Summary
The mesenchymal subtype of glioblastoma is thought to be determined by both cancer cell intrinsic alterations and extrinsic cellular interactions, but remains poorly understood. Here, we dissect glioblastoma-to-microenvironment interactions by single-cell RNA-sequencing analysis of human tumors and model systems, combined with functional experiments. We demonstrate that macrophages induce a transition of glioblastoma cells into mesenchymal-like (MES-like) states. This effect is mediated, both in vitro and in vivo, by macrophage-derived Oncostatin M (OSM) that interacts with its receptors (OSMR or LIFR) in complex with GP130 on glioblastoma cells and activates STAT3. We show that MES-like glioblastoma states are also associated with increased expression of a mesenchymal program in macrophages and with increased cytotoxicity of T cells, highlighting extensive alterations of the immune microenvironment with potential therapeutic implications.
Keywords: Glioblastoma, GBM, scRNA-seq, Mesenchymal, Tumor microenvironment, Macrophage, OSM
Introduction
Glioblastoma, isocitrate dehydrogenase (IDH)-wildtype, is a universally lethal form of brain cancer with no effective treatment available (Louis et al., 2016; Stupp et al., 2005; Wen et al., 2020). Despite extensive efforts to translate the basic glioblastoma biology to patients, molecularly targeted therapies have largely failed in trials over the past decade (Gilbert et al., 2014; Wen et al., 2020). Intra-tumoral heterogeneity in glioblastoma may confer resistance to targeted therapies (Patel et al., 2014; Sottoriva et al., 2013), such as receptor tyrosine kinase inhibitors (Furnari et al., 2015; Koga et al., 2019) or an integrin antagonist that is effective only for a subset of glioblastoma cells (Cosset et al., 2017). Moreover, multiple studies demonstrated spatio-temporal heterogeneity among transcriptionally-defined tumor subtypes, such as The Cancer Genome Atlas (TCGA) Proneural (TCGA-PN), Mesenchymal (TCGA-MES) and Classical (TCGA-CL) (Rooj et al., 2017; Verhaak et al., 2010; Wang et al., 2017), which may provide further mechanisms to tolerate therapeutics (Shibue and Weinberg, 2017). Thus, understanding cellular diversity and plasticity in glioblastoma is essential both for predicting cellular behaviors under treatment and for discovering key regulators to manage such heterogeneity.
Single-cell RNA-sequencing (scRNA-seq) and advanced computational analyses now allow to comprehensively identify recurrent cellular states across tumors that are affected by both cell-intrinsic and -extrinsic factors (Tanay and Regev, 2017; Tirosh and Suva, 2019). By applying scRNA-seq to glioblastoma patient samples, we previously found that the diverse malignant cells in glioblastoma converge to a limited set of four cellular states: neural-progenitor-like (NPC-like), oligodendrocyte-progenitor-like (OPC-like), astrocyte-like (AC-like), and mesenchymal-like (MES-like), with potential for state plasticity (Neftel et al., 2019). We also delineated the expression programs of infiltrating macrophages and resident microglia (that we collectively refer to as macrophages) (Neftel et al., 2019; Venteicher et al., 2017). While these cellular states provide cellular building blocks to guide our understanding of tumor heterogeneity, much remains to be determined regarding their functional impact and therapeutic sensitivity as well as the mechanisms underlying their emergence and plasticity. The gene expression programs of NPC-like, OPC-like and AC-like states are anchored in neurodevelopment and the frequency of these states in a tumor is influenced by genetic drivers of glioblastoma, such as CDK4, PDGFRA and EGFR, providing insights into the rules governing their emergence (Neftel et al., 2019). In contrast, the MES-like state appears to have only limited similarity to cell types detected in the physiological healthy human brain and it is only partially associated with genetic alterations of glioblastoma and thus remains poorly understood. Prior work showed that the bulk TCGA-MES subtype is correlated with abundance of macrophages and that NF1 mutations or deletions, enriched in TCGA-MES tumors, increase the recruitment of macrophages (Wang et al., 2017). Thus, TCGA-MES is in part associated with increased abundance of macrophages in the tumor, which might also explain its predominance in recurrent settings (Ozawa et al., 2014). In spite of these observations, what drives the emergence of a MES-like cancer state in glioblastoma has yet to be addressed, and any potential role(s) of cell-extrinsic factors on either the cancer or immune cells remain to be elucidated.
Here, we leverage a mouse model and gliomasphere models recapitulating glioblastoma cellular states to demonstrate that macrophages induce the MES-like glioblastoma cell state in vivo and in vitro. Analysis of ligand-receptor pairs coupled to functional approaches implicate macrophage-derived Oncostatin M (OSM) and its receptors (OSMR or LIFR) in complex with GP130 on glioblastoma cells, in activating STAT3 signaling to induce a MES-like state. Surprisingly, we find MES-like states both in malignant cells and in macrophages. We then show that glioblastoma MES-like states are associated with increased abundance and cytotoxicity of tumor infiltrating T-cells. Our work describes a functional interaction between immune cells and the cellular states that drive glioblastoma.
Results
Macrophages induce transition of cancer cells into a MES-like state
To better understand the composition of bulk subtypes of glioblastoma (as defined by TCGA), we leveraged the scRNA-seq datasets and examined the average expression of signatures of the three bulk subtypes within cell types and cellular states. This analysis demonstrated that the TCGA-MES signature is highly expressed in MES-like cancer cells, but even more so in immune cells, including macrophages, microglia and, to a more limited degree, T-cells (Figures 1A and S1A). This supports previous studies that suggested an association between macrophages and mesenchymal programs in glioblastoma (Bhat et al., 2013; Wang et al., 2017), yet whether this association is causal remains unclear.
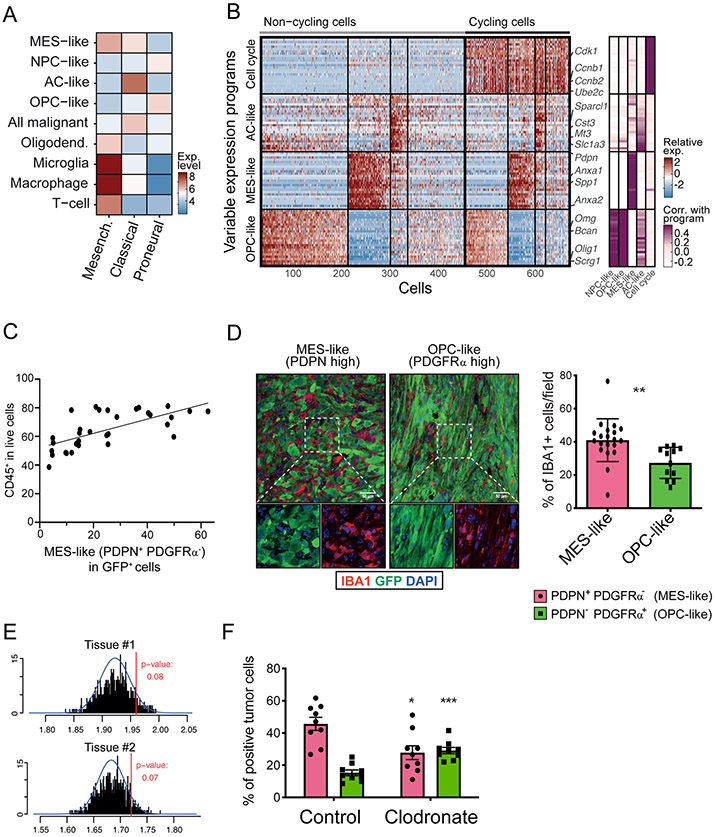
Figure 1: Macrophages induce transition of cancer cells into a MES-like state.
(A) Heatmap shows the average expression of the three TCGA signatures (Mesenchymal, Classical, and Proneural) in different cell types in human glioblastoma scRNA-seq, including the four GBM cell states: neural-progenitor-like (NPC-like), oligodendrocyte-progenitor-like (OPC-like), astrocyte-like (AC-like), and mesenchymal-like (MES-like). (B) Programs of heterogeneity identified using NMF in a mouse model of glioblastoma. Left panel: heatmap shows relative expression of genes from four programs across all cells. Cells are ordered in three subsequent steps by their score for the cell cycle, OPC-like, MES-like and AC-like programs; first, cells are separated into cycling and non-cycling; Second, cells within each group are further separated by the identity of the highest-scoring program, or defined as having no high-scoring program; Third, the cells with the same highest-scoring program are further sorted by their score for that program. Selected genes are indicated. Right panel: gene’s correlations to the corresponding human glioblastoma programs. (C) Quantification of the fraction of immune cells (CD45+) cells and MES-like (PDPN+ PDGFRα−) glioblastoma (GFP+) cells in each tumor (32 mouse glioblastoma samples). Pearson correlation is indicated between percentage of CD45+ cells and that of PDPN+ PDGFRα− cells, Pearson’r=0.6747, p<0.0001. (D) Left: Representative immunofluorescence (IF) staining of a MES-like, defined as PDPN+ PDGFRα− (left), and an OPC-like, defined as PDPN− PDGFRα+(right) tumor fields, stained for markers for macrophages (IBA1, red), cancer cells (GFP, green) and nuclei (DAPI, blue). See Figure S1D for details of PDPN and PDGFRα characterization. Scale bar, 50 μm. Right: Quantification of the fraction of macrophages (IBA1+) in tumor fields defined as being MES-high (based on PDPN staining) or OPC-high (based on PDGFRα staining). 8 mouse glioblastoma tissues were subjected to the analysis (PDPN; 20 regions, 1,4181 cells analyzed, PDGFRα; 12 regions, 6506 cells analyzed). Error-bars indicate Standard deviation (SD), and the difference between averages is significant by t-test (** denotes p<0.001). (E) Histogram showing observed mean number of macrophages in 30 nearest cells to each MES-like glioblastoma cell (red line) and distribution expected at random (black lines). Blue line shows the fitted normal distribution. Tumor section 1 shown on top and tumor section 2 shown on bottom (F) Fraction of MES-like (PDPN+ PDGFRα−) and OPC-like (PDPN− PDGFRα+) cells by flow cytometry, in liposome-PBS (control)-treated mice and mice injected with liposome-clodronate. Error bars indicate standard error of the mean (n=9 for each group), and the difference between averages is significant by t-test (PDPN group; * denotes p<0.0077, PDGFRα group;*** denotes p=0.0001).
To determine whether macrophages cause a MES-like state of glioblastoma cells in vivo, we utilized a mouse glioblastoma model induced by lentiviruses harboring GFP, HrasG12V and sh-p53, and a derivative tumor transplantation model (Friedmann-Morvinski et al., 2012). We profiled GFP+ cells of the lentiviral model by scRNA-seq, and applied non-negative matrix factorization (NMF) to identify variable expression programs. The NMF programs were highly consistent (albeit not identical) with those we previously observed in glioblastoma patients (Neftel et al., 2019), particularly for the MES-like and OPC-like states, and to a more limited extent for the AC-like state (Figures 1B and S1B-D; Table S1). Similar classification of cellular states was identified by an alternative approach using Louvain clustering (Figure S1E). Importantly, the co-existence of these distinct cellular states was consistently observed at multiple time points after lentiviral-induced transformation (Figure S1B). Thus, while this model has limitations in its genetic faithfulness to human glioblastoma, and shows minor species-specific differences in how cell states are encoded, these results establish its relevance for studying heterogeneity of cellular states in glioblastoma, especially the MES-like state.
Using established markers for the mouse MES-like (PDPN) and mouse OPC-like (PDGFRα) glioblastoma cellular states (Figures S1F-G), we found that the fraction of MES-like glioblastoma cells (PDPN+ PDGFRα− GFP+) is proportional to the abundance of immune cell (CD45+) (Figures 1C, S1H; n=32 mice, p<10−4). Immunohistochemical analysis using the pan-macrophage marker IBA1/AIF1 further showed that macrophages are enriched in the vicinity of MES-like glioblastoma cells, compared to OPC-like cells (Figures 1D and S1I; p<0.01, t-test). To further support such association, we selected the patient sample MGH157 as representative of a tumor that spans, by scRNA-seq, the spectrum of cell states and cell types of glioblastoma. We measured a panel of 135 genes by Multiplexed Error-Robust FISH (MERFISH, Methods) that allowed us to identify MES-like and macrophage cells (Chen et al., 2015). We probed two tissue sections from MGH157 that supported an enrichment of macrophages among MES-like glioblastoma cells in situ (Figures 1E and S1J-L). Thus, both in mouse models and in human samples MES-like glioblastoma states are enriched in the vicinity of macrophages.
