Abstract
Background
The prevalence of non-tuberculous mycobacteria (NTM) has been increasing worldwide in both developed and developing countries. NTM infection is clinically indistinguishable from tuberculosis and therefore poses significant challenges in patient management, especially in patients chronically treated for pulmonary TB. In this study, we evaluated a new highly sensitive Multiplex MTB/NTM assay that can differentiate M. tuberculosis complex (MTBC) from all NTM, including the treatable and most common NTM, M. avium complex (MAC).
Methods
We developed and optimized a new open- Multiplex MTB/NTM assay with two gene-targets for MTBC (IS6110/senX3-regX3) and two targets for MAC (IS1311/DT1) with samples spiked with stored strains and testing 20 replicates. Patients with presumptive TB and NTM were enrolled at the Respiratory Disease Department of The University Teaching Hospital of Point G, in Mali.
Findings
In the development stage, the new assay showed a high analytic performance with 100% detections of MTBC and MAC at only 5 colony forming units (CFUs). Overall, without the treatment failure cases, the Multiplex assay and the Xpert showed a sensitivity, specificity, PPV and NPV of 83·3% [66·4-92·6], 96·6% [88·6-99·0], 92·5% [82·3-96·5] and 92·2% [82·7-96·5] and the Xpert had values of 96·7% [83·3-99·4], 80·0% [68·2-88·1], 70·7 [55·5-82·3] and 97·9% [89·3-99·6], respectively. The Multiplex assay successfully detected all (5/5) the MAC cases.
Interpretation
Our new Multiplex assay demonstrates better specificity than Xpert for all group studied, in addition to detecting potential NTM cases. The assay could therefore complement the widely used Xpert assay and enhance discrimination of TB and NTM infections.
Funding
This work was supported by the National Institutes of Health (R03AI137674, U54EB027049, D43TW010350 and UM1AI069471) and Northwestern University's Institute for Global Health Catalyzer Fund.
Keywords: Tuberculosis, Nontuberculous mycobacteria, Simultaneous diagnosis
Research in context.
Evidence before this study
Tuberculosis (TB) is still a major public health problem in low resource settings. The prevalence of non-tuberculous mycobacteria (NTM) has also been increasing worldwide in both developed and developing countries. NTM infection is clinically indistinguishable from TB and poses significant challenges to patient management. The sputum smear microscopy, a widely used diagnostic test for TB, for more than a century, fails to detect up to half of the TB cases, and is unable to differentiate between mycobacterial species. The World Health Organization recommends use of Xpert MTB/RIF (Xpert) assay; however, Xpert is less sensitive than sputum culture and does not detect NTM infections.
Added value of this study
In this study, we developed and tested a new open-platform Multiplex MTB/NTM assay to simultaneously detect MTB, Mycobacterium avium complex (MAC) and other NTM infections. The new assay has demonstrated a complementarity with the Xpert assay, with a sensitivity and specificity of 83·3% [66·4-92·6] and 96·6% [88·6-99·0] for the Multiplex compared with 96·7% [83·3-99·4] and 80% [55·5-82·3] for the Xpert assay. In addition, the Multiplex MTB/NTM assay offers an extra value of detecting MAC and other NTM infections, an option that is not available with the Xpert assay. The Xpert assay, however, has the advantage of providing a rifampin resistance profile. The two assays are therefore highly complementary in patients with TB/NTM-like symptoms.
Implications of all the available evidence
Evidence shows that our new Multiplex MTB/NTM assay has good analytic and clinical performance for detection of TB and NTM infections. This assay complements current diagnostic tools for patients with TB/NTM infection-like symptoms, saves time and resources by its multi-detection nature and, more importantly, by detecting MAC and others NTM infections, will reduce mortality in TB endemic countries and worldwide.
Alt-text: Unlabelled box
1. Introduction
Worldwide, tuberculosis (TB) is the leading single pathogen affecting humans, with deaths greater even than from HIV/AIDS [1]. The genus Mycobacterium includes nearly 200 recognized tuberculous and non-tuberculous mycobacteria (NTM). Some NTM are either strictly or potentially pathogenic, while others are opportunistic or saprophyte non-pathogenic and associated with asymptomatic carriage.[2] The Mycobacterium tuberculosis complex (MTBC) species including Mycobacterium (M.) tuberculosis, M. africanum, M. bovis, M. cannetti, M. caprae, M. microti, and M. pinnipedi are known to cause all type of TB infections, although substantial differences in virulence have been reported between species.[3] Nontuberculous mycobacteria (NTM) represent a major public health threat due to the wide variety of species and the increasing number of people at risk, including the elderly, those with a history of lung disease, and immunocompromised individuals.[4] There are over 140 NTM species reported in the literature, of which 25 species have been strongly associated with NTM diseases.[5] Mycobacterium avium complex (MAC) is the most frequently associated with pulmonary disease, causing up to half of NTM cases worldwide.[6], [7], [8]
In developing countries where TB is endemic, our group and others have shown that failure to recognize NTM infections commonly leads to misdiagnosis of lung disease presenting with non-specific symptoms like coughs, fever and weight loss.[9,10] This results in inappropriate empiric treatment that is lengthy, expensive, toxic, and may result in the development TB drug resistance.[9,11] In Mali, we recently reported that NTM infections was identified in 18% of clinically classified chronic TB cases.[9,12] We also reported that 12% of all patients in Mali with presumptive TB are in fact infected with NTM.[9] Nearly 73% of the NTM cases, are caused by Mycobacterium avium complex (MAC).[9] The same epidemiological pattern has been reported in others TB endemic setting.[7]
Sputum smear microscopy (SSM), the most widely used TB diagnostic tool for more than a century and is relatively easy to perform. However, has only 50-80% sensitivity and cannot differentiate between mycobacterial species. While sputum culture (the gold standard) is very sensitive, it is time-consuming, requiring 10-42 days and needs a sophisticated infrastructure that is not typically available in the resource-limited settings where TB is endemic.[13,14] In light of these factors, the World Health Organization (WHO) endorsed use of the Xpert MTB/RIF (Xpert) molecular assay in developing countries. However, while Xpert provide essential information on rifampin resistance, it is less sensitive than TB sputum culture and cannot distinguish NTM infections.[15] Furthermore, the Xpert assay has limited performance for TB smear negatives and paediatric specimens (around 67% sensitivity).[16] Additionally, 57% of TB cases are bacteriologically-confirmed; the remaining are usually treated empirically.[17] These cases include Xpert-negative patients and patients with paucibacillary TB, such as children and people living with HIV (most of whom are sputum smear negatives).[18,19] This study evaluated the performance of a new highly sensitive quantitative polymerase chain reaction (qPCR) assay for simultaneous detection of MTBC and NTM in patients with clinically presumptive pulmonary tuberculosis. We hypothesized that the Multiplex assay may complement the current algorithm of diagnostic of patients with TB/NTM infection-like symptoms.
