Borchers et al. review regulatory principles in the activation of endocytic Rab GTPases and how they apply to other Rab and Arf GTPases.
Abstract
The eukaryotic endomembrane system consists of multiple interconnected organelles. Rab GTPases are organelle-specific markers that give identity to these membranes by recruiting transport and trafficking proteins. During transport processes or along organelle maturation, one Rab is replaced by another, a process termed Rab cascade, which requires at its center a Rab-specific guanine nucleotide exchange factor (GEF). The endolysosomal system serves here as a prime example for a Rab cascade. Along with endosomal maturation, the endosomal Rab5 recruits and activates the Rab7-specific GEF Mon1-Ccz1, resulting in Rab7 activation on endosomes and subsequent fusion of endosomes with lysosomes. In this review, we focus on the current idea of Mon1-Ccz1 recruitment and activation in the endolysosomal and autophagic pathway. We compare identified principles to other GTPase cascades on endomembranes, highlight the importance of regulation, and evaluate in this context the strength and relevance of recent developments in in vitro analyses to understand the underlying foundation of organelle biogenesis and maturation.
Membrane identity in the endomembrane system
One key feature of eukaryotic cells is the presence of membrane-enclosed organelles, which constantly exchange proteins, lipids, or metabolites via vesicular transport or membrane contact sites (MCSs). Along the endomembrane system, vesicular trafficking requires vesicle budding from the donor membrane and directed transport toward and fusion with the acceptor compartment. The resulting trafficking routes form a regulated network that connects not only the internal organelles, but also the interior and exterior of the cell.
The specific identity of organelles within the endomembrane system is defined by the lipid and protein composition of their membranes. This includes signaling lipids such as phosphoinositides (PIPs) and small GTPases of the Ras superfamily of small G proteins, namely of the Rab, Arf, and Arl families, which act as binding platforms for accessory proteins involved in multiple membrane trafficking processes (Balla, 2013).
Rab GTPases, like other small GTPases, are key regulatory proteins that switch between an inactive GDP-bound (Rab-GDP) and an active GTP-bound (Rab-GTP) state (Barr, 2013; Goody et al., 2017; Hutagalung and Novick, 2011). Rabs are posttranslationally modified by the addition of geranylgeranyl moieties to C-terminal cysteine residues, which allow their reversible membrane association. Within the cytosol, Rab-GDP is kept soluble by binding to the chaperone-like GDP dissociation inhibitor (GDI). At the target membrane, an organelle-specific guanine nucleotide exchange factor (GEF) activates the Rab after its previous release from GDI, a process possibly supported by other factors (Dirac-Svejstrup et al., 1997). GTP binding stabilizes two loops in the Rab GTPase domain, which allows recruitment and binding of various so-called effector proteins to the Rab-GTP on the membrane. Rab GTPases are inefficient enzymes with a low intrinsic GTP hydrolysis rate and thus depend on a GTPase-activating protein (GAP) to hydrolyze bound GTP. GDI then extracts the Rab-GDP and keeps it soluble in the cytosol until the next activation cycle (Barr, 2013; Goody et al., 2017; Hutagalung and Novick, 2011). In addition to their conserved GTPase domain, Rabs contain a hypervariable C-terminal domain (HVD), which supports GEF recognition and therefore correct localization of the Rab (Thomas et al., 2018)
Among various other functions, Rab GTPases are critical for the fusion of vesicles with the acceptor membrane by recruiting tethering proteins, which bring the two membranes into close proximity. Tethers, together with Sec1/Munc18 proteins, promote the folding of membrane-bound SNAREs at the vesicle and the target membrane into tetrameric coiled-coil complexes. This process further reduces the distance between the membranes, bypasses the hydration layer on membranes, and results in mixing of lipid bilayers and consequently membrane fusion (Wickner and Rizo, 2017; Ungermann and Kümmel, 2019).
Organization and function of the endolysosomal pathway
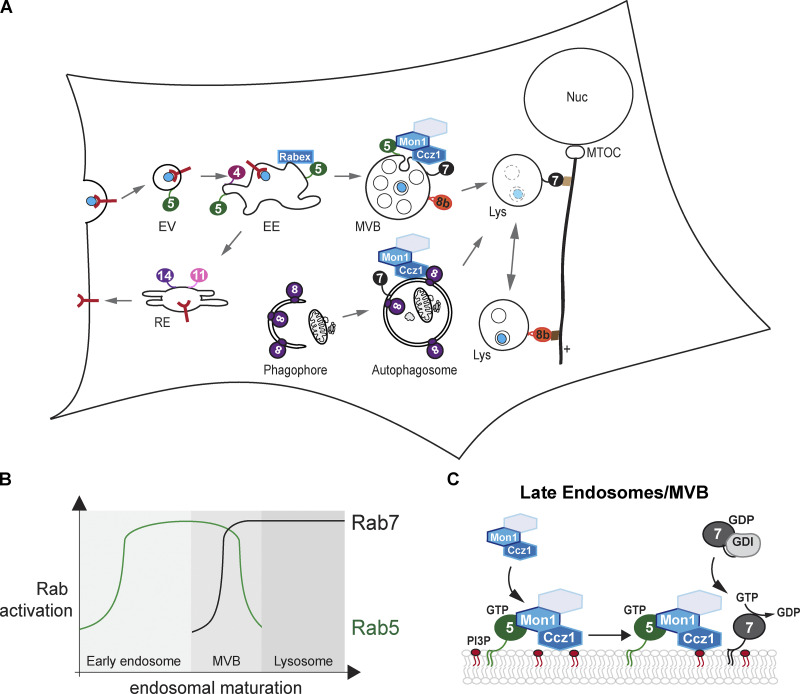
Endocytosis allows the rapid adaptation of plasma membrane composition in response to changing environmental conditions by the uptake of membrane proteins from the plasma membrane, which are either transported to and finally degraded in the lysosome or sorted back to the plasma membrane, e.g., receptors after releasing their cargo within the endosomal lumen (Sardana and Emr, 2021). A third fate of endocytosed cargo is trafficking to the Golgi (Laidlaw and MacDonald, 2018). In addition, various kinds of endocytosis allow the uptake of very large particles such as bacteria during phagocytosis or fluids during pinocytosis (Huotari and Helenius, 2011; Babst, 2014). The endocytic pathway is also involved in the quality control system of plasma membrane proteins and allows degradation of damaged cell surface proteins as well as the down-regulation of nutrient transporters and receptors (Sardana and Emr, 2021). During endocytosis, membrane proteins marked by ubiquitination are incorporated into endocytic vesicles, which pinch off the plasma membrane and fuse with the tubular-shaped early endosome (EE) in the cell periphery (Fig. 1 A). The EE serves as a sorting station, at which membrane proteins are either sorted into tubular structures and brought to the recycling endosome (RE) or get incorporated into intraluminal vesicles (ILVs) with the help of four endosomal sorting complexes required for transport (ESCRTs; Sardana and Emr, 2021). A prerequisite for the degradation of cargo in the lysosome is the maturation of EEs into late endosomes (LEs) by changing the organelle surface composition, including specific Rab GTPases and PIPs, and organelle shape. The LE is eventually spherically shaped, containing multiple ILVs and a more acidified lumen. Therefore, it is also called Multivesicular Body (MVB). Upon fusion with the lysosome, ILVs and their content are degraded into precursor molecules, which are reused by the cell (Fig. 1 A; Sardana and Emr, 2021; Huotari and Helenius, 2011).
Figure 1.
Rab GTPases in the endolysosomal pathway.(A) Localization of key Rab GTPases along the endolysosomal pathway. Endocytic vesicles containing cargo (blue dot) or receptor proteins (red) are substrates of endocytosis. Endocytic vesicles (EV) fuse with the EE. Rabs are shown by numbers: Rab5 (green) on early EE is replaced by Rab7 (black) on multivesicular bodies (MVBs). GEFs are shown in blue. Positioning of lysosomes (Lys) depends on binding to motor proteins by either Arl8b (orange, 8b) or Rab7. Recycling occurs via REs involving Rab4, Rab11, and Rab14. MTOC, microtubule organizing center; Nuc, nucleus. (B) Spatiotemporal Rab5-to-Rab7 transition during endosomal maturation. Rab5 (green graph) is rapidly recruited to EE and replaced by Rab7. (C) Model of Rab7 GEF recruitment and activation on endosomes. Mon1-Ccz1 (or the trimeric complex additionally containing Rmc1/C18orf8/Bulli, as indicated by the unlabeled hexagon) requires Rab5-GTP for activation to promote Rab7 recruitment. For details, see text.
Central functions of Rab5 and Rab7
Along the endolysosomal system, several Rabs coordinate sorting and recycling processes at the EE and LE. Early endosomal Rab5 and late endosomal Rab7 are here the key Rabs conserved among species. Their spatiotemporal activation and therefore functions are tightly coordinated on the level of the MVB/LE (Fig. 1 B).
In yeast, the Rab5-like GTPases Vps21, Ypt52, Ypt52, and Ypt10 and the Rab7-like Ypt7 structure the endocytic pathway (Singer-Krüger et al., 1994; Wichmann et al., 1992). In mammalian cells, Rab5 (with Rab5a, b, and c isoforms having nonredundant functions in the endocytic network; Chen et al., 2014, 2009) and Rab7 (with Rab7a and b isoforms, of which Rab7a is the main actor in transport processes along the endocytic pathway [Guerra and Bucci, 2016], whereas Rab7b has a role in the transport from endosome to the Golgi [Kjos et al., 2017; Progida et al., 2010]) are present (Wandinger-Ness and Zerial, 2014). While the overall organization of the endocytic pathway into EE and LE is conserved, yeast seems to have a more ancestral minimal endomembrane system, where the trans-Golgi network acts as EE and RE (Day et al., 2018). In mammalian cells, the more complex endolysosomal system depends on additional Rabs. Rab4 is involved in protein sorting at the EE, activation of Rab5, and recycling of cargo back to the plasma membrane (Kälin et al., 2015; Wandinger-Ness and Zerial, 2014; de Renzis et al., 2002), whereas Rab11 and Rab14 function at REs (Fig. 1 A; Linford et al., 2012; Takahashi et al., 2012). Furthermore, Rab9 is required for retrograde transport between LEs and the trans-Golgi network (Lombardi et al., 1993), and Rab32 and Rab38 function in the biogenesis of lysosome-related organelles (Bowman et al., 2019; Gerondopoulos et al., 2012; Wasmeier et al., 2006).
During endosomal maturation, Rab5 is exchanged for Rab7 (Rink et al., 2005; Poteryaev et al., 2010). This Rab switch is highly conserved and a prime example of coordinated Rab turnover during organelle maturation. The rapid transition from Rab5 to Rab7 was explained by a so-called cutout switch, where activation of Rab5 fosters at a threshold value activation of Rab7, which in turn suppresses further Rab5 activation (Fig. 1 B; Del Conte-Zerial et al., 2008). Such a principle may apply to most Rab cascades (Barr, 2013).
Rab5 has multiple functions on EEs (Wandinger-Ness and Zerial, 2014). It interacts with a number of effectors such as the lipid kinase Vps34, Rabaptin-5, which is found in complex with the Rab5-GEF Rabex5, Rabenosyn-5, and tethers such as the class C core vacuole/endosome tethering (CORVET) complex or EEA1. Therefore, Rab5 is critical for the homotypic fusion of EEs (Gorvel et al., 1991; Ohya et al., 2009; Christoforidis et al., 1999a, b; Perini et al., 2014; Marat and Haucke, 2016). Vps34 was initially identified in yeast (Schu et al., 1993) and exists in two heterotetrametric complexes, which differ by just one subunit (Kihara et al., 2001). Complex I resides on autophagosomes, whereas complex II functions on endosomes (Fig. 2 D). Both complexes generate a local pool of phosphatidylinositol-3-phosphate (PI3P), to which several effectors bind, including the early endosomal tether EEA1 and ESCRTs (Wallroth and Haucke, 2018). Recent structural insights revealed that Rab5 recruits and activates endosomal complex II, whereas Rab1 acts similarly on autophagosomal complex I (Tremel et al., 2021). This explains how Rab5-GTP promotes the formation of a local endosomal PI3P pool (Franke et al., 2019). Interestingly, Caenorhabditis elegans VPS-34 can recruit the Rab5 GAP TBC-2 to endosomal membranes, suggesting a possible link between PI3P generation and Rab5 inactivation (Law et al., 2017).
