Abstract
The present study aimed to provide evidence for the genetic heterogeneity of familial autism spectrum disorder (ASD), which might help to improve our understanding of the complex polygenic basis of this disease. Whole-exome sequencing (WES) was performed on two autistic children in a family pedigree, and reasonable conditions were set for preliminarily screening variant annotations. Sanger sequencing was used to verify the preliminarily screened variants and to determine the possible sources. In addition, autism-related genes were screened according to autism databases, and their variants were compared between two autistic children. The results showed that there were 21 genes respectively for autistic children IV2 and IV4, preliminarily screened from all variants based on the harmfulness (high) and quality (high or medium) of the variants, as well as the association between mutant genes and autism in human gene mutation database. Furthermore, candidate autism-related genes were screened according to the evidence score of >4 in the Autism KnowledgeBase (AutismKB) database or ≥3 in the AutDB database. A total of 11 and 10 candidate autism-related genes were identified in the autistic children IV2 and IV4, respectively. Candidate genes with an evidence score of >16 in AutismKB were credible autism-related genes, which included LAMC3, JMJD1C and CACNA1H in child IV2, as well as SCN1A, SETD5, CHD7 and KCNMA1 in child IV4. Other than the c.G1499A mutation of SCN1A, which is known to be associated with Dravet syndrome, the specific missense variant loci of other six highly credible putative autism-related genes were reported for the first time, to the best of the authors' knowledge, in the present study. These credible autism-related variants were inherited not only from immediate family members but also from extended family members. In summary, the present study established a reasonable and feasible method for screening credible autism-related genes from WES results, which by be worth extending into clinical practice. The different credible autism-related genes between the two autistic children indicated a complex polygenic architecture of ASD, which may assist in the early diagnosis of this disease.
Keywords: autism spectrum disorder, family pedigree, whole-exome sequencing, gene variation, genetic heterogeneity
Introduction
Autism spectrum disorder (ASD) is characterized by persistent deficits in language communication and social interaction, as well as the occurrence of stereotyped and repetitive behaviors (1). It is a group of neurodevelopmental disorders with high clinical and etiologic heterogeneity (1,2). The prevalence rate of autism is 1–2% worldwide according to the Autism KnowledgeBase (AutismKB 2.0) database (3). Autism is usually caused by genetic and environmental factors, of which genetic factors account for a larger proportion (1,3,4). Over the last decade, the genetic etiology of autism has been studied using classic cytogenetics, whole-genome linkage analysis, candidate gene resequencing and genetic association analysis, whole-genome association analysis, whole-genome copy number variation, whole-exome and whole-genome sequencing approaches (2,3). Marked progress has been made in the study of candidate genes associated with autism. For instance, 228 highly credible autism-related genes (30 syndrome genes and 198 non-syndrome genes) were identified and deposited in the Autism KnowledgeBase (AutismKB) database (3,4), including PTEN, NRXN 1, RELN, SHANK3, POGZ and CHD8. However, there is a lack of repeatability in different genetic studies (5–8) and it is not clear how many genes are associated with autism. General studies have shown that the genetic basis of autism has obvious heterogeneity (1,9). A number of autism-related genes can lead to a variety of phenotypes and diseases and the number of genes involved in autism is quite large, which brings great challenges to the genetic research of autism (6). Except for a small number of autistic patients with single-gene defects or dominant chromosomal abnormalities, most of the patients have not undergone prenatal diagnosis. Thus, it is necessary to understand the complex polygenic background of autism and to implement prenatal diagnosis of this disease (10,11).
Whole-exome sequencing (WES) is an advanced genomic technique that sequences all exonic regions in the entire genome captured through target enrichment. Comprehensive analysis of the whole exome aids in the identification of disease-related coding region mutations to determine the impact of these DNA mutations on the pathogenesis of a disease (12). There are a few reports on how to preliminarily screen for gene mutations of familial autism based on a large number of mutations annotated from the WES results and how to further identify highly credible autism-related genes (13–18). However, the screening conditions and parameters used are not consistent. WES has not been applied in the prenatal diagnosis of ASD.
In the present study, two autistic children were subjected to WES and reasonable conditions were established for the preliminary screening of variant annotations and the identification of credible autism-related genes. The present study aimed to compare and evaluate the genetic heterogeneity of credible autism-related genes between two autistic children in the same pedigree to provide more evidence for the genetic risk and complex polygenic background of familiar ASD.
Materials and methods
Autism pedigree and peripheral blood samples
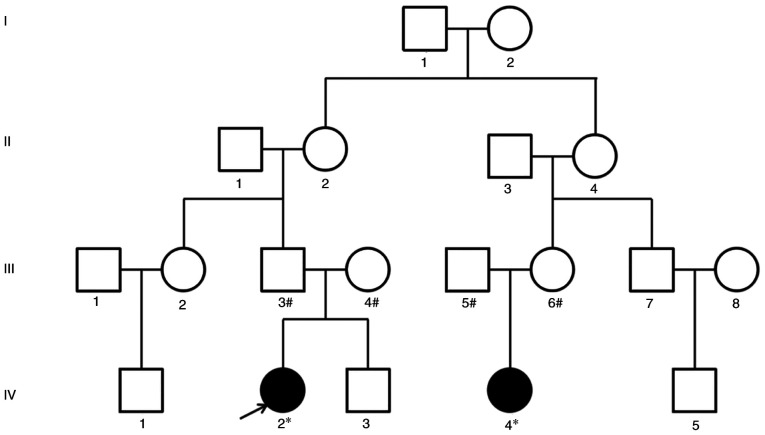
A total of two autistic children (IV2 and IV4) in a family pedigree were recruited between January 2019 and December 2020 for the present study (Fig. 1). They were diagnosed at the Second Affiliated Hospital of Wenzhou Medical University (Wenzhou, China) and identified as ASD according to Wenzhou Medical Identification Committee of Sick and Disabled Children. The diagnosis of ASD was confirmed by the Autism Diagnostic Observation Schedule (ADOS) (19) and expert clinical judgment. IV2 and IV4 children were found to have ASD characteristics at age 2 and 3 and diagnosed as autistic children at age 3 and 4, respectively. The scores of ADOS (model 1) were 10 and 8, respectively. IV2 was associated with mild cerebral palsy and mild intellectual disability, whereas IV4 was associated with epilepsy, brain development delay and mild intellectual disability. The parents of IV2 and IV4 were without clinical autism symptoms and the marriage type was not consanguineous. Psychological training, such as training in language communication and social interaction, was performed at the Mental Health Center, Wenzhou Medical University (Wenzhou, China), which is one of the author-affiliated institutes associated with this study. The present study was approved by the Ethics Committee of Wenzhou Hospital of Integrated Traditional Chinese and Western Medicine (approval number: 2018-57; Wenzhou, China); the Ethics Committee of this hospital was shared with the Wenzhou Maternal and Child Health Guidance Center (Wenzhou, China). Written informed consent forms was obtained from the parents of the children, for both their participation and the participation of the children. The clinical data and 2–3 ml/each peripheral blood samples of two autistic children and their parents were provided by Wenzhou Maternal and Child Health Guidance Center. The peripheral blood samples of IV2 and IV4 autistic children were collected at the age of 7 and 5 years, respectively.