2. Methods
2.1. Ethics
The study was approved by the Ethics Committee of the University of Sciences, Technics and Technologies of Bamako (USTTB), under the study number 2017/161CE-FMPOS and the Institutional Review Board of Northwestern University (NU) under study STU00206642. Informed consent was obtained from all participants.
2.2. Participants and study design
The Multiplex MTB/NTM assay was first developed at the Center for Innovation in Global Health Technologies (CIGHT) at NU, including primer design, selection and optimization. A clinical feasibility study was conducted at the University Clinical Research Center (UCRC) in Mali from April 2017 to March 2020. Clinically presumptive TB patients with at least 18 years old were from the six referral health centres in Bamako, Mali. Patients were referred to the Respiratory Disease Department at the University Teaching Hospital of Point-G where samples were obtained. Concentrated smear microscopy and sputum culture were performed in the UCRC Biosafety Level-3 (BSL-3) TB Laboratory and, the Multiplex RT-PCR MTB/NTM assay and Xpert assays were performed in UCRC molecular biology Laboratory.
2.3. Multiplex assay development
2.2.1. qPCR MTB/NTM Primers’ selection and assay design
Two M. tuberculosis target regions that we previously validated and have shown a clinical sensitivity of 95% or more, and a specificity of 100% compared to sputum culture and the Xpert assay.[20,21] These two target regions are preferred because the multi-copy of the Insertion Sequence (IS) 6110 and single-copy of target senX3-regX3 are highly conserved amongst MTBC species, but, more importantly, are not present in NTM species.[20,21] To add the MAC detection panel to the assay, we selected the IS1311 gene that has also been previously shown to be present in all MAC species except the M. intracellulare.[22,23] A second MAC target, DT1 gene, was added in which M. intracellulare (does not have the IS1311), M. avium subsp. avium (serotypes 1, 2 and 3) and M. avium subsp. silvaticuma are present.[23] The combination of these two gene target regions has been shown to cover and identify all MAC species but none of the MTBC species.[22,23] The Pan-Mycobacterium 16S rRNA gene that is present in all mycobacterial species was included as a third panel to identify Mycobacterium that are not MAC or MTBC. The negative control was composed of the reaction mixture only without any additional nucleic acid. The cotJC gene DNA fragments (Synthetic gBlocks) from Integrated DNA Technologies (IDT, Coralville, IA, USA) was added as the extraction and amplification controls, as previously shown.[20] In addition, a pre-extracted DNA mixture from MTBC and MAC strains also served as the assay reaction positive controls. All the designed primers (at least five for each gene) were crosschecked with related organisms that we had previously stored (clinical isolates), and the best ones in Multiplex format were selected for the final assay design and the analytic performance evaluation.
2.2.2. Results interpretation
A test was considered positive for MTBC if it was positive for ISI6110/senX3-regX3 genes, Pan-Mycobacterium, and for the positive controls (extraction/tube and assay controls), but negative for the negative control and MAC (Table 1). A test was considered positive for MAC if it was positive for IS1311/DT1 genes and positive for the Pan-Mycobacterium and positive controls, but negative for the negative control and MTBC. A test was considered positive for the “other NTM” panel if it was negative for MTBC, MAC and the negative control, but positive for Pan-Mycobacterium and positive controls. A negative test result was one negative for all the targets except the positive controls only. Any other combination of positive and negative results was considered inconclusive (Table 1).
Table 1.
Interpretation Criteria of the Multiplex MTB/NTN Assay.
| Identification | Criteria |
|||||
|---|---|---|---|---|---|---|
| ISI6110/senX3-regX3 | IS1311 +DT1 | Pan-Mycobacterium | Extraction control: cotJC gene | Positive controls MTBC/MAC | Negative control | |
| M. tuberculosis complex | + | - | + | + | + | - |
| M. avium complex | - | + | + | + | + | - |
| Other NTM | - | - | + | + | + | - |
| Negative results | - | - | - | + | + | - |
2.2.3. Analytic evaluation of the multiplex assay
This evaluation was conducted at CIGHT in Evanston using a Qiagen Rotor-Gene Q qPCR instrument. Stored uninfected sputum samples were spiked with mycobacterial DNA for MTB, MAC and other NTM, and were used to assess the designed primers and optimize the Multiplex assay using different concentrations corresponding to 50 CFUs/mL, 20 CFUs/mL, 10 CFUs/mL, 5 CFUs/mL and 1 CFU/mL in order to determine the limits of detection (Table 2). Twenty (20) samples were used for each dilution to determine the analytical performance and the detection limits of the assay. Further optimizations were performed in Mali on the Applied Biosystems™ 7500 Fast Dx Real-Time PCR Instrument using clinical isolates that were molecularly confirmed by AccuprobeTM (GenProbe Inc, San Diego, CA, USA) prior to the start of the clinical feasibility evaluation. The fully characterized isolates included M. tuberculosis H37Rv, M. avium, M. scrofulaceum, M. smegmatis, M. intracellulare, M. fortuitum, M. gordonae, M. kansasii, M. bovis, M. chelonae, M. palustre, M. simiae, M. cubicae.
Table 2.