To interrogate whether this association is causal, we injected our mouse transplant model with the macrophage-depleting agent clodronate. We observed a reduced percentage of PDPN+ PDGFRα− GFP+ MES-like glioblastoma cells upon clodronate treatment (Figure 1F and S1M-N; 39.1% decrease, n=9 mice, p<0.01, t-test), supporting a causal effect of macrophages on the fraction of glioblastoma cells expressing the MES-like state.
Macrophage-secreted ligands drive the MES-like state
To explore potential mechanisms underlying macrophage induction of the MES-like state in cancer cells, we examined putative ligand-receptor interactions predicted by the glioblastoma scRNA-seq datasets (Neftel et al., 2019). We identified five ligands (AREG, GAS6, HBEGF, OSM, PDGFB) that are highly expressed by macrophages and for which a corresponding receptor (AXL, EGFR, OSMR, PDGFRB) is highly expressed by the malignant cells (Figures 2A and S2A). Similarly, we identified potential ligand-receptor interactions of malignant cells with T-cells and with oligodendrocytes (Figure S2A) but focused follow-up experiments on macrophages due to their higher abundance in glioblastoma and the association with MES-like cells demonstrated above. Notably, three of the four receptors for macrophage-derived ligands (AXL, OSMR, PDGFRB) are preferentially expressed by MES-like cancer cells, compared to the other malignant states, in human tumor scRNA-seq (Neftel et al., 2019) and in TCGA datasets (Figures 2B and S2B). In addition, three of the macrophage-derived ligands (OSM, HBEGF, AREG) are more highly expressed in glioblastoma-associated macrophages compared to their counterpart in healthy brain in both human and mouse scRNA-seq datasets (Geirsdottir et al., 2019; Li et al., 2019; Masuda et al., 2019; Zhong et al., 2018) (Figure S2C).
Figure 2: Macrophage-secreted ligands as a driver of MES-like state.
(A) Average expression of each gene (dot) in glioblastoma malignant cells (x-axis) and macrophage cells (y-axis). Ligands expressed by macrophages (pink) with corresponding receptors expressed by malignant cells (cyan) are colored and labeled. (B) Correlation between the expression of receptors shown in (A) and the scores for 4 human glioblastoma cellular states (MES-like, AC-like, NPC-like, OPC-like) across the malignant cells in the human GBM scRNA-seq dataset (top) and across TCGA bulk-RNA seq (bottom). Error bars correspond to standard error, calculated by bootstrapping 500 cells (top) or 130 tumors (bottom) with 1000 iterations. (C) Fraction of MES-like (PDPN+ PDGFRα−) and OPC-high (PDPN− PDGFRα+) cells by flow cytometry in tumors of mice injected with cells overexpressing AREG, GAS6, HBEGF, OSM, or Luciferase as a control. Error bars indicate standard error of the mean (n=3 for each group), and the difference between averages is significant by t-test (* denotes p<0.05).
See also Figure S2.
To test the impact of macrophage-derived ligands on glioblastoma cellular states in vivo, we engineered glioblastoma mouse cells to overexpress each of the ligands (Figure S2D), injected them into syngeneic mice brains and compared the cellular state-distribution of the resulting tumors to those derived from control settings by flow cytometry (Methods). Overexpression of two of the five tested ligands (Osm, Hbegf) led to increased abundance of MES-like (PDPN+ PDGFRα−) cells (Figure 2C; n=3 mice, p<0.05, t-test), whereas Areg and Gas6 did not. Pdgfb overexpression greatly delayed tumor growth, therefore impeding our capacity to evaluate its impact on glioblastoma cells in vivo. Thus, Osm and Hbegf are ligands expressed by macrophages that induce an in vivo enrichment of the MES-like glioblastoma state in a mouse model.
OSM-induced transition to MES-like state is recapitulated in human cells in vitro
Next, we asked whether the induction of MES-like cells by macrophage-secreted ligands can also be recapitulated in human cells. We thus turned to examine the in vitro effect of the five ligands on primary human gliomasphere models. To identify a model that faithfully recapitulates the cellular states of glioblastoma, we profiled by scRNA-seq four glioblastoma gliomasphere models (MGG23, MGG75, MGH143, MGG18), and identified variable expression programs by NMF. The gliomaspheres partially recapitulated the cellular states observed in patient tumors (Figures 3A,B, S3A,B). In particular, the programs identified in MGG23 and MGG75 displayed strong similarities to the NPC-like, MES-like, and AC-like states, supporting their use as model systems (Table S2). We further confirmed the utility of CD44 and CD24 as efficient markers for the isolation and quantification of MES-like and NPC-like cells in gliomaspheres models, respectively (Figure S3C), consistent with our prior work (Neftel et al., 2019). Thus, human in vitro gliomaspheres partially recapitulate the heterogeneity of glioblastoma seen in patients, and represent additional models of the associated cellular states, complementary to the mouse model, with species-specific differences in markers for cellular state isolation.
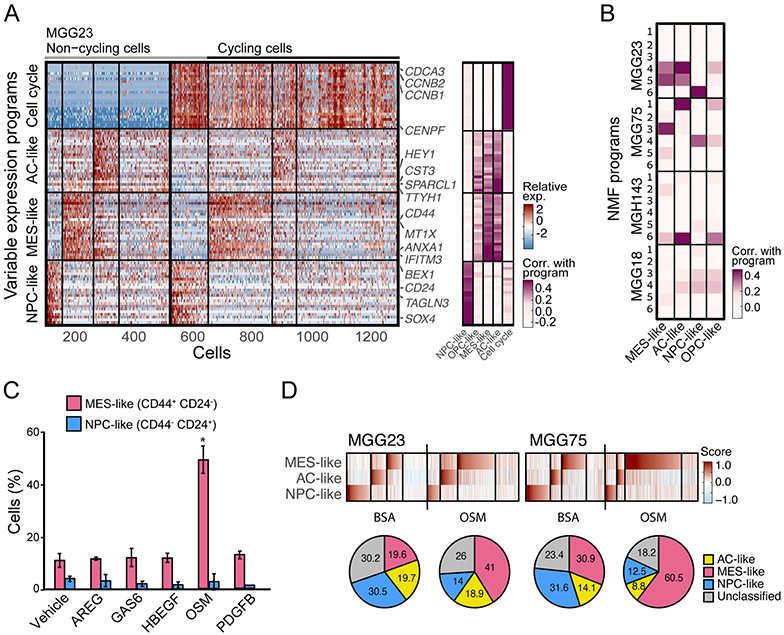
Figure 3: OSM-induced transition to MES-like state is recapitulated in human cells in vitro.
(A) Programs of heterogeneity identified using NMF in MGG23, a glioblastoma cell line grown as gliomaspheres. Right panel: heatmap shows relative expression of genes from four programs across all cells. Cells are ordered in three subsequent steps by their score for the cell cycle, NPC-like, MES-like and AC-like programs; first, cells are separated into cycling and non-cycling; Second, cells within each group are further separated by the identity of the highest-scoring program, or defined as having no high-scoring program; Third, the cells with the same highest-scoring program are further sorted by their score for that program Selected genes are indicated and labeled. Left panel: gene’s correlations with the corresponding human in vivo glioblastoma programs. (B) Comparison of the programs heterogeneity identified using NMF from the 4 cell lines profiled: MGG23, MGG75, MGH143, and MGG18. Heatmap shows each NMF program (6 per cell-line) correlations with the corresponding human in vivo glioblastoma programs (MES-like, AC-like, NPC-like, and OPC-like) (C) Fraction of MES-like (CD44+ CD24−) and NPC-like (CD44− CD24+) cells by flow cytometry, in MGG23 cells treated with recombinant AREG, GAS6, HBEGF, OSM, PDGFB, or control. Error bars indicate standard deviation (n=3 for each group), and the difference between averages is significant by t-test (* denotes p=0.029). (D) ScRNA-seq of MGG23 (left) and MGG75 (right) after treatment with OSM or BSA. Heatmaps show cells from each experiment, ordered by their score for the different glioblastoma states. Pie charts below show the relative proportion of each cell state in corresponding heatmap.
We treated MGG23 cells with the five macrophage-derived ligands for 24 hours and assessed the proportion of glioblastoma cellular states by CD24 and CD44 expression, measured by flow cytometry. Consistent with our observations in the mouse model, there was an increase of CD44+ MES-like cells upon treatment with OSM (Figure 3C; p<0.05, t-test). In contrast, the effect of HBEGF was not recapitulated in vitro, possibly reflecting a dependence on the TME, a long-term effect on cell growth, a model-specific effect or cross-species differences. We confirmed by RNA-seq that OSM globally upregulated the MES-like program, rather than only the marker gene CD44, while also downregulating the NPC-like program (Figure S3D). Similarly, OSM treatment increased the frequency of MES-like cells in a second glioblastoma model (MGG75; Figure S3E).
To further validate this result, we used scRNA-seq to profile MGG23 and MGG75 cells after treatment with OSM, as well as GFP+ glioblastoma cells from the mouse lentiviral model after in vivo treatment with recombinant OSM. As expected, OSM treatment led to an increase in the proportion of MES-like cells (Figures 3D and S3F). This came at the expense of reduced proportion of either OPC-like cells (in the mouse model) or NPC-like cells (in the gliomaspheres), while the proportion of AC-like cells was only mildly affected in all model systems. To further examine this potential interplay between states, we next examined bulk transcriptome data of 91 matched pairs of primary and recurrent glioblastoma samples (Wang et al., 2017) and found similar results. Specifically, we observed an inverse correlation between the difference in expression of MES-like or macrophage genes and the expression of OPC-like or NPC-like genes (but not of AC-like genes) between diagnosis vs. recurrence samples (Figure S3G-H). Taken together, these results suggest that the transition to MES-like cells, both upon experimental manipulation and in patients’ disease progression, occurs mainly from NPC-like and OPC-like states.
The MES-like state is induced by STAT3 through OSMR/LIFR-GP130 complexes
OSM is a pleiotropic cytokine of the interleukin-6 family that binds either OSMR or LIFR, in a complex with GP130 (Jahani-Asl et al., 2016; Jones and Jenkins, 2018). Due to the potential signaling of OSM via LIFR and the high similarity of OSM with the cytokine LIF, we also tested the effect of LIF on MGG23 cells. LIF also induced the MES-like program, albeit to a lesser extent and more transiently than OSM (Figures S3D and S4A). To further understand the mechanism by which OSM and LIF induce the MES-like program in human glioblastoma cells, we used CRISPR/Cas9-mediated genome editing to knockout (KO) OSMR, LIFR, OSMR+LIFR or IL6ST (encoding GP130) in MGG23 cells (Figures S4B-S4G, Methods).
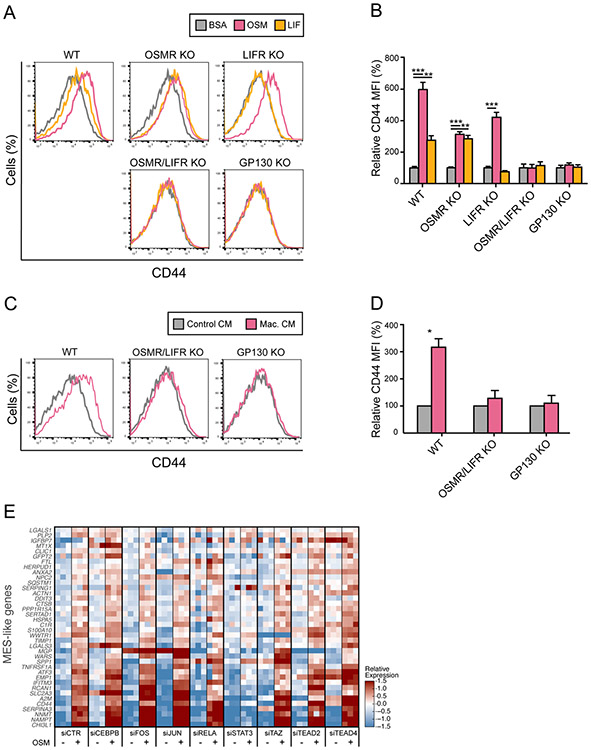
The induction of CD44 upon treatment with OSM was partially decreased in OSMR KO cells and completely abolished by double KO of OSMR and LIFR or by KO of IL6ST (Figures 4A-B). These results suggest that OSM-mediated induction of the MES-like program is dependent on either OSMR or LIFR (with a stronger effect of OSMR), in a complex with GP130. In contrast, the induction of CD44 upon treatment with LIF was abolished with either LIFR or IL6ST KO, but not by OSMR KO (Figures 4A-B), suggesting that the effect of LIF on the MES-like state is mediated by LIFR/GP130 heterodimeric complexes. A similar induction of CD44 was observed with conditioned media from human macrophages or microglia, or by co-culturing of MGG23 with human macrophages (Figures 4C-D and S4H-I). Also in these contexts, the double KO of OSMR and LIFR or the KO of IL6ST abolished the induction of CD44, mirroring the results with OSM treatments. These results suggest that OSM alone is largely sufficient to reproduce the effect of macrophages in inducing the MES-like state of MGG23 cells.
