Abstract
Modern-day drug discovery is now blessed with a wide range of high-throughput hit identification (hit-ID) strategies that have been successfully validated in recent years, with particular success coming from high-throughput screening, fragment-based lead discovery, and DNA-encoded library screening. As screening efficiency and throughput increases, this enables the viable exploration of increasingly complex three-dimensional (3D) chemical structure space, with a realistic chance of identifying highly specific hit ligands with increased target specificity and reduced attrition rates in preclinical and clinical development. This minireview will explore the impact of an improved design of multifunctionalized, sp3-rich, stereodefined scaffolds on the (virtual) exploration of 3D chemical space and the specific requirements for different hit-ID technologies.
Keywords: Drug discovery, chirality, three-dimensional, hit identification
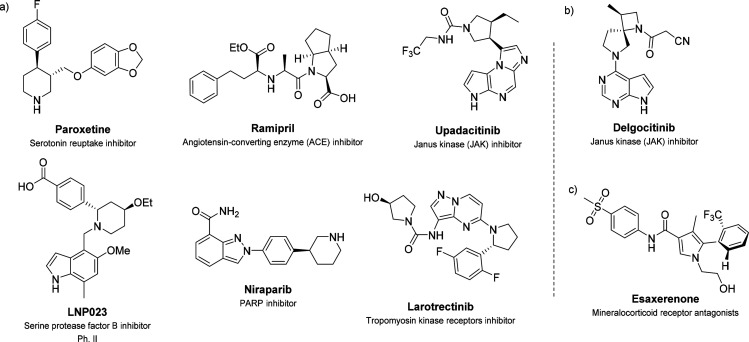
Chirality plays a fundamental role in the binding affinity and interactions between the drug and its target, thus shaping the drug’s pharmacology. For this reason, in 1992 the Food & Drug Administration (FDA) outlined a series of guidelines for the pharmaceutical development of single enantiomers and racemates.1 Since then, most drugs in the market are chiral, and the number of single-enantiomer and single-diastereomer drugs has consistently increased.2,3 Examples of chiral FDA-approved drugs and recent disclosures in clinical trials are shown in Figure 1a. Atropisomerism is a type of chirality that originates from a restricted bond rotation usually within an sp2-sp2 bond. An analysis of FDA-approved small-molecule drugs since 2011 revealed that 30% of these compounds have at least one atropisomeric axis,4 even though they exist as rapidly interconverting isomers.
Figure 1.
(a) Examples of FDA-approved chiral small-molecule drugs. (b) Structure of Delgocitinib. (c) Structure of the active atropisomer of Esaxerenone.
Chirality is an innate property of some molecules derived from an absence of an internal plane of symmetry. Many naturally occurring molecules (amino acids, sugars) and biologically relevant molecules (DNA, RNA, proteins) are chiral. Drugs interact in a chiral environment, and biological targets recognize their ligands in a three-dimensional (3D) fashion. It is logical that molecules with a higher degree of 3D shape will interact with their targets with an increased affinity and higher levels of specificity, thus making more efficacious and safer drugs. For the scope of this review, we define 3D molecules as any organic compound that deviates from linear (one-dimensional (1D)) or planar (two-dimensional (2D)) shape. The molecular shape can be compared by analyzing a set through the method of Sauer and Schwartz5 by normalizing the principle moment of inertia (PMI) values and plotting their ratios on a triangular graph in which each corner represents a linear, planar, or spherical shape.
Many of the drug-like small molecules developed to date have a higher sp2 fraction, while natural products have a higher sp3 fraction (Fsp3) and a larger number of chiral centers. A higher degree of Fsp3, defined as the fraction of sp3-hybridized carbons, has been suggested to have a positive correlation with better drug-like properties such as higher solubility6 and fewer off-target hits (lower promiscuity).7 A high Fsp3 value does not necessarily correlate to an enhanced 3D shape; fully sp2 compounds can sometimes have a high 3D character.
The concept of molecular complexity was introduced by Hann et al. using a simplified model of receptor–ligand interactions.8 The authors reported how more complex ligands have less chance to positively interact with a random target. Saturation can be used as a descriptor for molecular complexity, since it allows access to more complex molecular structures with only a small gain in molecular weight. Saturation also allows a higher number of isomers compared to isomers of 2D molecules (e.g., piperidine vs pyridine ring), which can sample a higher fraction of the chemical space. Increasing the Fsp3 character can also obtain better ligand/receptor interactions through the installment of out-of-plane substituents that are not accessible from flat sp2 molecules. In this context, stereodefined nonplanar molecules give access to the exact exit vectors needed for positive interactions, thus improving selectivity and potency. At the same time, an increase in 3D character and complexity will reduce the chances of binding to a specific target. This has been reported in a publication by Astex in the context of fragment screening, where the authors showed a lower hit rate for 3D-rich fragments.9
An analysis of the change in saturation and number of stereocenters during the drug discovery process showed that there is an average increase of 31% in Fsp3 and a 21% increase in the number of stereocenters from discovery compounds to drugs.10 Similarly, an analysis of attrition in the development phase showed that compounds with a higher sp3 fraction have a better chance to become drugs.8 As an example, the JAK inhibitor Delgocitinib (Figure 1b) has been recently approved in Japan to treat inflammatory skin disorders.11 In the modification of the head region, the authors decided to explore 3D-rich structures resulting in the discovery of a highly sp3-rich kinase inhibitor.
Synthesis and the structure–activity relationship (SAR) exploration of complex molecular architectures requires the development of synthetic methods and efficient approaches to regulate stereoisomerism and regioisomerism.
Chiral products can be accessed by an asymmetric synthesis, chiral pool synthesis, or resolution of racemic mixtures. The rapid development of asymmetric synthesis methodologies and chiral purification techniques has paved the way for the commercialization of sp3-rich and enantiomerically pure scaffolds, but they are still underrepresented in the commercial market landscape. Their accessibility on a large scale is still a challenge due to the high costs of chiral catalysts and time-consuming purifications to obtain a chirally pure compound.
The substrate specificity of most of the asymmetric transformations means that specific chiral ligands need to be developed and tested. A resolution of racemic mixtures is usually cheaper, but at least half of the material is discarded if the unwanted enantiomer cannot be racemized. A chiral pool synthesis is limited by the substrate availability.
The isolation of the preferred enantiomer can be achieved by a diastereomeric crystallization12 (classical resolution), kinetic resolution13 (chemical or enzymatic), preferential crystallization,14 or chromatographic resolution.15 The structure and absolute configuration of the desired enantiomer needs to be fully characterized, which can be a nontrivial task. The main techniques for the determination of an absolute configuration in use in industry and academia are X-ray crystallography and Mosher’s method (NMR), but recently vibrational circular dichroism (VCD) is gaining momentum due to the low amount of sample required and its complementarity with the previous techniques.16
While developing an enantiomerically pure drug, it should be confirmed that no change in purity or enantiomeric ratio occurs during the shelf life and, for therapeutic use, it is pivotal to characterize the enantiomeric purity and how each enantiomer is metabolized by the human body.17
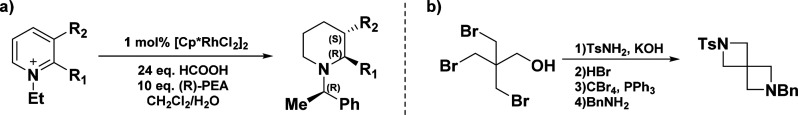
In the past decades, these challenges and the explosion of cross-couplings as robust synthetic methodologies have favored the use of easy-to-access 2D molecules while hampering the use of stereodefined building blocks (BBs) in drug discovery. Recently the trend has inverted, and more pharmaceutical companies are investing in improving their internal high-throughput screening (HTS) and fragment libraries.18 Increasing and improving the quality of in-house building block collections is an overlooked strategy according to scientists from AstraZeneca, who outlined the importance of saturated and chiral BBs in a recent publication.19 As a consequence, numerous contract research organizations (CROs) have focused in the development and optimization of chemical processes for the synthesis of 3D-rich libraries,20 making these attractive BBs available for screening. For example, the enantioselective transamination of substituted pyridiniums to the corresponding piperidines developed in Xiao’s group in 201421 (Figure 2a) is the founding technology of Liverpool ChiroChem (LCC).
