Summary
This protocol determines the fraction of a bacterial population that is viable and culturable, viable and non-culturable, or non-viable (dead). Each population is detected by isolating colonies on agar plates, performing direct counts, and staining for live or dead cells. Its application is limited to bacteria that are stainable and when permissible growth conditions are known. The quantitative data extracted allow for the detection of a viable but non-culturable (alive and non-dividing) population from a liquid culture.
For complete details on the use and execution of this protocol, please refer to Stott et al. (2015).
Subject areas: Cell Biology, Cell-based Assays, Microbiology, Microscopy
Graphical abstract

Highlights
-
•
Liquid cultures are spread onto agar plates to quantify CFU/mL
-
•
Direct counting is performed to determine the total number of bacteria per mL
-
•
Fluorescence microscopy is used to distinguish between live and dead bacteria
-
•
In sum, the protocol can reveal VBNC populations in a culture
This protocol determines the fraction of a bacterial population that is viable and culturable, viable and non-culturable, or non-viable (dead). Each population is detected by isolating colonies on agar plates, performing direct counts, and staining for live or dead cells. Its application is limited to bacteria that are stainable and when permissible growth conditions are known. The quantitative data extracted allow for the detection of a viable but non-culturable (alive and non-dividing) population from a liquid culture.
Before you begin
This assay was previously applied to the aquatic Gram-negative bacterium Caulobacter crescentus (Stott et al., 2015). The techniques outlined below are compatible with the Gram-negative bacterium Escherichia coli and the Gram-positive bacterium Bacillus subtilis. It is effective for bacteria that can be readily grown in lab and fluoresce with differential stains for cells that are alive or dead. This assay may not be effective for dense cluster-forming, or biofilm-forming, bacteria where it can be difficult to identify single cells or for the differential stains to penetrate.
Before initiating the assay, the experimentalist will prepare a 5× fixative and 2× differential stains. In addition, a liquid culture will be grown up to 1) optimize stain fluorescence and 2) identify an appropriate dilution for “Isolating Colonies on a Plate.”
On the day of the assay, exponential-phase cultures are diluted to a 0.100 working optical density. Thereafter, fresh agarose pads are made just prior to starting the assay. The assay is initiated by preserving an aliquot of culture with 5× fixative for direct counts to be performed at the end, plating for colony forming units at an appropriate dilution (isolating colonies on a plate) and using stains for distinguishing between live and dead cells. Finally, an aliquot of fixative-preserved culture is transferred to a Petroff-Hauser counting chamber for direct counts.
Preparation of fixative
Timing: 1 h
A concentrated 5× fixative (a solution of 150 mM sodium phosphate buffer, pH 7.5, containing 2.0% paraformaldehyde) is prepared prior to beginning the assay. Fixatives prevent autolysis and degradation of cell morphology (Howat and Wilson, 2014). The use of fixative reduces the number of experimentalists needed to conduct the assay by allowing downstream bacterial counts to be performed at a later time. Prepare 5× fixative as follows:
-
1.
Add 1.25 mL of 16% paraformaldehyde to 3 mL of 500 mM sodium phosphate dibasic anhydrous (Na2HPO4) buffer in a sterile 15 mL conical centrifuge tube.
CRITICAL: Paraformaldehyde is a potential carcinogen. Wear personal protective equipment and work under a chemical hood.
-
2.
Raise the volume to 8 mL by adding 3.75 mL of sterile deionized water.
-
3.
Use a dropper to adjust the pH to 7.5 by adding 1 M hydrochloric acid.
-
4.
Raise the volume to 10 mL with sterile deionized water and aliquot 1–1.5 mL volumes into sterile microcentrifuge tubes.
-
5.
Use on the same day or store at −20°C for up to a year.
Note: If fixative is frozen, prior to beginning the assay, thaw it using a 50°C heating block or water bath and finger vortex (hold in one hand and flick at the base with the other hand).
Alternatives: A longer preserving fixative may be available for the model organism to be studied (Chao and Zhang, 2011).
Preparation of 1% agarose pads
Timing: 30 min
-
6.
Clean microscope slides with glass wipes using 80% ethanol to remove debris.
