Abstract
Objective
Cancer-associated cachexia is a devastating pathological disorder characterized by skeletal muscle wasting and fat storage depletion. Circular RNA, a newly discovered class of noncoding RNAs with important roles in regulating lipid metabolism, has not been fully understood in the pathology of cachexia. We aimed to identify circular RNAs that are upregulated in adipose tissues from cachectic patients and explore their function and mechanism in lipid metabolism.
Methods
Whole transcriptome RNA sequencing was used to screen for differentially expressed circRNAs. Quantitative reverse transcription PCR was applied to detect the expression level of circPTK2 in adipose tissues. The diagnostic value of circPTK2 was evaluated in adipose tissues from patients with and without cachexia. Then, function experiments in vitro and in vivo were performed to evaluate the effects of circPTK2 on lipolysis and adipogenesis. Mechanistically, luciferase reporter assay, RNA immunoprecipitation, and fluorescent in situ hybridization were performed to confirm the interaction between circPTK2 and miR-182-5p in adipocytes.
Results
We detected 66 differentially expressed circular RNA candidates and proved that circPTK2 was upregulated in adipose tissues from cachectic patients. Then we identified that circPTK2 was closely related to the pathological process of cachexia and could be used as a diagnostic marker. Mechanistically, circPTK2 bound competitively to miR-182-5p and abrogated the suppression on its target gene JAZF1, which finally led to promotion of lipolysis and inhibition of adipogenesis. In vivo experiments demonstrated that overexpression of circPTK2 inhibited adipogenesis and enhanced lipolysis.
Conclusions
Our findings reveal the novel role of circPTK2 in promoting lipolysis and reducing adipogenesis via a ceRNA mechanism and provide a potential diagnostic biomarker and therapeutic target for cancer-associated cachexia.
Keywords: Cancer-associated cachexia, Circular RNAs, MicroRNAs, Adipose tissue wasting, Biomarkers
Highlights
-
•
A novel noncoding RNA termed circPTK2 was highly expressed in adipose tissues of patients with cancer-associated cachexia.
-
•
CircPTK2 was proven to be a potential diagnostic biomarker for cancer-associated cachexia.
-
•
CircPTK2 induced lipolysis and suppressed adipogenesis by sponging miR-182-5p to regulate JAZF1 expression.
-
•
A recombinant adeno-associated virus containing tissue-specific promoter was constructed and utilized in the animal experiment.
1. Introduction
Cancer-associated cachexia (CAC) is a devastating and systemic syndrome characterized by progressive loss of body weight due mainly to skeletal muscle atrophy from cancer growth, with or without loss of adipose mass [1]. CAC occurs in 50–80% of advanced cancer patients (up to 80–90% for gastric and pancreatic cancers) and eventually accounts for more than 20% of all cancer-related deaths [2]. CAC cannot be fully reversed by oral or intravenous nutritional support, and it diminishes the quality of life for patients. Although skeletal muscle atrophy due to increased protein breakdown is known as the major hallmark of CAC, depletion and remodeling of adipose tissue also plays a crucial role in cachectic patients [3]. The mechanism that drives adipose tissue depletion is multifactorial. Lipolysis is activated, accompanied by elevated total energy expenditure and suppressed adipogenesis, eventually reducing the fat mass [4]. Several studies have revealed that adipose tissue wasting typically occurs before the appearance of other classic cachexia signs [5,6]. Therefore, it is essential to improve our understanding regarding the molecular mechanisms involved in the onset and progression of CAC, as well as the pathophysiology of adipose tissue depletion, which represents an early clinical feature of cachexia.
Circular RNAs (circRNAs) have recently been identified as endogenous noncoding RNA molecules, characterized by a covalently closed loop structure without a 5′ cap and a 3′ poly A tail [7]. Unlike linear RNAs, circRNAs usually originate from back-splicing sites of exons or introns. With the development of high-throughput sequencing, numerous circRNAs have been identified. Most of them have been proven to be endogenous, abundant, and conserved in mammalian cells, suggesting specific roles in cellular physiology [8]. Currently, only a few circRNAs have been characterized functionally and mechanistically, but many of them have been demonstrated to have essential roles in the carcinogenesis and metastasis of tumors [[9], [10], [11]]. There are few studies on the role of specific circRNAs in adipose tissue of patients with malignant tumor, and methods of how these circRNAs regulate adipogenesis and lipolysis remain largely unknown.
JAZF1 (juxtaposed with another zinc finger gene 1), aka TIP27 or ZNF802, was first identified as a novel TAK1-interacting protein in 2004 [12]. JAZF1 encodes a 27kd nuclear protein containing three putative zinc finger motifs and is expressed in multiple tissues in mice and humans, especially in adipose tissue and testes [12]. A range of studies have proven that JAZF1 is associated with glucose metabolism, insulin sensitivity, and cell differentiation [[13], [14], [15], [16]]. In rats fed a high-fat diet, hypothalamus JAZF1 may decrease body weight and food intake and inhibit hepatic glucose production by increasing hepatic insulin signaling [17]. Another study showed that JAZF1-knockout mice had lower levels of serum insulin, pancreatic insulin expression, and decreased pancreatic β-cell size, which resulted in defects in the glucose homeostasis [18]. Few studies, however, have focused on the relationship between JAZF1 and lipid metabolism. Ling Li [19] and Guangfeng Ming [15] have demonstrated that overexpression of JAZF1 in 3T3-L1 cells contributes to decreased adipogenesis and increased lipolysis, but the molecular mechanism by which JAZF1 acts in adipocytes has not yet been clarified.
In this study, we used RNA sequencing to identify a novel circRNA derived from PTK2, termed circPTK2, which was significantly overexpressed in adipose tissues of cachectic patients compared to those of non-cachectic patients. Mechanistically, the elevated circPTK2 level upregulates JAZF1 expression to promote lipolysis and suppress adipogenesis by sponging miR-182-5p. Our findings suggest that circPTK2 functions as a promoter in CAC progression and may be a potential target in cachexia-related diagnosis and treatment.
2. Materials and methods
2.1. Human tissue specimens
In total, 90 patients with gastrointestinal tumors who received radical gastrectomy at the Department of Surgery, Zhongshan Hospital, Fudan University were enrolled in the study. A total of 42 patients with greater than 5% weight loss over the past six months were diagnosed with CAC; the rest did not reveal significant recent weight loss. The CT image of third lumbar vertebrae were used as a landmark to measure the skeletal muscle area (cm2) and adipose area (cm2). Data were shown in Additional file: Table S1. The patients did not receive radiotherapy or chemotherapy before surgery. All patients underwent total resection and were verified histologically and pathologically from 2018.06 to 2020.06. Subcutaneous adipose tissues from the abdominal incision site were collected. All samples were gathered and stored in liquid nitrogen at −80 °C. This study was approved by the Ethics Committee of Zhongshan Hospital, Fudan University (Approval No. B2019-193R). Written informed consents were collected from all patients.
2.2. Cell culture and differentiation
The mouse immortalized subcutaneous white preadipocytes were kindly provided by Professor Qiurong Ding from the Metabolism and Food Safety, Shanghai Institute of Nutrition and Health, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. This cell line has been described previously and used in several studies to assess the effects of different factors on adipose differentiation and function [20,21]. Preadipocytes, HEK293T cells, and C26 cells were cultured in high-glucose Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 mg/mL streptomycin. All cells were kept in an atmosphere of 5% CO2 at 37 °C. Differentiation of preadipocytes was initiated by an induction medium (0.5 mM isobutyl-1-methylxanthine (IBMX), 5 mM dexamethasone, 1 μM rosiglitazone, 5 μg/mL insulin) and replaced with a maintenance medium (5 μg/mL Insulin) after 2 days for further differentiation.
2.3. RNA sequencing (RNA-Seq)
The total RNA was extracted from six selected subcutaneous adipose tissues from patients with or without CAC. rRNA was removed from the total RNA with a ribosomal RNA removal kit. The libraries were then constructed using TruSeq Stranded Total RNA with Ribo-Zero Gold according to the manufacturer's instructions. Afterwards, quality inspection of the library was performed with the Agilent 2100 Bioanalyzer, including inspection of the total and effective concentrations. These libraries were sequenced on the Illumina sequencing platform HiSeqTM 2500. The raw sequencing dataset that supported the results of this study was deposited in the NCBI GEO database. The data are accessible through GEO: GSE174128. Differentially expressed transcripts between the two groups were identified when fold change was ≥2 and the p value was <0.05.
2.4. Ribonuclease R (RNase R) treatment
To measure the stability of hsa_circ_0005982, 2 μg total RNA was first treated with 3U/μg RNase R at 37 °C. 30 min later, the expression levels of hsa_circ_0005982 and linear PTK2 were detected using RT-PCR.
2.5. RNA isolation and qRT-PCR
Total RNA was isolated using TRIzol according to the manufacturer's protocol. For mRNA and circRNA, cDNA was synthesized using the FastKing RT Kit (Tiangen, Beijing, China). The abundance of circRNA was determined by divergent primers. Expression levels were normalized to the expression of GAPDH. The expression of miRNAs was detected using miRcute Plus miRNA First-Strand cDNA Kit (Tiangen, Beijing, China). U6 acted as a normalized control. qRT-PCR was performed according to the manufacturer's instructions and the relative fold change was calculated by the 2-△△Ct method. Primers used for real–time reactions were designed and synthesized by Sangon Biotech (Shanghai, China) and are listed in Additional file: Table S2. All experiments were repeated at least three times.