Figure 4: OSM-induced transition of MES-like state is mediated by OSMR/LIFR.
(A) Flow cytometry analysis of CD44 expression on MGG23 cells treated with OSM, LIF, or BSA, in wild-type cells (WT) and CRISPR knock-out of OSMR (OSMR KO), LIFR (LIFR KO), both OSMR and LIFR (OSMR/LIFR KO), and IL6ST (GP130 KO). (B) Quantification of mean fluorescence intensity (MFI) values in (A) reflecting CD44 expression on MGG23 cells treated with OSM, LIF or BSA, in WT, OSMR KO, LIFR KO, OSMR/LIFR KO and GP130 KO group, normalized by an averaged MFI of BSA-treated cells. Error bars indicate standard deviation (n=3 for each group), and the difference between averages is significant by t-test (** denotes p<0.001, *** denotes p<0.0001). (C) Flow cytometry analysis of CD44 expression on MGG23 cells treated with conditioned medium (CM) from human macrophage culture, in WT, OSMR/LIFR KO and GP130 KO group. (D) Quantification of MFI values in (C) reflecting CD44 expression on MGG23 cells treated with macrophage CM or control CM, in WT, OSMR/LIFR KO and GP130 KO group, normalized by an averaged MFI of control CM-treated cells. Error bars indicate standard deviation of means from three independent experiments, and the difference between averages is significant by t-test (* denotes p=0.0059). (E) MGG23 cells after knockdown of 8 different TFs (CEBPB, FOS, JUN, RELA, STAT3, WWTR1(TAZ), TEAD2, TEAD4) using siRNA were profiled by RNA-seq. Heatmap of relative expression of MES-like genes in bulk RNA-seq of MGG23 cells after knockdown of TFs and treatment with OSM or BSA, centered across all samples.
See also Figure S4.
OSMR is a co-receptor for EGFR and OSM has been shown to enhance EGFR signaling in glioblastoma (Jahani-Asl et al., 2016). However, several observations indicated that EGFR is not mediating the effect of OSM on the MES-like program (Figure S4J-L). First, we did not observe an impact of OSM on the phosphorylation status of EGFR and the downstream kinases, and in fact found decreased EGFR protein levels upon OSM treatment. Second, knockdown of EGFR did not affect the OSM-induction of CD44. To investigate the underlying mechanism for induction of the MES-like state by OSM, we examined the effects of siRNAs targeting eight transcription factors that were previously proposed to regulate the mesenchymal program in glioblastoma (CEBPB, STAT3, RELA, TAZ, FOS, JUN, TEAD2, TEAD4) (Bhat et al., 2013; Bhat et al., 2011; Carro et al., 2010; de Souza et al., 2018). We profiled by bulk RNA-seq MGG23 cells depleted of each transcription factor (Figure S4M) after treatment with OSM or BSA. Depletion of STAT3 abolished the OSM-mediated induction of MES-like genes such that expression of those genes was similar after OSM and BSA treatment (Figure 4E). Interestingly, while depletion of JUN and TAZ did not affect MES-like genes, they may play a significant role in the concomitant downregulation of the NPC-like program (Figure S4N). Thus, OSM-mediated induction of the MES-like program is primarily mediated through STAT3, while JUN, TAZ and potentially other factors may be needed for the repression of alternative programs.
A mesenchymal state of glioblastoma-associated macrophages
While macrophages induce a state transition in glioblastoma cells, they may also be reciprocally affected by this interaction or by other elements in the glioblastoma microenvironment. To explore this possibility, we compared scRNA-seq data of macrophages from glioblastoma with those from other glioma classes in which MES-like malignant cells were not detected by scRNA-seq, including IDH-mutant astrocytoma, oligodendroglioma, and H3K27M glioma (all profiled by the same lab and with the same scRNA-seq protocol) (Filbin et al., 2018; Tirosh et al., 2016; Venteicher et al., 2017). The first two principal components (PC1 and PC2) of a principal component analysis (PCA) highlighted the differences between macrophages of glioblastoma from those of other gliomas (Figure 5A, Methods). Specifically, PC1 reflects a macrophage-to-microglia axis, with macrophage-like genes (e.g. CD163) and microglia-like genes (e.g. P2RY12) associated with PC1low and PC1high cells, respectively (Table S3), consistent with an increased proportion of infiltrating macrophages in glioblastoma compared to other gliomas.
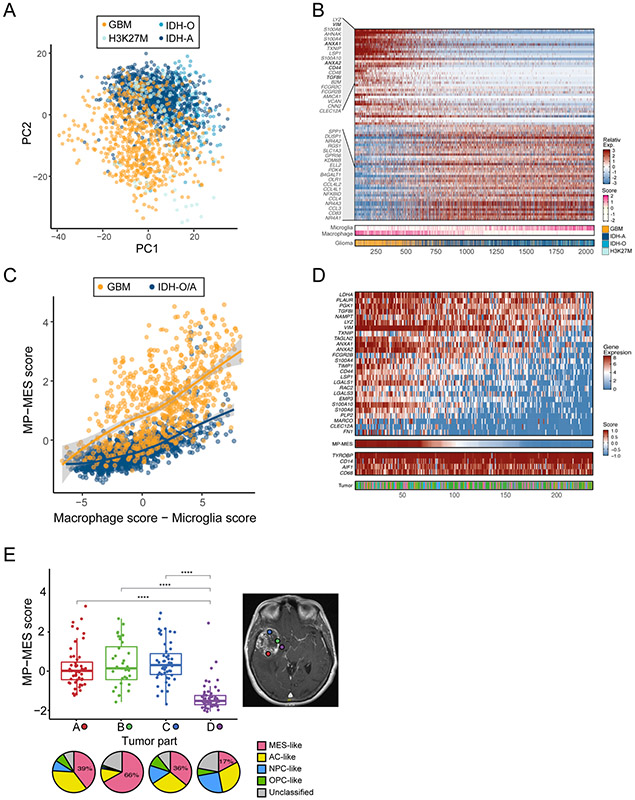
Figure 5: A mesenchymal state of glioblastoma-associated macrophages.
(A) PCA plot of myeloid cells from four glioma types (GBM for glioblastoma, IDH-mutant Astrocytoma and Oligodendroglioma, and H3K27M gliomas). (B) Heatmap of genes with top (positive and negative) loading scores in PC2, of which the top 40 are labeled. Cells are ordered by their expression of PC2-low genes. Lower panels indicate the glioma type and the score for the microglia and macrophage programs. (C) Myeloid cells MP-MES scores (Y-axis) and macrophage vs. microglia scores (X-axis). Colors distinguish GBM-derived cells from those of IDH-mutant tumors (Oligodendroglioma and Astrocytoma; IDH-O/A). Line indicates a LOESS regression. (D) The main heatmap (top) shows expression of the MP-MES program genes in macrophage-like cells in glioblastoma. Cells are ordered by their MP-MES scores, as shown in the lower panel. Additional lower panels show the expression of 4 housekeeping macrophage genes, and the tumor of origin (bottom). (E) Boxplots (the line in the box shows the median, the upper and lower borders of the box indicate the upper and lower quartile, lines below and above the box indicate the 5th and 95th percentiles, and all data points are shown) depict the MP-MES scores of macrophages from 4 different locations of glioblastoma tumor MGH105 (labeled by color on MRI, right). Pie charts below show the assignment of the malignant cells from each location to four states. Location D had the smallest percentage of MES-like malignant cells and a significantly lower macrophage MP-MES scores compared to locations A, B and C (**** denotes p<10−12 in three comparisons by t-test).
Surprisingly, among the top genes associated with PC1low cells, and even more notably with PC2low cells, and hence higher in macrophages of glioblastoma than those of other gliomas, were many of the same genes defining the MES-like state of cancer cells (e.g. VIM, CD44, ANXA1). This indicates that glioblastoma-macrophages display a different program than macrophages in other gliomas and defines a novel macrophage-mesenchymal (MP-MES) program (Figure 5B). MP-MES genes do not overlap with M1-like and M2-like macrophage polarization markers (Beyer et al., 2012; Gabrusiewicz et al., 2016; Mantovani et al., 2017), and combined analysis of all of those genes suggests that MP-MES is mutually exclusive from M1-like and M2-like macrophage states (Figure S5A). MP-MES is enriched among infiltrating macrophages, compared to resident microglia, and is partially correlated with the microglia-to-macrophage scores (Figures S5B-S5D). However, when comparing cells of the same macrophage score, we observed considerably higher expression of the MP-MES signature in glioblastoma compared to other gliomas (Figure 5C).
Notably, high MP-MES is not seen in all glioblastoma macrophages. Instead, we observe considerable variability in MP-MES scores of macrophages within each glioblastoma from our patient dataset (Figure 5D), as well as in our mouse model of glioblastoma (Figure S5E). This variability in MP-MES is correlated with the MES-like glioblastoma cells, both across spatial regions of the same tumor (Figures 5E and S5F), and across different tumors (Figure S5G).
These results suggest a connection between the mesenchymal-like states of macrophages and glioblastoma cells, such that either MES-like glioblastoma cells induce the MP-MES, or both states may commonly be induced by shared environmental factor(s), such as hypoxia (Figure S5H). To further examine this association, we leveraged the in vivo mouse model described above, in which glioblastoma cells were engineered to overexpress OSM. Macrophages isolated from this model had an increased proportion of MP-MES cells compared to macrophages from control mice (Figure S5I, p<0.005, Binomial test). The MP-MES state may be induced by ligands produced by the MES-like cancer cells, and our analysis identified 6 potential ligands (CSF1, CSF3, CX3CL1, TGFB2, TGFB3, CCL7) that are highly expressed in the MES-like cancer cells and for which a corresponding receptor (CSF1R, CSF3R, CX3CR1, TGFBR2, CCR2, CCR5) is highly expressed in the macrophages (Figures S5J-K).
MES-like states may be associated with T-cell activation
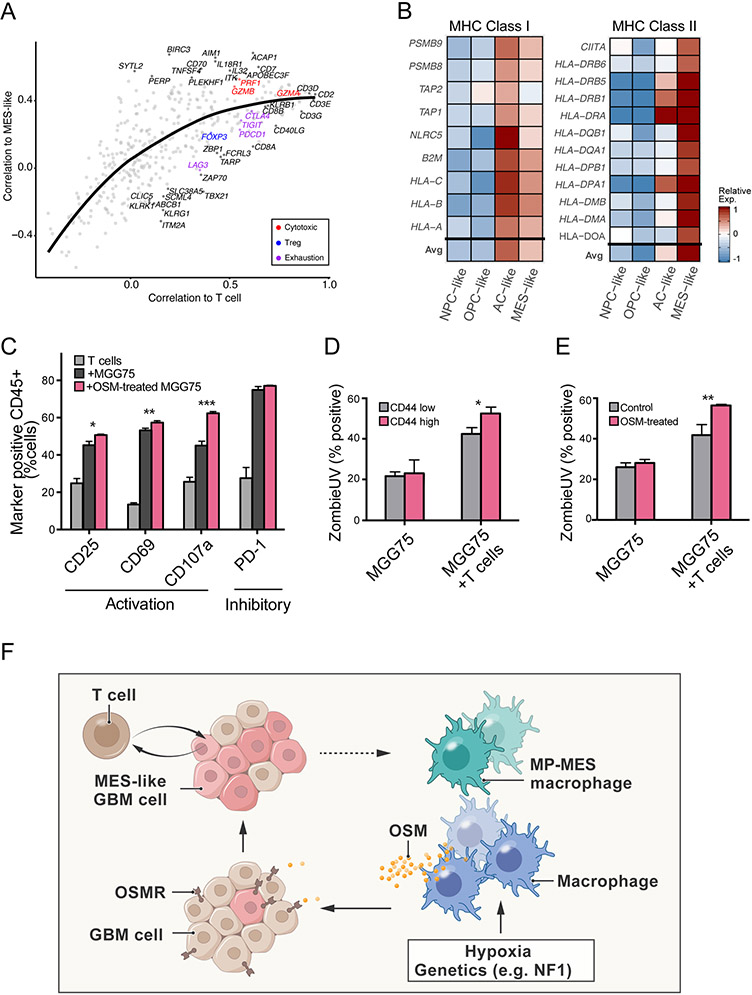
Glioblastoma-associated macrophages are typically thought of as being immune-suppressive (Grabowski et al., 2020). Moreover, mesenchymal programs have recently been shown to suppress the activity of T-cells in epithelial cancers and to be associated with poor prognosis in glioblastoma (Mariathasan et al., 2018; Terry et al., 2017). However, our analysis pointed towards an enrichment of T-cells within TCGA-MES tumors (Figure 1A). T-cell enrichment might be associated with increased or decreased anti-tumor activity, depending on the states of the T-cells. In particular, regulatory T-cells (Tregs) could indicate suppression, while effector CD8+ T-cells might indicate increased cytotoxic activity.
