ABSTRACT
The human YTH domain family (YTHDF) proteins are RNA-binding proteins that recognize N6-methyladenosine (m6A), facilitating various biological processes via m6A RNA modification. How these molecules associate with non–small-cell lung cancer (NSCLC) molecular mechanisms remain unclear. The protein expression levels of YTHDF1 and YTHDF2 in 603 cases of resected NSCLC were evaluated using immunohistochemistry. We analyzed the associations of these attributes with patient characteristics and survival. We also assessed four subsets of lymphocytes (PD-1+, CD8+, Foxp3+, and CD45RO+) as tumor-infiltrating lymphocytes (TILs) in the tumor nest and in the surrounding stroma separately. In addition, we investigated differentially expressed genes and the expression of PD-L1 in YTHDF1– and YTHDF2-deprived lung cancer cells. The expressions of both YTHDF1 and YTHDF2 were less in the advanced-stage tumors than in the early-stage tumors. The expressions of both YTHDF1 and YTHDF2 were also independent favorable prognostic factors for recurrence-free survival (HR, 0.745; 95% CI, 0.562–0.984 for YTHDF1; HR, 0.683; 95% CI, 0.503–0.928 for YTHDF2). The TIL densities of almost all four lymphocyte subsets in the stroma were significantly higher in the tumors with high YTHDF1 and YTHDF2 expression. In vitro, YTHDF1 and YTHDF2 knockdown in cells upregulated tumor PD-L1 expression and altered multiple immune-related genes. High expressions of both YTHDF1 and YTHDF2 are associated with a favorable prognostic outcome of NSCLC patients, a greater amount of TILs, and downregulation of PD-L1. YTHDF1 and YTHDF2 could be novel prognostic and druggable targets related to the tumor-immune microenvironment in lung cancers.
KEYWORDS: Lung cancer, ythdf1, ythdf2, pd-l1, tumor-immune microenvironment, m6A
Introduction
N6-methyladenosine (m6A) is the most prevalent internal modification in messenger RNA.1 In recent years, the significance of m6A in the context of various physiological functions such as obesity,2 synaptic signaling,3 sperm development,4 stem cell differentiation,5 circadian rhythms,6 and immunoregulation7 has been clarified, and the relationship between m6A and the pathogenesis of various diseases, including malignant disease, has been an enthusiastic topic of biomedical investigations.8,9 In cells, m6A is regulated by methyltransferase complex (writer) and demethylase (eraser), and m6A is recognized by RNA-binding proteins (reader). The fate of the subsequent m6A-modified mRNA is determined by the reader protein, which binds to the target mRNA.1,10
The human YTH domain family (YTHDF) proteins are RNA-binding proteins that directly recognize m6A. YTHDF1 actively promotes protein synthesis by interacting with initiation factors and ribosomes to facilitate translation initiation.11 YTHDF2 recognizes m6A and reduces the stability of target mRNA transcripts.12,13 YTHDF1 and YTHDF2 share 50% of their targets, and, through translation promotion or the alteration of mRNA stability, the m6A-modified target recognized by YTHDF1 or YTHDF2 regulates protein synthesis.11 To date, YTHDF1 is known to have an oncogenic role by promoting the translation of oncogenes among several solid cancers.14–17 On the contrary, studies have also reported that YTHDF1 has an antitumor effect through its promotion of translation of tumor suppressor gene in ocular melanoma.18 Similarly, YTHDF2 has an oncogenic role via the regulation of miR-493-3p in prostate cancer and 6-phosphogluconate dehydrogenase in non–small-cell lung cancer (NSCLC).19,20 On the other hand, YTHDF2 also has an antitumor effect in leukemic cells via the attenuation of the stability of MYC and CEBPA transcripts.21 Given these inconsistent findings, we can say it is still controversial as to how these m6A reader proteins, YTHDF1 and YTHDF2, affect oncogenic or anti-oncogenic function in defined clinical settings. They may work differently depending on whether the target genes are oncogenic or tumor suppressive. In lung cancer, there are several pathways involved in cancer development that vary based on the etiology, histopathological entity, and somatic mutational spectrum, it remains largely unknown how YTHDF1 and YTHDF2 are committed in these heterogeneous carcinogeneses of lung cancer.
Over the past decades, the NSCLC treatment strategy has dramatically changed, and immunotherapy has become one of the most common methods of treatment. The PD-1/PD-L1 axis has been deemed the main target of immunotherapy; however, the tumor-immune microenvironment, which is composed of various immune cells,22,23 is also now an area of focus and has been studied intensively as a secondary target of immunotherapy.24–27 Yang et al. reported that FTO, one of the m6A demethylases, was associated with several cell-intrinsic oncogenes.28 The combination of FTO inhibitors overcame the resistance of PD-L1 immunotherapy in melanoma. Thus, the association of m6A-related enzymes with tumor-immune systems might be important, and more evidence is still needed to enable comprehensive understanding of this association.
In this study, we first explored the clinical significance of the expressions of YTHDF1 and YTHDF2 using a relatively large number of resected NSCLC specimens, along with the association between these molecules and the tumor-immune microenvironment. Second, we investigated the effect of forced downregulation of YTHDF1 and YTHDF2 in lung cancer cell lines in terms of the effect on the alteration of PD-L1 and immunologic gene expression.
Materials and methods
Specimen preparation
Using immunohistochemistry, we evaluated the protein expression levels of both YTHDF1 and YTHDF2 among 603 resected NSCLCs, including 392 adenocarcinomas and 170 squamous cell carcinomas. The patients enrolled in this study were admitted to Hamamatsu University School of Medicine and Seirei-Mikatahara General Hospital between January 1990 and April 2014. This study was approved by the ethics committees of Hamamatsu University School of Medicine and Seirei-Mikatahara General Hospital and was carried out in accordance with approved guidelines. Because this study was based on reviews of previously acquired clinical records and archived data, the need for written informed consent was waived. All analyses were conducted in compliance with the ethical standards of the Helsinki Declaration.
All specimens were formalin fixed, paraffin embedded, and punched out from distinct tumor areas using 3-mm-diameter cylinders (Azumaya, Tokyo, Japan) by senior pathologists. The tissue cores were aligned on tissue microarrays (TMAs) as previously described.29–31 In cases with insufficient number of tumor cells in the core, another core with a higher number of tumor cells was chosen. We retrospectively collected clinicopathological data, such as age, sex, smoking status, adjuvant chemotherapy, and patient prognosis status, from the patients’ medical records. Tumors were classified into pathological stages I–III according to 8th edition of the TNM classification and histologically classified by three senior pathologists (KY, Y Inoue, and HS) based on the World Health Organization classification and the 2015 International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society ERS adenocarcinoma subclassification.32–34 Before conducting the TMA analysis, we confirmed the presence of a sufficient number of tumor cells in the TMA cores using hematoxylin–eosin staining of tissue sections.
Immunohistochemical analysis
TMA sections were assessed immunohistochemically using a previously published method.35 Paraffin sections were incubated with primary antibody, followed by incubation with Histofine Simple Stain MAX PO (Nichirei, Tokyo, Japan). The visualization of the antigen–antibody complex was performed using 3,3′-diaminobenzidine tetrahydrochloride. We used the following commercially available antibodies in recommended titrations: anti-YTHDF1 (1:100, 17479-1-AP, Proteintech, Rosemont, IL, USA), YTHDF2 (1:400, ab170118, Abcam, Cambridge, UK), PD-1 (1:50, EH33, Cell Signaling Technology [CST], Danvers, MA, USA), CD8 (1:200, C8/144B, Nichirei, Tokyo, Japan), Foxp3 (1:100, 22510, Abcam), CD45RO (1:600, UCHL1, CST), PD-L1 (1:100, E1L3N; CST), epidermal growth factor receptor (EGFR) E746-A750 deletion specific (1:100, D6B6, CST), and EGFR L858R mutant specific (1:100, 43B2, CST). Two observers (KT and KY) independently assessed the protein expressions levels in the tumor areas based on the H-score, which was derived from the intensity (0, absent; 1, weak; 2, moderate; and 3, strong; representative images are shown in Figure 1a). The area percentages of cells (0%–100%) were multiplied by the intensity values to obtain calculated values from 0 to 300. We also assessed the four subsets of tumor-infiltrating lymphocytes (TILs), such as PD-1+ immune cells, CD8+ cytotoxic T cells, Foxp3+ regulatory T cells, and CD45RO+ memory T cells, within the tumor nest and surrounding stroma separately, as previously described.36,37 TILs were counted under high-power fields (×200). Representative images are shown in Figure 1b. We calculated the average TIL number in the three areas and retrospectively explored their associations with patient characteristics and patient survival. The high expression levels of YTHDF1 or YTHDF2 were defined as cases with scores greater than 118 of the H-score for YTHDF1 and greater than 117 of the H-score for YTHDF2. We determined the scores using the minimum P-value method for overall survival (OS) based on each expression. PD-L1 positivity and EGFR mutant statuses were analyzed using a previous protocol.29,38,39
Figure 1.
