Abstract
Viruses of the subfamily Orthocoronavirinae can cause mild to severe disease in people, including COVID-19, MERS and SARS. Their most common natural hosts are bat and bird species, which are mostly split across four virus genera. Molecular clock analyses of orthocoronaviruses suggested the most recent common ancestor of these viruses might have emerged either around 10,000 years ago or, using models accounting for selection, many millions of years. Here, we reassess the evolutionary history of these viruses. We present time-aware phylogenetic analyses of a RNA-dependent RNA polymerase locus from 123 orthocoronaviruses isolated from birds and bats, including those in New Zealand, which were geographically isolated from other bats around 35 million years ago. We used this age, as well as the age of the avian-mammals split, to calibrate the molecular clocks, under the assumption that these ages are applicable to the analyzed viruses. We found that the time to the most recent ancestor common for all orthocoronaviruses is likely 150 or more million years, supporting clock analyses that account for selection.
Keywords: Coronavirus, Evolution, Bats, Biogeography
1. Introduction
Orthocoronaviruses (family Coronaviridae, subfamily Orthocoronavirinae) are infectious agents of birds and mammals. In humans, they can cause mild illness and commonly cause colds, but emergent viruses can cause more severe disease, with Severe Acute Respiratory Syndrome (SARS) (Peiris et al., 2003), Middle Eastern Respiratory Syndrome (MERS) (Zaki et al., 2012) and Coronavirus disease 2019 (COVID-19) (Huang et al., 2020; Wang et al., 2020; Wu et al., 2020) all causing more severe disease and death in varying proportions of cases. The Coronaviridae were recently reclassified, specifically the subfamily Coronavirinae was renamed to Orthocoronavirinae, with many species formally recognized (Gorbalenya et al., 2020). Orthocoronaviruses are positive-sense RNA viruses and are classified into Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus genera (Gorbalenya et al., 2020; de Groot RJ et al., 2011). Alphacoronaviruses and betacoronaviruses are only found in mammals, whereas gammacoronaviruses and deltacoronaviruses mainly infect birds. SARS (Peiris et al., 2003), in particular, initiated the discovery of novel orthocoronaviruses in humans, domesticated animals, and wildlife (Poon et al., 2005; Snijder et al., 2003; Wevers and van der Hoek, 2009; Zaki et al., 2012). Bats and birds host the greatest diversity of these viruses and are the likely natural ‘ancestral’ reservoirs of the viruses (Wertheim et al., 2013; Wong et al., 2019; Zhou et al., 2021). Previous studies have identified both evidence for possible orthocoronavirus – host codivergence and coevolution as well as recent cross-species transmission events (Leopardi et al., 2018; Zhang et al., 2020).
Molecular clock analyses of the RNA-dependent RNA polymerase (RdRp) gene and five other genomic regions using different models suggest a time of most recent common ancestor (tMRCA) for the orthocoronaviruses of either around 10,000 years ago (Woo et al., 2012) or 293 (95% CI, 190 to 489) million years ago (Wertheim et al., 2013). The large difference between these approaches was due to the first model using viral isolation (tip) dates and substitution models including the general time-reversible substitution model with a four-bin gamma rate distribution (GTR + Γ4), possibly in the absence of a temporal signal (Rieux and Balloux, 2016), and a second accounting for purifying selection. This was achieved through the development of models in HyPhy that model the substitutions using GTR + Γ4 and a branch site random effects likelihood (BS- REL) model (Pond et al., 2011) to account for variation in selection pressure across codon sites and phylogenetic lineages (Wertheim et al., 2013). These models reduce the effect of purifying selection that prevents the estimation of ages (Wertheim and Pond, 2011) through maintaining evidence of sequence homology after saturation at synonymous sites. For eukaryotic organisms, recent advances in phylogenetic analyses have allowed the use of fossils to calibrate these clocks (Gavryushkina et al., 2017; Heath, 2012), using fossil dates to calibrate nodes in the phylogenetic tree. Viruses are not fossilized and so tip calibration is usually used. However, endogenous viral elements and host species divergence ages have been used to estimate the age of other viruses, including single stranded, non-retroviral RNA viruses (Supplementary Table S1) (Belyi et al., 2010; Gifford et al., 2008; Gilbert and Feschotte, 2010; Han and Worobey, 2012; McGeoch and Cook, 1994; McGeoch et al., 1995; Suh et al., 2013, 2014; Taylor et al., 2010; Thézé et al., 2011). For example, the divergence of mammal hosts around 39–52 million years ago years ago with related endogenous filovirus elements lead to the estimation that filoviruses may be tens of millions years old, rather than the 10,000 years estimated by tip dates (Taylor et al., 2010).
Here we take advantage of biogeographic theory and mammalian speciation, including the unique features of phylogeography in New Zealand, to calibrate modeling approaches to estimate the age of bat and then representative bat and bird orthocoronaviruses. Specially, Alphacoronavirus RdRp RNA has been discovered in Mystacina tuberculata bats in New Zealand (Hall et al., 2014; Wang et al., 2015). New Zealand is estimated to have separated from other landmasses some 84 million years ago (Mortimer et al., 2017) and bats were, until the arrival of humans just 700 years ago between 1320 and 1350 (Walter et al., 2017), the only non-marine mammal present on the continent for nearly 35 million years. We assume there is an ancient, coevolutionary relationship between orthocoronaviruses and their bat or bird hosts for this analysis (Gorbalenya, 2008; Gorbalenya et al., 2006; Wertheim et al., 2013).
2. Materials and methods
2.1. Sequence data sets
Orthocoronavirus genomes (n = 123; Table 1 ) from all four genera were downloaded from GenBank in March 2020. Because only fragments of the RdRp gene (561 bp) were available from the New Zealand bats, we limited our analyses to this genomic region. This has additional benefits because this region is apparently free of recombinant sequences (Wertheim et al., 2013) and is relatively conserved (Ziebuhr et al., 2001). However, we tested nucleotide sequences for evidence of recombination using DualBrothers (Minin et al., 2005). A total of 105 sequences from bat hosts were used for the initial analyses, and a further 18 sequences from bird hosts were included in subsequent tests (Table 1). All nucleotide sequences were aligned at the amino acid level using MAFFT version 7 employing the E-INS-i algorithm (Katoh and Standley, 2013).
Table 1.