To distinguish between these possibilities, we further examined the correlation of T cell-specific genes with the MES-like program across the TCGA bulk RNA-seq data. Genes with higher correlations to the overall abundance of T-cells (as estimated by CD2, CD3D, CD3E and CD3G) also had higher correlations to the MES-like scores of bulk tumors, reflecting the overall association between T-cell abundance and MES-like states (Figures 6A). We estimated this expected trend with a LOESS regression and examined which of the T-cell genes have higher or lower correlations with the MES-like score compared to the regression. We found that cytotoxicity markers (GZMB, PRF1) had higher correlations, while the Treg marker FOXP3 and exhaustion markers (LAG3, PDCD1, TIGIT) had lower correlations with the MES-like scores (Figures 6A and S6A-S6B). The correlations of additional genes are shown in Table S4 although we note that this analysis had to be limited to a subset of T-cell genes with a restricted expression pattern.
Figure 6: MES-like states may be associated with T-cell activation.
(A) Scatter plot of 406 T-cell specific genes. Correlation of each gene’s expression to estimated T cell abundance, defined by genes (CD2, CD3D, CD3E, CD3G) (x-axis) and the correlation of each gene’s expression to the MES-like score (y-axis) in TCGA bulk RNA-seq. Line indicates a LOESS regression. Colors distinguish marker genes for T cell subtypes (Cytotoxic, Treg, Exhaustion). (B) Heatmap shows the relative average expression of MHC class I (left) and MHC class II (right) genes in simulated bulk profiles of NPC-like, OPC-like, AC-like, and MES-like cells. (C) Quantification of T cell activation markers (CD25 and CD69), a marker of T cell degranulation (CD107a) and an exhaustion marker PD-1 positive cells in CD45+ cells after 24 hours co-culture of engineered T cells with NY-ESO-1 expressing MGG75 cells pretreated with or without 20ng/mL of OSM. Error bars indicate standard deviation (n=3 for each group), and the difference between averages is significant by t-test (*p=0.011, **p=0.0083, ***p=0.0003). (D) Cellular viability of MGG75-NY-ESO-1 cells co-cultured with or without NY-ESO-1 TCR T cells. ZombieUV dye incorporation into CD44low and CD44high MGG75-NY-ESO-1 cells were measured by flow cytometry analysis after an 8 hours co-culture. Error bars indicate standard deviation (n=3 for each group), and the difference between averages is significant by t-test (*p=0.0124). (E) Cellular viability of NY-ESO-1 MGG75 cells pre-treated with or without 20 ng/mL of OSM for 24 hours, followed by co-culture with or without NY-ESO-1 TCR T cells. ZombieUV dye incorporation into CD45 negative MGG75-NY-ESO-1 cells were measured by flow cytometry analysis after an 8 hours co-culture. Error bars indicate standard deviation (n=3 for each group), and the difference between averages is significant by t-test (**p=0.00829). (F) Scheme explaining the establishment of a TCGA-MES tumor. An abundance of macrophages could be driven by tumor genetics, such as alterations of NF1 that promote macrophage recruitment, or by hypoxia and other aspects of the glioma microenvironment. Macrophage-derived OSM then induces a MES-like state of the glioblastoma cells. The MES-like glioblastoma cells and/or the associated microenvironment also facilitate a transition of macrophages into the MP-MES state through mechanisms that remain unclear.
Thus, TCGA-MES tumors are enriched with T-cells and specifically, with cytotoxic T-cells. We sought for potential mechanisms for this enrichment by evaluating the expression in MES-like cells of genes that might recruit or activate T cells. Cytokines and chemokines were generally not upregulated in MES-like cells (data not shown). However, MHC-I and MHC-II genes were expressed significantly higher in MES-like glioblastoma cells (as well as in AC-like cells) than in NPC-like and OPC-like cells (Figure 6B). Accordingly, MGG23 cells treated with OSM expressed significantly higher levels of MHC-I and MHC-II genes compared to controls (Figure S6C). Expression of MHC-II genes in cancer cells, such as melanoma, is associated with better response to immunotherapy (Rodig et al., 2018). These results, along with the enrichment of cytotoxic T-cells in TCGA-MES tumors, raise the hypothesis that MES-like states might be more efficiently killed by T cells.
To test the impact of glioblastoma cellular state on T-cell mediated tumor killing, we established a co-culture system between human T cells and the MGG75 cell line (Mathewson et al., 2021). We engineered MGG75 to express NY-ESO-1 and primary human T cells to express an HLA-A2*02:01-restricted, NY-ESO-1 antigen specific T cell receptor (TCR) (Rapoport et al., 2015), matching the HLA subtype of MGG75 (Methods). Upon co-culture, engineered MGG75 cells efficiently activated the engineered T cells, as measured by T cell activation (CD25, CD69), degranulation (CD107a) and exhaustion (PD-1) markers (Figure 6C). We then assessed state-specific T cell-mediated killing and found that MES-like cells (CD44high) were more efficiently killed by the T cells than CD44low cells in an 8 hours co-culture assay (Figure 6D, p<0.05, t-test). As a second approach to examine the differential T-cell response to MES-like cells, we treated MGG75 cells with OSM prior to co-culturing, and once again observed increased T-cell activation and T-cell mediated killing (Figures 6C, 6E, S6D, S6E and S3E; Methods).
Finally, we searched for evidence of state-specific immune response in clinical settings. To that end, we explored a recently published bulk RNA-seq dataset of PD-1-inhibitor treated glioblastoma (Zhao et al., 2019). The small number of patients (n=17) examined in this work and the limited significance of the reported responses limit the statistical power of our analysis and its interpretation. Nonetheless, we noticed that most responders, but not most non-responders, displayed lower MES-like scores after treatment (Figure S6F; p=0.1167). This preliminary observation is consistent with the hypothesis that T-cell mediated killing was biased towards MES-like cells, which might have therefore decreased in abundance in responders following PD-1 inhibition. Taken together, our results suggest that T-cell mediated killing of glioblastoma cells might be more efficient against MES-like states, and that inducing the transition of glioblastoma cells towards MES-like states by OSM could facilitate this process.
Discussion
Of the four recurrent cellular states described in glioblastoma, three states recapitulate neurodevelopment, while the origin of the fourth state - resembling mesenchymal cells - remains poorly understood. Here, we show that the MES-like state is not only correlated with macrophage abundance but is directly induced by them, specifically through macrophage-secreted OSM that interacts with OSMR or LIFR (in complex with GP130) on glioblastoma cells, thereby activating STAT3. Notably, MES-like glioblastoma cells were also observed in the absence of macrophages in spheroid in vitro models, indicating that while macrophages significantly increase the frequency of MES-like glioblastoma cells, their presence is not strictly required.
The establishment of a TCGA-MES tumor - in which macrophages are abundant and both the macrophages and the glioblastoma cells acquire a MES-like state - could be described as the combined result of several processes (Figure 6F). First, an abundance of macrophages could be driven by tumor genetics, such as alterations of NF1 that promote macrophage recruitment, or by hypoxia and other aspects of the glioma microenvironment. Macrophage-derived OSM then induces a MES-like state of the glioblastoma cells. Additional extrinsic factors (e.g. hypoxia) and intrinsic mechanisms may also contribute to the induction of MES-like states (e.g. a MES-like state is also found in vitro in the absence of macrophages). This is consistent with the occurrence of hypoxia-dependent and hypoxia-independent MES-like states, as previously described (Neftel et al., 2019).
The MES-like glioblastoma cells and/or the associated microenvironment may also facilitate a transition of macrophages into a MP-MES state, and further studies are required to elucidate its origin and consequences. We note that CSF1R and CSF3R have relatively lower expression in MP-MES macrophages compared to other macrophages and microglia, which may reduce the effectiveness of CSFR inhibitors in targeting these macrophages, possibly related to CSFR inhibitors failing in clinical trials (Butowski et al., 2016).
Interestingly, OSM is also produced by inflammatory monocytes in the gut where it activates inflammatory fibroblasts (West et al., 2017). This suggests a coherent function of OSM in supporting mesenchymal cells across biological contexts. Since OSM is secreted at high levels by various types of macrophages, this could reflect a common mechanism by which macrophages influence tumors and could partially account for the negative prognostic value of macrophages. Consistent with this possibility, additional studies suggested that OSM-OSMR may promote an epithelial-mesenchymal transition (EMT) in various carcinomas, such as breast, gastric, pancreatic and cervical cancers (Junk et al., 2017; Kucia-Tran et al., 2016; Smigiel et al., 2017; West et al., 2017). Thus, inhibition of OSM signaling may reflect a relevant therapeutic strategy for glioblastoma as well as for carcinomas, by preventing the aggressiveness of mesenchymal-like cancer cells.
However, at least in glioblastoma, MES-like states of macrophages and cancer cells may also represent a therapeutic opportunity, as they are associated with high levels of MHC-I and MHC-II and an abundance of T-cells, skewed towards a cytotoxic state, which might influence the response to immunotherapies. It is conceivable that different treatment modalities exert different pressures on glioblastoma cellular states and that the worse prognosis of TCGA-MES tumors to standard-of-care therapy might be contrasted with a better response to immunotherapies. Such an effect is hinted by our preliminary analysis of patients’ response to checkpoint-blockade (although larger cohorts would be needed to reach more conclusive results) and by our in vitro co-culture experiments, demonstrating increased T-cell killing of MES-like cells. Accordingly, the ability to induce MES-like states by OSM or other treatments may present a new therapeutic option when coupled with immunotherapies. Future work is required to fully assess the impact of diverse treatment modalities on the spectrum of cellular states that drive glioblastoma.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Itay Tirosh (Itayt@weizmann.ac.il).
Material availability
Unique reagents generated in this study are available with an MTA.
Data and Code Availability
Data generated for this study are available through the Gene Expression Omnibus (GEO: GSE168004). The Code supporting the current study is available from the corresponding authors on request. Additional datasets used for this study were obtained from the following studies: GSE GSE131928 (Neftel et al., 2019), GSE GSE134707 (Geirsdottir et al., 2019), GSE GSE124335 (Masuda et al., 2019), GSE GSE104276 (Zhong et al., 2018), GSE GSE89567 (Venteicher et al., 2017), GSE GSE70630 (Tirosh et al., 2016), GSE GSE102130 (Filbin et al., 2018), SRA PRJNA482620 (Zhao et al., 2019) and (Wang et al., 2017).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse model
All mice were maintained under pathogen-free conditions, and all procedures performed in this study were approved by the Institutional Animal Care and Use Committee at the Salk Institute and at Massachusetts General Hospital. Lentivirus prepared as described previously (Hara and Verma, 2019) or lentiviral model derived cells were stereotactically injected into the hippocampus of 6- to 16-week-old hGFAP-Cre, GFAP-FLPo or C57BL/6J mice (The Jackson Laboratory, Bar Harbor, ME). Briefly, lentiviruses (GFAP-cre model; 1-3 x 105 IFU, GFAP-FLPo model; 4-10 x 105 IFU) or cells (105 cells) suspended in 1 uL PBS were loaded on a Hamilton syringe with 33-gauge or 26-gauge needle, and injected at a speed of 0.1uL/30sec-1min using the following coordinate: 2.0 mm posterior, 1.5 mm lateral, and 2.3 mm dorsal to the bregma. Upon completing injection, the needle was left in place for 3 min, then withdrawn slowly in 2 min to help reduce virus reflux.