Analytic Performance and Limits of Detection of the New Multiplex MTB/NTM Assay.
| CFU/mL | Detection of MTBC and MAC |
||
|---|---|---|---|
| ISI6110 + senX3-regX3 for MTBC | IS1311 + DT1 for MAC | Pan-Mycobacterium for all Mycobacteria | |
| 50 | 20/20 | 20/20 | 20/20 |
| 20 | 20/20 | 20/20 | 15/20 |
| 10 | 20/20 | 20/20 | 12/20 |
| 5 | 20/20 | 20/20 | 0/20 |
| 1 | 0/20 | 0/20 | 0/20 |
MTBC: Mycobacterium tuberculosis complex, MAC: Mycobacterium avium complex
2.4. Clinical Feasibility Study: Data Collection and Management
After signing an IRB-approved informed consent form, new clinically TB presumptive patients which are 18 years old and more. To be enrolled patient can be chronically and empirically treated for TB with for more than six months with first-line drugs, and clinically presumptive NTM patients, were enrolled in the study. The control subjects were recruited from the same health care centres as tested patients and 5 mL sputum was collected from each as for the other groups. These controls were symptomatic patients presenting with presumptive TB but ended up being sputum culture negative. Two sputum samples and blood samples for HIV testing were obtained from each participant. A standardized questionnaire was used to collect participant's socio-demographic and clinical information. All data including laboratory results were recorded into a secure web-based data management platform, REDCap.
2.5. Statistics
The data were exported to and analysed with Stata 17® and Epi info version 7.2. The sensitivity, specificity, VPP, and VPN and their confidence intervals were determined using Wilson Score. The study was designed to be a “clinical feasibility study” and the early diagnostic performance between the new Multiplex and the Xpert MTB/RIF was measured. The full clinical evaluation study is expected to be next. The patients that were missing data from the multiplex, Xpert MTB/RIF or sputum culture for contamination or indeterminate results (6 cases) were not considered for data analysis.
2.6. Sample size determination
This was a pilot early clinical feasibility study to evaluate the potential performance of the new designed assay. As such, a sample size calculation was not needed, but most feasibility studies range between 10 and 300 participants.[24] In this particular study, a total of 190 participants were enrolled in 4 disease groups and a control group (Table 3). This first step of development will allow an improved next version that will move to a full automation and a formal clinical validation study with bigger sample size in different study settings.
Table 3.
Clinical and Sociodemographic Characteristics of the Study Participants.
| Participant Characteristics | Control n= 47 |
Naive n=30 |
HIV/TB n=36 |
Rx Failure n= 70 |
NTM n= 7 |
TOTAL N= 190 |
|---|---|---|---|---|---|---|
| Gender | ||||||
| Male n (%) | 32 (23·5) | 23 (16·9) | 19 (14·0) | 57 (41·9) | 5 (3·7) | 136 (71·6) |
| Female n (%) | 15 (27·8) | 7 (13·0) | 17 (31·5) | 13 (24·1) | 2 (3·7) | 54 (28·4) |
| HIV Status | ||||||
| Positive n (%) | 8 (11·9) | 14 (20·9) | 33 (49·3) | 11 (16·4) | 1 (1·5) | 67 (35·3) |
| Negative n (%) | 39 (31·7) | 16 (13·0) | 3 (2·4) | 59 (48·0) | 6 (4·9) | 123 (64·8) |
| Age | ||||||
| < 35 n (%) | 15 (22·4) | 15 (22·4) | 11 (16·4) | 26 (38·8) | 0 (0·0) | 67 (35·3) |
| 35 to 55 n (%) | 29 (28·7) | 11 (10·9) | 21 (20·8) | 33 (32·7) | 7 (6·9) | 101 (53·2) |
| >56 n (%) | 9 (40·9) | 4 (18·2) | 4 (18·2) | 5 (22·7) | 0 (0·0) | 22 (11·5) |
| Marital status | ||||||
| Married n (%) | 34 (24·4) | 26 (17·9) | 32 (22·1) | 47 (32·4) | 6 (4·1) | 145 (76·3) |
| Single n (%) | 11 (28·9) | 3 (7·9) | 3 (7·9) | 21 (55·3) | 0 (0·0) | 38 (20·0) |
| Others n (%) | 2 (28·6) | 1 (14·3) | 1 (14·3) | 2 (28·6) | 1 (14·3) | 7 (3·7) |
| Smoking | ||||||
| Yes n (%) | 4 (11·1) | 5 (13·9) | 3 (8·3) | 24 (66·7) | 0 (0·0) | 36 (18·9) |
| No n (%) | 43 (27·9) | 25 (16·2) | 33 (21·4) | 46 (29·9) | 7 (4·5) | 154 (81·1) |
| Sputum Culture n (%) | ||||||
| Positive n (%) | 0 (0·0) | 17 (56·7) | 8 (22·2) | 23 (32·9) | 7 (100) | 55 (28·9) |
| Negative n (%) | 42 (89·4) | 11 (36·7) | 28 (77·8) | 45 (64·3) | 0 (0·0) | 126 (66·3) |
| Contaminated n (%) | 5 (10·6) | 2 (6·6) | 0 (0·0) | 1 (2·8) | 0 (0·0) | 8 (4·8) |
2.7. TB Sputum smear microscopy, mycobacterial culture and identification
Part of each sputum samples received was decontaminated using the modified Petroff's method with N-acetyl-l-cysteine–sodium hydroxide (NALC-NaOH) by mixing 5 mL of sputum with an equal volume of NALC- NaOH in a 50 ml round bottom processing tube, which was then vortexed periodically for 15 minutes. This mixture was neutralized with a phosphate buffer (pH 6·8) to a maximum total volume of 50 ml and centrifuged for 8000 g for 10 minutes. The supernatant was discarded aseptically. The pellet was used for Acid Fast Bacilli (AFB) smears after staining with Auramine/Rhodamine and re-suspension of the pellet in 2 mL solution of the phosphate buffer; 0·5 mL of the suspension was used for the BACTECTM Mycobacteria Growth Indicator Tube (MGIT) liquid medium inoculation and 0·1 ml of the suspension was used for the solid media 7H11 Middlebrook inoculation under sterile conditions. The inoculated MGIT-960 tubes were loaded in the BACTECTM 960 system, and continuously monitored for positivity during over a 42-day period. The 7H11 solid media were also incubated at 37°C for 6 weeks and checked every week for bacterial growth. Positive results for the BACTECTM MGIT-960 tubes and colonies on 7H11 Middlebrook agar were confirmed by the presence of AFB on microscopic examination of a smears made from the cultures. Positive AFB culture was identified phenotypically and confirmed by the TB Capilia test, a validated immune-chromatographic test for the MTB complex members’ identification.[25] If an AFB positive culture was found to be negative for the Capilia test, it was consider to be a possible case of NTM infection and was investigated further using the GeneProbe Accuprobe molecular confirmatory assay (GenProbe Inc, San Diego, CA, USA) for NTM identification. All laboratory members performing the control tests were blinded to the PCR results and vice-versa.
