Abstract
Candida albicans is the main fungal species associated with the development of oral candidiasis. Currently, therapeutic options for these infections are limited by the adverse effects of antifungal drugs and by the emergence of drug resistant strains. Thus, the development of new antifungal agents is needed for the prevention and treatment of oral Candida infections. Caffeic acid phenethyl ester (CAPE) is a natural compound from propolis polyphenolic groups that exhibits many pharmacological properties. In this study, we investigated whether CAPE can have antifungal and immunomodulatory effects on oral candidiasis. Preliminary tests to assess the antifungal activity of CAPE were performed using the Minimum Inhibitory Concentration (MIC) assay that demonstrated inhibition in a range from 16 to 32 μg/mL, confirming its antifungal activity on several C. albicans strains isolated from the oral cavity. Subsequently, we analyzed Candida spp biofilms formed in vitro, in which CAPE treatment at 5 x MIC caused a reduction of 68.5% in the total biomass and ~2.60 Log in the viable cell count (CFU/mL) in relation to the untreated biofilm (p<0.0001). Next, RNA was extracted from untreated and CAPE-treated biofilms and analyzed by real-time qPCR. A series of genes analyzed (ALS1, ECE1, EPA1, HWP1, YWP1, BCR1, BGR1, CPH1, EFG1, NDT80, ROB1, TEC1, UME6, SAP2, SAP5, PBL2, and LIP9) were downregulated by CAPE compared to the untreated control group (p<0.0001). In in vivo studies using Galleria mellonella, the treatment with CAPE prolonged survival of larvae infected by C. albicans by 44.5% (p < 0.05) and accompanied by a 2.07-fold increase in the number of hemocytes. Flow cytometry revealed the most prominent increases were in types P2 and P3 hemocytes, granular cells, which phagocytize pathogens. In addition, CAPE treatment decreased the fungal load in the hemolymph and stimulated the expression of antifungal peptide genes such as galiomicin and gallerimycin. The antifungal and immunomodulatory activities observed in G. mellonella were extended to a murine model of oral candidiasis, in which CAPE decreased the levels of C. albicans colonization (~2 log CFU/mL) in relation to the untreated control group. In addition, CAPE treatment significantly reduced pseudomembranous lesions, invasion of hyphae on epithelium surfaces, tissue damage and inflammatory infiltrate (p < 0.05). CAPE was also able to increase the expression of β-defensin 3 compared to the infected and untreated group by 3.91-fold (p < 0.0001). Taken together, these results show that CAPE has both antifungal and immunomodulatory effects, making it a promising natural antifungal agent for the treatment and prevention of candidiasis and shows impact to oral candidiasis.
Keywords: CAPE (caffeic acid phenethyl ester), Candida albicans, Biofilms, Galleria mellonella, gene expression, oral candidiasis, β-defensin 3
Introduction
Candida albicans is the most prevalent fungus in the human oral mycobiome (Bandara et al., 2019) and can be a harmless commensal organism or, upon appropriate conditions, transition to a pathogen. It possess several virulence factors that can lead to overgrowth and invasion of host tissues, causing a range of intraoral, pharyngeal, and perioral manifestations that determine different forms of oral candidiasis (Lewis and Williams, 2017; Hellstein and Marek, 2019; Ponde et al., 2021). Some of the most important virulence factors of C. albicans are the expressions of adhesins, the secretion of hydrolytic enzymes, its ability to transition from yeasts to hyphal and the high metabolic adaptability (Ciurea et al., 2020). Perhaps one of the most significant transition events is the formation of biofilms, cellular communities that can adhere and proliferate in abiotic and biotic surfaces. Oral Candida biofilms are more resistant to standard antifungal treatments and present a higher relapse rates, which turn these infections into a considerable clinical challenge in dental practice that are difficult to eradicate (Ponde et al., 2021).
In addition to therapeutic challenges posed by biofilm drug recalcitrance, the current therapeutic options for oral candidiasis are limited due to few availability antifungal classes, undesirable side effects, and antifungal resistance (Campoy and Adrio, 2017; Scorzoni et al., 2021). Thus, the search for new compounds that can control Candida spp. infections and avoid the aforementioned issues had pointed to natural compounds that display antifungal efficacy comparable to or stronger than drugs currently available for clinical use (Ferreira et al., 2021; Scorzoni et al., 2021). In this context, caffeic acid phenethyl ester (CAPE) stands out as a potent and promising agent that appears to modulate host responses (Breger et al., 2007) and may have a better impact on treating oral candidiasis.
CAPE is one of the bioactive compounds of propolis that has several biological and pharmacological properties, including antioxidant, anti-inflammatory, anticarcinogenic, anti-viral, antifungal and immunomodulatory activities (Breger et al., 2007; Chan et al., 2013; Armutcu et al., 2015; Coleman et al., 2016). CAPE modulates the transcription factors NF-kβ showing an anti-inflammatory activities (Armutcu et al., 2015; Olgierd et al., 2021). The NF-κβ pathway is involved in the transcription of many cytokines, chemokines, enzymes, antiapoptotic and cell growth factors (Armutcu et al., 2015; Olgierd et al., 2021), signaling involved in a wide range of physiological processes throughout the body, including immune responses, cell proliferation and inflammation (Natarajan et al., 1996).
Thus, the aim of this study was to evaluate the antifungal and immunomodulatory effects of CAPE on C. albicans using in vitro and in vivo models. In vitro assessment focused on the fungal biofilm morphology that contributes to C. albicans virulence and is challenging to treat. CAPE efficacy was further assessed in vivo using G. mellonella and a murine oral candidiasis model.
Materials and Methods
Microorganisms and Growth Conditions
Forty oral clinical strains of Candida albicans and a reference strain (ATCC18804) from the fungal collection of the Oral Microbiology and Immunology Laboratory at the Institute of Science and Technology of São José dos Campos/UNESP were used in this study (Junqueira et al., 2012). The strains were cultured in yeast extract, peptone, dextrose (YPD broth, Difco, Detroit, USA) for 24 h at 37°C.
Antifungal Activity and Determination of Minimum Inhibitory Concentration (MIC)
All clinical C. albicans strains were subjected to broth microdilution assays to determine the MIC values for CAPE, amphotericin B and fluconazole. Quality control was performed using the strain C. parapsilosis ATCC 22019.
Investigational compounds CAPE, amphotericin B and fluconazole were purchased from Sigma/Aldrich and diluted in DMSO at a concentration of 10 mg/mL. MICs were determined according to Clinical and Laboratory Standards Institute, document M60 (Clinical Laboratory Standard Institute, 2017) using round-bottomed 96-well microtiter plates, containing Candida suspensions prepared in physiological solution and then diluted into RPMI 1640 at final concentrations of 0.5-2.5 x103 CFU/mL. Inoculated plates were incubated for 24-48 h at 37°C. Fluconazole and amphotericin B were used as positive controls with final concentrations ranging from 64 to 0.125 μg/mL. The concentration tested for CAPE ranged from 128 to 2 µg/mL. The breakpoints used followed the CLSI guidelines (Fluconazole: ≤ 2.0 μg/mL for susceptible, ≥ 8 μg/mL for resistant; Amphotericin B: >1 μg/mL for resistant) (CLSI, 2017). MIC values for fluconazole and amphotericin B were defined as the lowest concentrations that inhibited 50% and 90% of fungal growth, respectively.
In Vitro Study in C. albicans Biofilms
The anti-biofilm activity of CAPE was tested against two C. albicans strains resistant to fluconazole (CA14 and CA70). C. albicans pre-biofilms were formed in 96-well flat bottom microtiter plates (TPP®, Trasadingen, Switzerland), following the methodology described by de Barros et al. (2018). Briefly, standardized suspensions of C. albicans (107 cells/mL) were added in microtiter plates containing yeast nitrogen base broth (YNB) (Difco, Detroit, USA) with 100 mM glucose and incubated for 48 h at 37°C with shaking at 75 rpm. After this period, each well was washed twice with PBS, and 200 µL of YNB containing different concentrations of CAPE (2 x MIC and 5 x MIC) were added to the plates and incubated for another 24 h in the same experimental conditions. After the treatment, each well was washed twice with PBS for subsequent analysis of viability cell and total biomass. Fluconazole served as a positive control, in concentrations corresponding to 2 x MIC and 5 x MIC.
Biofilm Analysis by Viability Assay
For the biofilm viability assay, 200 µL of PBS was added to each well and the biofilms were disrupted using a ultrasonic homogenizer (Sonopuls HD 2200, Bandelin Electronic, Berlin, Germany) at 50 W for 30 s (de Barros et al., 2018). Serial dilutions were prepared and 10 µL aliquots of the dilutions were plated on Sabouraud Dextrose agar supplemented with chloramphenicol. The plates were incubated at 37°C for 48 h and the number of colony-forming units (CFU/mL) was counted. Three biological replicates were performed, using five biofilms per group.
Biofilm Analysis by Biomass Quantification
Biofilm biomass was quantified by crystal violet (CV) staining as described by (Rossoni et al., 2018). After washing the wells, they were incubated for 15 min with 100 μL of 99% methanol (Sigma-Aldrich, St. Louis, MO, USA) followed by 20 min of 1% CV solution and washed with 0.85% NaCl. Bound CV was released by adding 150 μL of 99% methanol and the absorbance was measured at 570 nm (AJX-1900 spectrophotometer, Micronal, São Paulo, Brazil) to determine the total amount of biofilm dyed with CV. The experiments were performed three times, using five biofilms per group.
Biofilm Analysis by Scanning Electron Microscopy (SEM)
Biofilms formed on resin discs were fixed in 1 mL of 2.5% glutaraldehyde for 1 h (de Barros et al., 2018). Specimens were dehydrated in 100% ethanol (Sigma-Aldrich, St. Louis, MO, USA) for 20 min. The plates were kept in an incubator at 37°C for 24 h to permit total drying of the specimens. After drying, the specimens were transferred to aluminum stubs and sputter coated with gold for 160 s at 40 mA (Denton Vacuum Desk II, Denton Vacuum LLC, Moorestown, NJ, USA). Images were acquired using a JEOL JSM-5600 scanning electron microscope (JEOL USA, Inc., Peabody, MA, USA).
Expression Analysis of C. albicans Biofilm Genes
Biofilms formed in 24-well microtiter plates, as described in section 2.3, were used to assess transcription of C. albicans pathogenicity-associated genes (de Barros et al., 2017). After incubation, 1 mL TRIzol® (Ambion, Carlsbad, CA, USA) was added to each well to extract RNA according to the manufacturer’s protocol. RNA concentration was measured using a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). Total extracted RNA (700 ng) was treated with RQ1 RNase-Free DNase (Promega Corporation, Madison, WI, USA) and transcribed to cDNA using the GoScript™ Reverse Transcription Mix, Random Primers (Promega Corporation, Madison, WI, USA) according to the manufacturer’s recommendations. Primers for the genes analyzed are listed in Table 1. cDNA was amplified to determine the relative quantification of ALS1, ECE1, EPA1, HWP1, YWP1, BCR1, BGR1, CPH1, EFG1, NDT80, ROB1, TEC1, UME6, SAP2, SAP5, PBL2 and LIP9 expression. Three reference genes, ACT1, PMA1 and RIP1, were tested in all experimental groups. The results were analyzed at http://www.leonxie.com/referencegene.phpe, and the selected reference gene was RIP1 (data not shown).
