Abstract
Background
Over the last two decades, humanity has observed the extraordinary anomaly caused by novel, weird coronavirus strains, such as severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS). As the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) virus has made its entry into the world, it has dramatically affected life in every domain by continuously producing new variants. The vaccine development is an ongoing process, although some vaccines got marketed. The big challenge is now whether the vaccine candidates can provide long-lasting protection or prevention against mutant variants.
Methods
The information was gathered from various journals, electronic searches via Internet-based information such as PubMed, Google Scholar, Science Direct, online electronic journals, WHO landscape, world meters, WHO website, and News.
Results
This review will present and discuss some coronavirus disease 19 (COVID-19) related aspects including: the pathophysiology, epidemiology, mutant variants vaccine candidates, vaccine efficacy, and management strategies. Due to the high death rate, continuous spread, an inadequate workforce, lack of required therapeutics, and incomplete understanding of the viral strain, it becomes crucial to build the knowledge of its biological characteristics and make available the rapid diagnostic and vital therapeutic machinery for the combat and management of an infection.
Conclusion
The data summarizes current research on the COVID 19 infection and therapeutic interventions, which will direct future decision-making on the effort-worthy phases of the COVID 19 and the development of critical therapeutics. The only possible solution is the vaccine development targeting against all variant strains to halt its progress; the identified theoretical and practical knowledge can eliminate the gaps to improve a better understanding of the novel coronavirus structure and its design of a vaccine. In addition, to that the long-lasting protection is another challenging objective that need to be looked into.
Abbreviations: ACE2, angiotensin-converting enzyme-2; ADRP, ADP ribose-1′-phosphatase; ARTES, Germany: based biotechnology company specialized in recombinant protein production and process development from microbial expression systems; BIOCAD, BIO computer aided design RNA; CanVirex AG, Swiss Biotech Association; CARDS, Covid 19 acute respiratory distress syndrome; CBC, complete blood count; CCHFV, Crimean-Congo hemorrhagic fever virus; CDC, center for disease control and prevention; CEA, carcinoembryonic antigen; CHIKV, chikungunya virus; CMV, cytomegalovirus; CNBG, China National Biotec Group; CNRS, centre national de la recherche scientifique; COPD, chronic obstructive pulmonary disease; COVID-19, corona virus disease 2019; CPAP, continuous positive airway pressure; CRP, C-reactive protein; DMVs, double-membrane vesicles; DWRAIR, Diseases/Walter Reed Army Institute of Research; DZIF, German Center for Infection Research; EBOV, Ebola virus; ECDC, European Centre for Disease Prevention and Control; ERGIC, endoplasmic reticulum- golgi intermediate compartment; ExoN, exoribonuclease; FiO2, fraction of inspired oxygen; GCIR, German Center for Infection Research; GMV, glycine mosaic virus; GLA, glucopyranosyl Lipid A; GPO, Government Pharmaceutical Organization; HeV, hepatitis virus E; HBV, hepatitis B virus; HFNC, high-flow nasal cannula; HLC, high lung compliance; IAVI, international AIDS vaccine initiative; IDIBAPS, Pi i Sunyer Biomedical Research Institute; IEM, Institute For Engineering in Medicine; InfA, influenza virus-A; INRAE, National Research Institute for Agriculture, Food and Environment; IPV, inactivated polio virus; IMV, Instruments de Médecine Vétérinaire; LASSA, lassa virus; LASV, lassa mammarenavirus; LinKinVax, French biotechnology startup that focuses on speeding up vaccine; LiteVax BV, spike-based (epitope screening); LLC, low lung compliance; LVVV, live viral vectored vaccine; MARV, Marburg virus; MDA5, melanoma differentiation associated protein; MERS-CoV, Middle East Respiratory Syndrome coronavirus; MIGAL, Galilee Research Institute Ltd; MMR, measles mumps rubella; MVA, modified vaccinia virus Ankara; NERVTAG, new and emerging respiratory virus Threats advisory group; NiV, Nippa virus; NIV, non-invasive ventilation; NLC, nanostructured Lipid Carriers; NORV, norovirus; NSCLC, non-small cell lung cancer; NSP, non-Structural proteins; O-MT, O-methyl transferase-2; OMV, outer membrane vesicle; ORFs, open reading frames; Osivax, clinical stage biotechnology company; P.C., preclinical; PaCO2, partial pressure of carbon dioxide; PaO2, partial pressure of oxygen; PEEP, positive end-expiratory pressure; PPI, proton pump inhibitors; RBD, receptor-binding domain; RVF, Rift Valley fever; RdRp, RNA dependent RNA polymerase; RTC, replication transcription complex; RTI, respiratory tract infections; SARS-CoV-2, severe acute respiratory syndrome coronavirus-2; SIG, SARS-COV-2 Interagency Group; SpO2, oxygen saturation; SRC VB VECTOR, State Research Centre of Virology and Biotechnology; TMPRSS2, transmembrane protein serine 2; TRSs, transcriptional regulatory sequences; USAMRIID/WARIAR, United States Army Medical Research Institute of Infectious; VEE, Venezuelan equine encephalitis; VLP, virus like particle; VOC, variant of concern; VOHC, variant of high consequences; VOI, variant of interest; VRI, Vaccine Research Institute; VSV, vesicular stomatitis virus
Keywords: SARS-COV-2, Epidemiology, Variant strains, Vaccine candidates, WHO landscape
Introduction
Andrews and Gledhill in 1951 screened a hepatitis virus from mice which are now known as a single-stranded RNA coronavirus. Its diseases and cause have been recognized in animals and humans for over 50 years. 229E and OC43 were the first coronaviruses to cause very mild infections like common colds in humans. Later on, severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) emerged from civet cats, camels, bats [1]. The family of coronavirus is called as Coronaviridae which comprises of two subfamilies, Coronaviridae and Letovirinae [2,3]. Pneumonia’s initial diagnosis and unknown cause made the cluster of patients in China admitted into hospitals in December 2019. The reports confirmed the potential coronavirus outbreak, of coronavirus disease 19 (COVID-19), and WHO gave it a name on February 11, 2020 [4]. In response to the outbreak, the center for disease control and prevention (CDC) conducted epidemiological and etiological investigations via Wuhan city's Health authorities. Nearly 1 billion cases per year and economic losses of hundreds of billions of dollars occur due to this zoonotic illness, demonstrating the importance of developing vaccine design strategies for virus families with pools of causing extensive zoonotic diseases. The transcriptional regulatory sequences (TRSs) mediates discontinuous transcription and transcribes sub genomic RNAs, which make up the structure of a virus particle [5,6]. Advances in understanding viral machinery, the role of various viral proteins, their genetic structure, host immune responses, and the ability to confer protection switch its way to developing a vaccine successfully. An additional challenge is conducting clinical trials in this pandemic loss by utilizing the current evidence and critical knowledge gaps to better understand the virus strategy to safeguard public health. This review, has focused and emphasized points on the COVID 19 pathophysiology, epidemiology, mutant variants, vaccine candidates, vaccine efficacy and strategies for disease management.
Pathophysiology
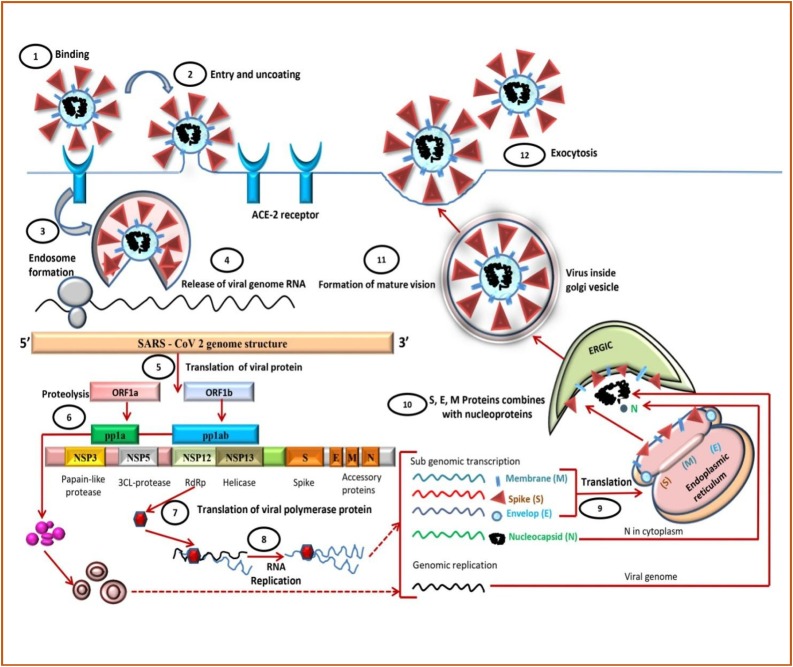
Coronavirus spike glycoprotein (S) with the expression of their subunits S1 and S2 binds to the host cell receptor's surface. S1 determines the cellular tropism and host range and helps attach the virus to the target cell. In contrast, the S2 subunit helps infusion with the host cells (mainly alveolar epithelial cells and small intestine enterocytes) with the aid of Angiotensin-Converting Enzyme-2 (ACE2) claw-like structure of the receptor by endocytosis [7,8]. When this S protein binds to the ACE-2 receptor, the transmembrane protein serine 2 (TMPRSS2), which is associated with the cell surface, mediates cleavage of S protein trimer, and the fusion mechanism is activated via endosomal acid proteases (cathepsin L) [[9], [10], [11]]. ACE2 is an 805 amino acid containing integral membrane protein consists of 3 domains that are a domain at the transmembrane side, a domain at the extracellular side and a domain at the cytoplasmic side, and the N-terminal peptide. The enzyme sheddase helps release soluble protein into the bloodstream by cleavage of the active carboxypeptidase domain from the transmembrane domain [12]. When there is a lysosome-mediated drop in pH, the endosome membrane fuses with the virus’s envelope and releases a nucleocapsid in the cytoplasm. The cellular proteases degrade the capsid, and the positive sense RNA viral genome ranges between 27−32 kbp is left free. This viral genomic RNA consists of at least 6 open reading frames (ORFs). ORF1a and ORF1b are present at the 5′end, constitute a common fraction of the whole genome length RNAs, and are translated into pp1a and pp1ab proteins. Protease helps in cleavage of those proteins in to 16 non-structural proteins (NSP1 –NSP16) [[13], [14], [15]]. Some of these NSPs, like NSP3, encode for papain-like proteases (PLP) and NSP5 codes for 3CL-proteases. Both of these proteins form a polypeptide “Replication transcription complex” (RTC) and helps in innate immune response blockage via genomic transcription and translation process. NSP15 encodes for RNA helicase, whereas NSP12 for RNA dependent RNA polymerase (RdRp) and synthesis of subgenomic RNAs (sgRNAs) stand genomic RNAs occurs [16] (Fig. 1 ).