2.6. Isolation of RNAs from nuclear and cytoplasmic fractions
The nuclear and cytoplasmic fractions were isolated using PARIS Kit (AM1921, Invitrogen, Grand Island, CA, USA) following the manufacturer's protocol. Briefly, adipocytes were collected and lysed with cell fractionation buffer, followed by centrifugation to separate the nuclear and cytoplasmic fractions. The supernatant containing the cytoplasmic fraction was collected. The cytoplasmic fraction and nuclear lysate were mixed with 2X Lysis/Binding Solution before 100% ethanol was added. The sample mixture was drawn through a filter cartridge, followed by washing with wash solution. The RNAs of nuclear and cytoplasmic fractions were eluted with elution solution, and 45S rRNA and 12S rRNA were employed as a positive control for nuclear and cytoplasmic fractions, respectively.
2.7. Plasmid and siRNA construction
The sequence of circPTK2 was amplified and cloned into a circRNA overexpression vector GV486 (GeneChem, Shanghai, China), which contains a front circular frame and back circular frame; an empty vector served as a control. The JAZF1 expression plasmid pcDNA3.1+/JAZF1 and empty plasmid pcDNA3.1+ were also constructed. A miR-182-5p mimic and inhibitor and specific siRNAs that targeted circPTK2 or JAZF1 were designed and synthesized by GenePharma (Shanghai, China). Cell transfection was conducted using Lipofectamine RNAiMAX Transfection Reagent kit (Invitrogen; 13778100) or Lipofectamine 2000 Transfection Reagent (Invitrogen; 11668027) according to the manufacturer's instruction.
2.8. AAV vector production
An AAV Helper-Free System (GeneChem, Shanghai, China) was utilized to transfect the targeted cells. In brief, the AAV Helper-Free System is constituted by three vectors. The viral vector contains a promoter (FABP4) and other elements required for gene expression, along with AAV inverted terminal repeats (ITRs). The pAAV-RC vector contains rep and cap genes encoding the replication protein and viral capsid protein of AAV. The pHelper vector contains a collection of adenovirus genes (VA, E2A, and E4) which are necessary to produce high-titer AAV. AAV concentration and purification were performed to increase the titer and avoid side effects on animals. Vector genome (vg) titers were obtained by qPCR using primers designed to selectively bind AAV9 ITRs. (Forward, 5′-ACGAGCTGTACAAGGCTAGCTAACTAGATTGGCCGCGTGGGGTGGC-3’. reverse, 5′-GTTCCTGCGGCCGCAGATCTGAACAAACGACCCAACACCCGTG-3′).
2.9. Oil Red O staining
Mature adipocytes were fixed with 4% formaldehyde for 30 min, then washed twice with PBS. They were then stained with 0.3% Oil Red O (ORO) solution and washed three times with distilled water. Images were captured under a light microscope.
2.10. Free fatty acid assay
FFA concentration in conditioned media were measured using a non-esterified free fatty acids assay kit (Jiancheng Bioengineering Institute, Nanjing, China). The procedure was conducted according to the manufacturer's instructions. Each experiment was repeated three times.
2.11. Luciferase reporter assay
Full length circPTK2, JAZF1–3′ UTR, and their corresponding mutant versions with mutant miR-182-5p binding sites were synthesized and cloned into the luciferase reporter vector psiCHECK2. All of these plasmids were validated by sequencing. The 293T cells were seeded into 24-well plates and co-transfected with corresponding plasmids and miRNA mimics using Lipofectamine 3000. After 48 h incubation, the cells were lysed and the relative luciferase activity was exanimated using a Dual Luciferase Assay Kit (Promega, WI, USA) in accordance with the manufacturer's protocol.
2.12. Fluorescence in situ hybridization (FISH)
FISH assays were performed to observe the locations of circPTK2 and miR-182-5p in mature adipocytes. Briefly, after prehybridization at 55 °C for 2 h, cell climbing pieces were hybridized with a specific Cy3-labeled circPTK2 probe (Cy3–5′-TGTCATATTATTGCAGCCTTTG-3′Cy3) and FITC-labeled miR-182-5p probe (FITC-5′AAACCGTTACCATCTTGAGTGTGGC-3′-FITC) (Geneseed, Guangzhou, China) at 37 °C overnight. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Slides were photographed with a confocal microscope (Leica, Wetzlar, Germany).
2.13. RNA immunoprecipitation (RIP)
RIP was conducted with a Magna RIP kit (Millipore, Billerica, MA, USA) following the manufacturer's instructions. Mature adipocytes were harvested and lysed in complete RIP lysis buffer. Then, cell lysates were incubated with magnetic beads conjugated with anti-Argonaute2 (AGO2, Abcam, ab57113) or a negative control IgG antibody (Millipore) on a rotator overnight at 4 °C. The beads were washed using washing buffer. Immunoprecipitated RNA and protein were then purified and enriched to detect circPTK2, miR-182-5p, and AGO2 by qRT-PCR or Western blotting.
2.14. Western blot
An equal amount of protein lysates was separated by 8–12% SDS-PAGE and transferred onto a PVDF membrane. After blocking, the membranes were incubated with primary antibody (1:1000 dilution) overnight at 4 °C, followed by an incubation with secondary antibody (1:5000 dilution) for 1 h at room temperature. Western blot analyses were performed with commercially available antibodies, ATGL (Abcam, ab109251), HSL (Cell signaling technology, #4107), AdipoQ (Cell signaling technology, #2789), CEBPα (Cell signaling technology, #2295), FABP4 (Cell signaling technology, #2120), and JAZF1 (Santa Cruz, sc-376503). All experiments were repeated at least three times.
2.15. Immunohistochemistry
The sections were deparaffinized, rehydrated, and incubated in 3% hydrogen peroxide at room temperature for 10 min. Antigens were retrieved in 0.01 mol/L citric buffer (pH 6.0) at 95–98 °C for 25 min. Slides were cooled down for 1 h before blocking with a preferred blocking solution for 30 min at room temperature. Staining with anti-JAZF1 was performed at 4 °C overnight.
2.16. Immunofluorescence
Adipose tissue sections were cut with a Leica CM3050 cryostat (Leica Biosystems, Buffalo Grove, IL, USA). Slides were then rinsed three times with PBS. Tissue sections on the slides were covered with 0.5% Triton X-100 and 1% BSA, each for 1 h. Anti-JAZF1 were diluted and incubated with the sections at 4 °C overnight. Slides were rinsed with PBS and incubated with secondary antibody at room temperature. Fluorescence images were taken by a Zeiss LSM 710 Spectral Confocal Laser Scanning Microscope.
2.17. In vivo experiments
Six-week-old male BALB/c mice were purchased from Shanghai SLAC Laboratory Animal Corp and maintained under SPF conditions in a controlled environment of 20–22 °C with a 12/12 h light/dark cycle. All the mice were randomly assigned to the AAV-circPTK2 group, AAV-NC group, CAC group, or NC group. Mice in the AAV-circPTK2 group were injected intravenously through the tail vein with 3 × 1011 vg/mouse AAV. Cachexia was induced in the CAC group by subcutaneous injection of 2 × 106 colon-26 adenocarcinoma (C26) cells into the right flanks of the mice. The littermate control mice received vehicles (AAV-NC or PBS) only. At 3 weeks post-injection, mice were injected with chloral hydrate (0.1 mL of 5% solution per 10g body weight) via intraperitoneal route. Subcutaneous adipose tissues were harvested and stored in liquid nitrogen and fixed in 4% paraformaldehyde. For this study, n = 6 mice were injected for each group. All animal studies were performed in accordance with the guidelines provided by the Shanghai Medical Experimental Animal Care Commission.
2.18. Statistical analysis
All statistical analyses were performed with SPSS 25.0 software. Continuous variables that obey a normal distribution were analyzed by Student's t-tests to compare the differences. Differences between groups were compared using analysis of variance (ANOVA). Correlation analysis was performed using Spearman's correlation coefficient method. Receiver operating characteristic (ROC) curve analysis was performed to estimate the diagnostic sensitivity and specificity. Unless otherwise specified, the results are presented as the mean ± standard deviation (SD). All statistical tests were two-sided, and p < 0.05 was considered statistically significant. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ns stands for not significant.