-
7.
Add 0.3 g of molecular-biology grade agarose into 30 mL of deionized water in a 125 mL Erlenmeyer flask.
-
8.
Microwave to dissolve agarose and air cool so that it is warm-to-hot when touched.
-
9.
Pipette 150 μL of liquid agarose, with steady control, onto a pre-cleaned microscope slide.
-
10.
Allow a moment for air cooling, then place a 22 X 22 mm coverslip parallel to the slide onto the agarose. This prevents an unleveled, and out-of-focus, plane of viewing.
-
11.
Store unused agarose solution at 4°C for a up to one month and re-microwave to liquefy agarose for future experimentation.
Note: Agarose pads left at room temperature for a prolonged period of time will dry out.
Preparation of stains to detect populations of live and dead cells
Timing: 30+ min
-
12.
Prepare a 0.85% NaCl (aq) solution using sterile deionized water.
-
13.Prepare Molecular Probes’ BacLight Bacterial stains, or Biotium’s Viability/Cytotoxicity Assay for Bacteria Live & Dead Cells, as follows:
-
a.For the Molecular Probes’ BacLight Bacterial stains: Pipette 1 mL of sterile deionized water into a sterile petri dish. Then take one manufacturer-provided plastic pipette containing either propidium iodide, or SYTO 9, powder and repeatedly draw in sterile deionized water to fully suspend the powder in solution. Then transfer all of the solution to a 15 mL conical centrifuge tube and raise the volume to 2.5 mL using sterile deionized water. Repeat this protocol with second powdered stain. Once both powders have been suspended, add 500 μL of each component to a black sterile microcentrifuge tube for a working 2× stock solution (Molecular Probes, 2004).
-
b.For the Biotium Viability/Cytotoxicity Assay: in a sterile microcentrifuge tube, combine 10 μL of DMAO and 20 μL volumes of EthD-III, mix thoroughly, then add 80 μL of 0.85% NaCl solution for a 100× dye solution of 110 μL (Biotium, 2020). Transfer 10 μL of 100× dye solution into a new microcentrifuge tube and add 490 μL of 0.85% NaCl (aq) solution for a 2× dye solution.
-
c.All solutions may be stored at −20°C, away from light, for up to 1 year.
-
a.
CRITICAL: Propidium iodide and SYTO 9 (Molecular Probes) and DMAO and EthD-III (Biotium) stains are known to bind nucleic acids. Wear personal protective equipment as they are potential mutagens (Biotium, 2020; Molecular Probes, 2004).
The Molecular Probes BacLight stains are compatible across numerous bacterial species such as Agrobacterium tumefaciens, Clostridium perfringens, Klebsiella pneumoniae, Pseudomonas aeruginosa, Salmonella oranienburg, Staphylococcus aureus and Streptococcus pyogenes (Hu et al., 2017; Lindbäck et al., 2010; Park and Kim, 2018; Robertson et al., 2019; Molecular Probes, 2004). The Biotium stains have been used with E. coli, Leptothrix, Lactobacillus fermentum, Lactobacillus reuteri, Pleurotus osreatus, and Salmonella enterica, (Adamski and Pietr, 2019; Bhave et al., 2015; Duplantis et al., 2015; Furutani et al., 2011; Hernandez et al., 2012; Kim et al., 2016; Rodes et al., 2013; Biotium, 2020).
Optimizing stain fluorescence for live and dead cells
Timing: 2–4 h
-
14.
Grow up 3 mL of culture to exponential phase.
-
15.
Dilute the culture down to an OD of 0.100.
-
16.
Aliquot 1 mL into a microcentrifuge tube.
-
17.
Centrifugate at 2,900 × g for 2 min.
CRITICAL: High speeds of centrifugation can presumably damage the cell membrane, resulting in a lower percentage of live cells.
-
18.
Carefully remove the supernatant and resuspend the pellet in 100 μL of 0.85% NaCl (aq) solution.
-
19.
Split this salt-suspended culture into two microcentrifuge tubes (50 μL each); one for experimental testing and the other for a control.
-
20.Set up the following in a separate microcentrifuge tube for the experimental culture:
-
a.Add 5 μL of salt-suspended culture.
