Abstract
Leptospirosis is a serious bacterial disease that affects both humans and animals. A wide range of symptoms have been described in humans; the disease in dogs is commonly associated with kidney and/or liver disease. In Malaysia, information about the common serovars infecting dogs is limited. Therefore, we investigated the occurrences of leptospirosis in 124 pet dogs diagnosed with kidney and/or liver disease. Blood, urine, abdominal effusion, and/or kidney and liver were collected from the dogs. Based on microscopic agglutination testing, 53 of 124 (42.7%) dogs were seropositive for leptospiral exposure. Sera were frequently positive to serovars Bataviae (n = 12), Javanica (n = 10), and Icterohaemorrhagiae (n = 10). Direct detection using PCR showed that 42 of 124 (33.9%) of the whole blood and 36 of 113 (31.9%) urine samples were positive for pathogenic Leptospira spp. By PCR, 2 of 23 (9.1%) kidney and 2 of 23 (9.1%) liver were positive for pathogenic Leptospira spp. Abdominal effusion from 4 dogs were PCR-positive for pathogenic Leptospira spp. The species detected were L. interrogans, L. borgpetersenii, L. kirschneri, and L. kmetyi by partial 16S rRNA sequencing. We further identified and characterized 11 Leptospira spp. isolates from 8 dogs as serovars Bataviae, Javanica, and Australis. The mortality rate of the Leptospira-infected dogs was high (18 of 53; 34%).
Keywords: canine, detection, isolation, Leptospira, Malaysia, serovars
Leptospirosis is a bacterial disease caused by pathogenic variants of Leptospira spp. that can affect almost all mammals.32,52 The disease is recognized as a widespread zoonosis and has emerged as a major public health issue in many developing countries.12 Transmission of the disease is strongly driven by environmental factors, such as high rainfall, flooding, natural disasters, uncontrolled urban expansion, and poor sanitation.29,42,50 Exposure to water and soil contaminated by the urine of infected animals is the most common route of transmission to humans and domestic animals,50 and rodents are considered the major source for human infection, a role likely attributed to the synanthropic behavior and widespread distribution of rats.12
Case reports of canine leptospirosis have been described worldwide.4,25,52 However, the actual role of dogs in the zoonotic transmission of leptospirosis remains poorly documented, and the overall contribution of dogs to the burden of human leptospirosis has yet to be determined.35 Alarmingly, asymptomatic urinary shedding of leptospires among dog populations has been widely reported.20,34,47 Thus, there is a possibility that dogs may contribute to the spread of pathogenic Leptospira spp. in the environment.
Re-emergence of clinical illness in dogs and humans has been documented, highlighting the importance of improving current diagnostic approaches and prevention strategies.22,52 Infected dogs could be asymptomatic with mild, self-limiting febrile illness, or manifest a broad spectrum of clinical signs, such as vomiting, depression, icterus, dehydration, diarrhea, and anorexia. Often these dogs are diagnosed with hepatic and/or renal failure accompanied by hemorrhagic and pulmonary disorders.2,24,45 Clinical and laboratory findings are usually nonspecific, and a definitive diagnosis requires additional confirmatory tests for the direct or indirect identification of the pathogen, such as darkfield microscopy, PCR, bacterial culture, and the microscopic agglutination test (MAT).44,52 For serodetection of acute leptospiral infection, MAT is still widely employed, despite the poor ability of the assay to predict the infecting serovar and an inability to distinguish between infection and vaccine-induced titers.33 Conversely, PCR has been used successfully to confirm leptospiral infection at the early stages of infection,18,19 and sequencing the PCR amplicon has enabled the identification of the different leptospiral species infecting dogs.40
In Malaysia, the seroprevalence of leptospirosis among dogs has been reported to be 3.1–22.2%,8,16,23,30,31 but most of these studies did not examine the seroprevalence of leptospirosis in dogs diagnosed with kidney and/or liver disease. The one study examining dogs with kidney disease reported a detection rate of 5.3%.30 Therefore, we investigated the diagnosis of leptospirosis in dogs with kidney and/or liver disease using multiple strategies based on serologic, molecular, and bacteriologic tests, along with characterization of the recovered leptospiral strains. The identification of the serovars circulating among dog populations remains the major strategy to support the development and commercialization of vaccines incorporating more specific serovar compositions, which would increase immunization effectiveness for local canine populations.
Materials and methods
Sample collection and inclusion criteria
We recruited dogs diagnosed with kidney and/or liver disease that had been presented to the University Veterinary Hospital (UVH) of Faculty of Veterinary Medicine (FVM), Universiti Putra Malaysia (UPM), or to private veterinary clinics within 10 km of UPM. Owner permission was obtained prior to sample collection. Ethical approval was obtained from the Institutional Animal Care and Use Committee (UPM/IACUC/AUP-R084/2016).
The selection criteria of the recruited dogs were: dogs with clinical signs of kidney and/or liver disease based on serum biochemistry profile with elevated parameters: urea > 7.5 mmol/L, creatinine > 176 µmol/L, alanine aminotransferase (ALT) > 90 U/L, and alkaline phosphatase (ALP) > 100 U/L. The signalment of each dog was recorded, and a history was taken based on information provided by the owners. Age, breed, and clinical illness characteristics were categorized according to published guidelines.17 The recruited pet dogs were manually restrained for venipuncture (serum and whole blood), and urine samples were collected either via cystocentesis or spontaneous micturition by experienced veterinarians. Blood samples collected using plain blood tubes (Vacutainer; Becton Dickinson) were allowed to clot at 4°C then centrifuged for 10 min at 2,130 × g (80-2 centrifuge; Jiangsu Jinyi). Sera were collected and stored at –20°C. Whole blood was collected in an EDTA tube (Vacutainer; Becton Dickinson), and urine samples were collected in sterile universal containers. Whole blood and urine samples were kept at 4°C until further analysis. Dogs euthanized or found dead as a result of the disease were autopsied, with permission. The kidney and liver were examined grossly and harvested. Samples of the kidney and liver were homogenized with liquid medium (Ellinghausen and McCullough modified by Johnson and Harris [EMJH]) for bacterial isolation. If there was abdominal effusion, the fluids were collected and stored at 4°C.