Fig. 1.
The life cycle of SARS-CoV-2 in the host cells.
Other NSPs protein functions are listed below in Table 1
Table 1.
Functions of non-structural proteins (NSP1-16).
| Nonstructural Proteins | Function | Reference |
|---|---|---|
| NSP1 | Innate immune response blockage by promotion of mRNA degradation | [[24], [25], [26], [27]] |
| NSP2 | Unknown function, binds to prohibiting proteins | [28,29] |
| NSP3 | Papain lyase, cytokine expression promotion, blocking host innate immune response, activity of ADRP | [[30], [31], [32], [33], [34], [35]] |
| NSP4 | Potential transmembrane scaffold protein, maintains DMVs proper structure | [36,37] |
| NSP5 | Main protease (Mpro), viral polyprotein cleavage | [38] |
| NSP6 | Potential transmembrane scaffold protein | [39] |
| NSP7 | NSP7 forms a hexadecameric complex with NSP8. For RNA polymerase, it acts as a processivity clamp | [40] |
| NSP8 | For RNA polymerase, it acts as a processivity clamp and also plays a role as a primase | [41,42] |
| NSP9 | Binding protein for RNA | [43] |
| NSP10 | It acts as a cofactor for NSP14 and 16 by forming a heterodimer with both of them and stimulating the activity of N viral exoribonuclease (ExoN) and O-methyl transferase | [44,45] |
| NSP12 | RdRp) | [46] |
| NSP13 | RNA Helicase, 5′ Triphosphatase | [47,48] |
| NSP14 | Shows ExoNactivity which is important for viral genome proofreading, addition of 5′ cap to viral RNAs by MTase. | [[49], [50], [51], [52]] |
| NSP15 | Endoribonuclease of virus | [53,54] |
| NSP16 | O-methyl transferase (2′-O-MT); viral RNA shielding from MDA5 recognition | [55,56] |
Four Structural proteins are coded by ORFs located on 3′ end:
-
1)
Membrane (M) shape the virions [17].
-
2)
Spike (S) recognizes the ACE2 receptor on the host cell surface [18].
-
3)
Envelope (E) helps in assembly and release of virions [19].
-
4)
Nucleocapsid (N) packages the genome in the virions, provides pathogenicity.
There are many other structural and accessory proteins that are specific to the different species. Membrane exocytosis helps budding of completed assembled SARS-COV-2 particles via endoplasmic reticulum in the endoplasmic reticulum-golgi intermediate compartment (ERGIC). It is currently known that interaction of mainly M protein with different structural proteins of virus E, S aids in the generation of the virion scaffold promoting assembly, budding, and release of mature virus particle by exocytosis. After the final phase of maturation, all the components of the virus fit together, the particle is infectious and ready to begin a new cycle [[20], [21], [22], [23]].
Epidemiology
Coronaviruses which mainly infect birds and mammals, comprise a large family (Coronaviridae) of giant enveloped positive-strand RNA viruses, which can be a cause of upper and lower respiratory tract infections (RTI) manifest as pneumonia, bronchitis, MERS, SARS, COVID 19. These three new coronaviruses caused respiratory disease outbreaks with their unique features, but SARS and MERS are less infectious and have significantly higher fatality rates than SARS-COV-2. The following Table 2 demonstrates the fundamental difference between the three of them [[57], [58], [59]].
Table 2.
Differences of SARS-COV, MERS CoV, SARS-COV-2.
| SARS-CoV | MERS-CoV | SARS-CoV-2 | |
|---|---|---|---|
| First notified | November 2002 in China’s Guangdong province | September 2012, in Saudi Arabia | December 2019 in Wuhan, China |
| Mode of transmission | Infected civets, droplets produced by sneezing, coughing, breathing, talking | Droplets from person to person (unclear from camels to humans) | Droplets by coughing, sneezing, talking |
| Mean incubation period | 4−5 days | 6−7 days | 1−14 days |
| Key symptoms | Dry cough at first, fever, malaise, body aches and pains, diarrhea (in the first or second week) | Fever, cough, shortness of breath, nausea/vomiting, diarrhea | Fever, dry cough, shortness of breath, fatigue |
| Treatment | No specific treatment | No specific treatment | No specific treatment |
| Mortality rate | 11% | 34% | 3−4% |
| Vaccine | No vaccine | No vaccine | No vaccine |
Origin
Disease spread globally over eight million after the emergence of many pneumonia-like cases in Wuhan, Hubei province China (Early December 2019).
Topological dispersal
On December 31, 2019, the Chinese government first confess the numerous cases of pneumonia of an unknown cause presumed to be a zoonotic illness that would later be called COVID 19. The first case of death was reported on January 11. The Wuhan city was locked down with over 11 million populations on January 23. A week later, thousands of cases were reported, and WHO immediately declared a global health emergency [60]. With the start of July 2020, there were approximately 8.6 million cases and 450,000 deaths due to COVID 19 [61].
Global response
Italy
Italy was the following unfortunate country after China experienced a significant outbreak and resulted in the highest death rate, 7.2%. Milan, the most dynamic city alone, represents 10% of the Italian economy, “The country’s economic engine” was drastically slowed after Italy experienced a surge in coronavirus cases. Italy went from discovering the first official COVID 19 case to the prohibition of all movements and non-essential business activities in a matter of weeks (February 21- March 22). Around 3,949,517 cases and 119,021 deaths took place until 25th of April 2021 [61].
France and Germany
The “quick and dirty Sunday morning” analysis confirmed the trend of viral proliferation in France and Germany exceeds more than in Italy, South Korea, and Japan with a similar temperature. The study also suggests that intense containment measures as in Italy, South Korea, and Japan may help to slowdown proliferation [62]. France is 4th country affected and around 5,473,579 cases and 102,713 deaths took place until 25th of April 2021. Germany 10th leading country in COVID 19 cases and around 3,277,661 cases and 82,117 deaths took place until 25th of April 2021 [61].
Japan
Japan, though with a more significant percentage of the population, experienced a low mortality rate compared to Italy mainly because of their cluster-based testing approach and adoption of the “3C” method (avoidance of closed spaces, crowded spaces, close contact) (Bloomberg, 2020). Around 556,999 cases and 9854 deaths were reported in Japan until 25th of April 2021 [61].
United States
U.S reached the highest count of reported cases worldwide from the first known patient in late January to August 28th where there are 6 million cases and nearly 2 lakh deaths. About 32,766,119 cases have been reported and 585,449 deaths had took place on 25th of April 2021 and US is at the forefront [61].
Brazil
The second highest number of cases is found in Brazil, especially the Amazon state country’s northwest, which has the highest mortality rate. Manaus, the state capital, and the bustling city make the potential hotspots for transmitting the virus (CDC, 2020).). Brazil is third affected country and around 14,238,110 cases and 386,623 deaths took place until 25th of April [61].
Russia
Russia was centered in Moscow’s city, accounted for the highest number of cases measuring 257.7 thousand, followed by Moscow Oblast with 67 thousand cases by August 24, 2020. It has registered ten folds of low mortality than Spain, Britain, Italy, and France despite having many cases. The country’s well-funded healthcare system is to be appreciated for managing better than those in the U.S and Western European countries. Mass testing (has nearly 6 million tests carried out so far) helps people identify and isolate more people affected by the virus. They quickly convert hospitals and clinics to virus treatment centers. Russia is also at top leading in COVID 19 cases. Around 4,753,789 cases and 107,900 deaths took place until 25th of April 2021 [61].