Figure 1.
Pedigree diagram of familial autism. *Underwent clinical assessment, whole-exome sequencing and Sanger sequencing; #underwent Sanger sequencing. Shaded and unshaded symbols indicate affected and unaffected individuals, respectively. Squares and circles represent males and females, respectively. The autistic patient indicated by an arrow is the proband, i.e. the autistic patient found first in this pedigree.
WES
WES of the autism samples (2 ml) was performed by iGeneTech Biotech (Beijing) Co. Ltd. Genomic DNA was extracted using a Blood Genomic DNA Isolation kit (magnetic bead method; Kangwei Biologic Co. Ltd.) in accordance with the manufacturer's protocols. The quality and quantity of DNA extracts were evaluated. DNA quality was examined using 1% agarose gel electrophoresis. The DNA concentrations were determined using a Qubit dsDNA BR assay kit (Thermo Fisher Scientific, Inc.). The whole exome liquid-phase capture technique was used to efficiently and specifically enrich all the human exonic regions. Briefly, fragmentation was carried out using a hydrodynamic shearing system (Covaris, Inc.) to generate 150–200 bp fragments. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities using the Fast Library Prep Kit [iGeneTech Biotech (Beijing) Co. Ltd.]. After adenylation of 3′ ends of DNA fragments, adapter oligonucleotides were ligated using the Fast Library Prep Kit. The Fast Library Prep Kit was used in accordance with the manufacturer's protocols. DNA fragments with ligated adapter molecules on both ends were selectively enriched by PCR. Subsequently, libraries were hybridized in liquid phase with a biotin-labeled probe, and magnetic beads with a streptomycin monolayer were employed to capture the exons of genes using the TargetSeq One Kit [iGeneTech Biotech (Beijing) Co. Ltd.], in accordance with the manufacturer's protocols. Captured libraries were enriched by PCR to prepare for sequencing. DNA library construction was performed using a Fast Library Preparation kit [iGeneTech Biotech (Beijing) Co. Ltd.]. Library quantification and quality assessment were then performed. Qubit 3.0 (Thermo Fisher Scientific, Inc.) was used for library quantification; library concentration >25 ng/µl was considered a qualified library. DNA quality was examined using Bioptic Qsep 100 DNA fragment analyzer (BiOptic, Inc.). The length of library fragments was generally between 270 and 350 bp. The Illumina NovaSeq6000 platform (Illumina, Inc.) was used for PE150 high-throughput double-terminal sequencing. The loading concentration of the final library was 450–600 pmol/l; library quantification was performed by qPCR. The raw WES data were obtained and mapped to the human reference genome (UCSC GRCh37/hg19, 2009). The type of analysis was single sample whole-exome analysis, which included quality evaluation of sequencing data as well as the detection and screening of variations. Based on the BAM results of alignment with the genome reference sequences, Samtools (http://www.htslib.org/) and GATK (https://gatk.broadinstitute.org/hc/en-us) were used to find single nucleotide polymorphisms (SNPs) and small insertions and deletions (InDel) of <50 bp. ANNOVAR (http://www.openbioinformatics.org/annovar/) software was used to annotate the SNP and InDel loci to determine the corresponding gene information, functional data and harmfulness. The annotations on SNP and InDel were carried out by iGeneTech Biotech (Beijing) Co. Ltd.
Screening for SNP and InDel annotations
A large number of genetic variants were detected. To identify the variants with significant functions, the following conditions were set: i) The variants could be divided into high level, possible high level, intermediate level and low level according to the priority of harmfulness. In the present study, the screening for the priority of variants was limited to high level only. The high priority of the variant indicated that it was located in the exonic region of the gene, had a minor allele frequency (MAF) of <0.01 in East Asian populations [i.e. the 1000 Genomes Project database (1000G), the Exome Aggregation Consortium (ExAC) and the Genome Aggregation Database (gnomAD)] (20) and could lead to an altered amino acid sequence. In addition, at least 2 of the 10 prediction software packages [including functional prediction software SIFT, Polyphen2_HDIV, Polyphen2_HVAR, LRT, MutationTaster, MutationAssessor, FATHMM, fathmm-MKL, total effect prediction software CADD, conservative prediction software GERP; all from dbNSFP version 3.5a (annotated using Annovar; updated on September 21, 2018)] predicted it as harmful. ii) The quality evaluation of the variants could be divided into high, medium and low quality. In the present study, the quality of the variant was limited to high or medium quality only. The high quality of the variant is denoted by its depth of ≥50 layers, genotype quality value of >30 and frequency deviation of <0.1 (heterozygous 0.4–0.6; homozygous 0.9–1.0) (20). The medium quality of the variant is denoted by its depth of ~20 layers, genotype quality value of >20 and frequency deviation of <0.2 (heterozygous 0.3–0.7; homozygous 0.8–1.0) (20). iii) The mutant genes were required to be associated with autism in the human gene mutation database (HGMD) (http://www.hgmd.cf.ac.uk/ac/index.php).
Sanger sequencing
All the preliminary screening variants of IV2 and IV4 were verified by Sanger sequencing. In addition, the parents of IV2 and IV4 were sequenced for these variants to determine the variant source. Conventional PCR amplification and Sanger sequencing were performed on all the exons where the variant loci of the corresponding gene were located; PCR primer synthesis and Sanger sequencing were conducted by Qingke Biology Co., Ltd. The primer sequences are provided in Table SI.