Serologic detection using MAT
Microscopic agglutination testing was performed with a panel of 20 leptospiral serovars (Table 1), comprised of 19 pathogenic species and 1 nonpathogenic strain (Patoc 1), all of which were available in the Bacteriology Laboratory of FVM, UPM. Endpoint titers were determined using serial 2-fold dilutions, and the last well with 50% agglutination was recorded. The cutoff for a positive MAT reaction was a titer ≥ 1:100, as defined in previous studies.10,37,39 The serovar with the highest MAT titer was recorded.
Table 1.
The panel of leptospiral serovars used in microscopic agglutination test for anti-leptospiral antibody detection.
| Species | Serovar | Strain |
|---|---|---|
| L. biflexa | Patoc | Patoc I |
| L. borgpetersenii | Ballum | Mus 127 |
| Hardjobovis | 117123 | |
| Javanica | Veldrat Bataviae 46 | |
| Tarassovi | Perepelitsin | |
| L. interrogans | Australis | Ballico |
| Autumnalis | Akiyami A | |
| Bataviae | Swart | |
| Canicola | Hond Utrecht IV | |
| Copenhageni | M20 | |
| Djasiman | Djasiman | |
| Hebdomadis | Hebdomadis | |
| Icterohaemorrhagiae | RGA | |
| Lai | Lai | |
| Pomona | Pomona | |
| Pyrogenes | Salinem | |
| L. kirschneri | Cynopteri | 3522C |
| Grippotyphosa | Moskva V | |
| L. kmetyi | Malaysia | Bejo-ISO9 |
| L. weilii | Celledoni | Celledoni |
Molecular detection using PCR and partial 16S rRNA sequencing
The whole blood, urine, abdominal effusion (if available), and samples of kidney and liver were used to detect leptospiral DNA. DNA extraction from the samples and the positive control of Leptospira interrogans serovar Canicola strain Hond Utrecht IV was performed (DNeasy blood and tissue kit; Qiagen). The protocols provided in the kit were used, namely: 1) non-nucleated protocol for whole blood, 2) cultured cell protocol for urine, abdominal effusion, and positive control, and 3) animal tissue protocol for kidney and liver samples. The end products (DNA template) were inspected for purity using 1.5% agarose gels.
Sets of primers that targeted the 16S rRNA and LipL32 genes1,46 (Table 2) were used; the LipL32 gene is present only in pathogenic Leptospira spp.54 The 25-μL reaction volume was optimized as follows: 12.5 μL of 2 × MyTaq Red Mix (Bioline), 2.5 μL of forward and reverse primer, and 10 μL of DNA template. Amplification was optimized and performed (Mastercycler Pro S; Eppendorf) with initial denaturation of 94°C for 5 min followed by 30 cycles of denaturation at 94°C for 1 min, primer annealing at 58°C for 45 s, and DNA extension at 72°C for 30 s before the final extension step at 72°C for 6 min. Amplicons were analyzed in Tris-borate-EDTA (TBE) buffer at 80 V for 1.5 h with 1.5% gel electrophoresis. The gel was pre-stained (SYBR Safe DNA gel stain; Invitrogen) and examined using gel documentation (AlphaImager; Alpha Innotech). Amplicons were identified by their band sizes at 541 bp (16S rRNA) and 756 bp (LipL32).
Table 2.
Primers used for amplification and sequencing of Leptospira spp.
After PCR analysis, amplicons were submitted to a commercial facility for DNA sequencing together with the relevant forward primer sequence. The sequencing data were compared to reference sequences deposited in GenBank using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/).
Isolation of Leptospira spp
Leptospira spp. were isolated based on the protocol of the World Organisation for Animal Health (OIE).56 Two drops of whole blood, urine, abdominal effusion, and ~1 mL of homogenized kidney and liver samples were inoculated into semisolid EMJH medium containing 200 µg/mL of 5-fluorouracil. Inoculation of all of the samples was performed within 2 h of collection. Primary cultures were kept in the incubator at 30°C for 12 wk. Cultures were checked fortnightly for leptospires using darkfield microscopy. If leptospires were observed within 12 wk, the positive cultures were transferred into liquid EMJH medium to enhance their growth and then filtered (0.45-µm filter; Millex) until pure isolates were obtained. Leptospira spp. isolates were maintained in liquid EMJH medium and further characterized through serologic and molecular tests. If negative for leptospires at 12 wk after final careful examination, the cultures were discarded.
Serologic characterization of Leptospira spp. isolates
The serovar of the isolates was determined by MAT using a panel of 18 hyperimmune sera, namely Australis, Autumnalis, Ballum, Bataviae, Celledoni, Cynopteri, Copenhageni, Djasiman, Hardjobovis, Hebdomadis, Icterohaemorrhagiae, Javanica, Lai, Malaysia, Patoc, Pomona, Pyrogenes, and Tarassovi. The isolates were assigned to a leptospiral serovar when they reacted specifically to the relevant hyperimmune serum. The hyperimmune sera were provided by Forensic and Scientific Services, Department of Health, Leptospirosis Reference Laboratory, Queensland, Australia.
Molecular characterization of Leptospira spp. isolates
Isolates were identified as either pathogenic or nonpathogenic Leptospira spp. PCR assays were performed using 16S rRNA and LipL32 primers as mentioned earlier. Partial 16S rRNA sequencing was performed with the relevant forward and reverse primer sequences (Table 2) for all isolates. The sequencing data obtained were compared with GenBank using BLAST. Sequences generated from the isolates, along with the representative sequences from genus Leptospira (pathogenic, intermediate, and saprophytic) and Leptonema illini strain Habaki (as an outgroup, obtained from GenBank) were aligned with OMEGA v.1.2.2 (Clustal; http://www.clustal.org/omega/). Sequencing data were then subjected to single and multiple pairwise alignments using CLUSTALW v.2.1 (Clustal; https://www.genome.jp/tools-bin/clustalw). The aligned sequencing data were analyzed by using phylogenetic analysis. The phylogenetic tree was generated using MEGA7 (https://www.megasoftware.net/) and was inferred by using the neighbor-joining method with 1,000 bootstraps based on a general time reversible model.26
Statistical analysis
Serologic detection, molecular detection, and isolation of Leptospira spp. were analyzed using descriptive statistics with 95% confidence intervals (CIs; SPSS Statistics v.23; IBM). Mortality rate was calculated based on molecular detection, and 95% CI was also applied.
