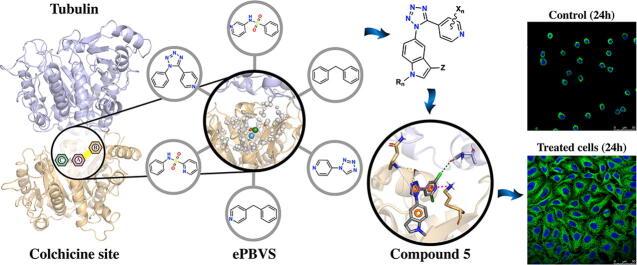
Graphical abstract
Keywords: Ensemble pharmacophore, Virtual screening, Tubulin, Colchicine, Drug design, Antimitotic
Highlights
-
•
Design of a new family of anti-tumor colchicine-site ligands.
-
•
Ensemble pharmacophore construction based on tubulin X-ray structures.
-
•
Virtual screening of a ZINC sublibrary by ensemble pharmacophores.
-
•
Favored structural elements combined in a novel compound family: indole, tetrazole and halogenated pyridines.
-
•
Diaryl tetrazole 5 satisfies the ensemble pharmacophore requirements and MD simulations and is a novel potent antiproliferative inhibitor of tubulin in vitro and in cells.
Abstract
Tubulin is a well-validated target for herbicides, fungicides, anti-parasitic, and anti-tumor drugs. Many of the non-cancer tubulin drugs bind to its colchicine site but no colchicine-site anticancer drug is available. The colchicine site is composed of three interconnected sub-pockets that fit their ligands and modify others’ preference, making the design of molecular hybrids (that bind to more than one sub-pocket) a difficult task. Taking advantage of the more than eighty published X-ray structures of tubulin in complex with ligands bound to the colchicine site, we generated an ensemble of pharmacophore representations that flexibly sample the interactional space between the ligands and target. We searched the ZINC database for scaffolds able to fit several of the subpockets, such as tetrazoles, sulfonamides and diarylmethanes, selected roughly ~8000 compounds with favorable predicted properties. A Flexi-pharma virtual screening, based on ensemble pharmacophore, was performed by two different methodologies. Combining the scaffolds that best fit the ensemble pharmacophore-representation, we designed a new family of ligands, resulting in a novel tubulin modulator. We synthesized tetrazole 5 and tested it as a tubulin inhibitor in vitro. In good agreement with the design principles, it demonstrated micromolar activity against in vitro tubulin polymerization and nanomolar anti-proliferative effect against human epithelioid carcinoma HeLa cells through microtubule disruption, as shown by immunofluorescence confocal microscopy. The integrative methodology succedes in the design of new scaffolds for flexible proteins with structural coupling between pockets, thus expanding the way in which computational methods can be used as significant tools in the drug design process.
1. Introduction
The microtubules of eukaryotic cells are hollow tubes formed by continuous assembly and disassembly of tubulin dimers engaged in complex polymerization and depolymerization equilibria. The microtubules are part of the cytoskeleton and play important roles in cell shape maintenance, intracellular transport of organelles and other cargoes, cell motility by cilia and flagella, form the mitotic spindle, and provide the scaffold for the chromosome movements during mitosis.
Drugs that alter the microtubule dynamics are usually referred to as antimitotics, as their more prominent but not only action is the perturbation of the mitotic events [1], [2]. Compounds acting on the microtubule dynamics usually bind to tubulin, and are important chemotherapeutic agents used as herbicides (e.g. oryzalin), fungicides (e.g. carbendazim), antiparasitic (e.g. mebendazole), and anticancer agents (e.g. paclitaxel or vincristine). The antitubulin drugs bind to at least seven distinct, well characterized binding sites, named after their most representative binders: the taxoids, the laulimalide/peloruside, the eribulin, the maytansine, the vinca minor alkaloids, the pironetin, and the colchicine binding sites [3]. Drugs binding at the first two sites promote tubulin polymerization (microtubule stabilizing agents), whereas binders of the other sites promote tubulin depolymerization (microtubule destabilizing agents). However, at the low concentrations relevant for the biological effects both classes disrupt tubulin polymerization dynamics, which is considered the actual mechanism of action for all of them.
In spite of the great success of taxanes and vinca minor alkaloids, they often show poor pharmacokinetics due to their large sizes and hydrophobic nature. Moreover, these suffer from resistance associated with efflux proteins such as the MDRs (multidrug resistance proteins), suggesting the need for new antitubulin agents with favorable profiles as an urgent need for cancer therapy [4]. Up until now drugs binding to the colchicine site have not succeeded in bringing representatives to the clinic, due to toxicity (e.g. colchicine), insufficient potency (e.g. ABT-751) or poor solubility (e.g. combretastatin A4). However, they are much smaller than taxoids and vinca alkaloids, many of them are not substrates of the MDR efflux pumps, and recently they have been shown to disrupt the tumor neovasculature [5], thus making them attractive for medicinal chemistry campaigns.
The colchicine site of tubulin is a hydrophobic domain located at the intra-dimer interface between α and β subunits [6]. Drugs binding to the colchicine site block the structural rearrangements needed to transform the curved tubulin structure observed in solution into the straight tubulin observed in the microtubules, thus shifting the equilibria towards unpolymerized tubulin and explaining their microtubule-destabilizing effect. In recent years, a vast number of colchicine-site inhibitors has been described, and many X-ray structures of tubulin in complex with colchicine-site ligands have been reported at good resolutions, mainly as a result of the methodological advances provided by the co-crystallization of the curved tubulin dimers with proteins of the stathmin-like family and tubulin tyrosine ligase [7].
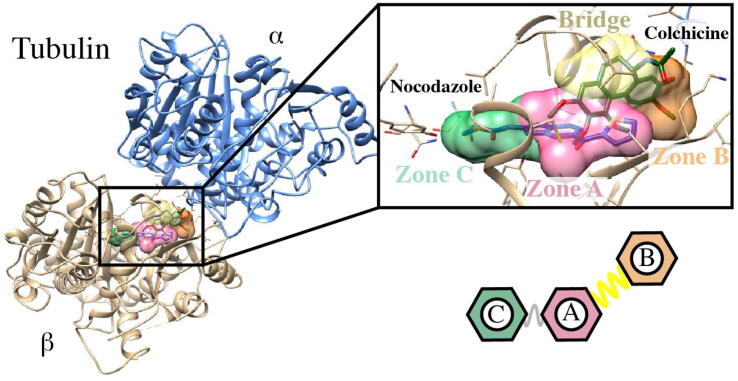
These structures have led to a better structural understanding of the colchicine site and its interactions with ligands. Based on the X-ray crystal structures, the colchicine domain has been divided in three sub-pockets called zones A-C (Fig. 1) arranged in a C-A-B disposition [3], [8], [9]. Zone A is the intermediate region of the domain that contacts only with the β subunit, between zones B and C. Zone B is close to the interfacial surface between the α and β subunits, thus making contacts with residues of both subunits. Zone C is buried deeper in the β subunit, further away from the interfacial surface. The connection between zones A and B (bridge) also projects towards the interfacial surface and can successfully allocate bridges of different sizes. Zones A and C are not so neatly divided and the connections are not included in a different pocket. Most known ligands bind to zones A and B, including the “classical” colchicine ligands such as colchicine, podophyllotoxin, and combretastatin A4, whereas a smaller group bind to zones A and C such as nocodazole, plinabulin, as well as TN16. Very few compounds bind to the tree zones (and only partially to one of them), such as ABT-751 and lexibulin, and only Bal27862 binds to zones B and C.
Fig. 1.
Tubulin structure and detail of the colchicine binding site located between tubulin α and β subunits with the Nocodazole and Colchicine ligands. A schematic of the colchicine site subpockets (A, B and C) is shown.
The early stages of antitubulin drug search were exclusively based on ligand-based drug design (LBDD) campaigns due to the scarcity of structural information. These were mainly based on blind screening or pharmacophoric models [10], which were in many cases only partially successful. Later, the accumulating breadth of information on tubulin binders, and binding conformations, has stimulated an intense search of new ligands applying structure-based drug design (SBDD) methodologies [3], [9]. Most of the approaches that target tubulin have applied pharmacophore-based or docking strategies based on a single target conformation, with few examples of ensemble approaches [11]. The employment of ensembles in SBDD enables taking into account the target’s flexibility, and having diverse representations of the active-site interactions. Different examples of pharmacophore-screening campaigns, which require less computational resources than docking simulations, make use of the target’s ensemble [12], [13]. These have been implemented to look for novel treatments against multiple pathogens [14], [15], and have been combined with other methodologies including 3D-QSAR [16] and conformations derived from molecular dynamics (MD) simulations [17].
In this work, we have taken advantage of the multiple structures of tubulin in complex with ligands bound to the colchicine binding site, retrieving more than 80 tubulin–compound complexes, that are used with the Flexi-pharma protocol [17] (originally developed for MD ensembles). We modeled the pharmacophore of each complex having multiple interaction configurations at this binding site. Then, we searched for tetrazoles, sulfonamides and diarylmethanes representative scaffolds (widely studied scaffolds to modulate tubulin) from the ZINC database (~8000) that matched the pharmacophores using the Flexi-pharma score. This screening was performed for all scaffolds using two different pharmacophore-matching programs. In this way, we found the scaffolds that best fit with the different zones (A, B or C) of the colchicine binding site on tubulin. With this information, we designed a novel tubulin modulator. The tetrazole-derivative compound 5 was synthesized and tested in vitro, demonstrating marked activity against tubulin polymerization, as well as antiproliferative effect in HeLa cells from human epithelioid carcinoma. The mechanism of action was confirmed by cell cycle studies and immunofluorescence confocal microscopy. This methodology is expected to design multiple compounds using scaffolds that best fit tubulin pharmacophores, thus, expanding the way in which computational methods are used as significant tools in the drug design process.