Orthocoronavirus sequences analyzed in this study.
| Genus and published virus namea | Host species | Accession | Year | Sampling country |
|---|---|---|---|---|
| Alphacoronavirus | ||||
| Miniopterus bat coronavirus 1 | Miniopterus spp. | AY864196 | 2004 | Hong Kong |
| Bat coronavirus HKU7 | Miniopterus magnater | DQ249226 | 2005 | Hong Kong |
| Scotophilus bat coronavirus 512 | Scotophilus kuhlii | DQ648858 | 2005 | China |
| Bat coronavirus HKU2 | Rhinolophus sinicus | EF203064 | 2006 | China |
| Bat coronavirus 1B | Miniopterus pusillus | EU420137 | 2006 | China |
| Bat coronavirus 1A | Miniopterus magnater | EU420138 | 2005 | China |
| Bat coronavirus HKU8 | Miniopterus pusillus | EU420139 | 2005 | China |
| Bat coronavirus | Miniopterus schreibersii | GU190243 | 2008 | Bulgaria |
| Bat coronavirus | Miniopterus schreibersii | GU190244 | 2008 | Bulgaria |
| Miniopterus bat coronavirus/Kenya/KY33/2006 | Miniopterus inflatus | HQ728485 | 2006 | Kenya |
| Coronavirus BtCoV/KP256/Art_jam/PAN/2010 | Artibeus jamaicensis | JQ731784 | 2010 | Panama |
| Coronavirus BtCoV/KP565/Art_jam/PAN/2010 | Artibeus jamaicensis | JQ731786 | 2010 | Panama |
| Bat coronavirus | Miniopterus schreibersii | KF294269 | 2012 | China |
| Bat coronavirus | Miniopterus schreibersii | KF294270 | 2012 | China |
| Bat coronavirus | Miniopterus schreibersii | KF294271 | 2012 | China |
| Bat coronavirus | Miniopterus schreibersii | KF294275 | 2012 | China |
| Mystacina coronavirus New Zealand/2013 | Mystacina tuberculata | KF515987 | 2013 | New Zealand |
| Mystacina coronavirus New Zealand/2013 | Mystacina tuberculata | KF515988 | 2013 | New Zealand |
| Mystacina coronavirus New Zealand/2013 | Mystacina tuberculata | KF515989 | 2013 | New Zealand |
| Mystacina coronavirus New Zealand/2013 | Mystacina tuberculata | KF515990 | 2013 | New Zealand |
| Alphacoronavirus BtCoV/MSTM2/Min_nat/RSA/2010 | Miniopterus cf. natalensis | KF843851 | 2010 | South Africa |
| Alphacoronavirus BtCoV/VH_NC2/Neo_cap/RSA/2012 | Neoromicia cf. capensis | KF843854 | 2010 | South Africa |
| Alphacoronavirus BtCoV/GrNC1/Neo_cap/RSA/2012 | Neoromicia cf. capensis | KF843855 | 2010 | South Africa |
| Alphacoronavirus BtCoV/GrNC2/Neo_cap/RSA/2012 | Neoromicia cf. capensis | KF843856 | 2010 | South Africa |
| BtRf-AlphaCoV/HuB2013 | Rhinolophus ferrumequinum | KJ473807 | 2013 | China |
| BtMs-AlphaCoV/GS2013 | Myotis sp. | KJ473810 | 2013 | China |
| 229E-related bat coronavirus | Hipposideros abae | KT253270 | 2010 | Ghana |
| 229E-related bat coronavirus | Hipposideros abae | KT253272 | 2010 | Ghana |
| 229E-related bat coronavirus | Hipposideros cf. ruber | KT253273 | 2010 | Ghana |
| 229E-related bat coronavirus | Hipposideros abae | KT253274 | 2010 | Ghana |
| 229E-related bat coronavirus | Hipposideros cf. ruber | KT253297 | 2011 | Ghana |
| 229E-related bat coronavirus | Hipposideros cf. ruber | KT253298 | 2011 | Ghana |
| Bat coronavirus MsBtCoV/4039 | Miniopterus schreibersii | KU343194 | 2012 | China |
| Bat coronavirus HpBtCoV/3723 | Hipposideros pomona | KU343196 | 2012 | China |
| Bat coronavirus RaBtCoV/4307-1 | Rhinolophus affinis | KU343197 | 2013 | China |
| Bat coronavirus | Myotis siligorensis | KY770850 | 2013 | China |
| Bat coronavirus | Myotis davidii | KY770851 | 2013 | China |
| Bat coronavirus | Hipposideros armiger | KY770852 | 2013 | China |
| Bat coronavirus | Rhinolophus pearsonii | KY770853 | 2013 | China |
| Bat coronavirus | Rhinolophus macrotis | KY770854 | 2013 | China |
| Bat coronavirus | Myotis davidii | KY770856 | 2013 | China |
| Bat coronavirus | Ia io | KY770857 | 2013 | China |
| Alphacoronavirus sp. | Neoromicia capensis | MF593271 | 2013 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG193605 | 2015 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG193610 | 2015 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG193611 | 2015 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG193616 | 2016 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG205585 | 2015 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG205586 | 2014 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG205590 | 2015 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG205592 | 2015 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG205598 | 2015 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG205599 | 2015 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG252860 | 2015 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG252863 | 2015 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG252866 | 2015 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG252870 | 2016 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG252871 | 2016 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG252874 | 2016 | South Africa |
| Bat alphacoronavirus | Neoromicia nanus | MG252875 | 2016 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG310222 | 2015 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG310232 | 2015 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG310235 | 2015 | South Africa |
| Bat alphacoronavirus | Miniopterus natalensis | MG310236 | 2016 | South Africa |
| Bat alphacoronavirus | Miniopterus natalensis | MG310239 | 2015 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG310241 | 2015 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG310242 | 2015 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG310246 | 2015 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG310247 | 2015 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG310253 | 2014 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG310257 | 2014 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG817491 | 2015 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG817498 | 2015 | South Africa |
| Bat alphacoronavirus | Neoromicia capensis | MG817499 | 2015 | South Africa |
| Alphacoronavirus sp. | Scotophilus kuhlii | MH687934 | 2014 | Vietnam |
| Alphacoronavirus sp. | Scotophilus kuhlii | MH687937 | 2014 | Vietnam |
| Alphacoronavirus sp. | Scotophilus kuhlii | MH687938 | 2014 | Vietnam |
| Alphacoronavirus sp. | Scotophilus kuhlii | MH687941 | 2014 | Vietnam |
| Alphacoronavirus sp. | Scotophilus kuhlii | MH687942 | 2014 | Vietnam |
| Alphacoronavirus sp. | Scotophilus kuhlii | MH687943 | 2014 | Vietnam |
| Alphacoronavirus sp. | Scotophilus kuhlii | MH687944 | 2014 | Vietnam |
| Alphacoronavirus sp. | Scotophilus kuhlii | MH687945 | 2014 | Vietnam |
| Alphacoronavirus sp. | Scotophilus kuhlii | MH687946 | 2014 | Vietnam |