Table 1.
Quantitative real-time PCR primers.
| Candida albicans | ||||
|---|---|---|---|---|
| Target gene | Oligonucleotide sequence 5’–3’* | Description | Amplicon size (bp)** | Reference |
| ACT1 |
F- GAAGCCCAATCCAAAAGA R- CTTCTGGAGCAACTCTCAATTC |
Structural constituent of cytoskeleton (Normalizing internal standard) |
130 bp | Nailis et al., 2010 |
| RIP1 |
F- TGTCACGGTTCCCATTATGATATTT R- TGGAATTTCCAAGTTCAATGGA |
Ubiquinol cytochrome-c reductase complex component (Normalizing internal standard) |
83 bp | Nailis et al., 2010 |
| PMA1 |
F- TTGCTTATGATAATGCTCCATACGA R- TACCCCACAATCTTGGCAAGT |
Plasma Membrane H (+) ATPase (Normalizing internal standard) |
66 bp | Nailis et al., 2010 |
| ALS1 |
F- CAACAGGCACCTCAGCATCTAC R- CTCCACCAGTAACAGATCCACTAGTAA |
Agglutinin-Like Sequence (cell adhesion molecule binding) |
88 bp | Nailis et al., 2010 |
| BCR1 |
F- AATGCCTGCAGGTTATTTGG R- TTTTAGGTGGTGGTGGCAAT |
Biofilm and Cell wall Regulator (transcription factor activity) |
112 bp | Hnisz et al., 2012 |
| BGR1 |
F- ACGATCAACCATTAGTGGAGG R- GAAGAAGTAGGTGTAGATGATCCAC |
Biofilm ReGulator (transcription factor activity) |
141 bp | Hnisz et al., 2012 |
| CPH1 |
F- ACGCAGCCACAAGCTCTACT R- GTTGTGTGTGGAGGTTGCAC |
Candida pseudohyphal regulator (transcription factor activity) |
119 bp | Sherry et al., 2014 |
| EFG1 |
F- CAGTATGGTCAGTATAATGCT R- TGTTGTTGCTGTTGGTATGGATATGATGATG |
Enhanced filamentous growth (transcription factor activity) |
222 bp | Hnisz et al., 2012 |
| ECE1 |
F- GTCGTCAGATTGCCAGAAATTG R- CTTGGCATTTTCGATGGATTGT |
Extent of Cell Elongation (Candidalysin) |
69 bp | Bassilana et al., 2005 |
| EPA1 |
F- ACCACCACCGGGTATACAAA R- GCCATCACATTTGGTGACAG |
Epithelial adhesion protein (adhesion) |
99 bp | Sherry et al., 2014 |
| HWP1 |
F- GAAACCTCACCAATTGCTCCAG R- GTAGAGACGACAGCACTAGATTCC |
Hyphal wall protein (cell adhesion molecule binding) |
92 bp | Hnisz et al., 2012 |
| NDT80 |
F-CTCAACAAGGCCCAACACCTC R- TTGACGTGGTTGTCTTGCTGG |
Meiosis-specific transcription factor | 121 bp | Hnisz et al., 2012 |
| ROB1 |
F- GAGGAATTTTACGCCCAACA R- TCTTGTGGTTGTGGTTCGTC |
Regulator of Biofilm | 130 bp | Hnisz et al., 2012 |
| TEC1 |
F- TGAGCAACAACAACAACAACCAC R- CTGGGTTGTTGTCATAGTGGCC |
TEA/ATTS transcription factor | 140 bp | Hnisz et al., 2012 |
| UME6 |
F- TCATTCAATCCTACTCGTCCACC R- CCAGATCCAGTAGCAGTGCTG |
Zn(II)2Cys6 transcription factor | 133 bp | Hnisz et al., 2012 |
| SAP2 |
F- GAATTAAGAATTAGTTTGGGTTCAGTTGA R-CCACAAGAACATCGACATTATCAGT |
Major secreted aspartyl proteinase | 122 bp | Nailis et al., 2010 |
| SAP5 |
F- CCAGCATCTTCCCGCACTT R- GCGTAAGAACCGTCACCATATTTAA |
Secreted Aspartyl Proteinase | 96 bp. | Nailis et al., 2010 |
| PLB2 |
F- TGAACCTTTGGGCGACAACT R- GCCGCGCTCGTTGTTAA |
PhosphoLipase B | 80 bp | Nailis et al., 2010 |
| LIP9 |
F- CGCAAGTTTGAAGTCAGGAAAA R- CCCACATTACAACTTTGGCATCT |
LIPase | 119 bp | Nailis et al., 2010 |
| YWP1 |
F- ACACCGGAAAATACCGTTGC R- ATGGCAGCTTTACCAGAACC |
Yeast-form wall protein (adhesion of symbiont to host) |
116 bp | Granger, 2012 |
| Galleria mellonella | ||||
| β -actin |
F- ACAGAGCGTGGCTACTCGTT R-GCCATCTCCTGCTCAAAGTC |
Structural constituent of cytoskeleton (Normalizing internal standard) |
104 bp | Rossoni et al., 2017 |
| Galiomicin |
F- TCCAGTCCGTTTTGTTGTTG R- CAGAGGTGTAATTCGTCGCA |
Antifungal peptide | 123 bp | Rossoni et al., 2017 |
| Gallerymicin |
F- GAAGATCGCTTTCATAGTCGC R- TACTCCTGCAGTTAGCAATGC |
Antifungal peptide | 175 bp | Bergin et al., 2006 |
| Mus Musculus | ||||
| GAPDH |
F- CGTCCCGTAGACAAAATGGT R- GAATTTGCCGTGAGTGGAGT |
Glyceraldehyde-3-phosphate dehydrogenase (Normalizing internal standard) |
177 bp | Tomalka et al., 2015 |
| β defesin3 |
F- GCATTGGCAACACTCGTCAGA R- CGGGATCTTGGTCTTCTCTATT |
Beta-defensin 3 | 85 bp | Tomalka et al., 2015 |
*F- Forward and R- Reverse.
**bp- base pairs.
For the qPCR reactions, a GoTaq® qPCR Master Mix kit (Promega Corporation, Madison, WI, USA) was used according to the manufacturer’s recommendations. The reactions were performed in triplicate in the StepOnePlus™ Real-Time PCR System (Applied Biosystems, Foster, CA, USA), consisting of 12.5 μL GoTaq® qPCR Master, 0.5 μL CXR, 0.4 μM primers (final concentration), and target cDNA, supplemented with RNAse-free ddH2O to a final volume of 25 μL. The cycling parameters for the amplification reactions were 95°C for 2 min; followed by 40 cycles of 95°C for 15 s and 60°C for 30 s. The level of gene expression was calculated by applying the 2-ΔΔCT method (Livak and Schmittgen, 2001).
In Vivo Study in G. mellonella Model
The antifungal and immunomodulatory effects of CAPE on G. mellonella model were assessed in larvae infected with the C. albicans 70 strain (resistant to fluconazole). For this study, the methodology described by Rossoni et al. (2019) was used with some modifications. G. mellonella in their final larval stage (250-350 mg) were stored in the dark and used within seven days from shipment. Groups inoculated with PBS and without any intervention (naïve) were included in the assays as controls.
G. mellonella larvae were infected with C. albicans and treated with CAPE 30 min after inoculation. At 24 h post-treatment, the experimental candidiasis was monitored by survival rate, hemocytes count, characterization of the hemocytes population by flow cytometry, fungal load in the hemolymph, and gene expression of antifungal peptides.
G. mellonella Survival
Initially, CAPE toxicity was evaluated. The compound was injected at varying concentrations (5-20 mg/kg) directly into G. mellonella hemolymph and monitored for 7 days to generate survival curves. To study the CAPE effects on candidiasis, larvae were infected with 5 x 107 cells/mL of C. albicans, and injected with CAPE (5 mg/kg) or fluconazole (5 mg/kg) after 30 min inoculation. Larvae were considered dead when they did not respond to touch. Sixteen larvae were used for each group.
G. mellonella Hemocyte Density
After 24 h of the treatments as described above, the hemolymph was collected into ice-cold sterile insect physiologic solution (IPS; 150 mM sodium chloride, 5 mM potassium chloride, 100 mM Tris hydrochloride, pH 6.9, with 10 mM EDTA, and 30 mM sodium citrate). After centrifugation, the supernatant was discarded, and the hemocytes were diluted in IPS medium and quantified using a hemocytometer. The results were averaged from four replicates.
Characterization of Hemocytes Sub-Populations in G. mellonella
The hemocytes sub-populations were analyzed by Fluorescence-Activated Cell Sorting (FACS). For this, hemolymph (150 mL) was extracted from larvae and diluted in ice cold PBS (800 μL) (Browne et al., 2015). Hemocytes were enumerated and the density was adjusted to 1 x 106 cells/mL. Cells were fixed in 4% formaldehyde (Sigma-Aldrich) in PBS for 10 min at 4°C. Hemocytes were washed in 1% BSA/PBS, collected through centrifugation at 1500 x g for 5 min at 4°C and resuspended in BSA/PBS at a density of 1 x 106 cells/mL. Cell populations were characterized using a FACSLyric™ flow cytometer (BD Biosciences) and differentiated based on side and forward scatter with a total of 10,000 events measured per sample using the BD FACSuit™ software. Hemocytes sub-populations were gated and analyzed using FlowJo™ Software.
Analysis of the Gene Expression of Antifungal Peptides in G. mellonella
The gene expression assay was performed as described by de Barros et al. (2019). Larvae were pulverized using a liquid nitrogen cooled mortar and pestle. RNA was extracted according to the manufacturer’s instruction using TRIzol (Ambion, Inc., Carlsbad, CA, USA). For cDNA preparation, the extracted total RNA (1000 ng) was transcribed applying reverse transcriptase and random primers according to the manufacturer’s instructions (GoScript Reverse Transcription Mix, Random Primers, Promega Corporation, Madison, WI, USA). Primers for the genes examined in this study were described in Table 1. The transcribed cDNAs were amplified for relative quantification of the expression of the genes encoding gallerimycin and galiomicin in relation to the concentration of the reference gene β-actin. Quantitative real-time PCR was conducted using the GoTaq qPCR Master Mix Kit (Promega Corporation, Madison, WI, USA) in the StepOnePlus™ apparatus (Applied Biosystems, Framingham, MA, USA). The level of gene expression was calculated by applying the 2-ΔΔCT method (Livak and Schmittgen, 2001).
Quantification of Candida in G. mellonella Hemolymph
For this study, the methodology described by de Barros et al. (2019) was used with some modifications. Fungal cells were extracted from G. mellonella hemolymph immediately after treatment (0 h) and 24 h after the larvae were treated with of CAPE or fluconazole. Five larvae were used in order to collect a pool of hemolymph. The experiment was performed in triplicate and a total of 15 larvae were used for each group and period. The extracted hemolymph was homogenized and aliquots of 0.1 mL of each dilution were transferred to Petri plates containing Sabouraud Dextrose agar supplemented with chloramphenicol. The plates were incubated for 48 h at 37°C and colonies were subsequently counted to calculate the CFU/mL numbers.