Screening of autism-related genes through autism databases
The AutDB autism database (http://www.mindspec.org/autdb.html; also known as SFARI Gene, http://gene.sfari.org) is updated in real time (21). By September 2020, it contained 1,237 genes related to autism, with an evidence score of 0–5. In the present study, the genes with evidence score of ≥3 in the AutDB database were selected as candidate autism-related genes. AutismKB Database is the world's largest database for the genetic evidence of ASD (3,4), with 1,379 genes and 11,669 single nucleotide variants or small InDels integrated in version 2.0 (2018). According to AutismKB, in addition to 99 syndrome genes, 1,280 non-syndrome genes with an evidence score of >4 were designated as candidate autism-related genes, of which 30 syndrome genes and 198 non-syndrome genes with an evidence score of >16 were designated as substantially credible autism-related genes. The criteria set by Yang et al (3) used for calculating the ‘evidence score’ of AutismKB v2.0 database were as follows: Next-generation sequencing (NGS) de novo mutation studies or low-scale gene studies, weight 10; NGS other studies, weight 8; genome-wide association studies, weight 4; Copy-number variation/structural variation studies, linkage analyses or low-scale genetic association studies, weight 2; and expression profiling or NGS mosaic mutation studies, weight 1.
Results
Preliminary screening of the WES results of autistic child IV2
Among all the detected variants in autistic child IV2, there were 21 variants (in 21 genes) with high harmful effects and high or medium quality, and their mutant genes were associated with autism in HGMD; further details are provided in Table I. Notably, the variant types of all genes were missense (nonsynonymous), except that GPR152 and OAS3 genes contained deletion (frameshift, InDel below 50 bp) and nonsense (stopgain) variants. All the variants were located in exons. The genotypes were all heterozygous and the variant frequency of each gene in this sample ranged from 0.32–0.59; that is, the minimum variant frequency (that of gene NRP2) was 0.32 and the maximum variant frequency (that of gene OAS3) was 0.59 in this sample. All of the mutant genes were located on autosomal chromosomes, except that ARHGEF6 was on the X chromosome.
Table I.
Preliminary screening of whole-exome sequencing results of autistic child IV2.
| Gene | Location | Transcript/Exon (cDNA change/AA change) | dbSNP150 ID | Variant frequency in IV2 |
|---|---|---|---|---|
| DNAH7 | 2q32.3 | NM018897/exon18 | rs769106832 | 0.52 |
| (c.G2323A/p.E775K) | ||||
| CAPN10 | 2q37.3 | NM023083/exon7 | rs552159586 | 0.48 |
| (c.T1145G/p.L382R) | ||||
| LPA | 6q25.3 | NM005577/exon34 (c.T5395A/p.C1799S) | rs189946882 | 0.42 |
| ABCB5 | 7p21.1 | NM178559/exon12 | rs80123476 | 0.47 |
| (c.G1234A/p.A412T) | ||||
| LAMB1 | 7q31.1 | NM002291/exon3 | N/A | 0.4 |
| (c.C38T/p.A13V) | ||||
| CTTNBP2 | 7q31.31 | NM033427/exon5 | N/A | 0.45 |
| (c.C2188G/p.L730V) | ||||
| ZNF79 | 9q33.3 | NM001286696/exon5 | rs182450731 | 0.56 |
| (c.T259C/p.W87R) | ||||
| LAMC3 | 9q34.12 | NM006059/exon15 | rs200121474 | 0.44 |
| (c.G2687A/p.R896Q) | ||||
| JMJD1C | 10q21.3 | NM001322254/exon6 | rs201464655 | 0.46 |
| (c.A1046C/p.D349A) | ||||
| SCUBE2 | 11p15.4 | NM001170690/exon7 | rs991815964 | 0.41 |
| (c.C838T/p.R280W) | ||||
| GPR152 | 11q13.2 | NM206997/exon1 | rs776403683 | 0.48 |
| (c.1149delG/p.Q383fs) | ||||
| OAS3 | 12q24.13 | NM006187/exon9 | N/A | 0.59 |
| (c.C1996T/p.Q666X) | ||||
| MYCBP2 | 13q22.3 | NM015057/exon50 | N/A | 0.41 |
| (c.C7394G/p.P2465R) | ||||
| CACNA1H | 16p13.3 | NM001005407/exon25 | N/A | 0.46 |
| (c.A4643G/p.N1548S) | ||||
| CHD3 | 17p13.1 | NM001005271/exon11 | rs747768901 | 0.49 |
| (c.G1969A/p.G657S) | ||||
| MYH4 | 17p13.1 | NM017533/exon4 | rs201896271 | 0.48 |
| (c.G331A/p.A111T | ||||
| ARHGEF6 | Xq26.3 | NM001306177/exon12 | rs201896882 | 0.44 |
| (c.G944A/p.R315Q) | ||||
| ATP2B4 | 1q32.1 | NM001001396/exon12 | rs138521935 | 0.4 |
| (c.G1996A/p.A666T) | ||||
| NRP2 | 2q33.3 | NM003872/exon9 | rs201900948 | 0.32 |
| (c.A1333C/p.I445L) | ||||
| CSMD1 | 8p23.2 | NM033225/exon41 | rs201454449 | 0.4 |
| (c.A6236T/p.Y2079F) | ||||
| SORCS1 | 10q25.1 | NM001013031/exon1 | rs118055903 | 0.38 |
| ′(c.C503T/p.T168M) |
Preliminary screening of the WES results in autistic child IV4
Among all the detected variants in autistic child IV4, there were 22 variants (in 21 genes) with high harmful effects and high or medium quality, and their mutant genes were associated with autism in HGMD; further details are given in Table II. All of the variants were missense type and were located in exonic regions. The genotypes were all heterozygous, and the variant frequency of each gene in this sample ranged from 0.32–0.57; that is, the minimum variant frequency (that of gene MTMR12) was 0.32 and the maximum variant frequency (that of gene SMARCC2) was 0.57 in this sample. The mutant genes were all located on autosomal chromosomes. Based on the comparative assessment of WES results in Tables I and II, the two autistic children IV2 and IV4 shared a rare variant (ATP2B4 c.G1996A).
Table II.
