Summary
Human rhinoviruses (RVs) are the primary aetiological agent of the common cold. Generally, the associated infection is mild and self‐limiting, but may also be associated with bronchiolitis in infants, pneumonia in the immunocompromised and exacerbation in patients with pulmonary conditions such as asthma or chronic obstructive pulmonary disease. Viral infection accounts for as many as two thirds of asthma exacerbations in children and more than half in adults. Allergy and asthma are major risk factors for more frequent and severe RV‐related illnesses. The prevalence of RV‐induced wheezing will likely continue to increase given that asthma affects a significant proportion of the population, with allergic asthma accounting for the majority. Several new respiratory viruses and their subgroups have been discovered, with various degrees of relevance. This review will focus on RV infection in the context of the epidemiologic evidence, genetic variability, pathobiology, clinical studies in the context of asthma, differences with other viruses including COVID‐19 and current treatment interventions.
Keywords: asthma exacerbations, influenza, rhinovirus, SARS‐Cov‐2
Abbreviations
- CDHR3
cadherin‐related family member 3
- CI
confidence interval
- COAST
Childhood Origins of Asthma
- COPD
chronic obstructive pulmonary disease
- COVID‐19
coronavirus disease 2019
- HIV
human immunodeficiency virus
- ICAM‐1
intercellular adhesion molecule 1
- ICS
inhaled corticosteroid
- IFN
interferon
- IL
interleukin
- LABA
long active beta agonist
- LDL
low‐density lipoprotein
- MRCA
Most Recent Common Ancestor
- Ne
effective population size
- OR
odds ratio
- PCR
polymerase chain reaction
- RSV
respiratory syncytial virus
- RV
rhinovirus
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
1. INTRODUCTION
The respiratory viruses that have been linked with asthma exacerbations include rhinovirus (RV), respiratory syncytial virus (RSV), enterovirus, influenza A and B, parainfluenza virus, adenovirus and coronavirus.1, 2, 3, 4, 5 It is relevant to distinguish the type of respiratory infection considering the ongoing concerns with the COVID‐19 virus pandemic. Common human coronaviruses—not to be confused with the novel coronavirus, or SARS‐CoV‐2, currently circulating—can cause mild to moderate upper‐respiratory tract illnesses, like the common cold. In fact, the majority of people will get infected with one or more of these viruses at some point in their lives; four common human coronaviruses cause 15%–30% of common colds.
Human RVs are small (27 nm), non‐enveloped, ssRNA viruses of the Picornaviridae family genus Enterovirus. The prevalence and spectrum of viral‐triggered exacerbations vary according to patient age and seasonality. Viral infection accounts for as many as 80% of exacerbations in children6 and more than 50% in adults.2 The prevalence of RV‐induced wheezing will likely continue to increase considering the prevalence of asthma, with an allergic component accounting for the majority of cases.7 It is important to emphasize that respiratory viruses act as triggers of wheezing. Despite improvements in asthma management and advances in therapeutics, the reported incidence of asthma exacerbations has not declined. Data from controlled clinical trials indicate that the development of asthma exacerbations in children as well as in adults are predictive of future exacerbations.2, 7, 8, 9, 10 Therefore, strategies enabling the management and control of viral‐induced events represent a priority to counter exacerbations linked to viral disease.
2. EPIDEMIOLOGY AND SEASONALITY
Children may be infected with RVs from 8 to 12 times per year, while adults may be infected 2–3 times per year, with peaks of infection observed throughout the year.11 While mild and self‐limiting in immunocompetent hosts, RV infection is associated with bronchiolitis in infants, pneumonia in the immunosuppressed and exacerbation of pre‐existing pulmonary conditions such as asthma or chronic obstructive pulmonary disease (COPD).12, 13 Bronchiolitis, acute wheezing illnesses and asthma are major clinical management challenges representing an unmet medical need.
RSV is the primary cause of bronchiolitis in infants less than 6 months of age. RV becomes more common later in infancy and is a much more common cause of wheezing in the second and third year of life than RSV.3, 4 The reasons for age‐specific manifestations and outcomes are poorly understood and may involve complex interactions between the host and intrinsic pathogenicity of the virus. Data have demonstrated that transient wheezing (from infancy up to age 3 years) may be linked to RSV infection.8 In addition to asthma exacerbations, severe respiratory illness induced by RSV or RV has been associated with subsequent development of asthma. In fact, RSV‐induced wheezing during infancy may affect respiratory health for years.14 There is evidence that RSV‐induced bronchiolitis can damage the airways and promote airway obstruction with recurrent wheezing.8, 14 While RV likely causes less structural damage, it remains a significant contributor to wheezing illnesses in young children. Because RV infections are common and a major cause of exacerbations in paediatric and adult patients with lung disease, interactions between viral virulence factors, personal risk factors (e.g., atopy, genetic susceptibility and age) and environmental exposures (e.g., allergen exposure and seasonality) promote more severe wheezing illnesses and the risk for progression to asthma.4, 5, 6 The prevalence of bronchiolitis is approximately 20%–30% in the first year and 10%–20% in the second year of life.15 Up to 50% of children have acute wheezing at least once before school age. Of these, about 35% will have recurrent wheezing. Once asthma is established, exposure to allergens and RV, with a potential synergistic effect, are important triggers of asthma exacerbation.16
A seasonal pattern of paediatric asthma exacerbation is well established.17, 18, 19 In particular, a peak in asthma exacerbations and related hospitalizations occurring in September has been observed in children in the Northern Hemisphere. There is considerable evidence to support a causal link between viral respiratory tract infection and asthma exacerbation in children, with respiratory viruses detected up to 80% of paediatric patients who experience asthma exacerbation.20, 21 A retrospective cohort study by Suruki et al. using US healthcare claims data reported the frequency and type of exacerbation in 734,114 paediatric patients with asthma.22 The investigators analysed the annual frequency of and seasonal trends for exacerbation in real‐world clinical practice. The mean annual exacerbation frequency was 1.4; 86% of these exacerbations were defined by systemic corticosteroid use. A consistent trend of increased exacerbation incidence in the fall and early winter was observed. In addition, a high proportion of asthma‐related hospitalizations occurred in patients of a younger age. This study further supports the epidemiological association of seasonal exacerbations linked to viral exposures that tend to occur between fall and winter. Figure 1 represents a simulated pattern of the expected exacerbation events based on prior reports.17, 19, 20, 21, 22
FIGURE 1.
