Abstract
Corona Virus Disease-2019 (COVID-19) is a global pandemic which is spreading briskly across the globe. The gold standard for the diagnosis of COVID-19 is viral nucleic acid detection with real-time polymerase chain reaction (RT-PCR). However, the sensitivity of RT-PCR in the diagnosis of early-stage COVID-19 is less. Recent research works have shown that computed tomography (CT) scans of the chest are effective for the early diagnosis of COVID-19. Convolutional neural networks (CNNs) are proven successful for diagnosing various lung diseases from CT scans. CNNs are composed of multiple layers which represent a hierarchy of features at each level. CNNs require a big number of labeled instances for training from scratch. In medical imaging tasks like the detection of COVID-19 where there is a difficulty in acquiring a large number of labeled CT scans, pre-trained CNNs trained on a huge number of natural images can be employed for extracting features. Feature representation of each CNN varies and an ensemble of features generated from various pre-trained CNNs can increase the diagnosis capability significantly. In this paper, features extracted from an ensemble of 5 different CNNs (MobilenetV2, Shufflenet, Xception, Darknet53 and EfficientnetB0) in combination with kernel support vector machine is used for the diagnosis of COVID-19 from CT scans. The method was tested using a public dataset and it attained an area under the receiver operating characteristic curve of 0.963, accuracy of 0.916, kappa score of 0.8305, F-score of 0.91, sensitivity of 0.917 and positive predictive value of 0.904 in the prediction of COVID-19.
Keywords: COVID-19, Computed tomography, CNN, KSVM, Computer-aided diagnosis
Introduction
On March 2019, the World Health Organization (WHO) has declared the Corona Virus Disease-2019 (COVID-19) a pandemic [17]. Early diagnosis and rapid isolation of the patients are crucial for effective treatment and for preventing the spread of the disease. Viral nucleic acid detection with real-time polymerase chain reaction (RT-PCR) is the gold standard for the detection of COVID-19. The availability of RT-PCR test kits is insufficient in most of the places where the disease has spread. Moreover, RT-PCR shows less sensitivity in the early diagnosis of COVID-19 [10]. Due to the shortage of RT-PCR test kits and the likelihood of false-negative RT-PCR findings during the early diagnosis, National Health Commission of China has endorsed the diagnosis of COVID-19 using chest CT findings [4]. COVID-19 in CT scans is characterized by ground glass opacity (GGO). GGO represents an area of hazy increased lung opacity through which underlying vessels are visible [14]. Manual analysis of a large number of CT scans by the radiologists is tedious. A computer-aided diagnosis mechanism could assist radiologists in the diagnosis of COVID-19 from CT scans.
There exist a number of computer-aided studies for the diagnosis of COVID-19 from CT scans. He et al. [13] proposed a method that combines transfer learning with self-supervised learning. Anwar and Zakir [3] and Sakagianni et al. [21] also used the public dataset created by [13] for their study. Anwar and Zakir [3] used EfficientNet to diagnose COVID-19 using CT scans. The model used three different learning approaches, namely, reduce on plateau, cyclic and constant learning rates. Sakagianni et al. [21] used auto Machine Learning Cloud Vision platform for the prediction of COVID-19. Abraham and Nair [2] proposed a multi-CNN architecture consisting of multiple pre-trained CNNs along with correlation-based feature selection (CFS) technique and Bayesnet classifier for the classification of COVID-19 and non-COVID-19 X-ray images. Pu et al. [19] developed three classifiers based on CNN for distinguishing COVID-19 and pneumonia. Mei et al. [15] used a combination of CNN, multi-layer perceptron (MLP) and clinical information to diagnose COVID-19 from CT scans. Xu et al. [28] used a local attention classification model for classifying the infected regions into COVID-19, influenza and irrelevant to infections. Wu et al. [27] used a deep learning- based multi-view fusion model for the detection of COVID-19. In addition to CT, few works have explored the use of X-rays [12] and cough audio recordings [9] for the computer-aided diagnosis of COVID-19.
All the above-mentioned methods which employed CT scans for the diagnosis of COVID-19 have used a single CNN for the diagnosis of COVID-19. Combination of features from multiple CNNs could produce better results than single CNNs. None of the state-of-the-art studies have explored the optimum combination of features from pre-trained CNNs along with an off-the-shelf classifier for the detection of COVID-19.