Preliminary screening of whole-exome sequencing results of autistic child IV4.
| Gene | Location | Transcript/Exon (cDNA change/AA change) | dbSNP150 ID | Variant frequency in IV4 |
|---|---|---|---|---|
| USH2A | 1q41 | NM007123/exon13 | rs145383772 | 0.45 |
| (c.G2792A/p.C931Y) | ||||
| SNTG2 | 2p25.3 | NM018968/exon3 | rs192264442 | 0.47 |
| (c.C229A/p.R77S) | ||||
| SCN1A | 2q24.3 | NM001165963/exon10 | rs200176684 | 0.45 |
| (c.G1499A/p.R500Q) | ||||
| SETD5 | 3p25.3 | NM001080517/exon20 | rs527600519 | 0.45 |
| c.G3325A/p.D1109N) | ||||
| CCDC14 | 3q21.1 | NM001308317/exon5 | rs116934420 | 0.51 |
| (c.A116G/p.H39R) | ||||
| ROS1 | 6q22.1 | NM002944/exon43 | rs538891848 | 0.47 |
| (c.C6764T/p.S2255L) | ||||
| ROS1 | 6q22.1 | NM002944/exon43 | rs547573917 | 0.47 |
| (c.T6763C/p.S2255P) | ||||
| SYNE1 | 6q25.2 | NM033071/exon72 | rs375752705 | 0.42 |
| (c.C11786A/p.A3929E) | ||||
| THSD7A | 7p21.3 | NM015204/exon24 | N/A | 0.56 |
| (c.G4540T/p.D1514Y) | ||||
| PLXNA4 | 7q32.3 | NM020911/exon31 | rs200763664 | 0.49 |
| (c.C5564T/p.S1855F) | ||||
| CHD7 | 8q12.2 | NM017780/exon3 | N/A | 0.57 |
| (c.G1892A/p.R631K) | ||||
| PKHD1L1 | 8q23.2 | NM177531/exon77 | rs146897796 | 0.41 |
| (c.C12673G/p.L4225V) | ||||
| KCNMA1 | 10q22.3 | NM001014797/exon1 | rs77602559 | 0.44 |
| (c.A34G/p.S12G) | ||||
| SLK | 10q24.33 | NM001304743/exon12 | rs554070501 | 0.47 |
| (c.G2774A/p.R925Q) | ||||
| NAV2 | 11p15.1 | NM001111018/exon5 | N/A | 0.46 |
| (c.T362C/p.F121S) | ||||
| NCAPD2 | 12p13.31 | NM014865/exon20 | rs200921592 | 0.49 |
| (c.C2548T/p.R850W) | ||||
| SMARCC2 | 12q13.2 | NM003075/exon26 | rs200833916 | 0.57 |
| (c.C2920A/p.P974T) | ||||
| GOLGA3 | 12q24.33 | NM001172557/exon12 | N/A | 0.5 |
| (c.C2492T/p.T831I) | ||||
| SPATA13 | 13q12.12 | NM001166271/exon2 | N/A | 0.44 |
| (c.T959C/p.V320A) | ||||
| ATP2B4 | 1q32.1 | NM001001396/exon12 | rs138521935 | 0.36 |
| (c.G1996A/p.A666T) | ||||
| MTMR12 | 5p13.3 | NM001040446/exon5 | rs768253146 | 0.32 |
| (c.C424T/p.H142Y) | ||||
| DNAH9 | 17p12 | NM004662/exon15 | N/A | 0.36 |
| (c.C2209T/p.P737S) |
Sanger sequencing to verify the preliminary screening variants in autistic children and to verify the sources of these variants
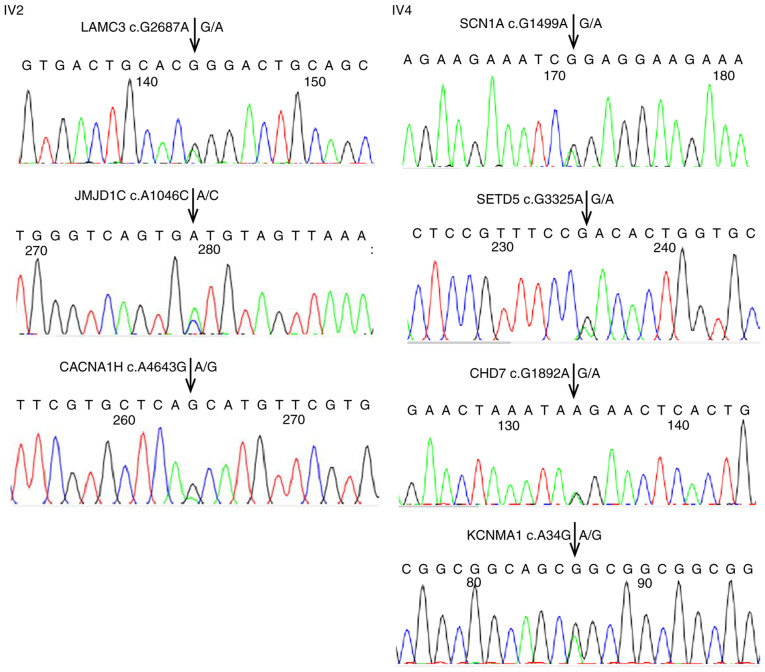
The 21 and 22 preliminary screening variants in autistic children IV2 and IV4, respectively, were verified by Sanger sequencing. The sequencing maps of three variants in IV2 and four variants in IV4 are provided in Fig. 2. As shown in Tables III and IV, these variants were ultimately identified as highly credible autism-related variants. The sequencing maps of the remaining variants will be provided upon reasonable request (see Availability of data and materials below). To determine the source of these preliminary screening variants, the sequencing data of the parents were analyzed. The results indicated that all the preliminary screening variants in the two autistic children were inherited from their parents, who were heterozygous for all of these variants, and there were no de novo variants (Tables III and IV). The sequencing maps of the parents will be provided upon reasonable request (see Availability of data and materials below).
Figure 2.
Key results of Sanger sequencing verification of preliminary screening gene mutations in autistic children IV2 and IV4. The arrows indicate the variant loci of genes.
Table III.
Further bioinformatics analysis of preliminary screening gene variations in autistic child IV2.
| Gene | Variation harmfulness | Variation quality | Variation source | Included in AutismKB 2.0/Evidence score | Included in AuthB/Evidence score |
|---|---|---|---|---|---|
| DNAH7 | High | High | Father | Yes/2 | No |
| CAPN10 | High | High | Mother | Yes/0 | No |
| LPA | High | High | Father | Yes/0 | No |
| ABCB5 | High | High | Father | No | No |
| LAMB1 a | High | High | Mother | Yes/16 | Yes/3 |
| CTTNBP2 a | High | High | Mother | Yes/2 | Yes/3 |
| ZNF79 | High | High | Father | Yes/2 | No |
| LAMC3 a | High | High | Father | Yes/38 | Yes/3 |
| JMJD1C a | High | High | Father | Yes/20 | Yes/3 |
| SCUBE2 | High | High | Father | Yes/0 | No |
| GPR152 | High | High | Father | Yes/0 | No |
| OAS3 | High | High | Mother | Yes/0 | No |
| MYCBP2 a | High | High | Mother | Yes/12 | No |
| CACNA1H a | High | High | Mother | Yes/22 | Yes/3 |
| CHD3 a | High | High | Mother | Yes/12 | Yes/3 |
| MYH4 a | High | High | Mother | Yes/12 | Yes/2 |
| ARHGEF6 a | High | High | Father | Yes/syndromic | No |
| ATP2B4 | High | Medium | Father | Yes/0 | No |
| NRP2 | High | Medium | Father | Yes/4 | Yes/0 |
| CSMD1 a | High | Medium | Father | Yes/14 | Yes/3 |
| SORCS1 a | High | Medium | Mother | Yes/15 | No |
Candidate autism-related genes; bold, highly credible autism-related genes. AutDB, Autism Database; AutismKB, Autism KnowledgeBase.