2.8. Method of the multiplex MTB/NTM sssay
2.8.1. Specific capture DNA extraction
Our new specific capture DNA extraction method included several steps as previously described.[20,21] gBlocks of the cotJC gene were used as the process control. Ten microliters of extraction control (DNA of CotJc gene) and 200µL of proteinase K were added to 1mL of sputum in a 1·8 mL microtube containing air dried 2% SDS (Thinning tube). This solution was incubated and shaken at 55°C in a shaking heat bloc for eight minutes. The temperature was then raised to 100°C for 10 minutes. The entire volume of the solution was then transferred into a Binding tube containing 5µL of capture probes, sodium salt and magnesium chloride. The capture probes are made of MTBC, MAC and Pan Mycobacteria-specific biotinylated oligonucleotides, which were added to the sputum. These oligonucleotides allow a streptavidin-coated paramagnetic particles (PMP) specific capture. The suspension was vortexed, incubating and shaken at 60°C for 20 minutes in the heat shaker bloc at 1,500 rpm. Twenty microliters of the PMP suspension were then added to each binding tube and mixed with a rotating tube holder for 10 minutes. This process links/captures the DNA to the magnetic beads. The mixture is placed in a magnetic rack in order for the captured DNA to remain linked to the magnetic beads and separated from the supernatant, which is then discarded. After repeating this step three times, the DNA is then separated from the magnetic beads by first adding 20µL of Buffer solution. That solution is incubated in shaking heat bloc at 70°C for 3 minutes. The eluted DNA containing solution was placed in a clean and DNase free microtube for the Multiplex Assay. The extracted samples were used, immediately, for PCR reaction or stored at -20°C for later use.
2.8.2. PCR Reaction
The PCR amplifications were performed as previously described.[20,21] The final optimization for the PCR reaction solution included adding 10% of glycerol, 1M of Bicine-KOH (pH= 8·0), 135mM of Potassium acetate, 20mM of Magnesium chloride, and 30mM Tris (pH=8·0) to 15ul per reaction with ultrapure water before adding 10ul of DNA sample for a total of 25ul. The two MTBC target genes are detected on FAM dye, the two MAC targets on HEX dye, the Pan-Mycobacterium target on TEX 615, and the cotJC extraction control on Cy5 dye (Supplementary Table 1). These dyes are on most qPCR instruments and were chosen for broad applicability of the assay).
2.9. Role of funding source
The funders had no role in study design, data collection, data analyses, interpretation, or writing of the manuscript.
3. Results
3.1. Analytic performance of the multiplex MTB/NTM
To determine the analytic performance of the newly designed Multiplex assay and its limit of detection, we selected the best performing dual-primers from a list of four primers for MTBC, MAC and Pan-Mycobacteria gene targets (Table 2). Each of the dual primers were tested on twenty (20) replicates of spiked sputum samples at each of the following concentrations: 50, 20, 10, 5 and 1 CFUs/mL. All tested 20 samples were successfully detected (100%) by the Multiplex assay down to 5 CFUs/mL for both MTBC and MAC spiked samples. The mycobacterial strains that were not MTBC or MAC were all detected by the Pan-Mycobacteria panel only at 50 CFUs/mL (20/20), 75% (15/20) at 20 CFUs/mL and 60% (12/20) at 10 CFUs/mL (Table 2). The new assay was successfully confirmed to detect all previously stored strains in Mali before beginning the clinical feasibility evaluation (Supplementary Table 2).
3.2. Clinical feasibility evaluation
3.2.1. Participants
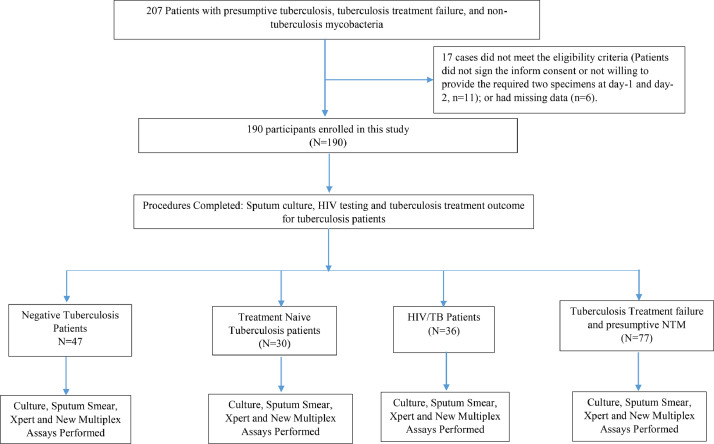
In order to conduct a clinical feasibility evaluation for the Multiplex MTB/NTM assay, a total of one hundred and ninety (190) participants with presumptive TB and/or NTM infections were enrolled (Fig. 1). Of the participants, 71·58% (136/190) were males and 28·42% (54/190) females, and 28·9% (55/190) were culture Positive and 66.3% (126/190) were culture Negative (Table 3). Participants average age was 41·5±13·8 years with a mean weight of 58·1±16·3 kg; 76·3% (145/190) of participants were married, 20% single, and 3·7% (7/190) other status (Table 3).
Fig. 1.
Study Participants’ Flow Chart.
Overall, the diagnostic performance of the Multiplex MTB/NTM assay was lower than the sputum culture but comparable to the Xpert assay for the different patient groups (TB culture negative control group, TB/HIV positive group, treatment failures and TB positive naive to treatment). The Xpert assay had greater sensitivity, but the Multiplex assay demonstrated higher specificity for all groups evaluated.
3.2.2. Multiplex assay performance for tuberculosis
The multiplex assay was able to detect M. tuberculosis at a Ct value of 23 for tuberculosis and 23-24 for MAC with ABI 7500 PCR Instrument (Supplementary Table 2). Overall, this assay had a sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) of 80·4% [66·8-89·3], 85·7% [77·7-91·1], 71·1% [57·7-81·6] and 90·9% [83·6-95·1] and the Xpert had value of 93·4% [82·85-97·7], 61·9% [52·3-70·6], 51·8% [41·2-62·2] and 95·5% [87·8-98·4], respectively (Table 4). The specificity calculation did not include the MAC cases detected. These results are impacted by the inclusion of treatment failure cases, for which molecular assays such as the Xpert assay are known to not perform well, yielding high rates of false positives, due to residual bacterial nucleic acid materials remaining in the lung and sputum samples after treatment.[26] While the bacteria will not grow on agar plates, they may yield a positive molecular assay. The rationale of having the treatment failure group was to increase the likelihood of detecting NTM cases.[9] When excluding treatment failure cases, the Multiplex assay had a Sensitivity, Specificity, PPV and NPV of 83·3% [66·4-92·6], 96·6% [88·6-99·0], 92·5% [82·3-96·5] and 92·2% [82·7-96·5] and the Xpert had values of 96·7% [83·3-99·4], 80·0% [68·2-88·1], 70·7 [55·5-82·3] and 97·9% [89·3-99·6], respectively (Table 4).