| Alphacoronavirus sp. | Scotophilus kuhlii | MH687948 | 2014 | Vietnam |
| Alphacoronavirus sp. | Scotophilus kuhlii | MH687954 | 2014 | Vietnam |
| Alphacoronavirus sp. | Scotophilus kuhlii | MH687955 | 2014 | Vietnam |
| Alphacoronavirus sp. | Scotophilus kuhlii | MH687956 | 2014 | Vietnam |
| Alphacoronavirus sp. | Scotophilus kuhlii | MH687957 | 2014 | Vietnam |
| Alphacoronavirus sp. | Scotophilus kuhlii | MH687958 | 2014 | Vietnam |
| Alphacoronavirus sp. | Scotophilus kuhlii | MH687960 | 2014 | Vietnam |
| Alphacoronavirus sp. | Scotophilus kuhlii | MH687961 | 2014 | Vietnam |
| Alphacoronavirus sp. | Scotophilus kuhlii | MH687965 | 2015 | Vietnam |
| Alphacoronavirus sp. | Scotophilus kuhlii | MH687966 | 2015 | Vietnam |
| Bat coronavirus | Pipistrellus pipistrellus | MH921428 | 2016 | China |
| Bat coronavirus | Pipistrellus pipistrellus | MH921429 | 2016 | China |
| Coronavirus BtSk-AlphaCoV/GX2018B | Scotophilus kuhlii | MK211370 | 2017 | China |
| Coronavirus BtSk-AlphaCoV/GX2018D | Scotophilus kuhlii | MK211372 | 2017 | China |
| Chalinolobus morio alphacoronavirus | Chalinolobus morio | MN602059 | 2016 | Australia |
| Chalinolobus morio alphacoronavirus | Chalinolobus morio | MN602060 | 2016 | Australia |
| Miniopterus pusillus bat coronavirus | Miniopterus pusillus | MN611518 | 2018 | China |
| Scotophilus kuhlii bat coronavirus 512-related |
Scotophilus kuhlii |
MN611521 |
2018 |
China |
| Betacoronavirus | ||||
| Bat SARS coronavirusc | Rhinolophus pearsoni | DQ071615 | 2004 | China |
| Bat coronavirus/133/2005 | Tylonycteris pachypus | DQ648794 | 2005 | China |
| Bat coronavirus HKU5 | Pipistrellus abramus | EF065512 | 2006 | China |
| Bat coronavirus HKU9 |
Rousettus leschenaulti |
HM211100 |
2006 |
China |
| Gammacoronavirus | ||||
| Infectious bronchitis virus | Gallus gallus | FJ904719 | 1991 | USA |
| Infectious bronchitis virus | Gallus gallus | FJ904721 | 1972 | USA |
| Turkey coronavirus | Meleagris gallopavo | GQ427173 | 2003 | USA |
| Turkey coronavirus | Meleagris gallopavo | GQ427175 | 1994 | USA |
| Turkey coronavirus | Meleagris gallopavo | GQ427176 | 1998 | USA |
| Infectious bronchitis virus | Gallus gallus | GQ504724 | 1941 | USA |
| Infectious bronchitis virus | Gallus gallus | GU393336 | 1954 | USA |
| Duck coronavirus | Duckf | JF705860 | 2004 | China |
| Infectious bronchitis virus |
Gallus gallus |
JF828980 |
2010 |
China |
| Deltacoronavirus | ||||
| Bulbul coronavirus HKU11 | Pycnonotus jocosus | FJ376619 | 2007 | China |
| Thrush coronavirus HKU12 | Turdus hortulorum | FJ376621 | 2007 | China |
| Munia coronavirus HKU13 | Lonshura striata | FJ376622 | 2007 | China |
| White-eye coronavirus HKU16 | Zosterops sp. | JQ065044 | 2007 | China |
| Sparrow coronavirus HKU17 | Passer montanus | JQ065045 | 2007 | China |
| Magpie robin coronavirus HKU18 | Copsychus saularis | JQ065046 | 2007 | China |
| Night-heron coronavirus HKU19 | Nycticorax nycticorax | JQ065047 | 2007 | China |
| Wigeon coronavirus HKU20 | Anas Penelope | JQ065048 | 2008 | China |
| Common-moorhen coronavirus HKU21 | Gallinula chloropus | JQ065049 | 2007 | China |
Species, strain/isolate names are in Supplemental Table S2.
2.2. Phylogenetic analyses
We chose to use amino acid sequences in our primary analyses but, due to the short length of the available sequences, we also re-ran analyses using the nucleotides. We used BEAST v1.10.4 (Suchard et al., 2018) and BEAST2 v2.6.3 (Bouckaert et al., 2019) to analyse the amino acid and nucleotide sequence alignments respectively. We did not use tip dates as the timescales mean all isolates are effectively contemporaneously sampled. We initially assumed a constant population size and a strict clock with an LG substitution model for amino acid sequences (only available in BEAST v1) and HKY substitution model as well as a four-bin gamma rate distribution (GTR + Γ4) with a proportion of invariant sites for nucleotides. After some initial tree topology checking to confirm that genera and the New Zealand and Australian bat-derived orthocoronaviruses were monophyletic, we put a calibration prior on the node that is the common ancestor of the New Zealand and Australian bat-derived orthocoronaviruses. To check the sensitivity of our assumptions we then also put a prior on the age of all the bird-derived viruses and subsequently only the bird derived orthocoronaviruses, so in total used two calibration points in three scenarios: a bat-only, a bird-bat, and a bird-only. The bat-only calibration was tested on two datasets, one complete (123 sequences from bat and bird hosts) and one using only bat hosts (105 sequences). The ages we used were 35 million years (2.5–97.5% quartiles = 30–42 MY) for the New Zealand bats, based on the estimated divergence time of Mysticina from other bats (Van den Bussche and Hoofer, 2000), and 150 million years (2.5–97.5% quartiles = 134–167 MY) for the age of birds (Hu et al., 2009). However, because New Zealand has one other living bat species, Chalinolobus tuberculatus, we also test the sensitivity of this divergence time, and use the age of C. tuberculatus, 17 million years (2.5–97.5% quartiles = 14–20 MY) (Dool et al., 2016). We identify calibrations with the taxa (e.g. bird, bat) and the age in superscriptx, e.g. bat35 is the 35MY calibration on the New Zealand-Australian bat virus ancestor node. Lastly, we also used a relaxed clock for each calibration scenario and general time-reversible substitution model with a four-bin gamma rate distribution (GTR + Γ4) for each nucleotide scenario. Strict clock models were run for 10 million MCMC samples with a 10% burnin and sampling every 1000, whereas the relaxed clocks require longer MCMC chains, so were run for 100 million and sampled every 10000. The analyses covered 186 amino acid and 561 nucleotide sites. All xml files are available at https://github.com/dtsh2/coronavirus_ancestry and can be replicated. Logs were visualized in Tracer 1.7.1 (Rambaut et al., 2018) and trees plotted in FigTree 1.4.4 (Rambaut, 2012). We checked for overall host-virus coevolution using parafit (Legendre et al., 2002) in the ape R package (Paradis and Schliep, 2019) using taxa (or sister taxa) from the TimeTree database (Hedges et al., 2015) and amino acid model with the bat35 and bird150 calibrations (see Supplementary Fig. S1). Further manipulation and visualization were performed in R v4.0.4 using beastie (du Plessis, 2020); ggplot2 (Wickham, 2016); ggmcmc (Fernandez-i-Marin, 2016); stringr (Wickham, 2019); and ggdistribute (Burling, 2018) packages.