Figure 2.
(a) Transamination of 2-substituted pyridiniums to piperidines. (b) Synthesis of 2,6-diazaspiro[3.3]heptane.
Despite piperidines being considered a privileged scaffold in medicinal chemistry and the considerable size of literature on their stereoselective synthesis,22 the offer of chirally pure, multifunctionalized novel derivatives remains still scarce.
Similarly, recent discoveries from Carreira’s group in the synthesis of spirocyclic building blocks are at the basis of Spirochem (Figure 2b), and the different synthetic methods developed for their synthesis has been reviewed elsewhere.23
The presence of this popular motif in medicinal chemistry has steadily increased in the past decade,24 and advances in the synthesis of structurally diverse spirocylces has paved the way for their exploitation in drug-discovery programs. Consequently, the synthesis of diverse fragment libraries based on the spirocycle moieties libraries has been recently reported.25 Notably, the influence of stereochemistry on different biological targets has been demonstrated by numerous studies.26
DNA-encoded libraries (DELs) are a set of drug-like small-molecule compounds attached to a DNA tag. Each tag is unique and provides the chemical information related to the compound to which each tag is coupled. The compounds are synthesized starting from selected sets of building blocks, which are combined in a mix-and-split manner to form libraries containing millions to billions of compounds. Hits from DEL campaigns have been successfully developed into clinical candidates, such as the Receptor Interacting Protein 1 (RIP1) kinase inhibitor GSK298277227 and the soluble epoxide hydrolase (sEH) inhibitor GSK2256294.28
The methodologies used to synthesize the library compounds are limited by the DNA stability and solubility. Continued efforts are being made to increase the range of on-DNA compatible chemistries,29 and consequently there is a need to expand the array of synthetic handles in the core scaffolds. The drug-discovery community has been trying to achieve diversity and complexity in DNA-encoded libraries through constantly increasing the toolbox of on-DNA chemical reactions. To preserve the integrity of the DNA barcode, chemists have focused on expanding the scope of DNA-compatible chemical transformations and DNA-protecting strategies. A review covering developments in DNA-compatible reactions has been recently published.30 Established DNA-compatible transformations require amines, carboxylic acids/esters, aldehydes, boronic acids/esters, aryl halides, azides, and alkynes as synthetic handles in the selected cores. Such methodologies are limited compared to conventional organic reactions under standard conditions, but the number of BBs commercially available for these transformations is big enough (thousands) to allow the creation of screening libraries with billions of compounds.
Many reports in the literature have described different approaches to select BBs for DEL technology and increase the sp3 fraction of the resulting drug-like compounds. When the library is built through an iteration of the same synthetic step (i.e., amidation31), the complexity of the drug-like compounds can be achieved by utilizing sets of BBs with a high diversity and high sp3 fraction. Thus, BB cores and the spatial distribution of the synthetic handles are the basis for diversity in designing a DEL library.
Recent reports in the literature showed how the stereochemical complexity of core scaffolds can be utilized to generate sp3-rich DELs with a higher scaffold diversity. A relevant example is the work of Clemons, Schreiber, and co-workers32 who used 2,3-disubstituted azetidines and pyrrolidines as core scaffolds for the generation of a relatively small DEL of just over 100 000 molecules. The implementation of all four stereoisomers of each heterocycle in the scaffold selection resulted in a collection that resembled diversity-oriented synthesis libraries more closely than other sources. In another example, Santini and Young reported the multigram synthesis of 20 2,3-disubstituted piperazines33 and 24 2,6-disubstituted piperazines34 in an enantiomerically pure form to be used as scaffolds for a library production. The stereochemical diversity of the core and the possibility to orthogonally functionalize the two nitrogens of the piperazine ring is the key to generate libraries with a high conformational diversity.
GlaxoSmithKline (GSK) also shared their principles for selecting BBs for DEL,35 where the authors highlighted the importance of nonplanar scaffolds to increase the coverage of chemical space. A different approach to this problem has been proposed by scientists from Pfizer, who managed to increase the coverage of 3D chemical space by developing a C–C bond-forming reaction of DNA-tagged reactants.36
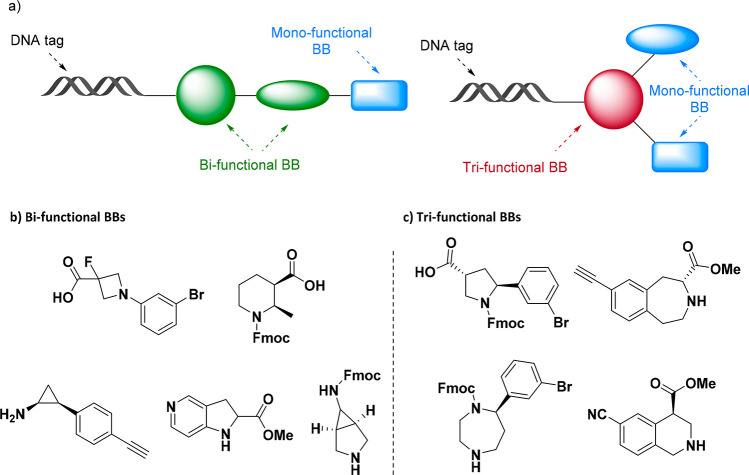
The design and selection of the starting set of BBs is crucial to ensure good predicted pharmacokinetic profiles of the drug-like library compounds. Molecular weight (MW) is the most important limiting factor for BB selection (usually <200 Da), and it is dependent on the number of cycles intended to synthesize the library. DELs are most commonly based on two or three cycles and can be built in a linear or branched fashion.37 Linear libraries require at least one bifunctional BB, while branched libraries need at least one trifunctional BB (Figure 3a). The commercial availability of BBs decreases drastically when increasing the number of synthetic handles. A recent study from Eli Lilly scientists38 on the availability of mono-, bi-, and trifunctionalized BBs for DEL showed that trifunctionalized BBs only cover 2.3% of the total. Limitations in the commercial availability of trifunctionalized BBs and the diversity of synthetic handles are currently affecting the way DELs are designed. There is a strong need for easy access to a variety of trifunctionalized compounds to use as core scaffolds that would diversify the binding mode and the overall coverage of chemical space of DELs.
Figure 3.
(a) Representation of branched and linear DEL compounds. (b) Bifunctional BBs. (c) Trifunctional BBs.
It is important that the reactivity of each BB is good enough to ensure high yields and improve the purification process between cycles. Moreover, synthetic handles in each BB need to have an orthogonal reactivity to minimize the byproduct formation and guarantee accuracy between the DNA tag and the corresponding chemical product. Examples of commercially available bi- and trifunctionalized BBs for DEL application are represented in Figure 3b,c.