India
India, among all other countries, has the lowest fatality rate (2.41%) as of July 23 in the World even though it stands third after the United States and Brazil for having a large number of cases in Asia. The first case of COVID 19 was reported in Kerala on January 30. By mid of May 2020, 6 major cities like Delhi, Mumbai, Kolkata, Chennai, Pune, and Ahmedabad accounted for around 50% of all cases reported in the country [65]. In India, there is progressive rise of COVID 19 and about 16,951,621 cases and 192,309 deaths are reported till 25th of April 2021 [61].
Etiology in different groups
The virus is transmitted via major routes such as droplets, contact, and aerosols. It has also been detected in the faecal samples of patients in the United States and China. Major risk factors include people more than 60 years of age, and even people underlying non-communicable diseases (NCDs): diabetes, hypertension, chronic lung disease, cerebrovascular disease, cardiac disease, chronic kidney failure, immune-suppression and cancer patient. Due to immunosuppressive state and many other physiological adaptive changes pregnant women are more susceptible to RTI but currently evidence of SARS-CoV-2 transmission through the placenta to the new-born has not been observed. In one of the studies, the samples of amniotic fluid, blood obtained from the umbilical cord, throat swab of neonates, and maternal milk were collected from new-born babies whose mothers were SARS-CoV-2 positive. Still, none of the neonates were found to be positive [66].
Mutations and severity of infection
Before SARS-CoV-2 other viruses also mutate themselves. There are different mutant variants of SARS-CoV-2 that have reported and increased the severity of infections. In UK, new and emerging respiratory virus threats advisory group (NERVTAG) published a paper with the result outcome from many preliminary analyses of B.1.1.7 [67]. One of the variants was detected in England and was highly transmissible and got dispersed to several other countries. This variant contains seventeen mutations in the genome in which 8 are on S protein which is main antigenic target of 3 SARS-CoV-2 vaccines that have been licensed in England [67]. The NERVTAG proposed that infections by B.1.1.7 have high chances of death, as compared to parent virus. The other highly infectious variant P.1 in Brazil was reported in the mid-2020. This variant has increased the rate of infections, severity of the disease and the Manaus city, in Brazil where the health department is totally in a collapsed position. Today Brazil is third leading country in the SARS-CoV-2 infection globally because of these variants [67].
The B.1.351 variant was first identified in South Africa in 2020. The vaccines manufactured by Moderna claimed on Jan 25, 2021 that our vaccine is effective against both B.1.1.7 and B.1.351 variants. However, the claim was based on an in vitro study. In addition to that, a South African variant was having decreased levels of neutralizing antibody titers. The company Pfizer has claimed that our vaccine will work against B.1.1.7 variant because of their investigations in laboratory, although these studies have not been peer reviewed [67]. As long as the variants emerged, the more havoc it will create, vaccines will not work and the severity of the disease will be more. In India, more than three hundred thousand cases hit each day as on 25th of April 2021 because of the double and triple mutant variants [61]. There should be big focus on the drug development against SARS-COV-2 rather than whole focus on the vaccines. Such as in cases of other viral diseases which have been controlled by drugs not by vaccines like HIV. Vaccines may be effective against a single variant but drugs might work against many variants as is evidenced by many antiviral drugs. There are three CDC established classifications for multiple variants of the virus in collaboration with SIG. They are: variant of concern (VOC), variant of interest (VOI), and variant of high consequences (VOHC).
VOHC’s
These variants have clear evidence that medical counter measures (MCMs) or measures for prevention reduced the effectiveness significantly compared to the previous variants. Fortunately, there are no SARS-CoV-2 variants observed so far at this high level of consequences [68].
VOC’s
There is a clear evidence of transmissibility, severity, and immunity which require efforts to control the virus spread, CDC reporting, health actions of public, test and research to evaluate the vaccine effectiveness [68]. List of variants are given in Table 3 .
Table 3.
Variants of concern (VOC).
| Additional mutations and lineage | First detected | Substitutions on spike protein | Impact on immunity evidence |
|---|---|---|---|
| B.1.1.7 (20I/501Y.V1) | United Kingdom (Sept 2020) | Δ69/70, Δ144, (E484K*), (S494P*), N501Y, A570D, D614G, P681H, T716I, S982A, D1118H (K1191N*) | Unclear [2] |
| B.1.1.7 + E484K | United Kingdom (Dec 2020) | E484K, N501Y, D614G | Neutralisation (v) [2,5] |
| B.1.351 (20H/501.V2) | South Africa (Sep 2020) | D80A, D215G, Δ241/242/243, K417N, E484K, N501Y, D614G, A701V | Escape (v) [7,8] |
| B.1.427 (20C/S:452R) | U.S.A (California) | L452R, D614G | Neutralisation (v) |
| B.1.429 (20C/S:452R) | U.S.A (California) | S13I, W152C, L452R, D614G | Neutralisation (v) |
| P.1 (20J/501Y.V3) | Japan/Brazil | L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I | Neutralisation (v) [11] |
* = Aminoacid substitution on the variants.
VOI’s
These variants have genetic markers which are specific which changes the binding of receptor, treatment efficacy reduction, has a clear evidence of transmissibility, severity, neutralization reduction by previously generated infection or vaccination. These variants require health actions of public, increased laboratory characterization, assessing how quickly the virus spreads by epidemiological investigations. B.1.617 and B.1.617.2 are classified as VOC by WHO and UK on 7th May 2021 respectively [2]. List of variants of interest are given in Table 4 .
Table 4.
Variants of interest (VOI) [2].
| Additional mutations and lineage | First detected | Substitutions on spike protein | Impact on immunity evidence |
|---|---|---|---|
| B.1.525 | Nigeria (Dec 2020) | E484K, D614G, Q677H | Neutralisation |
| B.1.427/B.1.429 | United States (Sep 2020) | L452R, D614G | Neutralisation |
| P.3 | The Philippines (Jan 2021) | E484K, N501Y, D614G | Neutralisation |
| B.1.616 | France (Feb 2021) | V483A, D614G, H655Y, G669S | |
| B.1.617 (20A) | India (Feb 2020) | L452R, E484Q, D614G | |
| B.1.617.1 (20A/S:154K) | India (Dec 2020) | (T95I), G142D, E154K, L452R, E484Q, D614G, P681R, Q1071H | Neutralisation |
| B.1.617.2 (20A/S:478K) | India (Dec 2020) | T19R, (G142D), Δ156, Δ157, R158G, L452R, T478K, D614G, P681R, D950N | Neutralisation |
| B.1.617.3 (20A) | India (Oct 2020)/(Feb 2021) | T19R, G142D, L452R, E484Q, D614G, P681R, D950N | Neutralisation |
| B.1.620 | Place not clear (Feb 2021) | S477N, E484K, D614G | Neutralisation |
| B.1.621 | Colombia (Jan 2020) | R346K, E484K, N501Y, D614G | Neutralisation |
Variants under monitoring
Through genomic variant rules-based screening, preliminary scientific evidence, epidemic intelligence these variants are detected as signals. There is a weak evidence which is not been assessed by European centre for disease prevention and control (ECDC) that they show similar properties as of VOC (Table 5 ).
Table 5.
Variants of monitoring.
| Additional mutations and lineage | First detected | Substitutions on spike protein | Impact on immunity evidence |
|---|---|---|---|
| B.1.214.2 | Place not clear (Dec 2020) | Q414K, N450K, ins214TDR, D614G | |
| A.23.1 + E484K | United Kingdom (Dec 2020) | E484K, Q613H | Neutralisation |
| A.27 | Place not clear (Dec 2020) | L452R, N501Y, H655Y | Neutralisation |
| A.28 | Place not clear (Dec 2020) | E484K, N501T, H655Y | Neutralisation |
| C.16 | Place not clear (Oct 2020) | L452R, D614G | Neutralisation |
| C.37 | Peru (Dec 2020) | L452Q, F490S, D614G | |
| B.1.351 + P384L | South Africa (Dec 2020) | P384L, K417N, E484K, N501Y, D614G | Escape |
| B.1.351 + E516Q | Place not clear (Jan 2021) | K417N, E484K, N501Y, E516Q, D614G | Escape |
| B.1.1.7 + L452R | United Kingdom (Jan 2021) | L452R, N501Y, D614G | Neutralisation |
| B.1.1.7 + S494P | United Kingdom (Jan 2021) | S494P, N501Y, D614G | Neutralisation |
| C.36 + L452R | Egypt (Dec 2020) | L452R, D614G | Neutralisation |
| AT.1 | Russia (Jan 2021) | E484K, D614G | Neutralisation |
| B.1.526 (20C/S:484K) | United States New York (Nov/Dec 2020) | (L5F*), T95I, D253G, (S477N*), (E484K*), D614G, (A701V*) | Neutralisation |
| B.1.526.1 (20C) | United States New York (Oct/Nov 2020) | D80G, Δ144, F157S, L452R, D614G, (T791I*), (T859N*), D950H | Neutralisation |
| B.1.526.2 | United States (Dec 2020) | S477N, D614G | |
| B.1.1.318 | Place not clear (Jan 2021) | E484K, D614G | Neutralisation |
| P.2 | Brazil (Jan 2021) | E484K, D614G | Neutralisation |
Detected in few sequences but not all.