-
b.Add 5 μL of the 2× stock fluorescent stains and mix by pipetting.
-
c.Allow for a 15-min incubation period (a 15-min incubation period is recommended by manufacturer, but we found this step unnecessary for Bacillus subtilis, Escherichia coli and Caulobacter crescentus). For more information, please refer to Supplemental File 1.
-
d.Take an agarose pad and remove the cover slip, then pipette 1 μL of this mix and allow drop to air-dry for immobilization. Then, replace cover slip.
-
a.
CRITICAL: Prepare only a single slide of samples at a time (two samples fit on each slide) as cells lose viability if left on an agarose pad for an extended period of time.
Note: Fluorescent cells will begin to lose fluorescence after 5 min under a cover slip.
Note: 1- to 2-h old agarose pads reduce air-drying time down to under a min. Fresher agarose pads may take up to 5 min for samples to air-dry.
-
21.Visualize cells at 1000× magnification via differential interference contrast (for DIC), or phase contrast, with fluorescence microscopy.
-
a.DIC images herein are acquired with a 120W metal halide lamp (X-Cite 120) light source at 100% light intensity (adjusted to 50% on a Zeiss Axio Imager.M1) and Hamamatsu ORCA-ER-1394 digital camera (set to auto-exposure).
-
b.Cells viewed under fluorescence microscopy are imaged with Cy3, or TexasRed, and FITC filters.
-
c.The excitation/emission spectra for the stains bound to DNA are 480/500 nm (FITC filter) for the SYTO9 stain, 490/635 nm (Cy3 or TexasRed filter) for propidium iodide (Molecular Probes, 2004), 503/530 nm (FITC filter) for the DMAO stain, and 532/625 nm (Cy3 or TexasRed filter) for EthD-III (Biotium, 2020).
-
a.
-
22.Prepare a negative control:
-
a.Add 200 μL of 70% isopropanol to the control microcentrifuge tube containing 50 μL of salt-suspended culture from step 19.
-
b.Centrifugate at 2,900 × g for 2 min.
-
c.Quickly remove all alcohol by pipetting and resuspend the pellet with 200 μL of 0.85% NaCl (aq) solution.Note: In some cases, spun-down cultures will adhere to the sidewall of the microcentrifuge tube. Pipette the salt solution repeatedly onto the side wall to suspend the cells back into solution.
-
d.Transfer 5 μL of salt-suspended culture to a new microcentrifuge tube.
-
e.Proceed from steps 20b-d and image cells (step 21) via differential interference contrast, or phase contrast, and fluorescence microscopy using Cy3 or Texas Red and FITC filters (as a reference, properly stained bacteria are displayed in Figure 1). Troubleshooting 1
-
a.
CRITICAL: Exposure time and power intensity will need to be optimized to produce fluorescence well below saturation while displaying a defined signal-to-background fluorescence intensity.
Note: An acceptable negative control will present red fluorescent cells under a Cy3 or TexasRed filter. No fluorescent cells should be detected under a FITC filter.
Figure 1.
