Abstract
Importation of exotic animals that may harbor infectious agents poses risks for native species with potentially severe impacts on animal health and animal production. Although the Asian water buffalo (Bubalus bubalis) population in Europe is steadily increasing, its susceptibility to viral infections and its role for interspecies transmission is largely unknown. To identify viral infections that are shared between exotic water buffaloes and native small ruminants, we collected blood samples from 3 Swiss farms on which water buffaloes were kept either without, or together with, sheep or goats. These samples were analyzed by next-generation sequencing (NGS) as well as by selected conventional tests, including PCR, ELISA, and in some cases a virus neutralization test. By NGS, a novel virus of the genus Gemykrogvirus (GyKV; Genomoviridae) was first detected in the buffaloes on one farm, and subsequently confirmed by PCR, and was also detected in the co-housed sheep. In contrast, this virus was not detected in buffaloes on the farms without sheep. Moreover, conventional methods identified a number of viral infections that were not shared between the exotic and the native animals, and provided evidence for potential roles of water buffaloes in the epidemiology of ruminant pestiviruses, especially bovine viral diarrhea virus, bluetongue virus, and possibly bovine alphaherpesvirus 2. Our results clearly indicate that water buffaloes are susceptible to interspecies viral transmission and may act as intermediate hosts, or even as reservoirs, for these viruses.
Keywords: buffalo, interspecies transmission, next-generation sequencing, small ruminants
The Asian water buffalo (Bubalus bubalis) was introduced to Europe several 100 y ago, and it is kept for milk and meat production.5 Compared to cattle, water buffalo farming in Switzerland is in its infancy. Since 1996, when the first Asian water buffaloes were introduced to Switzerland, water buffalo farming has been gaining in importance. There are now 2,400 water buffaloes on farms in all parts of Switzerland (Schwermer HP, pers. comm., 2020 Oct 18). Despite various advantages of water buffalo farming compared to cattle farming, such as a 5-fold higher milk price and more efficient conversion of poor-quality roughage into milk and meat,5 the intermixing of exotic cloven-hoofed species with native livestock entails risks of infection.
It is well known that certain disease agents may go unnoticed in one species, but cause severe and, in some cases, lethal illness in another. Examples are malignant catarrhal fever (MCF) viruses (genus Macavirus), which cause systemic infection with high lethality in cattle and deer, while remaining subclinical in sheep, goats, and other reservoir hosts10,35; bluetongue virus (BTV), which most severely affects sheep while causing rather mild infections in cattle23; or infection with foot-and-mouth disease virus (FMDV), in which clinical signs are overt in cattle and swine but mild or nonexistent in small ruminants.18
The species-associated difference in the clinical manifestations of viral infections harbors the risk of unnoticed introduction and spread of notifiable diseases that can have severe impacts on animal health and cause considerable economic losses as a result of animal deaths, control measures of epizootics, and trade restrictions. In Switzerland, where co-housing of various ruminant species and alpine farming is very common, the risk of interspecies transmission and rapid spread of viral infections is further enhanced.
Moreover, as a consequence of ongoing worldwide climate change, exotic arthropods are spreading north.34 Arthropods frequently act as vectors for viruses and may therefore either introduce exotic disease agents or contribute to the distribution of established or novel disease agents. In fact, the 2 most recent emerging viral diseases in farm animals in Switzerland, and probably all of Europe, are caused by arboviruses, namely BTV and Schmallenberg virus (SBV; Schmallenberg orthobunyavirus).32
Our aim was to identify viral infections that are shared between water buffaloes and native small ruminants (i.e., sheep and goats) by a variety of test methods. These species are of special interest because they may act as intermediate hosts or reservoirs for viruses predominantly affecting cattle, and because small ruminants are not included in eradication or monitoring programs of notifiable diseases in bovine species. Next-generation sequencing (NGS) has gained importance in viral identification and is increasingly used as a testing tool. Compared to conventional tests, high-throughput NGS technology allows the detection of an unlimited number of known as well as unknown and potentially emerging viruses in a nonspecific manner. Culture- and primer-independent NGS generates a wealth of sequencing data that may include all viruses present in a single sample. In addition to NGS, which we used on blood samples of water buffaloes only, we applied selected conventional assays targeting major viral infections in ruminants on blood samples of water buffaloes as well as co-housed small ruminants. These tests included conventional PCR and real-time PCR (rtPCR) assays as well as serologic assays, namely ELISA, and in some cases virus neutralization tests (VNTs). This procedure allowed insight into the blood-associated virome of Swiss water buffaloes as well as a comparison of the NGS and conventional assays, both of which are useful for assessing the risk of interspecies viral transmission. These data may have important implications for eradication or monitoring programs of notifiable diseases in ruminant species.
Materials and methods
Our study was carried out with written consent of animal owners and in strict accordance with the Swiss regulations for animal experimentation. The protocol for our study was approved by the Cantonal Veterinary Office Zurich, ZH, Switzerland (Permit 102/2012).
Farms and animals
We investigated 3 Swiss farms housing water buffaloes. Farm 1 was located in the central part of Switzerland. The animal stock consisted of 21 water buffaloes and 30 cattle. The 2 species were housed in the same barn separated by a feed trough, having direct contact only in the courtyard. Both cattle and water buffaloes spent the summer on an alpine pasture. Farm 2 was also situated in central Switzerland and housed 33 water buffaloes and 20 sheep. The buffaloes were kept in 2 barns, each with access to a courtyard. The sheep were separated from the buffaloes by a wooden wall; the 2 species did not share a pasture. The water buffaloes spent the summer on an alpine pasture. Farm 3 was located in northwestern Switzerland. It housed 5 water buffaloes, 65 cattle, and 7 goats. The adult buffaloes were housed with the cattle in one barn, buffalo calves were kept in another premises, separated from the goats by a wooden wall. Buffaloes and goats were always pastured separately.
Sample collection
In May and June 2013, EDTA blood samples were taken from water buffaloes and from co-housed small ruminants by the private veterinarian of the respective farm. Animals tested included 17 water buffaloes from farm 1, 26 water buffaloes and 19 sheep from farm 2, and 5 water buffaloes and 7 goats from farm 3. None of the animals had been reported to show any clinical signs at the time of sampling. Cattle were not included in the analysis given that our focus was on viral transmission between water buffaloes and small ruminants.
Sample preparation
Plasma was separated by centrifugation of the blood samples at room temperature (868 × g for 10 min). One part of the plasma was used for conventional tests, the other for NGS. Buffy coat cells were isolated using an in-house NaH4Cl lysis buffer.35 The buffy coat from each sample was aliquoted for NGS and for conventional test methods. Buffy coats and plasma samples were stored at −20°C until further processed.
Enrichment for viral particles and nucleic acid extraction for NGS
Only blood samples of the 48 water buffaloes were subjected to NGS given that our aim was to detect potentially novel viruses primarily in this exotic species. After thawing, 3–4 mL of plasma was centrifuged for 30 min at 3,000 × g and the supernatant filtered using a 0.45-µm syringe filter (13 mm Whatman Puradisc; GE Healthcare). To pellet any viral particles, the filtrate was centrifuged at 120,000 × g for 6 h at 16°C (AH650 swing out rotor and matching buckets, Beckmann ultra-clear 5-mL tubes, Sorvall Wx Ultra 80 ultra-centrifuge; Thermo Fisher). If the volume of plasma was < 5 mL, the volume was topped-up to 5 mL by adding nuclease-free water. After centrifugation, the supernatant was carefully removed and the (invisible) pellet resuspended using 200 µL of phosphate-buffered saline (PBS). The water buffalo buffy coat samples were thawed and 200 µL of nuclease-free water added. The pellet was homogenized (QIAshredder column; Qiagen) and was centrifuged for 2 min at full speed in a benchtop centrifuge. The flow-through was filtered using a 0.45-µm syringe filter (GE Healthcare) to remove larger particles. The filtrate (125 µL) was mixed with RNase A (Sigma) at a final concentration of 150 µg/mL with Benzonase (1 U/µL; Merck) to remove free nucleic acid not protected by a viral capsid, and incubated at 45°C for 45 min followed by 1 h at 37°C. The buffy coat (130 µL) and plasma (200 µL) preparations were thereafter pipetted together, and the nucleases immediately inactivated by adding 3 volumes of purification reagent (peqGold TriFast FL; VWR).