3. Results
3.1. Assumptions and model adequacy
We found no support for recombination across the RdRp fragment used (see Supplementary Fig. S2). As anticipated, the limited length of amino acid sequences did not provide information for the modelling of some parameter and as a result several of the relaxed clock models returned effective samples sizes (ESS) of <200. However, crucially the ESS for tree heights (ages) was >1179 (range 1179–5036). Using nucleotide sequences, all chains converged with ESS all >200, with all ESS for tree heights (ages) > 479 (range 479–5775). Overall, the nucleotide tree topology was well supported, with high posterior support for many nodes. There was support for coevolution between viruses and hosts over the whole tree using parafit (p value = 0.03, Supplementary Fig. S3). The posterior distributions of the HKY and GTR were essentially the same and, as expected there is increasing uncertainty of tMRCA estimates and lower node support with deeper nodes (see Supplementary Table S3, Supplementary Figs. S4–S33). Herein only the amino acid sequences using the 35 MY bat prior (bat35) are discussed, but all the results can be seen in Table 2 and the supplementary information.
TABLE 2.
Orthocoronavirus ages by different models in this study using amino acid sequences. Calibration confidence intervals at 2.5–97.5% are 14–20 MY for Bat-17 MYA, 30–42 MY for Bat-35 MYA and 134–167 MY for Bird-150 MYA. Mean and 95% highest posterior density (HPD) only are shown for the substitution rate (per site per million years).
| Orthocoronaviruses | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hosts | Bat | Bat & Bird | Bat & Bird | Bat | Bat & Bird | Bat & Bird | Bat & Bird | Bat | Bat & Bird | Bat & Bird | Bat | Bat & Bird | Bat & Bird | Bat & Bird |
| Model | ||||||||||||||
| Clock | Relaxeda | Relaxed | Relaxed | Relaxeda | Relaxeda | Relaxeda | Relaxed | Strict | Strict | Strict | Strict | Strict | Strict | Strict |
| Site model | LG | LG | LG | LG | LG | LG | LG | LG | LG | LG | LG | LG | LG | LG |
| Calibration | Bat-17 MYA | Bat-17 MYA | Bat-17 MYA + Bird-150 MYA | Bat-35 MYA | Bat-35 MYA | Bat-35 MYA + Bird-150 MYA | Bird-150 MYA | Bat-17 MYA | Bat-17 MYA | Bat-17 MYA + Bird-150 MYA | Bat-35 MYA | Bat-35 MYA | Bat-35 MYA + Bird-150 MYA | Bird-150 MYA |
| Age | ||||||||||||||
| Mean | 178.5 | 239.2 | 176.9 | 347.6 | 492 | 234.7 | 168 | 213.7 | 264.9 | 167.8 | 417.8 | 531.7 | 185.9 | 163.4 |
| Median | 160.4 | 223.4 | 170 | 305.8 | 465.2 | 218.4 | 162.8 | 198 | 246 | 166.4 | 391.3 | 493.2 | 184.4 | 162 |
| 95% HPD | 21.2–375.9 | 85.6–423.9 | 136–231 | 38.2–748.1 | 221.5–832.4 | 141.5–361.7 | 134.4–210 | 90.7–363.4 | 123.8–446.7 | 143.1–197.2 | 190.9–708 | 240.7–899.1 | 153.9–219.8 | 139–191.5 |
| Range | 14.9–1565.5 | 34.4–1077.3 | 126–765 | 31.2–3176.9 | 92.2–1504.1 | 136.2–792.2 | 129.1–454.8 | 68.5–942.8 | 72.2–760.4 | 121.5–240.3 | 106.8–1265.2 | 195.5–2897.1 | 139.1–262.8 | 124.8–240 |
| Bat Clade Age | ||||||||||||||
| Mean | 178.5 | 171.3 | 139.7 | 347.6 | 358.9 | 171.1 | 131 | 213.7 | 199.8 | 139.7 | 417.8 | 399.3 | 160 | 134.7 |
| Median | 160.4 | 160.3 | 137 | 305.8 | 340.6 | 165.5 | 129.4 | 198 | 186.4 | 138.4 | 391.3 | 367.2 | 158.9 | 133.2 |
| 95% HPD | 21.2–375.9 | 61.9–306.1 | 97–187 | 38.2–748.1 | 164.5–603.1 | 93.4–259.8 | 89.8–173.4 | 90.7–363.4 | 92.3–338.1 | 108.1–172.6 | 190.9–708 | 185.5–687.6 | 123.9–198 | 105.5–167.8 |
| Range | 14.9–1565.5 | 23.4–711.8 | 68–352 | 31.2–3176.9 | 63.6–1127.4 | 54.6–560.5 | 67.6–350.4 | 68.5–942.8 | 85.8–829.2 | 88.5–215.2 | 106.8–1265.2 | 134.9–2095 | 101.2–236.5 | 88.1–218.5 |
| Substitution rate | ||||||||||||||
| Mean | 2.41E-3 | 1.81E-3 | 2.20E-3 | 1.18E-3 | 9.02E-4 | 1.81E-3 | 2.36E-3 | 1.52E-3 | 1.63E-3 | 2.20E-3 | 7.71E-4 | 8.13E-4 | 1.91E-3 | 2.30E-3 |
| 95% HPD | 5.84E-4 –5.49E-3 | 8.29E-4 –2.94E-3 | 1.65E-3 –2.75E-3 | 2.50E-4 –2.79E-3 | 3.71E-4 –1.46E-3 | 1.04E-3 –2.69E-3 | 1.67E-3 -2.99E-3 | 6.79E-4 -2.51E-3 | 7.17E-4 -2.62E-3 | 1.79E-3 -2.60E-3 | 3.41E-4 -1.27E-3 | 3.63E-4 -1.32E-3 | 1.55E-3 -2.27E-3 | 1.86E-3 -2.75E-3 |
Low ESS (<200) for some parameters, see text.
