Important Compound Classes
Title
P300/CBP HAT Inhibitors and Methods for their Use
Patent Publication Number
WO 2020/176558 A1
Publication Date
September 3, 2020
Priority Application
US 62/811,143 and US 62/879,852
Priority Date
February 27, 2019 and July 29, 2019
Inventors
Wilson, J. E.
Assignee Company
Constellation Pharmaceuticals Inc., USA
Disease Area
Cancer, cardiac diseases, metabolic diseases, fibrotic diseases, inflammatory diseases, viral infections, and neurological disorders
Biological Target
Histone acetyltransferase (HAT)
Summary
Chromatin is a complex combination of DNA and protein that makes up chromosomes. It is found inside the nuclei of eukaryotic cells and is divided between heterochromatin (condensed) and euchromatin (extended) forms. The major components or chromatin are DNA and proteins. Histones are the chief protein components of chromatin, acting as spools around which DNA winds. The functions of chromatin are to package DNA into a smaller volume to fit in the cell to strengthen the DNA to allow mitosis and meiosis and to serve as a mechanism to control expression and DNA replication. The chromatin structure is controlled by a series of post-translational modifications to histone proteins, notably histones H3 and H4, and most commonly within the “histone tails” which extend beyond the core nucleosome structure. Histone tails tend to be free for protein–protein interaction and are also the portion of the histone most prone to post-translational modification. These modifications include acetylation, methylation, phosphorylation, ubiquitinylation, and SUMOylation. These epigenetic marks are written and erased by specific enzymes that place the tags on specific residues within the histone tail, thereby forming an epigenetic code, which is then interpreted by the cell to allow gene specific regulation of chromatin structure and thereby transcription.
Covalent modification of histones is a fundamental mechanism of control of gene expression and is one of the major epigenetic mechanisms at play in eukaryotic cells. Distinct classes of enzymes, namely, histone acetyltransferases (HATs) and histone deacetylases (HDACs), acetylate or deacetylate specific histone lysine residues.
Histone acetyltransferases (HATs) catalyze the acetylation (transfer of an acetyl group) on a ε-amino group of a target lysine side chain within a substrate histone and HDACs catalyze the removal of acetyl group from lysine residues. Subsequently, acetylated core histones were shown to preferentially associate with transcriptionally active chromatin. HATs are categorized into four major families based on primary sequence homology, shared structural features and functional roles: Gen5/PCAF (General control nonrepressed protein 5 and p300 and CBP associated factor); MYST (named for founding members MOZ, Ybf2/Sas3, Sas2, and Tip60); p300/CBP (protein of 300 kDa and CREB Binding Protein); and Rtt109 (Regulator of Tyl Transposition gene production 109).
The present application describes a series of novel histone acetyltransferase (HAT) inhibitors for the treatment of diseases including, cancer, cardiac diseases, metabolic diseases, fibrotic diseases, inflammatory diseases, viral infections, and neurological disorders. Further, the application discloses compounds, their preparation, use, pharmaceutical composition, and treatment.
Definitions
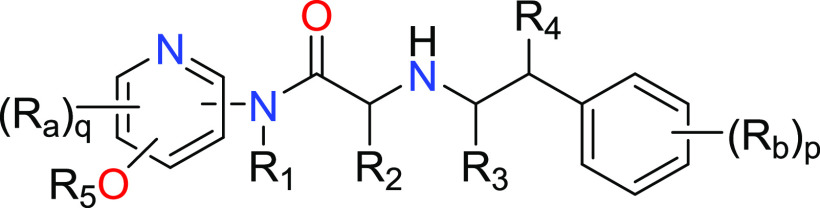
R1, R3 and R4 = H or C1–4alkyl;
R2 = phenyl or 5- to 6-membered heteroaryl, each of which is optionally substituted with 1 to 3 groups selected from Rc;
R5 = C1-6alkyl substituted with 4- to 6-membered heterocyclyl or 5- to 6-membered heteroaryl, wherein each of said 4- to 6-membered heterocyclyl or 5- to 6-membered heteroaryl are optionally substituted with 1 to 3 groups selected from Rd;
Ra, Rb, Rc and Rd = halo, CN, oxo, NO2, C1–6alkyl, C2–6alkenyl, C1–6alkoxy, halo(C1–6alkoxy), halo(C1–6alkyl), −C1–6alkylORe, −C(O)Rf, −C(O)OR, −C1–6alkylC(O)ORe, −C(O)N(Re)2, −C(O)NReC1–6alkylORe, −OC1–6alkylN(Re)2, −C1–6alkylC(O)N(Re)2, −C1–6alkylN(Re)2, −N(Re)2, −C(O)NReC1–6alkylN(Re)2, −NReC1–6alkylN(Re)2, −NReC1–6alkylORe, −SORe, −S(O)2Re, −SON(Re)2, −SO2N(Re)2, −O(C3-C6)cycloalkyl, −O–C1–4alkylaryl, −C1–6alkyl(C3–6)cycloalkyl, −C1–6alkylaryl, −-C1–6alkylheteroaryl, −C1–6alkylhetercyclyl, (C3–C6)cycloalkyl, heterocyclyl, heteroaryl or aryl;
q = 0, 1, or 2; and p = 0, 1, 2, or 3.