Cell culture
Mouse glioblastoma cells were maintained in 1 x N2-supplemented (Thermo Fisher Scientific) DMEM/Hams F12 (Corning) containing 20 ng/ml of human fibroblast growth factor (FGF)-2 (Peprotech), 20 ng/ml of human epidermal growth factor (EGF) (Peprotech), 2 mM of L-Glutamine (Corning), 1 x Antibiotic-Antimycotic (Thermo Fisher Scientific) and 40 mg/ml of heparin (Sigma). Patient derived human glioblastoma cells (MGG23, MGG75, MGH143, MGG18) (Neftel et al., 2019; Wakimoto et al., 2012) were maintained in Neurobasal Medium (Thermo Fisher Scientific) supplemented with 1/2 x N2 and 1 x B27 (Thermo Fisher Scientific), 1% Penicillin/Streptomycin (Thermo Fisher Scientific), 1.5 x Glutamax (Thermo Fisher Scientific), 20 ng/mL of EGF and 20 ng/mL of FGF2 (Shenandoah Biotechnology). The HLA-A*02:01 genotype was confirmed in MGG75 cells by performing Sanger sequencing on genomic DNA amplified with primers for forward: ACCGTCCAGAGGATGTATGG and reverse: CCAGGTAGGCTCTCAACTGC. Sequences were aligned to homo sapiens MHC class I antigen (HLA-A) gene, HLA-A*02010101 allele (GenBank: GQ996941.1).
Human Samples
Adult patients at Massachusetts General Hospital (MGH) provided preoperative informed consent to take part in the study in all cases after the Institutional Review Board Protocols DF/HCC 10-417. For tissue preparation for MERFISH, flash frozen glioblastoma was stored at −80 °C until cutting. Frozen tissue was sectioned at −18 °C on a cryostat (Leica CM3050 S). Monocyte-derived macrophages and iPSC-derived microglia were obtained from PromoCell and Applied StemCell, respectively. T cells were purified from PBMCs obtained from fresh leukaphoresis blood collars procured from the Brigham and Women’s Hospital blood bank.
METHOD DETAILS
Flow Cytometry
All mice were perfused with ice-cold PBS after euthanasia to reduce the influence of peripheral blood derived cells. The collected brains were mechanically and enzymatically dissociated using a papain-based neural tissue dissociation kit (Miltenyi Biotec) supplemented with 0.1 % typeI collagenase (Thermo Fisher Scientific) / PBS. The dissociated cells were first stained with calcein Blue AM (BD Biosciences) and Zombie NIR (BioLegend) for 25 min at 4 °C, and with anti-mouse CD16/32 (BD Biosciences) for 5 min. After washing cells with ice-cold 1% BSA / PBS, the cells were then stained with a combination of following antibodies; PerCP anti-mouse CD45 (clone 30-F11, BD Biosciences), phycoerythrin (PE) anti-mPdgfra (clone APA5, BioLegend), Allophycocyanin (APC) anti-mPodoplanin (clone 8.1.1, BioLegend), and APC anti-CD11b (clone M1/70, BioLegend), for 30 min at 4 °C. Note that CD44 antibody was not efficient in isolating the mouse MES-like population. We thus leveraged a data-driven approach to identify other markers based on the scRNA-seq mouse data, and found that PDPN and PDGFRα are highly efficient markers for isolating the mouse MES-like state and OPC-like, respectively, as validated by RNA-seq following FACS sorting. Sorting was performed with Becton Dickinson Influx cytometer (Becton Dickinson). Side scatter (SSC) width versus forward scatter (FSC) area, and Trigger Pulse Width versus FSC criteria were used to discriminate doublets and gate only singleton cells. Viable single cells were identified as calcein blue AM positive and Zombine NIR negative to low cells. Note that macrophages tend to uptake the dye to be Zombie NIR low population. We sorted viable CD45 negative/ GFP positive or CD45 positive/ CD11b positive single-cells into 96-well plates containing 10uL of TCL buffer (QIAGEN) with 1% beta-mercaptoethanol (Sigma Aldrich). Plates were frozen immediately after sorting and stored at −80 °C prior to whole transcriptome amplification, library preparation and sequencing.
ScRNA-seq
For the mouse model, libraries from isolated single cells were generated based on the Smart-seq2 protocol (Picelli et al., 2014) with the following modifications. RNA from single cells was first purified with Agencourt RNAClean XP beads (Beckman Coulter) before oligo-dT primed reverse transcription with Maxima reverse transcriptase (Thermo Scientific) and locked TSO oligonucleotide, which was followed by 20 cycle PCR amplification using KAPA HiFi HotStart ReadyMix (Roche) with subsequent Agencourt AMPure XP bead purification as described. Libraries were tagmented using the Nextera XT Library Prep kit (Illumina) with custom barcode adapters (sequences available upon request). Libraries from 768 cells with unique barcodes were combined and sequenced with paired-end, 38-base reads, using a NextSeq 500 sequencer (Illumina). For the cell lines, scRNA-seq libraries were generated using the Chromium Next GEM Single Cell 3′ GEM, Library & Gel Bead Kit v3.1, Chromium Single Cell 3′ Feature Barcode Library Kit, Chromium Next GEM Chip G, and 10x Chromium Controller (10x Genomics) according to the 10X Single Cell 3’ v3.1 with Feature Barcoding technology for Cell Surface Protein protocol. The cell lines were processed as a pool using TotalSeq-B/Hashtag antibodies (BioLegend), following the manufacturer’s recommendation. Each of the dissociated cells was first stained with Human TruStain FcX (BioLegend) in Cell Staining Buffer (BioLegend) for 10 min at 4 °C, and then stained with barcoded TotalSeq-B antibodies targeting human CD298 and β2 microglobulin in a separated tube for 30 min at 4 °C. We generated a single cell suspension of the pool in 0.04 % PBS-BSA and loaded approximately 9,900 single cells to the Chromium Controller with a targeted recovery of 6,000 cells. Single cells, reagents and single gel beads containing barcoded oligonucleotides were encapsulated into nanoliter-sized droplets and subjected to reverse transcription. Droplets were broken and the barcoded cDNAs were purified with DynaBeads and amplified by 12 cycles of PCR (98°C for 3 min; [98°C for 15 s, 63°C for 20 s, 72°C for 1 min] x 12; 72°C for 1 min). 3’ gene expression cDNA and TotalSeq antibody derived barcode cDNA were size-selected and separated with SPRIselect Reagent (Beckman Coulter), and then fragmented, end-repaired, ligated with index adaptors. Quality control of the resulting libraries was performed with the Bioanalyzer High Sensitivity DNA Analysis (Agilent). The constructed gene expression library and barcode library were combined at 4:1 ratio and sequenced with paired-end, 28 and 55-base reads, using a NextSeq 500 sequencer (Illumina).
Immunostaining
GFAP-cre or GFAP-FLPo mice injected with lentiviruses showing clinical symptoms were perfused with ice-cold PBS and followed by with 2% PFA/PBS after euthanasia. To avoid over-fixation, particularly for PDGFRα antigen, additional tissue fixation was performed in 2% PFA/PBS at 4°C and limited to 12-16 hours. Fixed tissues were washed with PBS for overnight and processed with vibratome into 100-200um thick brain sections and stored in tissue freezing media at serially assigned manners (25% Glycerol, 30% Ethylene glycol, 1.38g/L NaH2PO4, 5.48g/L Na2HPO4). Antibody staining of serial sections was performed overnight, and the primary antibodies used in this study as follows; anti-Iba1/ AIF1 (GTX100042; GeneTex), anti-Podoplanin (ab11936; Abcam), anti-Pdgfra (AF1062, R&D Systems). The secondary antibodies were as follows: Alexa Fluor 568 anti-rabbit IgG, Alexa Fluor 647 anti-goat IgG, Alexa Fluor 647 anti-hamster IgG (Thermo Fisher Scientific). The nucleus was stained with DAPI. The images were acquired with laser scanning microscopy, LSM 710, 810 or 880 with Airyscan (Zeiss) equipped with the laser lines (405, 458, 488, 514, 561, 594 and 633nm). In serial sections, tissues area which are positive for PDPN and GFP but negative for PDGFRα were defined as MES-like, and positive for PDGFRα and GFP but negative or weakly positive for PDPN were defined as OPC-like.
Multiplexed error-robust fluorescence in situ hybridization (MERFISH)
In order to distinguish among different malignant profiles and between glioblastoma and non-glioblastoma cell types present within the tumors using MERFISH, we designed a panel of 135 genes using mutual information analysis as reported previously (Li et al., 2017) based on previously published glioblastoma scRNA-seq data with corresponding cell-type and cell-state annotations from (Neftel et al., 2019). MERFISH encoding probes for the 135 genes were then designed as previously described (Moffitt et al., 2016) from a 16-bit, HammingDistance-4, Hamming-Weight-4 encoding scheme. We included 5 extra barcodes as “blank” barcodes, which were not assigned to any genes, to provide a measure of the false-positive rate in MERFISH as described previously (Moffitt et al., 2018). For the 135 gene panel used in this study, 12944 encoding probes were designed in total, with each gene targeted by 96 unique probes (except for PCNA, which was targeted by 80 probes). Each encoding probe contained a binding site for 2 of the 4 readout probes used to encode the genes identity. Each readout probes were conjugated to one of the two dye molecules (Alexa750, Cy5) via a disulfide linkage, as described previously (Moffitt et al., 2016) . For tissue preparation, two adjacent 12 μm thick tumor sections were mounted onto coverslips for MERFISH while another 50 μm thick tumor sections immediately anterior or posterior of the slices used for MERFISH were set aside for bulk RNA-seq. The tissue slices were mounted on prepared coverslips and stained with the MERFISH probe set as described previously (Moffitt et al., 2018; Moffitt et al., 2016).For MERFISH imaging, we used a home-built imaging platform as previously described (Xia et al., 2019). All MERFISH image analysis was performed using MERlin (Xia et al., 2019), a Python-based MERFISH analysis pipeline (https://github.com/ZhuangLab/MERlin), resulting in cell-by-gene matrices along with cellular spatial positional information.
RNA in situ hybridization
Paraffin-embedded tissue sections were mounted on glass slides and stored at −80 °C. Slides were stained using the RNAscope 2.5 HD Duplex Detection Kit (Advanced Cell Diagnostics, #322430), as previously described (Filbin et al., 2018). Briefly, slides were baked for 1 h at 60 °C, deparaffinized and dehydrated with xylene and ethanol. The tissue was pretreated with RNAscope Hydrogen Peroxide (Cat. No. 322335) for 10 min at room temperature and RNAscope Target Retrieval Reagent (Cat. No. 322000) for 15 min at 98 °C. RNAscope Protease Plus (Cat. No. 322331) was then applied to the tissue for 30 min at 40 °C. Hybridization probes were prepared by diluting the C2 probe (red) 1:50 into the C1 probe (green). Advanced Cell Technologies RNAscope Target Probes used included Hs-CD24 (#313021-C2), Hs-CD44 (#311271-C2), Hs-CD14 (#418801-C1). Probes were added to the tissue and hybridized for 2 h at 40 °C. A series of 10 amplification steps were performed using instructions and reagents provided in the RNAscope 2.5 HD Duplex Detection Kit. Tissue was counterstained with Gill’s hematoxylin for 25 s at room temperature followed by mounting with VectaMount mounting media (Vector Laboratories).
Macrophage depletion
Macrophage depletion was described previously (Saha et al., 2017). Lentiviral model derived mouse glioblastoma cells (105 cells) suspended in 1uL PBS were stereotactically injected into the hippocampus of age-matched C57BL/6J female animals. The mice were randomly separated into two groups for the treatment with either liposome-PBS (vehicle) or with liposome-clodronate (Encapsula NanoSciences). 50 mg per kg body weight (BW) of liposome-clodronate or an equal volume per BW of liposome-PBS were given to the animals intraperitoneally at day 4, followed by the treatment with a reduced dosing 25 mg/kg, to avoid an adverse effect, at day 7, 10, 13, 20, and 27. In this condition, the liposome-clodronate treatment improved the median survival of animals; liposome-PBS, 31days vs liposome-clodronate, 44 days. The mice were euthanized at day 28 for the flow cytometry analysis to evaluate the fraction of GFP+, PDPN+, PDGFRα− MES-like and GFP+, PDPN−, PDGFRα+ OPC-like cells and for immunofluorescence to evaluate macrophage depletion in the tissues. To assess systemic depletion of macrophages, peripheral blood and spleens were harvested from animals treated either with liposome-PBS or liposome-clodronate and subjected to flow cytometric analysis of monocytes; CD45+, CD115+, CD11b and T-cells; CD45+, CD3+. To evaluate the impact of liposome clodronate treatment on glioblastoma cells, mouse glioblastoma cells were treated with 7.5ug/mL of liposome-clodronate or an equal volume of liposome-PBS and subjected to flow cytometric analysis.