In Vivo Study in Murine OralCandidiasis Model
Adult 90-days-old male Swiss mice, weighing approximately 30g, from the Central Animal Care Facility of UNESP (Botucatu, SP, Brazil) were used in the study with approval from the Ethics Committee on the Use of Animals of the ICT/UNESP under protocol 019/2019-CEUA-ICT/UNESP. A total of 35 mice were divided into the following groups: mice not infected by C. albicans and untreated (n = 3) (control group), mice not infected by C. albicans and treated with CAPE (n = 8), mice infected by C. albicans and untreated (n = 8), mice infected by C. albicans and treated with nystatin (n = 8), and mice infected by C. albicans and treated with CAPE (n = 8). The control group was used as parameter to evaluate histological alterations and baseline expression of the genes investigated.
Oral candidiasis was induced as previously described by de Camargo Ribeiro et al. (2020). Animals were immunosuppressed using prednisolone (Depo-Medrol, Laboratórios Pfizer Ltda., Guarulhos, Brazil) on alternating days with Candida inoculations. To decrease normal oral bacteria, tetracycline hydrochloride (0.83mg/mL, Terramycin, Pfizer) was administered in the drinking water during the experiment period. For C. albicans infection, animals were anesthetized with intraperitoneal injections of ketamine (100 mg/kg) and xylazine (10 mg/kg) and inoculated with a C. albicans suspension (109 cells/mL) using a sterile swab. After 24 h of the second infection, 200 μL of the CAPE at a dose of 15 mg/kg or nystatin (40,000 IU/kg in 0.2 ml PBS) (Matsubara et al., 2012) were delivered into the oral cavity using a pipette.
Recovery of C. albicans From the Oral Cavity of Mice
After 48 h of treatment, saliva samples were recovered by mouthwash using 100 µL of sterile PBS. The samples were diluted and plated on Sabouraud Dextrose (SD) agar with chloramphenicol to determine the number of CFU/mL.
Macroscopic Analysis of Oral Candidiasis
After 48 h of treatment, euthanasia was performed and the dorsum tongue of the mice were analyzed on a stereomicroscopy (Zeiss, Göttingen, Germany) and scored between 0-4 for the presence of lesions. The scoring system was as follows: 0) absence of lesions; 1) white plaques on less than 20% of the tongue surface; 2) white plates covering 21-90% of the surface; 3) white plaques by more than 91%; 4) thick lesions with pseudomembrane in more than 91% (Takakura et al., 2003; de Camargo Ribeiro et al., 2020).
Microscopic Analysis of Oral Candidiasis
For the microscopic analysis, the tongues were removed, sagittally sectioned and fixed in 10% formalin for 24 h. After paraffin embedding, 5 μm serial sections were obtained, and stained with Hematoxylin-Eosin (HE) and Schiff’s Periodic Acid (PAS) according to laboratory standard protocols. On the histologic sections it was evaluated the presence of yeasts, hyphae and candidiasis lesions. The intensity of candidiasis lesions was evaluated following the study of Junqueira et al. (2005), considering epithelial alterations and inflammatory response of the conjunctive tissue.
Analysis of β-defensin 3 Gene Expression in the Tongue of Mice
The analysis of β-defensin 3 expression assay was performed as described by Tomalka et al. (2015). Frozen tongue was homogenized in 1 mL of TRIzol (Ambion, Inc., Carlsbad, CA, USA) in a porcelain mortar using a pestle and RNA extraction was made as recommended by the manufacturer. Total extracted RNA (1000 ng) was treated with RQ1 RNase-Free DNase (Promega Corporation, Madison, WI, USA) and transcribed to cDNA using the GoScript™ Reverse Transcription Mix, Random Primers (Promega Corporation, Madison, WI, USA). Relative quantification of gene expression (β-defensin 3) was determined by real-time PCR with SYBR green (GoTaq qPCR Master Mix Kit, Promega Corporation, Madison, WI, USA) normalized to GAPDH. These primers were described in Table 1. The level of gene expression was calculated by applying the 2-ΔΔCT method (Livak and Schmittgen, 2001).
Statistical Analysis
For statistical analysis level of significance of 5% was adopted. Analysis of variance (ANOVA), Tukey test, Student’s t-test, and Kruskal-Wallis were used depending on data distribution and groups, for each experimental assay. All analysis were performed using the GraphPad Prism 6 Program (GraphPad Software, Inc., La Jolla, CA, USA).
Results
Antifungal Activity of CAPE on C. albicans Clinical Strains
The antifungal effects of CAPE were evaluated on C. albicans growth by the minimum inhibitory concentration (MIC). Among the 40 clinical strains of C. albicans tested, 5 (12.5%) and 8 (20%) showed resistance to amphotericin B and fluconazole, respectively (Table 2). These strains showed susceptibility profile ranging from 0.0625 to 2 µg/mL for amphotericin B and 0.5 to ≥ 64 µg/mL for fluconazole. The overall susceptibility to CAPE ranged from 16 to 64 µg/mL (Table 2). C. albicans strains (CA70 and CA14) with resistance to fluconazole were selected for subsequent assays.
Table 2.
Minimum Inhibitory Concentration (MIC) of CAPE against Candida albicans isolates.
| Amphotericin B (µg/mL) | Fluconazole (µg/mL) | CAPE (µg/mL) | |
|---|---|---|---|
| C. albicans ATCC 18804 | 0.125 | 2 | 32 |
| C. albicans SC5314 | 0.0625 | 1 | 32 |
| C. albicans 3 | 0.5 | 1 | 64 |
| C. albicans 5 | 0.5 | 4 | 64 |
| C. albicans 14 | 1 | 64 | 32 |
| C. albicans 17 | 0.25 | 32 | 32 |
| C. albicans 21 | 1 | 0.5 | 64 |
| C. albicans 23 | 0.25 | 1 | 32 |
| C. albicans 26 | 0.0625 | 1 | 32 |
| C. albicans 28 | 1 | 1 | 64 |
| C. albicans 31 | 0.25 | 0.5 | 16 |
| C. albicans 38 | 1 | 1 | 64 |
| C. albicans 40 | 0.25 | 2 | >64 |
| C. albicans 52 | 1 | 1 | 64 |
| C. albicans 53 | 0.25 | 4 | 64 |
| C. albicans 60 | 1 | 1 | 16 |
| C. albicans 61 | 0.25 | 16 | 32 |
| C. albicans 70 | 0.625 | >64 | 16 |
| C. albicans 73 | 0.625 | >64 | 64 |
| C. albicans 4S | 2 | 8 | 16 |
| C. albicans 24S | 1 | 2 | 64 |
| C. albicans 31S | 0.5 | 4 | 32 |
| C. albicans 36S | 1 | 2 | 32 |
| C. albicans 39S | 0.5 | 16 | 32 |
| C. albicans 57S | 2 | 2 | 16 |
| C. albicans 65S | 1 | 1 | 32 |
| C. albicans 66S | 2 | 2 | 16 |
| C. albicans 69S | 0.25 | 1 | 32 |
| C. albicans 75S | 2 | 4 | 32 |
| C. albicans 97S | 1 | 32 | 32 |
| C. albicans 109S | 0.25 | 1 | 64 |
| C. albicans 110S | 0.5 | 2 | 32 |
| C. albicans 115S | 0.25 | 4 | 32 |
| C. albicans 137S | 1 | 0.5 | 32 |
| C. albicans 160S | 0.625 | 2 | 16 |
| C. albicans 165S | 0.5 | 4 | 16 |
| C. albicans 193S | 1 | 1 | 64 |
| C. albicans 194S | 0.25 | 1 | 16 |
| C. albicans 197S | 1 | 1 | 64 |
| C. albicans 205S | 2 | 0.5 | 16 |
| C. albicans 228S | 1 | 0.5 | 32 |
| C. albicans 230S | 0.5 | 1 | 64 |
| C. parapsilosis ATCC 22019 | 0.25 | 1 | 32 |
Effects of CAPE on C. albicans Biofilms
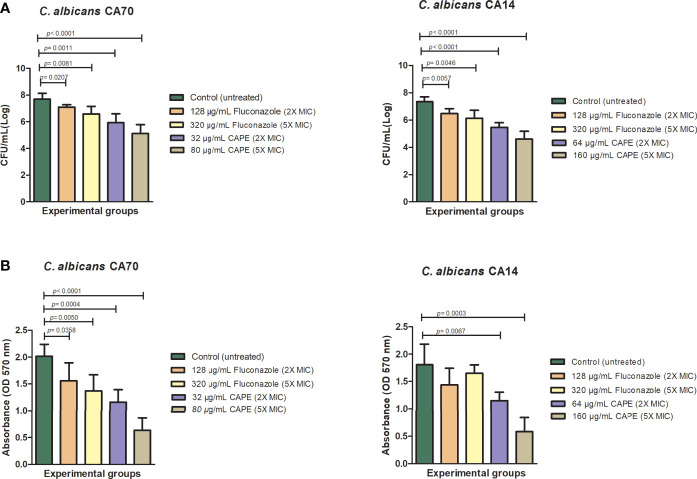
Next, we investigated the activity of CAPE on C. albicans biofilms since this virulene factor has an important role in the recurrent and chronic infections. CAPE at 2 x MIC (32 µg/mL for CA70 and 64 µg/mL for CA14) and 5 x MIC (80 µg/mL for CA70 and 160 µg/mL for CA14) concentrations significantly reduced the viable cells number (p<0.0001) and the quantity of total biomass (p<0.0001) for both C. albicans strains. The higher fungal CFU/mL reductions observed corresponded to 2.57 and 2.74 log respectively for the strains CA70 and CA14, when the biofilms were treated with CAPE at 5 x MIC in relation to the untreated group. Significant reductions were also found in the treatment with fluconazole, however this antifungal was used in concentration of 320 µg/mL, which was 2 to 4 times higher than the CAPE concentrations (Figure 1A). In relation to the biofilm biomass, the treatment with CAPE and fluconazole at 5 x MIC caused a reduction of 68.5 and 32%, respectively, in relation to the untreated control group (Figure 1B ).
Figure 1.
Analysis of in vitro C. albicans biofilms. Mature biofilms (48 h) formed in 96-wells plates for clinical strains of fluconazole-resistant C. albicans were treated with different concentrations of fluconazole and CAPE (2 x MIC and 5 x MIC). (A) The viable C. albicans cells number are shown in CFU per milliliter count. (B) Biofilm biomass analysis expressed in optical density (570nm). The treatments were compared to the control group using Student’s t test (p < 0.05).
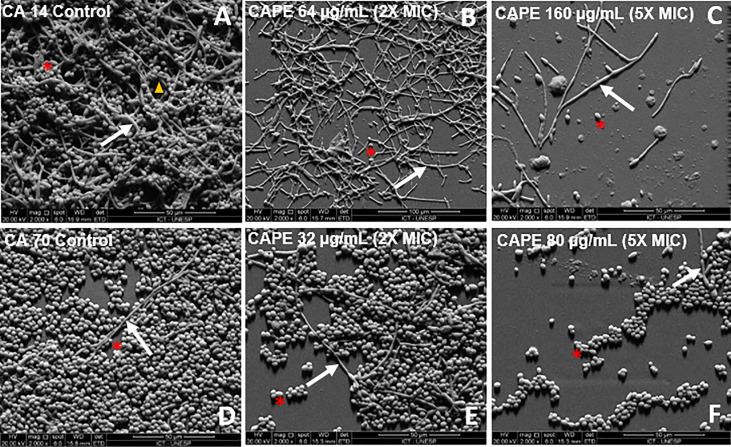
The effects of CAPE on C. albicans biofilms were visually confirmed by SEM. The SEM images revealed a dense biofilm, formed by several layers of hyphae associated with yeasts in the untreated control group (Figures 2A, D). However, the CAPE-treated groups exhibited biofilms formed by a single dispersed layer with fewer yeasts and hyphae (Figures 2B, C, E, F). These observations are consistent with the inhibition of biofilm observed in the viable cells count and total biomass assays.