Biomarkers helpful in assessing the clinical trend are HFNC (high-flow nasal cannula), CPAP (continuous positive airway pressure), NIV (noninvasive ventilation), PEEP (positive end-expiratory pressure), FiO2 (fraction of inspired oxygen), PaO2 (partial pressure of arterial blood), ARDS (acute respiratory distress syndrome), HLC (High Lung Compliance), LLC (Low lung Compliance) [68].
Management of COVID 19
Breathing movement: oxygen support: 5–15% of patients with COVID 19 require intensive care and ventilatory support as it primarily injures the vascular endothelium as it is a systemic disease, and a patient with COVID 19 ARDS that is covid 19 acute respiratory distress syndrome (CARDS) can develop multiorgan failure if not managed expertly. There are two types of SARS-COV-2 phenotypes for Respiratory support management. Target SpO2: 92–96% (82–92% in COPD patients), PaO2 ≥ 8 kPa = 60 mm Hg, PaCO2 < 6 kPa or pH > 7.3, FiO2 ≤ 0.4, Morphine sulphate, Midazolam, Benzodiazepine is given as per the severity of disease. Awake proning, monitoring C- reactive protein (CRP), CBC examination, chest x-ray [69]. Table 6 , shown, demonstrates the differences between different phenotypes [70,71].
Table 6.
Effect and management of corona in different phenotypes.
| L-Phenotype | H-Phenotype |
|---|---|
| Low elastance (HLC) | High elastance (LLC) |
| Low ventilation perfusion ratio | High shunt from right to left |
| The low weight of lung | The high weight of lung |
| Low recruitability | High recruitability |
| PaO2/FiO2 = 95 mmHg | PaO2/FiO2 = 84 mmHg |
| Non-intubated patients | Severe ARDS treated patients |
| -HFNC, CPAP, NIV | -PEEP is higher, |
| Intubated patients: | -Prone position lying |
| -Volumes greater than 6 mg/kg (up to 8−9 mg/kg) can be ventilated for hypercapnic patients | -Extracorporeal support |
| -PEEP reduced 8−10 cm H2O | |
| -Prone positioning only as a rescue maneuver |
Cough management
Sixty percent of COVID 19 positive cases report dry cough with zero phlegm production due to inflammation or irritation in your respiratory tract, which can get better with steam, humidifiers, honey consumption, cough suppressants, saltwater gargle, codeine phosphate, morphine sulfate as per the severity of infection [72,73].
Fever management
Fever after five days of infection is the most common in 99% of cases as your body’s normal immune response tries to kill a virus. Advising the patients to consume a large number of fluids and to take paracetamol or ibuprofen can help to manage fever. Still, the use of ibuprofen use has marked a question of concern as the adaptive immune response will interfere with the release of prostaglandins, suppressing the fever response also leads to an increase in activity of lymphocyte, hyperemic response, and organ tissues oxygenation, causing failure of multiple organs [74,75].
GI disturbances
Pain in the abdomen (3.6%), looseness of the bowels (10.1%), Emesis (3.6%) is joint in COVID 19 patients due to its ACE2 receptor, which is expressed highly in gastrointestinal intestinal epithelial cells. Also, its viral RNA has been found in stool specimens of patients. A cohort endoscopy study of 95 COVID 19 patients reported six additional cases and identified viral RNA in the stomach, esophagus, intestinal duodenum, and rectum from 2 severe cases. Treatment relies on supportive care, antiemetics, proton pump inhibitors (PPI), antidiarrheals, promethazine, ondansetron, metoclopramide, adequate hydration, fresh ginger boiled in water added with honey can also treat nausea and weakness can reduce vomiting. If loose motions persist, stop the diet and consume coconut water (John Wiley, 2020), (Lipi Roy, 2020).
Vaccines
Acute viral infections remain a leading cause of morbidity andmortality. This novel coronavirus pandemic has triggered unprecedented global health researchers and scientists to find a safe, effective vaccine against this virus. Extensive research can be done by gaining the knowledge from SARS and MERS vaccines development strategies and knowing the key targets such as receptor-binding domain (RBD) of spike protein, nucleotide identification, immunization route, suitable animal model utilization, production facility scalability, are some of the essential parameters to be considered [78]. There are seven COVID 19 vaccines in the third phase of clinical trials. Out of which four are from China, two of the candidates are from China National Biotec Group (CNBG) [79]. On August 27, Sputnik V advanced Russia vaccine trials of Sputnik V announced COVID 19 vaccine trials for over six months in forty thousand volunteers (Table 8).
Table 8.
Draft of COVID 19 vaccine candidates- WHO landscape 2020.
| Description of vaccine | Type of vaccine candidate | The target for coronavirus | Non-corona virus candidate’s same platform | Developers |
|---|---|---|---|---|
| A vaccine based on DNA | Vaccine inserts compatible with multiple delivery systems are engineered | SARS-CoV-2 and Sarbeco-CoV | University of Cambridge + DIOSynVax Ltd | |
| DNA Vaccine | SARS-CoV2 | University of Ege | ||
| Plasmid DNA vaccine N and RBD | Nottingham/Nottingham Trent University/Scancell | |||
| DNA with electroporation | Cobra Biologics/Karolinska Institute | |||
| DNA with electroporation | Vaccine Research Center Chula | |||
| DNA | Evvivax/Applied DNA Sciences/Takis | |||
| Needle-free delivery for DNA with plasmid | SARS | PharmaJet/Immunomic Therapeutics, Inc./EpiVax, Inc. | ||
| S, S1, S2, RBD and N, DNA plasmid vaccine | Egypt, National Research Centre | |||
| DNA vaccine | BioNet Asia | |||
| ms DNA vaccine | Waterloo University/MediphageBioceuticals | |||
| DNA Vaccine | Santos Pharmaceuticals | |||
| S-gene containing DNA plasmids | Biosun Pharmed | |||
| Plasmid vaccine with DNA | Bangladesh, Globe Biotech Limited | |||
| Nanostructured RBD with plasmid DNA | Slovenia, National Institute of Chemistry | |||
| Vaccine encoding RBD with plasmid DNA | Norway, Vaccibody, Oslo Research Park | |||
| Immunostimulatory DNA sequences | Inserm | |||
| Inactivated virus (IV) | CpG 1018 + Inactivated, egg-based, Membrane expressing whole Chimeric Newcastle Disease Virus (NDV) - SARS - CoV- 2, S protein (Lexapro) anchored Pre-fusion-stabilized trimeric | PATH/Dynavax/Institute of Vaccines and Medical Biologicals (IVAC; Vietnam) | ||
| CpG 1018 + Inactivated, egg-based, Membrane expressing whole Chimeric Newcastle Disease Virus (NDV) - SARS - CoV- 2, S protein (Lexapro) anchored Pre-fusion-stabilized trimeric | PATH/Dynavax/GPO; Thailand) | |||
| CpG 1018 + Inactivated, egg-based, Membrane expressing whole Chimeric Newcastle Disease Virus (NDV) - SARS - CoV- 2, S protein (Lexapro) anchored Pre-fusion-stabilized trimeric | PATH/Dynavax/Institute Butantan (Brazil) | |||
| Alum + Inactivated | J.E., ZIKA | KM Biologics | ||
| Inactivated | University of Selcuk | |||
| NIBIOHN/BIKEN/University of Osaka | ||||
| CpG 1018 + Inactivated | Dynavax/Sinovac | |||
| CpG 1018 + Inactivated | Dynavax/Valneva | |||
| The whole virus Inactivated | Egypt, National Research Centre | |||
| Inactivated | Kocak Farma Ilac ve Kimya San. A. S | |||
| Alum + Inactivated | Shifa Pharmed | |||
| Inactivated | MMR, IPV | Milad Pharmaceutics Co. | ||
| Inactivated | MMR, IPV | Zista Kian Azuma Co. | ||
| Live attenuated virus (LAV) | Live attenuated vaccines, which are codon deoptimized | AcıbademLabmed Health Services A.S./Mehmet Ali Aydinlar University | ||
| Live attenuated vaccines, which are codon deoptimized | University of Griffith/Indian Immunologicals Ltd | |||
| Live attenuated bacterial vector (LABV) | Live attenuated vaccine bacterial (pertussis) | Institut Pasteur Lille | ||
| Live attenuated bacterial vector | TheRex, ALtraBio | |||
| Viral vector (non-replicating) | Sendai virus vector | I.D. Pharma | ||
| Adenovirus-based | University of Ankara | |||
| (AAV SARS-COV-2) Adeno-associated virus vector | AveXis/Massachusetts General Hospital/Massachusetts Eye and Ear | |||
| VLP encoded by MVA | MARV, HIV, EBOV, LASV | BravoVax/GeoVax | ||
| MVA-S encoded | Multiple candidates | IDT Biologika GmbH/GCIR-DZIF | ||
| MVA-S | Spain, IDIBAPS-Hospital Clinic | |||
| Adeno5-based | University of Erciyes | |||
| (GREVAX™ platform) Ad5 S | MERS | Greffex | ||
| Oral Ad5 S | HSV-2, VZV, Zika and Norovirus | Stabilitech Biopharma Ltd | ||
| HLA-matched peptides + adenovirus-based | Pan-Corona | Valo Therapeutics Ltd | ||
| Structural proteins expressing MVA | SARS-CoV2 | Multiple candidates | Spain, Centro Nacional Biotecnología | |
| Spike protein expressing vaccine Parainfluenza virus 5 (PIV5)-based | MERS | Lowa University/Georgia University | ||