Table IV.
Further bioinformatics analysis of preliminary screening gene variations in autistic child IV4.
| Gene | Variation harmfulness | Variation quality | Variation source | Included in AutismKB 2.0/Evidence score | Included in AuthB/Evidence score |
|---|---|---|---|---|---|
| USH2A a | High | High | Father | Yes/10 | Yes/3 |
| SNTG2 a | High | High | Father | Yes/14 | Yes/2 |
| SCN1A a | High | High | Mother | Yes/20/syndromic | Yes/5 |
| SETD5 a | High | High | Mother | Yes/22 | Yes/5 |
| CCDC14 | High | High | Father | Yes/0 | No |
| ROS1 | High | High | Mother | Yes/0 | No |
| ROS1 | High | High | Mother | Yes/0 | No |
| SYNE1 a | High | High | Father | Yes/11 | Yes/3 |
| THSD7A | High | High | Father | Yes/0 | No |
| PLXNA4 a | High | High | Father | Yes/11 | Yes/2 |
| CHD7 a | High | High | Father | Yes/18/syndromic | Yes/3 |
| PKHD1L1 | High | High | Mother | Yes/0 | No |
| KCNMA1 a | High | High | Mother | Yes/34 | Yes/3 |
| SLK | High | High | Mother | Yes/0 | No |
| NAV2 a | High | High | Father | Yes/12 | Yes/3 |
| NCAPD2 | High | High | Mother | Yes/2 | No |
| SMARCC2 a | High | High | Mother | Yes/0 | Yes/4 |
| GOLGA3 | High | High | Mother | Yes/2 | No |
| SPATA13 | High | High | Father | Yes/3 | No |
| ATP2B4 | High | Medium | Mother | Yes/0 | No |
| MTMR12 | High | Medium | Father | Yes/0 | No |
| DNAH9 | High | Medium | Father | Yes/0 | No |
Candidate autism-related genes; bold, highly credible autism-related genes. AutDB, Autism Database; AutismKB, Autism KnowledgeBase.
Bioinformatics analysis of preliminary screening variants in autistic child IV2
A total of 11 candidate autism-related genes (indicated with a in Table III) in autistic child IV2 were screened according to the evidence score of >4 in AutismKB or ≥3 in AutDB. The candidate genes with an evidence score of >16 in AutismKB were regarded as highly credible autism-related genes, which included LAMC3, JMJD1C and CACNA1H (indicated in bold in Table III).
Based on the possible autism-related gene functions in Gene Ontology (GO; updated on January 1, 2018) database (https://www.uniprot.org/help/gene_ontology), LAMC3 was associated with astrocyte development, JMJD1C was related to transcriptional regulation and histone demethylase activity (that is, chromatin remodeling) and CACNA1H was involved in neuronal action potential and membrane potential regulation (that is, synaptic function).
The specific missense variant loci of these three genes were reported for the first time in the present study, to the best of our knowledge. None of the loci were found in AutismKB 2.0 autism database (version 2018), AutDB autism database (version 2020) or HGMD database (version 2019). The three gene variants were located outside the repeat region. The MAFs of LAMC3 c.G2687A were 0.001, 0.01 and 0.008 in East Asian populations (1000G, ExAC and gnomAD, respectively). However, CACNA1H c.A1046C and JMJD1C c.A4643G were not detected in these populations. According to the HGMD database, 40.91% (9/22), 92.00% (23/25) and 92.73% (51/55) of the missense variants in LAMC3, JMJD1C and CACNA1H genes, respectively, were pathogenic. The functional effects of the variants in these three genes, as predicted by 10 software packages are listed in Table V. Notably, eight of the 10 packages predicted LAMC3 c.G2687A as harmful (i.e. very high harmfulness), eight of the 10 packages are predicted CACNA1H c.A1046C as harmful (i.e. very high harmfulness) and two of 10 software predicted JMJD1C c.A4643G as harmful (i.e. high harmfulness).
Table V.
Functional impact of highly credible autism-related gene mutations in autistic children IV2 and IV4 as predicted by 10 software programs.
| Software | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||
| Gene | SIFTa | Polyphen2 HDIVb | Polyphen2 HVARc | LRTd | Mutation Tastere | Mutation Assessorf | FATHMMg | Fathmm MKLh | CADDi | GERPj |
| LAMC3 | D | D | P | D | D | L | T | D | 25.3 | 5.12 |
| JMJD1C | D | B | B | N | N | L | T | D | 4.9 | 4.91 |
| CACNA1H | D | D | D | D | D | N/A | D | D | 25.6 | 4 |
| SCN1A | D | B | B | N | D | M | D | D | 23.8 | 5.28 |
| SETD5 | D | D | D | N | D | M | D | D | 23.3 | 6.07 |
| CHD7 | T | P | P | D | D | L | T | D | 17.7 | 5.23 |
| KCNMA1 | D | B | B | N/A | N | N | D | T | 12.1 | −1.58 |
SIFT: D, deleterious (≤0.05); T, tolerated (>0.05).
Polyphen2 HDIV: D, probably damaging (≥0.957); P, possibly damaging (0.956–0.453); B, benign (≤0.452).
Polyphen2 HVAR: D, probably damaging (≥0.909), P, possibly damaging (0.908–0.447), B, benign (≤0.446).
LRT: D, deleterious; N, neutral.
MutationTaster: D, disease causing; N, polymorphism.
MutationAssessor: M, medium harmful; L, low harmful; N, neutral.
FATHMM: D, deleterious; T, tolerated.
Fathmm-MKL coding: D, deleterious; T, tolerated.
CADD: >15 is harmful.
GERP: >5 is harmful. SIFT, sorting intolerant from tolerant; Polyphen2, Polymorphism Phenotyping v2; LRT, Likelihood Ratio Test; FATHMM, Functional Analysis Through Hidden Markov Models; CADD, Combined Annotation Dependent Depletion; GERP, Genomic Evolutionary Rate Profiling.