Figure 2.
(A–F) Scanning electron microscopy images of biofilms formed in vitro on resin surfaces, as follows: biofilms formed by only C. albicans (CA14) (A) and CA70 (D), cells treated with CAPE (2 x MIC) (B, E), cells treated with CAPE (5 x MIC) (C, F). It is possible to observe the presence of channel water (→), yeasts (*), and hyphae (▲) of C. albicans. Original magnification 2000x.
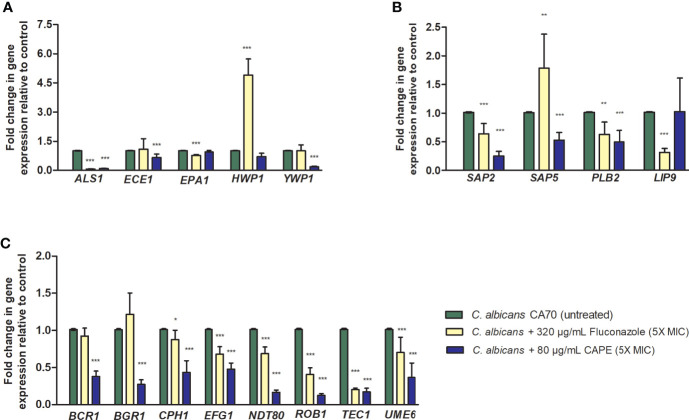
We also studied whether the inhibitory effect of CAPE on biofilm is related to changes in gene expression. To this end, the RNA was extracted from untreated and CAPE-treated biofilms and analyzed by real-time qPCR. Data obtained from real-time qPCR revealed a variety of C. albicans genes whose expression was affected differently upon CAPE treatment. CAPE at 5 x MIC downregulated the expression of genes involved in maintenance, development, and maturation of biofilms, such as transcriptional regulators: BCR1, BGR1, CPH1, NDT80, ROB1 and TEC1 (2.6, 3.7, 2.32, 6.25, 8.33, 5.88-fold); adhesion: ALS1, EPA1 and YWP1 (11.1, 1.06, 5.55-fold); and filamentation: ECE1, HWP1, EFG1 and UME6 (1.53, 1.4, 2.08, 2.78-fold) when compared to control (Figure 3). Importantly, genes involved in C. albicans host invasion (SAP2, SAP5, PBL2 and LIP9) were also downregulated 4.0, 1.92, 2.04, 0.98-fold, respectively, compared to the control group (p < 0.0001) (Figure 3). Conversely, HWP1 and SAP5 were upregulated with fluconazole treatment, increasing expression by 4.9 and 1.92-fold, respectively (p < 0.001) (Figure 3).
Figure 3.
Relative expression of C. albicans genes. (A) Relative quantification of ALS1, ECE1, EPA1, HWP1, YWP1 genes for the non-treated control group (PBS) and treated groups with fluconazole and CAPE (5 x MIC). (B) Relative quantification of SAP2, SAP5, PBL2 and LIP9 genes. (C) Relative quantification of BCR1, BGR1, CPH1, EFG1, NDT80, ROB1, TEC1 and UME6 genes. Normalization was done using RIP1 gene and values expressed as the mean and SD. The treatments were compared to the control group using Student’s t-test (*p < 0.05, **p < 0.01 and ***p < 0.0001).
Effects of CAPE on Experimental Candidiasis in G. mellonella Model
Initially, the G. mellonella model was used to evaluate the toxicity of CAPE in single doses of 5, 10, 15 and 20 mg/kg. CAPE did not exert toxic effects on the larvae at the tested concentrations (Figure 4A). Next, G. mellonella model was used to study the efficacy of treatment with CAPE in larvae infected by C. albicans. In the untreated group, infection with C. albicans caused death in 100% of the larvae within 3 days. When the larvae were treated with CAPE (5 mg/kg) after C. albicans infection, the survival rate of G. mellonella larvae was significantly increased by 44.5% (p < 0.05). Conversely, treatment with fluconazole at a dose of 5 mg/kg killed 100% of the larvae within 6 days (Figure 4B). These results indicated that CAPE had in vivo efficacy in treating C. albicans infections without toxicity to the host at the delivered therapeutics concentration.
Figure 4.
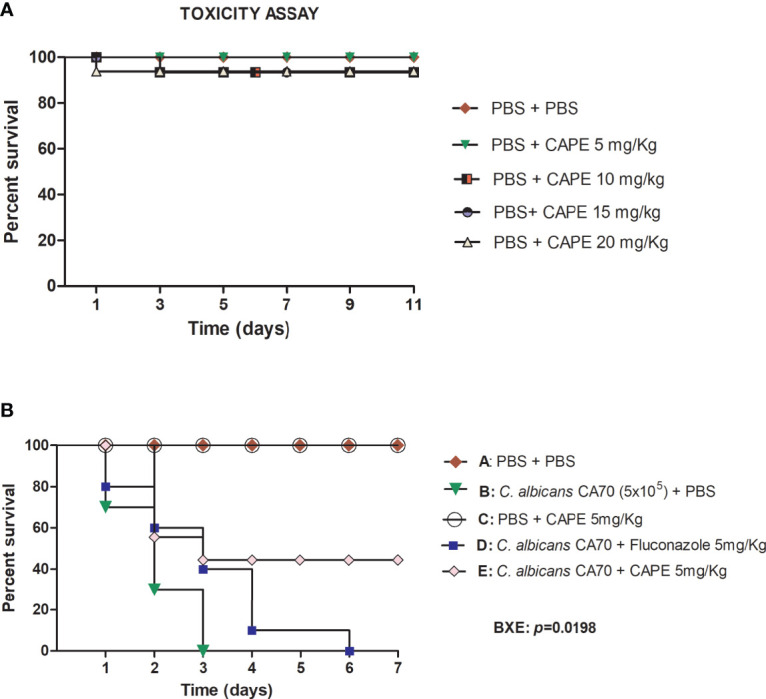
(A) Toxicity evaluation of CAPE in G. mellonella model. G. mellonella larvae were injected with serial concentrations of CAPE and no death was observed at the concentrations used. (B) CAPE prolonged the survival of G. mellonella larvae infected with C. albicans (CA70). Significant differences were observed in survival between the “CA70 + CAPE (5 mg/Kg) group” and “CA70 + PBS group”. Kaplan-Meier test, p ≤ 0.05.
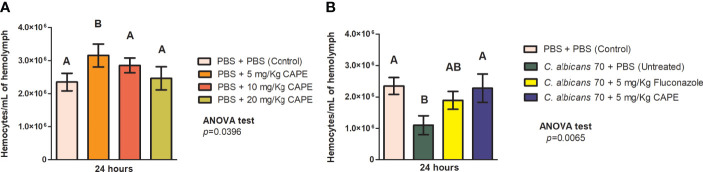
To investigate the effects of CAPE on immune system of G. mellonella larvae, we evaluated the hemocytes quantity in the hemolymph of G. mellonella with or without Candida infection. Analyzing the larvae not infected by C. albicans, we observed a significant increase in the number of hemocytes in the group treated with CAPE at 5 mg/kg (1.3-fold increase) compared to the PBS control group (Figure 5A). When the larvae were infected by C. albicans and untreated, we found hemocytes reductions (2.2-fold decrease) compared to the control group. Larvae infected and treated with CAPE exhibited an increase in the number of hemocytes compared to the infected and untreated group. The treatment with fluconazole also led to an increase in the hemocytes quantity (1.71-fold increase) although it was less pronounced than the treatment with CAPE (2.07-fold increase) (Figure 5B). These data suggested that CAPE can influence the immune system, acting as an immunomodulatory natural compound.
Figure 5.
CAPE modulates the immune system of G. mellonella. (A) G. mellonella larvae were injected with serial concentrations of CAPE increased the hemocyte number compared to a PBS control (PBS + PBS). (B) The group of CA70 + CAPE (5 mg/Kg) increased significantly the hemocyte number compared to CA70 + PBS (untreated). CA70 + PBS group showed a reduction of hemocyte quantity in relation to the PBS control group, but when the larvae were treated with CAPE, the hemocyte quantity was very similar to the values found in the PBS control group. The number of hemocytes slightly increased in the group of CA70 + fluconazole (5 mg/Kg) but did not show differences in relation to the CA70 + PBS group. Different letters A and B represent statistically significant differences among the groups. ANOVA and Tukey Tests (p < 0.05 was considered significant).
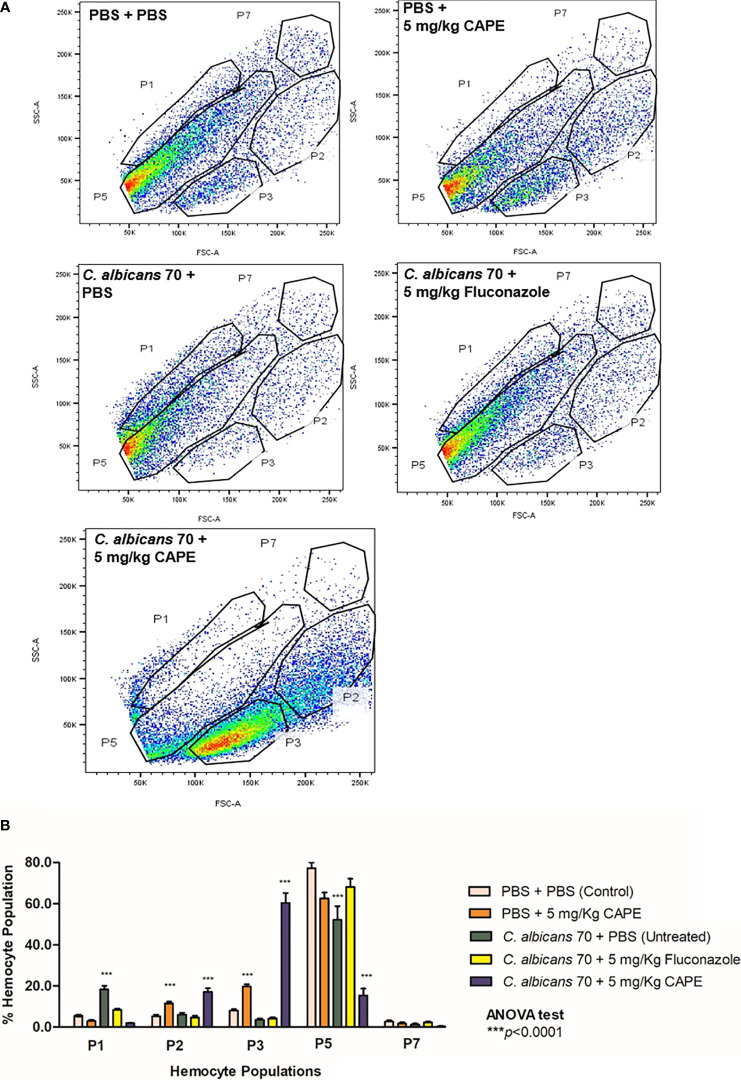
To explore the immunomodulatory action of CAPE on the cellular immune response of G. mellonella, we characterized the hemocyte population by flow cytometry. The hemocyte population was differentiated based on the size and granularity of cells, and at least 5 distinct sub-populations, labeled P1, P2, P3, P5 and P7, were visible (Figure 6A). Larvae infected with C. albicans showed a significant increase (3.33- fold) in the relative abundance of P1 hemocytes (small and granular cells) compared to control group (PBS +PBS) (p < 0.0001). There was a significant increase in hemocyte P5 (very large and nongranular cells) in all groups, except in larvae infected and treated by CAPE. In relation to P7 sub-population, the relative abundance was similar among the groups studied. Interestingly, the sub-populations of P2 and P3 (medium size and granular cells), that are considered important phagocytic cells against pathogens, were significantly increased by the treatment with CAPE in both infected and not infected larvae. When CAPE was administered in G. mellonella larvae not infected by C. albicans, the P2 and P3 sub-populations increased by 2.2 and 2.47 -fold, respectively, in comparison to control group. In larvae infected by C. albicans, the treatment with CAPE reached greater increases in P2 and P3 sub-populations with 2.85 and 17.20-fold, respectively, in relation to infected larvae and untreated (Figure 6B). Therefore, CAPE showed a promising action for recruiting phagocytic cells against C. albicans.