RNA was extracted following the manufacturer’s instructions with the exception of adding 40 µg of UltraPure glycogen (Thermo Fisher) to the aqueous phase to enhance RNA precipitation. DNA was extracted from the mid- and bottom layer using a DNA back extraction buffer consisting of 4 M guanidine thiocyanate, 50 mM sodium citrate, 1 M Tris (free base), pH 8.5–9 as recommended in the TRIzol manual for DNA extraction (Thermo Fisher). The extracted RNA and DNA were combined and stored at −80°C if not processed immediately. A bovine EDTA blood sample was spiked with known RNA (bovine viral diarrhea virus, BVDV; genus Pestivirus) and DNA (bovine alphaherpesvirus 1, BoHV1) viruses for positive control, and was used non-spiked as negative control. Both samples were prepared as described for the buffalo samples. The water buffalo samples were not grouped according to farm of origin but in randomized batches for sample preparation, and the DNA was again randomized for library preparation to avoid allocation bias.
Sequence-independent single primer amplification for NGS
Because of the nuclease treatment, the total nucleic acid concentration of the samples was too low for library preparation and therefore had to be amplified. In addition, RNA and single-stranded DNA (ssDNA) needed to be converted to double-stranded DNA (dsDNA). For cDNA synthesis, the RevertAid H minus first strand cDNA synthesis kit (Thermo Fisher) was used, primed by a random tagged primer consisting of random hexamers and a 20-nucleotide (nt) tag (5′-GTTGGAGCTCTGCAGTCATC-NNNNNN-3′). The cDNA was subjected to 2 rounds of second-strand synthesis by Klenow fragment exo– (Thermo Fisher), again under addition of the random tagged primer. The dsDNA was purified (PureLink PCR micro kit; Thermo Fisher) and the eluate subjected to 25 amplification cycles (HotStarTaq DNA polymerase; Qiagen) using the tag as single primer (5′-GTTGGAGCTCTGCAGTCATC-3′). The amplified product was purified (PureLink PCR micro kit) and stored at −80°C until library preparation.
Library preparation and sequencing
The DNA concentration of the amplified product was determined (DNA high sensitivity assay, Qubit 2.0 fluorometer; Thermo Fisher). DNA (100 ng) in 58 µL total volume was further processed. The DNA was sheared into 500-bp fragments (Focused-ultrasonicator E220; Covaris). The library was prepared (NEBNext ultra DNA library prep kit for Illumina, NEBNext multiplex oligos for Illumina; New England Biolabs) according to the manufacturer’s instructions but without size selection and using 6 instead of 8 amplification cycles. The size and molarity of the libraries were checked (Agilent TapeStation 2200, D1000 screen tape; Agilent). Libraries were equimolarly mixed to the concentration of the lowest library prior to a paired-end sequencing run (2 × 150 bp, NextSeq machine at mid-output; Illumina) at the Functional Genomics Center Zurich (FGCZ).
Analysis of NGS data
After the raw data were quality checked, trimmed, and de-multiplexed at the FGCZ, we performed reference-based alignment (Lasergene SeqMan NGen software; DNASTAR). Reads of the single libraries were screened against the NCBI RefSeq database (https://www.ncbi.nlm.nih.gov/refseq/) using the metagenomic pipeline of the program including host removal (bovine genome). All alignments were visually controlled for coverage and false matches (e.g., clonal alignments resulting from low-complexity reads were omitted from further analysis). Contigs of true matches were extracted and saved in FASTA format for further analysis.
GyKV full-genome Sanger sequencing
NGS revealed reads for a novel virus of genus Gemykrogvirus (GyKV). However, the reads only covered 18% of the genome. To determine the remaining genome sequence, primers for 2 overlapping PCR products that should cover the whole circular genome of ~ 2,200 nt were designed (Clone Manager v.9; Sci Ed Software). Design was based on the contigs composed of the NGS reads. DNA from animal F2_WB18 served as template for the 2 PCR assays (Suppl. Table 1). HotStarTaq DNA polymerase (Qiagen) was used according to the manufacturer’s instructions using a 200 nM final concentration of primers 1f (5′-TTAGCGAAGTGTGGGTCCTC-3′) and 1r (5′-CGGCTACTGCGTTCGATTAC-3′) for the first PCR that resulted in a 792-nt amplicon, and of primers 2f (5′-GTGGTCAAGTCGGATGTCTC-3′) and 2r (5′-AGCACGCCTACTTCAACCTC-3′) for the second PCR that resulted in a 1,667-nt product. Bands were visualized on 1.5% agarose gel, and products of the correct size were excised and extracted (QIAquick gel extraction kit; Qiagen). Purified amplicons were sent to Microsynth (Balgach, Switzerland) for bidirectional sequencing. The full genome of the bubaline-associated gemykrogvirus (BuGyKV) F18_L28 was assembled in silico using Clone Manager v.9 software and is available in GenBank (MT553114). Detailed examination of the genome and determination of the open reading frames (ORFs) was performed using Clone Manager v.9.
Extraction of nucleic acids for conventional tests
Because only one buffy coat was available for conventional testing, nucleic acids were extracted using the QIAamp DNA mini kit (Qiagen), which in preliminary tests copurified RNA and yielded reverse-transcription PCR (RT-PCR) results comparable to extraction with the QIAamp RNA blood mini kit (Qiagen; data not shown). The extractions were performed according to the manufacturer’s instructions, and extracted nucleic acids were stored at −20°C or used directly for PCR.
Conventional tests
Various PCR assays were used for detection of viral nucleic acids in the buffy coat cells. ELISA and, in some cases, VNT were used to detect virus-specific antibodies in the plasma samples (Table 1).
Table 1.
Overview of the conventional tests used for virus or antibody detection in water buffaloes and small ruminants in Switzerland.
| Virus | Method | Reference |
|---|---|---|
| Herpesviruses | Panherpesvirus nPCR | Ehlers et al.11 and VanDevanter et al.38 |
| OvHV2 | rtPCR | Hüssy et al.16 and Stahel et al.35 |
| CpHV2 | rtPCR | Cunha et al.8 |
| BoHV1 | rtPCR | Abril et al.1 |
| ELISA | IBR gB X3 (Idexx) | |
| BoHV5 | rtPCR | Abril et al.1 |
| BuHV1 | ELISA | Nogarol et al.28; Eradikit BuHV-1 (IN3 diagnostic) |
| CpHV1 | ELISA | Bertolotti et al.4; Eradikit CpHV1(IN3 diagnostic) |
| BoHV2 | ELISA | ID screen BHV2 indirect (IDvet) |
| VNT | World Organisation for Animal Health42 | |
| BVDV, BDV | Panpestivirus RT-PCR | Vilcek40 |
| ELISA | Canal et al.7 | |
| VNT | Kaiser et al.17 | |
| BLV | ELISA | Leukosis serum X2 test kit (Idexx) |
| BTV | ELISA | Bluetongue virus antibody test kit (VMRD) |
| SBV | ELISA | Svanovir SBV-Ab (Boehringer Ingelheim Svanova) |
| BuGyKV | rtPCR | In-house |
BDV = border disease virus; BLV = bovine leukemia virus; BoHV1 = bovine alphaherpesvirus 1; BoHV2 = bovine alphaherpesvirus 2; BoHV5 = bovine alphaherpesvirus 5; BTV = bluetongue virus; BuGyKV = bubaline-associated gemykrogvirus; BuHV1 = bubaline alphaherpesvirus 1; BVDV = bovine viral diarrhea virus; CpHV1 = caprine alphaherpesvirus 1; CpHV2 = caprine gammaherpesvirus 2; nPCR = nested PCR; OvHV2 = ovine gammaherpesvirus 2; rtPCR = real-time PCR; RT-PCR = reverse-transcription PCR; SBV = Schmallenberg virus; VNT = virus neutralization test.