| S1 containing Recombinant deactivated rabies virus | CCHFV, LASSA, EBOV, MERS, NiV, HeV | Thomas Jefferson University/Bharat Biotech | ||
| H1N1 vector H1N1 vector | Egypt, National Research Centre | |||
| S expressing Newcastle disease virus | Mount Sinai, Icahn School of Medicine | |||
| Lentiviral Vector | Institut Pasteur -Theravectys | |||
| Lentiviral Vector | AIOVA | |||
| Retro-VLP Particles Lentiviral Vector | University of Sorbonne | |||
| intranasal administration Ad 5 vector | Eastern Finland University and Helsinki University | |||
| TBD | VEE, HBV, RVF, EBOV, LASV, CHIKV, NORV, InfA | Vaxart | ||
| Protein subunit (P.S.) | Mannose-conjugated chitosan nanoparticle delivered via RBD protein | University of Kazakh National Agrarian/University of Ohio State | ||
| Essai O/W 1,849,101 adjuvant with recombinant spike protein | University of Kazakh National Agrarian | |||
| Peptides | Neo7Logic | |||
| Essai O/W 1,849,101 adjuvant with recombinant spike protein | National Scientific Center for Especially Dangerous Infections/Kazakhstan, University of Kazakh National Agrarian | |||
| Recombinant spike protein | Colloids and Interfaces Max-Planck-Institute | |||
| FAR-Squalene adjuvant + RBD protein (baculovirus production) | Universidad Peruana Cayetano Heredia (UPCH)/FarmacológicosVeterinarios SAC (FARVET SAC) | |||
| Protein Subunit | Rep of Kazakhstan, Research Institute for Biological Safety Problems | |||
| RBD-protein | Mynvax | |||
| Recombinant S protein | Biomedicine and Genome Center Izmir | |||
| Novel adjuvant + Peptide | HIV, Malaria, Zika, NSCLC | University of Bogazici | ||
| 3M052 adjs./S subunit intranasal liposomal formulation with GLA | University of Virginia | |||
| Adjuvant, E coli-based Expression + S-Protein (Subunit) | Nigeria, Oyo State, Ogbomoso, Trinity Immonoefficient and Ogbomoso Laboratory, Helix Biogen Consult | |||
| S, N, M and S1 protein subunit | Egypt, National Research Centre | |||
| Protein Subunit | Argentina, San Martin and CONICET University | |||
| Adj. + RBD protein fused with Fc of IgG | Thailand, GPO/University of Chulalongkorn | |||
| Capsid-like Particle | AdaptVac (PREVENT - n CoV consortium) | |||
| VLPs Drosophila S2 insect cell expression system | ExpreS2ion | |||
| LNP formulated peptide antigens | IMV Inc | |||
| S Protein | USAMRIID/WRAIR | |||
| Adjuvant + Sprotein | Influenza | UMN Pharma/Shionogi/National Institute of Infectious Disease, Japan | ||
| Adjuvant + VLP-recombinant protein | BIKEN/University of Osaka/National Institutes of Biomedical Innovation, Japan | |||
| S1 subunit microneedle arrays | MERS | Pittsburgh University | ||
| Peptide | Vaxil Bio | |||
| Adjuvanted protein subunit (RBD) | Biological E Ltd | |||
| Peptide | Breast CA vaccine, HPV therapeutic vaccine, ZIKA, Ebola, Marburg, HIV, Influenza | Flow Pharma Inc | ||
| S protein | A.J. Vaccines | |||
| Ii-Key peptide | SARS-CoV, Influenza, HIV | EpiVax/Generex | ||
| S protein | H7N9 | Georgia University/EpiVax | ||
| EPV-CoV-19 protein subunit | EpiVax | |||
| gp-96 backbone | HIV, Malaria, Zika, NSCLC | Miami University/Heat Biologics | ||
| vaccine subunit | Koltsovo, Rospotrebnadzor, FBRI SRC VB VECTOR | |||
| RBD protein or S1 | SARS | Baylor College of Medicine | ||
| Plant produced protein subunit | CC-Pharming/iBio | |||
| Nanoparticles (based on S-protein and other epitopes), recombinant protein | Saint-Petersburg scientific reseacrh institute of vaccines | |||
| Truncated S (spike) proteins SARS-COV-2 XWG-03 | HPV | GSK/University of Xiamen/Innovax | ||
| Microsphere adjuvanted peptide | VIDO-Intervac, Saskatchewan University | |||
| S and M proteins synthetic Long Peptide Vaccine candidate | OncoGen | |||
| S and N proteins Oral E. coli-based protein expression system | MIGAL Galilee Research Institute | |||
| Nanoparticle vaccine | Lake Pharma, Inc. | |||
| (RBD-Fc + Adjuvant) plant-based vaccine | Chula Vaccine Research Center/BaiyaPhytopharm | |||
| vaccine based on OMV | Flu A, Plague | Quadram Institute Biosciences | ||
| vaccine based on OMV | Trento University/BiOMViSSrI | |||
| tobacco mosaic virus (TMV) structurally modified spherical particles | Rotavirus, Rubella | University of Lomonosov Moscow State | ||
| Spike-based | Hepatitis C | Alberta University | ||
| S1-Fc fusion recombinant protein | AnyGo Technology | |||
| Recombinant protein | Yisheng Biopharma | |||
| (Insect cell line baculovirus expression system) Recombinant S protein in IC-BEVS | Bristol University U.K. and Vietnam, Vabiotech | |||
| Heat stable, orally delivered subunit | Applied Biotechnology Institute, Inc. | |||
| Spike protein peptides | Axon Neuroscience S.E. | |||
| Protein Subunit | G.C. Pharma, MOGAM Institute for Biomedical Research | |||
| RBD-based | Tel Aviv University/Neovii | |||
| OMVsubunit | Epivax/Intravacc | |||
| (Epitope screening) Spike-based | LiteVax BV ImmunoPrecise | |||
| Spike-based | University of Ankara, Middle East Technical University, Nanografi Nano Technology | |||
| Adjuvant with a recombinant spike | Iran | |||
| BEVS produced recombinant S protein | University of Tampere | |||
| Nanoformulated protein subunit | INRAE, CEA, Vaxinano | |||
| Adenoviral Carrier protein subunit | CNRS, CEA | |||
| DC-targeted epitopes | VRI, LinkinVax | |||
| Bacterial vector replicating | Protein expression system of RBD based on Oral Salmonella enteritidis (3934Vac) | Universidad Peruana Cayetano Heredia (UPCH)/FarmacológicosVeterinarios SAC (FARVET SAC) | ||
| Viral vector Replicating | YF17D Vector | K.U. Leuven | ||
| Measles Vector | Cadila Healthcare Limited | |||
| Measles Vector | Koltsovo, Rospotrebnadzor, FBRI SRC VB VECTOR | |||
| S, N targets measles virus | CHIKV, H7N9, Zika | CanVirex AG/DZIF - German Center for Infection Research | ||
| S protein expressing horsepox vector | Monkeypox, Smallpox | Southern Research/Tonix Pharma | ||
| (Intranasal) Atenuated influenza virus backbone LVVV | Influenza | IEM and BIOCAD | ||
| (Intranasal) Influenza A virus, recombinant vaccine | Influenza | Koltsovo, Rospotrebnadzor, FBRI SRC VB VECTOR | ||
| Expressing antigenic portion of the Spike protein attenuated influenza | Influenza | Fundação Oswaldo Cruz and Instituto Buntantan | ||
| RBD expressing influenza vector | Hong Kong University | |||
| SARS-CoV-2 Spike (S) glycoprotein delivered by Replication-competent VSV chimeric virus technology (VSVΔG) | Lassa, Marburg, Ebola | Merck/IAVI | ||
| DC-targeting replicating VSV vector | Manitoba University | |||
| VSV-S | MERS, HIV | Western Ontario University | ||
| VSV-S | Aurobindo | |||
| VSV vector | FBRI SRC VB VECTOR, Rospotrebnadzor, Koltsovo | |||
| Influenza vector M2-deficient single replication (M2SR) | Influenza | Bharat Biotech/FluGen/UW-Madison | ||
| (NDV-SARS-CoV-2/Spike) Newcastle disease virus vector | Utrecht University/Intravacc/Wageningen Bio veterinary Research | |||
| (APMV) Avian paramyxovirus vector | SARS-CoV3 | University of Lancaster, UK | ||
| RBD expressing Intranasal Newcastle disease virus vector (rNDV-FARVET) | Universidad Peruana Cayetano Heredia (UPCH)/FarmacológicosVeterinarios SAC (FARVET SAC) | |||
| Vaccine based on RNA | NLC saRNA formulated | SARS-CoV2 | Amyris, Inc. Infectious Disease Research Institute | |
| S encoding LNP-encapsulated Mrna | Max-Planck-Institute of Colloids and Interfaces | |||
| Self-amplifying RNA | Gennova | |||
| mRNA | Live, attenuated virus (LAV) | University of Selcuk | ||
| LNP-mRNA | Sanofi Pasteur/Translate Bio | |||
| LNP-mRNA | Precision Nanosystems/CanSino Biologics | |||
| Cocktail encoding VLP LNP-encapsulated Mrna | Live, attenuated bacterial vector (LABV) | RNA cure Biopharma/Shanghai Jiao Tong University/Fudan University | ||
| RBD encoding LNP-encapsulated mRNA | RNA cure Biopharma/Shanghai Jiao Tong University/Fudan University | |||
| SARS-CoV-2 derived replicating Defective RNAs | Centro Nacional Biotecnologia (CNB-CSIC), Spain | |||
| mRNA encapsulated LNP | MERS | Daiichi-Sankyo/Tokyo University | ||
| mRNA encapsulated Liposome | BIOCAD | |||
| mRNA candidates | RNAimmune, Inc. | |||
| mRNA | Koltsovo, Rospotrebnadzor, FBRI SRC VB VECTOR | |||
| mRNA | Stermina/University of Tongji/China CDC | |||
| Intranasal delivery system mRNA | eTheRNA | |||
| mRNA | Greenlight Biosciences | |||
| mRNA | IDIBAPS-Hospital Clinic, Spain | |||
| mRNA | Providence Therapeutics | |||
| mRNA | Cell Tech Pharmed | |||
| mRNA | ReNAP Co | |||
| LNP-encapsulated mRNA D614G variant | Globe Biotech Ltd | |||
| Encapsulated mRNA | CEA | |||