2. Materials and methods
2.1. Structural and computational studies
2.1.1. Selection of tubulin crystallographic structures
Crystal structures of tubulin with a co-crystallized ligand at the colchicine site were retrieved from the Protein Data Bank (PDB). Only the tubulin α and β subunits were conserved, as well as the modulators at the colchicine site, ions, and phosphorylated nucleotides. The structures were aligned against the tubulin-colchicine complex (PDB code: 4O2B) using the α-subunit as reference.
2.1.2. Selection of scaffolds and library construction
For the selection of reference molecules, three main families of scaffolds were included: tetrazoles, diarylmethanes and sulfonamides, given previous studies for the discovery of tubulin polymer-formation inhibitors [18], [19]. From these, a set of molecules containing the selected scaffolds were obtained to further perform the pharmacophore-based virtual screening. The dataset was downloaded from the ZINC database, by searching compounds containing the scaffolds (Fig. S3). For those scaffolds reporting more than 1000 compounds in the database, a fixed number of random molecules containing the scaffold was selected randomly as a representative group of the chemical space. A total of 9819 molecules was initially selected. Then, a filter was applied to avoid high hydrophobicity and large structures. Specifically, molecules with molecular weight lower than 400 g/mol, and Crippen LogP [20] lower than 4.0 were filtered. The properties were calculated using RDKit version 2020.09.3 (www.rdkit.org). From this step, a total of 2713 tetrazoles, 2966 diarylmethanes, and 3239 sulfonamides were prioritized for the Flexi-pharma strategy. All ligands were then prepared using the LigPrep and Epik modules of Maestro Suite [21]. Original chirality was retained. The force field, OPLS3 [22] has been used to define all the generated compounds.
2.1.3. Pharmacophore identification
Two different software were used to identify and characterize the pharmacophores from tubulin-ligand pre-aligned complexes, LigandScout [23] and Phase [24]. Briefly, with either software, each ligand and its macromolecular environment was extracted and interpreted from every pre-aligned tubulin-ligand complex. Using the default parameters of both software, 3D models of pharmacophores of each complex were detected. Subsequently, the obtained models were manually edited to eliminate non-relevant protein–ligand interactions, such as interactions with water molecules other than water-bridge interactions. To consider the steric characteristics of the binding site, excluded volume coats for all structure-based pharmacophores models were conserved. Finally, the pharmacophores were created and saved in two different formats in order to perform the screening analysis with two different software (see below).
2.1.4. Flexi-pharma using crystallographic ensemble
The Phase and Pharmer software were used to virtually screen the selected ZINC scaffolds against the tubulin pharmacophores. A hit is considered when a compound matches at least 3 pharmacophoric features from each pharmacophore. With Phase, multiple conformers per ligand (max 50) were generated during the search, partial matches of more features were preferred, and all other parameters were maintained as default, including the phase screen score as a scoring function. Only pharmacophores without water were considered. For Pharmer [25], we created subsets of pharmacophores obtained from all possible combinations of 3 pharmacophoric features, identified from each crystallographic complex (with or without water), together with the volume exclusion features. Pharmer was used to assess if a molecule matched at least one of the subset pharmacophores. For both programs, the molecule scoring criteria was the normalized number of matches. This is the same as the Flexi-pharma [17] method when the number of pharmacophores is equal to the number of structures. The molecules with higher scores are considered more active.
2.1.5. Molecular dynamics simulations (MDs)
MD simulations were run using the tubulin crystal (PDB code: 5Z4U) with the compound 5 at the colchicine binding site. The complex was obtained from the ePBVS step. The co-crystallized GTP cofactor and binding site ions were conserved for MDs and further analysis. The tubulin-ligand complex was solvated by an orthorhombic box of SPC water model. Chloride ions were added to neutralize the systems and then the ion concentration was set to 0.15 M NaCl. We performed two MDs for each tubulin–ligand complex. Prior equilibrium simulations were performed, the systems were relaxed using the default Desmond's relaxation protocol. Briefly, the first stage consists of a 100 ps brownian dynamics simulation in an NVT ensemble at 10 K, applying restrictions on the heavy atoms of the protein. The second stage corresponds to 12 ps simulation in a NVT ensemble at 10 K, applying restrictions to the solute heavy atoms. The third stage corresponds to 12 ps simulation in a NPT ensemble at 10 K maintaining the restrictions on the solute heavy atoms. The fourth and final relaxation stage consisted of heating from 10 K to 310 K during 36 ps in a NPT ensemble. Then, the system was equilibrated for 25 ns in a NPT ensemble at 310 K with the application of a restraint spring constant of 5 kcal × mol−1 × Å−2 to the protein backbone and ligands atoms at the colchicine site. After the equilibration, a 500 ns MDs in the NPT ensemble was performed without restrictions. In both equilibrium and production MDs, temperature and pressure were kept constant at 310 K and 1.01325 bar respectively by coupling to a Nose-Hoover Chain thermostat [26] and Martyna-Tobias-Klein barostat [27] with an integration time step of 2 fs. The simulations were performed with Desmond [28] and the OPLS3 force field [22]. As control, two extra MDs using the same protocol described above were performed with the co-crystalized ligand 96C (from 5Z4U structure) and without ligand. The simulations were analyzed with Desmond, KNIME, Schrödinger and in-house scripts. Visualization was carried-out with VMD [29] and Pymol [30].
2.2. Chemistry
2.2.1. General chemical techniques
Reagents were used as purchased without further purification. Solvents for synthesis (dichloromethane and acetonitrile) were dried and freshly distilled before use according to procedures described in the literature. TLC was performed on pre-coated silica gel polyester plates (0.25 mm thickness) with a UV fluorescence indicator 254 (Polychrome SI F254). Chromatographic purification was performed on silica gel columns by flash (Kieselgel 40, 0.040–0.063; Merck) chromatography. Melting point was determined on a Büchi 510. 1H NMR and 13C NMR spectra were recorded in CDCl3 or DMSO‑d6 on a Varian Mercury spectrometer operating at 400/100 MHz. Chemical shifts (δ) are given in ppm downfield from tetramethylsilane and coupling constants (J values) are in Hertz. IR spectra were run on a Nicolet Impact 410 Spectrophotometer. For FAB-HRMS analyses, a VG-TS250 apparatus (70 eV) was used.
2.2.2. Chemical synthesis
2.2.2.1. 2,6-dichloro-N-(1-methyl-1H-indol-5-yl)pyridine-4-carboxamide (4)
A solution of 2,6-dichloropyridine-4-carboxylic acid (770 mg, 4 mmol), DMAP (489 mg, 4 mmol) and EDC (765 mg, 4 mmol) in dry DCM (25 mL) was stirred for 1 h. Then, N-methyl-5-aminoindole (390 mg, 2.65 mmol) was added and the mixture was refluxed under N2 atmosphere for 24 h. Then the mixture was poured into cold water and the organic layer was washed with 1 M HCl, 5% NaHCO3 and NaCl saturated aqueous solution until neutral pH, dried over anhydrous Na2SO4, filtered, and the solvent was evaporated under vacuum. The crude was purified by column chromatography using 1:3–1:1 EtOAc-Hexane to obtain 687 mg of pure product (2.14 mmol, 78 %). 1H NMR δ (ppm, 400 MHz, DMSO‑d6): 10.47 (1H, s), 8.01 (2H, s), 7.99 (1H, s), 7.42 (1H, d, J = 1.2), 7.31 (1H, d, J = 3.2), 6.41 (1H, d, J = 3.2), 3.77 (3H, s). 13C NMR δ (ppm, 100 MHz, DMSO‑d6): 161.1 (CO), 150.2 (2C), 149.1 (C), 134.3 (C), 130.9 (CH), 130.6 (C), 128.2 (C), 122.3 (CH), 116.1 (CH), 112.9 (CH), 110.1 (CH), 100.9 (CH), 33.0 (CH3). IR (KBr): 3252, 1738, 1651, 1604, 1548, 1493 cm−1. HRMS (C15H11N3ONaCl2) calculated (M + Na+) 342.0168, found 342.0171.
2.2.2.2. 5-(5-(2,6-dichloropyridin-4-yl)-1H-tetrazol-1-yl)-1-methyl-1H-indole (5)
A solution of compound 4 (352 mg, 1.1 mmol) and sodium azide (572 mg, 8.8 mmol) in acetonitrile (7 mL) was prepared in a sealed tube. Silicon tetrachloride (1 mL, 8.8 mmol) was dropwised and the mixture was stirred at 90 °C for 72 h. Then it was cooled down in an ice bath and the reaction was quenched by dilution with DCM and 5% NaHCO3, and vigorously stirred for 30 min. The aqueous layer was extracted with EtOAc and the organic layers were washed with NaCl saturated aqueous solution, then dried over anhydrous Na2SO4, filtered and evaporated under vacuum. The product was crystallized in methanol to obtain 349 mg (1.01 mmol, 92 %). 1H NMR δ (ppm, 400 MHz, CDCl3): 7,65 (1H, d, J = 2.1), 7.48 (1H, d, J = 8.8), 7.44 (1H, s), 7.43 (1H, s), 7.26 (1H, d, J = 3.2), 7.12 (1H, dd, J = 8.8; J = 2.1), 6.60 (1H, d, J = 3.2), 3.90 (3H, s). 13C NMR δ (ppm, 100 MHz, DMSO‑d6): 151.0 (2C), 149.5 (C), 131.6 (CH), 136.8 (C), 136.2 (C), 128.3 (C), 128.3 (C), 124.8 (C), 121.3 (CH), 121.2 (CH), 117.8 (CH), 117.6 (CH), 110.4 (CH), 101.8 (CH), 32.9 (CH3). IR (KBr): 1600, 1540, 1492, 1241, 1167 cm−1. M. p.: 88–89 °C. HRMS (C15H10N6NaCl2) calculated (M + Na+) 367.0241, found 367.0236.