Table 4.
Multiplex and Xpert MTB/RIF diagnostic performances relative to sputum culture.
| Multiplex RT-PCR | Xpert MTB-RIF | ||
|
All participants (N=190) |
|||
| Sensitivity (%) [CI] | 80·4 [66·8-89·3] | 93·4 [82·85-97·7] | |
| Specificity (%) [CI] | 85·7 [77·7-91·1] | 61·9 [52·3-70·6] | |
| PPV (%) [CI] | 71·1 [57·7-81·6] | 51·8 [41·2-62·2] | |
| NPV (%) [CI] | 90·9 [83·6-95·1] | 95·5 [87·8-98·4] | |
| TB Patient Naïve to treatment (N=30) | Sensitivity (%) [CI] | 82·4 [58·9–93·8] | 94·1 [73·0–98·9] |
| Specificity (%) [CI] | 90·9 [62·2–98·3] | 81·8 [52·3–94·8] | |
| PPV (%) [CI] | 93·3 [70·1–98·8] | 88·9 [67·2–96·9] | |
| NPV (%) [CI] | 76·9 [49·7–91·8] | 90.0 [59·5–98·2] | |
| HIV Patient (N=36) | Sensitivity (%) [CI] | 71·4 [52·9–84·7] | 96·4 [82·2–99·3] |
| Specificity (%) [CI] | 96·2 [81·1–99·3] | 76.9 [57·9–88·9] | |
| PPV (%) [CI] | 95·2 [77·3–99·1] | 81·8 [65·6–91·3] | |
| NPV (%) [CI] | 75·8 [58·9 – 87·1] | 95·2 [77·3–99·1] | |
| Treatment failure, retreatment and NTM suspected patients (N=77) | Sensitivity (%) [CI] | 75·0 [50·5-89·8] | 87·5 [63·9-96·5] |
| Specificity (%) [CI] | 71·1 [56·6-82·2] | 37·7 [25·1-52·3] | |
| PPV (%) [CI] | 48·0 [30·0-66·5] | 33·3 [21·0-48·4] | |
| NPV (%) [CI] | 88·8 [74·6-95·5] | 89·4 [68·6-97·0] | |
| All (without treatment failure participants) (N=113) | Sensitivity (%) [CI] | 83·3 [66·4-92·6] | 96·7 [83·3-99·4] |
| Specificity (%) [CI] | 96·6 [88·6-99·0] | 80·0 [68·2-88·1] | |
| PPV (%) [CI] | 92·5 [82·3-96·5] | 70·7 [55·5-82·3] | |
| NPV (%) [CI] | 92·2 [82·7-96·5] | 97·9 [89·3-99·6] | |
CI: confidence Interval; PPV: Positive Predictive Value; NPV: Negative Predictive Value
3.2.3. Multiplex assay performance for tuberculosis naive to treatment
The performance of the Multiplex assay for this group had a sensitivity, specificity PPV, NPV of 82·4%, 90·9%, 93·3% and 76·9%) and the Xpert had a value of 94·1%, 81·8%, 88·9%, 90·0%, respectively (Table 4).
3.2.4. Multiplex assay performance for tuberculosis and HIV co-infected
Similar trends were seen in this group with better sensitivity for Xpert but increased specificity with the Multiplex assay, with a sensitivity and specificity of 71·4% and 96·2% for the Multiplex and 96·4 and 76·9 for the Xpert, respectively (Table 4).
3.2.5. Multiplex assay performance for tuberculosis treatment failure cases
The rationale for including this group, although historically performing less accurately under molecular assays, was to increase the likelihood of finding NTM cases given that the Multiplex has a panel for NTM detections. As expected, the sensitivity and specificity for this group were worse for both the Multiplex and Xpert at 75·0% and 71·1%, and 87·5% and 37·7%, respectively (Table 4).
4.2.6. Multiplex assay performance for nontuberculous mycobacterial infections
Of the 190 participants, there were seven presumptive NTM infection cases, but five (5) were confirmed to be positive by sputum culture. All five (5) were MAC cases (Table 5). These cases were all confirmed indirectly (post sputum culture) by Accuprobe assay. We should note that all five cases fulfilled the American Thoracic Society (ATS)’s definition of MAC disease, with MAC detected in at least two sputum samples collected at different times, a presence of TB/NTM infection-like respiratory symptoms (fever, cough, weight lost, night sweats) and M. tuberculosis not detected in the sputum samples.[27] The Multiplex assay in combination with the Xpert assay, could serve as a complement to the current diagnostic algorithm for patients with TB/NTM infection-like symptoms. For the pan-mycobacteria panel and “Other-NTM”, the clinical context is part of the diagnostic process, as patients have to fulfil the American Thoracic Society (ATS) diagnostic criteria for NTM disease described above.[27]
Table 5.