Figure 2.
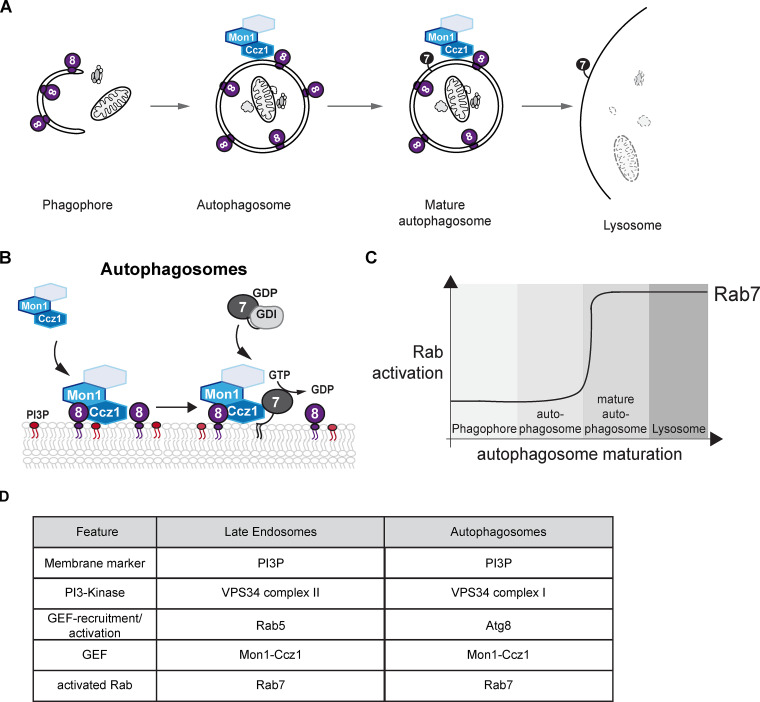
Rab7 activation on autophagosomes.(A and B) Atg8-dependent Mon1-Ccz1 recruitment and activation. Atg8 (violet) recruits Mon1-Ccz1 (and likely also the trimeric GEF complex in higher eukaryotes, as indicated by the unlabeled hexagon) and allows fusion with lysosome. (C) Model of spatiotemporal Rab7 activation on autophagosomes. Maturation is prerequisite for successful fusion. (D) Comparison of proteins involved in maturation of LEs and autophagosomes.
Rab7 is a key component in the late endocytic pathway (Langemeyer et al., 2018a). It is found on LEs, lysosomes, and autophagosomes and is required for the biogenesis and positioning of LEs and lysosomes, for MCSs of lysosomes with other organelles, and for the fusion of endosomes and autophagosomes with lysosomes (Fig. 1 A; Guerra and Bucci, 2016; McEwan et al., 2015; Ballabio and Bonifacino, 2020; Cabukusta and Neefjes, 2018). Even though both the metazoan Rab7 and yeast Ypt7 are activated by the homologous Mon1-Ccz1 GEF complex and are required for endosomal maturation, their function on LEs and lysosomes is not entirely conserved. In yeast, active Ypt7 directly binds the hexameric homotypic fusion and vacuole protein sorting (HOPS) tethering complex and mediates SNARE-dependent fusion of LEs or autophagosomes with vacuoles as well as homotypic vacuole fusion (Wickner and Rizo, 2017; Gao et al., 2018a, b). In higher eukaryotes, HOPS also promotes fusion between LEs and lysosomes, yet apparently does not directly interact with Rab7, but rather with the GTPases Rab2 and Arl8b (Gillingham et al., 2014; Fujita et al., 2017; Lőrincz et al., 2017; Khatter et al., 2015). How Rab7 contributes to fusion at the lysosome is still unclear. Rab7 interacts with several proteins on lysosomes, including the cholesterol sensor ORPL1 and the dynein-interacting lysosomal RILP (Jordens et al., 2001; Cantalupo et al., 2001; Rocha et al., 2009). Both proteins also bind HOPS (van der Kant et al., 2015, 2013), as does another multivalent adaptor protein, PLEKHM1 (McEwan et al., 2015), which binds both Arl8b and Rab7 (Marwaha et al., 2017). Interestingly, Arl8b in complex with its effector SKIP also binds TBC1D15, a Rab7 GAP, which may displace Rab7 from LEs before their fusion with lysosomes (Jongsma et al., 2020). It is thus possible that fusion of LEs and autophagosomes with lysosomes requires a complex coordination of the three GTPases, Rab7, Arl8b, and Rab2, with the HOPS complex and other effectors. Some of this complexity may be explained by a second function of Rab7 and Arl8b in binding adapters of the kinesin or dynein motor protein family, which connect LEs and lysosomes to the microtubule network. Thereby Rab7 and Arl8b control the positioning of these organelles to the periphery or perinuclear area via the microtubule network, which has functional implications (Fig. 1 A; Cabukusta and Neefjes, 2018; Bonifacino and Neefjes, 2017). Perinuclear lysosomes are the main places for degradation of cargo delivered by endosomes and autophagosomes, whereas peripheral lysosomes are involved in the regulation of mammalian target of rapamycin complex1 (mTORC1), the master regulator switching between cell growth and autophagy (Johnson et al., 2016; Korolchuk et al., 2011). This also may be connected to the role of lysosomes in lipid homeostasis, as Rab7 seems to control cholesterol export via the lysosomal NPC1 (van den Boomen et al., 2020; Shin and Zoncu, 2020; Castellano et al., 2017). How far the acidification state of perinuclear and peripheral lysosomes also affects their Rab7 and Arl8b mediated localization is still under debate (Ponsford et al., 2021). Thus, it is likely that Rab7 coordinates LE and lysosomal transport and fusion activity in coordination with endosomal biogenesis and cellular metabolism.
GEF function and regulation in endosomal maturation
The heterodimeric complex Mon1-Ccz1 was identified as the GEF for Ypt7 in yeast and for Rab7 in higher eukaryotes (Nordmann et al., 2010; Gerondopoulos et al., 2012). The Mon1-Ccz1 complex is an effector of Rab5 (Kinchen and Ravichandran, 2010; Langemeyer et al., 2020; Cui et al., 2014; Li et al., 2015; Poteryaev et al., 2010; Singh et al., 2014), suggesting a direct link to endosomal maturation and Rab turnover (Fig. 1 B). Structural analyses uncovered how the two central longin domains in Mon1 and Ccz1 displace the bound nucleotide from Ypt7 (Kiontke et al., 2017). Unlike yeast, the metazoan Mon1-Ccz1 complex contains a third subunit termed RMC1 or C18orf8 in mammals and Bulli in Drosophila (Vaites et al., 2017; Dehnen et al., 2020; van den Boomen et al., 2020). Even though loss of this subunit impairs endosomal and autophagosomal biogenesis, this subunit does not affect GEF activity toward Rab7 in vitro (Dehnen et al., 2020; Langemeyer et al., 2020), indicating that the general GEF mechanism is conserved across species. As Rab7 is required on LEs, autophagosomes, and lysosomes, spatial recruitment and activity of the Rab7 GEF must be tightly regulated.
Rab5 activates the Mon1-Ccz1 GEF complex
During endosomal maturation, the Mon1-Ccz1 complex is recruited to Rab5- and PI3P-positive endosomes and activates Rab7 for subsequent fusion of endosomes with lysosomes (Nordmann et al., 2010; Poteryaev et al., 2010; Cabrera and Ungermann, 2013; Cabrera et al., 2014; Singh et al., 2014; Fig. 1 C). However, it was postulated that (but remained unclear how) Rab5 affects Rab7 GEF activity. The activity of GEFs is in the simplest way determined in solution, where the respective Rab, which has been loaded with a fluorescent- or radioactive-labeled nucleotide, is incubated with the GEF (Schoebel et al., 2009; Bergbrede et al., 2009). GDP or GTP addition then triggers displacement of the bound nucleotide, which results in a decrease of fluorescence or increase of radioactive signal in solution. Such in-solution assays can uncover the Rab specificity of GEFs yet cannot recapitulate the membrane context and potential regulating factors. Recent approaches therefore used liposomes and prenylated Rab:GDI complexes to address the role of membrane lipids and proteins in GEF activation (Thomas and Fromme, 2016; Thomas et al., 2018; Langemeyer et al., 2020, 2018b; Cezanne et al., 2020; Bezeljak et al., 2020). Details of these reconstituted systems are discussed below. In yeast, prenylated, membrane-bound, and GTP-loaded Rab5-like Vps21 was surprisingly inefficient as a single factor to recruit Mon1-Ccz1 to membranes, whereas addition of PIPs together with Vps21 enhanced recruitment (Langemeyer et al., 2020). However, activity of both the yeast and metazoan Rab7 GEF complexes showed a striking dependence on membrane-bound Rab5-GTP in the GEF assay, whereas PIPs alone were not sufficient to drive GEF activation. These observations demonstrate that the Mon1-Ccz1 complex depends on membrane-bound Rab5 for its Rab7 GEF activity, which nicely explains some of the previous in vivo observations on endosomal Rab5-to-Rab7 exchange (Poteryaev et al., 2010; Rink et al., 2005).
This Rab exchange, which occurs similarly on phagosomes (Jeschke and Haas, 2016), is in vivo likely regulated in space and time. Time-lapse microscopy studies revealed that levels of fluorescently labeled Rab5 decreased, while fluorescently labeled Rab7 increased on the surface of a tracked endosome (Poteryaev et al., 2010; Yasuda et al., 2016). Analysis of the spatiotemporal Rab5-to-Rab7 transition in mammalian cells revealed that Rab5-positive endosomes can separate from Rab7-positive membranes, suggesting that a stepwise maturation process also occurs in some cells (Skjeldal et al., 2021). However, in all cases, only some insights on Mon1-Ccz1 regulation are presently available. Phosphorylation is one potential regulatory mechanism in GEF regulation (Kulasekaran et al., 2015). Indeed, yeast Mon1-Ccz1 is a substrate of the vacuolar casein kinase 1 Yck3 (Lawrence et al., 2014). When added to the Rab5-dependent GEF assay, Yck3-mediated phosphorylation inhibited Mon1-Ccz1 GEF activity, presumably by blocking the Rab5 interaction (Langemeyer et al., 2020). How the kinase is in turn regulated and whether this is the only mechanism of Mon1-Ccz1 GEF control is currently unknown.
Rab7 activation and function in autophagy
The lysosome is also the destination of the autophagic catabolic pathway. During autophagy, portions of the cytosol, specific organelles, aggregates, or pathogens are engulfed into a double-layered membrane, which upon closure fuses with the lysosome for degradation and reuse of its content (Fig. 2 A; Zhao and Zhang, 2019; Nakatogawa, 2020). Autophagy is a versatile pathway required for adaptation of a cell’s organelle repertoire and quality control.
Rab7 is found not just on LEs, but also on autophagosomes (Hegedűs et al., 2016; Gao et al., 2018a), although its precise function seems to differ between organisms (Kuchitsu and Fukuda, 2018). In yeast, the Rab7-homologue Ypt7 mediates HOPS-dependent fusion of autophagosomes with vacuoles (Gao et al., 2018a). In metazoan cells, Rab7 and its effectors PLEKHM1 and WDR91 are required for autolysosome/amphisome-lysosome fusion, yet Rab7 does not seem to directly bind HOPS during fusion of autophagosomes with lysosomes (Xing et al., 2021; McEwan et al., 2015; Gutierrez et al., 2004; Kuchitsu and Fukuda, 2018).