Results
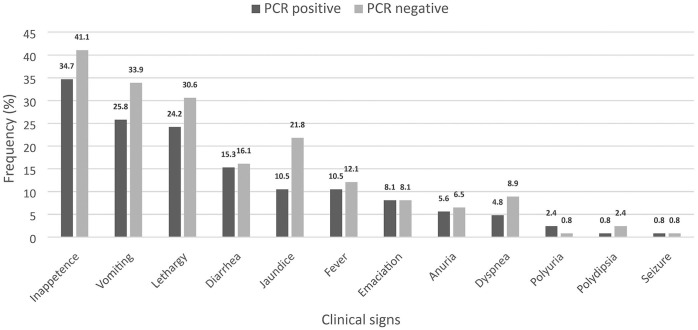
Of the 124 dogs recruited (Table 3), 68 dogs were diagnosed with both kidney and liver disease, 34 dogs had kidney disease (urea and/or creatinine elevated), and 22 dogs had liver disease (ALT and/or ALP elevated). The most common clinical signs observed in the dogs were inappetence, followed by vomiting, lethargy, jaundice, and diarrhea (Fig. 1). All of the dogs had at least 2 clinical signs.
Table 3.
Demographic data of the dogs diagnosed with kidney and/or liver disease (n = 124).
| Characteristic | No. of dogs (%) |
|---|---|
| Age | |
| Young (≤ 1 y) | 13 (10.5) |
| Adult (1 to < 6 y) | 48 (38.7) |
| Senior (≥ 6 y) | 63 (50.8) |
| Sex | |
| Male | 76 (61.3) |
| Female | 48 (38.7) |
| Vaccination status | |
| Vaccinated | 52 (41.9) |
| Nonvaccinated | 72 (58.1) |
| Management | |
| Indoor | 51 (41.1) |
| Outdoor | 73 (58.9) |
| Breed | |
| Small | 28 (22.6) |
| Medium | 65 (52.4) |
| Large | 31 (25.0) |
| Type of household | |
| Single | 68 (54.8) |
| Multiple | 56 (45.2) |
| Rat exposure | |
| Exposed | 78 (62.9) |
| Not exposed | 46 (37.1) |
| Clinical illness | |
| Acute (≤ 7 d) | 102 (82.3) |
| Chronic (> 7 d) | 22 (17.7) |
Figure 1.
Frequency of clinical signs observed in the dogs diagnosed with kidney and/or liver disease with leptospirosis (black bar) and without leptospirosis (gray bar) that had been presented to the hospital or private clinics (n = 124). Leptospirosis was confirmed by PCR tested in whole blood, urine, abdominal effusion, or kidney and liver samples.
We collected 124 sera, 124 whole blood, and 113 urine samples. During the study, 30 of 124 dogs did not survive despite treatment and supportive care; 23 kidneys and 23 livers were sampled after postmortem examination. Abdominal effusion samples were collected from 4 dogs. The majority of the dogs diagnosed with kidney and/or liver disease were older, male, nonvaccinated, outdoor, and medium-breed dogs (Table 3). Vaccinated dogs had been vaccinated with a commercial tetravalent vaccine (Vanguard Plus 5 L4; Zoetis) that included serovars Canicola, Grippotyphosa, Icterohaemorrhagiae, and Pomona.
Serologic detection
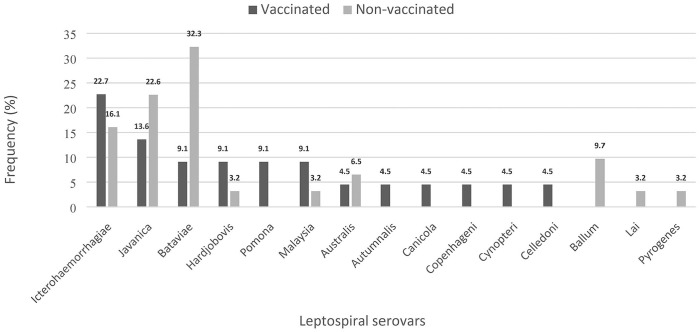
Of 124 sera tested using MAT, 53 (42.7%; 95% CI: 34.0–51.4%) dogs were seropositive with titers of 1:100 to 1:800 (Fig. 2). Sera were most frequently positive to serovars Bataviae (n = 12), Javanica (n = 10), and Icterohaemorrhagiae (n = 10), followed by Australis (n = 3), Ballum (n = 3), Hardjobovis (n = 3), Malaysia (n = 3), and Pomona (n = 2). The least frequent leptospiral serovars observed based on seropositivity were Autumnalis, Canicola, Celledoni, Copenhageni, Cynopteri, Lai, and Pyrogenes. All of the tested sera were negative for serovars Djasiman, Grippotyphosa, Hebdomadis, Patoc, and Tarassovi. In vaccinated dogs, seropositivity was highest against serovar Icterohaemorrhagiae (n = 5); in nonvaccinated dogs, seropositivity was highest against serovars Bataviae (n = 10) and Javanica (n = 7; Fig. 3).
Figure 2.