3.2. Time to the most recent common ancestor
The strict clock analyses using the bat17, bat35, bat17 and bird150, bat35 and bird150 or bird150 only calibration points and LG model, estimated bat orthocoronaviruses to be somewhere from 133 to 391 million years old (median values). These values overlapped with the estimates from all other models for the relaxed clocks (Fig. 1 ). The relaxed clock models had great uncertainty, for example for the bat35 only calibration with both bat and bird viruses the estimate was 305MY (38–748 95% HPD). The strict clock estimates were 391MY (190–708 95% HPD). All other values are in Table 2. Overall, our calibrations led to the substitution rates ranging from means of to per MY (see Table 2, Supplementary Table S4 and Fig. S34).
Fig. 1.
Crown date estimates for all amino acid models. A 35 MY bat calibration prior on the New Zealand and Australian bat derived virus clade was used. The present (solid), 35MY calibration (dashed), 50MY (approximate age of bats, dotted) and 150MY (approximate age of bats, dash-dot) are shown by vertical bars.
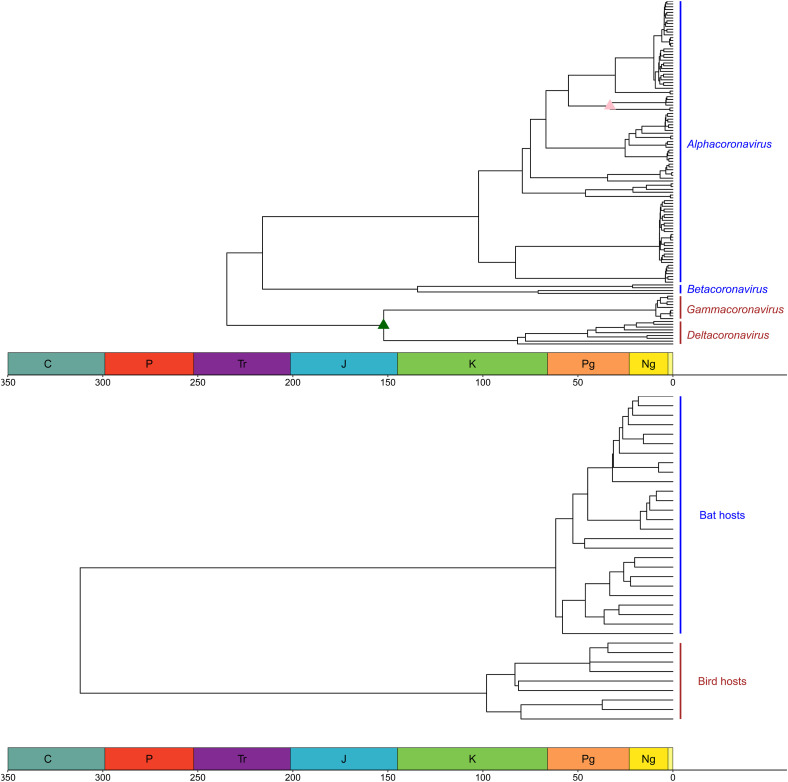
For all analyses, the single younger bat calibration point (17 MY) led to less uncertainty and younger tMRCA estimates if used alone. For the strict clock analyses the differences between estimates when the bird calibration point was used led to non-overlapping 95% HPD. However, in all analyses the estimates for the tMRCA for bat orthocoronaviruses is older than bats themselves (around 50MY, Fig. 2 ) and always includes the estimated age of birds (150MY), the only exception being the most uncertain strict clock analysis mentioned above with only the 35MY NZ-Australian bat clade prior (bat35), that estimates the viruses to be older. In all cases the estimated orthocoronavirus crown tMRCA is similar to those of bat orthocoronaviruses. The calibration points force the nodes to be monophyletic and in most analyses the maximum clade credibility (MCC) trees had bat (alpha- and betacoronaviruses) with one common ancestor and bird (gamma- and deltacoronaviruses) with another. However, with only the bat calibration point, both the relaxed and strict clocks placed the bird virus ancestors as ancestral to bat viruses, with the switch only once occurring when both calibration points were used using the relaxed clock and GTR+Γ4 model. All 14 amino acid and 16 nucleotide trees with the 35 million year old prior and their node support, 95% HPD and virus names are provided in the supplementary information.
Fig. 2.
Phylogeny of orthocoronaviruses and their hosts. Amino acid alignments with a relaxed molecular clock, 35MY bat calibration prior (pink triangle) on the New Zealand and Australian bat derived virus clade and 150MY bird calibration prior on delta- and gammacoronaviruses were used (dark green triangle). Host ages are provided for comparison from http://www.timetree.org/(Hedges et al., 2015) (see Supplementary Fig. S1). Bat viruses and bat hosts (blue) and bird viruses and bird hosts (brown) names are coloured.
4. Discussion
Our results support previous findings that orthocoronaviruses evolved millions of years ago (Wertheim et al., 2013). Increasingly analyses suggest that many viruses are ancient. Amniotes (reptiles, birds, and mammals) are estimated from fossil and molecular evidence to be around 325 million years old (Blair and Hedges, 2005; Shedlock and Edwards, 2009), whereas birds share an ancestor around 150 million years and bats around 50 million years ago (Simmons et al., 2008; Teeling et al., 2005). Our estimated tMRCA dates for orthocoronaviruses of somewhere from 133 to 391 million years ago (median estimates), older than bats and (mostly) birds ancestors, suggesting these viruses evolved prior to bat and possibly bird ancestors among earlier Amniotes. There is, of course, great uncertainty in our estimates (Table 2, Fig. 1), but our results were robust to changes in the use of calibration point position. While the values and uncertainty changed, the use of calibrations either closer to tree tips (bat17, bat35) or deeper in the tree (bird150) nearer the crown of all orthocoronaviruses all led to ages older than bats and, mostly, birds. The family Coronaviridae includes Letovirinae subfamily viruses from a frog (Bukhari et al., 2018) and metatranscriptomic sequencing has identified distinct and diverse Coronaviridae phylogenetic groups among jawless and bony fish, providing further evidence that these viruses have evolved with vertebrate hosts over millions of years (Miller et al., 2021; Mordecai et al., 2019).