Given the striking Rab5 dependence on endosomes in Mon1-Ccz1 activation, the question arises, how does Mon1-Ccz1-mediated Rab7 activation happen on autophagosomes? Some data suggest that yeast and metazoan Rab5 is directly involved in the autophagy process such as autophagosome closure (Ravikumar et al., 2008; Bridges et al., 2012; Zhou et al., 2019, 2017), whereas others do not find direct evidence, for instance in Drosophila (Hegedűs et al., 2016). Studies in yeast revealed that the LC3–like Atg8 protein directly binds and recruits Mon1-Ccz1 to the autophagosomal membrane during starvation, which results in Ypt7 activation as a prerequisite of HOPS-dependent fusion with the vacuole (Gao et al., 2018a; Fig. 2 B). Tight regulation of Mon1-Ccz1 GEF-activity is apparently mandatory to avoid fusion of premature autophagosomes with the vacuole (Fig. 2 C). How Mon1-Ccz1 localization to either endosomes or autophagosomes is coordinated (also with regard to similarities in organelle features; Fig. 2 D) and whether Atg8/LC3 also regulates the activity of the GEF complex are not yet known.
Of note, an endosomal-like Rab5-to-Rab7 cascade also occurs on the mitochondrial outer membrane during mitophagy in metazoan cells, a selective pathway to degrade damaged mitochondria (Yamano et al., 2018). Here, Rab5 is activated by a mitochondrially localized Rab5 GEF, followed by Mon1-Ccz1 recruitment and Rab7A activation, which then orchestrates the subsequent mitophagy process. How this process is coupled to autophagosome maturation, and whether Rab7 is then again needed on the formed autophagosome, has not been addressed so far.
These data nevertheless demonstrate the adjustable recruitment of Mon1-Ccz1 during endosomal maturation and autophagosome formation and even to the mitochondrial surface. Targeting of the Mon1-Ccz1 complex is likely coordinated between all these processes.
A role for ER-endosome MCSs in endosome maturation
Endosomes form MCSs with the ER. Such contact sites have multiple roles ranging from lipid transport to ion exchange (Scorrano et al., 2019; Reinisch and Prinz, 2021). The endosome-ER contact depends on Rab7 and contributes to transport and positioning of endosomes, supports endosomal fission, and facilitates endocytic cargo transport and cholesterol transfer between LEs and the ER (Rocha et al., 2009; Friedman et al., 2013; Rowland et al., 2014; Raiborg et al., 2015; Jongsma et al., 2016). Rab7 activation via the Mon1-Ccz1 complex is required for cholesterol export from the lysosome, likely in the context of MCSs. Rab7 binds to the NPC1 cholesterol transporter and may thus promote cholesterol export only at MCSs with the ER or other organelles (van den Boomen et al., 2020). The ER is also involved in endosome maturation, which requires an MCS between Reticulon-3L on the ER and endosomal Rab9. In fact, Rab9 is recruited shortly before the Rab5-to-Rab7 transition (Wu and Voeltz, 2021; Kucera et al., 2016). How Rab9 activation and MCS formation are coordinated with endosomal maturation has not yet been revealed. It is likely that the spatial positioning of endosomes (Fig. 1 A), their acidification, and TORC1 activity also contribute to this process (Bonifacino and Neefjes, 2017; Johnson et al., 2016).
Retromer opposes Rab7 activation
Retromer is a conserved heteropentameric complex that mediates the formation of vesicular carriers at the endosome and thus allows the transport of receptors back to the Golgi or plasma membrane. The complex consists of a trimeric core (Vps35, Vps26, and Vps29), which binds either a SNX1-SNX4 heterodimer or a SNX3 monomer (Simonetti and Cullen, 2018; Leneva et al., 2021; Kovtun et al., 2018). Retromer is an effector of Rab7, but also recruits the Rab7 GAP TBC1D5 in metazoan cells (Rojas et al., 2008; Kvainickas et al., 2019; Jimenez-Orgaz et al., 2018; Distefano et al., 2018; Seaman et al., 2009). This dual function of retromer may facilitate the formation of endosomal tubules after the Rab5-to-Rab7 transition, and these tubules eventually lose Rab7 once scission has occurred (Jongsma et al., 2020).
It is not yet clear how conserved the Rab7-retromer-GAP connection is. Yeast retromer is also an effector of the Rab7-like Ypt7 and coordinates protein recycling at the endosome (Liu et al., 2012; Balderhaar et al., 2010), yet a role of a Rab7 GAP has not been described. However, yeast retromer also binds to the Rab5 GEFs Vps9 and Muk1 (Bean et al., 2015), which suggests that both Rab5 and Rab7 function contribute to efficient tubule formation at the endosome. Whether and how the Rab7 GEF Mon1-Ccz1 is functionally coordinated with retromer will be a topic of future studies.
GEF regulation along the endomembrane system
In the previous section, we focused mainly on the role of the Rab7 GEF in the context of endosome and autophagosome maturation. However, the timing of GEF activation and the subsequent recruitment of their target Rabs is critical for all membrane trafficking processes along the endomembrane system to guarantee maintenance of intracellular organelle organization. Rabs in turn interact with effectors, and effectors such as the lysosomal HOPS complex not only bind SNAREs but also catalyze their assembly and thus drive membrane fusion (Fig. 3 A). The spatiotemporal regulation of GEF activation is therefore at the heart of organelle biogenesis and maturation, and thus membrane trafficking. Within this section, we will now broaden our view by comparing different regulatory principles of GEFs.
Figure 3.
Regulatory mechanisms influence the activity of GEFs.(A) Hierarchical cascade of factors controlling membrane fusion. GEFs integrate various signals and initiate a cascade of protein activities, finally leading to membrane fusion. Signaling lipids, the presence of cargo proteins, upstream GTPases, and kinases influence the activity of GEFs and therefore determine Rab GTPase activation. Consequently, effector proteins such as tethering factors are recruited. This ultimately leads to SNARE-mediated lipid bilayer mixing and membrane fusion. (B) A Rab cascade in yeast exocytosis. Active Ypt32 and PI4P (yellow) on late Golgi compartments and secretory vesicles recruit the GEF Sec2, which in turn promotes activation and stable membrane insertion of the Rab Sec4. (C) Mon1-Ccz1 regulation by phosphorylation. Mon1-Ccz1 is recruited to and activated on LEs by coincidence detection of membrane-associated Rab5 and PI3P (red, Fig. 1 C) and promotes stable membrane insertion of Rab7. This process is terminated by Mon1-Ccz1 phosphorylation by the type I casein kinase Yck3 in yeast (orange). (D) A positive feedback loop of GEF activation on endocytic vesicles and EEs. The Rab5 GEF Rabex-5 binds ubiquitinated cargo on endocytic vesicles and is autoinhibited. Rab5 recruits Rabaptin-5, which binds Rabex-5 and releases the GEF from autoinhibition, generating a positive feedback loop. (E) Membrane factors determine GEF activity of TRAPPII at the trans-Golgi. TRAPPII activity for the Rab Ypt32 requires membrane-associated Arf1 and PI4P. (F) The length of the hypervariable domain of Golgi Rabs defines the substrate specificity for TRAPP complexes. The yeast Rab GTPases Ypt1 and Ypt32 differ in the length of their C-terminal HVD (box). TRAPPII and TRAPPIII complexes have the same active site, which is positioned away from the membrane, and thus discriminate Rab accessibility. (G) Phosphorylation as a mechanism to promote GEF activity. DENND1 GEF activity is autoinhibited, which is released by Akt-mediated phosphorylation. For details, see text.
A Rab cascade in exocytosis
Another well-characterized Rab cascade is involved in the exocytic transport of secretory vesicles from the trans-Golgi network to the plasma membrane. At the trans-Golgi, the GEF transport protein particle II (TRAPPII) activates the Rab GTPase Ypt32, which then recruits the GEF Sec2 to secretory vesicles. Sec2 in turn activates the Rab Sec4, which binds the Sec15 subunit of the Exocyst tethering complex and allows vesicles to dock and fuse with the plasma membrane (Fig. 3 B; Walch-Solimena et al., 1997; Ortiz et al., 2002; Dong et al., 2007; Itzen et al., 2007). This cascade is conserved in humans. During ciliogenesis at the plasma membrane, the Ypt32 homologue Rab11 recruits the GEF Rabin 8, which in turn activates the human Sec4 homologue Rab8, a process regulated by phosphorylation (Hattula et al., 2002; Wang et al., 2015; Knödler et al., 2010). Interestingly, yeast Sec2 not only is a GEF, but also interacts with the Sec4 effector Sec15 (Medkova et al., 2006), a principle also observed in the endocytic Rab5 activation cycle, where the GEF Rabex5 interacts with the Rab5 effector Rabaptin-5. This dual role may also apply to Mon1-Ccz1, as the Mon1 homologue in C. elegans, SAND1, and yeast Mon1-Ccz1 can bind the HOPS tethering complex (Poteryaev et al., 2010; Nordmann et al., 2010).
At the Golgi, phosphatidylinositol-4-phosphate (PI4P) contributes to directionality and spatiotemporal regulation of the exocytic Rab cascade. Sec2 binds both Ypt32 and PI4P on secretory vesicles via two binding sites, a process called coincidence detection. However, PI4P binding inhibits the interaction of Sec2 with Sec15. As vesicles reach the cell periphery, PI4P levels drop by the activity of Osh4, a lipid transporter, which allows Sec2 to bind the Exocyst subunit rather than Ypt32 (Ling et al., 2014; Mizuno-Yamasaki et al., 2010). In addition, Sec2 is phosphorylated by the plasma membrane–localized casein kinases Yck1 and Yck2 (Stalder et al., 2013; Stalder and Novick, 2016), resulting in effector recruitment rather than further Rab activation.
Such a regulation may also apply to yeast Mon1-Ccz1. Anionic phospholipids and PI3P support Mon1-Ccz1 recruitment to liposomes and vacuoles (Langemeyer et al., 2020; Cabrera et al., 2014; Lawrence et al., 2014), whereas phosphorylation of the complex by the casein kinase Yck3 inhibits the binding of Mon1-Ccz1 to the Rab5-like Ypt10 and consequently reduces its GEF activity toward Rab7 (Fig. 3 C; Langemeyer et al., 2020). These observations suggest that the phosphorylation of GEFs by kinases may be a general regulatory principle in Rab cascades.
Autoinhibition controls the Rab5 GEF
Another widely used regulatory mechanism is the autoinhibition of GEFs to control their activity. This has been analyzed in detail for the early endosomal Rab5-specific GEF Rabex-5, which interacts with the Rab5-effector Rabaptin-5 (Horiuchi et al., 1997). One factor for Rabex-5 recruitment to endocytic vesicles are ubiquitinated cargo proteins at the plasma membrane (Fig. 3 D; Mattera et al., 2006; Lee et al., 2006). Yet, isolated Rabex-5 has only low GEF activity in vitro (Delprato and Lambright, 2007). Structural analysis revealed that binding of Rabaptin-5 to Rabex-5 causes a rearrangement in the Rabex-5 C-terminus, which releases the GEF from autoinhibition and therefore facilitates nucleotide exchange of Rab5 (Delprato and Lambright, 2007; Zhang et al., 2014). On endosomes, increasing amounts of Rab5-GTP further promotes recruitment of the Rabex-5–Rabaptin-5 complex, resulting in a positive feedback loop of Rab5 activation and GEF recruitment (Lippé et al., 2001). Overall, Rabex-5 GEF activity is regulated by autoinhibition, a feedback loop with the Rab5 effector protein Rabaptin-5, and ubiquitinated cargo, which guarantees precise timing in establishing a Rab5-positive endosome. Of note, the Mon1 subunit of the Rab7 GEF can displace Rabex-5 from endosomal membranes (Poteryaev et al., 2010), which suggests a negative feedback loop of the Rab5 activation cascade once the next GEF is present.