3. Results
3.1. CircPTK2 is upregulated in adipose tissues of cachectic patients
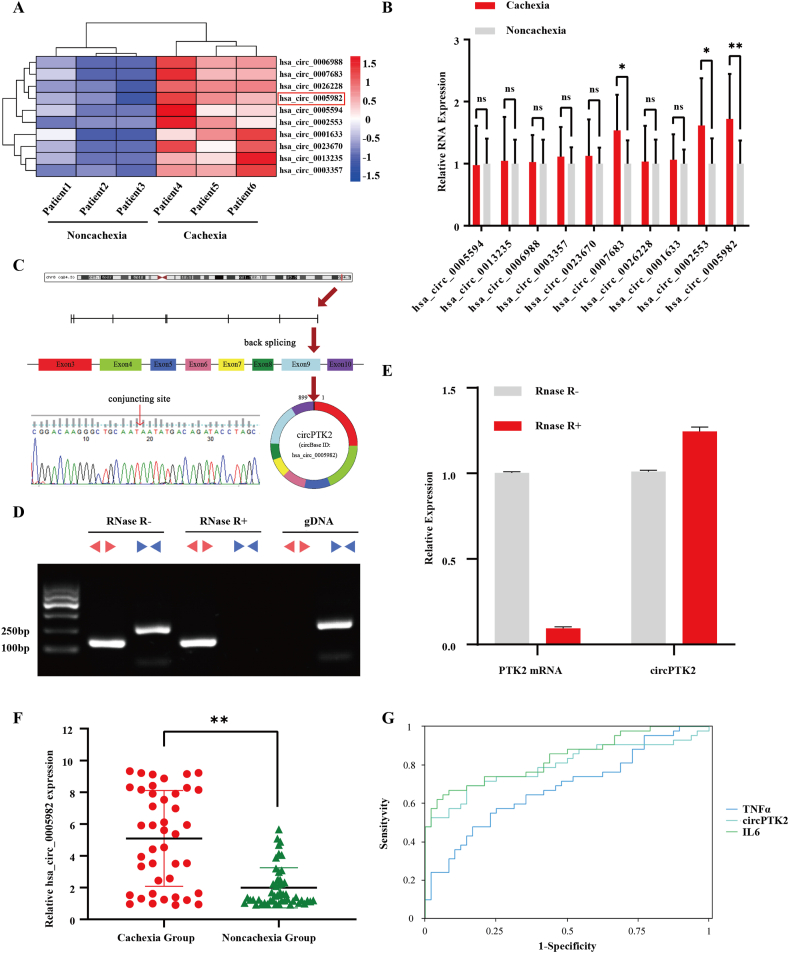
To investigate potential adipose-related circRNAs in CAC, we performed whole transcriptome RNA sequencing of subcutaneous white adipose tissue (WAT) from patients with and without CAC. Differentially expressed circRNAs were found between the two groups. Among 66 differentially expressed circRNAs, 29 circRNAs were upregulated, whereas 37 circRNAs were downregulated in the cachexia group (Fig. S1A, fold changes ≥2 and p < 0.05). We then filtered the 10 most upregulated circRNAs and verified them by qPCR in 10 pairs of adipose tissues from patients with and without cachexia (Figure 1A,B). Hsa_circ_0005982 (chr8: 141828375–141900868) was one of the most upregulated circRNAs. Using the human reference genome (GRCh37/hg19), we identified that hsa_circ_0005982 is derived from exons of the PTK2 gene, which is located on chromosome 8q24.3. Sanger sequencing of the amplified PCR products confirmed the back-splicing sites of hsa_circ_0005982 (Figure 1C). To evaluate its resistance to RNase R digestion, total RNA was treated with RNase R, and this treatment's efficacy was illustrated using linear isoform levels. In addition, convergent and divergent primers were designed to amplify hsa_circ_0005982 using cDNA and genomic DNA (gDNA) from adipocytes. The results showed that hsa_circ_0005982 could be detected by the divergent primer in cDNA but not gDNA, thus excluding the possibility that the back-splicing site was generated by trans-splicing or genomic rearrangement (Figure 1D). Furthermore, hsa_circ_0005982 was more resistant to RNase R than linear PTK2 mRNA, demonstrating the circular structure of circPTK2 in adipocytes (Figure 1E). We then searched circBank (ID: hsa_circPTK2_024) and circAtlas (ID: hsa_PTK2_0017) and found that mmu_circ_0005773 is the homologous RNA of hsa_circ_0005982. To further verify the identity, we resorted to NCBI BLAST and proved that the sequence of mmu_circ_0005773 has a 100% query cover and a 96.05% identity compared with hsa_circ_0005982 (Figs. S1B and S1C). This convinced us that both circRNAs are homologous, so we used circPTK2 as their general term in the following articles.
Figure 1.
CircPTK2 is upregulated in patients with CAC and is correlated with cachexia-related clinical characteristics. (A) Heatmap of the top 10 differentially expressed circRNAs between patients with and without CAC. (B) The expression of the top 10 differentially expressed circRNAs verified in adipose tissues by qPCR. (C) Genomic loci of the circPTK2 gene. The back-splicing junction was validated by Sanger sequencing. (D) RT-PCR analysis of PTK2 mRNA and circPTK2 expression after RNase R treatment. Divergent primers detected circPTK2 from cDNA, but not from gDNA. (E) qRT-PCR was used to determine the abundance of circPTK2 and PTK2 mRNA after treatment with RNase R in adipose tissue. (F) Relative expression of circPTK2 in adipose tissues from patients with (n = 42) or without (n = 48) CAC. (G) The ROC curve including IL6, TNFα, and circPTK2 in distinguishing cachexia and normal patients.
We also explored the clinical significance of circPTK2 in adipose tissues from patients diagnosed with gastrointestinal neoplasm. A total of 90 patients were enrolled and their serum determinations were tested before surgery (Table 1). Among the 90 adipose tissue samples, the qRT-PCR assay showed that circPTK2 was significantly upregulated in 42 samples from cachectic patients (p < 0.05, Figure 1F). Correlation analysis was performed between circPTK2 expression and cachexia-related clinicopathological characteristics. The results of the Spearman's correlation analysis showed that the expression level of circPTK2 was negatively correlated with BMI, prealbumin, and apoE and positively correlated with weight loss, IL6, and free fatty acid (FFA) (Table 2). These results suggested that circPTK2 was closely related to the pathological process of cachexia in gastrointestinal neoplasms. ROC curve was constructed to assess whether circPTK2 expression could be used as a potential diagnostic marker for CAC. The analysis indicated that the AUC of circPTK2 was 0.796 (95% CI: 0.698–0.895, p < 0.001); the AUCs of IL6 and TNFα were 0.839 (95% CI: 0.756–0.922, p < 0.001) and 0.682 (95% CI: 0.571–0.793, p < 0.001) (Figure 1G). The specificity and sensitivity for CAC of circPTK2 were 0.854 and 0.690, respectively. A circPTK2 expression level of 3.198 was the best cut-off value. These results indicated a moderately predictive value of circPTK2 to distinguish individuals with cachexia from controls.
Table 1.
Clinical Characteristics of 90 Participants.
| Clinical characteristics | Cachexia (n=42) | Noncachexia (n=48) | T/x2 | p Value |
|---|---|---|---|---|
| Age | 65.40±7.53 | 61.77±11.41 | 1.755 | 0.056 |
| BMI | 20.76±1.74 | 24.14±1.92 | 8.692 | 0.049∗ |
| Weight Loss | 6.95±2.98 | 0.91±0.53 | 15.026 | <0.001∗ |
| Disease stage(III/IV) | 36/42 | 16/48 | 7.174 | <0.001∗ |
| IL6(mmol/L) | 9.59±5.58 | 4.32±1.60 | 6.245 | <0.001∗ |
| TNFa(mmol/L) | 13.89±6.36 | 10.10±3.86 | 3.460 | 0.001∗ |
| Alb (g/L) | 38.33±3.93 | 41.81±5.11 | 3.577 | 0.116 |
| PAb(mg/L) | 196.0±61.81 | 252.33±29.84 | 5.613 | <0.001∗ |
| FAA(mmol/L) | 0.57±0.15 | 0.33±0.10 | 8.761 | <0.012∗ |
| TC(mmol/L) | 4.22±1.07 | 4.31±0.81 | 0.925 | 0.275 |
| TG(mmol/L) | 1.10±0.42 | 1.54±0.84 | 3.076 | 0.015∗ |
| LDL(mmol/L) | 2.49±0.97 | 2.46±0.67 | 0.123 | 0.155 |
| HDL(mmol/L) | 1.16±0.36 | 1.17±0.30 | 0.229 | 0.212 |
| ApoA(g/L) | 1.13±0.23 | 1.28±0.21 | 3.144 | 0.218 |
| ApoB(g/L) | 0.79±0.26 | 0.78±0.18 | 0.043 | 0.058 |
| ApoE(mg/L) | 39.50±17.78 | 44.85±10.55 | 1.762 | 0.002∗ |
| HB(g/L) | 125.64±23.28 | 127.77±16.96 | 2.848 | 0.282 |
| WBC(10ˆ9/L) | 6.02±1.83 | 6.15±1.88 | 0.333 | 0.790 |
| PLT(10ˆ9/L) | 224.83±80.65 | 232.68±99.17 | 0.408 | 0.612 |
| circPTK2 | 5.09±3.01 | 1.98±1.26 | 6.525 | 0.001∗ |
BMI Body mass index, HB Hemoglobin, WBC White blood cell count, PLT Platelet, ALB Albumin, PAb Prealbumin, TC Total cholesterol, TG Tri-glyceride, LDL Low-density lipoprotein, HDL High-density lipoprotein, ApoA Apolipoprotein A, ApoB Apolipoprotein B, ApoE Apolipoprotein E, FFA Free fatty acid, IL-6 Interleukin 6, TNF-α Tumor Necrosis Factor-α.
Table 2.
Correlations Between circPTK2 Expression and Clinical Characteristics in 90 Patients.
| Clinical characteristics | hsa_circ_0005982 |
|
|---|---|---|
| r | P | |
| Age | 0.181 | 0.087 |
| BMI | -0.526∗ | 0.001∗ |
| WL | 0.504 | 0.001∗ |
| HB | -0.127 | 0.233 |
| WBC | 0.031 | 0.773 |
| PLT | -0.192 | 0.069 |
| Alb | -0.301 | 0.063 |
| PAb | -0.635∗ | <0.001∗ |
| TC | -0.240 | 0.098 |
| TG | -0.064 | 0.547 |
| LDL | -0.118 | 0.269 |
| HDL | -0.179 | 0.092 |
| FFA | 0.604∗ | <0.001∗ |
| ApoA | -0.305 | 0.204 |
| ApoB | -0.032 | 0.765 |
| ApoE | -0.655∗ | 0.012∗ |
| TNFα | 0.366 | 0.015 |
| IL6 | 0.712∗ | <0.001∗ |
BMI Body mass index, WL Weight loss, HB Hemoglobin, WBC White blood cell count, PLT Platelet, Alb Albumin, PAb Prealbumin, TC Total cholesterol, TG Tri-glyceride, LDL Low-density lipoprotein, HDL High-density lipoprotein, ApoA Apolipoprotein A, ApoB Apolipoprotein B, ApoE Apolipoprotein E, FFA Free fatty acid, IL-6 Interleukin 6, TNF-α Tumor Necrosis Factor-α.