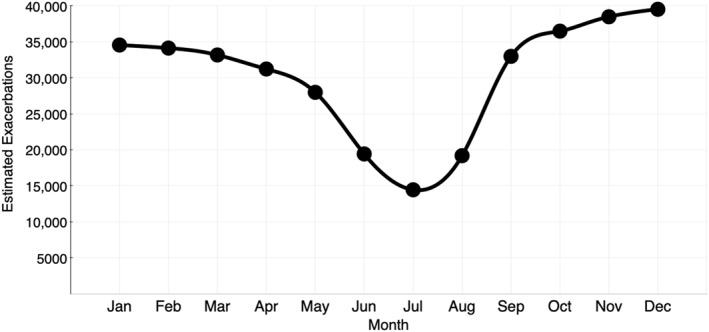
Estimated seasonality pattern related to asthma exacerbations by month. Expected seasonal increase in exacerbations in the fall and early winter compared with the summer months
3. RV STRAINS
More than 160 strains of RV have been identified and classified into three genetic clades (A, B and C) according to sequence similarity, including 80 RV‐A, 32 RV‐B and 65 RV‐C genotypes23, 24 while cross‐protection appears to be limited. Molecular epidemiologic studies suggest that the dominant species are RV‐A and RV‐C, while RV‐B is rarely detected. RV‐A and RV‐C are not only associated with wheezing illnesses in early childhood, but also these viruses are more often associated with exacerbations of asthma compared with RV‐B. RV‐C might be more strongly associated with more severe exacerbations, including those requiring hospitalization.25, 26 This could be due to faster replication rate and induction of more robust cellular responses, based on data from cultures of differentiated airway epithelial cells. In cohort studies, RV‐B infections do not increase the risk for exacerbations, but they might slightly increase the risk of exacerbation in children with severe asthma.
Competition assays for cellular binding sites have further grouped RVs into major or minor group viruses depending on the use of intercellular adhesion molecule 1 (ICAM‐1), low‐density lipoprotein (LDL) and cadherin‐related family member 3 (CDHR3) as receptors.24 While these advances in the understanding of the virus are encouraging, the unique structural and genetic variability of RVs has inhibited efforts to develop effective therapies.
4. GENETIC VARIATION
To build a context of the genetic variation of RV, it is important to understand how it fits in the world of genetic variation among other viruses. For example, the human immunodeficiency virus (HIV)27 exhibits one of the fastest evolving genomes ever observed.28 Within a single patient, the envelope gene evolves at an astounding rate of 1% per year.29 After approximately 4 years of within‐patient evolution, the viral population no longer has coalescent events that reach back to the infection time frame; this means that none of the nodes that existed near the time of infection are present. Serially sampled populations of viruses tend to cluster temporally, giving rise to a ‘ladder’ like phylogeny.29 Interestingly, among patients with influenza phylogenetic trees are ladder like as well.30, 31 This means that early sequences sampled in a particular calendar time tend to cluster together with a Most Recent Common Ancestor (MRCA), reaching back about 5–10 years. In both, the HIV within‐patient and the influenza among‐patient examples, the ladder shapes of the phylogenies are thought to be driven in part by diversified selection in the host's immune response to the virus (Figure 2). By exploiting the calendar sampling time, it is possible, with modest molecular clock assumptions, to estimate the timing of ancestral events that occur on the phylogeny.32 One of the most important demographic factors is the effective population size.33 In both cases, among patient influenza and within patient HIV, the total mutation at any one slice of time reaches back about 5 and 3 years, respectively.30, 34
FIGURE 2.
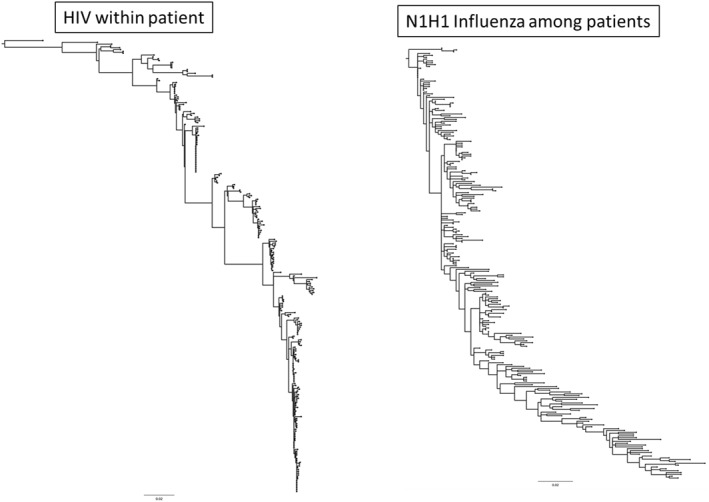
Tree pattern of changes in sequences collected overtime. The tree on the left is a set of sequences collected from a single individual approximately every 6 months who was not on effective therapy. The tree on the right is a selection of N1H1 Influenza viruses where the earliest sequences are from the 1918 Influenza Pandemic and the most recent are from the 2019 to 2020 flu season. Note that both tree shapes appear to shift from one population to the next suggesting that the effective population size at any one time is relatively small. Each of these trees are rooted by the earliest time points
The concept of effective population size (or Ne) is central to population genetics. Recombination events can drive the appearance of extremely high Ne in a phylogenetic context (Figure 3).35 It measures the time that a population can reach into the past with current variation. Thus, the time to the MRCA is directly proportional to the amount of genetic diversity with high diversity reaching further back in evolutionary history than low genetic variation. Unlike, influenza or within patient HIV, RV has deep evolutionary branches (Figure 3). The amount of genetic variation circulating at any one time is enormous and can reach back hundreds of years rather than just a few. The current circulating strains of RV‐A comprise an effective population size nearly two orders of magnitude larger than that of the 2019–2022 influenza H1N1 season and what can be found about 5 years post‐HIV infection without effective therapy. This suggests that at any given time there is very large pool of RV‐infected individuals unlike influenza where there is a yearly bottleneck before the next seasonal outbreak that drives variation from the population. Although RV is seasonal, it must maintain its genetic variation by a continuously infecting many people throughout time.