| Protein subunit | SARS-CoV-2 S, M, N and NSPs targets to induce T cell responses (CD8) | OSE immunotherapeutics | ||
| sVirus like particle | VLP | Max Planck Institute for Dynamics of Complex Technical Systems | ||
| Virus-like particle-based Dendritic Cell (D.C.)-targeting vaccine | Manitoba University | |||
| VLP | University of BezmialemVakif | |||
| VLP | Middle East Technical University | |||
| (eVLP) Enveloped Virus-Like Particle | SARS-CoV-2, SARS-CoV, & MERS-CoV | CMV, GBM, Zika | VBI Vaccines Inc. | |
| HIV VLPs integrated by S protein | SARS-CoV2 | Grifols/Barcelona Supercomputing/IRTA--CReSA/IrsiCaixa AIDS Research | ||
| Adjuvant + VLP | GPO/Siriraj Hospital/University of Mahidol | |||
| Baculovirus vehicles, Virus-like particles and lentivirus | Malaria | Oncoimmunology group, Navarrabiomed | ||
| RBD displayed on virus-like particles | Saiba GmbH | |||
| Multiepitope display ADDomerTM | Bristol University’s Max Planck Centre and Imophoron Ltd | |||
| Unknown | Doherty Institute | |||
| VLP | SARS-CoV1, SARS-CoV2 | OSIVAX | ||
| eVLP | SARS-CoV2 | Malaria | ARTES BiotechnologARTESBiotechnolog | |
| Whole virus/VLPs peptides | Sao Paulo University | |||
| VLPs produced in BEVS | University of Tampere | |||
| VLP derived by plant | University of Shiraz |
COVID 19 vaccine efficacy
The vaccines were marketed in short span of time after the deadly pandemic. The most important thing was whether these vaccines have good efficacy in neutralising the SARS-CoV-2 virus or there is long term protection by generating memory B-cells and T-cells. In Table 7 we have discussed various marketed vaccines and their efficacy like the dosage regimen, antibody response, T cell response, and effectiveness. There are many vaccines which have received emergency approval in many countries. As of April 2021, 28 vaccines which have entered phase III clinical trials, and other 5 reported showed efficacy in the submitted reports to the peer-reviewed regulatory authorities for their emergency use through literature and/or through detailed publicly reports are available. For minimal protection 2 doses of vaccines are required for most of the them. Only 2 mRNA vaccines have shown efficacy at first dose after the detection of moderate TH1 cells and non neutralizing antibodies (Nabs). Induction of antibody dependent effector mechanisms, T-cell response, virus neutralization suggests that T cells, innate immune mechanisms, NAbs low levels and other immune effector mechanisms are involved which helps in easy identification of protection mechanism and further understanding of immune system involvement for further development of vaccines.
Table 7.
Overview of efficacy of various marketed COVID 19 vaccine in human subjects.
| Dose regimen | Formulation | Effective against (Phase III trails) | Post implementation effectiveness | Response in humans for antibodies | Response in humans for T cells | References | |
|---|---|---|---|---|---|---|---|
| mRNA Pfizer/BioNtech (BNT162b2) |
30 g Mrna, 2 doses, 21 days apart | To lock protein in the pre – fusion state “S” subunit is modified by two mutations of proline by forming lipid nanoparticle | After 2 doses – 95% After 1 dose – 52% Data review suggestion 14 days after 1st dose – 93% 6 months post 2nd dose – 91% |
Symptomatic infection - 1 dose – 94−96%2 doses – 46 –80% Asymptomatic infection : 1 dose – 79% 2 doses 90% Hospitalization: 1 dose – 71−85% 2 doses - 87% Any infection: 1 dose – 46–72% 2 doses 86−92% |
After 2nd dose S1- binding antibody increases. Nab was present in significant amount after 2nd dose | After 2nd dose increase in antigen-specific IFNγ+ CD4+ and CD8+ T cells, more IFNγ and IL-2 secretion than IL- 4, TH1 cell polarization | [80] |
| Moderna (Mrna – 1273) | 100 g Mrna, 2 doses, 28 days apart | To lock protein in the pre – fusion state “S” subunit is modified by two mutations of proline by forming lipid nanoparticle | After 1 dose – 95% After 2 doses – 92% |
Symptomatic infection - 1 dose – 80% 2 doses – 90% |
After 14 days S - binding antibody detected and its levels increases slightly by 28 days and marked increase after 2nd dose. Nab levels are minimum after 1st dose reaches at peak after 2nd dose 14 days | After 1st dose small increases in TNF and IL-2-secreting cells. After 2nd dose Significant increases in CD4+ T cells secreting TH1 type cytokines (TNF > IL-2 > IFNγ). Minimum change in TH2 cell, CD8+ responses |
|
| Viral vector Oxford University/Astra-Zeneca (ChAdOx1 nCoV-19) |
2.5–5 × 1010 viral particles, 2 doses, ≥28 days apart | Simian adenovirus vector recombinant replication – deficient full – length S protein with a Tpa leader sequence | After 1 dose – 76% After 2 doses – 62–67% Low dose followed by high dose – 90% ≥12-week interval, (81%), <6-week interval (55%) |
Hospitalization : After 1st dose 80–94% | After 14 days S - binding antibody detected and its levels increases slightly by 28 days and marked increase after 2nd dose 14 days. More IgG3 and IgG1. Nab detected after 1st dose and increases after 14 days of 2nd dose. After 28–56 days of single dose and peak IgM and IgA responses at day 14 or 28 |
After 1st dose 14 days Peak T cell responses which is higher after 28 days 2nd dose. At 14 day there is increase in TNF and IFNγ production by CD4+ T cells | |
| Gamaleya Research Institute (Gam-COVID-Vac) | 1011 viral particles, 2 doses, 21 days apart | Dose 1 human adenovirus 26 replication-deficient, recombinant | After dose 1 74% After dose 2 91% |
– | After 14 days of 1st dose - NAb (61)% S – binding antibody (85−89%) are detected. After 14 days of 2nd dose- binding antibody (98%) and neutralizing antibody (95%) |
After 1st dose 14 days CD4+ and CD8+ T cells are observed. After 2nd dose 7 days S-specific IFNγ responses are observed |
|
| Janssen (Ad26.COV2.S) | 5 × 1010 viral particles, 1 dose | S protein two amino acid changes at S1/S2 junction that remove cleavage of furin and 2 proline substitutions for replication-deficient recombinant Human adenovirus 26 | After 1st dose 67% | _ | After 28 days of vaccination S-binding and neutralizing antibody are present and their levels remain after 84 days of post vaccination. | At 14 and 28 days of post vaccination CD4+ and CD8+ T cells are present IFNγ and/or IL-2 secretion suggesting TH1 cell | |
| CanSino Biologics (Ad5-nCoV) | 1 dose 5 × 1010 viral particles | Simian adenovirus vector recombinant replication – deficient full – length S protein with a Tpa leader sequence | After 1st dose 66% Decreases to 50% at 5–6 months |
After 1st dose Hospitalization (80−94%) | RBD binding antibodies are observed after 14 days of vaccination (44%). Anti RBD binding antibodies after 28 days of vaccination, Nab’s (47−50%) | After 28 days 78−88% had T cell response based on IFNγ ELIspot After 14 day peak T cell responses were observed |
|
| Protein subunit Novavax (NVX – CoV2373 |
(5 μg protein, 2 doses, 21 days apart | Full-length S protein with mutations at S1/S2 cleavage | After 2nd dose 7 days 90% | After 1st dose 21 days S-binding antibody and Nab is detected After 2nd dose- 7 days significant increase is observed | After 2nd dose 7 days CD4+ T cell responses are observed. Based on IL-2 and TNF production TH1 cell phenotype; minimal TH2 cell responses are measured | ||
| Whole - cellinactivated virus Bharatth Biotech (BBV152) |
6 μg protein, 2 doses, 28 days apart | Grow SARS – CoV- 2 in Vero cells adsorbed on to aluminium hydroxide and imidazoquinoline molecule after inactivation with β-propiolactone | After 2 doses 78% | After 1st dose anti – S binding antibodies are observerd (65%), NAbs (48%) After 2nd dose 14 day (98%) and 97% respectively |
Strong bias towards TH1 cell phenotype (IFNγ and TNF), with minimal TH2 cell responses After 2nd dose 76 day CD4+CD45RO+ memory T cells increases |
Conclusion
SARC-CoV-2 is a novel strain of coronavirus that is liable for causing the global pandemic. This has challenged all the crucial factors like the global economy, medical infrastructure, and public work life, particularly the variant strains are causing havoc. The impact of this pandemic is so severe that it has shaken most countries' economies. Since the tremendous advancement has been achieved in understanding the condition, shortly there seems a strong possibility of some of the therapeutic interventions to combat the SARS-COV-2 pandemic; till then, the only trusted intervention which is currently viable and proven to control is the following of strict quarantine measures, but to reach that intervention to all the affected groups there is a need of a more extensive set of randomized trials and fast testing of the condition to combat the disease effectively. However, this comprehensive review can provide some of the references for the follow-up medical studies. The spread of coronavirus trajectory across the World is difficult to predict as one country’s problems will become global. The only possible solution is the vaccine development targeting against all variant strains to halt its progress, the identified theoretical and practical knowledge, current evidence, international alliances, initiatives, and ideas based on the values of cooperation, inclusiveness, and equity can eliminate the gaps to improve better understanding of the novel coronavirus structure and its design of a vaccine.