Gram-positive and Gram-negative model organisms are stainable with the Biotium and Molecular Probes BacLight stains
Both set of stains produce a GFP signal in live Caulobacter crescentus NA1000 (Gram-negative; PYE broth, 28°C), Escherichia coli MG1655 (Gram-negative; LB broth, 37°C) and Bacillus subtilis PY79 (Gram-positive; LB broth, 37°C) whereas isopropanol-treated (dead) cells produce a strong Texas Red signal with minimal-to-no GFP fluorescence. (A) Biotium stains (B) Molecular Probes BacLight stains. Scale bars: 5 μM.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Bacillus subtilis PY79 | Gift from Lucy Shapiro | Wild type |
| Caulobacter crescentus NA1000 (CB15N) | Gift from Lucy Shapiro | Wild type |
| Caulobacter crescentus SM1309 (NA1000 PvanA::fabH ΔfabH ΔspoT) | Stott et al. 2015 | Mutant |
| Escherichia coli MG1655 (F- λ-rph-1) | Gift from Lucy Shapiro | Wild type |
| Critical commercial assays | ||
| Molecular Probes LIVE/DEAD BacLight Bacterial Viability Kit | Fisher Scientific | Cat#L13152 |
| Biotium Viability/Cytoxicity Assay Kit for Bacteria Live & Dead Cells | Fisher Scientific | Cat#NC1643156 |
| Chemicals, peptides, and recombinant proteins | ||
| Paraformaldehyde, 16% | Fisher Scientific | Cat#18505; CAS:50-00-0 |
| Sodium phosphate dibasic anhydrous | Fisher Scientific | Cat#S379-212; CAS:7558-79-4 |
| Agarose | Fisher Scientific | Cat#BP160-100; CAS:9012-36-6 |
| Isopropanol | Fisher Scientific | Cat#A417-1; CAS:67-63-0 |
| Vanillic acid | Sigma-Aldrich | Cat#94770; CAS:121-34-6 |
| Other | ||
| Plain Microscope Slides Precleaned | Fisher Scientific | Cat#12-550-A3 |
| Premium Cover Glass | Fisher Scientific | Cat#12548B |
| Axio Imager MOT M1 (fluorescent microscope) | Carl Zeiss Microscopy | Cat#4300049901000000 |
| C4742-80-12AG ORCA-ER-1394 (microscope camera) | Carl Zeiss Microscopy | Cat# 4108301700000000 |
Step-by-step method details
All data is acquired in triplicate in this assay, using three independent cultures per condition.
Isolating colonies on a plate
Timing: 30 min
Determine the colony forming units per milliliter of a 0.100 optical density culture, grown to exponential-phase, by performing a set of serial dilutions and subsequently plating dilutions onto agar media. This optical density was selected because it facilitates quick enumeration of total cell numbers per milliliter using a Petroff-Hauser counting chamber.
-
1.
Grow up bacterial cultures in 3 mL of liquid media.
-
2.Standardize exponential-phase cultures to an OD of 0.100 with the media they are grown in.
-
a.Add 1.0 mL of the 0.100 OD culture to a microcentrifuge tube for use in “Staining to detect live or dead cells”, step 11.
-
a.
Note: When diluting cultures, pipette immediately below the meniscus to avoid transferring bacteria from outside of the pipette tip.
-
3.
For easy counting of CFU, spread 10−5, 10−6, or 10−7 dilutions onto agar growth media from the standardized culture.
-
4.
Incubate at optimal growth temperature.
-
5.
Once colonies surface, calculate colony forming units (CFU) per milliliter by multiplying CFU by the appropriate dilution factor.
Note: A desired CFU count is between 25 to 250 as per standard microbiological laboratory practices.
Direct cell counts
Timing: 3–4 h
Standardized cultures are preserved with fixative to simultaneously prevent culture growth and cell lysis for an accurate direct count determination after other steps of this procedure have been completed. Enumerations of fixed cultures are performed with a Petroff-Hausser counting chamber to determine the cells per milliliter within cultures.
-
6.
Add 20 μL of 5× fixative to 80 μL of standardized 0.100 OD cultures (“Isolating Colonies on a Plate,” Step 2) into a microcentrifuge tube. Mix well by finger vortexing.
Pause point: Once fixative is applied to the culture, store at 4°C.
Note: Fixative will instantaneously preserve the culture once applied and may be stored at 4°C for up to one week.
CRITICAL: For an accurate representation of the percentage of live cells within a culture, perform the direct counts at a later time, or date, after all fluorescent stain imaging is completed.
-
7.
Resuspend the fixed cells (using a micropipettor) to ensure all cells are completely suspended (no pellet or clumps should be visible). Apply 20 μL of the fixative-culture mixture to a Petroff-Hausser counting chamber and apply the manufacturer-provided re-enforced cover slip.
-
8.
Allow 2 min to elapse for reduced capillary motion.
CRITICAL: Failure to suspend an observable pellet or dissociate clumping will affect the repeatability and accuracy of the direct counts.
-
9.