2.3. Biology
2.3.1. Inhibition of tubulin polymerization
Bovine brain tubulin was isolated as previously described [31]. Tubulin polymerization assays were carried out with 1.5 mg/mL protein at pH 6.7 in assay buffer containing 0.1 M 2-(N-morpholino)ethanesulfonic acid buffer, 1.5 mM GTP, 1 mM EGTA, 1 mM 2-mercaptoethanol, 1 mM MgCl2, and the required ligand concentration. Samples were incubated 30 min at 20 °C, followed by cooling on ice for 10 min. Tubulin polymerization was assessed by the UV absorbance increase at 450 nm due to the turbidity caused by a temperature shift from 4 °C to 37 °C. When a stable plateau was reached and maintained for at least 20 min, the temperature was switched back to 4 °C to ascertain the return to the initial absorption values, to confirm the reversibility of the process. The degree of tubulin assembly for each experiment was calculated as the difference in amplitude between the stable plateau and the initial baseline of the curves. Control experiments in identical conditions but the absence of ligand were taken as 100% tubulin polymerization. The IC50 value of tubulin polymerization was determined by measuring the tubulin polymerization inhibitory activity at different ligand concentrations. The obtained values of the mole ratio of total ligand to total tubulin in solution were fitted to mono exponential curves and the IC50 values of tubulin polymerization inhibition calculated from the best-fitting curves.
2.3.2. Cell culture
HeLa (human cervical carcinoma) cell line was grown at 37 °C in humidified 95% air and 5% CO2 in DMEM culture medium containing 10% (v/v) heat inactivated fetal bovine serum (FBS), 2 mM l-glutamine, 100 U/mL penicillin, and 100 mg/mL streptomycin. Cells were periodically tested for mycoplasma infection and found to be negative.
2.3.3. Cell growth inhibition assay
The effect of the compounds on the proliferation of human tumor cell lines was determined using the XTT (sodium 30-[1-(phenylaminocarbonyl)-3,4-tetrazolium]-bis(4-methoxy-6-nitro)-benzenesulfonic acid hydrate) cell proliferation kit (Roche Molecular Biochemicals, Mannheim, Germany) according to the manufacturer’s instructions as previously described [32]. Cells were incubated for 72 h in 96-well flat-bottomed microtiter plates (15,000 cells/mL, 100 µL/well) at 37 °C in a humidified atmosphere of air with 5% CO2 in culture medium containing 10% heat-inactivated FBS in the absence (control) and the presence of compound 5 at concentrations ranging from 10-5 to 10-10 M. After incubation, the XTT assay was performed. Each experiment was repeated three times and measurements were performed in triplicate. The IC50 (50% inhibitory concentration) value, defined as the drug concentration required to cause 50% inhibition in cellular proliferation with respect to the untreated controls, was determined for each compound by nonlinear curve fitting of the experimental data.
2.3.4. Cell cycle analysis
For cell cycle analyses, untreated and drug-treated cells (50000 cells/mL) were centrifuged and fixed overnight in 70% ethanol at 4 °C. Then cells were washed three times with PBS, incubated for 1 h with 1 mg/mL RNase A and 20 mg/mL propidium iodide at room temperature, and analyzed with a BD Accuri™-C6 Plus flow cytometer and the C6 (version 1.0.264.21) software (BD Biosciences) to determine cell cycle distribution and compared to control cells.
2.3.5. Confocal microscopy
HeLa cells were grown on 0.01% poly-l-lysine coated coverslips and after drug treatment, the coverslips were washed three times with PBS buffer, fixed with 4% formaldehyde in PBS buffer for 20 min, and permeabilized with 0.5% Triton X-100 as previously described [33]. Coverslips were incubated with a specific Ab-1, anti α-tubulin mouse monoclonal antibody (diluted 1:150 in PBS, Calbiochem, San Diego, CA) for 1 h, washed four times with PBS, and then incubated with donkey anti-mouse IgH (H + L) highly cross-adsorbed secondary Antibody, Alexa Fluor 488 (diluted 1:100 in PBS, Thermofisher Scientific) for 1 h at 4 °C. After four washes with PBS, a drop of ProLongTM Gold antifade reagent with DAPI (blue fluorescence) was added to stain nuclei. The samples were kept at 4 °C in a humidified chamber in the dark and analyzed in a Leica TCS SP5 confocal microscope. Negative controls lacking the primary antibody and alternatively, with an irrelevant primary antibody showed no fluorescence.
3. Results
We designed a systematic pipeline that allowed us to structurally explore the pharmacophores described in multiple tubulin crystallographic structures, and then search for molecular scaffolds regarding the explored pharmacophores. With this information, new chemical entities were designed and studied through MD simulations. Then, the lead compound was synthesized and evaluated for its biological activity against tubulin. The effect on tubulin polymerization was determined in vitro by measuring the degree of inhibition of polymerization of microtubular protein and in cells by confocal microscopy studies. The cytotoxic activity and effect on the cell cycle distribution profile was also measured. Our findings proved to be in concordance with tubulin inhibition by binding at the colchicine site, thus validating the virtual-screening and design.
3.1. Exploring tubulin crystallographic structures
For the selection of the tubulin crystallographic structures, tubulin crystals deposited in the PDB database were explored. The structures that had a crystallized ligand in the colchicine binding site (located between tubulin α and β subunits) were selected. This resulted in 81 structures, out of which 14 have colchicine as a co-crystallized ligand (Table S1). The structures were prepared with Maestro [21], leaving only α and β subunits, as well as the co-crystallized ligands at the colchicine site, ions, metals, and phosphorylated nucleotides. The colchicine binding site is composed of 3 zones denoted as A, B, and C (Fig. 1). We studied the binding site occupation by the co-crystallized ligands and found that 73% of the compounds interact with tubulin at A and B zones, 17% at A and C zones, and 10% at A, B and C zones (Table S1).
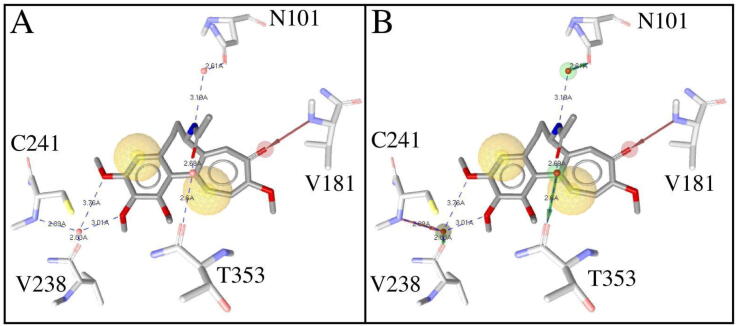
All tubulin PDB structures were pre-aligned against the α-subunit of the tubulin-colchicine complex (PDB code: 4O2B) using PyMol [30]. Then, for each complex pharmacophores representing tubulin–ligand interactions derived from each compound and its surrounding amino acid residues were identified using Phase [24] and LigandScout [23]. In cases where water-bridge interactions between the ligand and the binding site residues in tubulin were present, two different pharmacophores were created, with and without water(s). For instance, the tubulin-colchicine complex (PDB code: 5ITZ) presents water-bridge interactions, so, when water-bridge interactions are not considered, colchicine interacts with tubulin through hydrophobic contacts and one hydrogen bond with V181 (Fig. 2A). In contrast, when waters are considered a water-bridge network extends the colchicine interactions to residues N101, V238, C241, and T353 (Fig. 2B). This is why in our analysis we took into account “extended” pharmacophores considering waters, and pharmacophores without waters. In the end, a total of 118 pharmacophores were identified, 81 without waters and 37 with waters.
Fig. 2.
Structure-based pharmacophores for tubulin-colchicine complex (PDB code: 5ITZ). Pharmacophores without (A) and with (B) water included. Hydrogen bond acceptor (red), hydrogen bond donor (green), and hydrophobic (yellow) pharmacophoric features are displayed to illustrate how colchicine interacts with tubulin. Distances between interacting atoms are shown as dotted lines. Waters are illustrated as red spheres. For better visualization, hydrogens are not displayed. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Our pharmacophore ensemble covers all the points proposed by Nguyen et al [10] and Wang et al [34] (Fig. S1A), but taking into account many individual pharmacophores instead of a single representative of the ensemble provides a more diffuse definition of the elements and the distances between them that in turn results in a less restrictive selection. The incorporation of bridging waters in the definitions of pharmacophores further extends the accepted elements.
Performing conformational screening using multiple pharmacophores of the crystallized ligands at the colchicine binding site allows us to expand the knowledge we may have regarding the unique key interactions of certain ligands at this site. To study how similar the pharmacophores (with and without waters) used in this study are, we employed the tool “align_hypoPair” (included in Schrödinger's Suite – www.schrodinger.com/kb/132) to align one pharmacophore to another. Alignment is done using least-squares fitting of the matching site features in the two pharmacophores, considering all possible mappings. The best RMSD was selected to compute a similarity for each alignment and thus build a similarity matrix (the lower the value the higher the similarity). Then, a distance matrix was calculated and sorted (Fig. S1B) to compare the differences between the pharmacophores used in this study. The results indicate that there is no major similarity between the pharmacophores, which is mainly due to the diversity of ligands used, and because of the inclusion of key waters for the interaction of these ligands at the colchicine binding site.