The contributions of the proposed ensemble method are listed below.
i. The proposed method explored the combination of various pre-trained CNNs to put forward a model that gives better performance than single pre-trained CNNs and transfer learning using end-to-end CNNs for the diagnosis of COVID-19 from CT scans. Based on empirical analysis, an ensemble of features generated using 5 pre-trained CNNs (MobilenetV2, Shufflenet, Xception, Darknet53 and EfficientnetB0) is proposed in this study for the detection of COVID-19 from CT scans. Most of the state-of-the-art methods employing CT images for the detection of COVID-19 have used features from a sole CNN.
ii. The proposed technique used an off-the-shelf classifier, kernel support vector machine (KSVM), for the classification of COVID-19 from CT scans. No existing techniques have employed the combination of the above-mentioned ensemble of CNNs with KSVM for the detection of COVID-19.
Materials and methods
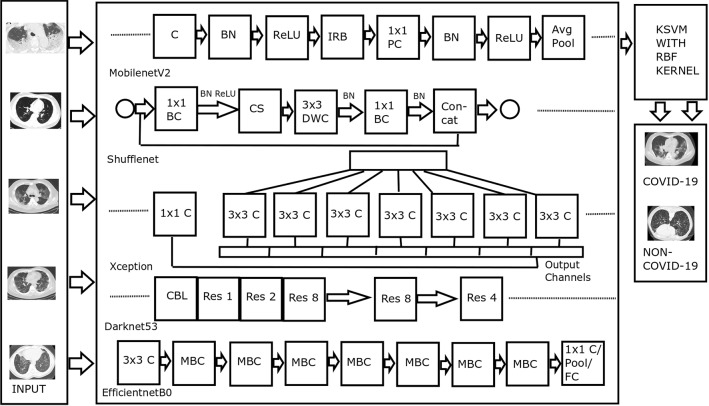
Figure 1 displays the architecture of the proposed method. The functions of various blocks of the architecture are described in the following subsections.
Fig. 1.
Architecture of the proposed method. Abbreviations—C: Convolution, BN: Batch Normalization, ReLU: Rectified Linear Unit, IRB: Inverted Residual Block, PC: Pointwise Convolution, Avg Pool: Average Pooling, BC: Batch Convolution, CS: Channel Shuffle, DWC: Depthwise Convolution, Res: Residual
Feature extraction using multiple CNNs
Convolutional neural networks produce powerful feature representations for computer vision tasks. Many research works like segmentation of liver tumor [30] and diagnosis of prostate cancer [1] have proven the effectiveness of CNN in medical applications. For most of the medical diagnostic tasks, training a network from scratch is difficult due to the non-availability of sufficiently large medical datasets. Off-the-shelf features extracted using pre-trained CNNs were found successful in medical diagnosis. Using a pre-trained network is much faster than training a network from scratch. Off-the-shelf features extracted using CNN could achieve superior results even without fine-tuning [23].
Combining features extracted from an ensemble of pre-trained networks could achieve better performance than features extracted using single CNNs. The proposed method combines the features generated from the last fully connected (FC) layer of an ensemble of various CNNs. The CNNs were pre-trained on Imagenet dataset [7] which contains millions of images. The pre-trained CNNs can classify objects into 1000 different categories. The number of layers, input size and number of parameters of the above-mentioned pre-trained networks are represented in Table 1.
Table 1.
Depth, input size and number of parameters of pre-trained CNNs
| CNN | Depth | Input size | Number of parameters (In Millions) |
|---|---|---|---|
| MobilenetV2 | 53 | 3.5 | |
| Shufflenet | 50 | 1.4 | |
| Xception | 71 | 22.9 | |
| Darknet53 | 53 | 41 | |
| EfficientnetB0 | 82 | 5.3 |
As the dataset contains 746 images, a feature vector of dimension is generated by each CNN (Activations from the last convolution layer of Darknet53 and last FC layer of other CNNs). Feature vectors produced by each CNN are concatenated to get a feature vector of size . Specimen features produced by the endmost convolutional layer of MobilenetV2, Shufflenet, Xception, Darknet53 and EfficientnetB0 are displayed in Fig 2.
Fig. 2.

Sample features. Two rows at the top represent sample features of a COVID-19 image, and the two rows at the bottom represent sample features of a non-COVID-19 CT image
The five pre-trained CNNs (MobilenetV2, Shufflenet, Xception, Darknet53 and EfficientnetB0) which achieved best performance in our study are described in the following sections.
MobilenetV2
The first layer of MobilenetV2 is a convolution layer that accepts input images of dimension [22]. The model introduced 19 Inverted Residual with Linear Bottleneck (IRB) layers which accepts a low- dimensional feature representation as input. Pointwise convolution operation is performed to increase the dimension of feature matrix to a high-dimensional one. A depth-wise convolution followed by a pointwise convolution is done subsequently.