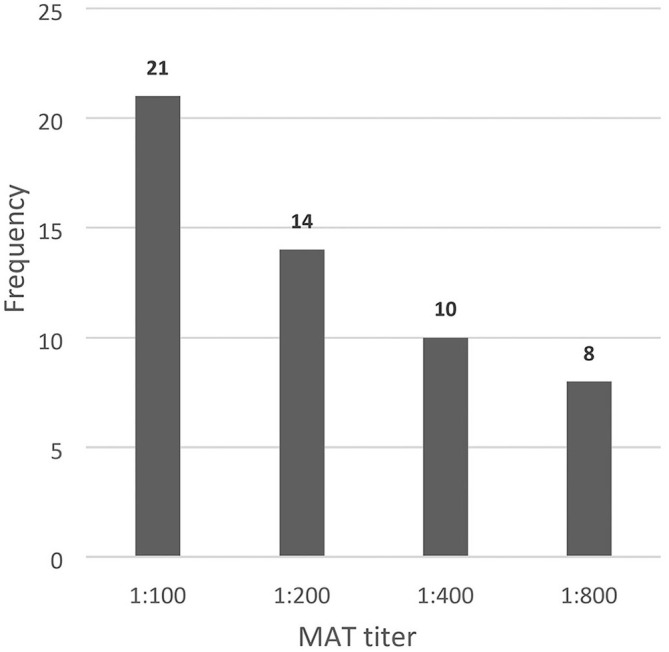
Frequency of microscopic agglutination test (MAT) titer from 53 seropositive dogs.
Figure 3.
Frequency of leptospiral serovars with highest titers detected in acute sera from seropositive vaccinated dogs (n = 22; black bar) and seropositive nonvaccinated dogs (n = 31; gray bar). Serovars Autumnalis, Copenhageni, Cynopteri, and Celledoni were each detected in a single vaccinated dog; serovars Ballum (3 dogs), Lai (1 dog), and Pyrogenes (1 dog) were detected in nonvaccinated dogs.
Molecular detection and partial 16S rRNA sequencing
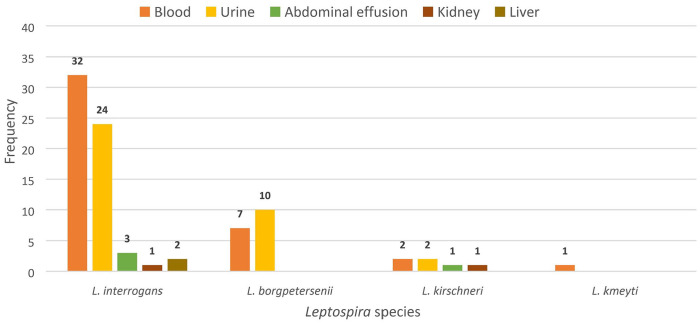
We found 53 of 124 (42.7%; 95% CI: 34.0–51.4%) dogs positive by PCR for leptospiral infection. The positive results were obtained from whole blood (42 samples), urine (36 samples), kidneys (2 samples), livers (2 samples), and abdominal effusions (4 samples). All of them were positive for pathogenic Leptospira spp., and 4 Leptospira spp. were detected using partial 16S rRNA sequencing. L. interrogans (n = 62) was the species detected most commonly, followed by L. borgpetersenii (n = 17) and L. kirschneri (n = 6). Only one sample (a blood sample) was positive for L. kmetyi (Fig. 4). Of the 53 PCR-positive dogs, 21 dogs were also seropositive by MAT. Furthermore, of the 53 PCR-positive dogs, 18 dogs had died, therefore the mortality rate of definitively diagnosed dogs was 34.0% (95% CI: 21.2–46.7%).
Figure 4.
Frequency of Leptospira species as determined by partial 16S rRNA sequencing in the samples obtained from 53 positive dogs.
Isolation and characterization of Leptospira spp
Leptospira spp. were successfully isolated from 8 of 124 dogs (6.5%; 95% CI: 2.1–10.8%); 3 dogs had been vaccinated, and 5 dogs had not been vaccinated. These 8 dogs were also positive by PCR direct detection. From these 8 dogs, 11 isolates were recovered from blood (3 isolates), urine (7 isolates), and abdominal effusion (1 isolate) samples. By MAT, 8 of the isolates were Bataviae, 2 isolates were Australis, and 1 isolate was Javanica. Molecular characterization using PCR revealed that all of the isolates were pathogenic. Further analysis of the partial 16S rRNA sequencing using BLAST confirmed that 10 isolates had 95% identity to L. interrogans, and 1 isolate had 95% identity to L. borgpetersenii. The sequences were submitted to GenBank (Table 4).
Table 4.
Serologic and molecular characterization results of the 8 Leptospira spp. isolates recovered from dog samples.
| Dog | Sample | Characterization | |
|---|---|---|---|
| Serologic | Molecular | ||
| 1 | Urine | Bataviae | L. interrogans UVH huseli (MF589180.1) |
| 2 | Blood | Bataviae | L. interrogans UVH khor_samei3 (MH745144.1) |
| Urine | Bataviae | L. interrogans UVH khor_samei1 (MH745142.1) | |
| Abdominal effusion | Bataviae | L. interrogans UVH khor_samei2 (MH745143.1) | |
| 3 | Urine | Bataviae | L. interrogans UVH khor_saron (MH799846.1) |
| 4 | Urine | Bataviae | L. interrogans PV huseli (MH799845.1) |
| 5 | Urine | Bataviae | L. interrogans UVH khor_savi (MH799847.1) |
| 6* | Blood | Javanica | L. borgpetersenii strain khorsayo (MN226648.1) |
| 7* | Urine | Australis | L. interrogans UVH khor_sapy1 (MN698724.1) |
| Blood | Australis | L. interrogans UVH khor_sapy3 (MN698723.1) | |
| 8* | Urine | Bataviae | L. interrogans khorgoh_jacky (MN226649.1) |
GenBank accessions in parentheses.
Dogs had been vaccinated annually with commercial tetravalent vaccine.
The phylogenetic tree of Leptospira spp. is composed of 3 distinct clades (pathogenic, intermediate, and saprophytic species), and all isolates obtained were placed within the pathogenic clade. Ten isolates were placed within species of L. interrogans; one isolate was placed within L. borgpetersenii. Three isolates, namely L. interrogans UVH khor_sapy1, L. interrogans UVH khor_sapy3, and L. borgpetersenii khorsayo, were somewhat divergent from the other strains within their respective species. A condensed tree with a 50.0% cutoff was created from the original tree to further investigate the significance of the divergence (Fig. 5). From the condensed tree, all 10 L. interrogans isolates were closely related (61%) with all of the included representative serovars of L. interrogans. In addition, L. interrogans UVH khor_sapy1 and L. interrogans UVH khor_sapy3, both isolates from the same dog, were 99% related. The L. borgpetersenii isolate was closely related (68%) to all of the representative serovars of L. borgpetersenii.