3.2. Exploring ligand scaffolds through pharmacophore virtual-screening
To validate the Flexi-pharma methodology using crystal structures, we screened the 81 co-crystalized ligands against the identified pharmacophores in three different setups: 1) pharmacophores without waters screened with Phase [24], 2) pharmacophores without waters screened with Pharmer [25], and 3) pharmacophores with waters screened with Pharmer. The total of screened pharmacophores in each setup varied due to the screening algorithm employed by each software (see Materials and Methods section). The results indicated that both software could reproduce the pharmacophore features with a high hit-match rate, 96% and 86% for Phase and Pharmer, respectively (Table S2). In addition, we also screened a dataset of 3354 decoys randomly extracted from DUD-E database (http://dude.docking.org/). Our results indicate that both Phase and Pharmer software presented higher prediction rates for positive controls (tubulin co-crystallized ligands) versus decoys (Fig. S2). The score distribution shows that for both programs the ligands have (on average) a higher score and better performance, indicating that it is possible to obtain an enrichment from the analysis.
3.2.1. Scaffold and compound libraries
Based on the structural characteristics of the colchicine site (Fig. 1), we searched the ZINC database [35] for several scaffolds that could fit colchicine binding site zones A, B, and C with aromatic and/or heteroaromatic rings. For the connections between the aryl rings, we have explored three structural options (sulfonamides, tetrazoles, or methenyles) that place the substituents at both ends (aryls) at different distances and geometries. This selection was made to also provide a straightforward synthetic access to compounds combining the preferred structural features found in the pharmacophoric search and not present in the searched database, therefore increasing originality, for the final validation of the methodology with experimental examples. In particular, we focused on searching the ZINC database for molecules containing these scaffolds that were divided into 20 subgroups (Fig. S3). The molecules from ZINC were then filtered by hydrophobicity and size (see Materials & Methods section), and the set was divided into different subgroups according to the scaffold used for the search. In the end, 8,918 molecules from ZINC database were included for the ePBVS as follows: Group 1: tetrazoles, divided into 9 subgroups with 2713 molecules; group 2: diarylmethane, divided into 4 subgroups with 2966 molecules; and group 3: sulfonamides, divided into 7 subgroups with 3239 molecules (Fig. S3).
3.2.2. Flexi-pharma virtual-screening using the crystallographic ensemble
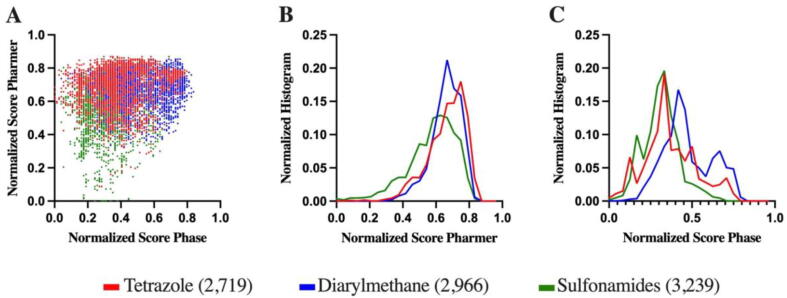
The 8918 molecules, representing 20 different scaffolds grouped as tetrazoles, diarylmethane and sulfonamides (Fig. S3) were screened with Phase and Pharmer against the tubulin pharmacophores previously created. For each molecule, a match (1) or mismatch (0) was determined in terms of interaction with each tubulin pharmacophore using both programs (see the Materials and Methods section). We used the frequency hits (normalized number of matches) for both software to score the compounds. Fig. 3A shows the frequency hits for the three groups of compounds screened against tubulin pharmacophores. Fig. S4 shows detailed frequency hits for each of the 20 subgroups. Interestingly, the results for the two programs are not completely correlated, validating the use of two different pharmacophore-screening programs to obtain complementary results. We found that with Pharmer the tetrazoles interact slightly more with a greater number of tubulin pharmacophores, while with Phase the ones that interact with more pharmacophores are diarylmethanes (Fig. 3B-C).
Fig. 3.
Frequency hits (normalized number of matches) for Pharmer and Phase: correlation plot (A), and normalized histograms for Pharmer (B) and Phase (C) for the three subgroups of compounds.
The colchicine-site occupation by the screened molecules was also monitored. The AB zones occupied 73% of the crystallographic structures selected in this study (Table S1), and exhibited the highest occupancy rate with both Pharmer and Phase (Fig. S5). For the virtual-screening done with Phase, the diarylmethanes mainly occupied the AB zones (101,434 hits), followed by AC (9452 hits), BC (946 hits), and ABC (48 hits) (Fig. S5A). Both sulfonamides and tetrazoles scaffolds presented similar occupations in the colchicine site. It is worth noting that a greater number of hits were identified in the Pharmer screening because 37 more pharmacophores were employed (those that include water). Again, the diarylmethane scaffold presented the highest occupation, mainly occupying the AB zones (221,649 hits), followed by AC (54,833 hits), ABC (23,206 hits), and BC (3239 hits) (Fig. S5B). The methodological design presented here allowed us to explore multiple tubulin structures and find potential molecular scaffolds to design new tubulin modulators.
3.3. Design of novel tubulin modulators
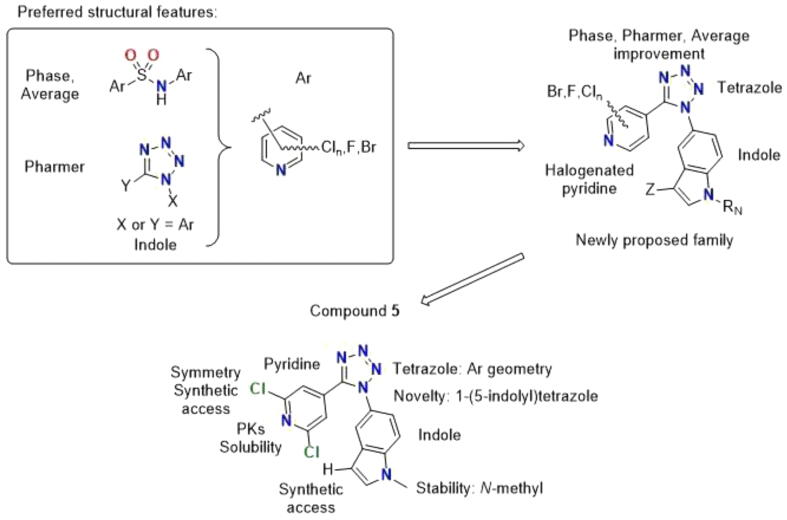
With the Flexi-pharma [17] virtual-screening results, we have analyzed the structures of the top-100 results for each program individually and for the average of its frequency-hit. The top-100 results for Phase (average frequency of 0.78) are also well scored by Pharmer (average frequency 0.72), but the top-100 results for Pharmer (average frequency of 0.85) are poorly scored by Phase (average frequency 0.49) (Table S3). The top-100 solutions for the average frequency score were also analyzed (Table S4), and we found that the top-100 ranked ZINC compounds presented similar Phase and Pharmer average frequencies, 0.76 and 0.79, respectively. We then analyzed the structural features selected by each scoring program to combine them in a new structural family not present in the ZINC database. We seeked to improve structural novelty and predicted activity while having synthetic accessibility, thus allowing easy access. Selecting a combination of features instead of ligands already represented in the database increases the ligand originality. The 2D-structures of Phase, Pharmer and average frequency score are shown in Tables S5, S6 and S7, respectively. Analyzing the results for the top-100 using the average frequency score (Table S4), it is observed that sulfonamides are the most favored scaffolds, representing 67%, with an average frequency score of 0.78, followed by tetrazoles and diarylmethanes, with 31% and 2%, respectively. Interestingly, the highly scored tetrazoles of Pharmer were mostly monoaryl tetrazoles (subgroup 8 – Fig. S3, Fig. S4D, and Table S3), and the lack of the second aryl group was considered the reason for the low scoring given by Phase. Therefore, the new family was designed to include diaryl tetrazoles. The second outstanding feature of the top ligands was the highly overrepresentation of halogenated pyridine residues in all the top subsets (Tables S5–S7), and we therefore decided to incorporate one such halogenated pyridine ring. For synthetic convenience and preferring more symmetric options, we opted for 4-pyridines with halogen atoms. Finally, the presence of the 5-indolyl substituent in the top-100 results for the average subset, despite its quite scarce presence in the ZINC database, prompted us to include it in the design to further increase the structural novelty. As a result of the above considerations, we have proposed a new family of halogenated 5-(5-(pyridin-4-yl)-1H-tetrazol-1-yl)-1H-indoles with different substituents (Fig. 4) in synthetically favored positions. From this new family, we selected compound 5 as the most synthetically accessible representative, we re-evaluated it with our computational model and we finally assayed it as a tubulin inhibitor as a proof of principle.
Fig. 4.
Design of the new family of colchicine site inhibitors based on the results of the Flexi-pharma virtual screening on the focussed ZINC sub-library. The most prominent structural features observed for the top-100 compounds from with Phase and Pharmer were combined in the new proposed family. Compound 5 was selected as an easily accessible representative due to the indicated reasons.