Clinical Performance of the Multiplex MTB/MTN in the Detection of Mycobacterium avium complex using Wilson confidence interval.
| Sputum Culture + Accuprobe MAC | Multiplex MTB/NTM |
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive (n) | 5 | 0 | 5 |
| Negative (n) | 0 | 185 | 185 |
| Sensitivity % (CI) | 100 [56•6-100] | ||
| Specificity % (CI) | 100 [97•9-100] | ||
| Positive Predictive Value % (CI) | 100 [56•6-100] | ||
| Positive Predictive Value % (CI) | 1000 [98•0-100] | ||
Mycobacterium avium complex (MAC)
4. Discussion
This new Multiplex molecular assay for TB, MAC and other NTM infections can significantly improve the management of patients presenting with TB/NTM infection-like symptoms. This is particularly important in the growing number of settings where these infections co-exist and are continuing to increase both in developing countries traditionally with endemic TB and in developed countries where MAC infections dominate among TB/NTM-like cases. Our Multiplex assay showed a great analytic and clinical performance similar to the Xpert assay, in addition to providing an option for simultaneous detection of mycobacterial infections other than TB. Some NTM stored samples previously identified were tested on the new primers and 100% concordance was found. This multiplex assay has shown a great analytical performance (5 CFUs/mL for TB and MAC and 20 CFUs/mL for other NTMs). The analytical performance for other NTMs (other than MAC) is lower at a detection limit of 20, but it is still at least as good as the analytic performance reported for the Xpert assay for tuberculosis (15.6 to 112.6 CFUs/mL).[28,29] The overall sensitivity, specificity, PPV and NPV of the Multiplex in cases without treatment failure were 83·3%, 96·6%, 92·5% and 92·2%, while the Xpert had value of 96·7%, 80·0%, 70·7% and 97·9%, respectively for the same patients compared to sputum culture (Table 4). The Multiplex assay with the Xpert assay, may be used as a complement to the current algorithm of diagnosis of patients with TB/NTM infection-like symptoms. The Multiplex and Xpert did not perform well in this study, because in big part of the inclusion of the treated group, in which residual DNA during and after treatment could lead to false positives.
Globally, NTM is not routinely diagnosed despite the availability of treatment, including the most prevalent, MAC infection. MAC is the leading cause of pulmonary NTM infections in both patients with or without HIV infection.[7,30] NTM infections are associated with substantial morbidity and mortality across the world.[31] While NTM pulmonary or systemic disease have been well characterized in developed countries,[32] it is largely undescribed and unappreciated in developing countries with a high incidence of both tuberculosis and HIV. This may be due to detection bias, because symptoms of NTM are often indistinguishable from TB and there is an absence in low and middle-income countries of appropriate diagnostic tools for NTM infections. A high proportion of TB patients are still empirically treated without any bacteriological confirmation.[17] SSM, in TB endemic settings, is often the only biological test available for TB.[13] It has a high detection limit [10,000 colony forming units per millilitre (CFU/mL)][33] and very poor sensitivity, identifying only 50-80% of all TB cases, particularly in settings where NTM is common.[9,34] Patients are generally presumed of potentially having NTM disease only after failure of toxic and lengthy TB treatment.[35]
Immunocompromised patients, including those with HIV infection, have worse outcomes for both TB and NTM. NTM and TB infections are often associated with immunocompromised patients (i.e., HIV infection); however, NTM infections are increasing in both prevalence and incidence in immunocompetent patients as well.[31,36,17,37,38] The ability to discriminate NTM from TB infection is thus of paramount importance in immunocompromised patients,[36] and highlights the need for new diagnostic tools to simultanously detect the two types of mycobacterial infections.
Treatments and outcomes may differ from one NTM species to another.[36] However, there is no available treatment for many species, but fortunately, MAC, the most frequent NTM disease, is treatable with two or three antimicrobials for at least 12 months, including macrolides (clarithromycin or azithromycin), ethambutol, and rifamycins (rifampin or rifabutin).[36] Physicians, in low resource settings, often initiate presumptive treatment, which may be toxic and time consuming, using precious resources that could have cured another patient for a more efficacious diagnostic test. Identifying these patients and ensuring appropriate treatment is critical in order to combat TB drug resistance and effectively treat those with NTM infection or TB. The NTM prevalence is not negligible in TB/NTM infection like patients and is increasing worldwide. In Mali and many other TB endemic countries, significant overlap exists between NTM diseases and TB.[2,3] In a recent study in Bamako, Mali, we found that 12% of presumptive TB cases were in fact infected by NTM, and, of those, 73% were infected by MAC.[9] Our most recent data from Mali also confirmed a 12% prevalence of NTM in all TB suspects (new and previously treated) seen at the major TB Referral Center at the Point-G Hospital of Bamako.[12] Similar data have been reported from Kenya, Burkina Faso and Nigeria.[39], [40], [41] Although it is not understood why MAC and other NTM diseases have been increasing worldwide including traditionally TB endemic setting,[7] environmental changes, aging, HIV/AIDS and use of immunosuppressors are known to be associated factors.[7,36]
The Multiplex had a better specificity than Xpert for all TB infection tested groups, although the Xpert had greater sensitivity. The improved specificity of the Multiplex is clinically relevant, as reflected in a better PPV. The new version of Xpert, the Xpert Ultra, has a 5% improvement compared to the old version but at the cost of the assay specificity.[26,42] Moreover, the new assay can detect and differentiate TB from NTM infection, while the Xpert includes drug resistance option. Thus, the new assay is not meant to replace the Xpert assay, but rather can serve as a complement by providing an additional fast and effective option, depending on geography, epidemiology and the patient's individual clinical context.[10] This will save resources as in the current practice, NTM patients often get tested for TB first (and sometimes even treated unnecessarily with TB drugs). The sputum culture for TB takes at least 8 weeks for negative samples before being tested for NTM. We found during the field clinical evaluation a slight decrease in the performance of the assay. An automated version of the assay in the next phase of development will ease the assay procedure and may help in performance consistency.
This study has some limitations. The assay is still at this point an open-system, usable on many PCR platforms but not yet automated to warranty consistency in performance. In addition, the project was not a clinical validation study but rather one of clinical feasibility to evaluate the early performance of the assay. Weaknesses of the assay from this early study will be recorded and corrected in the next phase of development. The Multiplex assay seems to have a great performance for MAC detection, but only five MAC cases were included in this study. Future clinical studies should include more MAC and NTM cases from different settings. Nevertheless, based on these encouraging results, the Multiplex assay should be adapted to a scalable, quick (less than 30 minutes) and easy point of care (POC) automated system that should be tested in various clinical and geographical settings in a multi-centric study. Many potential options are available for automation such as making cartridges for the assay for Cepheid's GeneXpert instrument or BD MAX™ PCR instruments. Nevertheless, this project has laid the framework and provided the rationale and justification for bigger trials of the assay, particularly in areas where TB and NTM disease are both prevalent.
This novel multiplex MTB/NTM assay has a great analytic and clinical performance for TB and NTM infections and will complement the current diagnostic algorithm and tools for patients with TB/NTM infection like symptoms. The employment of this multiplex assay will save time and resources by testing for more than one analyte, and more importantly, it has the potential to save lives worldwide, wherever TB and NTM coexist.
