Biol. 80 241–253. 10.1007/s11103-012-9941-y [DOI] [PubMed] [Google Scholar]
- Shimono M., Sugano S., Nakayama A., Jiang C. J., Ono K., Toki S., et al. (2007). Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 19 2064–2076. 10.1105/tpc.106.046250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R., Berg R. H., Schachtman D. P. (2005). Reactive oxygen species and root hairs in Arabidopsis root response to nitrogen, phosphorus and potassium deficiency. Plant Cell Physiol. 46 1350–1357. 10.1093/pcp/pci145 [DOI] [PubMed] [Google Scholar]
- Singh R., Dangol S., Chen Y., Choi J., Cho Y. S., Lee J. E., et al. (2016). Magnaporthe oryzae effector Avr-pii helps to establish compatibility by inhibition of the rice NADP-malic enzyme resulting in disruption of oxidative burst and host innate immunity. Mol. Cells 39 426–438. 10.14348/molcells.2016.0094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Jwa N. S. (2013). The rice MAPKK-MAPK interactome: the biological significance of MAPK components in hormone signal transduction. Plant Cell Rep. 32 923–931. 10.1007/s00299-013-1437-y [DOI] [PubMed] [Google Scholar]
- Singh R., Lee M. O., Lee J. E., Choi J., Park J. H., Kim E. H., et al. (2012). Rice mitogen-activated protein kinase interactome analysis using the yeast two-hybrid system. Plant Physiol. 160 477–487. 10.1104/pp.112.200071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockwell B. R., Friedmann Angeli J. P., Bayir H., Bush A. I., Conrad M., Dixon S. J., et al. (2017). Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell 171 273–285. 10.1016/j.cell.2017.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J., Yang L., Zhu Q., Wu H., He Y., Liu Y., et al. (2018). Active photosynthetic inhibition mediated by MPK3/MPK6 is critical to effector-triggered immunity. PLoS Biol. 16:e2004122. 10.1371/journal.pbio.2004122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Ishihama N., Yoshioka H., Huser A., O’Connell R., Tsuji G., et al. (2009). The Colletotrichum orbiculare SSD1 mutant enhances Nicotiana benthamiana basal resistance by activating a mitogen-activated protein kinase pathway. Plant Cell 21 2517–2526. 10.1105/tpc.109.068023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thulasi Devendrakumar K., Li X., Zhang Y. (2018). MAP kinase signalling: interplays between plant PAMP- and effector-triggered immunity. Cell. Mol. Life Sci. 75 2981–2989. 10.1007/s00018-018-2839-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M. A., Jones J. D. G., Dangl J. L. (2006). Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141 373–378. 10.1104/pp.106.079467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Sato M., Stoddard T., Glazebrook J., Katagiri F. (2009). Network properties of robust immunity in plants. PLoS Genet. 5:e1000772. 10.1371/journal.pgen.1000772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno Y., Yoshida R., Kishi-Kaboshi M., Matsushita A., Jiang C. J., Goto S., et al. (2015). Abiotic stresses antagonize the rice defence pathway through the tyrosine-dephosphorylation of OsMPK6. PLoS Pathog. 11:e1005231. 10.1371/journal.ppat.1005231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursini F., Maiorino M., Valente M., Ferri L., Gregolin C. (1982). Purifcation from pig liver of a protein which protects liposomes and biomembranes from peroxidative degradation and exhibits glutathione peroxidase activity on phosphatidylcholine hydroperoxides. Biochim. Biophys. Acta 710 197–211. 10.1016/0005-2760(82)90150-3 [DOI] [PubMed] [Google Scholar]
- Wang H., Hao J., Chen X., Hao Z., Wang X., Lou Y., et al. (2007). Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol. Boil. 65 799–815. 10.1007/s11103-007-9244-x [DOI] [PubMed] [Google Scholar]
- Wong H. L., Pinontoan R., Hayashi K., Tabata R., Yaeno T., Hasegawa K., et al. (2007). Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell 19 4022–4034. 10.1105/tpc.107.055624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X. Z., Xue Y. J., Zhou J. J., Zhang B., Chang H., Takano M. (2011). Phytochromes regulate SA and JA signaling pathways in rice and are required for developmentally controlled resistance to Magnaporthe grisea. Mol. Plant 4 688–696. 10.1093/mp/ssr005 [DOI] [PubMed] [Google Scholar]