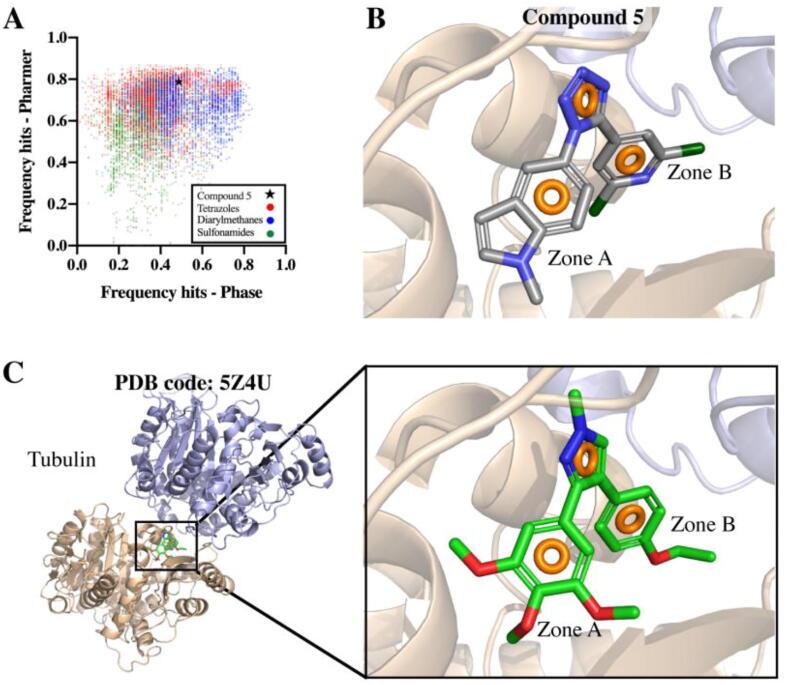
To corroborate that compound 5 indeed matches with the appropriate pharmacophores, thus interacting properly with tubulin at the colchicine site, a Flexi-pharma virtual-screening for this compound was performed against all tubulin pharmacophores using Phase and Pharmer. The results indicated that this compound indeed presents good frequency-hits with both software (Phase: 0.49, Pharmer: 0.79) (Fig. 5A). Additionally, it was observed that compound 5 interacts with tubulin through a 3-point pharmacophore, integrated by three aromatic rings, two of them located at A and B zones (Fig. 5B). These results are in agreement with the 3-point pharmacophore present in PDB 5Z4U crystallographic structure (Fig. 5C).
Fig. 5.
Flexi-pharma virtual-screening of compound 5 against tubulin multiple pharmacophores. A. Pharmer vs. Phase frequency hits including screened scaffolds. B. Compound 5 interacting with tubulin. C. Tubulin-ligand complex (PDB code: 5Z4U). Aromatic rings pharmacophoric features are displayed as orange rings. Tubulin α and β subunits are shown as sand and blue cartoons, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
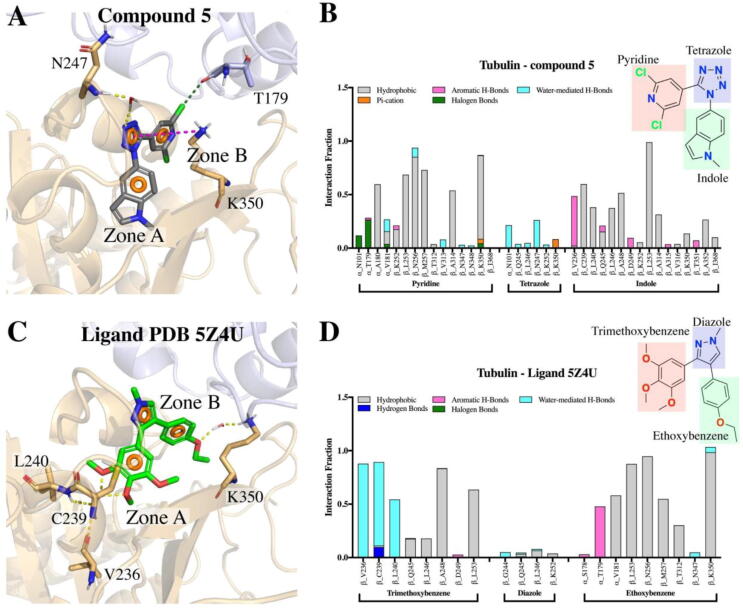
Using molecular dynamics simulations (MDs), we studied how compound 5 interacts at the colchicine site between the α and β subunits of tubulin. We analyzed both complexes, tubulin-compound 5 and 5Z4U crystal structure through 500 ns-MDs (Fig. 5B-C). To analyze the stability of tubulin throughout the simulation, the RMSD of the protein backbone was monitored in both systems, and it was observed that no major changes occurred within 500 ns of simulation (Figs. S6A and S6D). It was also observed that both ligands remained stable at the colchicine site during the simulation (Fig. S6A-E). Both ligands interact at Zones A-B mainly through hydrophobic interactions (Fig. 6). Pyridine, tetrazole and indole moieties of compound 5 remain interacting in zones A, bridge and B throughout the simulation (Fig. 6A), whereas 5Z4U ligand moieties trimethoxybenzene, diazole and ethoxybenzene interact at zones A, bridge and B, respectively (Fig. 6C).
Fig. 6.
Contacts of compound 5 (A-B) and ligand 5Z4U (C-D) with residues of the colchicine binding site during the 500 ns MDs and the nature of the chemical interactions. Hydrogen bonds are represented as dotted yellow lines. Interactions between the tubulin residues and analyzed ligands are categorized into five types: hydrophobic, hydrogen bonds, aromatic hydrogen bonds, halogen bonds and water-mediated hydrogen bonds (water bridges). The stacked bar charts are normalized over the course of the unrestrained MDs. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The principal interactions of compound 5 are hydrophobic, mainly between pyridine and indole moieties with residues α_A180, β_C239, β_L240, β_L246, β_A248, β_L253, β_N256, β_M257, β_A314, and β_K350. The tetrazole interacts through water-mediated hydrogen bonds (water bridges) with α_N101 and β_N247 residues. The compound 5 interaction pattern is similar to the ligand from the 5Z4U crystal structure, where the substituted phenyls are the ones that interact mainly with tubulin. In fact, the structural similarity of compound 5 is much greater to the 5Z4U-ligand than to other tubulin inhibitors that interact in different binding sites, which strengthens our hypothesis that compound 5 is indeed an inhibitor of the colchicine binding site (Table S8). The main interaction differences between both ligands were observed in the halogen bonds that compound 5 establish during simulation (Figs. 6, S6C and S6F), since compound 5, having two chlorines in the pyridine moiety establishes halogen bonds throughout the simulation, mainly with α_N101 and α_T179 (Fig. 6A-B). This interaction is not present for ligand 5Z4U, since there are no halogens in its structure (Figs. 5C and 6D). Additionally, compound 5 also presented a few Pi-cation interactions with residue K350 during the simulation (Fig. 6A-B and S6C). Finally, the trimethoxybenzene moiety of ligand 5Z4U, due its hydrophilic nature, is able to establish water-mediated hydrogen bonds with residues β_V236, β_C239 and β_L240.
The physicochemical descriptors of compound 5 were calculated (Table 1). It is worth noting here that compound 5 has a MW < 500 g/mol, which is required of compounds to be potentially useful as drugs. All the calculated physicochemical descriptors and pharmacokinetics properties are in the defined acceptable ranges, meeting the criteria of drug-likeness according to Lipinski’s rule of five. The calculated properties to predict drug-likeness according to Ghose, Veber, Egan, and Muegge rules [36] (Table 2) show that compound 5 does not present any violation. 5 has good predicted solubility values above micromolar, much better than combretastatin A4 and even an order of magnitude better than ABT-751, probably due to its increased TPSA of 61 Å2. This value, combined with a clogP close to 3 is a good toxicity predictor [37].
Table 1.
Physicochemical and pharmacokinetic descriptors of compound 5 calculated with SwissADME [36].
| Physicochemical Properties | Lipophilicity | Water Solubility (mol/L) | Pharmacokinetics | ||||
|---|---|---|---|---|---|---|---|
| MW1 | 345.19 | Log Po/w (iLOGP) | 2.93 | ESOL | 1.26E-05 | GI abs6 | High |
| RB2 | 2 | Log Po/w (XLOGP3) | 3.82 | Ali | 1.57E-05 | BBB7 | Yes |
| HB-A3 | 4 | Log Po/w (WLOGP) | 3.52 | SILICOS-IT | 1.07E-06 | log Kp (cm/s)8 | −5.69 |
| HB-D4 | 0 | Log Po/w (MLOGP) | 3.23 | ||||
| TPSA5 | 61.43 | Log Po/w (SILICOS-IT) | 2.6 | ||||
| Average Log Po/w | 3.22 | ||||||
Table 2.
Drug-likeness properties of the compound 5 calculated with SwissADME [36].
| Lipinski # violations1 | Ghose # violations2 | Veber # violations3 | Egan # violations4 | Muegge # violations5 | Bioavailability Score6 |
|---|---|---|---|---|---|
| 0 | 0 | 0 | 0 | 0 | 0.55 |
Lipinski (Pfizer) filter [29]: MW ≤ 500; MLOGP ≤ 4.15; N or O ≤ 10; NH or OH ≤ 5.
Ghose filter [30]: 160 ≤ MW ≤ 480; −0.4 ≤ WLOGP ≤ 5.6; 40 ≤ MR ≤ 130; 20 ≤ atoms ≤ 70.
Veber (GSK) filter [31]: Rotatable bonds ≤ 10; TPSA ≤ 140.
Egan (Pharmacia) filter [32]: WLOGP ≤ 5.88; TPSA ≤ 131.6.
Muegge (Bayer) filter [33]: 200 ≤ MW ≤ 600; −2 ≤ XLOGP ≤ 5; TPSA ≤ 150; Number of rings ≤7; Number of carbon atoms >4; Number of heteroatoms >1; Number of rotatable bonds ≤15.