3.2. CircPTK2 promotes lipolysis and suppresses adipogenesis in adipocytes
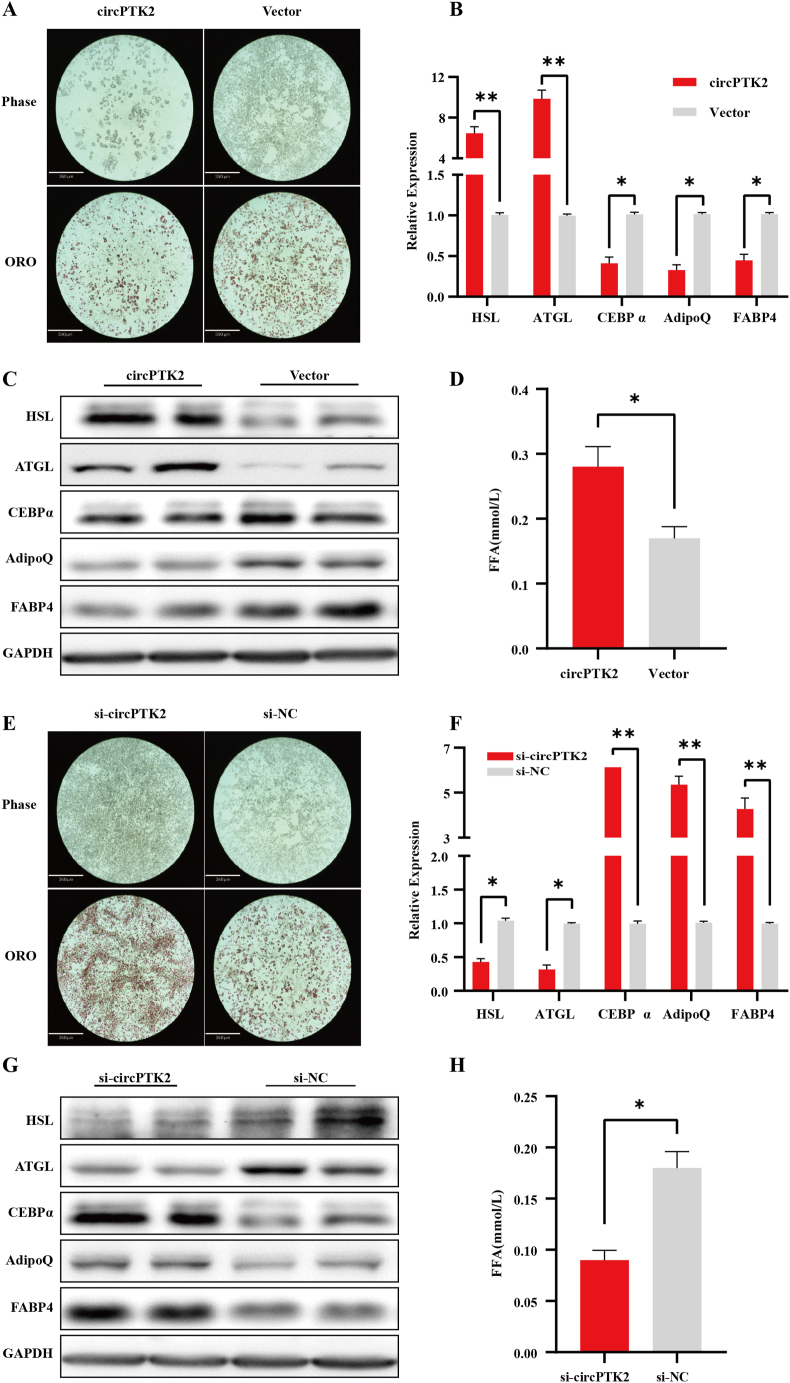
To investigate the role of circPTK2 in adipocyte metabolism, circPTK2 was upregulated by transfecting preadipocyte with circPTK2 overexpression vectors, followed by induction and differentiation. The qPCR results showed that circPTK2 expression had increased three-fold by differentiation day 6 in the circPTK2 overexpression group compared to the control group; PTK2 expression had no significant difference between the two groups (Fig. S2A). CircPTK2 overexpression promoted lipolysis and inhibited adipogenesis on day 6, as demonstrated by the decreased Oil Red O staining (Figure 2A). The expression of adipose-related markers, including adipose triglyceride lipase (ATGL), hormone-sensitive lipase (HSL), adiponectin (AdipoQ), fatty acid binding protein 4 (FABP4), and CCAAT/enhancer-binding protein alpha (CEBPα) was also analyzed by qPCR and western blotting (Figure 2B,C). In addition, FFA released in culture medium was increased by circPTK2 overexpression (Figure 2D), indicative of the involvement of the increased lipolysis. To further demonstrate whether circPTK2 was required for adipocyte metabolism, adipocytes were transfected with three independent siRNAs specifically targeting the back-splicing junction of circPTK2 but not changing the linear mRNA expression, followed by induction of differentiation. Approximately 70% knockdown was achieved by one of the siRNAs (Fig. S2B). CircPTK2 knockdown resulted in reduced lipolysis and enhanced adipogenesis according to the Oil Red O staining (Figure 2E). Lipolysis markers HSL and ATGL were also simultaneously downregulated with the knockdown, while the adipogenesis markers were upregulated (Figure 2F,G). FFA released in culture medium was decreased by circPTK2 knockdown (Figure 2H), which indicated the involvement of circPTK2 in lipolysis.
Figure 2.
CircPTK2 promotes lipolysis and suppresses adipogenesis in adipocytes. (A) Oil Red O staining of lipid accumulation in adipocytes (6d) without/with overexpression of circPTK2 (Scale bar: 360 μm). (B–C) qPCR and Western blot analysis of the expression of adipose-related markers in adipocytes (6d) without/with overexpression of circPTK2. (D) Concentration of FFA released in culture medium by adipocytes without/with overexpression of circPTK2. (E) Oil Red O staining of lipid accumulation in adipocytes (6d) without/with knockdown of circPTK2 (Scale bar: 360 μm). (F–G) qPCR and Western blot analysis of the expression of adipose-related markers in adipocytes (6d) without/with knockdown of circPTK2. (H) Concentration of FFA released in culture medium by adipocytes without/with knockdown of circPTK2.
3.3. CircPTK2 acts as a sponge for miR-182-5p
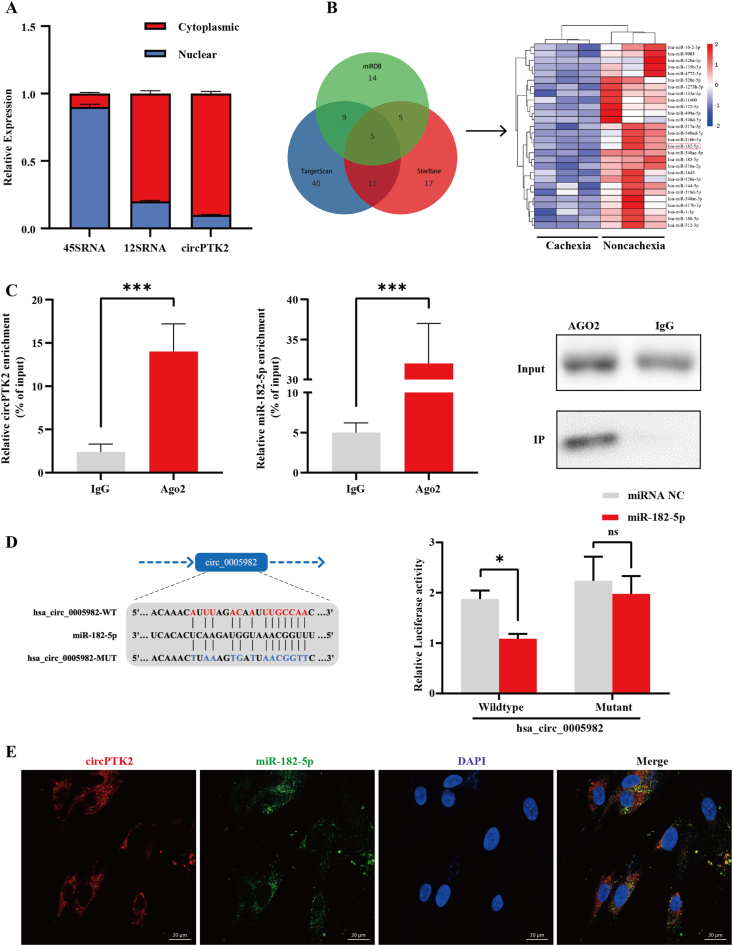
To observe the cellular distribution of circPTK2, we performed a qPCR assay for nuclear and cytoplasmic circPTK2 expression. The results from nuclear and cytoplasmic fractions indicated that circPTK2 was predominantly localized in the cytoplasm of adipocytes (Figure 3A). Studies have reported that circRNAs in cytoplasm can function as miRNA sponges to regulate gene expression [7,22]. To explore potential miRNAs that interact with circPTK2, we analyzed the results of the RNA sequencing mentioned above to evaluate the miRNA expression profiles. The results revealed 70 upregulated and 28 downregulated miRNAs in the CAC group (fold changes ≥ 2 and p < 0.05). Subjecting differentially expressed miRNAs to the bioinformatic algorithms miRDB, TargetScan, and StarBase to predict miRNAs revealed that miR-182-5p may contain putative targeting sites for the circPTK2 region (Figure 3B). Given that circPTK2 was enriched in the cytoplasm, AGO2 interacting with both circRNAs and miRNAs in the context of the ceRNA mechanism may be a prevalent phenomenon [23]. RNA immunoprecipitation (RIP) analysis revealed that circPTK2 and miR-182-5p were significantly enriched by the AGO2 antibody (Figure 3C, p < 0.001), validating the direct binding of circPTK2 with miR-182-5p. This result suggested that circPTK2 may possess miRNA-related functions. We then performed dual luciferase assays to further confirm the binding capability of miR-182-5p to circPTK2. A miR-182-5p mimic was co-transfected with wild- and mutant-type circPTK2 luciferase reporters into 293T cells, respectively. The results demonstrated that the luciferase activity was reduced by approximately 40% after co-transfection of the wild-type circPTK2 reporter plasmid (Luc-circ circPTK2-WT) with miR-182-5p mimic (Figure 3D). Moreover, the double FISH assay indicated circPTK2 and miR-182-5p colocalization in the cytoplasm of adipocytes, supporting the direct interaction of circPTK2 with miR-182-5p (Figure 3E). All of these findings have demonstrated that circPTK2 can bind directly to miR-182-5p.