Figure 5.
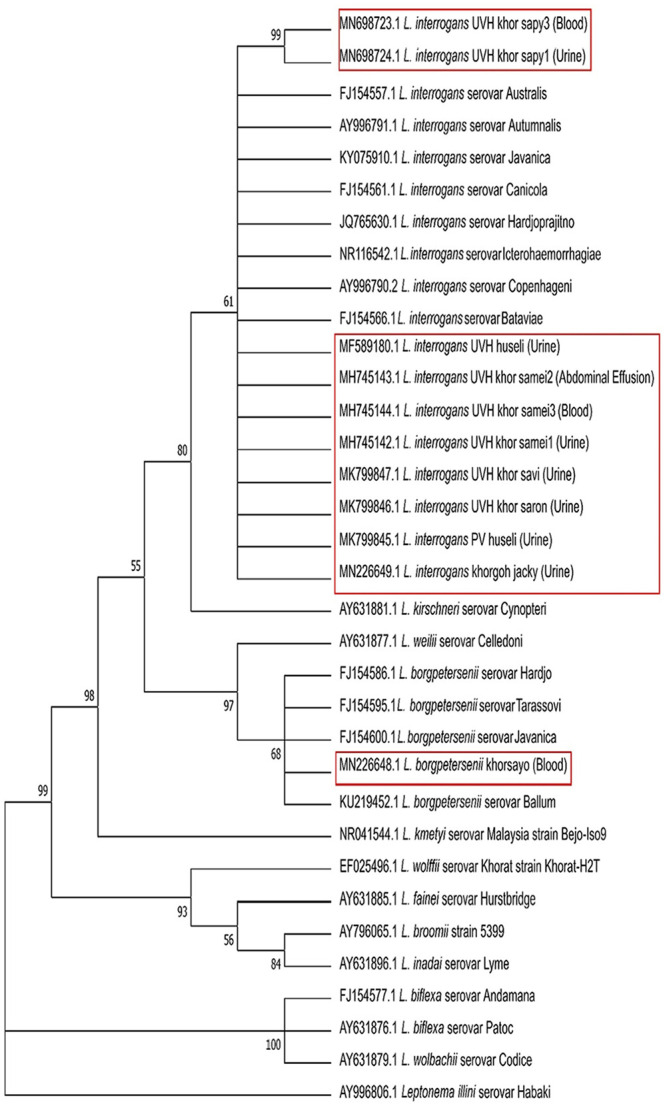
Condensed tree generated from the original phylogenetic tree with a 50.0% cutoff showing the evolutionary relationships of taxa based on partial 16S rRNA gene from Leptospira spp. isolates (in red boxes) as well as representative Leptospira spp., including pathogenic, intermediate, and saprophytic species. Leptonema illini serovar Habaki was used as an outgroup.
Discussion
Although the MAT is the cornerstone of serodetection of leptospirosis, the antigenic heterogeneity of Leptospira spp. means that many serovars are required as antigens.9 The overall serologic detection of leptospiral infection in dogs with kidney and/or liver disease in our study was 42.7%, which is higher16,23,30,31 and lower39 compared to previous studies, likely as a result of our specific selection criteria for recruited dogs. Previous studies investigated a smaller population of unhealthy dogs,39 or pet dogs only,30 or a larger population of apparent healthy shelter and working dogs16; some studies investigated healthy dogs from a single location.23,31
The MAT titer was 1:100 in 21 dogs at the time they were clinically ill and subsequently diagnosed with leptospirosis. Titers this low may be interpreted by some as inconsistent with a diagnosis of leptospirosis and may be dismissed as consistent with previous vaccination or exposure, delaying appropriate therapy and zoonotic preventive measures. A previous study also reported that an antibody titer of 1:100 associated with a clinically compatible illness was regarded as a probable case of leptospirosis.11 It is recommended that the confirmation of leptospiral infection by MAT should be based on testing paired serum samples, collected 14 d apart, to detect at least a 4-fold increase in agglutinating antibodies against Leptospira.52 In our study, paired serum samples were not collected because of the high mortality rate, and PCR was performed simultaneously to confirm leptospirosis. A total of 3 dogs with a titer level of 1:100 did not survive despite being treated in our study. On the other hand, 6 dogs with a high titer (1:800) did survive after being treated. Hence, the titer levels may not correlate with the risk of mortality. The immunity level of the individual dog, the infecting serovar, and the treatment initiated might all have played important roles in the survivability of infected dogs.
The panel of leptospiral serovars that we used was based on the serovars that have been reported in the previous serosurveys in Malaysia for humans, rats, and dogs.8,16,23,30,31,48 Incorporating known leptospiral serovars that have been reported previously is highly recommended in the diagnostic workflow to improve the detection rate in dogs. Limited local reports had found both serovars Bataviae and Javanica in dogs,16,31 and the high serodetection rate of both serovars in our study was likely linked to direct contact with rats, based on the history provided by the pet owners. This is not surprising given that serovars Bataviae and Javanica have been reported to be circulating among the urban rat populations in Peninsular Malaysia.6 Serovars Australis, Canicola, Icterohaemorrhagiae, and Pomona had been detected in rats within Kuala Lumpur,15 matching the findings of our study. Serovars Copenhageni and Hardjobovis were commonly reported in working dogs from livestock farms in New Zealand,21 but were infrequently detected in our study, reflecting a differing local endemicity.
Direct PCR on specimens enables rapid and direct diagnosis, in both early and convalescent stages of infection.9 In our study, the overall molecular detection of leptospiral infection in dogs diagnosed with kidney and/or liver disease was 42.7%. These results were consistent with a previous study, which reported 42.4% (14 of 33; 95% CI: 25.6–59.3%), in a smaller, but similar, target population of dogs.39 In contrast, lower molecular detection rates of 19.8% (26 of 131; 95% CI: 13.0–26.7%) and 1.0% (1 of 106; 95% CI: 0.0–2.8%) have been reported in 2 other studies.28,49 Although these latter 2 studies were of a similar size to our study, the target populations were apparently healthy dogs, which could explain the lower detection rate.