12S rRNA rtPCR
Amplification of the 12S rRNA reference gene served as internal control to confirm successful DNA extraction and sensitivity of the PCR reaction.35 Duplicates of the samples were tested in 10-fold dilution. PCR conditions corresponded to those used for ovine gammaherpesvirus 2 (OvHV2) detection.
Panherpesvirus nested PCR
Panherpesvirus nested PCR (nPCR) was performed as described previously,11,38 with slight modifications: for first-round PCR, 5 µL of extracted undiluted and 10-fold diluted sample DNA was used as template; for second-round PCR, 1 μL of the product from first-round PCR was used. In both PCR runs, the final mixture had a volume of 25 µL, containing: 2.5 µL of PCR buffer (10×; Qiagen); final concentrations of 200 µM for each dNTP and 1 µM for each degenerate primer38; 2 units of HotStarTaq DNA polymerase (5 U/µL); and topped up with diethylpyrocarbonate (DEPC)-treated water. Thermal cycling (Peltier thermal cycler-200; MJ Research) used conditions described previously,11 except for 12 min of initial denaturation at 95°C. Products were analyzed on 2% agarose gel, and bands of the expected size (215–235 bp) were extracted (QIAquick gel extraction kit) according to the manufacturer’s instructions. Gel-extracted DNA was subsequently amplified in a third sequencing PCR using non-degenerate primers.38 The reaction mixture for sequencing PCR and cycling conditions corresponded to those described for second-round PCR with the exception of a 200-nM final concentration of each sequencing primer and 1 unit of HotStarTaq DNA polymerase (5 U/µL). Products were purified (QIAquick PCR purification kit) and sent to Microsynth for bidirectional sequencing. Sequences were assembled by SeqMan Pro (DNASTAR), screened by NCBI BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and phylogenetically analyzed using MEGAX.21
OvHV2 rtPCR
For detection of OvHV2, a rtPCR was performed using published primers and probes.35 PCR reaction mixture and cycling conditions have been described previously.16 The PCR was run on a 7900HT fast real-time PCR system (Applied Biosystems) with 9600 emulation ramping. Given the limited amount of DNA available, duplicates of the samples were tested in 2-fold and 10-fold dilutions.
CpHV2 rtPCR
For detection of caprine gammaherpesvirus 2 (CpHV2), primer and probes were used as described previously,8 with the exception that the upstream primer contained a guanine instead of an adenine base in position 9, which is supposed to raise specificity for CpHV2 (data not shown). Further, the probe was labeled with TET instead of HEX at the 5′-end. The reaction mixture had a final volume of 25 µL and was composed of: 12.5 µL of TaqMan universal PCR master mix (Applied Biosystems); 6.8 µL of DEPC-treated water; and final concentrations of 200 nM of each primer and 80 nM probe; to which 5 µL of undiluted or 10-fold diluted template DNA were added. Samples were tested in duplicate. The cycle protocol corresponded to the one used for OvHV2 detection, with the ramping of the cycler set to standard rate.
BoHV1 and BoHV5 rtPCR
For detection of BoHV1 and bovine alphaherpesvirus 5 (BoHV5), 2 rtPCR assays were performed.1 Only the water buffalo samples were tested, undiluted and in 10-fold dilution. The PCR was run on a 7900HT fast real-time PCR system with 9,600 emulation ramping. While the BoHV1 rtPCR is able to also amplify BoHV5 DNA, although less efficiently, the BoHV5 rtPCR amplifies BoHV5 DNA specifically, allowing discrimination of the 2 viruses in positive samples.
BoHV1, BuHV1, CpHV1, BoHV2, BLV, BTV, and SBV ELISAs
ELISA for antibody detection of BoHV1, bubaline alphaherpesvirus 1 (BuHV1),28 caprine alphaherpesvirus 1 (CpHV1),4 bovine alphaherpesvirus 2 (BoHV2), bovine leukemia virus (BLV), BTV (24 different serotypes), and SBV were performed as per the manufacturer’s instructions (Table 1).
BoHV2 VNT
Samples exceeding the negative cutoff in the BoHV2 ELISA were subjected to VNT consecutively, as described for BoHV1.42 In short, plasma samples were complement-inactivated at 56°C for 30 min, diluted in 2-fold dilution steps, and then mixed with equal volumes of a working solution (2,000 TCID50/mL) of BoHV2 strain V766. After incubation for 24 h at 37°C and 5% CO2, 100 µL of each plasma–virus mixture were distributed in 4 wells coated with Madin–Darby bovine kidney (MDBK) cells, yielding ~ 100 TCID50 of BoHV2 per well. Medium and cell controls as well as viral back titration were included. The plate was incubated for 3 d at 37°C. Wells displaying cytopathic effects (CPE) typical for BoHV2 were identified, and antibody titers were calculated according to Reed and Muench. The VNT was valid if the titration of the working suspension yielded a virus concentration of 600–6,000 TCID50/mL to result in 30–300 TCID50 per well.
Panpestivirus RT-PCR
A panpestivirus RT-PCR40 was performed (OneStep RT-PCR kit; Qiagen) according to the manufacturer’s instructions. The reaction had a final reaction volume of 20 µL and contained 2 µL of RNA. Each sample was tested undiluted and in 10-fold dilution. PCR products had an expected size of 244–247 bp and were analyzed on 1.5% agarose gel. Gel extraction and sequencing were carried out as described for the panherpesvirus nPCR.
BVDV/BDV biphasic indirect ELISA and VNT
For serologic analysis of the ruminant pestiviruses BVDV and border disease virus (BDV; Pestivirus D), a biphasic indirect ELISA was performed7 with slight modifications of incubation periods at 60 min instead of 90 min. For water buffalo samples, goat anti-bovine IgG was used as conjugate; peroxidase-labeled protein G was used for small ruminant samples. Sample-to-positive control values of < 20% were considered seronegative; samples with reactivities of ≥ 30% were considered seropositive. Ratios in between were considered doubtful. Given the marked cross-reactions among pestiviruses, the ELISA allows no distinction between BVDV and BDV antibodies. Antibody specificity of samples exceeding the negative cutoff of 20% in the ELISA was therefore determined by VNT, by which each plasma sample was tested against 3 pestiviral strains: the cytopathogenic (cp) BVDV1a R1935/72 (Oregon C24V), the non-cytopathogenic (ncp) BVDV1h (CH-04-01b), and BDV Swiss a R9336/11 (ncp).17 To relate the antibodies to either BVDV or BDV, the titer ratios (BVDV/BDV) of BVDV1a and BDV, as well as BVDV1h and BDV, were calculated for each sample. Ratios ≥ 4 indicated seropositivity against BVDV (1a or 1h); ratios ≤ 0.25 indicated antibody specificity for BDV. For ratios in between (0.25 < ratio < 4), antibody specificity could not be determined.17
BuGyKV rtPCR
NGS revealed reads for a novel virus of the genus Gemykrogvirus in 15 water buffalo samples. To confirm the presence of BuGyKV among water buffaloes and to also address its prevalence in the small ruminants, we designed a specific rtPCR targeting a 60-bp sequence located in the small intergenic region between the replication (Rep) and the capsid (Cap) genes (Fig. 1). The full genome sequence gained from sample F2_WB18 (Suppl. Table 1) by NGS and Sanger sequencing was used as reference genome (MT553114). The reaction mixture had a final volume of 20 µL and consisted of: 10 µL of TaqMan universal PCR master mix; 5 µL of DEPC-treated water, final concentrations of 500 nM of each primer (forward 5′-GATCGTTCGCTTCTTTCGGTAT-3′, reverse 5′-TGGCTAGGCGCACAAAAAC-3′) and 250 nM probe (5′-6FAM-TGTCCGTGATGACAAAT-MGB-3′); to which 2 µL of DNA were added. If no DNA and little plasma was left from a sample, plasma was used directly, after heating it to 95°C for 3 min and then cooling it on ice for 5 min. PCR was run on a QuantStudio flex real-time PCR system (Applied Biosystems). Cycling was carried out according to the manufacturer’s instructions (TaqMan universal PCR master mix).