Shufflenet
Shufflenet architecture introduced by Zhang et al. [31] consists of pointwise batch convolution operation and channel shuffle (CS) operation. Pointwise batch convolution reduces the computation complexity of dense convolution with a cost of weakened representation. Weakened representation caused by pointwise batch convolution can be overcome by using channel shuffle operation which secures input from different batches by dividing the channels in each batch into various groups.
Xception
Xception [6] is a pre-trained CNN model based on Inception, where stacks of depthwise separable convolution layers with residual connections substitute inception modules. A depthwise separable convolution is composed of a depthwise convolution which is a spatial convolution operation done separately over each input channel, followed by a convolution operation. The Xception architecture consists of 36 convolutional layers which are grouped into 14 modules, all of which except the first and last modules have linear residual connections among them.
Darknet53
The basic building block of Darknet53 is CBL (Convolution, Batch normalization, Leaky ReLU) layer [20]. Darknet53 is a pre-trained CNN mainly composed of a series of and convolutional layers. The network consists of 5 residual layers which are used to solve the gradient vanishing problem. The residual layers are indicated as Res n, where n is the number of residual blocks in a residual layer. The different values of n are 1, 2, 4 and 8.
EfficientnetB0
The main building block for EfficientNetB0 is the Mobile inverted Bottleneck MBC to which squeeze and excitation optimization is added [24]. MBC blocks consist of a layer that performs expansion followed by compression of the channels. Direct connections are used among bottlenecks which connect much lesser channels compared to expansion layers.
KSVM
Support vector machine (SVM) proposed by Vapnik et al. [25] is a classifier that determines the optimum hyperplane which separates the positive and negative instances of a training data. A training data of n observations can be defined as
| 1 |
where is an instance of training data and is the corresponding label.
A hyperplane in the feature space can be represented as
| 2 |
where w represents the weight matrix and b is the bias term. Distance of an instance from the hyperplane plane (w, b) is defined as
| 3 |
SVM model proposes a maximum margin separating and , which can be obtained by solving the optimization problem
| 4 |
conditional on
| 5 |
By introducing a mapping function , the vector x in the original low-dimensional space can be transformed into of high-dimensional space. A kernel function is described as
| 6 |
The optimization problem defined in Eq. 4 becomes
| 7 |
conditional on
where C is a regularization parameter and is a slack variable used to solve overfitting.
The optimization problem in Eq. 7 can be solved in dual form as given below.
| 8 |
conditional on
,
where represents the weight of and
, a matrix of dimension .
The proposed method uses RBF kernel in association with KSVM classifier [5]. RBF kernel is described as
| 9 |
where and is a free parameter. The decision function for the KSVM is specified as
| 10 |
Results and discussion
A publicly available dataset consisting of 349 CT scans diagnosed with COVID-19 and 397 CT scans confirmed as non-COVID was used for the study [33]. The area under the receiver operating characteristic curve (AUROC), accuracy, kappa score, positive predictive value (PPV), sensitivity and F-score were used as the performance metrics. MATLAB 2020b with default parameter settings was used for feature extraction using the CNNs. The classification was performed using LibSVM package of Weka 3.6. Parameter settings of the classifier are given in Table 2.
Table 2.
Parameter Settings
| Parameter | Value |
|---|---|
| Cost | 4.5 |
| Degree | 3 |
| eps | 0.001 |
| Kernel | Radial Basis Function (RBF) |
| Loss | 0.1 |
A 10-fold cross-validation (CV) was performed on the dataset, and the average results of 10-fold were taken into account. Using the same training and test set will give more accurate results when the performance of different models is compared. A seed value of 1 was used for generating the folds of 10-fold cross-validation so that the same data partitions were created for each of the experiments. All experiments were performed on the same training and test sets so that the results are comparable. The proposed method achieved an accuracy of 91.6% and AUROC of 0.963. Figure 3 represents the confusion matrix of the proposed method. Figure 4 displays the AUROC achieved by the proposed method. The p value based on Chi-square statistic was computed to determine the statistical significance of the proposed method. Level of significance was set at 0.05. The p value achieved was less than 0.00001, significant at .
Fig. 3.