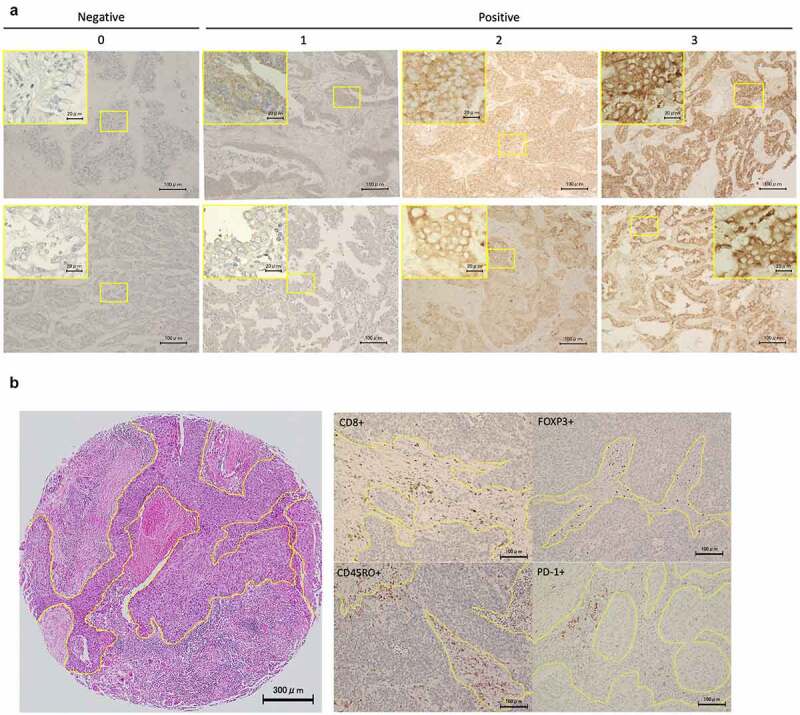
Representative images of YTHDF1, YTHDF2, and TIL expression in immunohistochemistry
(a) Representative images of immunohistochemistry of YTHDF1 and YTHDF2 (×20 magnification, inset ×60 magnification). The yellow square indicates the position of the inset. Staining intensity was categorized as 0 (absent), 1 (weak), 2 (moderate), or 3 (strong). (b) Representative images of tumor-infiltrating lymphocytes such as FOXP3+, PD-1+, CD8+, and CD45RO+ immune cells (×20 magnification).
Online data analysis
We used two online cohorts to validate the mRNA expression and survival in patients with lung cancer: The Cancer Genome Atlas (TCGA) data set and the Kaplan–Maier plotter. We collected the mRNA expression data for lung adenocarcinoma (TCGA ID: LUAD) and lung squamous cell carcinoma (TCGA ID: LUSC) from the TCGA data portal in the form of RNA-seq by expectation–maximization. Clinical information was downloaded from the cBioPortal (http://www.cbioportal.org/) on March 15, 2019.40 The high level of mRNA expression of YTHDF1 and YTHDF2 was defined by the value derived from the minimum P-value method for OS. Survival results were produced from the Kaplan–Meier Plotter (lung; http://kmplot.com/analysis/index.php?p=service&cancer=lung).41,42 These data were downloaded on October 12, 2019. We defined our cutoffs of YTHDF1 and YTHDF2 by the median value. We performed gene set enrichment analysis (GSEA) to examine the regulated gene set stratified by YTHDF1 and YTHDF2 expressions using TCGA data (http:/software.broadinstitute. org/gsea/omdex.jsp). Normalized enrichment scores >1.5 and a false discover rate (FDR) <0.05 were used as cutoffs.
Cell line and transient knockdown with siRNA
The human lung cancer cell lines H1299 and A549 were obtained from Health Science Research Resources Bank (Osaka, Japan) or the American Type Culture Collection (Manassas, VA, USA). We purchased PC9 lung cancer cells from RIKEN BRC (Tsukuba, Japan). All lung cancer cell lines were cultured in RPMI1640 medium (R8758, Thermo Fisher Scientific, Waltham, MA, USA) containing 10% (vol/vol) fetal bovine serum (FBS) and 100 IU/mL penicillin G and 100 mg/mL streptomycin. Cells were maintained in a 5% CO2 and 95% air incubator at 37°C. Silencer Select Pre-designed siRNA for YTHDF1 (s29743, s29744, Invitrogen, Carlsbad, CA, USA) and YTHDF2 (s28147, s28148, Invitrogen) and Silencer Select Negative control (4390843, Invitrogen) were purchased for a transient knockdown. Lung cancer cells were cultured for 24 hours before transfection and transfected with final concentrations of 15 nM of siRNAs using Opti-MEM (31985070, Gibco, Waltham, MA, USA) and Lipofectamine 2000 (11668019, Thermo Fisher). Forty-eight hours after transfection, the cells were used for further assays.
Cell viability assay
Cells were seeded in 96-well plates at 3000 cells per well after 48 hours of knockdown. Cell proliferation was monitored using the Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan) according to the manufacturer’s protocol. After incubation, the cells were further incubated with 10% CCK-8 for 1 hour, and we assessed the absorbance at 450 nm in each well by spectrophotometry (Synergy HT, BioTek) every 24 hours.
Transwell migration assay
Cell migration was evaluated using 24-well plates with cell culture inserts (353097, Falcon) containing a filter with 8 μm-diameter pores. Briefly, after serum starvation for 24 h with 0.1% FBS-containing RPMI1640 medium, 1 × 105 cells resuspended in 500 μL of RPMI1640 medium (Gibco, USA) were seeded into the upper chamber, after which the RPMI1640 medium with 10% FBS was placed in the lower compartment of the chamber. After incubation for 16 h, the upper surface of the membrane was wiped with a cotton-tipped applicator whereas the migrating cells on the lower surface were fixed with cold methanol and stained with 0.5% crystal violet. Migrating cells were counted automatically in three random microscopic fields using the Hybrid Cell Count function of the BZ-II Analyzer (Keyence, Osaka, Japan).
Cell cycle analysis
Cell Cycle Assay Solution Blue (C549, Dojindo) was used according to the manufacturer’s instructions to analyze cell cycle. Briefly, treated cells were synchronized at the G1 phase by serum starvation with 0.1% FBS-containing medium for 48 h. After 24–36 h after release from serum starvation, the treated cells were collected, washed with phosphate-buffered saline (PBS), and incubated with 5 μL cell cycle assay solution for 15 min at 37°C. Thereafter, DNA content was determined by measuring staining intensity using a Gallios flow cytometer (Beckman Coulter, Miami, FL, USA). Results were analyzed using the FlowJo software (Becton, Dickinson and Company, Franklin Lakes, NJ, USA).
Apoptosis assay
The annexin V-FITC apoptosis detection kit (15342–54, Nacalai, Kyoto, Japan) was used to detect apoptosis by measuring annexin V and propidium iodide-positive cells, according to the manufacturer’s instructions. Briefly, cells were incubated for 96 h after siRNA transfection. The treated cells were collected, washed with PBS, and incubated with 5 μL of annexin V-FITC solution and 5 μL of propidium iodide solution for 15 min at room temperature. Thereafter, the number of apoptotic cells was determined using a Gallios flow cytometer with the FlowJo software.
Western blotting
Total protein lysates were extracted from whole-cell lines in phosphate-buffered saline. The Pierce BCA Protein Assay Kit (Cat#23225, Thermo Fisher) was used to determine concentration. All proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and the proteins in the gel were then transferred to polyvinylidene difluoride blotting membrane (A29532146, P 0.45 GE Healthcare Life Science, Chicago, IL, USA). Trans-Blot Turbo Cassette (Bio-Rad, Hercules, CA, USA) was used for blotting. For blocking, 5% skimmed milk was used. Primary antibodies including anti-YTHDF1 (1:1000 dilution, 17479-1-AP, Proteintech), YTHDF2 (1:1000 dilution, ab170118, Abcam), PD-L1 (1:1000 dilution, A1935; ABclonal, Tokyo, Japan), and GAPDH (1:1000 dilution, Ab8245; Abcam) were incubated overnight at 4°C. Secondary antibodies for rabbit (1:20,000 dilution, NA9340; GE Healthcare Life Science) or mouse (1:20,000 dilution, NA9310; GE Healthcare Life Science) were incubated at room temperature for 1 hour. To visualize the band, we used enhanced chemiluminescence (Pierce ECL Plus Substrate, Thermo Scientific). The photo was detected by a chemi-doc (Bio-Rad). Signal intensity was quantified using Image Labo Software (Bio-Rad).