Figure 1.
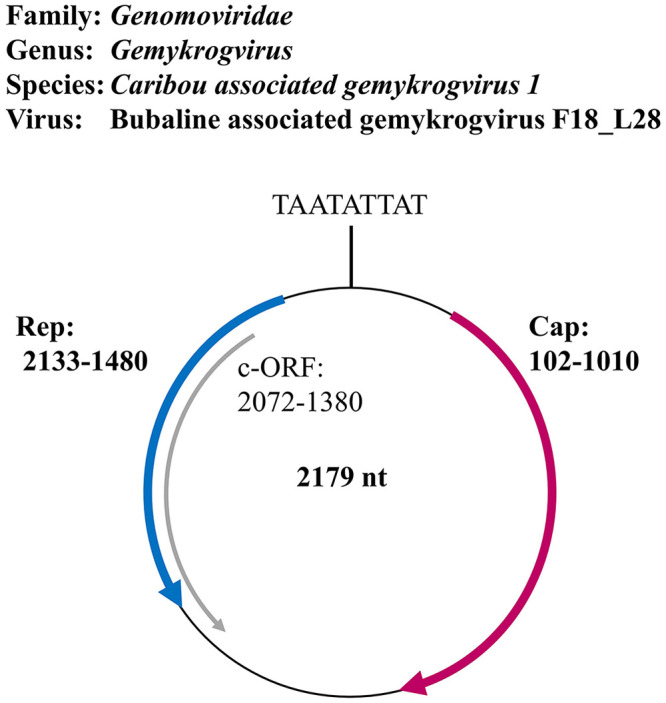
Taxonomic affiliation and genome organization of the gemykrogvirus (GyKV) detected in blood samples of water buffaloes (bubaline-associated gemykrogvirus, BuGyKV). The conserved nonamer in the hairpin structure as well as the 3 open reading frames (ORFs) are depicted; 1 coding in sense direction (red) for a putative capsid protein (Cap), and 2 coding in anti-sense direction for a putative replicase-associated protein (Rep, blue) and an unknown putative protein (c-ORF, gray). The genome numbering starts with the conserved nonamer sequence.
Phylogenetic analysis of GyKV and gammaherpesviruses
To determine the taxonomic affiliation of the novel GyKV, the full-length sequence was compared to representatives of all 73 official species of the 9 genera of the family Genomoviridae using SDT v.1.227 after MUSCLE alignment. Taxonomy and reference genomes as well as the species demarcation cutoff of 78%39 were chosen according to the International Committee on Taxonomy of Viruses (ICTV, status July 2020; https://talk.ictvonline.org/taxonomy/). In addition, a phylogenetic tree was constructed in MEGAX21 using the same 73 full-length reference genomes after MUSCLE alignment and 100 bootstraps. The evolutionary history was inferred by using the maximum likelihood method and Tamura–Nei model. The tree with the highest log likelihood was selected. The same methods (pairwise comparison and maximum likelihood tree) were used to analyze the sequences resulting from the panherpesvirus nPCR of water buffaloes, sheep, and goats. As reference genomes, one representative—if available, the RefSeq genome—of each species belonging to the 4 official genera of the subfamily Gammaherpesvirinae was chosen and shortened to the 166–178-nt sequence of the PCR product (primer binding sites were omitted). Taxonomy and nomenclature are according to the latest ICTV report (status October 9, 2020).
Results
Detection of a novel virus of genus Gemykrogvirus in water buffaloes and sheep
NGS of the 48 water buffalo samples and 2 control samples resulted in 313,632,454 quality-controlled total reads that were screened to the RefSeq database. The average read count per sample was 6,272,649. The positive control showed efficient recovery of the spiked-in RNA and DNA viruses; the negative control did not provide evidence for contamination (data not shown). Upon visual control of the single alignments regarding genome coverage and contig composition, 2 reference sequences proved to be valid matches. Reads of 15 of 48 libraries matched to these reference sequences: 398 reads to the caribou feces–associated gemycircularvirus (NC_024909.1) and 170 reads to the related HCBI9.212 (NC_024690.1; Suppl. Table 1), which is a circular ssDNA virus detected in bovine serum. Interestingly, the reads were only found in samples of water buffaloes from farm 2. The highest number of matching reads was found in sample F2_WB18 (library 28) with 99 reads for caribou feces–associated gemycircularvirus and 40 for HCBI9.212 (Suppl. Table 1). Genome coverage was 18% and 15%, respectively. Based on the contigs, primers were designed to cover the remainder of the genome and to validate the sequence of the contigs. Indeed, the Sanger sequences confirmed the NGS contigs and covered the whole genome. The final sequence has a length of 2,179 nt, and BLAST analysis showed it to be most closely related to members of family Genomoviridae (Fig. 1). The genome encodes for 2 major ORFs: a 909-nt sequence in positive-sense direction relative to the conserved nonamer, coding for a 302 amino acid protein closely related (up to 91.4%) to Cap proteins of other Genomoviridae, and a second ORF in anti-sense direction coding for a putative Rep protein of 217 amino acids, up to 96.6% identical to Rep proteins of other Genomoviridae. Another ORF in anti-sense (Fig. 1) had 60% identity to putative proteins of genomoviruses associated with mice and giant pandas.
To allocate genomoviruses to 1 of the 9 official genera, the sequence of the conserved nonamer in the stem loop as well as several Rep motifs can be considered.39 These analyses indicated that our sequence belonged to genus Gemykrogvirus (data not shown). To prove the genus allocation and determine species allocation, we performed a pairwise comparison including one full-length representative of each of the 73 species belonging to family Genomoviridae with the species demarcation cutoff of 78% applied39 (Suppl. Table 2). With 84.3% pairwise identity, the genome was shown to belong to caribou associated gemykrogvirus 1 (strain caribou feces–associated gemycircularvirus, NC_024909.1; Gemykrogvirus carib1) within genus Gemykrogvirus. All other values were below 78%; the bovine associated gemykrogvirus 1 (strain HCBI9.212, NC_024690.1; Gemykrogvirus bovas1) was the next related species with 76.7% identity. Furthermore, a phylogenetic maximum likelihood tree using the same 73 reference sequences as for the pairwise comparison confirmed the allocation with high confidence (Fig. 2). Therefore, the novel virus was named bubaline-associated gemykrogvirus F18_L28 (BuGyKV).
Figure 2.