Cyclic saturated N-heterocycles are optimal starting points for the design of bi- and trifunctional BBs. The endocyclic nitrogen can be used as a synthetic handle, and the cyclic core can be decorated with additional functionalities to obtain a low-molecular-weight, rigid, sp3-rich building block. Many major BB providers have developed collections to be employed in DNA-encoded library technologies that exploit these scaffolds.
Access to single enantiomers is fundamental when using a technology capable of generating billions of compounds by a split-and-pool strategy, especially when multiple BBs are chiral. The inclusion of both enantiomers in the library design will ensure a thorough exploration of potential chiral space. Finally, each BB needs to be available in gram quantities to facilitate a rapid off-DNA synthesis of virtual hits to confirm their biological activity.
Fragment library screening is employed to identify low-molecular-weight molecules (MW < 300 Da) that bind to biologically important targets.
Fragment-based lead discovery (FBLD) offers multiple advantages compared to normal high-throughput screening; the average library size is in the order of a few thousand fragments compared to millions of compounds for HTS. The technique is based on the principle that low-MW compounds can sample a wider range of chemical space than is possible for higher-MW compounds. Moreover, there’s a high potential for fragments to be optimized through a fragment growth and fragment merging, for example. Currently, several compounds derived from a fragment-based approach have entered different stages of clinical trials, and some have been approved by the FDA, such as Vemurafenib39 developed by Zelboraf and Plexxikon/Roche by fragment growing.
A noncomprehensive list of screening methods for fragment libraries includes X-ray crystallography40 and NMR,41 alongside other biophysical methods such as surface plasmon resonance (SPR),42 isothermal titration calorimetry (ITC),43 and mass spectrometry (mass-spec),44 which are very sensitive and can therefore detect weak bindings. NMR and X-ray crystallography can give structural information together with binding and, therefore, facilitate the optimization of binding fragments at the same time (structure-based design). The throughput of techniques such as SPR is also rapidly increasing, which allows larger fragment libraries to be screened45 with a better depth and breadth to the pharmacophore diversity.
There are specific challenges related to FBLD. First of all, highly sensitive methods are required to detect binding in the millimolar region. Fragment hits with low affinity binding (usually mM potencies) will need optimization, but thanks to countless possibilities for fragment expansion and the creativity of medicinal chemists, it is nearly always possible to obtain nanomolar leads form millimolar fragment hits.46 Fragments need to be soluble at the high concentrations required for screening; therefore, measured water and/or dimethyl sulfoxide (DMSO) solubility is an added value for fragment libraries. An aggregation of fragments at high concentrations should also be considered, as it can lead to false negatives.47
Despite a low affinity binding, fragments may often have a high ligand efficiency (free energy per heavy atom) compared to lead-like and drug-like HTS hits, which is beneficial for the following optimization.
Even though the principles for the design of a fragment library depend strongly on the biophysical method of screening, some common features can be found: diversity, chemically expandable fragments, availability of materials for hit validation, access to structurally related analogues, and exclusion of unwanted functionalities (pan-assay interference compounds (PAINS) or functionalities that might react with the target protein).
The molecular complexity of the fragments needs to be carefully modulated. The probability of a ligand matching a target decreases as the complexity of the ligand increases,48 but some degree of complexity is needed to achieve a detectable binding and a satisfactory sampling of the chemical space. It is known that, even though the hit rate of fragments is high, the binding affinity is usually low; therefore, the complexity of a fragment can only be decreased as long as the binding is still detectable. Increasing the number of diversely shaped fragments (i.e., moving away from flat, sp2-rich compounds) could be a method to introduce complexity and diversity into fragment libraries and explore new areas of chemical space. A higher degree of shape diversity has been correlated to a broader range of biological activities and, thus, a higher chance to engage challenging targets.49
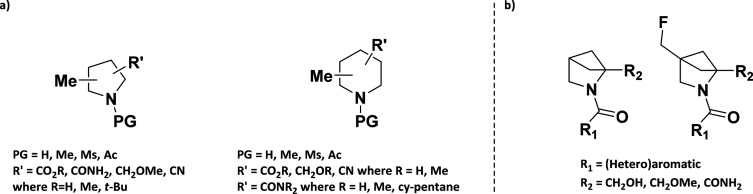
A recent paper from O’Brien’s group50 described the design and synthesis of a small collection of fragments to be used in addition to existing libraries to improve diversity and three-dimensionality. This collection shows how simple derivatives of privileged scaffolds (disubstituted piperidines and pyrrolidines in this specific example, Figure 4a) can increase the shape diversity of existing commercial libraries and explore under-represented areas of chemical space. Recent reports from the University of Sussex in collaboration with Photodiversity and Abbvie51 showed how carefully designed fragments, characterized by a stereodefined heterocyclic saturated core decorated with a different range of pharmacophores (Figure 4b), can effectively sample the chemical space while maintaining desirable features for drug discovery programs like a high solubility, 3D shape, high sp3 fraction, and low number of rotatable bonds.
Figure 4.
(a) Representation of sp3-rich fragments included in the O’Brien collection. (b) General structures of 3D-fragments reported by Cox et al.
A fragment library design should focus on the balance of simplicity, novelty, and diversity. The inclusion of annulated, spiro, and bridged compounds to novel variants of piperidine- and pyrrolidine-based scaffolds will ensure a low number of rotatable bonds and a desirable sp3 character for optimal physicochemical properties. The spatial distribution of the exit vectors and the pharmacophore diversity should be maximized to improve the probability of positive interactions with the target. A designed diversification of the synthetic handles combined with stereodefined structures allows for fragment growth/merging with specific geometry. Sets of fragments such as these could be exploited as an sp3-rich collection to add value and diversity to any existing fragment library. The impact and importance of 3D-rich fragment libraries exhibiting high Fsp3 and molecular complexity has been recently evaluated.52
Recently the importance of novelty in the design of fragment libraries has been challenged,53 shifting the focus on diversity. It has been shown how different FBLD campaigns resulted in very similar (or even identical) fragments being selected for different biological targets.54 Even when the binding mode was the same, each group went on to develop the fragment in a unique way, which resulted in distinctive lead-like compounds with no intellectual property overlap. These examples illustrate how selectivity and potency can arise from common promiscuous fragments.
We believe that a stronger focus on pharmacophore diversity and potential growth vectors of the fragments will maximize the possibilities for their expansion. New synthetic strategies can play an important role in developing lead-like compounds from fragment hits. For example, the development of C–H activation methodologies can facilitate the functionalization of fragments in specific directions not yet accessible. Mastering the challenge of balancing complexity and Fsp3 character will deliver fragment libraries with a satisfactory hit rate and developability.
HTS is a method used in drug discovery that allows testing of millions of chemical substances against individual proteins (or a family of related proteins, target-based) or cellular systems (phenotypic-based) to identify biologically relevant molecules. The primary goal is to identify lead-like compounds that can be further optimized. Since the chemical space is practically infinite, the design, synthesis, and maintenance of compound libraries requires significant resources and constant upgrading to ensure competitiveness and novelty.55
HTS libraries are usually designed to contain a significant structural diversity56 even though having several examples of the same scaffold is useful to detect real hits, since it is more likely to have a real or validated hit when hits emerge from different compounds containing the same scaffolds.57
Target-oriented libraries are a collection of compounds that are specifically designed for a certain target (or family). It has been reported that focused libraries can highly increase the number of hits.58
There are different approaches to design targeted screening libraries. If a crystal structure of the target protein (or homologous) is available, the corresponding screening library can be built from a structure-based approach by docking sets of compounds into the active site of the protein. A different approach is to design the focused screening library from the information generated by known binders. For certain gene families (kinases, G-protein coupled receptor (GPCR), etc.) there are hundreds of known active compounds (ligand-based approach).