Quantitative real-time polymerase chain reaction (qRT-PCR) analysis
Cells were incubated after 48 hours and transfected by siRNA. We subsequently extracted total RNA using RNeasy Plus Mini Kit (#74136, Qiagen, Hilden, Germany) according to the manufacturer’s instructions. We calculated the total RNA concentration using Nanodrop (NanoDrop1000, Thermo Scientific). We synthesized cDNA from 2 μg of total RNA with ReverTra Ace qPCR RT Master Mix (FSQ-201, TOYOBO, Osaka, Japan) according to the manufacturer’s instructions. qPCR were performed in triplicate on a StepOne Plus Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific) using a THUNDERBIRD qPCR Mix (QPS-201, TOYOBO). We calculated the relative RNA expression levels using the ΔΔCt method, with the levels normalized to GAPDH mRNA. All amplicons were confirmed as a single product by agarose gel visualization and/or melting curve analysis. Table S1 lists the applied primer sequences.
Flow cytometric analysis
Cells were incubated after 48 hours of transfection by siRNA. Recombinant human interferon-γ (AF-300-02, PeproTech, Cranbury, NJ, USA) was added to a final concentration of 10 ng/mL for the last 24 hours of incubation. Then, PC9 cells were incubated with Human TruStain FcX (cat422302, Biolegend, San Diego, CA, USA) for 10 minutes at 4°C and washed with phosphate-buffered saline containing 1% FBS. The cells were incubated with 5 μL of PE anti-human CD274 (B7-H1, PD-L1) antibody (cat #329706, Biolegend) or the corresponding isotype control (cat #400314, Biolegend) at 4°C for 30 minutes. A Gallios flow cytometer and the FlowJo software were used for data analysis.
Multiplex RNA expression analysis with the Nanostring nCounter
A total of 100 ng of RNA was hybridized using the PanCancer IO 360 Gene Expression Panel on the nCounter system (NanoString Technologies, Seattle, WA), following the manufacturer’s recommendations. The PanCancer IO 360 Gene Expression Panel is designed to quantitate 770 target genes, 40 housekeeping genes, and additional positive and negative controls. For downstream analysis, absolute read counts of all panel genes were extracted from the nSolver software (NanoString Technologies, Seattle, WA). Target genes were normalized to 40 reference genes and fold changes, and associated statistics were calculated with the R-based NanoStringDiff package. We used differential expression analysis to calculate the t-statistic for each gene against each covariate in the model. We excluded genes with a low-expression value of <10.0 from the analysis. The raw data have been deposited in the Gene Expression Omnibus at the National Center for Biotechnology Information (accession No. GSE171634). Processed data are shown in the Supplemental Online Material.
Statistical analysis
Discrete variables are expressed as counts (percentage), and continuous variables are expressed as the mean ± standard deviation or median (range), unless otherwise specified. We used the Mann–Whitney U test or Student’s t test for continuous variables. Categorical data were compared between groups using the Fisher exact test for independence. We used the Wilcoxon matched-pairs signed-rank test for the analysis of the comparison between the two corresponding groups. We used Spearman’s rank-order correlation coefficient for correlation analysis. Survival was analyzed by the Kaplan–Meier curves with the log-rank test. OS was defined as the time from baseline to the date of death, and recurrence-free survival (RFS) was defined as the time from baseline to the recurrence date. We applied univariate and multivariate Cox proportional hazards models to generate the hazard ratios (HRs) of death, with adjustments for other potential confounding factors. Cell proliferation assays were analyzed using a two-way analysis of variance. Due to the exploratory nature of this study, none of the P values were adjusted for multiple testing.43 Statistical analyses were performed using GraphPad Prism version 8 (GraphPad Software, San Diego, CA, USA) and EZR software (Saitama Medical Center, Jichi Medical University, Saitama, Japan). A P value less than 0.05 was considered statistically significant.
Results
Levels of YTHDF1 and YTHDF2 expression are elevated in NSCLC
We first compared the expression levels of YTHDF1, YTHDF2, and YTHDF3 mRNA in lung cancer tissues and their corresponding normal lung tissues in the publicly available TCGA database (n = 109). We found that the expression levels of YTHDF1 and YTHDF2 were significantly higher in tumor tissues as compared with normal lung tissues (P < .0001 for YTHDF1, P = .0003 for YTHDF2). On the other hand, there was no significant difference in the YTHDF3 expression in lung cancer tissues (P = .188; Figure 2a).
Figure 2.
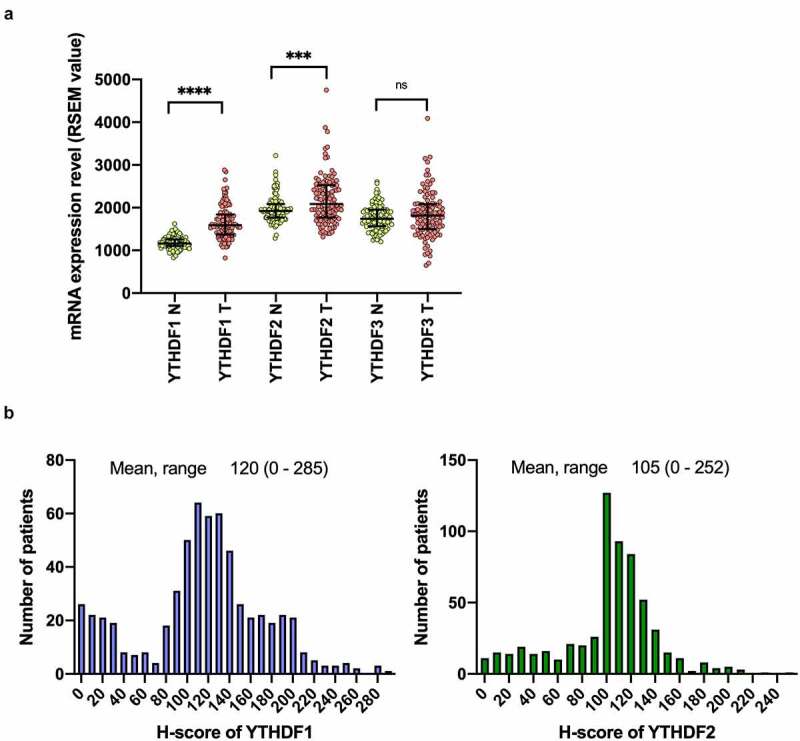
Level of YTHDF1 and YTHDF2 expression is elevated in NSCLC
(a) Comparison of the expressions of YTHDF 1, YTHDF2, and YTHDF3 in tumor and nontumor lung tissue in patients with non–small-cell lung cancer (N: nontumor tissue, T: tumor tissue). ***P < .001 and ****P < .0001, Mann–Whitney U test). Results were presented as means ± SD. (b) The histograms show the distribution of the H-scores for YTHDF1 and YTHDF2 evaluated by immunohistochemistry.
We further analyzed the expressions of YTHDF1 and YTHDF2 using our own cohort of lung cancer patients. The median age of the patients was 68 years (range, 23–88 years). Among the patients, 414 (68.7%) were male and 411 (68.2%) had a smoking history. The median H-score was 120 (0–285) for YTHDF1 and 105 (0–252) for YTHDF2 (Figure 2b). The expression levels of YTHDF1 and YTHDF2 were statistically more abundant in adenocarcinomas than in other histologies (P = .007 for YTHDF1, P = .011 for YTHDF2). In contrast, there was a reduced expression of YTHDF1 and YTHDF2 in the advanced pathological stages (P = .010 for YTHDF1, P = .047 for YTHDF2) and in the subsets with worse nodal metastases (P = .010 for YTHDF1, P = .004 for YTHDF2). As revealed by immunohistochemistry, we observed significantly higher YTHDF1 expression in patients with EGFR mutations (P < .001; Table 1). There was also a weak positive correlation between the expression levels of YTHDF1 and YTHDF2 (r = 0.325, P < .0001; Figure S1).
Table 1.