Among 53 positive dogs, 17 dogs (13 with kidney and liver disease, 2 with kidney disease only, 2 with liver disease only) were in the leptospiremia phase, given that Leptospira spp. were detected only from whole blood; 11 dogs (6 with kidney and liver disease, 5 with kidney disease only) were in the leptospiruric phase, with positive detection only from urine samples. In another 25 dogs, Leptospira spp. were detected in both whole blood and urine samples, suggesting these dogs were in the period of active infection and actively shedding.
In addition, among these 53 positive dogs, 21 dogs were also seropositive by MAT. Of the 21 PCR-MAT–positive dogs, Leptospira spp. were detected in blood from 6 dogs (5 with kidney and liver disease, 1 with liver disease only), in urine from 5 dogs (2 with kidney and liver disease, 3 with kidney disease only), and both samples from 10 dogs (6 with kidney and liver disease, 3 with kidney disease only, 1 with liver disease only), which suggested that the dogs were either in active infection or in the recovery phase.
Dogs with clinical suspicion of leptospirosis, especially kidney disease, should have a positive PCR result on urine.18,19,57 However, using the PCR method, we found that blood samples were more commonly positive than were urine samples. This finding could be related to samples obtained at different phases of infection, the biological feature of urine itself, or the number of organisms being below the limit of detection of the PCR assay. There are limitations to PCR with regards to urine samples given that urea may degrade polymerase, affecting the sensitivity of the assay or even causing false-negative results.5,51 Furthermore, the optimal time for detection of Leptospira spp. in the urine of infected dogs was reported as ≥7 d of clinical illness.9,27,36 Therefore, the detection of leptospiral DNA from blood rather than urine should be considered a more reliable strategy to diagnose leptospirosis in dogs presented with both kidney and liver disease.
Only 2 kidneys and 2 livers were positive for pathogenic Leptospira spp. Low detection from tissue samples could be because of PCR inhibitors, such as hemoglobin and hormones.51 The abdominal effusion samples obtained from 4 different dogs were all positive for pathogenic Leptospira spp., suggesting that abdominal effusions could be the preferred samples for molecular investigation. Further studies are required on this issue.
In our study, of 53 positive dogs, 41 were exposed to rats and 32 were kept outdoors, suggesting that the dogs could have been infected through environmental exposure. The partial 16S rRNA sequencing performed after direct PCR revealed that L. interrogans were detected in all types of samples (blood, urine, abdominal effusion, kidney, liver); L. kirschneri was detected in all sample types except liver. The detection of L. interrogans and L. kirschneri were expected in our study; both species had been commonly associated with canine leptospirosis.18,19,38,52 The detection of L. borgpetersenii was linked to contacts with rats, given that this species is commonly shed by rats.6,7,55 A study in Germany reported that 3 of 200 healthy dogs shed leptospires and the species involved were L. interrogans (n = 2) and L. borgpetersenii (n = 1).34 Even though L. borgpetersenii is not common in dogs, this species remains a concern to canine leptospirosis.34 L. kmetyi was detected from one blood sample and this could be associated with environmental exposure; L. kmetyi has been isolated from the environment in Malaysia.53,58
Eight of 53 positive dogs were positive for leptospiral isolation. Leptospira spp. isolates were recovered from whole blood (n = 3), urine (n = 7), and abdominal effusion (n = 1) samples. In comparison, previous studies only recovered leptospires from urine samples of diseased, stray, and farm dogs.8,38,41 Surprisingly, we successfully cultured Leptospira sp. from abdominal effusion. Further serologic and molecular characterization of the 11 isolates revealed that all isolates were pathogenic, a finding supported by the phylogenetic analysis that showed that all the isolates were located within the pathogenic clade. Phylogenetic analysis concurred with the findings of previous reports that the pathogenic, intermediate, and saprophytic species each formed a clade.3,43
The isolation of serovars Bataviae, Javanica, and Australis is important given their absence in commercial vaccines. In Malaysia, bivalent (Canicola and Icterohaemorrhagiae) and tetravalent vaccines (Canicola, Grippotyphosa, Icterohaemorrhagiae, and Pomona) were adopted based on the availability of imported vaccine and the guidelines of the World Small Animal Veterinary Association.13 However, vaccination does not provide cross-protection toward nonvaccinal serovars,14 perhaps explaining the recovery of nonvaccinal serovars namely Bataviae, Javanica, and Australis from 3 vaccinated dogs. Therefore, proper characterization of leptospiral isolates is critical for the provision of evidence-based knowledge to support the development and commercialization of multivalent vaccines containing serovars that are circulating among local populations.
Our study has shown that at least 1 in 3 dogs with kidney and/or liver disease in Malaysia have the potential to have leptospirosis. Veterinarians examining dogs with these conditions should consider leptospirosis as a differential diagnosis.
Acknowledgments
We thank the small animal veterinarians at University Veterinary Hospital (UVH), and the staff of the Bacteriology Laboratory, Faculty of Veterinary Medicine (FVM), UPM for their valuable help in taking the samples from the recruited dogs as well as laboratory assistance. We also thank the private veterinary clinics near UPM (St. Angel Animal Medical Centre and J Avenue Veterinary Clinic) for providing samples.
Footnotes
Declaration of conflicting interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: Our research was supported by Ministry of Higher Education (MOHE), Malaysia under the Fundamental Research Grant Scheme (FRGS); FRGS/1/2016/SKK02/UPM/02/1/5524930.
ORCID iD: Kuan H. Khor  https://orcid.org/0000-0003-1500-9705
https://orcid.org/0000-0003-1500-9705
Contributor Information
Sabri A. Rahman, Departments of Veterinary Clinical Studies, Faculty of Veterinary Medicine, Universiti Putra Malaysia, Serdang, Selangor, Malaysia
Kuan H. Khor, Departments of Veterinary Clinical Studies, Faculty of Veterinary Medicine, Universiti Putra Malaysia, Serdang, Selangor, Malaysia.
Siti Khairani-Bejo, Veterinary Pathology and Microbiology, Faculty of Veterinary Medicine, Universiti Putra Malaysia, Serdang, Selangor, Malaysia.