Figure 6.
FACS analysis of larval hemocyte population. (A) Hemocytes were extracted from larvae and differentiated by FACS based on cell size (x-axis) and granularity (y-axis). Representative images of hemocytes in the P1, P2, P3, P5, and P7 sub-populations are presented in each group. (B) Fluctuations in hemocyte sub-populations in larvae for the control group (PBS), group infected only CA70 (untreated) and groups infected and treated with fluconazole and CAPE (5 mg/Kg). The relative proportion of hemocyte sub-populations in larvae was measured by FACS analysis (p is relative to the hemocyte sub-population in the control (PBS) in relation to others groups). ANOVA test (***p < 0.0001).
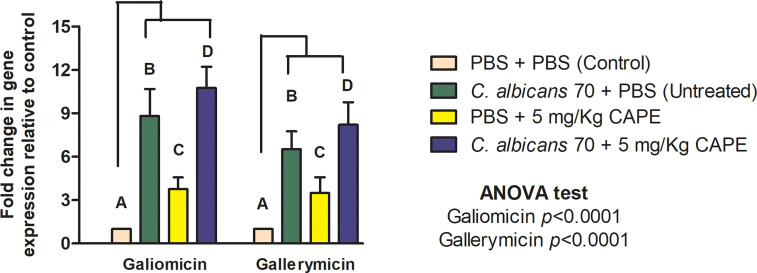
Subsequently, we investigated the immunomodulatory effects of CAPE on humoral immune response of G. mellonella by analyzing the gene expression of antifungal peptides (galiomycin and galeriomycin). We found that CAPE was able to increase the expression of both galiomicin and galeriomycin peptides. The group infected with C. albicans and treated with CAPE had a statistically significant increase (p < 0.0001) in relation to the group infected and untreated (galiomicin: 1.93-fold increase; galeriomycin: 1.7-fold increase). CAPE induced an increase in the gene expression of galiomycin and galleriomycin by 3.75 and 3.48-fold, respectively, compared to the control group formed by consecutive PBS injections (Figure 7).
Figure 7.
CAPE increased the expression of antimicrobial peptides genes of G. mellonella at 24h after treatment of CAPE. Relative quantification (log) of galiomicin and gallerimycin for the groups inoculated with PBS (Control), infected with CA70 only (untreated), treated with CAPE, and infected and treated with CAPE [CA70 + CAPE (5 mg/Kg)]. Gene expression was represented as mean and SD. Normalization was done using β-actin gene. Different letters A, B, C and D represent statistically significant differences among the groups. ANOVA and Tukey Tests (p < 0.05).
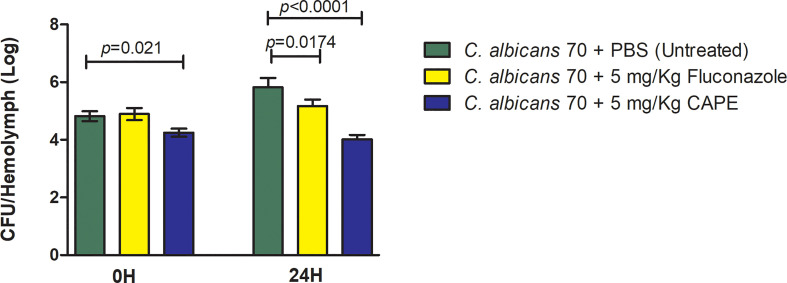
Finally, the antifungal effect of CAPE on experimental candidiasis in G. mellonella model was evaluated by the fungal load analysis in the hemolymph. The immediate action of CAPE on infected larvae caused a reduction of 0.58 Log in the CFU/mL in the group infected and untreated (p = 0.021). After 24 h, the CAPE treatment was more effective, further reducing the fungal load by 1.80 log compared to the infected and untreated (p < 0.0001). In the treatment with fluconazole, only slight reductions in the fungal load were obtained in comparison to the control in 24 h (Figure 8). Based on the results obtained in the studies with G. mellonella model, CAPE proved to be effective to treat experimental candidiasis by both antifungal activity and immunomodulatory action.
Figure 8.
CAPE decreased the number of fungal cells in G. mellonella hemolymph. Mean and SD of C. albicans counts (CFU/larvae) in the hemolymph of G. mellonella at 0 h and 24 h after treatment of CAPE. The following groups were compared at each time of infection: CA70 + PBS (untreated), CA70 + fluconazole and CA70 + CAPE (5 mg/Kg). Treatment with CAPE was able to significantly reduce fungal burden in times evaluated, and fluconazole only after 24 hours. Student’s t-test, p < 0.05.
Evaluation of the Treatment With CAPE on Oral Candidiasis in Mice
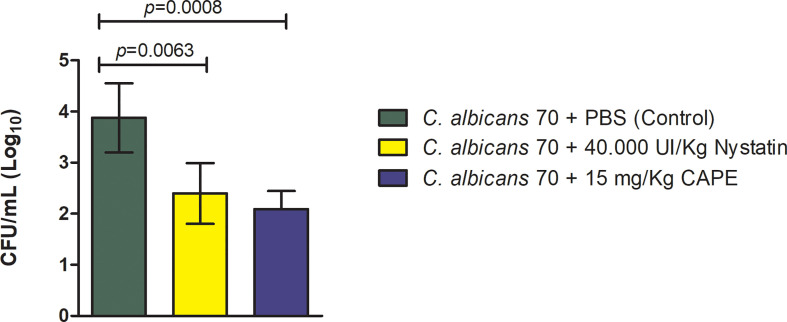
With antifungal and immunomodulatory CAPE activity demonstrated in the results in G. mellonella model, we employed a more refined model of oral candidiasis in mice to continue our investigation of CAPE treatment. C. albicans cells were recovered from the oral cavity of all infected mice. The CFU/mL counts were 3.97 ± 0.67 (Log10) for the infected and untreated group, 2.39 ± 0.59 (Log10) for the group treated with nystatin and 2.0 ± 0.35 (Log10) for the group treated with CAPE. Both treatments were capable of reducing C. albicans colonization with a statistically significant difference in relation to the untreated group (Figure 9).
Figure 9.
Recovery of C. albicans from the oral cavity of mice. C. albicans CFU/mL recovered from the oral cavity of mice infected by CA70 and treated with PBS (control group), with nystatin and CAPE (Student’s t test, p < 0.05).
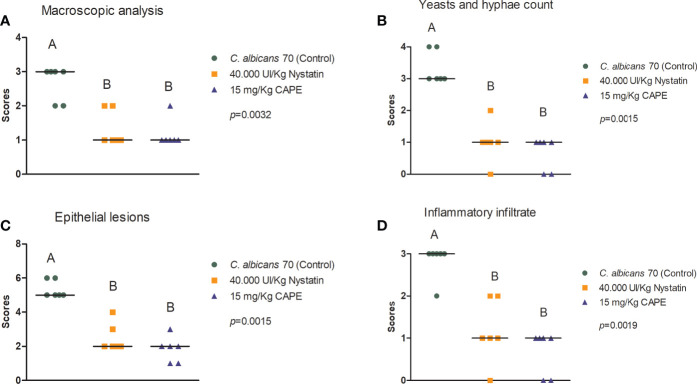
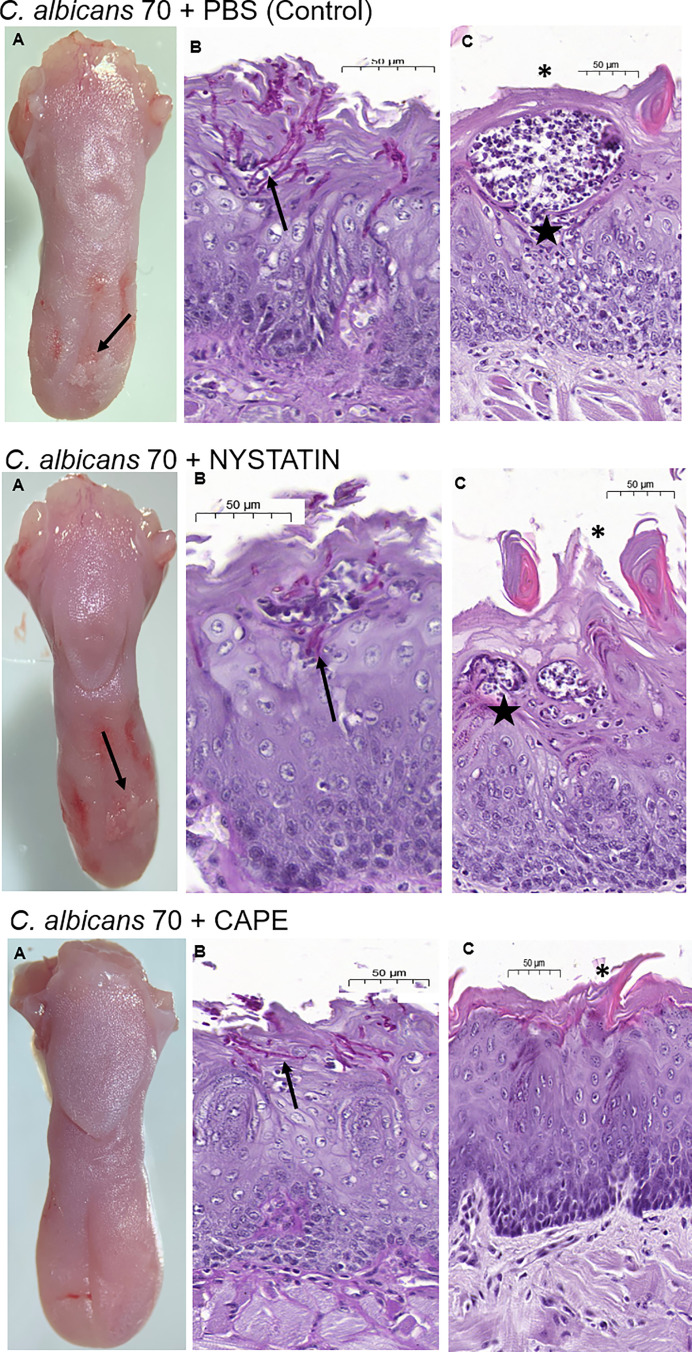
Macroscopic analysis of the dorsum tongue revealed the formation of candidiasis lesions characterized by whitish regions with the presence of pseudomembrane. Promisingly, these lesions were significantly reduced in the groups treated with nystatin or CAPE compared to infected and untreated mice (Figure 10A). These findings were confirmed in the microscopic analysis, in which the group treated with CAPE or nystatin presented less quantity of yeasts and hyphae in the keratin layer (Figure 10B), of epithelial lesions such as microabscesses, exocytosis, spongiosis and loss of filiform papillae (Figure 10C), and of inflammatory infiltrate than infected and untreated group (Figure 10D). In general, CAPE was capable of decreasing the macroscopic and microscopic lesions of oral candidiasis (Figure 11).