Comparison of clinical characteristics based on YTHDF1 and YTHDF2 expression in the tumor
| YTHDF1 |
YTHDF2 |
||||||
|---|---|---|---|---|---|---|---|
| Total | Low | High | Low | High | |||
| N = 603 | N = 291 | N = 312 | P-value | N = 404 | N = 199 | P-value | |
| Age | |||||||
| Mean, (range) | 68 (23–88) | 68 (23–88) | 68 (39–88) | N.S. | 68 (23–88) | 68 (33–88) | N.S. |
| Sex, n, (%) | |||||||
| Male | 414 (68.7) | 205 (70.4) | 209 (67.0) | N.S. | 283 (70.0) | 131 (65.8) | N.S. |
| Female | 189 (31.3) | 86 (29.6) | 103 (33.0) | 121 (33.0) | 68 (34.2) | ||
| Smoking status, n, (%) | |||||||
| Never | 179 (29.7) | 79 (27.1) | 100 (32.1) | N.S. | 115 (28.5) | 64 (32.2) | N.S. |
| Ever | 411 (68.2) | 204 (70.1) | 207 (66.3) | 277 (68.6) | 134 (67.3) | ||
| Unknown | 13 (2.1) | 8 (2.7) | 5 (1.6) | 12 (3.0) | 1 (0.5) | ||
| Histology, n, (%) | |||||||
| Adenocarcinoma | 392 (65.0) | 171 (58.8) | 221 (70.8) | .007 | 250 (61.9) | 142 (71.4) | .012 |
| Squamous cell carcinoma | 170 (27.0) | 98 (33.7) | 72 (23.1) | 129 (32.0) | 41 (20.6) | ||
| Others | 41 (68.0) | 22 (7.6) | 19 (6.1) | 25 (6.2) | 16 (8.0) | ||
| Tumor status, n, (%) | |||||||
| 1 | 236 (39.1) | 106 (36.4) | 130 (45.2) | N.S. | 148 (36.6) | 88 (44.2) | N.S. |
| 2 | 267 (44.3) | 126 (43.3) | 141 (45.2) | 186 (46.0) | 81 (40.7) | ||
| 3 | 63 (10.5) | 38 (13.1) | 25 (8.0) | 42 (10.4) | 21 (10.6) | ||
| 4 | 37 (6.1) | 21 (7.2) | 16 (5.1) | 28 (6.9) | 9 (4.5) | ||
| Node metastasis, n, (%) | |||||||
| 0 | 445 (73.8) | 198 (68.0) | 247 (79.2) | .007 | 285 (70.5) | 160 (80.4) | .018 |
| 1 | 70 (11.6) | 41 (14.1) | 29 (9.3) | 57 (14.1) | 13 (6.5) | ||
| 2 | 81 (13.4) | 46 (15.8) | 35 (11.2) | 56 (13.9) | 25 (12.6) | ||
| 3 | 7 (1.2) | 6 (2.1) | 1 (0.3) | 6 (1.5) | 1 (0.5) | ||
| Pathological stage, n, (%) | |||||||
| I | 378 (62.7) | 164 (56.4) | 214 (68.6) | .010 | 241 (59.7) | 137 (68.8) | .089 |
| II | 106 (17.6) | 56 (19.2) | 50 (16.0) | 78 (19.3) | 28 (14.1) | ||
| III | 119 (19.7) | 71 (24.4) | 48 (15.4) | 85 (21.0) | 34 (17.1) | ||
| Adjuvant chemotherapy, n, (%) | |||||||
| Yes | 254 (42.1) | 117 (40.2) | 137 (43.9) | N.S. | 160 (39.6) | 94 (47.2) | N.S. |
| No | 349 (57.9) | 174 (59.8) | 175 (56.1) | 244 (60.4) | 105 (71.4) | ||
| EGFR mutation, n, (%) | |||||||
| Wild type | 478 (79.3) | 251 (86.3) | 227 (72.8) | <.001 | 329 (81.4) | 149 (74.9) | .069 |
| Mutant | 125 (20.7) | 40 (13.7) | 85 (27.2) | 75 (18.6) | 50 (25.1) | ||
| PD-L1%TPS | |||||||
| ≧1% | 223 (37.0) | 112 (38.49) | 111 (35.58) | N.S. | 141 (34.90) | 82 (41.21) | N.S. |
| <1% | 380 (63.0) | 179 (61.51) | 201 (64.42) | 263 (65.10) | 117 (58.79) | ||
YTHDF1 and YTHDF2 are favorable prognostic factors in NSCLC
Figure 3 presents the Kaplan–Meier curves, based on the YTHDF1 and YTHDF2 expressions in our own cohort. Tumors with high levels of YTHDF1 and YTHDF2 expression showed significant favorable prognoses for OS (log-rank test, P = .002 for YTHDF1, P = .009 for YTHDF2; Figure 3a, b). Similarly, tumors with high YTHDF1 and YTHDF2 expressions showed significantly favorable RFS (P = .001 for YTHDF1, P = .005 for YTHDF2; Figure 3c, d). Further analysis of our cohort which was divided into two groups, one as the discovery set (Seireimikatahara Hospital [Mikatahara] cohort) and one as the validation set (Hamamatsu University School of Medicine [HUSM] cohort), showed a consistent trend that YTHDF1 and YTHDF2 expression was associated with favorable prognosis (P = .04 and P = .09 for YTHDF1 and YTHDF2, respectively, in the Mikatahara cohort; P = .02 and P = .04 for YTHDF1 and YTHDF2, respectively, in the HUSM cohort; Figure S2A–E). Multivariate Cox regression analyses adjusted for sex, smoking status, histology, stage, EGFR mutation status, and adjuvant chemotherapy demonstrated that YTHDF1 and YTHDF2 were independent favorable prognostic factors for RFS (HR, 0.745; 95% confidence interval [CI], 0.562–0.984 for YTHDF1; HR, 0.683; 95% CI, 0.503–0.928 for YTHDF2; Table 2, Table 3).
Figure 3.
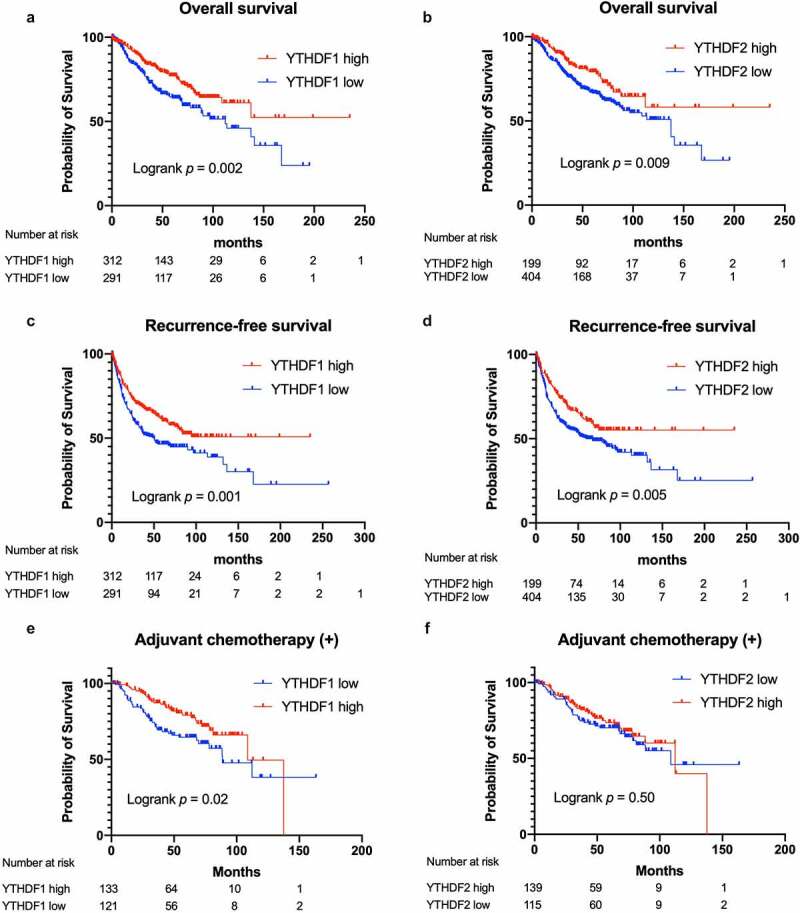
Survival curves according to YTHDF1 and YTHDF2 expression in patients with non–small-cell lung cancer (NSCLC)
Kaplan–Meier survival curves for overall survival (OS) according to the expression of YTHDF1 (a) and YTHDF2 (b) (n = 603, log-rank test). Kaplan–Meier survival curves for recurrence-free survival (RFS) according to YTHDF1 (c) and YTHDF2 (d) expression (n = 603, log-rank test). Kaplan–Meier survival curves for OS according to YTHDF1 (e) and YTHDF2 (f) expression in the population who received adjuvant chemotherapy (n = 254, log-rank test).
Table 2.