Author’s contribution
TF and JAM contributed equally in data collection and drafting the article. AHM, TA and KA contributed in data analysis and interpretation, and drafting the article. SA supervision and critical revision. All authors approved the final version of the article.
Funding
No funding sources.
Competing interests
None declared.
Ethical approval
Not required.
Acknowledgement
The authors would like to thank National Institute of Pharmaceutical Education and Research (NIPER), Guwahati and Hyderabad for fellowships.
References
- 1.Malik JA, Maqbool M. COVID-19: an overview of current scenario n.d. 10.5667/CellMed.2020.0021. [DOI]
- 2.Isaacs A., Gledhill A.W., Andrewes C.H. Influenza A viruses; laboratory studies, with special reference to European outbreak of 1950-1. Bull World Health Organ. 1952;6:287–315. [PMC free article] [PubMed] [Google Scholar]
- 3.Vassilara F., Spyridaki A., Pothitos G., Deliveliotou A., Papadopoulos A. A rare case of human coronavirus 229E associated with acute respiratory distress syndrome in a healthy adult. Case Rep Infect Dis. 2018;2018:1–4. doi: 10.1155/2018/6796839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization Coronavirus press conference 11 February, 2020 Speaker. J Chem Inf Model. 2019;53:1689–1699. doi: 10.1017/CBO9781107415324.004. [DOI] [Google Scholar]
- 5.Alonso S., Izeta A., Sola I., Enjuanes L. Transcription regulatory sequences and mRNA expression levels in the coronavirus transmissible gastroenteritis virus. J Virol. 2002;76:1293–1308. doi: 10.1128/jvi.76.3.1293-1308.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di H., McIntyre A.A., Brinton M.A. New insights about the regulation of Nidovirus subgenomic mRNA synthesis. Virology. 2018;517:38–43. doi: 10.1016/j.virol.2018.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L., et al. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016 doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV-A target for vaccine and therapeutic development. Nat Rev Microbiol. 2009 doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alanagreh L., Alzoughool F., Atoum M. The human coronavirus disease covid-19: its origin, characteristics, and insights into potential drugs and its mechanisms. Pathogens. 2020 doi: 10.3390/pathogens9050331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inhibitor P., Hoffmann M., Kleine-weber H., Schroeder S., Mu M.A., Drosten C., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven article SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou P., Yang X., Wang X., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;2019 doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu M., Wang T., Zhou Y., Zhao Y., Zhang Y. Potential role of ACE2 in coronavirus disease 2019 (COVID-19) prevention and management. J Transl Int Med. 2020;8 doi: 10.2478/jtim-2020-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui J. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masters PS. The molecular biology of coronaviruses n.d.;66. 10.1016/S0065-3527(06)66005-3. [DOI]
- 16.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neuman B.W., Kiss G., Kunding A.H., Bhella D., Baksh M.F., Connelly S., et al. A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol. 2011 doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Demogines A., Farzan M., Sawyer S.L., Demogines A., Farzan M., Sawyer S.L. Evidence for ACE2-utilizing coronaviruses (CoVs) related to severe acute respiratory syndrome CoV in bats. J Virol. 2012;86 doi: 10.1128/JVI.00311-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almaza F., Dediego M.L., Enrique A., Rejas T., Lamirande E., Roberts A., et al. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J Virol. 2007;81:1701–1713. doi: 10.1128/JVI.01467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fehr A.R., Perlman S. Chapter 1 coronaviruses: an overview of their replication and pathogenesis. Coronaviruses. 2015;1282 doi: 10.1007/978-1-4939-2438-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui L., Wang H., Ji Y., Yang J., Xu S., Huang X., Wang Z., Qin L., Tien P., Zhou X., Guo D., Chen Y. The nucleocapsid protein of coronaviruses acts as a viral suppressor of RNA silencing in mammalian cells. J Virol. 2015;89(17):9029–9043. doi: 10.1128/JVI.01331-15. Epub 2015 Jun 17. PMID: 26085159; PMCID: PMC4524063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wit E. De, Doremalen N. Van, Falzarano D., Munster V.J. REVIEWS SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim Y.X., Ng Y.L., Tam J.P., Liu D.X. Human coronaviruses: a review of virus – host interactions. Diseases. 2016 doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang C., Lokugamage K.G., Rozovics J.M., Narayanan K., Semler B.L., Makino S., et al. Alphacoronavirus transmissible gastroenteritis virus nsp1 protein suppresses protein translation in mammalian cells and in cell-free HeLa cell extracts but not in rabbit reticulocyte lysate alphacoronavirus transmissible gastroenteritis virus nsp1 protein. J Virol. 2011 doi: 10.1128/JVI.01806-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamitani W., Huang C., Narayanan K., Lokugamage K.G., Makino S. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat Struct Mol Biol. 2009;16:1134–1140. doi: 10.1038/nsmb.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamitani W., Narayanan K., Huang C., Lokugamage K., Ikegami T., Ito N., et al. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc Natl Acad Sci U S A. 2006 doi: 10.1073/pnas.0603144103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka T., Kamitani W., Dediego M.L., Enjuanes L., Matsuura Y. Severe acute respiratory syndrome coronavirus nsp1 facilitates efficient propagation in cells through a specific translational shutoff of host mRNA. J Virol. 2012;86:11128–11137. doi: 10.1128/JVI.01700-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graham R.L., Sims A.C., Brockway S.M., Baric R.S., Denison M.R. The nsp2 replicase proteins of murine hepatitis virus and severe acute respiratory syndrome coronavirus are dispensable for viral replication. J Virol. 2005;79:13399–13411. doi: 10.1128/JVI.79.21.13399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cornillez-ty C.T., Liao L., Iii J.R.Y., Kuhn P., Buchmeier M.J. Severe acute respiratory syndrome coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling. J Virol. 2009;83:10314–10318. doi: 10.1128/JVI.00842-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chatterjee A., Johnson M.A., Serrano P., Pedrini B., Joseph J.S., Neuman B.W., et al. Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold. J Virol. 2009;83:1823–1836. doi: 10.1128/JVI.01781-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heinonen M., Egloff M., Frangeul A., Gruez A., Cambillau C., Ziebuhr J., et al. Structural and functional basis for ADP-ribose and poly (ADP-ribose) binding by viral macro domains. J Virol. 2006;80:8493–8502. doi: 10.1128/JVI.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eriksson K.K., Cervantes-barraga L., Ludewig B., Thiel V. Mouse hepatitis virus liver pathology is dependent on ADP-ribose-1″-phosphatase, a viral function conserved in the alpha-like supergroup. J Virol. 2008;82:12325–12334. doi: 10.1128/JVI.02082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Signaling N.-B., Frieman M., Ratia K., Johnston R.E., Mesecar A.D., Baric R.S. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J Virol. 2009;83:6689–6705. doi: 10.1128/JVI.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neuman B.W., Joseph J.S., Saikatendu K.S., Serrano P., Chatterjee A., Johnson M.A., et al. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J Virol. 2008;82:5279–5294. doi: 10.1128/JVI.02631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Serrano P., Johnson M.A., Almeida M.S., Horst R., Herrmann T., Joseph J.S., et al. Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus. J Virol. 