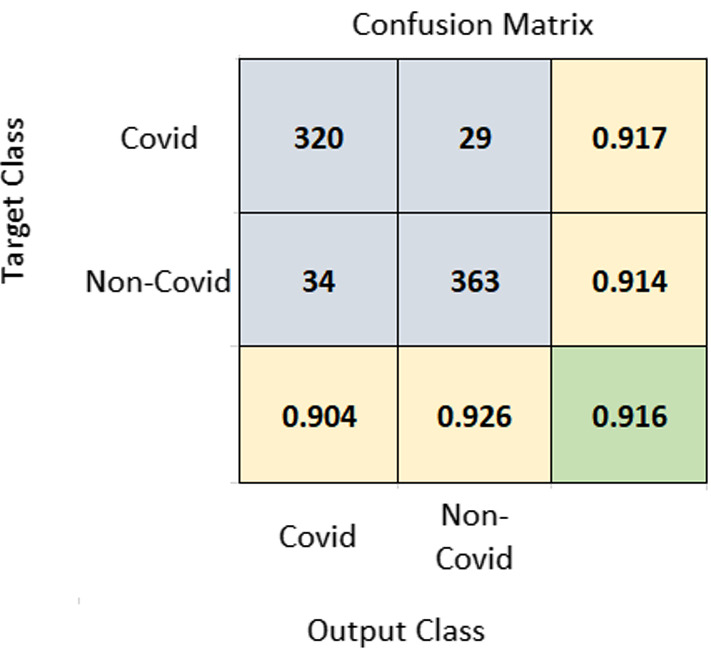
Confusion matrix of COVID-19 vs. non-COVID-19 classification. The diagonal element at the bottom shown in green color corresponds to accuracy. Rightmost column values represent sensitivity and bottom-most row elements represent PPV
Fig. 4.
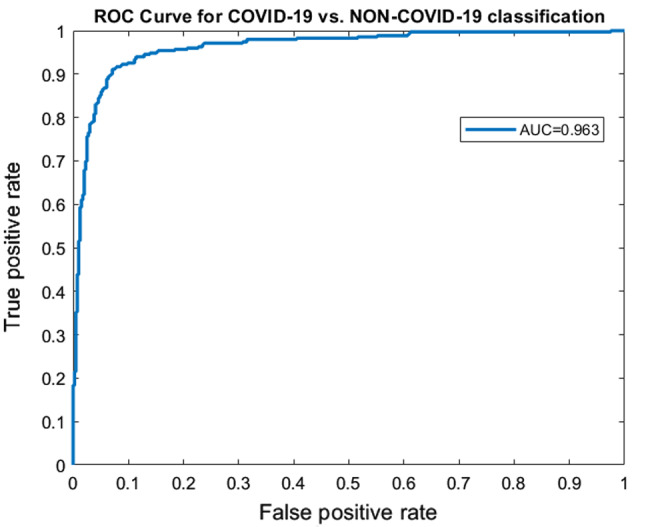
ROC curves corresponding to COVID-19 vs. non-COVID-19 classification
Two more types of validation were performed to ensure the performance of the method further. Partitions were created using different seed values from 1 to 10 and repeated the experiments 10 times. Ten-fold CV was performed using the training sets and test sets created using each seed value. The average PPV, sensitivity, F-score, kappa score, AUROC and accuracy achieved on repeating the experiments 10 times were 0.9033, 0.9194, 0.9112, 0.8319, 0.9622 and 0.9155, respectively. We also performed validation by dividing the data into a 70% training set and a 30% test set. This type of validation achieved a PPV, sensitivity, F-score, kappa score, AUROC and accuracy of 0.933, 0.866, 0.898, 0.959 and 0.9018, respectively. The results achieved using the three types of validation were comparable.
Table 3 represents the results achieved using various combinations of pre-trained CNNs. Ensemble of 4 pre-trained CNNs consisting of Darknet53, Shufflenet, Xception and EfficientnetB0 also achieved an AUROC of 0.963. However, the proposed 5 CNNs achieved a slightly better accuracy, sensitivity, kappa score and F-score. The combinations of various 4 CNNs selected from the 5 networks of the proposed ensemble also achieved good performance, but slightly lower than the proposed architecture. The results show that ensemble of CNNs, selected based on empirical analysis, could perform better than single CNNs.
Table 3.