Univariate and multivariate Cox hazards model analyses for overall survival of patients with non–small-cell lung cancer
| Variable | Per unit for HR | univariate |
multivariate |
||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | ||
| Age | 1-year | 1.022 | 1.006–1.038 | .008 | 1.038 | 1.018–1.058 | <.001 |
| Sex | Male/female | 2.286 | 1.563–3.345 | <.001 | 0.699 | 0.375–1.305 | .261 |
| Smoking status | Ever/never | 3.394 | 2.291–5.029 | <.001 | 2.771 | 1.371–5.598 | .004 |
| Histology | Adenocarcinoma/the other histologies | 2.108 | 1.522–2.918 | <.001 | 1.052 | 0.718–1.539 | .794 |
| Pathological stage | 1-stage | 2.053 | 1.734–2.432 | <.001 | 2.108 | 1.723–2.58 | <.001 |
| EGFR mutation | Mutant/wild type | 0.61 | 0.399–0.933 | .027 | 0.839 | 0.502–1.401 | .503 |
| chemotherapy | Yes/no | 0.994 | 0.731–1.350 | .967 | |||
| YTHDF1 | High/low | 0.631 | 0.464–0.859 | .003 | 0.729 | 0.513–1.035 | .077 |
| YTHDF2 | High/low | 0.677 | 0.498–0.919 | .012 | 0.675 | 0.456–1.002 | .051 |
Table 3.
Univariate and multivariate Cox hazards model analyses for recurrence-free survival of patients with non–small-cell lung cancer
| Variable | Per unit for HR | univariate |
multivariate |
||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | ||
| Age | 1-year | 1.003 | 0.991–1.015 | .632 | |||
| Sex | Male/female | 1.663 | 1.248–2.215 | <.001 | 0.787 | 0.504–1.228 | .291 |
| Smoking status | Ever/never | 1.976 | 1.457–2.680 | <.001 | 1.746 | 1.069–2.854 | .026 |
| Histology | Adenocarcinoma/the other histologies | 1.763 | 1.350–2.303 | <.001 | 1.123 | 0.818–1.54 | .473 |
| Pathological stage | 1-stage | 2.433 | 2.109–2.807 | <.001 | 2.293 | 1.93–2.72 | <.001 |
| EGFR mutation | Mutant/wild type | 0.696 | 0.501–0.966 | .03 | 0.917 | 0.628–1.339 | .654 |
| Adjuvant chemotherapy | Yes/no | 1.403 | 1.095–1.798 | .007 | 1.084 | 0.808–1.454 | .588 |
| YTHDF1 | High/low | 0.664 | 0.518–0.850 | .001 | 0.745 | 0.564–0.984 | .038 |
| YTHDF2 | High/low | 0.67 | 0.508–0.885 | .005 | 0.683 | 0.503–0.928 | .014 |
We also analyzed the prognostic association of YTHDF1 and YTHDF2 using the TCGA data set (n = 974) and Kaplan–Meier plotter data set (n = 1926). The result of the TCGA data set demonstrated that high YTHDF2 mRNA expression was significantly associated with a favorable prognosis (log-rank test, P = .0009; Figure S3A). High YTHDF1 mRNA expression tended to have a favorable prognosis, although this was not statistically significant (log-rank test, P = .1185; Figure S3B). The result of the Kaplan–Meier plotter data set also showed that the tumors with high YTHDF1 and YTHDF2 expressions showed significant favorable survival (log-rank test, P = .000019 for YTHDF1, P = .000051 for YTHDF2; Figure S3C, S3D). Moreover, we performed prognostic analysis in the population that received adjuvant chemotherapy to investigate whether the expression of YTHDF1 and YTHDF2 might have predictive significance for response to adjuvant chemotherapy. Interestingly, the group with low YTHDF1 had poor prognosis compared with the YTHDF1-high group, but there was no significant difference in prognosis dependent on YTHDF2 expression (log-rank test, P = .02 for YTHDF1, P = .50 for YTHDF2; Figure 3e, 3f). These results indicated that YTHDF1 and YTHDF2 were favorable prognostic factors in NSCLC, which might be partly due to better response to adjuvant chemotherapy in YTHDF1.
Expression of YTHDF1 and YTHDF2 is associated with TILs in the stroma in non–small-cell lung cancer
Because the expression levels of YTHDF1 and YTHDF2 were high in cancerous tissues, we investigated the surrounding tumor environment to elucidate the mechanism underlying the role of YTHDF1 and YTHDF2 in better clinical outcomes. Hence, we evaluated CD8+, FOXP3+, PD-1+, and CD45RO+ immune cells on behalf of TILs among tumor nests and tumor-surrounding stroma. All subsets of TILs showed significant positive correlations between tumor nests and tumor surrounding stroma (r = 0.266 for PD-1+, r = 0.385 for FOXP3+, r = 0.415 for CD45RO+, r = 0.581 for CD8+; Figure S4A–D). In the tumor surrounding stroma, the number of almost all subsets of TILs was significantly higher in high YTHDF1 and YTHDF2 tumors (P < .001 for Foxp3+, P = .01 for PD-1+, and P = .005 for CD45RO+ TILs in YTHDF1; P < .001 for all subsets of TILs in YTHDF2; Figure 4a, 4b). In the tumor nests, the numbers of Foxp3+ and PD-1+ TILs were significantly higher in high YTHDF2 tumors (P < .001 for Foxp3+, P = .028 for PD-1+); however, there were no significant differences in TILs based on YTHDF1 expression (Figure 4c, 4d). These observations suggest that the high expression of YTHDF1 and YTHDF2 is associated with a higher number of TILs in the stroma.
Figure 4.
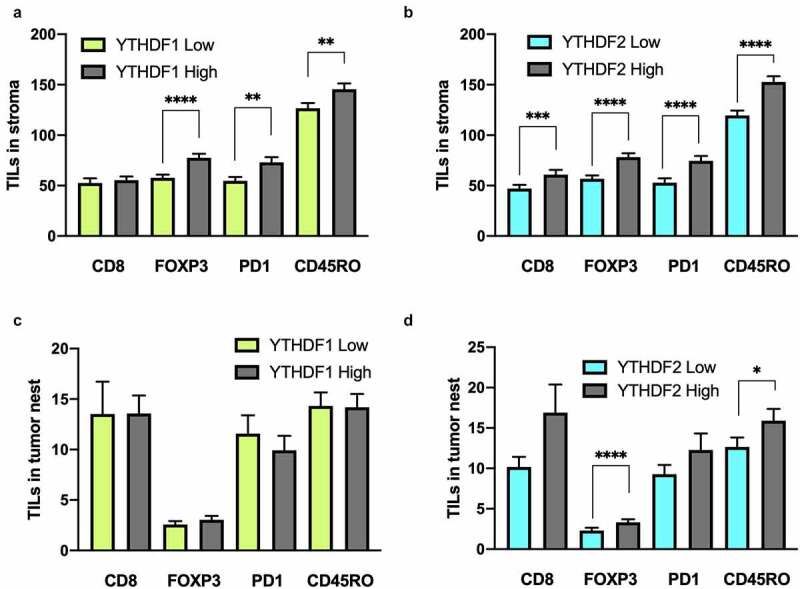
Expression of YTHDF1 and YTHDF2 is associated with tumor-infiltrating lymphocytes (TILs) in the stroma in patients with non–small-cell lung cancer
(a) Number of TILs in tumor nests according to the expression of YTHDF1 (Mann–Whitney U test). (b) Number of TILs in tumor nests according to the expression of YTHDF2 (*P < .05 and ****P < .0001, Mann–Whitney U test). (c) Number of TILs in stroma according to the expression of YTHDF1 (**P < .01 and ****P < .0001, Mann–Whitney U test). (d) Number of TILs in stroma according to the expression of YTHDF2 (***P < .001 and ****P < .0001, Mann–Whitney U test). All results were presented as means ± SEM.
YTHDF1 and YTHDF2 knockdown suppresses cell proliferation in lung cancer cells
To investigate the impact of YTHDF1 and YTHDF2 deficiency in lung cancer cells, we used small interfering RNA (siRNA) to knock down YTHDF1 and YTHDF2 in lung cancer cells. Knockdown efficacy was confirmed by qRT-PCR analysis (Figure 5a, 5b). YTHDF1 knockdown significantly suppressed the cell proliferation of PC9, A549, and H1299 cells. YTHDF2 knockdown also significantly suppressed the cell proliferation of PC9, A549, and H1299 cells (Figure 5c). We next assessed the migration ability of YTHDF1 – and 2-knockdown cells using a transwell migration assay, which revealed that there was no significant reduction in the migration of PC9, A549, and H1299 cells with YTHDF1 – or YTHDF2-knockdown (Figure S5). Subsequently, flow cytometric analysis for apoptosis showed that YTHDF1 and YTHDF2 knockdown increased the number of apoptotic PC9 and H1299 cells (Figure 5d). These results showed that YTHDF1 and YTHDF2 facilitated cell proliferation and partly reduced apoptosis in vitro. However, these observations were inconsistent with the favorable prognosis in NSCLC patients with high expression of YTHDF1 and YTHDF2.
Figure 5.