Here we also assumed these viruses were exclusively bat and bird viruses. It is feasible that other hosts (for example porcine, rodent, etc.) will come to light since host switching does occur, as evidenced by the recent emergence of orthocoronaviruses causing SARS, MERS and COVID-19 in people and swine acute diarrhoea syndrome in pigs (Zhou et al., 2018), and the widespread distribution of other now endemic orthocoronaviruses, such as HCoV-229E. We excluded other viruses because of this, but our results may have influenced by such host switches among bat taxa. The monophyletic relationships of orthocoronaviruses in bats (i.e., alphacoronaviruses and betacoronaviruses) and birds (gammacoronaviruses and deltacoronaviruses), however, suggest these relationships are old and real (Chu et al., 2011; Wong et al., 2019; Woo et al., 2012). The sequencing of additional and/or more complete genomes of bat-derived orthocoronaviruses from islands, especially in the Pacific, may help support these findings.
It is most likely orthocoronaviruses arrived in New Zealand with the colonizing Mysticina (~35MYA) or Chalinolobus (~17MYA) bats, though the arrival through other non-bat species such as marine mammals or with humans is possible. Given the isolated population of Mysticina on a New Zealand South Island island in the Pacific Ocean the alphacoronavirus RdRp RNA was isolated from and the widespread distribution of bat orthocoronaviruses globally, this later arrival seems unlikely. Further, more recent host switching from non-bat (e.g. rodent) hosts is possible but highly unlikely given the close association of New Zealand bat orthocoronaviruses with Australian bat alphacoronaviruses and the very recent arrival of the first rodents to New Zealand. Rodents (specifically the Polynesian rat (Rattus exulans), known to Māori as kiore) only arrived in New Zealand from Polynesia with Māori, not via Australia, approximately 700 years ago, and other rodents only with Europeans. Therefore, it is more likely our results support those using the BS-REL tree calibration and that orthocoronaviruses have been infecting bats and birds for millions of years and, possibly, since their Amniote ancestors diverged approximately 300 million years ago.
There remain other issues to resolve for orthocoronaviruses. For example, while the partial RdRp gene we used was likely free of recent recombination, recombination among orthocoronaviruses is common and can even include genetic material from unrelated viruses, impacting any estimates relating to ancestry (Paskey et al., 2020). By using calibration points so far in the past, we automatically reduce the substitution rate (range to per MY). These rates had previously been estimated to be 10−5 to 10−6 mutations per site per replication (Eckerle et al., 2010) or around 10−3 substitutions per site per year (Hon et al., 2008; Wertheim et al., 2013). When clocks become unreliable, therefore, is unknown, though clearly analyses of more recent ancestries like SARS-coronaviruses are most reliable of all (Boni et al., 2020). Time dependent rates of molecular evolution have been observed from a wide range of taxa with short-term substitution rates exceeding long-term by an order of magnitude or more (Ho et al., 2011). Rates of molecular evolution in RNA viruses may span several orders of magnitude (Duffy et al., 2008) with this rapid accumulation of genetic differences in the short-term primarily due to the fidelity of the polymerase that is used during replication, though genome size and replication speed are also important factors. In the longer term, it is thought that strong purifying selection constrains the substitution rate (Duchêne et al., 2014; Holmes, 2003). Together, the evidence supports time varying rates of evolution and so substitution rates (including ours) need to be used and interpreted with caution (Duchêne et al., 2014).
In summary, our analyses using Bayesian evolutionary and tree analyses and mammalian tMRCA estimates have allowed us to make inferences about the age of orthocoronaviruses and support our intuition that orthocoronaviruses probably have an evolutionary history that matches their vertebrate hosts.
Funding
This work was supported by Royal Society Te Apārangi Rutherford Discovery Fellowship RDF-MAU1701
CRediT authorship contribution statement
David T.S. Hayman: Conceptualization, Methodology, Data curation, Formal analysis, Writing – original draft, preparation. Matthew A. Knox: Data curation, Formal analysis, Writing – review & editing.
Declaration of competing interest
☒ The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
☐The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Acknowledgements
Thanks to Prof Eddie Holmes, for his very helpful discussions and great generosity. We also thank two anonymous reviewers, and Virology editor, Prof Gorbalenya, who was exceptionally helpful.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2021.08.004.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Belyi V.A., Levine A.J., Skalka A.M. Unexpected inheritance: multiple integrations of ancient bornavirus and ebolavirus/marburgvirus sequences in vertebrate genomes. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair J.E., Hedges S.B. Molecular phylogeny and divergence times of deuterostome animals. Mol. Biol. Evol. 2005;22:2275–2284. doi: 10.1093/molbev/msi225. [DOI] [PubMed] [Google Scholar]
- Boni M.F., Lemey P., Jiang X., Lam T.T.-Y., Perry B., Castoe T., Rambaut A., Robertson D.L. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat. Microbiol. 2020;5:1408–1417. doi: 10.1038/s41564-020-0771-4. 11th ed. [DOI] [PubMed] [Google Scholar]
- Bouckaert R., Vaughan T.G., Barido-Sottani J., Duchene S., Fourment M., Gavryushkina A., Heled J., Jones G., Kuhnert D., De Maio N., Matschiner M., Mendes F.K., Muller N.F., Ogilvie H.A., du Plessis L., Popinga A., Rambaut A., Rasmussen D., Siveroni I., Suchard M.A., Wu C.H., Xie D., Zhang C., Stadler T., Drummond A.J. Beast 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019;15 doi: 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari K., Mulley G., Gulyaeva A.A., Zhao L., Shu G., Jiang J., Neuman B.W. Description and initial characterization of metatranscriptomic nidovirus-like genomes from the proposed new family Abyssoviridae, and from a sister group to the Coronavirinae, the proposed genus Alphaletovirus. Virology. 2018;524:160–171. doi: 10.1016/j.virol.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burling J. 2018. Ggdistribute: A Ggplot2 Extension for Plotting Unimodal Distributions.https://github.com/iamamutt/ggdistribute/ R package version 1.0.1. [Google Scholar]