Abbott Bioavailability Score [40].
3.4. Synthesis and biological evaluation as antitumor antimitotic agents
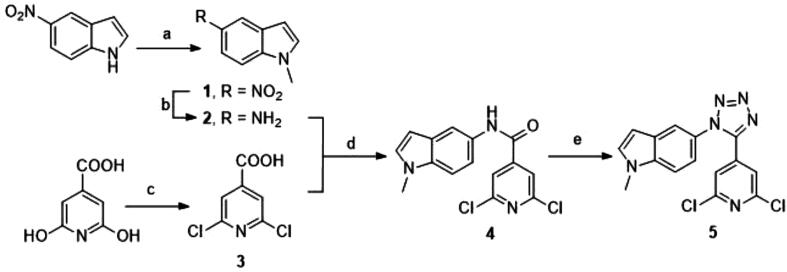
3.4.1. Chemical synthesis
The selected tetrazole derivative 5 was prepared as outlined in Scheme 1. 5-nitroindole was N-methylated under phase transfer basic catalysis treatment to obtain N-methyl-5-nitroindole (1). Heterogeneous catalytic hydrogenation was performed for the reduction of nitro group for the required N-methyl-5-aminoindole (2) [41]. On the other hand, 2,6-dichloroisonicotinic acid (3) was prepared from citrazinic acid by treatment with neat phosphorous oxychloride [42]. Amide coupling reaction between the amino group of 2 and the carboxylic acid derivative 3 was achieved with 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) as carboxyl activating agent to obtain compound 4. The tetrachlorosilane-azide [43] system was employed to transform the amido group into the 1,5-disubstituted tetrazole ring to synthesize the target compound 5 (Scheme 1).
Scheme 1.
Synthesis of tetrazole derivative 5. Reagents and conditions: (a) MeI, NaOH, n-Bu4NHSO4, dry DCM, rt, 24 h; (b) H2, Pd/C, MeOH, DCM, rt, 24 h; (c) POCl3, Me4NBr, 90–140 °C, 24 h; (d) EDC, DMAP, dry DCM, reflux, 24 h; (e) NaN3, SiCl4, MeCN, reflux, 24 h.
3.4.2. Biological assays
In order to study the effect of compound 5 on tubulin, the in vitro inhibitory activity on the microtubular protein polymerization was performed. Tubulin was isolated and purified from calf brains following a modified Shelanski procedure [44], [45]. The tubulin polymerization inhibition IC50 value obtained for 5 was 2.8 µM, similar to the reference compound combretastatin A-4 (3 µM) [46] thus confirming the interaction with tubulin.
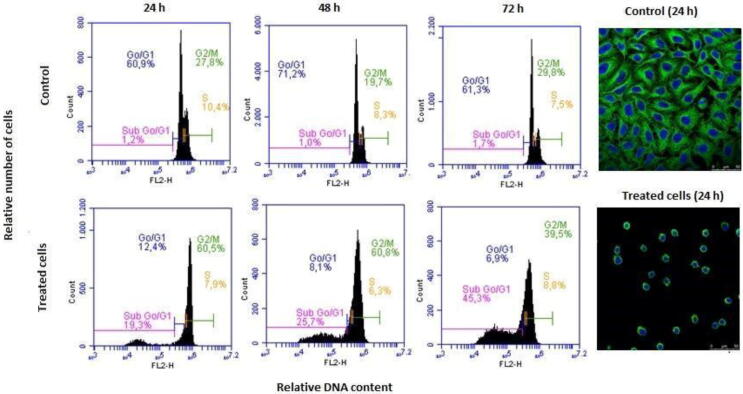
The cell proliferation inhibitory activity of the tetrazole derivative 5 was assayed by the XTT method against HeLa cells from human cervix epithelioid carcinoma, following previously described procedures [32], [46]. A preliminary evaluation at 1 µM (three independent assays in at least two independent assays) showed more than 50% growth inhibition, and thereafter it was evaluated at concentrations ranging from 10 µM to 0.1 nM. The compound exhibited a remarkable cytotoxic effect, with an IC50 value of 45 nM. In order to elucidate the mechanism of action of the compound in HeLa cells, the effect on the cell cycle progression was evaluated by flow cytometry at different time points (24, 48 and 72 h) after drug treatment. The fluorescent dye propidium iodide (PI) was used for DNA quantification. After 24 h, compound 5 promoted a significant increase in the percentage of cells in the G2/M phase as compared with the untreated sample (60.5% vs. 27.8%, respectively), and the G2/M arrest was maintained 48 h after treatment. This effect was accompanied by the appearance of the Sub G0/G1 fraction, an hypodiploid peak which corresponds to apoptotic cells that undergo DNA condensation and fragmentation. The Sub G0/G1 region represented 19.3% and 25.7 % of cell population after 24 h and 48 h, respectively. At a later time point of 72 h the G2/M peak decreased to 39.5% while more than 45% of the cells were suffering apoptosis. The observed mitotic arrest followed by the induction of apoptosis is consistent with the mechanism of action of tubulin binding drugs.
The alteration of tubulin cytoskeleton in HeLa cells was assessed by confocal immunofluorescence microscopy as described [46]. The experiments were performed 24 h after cell treatment when the maximum antimitotic effect can be observed having minimal cell death. The compound 5 promoted a microtubule network disruption in HeLa cells whereas in the micrograph for the untreated sample it is possible to distinguish the organized tubulin fibers.
4. Discussion
Ligands binding at the colchicine site of tubulin perturb tubulin polymerization and block the mitosis of eukaryotic cells, thus causing cell death [47]. As a consequence, they have great potential as chemotherapeutic agents against eukaryotic cells, such as those of cancer, parasites, plants, and fungi [1]. The blind search for ligands directed toward the colchicine site has yielded a plethora of chemical structures that have demonstrated that active ligands of different sizes and with great chemical diversity can be found [3]. It also exhibits surprisingly high class-dependent structure–activity relationships. As a result, attempts to combine ligands from different structural classes have been often unsuccessful [9].
Recent increase in tubulin 3D-structures (deposited in the PDB) in complexes with ligands bound to the colchicine site has provided structural insights related to tubulin inhibition. The colchicine site is subdivided in three different pockets (zones A-C) variably interconnected, thus different binders induce on the neighbor sites subtle variations that limit their access [8], [9]. This site configuration imposes severe restrictions to the ligands binding to them and to the allowed linkages between pockets. This structural complexity and variability requires an exhaustive sampling of the protein flexibility in the application of structure-based drug design approaches that is not usually applied to reduce computational and analysis resources. The application of ensemble methodologies seems to be perfectly suited to this problem, but the mere combination of too explicit rigid snapshots, such as the structures used for ensemble docking, cannot efficiently reproduce the structural variations observed upon different ligand binding. Therefore, a less explicit approach, such as the pharmacophore ensemble here proposed, should provide a more flexible alternative for the design of new colchicine-site inhibitors.
Here, we explored 81 tubulin-ligand complexes (Table S1) along with the structural diversity of interactions presented at the colchicine binding site. To this effect we investigated how ligands interact with tubulin and extracted the pharmacophores describing such interactions, in each crystal structure examined, also taking into account water-mediated interactions [23]. It is important to highlight that we were able to perform this broad sampling due to the extensive tubulin structural information available in databases, and we recommend (when available data permit) using similar methodologies that enable sampling the multiple interaction options at a given binding site.
A representative dataset of scaffolds of interest was extracted from the ZINC database [35], resulting in ~9000 compounds representing tetrazoles, diarylmethanes and sulfonamides, then the dataset was screened against all tubulin-pharmacophores through a Flexi-pharma approach. This extensive sampling of thousands of compounds with structural diversity was performed in multiple tubulin-pharmacophores with two different software, and allowed us to determine which scaffolds best fit the studied tubulin-pharmacophores (Fig. 3 and Fig S4). We were also able to observe in which areas of the colchicine site (Fig. 1) the screened compounds interact most frequently (Fig. S5). The major interaction pockets were the AB zones, where mainly diarylmethanes and tetrazoles fit with the pharmacophores describing the interaction of multiple ligands with tubulin. To determine whether or not a compound was a good ligand for a given pharmacophore we used the number of matches as a criterion. When the compound matches at least 3 of the total pharmacophoric features described for a pharmacophore it is considered a match, and otherwise it is considered a mismatch. In this simple approach, and using two different software (Phase and Pharmer) employing different screening algorithms, we were able to calculate the frequency with which a given compound interacts with multiple tubulin pharmacophores (Fig. 3). Then, focusing on the top-100 results (Tables S3–S7) we found that diaryltetrazoles as well as halogenated pyridine were the scaffolds that best interact with the multiple conformations adopted by the colchicine site, described by the pharmacophores screened.
To prove that the identified scaffolds effectively inhibit tubulin, we proceeded to use them in the design of a new series of compounds (Fig. 4). According to synthetic accessibility in this work we proceeded to test compound 5 as a proof of concept. So, before synthesizing it we performed molecular dynamics simulations (500 ns) and studied how this compound potentially interacts with tubulin, comparing it with a known colchicine-site ligand with a similar chemical structure (PDB code 5X4U). Our results confirm that both ligands exhibit the same pharmacophore and interact very similarly with tubulin at the colchicine binding site, presenting mainly hydrophobic interactions (Fig. 6). Furthermore, compound 5 showed favourable predicted pharmacokinetic properties that make it a good starting point for new drug development projects.
The proposed tetrazole derivative 5 was obtained by a short synthetic route (1 to 5) in good overall yield. Amide 4 was prepared by amide coupling reaction between N-methyl-5-aminoindole (2) and 2,6-dichloroisonicotinic acid (3), and subsequently transformed in the tetrazole by treatment with tetrachlorosilane and sodium azide (Scheme 1).