Figure 3.
CircPTK2 acts as a sponge for miR-182-5p. (A) qPCR analyses of nuclear (Nuc) and cytoplasmic (Cyt) fractions from differentiated adipocytes. 12S rRNA served as positive controls in cytoplasm; 45S rRNA served as positive controls in nuclei. (B) Schematic illustration exhibiting the heatmap of 28 significantly downregulated miRNAs and the Venn diagram of circPTK2-targeted miRNAs, predicted by TargetScan, StarBase, and miRDB databases. (C) RNA immunoprecipitation (RIP) analysis of circPTK2 and miR-182-5p in adipocytes using antibodies against AGO2. Western blotting analysis of immunoprecipitated AGO2 protein is shown. (D) Schematic of the predicted miR-182-5p binding site on circPTK2. (E) Luciferase activity of wild-type or mutated circPTK2 in 293T cells after co-transfection with miR-182-5p or miRNA control. (F) Colocalization between miR-182-5p and circPTK2 was observed by RNA FISH in adipocytes. The nuclei were stained with DAPI (Scale bar: 20 μm).
3.4. JAZF1 is a downstream target of miR-134-5p and circPTK2 and is involved in lipolysis and adipogenesis
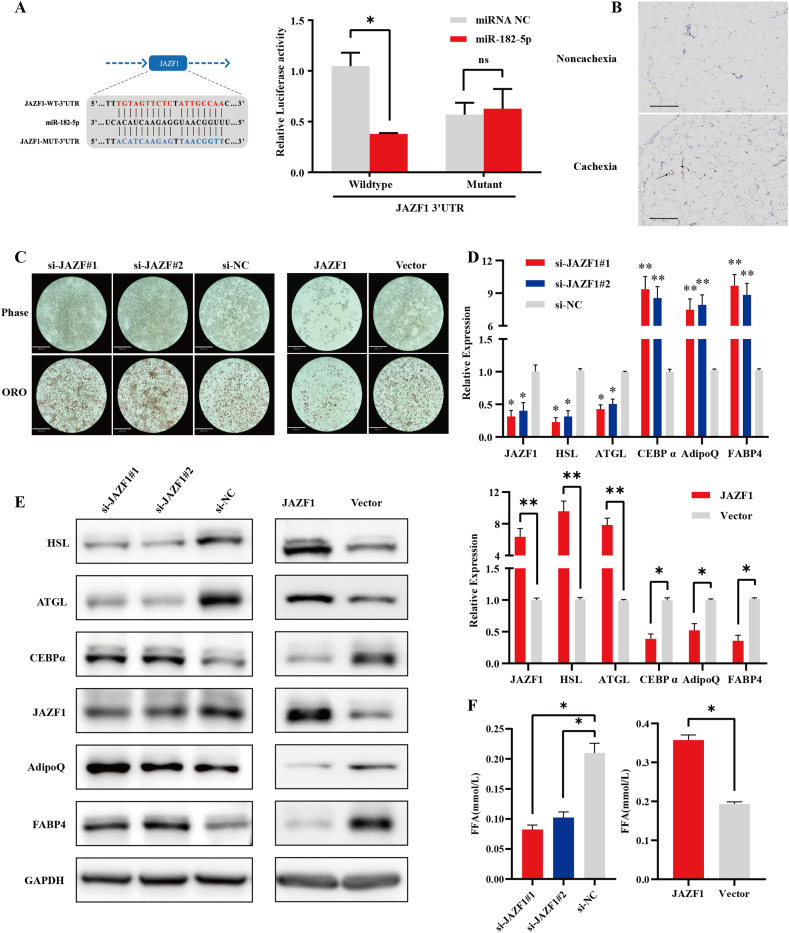
We employed the TargetScan algorithm to explore the downstream targets of circPTK2 and miR-182-5p. JAZF1 was predicted as a putative target gene of miR-182-5p. To confirm this, we constructed a luciferase reporter vector with the wild-type or mutated JAZF1 3′ UTR binding site for miR-182-5p. The luciferase activities of the JAZF1 3′ UTR wild-type reporter were significantly reduced in 293T cells transfected with miR-182-5p mimic (Figure 4A). However, no significant difference in luciferase activity was noted for scrambled control and miR-182-5p mimic when transfected with the mutated JAZF1 3′ UTR reporter. The qPCR and western blotting analysis revealed that miR-182-5p mimics significantly reduced JAZF1 mRNA and protein levels, while miR-182-5p inhibitor significantly increased JAZF1 mRNA and protein levels (Figs. S3A and S3B). Similarly, circPTK2 upregulation significantly increased JAZF1 expression, whereas circPTK2 downregulation significantly reduced JAZF1 expression (Figs. S3C and S3D). These data indicate that JAZF1 can be regulated by miR-182-5p and circPTK2.
Figure 4.
JAZF1 is a downstream target of miR-134-5p and circPTK2 and is involved in lipolysis and adipogenesis. (A) Schematic of the predicted miR-182-5p binding site in the JAZF1 3′ UTR. Luciferase activity of wild-type or mutated JAZF1 3′ UTR in 293T cells after co-transfection with miR-182-5p or miRNA control. (B) Representative IHC staining of JAZF1 expression in adipose tissues from patients without/with CAC (Scale bar: 150 μm). (C) Oil Red O staining of lipid accumulation in adipocytes (6d) with knockdown/overexpression of JAZF1 (Scale bar: 360 μm). (D) qPCR results of the expression of adipose-related markers in adipocytes (6d) with knockdown/overexpression of JAZF1. (E) Western blot analyses of the expression of adipose-related markers in adipocytes (6d) with knockdown/overexpression of JAZF1. (F) Concentration of FFA released in culture medium by adipocytes with knockdown/overexpression of JAZF1.
To identify the effects of JAZF1 on adipose phenotypes, IHC was performed in the adipose tissues of patients with and without cachexia. Through IHC analysis, we presented that the JAZF1 level was significantly upregulated in the adipose tissues of cachectic patients (Figure 4B). To investigate the specific function of JAZF1 in adipocytes, we constructed a JAZF1 overexpression vector, designed and synthesized siRNAs targeting JAZF1, and then transfected them into adipocytes. After 8 days of induction and differentiation, the cells were collected and subjected to Oil Red O Staining, qPCR, and Western blot. With Oil Red O staining, the amount of lipid droplets significantly increased when JAZF1 was downregulated. By contrast, the amount of lipid droplets significantly decreased when JAZF1 was overexpressed (Figure 4C). The qPCR results showed that the expression of JAZF1 and lipolysis markers were significantly suppressed after transfection with siRNAs; the adipogenesis markers revealed the opposite results. The expression of JAZF1 and lipolysis markers were significantly upregulated after JAZF1 overexpression vector transfection. Conversely, the adipogenesis markers were downregulated compared to the control group (Figure 4D). Western blotting analysis further confirmed these results (Figure 4E). The FFA content in the culture medium significantly decreased when JAZF1 was downregulated and increased when JAZF1 expression was overexpressed (Figure 4F).