- Yang K. Y., Liu Y., Zhang S. (2001). Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc. Natl. Acad Sci. 98 741–746. 10.1073/pnas.98.2.741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. S., Stockwell B. R. (2016). Ferroptosis: death by lipid peroxidation. Trends Cell Biol. 26 165–176. 10.1016/j.tcb.2015.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Ma H., Hong H., Yao W., Xie W., Xiao J., et al. (2015). Transcriptome-based analysis of mitogen-activated protein kinase cascades in the rice response to Xanthomonas oryzae infection. Rice 8:4. 10.1186/s12284-014-0038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka H., Numata N., Nakajima K., Katou S., Kawakita K., Rowland O., et al. (2003). Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 15 706–718. 10.1105/tpc.008680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Zhou R. G., Gao Y. J., Zheng S. Z., Xu P., Zhang S. Q., et al. (2009). Molecular and genetic evidence for the key role of AtCaM3 in heat-shock signal transduction in Arabidopsis. Plant Physiol. 149 1773–1784. 10.1104/pp.108.133744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C. (2008). Pattern-recognition receptors in plant innate immunity. Curr. Opin. Immunol. 20 10–16. 10.1016/j.coi.2007.11.003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amino acid sequence alignment and phylogenetic tree of rice MAPKKs (OsMEKs) with Arabidopsis MAPKKs. (A) Amino acid sequence alignment of OsMEKs with Arabidopsis MAPKKs. Rice MAPKKs are aligned with Arabidopsis MAPKKs, which are categorized into four groups (Group A–D) using Clustal Omega (EMBL-EBI). The MAPKK active site [D(I/L/V)K] and conserved domain [S/T-X-S/T] are located between kinase subdomains VII and VIII. (B) Phylogenetic tree of OsMEKs with Arabidopsis MAPKKs was constructed using the neighbor-joining method based on Molecular Evolutionary Genetics Analysis Version 7.0 (MEGA7) (Kumar et al., 2016). Accession numbers of the plant MAPKKs are OsMEK1 (Os01g32660), OsMEK2 (Os06g05520), OsMEK3 (Os03g12390), OsMEK4 (Os02g46760), OsMEK5 (Os06g09190), OsMEK6 (Os02g54600), OsMEK7 (Os06g09180), OsMEK8 (Os06g27890), OsWNK1 (Os07g38530), AtMKK1 (At4g26070), AtMKK2 (At4g29810), AtMEK3 (NP_198860), AtMEK4 (At1g51660), AtMEK5 (At3g21220), AtMKK6 (At5g56580), AtMKK9 (At1g73500), and AtWNK9 (At3g04910).
Amino acid sequence alignment of rice MAPKKs (OsMEKs) with Arabidopsis, tomato, and tobacco MEKs. Amino acid sequence of OsMEK2 was aligned with MAP kinase kinases of Arabidopsis, tomato, and tobacco using CLUSTAL OMEGA (EMBL-EBI). Accession numbers of the aligned plant MAPKKs are OsMEK1 (Os01g32660), OsMEK2 (Os06g05520), OsMEK3 (Os03g12390), OsMEK6 (Os02g54600), OsMEK8 (Os06g27890), AtMKK1 (At4g26070), AtMKK2 (At4g29810), AtMEK3 (NP_198860), AtMEK4 (At1g51660), AtMEK5 (At3g21220), SlMKK2 (NP_001234588), NtMEK2 (AF325168), and NbMEK2 (LOC107818847). Asterisks and dots at the bottom of sequences indicate identical and similar amino acids, respectively. Domain numbers (I∼XI) on the top of sequences indicate conserved subdomains. The conserved consensus motif (GXGXXG) in conserved subdomain I is boxed. The active motif [D(I/L/V)K] and the conserved consensus motif [S/TXXXXXS/T, serine(S)/threonine (T) residues] between conserved subdomains VII and VIII are shown in yellow.
Circular phylogenetic tree of rice MAPKKs (OsMEKs) with Arabidopsis, tomato, and tobacco MAPKs. Phylogenetic tree was constructed using the neighbor-joining method based on Molecular Evolutionary Genetics Analysis Version 7.0 (MEGA7) (Kumar et al., 2016). Accession numbers of the plant MAPKKs are OsMEK1 (Os01g32660), OsMEK2 (Os06g05520), OsMEK3 (Os03g12390), OsMPK8a (Os06g27890), AtMEK3 (NP_198860), AtMEK4 (At1g51660), AtMEK5 (At3g21220), AtMKK1 (At4g26070), AtMKK2 (At4g29810), SlMKK1 (NP_001234744), SlMKK2 (NP_001234588), NtMEK2 (AF325168), NtNPK2 (BAA06731), and NbMEK2 (LOC107818847).
Nucleotide sequences and deduced amino acid sequences of rice MAP kinase kinase 2 (OsMEK2) genomic DNA. Small letters represent nucleotide sequences of exons and introns. Capital letters represent deduced amino acid sequences of exons. Initiation and termination codons of the OsMEK2 coding region are represented by asterisks. Exon-intron splice junctions (gt/ag) are represented by bold letters. Numbers at the right refer to nucleotide and amino acid (in parentheses) residue positions in the respective sequence.