View chamber under a table-top phase-contrast microscope at 400× magnification perform total cell counts (as a reference, boxes to be counted are highlighted in Figure 2) Troubleshooting 2
-
10.Once bacteria are counted, adjust the average number of cells in each of the 5 diagonal quadrants to account for the 5× fixative dilution by multiplying with 1.25 (100/80=1.25), calculated as follows:
-
a.Cells per milliliter (refer to Figure 4 for a reference range)
-
b.Percentage of cells that form coloniesTroubleshooting 3Note: Cells touching the left and top edges in each sub-quadrant are counted whereas the right and bottom edges are not.Note: A cell with a clear septum is counted as two cells.
-
a.
Figure 2.
Pictogram of petroff-hausser counting chamber centric counting area
Bacterial counts are performed on the 5 highlighted diagonal boxes. Within each box lies a grid composed of 16 smaller sub-quadrants where bacteria are counted.
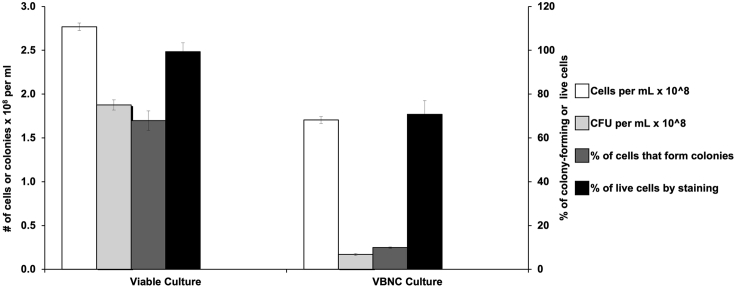
Figure 4.
A graphical representation of data from direct counts, CFU and LIVE/DEAD staining in Caulobacter crescentus mutants using the Molecular Probes BacLight stains
A viable but non-culturable population can be observed when there is a large difference between the percentage of live cells by staining to a percentage of lives cells that form colonies. SM1309 (Caulobacter crescentus NA1000 PvanA::fabH ΔfabH ΔspoT) was initially grown in PYE broth supplemented with 0.5 mM vanillate, washed 3 times in PYE to remove vanillate, and split into PYE broth with, or without, vanillate. The culture with vanillate remained in exponential-phase until assayed (viable culture); the culture in the absence of vanillate depleted FabH (VBNC culture) and growth arrested by 22 h. Data was collected after 24 h of incubation at 28°C. Scales for cells or colonies per milliliter are placed on the left y-axis and percentage of colony forming or living cells are placed on the right y-axis. Data, from triplicate independent experiments, is presented as the mean ± SEM.
Staining to detect live or dead cells
Timing: 2–4 h
Normalized cultures are treated with stains for the detection of live and dead cells and visualized by fluorescence microscopy to enumerate the percentage of living cells.
-
11.
Centrifuge the remainder of the standardized 1.0 mL of the 0.100 OD culture prepared in “Isolating Colonies on a Plate,” step 2 at 2,900 × g for 2 min.
-
12.
Proceed from “Optimizing Stain Fluorescence for live and dead cells” steps 17–22.
Note: For step 21 photograph at least several hundred cells via differential interference contrast, or phase contrast, and fluorescence microscopy using Cy3 or Texas Red and FITC filters across triplicate cultures.
-
13.From the array of experimental images captured, calculate:
-
a.The percentage of live cells by stainingTroubleshooting 4
-
a.
Note: In predivisional cells, if a clear septum is visible, count them as two entities (Stott et al., 2015). Some compartmentalized late predivisional cells stain green in one compartment (SYTO9) and red in the other (propidium iodide).
Expected outcomes
DIC and fluorescence microscopy images from staining for live and dead cells in liquid cultures are shown in Figures 3A and 3B. The figure displays sampled images of viable and viable but non-culturable (VBNC) Caulobacter crescentus populations as living, as indicated by green fluorescence with the both the Biotium and BacLight stains. Viable cells will form colonies on agar media whereas VBNC cells will not. In contrast, dead, isopropanol-treated cells fluoresce red with both sets of stains.
Figure 3.