Figure 10.
Macroscopic and histological assessment of candidiasis lesions formed on mice’s dorsum tongue. Mice were treated with PBS (control group), nystatin or CAPE (A–D). Scores and median of macroscopic lesions quantification (A), Yeasts count and hyphae in PAS stained sections (B), and epithelial lesions (C) and inflammatory infiltrate (D) determined in H&E slides (Kruskal-Wallis and Dunn’s posttest; statistically significant differences between groups are represented by different letters).
Figure 11.
Analysis of the dorsum tongue of mice infected with C. albicans. The mice were treated with PBS (control group, top row), nystatin (middle row) and CAPE (bottom row). (A) White patches of candidiasis (→) can be observed in the macroscopic analysis of the untreated group and the one treated with nystatin. (B) PAS sections, showing hyphae and yeast in the epithelium-keratinized layer (→). (C) Images of histological cuts stained by H&E, where intraepithelial microabscesses (☆) and loss of filiform papillae were observed (*).
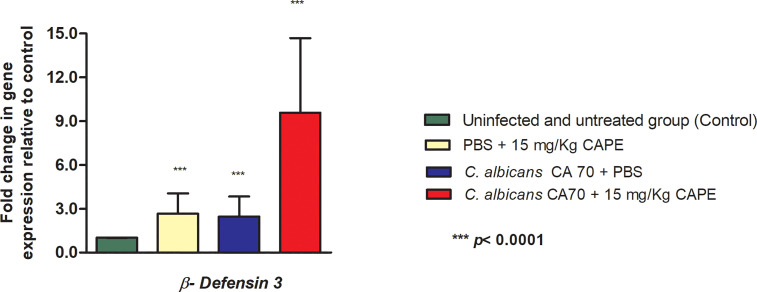
The immunomodulatory effect of CAPE was also evaluated in the oral cavity of mice by gene expression analysis of β-defensin 3. In experimental candidiasis in mice treated with CAPE, this gene showed an increase of 3.91-fold compared to the infected and untreated group. Furthermore, uninfected and CAPE-treated mice, the expression of the β-defensin 3 gene was increased by 2.66-fold compared to the control group (uninfected and untreated) (Figure 12).
Figure 12.
CAPE increased the expression of β-defensin 3 gene. Relative quantification (log) of β-defensin 3 gene for the uninfected and untreated group (control), infected with CA70 only, treated with CAPE, and infected with CA70 + CAPE (15 mg/Kg). Gene expression was represented as mean and SD. Normalization was done using β-actin gene. ANOVA and Tukey Test (***p < 0.0001).
Taken together, the study using the oral candidiasis mouse model demonstrated that CAPE was able to reduce the oral colonization by C. albicans, to decrease macroscopic and microscopic of candidiasis lesions, and to stimulate the innate defense mechanism associated to β-defensins.
Discussion
In this study, we investigated the antifungal action of CAPE as a new natural source for the development of therapeutic agents. Mature biofilm was disrupted by the presence of CAPE. Interestingly, the most pronounced effects were observed when CAPE exerted immunomodulatory protection against C. albicans in two in vivo infection models: G. mellonella and oropharyngeal candidiasis of immunosuppressed mice. Delivery of the natural compound was found to elevate antifungal peptide expression in the G. mellonella model that resulted in prolonged survival. In the murine oral candidiasis model, a significant reduction in fungal cells at the infection site was observed that resulted in reduced lesions and tissue inflammation.
In planktonic assays to determine the antifungal activity of CAPE, we found MIC results ranged from 16 to 64 μg/mL among the 40 clinical C. albicans isolates from oral candidiasis. Although inhibition was not as efficacious as standard therapeutic agents, interestingly, CAPE was active against both fluconazole-sensitive and fluconazole-resistant strains, in agreement with some studies that found the same range of MIC (Breger et al., 2007; Coleman et al., 2016; Sun et al., 2018). Sun et al. (2018) found the same MIC values for CAPE (32 to 64 μg/mL) when testing clinical isolates of C. albicans, sensitive and resistant to fluconazole, from Shandong Provincial Hospital Qianfoshan. They observed a synergically action of CAPE and fluconazole against resistant clinical isolates of C. albicans. Additionally, the CAPE-Fluconazole combination increased the longevity and decreased fungal burden in C. elegans when compared with the compounds alone. Some limitations of our study is related to susceptibility of these strains to other antifungals, including echinocandins. In addition, the MIC values for fluconazole, amphotericin B and CAPE that inhibited 50% of growth have not been evaluated.
Since C. albicans can be found in both planktonic and biofilm forms during infection, the antifungal effects of CAPE were tested on C. albicans biofilms. The elevate resistance to antifungal drugs, the ability to withstand host immune defenses, and the role as a reservoir for continuing infections, makes Candida biofilms a crucial factor for the increase morbidity and mortality of patients with C. albicans infections (Liu et al., 2017). In our study, biofilms treated with CAPE at a concentration of 5 x MIC showed significant reductions in the viable cell count (CFU/mL) and in the total biomass. Biofilms treated with CAPE reached reductions of up to 68.5% in the amount of total biomass in relation to the untreated control group. Likewise, some studies have shown the antifungal effect of CAPE on C. albicans biofilms as an alternative in the treatment or prevention of this infection (De Vita et al., 2014; Freires et al., 2016). The reduction in the biomass of the biofilm obtained in this study were superior to those found by (Breger et al., 2007), who tested the effects of CAPE on mature biofilms formed by the C. albicans DAY185 in silicone squares. They found a 50% reduction in the dry weight of CAPE-treated biofilms compared to untreated control.
Although fluconazole has few side effects, its extensive application has led to the frequent emergence of resistance compromised the successful treatment of oral candidiasis. Treatment with fluconazole at concentration of 5x MIC (320 μg/mL) was not so effective as CAPE in reducing the total biomass of C. albicans biofilms (only 32% of reduction). This result was expected since the C. albicans strain employed in our study showed resistance to fluconazole in MIC assays. Several studies have reported low susceptibility of biofilm-producing C. albicans strains to fluconazole, a front-line antifungal drug for the treatment of oral candidiasis (Mukherjee et al., 2003; Uppuluri et al., 2008; Jia et al., 2016; Lu et al., 2019). Fluconazole showed an insignificant inhibition (< 20%) of C. albicans biofilms at concentration of 1024 μg/mL (Jia et al., 2016).
Microscopic images confirmed CAPE impacted C. albicans biofilm, reducing the number of recovered fungal cells and overall biomass. Importantly, this analysis showed that treatment with CAPE inhibited the hyphae formation that is another crucial virulence factor of C. albicans species. Here, we also analyzed the gene expressions of C. albicans in biofilms employing several genes associated with different virulence mechanisms. Genes related to surface adhesins, such as ALS1 (agglutinin-like sequence) and YWP1 (yeast cell wall protein) were suppressed by CAPE treatment in biofilms. It is known that the agglutinin-like sequence (ALS) genes play an important role in C. albicans adhesion and biofilm formation on biotic and abiotic surfaces (Hosseini et al., 2019; Jordão et al., 2021). YWP1 might be involved in C. albicans yeast dispersion, once it was found that YWP1-deficient blastoconidia exhibited increase adhesiveness and biofilm formation (Granger et al., 2005). Therefore, the decrease in the expression of ALS1 and YWP1 represent a promising action of CAPE in control to prevent C. albicans biofilm formations.
Gene expression analysis revealed a decrease in the expression of an integrated network of transcription factors (BCR1, BGR1, CPH1, EFG1, NDT80, ROB1, TEC1 and UME6) that play a role in regulating biofilm-related processes. Nobile et al. (2012) have provided a portrait of a six-transcription factors network (BRG1, NDT80, ROB1, TEC1, BCR1 and EFG1) involved in a variety of phenotypes with relevance to C. albicans biology and pathobiology. Their data indicated that the transcriptional circuitry for biofilm formation is highly integrated and regulates the expression of key biofilm effector genes. Apart from biofilm formation, the secreted virulence factors of C. albicans, such as proteases and phospholipases, contribute to elevate its pathogenicity. These hydrolases degrade surface membrane proteins, thereby facilitating the invasion of C. albicans into host tissue (Mayer et al., 2013). Overall, our results indicate that CAPE treatment may affect biofilm formation by reducing the levels of adhesins and by restricting the long-term maintenance of biofilm through transcriptional regulator factors. In addition, CAPE can impair C. albicans invasion into host tissues by inhibiting the expression of proteases and phospholipases. Thereby CAPE exerts its anti-biofilm activity towards C. albicans biofilm through a multi-target mode of action, which differs from commonly used antifungals.
Assessment of the impact of CAPE during an active infection using in vivo studies immunomodulatory effects. G. mellonella is a widely studied infection model, which provides the possibility of evaluating the efficacy of new antimicrobial compounds and immune host responses. G. mellonella larvae are easy to use, inexpensive to purchase, and free from the legal/ethical restriction that are applied to the use of mammals (Kavanagh and Sheehan, 2018; Pereira et al., 2018). The use of G. mellonella larvae in this capacity will not completely replace the need to use of mammalian models in this role, but judicious use of larvae can accelerate the identification of potential therapeutic doses for use in mammals and allow the rapid initial evaluation of antifungal agents (Kavanagh and Sheehan, 2018). In addition, the larvae immune system presents structural and functional similarities to the innate immune response of mammals, which are composed of cellular (hemocytes) and humoral defenses (antimicrobial peptides) (Pereira et al., 2018; de Barros et al., 2019). The cellular branch of G. mellonella immunity is mediated by hemocytes. G. mellonella possesses five types of hemocytes: prohemocytes, granulocytes, plasmatocytes, oenocytoids and spherulocytes (Browne et al., 2015; Wojda et al., 2020). To the best of our knowledge, this is the first article in the literature that investigated the anti-C. albicans properties of CAPE using G. mellonella. We found that the treatment with CAPE in G. mellonella larvae infected by C. albicans increased the survival rate by killing C. albicans cells in hemolymph and also by stimulating the immune system.