FIGURE 3.
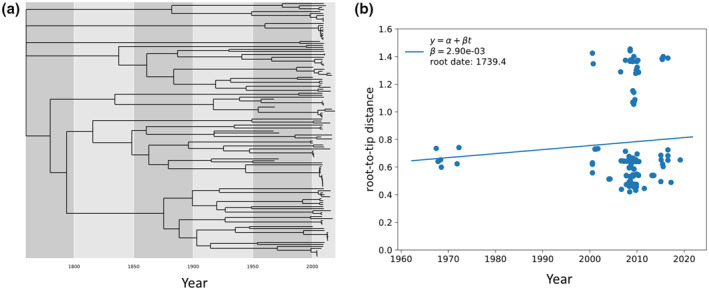
Evolutionary rate based off dated tips of rhinovirus A with an estimated divergence rate. Rhinovirus A has very deep evolutionary nodes, indicating that the population has a very high Ne. (a) Dated tip phylogeny can be used to estimate the dates of all the nodes on a tree including the date of the Most Recent Common Ancestor (MRCA). (b) These dates can be used to assess the maximum likelihood estimate of the divergence rate (2.90e‐03). Importantly, recombination events can drive the appearance of extremely high Ne in a phylogenetic context
There is a relationship between Ne and the difficulty in developing a vaccine, such that, the larger the Ne the harder it is to generate a durable immune response with a vaccine. As such, some of the challenges in developing a RV vaccine are not simply due to the mutation rate. Influenza has a very similar mutation rate, yet yearly vaccines are possible because the Ne is sufficiently small. However, RV vaccine efforts do not enjoy these low Ne and thus vaccine efforts have largely failed in part due to the vast variation of the current circulating strains through human populations.
The plasticity of viral genomes allows for the generation of enormous numbers of viable mutants, resulting in circulating sequences that can differ by more than 30% in the maximally variable genes of viruses. Since the genetic diversity of viruses, RV in particular, will continue to increase, it is critical to understand the genetic variation in a phylogenetic sense in an effort to develop effective antiviral or vaccine strategies.
5. MECHANISMS AND PATHOBIOLOGY
RV is transmitted mainly through direct contact with aerosolized particles and replicate in ciliated epithelial cells of the upper airways and in medium‐to large‐sized lower airways.36 Viruses attach to unique cellular receptors: ICAM‐1 used by RV‐B and most RV‐As, LDL receptor used by some RV‐As, and cadherin‐related family member 3 (CDHR3) used by RV‐C.37 After RV attachment, infected cells recognize RV pathogen‐associated molecular patterns through interaction with two different families of pattern recognition receptors: Toll‐like receptor (TLR) 2, TLR3, TLR7 and TLR8 and retinoic acid‐inducible gene I‐like (RIG‐1) receptors.38, 39 These receptors activate transcription factors (e.g., interferon regulatory transcription factor 7 and nuclear factor kB) that promote the expression of type I and type III interferons and several inflammatory cytokine genes.40 Early innate immune responses, such as type 1 interferon release, occur rapidly after infection of the epithelium. RV induce production of cytokines (IL‐1, IFN‐γ, TNF, IL‐6, IL‐12 and IL‐18), chemokines (CCL3, CCL5, CXCL8 and CXCL10), and growth factors that attract and activate and granulocytes, dendritic cells and monocytes at the site of infection.40 , 41 Figure 4 illustrates the combined effects of the virus and the inflammatory response leading to epithelial damage and sloughing, mucus production and ultimately airway obstruction.42, 43, 44 There is evidence that viruses and bacteria interact in patients with respiratory illnesses; viral infection may be associated with transient detection of common bacterial pathogens such as Moraxella catarrhalis, Streptococcus pneumoniae and Haemophlius influenzae.45
FIGURE 4.
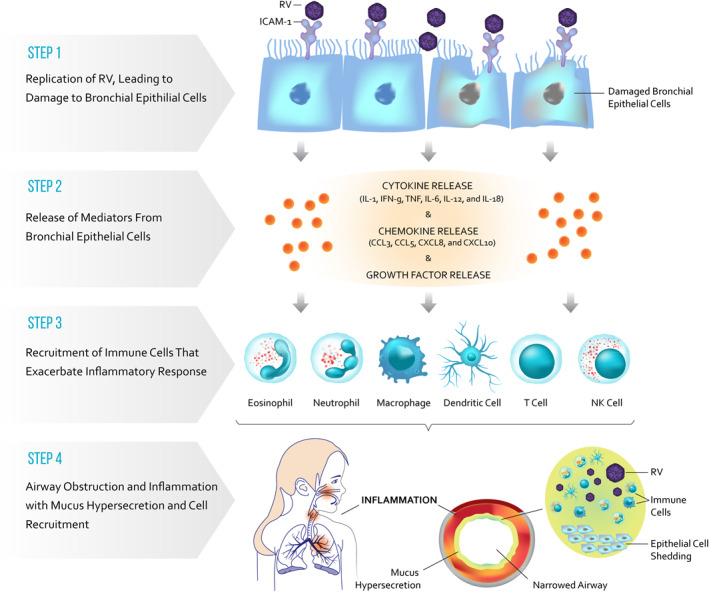
Inflammatory response following viral infection. Rhinovirus (RV) is transmitted mainly through direct contact and aerosolized particles and replicates in ciliated epithelial cells of the upper and lower airways. The viruses attach to unique cellular receptors. After attachment, infected cells recognize RV pathogen‐associated molecular patterns through interaction with two different families of pattern recognition receptors, that is, Toll‐like receptors. These receptors activate transcription factors that promote the expression of type I and type III interferons and several inflammatory cytokine genes. Early innate immune responses, such as type I interferons, occur rapidly after infection. RV induce cytokines, chemokines and growth factors that activate and attract granulocytes, dendritic cells and monocytes at the site of infection and trigger an inflammatory response and induce an asthma exacerbation
Disrupted airway epithelium favours RV replication by allowing access to deeper layers in tissue in which RV replicates most actively and by increasing the number of ICAM1 receptors as shown in in vitro studies.46 The damaged barrier function of the airway epithelium can also enhance engagement of aeroallergens or bacterial pathogens through the airway wall.47 RVs also may contribute to airway remodelling by inducing vascular endothelial growth factor, TGF‐β, and other mediators into airway smooth muscle cells.41, 48 It is possible that these effects are more pronounced in early life.6, 10, 14, 49 Thus, repeated RV infections that extend to the lower airways may cause damage that subsequently leads to remodelling of the airways.