Biotium and Molecular Probes BacLight stains of Caulobacter crescentus SM1309 (NA1000 PvanA::fabH ΔfabH ΔspoT) cultures that form viable but non-culturable cells when depleted of the ß-ketoacyl-acyl carrier protein synthase FabH, resulting in fatty acid starvation
Regions of interest on agarose pads are visualized using differential interface contrast microscopy. Under a FITC (GFP) filter, live cells will fluoresce green. Isopropanol-treated, dead cells will fluoresce red under a Cy3 (TexasRed) filter. Viable cells (labeled C. crescentus) were grown in the presence of vanillate whereas viable but non-culturable cells were grown in the absence of vanillate (FabH depletion conditions) for 24 h (PYE broth, 28°C). Only viable, but not viable but non-culturable, cells will form colonies on agar growth media. Isopropanol kills both viable and viable but non-culturable cells. (A) Biotium stains (B) Molecular Probes BacLight stains. Scale bars = 5 μM.
Results from a Caulobacter crescentus viable but non-culturable assay are shown in Figure 4. This strain lacks (p)ppGpp (due to a non-polar deletion in spoT) and has the expression of the FabH –ketoacyl-acyl carrier protein synthase enzyme controlled by a vanillate-inducible promoter. When this strain is grown in the presence of 0.5 mM vanillate, FabH is expressed and the majority of cells are viable. When the strain is grown in the absence of vanillate, the cells become starved for fatty acids. This eventually arrests growth and the majority of cells enter a VBNC state (Stott et al., 2015). As seen in Figure 4, in viable cultures, cell number per mL (from direct counts) correlate with CFU per mL, with a high percentage of cells forming colonies and most cells staining as live. In contrast, under VBNC conditions, cell number per mL is not correlated with CFU per mL, resulting in a low percentage of cells forming colonies despite the majority of cells staining as live. Please note that we represented CFU per milliliter and cells per milliliter on one y-axis and the percentage of cells that form colonies and percentage of live cells by staining on the other y-axis. Since the direct counts, CFU/mL and fluorescent staining were performed from the same culture, the percentage of VBNC cells can be calculated by the percentage of live cells that do not form colonies (as in Supplemental File 2).
Limitations
The protocol herein is limited to known model organisms whose permissive growth conditions are known and stainable by the Molecular Probes and Biotium stains. This assay may not be effective on bacteria that form dense clusters, or biofilms, where it can be difficult to identify single cells or for the stains to penetrate.
Troubleshooting
Problem 1
LIVE/DEAD staining of the negative control is producing fluorescent green (live) cells (step 22 in “before you begin”).
Potential solution 1
The manufacturer recommends adjusting the ratio of individual stains to optimize their fluorescence.
Potential solution 2
The microscope imaging program (set to auto-exposure) may be incorrectly fluorescing dead cells as green. Troubleshooting could include (1) observing cells in real-time through the ocular lens under Cy3, or TexasRed, and FITC filters to disgtinguish between bright-fluorescing red cells and background-fluorescing green cells, respectively; or (2) observing cells in real-time through the microscopes camera on the attached computer monitor under Cy3, or TexasRed, and FITC filters. The camera detector should detect bright-fluorescing red cells but not background-fluorescing green cells
If both these conditions are met, suggesting that the hardware is functioning properly, then the imaging program (of the user’s choice) could be scoring dead cells as alive. Precisely, the software could be inappropriately displaying the scale of the image pixel intensity histogram. Adjusting the histogram to a linear scaling can correct this issue during or after image acquisition. Optimizing exposure to a maximum signal-to-background ratio and normalizing fluorescence intensity when comparing controls and experimental images can also be helpful.
Problem 2
Excess debris is present in fixed cell mixture under the Petroff-Hausser counting chamber making it difficult to count cells (step 9, “step-by-step method details”).
Potential solution
Prior to fixing cultures in “Direct Cell Counts,” warm fixative in a 50°C water bath, or heat block, and vortex lightly, repeatedly, to clear debris.
Problem 3
The total cells per mL calculated are considerably lower than the CFU per mL (step 10, “step-by-step method details”).
Potential solution 1
Test fixative for preservation efficacy over time. Preserve an aliquot of culture with fixative and perform Petroff-Hausser counting chamber counts to enumerate the population over time. A decreasing total population in a fixed culture over time indicates low fixative efficacy (Günther et al., 2008).