The number of hemocytes was increased after treatment with CAPE, mainly the P2 and P3 subpopulations that are granular cells with function in phagocyting pathogens (Araújo et al., 2008; Browne et al., 2015). The increase relative proportion of P2 and P3 hemocytes with the CAPE treatment corresponded to a decrease in the proportion of P5 hemocytes. P5 hemocytes demonstrate globular inclusions and resembled adipohemocytes, which stores energy in the form of lipids and glycogen (Araújo et al., 2008; Browne et al., 2015). The reduction in P5 cells in larval hemocyte population treated with CAPE may be an indication that energy reserves in the form of lipids were used to maintain their survival against infection by C. albicans (Browne et al., 2015).
In addition to modulating hemocyte responses, antifungal peptide expression was also altered. The immune response of G. mellonella against pathogens is comprised of a range of antimicrobial peptides (AMPs), which are produced according to the aggressor agent type. Thus, we further explored alterations in the immune response examining the expression of antifungal peptides such as galiomicin, a defensin, and gallerymicin, a cysteine-rich peptide. We observed an increased expression of both AMPs, indicating that CAPE also modulated the humoral immune response of G. mellonella larvae, which may have contributed to protect larvae from C. albicans infection. Taken together, these findings indicate that CAPE is capable of stimulating the cellular and humoral immune responses of the larvae and consequently prevent C. albicans infection.
Finally, the effects of CAPE were evaluated in an oral candidiasis murine model which mimics the ones observed in the human oral cavity, such as salivary flow, pH variations, the presence of teeth, mucous membranes characteristics and immune response (Sparber and LeibundGut-Landmann, 2015; Borman, 2018). We verified that the treatment with CAPE in mice infected by C. albicans was able to reduce the fungal burden, hyphae invasion and inflammation in the oral tissues compared to untreated animals. A single treatment with CAPE was able to reduce the fungal load in the mouth of mice with oral candidiasis by 1.97 log (CFU/mL).
To investigate the possible mechanisms related to the preventive effects of CAPE on oral candidiasis in mice, we analyzed the gene expression of antimicrobial peptides with local action in the immune defense of oral cavity. Epithelial cells of mice release small anti-microbial peptides (AMPs), especially in saliva, which are capable of exerting direct effects against pathogens, including the family of defensins (Tomalka et al., 2015). Certain murine defensins have shown antifungal properties, such as β-defensin 3 (mBD3), which induces perforations in the fungal wall, and consequently, cell lysis (Jiang et al., 2012; Tomalka et al., 2015). The mBD3 is essential to prevent oropharyngeal candidiasis in mice (Conti et al., 2016), although it has no direct ortholog in humans (Ganz, 2003; Zhou et al., 2020). In this context, CAPE treatment exhibited potent anti-C. albicans activity in oral cavity of mice, as it was able to stimulate the expression of the β-defensin 3 gene, favoring the reduction of candidiasis lesions.
In summary, CAPE exhibited strong antifungal activity against a large number of C. albicans oral cavity isolates, including drug resistant strains. CAPE also showed activity against C. albicans biofilms, leading to inhibition of viable cells, decrease of the total biomass, reduction of filamentation and downregulation of genes associated with adherence, transcriptional factors and enzymes production. In in vivo study, CAPE protected G. mellonella larvae from candidiasis due to both antifungal and immunomodulatory properties. The treatment with CAPE increased the larvae survival by killing the fungal cells and by stimulating the humoral and cellular immune responses. In addition, CAPE was able to decrease oral candidiasis in mice with significant reductions of the macroscopic lesions, penetration of hyphae into oral tissues, epithelial damages and inflammatory infiltrate, as well as with an increase in the expression of β-defensin 3 gene. The treatment with CAPE in G. mellonella and mouse models was not harmful to the hosts. Thus, CAPE is a promising natural compound that should be explored further for its potential in prevention and treatment of oral candidiasis.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by Ethics Committee on the Use of Animals of the ICT/UNESP under protocol 019/2019-CEUA-ICT UNESP.
Author Contributions
Conceived and designed the experiments: JJ and EM. Performed the experiments: PB. Contributed to FACS analysis: VK and FL. Contributed to the experiments in oral candidiasis model: RR and MG. Contributed with reagents/materials/analysis tools in RIH/Brown University: BF and EM. Analyzed the data: PB and JJ. Wrote and revised the paper: PB, BF, EM, and JJ. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
National Council for Scientific Development (CNPq): Researcher on Productivity PQ-1C for JCJ (306330/2018-0). São Paulo Research Foundation (FAPESP): scholarship for PPB (2017/02652-6) and abroad (2019/05664-0). Grant 2019/10097-8 (Multi-user equipment FACs Lyric), São Paulo Research Foundation (FAPESP).
References
- (2017). Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts. 4th ed. CLSI standard M27 (Wayne, PA: Clinical and Laboratory Standards Institute; ). [Google Scholar]
- Araújo H. C. R., Cavalcanti M. G. S., Santos S. S., Alves L. C., Brayner F. A. (2008). Hemocytes Ultrastructure of Aedes aegypti (Diptera: Culicidae). Micron. 39, 184–189. 10.1016/j.micron.2007.01.003 [DOI] [PubMed] [Google Scholar]
- Armutcu F., Akyol S., Ustunsoy S., Turan F. F. (2015). Therapeutic Potential of Caffeic Acid Phenethyl Ester and Its Anti-Inflammatory and Immunomodulatory Effects (Review). Exp. Ther. Med. 9, 1582–1588. 10.3892/etm.2015.2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandara H. M. H. N., Panduwawala C. P., Samaranayake L. P. (2019). Biodiversity of the Human Oral Mycobiome in Health and Disease. Oral. Dis. 25, 363–371. 10.1111/odi.12899 [DOI] [PubMed] [Google Scholar]
- Bassilana M., Hopkins J., Arkowitz R. A. (2005). Regulation of the Cdc42/Cdc24 GTPase Module During Candida Albicans Hyphal Growth. Eukaryot. Cell 4 (3), 588–603. 10.1128/EC.4.3.588-603.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassilana D., Hopkins L., Arkowitz J., Clynes M., Kavanagh K. (2006). Pre-Exposure to Yeast Protects Larvae of Galleria Mellonella from A Subsequent Lethal Infection by Candida Albicans and Is Mediated by the Increased Expression of Antimicrobial Peptides. Microbes Infect. 8(8), 2105–12. 10.1016/j.micinf.2006.03.005 [DOI] [PubMed] [Google Scholar]
- Bergin D., Murphy L., Keenan J., Clynes M., Kavanagh K. (2006). Pre-Exposure to Yeast Protects Larvae of Galleria mellonella From A Subsequent Lethal Infection by Candida albicans and Is Mediated by the Increased Expression of Antimicrobial Peptides. Microbes Infect. 8 (8), 2105–2112. 10.1016/j.micinf.2006.03.005 [DOI] [PubMed] [Google Scholar]
- Borman A. M. (2018). Of Mice and Men and Larvae: Galleria Mellonella to Model the Early Host-Pathogen Interactions After Fungal Infection. Virulence 9, 9–12. 10.1080/21505594.2017.1382799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breger J., Fuchs B. B., Aperis G., Moy T. I., Ausubel F. M., Mylonakis E. (2007). Antifungal Chemical Compounds Identified Using a C. elegans Pathogenicity Assay. PloS Pathog. 3, 0168–0178. 10.1371/journal.ppat.0030018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne N., Surlis C., Maher A., Gallagher C., Carolan J. C., Clynes M., et al. (2015). Prolonged Pre-Incubation Increases the Susceptibility of Galleria Mellonella Larvae to Bacterial and Fungal Infection. Virulence 6, 458–465. 10.1080/21505594.2015.1021540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campoy S., Adrio J. L. (2017). Antifungals. Biochem. Pharmacol. 133, 86–96. 10.1016/j.bcp.2016.11.019 [DOI] [PubMed] [Google Scholar]
- Chan G. C. F., Cheung K. W., Sze D. M. Y. (2013). The Immunomodulatory and Anticancer Properties of Propolis. Clin. Rev. Allergy Immunol. 44, 262–273. 10.1007/s12016-012-8322-2 [DOI] [PubMed] [Google Scholar]
- Ciurea C. N., Kosovski I. B., Mare A. D., Toma F., Pintea-Simon I. A., Man A. (2020). Candida and Candidiasis—Opportunism Versus Pathogenicity: A Review of the Virulence Traits. Microorganisms 8, 1–17. 10.3390/microorganisms8060857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman J. J., Komura T., Munro J., Wu M. P., Busanelli R. R., Koehler A. N., et al. (2016). The Immunomodulatory Activity of Caffeic Acid Phenethyl Ester in Caenorhabditis Elegans is Mediated by the CED-10 (Rac-1)/PAK1 Pathway. Future Med. Chem. 8, 2033–2046. 10.4155/fmc-2016-0085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti H. R., Bruno V. M., Childs E. E., Daugherty S., Hunter J. P., Mengesha B. G., et al. (2016). IL-17 Receptor Signaling in Oral Epithelial Cells Is Critical for Protection Against Oropharyngeal Candidiasis. Cell Host Microbe 20, 606–617. 10.1016/j.chom.2016.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Barros P. P., Rossoni R. D., De Camargo Ribeiro F., Junqueira J. C., Jorge A. O. C. (2017). Temporal Profile of Biofilm Formation, Gene Expression and Virulence Analysis in Candida albicans Strains. Mycopathologia 182, 285–295. 10.1007/s11046-016-0088-2 [DOI] [PubMed] [Google Scholar]
- de Barros P. P., Rossoni R. D., Freire F., Ribeiro F. de C., Lopes L.A.d. C., Junqueira J. C., et al. (2018). Candida Tropicalis Affects the Virulence Profile of Candida albicans: An In Vitro and In Vivo Study. Pathog. Dis. 76:1–9. 10.1093/femspd/fty014 [DOI] [PubMed] [Google Scholar]
- de Barros P. P., Rossoni R. D., Ribeiro F., de C., Silva M. P., de Souza C. M., et al. (2019). Two Sporulated Bacillus Enhance Immunity in Galleria Mellonella Protecting Against Candida albicans. Microb. Pathog. 132, 335–342. 10.1016/j.micpath.2019.05.023 [DOI] [PubMed] [Google Scholar]
- de Camargo Ribeiro F., Junqueira J. C., dos Santos J. D., de Barros P. P., Rossoni R. D., Shukla S., et al. (2020). Development of Probiotic Formulations for Oral Candidiasis Prevention: Gellan Gum as a Carrier to Deliver Lactobacillus paracasei 28.4. Antimicrob. Agents Chemother. 64, 1–16. 10.1128/AAC.02323-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vita D., Friggeri L., D’Auria F. D., Pandolfi F., Piccoli F., Panella S., et al. (2014). Activity of Caffeic Acid Derivatives Against Candida albicans Biofilm. Bioorg. Med. Chem. Lett. 24, 1502–1505. 10.1016/j.bmcl.2014.02.005 [DOI] [PubMed] [Google Scholar]