- Chu D.K., Leung C.Y., Gilbert M., Joyner P.H., Ng E.M., Tsemay M.T., Guan Y., Peiris J.S., Poon L.L. Avian coronavirus in wild aquatic birds. J. Virol. 2011;85:12815–12820. doi: 10.1128/JVI.05838-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Baker S.C., Baric R., Enjuanes L., Gorbalenya A.E., Holmes K.V., Perlman S., Poon L., Rottier P.J.M., Talbot P.J., Woo P.C.Y., J Z. In: Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses. King A.M.Q., Adams M.J., Carstens E.B., EJ L., editors. Academic Press, Ltd.; London, United Kingdom: 2011. Family Coronaviridae; pp. 806–828. [Google Scholar]
- Dool S.E., O'Donnell C.F.J., Monks J.M., Puechmaille S.J., Kerth G. Phylogeographic-based conservation implications for the New Zealand long-tailed bat, (Chalinolobus tuberculatus): identification of a single ESU and a candidate population for genetic rescue. Conserv. Genet. 2016;17:1067–1079. [Google Scholar]
- du Plessis L. 2020. Beastio: Functions for Pre- and Post-processing of BEAST and BEAST2 Files. R package version 0.2.5. [Google Scholar]
- Duchêne S., Holmes E.C., Ho S.Y. vol. 281. 2014. Analyses of evolutionary dynamics in viruses are hindered by a time-dependent bias in rate estimates; p. 20140732. (Proceedings of the Royal Society B: Biological Sciences). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S., Shackelton L.A., Holmes E.C. Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 2008;9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- Eckerle L.D., Becker M.M., Halpin R.A., Li K., Venter E., Lu X.T., Scherbakova S., Graham R.L., Baric R.S., Stockwell T.B., Spiro D.J., Denison M.R. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-i-Marin X. Ggmcmc: analysis of MCMC samples and bayesian inference. J Stat Softw. 2016;70 [Google Scholar]
- Gavryushkina A., Heath T.A., Ksepka D.T., Stadler T., Welch D., Drummond A.J. Bayesian total-evidence dating reveals the recent crown radiation of penguins. Syst. Biol. 2017;66:57–73. doi: 10.1093/sysbio/syw060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford R.J., Katzourakis A., Tristem M., Pybus O.G., Winters M., Shafer R.W. A transitional endogenous lentivirus from the genome of a basal primate and implications for lentivirus evolution. Proc. Natl. Acad. Sci. Unit. States Am. 2008;105:20362–20367. doi: 10.1073/pnas.0807873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C., Feschotte C. Genomic fossils calibrate the long-term evolution of hepadnaviruses. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. In: Nidoviruses. Perlman S., Gallagher T., EJ S., editors. ASM Press; Washington, DC: 2008. Genomics and evolution of the Nidovirales; pp. 15–28. [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J., Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R.J., Wang J., Peacey M., Moore N.E., McInnes K., Tompkins D.M. New alphacoronavirus in Mystacina tuberculata bats, New Zealand. Emerg. Infect. Dis. 2014;20:697–700. doi: 10.3201/eid2004.131441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G.-Z., Worobey M. An endogenous foamy-like viral element in the coelacanth genome. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath T.A. A hierarchical bayesian model for calibrating estimates of species divergence times. Syst. Biol. 2012;61:793–809. doi: 10.1093/sysbio/sys032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges S.B., Marin J., Suleski M., Paymer M., Kumar S. Tree of life reveals clock-like speciation and diversification. Mol. Biol. Evol. 2015;32:835–845. doi: 10.1093/molbev/msv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S.Y.W., Lanfear R., Bromham L., Phillips M.J., Soubrier J., Rodrigo A.G., Cooper A. Time-dependent rates of molecular evolution. Mol. Ecol. 2011;20:3087–3101. doi: 10.1111/j.1365-294X.2011.05178.x. [DOI] [PubMed] [Google Scholar]
- Holmes E.C. Patterns of intra- and interhost Nonsynonymous variation reveal strong purifying selection in dengue virus. J. Virol. 2003;77:11296–11298. doi: 10.1128/JVI.77.20.11296-11298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon C.C., Lam T.Y., Shi Z.L., Drummond A.J., Yip C.W., Zeng F., Lam P.Y., Leung F.C.C. Evidence of the recombinant origin of a bat severe acute respiratory syndrome (SARS)-like coronavirus and its implications on the direct ancestor of SARS coronavirus. J. Virol. 2008;82:1819–1826. doi: 10.1128/JVI.01926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D.Y., Hou L.H., Zhang L.J., Xu X. A pre-Archaeopteryx troodontid theropod from China with long feathers on the metatarsus. Nature. 2009;461:640–643. doi: 10.1038/nature08322. [DOI] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:496. doi: 10.1016/S0140-6736(20)30183-5. vol 395, pg 497, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre P., Desdevises Y., Bazin E. A statistical test for host–parasite coevolution. Syst. Biol. 2002;51:217–234. doi: 10.1080/10635150252899734. [DOI] [PubMed] [Google Scholar]
- Leopardi S., Holmes E.C., Gastaldelli M., Tassoni L., Priori P., Scaravelli D., Zamperin G., De Benedictis P. Interplay between co-divergence and cross-species transmission in the evolutionary history of bat coronaviruses. Infect. Genet. Evol. 2018;58:279–289. doi: 10.1016/j.meegid.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch D.J., Cook S. Molecular phylogeny of the alphaherpesvirinae subfamily and a proposed evolutionary timescale. J. Mol. Biol. 1994;238:9–22. doi: 10.1006/jmbi.1994.1264. [DOI] [PubMed] [Google Scholar]
- McGeoch D.J., Cook S., Dolan A., Jamieson F.E., Telford E.A. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J. Mol. Biol. 1995;247:443–458. doi: 10.1006/jmbi.1995.0152. [DOI] [PubMed] [Google Scholar]
- Miller A.K., Mifsud J.C.O., Costa V.A., Grimwood R.M., Kitson J., Baker C., Brosnahan C.L., Pande A., Holmes E.C., Gemmell N.J., Geoghegan J.L. Slippery when wet: cross-species transmission of divergent coronaviruses in bony and jawless fish and the evolutionary history of the Coronaviridae. Virus Evolution. 2021 doi: 10.1093/ve/veab050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minin V.N., Dorman K.S., Fang F., Suchard M.A. Dual multiple change-point model leads to more accurate recombination detection. Bioinformatics. 2005;21:3034–3042. doi: 10.1093/bioinformatics/bti459. [DOI] [PubMed] [Google Scholar]