Plasmids
Mouse Areg, Gas6, Hb-egf, Osm and Pdgfb cDNA were synthesized by Integrated DNA Technologies, and cloned into pLenti-PGK-puro-EF1-Luc vector. These macrophage-derived ligands were expressed in mouse glioblastoma cells with lentiviral infection followed by puromycin selection of transduced cells. The overexpression of each of the ligands were confirmed by qRT-PCR, as well as ELISA of secreted OSM protein (R&D systems). The following specific primers were used: mouse beta-actin forward; GGCTGTATTCCCCTCCATCG, reverse; CCAGTTGGTAACAATGCCATGT, mouse Areg forward; GGTCTTAGGCTCAGGCCATTA, reverse; CGCTTATGGTGGAAACCTCTC, mouse Gas6 forward; TGCTGGCTTCCGAGTCTTC, reverse; CGGGGTCGTTCTCGAACAC, mouse Hbegf forward; CGGGGAGTGCAGATACCTG, reverse; TTCTCCACTGGTAGAGTCAGC, mouse Osm forward; CCCGGCACAATATCCTCGG, reverse; TCTGGTGTTGTAGTGGACCGT, mouse Pdgfb forward; AAGTGTGAGACAATAGTGACCCC, and reverse; CATGGGTGTGCTTAAACTTTCG. The NY-ESO-1 construct was generated by linking the NY-ESO-1 cDNA (Accession number: NM_139250.2) encoding the relevant NY-ESO-1 protein (processed peptide SLLMWITQC) to luciferase via a T2A peptide upstream of an IRES followed by the non-functional human nerve growth factor receptor (NGFR) extracellular domain as a selection marker. The NY-ESO-1 [1G4] TCR cDNA (Robbins et al., 2008) GeneArt, Thermo Fisher Scientific) was cloned into the pHAGE-MCS lentiviral vector.
Cytokine treatment
For in vitro cytokine experiments, human glioblastoma cells were treated with 20 ng/mL of human AREG, HB-EGF, OSM (PeproTech), GAS6 (Sino Biological), and PDGF-BB (Miltenyi Biotec) for 24 hours, and glioma spheres were dissociated with TrypLE (Thermo Fisher Scientific) to prepare single cell suspension. The dissociated cells were stained with calcein Blue AM, Zombie NIR and antibodies; anti-CD44 (clone REA690, Miltenyi Biotec) and anti-CD24 (clone REA832, Miltenyi Biotec), and analyzed with BD LSR II (Becton Dickinson) or CytoFLEX (Beckman Coulter). For in vivo treatment of OSM, lentiviruses were co-injected with 20 ng of recombinant OSM (R&D Systems) and the mice were collected 7 days after the injection for single-cell isolation.
Gene knockout in gliomaspheres
To knockout genes in glioma sphere culture models, cells were transduced with multiple sgRNAs-CAS9 protein complexes (Gene Knockout Kit v2, Synthego) using Neon transfection system (10uL kit, Thermo Fisher Scientific). Briefly, 1 x 105 cells suspended in 10uL of resuspension buffer R were mixed with pre-assembled ribonucleoprotein (RNP) complexes (6:1 sgRNA to Cas9 ratio), and electroporated with double pulses (1200mV, 20ms). To evaluate knockout efficiency, genomic DNA was extracted from cells using QuickExtract DNA Extract Solution (Lucigen). Prior to Sanger sequencing, the genomic regions targeted by sgRNAs were amplified by polymerase chain reaction (PCR) using Q5 high fidelity polymerase (New England Biolabs) and following primers, OSMR Fw: CACTGGCACATATCGTGGAC and Rv: TTCCAGTGCCAAGTTTCACA, LIFR Fw: CCAGGGAAGCTTGAGTTTGA and Rv: TCTTGATTGTGCTGGTGGTT, IL6ST (GP130) Fw: CCATAATCCAACCACACTTAACA and Rv: CCCAGAAGAGGTTATCAAGCA. Editing efficiency was determined with Inference of CRISPR Edits (ICE) tool (Synthego), and we observed 80-90% KO score defined by frameshifts or deletions of more than 21bp. Edited cells were further isolated by FACS sorting with antibodies; APC anti-human OSMR (clone AN-V2, Thermo Fisher Scientific), APC anti-human LIFR (clone 32953, R&D systems), APC anti-human GP130 (clone 2E1B02, BioLegend), APC mouse IgG1 (clone MOPC-21, BioLegend), APC mouse IgG2a (clone MOPC-173, BioLegend).
SiRNA knockdown
siGENOME siRNA pools were purchased from Dharmacon, and introduced into glioma spheres using Neon transfection system. Briefly, 1 x 105 cells suspended in 10uL of resuspension buffer R were mixed with 100 pmol of siRNAs, and electroporated with double pulses (1200mV, 20ms). 24 hours later, the cells were dissociated and seeded into ultra-low attachment plates (Corning), and then harvested at 72 hours after electroporation to evaluate knockdown efficiency by qRT-PCR or western blot, or treated with 20 ng/mL of OSM or BSA for another 24 hours. For RNA-seq readout, 1,000 cells were lysed in 10uL of buffer TCL (Qiagen) with 1 % 2-Mercaptoethanol, and processed with the Smart-Seq2 protocol described above with a modification of reduced PCR cycle (20 cycle to 9 cycles).
Western blotting
The cells were lysed with NP-40 lysis buffer and centrifuged at 15,000 rpm for 15 min at 4 °C. The supernatants were collected, and total protein content was measured using the Bradford assay (Bio-Rad Laboratories). Lysates were separated with Bolt 4 to 12 % Bis-Tris polyacrylamide gels (Thermo Fisher Scientific), transferred to membrane filters, and subjected to immunoblotting using anti-GP130 (1:2000, Cell Signaling, #3732), anti-LIFR (1:1000, Santa Cruz Biotechnology, A-10), anti-EGFR (1:1000, Cell Signaling, #2232), anti-p-EGFR-Tyr1068 (1:1000, Cell Signaling, #2234), anti-AKT (1:1000, Cell Signaling, #4691), anti-p-AKT-Ser473 (1:1000, Cell Signaling, #4060), anti-ERK1/2 (1:1000, Cell Signaling, #9102), anti-p-ERK1/2-Thr202/Tyr204 (1:1000, Cell Signaling, #9101), anti-MEK1/2 (1:1000, Cell Signaling, #8727), anti-p-MEK1/2-Ser217/221 (1:1000, Cell Signaling, #9154), anti-Stat3 (1:1000, Cell Signaling, #4904), anti-p-Stat3-Tyr705 (1:1000, Cell Signaling, #9145), anti-Jak1 (1:1000, Cell Signaling, #3344), anti-p-Jak1-Tyr1034/1035 (1:1000, Cell Signaling, #74129), anti-Jak2 (1:1000, Cell Signaling, #3230), anti-p-Jak2-Tyr1008 (1:1000, Cell Signaling, #8082), anti-Jak3 (1:1000, Cell Signaling, #8827), anti-p-Jak3-Tyr980/981 (1:1000, Cell Signaling, #5031), and anti-beta-tubulin mouse antibody (1:5000, Sigma).
Human macrophage co-culture
Human monocyte-derived macrophages (C12915, PromoCell) or iPSC-derived human microglia (ASE-9601, Applied StemCell,) were cultured at 5 x 105 cells/mL in DMEM (Corning) supplemented with 20 ng/mL of M-CSF (Peprotech) and 10 % FBS. To obtain conditioned medium, MGG23 cells were cultured with or without macrophages for 72 hours in EGF/FGF/M-CSF-containing serum-free medium, and the supernatant was harvested by centrifugation at 2000 rpm for 15 min and stored at 4 °C or at −80 °C for a long-term storage. The glioma sphere pellets from the co-culture supernatant were further dissociated with TrypLE to prepare single-cell suspension for flowcytometry analysis. CD45 expression was used to discriminate glioblastoma cells from macrophages. MGG23 cells were then incubated with control or macrophage conditioned medium additionally supplemented with EGF/FGF for 24 hours before flow-cytometric analysis.
Primary human T cell isolation
Human T cells were purified from PBMCs obtained from fresh leukaphoresis blood collars procured from the Brigham and Women’s Hospital blood bank and separated using SepMate PBMC isolation tubes (Stemcell Technologies) (Mathewson et al., 2021). T cells were isolated by utilization of the Human T cell kit (EasySep, Stemcell Technologies). Human T cells were maintained in RPMI-1640 supplemented with 9% fetal bovine serum (FBS), 1% human serum, 50 units/ml penicillin/streptomycin, 5 mM HEPES, 2 mM Glutamax, 5 mM non-essential amino acids, 5 mM sodium pyruvate, 50 μM β-mercaptoethanol, and 30 units/ml of recombinant human IL-2 (Peprotech). T cells were activated with human dynabeads (Life Technologies) at a ratio of 1:1 in the presence of IL-2 for 3 days followed by the inactivation of the endogenous T cell receptor ɑ constant (TRAC) locus using a ribonucleoprotein (RNP) complex of Cas9 protein and a gRNA encoding 5′- TGTCTATAGGTCTTGGGAC-3’. The RNP was electroporated into the stimulated primary human T cells using an Amaxa 4-D nucleofector. The T cells were next transduced with lentivirus to induce expression of the NY-ESO-1 TCR (Mathewson et al., 2021). The T cells were cultured in IL-2 and rested for 3 days prior to the co-culture assay by removing the magnetic dynabeads.
Human T cell co-culture
As recently described (Mathewson et al., 2021), MGG75 cells were pretreated with 20 ng/mL of OSM or vehicle control for 24 hours prior to co-culture experiment. The next day, the MGG75 cells were collected and dissociated into a single-cell suspension and washed two times to remove OSM. The MGG75 cells were pre-incubated in ultra-low attachment 96-well plates (25,000 cells per well) for 3 hours, then NY-ESO-1 TCR+ T cells were seeded at a 1:1 ratio and cultured for 8 hours (killing assay) or 24 hours (flow cytometry). Cells were stained with Zombie UV (BioLegend) for 10 minutes, and subsequently stained with antibodies against PE anti-human CD44, APC anti-human CD3 (clone HIT3a, BioLegend), BV785 anti-human CD25 (clone BC96, BioLegend), BV421 anti-human CD69 (clone FN50, BioLegend), and PE/Dazzle 594 anti-human PD-1 (clone EH12.2H7, BioLegend).
QUANTIFICATION AND STATISTICAL ANALYSIS
ScRNA-seq data processing
For the mouse model, as quality control, we excluded sequenced cells with fewer than 3,000 detected genes. Among the remaining cells, we detected on average 5,855 genes per cell. Expression levels were quantified as E(i,j) = log2(TPM(i,j)/10+1), where TPM(i,j) refers to transcript-per-million for gene i in cell j, as quantified by RSEM (Li and Dewey, 2011). The average number of transcripts detected per cell was less than 100,000, thus TPM values were divided by 10, to avoid inflating the differences between detected (E(i,j) > 0) and non-detected (E(i,j) = 0) genes, as previously described (Tirosh et al., 2016). For the remaining cells, we calculated Eavg(i) = log2(mean(TPMi,1…n)+1) for each gene, and excluded genes with Eavg < 4. For the cell lines, cell barcode filtering, alignment of reads, and UMI counting were performed using CellRanger 3.0.1 (10x Genomics). Expression levels were quantified as E(i,j) = log2(1 + CPM(i,j)/10), where CPM(i,j) refers to 106*UMI(i,j)/sum[UMI 1..n,j], for gene i in sample j, with n being the total number of analyzed genes. CPM values were divided by 10, as described above for TPM values. Cells with a number of detected genes less than 2,000 were excluded, and we detected an average of 2,885 genes detected per retained cells. For the remaining cells, we assigned cells according to their cell-line (described below), and for each cell line, calculated the Eavg(i) = log2(mean(CPMi,1…n)+1) for each gene, and excluded genes with Eavg < 4. For both systems, we defined relative expression over the remaining cells and genes, by centering the expression levels per gene, Erel(i,j) = Ei,j - mean[Ei,1…n].
Cell line assignment
We used a gene expression-based method to assign cells from the pool to individual cell lines, similar to the method previously described (Kinker et al., 2020). First, we subsetted the gene expression matrix of all of the single cells from the pool to only include cells with at least 2,000 genes detected, and the top 7,000 most highly expressed genes. Next, we used hierarchical clustering of the all of retained single cells and genes, using one minus the Pearson correlation (across all analyzed genes) as the distance metric. This separated the cells of the pool into 5 clusters, each representing a separate cell line. Finally, for each cluster, we identified the reference cell line from the pool with the most similar bulk RNA-seq gene expression profile (Pearson correlation similarity), and in this way, assigned the cells of that cluster to that cell line.