In addition to LAMC3 and JMJD1C inherited from the father III3 (descended from I1 and I2), there was also the cumulative effect of CACNA1H inherited from the mother III4 (not descended from I1 and I2), i.e. there was a cumulative effect of CACNA1H with LAMC3 and JMJD1C. The results of the present study could provide guidance for the prenatal diagnosis of ASD. If the father III3 and mother III4 of autistic child IV2 give birth again, prenatal genetic diagnosis should be conducted on these three highly credible autism-related genes.
Bioinformatics analysis of preliminary screening variants in autistic child IV4
A total of 10 candidate autism-related genes (indicated with a in Table IV) in autistic child IV4 were screened according to the evidence score of >4 in the AutismKB or at ≥3 in the AutDB. The candidate genes with an evidence score of >16 in the AutismKB were considered as highly credible autism-related genes, which included SCN1A, SETD5, CHD7 and KCNMA1 (indicated in bold in Table IV).
Based on the possible autism-related gene functions in GO database, SCN1A was related to axon initiation segment and neuronal action potential transmission (synaptic function), SETD5 was involved in transcriptional regulation and histone acetylation regulation (i.e. chromatin remodeling), CHD7 was associated with central nervous system development (i.e. synaptic function) and transcriptional positive regulation, and KCNMA1 was responsible for the regulation of membrane potential (i.e. synaptic function).
SCN1A c.G1499A has been reported to be associated with Dravet syndrome in the HGMD database (updated in 2019), whereas the missense variant loci of the remaining three genes were first discovered in the present study, to the best of our knowledge, and were not found in the AutismKB 2.0 database (version 2018), the AutDB database (version 2020) or the HGMD database (version 2019).
All variants in these four genes were located outside the repeat region. The MAFs of SCN1A c.G1499A, SETD5 c.G3325A and KCNMA1 c.A34G were (0.003, 0 and 0.0016), (0.001, 0 and 0.0032) and (0.001, 0 and 0.0014) in East Asian populations (1000G, ExAC and gnomAD), respectively; whereas CHD7 c.G1892A was not found in these populations. According to the HGMD database, 51.62% (860/1666), 18.75% (6/32), 20.14% (171/849) and 66.67% (6/9) of the missense variants in SCN1A, SETD5, CHD7 and KCNMA1 genes, respectively, were pathogenic. The functional effects of the variants in these four genes predicted by 10 software are listed in Table V. Notably, seven of the 10 software packages predicted that SCN1A c.G1499A was harmful (i.e. very high harmfulness), nine of the 10 software packages predicted that SETD5 c.G3325A was harmful (i.e. very high harmfulness), seven of the 10 software packages predicted that CHD7 c.G1892A was harmful (i.e. very high harmfulness) and two of the 10 software packages predicted that KCNMA1 c.A34G was harmful (i.e. high harmfulness).
In addition to SCN1A, SETD5 and KCNMA1 inherited from the mother III6 (descended from I1 and I2), there was also the cumulative effect of CHD7 inherited from the father III5 (not descended from I1 and I2), i.e. there was a cumulative effect of CHD7 with SCN1A, SETD5 and KCNMA1. The results of the present study may provide guidance for the prenatal diagnosis of ASD. If the father III5 and the mother III6 of child IV4 give birth again, prenatal genetic diagnosis should be performed on these four highly credible autism-related genes.
The filtering procedure and number of the remaining variants in IV2 and IV4 autistic children are summarized in Table VI.
Table VI.
Filtering procedure and number of remaining variants of autistic children IV2 and IV4 for each step.
| Variant | ||
|---|---|---|
|
|
||
| Filtering procedure | IV2 | IV4 |
| Total variants | 252,419 | 176,044 |
| Excluding variants if QD<2.0 or FS >60.0 or MQ<40.0 or DP<4 | 135,315 | 121,156 |
| Excluding variants if harmfulness of variant (priority) was low | 3,237 | 2,105 |
| Retaining variants if genes involved in autism in HGMD | 145 | 84 |
| Retaining variants if harmfulness of variant was high and if quality of variant was high or medium (preliminary screening gene variants in Tables I and II) | 21 | 22 |
| Following confirmation by Sanger sequencing; retaining variants if evidence score of gene was >4 in AutismKB or at ≥3 in AutDB (candidate autism–related gene variants in Tables III and IV) | 11 | 10 |
| Retaining variants if evidence score of gene was >16 in AutismKB (highly credible autism-related gene variants in Tables III and IV) | 3 | 4 |
AutDB, Autism Database; AutismKB, Autism KnowledgeBase; DP, read depth; FS, Phredscaled P-value using Fisher's exact test to detect strand bias; MQ, Root Mean Square of the mapping quality; QD, variant confidence/quality by depth.
Discussion
De novo mutations and genetic variations both have important effects on autism. In recent years, remarkable progress has been made towards understanding the genetic risks of autism using WES (22,23). For example, some de novo mutations were found by comparing the WESs of sporadic cases with those of their parents (17,24–29). It has been suggested that these de novo mutations can increase the risk of autism but may not lead to autism. Therefore, it is necessary to perform a WES study based on the family pedigree of ASD patients to explore possible hereditary mutations (14–17).
The present study set the reasonable conditions of high or medium variant quality, such as depth, genotype quality value and frequency deviation, to ensure the reliability of the WES results. Additionally, to ensure that the selected variants were of high pathogenicity it set the conditions that the harmfulness of the variant was high, the variant was located in the exonic region, the MAF value was low in three East Asian populations, the variant resulted in amino acid changes and that various software packages predicted the variant as harmful. The present study also set reasonable conditions for the preliminary screening of mutant genes related to autism in the HGMD database and further selected the genes with high evidence score in the AutismKB and the AutDB autism databases to ensure that the identified autism-related genes were highly credible.
The results of the present study showed that all the preliminary screening genes, except for ATP2B4, were different between two autistic children in the same pedigree and the highly credible autism-related genes were completely different between two autistic children, including LAMC3, JMJD1C and CACNA1H in autistic child IV2 and SCN1A, SETD5, CHD7 and KCNMA1 in autistic child IV4. These data indicated that familial ASD patients exhibited a high degree of genetic heterogeneity. The two autistic children in this pedigree were fourth-degree relatives, which might contribute to a high degree of genetic heterogeneity. A previous study showed that genetic heterogeneity exists in the first-degree relatives of autistic patients and even identical twins (1). High clinical heterogeneity might also be the main cause of high genetic heterogeneity observed in the present study (30). Apart from the core symptoms, the vast majority of autism cases are accompanied by other concurrent symptoms, including abnormal head circumference, sleep disorders, epilepsy, gastrointestinal diseases, immune disorders and metabolic disorders (31). Autistic child IV2 in the present study pedigree was accompanied by mild cerebral palsy and mild intellectual disability, while IV4 was accompanied by epilepsy, brain growth delay and mild intellectual disability, which may be partly attributed to the heterogeneous variations of autism risk genes.