Eosinophils exert prominent cytotoxic properties that damage the respiratory mucosa and attenuate lung function during stable asthma and during exacerbation. Using an experimental human exposure model of mild asthma, Sabogal Piñeros et al. reported that RV16‐inoculation induced loss of asthma control with a strong correlation with CD69 expression by eosinophils.50 Interestingly, eosinophils from patients with asthma displayed a reduced capacity to bind the virus, suggesting that human eosinophils may be important scavengers of virus in the respiratory mucosa, preventing viral propagation. More recently, a study51 in children with asthma using peripheral blood mononuclear cells stimulated with peptide formulations to induce species‐specific responses to RV‐A and RV‐C showed that responses to RV‐A have higher expression of IFNγ and STAT1 compared with RV‐C, and significant expression of CXCL9, 10 and 11 was not found for RV‐C. In contrast, RV‐C induced higher expression of CCL24 (eotaxin‐2) than RV‐A in the responses of children with and without asthma. Upstream regulator analysis showed both RV‐A and RV‐C induced predominant Th1 and inflammatory cytokine expression. The responses of children with asthma compared with those without asthma were lower for both RV‐A and RV‐C while retaining the pattern of gene expression and upstream regulators characteristic of each species. Notably, all groups showed activation of the IL‐17A pathway. Overall, the study showed that RV‐A and RV‐C induce qualitatively different T‐cell responses providing a possible mechanism to explain why RV‐C tends to be associated with more severe symptoms of infection and asthma exacerbations.
6. CLINICAL STUDIES
Most RV infections cause common cold symptoms with damage to the epithelial barrier integrity, causing an increased translocation of pathogens and complications of respiratory diseases. Up to 35% of asymptomatic subjects may test positive for RV, but the virus does not cause chronic infection or colonization in healthy subjects.11 However, both symptomatic and asymptomatic infection can induce systemic immune responses in young wheezing children. The Childhood Origins of Asthma study demonstrated that the risk for development of asthma by age 6 years was increased if children had wheezing with RV (odds ratio [OR], 9.8; 95% CI, 4.3, 22.0) during the first 3 years of life. Notably, 88% of children with RV‐induced wheezing in the third year of life had asthma by age 6 years.6, 52 Although RV‐induced wheezing was an independent asthma risk factor, allergen sensitization significantly increases the RV‐associated risk of asthma.5, 6, 7
A study by Tan et al. reported a viral detection rate of 59% in patients treated for life‐threatening asthma in the intensive care unit.9 This study evaluated the prevalence and spectrum of respiratory viruses in adult patients hospitalized for life‐threatening asthma, severe asthma and COPD. RV was the most common virus in near‐fatal severe attacks, and coinfection with adenovirus was detected. These data highlight the importance of viral infection in the burden of respiratory disease‐related morbidity and mortality.
Prazma et al. investigated the frequency of asthma exacerbation and respiratory tract infection occurring during the fall season in a paediatric population using an at‐home mucus collection methodology during a 16‐week, randomized, double‐blind study.52 Children, 4–11 years of age with a clinical diagnosis of asthma treated with inhaled corticosteroids (ICS), a morning peak expiratory flow ≥70% predicted, and a history of ≥1 asthma exacerbation during the previous respiratory viral season were eligible for enrolment. Mucus samples obtained during symptomatic periods were analysed for common respiratory viruses by multiplex polymerase chain reaction. Table 1 illustrates the type of viruses identified. Notably, 80% of the samples tested positive for RV. Of the 537 mucus samples collected, 64% tested positive for viruses but less than 10% of patients presented with asthma exacerbation.53 Noteworthy, all patients were treated during the study with either ICS alone or ICS/LABA combination, prior attending school. It is possible that this continuous use and adherence to treatment may have mitigated the manifestation of exacerbations. In contrast, Korean investigators demonstrated an association between RV infection and asthma exacerbations in hospitalized children [OR] 3.9 (95% CI, 1.4, 10.5); this effect was further enhanced in atopic versus non‐atopic patients, OR 8.3 (95% CI, 1.5,43.3) versus 2.4 (95% CI, 0.5, 10.7), respectively.54
TABLE 1.
Virology from mucus sample collection in children 4–11 years of age with asthma
| Samples containing virus | Total (N = 344) |
|---|---|
| Number of viruses per sample, n (%) | |
| Positive for 1 virus | 320 (93) |
| Positive for 2 viruses | 23 (7) |
| Positive for 3 viruses | 1 (<1) |
| Virus type, n (%) | |
| Rhinovirus | 276 (80) |
| Parainfluenza virus 2 | 29 (8) |
| Coronavirus NL63 | 14 (4) |
| Coronavirus OC43 | 15 (4) |
| Other: Enterovirus, bocavirus, adenovirus C, parainfluenza 4b, parainfluenza 1 and influenza B | 35 (10) |
Note: Modified from Prazma et al. Resp Med 2015.
Respiratory symptoms typically develop 1–2 days after inoculation in studies, and uncomplicated RV symptoms usually peak 2–4 days after inoculation. The median duration of RV colds is 1 week, but up to 25% last more than 2 weeks.3 During illness caused by RV, viral shedding occurs naturally for up to 21 days, but predominantly over an initial 3‐ to 4‐day period.
Recent data suggest that people who present with symptoms of respiratory illness at an emergency department, and who are subsequently diagnosed with a common respiratory virus, are in fact co‐infected with the COVID‐19 virus.55 In a single‐centre analysis, 562 individuals were tested for COVID‐19; 49 tested positive for infection with SARS‐CoV‐2. Of these 562 individuals, 517 were also tested for the presence of other common respiratory viruses; 127 tested positive for at least one other respiratory virus. Specifically, the top six viruses identified were RV/enterovirus, 46 (36.2%); influenza A, 21 (16.5%); meta‐pneumovirus 21 (16.5%); RSV, 14 (11%); coronavirus, 10 (7.8%), and parainfluenza 1, 4(3.1%).6 Of the individuals tested for both SARS‐CoV‐2 and other respiratory viruses, 11 of 49 (22.4%) confirmed COVID‐19 cases and 11 of 127 (8.7%) with other respiratory viruses—were found to be co‐infected. These findings suggest that about 1 in 5 people with SARS‐CoV‐2 are also infected with other respiratory viruses.