The first step to check the success of the design strategy was to ascertain if compound 5 was able to inhibit tubulin polymerization (TPI). Microtubular protein was isolated from calf brain and its polymerization in vitro was monitored by the increase in turbidity detected as a reduced UV transmission upon a temperature change from 4 °C (soluble dimers) to 37 °C (microtubule formation, that cause the increase in sample turbidity). Inhibitors of tubulin polymerization decrease the amplitude of the increment and the percentage of reduction compared to an untreated sample gives the tubulin polymerization inhibitory activity at the tested concentration. The TPI IC50 is the compound concentration that reduces the turbidity measurement to 50% with respect to the control, and can be used to compare the inhibitory potency of different compounds. Compound 5 was found to be a potent tubulin polymerization inhibitor, having a micromolar IC50 value (2.8 µM) comparable to reference compounds combretastatin A-4 (3 µM) and ABT-751 (4 µM) [46], thus confirming the proposed hypothesis of interaction with tubulin.
Cell-permeable compounds able to alter the tubulin polymerization-depolymerization equilibria cause changes in the cells’ cytoskeleton that result in a disruption of the mitotic events as the more remarkable effect, due to the more dynamic character of these microtubules compared to those of the interphase. As a result of the treatment with such tubulin inhibitors, cell proliferation is reduced, cultured cells arrest in mitosis and accumulate in the G2/M phase of the cell-cycle compared to other phases, and cells eventually die [48]. We therefore assessed the effect of compound 5 on the proliferation of the cervix epithelioid carcinoma cell line HeLa by the XTT method that measures the metabolic activity of live cells. The compound exhibited an anti-proliferative IC50 value in the double-digit nanomolar range (45 nM), which is between those of combretastatin A-4 (2 nM) and ABT-751 (388 nM), two tubulin inhibitors in clinical trials [49]. To further confirm the anti-tubulin effect of 5 in cells, the cell cycle profile of treated cells was compared with untreated controls, and was analyzed by DNA staining and flow cytometric quantification. These studies revealed that the cytotoxic activity of compound 5 was achieved as a result of the mitotic arrest observed 24 and 48 h after drug treatment. Over 60% of cell population was accumulated in the G2/M phase 48 h after treatment whereas at a time point of 72 h the mitotic arrest led to cell death as demonstrated by the increase in the subG0/G1 region, up to 45.3%, which represents hypodiploid apoptotic cells (Fig. 7). To further ascertain that the effects are due to microtubule inhibition, immunofluorescence microscopy experiments in HeLa cells, performed by fluorescent labelling of tubulin 24 h after the drug treatment, revealed a complete microtubule network disorganization in comparison with the control sample, where the tubulin fibers were perfectly visible (Fig. 7). This observation is in agreement with inhibition of tubulin polymerization upon ligand interaction triggering cell cycle arrest at G2/M phase and ensuing apoptosis after 72 h. All these results are in good agreement with the design objectives of achieving easily accessible tubulin inhibitors with adequate pharmacokinetics for acting inside cells.
Fig. 7.
Effects of compound 5 on the microtubule network and cell cycle in HeLa cells. Cells were incubated in the absence (control) or in the presence of 100 nM of compound 5 for 24, 48 or 72 h and analyzed by flow cytometry (left) and by confocal microscopy (right).
The use of the Flexi-pharma methodology described here allowed us not only to identify the scaffolds to be included in the new microtubule polymerization inhibitors to be designed, but also helped us to predict how they should be interconnected and where they would interact with tubulin. It allowed us to save time and resources that we would have had to invest if we had used a different sampling method, such as molecular docking, which has proven to be ineffective when the flexibility of the receptor is not taken into account [50], [51]. So, it is always recommended to couple docking to other methods such as molecular dynamics and free energy calculations. When attempting to explore more than 80 structures of the same target, each with ~9000 different compounds (as we did in this study) a methodology such as Flexi-pharma is effective and relatively fast. Finally, having identified, designed, synthesized and proven that compound 5 is indeed a potent inhibitor of tubulin polymerization, in future work we will modify this compound by medicinal chemistry to obtain better biological results and thus enhance as potential antimitotic tubulin inhibitors.
5. Conclusions
In this work, we used a pharmacophore-screening methodology for identifying key components inside the colchicine-binding site of tubulin. Due to the large availability of X-ray crystal structures in complex with multiple ligands in different subpockets, we could generate an ensemble of pharmacophores to take into account the flexibility and diversity of this binding site. The Flexi-pharma campaign allowed us to identify several characteristics to design a novel family of colchicine-site inhibitors. In particular, diaryltetrazoles and halogenated pyridine were the scaffolds that lead to the easily synthesizable tetrazole-derivative 5. This compound was shown to be a potent tubulin inhibitor in vitro, and presented anti-proliferative activity, through microtubule disruption, against human epithelioid carcinoma HeLa cells. Overall, our results show that sampling the flexibility of a given binding site prior to evaluating potential ligands by virtual screening, using methodologies such as Flexi-pharma, enables the efficient design of new active compounds due to the recognition of key interactions and molecular determinants for the modulation of a specific target.
CRediT authorship contribution statement
Laura Gallego-Yerga: Methodology, Investigation, Validation, Writing – original draft. Rodrigo Ochoa: Methodology, Investigation, Data curation, Writing – original draft. Isaías Lans: Methodology, Investigation. Carlos Peña-Varas: Methodology, Data curation, Visualization. Melissa Alegría-Arcos: Methodology, Data curation, Visualization. Pilar Cossio: Methodology, Supervision, Writing – original draft, Writing - review & editing, Funding acquisition. David Ramírez: Methodology, Investigation, Writing – original draft, Writing - review & editing, Funding acquisition, Supervision. Rafael Peláez: Methodology, Investigation, Writing – original draft, Writing - review & editing, Funding acquisition, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors acknowledge the support of the Enlighten Your Research LatinAmerica2Europe (EYR-LA2EU) programme. I.L, R.O. and P.C. were also supported by MinCiencias, University of Antioquia, Ruta N, Colombia and the Max Planck Society, Germany. L.G-Y. and R.P-L. acknowledge the support by the Consejería de Educación de la Junta de Castilla y León (SA262P18 and SA116P20) and the Spanish Ministry of Science, Innovation and Universities (RTI2018-099474-BI00), co-funded by the EU's European Regional Development Fund-FEDER. D.R. was supported by the CONICYT-PCI grant N° REDES190074 and FONDECYT grant N° 11180604. This work has been partially carried out on the ACME cluster, which is owned by CIEMAT and funded by the Spanish Ministry of Economy and Competitiveness project CODEC2 (TIN2015-63562-R) with FEDER funds as well as supported by the CYTED co-founded RICAP Network (517RT0529).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.07.039.
Contributor Information
David Ramírez, Email: david.ramirez@uautonoma.cl.