Avirulent Magnaporthe oryzae 007 infection causes susceptible responses in ΔOsmek2 knock-out lines, but resistant responses in the wild-type rice DJ and 35S:OsMEK2 overexpression lines. Rice leaf sheaths were inoculated with a conidial suspension (4 × 105 conidia mL–1). (A) Images of rice sheath epidermal cells infected with M. oryzae 007 (48 hpi). M. oryzae 007 grew well and produced invasive hyphae in the ΔOsmek2 #2 and #4 knock-out plants, but induced hypersensitive cell death in wild-type (WT) rice DJ and 35S:OsMEK2 #4 and #6 overexpression plants. Images were captured using a fluorescence microscope. hpi, hours post-inoculation. Scale bars = 20 μm. (B) Quantification of invasive hyphae (IH) and cell death (CD) in rice sheath cells infected with M. oryzae 007 (48 hpi). Results are presented as mean values ± SD; n = 4 leaf sheaths from different plants. Different letters above the bars indicate significantly different means (P < 0.05), as analyzed by Fisher’s protected least significant difference (LSD) test. IH, invasive hyphae; CD, cell death.
Quantitative real-time RT-PCR analysis of time-course expression of defense-response genes OsPR-1b, OsPAL1, OsAPX1, and OsAPX2 in wild-type (WT) rice (cultivar DJ), ΔOsmek2 #2, and 35S:OsMEK2 #4 plants during Magnaporthe oryzae 007 infection. Leaf sheaths of wild-type (cultivar DJ), ΔOsmek2 #2, and 35S:OsMEK2 #4 rice plants were sampled at different time points after inoculation, followed by total RNA extraction. Relative gene expression levels of defense-responsive genes OsPR-1b (Os01g28450), OsPAL1 (Os04g43760), OsAPX1 (Os0.g17690), and OsAPX2 (Os07g49400) at each time point were obtained by normalizing the gene expression with respect to expression of the internal control 18S rRNA gene. Data represent the means ± SD from three independent experiments. Different letters above the bars indicate significantly different means (P < 0.05) as analyzed by Fisher’s protected LSD test.
Images of Prussian blue-stained and DAB-stained leaf sheath cells of wild-type (WT) rice (cultivar DJ) at different time points after erastin treatment. Rice DJ leaf sheaths were treated with mock (water) and 10 μM erastin. Images were taken using a fluorescence microscope (Zeiss equipped with Axioplan 2). Prussian blue and DAB staining in erastin-treated rice leaf sheaths did not detect ferric ion and ROS accumulation, respectively, at different time points after treatment. Experiments were repeated three times with similar results. hpt, hours post-treatment. Scale bars = 20 μm.
Nucleotide sequences and deduced amino acid sequences of rice MAP kinase 1 (OsMPK1) genomic DNA. Small letters represent nucleotide sequences of exons and introns. Capital letters represent deduced amino acid sequences of exons. Initiation and termination codons of the OsMPK1 coding region are represented by asterisks. Numbers at the right refer to nucleotide and amino acid (in parentheses) residue positions in the respective sequence.
Amino acid sequence alignment of rice MAP kinase 1 (OsMPK1) with other plant MPKs. Amino acid sequence of OsMPK1 was aligned with plant MAP kinases using CLUSTAL OMEGA (EMBL-EBI). Accession numbers of the aligned plant MPKs are OsMPK1 (Os06g06090), OsMPK6 (Os10g38950), AtMPK6 (At2g43790), AtMPK3 (At3g45640), AtMPK4 (At4g01370), AtMPK11 (At1g01560), NbMKK1 (BAE95414), LeMPK1 (NP_001234011), LeMPK2 (NP_001234355), LeMPK3 (NP_001234360), and ZmMPK5 (PW05027).
Phylogenetic tree of rice MAP kinase 1 (OsMPK1) with plant MPKs. Accession numbers: MPKs are OsMPK1 (Os06g06090), OsMPK6 (Os10g38950), AtMPK6 (At2g43790), AtMPK3 (At3g45640), AtMPK4 (At4g01370), AtMPK11 (At1g01560), NbMKK1 (BAE95414), LeMPK1 (NP_001234011), LeMPK2 (NP_001234355), LeMPK3 (NP_001234360), and ZmMPK5 (PW05027).
Transcriptional analysis of OsMPK1 expression in leaf sheath epidermal cells of wild-type (WT) rice (cultivar NB) and 35S:OsMPK1 plants using qRT-PCR. Relative gene expression of OsMPK1 was obtained by normalizing with respect to the expression of the internal control OsUbiquitin gene. Data represent the means ± SD from three independent experiments. Asterisks indicate statistically significant differences (Student’s t-test, P < 0.01).
Primers used in this study.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.