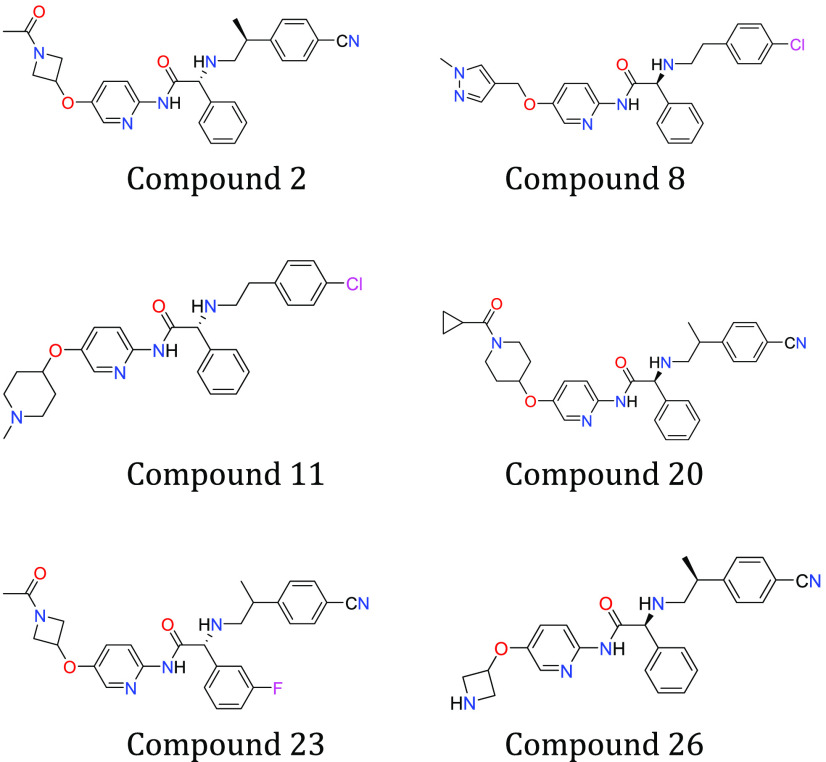
Key Structures
Biological Assay
The p300/CBP HAT scintillation proximity assay (SPA) was performed. The compounds described in this application were tested for their ability to inhibit HAT. The HAT IC50 (μM) are shown in the following Table.
Biological Data
The table below shows representative
compounds were tested for HAT inhibition. The biological data obtained
from testing representative examples are listed in the following table.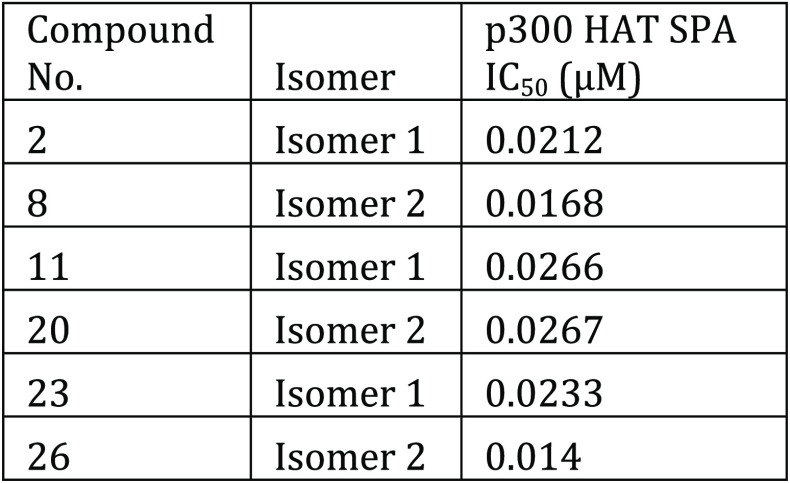
Claims
Total claims: 44
Compound claims: 25
Pharmaceutical composition claims: 17
Method of treatment claims: 1
Use of compound claims: 1
Recent Review Articles
-
1.
He Z.; Wei B.; Zhang X.; Gong Y.; Ma L.; Zhao W.. Eur. J. Med. Chem. 2021, 209, 112861.
-
2.
Gomathi K.; Akshaya N.; Srinaath N.; Rohini M.; Selvamurugan N.. Int. J. Biol. Macromol. 2021, 170, 326.
-
3.
Wang Y.; Xie Q.; Tan H.; Liao M.; Zhu S.; Zheng L.; Huang H.; Liu B.. Pharmacol. Res. 2021, in press.
-
4.
Burns A. M.; Graff J.. Curr. Opin. Neurobiol. 2021, 67, 75.
-
5.
Fioravanti R.; Mautone N.; Rovere A.; Rotili D.; Mai A.. Curr. Opin. Chem. Biol. 2020, 57, 65.
The author declares no competing financial interest.
Special Issue
Published as part of the ACS Medicinal Chemistry Letters special issue “Epigenetics 2022”.