Regarding the functions of seven highly credible autism-related genes in autistic children, JMJD1C and SETD5 were associated with transcriptional regulation and chromatin remodeling, CACNA1H, SCN1A and KCNMA1 were related to synaptic function, CHD7 was involved in synaptic function and transcriptional regulation and LAMC3 was related to astrocytes. There are three key pathways by which gene mutations affect nerve development in patients with ASD: i) Chromatin remodeling, ii) transcriptional regulation and splicing regulation and iii) synapse formation and function (32). Chromatin remodeling controls potential events in the formation of neural connections, including neurogenesis and neural differentiation and is dependent on epigenetic mechanisms (32). Among the top autism candidate genes, some are related to transcriptional regulation and alternative splicing (32). Mutations in synapse-related genes can affect multiple components in the synaptic network, from receptors, ion channels to scaffold proteins, thus resulting in an impaired synaptic function (32). Synaptic defects and neuroelectrophysiological impairments have been reported in experimental ASD models (32). Furthermore, astrocytes serve an important role in regulating synaptic connections (33).
Genetic studies on autism have focused on both ends of the frequency spectrum of genetic variation; namely, rare and common variations (34). All of the seven highly credible autism-related gene variants reported in the present study belonged to rare variations and the corresponding genes were included in the AutismKB and AutDB autism databases. According to the AutDB database, CHD7, SCN1A and SETD5 genes not only belong to rare variations but are also associated with CHARGE, Dravet and MRD23 syndrome (OMIM#214800, 607208 and 615761, respectively). It has been suggested that the etiology of autism can be caused by: i) Single gene defects, ii) a combination of two or more rare mutations, each with high pathogenic effect, iii) a rare variant with high pathogenic effect, on the background of common variants with low pathogenic effects, iv) multiple common variants and v) complex genetic mechanism, including common susceptibility variants, environmental risk factors and epigenetic variations (2,30). In the present study, the highly credible autism-related genes of autistic child IV2 (or IV4) were inherited not only from III3 (or III6) (immediate family members) but also from III4 (or III5) (extended family members), which could provide more evidence for the irregular inheritance pattern of familial ASD. The results demonstrated that a number of autism-related genes may be involved in the pathogenesis of familial ASD, and each gene probably has different degrees of contribution to this disease.
In summary, the present study provided a reasonable and feasible method for screening highly reliable autism-related genes from WES results, which has high application value in clinical practice. The analysis and comparison of the WES results indicated that several highly credible autism-related genes were identified such as LAMC3, JMJD1C and CACNA1H in autistic child IV2 and SCN1A, SETD5, CHD7 and KCNMA1 in autistic child IV4. These candidate genes were completely different between the two autistic children, suggesting a high degree of genetic heterogeneity. Except for SCN1A c.G1499A, the specific missense variant loci of the remaining six highly credible autism-related genes were reported for the first time in the present study. Some of the highly credible autism-related genes were originated from the extended family members of the pedigree. The findings provided strong evidence for the complex polygenic architecture of ASD, which may help in the early detection of this disease.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by grants from The Wenzhou Science and Technology Bureau of China (grant no. Y20180787), The Natural Science Foundation of Zhejiang Province (grant no. LY19C070001) and Horizontal Scientific Research Project of Wenzhou Medical University (grant no. KJHX1710).
Funding
The present study was supported by grants from The Wenzhou Science and Technology Bureau of China (grant no. Y20180787), The Natural Science Foundation of Zhejiang Province (grant no. LY19C070001) and Horizontal Scientific Research Project of Wenzhou Medical University (grant no. KJHX1710).
Availability of data and materials
The datasets generated during the current study are available in the NCBI sequence Read Archive (SRA) repository, https://www.ncbi.nlm.nih.gov/sra/PRJNA733596; BioProject accession: PRJNA733596. Other datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LS collected and analyzed the WES data, and wrote and revised the paper. PL, TZ, ML, SZ, YTH and YWH collected the samples, and clinical, WES and Sanger sequencing data, and analyzed the data. PL, TZ and ML confirm the authenticity of all the raw data. HL designed the research, analyzed the data, and wrote and revised the paper. All of the authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was given ethics approval by the Ethics Committee of Wenzhou Hospital of Integrated Traditional Chinese and Western Medicine (Wenzhou, China; approval no. 2018-57); the Ethics Committee of this hospital was shared with the Wenzhou Maternal and Child Health Guidance Center. Written informed consent was obtained from the parents of the children, for both their participation and the participation of the children.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jeste SS, Geschwind DH. Disentangling the heterogeneity of autism spectrum disorder through genetic findings. Nat Rev Neurol. 2014;10:74–81. doi: 10.1038/nrneurol.2013.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoon SH, Choi J, Lee WJ, Do JT. Genetic and epigenetic etiology underlying autism spectrum disorder. J Clin Med. 2020;9:966. doi: 10.3390/jcm9040966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang C, Li J, Wu Q, Yang X, Huang AY, Zhang J, Ye AY, Dou Y, Yan L, Zhou WZ, et al. AutismKB 2.0: A knowledgebase for the genetic evidence of autism spectrum disorder. Database. 2018;2018:bay106. doi: 10.1093/database/bay106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu LM, Li JR, Huang Y, Zhao M, Tang X, Wei L. AutismKB: An evidence-based knowledgebase of autism genetics. Nucleic Acids Res. 2012;40:D1016–D1022. doi: 10.1093/nar/gkr1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Betancur C. Etiological heterogeneity in autism spectrum disorders: More than 100 genetic and genomic disorders and still counting. Brain Res. 2011;1380:42–77. doi: 10.1016/j.brainres.2010.11.078. [DOI] [PubMed] [Google Scholar]