Contributors
YSS, MAB, BD, SMM, BT, RLM, JLH, FS, OK, MT, MG, BB, SD, IBD, DD, BPPD, IC, IK, ED, KO, BK, MM designed the study; MAB, YSS, MM, SMM set up and optimized the assay; FS, OK, MT, MG, BB, SD, IBD, DD, BPPD, IC, IK, ED, KO and BK enrolled, conducted the lab assays, collected and entered the data; YSS, IC, BK, AK and MM analysed the data; YSS, OK and FS verified the underlying data; YSS, BB, OK, IC, BPP, MM, SD drafted the manuscript; MM, CJA, BD, GT, YT, SD, BT, JLH, RLM, SD, SMM edited the manuscript. All authors read and approved the final version of the manuscript.
Declaration of Competing Interest
The authors have no conflicts of interest to report.
Acknowledgments
Acknowledgments
This work was supported by the Institute of Global Health of Northwestern University (Catalyzer Fund) and the U.S. National Institutes of Health (R03AI137674, U54EB027049, D43TW010350 and UM1AI069471). We would like to thank all the study participants and the TB unit of the Referral Health Center 4, 5 and 6 of Bamako, the Pulmonary Department of Teaching Hospital of Point G, and the National Institute of Public (INSP) for contributing to patient recruitment and follow-ups during the study. We also thank the team of the University Clinical Research Center (UCRC) of the University of Sciences, techniques, Technologies of Bamako (USTTB) for their valuable scientific and technical assistance during the study.
Data sharing statement
All data and code used for producing the results are freely available for others upon request. Request can be sent to these emails: mamoudou.maiga@northwestern.edu and/or sadio@icermali.org.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103527.
Contributor Information
Sally M McFall, Email: s-mcfall@northwestern.edu.
Mamoudou Maiga, Email: mamoudou.maiga@northwestern.edu.
Appendix. Supplementary materials
References
- 1.WHO. Global tuberculosis report, 2019.
- 2.Forbes B.A., Hall G.S., Miller M.B. Practice guidelines for clinical microbiology laboratories: mycobacteria. Clin Microbiol Rev. 2018;31(2) doi: 10.1128/CMR.00038-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forrellad M.A., Klepp L.I., Gioffre A. Virulence factors of the Mycobacterium tuberculosis complex. Virulence. 2013;4(1):3–66. doi: 10.4161/viru.22329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeon D. Infection source and epidemiology of nontuberculous mycobacterial lung disease. Tuberculosis Respir Dis. 2019;82(2):94–101. doi: 10.4046/trd.2018.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Ingen J. Diagnosis of nontuberculous mycobacterial infections. Semin Respir Crit Care Med. 2013;34(1):103–109. doi: 10.1055/s-0033-1333569. [DOI] [PubMed] [Google Scholar]
- 6.Kone B., Sarro Y.S., Maiga M. Clinical characteristics of non-tuberculous mycobacterial pulmonary infections in Bamako, Mali. Epidemiol Infect. 2018;146(3):354–358. doi: 10.1017/S0950268817003090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishiuchi Y., Iwamoto T., Maruyama F. Infection Sources of a Common Non-tuberculous Mycobacterial Pathogen, Mycobacterium avium Complex. Front Med (Lausanne) 2017;4:27. doi: 10.3389/fmed.2017.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan Y., Su B., Shu W. Epidemiology of pulmonary disease due to nontuberculous mycobacteria in Southern China, 2013-2016. BMC Pulmonary Med. 2018;18(1):168. doi: 10.1186/s12890-018-0728-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maiga M., Siddiqui S., Diallo S. Failure to recognize nontuberculous mycobacteria leads to misdiagnosis of chronic pulmonary tuberculosis. PLoS One. 2012;7(5):e36902. doi: 10.1371/journal.pone.0036902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarro Y.D., Kone B., Diarra B. Simultaneous diagnosis of tuberculous and non-tuberculous mycobacterial diseases: time for a better patient management. Clin Microbiol Infect Dis. 2018;3(3) doi: 10.15761/CMID.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diarra B., Toloba Y., Konate B. Extensively drug resistant tuberculosis in Mali: a case report. BMC Res Notes. 2017;10(1):561. doi: 10.1186/s13104-017-2890-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kone B., Sarro Y.S., Sanogo M. Clinical Characteristics of Nontuberculous Mycobacterial Pulmonary Infections in Bamako, Mali. Semin Respir Crit Care Med (Under Review) 2017 [Google Scholar]
- 13.Maiga M., Abaza A., Bishai W.R. Current tuberculosis diagnostic tools & role of urease breath test. Indian J Med Res. 2012;135(5):731–736. [PMC free article] [PubMed] [Google Scholar]
- 14.Hatta M., Sultan A.R., Tandirogang N., Masjudi Yadi. Detection and identification of mycobacteria in sputum from suspected tuberculosis patients. BMC Res Notes. 2010;3:72. doi: 10.1186/1756-0500-3-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.WHO. Global tuberculosis report, 2016.
- 16.Steingart K.R., Schiller I., Horne D.J., Pai M., Boehme C.C., Dendukuri N. Xpert(R) MTB/RIF assay for pulmonary tuberculosis and rifampicin resistance in adults. Cochrane Database Syst Rev. 2014;(1) doi: 10.1002/14651858.CD009593.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.WHO: World Health Organization. Global tuberculosis report 2016. 2016.