Regulation of Arf1 GEFs at different Golgi subcompartments
These key principles of GEF regulation in GTPase cascades are also found for Arf GTPases. Arf GTPases are soluble in their GDP-bound state by shielding their N-terminal myristate anchor in a hydrophobic pocket. Like Rabs, Arf GTPases are activated by specific GEFs, and their inactivation requires a specific GAP (Sztul et al., 2019). However, this review only highlights some key findings in the regulation of Rab GEFs and does not address regulation of the corresponding GAPs. Once activated, Arfs insert their lipid anchor and an adjacent amphipathic helix into membranes and are then able to bind effector proteins (Sztul et al., 2019). One of the best-studied Arf-GEFs is Sec7, which activates Arf1, an Arf GTPase involved in intra-Golgi trafficking (Achstetter et al., 1988). Studies on yeast Sec7 revealed that the protein is autoinhibited in solution and depends on three small GTPases—Arf1, the Rab Ypt1, and the Arf-like Arl1—for recruitment to the Golgi, a process supported by anionic lipids found in the late Golgi compartment. Importantly, the late Golgi Rabs Ypt31/32 strongly stimulate GEF activity (McDonold and Fromme, 2014; Richardson et al., 2012, 2016), indicating allosteric activation, as observed for Rab5-dependent Mon1-Ccz1 activation (Langemeyer et al., 2020). In this process, Sec7 dimerizes and promotes Arf1 recruitment and thus establishes a positive feedback loop. Interestingly, membrane binding of two additional Arf1 GEFs of the early Golgi, Gea1/2, depends on Rab1/Ypt1 and neutral membranes. Under these conditions, Gea1/2 is released from autoinhibition, although no positive feedback loop was observed (Gustafson and Fromme, 2017). Thus, Arf GEF regulation and Arf activation are tightly linked to multiple small GTPases and the membrane environment to establish Golgi compartments.
Regulation and specificity of TRAPP complexes at the Golgi
Arf1 activation is also linked to the activation of Golgi-specific Rabs. Arf1-GTP binds to the highly conserved TRAPP GEF complexes at the Golgi (Fig. 3 E). Yeast and mammalian cells contain two TRAPP complexes. In yeast, both complexes share seven core components. TRAPPIII in addition contains Trs85, while accessory TRAPPII subunits are instead Trs130, Trs120, Trs65, and Tca17. Metazoan TRAPP complexes contain additional subunits (Lipatova and Segev, 2019).
Interestingly, both complexes share the same catalytic site for Rab1/Ypt1 and Rab11/Ypt32. However, TRAPPIII provides GEF activity toward Rab1/Ypt1. Initially, it was proposed that TRAPPII can activate both Rab1/Ypt1 and Rab11/Ypt32 (Thomas et al., 2019, 2018; Thomas and Fromme, 2016; Riedel et al., 2018); however, it was recently shown that the TRAPPII complex is specific for Rab11/Ypt32 (Riedel et al., 2018; Thomas et al., 2019). Reconstitution of GEF activity on liposomes helped here to unravel TRAPP complex substrate specificity, since in solution assays are not adequate to address some of the features important for specific interactions: Rab11/Ypt32 has a longer HVD between the prenyl anchor and the GTPase domain compared with Rab1/Ypt1 (Fig. 3 F, box). The HVD not only binds TRAPPII but also stretches a longer distance from the membrane (Fig. 3 F). Thereby it allows Rab11/Ypt32, but not Rab1/Ypt1, to reach the active site of membrane-bound TRAPPII. Thus, substrate specificity is controlled by the distance of the GTPase domain from the membrane surface, since the active site seems to be located on the opposing site of the complex from the site of membrane interaction (Fig. 3 F; Thomas et al., 2019). The smaller TRAPPIII has its active site closer to the membrane, binds Ypt1 via its shorter HVD, and facilitates its activation, while Ypt32 with its longer HVD may be positioned too far away from the active site. In addition, both complexes require their respective membrane environment for optimal activity, indicating how Arf and Rab GEFs cooperate in Golgi biogenesis.
The GEF DENND1 requires Arf5 for Rab35 activation
Recently, another example of Arf-mediated Rab activation was reported (Kulasekaran et al., 2021). Rab35, an endocytic Rab found at the plasma membrane and REs (Sato et al., 2008; Kouranti et al., 2006), is involved in cell adhesion and cell migration by controlling the trafficking of β1-integrin and the EGF receptor (Klinkert and Echard, 2016; Allaire et al., 2013). Arf5 binds the Rab35 GEF DENND1 and stimulates its GEF activity, with dysregulation of this cascade linked to glioblastoma growth (Kulasekaran et al., 2021). DENND1 GEF activity is initially autoinhibited and relieved by phosphorylation via the central Akt kinase (Fig. 3 G; Kulasekaran et al., 2015). Similarly, another DENN-domain containing GEF, DENND3, is phosphorylated by the autophagy-specific ULK kinase and then activates Rab12, a small GTPase involved in autophagosome trafficking (Xu et al., 2015). Thus, it seems that Rab GEF activation is more generally linked to other trafficking proteins, such as Arfs, and controlled by kinases and likely also phosphatases.
Lessons from reconstitution
Organelle biogenesis and maintenance in the endomembrane system are tightly linked to the correct spatial and temporal activation of Rab GTPases. A small yeast cell gets by with 11 Rabs, while human cells encode >60 (Hutagalung and Novick, 2011). Rab activation, and therefore membrane identity, of each organelle depends on the cognate GEF. This puts GEFs into the driver’s seat of any Rab-directed function at cellular membranes. It seems that GEFs integrate, by several regulatory loops, incoming signals from various sources such as membrane composition, cargo proteins, upstream GTPases, or kinases/phosphatases (Fig. 3 A). Yet our insights on the specific membrane targeting and regulation of GEFs remain incomplete for want of available experimental approaches. We briefly discuss here how recent advances on the reconstitution of GEF-mediated Rab activation at model membranes have advanced our understanding of organelle maturation and biogenesis.
Reconstitution of any reaction to uncover the essential constituents is limited by the available tools. GEFs, Rabs, Sec18/Munc1 proteins, tethering factors, and SNAREs are for instance required for membrane fusion (Fig. 3 A). Initial assays focused on SNAREs and revealed their important but rather inefficient fusogenicity (Weber et al., 1998). Further analyses uncovered critical activation steps for SNAREs (Malsam et al., 2012; Pobbati et al., 2006; Südhof and Rothman, 2009; Jahn and Scheller, 2006), yet fusion at physiological SNARE concentrations in various in vitro systems does not occur, unless assisted by chaperoning Sec1/Munc18 proteins and tethering factors (Bharat et al., 2014; Lai et al., 2017; Mima and Wickner, 2009; Ohya et al., 2009; Wickner and Rizo, 2017). Most tethers again depend on Rabs for their localization, and Rab localization to membranes requires a GEF (Cabrera and Ungermann, 2013), whose activity can be a limiting factor for fusion (Langemeyer et al., 2020, 2018b). The long avenue of understanding the mechanism and regulation of membrane fusion exemplifies the challenges in dissecting the complexity of a cellular reaction, but also demonstrates the powerful insights obtained from reconstitution of these processes.
GEFs determine the localization of the corresponding Rab, and consequently, Rabs follow their GEF if they are mistargeted (Gerondopoulos et al., 2012; Blümer et al., 2013; Cabrera and Ungermann, 2013). However, these anchor-away approaches completely bypass the tight cellular regulation of GEF activation by the mistargeting and additional overexpression of the GEF protein and may allow only statements about GEF/substrate specificity. The spatiotemporal activation of each GEF at the right organelle is vital for the timing of all downstream reactions. GEFs are recruited to membranes by coincidence detection, which includes membrane lipids such as PIPs, membrane packaging defects, and peripheral membrane proteins such as upstream Rabs or other small GTPases. This recruitment is often accompanied by the release from autoinhibition, which may be triggered or inhibited by other regulatory processes such as phosphorylation. It comes as no surprise that pathogens such as Legionella and Salmonella take advantage of the central function of GEFs to establish and nourish their intracellular organellar niche by manipulating small GTPase activity (Spanò and Galán, 2018).
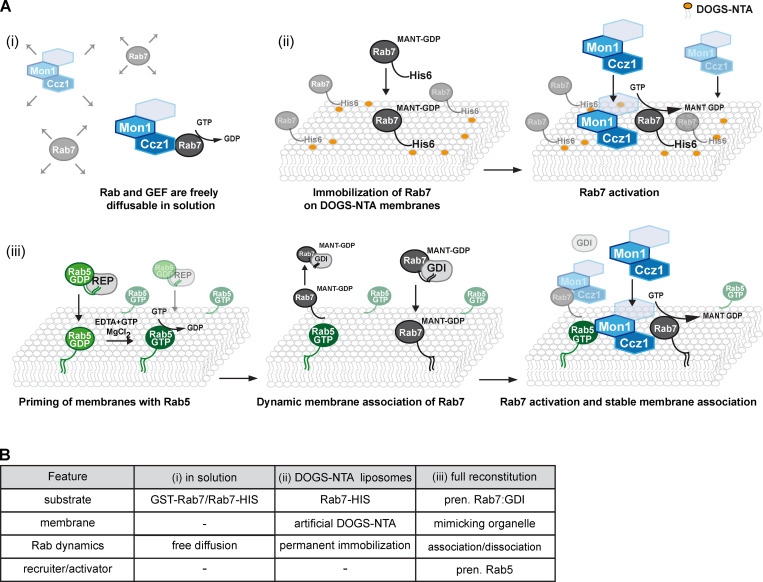
To understand the specificity of Rab GEFs (and GAPs), mostly very simplified systems were used. Most GEF assays analyze soluble Rabs loaded with fluorescent 2′-O-(N-methylanthraniloyl) (MANT)-nucleotide or radioactively labeled GTP/GDP and soluble GEF in a test tube, where nucleotide exchange activity is observed upon addition of unlabeled nucleotide (Fig. 4 A). This strategy allows the identification of substrate (Rab) specificity of GEFs, but could also lead to misleading results, as pointed out earlier on the example of the TRAPP complexes and Rab1/Ypt1 or Rab11/Ypt32. In addition, GEF-Rab pairs negatively regulated by one of the above principles could easily be missed.
Figure 4.
Approaches to determine GEF activity in vitro. Methods to determine GEF activity for Mon1-Ccz1. In all approaches, Rab7 is preloaded with fluorescent MANT-GDP. Fluorescence decreases upon GEF-mediated nucleotide exchange. (A) GEF assays. (Ai) In-solution Rab GEF assay. Mon1-Ccz1 (blue, Bulli/Rmc1/C18orf8 subunit, indicated by unlabeled hexagon) and Rab7 (gray) are freely diffusible in the test tube, which results in random collision and Rab activation. (Aii) GEF-mediated activation of artificially recruited Rab7 on liposomes. Rab7 with a C-terminal 6xHis-tag is permanently immobilized on membranes containing the cationic lipid DOGS-NTA. Mon1-Ccz1 unspecifically binds to this membrane surface and activates Rab7. Diffusion is limited to the membrane surface, thus increasing chances of interactions. (Aiii) Reconstitution of Rab5-mediated Rab7 activation by Mon1-Ccz1 on liposomes. Chemically activated, prenylated Rab5 (green), delivered to the membrane by the Rab Escort Protein (REP), allows Mon1-Ccz1 recruitment and Rab7 activation from the GDI complex (see text for further details). (B) Summary of Ai–Aiii. pren., prenylation.
As Rabs and GEFs function on membranes, we and others adopted strategies for measuring Rab activation by GEFs on membranes (Fig. 4 B). In a first approach, Rab and other small GTPases (Sot et al., 2013; Schmitt et al., 1994) were immobilized with C-terminal hexahistidine tags on liposomes containing the polycationic lipid 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (DOGS-NTA) and observed higher activity of the added GEF (Cabrera et al., 2014; Thomas and Fromme, 2016). A drawback of this technique is the artificial membrane composition. To avoid potential artifacts of unnaturally charged membranes and permanently membrane-bound Rab, recent studies relied on prenylated Rabs in complex with GDI. Reflecting the natural source of the cytoplasmic Rab pool, this complex was used as a GEF substrate in the presence of liposomes mimicking the natural membrane composition (Cezanne et al., 2020; Bezeljak et al., 2020; Langemeyer et al., 2020, 2018b; Thomas et al., 2018, 2019; Thomas and Fromme, 2016).