3.5. CircPTK2 promotes lipolysis and suppresses adipogenesis by relieving the suppression effects of miR-182-5p on JAZF1
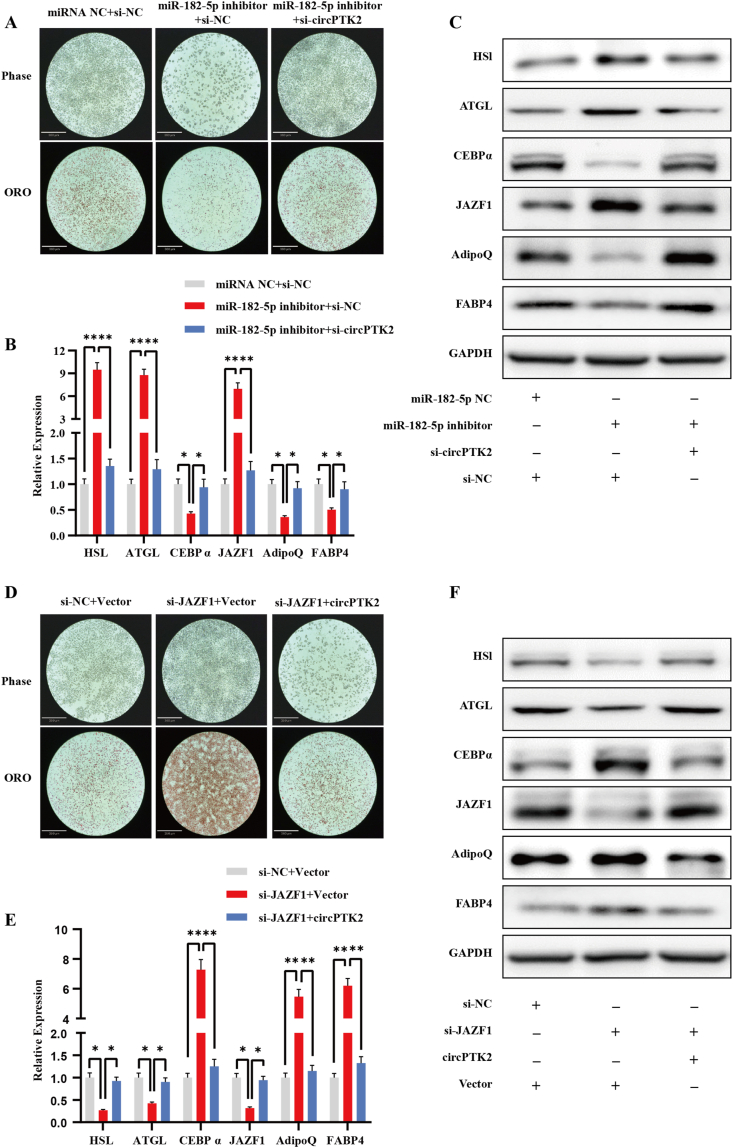
To confirm whether circPTK2 exerts its effect on adipocytes through miR-182-5p, Oil Red O staining, qPCR, and Western blotting were performed after miR-182-5p inhibitor transfection to knockdown miR-182-5p expression. We observed that miR-182-5p downregulation significantly promoted lipolysis and suppressed adipogenesis. In addition, when co-transfecting miR-182-5p inhibitor with circPTK2 siRNA, the promotion of lipolysis and suppression of adipogenesis from the miR-182-5p inhibitor were abrogated by circPTK2 siRNA (Figure 5A–C). Therefore, these findings suggest that circPTK2 regulates JAZF1 expression and regulates adipogenesis and lipolysis through miR-182-5p. We then investigated whether JAZF1 is essential for regulation of the circPTK2/miR-182-5p axis in adipocytes. The results showed that circPTK2 overexpression substantially rescued the reduced lipolysis and enhanced adipogenesis caused by JAZF1 depletion (Figure 5D–F). Taken together, these results reveal that circPTK2 served as a sponge for miR-182-5p to regulate JAZF1 expression and regulate adipogenesis and lipolysis by a ceRNA mechanism.
Figure 5.
CircPTK2 promotes lipolysis and suppresses adipogenesis by relieving the suppression effects of miR-182-5p on JAZF1. (A–C) Oil Red O staining, qRT-PCR, and Western blotting were performed in mature adipocytes treated with negative control, miR-182-5p inhibitor, and circPTK2 siRNA + miR-182-5p inhibitor (Scale bar: 360 μm). (D–F) Oil Red O staining, qPCR, and Western blotting were performed on mature adipocytes treated with negative control, JAZF siRNA, and circPTK2 overexpression vector + JAZF1 siRNA (Scale bar: 360 μm).
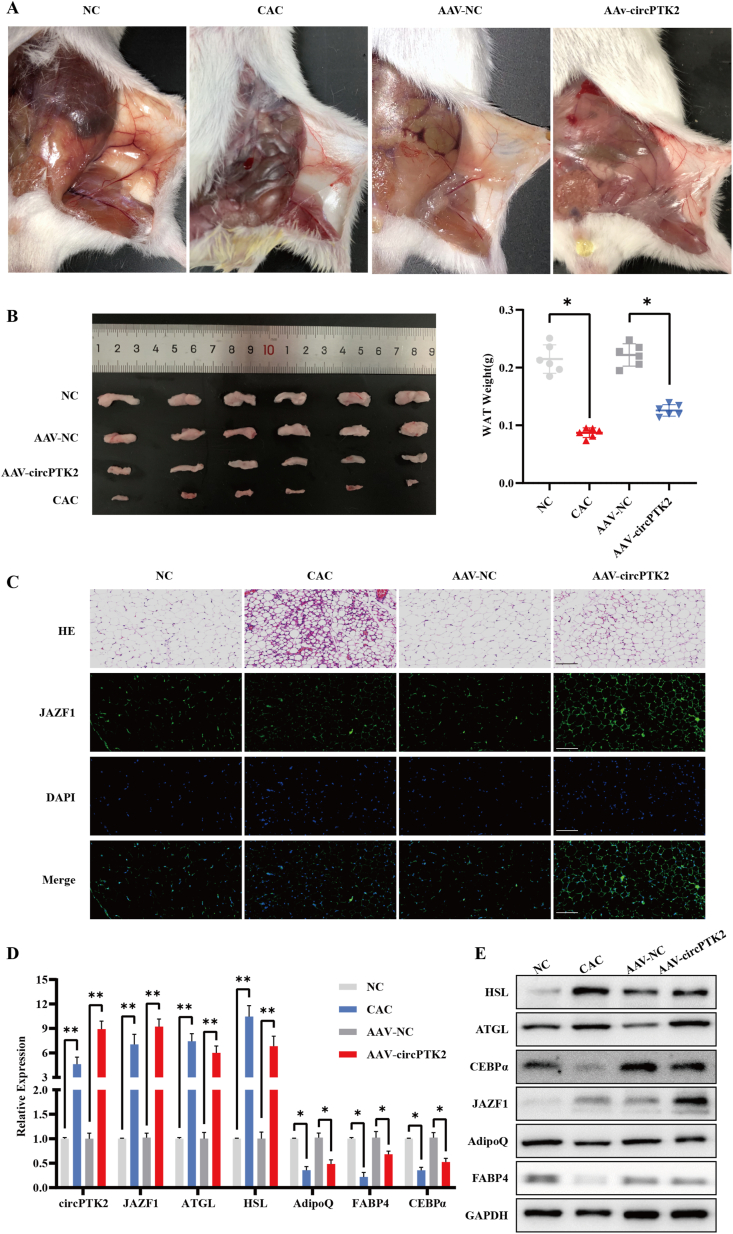
3.6. CircPTK2 promotes lipolysis and suppresses adipogenesis in vivo
To further investigate the effects of circPTK2 on adipose tissue in an animal model, a recombinant adeno-associated virus (rAAV) containing circPTK2 overexpression plasmid (AAV-circPTK2) was constructed and injected into the tail vein of BALB/c mice. The tissue-specific promoter was combined into the AAV to regulate circPTK2 expression in adipose tissues, specifically. C26 cells were injected into the right flanks of mice to construct a CAC mouse model in another group. Mice treated with C26 cells or AAV-circPTK2 revealed thinner subcutaneous WAT than those injected with PBS or AAV-negative control (Figure 6A). After dissection, the harvested subcutaneous WAT was observed and measured between the groups (Figure 6B). Immunofluorescent (IF) staining confirmed the increasing expression of JAZF1 in mice injected with C26 cells and AAV-circPTK2 (Figure 6C). We then evaluated circPTK2 expression in adipose tissues by qPCR and found 9-fold higher expression in AAV-circPTK2–injected mice and 5-fold higher expression in CAC mice, compared to the control groups. In addition, qPCR results showed an upregulation in HSL, ATGL, and JAZF1 and downregulation in AdipoQ, CEBPα, and FABP4 (Figure 6E). Western blotting analysis confirmed these results (Figure 6F). Altogether, in vivo experiments demonstrated that overexpression of circPTK2 dramatically inhibits adipogenesis and enhances lipolysis.
Figure 6.
CircPTK2 promotes lipolysis and suppresses adipogenesis in vivo. (A) Representative images of subcutaneous adipose tissues in control mice, CAC mice, AAV-NC mice, and AAV-circPTK2 mice. (B) Comparison of harvested subcutaneous WAT among each group. (C) Representative immunofluorescent staining of JAZF1 in WAT from each group. DAPI shown in blue. (Scale bar: 120 μm). (D–E) The indicated RNA and protein levels in adipose tissues from different groups were analyzed via qPCR and Western blotting.
4. Discussion
In this study, we identified circPTK2 as a significantly upregulated circRNA in the subcutaneous adipose tissues of patients with CAC. As an innovative first step, we recurred to the whole transcriptome sequence and explored the function and mechanism of circRNA in adipose tissues from cachectic patients. Overexpression of circPTK2 promoted lipolysis and suppressed adipogenesis by competitively binding to miR-182-5p and then relieving the inhibitory effects on JAZF1. Our study revealed the vital roles and mechanisms of circPTK2 in lipolysis and adipogenesis and highlighted the diagnostic and therapeutic importance of circPTK2 in cachexia.
Previous studies have clarified the significance of circRNAs in adipose tissue, especially in regulating the proliferation and differentiation of adipocytes [24,25]; however, most of the research on circRNAs has focused on tumorigenesis and metastasis. Interestingly, we identified three circRNAs also termed circPTK2 (hsa_circ_0005273, hsa_circ_0008305, and hsa_circ_0003221), derived from the same pre-mRNA PTK2, which were found to regulate proliferation, migration, and metastasis of multiple cancer cells. Hsa_circ_0003221 was reported to be upregulated in tissues and blood samples from patients with bladder cancer and enhance the proliferation and migration of bladder cancer cells [26]. Hsa_circ_0008305 was proven to inhibit TGF-β-induced epithelial–mesenchymal transition and metastasis by controlling TIF1γ in non–small cell lung cancer [27]. Hsa_circ_0005273 was seen to promote epithelial–mesenchymal transition in vitro and in vivo by binding to vimentin in colorectal cancer [28]. We explored and proved the novel roles and functions of circPTK2 in adipose tissues from mice and patients with CAC. It is worth noting that these circRNAs possess distinctive functions, as they have different mechanisms depending on the cellular context or downstream target molecules.