Bayer recently renewed their internal HTS library by adding 500 000 novel compounds. In the resulting publication, their approach to novel and highly attractive lead-like structures was described.59 As their previous library was biased toward sp2-rich molecules arising from flat (hetero)aromatic scaffolds (average Fsp3 ≈ 0.3), they established Fsp3 > 0.4 as a goal for the new set of compounds to increase the diversity through the introduction of saturated and 3D-shaped molecules. This enhancement of their library has shown a positive impact on their newest screens.
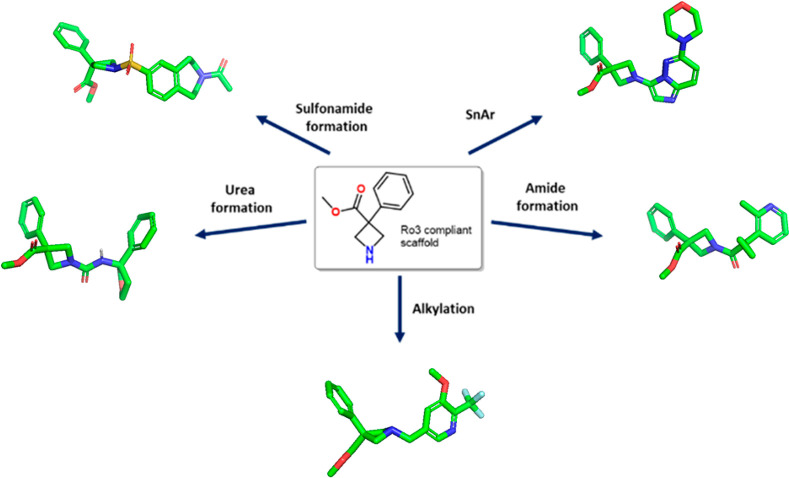
Hit identification/expansion can also be accelerated by advances in computational screening. Key to the success of virtual screening is the quality of the virtual collections. Virtual libraries (VLs) can be divided into four categories, namely, (a) internal collections; (b) collections from chemical vendors; (c) public collections, and (d) collections of compounds that theoretically exist, and the screening workflow has been reviewed recently.60 Recent computational advances have allowed significant improvements in both the size and quality of the virtual libraries that can be screened. Examples of lead-like libraries for virtual screening from commercial vendors include Enamine’s REAL database,61 Mcule’s Ultimate,62 WuXi’s Galaxi,63 and LCC’s 3Discovery.64 Cyclic Ro3-compliant amines can be strategically decorated to access a 3D lead-like chemical space in one step. A schematic representation of lead-like compounds derived from a heterocyclic scaffold is shown in Figure 5.
Figure 5.
Schematic representation of 3D lead-like compounds.
Concerning VLs from chemical vendors, any virtual hit from the collections will have to be validated experimentally, and therefore there is a strong advantage in screening VLs whose compounds can be synthesized rapidly and from in-house scaffolds and reagents. To ensure the delivery of such chemical entities in a reasonable lead-time and price, the established commercial model is based on a free screening access and make-on-demand hit validation.
Recent literature has highlighted the impact of chirality in virtual screenings. Of interest, an enantiomer-based virtual screening approach has been used by Wu and co-workers65 to identify bioactive compounds to human AChE, while a general outlook on the importance of absolute stereochemistry in virtual screening has been published elsewhere.66,67 The development of new tools to generate and screen 3D structures will facilitate the rational design and help the initial discovery phase and optimization of such challenging molecules.
Given the advantages of FBLD discussed in the previous chapter, screening electrophilic fragments has recently gained momentum as an alternative methodology to generate new chemical probes and validate new biological targets.68 An overview on available warheads and principles for a covalent library design has been published elsewhere69 and is out of the scope of this manuscript.
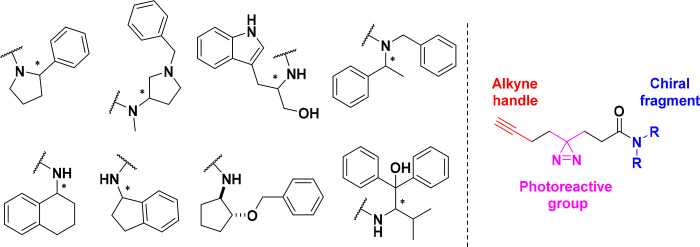
Author: In 2017 Cravatt’s group introduced a platform to perform FBLD directly in human cells by combining a library of fully functionalized fragment (FFF) probes with quantitative chemical proteomics70 to screen and validate the fraction of the human proteome that can be targeted by small molecules. A drawback of this revolutionary approach was the fact that different fragments have different physicochemical properties and, consequently, a different protein-binding potential, which can mislead the study of a structure–activity relationship. A brilliant solution to this came with the introduction of enantioprobes, pairs of FFF differing only in an absolute configuration. Numerous stereoselective protein-fragment interactions were identified in an unbiased way across a diverse set of protein classes.71
Currently, the published set of enantioprobes is commercialized by Sigma-Aldrich,72 and this technology is at the foundation of Vividion Therapeutics and Jnana, biotechnology companies for the discovery and development of small-molecule medicines.
Although a small set of enantioprobes (eight pairs, Figure 6) was used, a substantial fraction of the protein targets showed stereoselective interactions. It is clear how a limited set of enantioprobes will result in undersampling the amount of human proteins that can show stereoselective interactions with small molecules. Therefore, future development should be focused on the expansion and diversity of the enantioprobe library. A similar strategy was used by researchers at GSK, who developed a fragment-screening platform named PhotoAffinity Bits (PhABits)73 based on a library of photoreactive fragments. After irradiation, the photoreactive tag can covalently bind to proteins and identify weak reversible interactions between the fragment and the target.
Figure 6.
Structures of enantioprobes used by Cravatt’s group.
Recently, new photoreactive cross-linking reagents have been developed by a number of vendors74 with differentiated synthetic handles, and the benefits of employing diazirines have been reviewed elsewhere.75,66 The diazirine photoaffinity labeling of biomolecules is widely recognized as a powerful method for challenging aspects of drug discovery such as target identification.76
In the past decades, the complexity of the challenges associated with the synthesis and characterization of stereodefined compounds have limited their exploitation in medicinal chemistry. Although many technologies are emerging for the development of new drugs, clearly the necessity for a high Fsp3, 3D, multifunctionalized compounds is widespread throughout the drug-discovery community. Chemistry CROs are responding by increasing the diversity and complexity of their BBs libraries, but this brings additional challenges for their purification and characterization, limiting the number of such compounds commercially available on a gram scale and within reasonable lead times. When chiral compounds are available, the offer of chirally pure forms drops drastically. For example, Merk (Sigma-Aldrich) has 9751 heterocyclic building blocks, of which only 245 are chirally pure.77
Since the publication of Lovering’s seminal paper, a lot of interest has been placed on adding a 3D character to screening libraries. The recent advances in multiple fields of chemical synthesis such as photochemistry, electrochemistry, and biocatalysis is paving the way for the proliferation of high-value stereodefined fragments and BBs in the market. Chirality is a crucial property of molecules for specific interactions with the biological target, and its exploitation in different approaches for hit identification has been discussed. The increasing number and commercial availability in a multigram scale of diverse, stereodefined, sp3-rich molecules and the easier access to their analogues will provide the basis for the enhancement of existing libraries for screening technologies. We anticipate that a widespread access to chirally pure BBs and a broader selection of synthetic handles and/or functionalities will meet the demand of modern-day medicinal chemistry and enable the design of screening libraries with optimal physicochemical properties and, eventually, the development of improved drug-like compounds. The development of new enantiomerically pure bi- and trifunctionalized scaffolds will boost the design and diversity of (virtual) screening libraries and allow a better sampling of the chemical space.