- 18.DB Jen.nifer L.Re.eda, Butzlera Matthew.A., McFall Sall.y M. 2017. XtracTB Assay, a Mycobacterium tuberculosis molecular screening test with sensitivity approaching culture. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiang S.S., Swanson D.S., Starke J.R. New Diagnostics for Childhood Tuberculosis. Infect Dis Clin North Am. 2015;29(3):477–502. doi: 10.1016/j.idc.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 20.Reed J.L., Basu D., Butzler M.A., McFall S.M. XtracTB Assay, a Mycobacterium tuberculosis molecular screening test with sensitivity approaching culture. Sci Rep. 2017;7(1):3653. doi: 10.1038/s41598-017-03930-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reed J.L., Walker Z.J., Basu D. Highly sensitive sequence specific qPCR detection of Mycobacterium tuberculosis complex in respiratory specimens. Tuberculosis (Edinb) 2016;101:114–124. doi: 10.1016/j.tube.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Kaur P., Filia G., Singh S.V., Patil P.K., Ravi Kumar G.V., Sandhu K.S. Molecular epidemiology of Mycobacterium avium subspecies paratuberculosis: IS900 PCR identification and IS1311 polymorphism analysis from ruminants in the Punjab region of India. Comp Immunol Microbiol Infect Dis. 2011;34(2):163–169. doi: 10.1016/j.cimid.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 23.Shin S.J., Lee B.S., Koh W.J. Efficient differentiation of Mycobacterium avium complex species and subspecies by use of five-target multiplex PCR. J Clin Microbiol. 2010;48(11):4057–4062. doi: 10.1128/JCM.00904-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Billingham S.A., Whitehead A.L., Julious S.A. An audit of sample sizes for pilot and feasibility trials being undertaken in the United Kingdom registered in the United Kingdom Clinical Research Network database. BMC Med Res Methodol. 2013;13:104. doi: 10.1186/1471-2288-13-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramos A., Carvalho T., Ribeiro M., Guimarães J.T. CapiliaTM TB-Neo assay: a new tool for rapid distinction between tuberculous and non-tuberculous mycobacteria. Int J Tuberculosis Lung Dis. 2016;20(6):753–756. doi: 10.5588/ijtld.15.0528. [DOI] [PubMed] [Google Scholar]
- 26.Mishra H., Reeve B.W.P., Palmer Z. Xpert MTB/RIF Ultra and Xpert MTB/RIF for diagnosis of tuberculosis in an HIV-endemic setting with a high burden of previous tuberculosis: a two-cohort diagnostic accuracy study. Lancet Respir Med. 2020;8(4):368–382. doi: 10.1016/S2213-2600(19)30370-4. [DOI] [PubMed] [Google Scholar]
- 27.Ghio A.J., Smith G.S., DeFlorio-Barker S. Application of diagnostic criteria for non-tuberculous mycobacterial disease to a case series of mycobacterial-positive isolates. J Clin Tuberc Other Mycobact Dis. 2019;17 doi: 10.1016/j.jctube.2019.100133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blakemore R., Story E., Helb D. Evaluation of the analytical performance of the Xpert MTB/RIF assay. J Clin Microbiol. 2010;48(7):2495–2501. doi: 10.1128/JCM.00128-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang M., Xue M., He JQ. Diagnostic accuracy of the new Xpert MTB/RIF Ultra for tuberculosis disease: a preliminary systematic review and meta-analysis. Int J Infect Dis. 2020;90:35–45. doi: 10.1016/j.ijid.2019.09.016. [DOI] [PubMed] [Google Scholar]
- 30.Porvaznik I., Solovic I., Mokry J. Non-Tuberculous Mycobacteria: classification, Diagnostics, and Therapy. Adv Exp Med Biol. 2017;944:19–25. doi: 10.1007/5584_2016_45. [DOI] [PubMed] [Google Scholar]
- 31.Griffith D.E., Aksamit T., Brown-Elliott B.A. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007;175(4):367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 32.Adjemian J., Olivier K.N., Seitz A.E., Holland S.M., Prevots D.R. Prevalence of nontuberculous mycobacterial lung disease in U.S. Medicare beneficiaries. Am J Respir Crit Care Med. 2012;185(8):881–886. doi: 10.1164/rccm.201111-2016OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diagnostic Standards and Classification of Tuberculosis in Adults and Children This official statement of the American Thoracic Society and the Centers for Disease Control and Prevention was adopted by the ATS Board of Directors, July 1999. This statement was endorsed by the Council of the Infectious Disease Society of America, September 1999. Am J Respir Crit Care Med. 2000;161(4 Pt 1):1376–1395. doi: 10.1164/ajrccm.161.4.16141. [DOI] [PubMed] [Google Scholar]
- 34.Ngabonziza J.C., Ssengooba W., Mutua F. Diagnostic performance of smear microscopy and incremental yield of Xpert in detection of pulmonary tuberculosis in Rwanda. BMC Infect Dis. 2016;16(1):660. doi: 10.1186/s12879-016-2009-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atif M., Sulaiman S.A., Shafie A.A., Ali I., Hassali M.A., Saleem F. WHO guidelines for treatment of tuberculosis: the missing links. Int J Clin Pharm. 2012;34(4):506–509. doi: 10.1007/s11096-012-9657-8. [DOI] [PubMed] [Google Scholar]
- 36.Ryu Y.J., Koh W.J., Daley C.L. Diagnosis and treatment of nontuberculous mycobacterial lung disease: clinicians' perspectives. Tuberc Respir Dis (Seoul) 2016;79(2):74–84. doi: 10.4046/trd.2016.79.2.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Getahun H., Harrington M., O’Brien R., Nunn P. Diagnosis of smear-negative pulmonary tuberculosis in people with HIV infection or AIDS in resource-constrained settings: informing urgent policy changes. Lancet. 2007;369(9578):2042–2049. doi: 10.1016/S0140-6736(07)60284-0. [DOI] [PubMed] [Google Scholar]
- 38.Pawlowski A., Jansson M., Skold M., Rottenberg M.E., Kallenius G. Tuberculosis and HIV co-infection. PLoS Pathog. 2012;8(2) doi: 10.1371/journal.ppat.1002464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nyamogoba H.D., Mbuthia G., Mining S. HIV co-infection with tuberculous and non-tuberculous mycobacteria in western Kenya: challenges in the diagnosis and management. Afr Health Sci. 2012;12(3):305–311. doi: 10.4314/ahs.v12i3.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zida S., Tarnagda Z., Kaboré A. Current status of atypical mycobacterial infections in Burkina Faso: results of a regional survey. Pan Afr Med J. 2014;17:188. doi: 10.11604/pamj.2014.17.188.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akanbi M.O., Achenbach C., Taiwo B. Evaluation of gene xpert for routine diagnosis of HIV-associated tuberculosis in Nigeria: a prospective cohort study. BMC Pulm Med. 2017;17(1):87. doi: 10.1186/s12890-017-0430-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dorman S.E., Schumacher S.G., Alland D. Xpert MTB/RIF Ultra for detection of Mycobacterium tuberculosis and rifampicin resistance: a prospective multicentre diagnostic accuracy study. Lancet Infect Dis. 2018;18(1):76–84. doi: 10.1016/S1473-3099(17)30691-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.