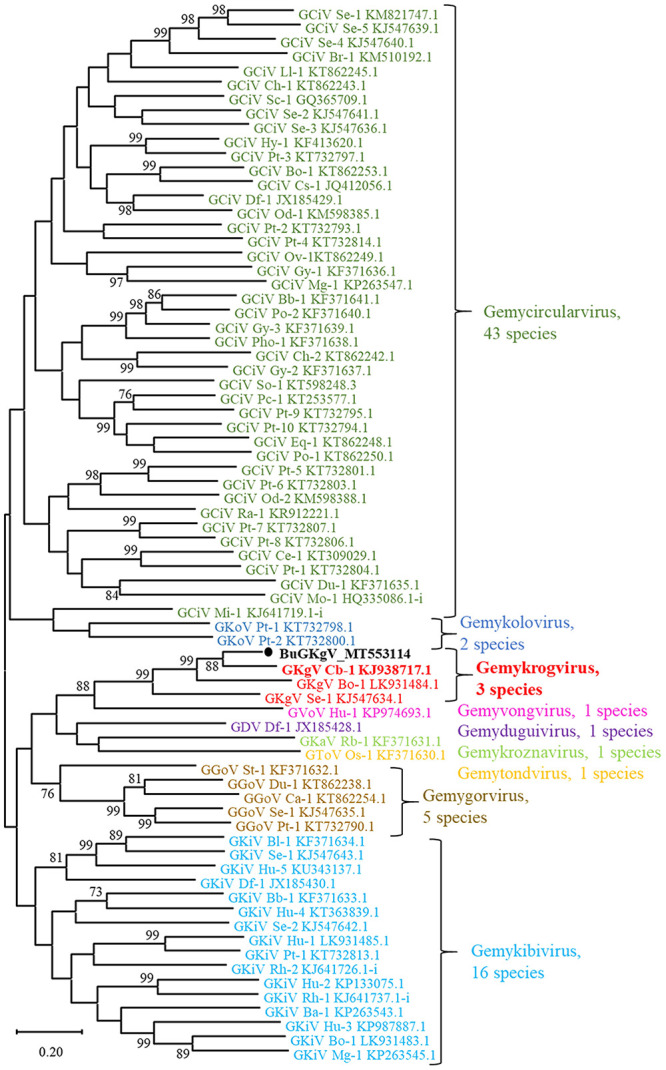
Phylogenetic tree of the family Genomoviridae. A maximum likelihood tree was constructed including full-length nucleotide sequences of representatives of all 73 official species of the family. The tree with the highest likelihood is presented, and bootstrap values > 70 are provided near the respective branching point. The 9 genera are distinguished by different colors. The bubaline-associated gemykrogvirus (BuGyKV) detected in the blood samples of water buffaloes is highlighted with a black dot. The representative of caribou associated gemykrogvirus 1 (GyKV Cb-1; Gemykrogvirus carib1) is highlighted in bold. Accessions of all reference genomes are provided after the genus and species abbreviation. The full names of the virus species and strains used as references as well as the values of the pairwise comparison are provided in Suppl. Table 2.
Re-assembling the reads of all libraries to the full-length BuGyKV revealed an average of 6 times more matching reads than against the caribou feces–associated gemycircularvirus and more positive samples (Suppl. Table 2). Interestingly, 21 reads from a single sample from farm 1 matched BuGyKV; all of the other samples were derived from farm 2 (Suppl. Table 1). By specific BuGyKV rtPCR, 22 of 48 water buffalo samples were found positive, with cycle threshold (Ct) values of 25.9–40.4 (Table 2, Suppl. Table 1). The 15 samples that had been positive by NGS were confirmed by rtPCR and had the lowest Ct values. Another 8 buffaloes were newly detected as carriers of BuGyKV DNA by rtPCR. In the sheep, which had not been tested by NGS, 11 BuGyKV-positive samples were detected, with Ct values of 35.0–39.8. All positive samples originated from farm 2, whereas the single NGS-positive sample from farm 1 could not be confirmed by specific rtPCR. All of the goats tested negative for BuGyKV (Table 2, Suppl. Table 1).
Table 2.
Viral infections detected on 3 farms by molecular (conventional or real-time RT-PCR) or serologic methods (ELISA, VNT).
| Targeted virus | Farm 1 (+ 30 cattle) |
Farm 2 |
Farm 3 (+ 65 cattle) |
||
|---|---|---|---|---|---|
| Water buffalo (n = 17) | Water buffalo (n = 26) | Sheep (n = 19) | Water buffalo (n = 5) | Goats (n = 7) | |
| Genome | |||||
| BuGyKV | ND | 22 | 11 | ND | ND |
| Herpesviruses | ND | 1 BoHV6 | 15 OvHV2; 3 unass. sheep LHV | ND | 1 unass. goat LHV |
| Antibodies | |||||
| BoHV2 | 1 | 1 | ND | ND | ND |
| Ruminant pestiviruses | 1 BVDV | 2 BVDV; 1 BVDV/BDV | 1 BDV | 1 BVDV | 1 BVDV |
| BTV | ND | ND | 1 | 3 | ND |
| SBV | ND | ND | 8 | ND | 2 |
Viruses shared between water buffaloes and small ruminants (BuGyKV, BVDV/BDV), and possibly cattle (BoHV2), are marked in bold.
BDV = border disease virus; BoHV2 = bovine alphaherpesvirus 2; BoHV6 = bovine gammaherpesvirus 6; BTV = bluetongue virus; BuGyKV = bubaline-associated gemykrogvirus; BVDV = bovine viral diarrhea virus; LHV = lymphotropic herpesvirus; ND = not detected; OvHV2 = ovine gammaherpesvirus 2; RT-PCR = reverse-transcription PCR; SBV = Schmallenberg virus; unass. = unassigned; VNT = virus neutralization test.
Detection of various herpesviruses in water buffaloes and small ruminants
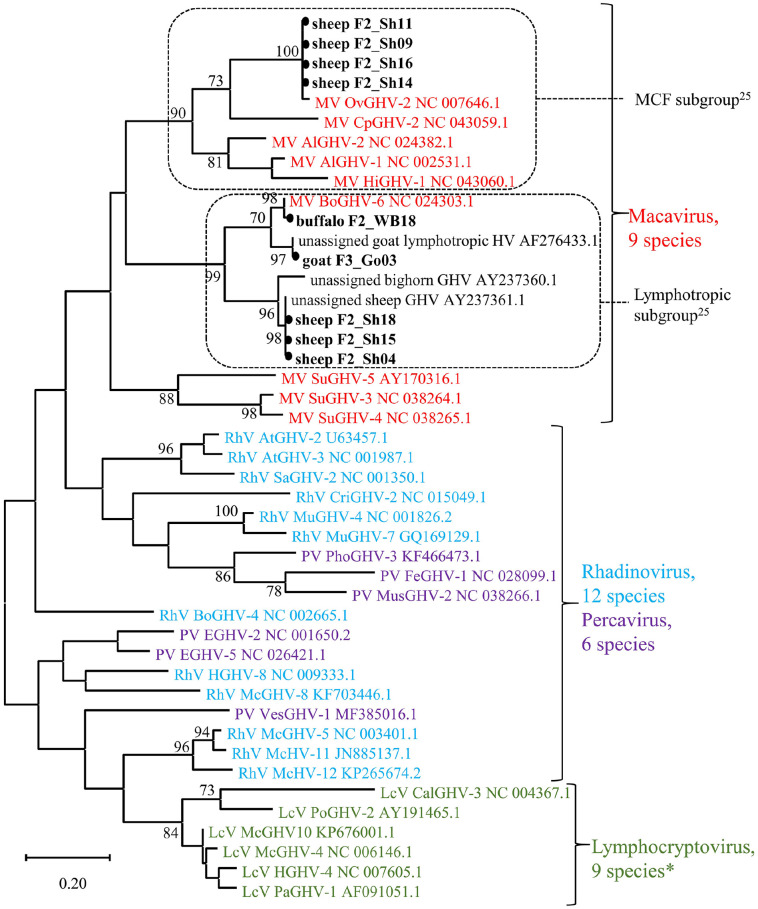
To search for a broad range of a priori unspecified herpesviruses, a panherpesvirus nPCR targeting a highly conserved region of the herpesviral DNA polymerase gene was performed.11,38 Thereafter, gel electrophoresis revealed discernible bands of the expected size (215–235 bp) in 9 of 74 samples. The positive samples originated from 1 water buffalo and 7 sheep from farm 2, and 1 goat from farm 3 (Table 2, Suppl. Table 1). BLAST analysis indicated that all viral sequences belonged to genus Macavirus within the subfamily Gammaherpesvirinae (Fig. 3). Four sheep viruses were identical to each other and nearly identical to OvHV2 within the MCF subgroup (Fig. 3; Suppl. Table 3).24 The other 3 sheep sequences were also identical to each other but clustered within the lymphotropic subgroup of the macaviruses,25 most closely to a yet unassigned sheep gammaherpesvirus (Fig. 3). In slight contrast, the sequence from the water buffalo could clearly be assigned to bovine gammaherpesvirus 6 (BoHV6; formerly bovine lymphotropic herpesvirus) and was closely related, although not identical, to the sequence from the goat (Fig. 3; Suppl. Table 3), which in turn was identical to an unassigned lymphotropic gammaherpesvirus (GHV) reported in goats (Fig. 3).25
Figure 3.