Performance of various CNNs and ensemble of CNNs in combination with KSVM
| Network | PPV | Sensitivity | F-Score | Kappa score | AUROC | Accuracy |
|---|---|---|---|---|---|---|
| MobilenetV2+Shufflenet+Xception+ | 0.904 | 0.917 | 0.91 | 0.8305 | 0.963 | 0.916 |
| Darknet53+EfficientnetB0 | ||||||
| MobilenetV2+Shufflenet+Xception+ | 0.892 | 0.90 | 0.896 | 0.8036 | 0.958 | 0.902 |
| Darknet53+Alexnet | ||||||
| MobilenetV2+Shufflenet+Xception+ | 0.892 | 0.897 | 0.894 | 0.8009 | 0.958 | 0.901 |
| Darknet53+Densenet201 | ||||||
| Darknet+Shufflenet+ | 0.906 | 0.911 | 0.909 | 0.8278 | 0.963 | 0.914 |
| Xception+EfficientnetB0 | ||||||
| MobilenetV2+Darknet+ | 0.90 | 0.90 | 0.90 | 0.8116 | 0.96 | 0.906 |
| Xception+EfficientnetB0 | ||||||
| MobilenetV2+Shufflenet+ | 0.905 | 0.903 | 0.904 | 0.8196 | 0.962 | 0.910 |
| Xception+EfficientnetB0 | ||||||
| MobilenetV2+Shufflenet+ | 0.892 | 0.9 | 0.896 | 0.8036 | 0.959 | 0.902 |
| Xception+Darknet53 | ||||||
| Alexnet+Googlenet+Resnet101 | 0.863 | 0.868 | 0.866 | 0.747 | 0.943 | 0.874 |
| Efficientnet+Shufflenet+MobilenetV2 | 0.904 | 0.891 | 0.898 | 0.8087 | 0.957 | 0.905 |
| Darknet53+Shufflenet+MobilenetV2 | 0.887 | 0.897 | 0.892 | 0.7955 | 0.953 | 0.898 |
| Darknet53+Shufflenet+Xception | 0.884 | 0.897 | 0.89 | 0.7929 | 0.956 | 0.897 |
| MobilenetV2+Shufflenet+Xception | 0.886 | 0.871 | 0.879 | 0.773 | 0.956 | 0.887 |
| MobilenetV2+Darknet53+Xception | 0.881 | 0.888 | 0.884 | 0.7821 | 0.954 | 0.891 |
| MobilenetV2+Shufflenet | 0.87 | 0.883 | 0.876 | 0.766 | 0.883 | 0.883 |
| MobilenetV2+Darknet53 | 0.871 | 0.888 | 0.879 | 0.7714 | 0.886 | 0.886 |
| Xception | 0.831 | 0.848 | 0.84 | 0.6962 | 0.916 | 0.849 |
| Darknet53 | 0.85 | 0.845 | 0.848 | 0.7145 | 0.93 | 0.857 |
| MobilenetV2 | 0.84 | 0.857 | 0.848 | 0.7123 | 0.926 | 0.857 |
| Shufflenet | 0.839 | 0.865 | 0.852 | 0.7148 | 0.926 | 0.858 |
| Alexnet | 0.826 | 0.805 | 0.816 | 0.6576 | 0.909 | 0.830 |
| Googlenet | 0.813 | 0.811 | 0.812 | 0.6473 | 0.904 | 0.824 |
| Resnet101 | 0.844 | 0.868 | 0.856 | 0.7259 | 0.928 | 0.863 |
| Densenet201 | 0.845 | 0.857 | 0.85 | 0.7176 | 0.93 | 0.859 |
| EfficientnetB0 | 0.87 | 0.86 | 0.865 | 0.7468 | 0.948 | 0.873 |
| Resnet18 | 0.843 | 0.828 | 0.835 | 0.7339 | 0.925 | 0.847 |
Performance of the proposed method (Ensemble of CNNs+KSVM) is represented in bold
Pre-trained networks can be used in two different ways for the classification task. In the proposed method, the pre-trained CNNs serve as a feature extractor, where the activations of a layer are used as features to train an off-the-shelf classifier like KSVM. Pre-trained CNNs can also be used for transfer learning where the layers of the pre-trained network are fine-tuned using the data at hand. Table 4 illustrates the performance of transfer learning using MobilenetV2, Shufflenet, Xception, Darknet53 and EfficientnetB0 with the proposed method. Number of epochs, mini batch size and learning rate were set as 5, 8 and 0.0001, respectively. The proposed method achieved better accuracy and kappa score than transfer learning using pre-trained CNNs. The execution time in minutes of transfer learning using MobilenetV2, Shufflenet, Xception, Darknet53 and EfficientnetB0 was 146.52, 70.08, 312.78, 657.35 and 298.99, respectively, whereas the proposed method took only 10.79 minutes for 10-fold CV. The time required for feature extraction and classification of a single instance was 24.315 seconds.
Table 4.