Importantly, common cold symptoms include stuffy nose, sneezing, sore throat, possibly cough and body aches. In contrast, SARS‐CoV‐2 symptoms include fever, dry cough, loss of taste or smell, body aches, fatigue, shortness of breath, possibly diarrhoea and vomiting. In more severe cases, the virus can cause pneumonia, severe acute respiratory distress syndrome, kidney failure and death. Thus, while there is the possibility of co‐infection and similarities exist in the presentation of these viral diseases, the symptoms can overlap. Table 2 contrasts the clinical features of the common cold, influenza and COVID‐19 infection. A recent case series of 393 consecutive confirmed COVID‐19 admissions in New York state documented a rate of asthma of 12.5%, slightly higher than the prevalence of current adult asthma (10.1%) in that state.56 This finding is consistent with another report from Ireland, where review of medical records of 193 consecutive admissions who were SARSCoV‐2‐positive found that 8.8% had a physician diagnosis of asthma which is slightly higher than the prevalence of current asthma of 7.0% in adults in Ireland.57 While these findings suggest that patients with asthma are not at a higher risk of SARS‐CoV‐2, recent data58 using air–liquid interface cultures from nasal tissues biopsied from 30 patients with asthma and infected with common RV strains (RV‐A16 and RV‐C15) suggest that RV infections are potential mechanisms of ACE2 overexpression (threefold increase). Thus, it is possible that RV infections play synergistic interactions with SARS‐CoV‐2 infection by priming the host to respond excessively to COVID‐19 infection.
TABLE 2.
Clinical features of common cold, influenza and COVID‐19
| Clinical features | Common cold | Influenza | COVID‐19 |
|---|---|---|---|
| Incubation period | 1–3 days | 1–4 days | 2–14 days |
| Symptoms | Gradual | Abrupt | Gradual |
| Fever | Sometimes | Common | Common |
| Cough | Common | Common | Common |
| Shortness of breath | Mild | Sometimes | Common |
| Fatigue | Sometimes | Common | Common |
| Body aches | Sometimes | Common | Common |
| Sore throat | Common | Sometimes | Sometimes |
| Nasal congestion | Common | Sometimes | Sometimes |
| Loss of smell | Sometimes | Sometimes | Common |
| Chills | Uncommon | Common | Sometimesa |
| Diarrhoea | Rare | Sometimes | Sometimes |
| Loss of appetite | Sometimes | Common | Sometimes |
Note: Modified from www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html.
Including repeated shaking with chills.
Muehling et al. performed a comprehensive analysis of Type 1 and Type 2 innate and adaptive responses in allergic asthmatics infected with RV.59 T cells were sampled for up to 11 weeks to capture steady state and post‐infection periods. T‐cell responses were analysed in parallel with nasal cytokines, upper and lower airway symptoms and lung function. The investigators reported that in uninfected asthmatics, higher numbers of circulating virus‐specific PD‐1+ Th1 cells, but not allergen‐specific Th2 cells, were linked to worse lung function. RV infection induced an amplified anti‐viral Th1 response in asthmatics versus controls, with allergen‐specific Th2 expansion, and production of Type 1 and 2 nasal cytokines. Notably, Th2 responses were absent in infected asthmatics who had normal lung function, and in those receiving the anti‐IgE monoclonal antibody omalizumab. Across all subjects, early induction of a minimal set of nasal cytokines that discriminated high responders included G‐CSF, IFN‐γ, TNF‐α and correlated with both egress of circulating virus‐specific Th1 cells and worse symptoms. These findings suggest that RV induces robust Th1 responses in allergic asthmatics that may promote disease, even after infection resolves.
7. TREATMENT APPROACHES
There are multiple on‐going efforts to address the unmet need in patients reactive to viral‐induced exacerbations. Here are some examples to illustrate these efforts:
Targeting ICAM‐1 in transgenic mice engineered to overexpress extracellular domains 1 and 2 of human ICAM‐1 has been shown to prevent the cellular entry of two major groups of RVs, RV16 and RV14.60 Reduced cellular inflammation, pro‐inflammatory cytokine production and virus load were also observed in this model. However, targeting and blockage of other receptors used by minor group RV, such as the LDL receptor, has been challenging.60 This is also the case in the context of the development of a universal anti‐RV antibody, which is problematic due to the antigenic diversity of circulating RV. In addition, there are significant challenges associated with the identification of new antigenic variants plus the fact that approximately 90% of RV serotypes cannot bind to the murine ICAM‐1 receptor.61 Ultimately, development of novel therapeutics that interferes with RV binding, entry and replication in the host cell could yield promising results.
In vitro studies have shown that exogenous delivery of interferons (IFN‐α, IFN‐β, IFN‐λ1 or IFN‐λ2) reduced RV1A viral copies in human primary bronchial epithelial cells (HPBECs).62 Interestingly, the addition of IFN‐β also suppressed RV16 and RV1B replication in HPBECs isolated from healthy and asthmatic individuals.63 A study by Djukanovic et al.64 evaluated the effect of inhaled IFN‐β in asthma patients at the onset of cold or flu symptoms. Although the trial did not meet its primary endpoint (asthma control), in a post‐hoc analysis the data suggest that inhaled IFN‐β is a potential treatment for virus‐induced deteriorations of asthma in difficult‐to‐treat patients. More recently Watson et al.65 reported in an in vitro model of IFN‐β the potential for intermittent prophylactic doses of exogenous IFN‐β to modulate viral infection. The findings showed that chronic dosing with IFN‐β was more effective than dosing after infection and it was not associated with induction of inflammatory mediators.