Potential solution 2
Pipetting error could occur at the dilution step; pipette immediately below the meniscus to avoid bacterial transfer from the outer part of the tip.
Problem 4
The percentage of live cells across triplicate data varies considerably (step 13, “step-by-step method details”).
Potential solution 1
Decrease the amount of time spent between applying the fluorescent stains to a prepared culture and imaging. Immediately after applying the stains, add to agarose pads on slides and proceed with imaging.
Potential solution 2
Reduce the speed and time spent under centrifugation prior to fluorescent stain treatment. We have found a reduction of these parameters yields considerably higher percentages of live cells across a variety of cultures tested.
Resource availability
Lead contact
Requests for information, resources, or reagents should be sent to the lead contact, Sean Murray (sean.murray@csun.edu).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
We thank previous and current lab members, especially Kristina Stott, Shannon Schreiner, and Bao Nguyen, for aiding in the development of this assay. This work was supported by NIH Grant GM121234 to S.R.M.
Author contributions
Conceptualization and methodology, I.S.A. and S.R.M.; investigation, I.S.A.; writing – original draft, I.S.A.; writing – reviewing and editing, I.S.A. and S.R.M.; funding acquisition, resources, and supervision, S.R.M.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2021.100738.
Contributor Information
Ignacio Servando Arvizu, Email: Ignacio.arvizu.36@my.csun.edu.
Sean Richard Murray, Email: sean.murray@csun.edu.
Supplemental information
Data from 15-min incubation vs immediate viewing of cells with stains for the detection of live or dead cells is shown. No significant differences (using paired two-tailed t-tests) were found between an immediate and 15-min incubation period with the BacLight or Biotium stains for detecting live and dead Caulobacter crescentus NA1000 (PYE broth, 28°C), Escherichia coli MG1655 (LB broth, 37°C), or Bacillus subtilis PY79 (LB broth, 37°C). The data is based off results with three independent cultures derived from single colonies for each bacterial species shown.
Viable but non-culturable data for Caulobacter crescentus SM1309 (NA1000 PvanA::fabH ΔfabH ΔspoT) was used to generate Figure 4. It includes cells per mL, CFU per mL, and staining for live or dead cells. The data were combined to detect a viable but non-culturable population.
Data and code availability
The dataset supporting the current study is available as Supplemental Files.
References
- Adamski M., Pietr S.J. Biodiversity of bacteria associated with eight pleurotus ostreatus (Fr.) P. kumm. strains from Poland, Japan and the USA. Pol. J. Microbiol. 2019;68:71–81. doi: 10.21307/pjm-2019-009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhave M., Hassanbhai A., Anand P., Luo K.Q., Teoh S.H. Effect of heat-inactivated clostridium sporogenes and its conditioned media on 3-dimensional colorectal cancer cell models. Sci Rep. 2015;5:15681. doi: 10.1038/srep15681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biotium Viability/cytotoxicity assay for bacteria live and dead cells manual. 2020. https://biotium.com/wp-content/uploads/2015/01/PI-30027.pdf
- Chao Y., Zhang T. Optimization of fixation methods for observation of bacterial cell morphology and surface ultrastructures by atomic force microscopy. Appl. Microbiol. Biotechnol. 2011;92:381–392. doi: 10.1007/s00253-011-3551-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplantis B.N., Puckett S.M., Rosey E.L., Ameiss K.A., Hartman A.D., Pearce S.C., Nano F.E. Temperature-sensitive salmonella enterica serovar enteritidis PT13a expressing essential proteins of psychrophilic bacteria. Appl. Environ. Microbiol. 2015;81:6757–6766. doi: 10.1128/AEM.01953-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani M., Suzuki T., Ishihara H., Hashimoto H., Kunoh H., Takada J. Assemblage of bacterial saccharic microfibrils in sheath skeleton formed by cultured Leptothrix sp. strain OUMS1. J. Marine Sci. Res. Dev. 2011;5:001. [Google Scholar]