It has been identified that circRNA participates in multiple regulatory mechanisms, such as ceRNAs, protein interaction, gene transcription, and translational regulation [29]. CircRNAs contain miRNA target-binding sites and may weaken the downregulation of target genes mediated by miRNA, widely known as the “miRNA sponge” effect [22]. In the current study, we used databases (miRDB, TargetScan, and StarBase) and RNA-seq to predict miRNAs that were potentially bound by circPTK2. The relationship between circPTK2 and miR-182-5p was further verified by luciferase reporter assay and RIP assay. Luciferase reporter assay results revealed that miR-182-5p did bind to circPTK2. Additionally, RIP assay indicated that miR-182-5p interacted with circPTK2. Then, we used FISH analysis to confirm that circPTK2 and miR-182-5p were predominantly distributed in the cytoplasm. Taken together, these results suggest that circPTK2 acted as a sponge to miR-182-5p. MiRNA is an important post-transcriptional regulator, resulting in a decrease in mRNA expression through direct base pairing with mRNA 3′-UTR target sites [30]. MiR-182-5p has been proven to regulate the key process in multiple cancers, such as breast cancer, bladder cancer, and colon cancer [[31], [32], [33], [34]], but our study is the first to demonstrate its acting as a functional target in the ceRNA network in adipocytes and cachexia.
Combining the results of miRNA target prediction with RNA-seq, we deduced that JAZF1 may be the potential target in this axis. All of the results pointed to the fact that circPTK2 serves as a ceRNA to contribute to cachexia progression through the miR-182-5p/JAZF1 axis. JAZF1, known as a regulatory gene in T2DM and prostate cancer, has previously proven to be a potential target for regulating lipid metabolism. Overexpression of JAZF1 in adipocytes and hepatocytes could reduce lipid synthesis and increase lipolysis [15,19]. However, the relationship between JAZF1 and non-coding RNAs has rarely been explored. It has been reported that miR-31-5p inhibits the proliferation and DNA synthesis of human spermatogonia stem cells and facilitates apoptosis of human spermatogonia stem cells by targeting JAZF1 [35]. In the current study, we showed that circPTK2 serves as a miR-182-5p sponge to decrease JAZF1 inhibition, resulting in activation of lipolysis and suppression of adipogenesis. To further demonstrate the function of circPTK2 in vivo, an AAV-containing circPTK2 overexpression plasmid was constructed and injected into the tail vein of BALB/c mice. One study has provided the first proof-of-principle that circRNAs can be overexpressed using AAV vectors in vivo [36]. To avoid influencing irrelevant organs and tissues in mice, the AAV was combined with a tissue-specific promotor to target the adipose tissues in particular. It turned out that AAV-circPTK2–injected mice revealed loss of subcutaneous adipose tissues, similar to the mice injected with C26 cells. The qPCR and Western blot analysis of harvested adipose tissues further confirmed our theory that circPTK2 inhibits adipogenesis and enhances lipolysis in adipose tissues.
CAC is outlined into three distinct stages: pre-cachexia, cachexia, and refractory cachexia [1]. Refractory cachexia is the stage that is most frequently recognized, as it is usually accompanied by a late stage of tumor progression [37]. Body weight loss and symptoms related to CAC could be easily ignored by patients themselves in the early stages. Hence, to heighten the awareness of conditions potentially inducing cachexia, it is essential to speed up the identification of novel diagnostic markers to improve the diagnosis of cachexia. The onset of cachexia has been proven to occur before evident weight loss and muscle wasting; thus, it is critical to search for adipose-related biomarkers to facilitate early diagnosis and an appropriate therapy response [38]. It has been suggested that non-coding RNAs, with their conserved structure and stable expression in the cytoplasm, are ideal biomarkers [39]. MiR-483-5p, miR-23a, miR-744, and miR-99b were downregulated, whereas miR-378 was significantly upregulated in abdominal subcutaneous adipose tissue from cachectic patients with gastrointestinal cancers when compared with patients without cachexia [40]. Our previous research revealed that CAAlnc1 suppresses adipogenesis by interacting with an RNA-binding protein required for adipogenesis (HuR), leading to loss of adipose tissue [41]. CircRNAs have also been demonstrated as novel regulators in lipid metabolism in adipose tissue. CircSAMD4A was found to promote preadipocyte adipogenesis and differentiation through the miR-138-5p/EZH2 pathway in mice [42]. In the tumor microenvironment, adipose tissue secretes adiponectin, leptin, and chemokines to regulate tumor behavior. Additionally, secretions from cancer cells could cause dysfunction of lipid metabolism. Circ-DB, secreted from adipocytes, regulates tumor growth and DNA damage via the suppression of miR-34a and the activation of deubiquitylation-related USP7 [43]. In contrast, ciRS-133, derived from gastric cancer cells and transported by exosomes into preadipocytes, resulted in the browning of preadipocytes by sponging miR-133 to activate PRDM16 [44]. In this way, we discovered that high circPTK2 expression was associated with cachexia-related clinical outcomes, indicating the prognostic importance of circPTK2 in CAC. To further advance current research with clinical utilization, we plan to detect the expression of multiple non-coding RNAs in plasma and tissues from cachexia patients and construct a diagnostic model. This model would be a practical tool for clinicians to improve early recognition of CAC.
5. Conclusions
In summary, we analyzed the RNA-seq and detected a novel circRNA (has_circ_ 0005982, derived from PTK2) that was overexpressed in adipose tissues from cancer cachexia patients. A high level of circPTK2 was positively correlated with cachexia-related laboratory indicators. CircPTK2 could competitively sponge miR-182-5p to block the suppression effect of miR-182-5p on JAZF1 and contribute to the suppression of adipogenesis and promotion of lipolysis. Our study results not only elucidate the potential mechanism by which circRNA regulates lipid metabolism in adipocytes, but also suggest that the circPTK2 could be a potential diagnostic biomarker for CAC.
Author contributions
Z.D., J.H. and G.W. conceived and designed the study. Z.D., D.S., F.Y., S.S., and X.S. performed the experiments. Z.D., D.S., S.L., and J.H. conducted the statistical analysis. Z.D. wrote the paper; Z.D., J.H., and G.W. revised the paper. All authors reviewed and approved this manuscript.
Data availability
Microarray data are deposited at the GEO database with the accession number GSE174128 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE174128).
Acknowledgments
We thank Prof. Ying Feng from the CAS Key Laboratory of Nutrition, Metabolism and Food Safety, Shanghai Institute of Nutrition and Health, University of Chinese Academy of Sciences, Chinese Academy of Sciences. She provided experiment equipment and offered helpful advice during the experiments. We also thank Prof. Qiurong Ding from CAS Key Laboratory of Nutrition, Metabolism and Food Safety, Shanghai Institute of Nutrition and Health, Shanghai Institutes for Biological Sciences, University of Chinese Academy of Sciences. She provided the immortalized subcutaneous mouse white preadipocytes. This work was supported by the Youth of National Natural Science Foundation of China (81803091) and Shanghai Natural Science Foundation Project (19ZR1409100).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101310.
Contributor Information
Jun Han, Email: hanjun198626@163.com.
Guohao Wu, Email: profwugh@163.com, wu.guohao@zs.hospital.sh.cn.