Seng F. Lau, Departments of Veterinary Clinical Studies, Faculty of Veterinary Medicine, Universiti Putra Malaysia, Serdang, Selangor, Malaysia
Mazlina Mazlan, Veterinary Pathology and Microbiology, Faculty of Veterinary Medicine, Universiti Putra Malaysia, Serdang, Selangor, Malaysia.
Azri Roslan, Veterinary Pathology and Microbiology, Faculty of Veterinary Medicine, Universiti Putra Malaysia, Serdang, Selangor, Malaysia.
Soon H. Goh, Departments of Veterinary Clinical Studies, Faculty of Veterinary Medicine, Universiti Putra Malaysia, Serdang, Selangor, Malaysia Faculty of Veterinary Medicine, Universiti Malaysia Kelantan, Kota Bharu, Kelantan, Malaysia.
References
- 1.Ahmed SA, et al. Rapid diagnosis of leptospirosis by multiplex PCR. Malays J Med Sci 2012;19:9–16. [PMC free article] [PubMed] [Google Scholar]
- 2.André-Fontaine G.Canine leptospirosis—do we have a problem? Vet Microbiol 2006;117:19–24. [DOI] [PubMed] [Google Scholar]
- 3.Azali MA, et al. Molecular characterization of Leptospira spp. in environmental samples from north-eastern Malaysia revealed a pathogenic strain, Leptospira alstonii. J Trop Med 2016;2016:2060241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azócar-Aedo L, Monti G.Meta-analyses of factors associated with leptospirosis in domestic dogs. Zoonoses Public Health 2016;63:328–336. [DOI] [PubMed] [Google Scholar]
- 5.Bal AE, et al. Detection of leptospires in urine by PCR for early diagnosis of leptospirosis. J Clin Microbiol 1994;32:1894–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benacer D, et al. Determination of Leptospira borgpetersenii serovar Javanica and Leptospira interrogans serovar Bataviae as the persistent Leptospira serovars circulating in the urban rat populations in Peninsular Malaysia. Parasit Vectors 2016;9:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benacer D, et al. Isolation and molecular characterization of Leptospira interrogans and Leptospira borgpetersenii isolates from the urban rat populations of Kuala Lumpur, Malaysia. Am J Trop Med Hyg 2013;88:704–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benacer D, et al. Serological and molecular identification of Leptospira spp. in swine and stray dogs from Malaysia. Trop Biomed 2017;34:89–97. [PubMed] [Google Scholar]
- 9.Budihal SV, Perwez K.Leptospirosis diagnosis: competency of various laboratory tests. J Clin Diagn Res 2014;8:199–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chadsuthi S, et al. Investigation on predominant Leptospira serovars and its distribution in humans and livestock in Thailand, 2010–2015. PLoS Negl Trop Dis 2017;11:e0005228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou YL, et al. Leptospirosis in Taiwan, 2001–2006. Emerg Infect Dis 2008;14:856–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costa F, et al. Global morbidity and mortality of leptospirosis: a systematic review. PLoS Negl Trop Dis 2015;9:e0003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Day MJ, et al. WSAVA Guidelines for the vaccination of dogs and cats. J Small Anim Pract 2016;57:E1–E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eric-Klaasen HL, Adler B.Recent advances in canine leptospirosis: focus on vaccine development. Vet Med (Auckl) 2015;6:245–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghada AK, et al. Serological and molecular prevalence of Leptospira infection in rat populations in Kuala Lumpur. Aust J Basic Appl Sci 2017;11:62–72. [Google Scholar]
- 16.Goh SH, et al. Risk factors and prediction of leptospiral seropositivity among dogs and dog handlers in Malaysia. Int J Environ Res Public Health 2019;16:1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greene CE, et al. Leptospirosis. In: Greene CE, ed. Infectious Diseases of the Dog and Cat. 4th ed.Saunders, 2012:431–447. [Google Scholar]
- 18.Harkin KR, et al. Clinical application of a polymerase chain reaction assay for diagnosis of leptospirosis in dogs. J Am Vet Med Assoc 2003;222:1224–1229. [DOI] [PubMed] [Google Scholar]
- 19.Harkin KR, et al. Comparison of polymerase chain reaction assay, bacteriologic culture, and serologic testing in assessment of prevalence of urinary shedding of leptospires in dogs. J Am Vet Med Assoc 2003;222:1230–1233. [DOI] [PubMed] [Google Scholar]
- 20.Harkin KR, Hays MP.Variable-number tandem-repeat analysis of leptospiral DNA isolated from canine urine samples molecularly confirmed to contain pathogenic leptospires. J Am Vet Med Assoc 2016;249:399–405. [DOI] [PubMed] [Google Scholar]
- 21.Harland AL, et al. A serological survey of leptospiral antibodies in dogs in New Zealand. N Z Vet J 2013;61:98–106. [DOI] [PubMed] [Google Scholar]
- 22.Hartskeerl RA, et al. Emergence, control and re-emerging leptospirosis: dynamics of infection in the changing world. Clin Microbiol Infect 2011;17:494–501. [DOI] [PubMed] [Google Scholar]
- 23.Khor KH, et al. Seroprevalence and molecular detection of leptospirosis from a dog shelter. Trop Biomed 2016;33:276–284. [PubMed] [Google Scholar]
- 24.Klopfleisch R, et al. An emerging pulmonary haemorrhagic syndrome in dogs: similar to the human leptospiral pulmonary haemorrhagic syndrome? Vet Med Int 2010;2010:928541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koizumi N, et al. Molecular and serological investigation of Leptospira and leptospirosis in dogs in Japan. J Med Microbiol 2013;62:630–636. [DOI] [PubMed] [Google Scholar]
- 26.Kumar S, et al. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 2016; 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lane AB, Dore MM.Leptospirosis: a clinical review of evidence-based diagnosis, treatment and prevention. World J Clin Infect Dis 2016;6:61–66. [Google Scholar]
- 28.Latosinski GS, et al. Serological and molecular detection of Leptospira spp. in dogs. Rev Soc Bras Med Trop 2018;51:364–367. [DOI] [PubMed] [Google Scholar]