- Günther S., Hübschmann T., Rudolf M., Eschenhagen M., Röske I., Harms H., Müller S. Fixation procedures for flow cytometric analysis of environmental bacteria. J. Microbiol. Methods. 2008;75:127–134. doi: 10.1016/j.mimet.2008.05.017. [DOI] [PubMed] [Google Scholar]
- Hernandez S.B., Cota I., Ducret A., Aussel L., Casadesus J. Adaptation and preadaptation of salmonella enterica to bile. PLoS Genet. 2012;8:e1002459. doi: 10.1371/journal.pgen.1002459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howat W.J., Wilson B.A. Tissue fixation and the effect of molecular fixatives on downstream staining procedures. Methods. 2014;70:12–19. doi: 10.1016/j.ymeth.2014.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Murata K., Zhang D. Applicability of LIVE/DEAD BacLight stain with glutaraldehyde fixation for the measurement of bacterial abundance and viability in rainwater. J. Environ. Sci. 2017;51:202–213. doi: 10.1016/j.jes.2016.05.030. [DOI] [PubMed] [Google Scholar]
- Kim S.E., Zhang C., Advincula A.A., Baer E., Pokorski J.K. Protein and bacterial antifouling behavior of melt-coextruded nanofiber mats. ACS Appl. Mater. Interfaces. 2016;8:8928–8938. doi: 10.1021/acsami.6b00093. [DOI] [PubMed] [Google Scholar]
- Lindbäck T., Rottenberg M.E., Roche S.M., Rørvik L.M. The ability to enter into an avirulent viable but non-culturable (VBNC) form is widespread among Listeria monocytogenes isolates from salmon, patients and environment. Vet. Res. 2010;41:8. doi: 10.1051/vetres/2009056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molecular Probes LIVE/DEAD® BacLight™ bacterial viability kits manual. 2004. https://www.thermofisher.com/document-connect/document-connect.html?url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FLSG%2Fmanuals%2Fmp07007.pdf&title=TElWRSYjNDc7REVBRCAmbHQ7aSZndDtCYWMmbHQ7L2kmZ3Q7TGlnaHQgQmFjdGVyaWFsIFZpYWJpbGl0eSBLaXRz
- Park S.Y., Kim C.G. A comparative study of three different viability tests for chemically or thermally inactivated Escherichia coli. Environ. Eng. Res. 2018;23:282–287. [Google Scholar]
- Robertson J., McGoverin C., Vanholsbeeck F., Swift S. Optimisation of the protocol for the LIVE/DEAD® BacLight™ bacterial viability kit for rapid determination of bacterial load. Front. Microbiol. 2019;10:801. doi: 10.3389/fmicb.2019.00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodes L., Coussa-Charley M., Marinescu D., Paul A., Fakhoury M., Abbasi S., Khan A., Tomaro-Duchesneau C., Prakash S. Design of a novel gut bacterial adhesion model for probiotic applications. Artif. Cells Nanomed. Biotechnol. 2013;41:116–124. doi: 10.3109/10731199.2012.712047. [DOI] [PubMed] [Google Scholar]
- Stott K.V., Wood S.M., Blair J.A., Nguyen B.T., Herrera A., Mora Y.G., Cuajungco M.P., Murray S.R. ppGpp modulates cell size and the initiation of DNA replication in Caulobacter crescentus in response to a block in lipid biosynthesis. Microbiology (Reading) 2015;161:553–564. doi: 10.1099/mic.0.000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data from 15-min incubation vs immediate viewing of cells with stains for the detection of live or dead cells is shown. No significant differences (using paired two-tailed t-tests) were found between an immediate and 15-min incubation period with the BacLight or Biotium stains for detecting live and dead Caulobacter crescentus NA1000 (PYE broth, 28°C), Escherichia coli MG1655 (LB broth, 37°C), or Bacillus subtilis PY79 (LB broth, 37°C). The data is based off results with three independent cultures derived from single colonies for each bacterial species shown.
Viable but non-culturable data for Caulobacter crescentus SM1309 (NA1000 PvanA::fabH ΔfabH ΔspoT) was used to generate Figure 4. It includes cells per mL, CFU per mL, and staining for live or dead cells. The data were combined to detect a viable but non-culturable population.
Data Availability Statement
The dataset supporting the current study is available as Supplemental Files.