Conflict of interest
The authors declare no competing interests.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Multimedia component 1
References
- 1.Fearon K., Strasser F., Anker S.D., Bosaeus I., Bruera E., Fainsinger R.L. Definition and classification of cancer cachexia: an international consensus. The Lancet Oncology. 2011;12:489–495. doi: 10.1016/S1470-2045(10)70218-7. [DOI] [PubMed] [Google Scholar]
- 2.Fearon K., Arends J., Baracos V. Understanding the mechanisms and treatment options in cancer cachexia. Nature Reviews Clinical Oncology. 2013;10:90–99. doi: 10.1038/nrclinonc.2012.209. [DOI] [PubMed] [Google Scholar]
- 3.Mannelli M., Gamberi T., Magherini F., Fiaschi T. The adipokines in cancer cachexia. International Journal of Molecular Sciences. 2020:21. doi: 10.3390/ijms21144860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daas S.I., Rizeq B.R., Nasrallah G.K. Adipose tissue dysfunction in cancer cachexia. Journal of Cellular Physiology. 2018;234:13–22. doi: 10.1002/jcp.26811. [DOI] [PubMed] [Google Scholar]
- 5.Das S.K., Hoefler G. The role of triglyceride lipases in cancer associated cachexia. Trends in Molecular Medicine. 2013;19:292–301. doi: 10.1016/j.molmed.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsoli M., Swarbrick M.M., Robertson G.R. Lipolytic and thermogenic depletion of adipose tissue in cancer cachexia. Seminars in Cell & Developmental Biology. 2016;54:68–81. doi: 10.1016/j.semcdb.2015.10.039. [DOI] [PubMed] [Google Scholar]
- 7.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 8.Rybak-Wolf A., Stottmeister C., Glažar P., Jens M., Pino N., Giusti S. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Molecular Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 9.Kristensen L.S., Hansen T.B., Venø M.T., Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang F., Fan M., Cai Y., Zhou X., Tai S., Yu Y. Circular RNA circRIMS1 acts as a sponge of miR-433-3p to promote bladder cancer progression by regulating CCAR1 expression. Molecular Therapy - Nucleic Acids. 2020;22:815–831. doi: 10.1016/j.omtn.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei C.Y., Zhu M.X., Lu N.H., Liu J.Q., Yang Y.W., Zhang Y. Circular RNA circ_0020710 drives tumor progression and immune evasion by regulating the miR-370-3p/CXCL12 axis in melanoma. Molecular Cancer. 2020;19:84. doi: 10.1186/s12943-020-01191-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakajima T., Fujino S., Nakanishi G., Kim Y.S., Jetten A.M. TIP27: a novel repressor of the nuclear orphan receptor TAK1/TR4. Nucleic Acids Research. 2004;32:4194–4204. doi: 10.1093/nar/gkh741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson J.A., Watson J.K., Nikolić M.Z., Rawlins E.L. Fank1 and Jazf1 promote multiciliated cell differentiation in the mouse airway epithelium. Biol Open. 2018;7 doi: 10.1242/bio.033944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meng F., Lin Y., Yang M., Li M., Yang G., Hao P. JAZF1 inhibits adipose tissue macrophages and adipose tissue inflammation in diet-induced diabetic mice. BioMed Research International. 2018;2018:4507659. doi: 10.1155/2018/4507659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ming G.F., Xiao D., Gong W.J., Liu H.X., Liu J., Zhou H.H. JAZF1 can regulate the expression of lipid metabolic genes and inhibit lipid accumulation in adipocytes. Biochemical and Biophysical Research Communications. 2014;445:673–680. doi: 10.1016/j.bbrc.2014.02.088. [DOI] [PubMed] [Google Scholar]
- 16.Yuan L., Luo X., Zeng M., Zhang Y., Yang M., Zhang L. Transcription factor TIP27 regulates glucose homeostasis and insulin sensitivity in a PI3-kinase/Akt-dependent manner in mice. International Journal of Obesity. 2015;39:949–958. doi: 10.1038/ijo.2015.5. [DOI] [PubMed] [Google Scholar]
- 17.Zhou M., Xu X., Wang H., Yang G., Yang M., Zhao X. Effect of central JAZF1 on glucose production is regulated by the PI3K-Akt-AMPK pathway. The FASEB Journal. 2020;34:7058–7074. doi: 10.1096/fj.201901836RR. [DOI] [PubMed] [Google Scholar]
- 18.Park S.J., Kwon W., Park S., Jeong J., Kim D., Jang S. Jazf1 acts as a regulator of insulin-producing β-cell differentiation in induced pluripotent stem cells and glucose homeostasis in mice. FEBS Journal. 2021;288:4412–4427. doi: 10.1111/febs.15751. [DOI] [PubMed] [Google Scholar]
- 19.Li L., Yang Y., Yang G., Lu C., Yang M., Liu H. The role of JAZF1 on lipid metabolism and related genes in vitro. Metabolism. 2011;60:523–530. doi: 10.1016/j.metabol.2010.04.021. [DOI] [PubMed] [Google Scholar]
- 20.Qiu Y., Sun Y., Xu D., Yang Y., Liu X., Wei Y. Screening of FDA-approved drugs identifies sutent as a modulator of UCP1 expression in brown adipose tissue. EBioMedicine. 2018;37:344–355. doi: 10.1016/j.ebiom.2018.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng Z., Wei Y., Zhang Y., Qiu Y., Liu X., Su L. Identification of a rhodanine derivative BML-260 as a potent stimulator of UCP1 expression. Theranostics. 2019;9:3501–3514. doi: 10.7150/thno.31951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nature Communications. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou W.Y., Cai Z.R., Liu J., Wang D.S., Ju H.Q., Xu R.H. Circular RNA: metabolism, functions and interactions with proteins. Molecular Cancer. 2020;19:172. doi: 10.1186/s12943-020-01286-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang R., Li H., Yang J., Shen X., Song C., Yang Z. circRNA profiling reveals an abundant circFUT10 that promotes adipocyte proliferation and inhibits adipocyte differentiation via sponging let-7. Molecular Therapy - Nucleic Acids. 2020;20:491–501. doi: 10.1016/j.omtn.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang Z., Zhang S., Jiang E., Wang X., Wang Z., Chen H. circFLT1 and lncCCPG1 sponges miR-93 to regulate the proliferation and differentiation of adipocytes by promoting lncSLC30A9 expression. Molecular Therapy - Nucleic Acids. 2020;22:484–499. doi: 10.1016/j.omtn.2020.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Z.Q., Yang M.G., Liu H.J., Su C.Q. Circular RNA hsa_circ_0003221 (circPTK2) promotes the proliferation and migration of bladder cancer cells. Journal of Cellular Biochemistry. 2018;119:3317–3325. doi: 10.1002/jcb.26492. [DOI] [PubMed] [Google Scholar]
- 27.Wang L., Tong X., Zhou Z., Wang S., Lei Z., Zhang T. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-β-induced epithelial-mesenchymal transition and metastasis by controlling TIF1γ in non-small cell lung cancer. Molecular Cancer. 2018;17:140. doi: 10.1186/s12943-018-0889-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang H., Li X., Meng Q., Sun H., Wu S., Hu W. CircPTK2 (hsa_circ_0005273) as a novel therapeutic target for metastatic colorectal cancer. Molecular Cancer. 2020;19:13. doi: 10.1186/s12943-020-1139-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kristensen L.S., Andersen M.S., Stagsted L.V.W., Ebbesen K.K., Hansen T.B., Kjems J. The biogenesis, biology and characterization of circular RNAs. Nature Reviews Genetics. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 30.Wu J., Qi X., Liu L., Hu X., Liu J., Yang J. Emerging epigenetic regulation of circular RNAs in human cancer. Molecular Therapy - Nucleic Acids. 2019;16:589–596. doi: 10.1016/j.omtn.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu S., Liu Y., Wang X., Wang J., Xi G. lncRNA SNHG10 promotes the proliferation and invasion of osteosarcoma via wnt/β-catenin signaling. Molecular Therapy - Nucleic Acids. 2020;22:957–970. doi: 10.1016/j.omtn.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu X., Chen H., Wu M., Peng S., Zhang L. Downregulation of miR-182-5p inhibits the proliferation and invasion of triple-negative breast cancer cells through regulating TLR4/NF-κB pathway activity by targeting FBXW7. Annals of Translational Medicine. 2020;8:995. doi: 10.21037/atm-20-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu C., Zhao K., Zhou N., Liu F., Xie F., Yu S. UBAC2 promotes bladder cancer proliferation through BCRC-3/miRNA-182-5p/p27 axis. Cell Death & Disease. 2020;11:733. doi: 10.1038/s41419-020-02935-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan S., Wang H., Chen X., Liang C., Shang W., Wang L. MiR-182-5p inhibits colon cancer tumorigenesis, angiogenesis, and lymphangiogenesis by directly downregulating VEGF-C. Cancer Letters. 2020;488:18–26. doi: 10.1016/j.canlet.2020.04.021. [DOI] [PubMed] [Google Scholar]
- 35.Fu H., Zhou F., Yuan Q., Zhang W., Qiu Q., Yu X. miRNA-31-5p mediates the proliferation and apoptosis of human spermatogonial stem cells via targeting JAZF1 and cyclin A2. Molecular Therapy - Nucleic Acids. 2019;14:90–100. doi: 10.1016/j.omtn.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meganck R.M., Borchardt E.K., Castellanos Rivera R.M., Scalabrino M.L., Wilusz J.E., Marzluff W.F. Tissue-Dependent expression and translation of circular RNAs with recombinant AAV vectors in vivo. Molecular Therapy - Nucleic Acids. 2018;13:89–98. doi: 10.1016/j.omtn.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruggeman A.R., Kamal A.H., LeBlanc T.W., Ma J.D., Baracos V.E., Roeland E.J. Cancer cachexia: beyond weight loss. J Oncol Pract. 2016;12:1163–1171. doi: 10.1200/JOP.2016.016832. [DOI] [PubMed] [Google Scholar]
- 38.Wyart E., Bindels L.B., Mina E., Menga A., Stanga S., Porporato P.E. Cachexia, a systemic disease beyond muscle atrophy. International Journal of Molecular Sciences. 2020:21. doi: 10.3390/ijms21228592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santos J.M.O., Peixoto da Silva S., Gil da Costa R.M., Medeiros R. The emerging role of MicroRNAs and other non-coding RNAs in cancer cachexia. Cancers. 2020;12 doi: 10.3390/cancers12041004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kulyté A., Lorente-Cebrián S., Gao H., Mejhert N., Agustsson T., Arner P. MicroRNA profiling links miR-378 to enhanced adipocyte lipolysis in human cancer cachexia. American Journal of Physiology. Endocrinology and Metabolism. 2014;306:E267–E274. doi: 10.1152/ajpendo.00249.2013. [DOI] [PubMed] [Google Scholar]
- 41.Shen L., Han J., Wang H., Meng Q., Chen L., Liu Y. Cachexia-related long noncoding RNA, CAAlnc1, suppresses adipogenesis by blocking the binding of HuR to adipogenic transcription factor mRNAs. International Journal of Cancer. 2019;145:1809–1821. doi: 10.1002/ijc.32236. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y., Liu H., Li Y., Mao R., Yang H., Zhang Y. Circular RNA SAMD4A controls adipogenesis in obesity through the miR-138-5p/EZH2 axis. Theranostics. 2020;10:4705–4719. doi: 10.7150/thno.42417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang H., Deng T., Ge S., Liu Y., Bai M., Zhu K. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2019;38:2844–2859. doi: 10.1038/s41388-018-0619-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Zhang H., Zhu L., Bai M., Liu Y., Zhan Y., Deng T. Exosomal circRNA derived from gastric tumor promotes white adipose browning by targeting the miR-133/PRDM16 pathway. International Journal of Cancer. 2019;144:2501–2515. doi: 10.1002/ijc.31977. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multimedia component 1
Data Availability Statement
Microarray data are deposited at the GEO database with the accession number GSE174128 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE174128).