Phylogenetic tree of the subfamily Gammaherpesvirinae. A maximum likelihood tree was constructed including the 9 sequences from our study as well as 1 representative of each species (n = 33) of the 4 official genera, except for 3 species in the genus Lymphocryptovirus (indicated by asterisk [*]), for which the polymerase coding region was not available. In addition, 3 unassigned viruses that by BLAST showed highest relatedness to the sheep and goat viruses were added. The 166–178-nt sequence of the polymerase coding region, flanked by the inner primer pair of the panherpesvirus nested PCR was compared. The tree with the highest likelihood is presented, and bootstrap values > 70 are provided near the respective branching point. The 4 genera are distinguished by different colors. Dashed frames indicate the subgroups within the genus Macavirus.25 The gammaherpesviruses detected in our study are highlighted with a black dot. Accessions of all reference genomes are provided after the genus and species abbreviation. The full name of the reference viruses as well as the values of the pairwise comparison are provided in Suppl. Table 3.
By specific rtPCR, OvHV2 was detected in 15 of 19 sheep samples (Table 2, Suppl. Table 1), simultaneously confirming the 4 OvHV2-positive results by panherpesvirus nPCR. In contrast, all water buffaloes and the goats tested OvHV2-negative. BoHV1, BoHV5, BuHV1, CpHV1, and CpHV2 were not detected in any of the samples tested by either PCR or ELISA (no plasma left from 1 buffalo on farm 1 for BuHV1 and CpHV1 ELISA). Interestingly, 1 water buffalo from farm 1 tested seropositive for BoHV2 antibodies in the ELISA; another buffalo from farm 2 gave a questionable result. In the VNT, seropositivity was confirmed in both cases, with plasma samples displaying neutralizing activity against BoHV2 at titers of 80 and 62, respectively (Table 2, Suppl. Table 1).
Detection of antibodies against ruminant pestiviruses in water buffaloes and small ruminants
Panpestivirus RT-PCR yielded negative results throughout (no sample material left from 1 sheep on farm 2). However, in the biphasic indirect ELISA for BVDV/BDV, 4 water buffaloes from farms 1 and 2, 1 sheep from farm 2, and 1 goat from farm 3 tested seropositive; another buffalo from farm 3 was classified as questionable (Table 2, Suppl. Table 1). In the VNT, each of the 7 samples had antibody titers > 8 against at least 1 virus strain, thus confirming their seropositivity (Table 3). Four buffalo samples and the goat sample had significantly higher titers against BVDV than against BDV (BVDV/BDV ratio ≥ 4). In contrast, the sheep sample showed a significantly higher level of neutralizing antibodies against BDV than against either of the 2 BVDV strains (BVDV/BDV ratio ≤ 0.25). One buffalo from farm 2 showed rather high neutralizing antibody titers against both BVDV (BVDV1h titer 190) and BDV (titer 95), providing a nonsignificant titer difference (0.25 < BVDV/BDV ratio < 4). Regarding the 2 BVDV strains, all of the samples displayed higher titers against BVDV1h than against BVDV1a (Table 3).
Table 3.
Virus neutralization test (VNT) titers and result interpretation of samples that exceeded the negative cutoff value of 20% in the pestivirus ELISA.17
| Farm | Species | ELISA (%) | VNT | Highest BVDV/BDV ratio* | Interpretation* | ||
|---|---|---|---|---|---|---|---|
| BVDV1a | BVDV1h | BDV Swiss a | |||||
| 1 | Buffalo | 41 | 14 | 80 | 8 | 10 | BVDV |
| 2 | Buffalo | 42 | 67 | 190 | 95 | 2 | Ambiguous |
| Buffalo | 74 | 95 | 538 | 80 | 6.37 | BVDV | |
| Buffalo | 57 | 190 | 1,076 | 160 | 6.37 | BVDV | |
| Sheep | 56 | 8 | 10 | 190 | 0.05 | BDV | |
| 3 | Buffalo | 21 | 40 | 48 | ≤ 7 | ≥ 6.86 | BVDV |
| Goat | 50 | 57 | 320 | 28 | 11.43 | BVDV | |
BDV = border disease virus; BVDV = bovine viral diarrhea virus.
The titer ratios (BVDV/BDV) of the higher BVDV titer (1 h) and BDV were calculated for each sample. Ratios ≥ 4 indicated seropositivity against BVDV (1 h), ratios ≤ 0.25 indicated antibody specificity for BDV. For ratios in between (0.25 < ratio < 4), the source of infection could not be determined.
Detection of BTV antibodies in water buffaloes and sheep
In the BTV competitive ELISA, 3 buffaloes on farm 3 and 1 sheep on farm 2 tested seropositive for BTV. In contrast, all of the water buffalo samples (no plasma left from 2 buffaloes on farm 1) had a seronegative result in the SBV ELISA. However, 8 sheep on farm 2 and 2 goats on farm 3 tested seropositive. All plasma samples tested negative in the BLV ELISA (Table 2, Suppl. Table 1).
Discussion
Modern NGS and conventional methods were shown to complement each other well in describing virus circulation among different animal species. However, our sample size is rather small and not representative of the Swiss water buffalo population; hence, results must be interpreted with caution and more extensive studies are needed to corroborate our findings.
The newly detected BuGyKV belongs to the group of circular Rep-encoding single-stranded (CRESS) DNA viruses. CRESS DNA viruses appear to be ubiquitous and, by metagenomic NGS analyses, have been found in a wide variety of blood and fecal samples from various animal species and humans, insects, sewage, in marine ecosystems, and even in air samples.13,19,22,30,41,44 With single exceptions, such as known porcine circoviruses and specific viruses infecting bacteria, plants, and fungi, neither the host nor the clinical significance of most CRESS DNA viruses are known, and they rarely grow in cell cultures.20 Remarkably, only animals from farm 2 were found positive for BuGyKV; a single sample from farm 1 positive by NGS, but not by rtPCR, may be attributed to low level contamination or index hopping during NGS.29
Detection of BuGyKV in water buffaloes and co-housed sheep of one farm indicates possible interspecies transmission. The frequency of detection of this novel virus in blood samples from buffaloes and sheep suggests that it was caught at the stage of simultaneous active viremia in both animal species. As herd-specific contamination during sample and library preparation in the lab can be excluded because of the randomized processing, the GyKV sequences are highly likely to originate from farm 2. Sheep may have been the original host of BuGyKV because sheep were kept solely on farm 2 but not on farms 1 and 3. There is also the remote possibility that the samples were contaminated with a farm-specific environmental virus during sampling. Serologic surveys for BuGyKV in water buffaloes, sheep, and other animal species would be highly desirable to clarify this issue.
After a national eradication program was launched in 2008, BVDV was successfully brought to the brink of extinction in Swiss cattle by 2013.33 Moreover, BDV has not been reported to circulate among water buffaloes and is thought to have a low prevalence in the Swiss sheep population.6 Hence, it was not unexpected that all of the animals tested negative for persistent pestivirus viremia by panpestivirus RT-PCR. However, seropositive animals were detected by ELISA and confirmed by VNT. Considering the ban of anti-pestivirus vaccines in Switzerland, these results strongly suggest that the water buffaloes on the 3 farms had been transiently infected with BVDV, in one case possibly also with BDV. In all BVDV seropositive animals, the highest antibody titers were measured against BVDV1h, which was the most frequent subgroup in Switzerland prior to eradication.36 This observation argues for a bovine source of transmission. Interestingly, 1 sheep on farm 2 was BDV seropositive. On the same farm, we also detected a water buffalo with high antibody titers against both BVDV and BDV, which could suggest a past infection with both pestiviruses. Although the low overall pestivirus seroprevalence on the 3 farms argued against recent presence of either BVDV or BDV persistently infected animals, these serologic observations strongly suggest that water buffaloes may be part of the epidemiology of ruminant pestiviruses.