Acknowledgments
The authors thank the colleagues at Liverpool Chirochem (LCC) for valuable discussion and comments during the preparation of this manuscript. In particular, we thank Prof. J. Xiao, Dr. J. Wu, and Dr. J. Ruan for their involvement in the development of the foundation technology at LCC.
Glossary
Abbreviations
- 2D
2-dimensional
- 3D
3-dimensional
- BB
Building block
- CRO
contract research organization
- DEL
DNA-encoded library
- DMSO
Dimethyl sulfoxide
- DNA
DNA
- FBLD
Fragment-based lead discovery
- FDA
Federal drug administration
- FFF
Fully functionalized fragment
- GPCR
G-protein coupled receptor
- HTS
High-throughput screening
- ITC
isothermal titration calorimetry
- NMR
Nuclear magnetic resonance
- PAINS
Pan-Assay Interference Compounds
- PMI
Principle moments of Inertia
- PROTAC
Proteolysis targeting chimera
- RNA
ribonucleic acid
- SPR
Surface plasmon resonance
- VCD
Vibrational circular dichroism
- VL
Virtual library
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
This work was supported by Liverpool ChiroChem Ltd. (LCC).
The authors declare the following competing financial interest(s): I.P.S. and P.C. are employees of Liverpool ChiroChem (LCC).
References
- Development of New Stereoisomeric Drugs ;Food and Drug Administration: Rockville, MD, 1992. [Google Scholar]
- Calcaterra A.; D’Acquarica I. The market of chiral drugs: Chiral switches versus de novo enantiomerically pure compounds. J. Pharm. Biomed. Anal. 2018, 147, 323–340. 10.1016/j.jpba.2017.07.008. [DOI] [PubMed] [Google Scholar]
- Agranat I.; Wainschtein S. R.; Zusman E. Z. The predicated demise of racemic new molecular entities is an exaggeration. Nat. Rev. Drug Discovery 2012, 11, 972–973. 10.1038/nrd3657-c1. [DOI] [PubMed] [Google Scholar]
- Toenjes S. T.; Gustafson J. L. Atropisomerism in medicinal chemistry: challenges and opportunities. Future Med. Chem. 2018, 10, 409–422. 10.4155/fmc-2017-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer W. H. B.; Schwarz M. K. Molecular shape diversity of combinatorial libraries: a prerequisite for broad bioactivity. J. Chem. Inf. Comput. Sci. 2003, 43, 987–1003. 10.1021/ci025599w. [DOI] [PubMed] [Google Scholar]
- Ishikawa M.; Hashimoto Y. Improvement in Aqueous Solubility in Small Molecule Drug Discovery Programs by Disruption of Molecular Planarity and Symmetry. J. Med. Chem. 2011, 54, 1539–1554. 10.1021/jm101356p. [DOI] [PubMed] [Google Scholar]
- Lovering F. Escape from Flatland 2: complexity and promiscuity. MedChemComm 2013, 4, 515–519. 10.1039/c2md20347b. [DOI] [Google Scholar]
- Hann M. M.; Leach A. R.; Harper G. Molecular Complexity and Its Impact on the Probability of Finding Leads for Drug Discovery. J. Chem. Inf. Comput. Sci. 2001, 41, 856–864. 10.1021/ci000403i. [DOI] [PubMed] [Google Scholar]
- Hall R. J.; Mortenson P. N.; Murray W. M. Efficient exploration of chemical space by fragment-based screening. Prog. Biophys. Mol. Biol. 2014, 116, 82–91. 10.1016/j.pbiomolbio.2014.09.007. [DOI] [PubMed] [Google Scholar]
- Lovering F.; Bikker J.; Humblet C. Escape from Flatland: Increasing Saturation as an Approach to Improving Clinical Success. J. Med. Chem. 2009, 52, 6752–6756. 10.1021/jm901241e. [DOI] [PubMed] [Google Scholar]
- Noji S.; Hara Y.; Miura T.; Yamanaka H.; Maeda K.; Hori A.; Yamamoto H.; Obika S.; Inoue M.; Hase Y.; Orita T.; Doi S.; Adachi T.; Tanimoto A.; Oki C.; Kimoto Y.; Ogawa Y.; Negoro T.; Hashimoto H.; Shiozaki M. Discovery of a Janus Kinase Inhibitor Bearing a Highly Three-Dimensional Spiro Scaffold: JTE-052 (Delgocitinib) as a New Dermatological Agent to Treat Inflammatory Skin Disorders. J. Med. Chem. 2020, 63, 7163–7185. 10.1021/acs.jmedchem.0c00450. [DOI] [PubMed] [Google Scholar]
- Faigl F.; Fogassy E.; Nógrádi M.; Pálovics E.; Schindler J. Strategies in optical resolution: a practical guide. Tetrahedron: Asymmetry 2008, 19, 519–536. 10.1016/j.tetasy.2008.02.004. [DOI] [Google Scholar]
- Kagan H. B.; Fiaud J. C.. Kinetic resolution. In Topics in Stereochemistry ;Eliel E. L., Wilen S. H., Eds.; John Wiley & Sons: New York, 1988; pp 249–330. [Google Scholar]
- Fogassy E.; Nógrádi M.; Kozma D.; Egri G.; Pálovics E.; Kiss V. Optical Resolution Methods. Org. Biomol. Chem. 2006, 4, 3011–3030. 10.1039/B603058K. [DOI] [PubMed] [Google Scholar]
- Andersson S.; Allenmark S. G. Preparative chiral chromatographic resolution of enantiomers in drug discovery. J. Biochem. Biophys. Methods 2002, 54, 11–23. 10.1016/S0165-022X(02)00126-4. [DOI] [PubMed] [Google Scholar]
- He Y.; Bo W.; Dukor R. K.; Nafie L. A. Determination of Absolute Configuration of Chiral Molecules Using Vibrational Optical Activity: A Review. Appl. Spectrosc. 2011, 65, 699–723. 10.1366/11-06321. [DOI] [PubMed] [Google Scholar]
- Kaminsky L. S.; Zhang Z. Y. Human P450 metabolism of warfarin. Pharmacol. Ther. 1997, 73, 67–74. 10.1016/S0163-7258(96)00140-4. [DOI] [PubMed] [Google Scholar]
- Schuffenhauer A.; Schneider N.; Hintermann S.; Auld D.; Blank J.; Cotesta S.; Engeloch C.; Fechner N.; Gaul C.; Giovannoni J.; Jansen J.; Joslin J.; Krastel P.; Lounkine E.; Manchester J.; Monovich L. G.; Pelliccioli A. P.; Schwarze M.; Shultz M.; Stiefl N.; Baeschlin D. K. Evolution of Novartis’ Small Molecule Screening Deck Design. J. Med. Chem. 2020, 63, 14425–14447. 10.1021/acs.jmedchem.0c01332. [DOI] [PubMed] [Google Scholar]
- Goldberg F. W.; Kettle J. G.; Kogej T.; Perry M. W. D.; Tomkinson N. P. Designing novel building blocks is an overlooked strategy to improve compound quality. Drug Discovery Today 2015, 20, 11–17. 10.1016/j.drudis.2014.09.023. [DOI] [PubMed] [Google Scholar]
- a Featured Products. Online at https://lifechemicals.com (accessed April 2021).; b Building Block Catalogue (accessed April 2021). Online at https://enamine.net.; c 3D-Rich Building Blocks. Online at https://www.liverpoolchirochem.com/chemical-space (accessed April 2021).