Characterization and comparison to human transcriptional heterogeneity
We analyzed the mouse model and each cell line separately to identify patterns of expression heterogeneity. NMF was applied to the relative expression values (Er), by transforming all negative values to zero, as previously described (Kinker et al., 2020; Puram et al., 2017). We performed NMF using the R package NMF with the number of factors k=6 and defined expression programs as the top 40 genes (by NMF score). Programs of expression heterogeneity in human GBM were previously defined- MES1-like, MES2- like, NPC1-like, NPC2-like, AC-like, OPC-like, G1S, and G2M (Neftel et al., 2019). For each NMF program found each of our model systems, we calculated the Jaccard similarity with each of the human programs, and found the program it was most similar to. Next, we calculated for each gene in each NMF program, the correlation of its expression with the average expression of the human program it was most correlated to across all of the cells in that model system, and selected the 25 highest correlated genes for each NMF program.
Definition of single-cell gene signature scores
Cells were scored to a gene signature as previously described (Neftel et al., 2019), using the R package scalop (https://github.com/jlaffy/scalop). Given a set of genes (Gj ) reflecting an expression signature of a specific cell type or biological function, we calculate for each cell i, a score, SCj (i), quantifying the relative expression of Gj in cell i, as the average relative expression (Er) of the genes in Gj , compared to the average relative expression of a control gene-set (Gj cont): SCj (i) = average[Er(Gj ,i)] – average[Er(Gj cont,i)]. The control gene-set is defined by first binning all analyzed genes into 30 bins of aggregate expression levels (Ea) and then, for each gene in the gene-set Gj, randomly selecting 100 genes from the same expression bin. In this way, the control gene-set has a comparable distribution of expression levels to that of Gj, and the control gene set is 100-fold larger, such that its average expression is analogous to averaging over 100 randomly selected gene-sets of the same size as the considered gene-set.
Assignment of cells to metaprograms
Malignant cells were first separated by cycling and non-cycling cells, based on their score for the G1S and G2M meta-modules. Next, cells were assigned to the meta-module with the highest score between the six meta-modules (MES1-like, MES2-like, NPC1-like, NPC2-like, AC-like, OPC-like) as previously described (Neftel et al., 2019), however cells with a score of less than 0.3 for any program or a difference of less than 0.1 between the highest and second-highest state scores were defined “unclassified”. For most analyses, we collapsed the MES1 and MES2 groups of cells into one group of MES-like cells, and similarly, the NPC1 and NPC2 cells into one group of NPC-like cells. Myeloid cells were assigned in a similar matter using gene-signatures for microglia and macrophages, as defined in (Venteicher et al., 2017).
Simulated bulk expression of glioblastoma cell types
ScRNA-seq dataset of glioblastoma was taken from our previous study (Neftel et al., 2019). Cell-types were already annotated in the dataset (malignant cells, myeloid cells, t-cells, oligodendrocytes). Malignant cells were assigned to the specific meta-programs and myeloid cells were assigned as microglia or macrophages as described in assignment of cells to metaprograms. Bulk expression levels for each gene j in cell i of a certain cell-type was quantified as Ei,j = log2( avg(TPMi,j) +1). Simulated bulk expression was used to examine the relative contribution of various cell types to the TCGA subtype signatures by calculating the average expression of each TCGA signature in each bulk cell-type. In addition, simulated bulk expression was used in the ligand-receptor analysis to find ligands highly expressed in non-malignant cell types with receptors highly expressed in the malignant cells.
Processing of bulk RNA-seq data
Reads were aligned to the GHCh38/hg38 human genome using Bowtie and expression values were quantified using RSEM. Data underwent quantile normalization using the R package preprocessCore. Data are presented as Ei,j = log2[(TPMi,j) + 1], where TPMi,j refers to transcript-per-million for gene i in sample j, as calculated by RSEM (Li and Dewey, 2011).
Bulk scores defined for TCGA samples
Expression data from TCGA samples was based on both the Agilent microarray and RNA-sequencing platforms and scoring for meta-modules was performed as previously described (Neftel et al., 2019). We first defined initial bulk scores by the average expression of a meta-program. Next, we calculated the correlation of each meta-program gene with the initial scores. Genes were excluded if their correlation was below 0.4 or if the correlation was higher for a different meta-program. The remaining genes were then used to define refined bulk scores.
MERFISH data analysis
For quality control, we first assessed the consistency of average log10 gene expression magnitudes between tissue sections as measured by MERFISH and bulk RNA-seq. Overall, we observed the high correlation between gene expression in both MERFISH sections and the corresponding bulk RNA-sequencing sections (Pearson Correlation R = 0.82 and 0.83) as well as between the two MERFISH sections (Pearson Correlation R = 0.87). We further confirm that on average, the blank barcodes were detected at lower levels compared to 133 out of the 135 real genes. To restrict our analysis to high quality cells and fields of view, we removed from downstream analysis cells with fewer than 10^1.5 total gene counts and cells with volumes less than 10^1.5 or greater than 10^3.5. To avoid edge effects, we further removed cells in fields of views along the edge of the tumor samples. Overall, 20244 cells (10354 in section 1 and 9890 in section 2) in total were retained for downstream analysis. Gene counts were then normalized by cell volume as described previously (Moffitt et al., 2018; Moffitt et al., 2016). To annotate cell-types and cell-states in MERFISH, we again leveraged previously published GBM scRNA-seq data with corresponding cell-type and cell-state annotations from Neftel et. al. scRNA-seq counts were CPM normalized and log normalized with a pseudocount of 1. Keeping only the 135 genes that were also measured in the MERFISH samples and using the previous cell-type annotations, we restricted analysis to 115 significantly differentially expressed genes between annotated groups based on Wilcoxon rank sum testing with a Benjamini-Hochberg multiple hypothesis correction (adjusted p-value < 0.05). We performed unified clustering analysis between the scRNA-seq data and the MERFISH data using Harmony (Korsunsky et al., 2019). We trained a linear discriminant analysis model to predict cell-types using the harmonized scRNA-seq data and corresponding cell-type annotations, and applied the trained model to transfer annotations to cells in the MERFISH data. We further confirm the reasonableness of cell-type annotations for MERFISH using clustering analysis on the harmonized expressions and evaluating for the expression of expected marker genes as compared to scRNA-seq data. We then used the transferred cell-type annotations to evaluate the spatial co-localization patterns in the MERFISH tissue sections. To assess the spatial co-localization between macrophages and MES-like glioblastoma cells, we computed the average number of macrophages in the 30 spatially closest cells to each MES-like cell. To calculate significance, we randomly permuted cell-type labels and computed average number of macrophages nearest neighbors as above to simulate the null distribution expected if there was no spatial association. We then fit a Gaussian distribution to the simulated null distribution and used it to compute a one-sided permutation P-value for the observed average number of macrophages nearest neighbors. We repeated this analysis to assess the spatial co-localization between oligodendrocytes and MES-like cells.
Inferring ligand-receptor interactions
Simulated bulk expression of each cell type in glioblastoma was used to infer ligand-receptor interactions. A list of putative ligand–receptor pairs was taken from a known study (Ramilowski et al., 2015). The list was subsetted to only include pairs that were considered “known” and for ligands which are known secreted factors. For each cell-type interaction tested, all ligands that had a higher average expression than 4 in the interacting cell-type (i.e. macrophages) and a corresponding receptor that had a higher average expression than 4 in the glioblastoma malignant cells were chosen as possible interaction axis. If more than one receptor met this requirement for a given ligand, such as the case with ligand GAS6 and receptors (AXL, TYRO3), the receptor that was most highly associated with the MES-like malignant state was chosen. Association of each receptor to the meta-programs was calculated by the correlation of each receptor’s expression across all malignant cells in the single cell RNA seq dataset, as well as the tumors of TCGA datasets, with the corresponding meta-program scores. The robustness of this correlation was tested by repeating this correlation 1000 times where 500 cells were sampled each time.
Comparing macrophages across glioma
ScRNA-seq dataset of gliomas was taken from our previous studies: IDH-wildtype glioblastoma (Neftel et al., 2019), IDH-mutant astrocytoma (Venteicher et al., 2017), IDH-mutant oligodendroglioma (Tirosh et al., 2016), and H3K27M gliomas (Filbin et al., 2018). PCA was performed over the relative expression matrix Erel of all microglia/macrophages, which were previously annotated in the datasets, from these gliomas including all genes with Ea > 4 (defined only on the basis of microglia/macrophage cells). The second principal component, PC2, separated the GBM myeloid cells from the other glioma myeloid cells. PC2 genes were defined as those with a loading score above 0.03 (PC2-high genes) or below −0.025 (PC2-low genes). We observed many MES-like genes in the PC2-low genes, and defined a MP-MES signature as genes that were PC2-low and MES-like. Because the MP-MES was more associated with macrophages than microglia, we repeated the PCA analysis in 2 parts (Figure S4 B): 1) cells that were confidently assigned as microglia from each glioma and 2) cells that were confidently assigned as macrophages from each glioma using previously published microglia and macrophage signatures (Venteicher et al., 2017). In addition, in order to compare the MP-MES score between GBM and the other gliomas, taking into account for the macrophage bias, we scored each cell for its macrophage, microglia, and MP-MES score and plotted the MP-MES score against the difference between the macrophage and microglia score.
Associations of MES-like state with T-cell and myeloid states in TCGA
To evaluate if the MES-state is associated with a specific subtype of T-cells, we used the bulk expression profiles from TCGA, separately analyzing the RNA-seq and the microarray datasets, both of which were log-transformed and normalized. First, we used our glioblastoma scRNA-seq data (Neftel et al., 2019) to compare the average expression of each gene across different cell types and identified genes that were at least 8-fold higher expressed in T-cells than in any other cell-type. Next, we scored each tumor in the TCGA dataset for the glioblastoma MES-like state, and for their total T-cell signal, defined by the average normalized expression of the genes CD2, CD3D, CD3E, and CD3G. Since the state of T-cells cannot be properly evaluated if the total T-cell signal is low, we restricted further analysis to 79 out of 179 tumors from RNA-seq dataset (and 202 out of 441 tumors for the microarray tumor) that had a total T-cell score above 4. For the scatter plot in Figure 6A, we then plotted each gene expression’s correlation to the MES-like score against its correlation to the total T-cell signal. For the histograms in Figure S6A, we generated a relative expression profile for each T-cell specific gene, by subtracting from its expression the median expression of all T-cell specific genes. We then calculated the Pearson correlation of each gene’s normalized expression to the MES-like score across all tumors. T-cell signatures were defined as follows: FOXP3 for T-regs, and PRF1, GZMB for Cytotoxic T-cells. For myeloid cells, the same analysis was repeated using myeloid-specific genes, and the macrophage/microglia signatures were defined as described above.
Analysis of bulk RNA-seq from PD-1 inhibitor trial
TPM matrix of bulk RNA seq of glioblastoma patients before and after treatment with PD-1 inhibitor was taken from a published study (Zhao et al., 2019). Each tumor was scored for the MES-like program as explained earlier. If multiple tumors were taken for the same patient before or after treatment, the average score was calculated. We compared the MES-like score for pre-treatment and post-treatment between patients that responded vs non-responders.
Comparing OSM expression in glioblastoma-associated myeloid cells with healthy microglia
Three datasets of scRNA-seq of healthy human microglia (Geirsdottir et al., 2019; Masuda et al., 2019; Zhong et al., 2018) and two datasets of healthy mouse microglia (Li et al., 2019; Masuda et al., 2019) were used to compare the relative expression of OSM in glioblastoma associated myeloid cells from our previously published scRNA-seq from 10x and SS2 platforms (Neftel et al., 2019). For each dataset simulated bulk expression was calculated for the myeloid cells. Next, for each comparison separately, simulated bulk expression of each gene in the healthy microglia simulated bulk (X) was plotted against its expression in the glioblastoma-associated macrophages simulated bulk (Y). Then, to overcome the differences in the platforms used for scRNA-seq, a line was fitted over the expression, which was defined in the following manner: for i=1..100 the point (Xi,Yi) such that Xi is the i'th percentile value of the X dataset and Yi is the i'th percentile value of the Y dataset. The Y distance of OSM from the line was calculated for each comparison between healthy microglia and glioblastoma myeloid cells, as well as control comparisons comparing: glioblastoma microglia vs glioblastoma macrophages, glioblastoma myeloid cells profiled from the 10x platform vs glioblastoma myeloid cells profiled by SS2 platform, and two healthy microglia datasets against each other.
Supplementary Material
Table S1: Mouse model NMF programs, related to Figure 1.
Table S2: MGG23 NMF programs, related to Figure 3.