To our knowledge, the 2 cases described here are the first tentative evidence for water buffaloes being susceptible to BoHV2 infection leading to the production of neutralizing antibodies. Possibly, BoHV2 can be shared between cattle and water buffaloes. BoHV2 is known to circulate in Swiss cattle,12,37 and unpublished data using ELISA indicate a seroprevalence of 28.5% (Bachofen C., pers. comm., 2020 Sept 10). Infections most often seem to take a subclinical course.37 Indeed, no clinical signs of herpes mammillitis were reported from the 2 farms on which the infected water buffaloes were housed. However, the potential role of water buffaloes in transmitting BoHV2 remains to be elucidated.
Various PCR tests confirmed the presence of at least 4 different gammaherpesviruses in water buffaloes, sheep, and a goat. However, none of these viruses had (yet) been transferred to the co-housed animal species on the same farm. BoHV6 infection has not been reported previously in Swiss water buffaloes, to our knowledge. Based on the short sequence from the panherpesvirus nPCR alone, it is difficult to conclude if this virus is identical to BoHV6 viruses detected in cattle, and hence the result of an interspecies transmission, or a water buffalo–specific variant. BoHV6 has been detected in cattle as well as water buffaloes suffering from various diseases, such as lymphoproliferative disease, abortion, and postpartum metritis.3,9,14,31 However, neither for this bovine lymphotropic herpesvirus, nor for the unclassified lymphotropic gammaherpesvirus found in small ruminants,24,25 has a causative association to a disease been firmly established, and was not seen in our study either. Interestingly, even though BoHV6 was detected by panherpesvirus nPCR in one water buffalo, no NGS reads matched this virus. We attribute this lack to the degradation of non-encapsidated nucleic acids during sample preparation. Latent herpesviruses may indeed be missed when enriching for viral particles. The absence of NGS reads also illustrates the seemingly trivial fact that sample-taking and processing may influence the outcome of downstream analyses.
Arboviruses seemed to circulate either only in water buffaloes or the co-housed small ruminants of the same farm, but not in both. Before sample taking in 2013, the only BTV outbreak in Switzerland occurred from 2007 to 2010 and was caused by BTV serotype 8.15 A vaccine against BTV8 was used to overcome this outbreak. Yet, none of the 3 BTV seropositive water buffaloes had a confirmed history of vaccination against BTV. Although maternal antibodies could not be excluded for 2 of the seropositive individuals (4-mo-old twins), the third seropositive buffalo (10 mo old), probably was too old for significant amounts of maternal antibodies to be present.23 Considering the prior serologic evidence that water buffaloes are susceptible to BTV infection,26 a natural infection with BTV seems most likely in this third water buffalo. Unfortunately, the volume of plasma was not sufficient for further serotyping of the BTV seropositive plasma samples. According to the farmers, none of the sampled animals had suffered from or was diagnosed with bluetongue disease. However, an inapparent BTV infection or a false-positive result cannot be fully excluded, and the potential role of water buffaloes in BTV epidemiology deserves further attention.
For SBV, only small ruminants, but no water buffaloes, tested seropositive. This fact may be attributed to differences in susceptibility and/or risk of Culicoides bites and susceptibility to the virus between the different species. In fact, lower seroprevalences in water buffaloes compared to other species and possible resistance of water buffaloes to SBV have been reported previously.2,43
Serologic results confirmed the absence of viruses that have been eradicated in Switzerland, namely BoHV1, the agent of infectious bovine rhinotracheitis and infectious pustular vulvovaginitis, and BLV, the agent of enzootic bovine leukosis. Interestingly, OvHV2, a very important MCF agent, was detected in co-housed sheep but apparently not (yet) transmitted to susceptible water buffaloes. The absence of such otherwise known and fatal interspecies transmission reminds us to remain cautious about uncontrolled co-housing of different animal species and ignoring seemingly harmless viral infections.
Supplemental Material
Supplemental material, sj-pdf-2-vdi-10.1177_10406387211027131 for Viral infections shared between water buffaloes and small ruminants in Switzerland by Julia Lechmann, Mathias Ackermann, Vanessa Kaiser and Claudia Bachofen in Journal of Veterinary Diagnostic Investigation
Supplemental material, sj-xlsx-1-vdi-10.1177_10406387211027131 for Viral infections shared between water buffaloes and small ruminants in Switzerland by Julia Lechmann, Mathias Ackermann, Vanessa Kaiser and Claudia Bachofen in Journal of Veterinary Diagnostic Investigation
Acknowledgments
We thank Eva-Maria Christen for organizing sample collection and sample preparation, Libby Nebel for providing material and support for pestivirus serology, and the Functional Genomics Center of the University of Zurich (FGCZ) for their support concerning NGS.
Footnotes
Declaration of conflicting interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: Our work was funded by the Federal Food Safety and Veterinary Office (FSVO; project 1.13.08).
ORCID iDs: Julia Lechmann  https://orcid.org/0000-0002-7389-846X
https://orcid.org/0000-0002-7389-846X
Mathias Ackermann  https://orcid.org/0000-0001-5268-1467
https://orcid.org/0000-0001-5268-1467
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Julia Lechmann, Institute of Virology, Vetsuisse Faculty, University of Zurich, Zurich, Switzerland.
Mathias Ackermann, Institute of Virology, Vetsuisse Faculty, University of Zurich, Zurich, Switzerland.
Vanessa Kaiser, Institute of Virology and Immunology, Vetsuisse Faculty, University of Bern, Bern, Switzerland; Current address: MSD Animal Health, Lucerne, Switzerland.
Claudia Bachofen, Institute of Virology, Vetsuisse Faculty, University of Zurich, Zurich, Switzerland.