Other approaches with small molecule ‘capsid binders’ that inhibit RV‐A and RV‐B binding and replication are not effective against RV‐C because of differences in capsid structure.66 While 3C protease inhibitors are effective in vitro, results in clinical trials have been disappointing.67 The large number of antigenically distinct RV types has been a barrier to vaccine development, although new approaches have identified some degree of cross‐reactivity among RV types.65 Pleconaril, for example, binds to hydrophobic pockets within viral capsids, altering binding of the viral pathogen to host cell receptors and blocking the uncoating process. Assessment of pleconaril on the effect on RV serotypes (RV‐2, 14, 16, 39 and A21) and 46 clinical isolates using in vitro cytopathic effect inhibition assays68 reported antiviral activity against the five serotypes (median EC50 of 0.02 μg/mL) and against the majority of the untyped clinical isolates. However, pleconaril failed the first clinical trial with an oral formulation that was given three times per day when treatment started 24 hours after challenge, in that only 1–1.5 days of reduction in symptom resolution time was demonstrated).69 Furthermore, drug‐resistant RV strains were identified in 24% of enrolled patients: 13% of patients were naturally resistant at baseline and 11% exhibited a reduced susceptibility by day 5 of treatment.70, 71 A phase II study, which used an intranasal formulation of pleconaril, failed to show significant results for RV‐positive participants either with or without asthma exacerbation.70 Overall, although capsid binders are attractive and potent early stage inhibitors of RV replication in vitro, problems with pharmacodynamics, in vivo efficacy, and resistance development have been reported. In addition, with the recent resolution of the capsid structure, it was demonstrated that RV‐C species lack the hydrophobic pocket, the binding target of capsid binders.
8. CONCLUSIONS
At present there are no licensed antibodies for clinical use in RV infection, underscoring the need for alternative therapeutic strategies. Many viruses, including RV, for which vaccines are not available, produce a significant impact on public health. Many of these viral targets could be classified as ‘difficult’ based on the fact that: (i) infection is not self‐limited, is associated with a high frequency of severe disease, and often leads to persistence; (ii) the virus has developed multiple mechanisms to alter and evade host immune responses, (iii) the host can be re‐infected; (iv) there is significant genetic variation; (v) the site of infection is the same as the major target organ for disease; (vi) there is integration of the viral genome or sequestration of the virus making it less accessible to immune effectors; (vii) animal models fail to recapitulate pathogenesis of human disease and (viii) there is a long delay between initiation of infection and onset of adaptive cellular immunity.72 Today we face global challenges with emerging viral diseases for which vaccines or antibody‐specific therapies would substantially benefit the public health. In particular, the current pandemic with COVID‐19 has tremendously accelerated the understanding of the science around viral disease, including the biology, diagnostics and antibody and vaccine development technologies. There is need for primary prevention through development of immuno‐prophylaxis interventions for patients with asthma and other respiratory diseases. In addition, drug‐discovery research must be accelerated to identify effective interventions that prevent RV infections and provide more practical approaches to disease prevention. Early detection of patients at risk as well as better understanding of the interactions between viral infection and the host is paramount. Designing novel treatments or preventive strategies for virus‐induced exacerbations across respiratory diseases could provide an invaluable therapeutic avenue for addressing a current unmet medical need.
CONFLICT OF INTEREST
The authors report no conflict of interest.
AUTHOR CONTRIBUTIONS
Hector Ortega: writing original draft, reviewing and editing; David Nickle: writing, reviewing and editing; Laura Carter: reviewing and editing.
ACKNOWLEDGEMENTS
The authors would like to thank Jill Luer, PharmD for her support reviewing and editing the manuscript.
Ortega H, Nickle D, Carter L. Rhinovirus and asthma: challenges and opportunities. Rev Med Virol. 2021;31(4):e2193. 10.1002/rmv.2193
DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.
REFERENCES
- 1.Meissner HC. Viral bronchiolitis in children. N Engl J Med. 2016;374:1793‐1794. [DOI] [PubMed] [Google Scholar]
- 2.Nicholson KG, Kent J, Ireland DC. Respiratory viruses and exacerbations of asthma in adults. Br Med J. 1993;307:982‐986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saraya T, Kurai D, Ishii H, et al. Epidemiology of virus‐induced asthma exacerbations: with special reference to the role of human rhinovirus. Front Microbiol. 2014;226:1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jartti T, Lehtinen P, Vuorinen T, Ruuskanen O. Bronchiolitis: age and previous wheezing episodes are linked to viral etiology and atopic characteristics. Pediatr Infect Dis J. 2009;28:311‐317. [DOI] [PubMed] [Google Scholar]
- 5.Kusel MM, de Klerk NH, Kebadze T, et al. Early‐life respiratory viral infections, atopic sensitization, and risk of subsequent development of persistent asthma. J Allergy Clin Immunol. 2007;119:1105‐1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson DJ, Gangnon RE, Evans MD, et al. Wheezing rhinovirus illnesses in early life predict asthma development in high‐risk children. Am J Respir Crit Care Med. 2008;178:667‐672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kantor DB, Stenquist N, McDonald MC, et al. Rhinovirus and serum IgE are associated with acute asthma exacerbation severity in children. J Allergy Clin Immunol. 2016;138:1467‐1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taussig LM, Wright AL, Holberg CJ, Halonen M, Morgan WJ, Martinez FD. Tucson children's respiratory study: 1980 to present. J Allergy Clin Immunol. 2003;111:661‐675. [DOI] [PubMed] [Google Scholar]
- 9.Tan WC, Xiang X, Qiu D, et al. Epidemiology of respiratory viruses in patients hospitalized with near‐fatal asthma, acute exacerbations of asthma, or chronic obstructive pulmonary disease. Am J Med. 2003;115:272‐277. [DOI] [PubMed] [Google Scholar]