- 29.Lau CL, et al. Climate change, flooding, urbanisation and leptospirosis: fuelling the fire? Trans R Soc Trop Med Hyg 2010;104:631–638. [DOI] [PubMed] [Google Scholar]
- 30.Lau SF, et al. Prevalence of leptospirosis in healthy dogs and dogs with kidney disease in Klang valley, Malaysia. Trop Biomed 2016;33:469–475. [PubMed] [Google Scholar]
- 31.Lau SF, et al. Seroprevalence of leptospirosis in working dogs. Top Compan Anim Med 2017;32:121–125. [DOI] [PubMed] [Google Scholar]
- 32.Levett PN.Leptospirosis. Clin Microbiol Rev 2001;14:296–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levett PN.Usefulness of serologic analysis as a predictor of the infecting serovar in patients with severe leptospirosis. Clin Infect Dis 2003;36:447–452. [DOI] [PubMed] [Google Scholar]
- 34.Llewellyn JR, et al. Urinary shedding of leptospires and presence of Leptospira antibodies in healthy dogs from Upper Bavaria. Berl Munch Tierarztl Wochenschr 2016;129:251–257. [PubMed] [Google Scholar]
- 35.Martins G, et al. The dog in the transmission of human leptospirosis under tropical conditions: victim or villain? Epidemiol Infect 2012;140:207–209. [DOI] [PubMed] [Google Scholar]
- 36.Mauro T, Harkin K.Persistent leptospiruria in five dogs despite antimicrobial treatment (2000–2007). J Am Anim Hosp Assoc 2019;55:42–47. [DOI] [PubMed] [Google Scholar]
- 37.Miller MD, et al. Variability in results of the microscopic agglutination test in dogs with clinical leptospirosis and dogs vaccinated against leptospirosis. J Vet Intern Med 2011;25:426–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miotto BA, et al. Development and validation of a modified TaqMan based real-time PCR assay targeting the lipl32 gene for detection of pathogenic Leptospira in canine urine samples. Braz J Microbiol 2018;49:584–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miotto BA, et al. Diagnosis of acute canine leptospirosis using multiple laboratory tests and characterization of the isolated strains. BMC Vet Res 2018;14:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miotto BA, et al. Molecular and serological characterization of the first Leptospira santarosai strain isolated from a dog. Acta Trop 2016;162:1–4. [DOI] [PubMed] [Google Scholar]
- 41.Miotto BA, et al. Prospective study of canine leptospirosis in shelter and stray dog populations: identification of chronic carriers and different Leptospira species infecting dogs. PLoS One 2018;13:e0200384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mohd Ali MR, et al. Isolation of Leptospira kmetyi from residential areas of patients with leptospirosis in Kelantan, Malaysia. J Infect Public Health 2018;11:578–580. [DOI] [PubMed] [Google Scholar]
- 43.Morey RE, et al. Species-specific identification of Leptospiraceae by 16S rRNA gene sequencing. J Clin Microbiol 2006;44:3510–3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Musso D, La-Scola B.Laboratory diagnosis of leptospirosis: a challenge. J Microbiol Immunol Infect 2013;46:245–252. [DOI] [PubMed] [Google Scholar]
- 45.Rissi DR, Brown CA.Diagnostic features in 10 naturally occurring cases of acute fatal canine leptospirosis. J Vet Diagn Invest 2014;26:799–804. [DOI] [PubMed] [Google Scholar]
- 46.Sabri AR, et al. Molecular detection of Leptospira sp. in cattle and goats in Kelantan, Malaysia after a massive flood using multiplex polymerase chain reaction. Trop Biomed 2019;36:165–171. [PubMed] [Google Scholar]
- 47.Samir A, et al. Leptospirosis in animals and human contacts in Egypt: broad range surveillance. Rev Soc Bras Med Trop 2015;48:272–277. [DOI] [PubMed] [Google Scholar]
- 48.Samsi NS, et al. Serodiagnosis of leptospirosis in domestic animals and humans. Malaysian J Vet Res 2013;4:21–26. [Google Scholar]
- 49.Santanna R, et al. High number of asymptomatic dogs as leptospiral carriers in an endemic area indicates a serious public health concern. Epidemiol Infect 2017;145:1852–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneider MC, et al. Leptospirosis in Rio Grande do Sul, Brazil: an ecosystem approach in the animal-human interface. PLoS Negl Trop Dis 2015;9:e0004095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schrader C, et al. PCR inhibitors—occurrence, properties, and removal. J Appl Microbiol 2012;113:1014–1026. [DOI] [PubMed] [Google Scholar]
- 52.Schuller S, et al. European consensus statement on leptospirosis in dogs and cats. J Small Anim Pract 2015;56:159–179. [DOI] [PubMed] [Google Scholar]
- 53.Slack AT, et al. Leptospira kmetyi sp. nov., isolated from an environmental source in Malaysia. Int J Syst Evol Microbiol 2009;59:705–708. [DOI] [PubMed] [Google Scholar]
- 54.Tansuphasiri U, et al. Duplex PCR-hybridization based detection of pathogenic Leptospira in environmental water samples obtained from endemic areas in northeast region of Thailand. Southeast Asian J Trop Med Public Health 2006;37:729–741. [PubMed] [Google Scholar]
- 55.Vedhagiri K, et al. Characterization of Leptospira borgpetersenii isolates from field rats (Rattus norvegicus) by 16s rRNA and lipl32 gene sequencing. Braz J Microbiol 2010;41:150–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.World Organisation for Animal Health (OIE). OIE manual of diagnostic tests and vaccines for terrestrial animals. OIE, 2004. https://www.oie.int/en/what-we-do/standards/codes-and-manuals/terrestrial-manual-online-access/
- 57.Yamaguchi T, et al. Characterizing interactions of Leptospira interrogans with proximal renal tubule epithelial cells. BMC Microbiol 2018;18:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yusof NY, et al. Complete genome sequence of Leptospira kmetyi LS 001/16, isolated from a soil sample associated with a leptospirosis patient in Kelantan, Malaysia. Microbiol Resour Announc 2019;8:e00015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]