2007;81:12049–12060. doi: 10.1128/jvi.00969-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clementz M.A., Kanjanahaluethai A., O’Brien T.E., Baker S.C. Mutation in murine coronavirus replication protein nsp4 alters assembly of double membrane vesicles. Virology. 2008;375:118–129. doi: 10.1016/j.virol.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gadlage M.J., Sparks J.S., Beachboard D.C., Cox R.G., Doyle J.D., Stobart C.C., et al. Murine hepatitis virus nonstructural protein 4 regulates virus-induced membrane modifications and replication complex function. J Virol. 2010;84:280–290. doi: 10.1128/jvi.01772-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu Y., Lu X., Denison M.R. Identification and characterization of a serine-like proteinase of the murine coronavirus MHV-A59. J Virol. 1995;69:3554–3559. doi: 10.1128/jvi.69.6.3554-3559.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oostra M., Hagemeijer M.C., van Gent M., Bekker C.P.J., te Lintelo E.G., Rottier P.J.M., et al. Topology and membrane anchoring of the coronavirus replication complex: not all hydrophobic domains of nsp3 and nsp6 are membrane spanning. J Virol. 2008;82:12392–12405. doi: 10.1128/jvi.01219-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhai Y., Sun F., Li X., Pang H., Xu X., Bartlam M., et al. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat Struct Mol Biol. 2005;12:980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhai Y., Sun F., Li X., Pang H., Xu X., Bartlam M., et al. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat Struct Mol Biol. 2005;12:980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Imbert I., Guillemot J.C., Bourhis J.M., Bussetta C., Coutard B., Egloff M.P., et al. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 2006;25:4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Egloff M.P., Ferron F., Campanacci V., Longhi S., Rancurel C., Dutartre H., et al. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc Natl Acad Sci U S A. 2004;101:3792–3796. doi: 10.1073/pnas.0307877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bouvet M., Debarnot C., Imbert I., Selisko B., Snijder E.J., Canard B., et al. In vitro reconstitution of sars-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6:1–13. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Decroly E., Debarnot C., Ferron F., Bouvet M., Coutard B., Imbert I., et al. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-o-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu X., Liu Y., Weiss S., Arnold E., Sara S.G. Molecular model of SARS coronavirus polymerase: implications for biochemical functions and drug design. Nucleic Acids Res. 2003;31 doi: 10.1093/nar/gkg916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ivanov K.A., Thiel V., Dobbe J.C., Meer Y. Van Der, Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ivanov K.A., Ziebuhr J. Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5 Ј-triphosphatase activities. J Virol. 2004;78:7833–7838. doi: 10.1128/JVI.78.14.7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eckerle L.D., Becker M.M., Halpin R.A., Li K., Venter E., Lu X., et al. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eckerle L.D., Lu X., Sperry S.M., Choi L., Denison M.R. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J Virol. 2007;81:12135–12144. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cambillau C., Minskaia E., Hertzig T., Gorbalenya A.E., Canard B., Ziebuhr J. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci U S A. 2006:10–15. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Y., Cai H., Xiang N., Tien P., Ahola T., Guo D. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc Natl Acad Sci U S A. 2009;106:3–8. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bhardwaj K., Sun J., Holzenburg A., Guarino L.A., Kao C.C. RNA recognition and cleavage by the SARS coronavirus endoribonuclease. J Mol Biol. 2020 doi: 10.1016/j.jmb.2006.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ivanov K.A., Hertzig T., Rozanov M., Bayer S., Thiel V., Gorbalenya A.E., et al. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc Natl Acad Sci U S A. 2004 doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Decroly E., Imbert I., Coutard B., Selisko B., Alvarez K., Gorbalenya A.E., et al. Coronavirus nonstructural protein 16 is a Cap-0 binding enzyme possessing (Nucleoside-2 Ј O) -methyltransferase activity. J Virol. 2008;82:8071–8084. doi: 10.1128/JVI.00407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Züst R., Cervantes-barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J., et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol. 2011;12:137–144. doi: 10.1038/ni.1979. Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gilbert G.L. Commentary: SARS, MERS and COVID-19—new threats; old lessons. Int J Epidemiol. 2020:1–3. doi: 10.1093/ije/dyaa061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petrosillo N., Viceconte G., Ergonul O., Ippolito G., Petersen E. COVID-19, SARS and MERS: are they closely related? Clin Microbiol Infect. 2020;26:729–734. doi: 10.1016/j.cmi.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rockx B., Kuiken T., Herfst S., Bestebroer T., Lamers M.M., Munnink B.B.O., et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;7314:1–10. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jennifer M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020 doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 61.COVID Live Update: 146,881,796 Cases and 3,107,317 Deaths from the Coronavirus - Worldometer n.d. https://www.worldometers.info/coronavirus/ (accessed April 25, 2021).
- 62.Puhani P.A. 2020. France and Germany exceed Italy, South Korea and Japan in temperature-adjusted corona proliferation a quick and dirty Sunday morning analysis. [Google Scholar]
- 65.March I. 2020. COVID-19 pandemic in India. 2019. [Google Scholar]
- 66.Kohler P.F., Farr R.S. Elevation of Cord over maternal IgG immunoglobulin: evidence for an active placental IgG transport. Nature. 1966;210:1070–1071. doi: 10.1038/2101070a0. [DOI] [PubMed] [Google Scholar]
- 67.Burki T. Understanding variants of SARS-CoV-2. Lancet. 2021 doi: 10.1016/S0140-6736(21)00298-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Who W . 2020. Clinical management of severe acute respiratory infection (SARI) when COVID-19 disease is suspected. 2019. [Google Scholar]
- 69.Möhlenkamp S., Thiele H. Ventilation of COVID-19 patients in intensive care units. Herz. 2020;45:329–331. doi: 10.1007/s00059-020-04923-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gattinoni L., Chiumello D., Caironi P., Busana M., Romitti F., Brazzi L., et al. COVID-19 pneumonia: different respiratory treatments for different phenotypes? Intensive Care Med. 2020;46:1099–1102. doi: 10.1007/s00134-020-06033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marini J.J., Hotchkiss J.R., Broccard A.F. Bench-to-bedside review: microvascular and airspace linkage in ventilator-induced lung injury. Crit Care. 2003;7:435–444. doi: 10.1186/cc2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao Y., Cao Y., Wang S., Cai K., Xu K. COVID-19 and gastrointestinal symptoms. Br J Surg. 2020;107(September (10)):e382–e383. doi: 10.1002/bjs.11821. Epub 2020 Aug 5. PMID: 32757447; PMCID: PMC7436706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Who W . 2020. Clinical management of severe acute respiratory infection (SARI) when COVID-19 disease is suspected. 2019. [Google Scholar]
- 74.Zhao Y., Cao Y., Wang S., Cai K., Xu K. COVID-19 and gastrointestinal symptoms. Br J Surg. 2020;2019:2019–2020. doi: 10.1002/bjs.11821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moore N., Carleton B., Blin P., Bosco P., Cecile L. Does ibuprofen worsen COVID 19? Drug Saf. 2020:19–22. doi: 10.1007/s40264-020-00953-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Malik J.A., Mulla A.H., Farooqi T., Pottoo F.H., Anwar S., Rengasamy K.R.R. Targets and strategies for vaccine development against SARS-CoV-2. Biomed Pharmacother. 2021;137 doi: 10.1016/j.biopha.2021.111254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dutta N.K., Mazumdar K., Gordy J.T. The nucleocapsid protein of SARS–CoV-2: a target for vaccine development. J Virol. 2020;94:1–2. doi: 10.1128/jvi.00647-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sadarangani M., Marchant A., Kollmann T.R. Immunological mechanisms of vaccine-induced protection against COVID-19 in humans. Nat Rev Immunol. 2021;21:1–10. doi: 10.1038/s41577-021-00578-z. [DOI] [PMC free article] [PubMed] [Google Scholar]