- 6.De Rubeis S, Buxbaum JD. Genetics and genomics of autism spectrum disorder: Embracing complexity. Hum Mol Genet. 2015;24:R24–R31. doi: 10.1093/hmg/ddv273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang T, Guo H, Xiong B, Stessman HA, Wu H, Coe BP, Turner TN, Liu Y, Zhao W, Hoekzema K, et al. De novo genic mutations among a Chinese autism spectrum disorder cohort. Nat Commun. 2016;7:13316. doi: 10.1038/ncomms13316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devlin B, Scherer SW. Genetic architecture in autism spectrum disorder. Curr Opin Genet Dev. 2012;22:229–237. doi: 10.1016/j.gde.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Vanya M, Szucs S, Szili K, Vetro A, Bartfai G. The possibility of prenatal diagnosis of autism spectrum disorder. Psychiatr Hung. 2015;30:303–307. (In Hu) [PubMed] [Google Scholar]
- 11.Jiang YH, Wang Y, Xiu X, Choy KW, Pursley AN, Cheung SW. Genetic diagnosis of autism spectrum disorders: The opportunity and challenge in the genomics era. Crit Rev Clin Lab Sci. 2014;51:249–262. doi: 10.3109/10408363.2014.910747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sener EF, Canatan H, Ozkul Y. Recent Advances in autism spectrum disorders: Applications of whole exome sequencing technology. Psychiatry Investig. 2016;13:255–264. doi: 10.4306/pi.2016.13.3.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapman NH, Nato AQ, Jr, Bernier R, Ankenman K, Sohi H, Munson J, Patowary A, Archer M, Blue EM, Webb SJ, et al. Whole exome sequencing in extended families with autism spectrum disorder implicates four candidate genes. Hum Genet. 2015;134:1055–1068. doi: 10.1007/s00439-015-1585-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Egawa J, Watanabe Y, Wang C, Inoue E, Sugimoto A, Sugiyama T, Igeta H, Nunokawa A, Shibuya M, Kushima I, et al. Novel rare missense variations and risk of autism spectrum disorder: Whole-exome sequencing in two families with affected siblings and a two-stage follow-up study in a Japanese population. PLoS One. 2015;10:e0119413. doi: 10.1371/journal.pone.0119413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue E, Watanabe Y, Egawa J, Sugimoto A, Nunokawa A, Shibuya M, Igeta H, Someya T. Rare heterozygous truncating variations and risk of autism spectrum disorder: Whole-exome sequencing of a multiplex family and follow-up study in a Japanese population. Psychiatry Clin Neurosci. 2015;69:472–476. doi: 10.1111/pcn.12274. [DOI] [PubMed] [Google Scholar]
- 16.Toma C, Torrico B, Hervás A, Valdés-Mas R, Tristán-Noguero A, Padillo V, Maristany M, Salgado M, Arenas C, Puente XS, et al. Exome sequencing in multiplex autism families suggests a major role for heterozygous truncating mutations. Mol Psychiatry. 2014;19:784–790. doi: 10.1038/mp.2013.106. [DOI] [PubMed] [Google Scholar]
- 17.Thongnak C, Hnoonual A, Tangviriyapaiboon D, Silvilairat S, Puangpetch A, Pasomsub E, Chantratita W, Limprasert P, Sukasem C. Whole-exome sequencing identifies one de novo variant in the FGD6 Gene in a Thai family with autism spectrum disorder. Int J Genomics. 2018;2018:8231547. doi: 10.1155/2018/8231547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al-Mubarak B, Abouelhoda M, Omar A, AlDhalaan H, Aldosari M, Nester M, Alshamrani HA, El-Kalioby M, Goljan E, Albar R, et al. Whole exome sequencing reveals inherited and de novo variants in autism spectrum disorder: A trio study from Saudi families. Sci Rep. 2017;7:5679. doi: 10.1038/s41598-017-06033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou B, Xu Q, Lu P, Xu X. Evaluation on reliability and validity of Chinese Version Autism Diagnostic Observation Schedule Module-1 and clinical application. Chin J Evid Based Pediatr. 2013;8:257–261. (In Chinese) [Google Scholar]
- 20.Do R, Stitziel NO, Won H, Jørgensen AB, Duga S, Merlini PA, Kiezun A, Farrall M, Goel A, Zuk O, et al. Multiple rare alleles at LDLR and APOA5 confer risk for earlyonset myocardial infarction. Nature. 2015;518:102–106. doi: 10.1038/nature13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basu SN, Kollu R, Banerjee-Basu S. AutDB: A gene reference resource for autism research. Nucleic Acids Res. 2009;37:D832–D836. doi: 10.1093/nar/gkn835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An JY, Peng M, Collins R, Grove J, Klei L, et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell. 2020;180:568–584.e23. doi: 10.1016/j.cell.2019.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feliciano P, Zhou X, Astrovskaya I, Turner TN, Wang T, Brueggeman L, Barnard R, Hsieh A, Snyder LG, Muzny DM, et al. Exome sequencing of 457 autism families recruited online provides evidence for autism risk genes. NPJ Genom Med. 2019;4:19. doi: 10.1038/s41525-019-0093-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, Karakoc E, Mackenzie AP, Ng SB, Baker C, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee YH, Narzisi G, Leotta A, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neale BM, Kou Y, Liu L, Ma'ayan A, Samocha KE, Sabo A, Lin CF, Stevens C, Wang LS, Makarov V, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iossifov I, O'Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sjaarda CP, Wood S, McNaughton AJM, Taylor S, Hudson ML, Liu X, Guerin A, Ayub M. Exome sequencing identifies de novo splicing variant in XRCC6 in sporadic case of autism. J Hum Genet. 2020;65:287–296. doi: 10.1038/s10038-019-0707-0. [DOI] [PubMed] [Google Scholar]
- 30.Guo H, Wang T, Wu H, Long M, Coe BP, Li H, Xun G, Ou J, Chen B, Duan G, et al. Inherited and multiple de novo mutations in autism/developmental delay risk genes suggest a multifactorial model. Mol Autism. 2018;9:64. doi: 10.1186/s13229-018-0247-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai MC, Lombardo MV, Baron-Cohen S. Autism. Lancet. 2014;383:896–910. doi: 10.1016/S0140-6736(13)61539-1. [DOI] [PubMed] [Google Scholar]
- 32.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petrelli F, Bezzi P. mGlu5-mediated signalling in developing astrocyte and the pathogenesis of autism spectrum disorders. Curr Opin Neurobiol. 2018;48:139–145. doi: 10.1016/j.conb.2017.12.014. [DOI] [PubMed] [Google Scholar]
- 34.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available in the NCBI sequence Read Archive (SRA) repository, https://www.ncbi.nlm.nih.gov/sra/PRJNA733596; BioProject accession: PRJNA733596. Other datasets used and analyzed during the current study are available from the corresponding author on reasonable request.