- a Wu J.; Tang W.; Pettman A.; Xiao J. Efficient and Chemoselective Reduction of Pyridines to Tetrahydropyridines and Piperidines via Rhodium-Catalyzed Transfer Hydrogenation. Adv. Synth. Catal. 2013, 355, 35–40. 10.1002/adsc.201201034. [DOI] [Google Scholar]; b Patent No. WO2015145143.
- A SciFinder search of “synthesis of chiral piperidines (1946–2020) gives 27 986 entries
- a Carreira E. M.; Fessard T. C. Four-Membered Ring-Containing Spirocycles: Synthetic Strategies and Opportunities. Chem. Rev. 2014, 114, 8257–8322. 10.1021/cr500127b. [DOI] [PubMed] [Google Scholar]; b Burkhard J.; Carreira E. M. 2,6-Diazaspiro[3.3]heptanes: Synthesis and Application in Pd-Catalyzed Aryl Amination Reactions. Org. Lett. 2008, 10, 3525–3526. 10.1021/ol801293f. [DOI] [PubMed] [Google Scholar]
- Hiesinger K.; Dar’in D.; Proschak E.; Krasavin M. Spirocyclic Scaffolds in Medicinal Chemistry. J. Med. Chem. 2021, 64, 150–183. 10.1021/acs.jmedchem.0c01473. [DOI] [PubMed] [Google Scholar]
- King T. A.; Stewart H. L.; Mortensen K. T.; North A. J. P.; Sore H. F.; Spring D. R. Cycloaddition Strategies for the Synthesis of Diverse Heterocyclic Spirocycles for Fragment-Based Drug Discovery. Eur. J. Org. Chem. 2019, 2019, 5219–5229. 10.1002/ejoc.201900847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Zou B.; Chan W. L.; Ding M.; Leong S. Y.; Nilar S.; Seah P. G.; Liu W.; Karuna R.; Blasco F.; Yip A.; Chao A.; Susila A.; Dong H.; Wang Q. Y.; Xu H. Y.; Chan K.; Wan K. F.; Gu F.; Diagana T. T.; Wagner T.; Dix I.; Shi P.-Y.; Smith P. W. Lead Optimization of Spiropyrazolopyridones: A New and Potent Class of Dengue Virus Inhibitors. ACS Med. Chem. Lett. 2015, 6, 344–348. 10.1021/ml500521r. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Tai V. W. F.; Garrido D.; Price D. J.; Maynard A.; Pouliot J. J.; Xiong Z.; Seal J. W.; Creech K. L.; Kryn L. H.; Baughman T. M.; Peat A. J. Design and synthesis of spirocyclic compounds as HCV replication inhibitors by targeting viral NS4B protein. Bioorg. Med. Chem. Lett. 2014, 24, 2288–2294. 10.1016/j.bmcl.2014.03.080. [DOI] [PubMed] [Google Scholar]; c Imaeda T.; Ono K.; Nakai K.; Hori Y.; Matsukawa J.; Takagi T.; Fujioka Y.; Tarui N.; Kondo M.; Imanishi A.; Inatomi N.; Kajino M.; Itoh F.; Nishida H. Discovery, synthesis, and structure-activity relations of 3,4-dihydro-1H-spiro(naphthalene-2,2′-piperidin)-1-ones as potassium-competitive acid blockers. Bioorg. Med. Chem. 2017, 25, 3719–3735. 10.1016/j.bmc.2017.05.012. [DOI] [PubMed] [Google Scholar]
- Harris P. A.; Berger S. B.; Jeong J. U.; Nagilla R.; Bandyopadhyay D.; Campobasso N.; Capriotti C. A.; Cox J. A.; Dare L.; Dong X.; Eidam P. M.; Finger J. N.; Hoffman S. J.; Kang J.; Kasparcova V.; King B. W.; Lehr R.; Lan Y.; Leister L. K.; Lich J. D.; MacDonald T. T.; Miller N. A.; Ouellette M. T.; Pao C. S.; Rahman A.; Reilly M. A.; Rendina A. R.; Rivera E. J.; Schaeffer M. C.; Sehon C. A.; Singhaus R. R.; Sun H. H.; Swift B. A.; Totoritis R. D.; Vossenkämper A.; Ward P.; Wisnoski D. D.; Zhang D.; Marquis R. W.; Gough P. J.; Bertin J. Discovery of a First-in-Class Receptor Interacting Protein 1 (RIP1) Kinase Specific Clinical Candidate (GSK2982772) for the Treatment of Inflammatory Diseases. J. Med. Chem. 2017, 60, 1247–1261. 10.1021/acs.jmedchem.6b01751. [DOI] [PubMed] [Google Scholar]
- Belyanskaya S. L.; Ding Y.; Callahan J. F.; Lazaar A. L.; Israel D. I. Discovering Drugs with DNA-Encoded Library Technology: From Concept to Clinic with an Inhibitor of Soluble Epoxide Hydrolase. ChemBioChem 2017, 18, 837–842. 10.1002/cbic.201700014. [DOI] [PubMed] [Google Scholar]
- Madsen D.; Azevedo C.; Micco I.; Petersen L. K.; Hansen N. J. V.. An overview of DNA-encoded libraries: A versatile tool for drug discovery. In Progress in Medicinal Chemistry ;Witty D. R., Brian Cox B., Eds.; Elsevier, 2020; pp 181–249. [DOI] [PubMed] [Google Scholar]
- Shi Y.; Wu Y.-R.; Yu J.-Q.; Zhang W.-N.; Zhuang C.-L. DNA-encoded libraries (DELs): a review of on-DNA chemistries and their output. RSC Adv. 2021, 11, 2359–2376. 10.1039/D0RA09889B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M. H.; Blakskjær P.; Petersen L. K.; Hansen T. H.; Højfeldt J. W.; Gothelf K. V.; Hansen N. J. V. A Yoctoliter-Scale DNA Reactor for Small-Molecule Evolution. J. Am. Chem. Soc. 2009, 131, 1322–1327. 10.1021/ja808558a. [DOI] [PubMed] [Google Scholar]
- Gerry C. J.; Wawer M. J.; Clemons P. A.; Schreiber S. L. DNA Barcoding a Complete Matrix of Stereoisomeric Small Molecules. J. Am. Chem. Soc. 2019, 141, 10225–10235. 10.1021/jacs.9b01203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy Guduru S. K.; Chamakuri S.; Raji I. O.; MacKenzie K. R.; Santini C.; Young D. W. Synthesis of Enantiomerically Pure 3-Substituted Piperazine-2-acetic Acid Esters as Intermediates for Library Production. J. Org. Chem. 2018, 83, 11777–11793. 10.1021/acs.joc.8b01708. [DOI] [PubMed] [Google Scholar]
- Chamakuri S.; Jain P.; Reddy Guduru S. K.; Arney J. W.; MacKenzie K. R.; Santini C.; Young D. W. Synthesis of Enantiomerically Pure 6-Substituted-Piperazine-2-Acetic Acid Esters as Intermediates for Library Production. J. Org. Chem. 2018, 83, 6541–6555. 10.1021/acs.joc.8b00854. [DOI] [PubMed] [Google Scholar]