- Ferreira E. D. S., Rosalen P. L., Benso B., De Cássia Orlandi Sardi J., Denny C., Alves De Sousa S., et al. (2021). The Use of Essential Oils and Their Isolated Compounds for the Treatment of Oral Candidiasis: A Literature Review. Evidence-Based Complement. Altern. Med. 2021, 1–16. 10.1155/2021/1059274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freires I. A., Queiroz V. C. P. P., Furletti V. F., Ikegaki M., de Alencar S. M., Duarte M. C. T., et al. (2016). Chemical Composition and Antifungal Potential of Brazilian Propolis Against Candida spp. J. Mycol. Med. 26, 122–132. 10.1016/j.mycmed.2016.01.003 [DOI] [PubMed] [Google Scholar]
- Ganz T. (2003). Defensins: Antimicrobial Peptides of Innate Immunity. Nat. Rev. Immunol. 3, 710–720. 10.1038/nri1180 [DOI] [PubMed] [Google Scholar]
- Granger B. L. (2012). Insight into the Antiadhesive Effect of Yeast Wall Protein 1 of Candida Albicans. Eukaryot Cell 11 (6), 795–805. 10.1128/EC.00026-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger B. L., Flenniken M. L., Davis D. A., Mitchell A. P., Cutler J. E. (2005). Yeast Wall Protein 1 of Candida albicans. Microbiology 151, 1631–1644. 10.1099/mic.0.27663-0 [DOI] [PubMed] [Google Scholar]
- Hellstein J. W., Marek C. L. (2019). Candidiasis: Red and White Manifestations in the Oral Cavity. Head Neck Pathol. 13, 25–32. 10.1007/s12105-019-01004-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D., Bardet A. F., Nobile C. J., Petryshyn A., Glaser W., Schöck U., et al. (2012). A Histone Deacetylase Adjusts Transcription Kinetics at Coding Sequences During Candida Albicans Morphogenesis. PLoS Genet. 8 (12), 1–13. 10.1371/journal.pgen.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini S. S., Ghaemi E., Noroozi A., Niknejad F. (2019). Zinc Oxide Nanoparticles Inhibition of Initial Adhesion and ALS1 and ALS3 Gene Expression in Candida albicans Strains From Urinary Tract Infections. Mycopathologia 184, 261–271. 10.1007/s11046-019-00327-w [DOI] [PubMed] [Google Scholar]
- Jiang Y., Yi X., Li M., Wang T., Qi T., She X. (2012). Antimicrobial Activities of Recombinant Mouse β-Defensin 3 and its Synergy With Antibiotics. J. Mater. Sci. Mater. Med. 23, 1723–1728. 10.1007/s10856-012-4645-z [DOI] [PubMed] [Google Scholar]
- Jia W., Zhang H., Li C., Li G., Liu X., Wei J. (2016). The Calcineruin Inhibitor Cyclosporine a Synergistically Enhances the Susceptibility of Candida albicans Biofilms to Fluconazole by Multiple Mechanisms. BMC Microbiol. 16, 113. 10.1186/s12866-016-0728-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordão C. C., Klein M. I., Carmello J. C., Dias L. M., Pavarina A. C. (2021). Consecutive Treatments With Photodynamic Therapy and Nystatin Altered the Expression of Virulence and Ergosterol Biosynthesis Genes of a Fluconazole-Resistant Candida albicans In Vivo . Photodiagnosis Photodyn. Ther. 33, 1–11. 10.1016/j.pdpdt.2020.102155 [DOI] [PubMed] [Google Scholar]
- Junqueira J. C., Colombo C. E. D., Da Silva Martins J., Ito C. Y. K., Carvalho Y. R., Jorge A. O. C. (2005). Experimental Candidosis and Recovery of Candida albicans From the Oral Cavity of Ovariectomized Rats. Microbiol. Immunol. 49, 199–207. 10.1111/j.1348-0421.2005.tb03721.x [DOI] [PubMed] [Google Scholar]
- Junqueira J. C., Vilela S. F. G., Rossoni R. D., Barbosa J. O., Costa A. C. B. P., Rasteiro V. M. C., et al. (2012). Colonização Oral Por Leveduras Em Pacientes HIV-Positivos No Brasil. Rev. Inst. Med. Trop. Sao Paulo 54, 17–24. 10.1590/S0036-46652012000100004 [DOI] [PubMed] [Google Scholar]
- Kavanagh K., Sheehan G. (2018). The Use of Galleria mellonella Larvae to Identify Novel Antimicrobial Agents Against Fungal Species of Medical Interest. J Fungi (Basel) 4 (3), 13. 10.3390/jof4030113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M. A. O., Williams D. W. (2017). Diagnosis and Management of Oral Candidosis. Br. Dent. J. 223, 675–681. 10.1038/sj.bdj.2017.886 [DOI] [PubMed] [Google Scholar]
- Liu X., Ma Z., Zhang J., Yang L. (2017). Antifungal Compounds Against Candida Infections From Traditional Chinese Medicine. BioMed. Res. Int. 2017, 1–12. 10.1155/2017/4614183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCT Method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lu M., Yang X., Yu C., Gong Y., Yuan L., Hao L., et al. (2019). Linezolid in Combination With Azoles Induced Synergistic Effects Against Candida albicansand Protected Galleria Mellonella Against Experimental Candidiasis. Front. Microbiol. 10, 3142. 10.3389/fmicb.2018.03142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara V. H., Silva E. G., Paula C. R., Ishikawa K. H., Nakamae A. E. M. (2012). Treatment With Probiotics in Experimental Oral Colonization by Candida albicans in Murine Model (DBA/2). Oral. Dis. 18, 260–264. 10.1111/j.1601-0825.2011.01868.x [DOI] [PubMed] [Google Scholar]
- Mayer F. L., Wilson D., Hube B. (2013). Candida albicans Pathogenicity Mechanisms. Virulence 4, 119–128. 10.4161/viru.22913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee P. K., Chandra J., Kuhn D. M., Ghannoum M. A. (2003). Mechanism of Fluconazole Resistance in Candida albicans Biofilms: Phase-Specific Role of Efflux Pumps and Membrane Sterols. Infect. Immun. 71, 4333–4340. 10.1128/IAI.71.8.4333-4340.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nailis H., Kucharíková S., Ricicová M., Van Dijck P., Deforce D., Nelis H., et al. (2010). Real-Time PCR Expression Profiling of Genes Encoding Potential Virulence Factors in Candida Albicans Biofilms: Identification of Model-Dependent and -Independent Gene Expression. BMC Microbiol. 10 (114), 1–11 10.1186/1471-2180-10-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan K., Singh S., Burke T. R., Grunberger D., Aggarwal B. B. (1996). Caffeic Acid Phenethyl Ester Is a Potent and Specific Inhibitor of Activation of Nuclear Transcription Factor NF-κB. Proc. Natl. Acad. Sci. U. S. A. 93, 9090–9095. 10.1073/pnas.93.17.9090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile C. J., Fox E. P., Nett J. E., Sorrells T. R., Mitrovich Q. M., Hernday A. D., et al. (2012). A Recently Evolved Transcriptional Network Controls Biofilm Development in Candida albicans. Cell 148, 126–138. 10.1016/j.cell.2011.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olgierd B., Kamila Ż., Anna B., Emilia M. (2021). The Pluripotent Activities of Caffeic Acid Phenethyl Ester. Molecules 26, 1335. 10.3390/molecules26051335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira T. C., Barros P.P. de, de Oliveira Fugisaki L. R., Rossoni R. D., Ribeiro F. de C., Menezes R. T. de, et al. (2018). Recent Advances in the Use of Galleria Mellonella Model to Study Immune Responses Against Human Pathogens. J. Fungi 4, 1–19. 10.3390/jof4040128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponde N. O., Lortal L., Ramage G., Naglik J. R., Richardson J. P. (2021). Candida albicans Biofilms and Polymicrobial Interactions. Crit. Rev. Microbiol. 47, 91–111. 10.1080/1040841X.2020.1843400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossoni R. D., de Barros P. P., de Alvarenga J. A., Ribeiro F. de C., Velloso M.d. S., Fuchs B. B., et al. (2018). Antifungal Activity of Clinical Lactobacillus Strains Against Candida albicans Biofilms: Identification of Potential Probiotic Candidates to Prevent Oral Candidiasis. Biofouling 34, 212–225. 10.1080/08927014.2018.1425402 [DOI] [PubMed] [Google Scholar]
- Rossoni R. D., de Barros P. P., Lopes L. A., das C., Ribeiro F. C., Nakatsuka T., et al. (2019). Effects of Surface Pre-Reacted Glass-Ionomer (S-PRG) Eluate on Candida spp.: Antifungal Activity, Anti-Biofilm Properties, and Protective Effects on Galleria Mellonella Against C. albicans Infection. Biofouling 35, 997–1006. 10.1080/08927014.2019.1686485 [DOI] [PubMed] [Google Scholar]
- Rossoni R. D., Fuchs B. B., de Barros P. P., Velloso M. D., Jorge A. O., Junqueira J. C., et al. (2017). Lactobacillus Paracasei Modulates the Immune System of Galleria Mellonella and Protects Against Candida Albicans Infection. PLoS ONE 12 (3), e0173332. 10.1371/journal.pone.0173332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorzoni L., Fuchs B. B., Junqueira J. C., Mylonakis E. (2021). Current and Promising Pharmacotherapeutic Options for Candidiasis. Expert Opin. Pharmacother 22 (7), 887–888. 10.1080/14656566.2021.1873951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry L., Rajendran R., Lappin D. F., Borghi E., Perdoni F., Falleni M. (2014). Biofilms Formed by Candida Albicans Bloodstream Isolates Display Phenotypic and Transcriptional Heterogeneity that Are Associated With Resistance and Pathogenicity. BMC Microbiol. 14 (182), 1–14. 10.1186/1471-2180-14-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparber F., LeibundGut-Landmann S. (2015). Interleukin 17-Mediated Host Defense Against Candida albicans. Pathogens 4, 606–619. 10.3390/pathogens4030606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Liao K., Hang C. (2018). Caffeic Acid Phenethyl Ester Synergistically Enhances the Antifungal Activity of Fluconazole Against Resistant Candida albicans. Phytomedicine 40, 55–58. 10.1016/j.phymed.2017.12.033 [DOI] [PubMed] [Google Scholar]
- Takakura N., Sato Y., Ishibashi H., Oshima H., Uchida K., Yamaguchi H., et al. (2003). A Novel Murine Model of Oral Candidiasis With Local Symptoms Characteristic of Oral Thrush. Microbiol. Immunol. 47, 321–326. 10.1111/j.1348-0421.2003.tb03403.x [DOI] [PubMed] [Google Scholar]
- Tomalka J., Azodi E., Narra H. P., Patel K., O’Neill S., Cardwell C., et al. (2015). β-Defensin 1 Plays a Role in Acute Mucosal Defense Against Candida albicans. J. Immunol. 194, 1788–1795. 10.4049/jimmunol.1203239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppuluri P., Nett J., Heitman J., Andes D. (2008). Synergistic Effect of Calcineurin Inhibitors and Fluconazole Against Candida albicans Biofilms. Antimicrob. Agents Chemother. 52, 1127–1132. 10.1128/AAC.01397-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojda I., Staniec B., Sułek M., Kordaczuk J. (2020). The Greater Wax Moth Galleria Mellonella: Biology and Use in Immune Studies. Pathog. Dis. 78, 1–15. 10.1093/femspd/ftaa057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Monin L., Gordon R., Aggor F. E. Y., Bechara R., Edwards T. N., et al. (2020). An IL-17F.S65L Knock-In Mouse Reveals Similarities and Differences in IL-17F Function in Oral Candidiasis: A New Tool to Understand IL-17F. J. Immunol. 205, 720–730. 10.4049/jimmunol.2000394 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.