Even though these observations are recent, the outcome and the understanding of GEF regulation is encouraging. For the Rab5 GEF complex consisting of Rabex5 and Rabaptin5, GEF-dependent Rab5 recruitment to membranes revealed a self-organizing system, nonlinear Rab5 patterning, and collective switching of the Rab5 population (Bezeljak et al., 2020; Cezanne et al., 2020). This is in agreement with mathematical modeling and predictions on bistability and ultrasensitivity of Rab networks (Del Conte-Zerial et al., 2008; Barr, 2013). For the Golgi-resident TRAPPII and TRAPPIII complexes, the membrane composition, the length of the Rab HVD, and the presence of membrane-bound Arf1 determined the GEF specificity for their Rabs (Fig. 3 F; Thomas et al., 2019, 2018; Thomas and Fromme, 2016; Riedel et al., 2018), which is nicely supported by recent structural analyses of yeast and metazoan TRAPPIII (Galindo et al., 2021; Joiner et al., 2021)
Our own data uncovered that the yeast and metazoan Mon1-Ccz1(-RMC1) complex required membrane-bound Rab5-GTP to activate Rab7 out of the GDI complex (Langemeyer et al., 2020). Surprisingly, Rab5-GTP not only determined membrane binding of Mon1-Ccz1, but also activated the GEF on membranes by a yet-unknown mechanism (Fig. 1 C). Phosphorylation of yeast Mon1-Ccz1 by the casein kinase Yck3 inhibited this activation, demonstrating possible regulation of GEF activity (Fig. 3 C). Importantly, this finding agrees with the observed Rab5-to-Rab7 switch in vivo (Poteryaev et al., 2010; Rink et al., 2005).
Taken together, the available tools open exciting avenues for our understanding of organelle maturation. Reconstitution will allow the investigation of an entire Rab cascade and its regulation by kinases or membrane lipids. It will be possible to determine the cross-talk with lipid kinases and observe possible regulatory loops between Rabs and PI kinases (Tremel et al., 2021). We are confident that such analyses, complemented by in vivo analyses of Rabs or other small GTPases and their GEFs, will clarify the underlying mechanism of organelle maturation and biogenesis along the endomembrane system of eukaryotic cells.
Acknowledgments
Funding of this work was provided by grants from the Deutsche Forschungsgemeinschaft (SFB 944, P11, and UN111/13-1 to C. Ungermann).
The authors declare no competing financial interests.
Author contributions: The intial draft was written by A.-C. Borchers and L. Langemeyer, revised by C. Ungermann, and was approved by all authors.
References
- Achstetter, T., Franzusoff A., Field C., and Schekman R.. 1988. SEC7 encodes an unusual, high molecular weight protein required for membrane traffic from the yeast Golgi apparatus. J. Biol. Chem. 263:11711–11717. 10.1016/S0021-9258(18)37842-6 [DOI] [PubMed] [Google Scholar]
- Allaire, P.D., Seyed Sadr M., Chaineau M., Seyed Sadr E., Konefal S., Fotouhi M., Maret D., Ritter B., Del Maestro R.F., and McPherson P.S.. 2013. Interplay between Rab35 and Arf6 controls cargo recycling to coordinate cell adhesion and migration. J. Cell Sci. 126:722–731. 10.1242/jcs.112375 [DOI] [PubMed] [Google Scholar]
- Babst, M.2014. Quality control: quality control at the plasma membrane: one mechanism does not fit all. J. Cell Biol. 205:11–20. 10.1083/jcb.201310113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balderhaar, H.J., Arlt H., Ostrowicz C., Bröcker C., Sündermann F., Brandt R., Babst M., and Ungermann C.. 2010. The Rab GTPase Ypt7 is linked to retromer-mediated receptor recycling and fusion at the yeast late endosome. J. Cell Sci. 123:4085–4094. 10.1242/jcs.071977 [DOI] [PubMed] [Google Scholar]
- Balla, T.2013. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93:1019–1137. 10.1152/physrev.00028.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballabio, A., and Bonifacino J.S.. 2020. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. Cell Biol. 21:101–118. 10.1038/s41580-019-0185-4 [DOI] [PubMed] [Google Scholar]
- Barr, F.A.2013. Review series: Rab GTPases and membrane identity: causal or inconsequential? J. Cell Biol. 202:191–199. 10.1083/jcb.201306010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean, B.D.M., Davey M., Snider J., Jessulat M., Deineko V., Tinney M., Stagljar I., Babu M., and Conibear E.. 2015. Rab5-family guanine nucleotide exchange factors bind retromer and promote its recruitment to endosomes. Mol. Biol. Cell. 26:1119–1128. 10.1091/mbc.E14-08-1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergbrede, T., Chuky N., Schoebel S., Blankenfeldt W., Geyer M., Fuchs E., Goody R.S., Barr F., and Alexandrov K.. 2009. Biophysical analysis of the interaction of Rab6a GTPase with its effector domains. J. Biol. Chem. 284:2628–2635. 10.1074/jbc.M806003200 [DOI] [PubMed] [Google Scholar]
- Bezeljak, U., Loya H., Kaczmarek B., Saunders T.E., and Loose M.. 2020. Stochastic activation and bistability in a Rab GTPase regulatory network. Proc. Natl. Acad. Sci. USA. 117:6540–6549. 10.1073/pnas.1921027117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharat, T.A.M., Malsam J., Hagen W.J.H., Scheutzow A., Söllner T.H., and Briggs J.A.G.. 2014. SNARE and regulatory proteins induce local membrane protrusions to prime docked vesicles for fast calcium-triggered fusion. EMBO Rep. 15:308–314. 10.1002/embr.201337807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blümer, J., Rey J., Dehmelt L., Mazel T., Wu Y.W., Bastiaens P., Goody R.S., and Itzen A.. 2013. RabGEFs are a major determinant for specific Rab membrane targeting. J. Cell Biol. 200:287–300. 10.1083/jcb.201209113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino, J.S., and Neefjes J.. 2017. Moving and positioning the endolysosomal system. Curr. Opin. Cell Biol. 47:1–8. 10.1016/j.ceb.2017.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, S.L., Bi-Karchin J., Le L., and Marks M.S.. 2019. The road to lysosome-related organelles: Insights from Hermansky-Pudlak syndrome and other rare diseases. Traffic. 20:404–435. 10.1111/tra.12646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges, D., Fisher K., Zolov S.N., Xiong T., Inoki K., Weisman L.S., and Saltiel A.R.. 2012. Rab5 proteins regulate activation and localization of target of rapamycin complex 1. J. Biol. Chem. 287:20913–20921. 10.1074/jbc.M111.334060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera, M., and Ungermann C.. 2013. Guanine nucleotide exchange factors (GEFs) have a critical but not exclusive role in organelle localization of Rab GTPases. J. Biol. Chem. 288:28704–28712. 10.1074/jbc.M113.488213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera, M., Nordmann M., Perz A., Schmedt D., Gerondopoulos A., Barr F., Piehler J., Engelbrecht-Vandré S., and Ungermann C.. 2014. The Mon1-Ccz1 GEF activates the Rab7 GTPase Ypt7 via a longin-fold-Rab interface and association with PI3P-positive membranes. J. Cell Sci. 127:1043–1051. 10.1242/jcs.140921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabukusta, B., and Neefjes J.. 2018. Mechanisms of lysosomal positioning and movement. Traffic. 19:761–769. 10.1111/tra.12587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantalupo, G., Alifano P., Roberti V., Bruni C.B., and Bucci C.. 2001. Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. EMBO J. 20:683–693. 10.1093/emboj/20.4.683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano, B.M., Thelen A.M., Moldavski O., Feltes M., van der Welle R.E.N., Mydock-McGrane L., Jiang X., van Eijkeren R.J., Davis O.B., Louie S.M., et al. 2017. Lysosomal cholesterol activates mTORC1 via an SLC38A9-Niemann-Pick C1 signaling complex. Science. 355:1306–1311. 10.1126/science.aag1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cezanne, A., Lauer J., Solomatina A., Sbalzarini I.F., and Zerial M.. 2020. A non-linear system patterns Rab5 GTPase on the membrane. eLife. 9:e54434. 10.7554/elife.54434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, P.-I., Kong C., Su X., and Stahl P.D.. 2009. Rab5 isoforms differentially regulate the trafficking and degradation of epidermal growth factor receptors. J. Biol. Chem. 284:30328–30338. 10.1074/jbc.M109.034546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, P.-I., Schauer K., Kong C., Harding A.R., Goud B., and Stahl P.D.. 2014. Rab5 isoforms orchestrate a “division of labor” in the endocytic network; Rab5C modulates Rac-mediated cell motility. PLoS One. 9:e90384. 10.1371/journal.pone.0090384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforidis, S., McBride H.M., Burgoyne R.D., and Zerial M.. 1999a. The Rab5 effector EEA1 is a core component of endosome docking. Nature. 397:621–625. 10.1038/17618 [DOI] [PubMed] [Google Scholar]
- Christoforidis, S., Miaczynska M., Ashman K., Wilm M., Zhao L., Yip S.-C., Waterfield M.D., Backer J.M., and Zerial M.. 1999b. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat. Cell Biol. 1:249–252. 10.1038/12075 [DOI] [PubMed] [Google Scholar]
- Cui, Y., Zhao Q., Gao C., Ding Y., Zeng Y., Ueda T., Nakano A., and Jiang L.. 2014. Activation of the Rab7 GTPase by the MON1-CCZ1 Complex Is Essential for PVC-to-Vacuole Trafficking and Plant Growth in Arabidopsis. Plant Cell. 26:2080–2097. 10.1105/tpc.114.123141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day, K.J., Casler J.C., and Glick B.S.. 2018. Budding Yeast Has a Minimal Endomembrane System. Dev. Cell. 44:56–72.e4. 10.1016/j.devcel.2017.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Renzis, S., Sönnichsen B., and Zerial M.. 2002. Divalent Rab effectors regulate the sub-compartmental organization and sorting of early endosomes. Nat. Cell Biol. 4:124–133. 10.1038/ncb744 [DOI] [PubMed] [Google Scholar]
- Dehnen, L., Janz M., Verma J.K., Psathaki O.E., Langemeyer L., Fröhlich F., Heinisch J.J., Meyer H., Ungermann C., and Paululat A.. 2020. A trimeric metazoan Rab7 GEF complex is crucial for endocytosis and scavenger function. J. Cell Sci. 133:jcs247080. 10.1242/jcs.247080 [DOI] [PubMed] [Google Scholar]
- Del Conte-Zerial, P., Brusch L., Rink J.C., Collinet C., Kalaidzidis Y., Zerial M., and Deutsch A.. 2008. Membrane identity and GTPase cascades regulated by toggle and cut-out switches. Mol. Syst. Biol. 4:206. 10.1038/msb.2008.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delprato, A., and Lambright D.G.. 2007. Structural basis for Rab GTPase activation by VPS9 domain exchange factors. Nat. Struct. Mol. Biol. 14:406–412. 10.1038/nsmb1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirac-Svejstrup, A.B., Sumizawa T., and Pfeffer S.R.. 1997. Identification of a GDI displacement factor that releases endosomal Rab GTPases from Rab-GDI. EMBO J. 16:465–472. 10.1093/emboj/16.3.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distefano, M.B., Haugen L.H., Wang Y., Perdreau-Dahl H., Kjos I., Jia D., Morth J.P., Neefjes J., Bakke O., and Progida C.. 2018. TBC1D5 controls the GTPase cycle of Rab7b. J. Cell Sci. 131:jcs.216630. doi:. 10.1242/jcs.216630 [DOI] [PubMed]