- Mordecai G.J., Miller K.M., Di Cicco E., Schulze A.D., Kaukinen K.H., Ming T.J., Li S., Tabata A., Teffer A., Patterson D.A. Endangered wild salmon infected by newly discovered viruses. Elife. 2019;8 doi: 10.7554/eLife.47615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer N., Campbell H.J., Tulloch A.J., King P.R., Stagpoole V.M., Wood R.A., Rattenbury M.S., Sutherland R., Adams C.J., Collot J., Seton M. Zealandia: earth's hidden continent. GSA Today (Geol. Soc. Am.) 2017;27:27–35. [Google Scholar]
- Paradis E., Schliep K. Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35:526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- Paskey A.C., Ng J.H.J., Rice G.K., Chia W.N., Philipson C.W., Foo R.J.H., Cer R.Z., Long K.A., Lueder M.R., Lim X.F., Frey K.G., Hamilton T., Anderson D.E., Laing E.D., Mendenhall I.H., Smith G.J., Wang L.-F., Bishop-Lilly K.A. Detection of recombinant rousettus bat coronavirus GCCDC1 in lesser dawn bats (eonycteris spelaea) in Singapore. Viruses. 2020;12:539. doi: 10.3390/v12050539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S.M., Lai S.T., Poon L.L.M., Guan Y., Yam L.Y.C., Lim W., Nicholls J., Yee W.K.S., Yan W.W., Cheung M.T., Cheng V.C.C., Chan K.H., Tsang D.N.C., Yung R.W.H., Ng T.K., Yuen K.Y., Grp S.S. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond S.L.K., Murrell B., Fourment M., Frost S.D.W., Delport W., Scheffler K. A random effects branch-site model for detecting episodic diversifying selection. Mol. Biol. Evol. 2011;28:3033–3043. doi: 10.1093/molbev/msr125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L.M., Chu D.K.W., Chan K.H., Wong O.K., Ellis T.M., Leung Y.H.C., Lau S.K.P., Woo P.C.Y., Suen K.Y., Yuen K.Y., Guan Y., Peiris J.S.M. Identification of a novel coronavirus in bats. J. Virol. 2005;79:2001–2009. doi: 10.1128/JVI.79.4.2001-2009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. University of Edinburgh, Institute of Evolutionary Biology; Edinburgh: 2012. FigTree V1. 4. Molecular Evolution, Phylogenetics and Epidemiology. [Google Scholar]
- Rambaut A., Drummond A.J., Xie D., Baele G., Suchard M.A. Posterior summarization in bayesian phylogenetics using tracer 1.7. Syst. Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieux A., Balloux F. Inferences from tip-calibrated phylogenies: a review and a practical guide. Mol. Ecol. 2016;25:1911–1924. doi: 10.1111/mec.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedlock A.M., Edwards S.V. In: The Timetree of Life. Hedges SB S.K., editor. Oxford University Press; Oxford, United Kingdom: 2009. Amniotes (amniota) pp. 375–379. [Google Scholar]
- Simmons N.B., Seymour K.L., Habersetzer J., Gunnell G.F. Primitive Early Eocene bat from Wyoming and the evolution of flight and echolocation. Nature. 2008;451 doi: 10.1038/nature06549. 818-U816. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L.M., Guan Y., Rozanov M., Spaan W.J.M., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard M.A., Lemey P., Baele G., Ayres D.L., Drummond A.J., Rambaut A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evolution. 2018;4 doi: 10.1093/ve/vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh A., Brosius J., Schmitz J., Kriegs J.O. The genome of a Mesozoic paleovirus reveals the evolution of hepatitis B viruses. Nat. Commun. 2013;4:1791. doi: 10.1038/ncomms2798. [DOI] [PubMed] [Google Scholar]
- Suh A., Weber C.C., Kehlmaier C., Braun E.L., Green R.E., Fritz U., Ray D.A., Ellegren H. Early mesozoic coexistence of amniotes and hepadnaviridae. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D.J., Leach R.W., Bruenn J. Filoviruses are ancient and integrated into mammalian genomes. BMC Evol. Biol. 2010;10:193. doi: 10.1186/1471-2148-10-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling E.C., Springer M.S., Madsen O., Bates P., O'brien S.J., Murphy W.J. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307:580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- Thézé J., Bézier A., Periquet G., Drezen J.-M., Herniou E.A. Paleozoic origin of insect large dsDNA viruses. Proc. Natl. Acad. Sci. Unit. States Am. 2011;108:15931–15935. doi: 10.1073/pnas.1105580108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Bussche R.A., Hoofer S.R. Further evidence for inclusion of the New Zealand short-tailed bat (Mystacina tuberculata) within Noctilionoidea. J. Mammal. 2000;81:865–874. [Google Scholar]
- Walter R., Buckley H., Jacomb C., Matisoo-Smith E. Mass migration and the polynesian settlement of New Zealand. J World Prehist. 2017;30:351–376. [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Moore N.E., Murray Z.L., McInnes K., White D.J., Tompkins D.M., Hall R.J. Discovery of novel virus sequences in an isolated and threatened bat species, the New Zealand lesser short-tailed bat (Mystacina tuberculata) The Journal of general virology. 2015;96:2442. doi: 10.1099/vir.0.000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim J.O., Chu D.K.W., Peiris J.S.M., Pond S.L.K., Poon L.L.M. A case for the ancient origin of coronaviruses. J. Virol. 2013;87:7039–7045. doi: 10.1128/JVI.03273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim J.O., Pond S.L.K. Purifying selection can obscure the ancient age of viral lineages. Mol. Biol. Evol. 2011;28:3355–3365. doi: 10.1093/molbev/msr170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wevers B.A., van der Hoek L. Recently discovered human coronaviruses. Clin. Lab. Med. 2009;29:715. doi: 10.1016/j.cll.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. Springer-Verlag; New York: 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- Wickham H. 2019. Stringr: Simple, Consistent Wrappers for Common String Operations. R Package Version 1.4.0.https://CRAN.R-project.org/package=stringr [Google Scholar]
- Wong A.C., Li X., Lau S.K., Woo P.C. Global epidemiology of bat coronaviruses. Viruses. 2019;11:174. doi: 10.3390/v11020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhang L., Liang J., Zhu C. Coevolution of coronavirus and paramyxovirus with their bat hosts in the same geographical areas. BMC Evolutionary Biology Pre-print in Review. 2020 [Google Scholar]
- Zhou H., Ji J., Chen X., Bi Y., Li J., Wang Q., Hu T., Song H., Zhao R., Chen Y., Cui M., Zhang Y., Hughes A.C., Holmes E.C., Shi W. 2021. Identification of Novel Bat Coronaviruses Sheds Light on the Evolutionary Origins of SARS-CoV-2 and Related Viruses. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Fan H., Lan T., Yang X.-L., Shi W.-F., Zhang W., Zhu Y., Zhang Y.-W., Xie Q.-M., Mani S. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Thiel V., Gorbalenya A.E. The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond. J. Biol. Chem. 2001;276:33220–33232. doi: 10.1074/jbc.M104097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.