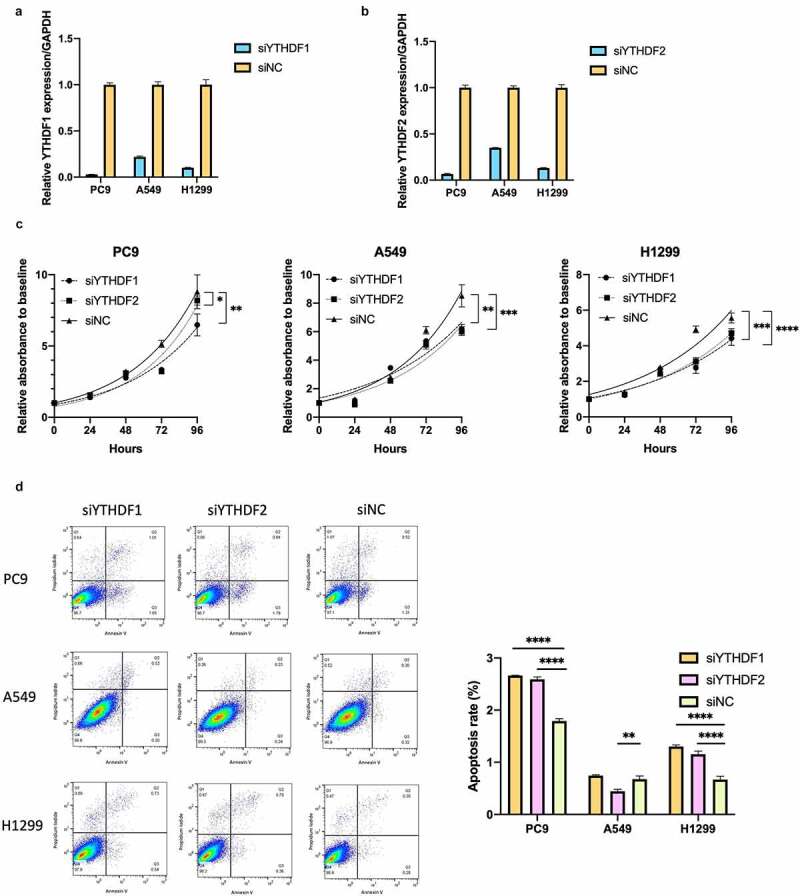
YTHDF1 and YTHDF2 knockdown suppresses cell proliferation and partially induces apoptosis in lung cancer cells
YTHDF1 and YTHDF2 mRNA expression were reduced by silencing YTHDF1 (a) and YTHDF2 (b) in PC9, A549, and H1299. Results were presented as means ± SD. (c) Effect of relative cell proliferation in PC9, A549, and H1299 transfected with siRNA of YTHDF1 or YTHDF2 and control (siNC) (n = 3 for each group, *P < .05, **P < .01, ***P < .001, ****P < .0001 by two-way analysis of variance). (d) The number of apoptotic cells was determined using flow cytometric analysis of PC9, A549, and H1299 cells transfected with siRNA for YTHDF1, YTHDF2, or siNC. Percentages of apoptotic cells are shown in a representative scatter plot. The apoptosis rates in cells with YTHDF1 or YTHDF2 knockdown were compared to those transfected with siNC and shown as a bar chart (n = 3, **P < .01 and ****P < .0001, Mann–Whitney U test). Results were presented as means ± SD.
YTHDF1 and YTHDF2 knockdown increased tumoral PD-L1 expression in lung cancer cells
We evaluated the changes in global RNA expression in the YTHDF1 and YTHDF2 knockdown PC9 cells using the PanCancer IO 360 Gene Expression Panel with nCounter analysis. We extracted total RNA from the PC9 cell line in which YTHDF1 and YTHDF2 expressions were silenced by siRNA and analyzed differential gene expression using nSolver 4.0. The threshold was determined with a fold change of 2.0. Five upregulated genes including CD274 and two downregulated genes overlapped between the knockdown of YTHDF1 siRNA no.1 (siYTHDF1#1) and the knockdown of YTHDF1 siRNA no.2 (siYTHDF1#2). Six upregulated genes including CD274 overlapped between the knockdown of YTHDF2 siRNA no.1 (siYTHDF2#1) and knockdown of YTHDF2 siRNA no.2 (siYTHDF2#2; Figure 6a). Based on the panel expression analysis, we next evaluated the YTHDF1 and YTHDF2 associations with tumoral PD-L1, which is encoded by CD274 and a ligand for PD-1 receptor and acts as a co-inhibitory molecule, which is also an important complementary marker for immune checkpoint inhibitors. The TCGA data set showed that the expression level of CD274 was significantly high in the low YTHDF2 group. Although the expression level of PD-L1 was not significantly high, there was a high tendency in the low YTHDF1 groups (Figure S6A, S6B). We also performed GSEA with a hallmark gene set using the TCGA data set stratified by the expression levels of YTHDF1 and YTHDF2. The GSEA revealed that the gene set of the inflammatory response and complements was significantly enriched in the low YTHDF1 and YTHDF2 groups (Figure S6C, S6D). These findings suggest that the expressions of YTHDF1 and YTHDF2 are significantly associated with the gene profiles linked to tumor immunity.
Figure 6.
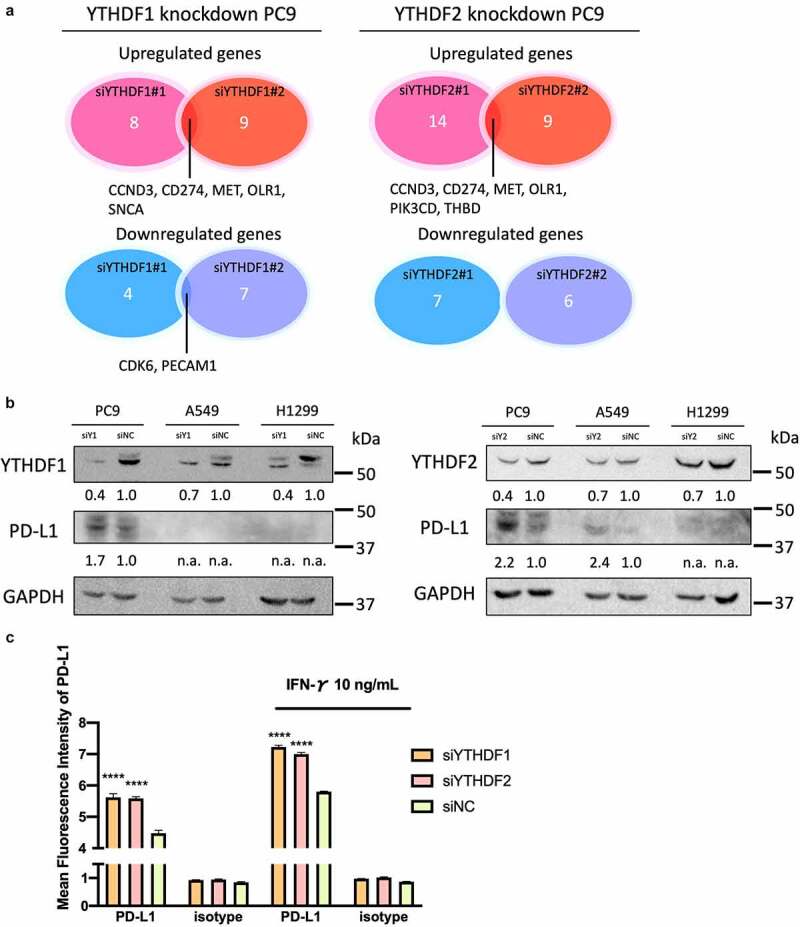
YTHDF1 and YTHDF2 knockdown increased tumoral PD-L1 expression in lung cancer cells
(a) Venn diagram indicating the number of common genes in different sequences of siRNA at least 2.0-fold alteration of RNA expression (n = 1). (b) Representative immunoblot analysis of YTHDF1 and YTHDF knockdown lung cancer cells. The protein expression of YTHDF1 or YTHDF2 and PD-L1 and GAPDH were shown. The score below the photo is a quantification of the relative intensity of the PD-L1 band to GAPDH. (c) PD-L1 cell surface expression in PC9 cells with YTHDF1 or YTHDF2 knockdown with or without interferon-γ stimulation determined by flow cytometric analysis (n = 3 per group, ****P < .0001, Student’s t test). Results were presented as means ± SD.