- Dong, G., Medkova M., Novick P., and Reinisch K.M.. 2007. A catalytic coiled coil: structural insights into the activation of the Rab GTPase Sec4p by Sec2p. Mol. Cell. 25:455–462. 10.1016/j.molcel.2007.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke, C., Repnik U., Segeletz S., Brouilly N., Kalaidzidis Y., Verbavatz J.M., and Zerial M.. 2019. Correlative single-molecule localization microscopy and electron tomography reveals endosome nanoscale domains. Traffic. 20:601–617. 10.1111/tra.12671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, J.R., Dibenedetto J.R., West M., Rowland A.A., and Voeltz G.K.. 2013. Endoplasmic reticulum-endosome contact increases as endosomes traffic and mature. Mol. Biol. Cell. 24:1030–1040. 10.1091/mbc.e12-10-0733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, N., Huang W., Lin T.-H., Groulx J.-F., Jean S., Nguyen J., Kuchitsu Y., Koyama-Honda I., Mizushima N., Fukuda M., and Kiger A.A.. 2017. Genetic screen in Drosophila muscle identifies autophagy-mediated T-tubule remodeling and a Rab2 role in autophagy. eLife. 6:e23367. 10.7554/elife.23367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo, A., Planelles-Herrero V.J., Degliesposti G., and Munro S.. 2021. Cryo-EM structure of metazoan TRAPPIII, the multi-subunit complex that activates the GTPase Rab1. EMBO J. 40:e107608. 10.15252/embj.2020107608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, J., Langemeyer L., Kümmel D., Reggiori F., and Ungermann C.. 2018a. Molecular mechanism to target the endosomal Mon1-Ccz1 GEF complex to the pre-autophagosomal structure. eLife. 7:e31145. 10.7554/elife.31145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, J., Reggiori F., and Ungermann C.. 2018b. A novel in vitro assay reveals SNARE topology and the role of Ykt6 in autophagosome fusion with vacuoles. J. Cell Biol. 217:3670–3682. 10.1083/jcb.201804039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerondopoulos, A., Langemeyer L., Liang J.-R., Linford A., and Barr F.A.. 2012. BLOC-3 mutated in Hermansky-Pudlak syndrome is a Rab32/38 guanine nucleotide exchange factor. Curr. Biol. 22:2135–2139. 10.1016/j.cub.2012.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham, A.K., Sinka R., Torres I.L., Lilley K.S., and Munro S.. 2014. Toward a comprehensive map of the effectors of rab GTPases. Dev. Cell. 31:358–373. 10.1016/j.devcel.2014.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goody, R.S., Müller M.P., and Wu Y.-W.. 2017. Mechanisms of action of Rab proteins, key regulators of intracellular vesicular transport. Biol. Chem. 398:565–575. 10.1515/hsz-2016-0274 [DOI] [PubMed] [Google Scholar]
- Gorvel, J.P., Chavrier P., Zerial M., and Gruenberg J.. 1991. rab5 controls early endosome fusion in vitro. Cell. 64:915–925. 10.1016/0092-8674(91)90316-Q [DOI] [PubMed] [Google Scholar]
- Guerra, F., and Bucci C.. 2016. Multiple Roles of the Small GTPase Rab7. Cells. 5:34. 10.3390/cells5030034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson, M.A., and Fromme J.C.. 2017. Regulation of Arf activation occurs via distinct mechanisms at early and late Golgi compartments. Mol. Biol. Cell. 28:3660–3671. 10.1091/mbc.e17-06-0370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez, M.G., Munafó D.B., Berón W., and Colombo M.I.. 2004. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J. Cell Sci. 117:2687–2697. 10.1242/jcs.01114 [DOI] [PubMed] [Google Scholar]
- Hattula, K., Furuhjelm J., Arffman A., and Peränen J.. 2002. A Rab8-specific GDP/GTP exchange factor is involved in actin remodeling and polarized membrane transport. Mol. Biol. Cell. 13:3268–3280. 10.1091/mbc.e02-03-0143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedűs, K., Takáts S., Boda A., Jipa A., Nagy P., Varga K., Kovács A.L., and Juhász G.. 2016. The Ccz1-Mon1-Rab7 module and Rab5 control distinct steps of autophagy. Mol. Biol. Cell. 27:3132–3142. 10.1091/mbc.e16-03-0205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi, H., Lippé R., McBride H.M., Rubino M., Woodman P., Stenmark H., Rybin V., Wilm M., Ashman K., Mann M., and Zerial M.. 1997. A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell. 90:1149–1159. 10.1016/S0092-8674(00)80380-3 [DOI] [PubMed] [Google Scholar]
- Huotari, J., and Helenius A.. 2011. Endosome maturation. EMBO J. 30:3481–3500. 10.1038/emboj.2011.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutagalung, A.H., and Novick P.J.. 2011. Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 91:119–149. 10.1152/physrev.00059.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzen, A., Rak A., and Goody R.S.. 2007. Sec2 is a highly efficient exchange factor for the Rab protein Sec4. J. Mol. Biol. 365:1359–1367. 10.1016/j.jmb.2006.10.096 [DOI] [PubMed] [Google Scholar]
- Jahn, R., and Scheller R.H.. 2006. SNAREs--engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7:631–643. 10.1038/nrm2002 [DOI] [PubMed] [Google Scholar]
- Jeschke, A., and Haas A.. 2016. Deciphering the roles of phosphoinositide lipids in phagolysosome biogenesis. Commun. Integr. Biol. 9:e1174798. 10.1080/19420889.2016.1174798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Orgaz, A., Kvainickas A., Nägele H., Denner J., Eimer S., Dengjel J., and Steinberg F.. 2018. Control of RAB7 activity and localization through the retromer-TBC1D5 complex enables RAB7-dependent mitophagy. EMBO J. 37:235–254. 10.15252/embj.201797128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, D.E., Ostrowski P., Jaumouillé V., and Grinstein S.. 2016. The position of lysosomes within the cell determines their luminal pH. J. Cell Biol. 212:677–692. 10.1083/jcb.201507112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joiner, A.M., Phillips B.P., Yugandhar K., Sanford E.J., Smolka M.B., Yu H., Miller E.A., and Fromme J.C.. 2021. Structural basis of TRAPPIII-mediated Rab1 activation. EMBO J. 40:e107607. 10.15252/embj.2020107607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongsma, M.L.M., Berlin I., Wijdeven R.H.M., Janssen L., Janssen G.M.C., Garstka M.A., Janssen H., Mensink M., van Veelen P.A., Spaapen R.M., and Neefjes J.. 2016. An ER-Associated Pathway Defines Endosomal Architecture for Controlled Cargo Transport. Cell. 166:152–166. 10.1016/j.cell.2016.05.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongsma, M.L., Bakker J., Cabukusta B., Liv N., van Elsland D., Fermie J., Akkermans J.L., Kuijl C., van der Zanden S.Y., Janssen L., et al. 2020. SKIP-HOPS recruits TBC1D15 for a Rab7-to-Arl8b identity switch to control late endosome transport. EMBO J. 39:e102301. 10.15252/embj.2019102301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordens, I., Fernandez-Borja M., Marsman M., Dusseljee S., Janssen L., Calafat J., Janssen H., Wubbolts R., and Neefjes J.. 2001. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr. Biol. 11:1680–1685. 10.1016/S0960-9822(01)00531-0 [DOI] [PubMed] [Google Scholar]
- Kälin, S., Hirschmann D.T., Buser D.P., and Spiess M.. 2015. Rabaptin5 is recruited to endosomes by Rab4 and Rabex5 to regulate endosome maturation. J. Cell Sci. 128:4126–4137. 10.1242/jcs.174664 [DOI] [PubMed] [Google Scholar]
- Khatter, D., Raina V.B., Dwivedi D., Sindhwani A., Bahl S., and Sharma M.. 2015. The small GTPase Arl8b regulates assembly of the mammalian HOPS complex on lysosomes. J. Cell Sci. 128:1746–1761. 10.1242/jcs.162651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara, A., Noda T., Ishihara N., and Ohsumi Y.. 2001. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152:519–530. 10.1083/jcb.152.3.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinchen, J.M., and Ravichandran K.S.. 2010. Identification of two evolutionarily conserved genes regulating processing of engulfed apoptotic cells. Nature. 464:778–782. 10.1038/nature08853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke, S., Langemeyer L., Kuhlee A., Schuback S., Raunser S., Ungermann C., and Kümmel D.. 2017. Architecture and mechanism of the late endosomal Rab7-like Ypt7 guanine nucleotide exchange factor complex Mon1-Ccz1. Nat. Commun. 8:14034. 10.1038/ncomms14034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjos, I., Borg Distefano M., Sætre F., Repnik U., Holland P., Jones A.T., Engedal N., Simonsen A., Bakke O., and Progida C.. 2017. Rab7b modulates autophagic flux by interacting with Atg4B. EMBO Rep. 18:1727–1739. 10.15252/embr.201744069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinkert, K., and Echard A.. 2016. Rab35 GTPase: A Central Regulator of Phosphoinositides and F-actin in Endocytic Recycling and Beyond. Traffic. 17:1063–1077. 10.1111/tra.12422 [DOI] [PubMed] [Google Scholar]
- Knödler, A., Feng S., Zhang J., Zhang X., Das A., Peränen J., and Guo W.. 2010. Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc. Natl. Acad. Sci. USA. 107:6346–6351. 10.1073/pnas.1002401107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk, V.I., Saiki S., Lichtenberg M., Siddiqi F.H., Roberts E.A., Imarisio S., Jahreiss L., Sarkar S., Futter M., Menzies F.M., et al. 2011. Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 13:453–460. 10.1038/ncb2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranti, I., Sachse M., Arouche N., Goud B., and Echard A.. 2006. Rab35 regulates an endocytic recycling pathway essential for the terminal steps of cytokinesis. Curr. Biol. 16:1719–1725. 10.1016/j.cub.2006.07.020 [DOI] [PubMed] [Google Scholar]
- Kovtun, O., Leneva N., Bykov Y.S., Ariotti N., Teasdale R.D., Schaffer M., Engel B.D., Owen D.J., Briggs J.A.G., and Collins B.M.. 2018. Structure of the membrane-assembled retromer coat determined by cryo-electron tomography. Nature. 561:561–564. 10.1038/s41586-018-0526-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucera, A., Borg Distefano M., Berg-Larsen A., Skjeldal F., Repnik U., Bakke O., and Progida C.. 2016. Spatiotemporal Resolution of Rab9 and CI-MPR Dynamics in the Endocytic Pathway. Traffic. 17:211–229. 10.1111/tra.12357 [DOI] [PubMed] [Google Scholar]
- Kuchitsu, Y., and Fukuda M.. 2018. Revisiting Rab7 Functions in Mammalian Autophagy: Rab7 Knockout Studies. Cells. 7:215. 10.3390/cells7110215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulasekaran, G., Nossova N., Marat A.L., Lund I., Cremer C., Ioannou M.S., and McPherson P.S.. 2015. Phosphorylation-dependent Regulation of Connecdenn/DENND1 Guanine Nucleotide Exchange Factors. J. Biol. Chem. 290:17999–18008. 10.1074/jbc.M115.636712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulasekaran, G., Chaineau M., Piscopo V.E.C., Verginelli F., Fotouhi M., Girard M., Tang Y., Dali R., Lo R., Stifani S., and McPherson P.S.. 2021. An Arf/Rab cascade controls the growth and invasiveness of glioblastoma. J. Cell Biol. 220:e202004229. 10.1083/jcb.202004229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvainickas, A., Nägele H., Qi W., Dokládal L., Jimenez-Orgaz A., Stehl L., Gangurde D., Zhao Q., Hu Z., Dengjel J., et al. 2019. Retromer and TBC1D5 maintain late endosomal RAB7 domains to enable amino acid-induced mTORC1 signaling. J. Cell Biol. 218:3019–3038. 10.1083/jcb.201812110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, Y., Choi U.B., Leitz J., Rhee H.J., Lee C., Altas B., Zhao M., Pfuetzner R.A., Wang A.L., Brose N., et al. 2017. Molecular Mechanisms of Synaptic Vesicle Priming by Munc13 and Munc18. Neuron. 95:591–607.e10. 10.1016/j.neuron.2017.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidlaw, K.M.E., and MacDonald C.. 2018. Endosomal trafficking of yeast membrane proteins. Biochem. Soc. Trans. 46:1551–1558. 10.1042/BST20180258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langemeyer, L., Fröhlich F., and Ungermann C.. 2018a. Rab GTPase Function in Endosome and Lysosome Biogenesis. Trends Cell Biol. 28:957–970. 10.1016/j.tcb.2018.06.007 [DOI] [PubMed] [Google Scholar]