Comparison of the performance of proposed method with transfer learning
| Network | PPV | Sensitivity | F-Score | Kappa score | AUROC | Accuracy |
|---|---|---|---|---|---|---|
| Proposed | 0.904 | 0.917 | 0.91 | 0.8305 | 0.963 | 0.916 |
| Xception | 0.9398 | 0.8241 | 0.8782 | 0.7571 | 0.9590 | 0.8780 |
| MobilenetV2 | 0.889 | 0.874 | 0.882 | 0.687 | 0.923 | 0.843 |
| Shufflenet | 0.796 | 0.883 | 0.837 | 0.679 | 0.927 | 0.839 |
| Darknet53 | 0.777 | 0.937 | 0.849 | 0.692 | 0.943 | 0.845 |
| EfficientnetB0 | 0.9427 | 0.8246 | 0.8797 | 0.7598 | 0.9659 | 0.879 |
Performance of the proposed method is indicated in bold
We selected KSVM as the classifier empirically, after experimenting with other major classifiers. The results attained using various major classifiers along with the proposed ensemble of CNNs are shown in Table 5. Even though all the classifiers produced statistically significant results at , their performance was considerably less compared to KSVM.
Table 5.
Performance of different classifiers when used along with the proposed ensemble of CNNs
| Classifier | PPV | Sensitivity | F-Score | Kappa score | AUROC | Accuracy |
|---|---|---|---|---|---|---|
| KSVM | 0.904 | 0.917 | 0.91 | 0.8305 | 0.963 | 0.916 |
| AdaBoostM1 | 0.691 | 0.691 | 0.691 | 0.4185 | 0.79 | 0.710 |
| Random Forest | 0.805 | 0.768 | 0.786 | 0.6059 | 0.89 | 0.804 |
| Naive Bayes | 0.729 | 0.639 | 0.681 | 0.433 | 0.801 | 0.7198 |
| KNN | 0.71 | 0.756 | 0.732 | 0.4825 | 0.7776 | 0.741 |
| Bayesnet | 0.726 | 0.668 | 0.696 | 0.4481 | 0.808 | 0. 727 |
Results achieved using KSVM are indicated in bold
Majority of the state-of-the-art techniques have used the public dataset used by the proposed method. Few other methods have used private datasets acquired from various hospitals. The results of different state-of-the-art methods are compared in Table 6. Methods by He et al. [13], Anwar and Zakir [3], Sakagianni et al. [21], Polsinelli et al. [18] and Yener and Oktay [29] employed the public dataset used in the proposed method. The proposed method and the method by Anwar and Zakir [3] used cross-validation, whereas the other methods which used the public dataset validated the performance using hold-out data. The proposed method achieved better results than the other methods that used the same dataset. The methods by Wu et al. [27], Pu et al. [19], Xu et al. [28], Mei et al. [15], Wang et al. [26], Di et al. [8], Gao et al. [11], Zhang et al. [32] and Ouyang et al. [16] were validated on other datasets which are not publicly available.
Table 6.
Performance achieved by different methods
| Method | Dataset | PPV | Sensitivity | F-Score | Kappa score | AUROC | Accuracy |
|---|---|---|---|---|---|---|---|
| Proposed | He et al. [33] | 0.904 | 0.917 | 0.91 | 0.8305 | 0.963 | 0.916 |
| He et al. [13] | He et al. [33] | – | – | 0.85 | – | 0.94 | 0.86 |
| Sakagianni et al. [21] | He et al. [33] | 0.8831 | 0.8831 | – | – | – | – |
| Anwar and Zakir [3] | He et al. [33] | – | – | – | – | 0.90 | 0.90 |
| Wu et al. [27] | Private | – | 0.811 | – | – | 0.819 | 0.76 |
| Pu et al. [19] | Private | – | - | – | – | 0.70 | – |
| Xu et al. [28] | Private | 0.813 | 0.867 | 0.839 | – | – | 0.867 |
| Mei et al. [15] | Private | – | 0.843 | – | – | 0.92 | – |
| Polsinelli et al. [18] | He et al. [33] | 0.8501 | 0.8755 | 0.8620 | – | – | 0.8503 |
| Wang et al. [26] | Private | – | 0.974 | – | – | 0.991 | – |
| Di et al. [8] | Private | 0.9065 | 0.9327 | – | – | – | 0.8979 |
| Gao et al. [11] | Private | – | 0.8914 | – | – | 0.9755 | 0.9599 |
| Zhang et al. [32] | Private | – | 0.9444 | – | – | – | 0.9403 |
| Yener and Oktay [29] | Kaggle, He et al. [33] | 0.90 | 0.94 | 0.92 | – | 0.91 | 0.91 |
| Ouyang et al. [16] | Private | – | 0.869 | 0.820 | – | 0.944 | 0.875 |
Performance attained by the proposed method (ensemble of CNNs+KSVM) is shown in bold
Conclusion
The experimental analysis performed in this work shows the effectiveness of ensemble of features extracted using MobilenetV2, Shufflenet, Xception, Darknet53 and EfficientnetB0 in predicting COVID-19 from CT scans. Features extracted using the aforementioned CNNs in combination with KSVM classifier achieved significant performance in the classification of COVID-19 vs. non-COVID-19. There can be combinations of CNNs that could provide better results. We leave it to the readers to experiment with more combinations of pre-trained CNNs to come up with improved results. A multi-class classification of CT scans into COVID-19, normal, viral pneumonia and bacterial pneumonia using an ensemble of CNNs is proposed as future research work.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Bejoy Abraham, Email: bjoyabraham@gmail.com.