- Arico-Muendel C. C. From haystack to needle: finding value with DNA encoded library technology at GSK. MedChemComm 2016, 7, 1898–1909. 10.1039/C6MD00341A. [DOI] [Google Scholar]
- Kölmel D. K.; Loach R. P.; Knauber T.; Flanagan M. E. Employing Photoredox Catalysis for DNA-Encoded Chemistry: Decarboxylative Alkylation of α-Amino Acids. ChemMedChem 2018, 13, 2159–2165. 10.1002/cmdc.201800492. [DOI] [PubMed] [Google Scholar]
- Satz A. L.Foundations of a DNA-Encoded Library (DEL). In A Handbook for DNA-Encoded Chemistry: Theory and Applications for Exploring Chemical Space and Drug Discovery ;Goodnow R. A., Eds.; Wiley, 2014; pp 99–121. [Google Scholar]
- Martín A.; Nicolaou C. A.; Toledo M. A. Navigating the DNA encoded libraries chemical space. Commun. Chem. 2020, 3, 127. 10.1038/s42004-020-00374-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag G.; Tsai J.; Zhang J.; Zhang C.; Ibrahim P.; Nolop K.; Hirth P. Vemurafenib: the first drug approved for BRAF-mutant cancer. Nat. Rev. Drug Discovery 2012, 11, 873–886. 10.1038/nrd3847. [DOI] [PubMed] [Google Scholar]
- Dias D. M.; Ciulli A. NMR approaches in structure-based lead discovery: recent developments and new frontiers for targeting multi-protein complexes. Prog. Biophys. Mol. Biol. 2014, 116, 92–100. 10.1016/j.pbiomolbio.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harner M. J.; Frank A. O.; Fesik S. W. Fragment-based drug discovery using NMR spectroscopy. J. Biomol. NMR 2013, 56, 65–75. 10.1007/s10858-013-9740-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navratilova I.; Hopkins A. L. Fragment Screening by Surface Plasmon Resonance. ACS Med. Chem. Lett. 2010, 1, 44–48. 10.1021/ml900002k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull W. B.; Daranas A. H. On the Value of c: Can Low Affinity Systems Be Studied by Isothermal Titration Calorimetry?. J. Am. Chem. Soc. 2003, 125, 14859–14866. 10.1021/ja036166s. [DOI] [PubMed] [Google Scholar]
- Vivat Hannah V.; Atmanene C; Zeyer D; Van Dorsselaer A; Sanglier-Cianferani S. Native MS: an ’ESI’ way to support structure- and fragment-based drug discovery. Future Med. Chem. 2010, 2, 35–50. 10.4155/fmc.09.141. [DOI] [PubMed] [Google Scholar]
- Neumann T.; Junker H.-D.; Schmidt K.; Sekul R. SPR-based Fragment Screening: Advantages and Applications. Curr. Top. Med. Chem. 2007, 7, 1630–1642. 10.2174/156802607782341073. [DOI] [PubMed] [Google Scholar]
- Murray C. W.; Rees D. C. The rise of fragment-based drug discovery. Nat. Chem. 2009, 1, 187–192. 10.1038/nchem.217. [DOI] [PubMed] [Google Scholar]
- Zega A. NMR Methods for Identification of False Positives in Biochemical Screens. J. Med. Chem. 2017, 60, 9437–9447. 10.1021/acs.jmedchem.6b01520. [DOI] [PubMed] [Google Scholar]
- Leach A. R.; Hann M. M. Molecular complexity and fragment-based drug discovery: ten years on. Curr. Opin. Chem. Biol. 2011, 15, 489–496. 10.1016/j.cbpa.2011.05.008. [DOI] [PubMed] [Google Scholar]
- Morley A. D.; Pugliese A.; Birchall K.; Bower J.; Brennan P.; Brown N.; Chapman T.; Drysdale M.; Gilbert I. H.; Hoelder S.; Jordan A.; Ley S. V.; Merritt A.; Miller D.; Swarbrick M. E.; Wyatt P. G. Fragment-based hit identification: thinking in 3D. Drug Discovery Today 2013, 18, 1221–1227. 10.1016/j.drudis.2013.07.011. [DOI] [PubMed] [Google Scholar]
- Downes T. D.; Jones S. P.; Klein H. F.; Wheldon M. C.; Atobe M.; Bond P. S.; Firth J. D.; Chan N. S.; Waddelove L.; Hubbard R. E.; Blakemore D. C.; De Fusco C.; Roughley S. D.; Vidler L. R.; Whatton M. A.; Woolford A. J.-A.; Wrigley G. L.; O’Brien P. Design and Synthesis of 56 Shape-Diverse 3D Fragments. Chem. - Eur. J. 2020, 26, 8969–8975. 10.1002/chem.202001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Cox B.; Zdorichenko V.; Cox P. B.; Booker-Milburn K. I.; Paumier R.; Elliott L. D.; Robertson-Ralph M.; Bloomfield G. Escaping from Flatland: Substituted Bridged Pyrrolidine Fragments with Inherent Three-Dimensional Character. ACS Med. Chem. Lett. 2020, 11, 1185–1190. 10.1021/acsmedchemlett.0c00039. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Garner P.; Cox P. B.; Rathnayake U.; Holloran N.; Erdman P. Design and Synthesis of Pyrrolidine-based Fragments That Sample Three-dimensional Molecular Space. ACS Med. Chem. Lett. 2019, 10, 811–815. 10.1021/acsmedchemlett.9b00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. A.; Nicolaou C. A.; Kirberger S. E.; Pandey A. K.; Hu H.; Pomerantz W. C. K. Evaluating the Advantages of Using 3D-Enriched Fragments for Targeting BET Bromodomains. ACS Med. Chem. Lett. 2019, 10, 1648–1654. 10.1021/acsmedchemlett.9b00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Fragments and Novelty. Online at https://www.cambridgemedchemconsulting.com/resources/hit_identification/fragmentNovelty.html (accessed April 2021).; b Fragments vs PKC-l: 7-azaindole strikes again. Online at http://practicalfragments.blogspot.com/2018/05/fragments-vs-pkc-7-azaindole-strikes.html (accessed April 2021).