To validate the results of the gene expression panel, we next evaluated the expression levels of CD274 in YTHDF1 – and YTHDF2-downregulated PC9 cells. The mRNA expression levels of CD274 were significantly upregulated in YTHDF1 and YTHDF2 knockdown PC9 cells (P < .0001 for si-YTHDF1#1 and #2, and P < .0001 for si-YTHDF2#1 and #2; Figure S6E, S6F). In addition, the protein expression levels of PD-L1 were upregulated in YTHDF1 and YTHDF2 knockdown cells (Figure 6b). Next, we evaluated the cell surface expression of PD-L1 among YTHDF1 – and YTHDF2-downregulated PC9 cells by flow cytometry. PD-L1 expression was upregulated on YTHDF1 knockdown cells (P = .002) and YTHDF2 knockdown cells (P = .0002). PD-L1 expression was also upregulated on YTHDF1 knockdown cells (P = .002) and YTHDF2 knockdown cells by interferon-γ stimulation (P = .0002; Figure 6c). These results indicate that the high expression of YTHDF1 and YTHDF2 is associated with the low expression of the co-inhibitor molecule PD-L1.
Discussion
In this study, we first demonstrated that YTHDF1 and YTHDF2 were significantly higher in tumor tissues as compared with normal tissue in lung cancer series and significantly associated with a better prognosis in patients with NSCLC. Mechanistically, we showed that most subsets of TIL densities were significantly higher in tumors with high expressions of YTHDF1 or YTHDF2 as compared with those with low expressions and tumoral PD-L1 expression was also upregulated in YTHDF1 – and YTHDF2-deficient lung cancer cells. Our results suggest that YTHDF1 and YTHDF2 inflamed the tumor-immune microenvironment in NSCLC.
The m6A reader proteins specifically recognize the m6A modification sites on mRNA, deciding the fate of affected mRNA. Although various types of m6A reader proteins have been reported, YTHDF1 and YTHDF2 are proteins that have been intensively studied. YTHDF1 is known to promote the translation of the target transcripts via the interaction with EIFs in some solid cancers.15,16 YTHDF2 is also a common m6A reader protein, which promotes RNA decay and regulates the target gene function via RNA instability. However, the exact roles and the underlying mechanisms of these reader proteins remain under investigation. Moreover, to draw conclusions regarding tumor aggressiveness or antitumor function, there is still room for discussion of m6A’s function against cancer.
A few studies have already been published regarding the clinical implications of YTHDF1 and YTHDF2 in NSCLC. YTHDF1, which is highly expressed in tumor cells, was reported to be associated with better prognosis because its depletion renders cancerous cells resistant to chemotherapy, even though YTHDF1 has some pro-tumorigenic nature via the modulation of cell cycle–related genes in vitro.44 These results were consistent with our in vitro study and patient prognostic profiles. As a hypothesis to explain these two conflicting events, i.e., YTHDF1, which is highly expressed in tumors, is associated with improved prognosis whereas the in vitro results are in contrast to its prognostic role in vivo, our experimental results have novel implications that tumor immunity including inhibitory co-stimulatory molecules may be indirectly mediated. On the other hand, YTHDF2, which is highly expressed in tumor cells, was reported to promote the translation of 6GPD mRNA, which led to the activation of the pentose phosphate pathway, resulting in the promotion of cell proliferation in lung cancer cells. However, there was no association with worse survival in patients with NSCLC.20 In this previous study, there was a discrepancy between the results of in vitro cell proliferation using lung cancer cells and the results of analysis of patient prognosis. Tumor-mediated immunity via inhibitory co-stimulatory molecules, as shown in the present study, may also provide an explanation. A recent study showed that the durable neoantigen-specific immunity is regulated by YTHDF1, and YTHDF1-deficient mice showed an elevated antigen-specific CD8 + T cell antitumor response in colon cancer.45 In contrast to previous reports, our study demonstrated that almost all subsets of TILs in the high YTHDF1 and YTHDF2 groups were also high in the tumor-associated stroma. This might be explained by the difference in target genes between different cancer types. In addition, PD-L1 was also upregulated in YTHDF1 – and YTHDF2-downregulated lung cancer cells. These results suggest that high expression of TILs and deficiency in the expression of the co-inhibitor molecule PD-L1 may be associated with better prognosis in NSCLC with high YTHDF1 and YTHDF2.
Our study had several limitations. First, our in vitro experiment could not emulate the environment around a tumor, which may not accurately reflect the in vivo observation. Because tumor microenvironment–mediated tumor immunity is involved in a cancer-promoting role, the results of in vitro experiments that lack a tumor microenvironment might differ from prognostic results using human samples. Second, this study did not scrutinize the underpinning molecular mechanisms, especially their association with m6A RNA modification. Indeed, YTHDF1 promotes the translation of the transcript through the target mRNA, whereas YTHDF2 destabilizes the mRNA. In this study, the expression of PD-L1 was upregulated by the knockdown of YTHDF1 and YTHDF2. This result contradicts that CD274 is a direct target for YTHDF1 and YTHDF2. To clarify the role of m6A in the machinery of YTHDF, further studies should be performed on tumor microenvironment–related genes and m6A-mediated regulation of transcripts and proteins by YTHDF perturbations.
In conclusion, high expressions of YTHDF1 and YTHDF2 were associated with an inflamed tumor microenvironment and better patient survival in NSCLC. These m6A reader proteins could be the novel prognostic and druggable targets related to the tumor-immune system in lung cancers.
Supplementary Material
Acknowledgments
We thank Mrs. Shiho Omori and Mr. Hisaki Igarashi (Hamamatsu University School of Medicine) for their technical assistance.
Funding Statement
This work was supported by grants from the Japanese Ministry of Health, Labor and Welfare (19–19, 10103838), the Japan Society for the Promotion of Science (JP22590356, JP23790396, JP22659072, JP24659161, JP26670187, JP16K15256), the Ministry of Education, Culture, Sports, Science and Technology (S-001), the National Cancer Center Research and Development Fund (25-A-1), Research on Global Health Issues from the Japanese Ministry of Health, Labor and Welfare, Japan Agency for Medical Research and Development (AMED, JP19ck0106264, JP20ck0106545), and Smoking Research Foundation.
Data availability:
All data and supplemental information within the article are available from the corresponding author upon reasonable request.
Disclosure statement:
The authors report no conflict of interest.
Authors’ contributions:
Study design: K.T., K.Y, H.S. Study conduct: K.T., K.Y. Data collection: K.Y., Yusuke.Inoue., T.W., H.Y., Yuji Iwashita, A.K., K.F., M.T., H.O., Data analysis: K.T., K.Y., Yuji Iwashita, Yusuke Inoue, K.S., H.S. Data interpretation: K.T., K.Y, Yuji Iwashita, K.F., T.S., H.S. Drafting the manuscript: K.T., Yuji.Iwashita., K.Y., H.S. Revising the manuscript content: K.Y., Yusuke Inoue, Yuji Iwashita, T.K., T.S., H.S. Approving final version of the manuscript: all authors.