Madhu S. Nair, Email: madhu_s_nair2001@yahoo.com
References
- 1.Abraham B, Nair MS. Computer-aided grading of prostate cancer from mri images using convolutional neural networks. J. Intell. Fuzzy Syst. 2019;36(3):2015–2024. doi: 10.3233/JIFS-169913. [DOI] [Google Scholar]
- 2.Abraham B, Nair MS. Computer-aided detection of Covid-19 from x-ray images using multi-cnn and bayesnet classifier. Biocybern. Biomed. Eng. 2020;40(4):1436–1445. doi: 10.1016/j.bbe.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anwar, T., Zakir, S.: Deep learning based diagnosis of Covid-19 using chest ct-scan images. TechRxiv (2020)
- 4.Bernheim, A., Mei, X., Huang, M., Yang, Y., Fayad, ZA., Zhang, N., Diao, K., Lin, B., Zhu, X., Li, K., et al.: Chest ct findings in coronavirus disease-19 (Covid-19): relationship to duration of infection. Radiology 200463 (2020) [DOI] [PMC free article] [PubMed]
- 5.Chang CC, Lin CJ. Libsvm: a library for support vector machines. ACM Trans. Intell. Syst. Technol. (TIST) 2011;2(3):1–27. doi: 10.1145/1961189.1961199. [DOI] [Google Scholar]
- 6.Chollet, F.: Xception: Deep learning with depthwise separable convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 1251–1258 (2017)
- 7.Deng, J., Dong, W., Socher, R., L,i LJ., Li, K., Fei-Fei, L.: Imagenet: a large-scale hierarchical image database. In: 2009 IEEE conference on computer vision and pattern recognition, Ieee, 248–255 (2009)
- 8.Di D, Shi F, Yan F, Xia L, Mo Z, Ding Z, Shan F, Song B, Li S, Wei Y, et al. Hypergraph learning for identification of Covid-19 with ct imaging. Med. Image Anal. 2020;68(101):910. doi: 10.1016/j.media.2020.101910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fakhry, A., Jiang, X., Xiao, J., Chaudhari, G., Han, A., Khanzada, A.: Virufy: a multi-branch deep learning network for automated detection of Covid-19. (2021) arXiv preprint arXiv:2103.01806
- 10.Fang Y, Zhang H, Xie J, Lin M, Ying L, Pang P, Ji W. Sensitivity of chest ct for Covid-19: comparison to rt-pcr. Radiology. 2020;296:E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao K, Su J, Jiang Z, Zeng LL, Feng Z, Shen H, Rong P, Xu X, Qin J, Yang Y, et al. Dual-branch combination network (dcn): towards accurate diagnosis and lesion segmentation of Covid-19 using ct images. Med. Image Anal. 2020;67(101):836. doi: 10.1016/j.media.2020.101836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gazda, M., Gazda, J., Plavka, J., Drotar, P.: Self-supervised deep convolutional neural network for chest x-ray classification. (2021) arXiv preprint arXiv:2103.03055
- 13.He, X., Yang, X., Zhang, S., Zhao, J., Zhang, Y., Xing, E., Xie, P.: Sample-efficient deep learning for Covid-19 diagnosis based on ct scans. (2020) medRxiv
- 14.Infante M, Lutman R, Imparato S, Di Rocco M, Ceresoli G, Torri V, Morenghi E, Minuti F, Cavuto S, Bottoni E, et al. Differential diagnosis and management of focal ground-glass opacities. Eur. Respir. J. 2009;33(4):821–827. doi: 10.1183/09031936.00047908. [DOI] [PubMed] [Google Scholar]
- 15.Mei X, Lee HC, Diao KY, Huang M, Lin B, Liu C, Xie Z, Ma Y, Robson PM, Chung M, et al. Artificial intelligence-enabled rapid diagnosis of patients with Covid-19. Nat. Med. 2020;26(8):1224–1228. doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ouyang X, Huo J, Xia L, Shan F, Liu J, Mo Z, Yan F, Ding Z, Yang Q, Song B, et al. Dual-sampling attention network for diagnosis of Covid-19 from community acquired pneumonia. IEEE Transa. Med. Imaging. 2020;38:2595–2605. doi: 10.1109/TMI.2020.2995508. [DOI] [PubMed] [Google Scholar]