- a Zhang C.; Ibrahim P. N.; Zhang J.; Burton E. A.; Habets G.; Zhang Y.; Powell B.; West B. L.; Matusow B.; Tsang G.; Shellooe R.; Carias H.; Nguyen H.; Marimuthu A.; Zhang K. Y. J.; Oh A.; Bremer R.; Hurt C. R.; Artis D. R.; Wu G.; Nespi M.; Spevak W.; Lin P.; Nolop K.; Hirth P.; Tesch G. H.; Bollag G. Design and pharmacology of a highly specific dual FMS and KIT kinase inhibitor. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 5689–5694. 10.1073/pnas.1219457110. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Kwiatkowski J.; Liu B.; Tee D. H. Y.; Chen G.; Ahmad N. H. B.; Wong Y. X.; Poh Z. Y.; Ang S. H.; Tan E. S. W.; Ong E. H.; Nurul Dinie; Poulsen A.; Pendharkar V.; Sangthongpitag K.; Lee M. A.; Sepramaniam S.; Ho S. Y.; Cherian J.; Hill J.; Keller T. H.; Hung A. W. Fragment-Based Drug Discovery of Potent Protein Kinase C Iota Inhibitors. J. Med. Chem. 2018, 61, 4386–4396. 10.1021/acs.jmedchem.8b00060. [DOI] [PubMed] [Google Scholar]
- Karawajczyk A.; Giordanetto F.; Benningshof J.; Hamza D.; Kalliokoski T.; Pouwer K.; Morgentin R.; Nelson A.; Müller G.; Piechot A.; Tzalis D. Expansion of chemical space for collaborative lead generation and drug discovery: the European Lead Factory Perspective. Drug Discovery Today 2015, 20, 1310–1316. 10.1016/j.drudis.2015.09.009. [DOI] [PubMed] [Google Scholar]
- There are multiple ways of analyzing structural diversity, for example, by shape analysis. SeeAkritopoulou-Zanze I.; Metz J. T.; Djuric S. W. Topography-biased compound library design: the shape of things to come?. Drug Discovery Today 2007, 12, 948–952. 10.1016/j.drudis.2007.08.017. [DOI] [PubMed] [Google Scholar]
- Dahlin J. L.; Walters M. A. The essential roles of chemistry in high-throughput screening triage. Future Med. Chem. 2014, 6, 1265–1290. 10.4155/fmc.14.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters W. P.; Namchuk M. Designing screens: how to make your hits a hit. Nat. Rev. Drug Discovery 2003, 2, 259–266. 10.1038/nrd1063. [DOI] [PubMed] [Google Scholar]
- Follmann M.; Briem H.; Steinmeyer A.; Hillisch A.; Schmitt M. H.; Haning H.; Meier H. An approach towards enhancement of a screening library: The Next Generation Library Initiative (NGLI) at Bayer — against all odds?. Drug Discovery Today 2019, 24, 668–672. 10.1016/j.drudis.2018.12.003. [DOI] [PubMed] [Google Scholar]
- Li Q.Virtual screening of small-molecule libraries. In Small Molecule Drug Discovery Methods, Molecules and Applications ;Trabocchi A., Lenci E., Eds.; Elsevier, 2020; pp 103–125. [Google Scholar]
- Real Space Navigator is just a click away. Online at https://www.biosolveit.de/REALSpaceNavigator/.
- Ultimate 100+ Million Compounds. Online at https://ultimate.mcule.com/.
- WuXi AppTec Research Service Division and BioSolveIT Introduce GalaXi, a Vast New Chemical Space of Tangible Molecules. Online at https://wxpress.wuxiapptec.com/wuxi-apptec-research-service-division-and-biosolveit-introduce-galaxi-a-vast-new-chemical-space-of-tangible-molecules/ (accessed April 2021).
- The PSL “Parallel Synthesis Laboratory”. Online at https://www.liverpoolchirochem.com/parallel-synthesis-laboratory (accessed April 2021).
- Zhang A.; Mu Y.; Wu F. An enantiomer-based virtual screening approach: Discovery of chiral organophosphates as acetyl cholinesterase inhibitors. Ecotoxicol. Environ. Saf. 2017, 138, 215–222. 10.1016/j.ecoenv.2016.12.035. [DOI] [PubMed] [Google Scholar]
- Guha R.; Van Drie J. H. Structure--activity landscape index: identifying and quantifying activity cliffs. J. Chem. Inf. Model. 2008, 48, 639–645. 10.1021/ci7004093. [DOI] [PubMed] [Google Scholar]
- a Watts K. S.; Dalal P.; Tebben A. J.; Cheney D. L.; Shelley J. C. Macrocycle Conformational Sampling with MacroModel. J. Chem. Inf. Model. 2014, 54, 2680–2696. 10.1021/ci5001696. [DOI] [PubMed] [Google Scholar]; b Drummond M. L.; Williams C. I. In Silico Modeling of PROTAC-Mediated Ternary Complexes: Validation and Application. J. Chem. Inf. Model. 2019, 59, 1634–1644. 10.1021/acs.jcim.8b00872. [DOI] [PubMed] [Google Scholar]
- Lu W.; Kostic M.; Zhang T.; Che J.; Patricelli M. P.; Jones L. H.; Chouchani E. T.; Gray N. S. Fragment-based covalent ligand discovery. RSC Chem. Biol. 2021, 2, 354. 10.1039/D0CB00222D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeley A.; Petri L.; Ábrányi-Balogh P.; Keserű G. M. Covalent fragment libraries in drug discovery. Drug Discovery Today 2020, 25, 983–996. 10.1016/j.drudis.2020.03.016. [DOI] [PubMed] [Google Scholar]
- Parker C. G.; Galmozzi A.; Wang Y.; Correia B. E.; Sasaki K.; Joslyn C. M.; Kim A. S.; Cavallaro C. L.; Lawrence R. M.; Johnson S. R.; Narvaiza I.; Saez E.; Cravatt B. F. Cell 2017, 168, 527–541. 10.1016/j.cell.2016.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Dix M. M.; Bianco G.; Remsberg J. R.; Lee H.-Y.; Kalocsay M.; Gygi S. P.; Forli S.; Vite G.; Lawrence R. M.; Parker C. G.; Cravatt B. F. Expedited mapping of the ligandable proteome using fully functionalized enantiomeric probe pairs. Nat. Chem. 2019, 11, 1113–1123. 10.1038/s41557-019-0351-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enantioprobe (S)-4. Online at https://www.sigmaaldrich.com/catalog/product/aldrich/913189?lang=en®ion=GB (accessed April 2021).
- Grant E. K.; Fallon D. J.; Hann M. M.; Fantom K. G. M.; Quinn C.; Zappacosta F.; Annan R. S.; Chung C.-W.; Bamborough P.; Dixon D. P.; Stacey P.; House D.; Patel V. K.; Tomkinson N. C. O.; Bush J. T. A Photoaffinity-Based Fragment-Screening Platform for Efficient Identification of Protein Ligands. Angew. Chem., Int. Ed. 2020, 59, 21096–21105. 10.1002/anie.202008361. [DOI] [PubMed] [Google Scholar]
- Martyloga O. V.; Myronenko A.; Tkachenko A. M.; Matvienko V. O.; Kuchkovska Y. O.; Grygorenko O. O. Multigram Synthesis of Functionalized Spirocyclic Diazirines. Eur. J. Org. Chem. 2019, 2019, 3744–3750. 10.1002/ejoc.201900485. [DOI] [Google Scholar]
- Ge S.-S.; Chen B.; Wu Y.-Y.; Long Q.-S.; Zhao Y.-L.; Wang P.-Y.; Yang S. Current advances of carbene-mediated photoaffinity labeling in medicinal chemistry. RSC Adv. 2018, 8, 29428–29454. 10.1039/C8RA03538E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser A. L.; Menzies S. K.; King E. F. B.; Tulloch L. B.; Gould E. R.; Zacharova M. K.; Smith T. K.; Florence G. J. Design and Synthesis of Broad Spectrum Trypanosomatid Selective Inhibitors. ACS Infect. Dis. 2018, 4, 560–567. 10.1021/acsinfecdis.7b00187. [DOI] [PubMed] [Google Scholar]
- Organic Building Blocks. Online at https://www.sigmaaldrich.com/chemistry/chemistry-products.html?TablePage=16270415 (accessed April 2021).