Notification of prior presentation:
The data in this article were presented at the poster session at the 111th AACR Virtual Annual Meeting II, June 22–24, 2020, https://www.aacr.org/meeting/aacr-annual-meeting-2020/.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
References
- 1.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(201–206):201–13. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 2.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(885–887):885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hess ME, Hess S, Meyer KD, Verhagen LA, Koch L, Bronneke HS, Dietrich MO, Jordan SD, Saletore Y, Elemento O, et al. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat Neurosci. 2013;16(1042–1048):1042–1048. doi: 10.1038/nn.3449. [DOI] [PubMed] [Google Scholar]
- 4.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(18–29):18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16(191–198):191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fustin JM, M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155(793–806):793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 7.Zhang C, Fu J, Zhou Y.. A review in research progress concerning m6A methylation and immunoregulation. Front Immunol. 2019;10(922). doi: 10.3389/fimmu.2019.00922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dang W, Xie Y, Cao P, Xin S, Wang J, Li S, Li Y, Lu J. N(6)-methyladenosine and viral infection. Front Microbiol. 2019;10(417). doi: 10.3389/fmicb.2019.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lan Q, Liu PY, Haase J, Bell JL, Huttelmaier S, Liu T. The critical role of RNA m6A methylation in cancer. Cancer Res. 2019;79(1285–1292):1285–1292. doi: 10.1158/0008-5472.CAN-18-2965. [DOI] [PubMed] [Google Scholar]
- 10.Patil DP, Pickering BF, Jaffrey SR. Reading m(6)A in the transcriptome: m(6) A-bindingProteins. Trends Cell Biol. 2018;28(113–127). doi: 10.1016/j.tcb.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161(1388–1399):1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(117–120):117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng X, Su R, Weng H, Huang H, Li Z, Chen J. RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018;28(507–517):507–517. doi: 10.1038/s41422-018-0034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bai Y, Yang C, Wu R, Huang L, Song S, Li W, Yan P, Lin C, Li D, Zhang Y. YTHDF1 regulates tumorigenicity and cancer stem cell-like activity in human colorectal carcinoma. Front Oncol. 2019;9(332). doi: 10.3389/fonc.2019.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orouji A, Peitsch WK, Orouji A, Houben R, Utikal J. Oncogenic role of an epigenetic reader of m(6)A RNA modification: YTHDF1 in merkel cell carcinoma. Cancers (Basel). 2020;12(1):202. doi: 10.3390/cancers12010202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y, Cheng C, Li L, Pi J, Si Y, et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020;48(3816–3831):3816–3831. doi: 10.1093/nar/gkaa048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao X, Chen Y, Mao Q, Jiang X, Jiang W, Chen J, Xu W, Zhong L, Sun X. Overexpression of YTHDF1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Biomark. 2018;21(859–868):859–868. doi: 10.3233/CBM-170791. [DOI] [PubMed] [Google Scholar]
- 18.Jia R, Chai P, Wang S, Sun B, Xu Y, Yang Y, Ge S, Jia R, Yang YG, Fan X. m(6)A modification suppresses ocular melanoma through modulating HINT2 mRNA translation. Mol Cancer. 2019;18(161). doi: 10.1186/s12943-019-1088-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Meng S, Xu M, Wang S, He L, Xu X, Wang X, Xie L. Downregulation of N(6)-methyladenosine binding YTHDF2 protein mediated by miR-493-3p suppresses prostate cancer by elevating N(6)-methyladenosine levels. Oncotarget. 2018;9(3752–3764). doi: 10.18632/oncotarget.23365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheng H, Li Z, Su S, Sun W, Zhang X, Li L, Li J, Liu S, Lu B, Zhang S, et al. YTH domain family 2 promotes lung cancer cell growth by facilitating 6-phosphogluconate dehydrogenase mRNA translation. Carcinogenesis. 2020;41(541–550):541–550. doi: 10.1093/carcin/bgz152. [DOI] [PubMed] [Google Scholar]
- 21.Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, Deng X, Wang Y, Weng X, Hu C, et al. R-2HG exhibits anti-tumor activity by targeting FTO/m(6)A/MYC/CEBPA signaling. Cell. 2018;172(1–2):90–105 e123. doi: 10.1016/j.cell.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, Coussens LM, Gabrilovich DI, Ostrand-Rosenberg S, Hedrick CC, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24(541–550):541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teng MW, Ngiow SF, Ribas A, Smyth MJ. Classifying cancers based on T-cell infiltration and PD-L1. Cancer Res. 2015;75(2139–2145):2139–2145. doi: 10.1158/0008-5472.CAN-15-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao R, Yuan L, Ma B, Wang G, Tian Y. Tumour microenvironment (TME) characterization identified prognosis and immunotherapy response in muscle-invasive bladder cancer (MIBC). Cancer Immunol Immunother. 2021;70(1–18):1–18. doi: 10.1007/s00262-020-02649-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Datta M, Coussens LM, Nishikawa H, Hodi FS, Jain RK. Reprogramming the tumor microenvironment to improve immunotherapy: Emerging strategies and combination therapies. Am Soc Clin Oncol Educ Book. 2019;39(165–174). doi: 10.1200/EDBK_237987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, Hodi FS, Martin-Algarra S, Mandal R, Sharfman WH, et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017;171(4):934–949 e916. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu F, Qin L, Liao Z, Song J, Yuan C, Liu Y, Wang Y, Xu H, Zhang Q, Pei Y, et al. Microenvironment characterization and multi-omics signatures related to prognosis and immunotherapy response of hepatocellular carcinoma. Exp Hematol Oncol. 2020;9(10). doi: 10.1186/s40164-020-00165-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang S, Wei J, Cui YH, Park G, Shah P, Deng Y, Aplin AE, Lu Z, Hwang S, He C, et al. m(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat Commun. 2019;10(2782). doi: 10.1038/s41467-019-10669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inoue Y, Yoshimura K, Mori K, Kurabe N, Kahyo T, Mori H, Kawase A, Tanahashi M, Ogawa H, Inui N, et al. Clinical significance of PD-L1 and PD-L2 copy number gains in non-small-cell lung cancer. Oncotarget. 2016;7(32113–32128):32113–32128. doi: 10.18632/oncotarget.8528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sugimura H. Detection of chromosome changes in pathology archives: an application of microwave-assisted fluorescence in situ hybridization to human carcinogenesis studies. Carcinogenesis. 2008;29(681–687):681–687. doi: 10.1093/carcin/bgn046. [DOI] [PubMed] [Google Scholar]
- 31.Sugimura H, Mori H, Nagura K, Kiyose S, Tao H, Isozaki M, Igarashi H, Shinmura K, Hasegawa A, Kitayama Y, et al. Fluorescence in situ hybridization analysis with a tissue microarray: ‘FISH and chips’ analysis of pathology archives. Pathol Int. 2010;60(543–550):543–550. doi: 10.1111/j.1440-1827.2010.02561.x. [DOI] [PubMed] [Google Scholar]
- 32.Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger K, Yatabe Y, Powell CA, Beer D, Riely G, Garg K, et al. International association for the study of lung cancer/american thoracic society/european respiratory society: international multidisciplinary classification of lung adenocarcinoma: executive summary. Proc Am Thorac Soc. 2011;8(381–385):381–385. doi: 10.1513/pats.201107-042ST. [DOI] [PubMed] [Google Scholar]
- 33.Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, Chirieac LR, Dacic S, Duhig E, Flieder DB, et al. The 2015 world health organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10(9):1243–1260. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 34.Goldstraw P, Chansky K, Crowley J, Rami-Porta R, Asamura H, Eberhardt WE, Nicholson AG, Groome P, Mitchell A, Bolejack V, et al. The IASLC lung cancer staging project: proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM classification for lung cancer. J Thorac Oncol. 2016;11(39–51):39–51. doi: 10.1016/j.jtho.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 35.Inoue Y, Matsuura S, Kurabe N, Kahyo T, Mori H, Kawase A, Karayama M, Inui N, Funai K, Shinmura K, et al. Clinicopathological and survival analysis of japanese patients with resected non-small-cell lung cancer harboring NKX2-1, SETDB1, MET, HER2, SOX2, FGFR1, or PIK3CA gene amplification. Journal of Thoracic Oncology. 2015;10(1590–1600):1590–1600. doi: 10.1097/jto.0000000000000685. [DOI] [PubMed] [Google Scholar]
- 36.Yoshimura K, Inoue Y, Tsuchiya K, Karayama M, Yamada H, Iwashita Y, Kawase A, Tanahashi M, Ogawa H, Inui N, et al. Elucidation of the relationships of MET protein expression and gene copy number status with PD-L1 expression and the immune microenvironment in non-small cell lung cancer. Lung Cancer. 2020;141(21–31):21–31. doi: 10.1016/j.lungcan.2020.01.005. [DOI] [PubMed] [Google Scholar]
- 37.Yoshimura K, Suzuki Y, Inoue Y, Tsuchiya K, Karayama M, Iwashita Y, Kahyo T, Kawase A, Tanahashi M, Ogawa H, et al. CD200 and CD200R1 are differentially expressed and have differential prognostic roles in non-small cell lung cancer. Oncoimmunology. 2020;9(1746554):1746554. doi: 10.1080/2162402X.2020.1746554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brevet M, Arcila M, Ladanyi M. Assessment of EGFR mutation status in lung adenocarcinoma by immunohistochemistry using antibodies specific to the two major forms of mutant EGFR. J Mol Diagn. 2010;12(169–176):169–176. doi: 10.2353/jmoldx.2010.090140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu J, Kane S, Wu J, Benedettini E, Li D, Reeves C, Innocenti G, Wetzel R, Crosby K, Becker A, et al. Mutation-specific antibodies for the detection of EGFR mutations in Non–Small-Cell lung cancer. Clin Cancer Res. 2009;15(3023–3028):3023–3028. doi: 10.1158/1078-0432.CCR-08-2739. [DOI] [PubMed] [Google Scholar]
- 40.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(e82241):e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(401–404):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(pl1):pl1–pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Althouse AD. Adjust for multiple comparisons? It’s not that simple. Ann Thorac Surg. 2016;101(1644–1645):1644–1645. doi: 10.1016/j.athoracsur.2015.11.024. [DOI] [PubMed] [Google Scholar]
- 44.Shi Y, Fan S, Wu M, Zuo Z, Li X, Jiang L, Shen Q, Xu P, Zeng L, Zhou Y, et al. YTHDF1 links hypoxia adaptation and non-small cell lung cancer progression. Nat Commun. 2019;10(4892). doi: 10.1038/s41467-019-12801-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han D, Liu J, Chen C, Dong L, Liu Y, Chang R, Huang X, Liu Y, Wang J, Dougherty U, et al. Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature. 2019;566(270–274):270–274. doi: 10.1038/s41586-019-0916-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and supplemental information within the article are available from the corresponding author upon reasonable request.