- Langemeyer, L., Perz A., Kümmel D., and Ungermann C.. 2018b. A guanine nucleotide exchange factor (GEF) limits Rab GTPase-driven membrane fusion. J. Biol. Chem. 293:731–739. 10.1074/jbc.M117.812941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langemeyer, L., Borchers A.-C., Herrmann E., Füllbrunn N., Han Y., Perz A., Auffarth K., Kümmel D., and Ungermann C.. 2020. A conserved and regulated mechanism drives endosomal Rab transition. eLife. 9:e56090. 10.7554/elife.56090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law, F., Seo J.H., Wang Z., DeLeon J.L., Bolis Y., Brown A., Zong W.-X., Du G., and Rocheleau C.E.. 2017. The VPS34 PI3K negatively regulates RAB-5 during endosome maturation. J. Cell Sci. 130:2007–2017. 10.1242/jcs.194746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, G., Brown C.C., Flood B.A., Karunakaran S., Cabrera M., Nordmann M., Ungermann C., and Fratti R.A.. 2014. Dynamic association of the PI3P-interacting Mon1-Ccz1 GEF with vacuoles is controlled through its phosphorylation by the type 1 casein kinase Yck3. Mol. Biol. Cell. 25:1608–1619. 10.1091/mbc.e13-08-0460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., Tsai Y.C., Mattera R., Smith W.J., Kostelansky M.S., Weissman A.M., Bonifacino J.S., and Hurley J.H.. 2006. Structural basis for ubiquitin recognition and autoubiquitination by Rabex-5. Nat. Struct. Mol. Biol. 13:264–271. 10.1038/nsmb1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leneva, N., Kovtun O., Morado D.R., Briggs J.A.G., and Owen D.J.. 2021. Architecture and mechanism of metazoan retromer:SNX3 tubular coat assembly. Sci. Adv. 7:eabf8598. 10.1126/sciadv.abf8598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Li B., Liu L., Chen H., Zhang H., Zheng X., and Zhang Z.. 2015. FgMon1, a guanine nucleotide exchange factor of FgRab7, is important for vacuole fusion, autophagy and plant infection in Fusarium graminearum. Sci. Rep. 5:18101. 10.1038/srep18101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linford, A., Yoshimura S., Bastos R.N., Langemeyer L., Gerondopoulos A., Rigden D.J., and Barr F.A.. 2012. Rab14 and Its Exchange Factor FAM116 Link Endocytic Recycling and Adherens Junction Stability in Migrating Cells. Dev. Cell. 22:952–966. 10.1016/j.devcel.2012.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling, Y., Hayano S., and Novick P.. 2014. Osh4p is needed to reduce the level of phosphatidylinositol-4-phosphate on secretory vesicles as they mature. Mol. Biol. Cell. 25:3389–3400. 10.1091/mbc.e14-06-1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatova, Z., and Segev N.. 2019. Ypt/Rab GTPases and their TRAPP GEFs at the Golgi. FEBS Lett. 593:2488–2500. 10.1002/1873-3468.13574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippé, R., Miaczynska M., Rybin V., Runge A., and Zerial M.. 2001. Functional synergy between Rab5 effector Rabaptin-5 and exchange factor Rabex-5 when physically associated in a complex. Mol. Biol. Cell. 12:2219–2228. 10.1091/mbc.12.7.2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, T.-T., Gomez T.S., Sackey B.K., Billadeau D.D., and Burd C.G.. 2012. Rab GTPase regulation of retromer-mediated cargo export during endosome maturation. Mol. Biol. Cell. 23:2505–2515. 10.1091/mbc.e11-11-0915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi, D., Soldati T., Riederer M.A., Goda Y., Zerial M., and Pfeffer S.R.. 1993. Rab9 functions in transport between late endosomes and the trans Golgi network. EMBO J. 12:677–682. 10.1002/j.1460-2075.1993.tb05701.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lőrincz, P., Tóth S., Benkő P., Lakatos Z., Boda A., Glatz G., Zobel M., Bisi S., Hegedűs K., Takáts S., et al. 2017. Rab2 promotes autophagic and endocytic lysosomal degradation. J. Cell Biol. 216:1937–1947. 10.1083/jcb.201611027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malsam, J., Parisotto D., Bharat T.A.M., Scheutzow A., Krause J.M., Briggs J.A.G., and Söllner T.H.. 2012. Complexin arrests a pool of docked vesicles for fast Ca2+-dependent release. EMBO J. 31:3270–3281. 10.1038/emboj.2012.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marat, A.L., and Haucke V.. 2016. Phosphatidylinositol 3-phosphates-at the interface between cell signalling and membrane traffic. EMBO J. 35:561–579. 10.15252/embj.201593564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marwaha, R., Arya S.B., Jagga D., Kaur H., Tuli A., and Sharma M.. 2017. The Rab7 effector PLEKHM1 binds Arl8b to promote cargo traffic to lysosomes. J. Cell Biol. 216:1051–1070. 10.1083/jcb.201607085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattera, R., Tsai Y.C., Weissman A.M., and Bonifacino J.S.. 2006. The Rab5 guanine nucleotide exchange factor Rabex-5 binds ubiquitin (Ub) and functions as a Ub ligase through an atypical Ub-interacting motif and a zinc finger domain. J. Biol. Chem. 281:6874–6883. 10.1074/jbc.M509939200 [DOI] [PubMed] [Google Scholar]
- McDonold, C.M., and Fromme J.C.. 2014. Four GTPases differentially regulate the Sec7 Arf-GEF to direct traffic at the trans-golgi network. Dev. Cell. 30:759–767. 10.1016/j.devcel.2014.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan, D.G., Popovic D., Gubas A., Terawaki S., Suzuki H., Stadel D., Coxon F.P., Miranda de Stegmann D., Bhogaraju S., Maddi K., et al. 2015. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol. Cell. 57:39–54. 10.1016/j.molcel.2014.11.006 [DOI] [PubMed] [Google Scholar]
- Medkova, M., France Y.E., Coleman J., and Novick P.. 2006. The rab exchange factor Sec2p reversibly associates with the exocyst. Mol. Biol. Cell. 17:2757–2769. 10.1091/mbc.e05-10-0917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mima, J., and Wickner W.. 2009. Complex lipid requirements for SNARE- and SNARE chaperone-dependent membrane fusion. J. Biol. Chem. 284:27114–27122. 10.1074/jbc.M109.010223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno-Yamasaki, E., Medkova M., Coleman J., and Novick P.. 2010. Phosphatidylinositol 4-phosphate controls both membrane recruitment and a regulatory switch of the Rab GEF Sec2p. Dev. Cell. 18:828–840. 10.1016/j.devcel.2010.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa, H.2020. Mechanisms governing autophagosome biogenesis. Nat. Rev. Mol. Cell Biol. 21:439–458. 10.1038/s41580-020-0241-0 [DOI] [PubMed] [Google Scholar]
- Nordmann, M., Cabrera M., Perz A., Bröcker C., Ostrowicz C., Engelbrecht-Vandré S., and Ungermann C.. 2010. The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr. Biol. 20:1654–1659. 10.1016/j.cub.2010.08.002 [DOI] [PubMed] [Google Scholar]
- Ohya, T., Miaczynska M., Coskun U., Lommer B., Runge A., Drechsel D., Kalaidzidis Y., and Zerial M.. 2009. Reconstitution of Rab- and SNARE-dependent membrane fusion by synthetic endosomes. Nature. 459:1091–1097. 10.1038/nature08107 [DOI] [PubMed] [Google Scholar]
- Ortiz, D., Medkova M., Walch-Solimena C., and Novick P.. 2002. Ypt32 recruits the Sec4p guanine nucleotide exchange factor, Sec2p, to secretory vesicles; evidence for a Rab cascade in yeast. J. Cell Biol. 157:1005–1015. 10.1083/jcb.200201003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perini, E.D., Schaefer R., Stöter M., Kalaidzidis Y., and Zerial M.. 2014. Mammalian CORVET is required for fusion and conversion of distinct early endosome subpopulations. Traffic. 15:1366–1389. 10.1111/tra.12232 [DOI] [PubMed] [Google Scholar]
- Pobbati, A.V., Stein A., and Fasshauer D.. 2006. N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science. 313:673–676. 10.1126/science.1129486 [DOI] [PubMed] [Google Scholar]
- Ponsford, A.H., Ryan T.A., Raimondi A., Cocucci E., Wycislo S.A., Fröhlich F., Swan L.E., and Stagi M.. 2021. Live imaging of intra-lysosome pH in cell lines and primary neuronal culture using a novel genetically encoded biosensor. Autophagy. 17:1500–1518. 10.1080/15548627.2020.1771858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteryaev, D., Datta S., Ackema K., Zerial M., and Spang A.. 2010. Identification of the switch in early-to-late endosome transition. Cell. 141:497–508. 10.1016/j.cell.2010.03.011 [DOI] [PubMed] [Google Scholar]
- Progida, C., Cogli L., Piro F., De Luca A., Bakke O., and Bucci C.. 2010. Rab7b controls trafficking from endosomes to the TGN. J. Cell Sci. 123:1480–1491. 10.1242/jcs.051474 [DOI] [PubMed] [Google Scholar]
- Raiborg, C., Wenzel E.M., and Stenmark H.. 2015. ER-endosome contact sites: molecular compositions and functions. EMBO J. 34:1848–1858. 10.15252/embj.201591481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar, B., Imarisio S., Sarkar S., O’Kane C.J., and Rubinsztein D.C.. 2008. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J. Cell Sci. 121:1649–1660. 10.1242/jcs.025726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinisch, K.M., and Prinz W.A.. 2021. Mechanisms of nonvesicular lipid transport. J. Cell Biol. 220:e202012058. 10.1083/jcb.202012058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, B.C., McDonold C.M., and Fromme J.C.. 2012. The Sec7 Arf-GEF is recruited to the trans-Golgi network by positive feedback. Dev. Cell. 22:799–810. 10.1016/j.devcel.2012.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, B.C., Halaby S.L., Gustafson M.A., and Fromme J.C.. 2016. The Sec7 N-terminal regulatory domains facilitate membrane-proximal activation of the Arf1 GTPase. eLife. 5:e12411. 10.7554/eLife.12411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel, F., Galindo A., Muschalik N., and Munro S.. 2018. The two TRAPP complexes of metazoans have distinct roles and act on different Rab GTPases. J. Cell Biol. 217:601–617. 10.1083/jcb.201705068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink, J., Ghigo E., Kalaidzidis Y., and Zerial M.. 2005. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 122:735–749. 10.1016/j.cell.2005.06.043 [DOI] [PubMed] [Google Scholar]
- Rocha, N., Kuijl C., van der Kant R., Janssen L., Houben D., Janssen H., Zwart W., and Neefjes J.. 2009. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J. Cell Biol. 185:1209–1225. 10.1083/jcb.200811005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas, R., van Vlijmen T., Mardones G.A., Prabhu Y., Rojas A.L., Mohammed S., Heck A.J.R., Raposo G., van der Sluijs P., and Bonifacino J.S.. 2008. Regulation of retromer recruitment to endosomes by sequential action of Rab5 and Rab7. J. Cell Biol. 183:513–526. 10.1083/jcb.200804048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland, A.A., Chitwood P.J., Phillips M.J., and Voeltz G.K.. 2014. ER contact sites define the position and timing of endosome fission. Cell. 159:1027–1041. 10.1016/j.cell.2014.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardana, R., and Emr S.D.. 2021. Membrane Protein Quality Control Mechanisms in the Endo-Lysosome System. Trends Cell Biol. 31:269–283. 10.1016/j.tcb.2020.11.011 [DOI] [PubMed] [Google Scholar]