- 10.Jackson DJ, Evans MD, Gangnon RE, et al. Evidence for a causal relationship between allergic sensitization and rhinovirus wheezing in early life. Am J Respir Crit Care Med. 2012;185:281‐285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Busse WW, LemanskeRF, Jr, Gern JE. Role of viral respiratory infections in asthma and asthma exacerbations. Lancet. 2004;376:826‐834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brzezińska‐Pawłowska OE, Rydzewska AD, Łuczyńska M, Majkowska‐Wojciechowska B, Kowalski ML, Makowska JS. Environmental factors affecting seasonality of ambulance emergency service visits for exacerbations of asthma and COPD. J Asthma. 2015:53:139‐145. [DOI] [PubMed] [Google Scholar]
- 13.Jartti T, Gern EJ. Role of viral infections in the development and exacerbation of asthma in children. J Allergy Clin Immunol. 2017;140:895‐906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez FD. Managing childhood asthma: challenge of preventing exacerbations. Pediatrics. 2009;123(suppl 3):S146‐S150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turunen R, Koistinen A, Vuorinen T, et al. The first wheezing episode: respiratory virus etiology, atopic characteristics, and illness severity. Pediatr Allergy Immunol. 2014;25:796‐803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zambrano JC, Carper HT, Rakes GP, et al. Experimental rhinovirus challenges in adults with mild asthma: response to infection in relation to IgE. J Allergy Clin Immunol. 2003;111:1008‐1016. [DOI] [PubMed] [Google Scholar]
- 17.Sears MR. Epidemiology of asthma exacerbations. J Allergy Clin Immunol. 2008;122(4):662–668. [DOI] [PubMed] [Google Scholar]
- 18.Teach SJ, Gergen PJ, Szefler SJ, et al. Seasonal risk factors for asthma exacerbations among inner‐city children. J Allergy Clin Immunol. 2015;135:1465‐1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnston NW, Johnston SL, Duncan JM, et al. The September epidemic of asthma exacerbations in children: a search for etiology. J Allergy Clin Immunol. 2005;115:132‐138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heinonen S, Jartti T, Garcia C, et al. Rhinovirus detection in symptomatic and asymptomatic children: value of host transcriptome analysis. Am J Respir Crit Care Med. 2016;193:772‐782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toivonen L, Schuez‐Havupalo L, Karppinen S, et al. Rhinovirus infections in the first 2 years of life. Pediatrics. 2016;138:1309‐1314. [DOI] [PubMed] [Google Scholar]
- 22.Suruki RY, Boudiaf N, Ortega H. Retrospective cohort analysis of healthcare claims in the United States characterising asthma exacerbations in paediatric patients. World Allergy Org J. 2016;9:18‐27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McIntyre CL, Knowles NJ, Simmonds P. Proposals for the classification of human rhinovirus species A, B and C into genotypically assigned types. J Gen Virol. 2013;94:1791‐1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobs SE, Lamson DM, St George K, Walsh TJ. Human rhinoviruses. Clin Microbiol Rev. 2013;26:135‐162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cox DW, Bizzintino J, Ferrari G, et al. Human rhinovirus species C infection in young children with acute wheeze is associated with increased acute respiratory hospital admissions. Am J Respir Crit Care Med. 2013;188:1358‐1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnston SL, Pattemore PK, Sanderson G, et al. Community study of role of viral infections in exacerbations of asthma in 9‐11 year‐old children. BMJ. 1995;310:1225‐1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.UNAIDS/WHO . AIDS Epidemic Update. Geneva, Switzerland: UNAIDS/WHO; 2002. [Google Scholar]
- 28.Belshaw R, Gardner A, Rambaut A, Pybus GO. Pacing a small cage: mutation and RNA viruses. Trends Ecol Evol. 2008;23:188‐193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shankarappa R, Margolick JB, Gange SJ, et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73:10489‐10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bedford T, Cobey S, Pascual M. Strength and tempo of selection revealed in viral gene genealogies. BMC Evol Biol. 2011;11:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fitch MW, Leiter JM, Li XQ, Palese P. Positive Darwinian evolution in human influenza A viruses. Proc Natl Acad Sci U S A. 1991;88:4270‐4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sagulenko P, Puller V, Neher AR. Tree time: maximum‐likelihood phylodynamic analysis. Virus Evol 2018;4:vex042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wright S. Evolution in Mendelian populations. Genetics. 1931;16:97‐159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drummond JA, Nicholls KG, Rodrigo GA, Solomon W. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics. 2002;161:1307‐1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kosakovsky LS, Posada D, Gravenor BM, et al. Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol. 2006;23:1891‐1901. [DOI] [PubMed] [Google Scholar]
- 36.Mosser AG, Vrtis R, Burchell L, et al. Quantitative and qualitative analysis of rhinovirus infection in bronchial tissues. Am J Respir Crit Care Med. 2005;171:645‐651. [DOI] [PubMed] [Google Scholar]
- 37.Bochkov YA, WattersK, Ashraf S, et al. Cadherin related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication. Proc Natl Acad Sci U S A. 2015;112:5485‐5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han M, Chung Y, Young Hong J, et al. Toll‐like receptor 2‐expressing macrophages are required and sufficient for rhinovirus induced airway inflammation. J Allergy Clin Immunol. 2016;138:1619‐1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slater L, Bartlett NW, Haas JJ, et al. Co‐ordinated role of TLR3, RIG‐I and MDA5 in the innate response to rhinovirus in bronchial epithelium. PLoS Pathog. 2010;6:e1001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bosco A, Wiehler S, Proud D. Interferon regulatory factor 7 regulates airway epithelial cell responses to human rhinovirus infection. BMC Genomics. 2016;17:76‐82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leigh R, Oyelusi W, Wiehler S, et al. Human rhinovirus infection enhances airway epithelial cell production of growth factors involved in air way remodeling. J Allergy Clin Immunol. 2008;121:1238‐1245. [DOI] [PubMed] [Google Scholar]
- 42.Rossi GA, Colin AA. Infantile respiratory syncytial virus and human rhinovirus infections: respective role in inception and persistence of wheezing. Eur Respir J. 2015;45:774‐789. [DOI] [PubMed] [Google Scholar]
- 43.Williams JV, Piedra PA, Englund JA. Respiratory syncytial virus, human metapneumovirus and parainfluenza viruses. In: Richman DD, Whitley RJ, Hayden FG, eds. Clinical Virology. 4th ed.Washington, DC: ASM Press; 2017:873‐902. [Google Scholar]