Rafael Peláez, Email: pelaez@usal.es.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Dumontet C., Jordan M.A. Microtubule-binding agents: a dynamic field of cancer therapeutics. Nat Rev Drug Discov. 2010;9(10):790–803. doi: 10.1038/nrd3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jordan M.A., Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer. 2004;4(4):253–265. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- 3.Vicente‐Blázquez A., González M., Álvarez R., del Mazo S., Medarde M., Peláez R. Antitubulin sulfonamides: The successful combination of an established drug class and a multifaceted target. Med Res Rev. 2019;39(3):775–830. doi: 10.1002/med.21541. [DOI] [PubMed] [Google Scholar]
- 4.Kavallaris M. Microtubules and resistance to tubulin-binding agents. Nat Rev Cancer. 2010;10(3):194–204. doi: 10.1038/nrc2803. [DOI] [PubMed] [Google Scholar]
- 5.Pérez-Pérez M.-J., Priego E.-M., Bueno O., Martins M.S., Canela M.-D., Liekens S. Blocking blood flow to solid tumors by destabilizing tubulin: an approach to targeting tumor growth. J Med Chem. 2016;59(19):8685–8711. doi: 10.1021/acs.jmedchem.6b00463. [DOI] [PubMed] [Google Scholar]
- 6.Ravelli R.B.G., Gigant B., Curmi P.A., Jourdain I., Lachkar S., Sobel A. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 2004;428(6979):198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- 7.Prota AE, Magiera MM, Kuijpers M, Bargsten K, Frey D, Wieser M, et al. Structural basis of tubulin tyrosination by tubulin tyrosine ligase. J Cell Biol 2013;200:259–70. [DOI] [PMC free article] [PubMed]
- 8.Massarotti A., Coluccia A., Silvestri R., Sorba G., Brancale A. The tubulin colchicine domain: a molecular modeling perspective. ChemMedChem. 2012;7(1):33–42. doi: 10.1002/cmdc.201100361. [DOI] [PubMed] [Google Scholar]
- 9.Álvarez R., Medarde M., Peláez R. New ligands of the tubulin colchicine site based on X-ray structures. Curr Top Med Chem. 2014;14:2231–2252. doi: 10.2174/1568026614666141130092637. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen T.L., McGrath C., Hermone A.R., Burnett J.C., Zaharevitz D.W., Day B.W. A common pharmacophore for a diverse set of colchicine site inhibitors using a structure-based approach. J Med Chem. 2005;48(19):6107–6116. doi: 10.1021/jm050502t. [DOI] [PubMed] [Google Scholar]
- 11.Zhou Y., Di B., Niu M.-M. Structure-based pharmacophore design and virtual screening for novel tubulin inhibitors with potential anticancer activity. Molecules. 2019;24(17):3181. doi: 10.3390/molecules24173181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wieder M., Garon A., Perricone U., Boresch S., Seidel T., Almerico A.M. Common hits approach: combining pharmacophore modeling and molecular dynamics simulations. J Chem Inf Model. 2017;57(2):365–385. doi: 10.1021/acs.jcim.6b00674. [DOI] [PubMed] [Google Scholar]
- 13.Polishchuk P., Kutlushina A., Bashirova D., Mokshyna O., Madzhidov T. Virtual screening using pharmacophore models retrieved from molecular dynamic simulations. Int J Mol Sci. 2019;20(23):5834. doi: 10.3390/ijms20235834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lone M.Y., Kumar S.P., Athar M., Jha P.C. Exploration of Mycobacterium tuberculosis structural proteome: an in-silico approach. J Theor Biol. 2018;439:14–23. doi: 10.1016/j.jtbi.2017.11.021. [DOI] [PubMed] [Google Scholar]
- 15.Manhas A., Lone M.Y., Jha P.C. In search of the representative pharmacophore hypotheses of the enzymatic proteome of Plasmodium falciparum: a multicomplex-based approach. Mol Divers. 2019;23(2):453–470. doi: 10.1007/s11030-018-9885-5. [DOI] [PubMed] [Google Scholar]
- 16.Lone M.Y., Athar M., Manhas A., Jha P.C., Bhatt S., Shah A. In silico exploration of vinca domain tubulin inhibitors: a combination of 3D-QSAR-based pharmacophore modeling, docking and molecular dynamics simulations. ChemSelect. 2017;2(33):10848–10853. doi: 10.1002/slct.201701971. [DOI] [Google Scholar]
- 17.Lans I., Palacio-Rodríguez K., Cavasotto C.N., Cossio P. Flexi-pharma: a molecule-ranking strategy for virtual screening using pharmacophores from ligand-free conformational ensembles. J Comput Aided Mol Des. 2020;34(10):1063–1077. doi: 10.1007/s10822-020-00329-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong M., Liu F., Zhou H., Zhai S., Yan B. Novel natural product- and privileged scaffold-based tubulin inhibitors targeting the colchicine binding site. Molecules. 2016;21:1375. doi: 10.3390/molecules21101375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu Y., Chen J., Xiao M., Li W., Miller D.D. An overview of tubulin inhibitors that interact with the colchicine binding site. Pharm Res. 2012;29(11):2943–2971. doi: 10.1007/s11095-012-0828-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mannhold R., van de Waterbeemd H. Substructure and whole molecule approaches for calculating log P. J Comput Aided Mol Des. 2001;15:337–354. doi: 10.1023/a:1011107422318. [DOI] [PubMed] [Google Scholar]
- 21.Schrödinger. Release 2020-1: Maestro, Schrödinger, LLC, New York, NY 2020.
- 22.Harder E., Damm W., Maple J., Wu C., Reboul M., Xiang J.Y. OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J Chem Theory Comput. 2016;12(1):281–296. doi: 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- 23.Wolber G., Langer T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J Chem Inf Model. 2005;45:160–169. doi: 10.1021/ci049885e. [DOI] [PubMed] [Google Scholar]
- 24.Dixon S.L., Smondyrev A.M., Rao S.N. PHASE: a novel approach to pharmacophore modeling and 3D database searching. Chem Biol Drug Des. 2006;67(5):370–372. doi: 10.1111/j.1747-0285.2006.00384.x. [DOI] [PubMed] [Google Scholar]
- 25.Koes D.R., Camacho C.J. Pharmer: efficient and exact pharmacophore search. J Chem Inf Model. 2011;51(6):1307–1314. doi: 10.1021/ci200097m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng A., Merz K.M. Application of the Nosé−Hoover chain algorithm to the study of protein dynamics. J Phys Chem. 1996;100(5):1927–1937. [Google Scholar]
- 27.Martyna G.J., Tobias D.J., Klein M.L. Constant pressure molecular dynamics algorithms. J Chem Phys. 1994;101(5):4177–4189. [Google Scholar]
- 28.Bowers KJ, Chow E, Xu H, Dror RO, Eastwood MP, Gregersen BA, et al. Scalable algorithms for molecular dynamics simulations on commodity clusters, 2006, p. 43–43.
- 29.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 30.DeLano WL. PyMOL. DeLano Scientific, San Carlos, CA 2002;700.
- 31.Álvarez R., Puebla P., Díaz J.F., Bento A.C., García-Navas R., de la Iglesia-Vicente J. Endowing indole-based tubulin inhibitors with an anchor for derivatization: highly potent 3-substituted indolephenstatins and indoleisocombretastatins. J Med Chem. 2013;56(7):2813–2827. doi: 10.1021/jm3015603. [DOI] [PubMed] [Google Scholar]
- 32.Jiménez C., Ellahioui Y., Álvarez R., Aramburu L., Riesco A., González M. Exploring the size adaptability of the B ring binding zone of the colchicine site of tubulin with para-nitrogen substituted isocombretastatins. Eur J Med Chem. 2015;100:210–222. doi: 10.1016/j.ejmech.2015.05.047. [DOI] [PubMed] [Google Scholar]
- 33.Maya A.B.S., Pérez-Melero C., Mateo C., Alonso D., Fernández J.L., Gajate C. Further naphthylcombretastatins. An investigation on the role of the naphthalene moiety. J Med Chem. 2005;48(2):556–568. doi: 10.1021/jm0310737. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y., Zhang H., Gigant B., Yu Y., Wu Y., Chen X. Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery. FEBS J. 2016;283(1):102–111. doi: 10.1111/febs.13555. [DOI] [PubMed] [Google Scholar]
- 35.Irwin J.J., Shoichet B.K. ZINC-a free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45(1):177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 2017;7:42717–42717. [DOI] [PMC free article] [PubMed]
- 37.Hughes J.D., Blagg J., Price D.A., Bailey S., DeCrescenzo G.A., Devraj R.V. Physiochemical drug properties associated with in vivo toxicological outcomes. Bioorg Med Chem Lett. 2008;18(17):4872–4875. doi: 10.1016/j.bmcl.2008.07.071. [DOI] [PubMed] [Google Scholar]
- 38.Ertl P., Rohde B., Selzer P. Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J Med Chem. 2000;43(20):3714–3717. doi: 10.1021/jm000942e. [DOI] [PubMed] [Google Scholar]
- 39.Potts R.O., Guy R.H. Predicting skin permeability. Pharm Res. 1992;9:663–669. doi: 10.1023/a:1015810312465. [DOI] [PubMed] [Google Scholar]
- 40.Martin Y.C. A bioavailability score. J Med Chem. 2005;48(9):3164–3170. doi: 10.1021/jm0492002. [DOI] [PubMed] [Google Scholar]
- 41.Esikov K.A., Zubarev V.Y., Malin A.A., Ostrovskii V.A. Use of the tetrachlorosilane-sodium azide system for synthesis of tetrazoles from carboxylic acid amides. Chem Heterocycl Compd. 2000;36(7):878–879. [Google Scholar]
- 42.Henegar K.E., Ashford S.W., Baughman T.A., Sih J.C., Gu R.-L. Practical Asymmetric Synthesis of (S)-4-Ethyl-7,8-dihydro-4-hydroxy-1H-pyrano[3,4-f]indolizine- 3,6,10(4H)-trione, a Key Intermediate for the Synthesis of Irinotecan and Other Camptothecin Analogs. J Org Chem. 1997;62(19):6588–6597. [Google Scholar]
- 43.Esikov K.A., Morozova S.E., Malin A.A., Ostrovskii V.A. Tetrachlorosilane-sodium azide system in the synthesis of tetrazole-containing amino acid derivatives. Russ J Org Chem. 2002;38:1370–1373. [Google Scholar]
- 44.Shelanski M.L., Gaskin F., Cantor C.R. Microtubule assembly in the absence of added nucleotides. Proc Natl Acad Sci U S A. 1973;70(3):765–768. doi: 10.1073/pnas.70.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dumortier C., Gorbunoff M.J., Andreu J.M., Engelborghs Y. Different kinetic pathways of the binding of two biphenyl analogues of colchicine to tubulin. Biochemistry. 1996;35(14):4387–4395. doi: 10.1021/bi952451f. [DOI] [PubMed] [Google Scholar]
- 46.González M., Ovejero-Sánchez M., Vicente-Blázquez A., Álvarez R., Herrero A.B., Medarde M. Microtubule destabilizing sulfonamides as an alternative to taxane-based chemotherapy. Int J Mol Sci. 2021;22(4):1907. doi: 10.3390/ijms22041907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhattacharyya B., Panda D., Gupta S., Banerjee M. Anti-mitotic activity of colchicine and the structural basis for its interaction with tubulin. Med Res Rev. 2008;28(1):155–183. doi: 10.1002/med.20097. [DOI] [PubMed] [Google Scholar]
- 48.Mollinedo F., Gajate C. Microtubules, microtubule-interfering agents and apoptosis. Apoptosis. 2003;8:413–450. doi: 10.1023/a:1025513106330. [DOI] [PubMed] [Google Scholar]
- 49.Home - ClinicalTrials.gov n.d. https://clinicaltrials.gov/ (accessed February 23, 2021).
- 50.Ramírez D, Caballero J. Is it reliable to take the molecular docking top scoring position as the best solution without considering available structural data? Molecules 2018;23:1038–1038. [DOI] [PMC free article] [PubMed]
- 51.Caballero J. The latest automated docking technologies for novel drug discovery. Expert Opin Drug Discov. 2020:1–21. doi: 10.1080/17460441.2021.1858793. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.