- Sato, M., Sato K., Liou W., Pant S., Harada A., and Grant B.D.. 2008. Regulation of endocytic recycling by C. elegans Rab35 and its regulator RME-4, a coated-pit protein. EMBO J. 27:1183–1196. 10.1038/emboj.2008.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt, L., Dietrich C., and Tampe R.. 1994. Synthesis and Characterization of Chelator-Lipids for Reversible Immobilization of Engineered Proteins at Self-Assembled Lipid Interfaces. J. Am. Chem. Soc. 116:8485–8491. 10.1021/ja00098a008 [DOI] [Google Scholar]
- Schoebel, S., Oesterlin L.K., Blankenfeldt W., Goody R.S., and Itzen A.. 2009. RabGDI displacement by DrrA from Legionella is a consequence of its guanine nucleotide exchange activity. Mol. Cell. 36:1060–1072. 10.1016/j.molcel.2009.11.014 [DOI] [PubMed] [Google Scholar]
- Schu, P.V., Takegawa K., Fry M.J., Stack J.H., Waterfield M.D., and Emr S.D.. 1993. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 260:88–91. 10.1126/science.8385367 [DOI] [PubMed] [Google Scholar]
- Scorrano, L., De Matteis M.A., Emr S., Giordano F., Hajnóczky G., Kornmann B., Lackner L.L., Levine T.P., Pellegrini L., Reinisch K., et al. 2019. Coming together to define membrane contact sites. Nat. Commun. 10:1287. 10.1038/s41467-019-09253-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman, M.N.J., Harbour M.E., Tattersall D., Read E., and Bright N.. 2009. Membrane recruitment of the cargo-selective retromer subcomplex is catalysed by the small GTPase Rab7 and inhibited by the Rab-GAP TBC1D5. J. Cell Sci. 122:2371–2382. 10.1242/jcs.048686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, H.R., and Zoncu R.. 2020. The Lysosome at the Intersection of Cellular Growth and Destruction. Dev. Cell. 54:226–238. 10.1016/j.devcel.2020.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti, B., and Cullen P.J.. 2018. Endosomal Sorting: Architecture of the Retromer Coat. Curr. Biol. 28:R1350–R1352. 10.1016/j.cub.2018.10.040 [DOI] [PubMed] [Google Scholar]
- Singer-Krüger, B., Stenmark H., Düsterhöft A., Philippsen P., Yoo J.S., Gallwitz D., and Zerial M.. 1994. Role of three rab5-like GTPases, Ypt51p, Ypt52p, and Ypt53p, in the endocytic and vacuolar protein sorting pathways of yeast. J. Cell Biol. 125:283–298. 10.1083/jcb.125.2.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, M.K., Krüger F., Beckmann H., Brumm S., Vermeer J.E.M., Munnik T., Mayer U., Stierhof Y.-D., Grefen C., Schumacher K., and Jürgens G.. 2014. Protein delivery to vacuole requires SAND protein-dependent Rab GTPase conversion for MVB-vacuole fusion. Curr. Biol. 24:1383–1389. 10.1016/j.cub.2014.05.005 [DOI] [PubMed] [Google Scholar]
- Skjeldal, F.M., Haugen L.H., Mateus D., Frei D.M., Rødseth A.V., Hu X., and Bakke O.. 2021. De novo formation of early endosomes during Rab5 to Rab7 transition. J. Cell Sci. 134:jcs.254185. doi:. 10.1242/jcs.254185 [DOI] [PMC free article] [PubMed]
- Sot, B., Behrmann E., Raunser S., and Wittinghofer A.. 2013. Ras GTPase activating (RasGAP) activity of the dual specificity GAP protein Rasal requires colocalization and C2 domain binding to lipid membranes. Proc. Natl. Acad. Sci. USA. 110:111–116. 10.1073/pnas.1201658110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanò, S., and Galán J.E.. 2018. Taking control: Hijacking of Rab GTPases by intracellular bacterial pathogens. Small GTPases. 9:182–191. 10.1080/21541248.2017.1336192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder, D., and Novick P.J.. 2016. The casein kinases Yck1p and Yck2p act in the secretory pathway, in part, by regulating the Rab exchange factor Sec2p. Mol. Biol. Cell. 27:686–701. 10.1091/mbc.E15-09-0651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder, D., Mizuno-Yamasaki E., Ghassemian M., and Novick P.J.. 2013. Phosphorylation of the Rab exchange factor Sec2p directs a switch in regulatory binding partners. Proc. Natl. Acad. Sci. USA. 110:19995–20002. 10.1073/pnas.1320029110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof, T.C., and Rothman J.E.. 2009. Membrane fusion: grappling with SNARE and SM proteins. Science. 323:474–477. 10.1126/science.1161748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztul, E., Chen P.-W., Casanova J.E., Cherfils J., Dacks J.B., Lambright D.G., Lee F.S., Randazzo P.A., Santy L.C., Schürmann A., et al. 2019. ARF GTPases and their GEFs and GAPs: concepts and challenges. Mol. Biol. Cell. 30:1249–1271. 10.1091/mbc.E18-12-0820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, S., Kubo K., Waguri S., Yabashi A., Shin H.-W., Katoh Y., and Nakayama K.. 2012. Rab11 regulates exocytosis of recycling vesicles at the plasma membrane. J. Cell Sci. 125:4049–4057. 10.1242/jcs.102913 [DOI] [PubMed] [Google Scholar]
- Thomas, L.L., and Fromme J.C.. 2016. GTPase cross talk regulates TRAPPII activation of Rab11 homologues during vesicle biogenesis. J. Cell Biol. 215:499–513. 10.1083/jcb.201608123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, L.L., Joiner A.M.N., and Fromme J.C.. 2018. The TRAPPIII complex activates the GTPase Ypt1 (Rab1) in the secretory pathway. J. Cell Biol. 217:283–298. 10.1083/jcb.201705214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, L.L., van der Vegt S.A., and Fromme J.C.. 2019. A Steric Gating Mechanism Dictates the Substrate Specificity of a Rab-GEF. Dev. Cell. 48:100–114.e9. 10.1016/j.devcel.2018.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremel, S., Ohashi Y., Morado D.R., Bertram J., Perisic O., Brandt L.T.L., von Wrisberg M.-K., Chen Z.A., Maslen S.L., Kovtun O., et al. 2021. Structural basis for VPS34 kinase activation by Rab1 and Rab5 on membranes. Nat. Commun. 12:1564. 10.1038/s41467-021-21695-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungermann, C., and Kümmel D.. 2019. Structure of membrane tethers and their role in fusion. Traffic. 20:479–490. 10.1111/tra.12655 [DOI] [PubMed] [Google Scholar]
- Vaites, L.P., Paulo J.A., Huttlin E.L., and Harper J.W.. 2017. Systematic Analysis of Human Cells Lacking ATG8 Proteins Uncovers Roles for GABARAPs and the CCZ1/MON1 Regulator C18orf8/RMC1 in Macroautophagic and Selective Autophagic Flux. Mol. Cell. Biol. 38:e00392-17. 10.1128/MCB.00392-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Boomen, D.J.H., Sienkiewicz A., Berlin I., Jongsma M.L.M., van Elsland D.M., Luzio J.P., Neefjes J.J.C., and Lehner P.J.. 2020. A trimeric Rab7 GEF controls NPC1-dependent lysosomal cholesterol export. Nat. Commun. 11:5559. 10.1038/s41467-020-19032-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kant, R., Fish A., Janssen L., Janssen H., Krom S., Ho N., Brummelkamp T., Carette J., Rocha N., and Neefjes J.. 2013. Late endosomal transport and tethering are coupled processes controlled by RILP and the cholesterol sensor ORP1L. J. Cell Sci. 126:3462–3474. 10.1242/jcs.129270 [DOI] [PubMed] [Google Scholar]
- van der Kant, R., Jonker C.T.H., Wijdeven R.H., Bakker J., Janssen L., Klumperman J., and Neefjes J.. 2015. Characterization of the Mammalian CORVET and HOPS Complexes and Their Modular Restructuring for Endosome Specificity. J. Biol. Chem. 290:30280–30290. 10.1074/jbc.M115.688440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Solimena, C., Collins R.N., and Novick P.J.. 1997. Sec2p mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post-Golgi vesicles. J. Cell Biol. 137:1495–1509. 10.1083/jcb.137.7.1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallroth, A., and Haucke V.. 2018. Phosphoinositide conversion in endocytosis and the endolysosomal system. J. Biol. Chem. 293:1526–1535. 10.1074/jbc.r117.000629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandinger-Ness, A., and Zerial M.. 2014. Rab proteins and the compartmentalization of the endosomal system. Cold Spring Harb. Perspect. Biol. 6:a022616. 10.1101/cshperspect.a022616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Ren J., Wu B., Feng S., Cai G., Tuluc F., Peränen J., and Guo W.. 2015. Activation of Rab8 guanine nucleotide exchange factor Rabin8 by ERK1/2 in response to EGF signaling. Proc. Natl. Acad. Sci. USA. 112:148–153. 10.1073/pnas.1412089112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmeier, C., Romao M., Plowright L., Bennett D.C., Raposo G., and Seabra M.C.. 2006. Rab38 and Rab32 control post-Golgi trafficking of melanogenic enzymes. J. Cell Biol. 175:271–281. 10.1083/jcb.200606050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, T., Zemelman B.V., McNew J.A., Westermann B., Gmachl M., Parlati F., Söllner T.H., and Rothman J.E.. 1998. SNAREpins: minimal machinery for membrane fusion. Cell. 92:759–772. 10.1016/S0092-8674(00)81404-X [DOI] [PubMed] [Google Scholar]
- Wichmann, H., Hengst L., and Gallwitz D.. 1992. Endocytosis in yeast: evidence for the involvement of a small GTP-binding protein (Ypt7p). Cell. 71:1131–1142. 10.1016/S0092-8674(05)80062-5 [DOI] [PubMed] [Google Scholar]
- Wickner, W., and Rizo J.. 2017. A cascade of multiple proteins and lipids catalyzes membrane fusion. Mol. Biol. Cell. 28:707–711. 10.1091/mbc.e16-07-0517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H., and Voeltz G.K.. 2021. Reticulon-3 Promotes Endosome Maturation at ER Membrane Contact Sites. Dev. Cell. 56:52–66.e7. 10.1016/j.devcel.2020.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, R., Zhou H., Jian Y., Li L., Wang M., Liu N., Yin Q., Liang Z., Guo W., and Yang C.. 2021. The Rab7 effector WDR91 promotes autophagy-lysosome degradation in neurons by regulating lysosome fusion. J. Cell Biol. 220:e202007061. 10.1083/jcb.202007061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J., Fotouhi M., and McPherson P.S.. 2015. Phosphorylation of the exchange factor DENND3 by ULK in response to starvation activates Rab12 and induces autophagy. EMBO Rep. 16:709–718. 10.15252/embr.201440006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano, K., Wang C., Sarraf S.A., Münch C., Kikuchi R., Noda N.N., Hizukuri Y., Kanemaki M.T., Harper W., Tanaka K., et al. 2018. Endosomal Rab cycles regulate Parkin-mediated mitophagy. eLife. 7:e31326. 10.7554/eLife.31326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda, S., Morishita S., Fujita A., Nanao T., Wada N., Waguri S., Schiavo G., Fukuda M., and Nakamura T.. 2016. Mon1-Ccz1 activates Rab7 only on late endosomes and dissociates from the lysosome in mammalian cells. J. Cell Sci. 129:329–340. 10.1242/jcs.178095 [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Zhang T., Wang S., Gong Z., Tang C., Chen J., and Ding J.. 2014. Molecular mechanism for Rabex-5 GEF activation by Rabaptin-5. eLife. 3:e02687. 10.7554/eLife.02687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y.G., and Zhang H.. 2019. Autophagosome maturation: An epic journey from the ER to lysosomes. J. Cell Biol. 218:757–770. 10.1083/jcb.201810099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, F., Zou S., Chen Y., Lipatova Z., Sun D., Zhu X., Li R., Wu Z., You W., Cong X., et al. 2017. A Rab5 GTPase module is important for autophagosome closure. PLoS Genet. 13:e1007020–e1007024. 10.1371/journal.pgen.1007020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, F., Wu Z., Zhao M., Murtazina R., Cai J., Zhang A., Li R., Sun D., Li W., Zhao L., et al. 2019. Rab5-dependent autophagosome closure by ESCRT. J. Cell Biol. 218:1908–1927. 10.1083/jcb.201811173 [DOI] [PMC free article] [PubMed] [Google Scholar]