References
- 1.Abril C, et al. Both viral and host factors contribute to neurovirulence of bovine herpesviruses 1 and 5 in interferon receptor-deficient mice. J Virol 2004;78:3644–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azkur AK, et al. Antibodies to Schmallenberg virus in domestic livestock in Turkey. Trop Anim Health Prod 2013; 45:1825–1828. [DOI] [PubMed] [Google Scholar]
- 3.Banks M, et al. Bovine lymphotropic herpesvirus and non-responsive post-partum metritis in dairy herds in the UK. Vet J 2008;176:248–250. [DOI] [PubMed] [Google Scholar]
- 4.Bertolotti L, et al. Characterization of caprine herpesvirus 1 (CpHV1) glycoprotein E and glycoprotein I ectodomains expressed in mammalian cells. Vet Microbiol 2013;164:222–228. [DOI] [PubMed] [Google Scholar]
- 5.Borghese A.Buffalo livestock and products in Europe. Buffalo Bull 2013;32:50–74. [Google Scholar]
- 6.Braun U, et al. Investigation of border disease and bovine virus diarrhoea in sheep from 76 mixed cattle and sheep farms in eastern Switzerland. Schweiz Arch Tierheilkd 2013;155:293–298. [DOI] [PubMed] [Google Scholar]
- 7.Canal CW, et al. Detection of antibodies to bovine viral diarrhoea virus (BVDV) and characterization of genomes of BVDV from Brazil. Vet Microbiol 1998;63:85–97. [DOI] [PubMed] [Google Scholar]
- 8.Cunha CW, et al. Development of a multiplex real-time PCR for detection and differentiation of malignant catarrhal fever viruses in clinical samples. J Clin Microbiol 2009;47:2586–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Oliveira CHS, et al. Bovine herpesvirus 6 in buffaloes (Bubalus bubalis) from the Amazon region, Brazil. Trop Anim Health Prod 2015;47:465–468. [DOI] [PubMed] [Google Scholar]
- 10.Dettwiler M, et al. A possible case of caprine-associated malignant catarrhal fever in a domestic water buffalo (Bubalus bubalis) in Switzerland. BMC Vet Res 2011;7:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehlers B, et al. Detection of new DNA polymerase genes of known and potentially novel herpesviruses by PCR with degenerate and deoxyinosine-substituted primers. Virus Genes 1999;18:211–220. [DOI] [PubMed] [Google Scholar]
- 12.Engels M, et al. Ein Virus sucht seine Krankheit: seroepizootologische Untersuchung über das Vorkommen der Bovinen Herpes Mammillitis in der Schweiz [Virus seeks its disease: a seroepizootiological study of the presence of bovine herpes mammillitis in Switzerland]. Schweiz Arch Tierheilkd 1979; 121:565–576.German. [PubMed] [Google Scholar]
- 13.Fahsbender E, et al. Discovery of a novel circular DNA virus in the Forbes sea star, Asterias forbesi. Arch Virol 2015;160: 2349–2351. [DOI] [PubMed] [Google Scholar]
- 14.Gagnon CA, et al. Quebec: detection of bovine lymphotropic herpesvirus DNA in tissues of a bovine aborted fetus. Can Vet J 2010;51:1021–1022. [PMC free article] [PubMed] [Google Scholar]
- 15.Hofmann M, et al. Blauzungenkrankheit erreicht die Schweiz [Bluetongue disease reaches Switzerland]. Schweiz Arch Tierheilkd 2008;150:49–56. German. [DOI] [PubMed] [Google Scholar]
- 16.Hüssy D, et al. Quantitative fluorogenic PCR assay for measuring ovine herpesvirus 2 replication in sheep. Clin Diagn Lab Immunol 2001;8:123–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaiser V, et al. Influence of border disease virus (BDV) on serological surveillance within the bovine virus diarrhea (BVD) eradication program in Switzerland. BMC Vet Res 2017;13:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitching RP, et al. Clinical variation in foot and mouth disease: sheep and goats. Rev Sci Tech 2002;21:505–512. [DOI] [PubMed] [Google Scholar]
- 19.Kraberger S, et al. Characterisation of a diverse range of circular replication-associated protein encoding DNA viruses recovered from a sewage treatment oxidation pond. Infect Genet Evol 2015;31:73–86. [DOI] [PubMed] [Google Scholar]
- 20.Krupovic M, et al. Cressdnaviricota: a virus phylum unifying seven families of Rep-encoding viruses with single-stranded, circular DNA genomes. J Virol 2020;94:e00582-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar S, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 2018;35: 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamberto I, et al. Mycovirus-like DNA virus sequences from cattle serum and human brain and serum samples from multiple sclerosis patients. Genome Announc 2014;2:e00848-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leemans J, et al. Interference of colostral antibodies with response to a bluetongue serotype 8 inactivated vaccine in lambs born from hyperimmune ewes. Vaccine 2013;31:1975–1980. [DOI] [PubMed] [Google Scholar]
- 24.Li H, et al. Recognition of another member of the malignant catarrhal fever virus group: an endemic gammaherpesvirus in domestic goats. J Gen Virol 2001;82:227–232. [DOI] [PubMed] [Google Scholar]
- 25.Li H, et al. A novel subgroup of rhadinoviruses in ruminants. J Gen Virol 2005;86:3021–3026. [DOI] [PubMed] [Google Scholar]
- 26.Maan S, et al. A comprehensive study on seroprevalence of bluetongue virus in Haryana state of India. Vet World 2017;10: 1464–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muhire BM, et al. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One 2014;9:e108277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nogarol C, et al. Expression and antigenic characterization of bubaline herpesvirus 1 (BuHV1) glycoprotein E and its potential application in the epidemiology and control of alphaherpesvirus infections in Mediterranean water buffalo. J Virol Methods 2014;207:16–21. [DOI] [PubMed] [Google Scholar]
- 29.Ros-Freixedes R, et al. Impact of index hopping and bias towards the reference allele on accuracy of genotype calls from low-coverage sequencing. Genet Sel Evol 2018;50:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosario K, et al. Diverse circular ssDNA viruses discovered in dragonflies (Odonata: Epiprocta). J Gen Virol 2012;93: 2668–2681. [DOI] [PubMed] [Google Scholar]
- 31.Rovnak J, et al. Detection of a novel bovine lymphotropic herpesvirus. J Virol 1998;72:4237–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schorer M, et al. Überwachung des Schmallenberg-Virus in der Schweiz [Monitoring of Schmallenberg virus in Switzerland]. Schweiz Arch Tierheilkd 2012;154:543–547. German. [DOI] [PubMed] [Google Scholar]
- 33.Schwermer H, et al. Data management systems for the bovine viral diarrhoea eradication programme in Switzerland. Rev Sci Tech 2013;32:741–750. [DOI] [PubMed] [Google Scholar]
- 34.Semenza JC, et al. Vector-borne diseases and climate change: a European perspective. FEMS Microbiol Lett 2018;365: fnx244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stahel AB, et al. Two different macaviruses, ovine herpesvirus-2 and caprine herpesvirus-2, behave differently in water buffaloes than in cattle or in their respective reservoir species. PLoS One 2013;8:e83695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stalder H, et al. Traces of history conserved over 600 years in the geographic distribution of genetic variants of an RNA virus: bovine viral diarrhea virus in Switzerland. PLoS One 2018;13:e0207604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Syring C, et al. Bovine herpes mammillitis in three dairy cows. Tierarztl Prax Ausg G Grosstiere Nutztiere 2010;38:317–320. [Google Scholar]
- 38.VanDevanter DR, et al. Detection and analysis of diverse herpesviral species by consensus primer PCR. J Clin Microbiol 1996;34:1666–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varsani A, Krupovic M.Sequence-based taxonomic framework for the classification of uncultured single-stranded DNA viruses of the family Genomoviridae. Virus Evol 2017;3:vew037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vilcek S.Vývoj PCR testov na detekciu BHV-1, BRSV a pestivírusov [Development of PCR tests for the detection of bovine herpesvirus-1, bovine respiratory syncytial viruses and pestiviruses]. Vet Med (Praha) 1994;39:687–700. Slovak. [PubMed] [Google Scholar]
- 41.Whon TW, et al. Metagenomic characterization of airborne viral DNA diversity in the near-surface atmosphere. J Virol 2012;86:8221–8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.World Organisation for Animal Health (OIE). Infectious bovine rhinotracheitis/infectious pustular vulvovaginitis. In: Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. 8th ed.OIE, 2018:1139–1157. [Google Scholar]
- 43.Zhai SL, et al. Preliminary serological evidence for Schmallenberg virus infection in China. Trop Anim Health Prod 2018;50:449–453. [DOI] [PubMed] [Google Scholar]
- 44.Zhao L, et al. Eukaryotic circular Rep-encoding single-stranded DNA (CRESS DNA) viruses: ubiquitous viruses with small genomes and a diverse host range. Adv Virus Res 2019;103:71–133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-2-vdi-10.1177_10406387211027131 for Viral infections shared between water buffaloes and small ruminants in Switzerland by Julia Lechmann, Mathias Ackermann, Vanessa Kaiser and Claudia Bachofen in Journal of Veterinary Diagnostic Investigation
Supplemental material, sj-xlsx-1-vdi-10.1177_10406387211027131 for Viral infections shared between water buffaloes and small ruminants in Switzerland by Julia Lechmann, Mathias Ackermann, Vanessa Kaiser and Claudia Bachofen in Journal of Veterinary Diagnostic Investigation