- 44.Tregoning JS, Schwarze J. Respiratory viral infections in infants: causes, clinical symptoms, virology, and immunology. Clin Microbiol Rev. 2010;23:74‐98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kloepfer KM, Lee WM, Pappas TE, et al. Detection of pathogenic bacteria during rhinovirus infection is associated with increased respiratory symptoms and asthma exacerbations. J Allergy Clin Immunol. 2014;133:1301‐1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jakiela B, Brockman‐Schneider R, Amineva S, Lee WM, Gern JE. Basal cells of differentiated bronchial epithelium are more susceptible to rhinovirus infection. Am J Respir Cell Mol Biol. 2008;38:517‐523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sajjan U, Wang Q, Zhao Y, Gruenert DC, Hershenson MB. Rhinovirus disrupts the barrier function of polarized airway epithelial cells. Am J Respir Crit Care Med. 2008;178:1271‐1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shariff S, Shelfoon C, Holden NS, et al. Human rhinovirus infection of epithelial cells modulates airway smooth muscle migration. Am J Respir Cell Mol Biol. 2017;56:796‐803. [DOI] [PubMed] [Google Scholar]
- 49.Hong JY, Bentley JK, Chung Y, et al. Neonatal rhinovirus Induces mucous metaplasia and airways hyperresponsiveness through IL‐25 and type 2 innate lymphoid cells. J Allergy Clin Immunol. 2014;134:429‐439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sabogal Piñeros YS, Bal SM, Dijkhuis A, et al. Eosinophils capture viruses, a capacity that is defective in asthma. Allergy. 2019;74:1898‐1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anderson D, Jones CA, Gaido MC, et al. Differential gene expression of lymphocytes stimulated with rhinovirus A and C in children with asthma. Am J Respir Crit Care Med. 2020;202:202‐209. [DOI] [PubMed] [Google Scholar]
- 52.LemanskeRF, Jr, Jackson DJ, Gangnon RE, et al. Rhinovirus illnesses during infancy predict subsequent childhood wheezing. J Allergy Clin Immunol. 2005;116:571‐577. [DOI] [PubMed] [Google Scholar]
- 53.Prazma MC, Gern EJ, Weinstein FS, et al. The association between seasonal asthma exacerbations and viral respiratory infections in a pediatric population receiving inhaled corticosteroid therapy with or without long‐acting beta‐adrenoceptor agonist: a randomized study. Resp Med. 2015;109:1280‐1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kwon J‐M, Shim WJ, Kim SD, et al. Prevalence of respiratory viral infection in children hospitalized for acute lower respiratory tract diseases, and association of rhinovirus and influenza virus with asthma exacerbations. Korean J Pediatr. 2014;57:29‐34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown I. Higher Co‐infection Rates in COVID19. Medium; 2020. https://medium.com/@nigam/higher-co-infection-rates-in-covid19-b2496508833355. [Google Scholar]
- 56.Goyal P, Choi JJ, Pinheiro LC, et al. Clinical characteristics of Covid‐19 in New York City [published online ahead of print April 17,2020]. N Engl J Med. 2020;382:2372‐2374. 10.1056/NEJMc2010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Butler WM, O'Reilly A, Dunican ME, et al. Prevalence of comorbid asthma in COVID‐19 patients. J Allergy Clin Immunol. 2020;146:P334‐P335. 10.1016/j.jaci.2020.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang HE, Willis LA, Romanoski EC, et al. RV infections in asthmatics increase ACE2 expression and cytokine pathways implicated in COVID‐19. Am J Resp Crit Care Med. 2020;202:753‐755. https://10.1164/rccm.202004-1343LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Traub S, Nikonova A, Carruthers A, et al. An anti‐human ICAM‐1 antibody inhibits rhinovirus‐induced exacerbations of lung inflammation. PLoS Pathog. 2013;9:e1003520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muehling LM, Heymann PW, Wright PW, et al. Human Th1 and Th2 cells targeting rhinovirus and allergen coordinately promote allergic asthma. J Allergy Clin Immunol. 2020;146:P555‐P570. 10.1016/j.jaci.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Casanova V, Sousa HF, Stevens C, Barlow GP. Antiviral therapeutic approaches for human rhinovirus infections. Future Virol. 2018;13:505‐518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gaajetaan GR, Geelen TH, Vernooy JH, et al. Interferon‐beta induces a long‐lasting antiviral state in human respiratory epithelial cells. J Infect. 2013;66:163‐169. [DOI] [PubMed] [Google Scholar]
- 63.Basta HA, Ashraf S, Sgro JY, Bochkov YA, Gern JE, Palmenberg AC. Modeling of the human rhinovirus C capsid suggests possible causes for antiviral drug resistance. Virology. 2014;448:82‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Djukonovic R, Harrison T, Johnston LS, et al. The Effect of inhaled IFN‐β on worsening of asthma symptoms caused by viral infections. Am J Respir Crit Care Med. 2014;190:145‐154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Watson A, Spalluto CM, McCrae C, et al. Dynamics of IFN‐β responses during respiratory viral infection: Insights for therapeutic strategies. Am J Respir Crit Care Med 2020;201:83‐94. [DOI] [PubMed] [Google Scholar]
- 66.Hayden FG, Turner RB, Gwaltney JM, et al. Phase II, randomized, double‐blind, placebo‐controlled studies of ruprintrivir nasal spray 2‐percent suspension for prevention and treatment of experimentally induced rhinovirus colds in healthy volunteers. Antimicrob Agents Chemother. 2003;47:3907‐3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Glanville N, McLean GR, Guy B, et al. Cross‐serotype immunity induced by immunization with a conserved rhinovirus capsid protein. PLoS Pathog. 2013;9:e1003669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kaiser L, Crump CE, Hayden FG. In vitro activity of pleconaril and AG7088 against selected serotypes and clinical isolates of human rhinoviruses. Antiviral Res. 2000;47:215‐220. [DOI] [PubMed] [Google Scholar]
- 69.Hayden FG, Coats T, Kim K, et al. Oral pleconaril treatment of picornavirus associated viral respiratory illness in adults: efficacy and tolerability in phase II clinical trials. Antivir Ther. 2002;7:53‐65. [PubMed] [Google Scholar]
- 70.Patick AK. Rhinovirus chemotherapy. Antiviral Res. 2006;71:391‐396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pevear DC, Hayden FG, Demenczuk TM, et al. Relationship of pleconaril susceptibility and clinical outcomes in treatment of common colds caused by rhinoviruses. Antimicrob Agents Chemother. 2005;49:4492‐4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Graham BS, Walker C. Meeting the challenge of vaccine design to control HIV and other difficult viruses. In: Kaufmann SHE, Rouse BT, Sacks DL, eds. The Immune Response to Infection. Washington, DC: ASM Press; 2011:559‐570. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.