- 17.Perc M, Gorišek Miksić N, Slavinec M, Stožer A. Forecasting Covid-19. Front. Phys. 2020;8:127. doi: 10.3389/fphy.2020.00127. [DOI] [Google Scholar]
- 18.Polsinelli, M., Cinque, L., Placidi, G.: A light cnn for detecting Covid-19 from ct scans of the chest. (2020) arXiv preprint arXiv:2004.12837 [DOI] [PMC free article] [PubMed]
- 19.Pu J, Leader J, Bandos A, Shi J, Du P, Yu J, Yang B, Ke S, Guo Y, Field JB, et al. Any unique image biomarkers associated with Covid-19? Eur. Radiol. 2020;30:6221–6227. doi: 10.1007/s00330-020-06956-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Redmon, J.: Darknet: open source neural networks in c (2013)
- 21.Sakagianni A, Feretzakis G, Kalles D, Koufopoulou C, Kaldis V. Setting up an easy-to-use machine learning pipeline for medical decision support: case study for Covid-19 diagnosis based on deep learning with ct scans. Stud. Health Technol. Inf. 2020;272:13–16. doi: 10.3233/SHTI200481. [DOI] [PubMed] [Google Scholar]
- 22.Sandler, M., Howard, A., Zhu, M., Zhmoginov, A., Chen, LC.: Mobilenetv2: inverted residuals and linear bottlenecks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 4510–4520 (2018)
- 23.Sharif Razavian, A., Azizpour, H., Sullivan, J., Carlsson, S.: Cnn features off-the-shelf: an astounding baseline for recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition workshops, 806–813 (2014)
- 24.Tan, M., Le, Q.: Efficientnet: rethinking model scaling for convolutional neural networks. In: International Conference on Machine Learning, PMLR, 6105–6114 (2019)
- 25.Vapnik V, Guyon I, Hastie T. Support vector machines. Mach. Learn. 1995;20(3):273–297. [Google Scholar]
- 26.Wang, B., Jin, S., Yan, Q., Xu, H., Luo, C, Wei, L., Zhao, W., Hou, X., Ma, W., Xu, Z., et al.: Ai-assisted ct imaging analysis for Covid-19 screening: building and deploying a medical ai system. Appl. Soft Comput. 106897 (2020) [DOI] [PMC free article] [PubMed]
- 27.Wu X, Hui H, Niu M, Li L, Wang L, He B, Yang X, Li L, Li H, Tian J, et al. Deep learning-based multi-view fusion model for screening 2019 novel coronavirus pneumonia: a multicentre study. Eur. J. Radiol. 2020;128:109041. doi: 10.1016/j.ejrad.2020.109041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu X, Jiang X, Ma C, Du P, Li X, Lv S, Yu L, Ni Q, Chen Y, Su J, et al. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering. 2020;6:1122–1129. doi: 10.1016/j.eng.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yener, FM., Oktay, AB.: Diagnosis of Covid-19 with a deep learning approach on chest ct slices. In: 2020 Medical Technologies Congress (TIPTEKNO), IEEE, 1–4 (2020)
- 30.Zhang, J., Xie, Y., Zhang, P., Chen, H., Xia, Y., Shen, C.: Light-weight hybrid convolutional network for liver tumor segmentation. In: IJCAI, 4271–4277 (2019)
- 31.Zhang, X., Zhou, X., Lin, M., Sun, J.: Shufflenet: an extremely efficient convolutional neural network for mobile devices. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 6848–6856 (2018)
- 32.Zhang, Y.D., Satapathy, S.C., Zhu, L.Y., Górriz, J.M., Wang, S.H.: A seven-layer convolutional neural network for chest ct based Covid-19 diagnosis using stochastic pooling. IEEE Sens. J. (2020) [DOI] [PMC free article] [PubMed]
- 33.Zhao, J., Zhang, Y., He, X., Xie, P.: COVID-CT-Dataset: a CT scan dataset about COVID-19. (2020) arXiv preprint arXiv:2003.13865