Table S3: PC1 and PC2 associated genes, related to Figure 5.
Table S4: Correlation of each T cell specific gene expression to total T cell signal or MES-like score, related to Figure 6.
Acknowledgements
This work was supported by a Broad Institute–Israel Science Foundation Collaborative Project Award (I.T. and M.L.S.), grants from the Mark Foundation Emerging Leader Award (M.L.S.), the Sontag Foundation Distinguished Scientist Award (M.L.S.), N.I.H. P50CA165962 (M.L.S.) and N.I.H. R37CA245523 (M.L.S). I.T. is the incumbent of the Dr. Celia Zwillenberg-Fridman and Dr. Lutz Zwillenberg Career Development Chair, and is supported by the Zuckerman STEM Leadership Program, the Mexican Friends New Generation, the Benoziyo Endowment Fund, and grants from the Human Frontiers Science Program. T.Hara was supported by Grant-in-Aid for JSPS Fellows from the Japan Society for the Promotion of Science, SENSHIN Medical Research Foundation, and Kanae foundation for the promotion of medical science. N.D.M. was supported by a postdoctoral fellowship from the American Cancer Society (PF-17-042-01-LIB) and the NIH education loan repayment program funded by the NCI (L30CA231679-01). G.S.K. was supported by fellowships from the Sao Paulo Research Foundation (FAPESP, 2014/27287-0 and 2017/24287-8). L.N.G.C. was supported by a grant from N.I.H. K12CA090354. S.W.E. and X.Z. were supported by N.I.H. and Howard Hughes Medical Institute. I.M.V. and T.Hunter were supported by a grant from N.I.H. R01CA195613. This work was also supported by the Flow Cytometry Core, the Advanced Biophotonics Core and the Next Generation Sequencing core of the Salk Institute, with funding from N.I.H. CCSG P30CA014195.
Footnotes
Declaration of interests
M.L.S. and K.W.W. are equity holders, scientific co-founders and advisory board members of Immunitas Therapeutics. I.T. is advisory board member of Immunitas Therapeutics. A.R. is a founder and equity holder of Celsius Therapeutics, an equity holder in Immunitas Therapeutics, and until July 31, 2020 was an SAB member for Thermo Fisher Scientific, Syros Pharmaceuticals, Asimov and Neogene Therapeutics. O.R.R. and A.R. are employees of Genentech since October 19, 2020 and August 1, 2020, respectively. O.R.R. is a co-inventor on patent applications filed by the Broad Institute for inventions related to single cell genomics. K.W.W. serves on the scientific advisory board of TCR2 Therapeutics, T-Scan Therapeutics, SQZ Biotech, Nextechinvest and receives sponsored research funding from Novartis. N.D.M. serves as a scientific advisor to Immunitas Therapeutics. X.Z. is a co-founder and consultant for Vizgen, Inc.
References
- Beyer M, Mallmann MR, Xue J, Staratschek-Jox A, Vorholt D, Krebs W, Sommer D, Sander J, Mertens C, Nino-Castro A, et al. (2012). High-resolution transcriptome of human macrophages. PLoS One 7, e45466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat KP, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F, Wani K, Heathcock L, James JD, Goodman LD, et al. (2013). Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer cell 24, 331–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat KP, Salazar KL, Balasubramaniyan V, Wani K, Heathcock L, Hollingsworth F, James JD, Gumin J, Diefes KL, Kim SH, et al. (2011). The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes & development 25, 2594–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butowski N, Colman H, De Groot JF, Omuro AM, Nayak L, Wen PY, Cloughesy TF, Marimuthu A, Haidar S, Perry A, et al. (2016). Orally administered colony stimulating factor 1 receptor inhibitor PLX3397 in recurrent glioblastoma: an Ivy Foundation Early Phase Clinical Trials Consortium phase II study. Neuro-oncology 18, 557–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, et al. (2010). The transcriptional network for mesenchymal transformation of brain tumours. Nature 463, 318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KH, Boettiger AN, Moffitt JR, Wang S, and Zhuang X (2015). RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosset E, Ilmjarv S, Dutoit V, Elliott K, von Schalscha T, Camargo MF, Reiss A, Moroishi T, Seguin L, Gomez G, et al. (2017). Glut3 Addiction Is a Druggable Vulnerability for a Molecularly Defined Subpopulation of Glioblastoma. Cancer cell 32, 856–868 e855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza CF, Sabedot TS, Malta TM, Stetson L, Morozova O, Sokolov A, Laird PW, Wiznerowicz M, Iavarone A, Snyder J, et al. (2018). A Distinct DNA Methylation Shift in a Subset of Glioma CpG Island Methylator Phenotypes during Tumor Recurrence. Cell reports 23, 637–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filbin MG, Tirosh I, Hovestadt V, Shaw ML, Escalante LE, Mathewson ND, Neftel C, Frank N, Pelton K, Hebert CM, et al. (2018). Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science 360, 331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann-Morvinski D, Bushong EA, Ke E, Soda Y, Marumoto T, Singer O, Ellisman MH, and Verma IM (2012). Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science 338, 1080–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari FB, Cloughesy TF, Cavenee WK, and Mischel PS (2015). Heterogeneity of epidermal growth factor receptor signalling networks in glioblastoma. In Nat Rev Cancer, (England), pp. 302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrusiewicz K, Rodriguez B, Wei J, Hashimoto Y, Healy LM, Maiti SN, Thomas G, Zhou S, Wang Q, Elakkad A, et al. (2016). Glioblastoma-infiltrated innate immune cells resemble M0 macrophage phenotype. JCI Insight 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geirsdottir L, David E, Keren-Shaul H, Weiner A, Bohlen SC, Neuber J, Balic A, Giladi A, Sheban F, Dutertre CA, et al. (2019). Cross-Species Single-Cell Analysis Reveals Divergence of the Primate Microglia Program. Cell 179, 1609–1622 e1616. [DOI] [PubMed] [Google Scholar]
- Gilbert MR, Dignam JJ, Armstrong TS, Wefel JS, Blumenthal DT, Vogelbaum MA, Colman H, Chakravarti A, Pugh S, Won M, et al. (2014). A randomized trial of bevacizumab for newly diagnosed glioblastoma. The New England journal of medicine 370, 699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski MM, Sankey EW, Ryan KJ, Chongsathidkiet P, Lorrey SJ, Wilkinson DS, and Fecci PE (2020). Immune suppression in gliomas. Journal of neuro-oncology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, and Verma IM (2019). Modeling Gliomas Using Two Recombinases. Cancer research 79, 3983–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahani-Asl A, Yin H, Soleimani VD, Haque T, Luchman HA, Chang NC, Sincennes MC, Puram SV, Scott AM, Lorimer IA, et al. (2016). Control of glioblastoma tumorigenesis by feed-forward cytokine signaling. Nat Neurosci 19, 798–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SA, and Jenkins BJ (2018). Recent insights into targeting the IL-6 cytokine family in inflammatory diseases and cancer. Nat Rev Immunol 18, 773–789. [DOI] [PubMed] [Google Scholar]
- Junk DJ, Bryson BL, Smigiel JM, Parameswaran N, Bartel CA, and Jackson MW (2017). Oncostatin M promotes cancer cell plasticity through cooperative STAT3-SMAD3 signaling. Oncogene 36, 4001–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinker GS, Greenwald AC, Tal R, Orlova Z, Cuoco MS, McFarland JM, Warren A, Rodman C, Roth JA, Bender SA, et al. (2020). Pan-cancer single cell RNA-seq uncovers recurring programs of cellular heterogeneity. bioRxiv 807552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga T, Chen CC, and Furnari FB (2019). When less is more: Gaining power through gene rearrangement of amplified EGFR. Oncotarget 10, 2116–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh PR, and Raychaudhuri S (2019). Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 16, 1289–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucia-Tran JA, Tulkki V, Smith S, Scarpini CG, Hughes K, Araujo AM, Yan KY, Botthof J, Perez-Gomez E, Quintanilla M, et al. (2016). Overexpression of the oncostatin-M receptor in cervical squamous cell carcinoma is associated with epithelial-mesenchymal transition and poor overall survival. Br J Cancer 115, 212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. In BMC Bioinformatics, (England), p. 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Horns F, Wu B, Xie Q, Li J, Li T, Luginbuhl DJ, Quake SR, and Luo L (2017). Classifying Drosophila Olfactory Projection Neuron Subtypes by Single-Cell RNA Sequencing. Cell 171, 1206–1220 e1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Cheng Z, Zhou L, Darmanis S, Neff NF, Okamoto J, Gulati G, Bennett ML, Sun LO, Clarke LE, et al. (2019). Developmental Heterogeneity of Microglia and Brain Myeloid Cells Revealed by Deep Single-Cell RNA Sequencing. Neuron 101, 207–223 e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN, Ohgaki H, Wiestler OD, and Cavenee WK (2016). WHO Classification of Tumors of the Central Nervous System (Revised 4th edition). IARC: Lyon. [Google Scholar]
- Mantovani A, Marchesi F, Malesci A, Laghi L, and Allavena P (2017). Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol 14, 399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, Kadel EE III, Koeppen H, Astarita JL, Cubas R, et al. (2018). TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554, 544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda T, Sankowski R, Staszewski O, Bottcher C, Amann L, Sagar, Scheiwe C, Nessler S, Kunz P, van Loo G, et al. (2019). Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature 566, 388–392. [DOI] [PubMed] [Google Scholar]
- Mathewson ND, Ashenberg O, Tirosh I, Gritsch S, Perez EM, Marx S, Jerby-Arnon L, Chanoch-Myers R, Hara T, Richman AR, et al. (2021). Inhibitory CD161 receptor identified in glioma-infiltrating T cells by single-cell analysis. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Bambah-Mukku D, Eichhorn SW, Vaughn E, Shekhar K, Perez JD, Rubinstein ND, Hao J, Regev A, Dulac C, and Zhuang X (2018). Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Hao J, Bambah-Mukku D, Lu T, Dulac C, and Zhuang X (2016). High-performance multiplexed fluorescence in situ hybridization in culture and tissue with matrix imprinting and clearing. Proceedings of the National Academy of Sciences of the United States of America 113, 14456–14461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, Richman AR, Silverbush D, Shaw ML, Hebert C, et al. (2019). An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 178, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa T, Riester M, Cheng YK, Huse JT, Squatrito M, Helmy K, Charles N, Michor F, and Holland EC (2014). Most Human Non-GCIMP Glioblastoma Subtypes Evolve from a Common Proneural-like Precursor Glioma. Cancer cell 26, 288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, et al. (2014). Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, and Sandberg R (2014). Full-length RNA-seq from single cells using Smart-seq2. In Nat Protoc, (England), pp. 171–181. [DOI] [PubMed] [Google Scholar]
- Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, Rodman C, Luo CL, Mroz EA, Emerick KS, et al. (2017). Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 171, 1611–1624 e1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramilowski JA, Goldberg T, Harshbarger J, Kloppmann E, Lizio M, Satagopam VP, Itoh M, Kawaji H, Carninci P, Rost B, and Forrest AR (2015). A draft network of ligand-receptor-mediated multicellular signalling in human. Nat Commun 6, 7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport AP, Stadtmauer EA, Binder-Scholl GK, Goloubeva O, Vogl DT, Lacey SF, Badros AZ, Garfall A, Weiss B, Finklestein J, et al. (2015). NY-ESO-1-specific TCR-engineered T cells mediate sustained antigen-specific antitumor effects in myeloma. Nat Med 21, 914–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins PF, Li YF, El-Gamil M, Zhao Y, Wargo JA, Zheng Z, Xu H, Morgan RA, Feldman SA, Johnson LA, et al. (2008). Single and dual amino acid substitutions in TCR CDRs can enhance antigen-specific T cell functions. J Immunol 180, 6116–6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodig SJ, Gusenleitner D, Jackson DG, Gjini E, Giobbie-Hurder A, Jin C, Chang H, Lovitch SB, Horak C, Weber JS, et al. (2018). MHC proteins confer differential sensitivity to CTLA-4 and PD-1 blockade in untreated metastatic melanoma. Sci Transl Med 10. [DOI] [PubMed] [Google Scholar]
- Rooj AK, Ricklefs F, Mineo M, Nakano I, Chiocca EA, Bronisz A, and Godlewski J (2017). MicroRNA-Mediated Dynamic Bidirectional Shift between the Subclasses of Glioblastoma Stem-like Cells. Cell reports 19, 2026–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha D, Martuza RL, and Rabkin SD (2017). Macrophage Polarization Contributes to Glioblastoma Eradication by Combination Immunovirotherapy and Immune Checkpoint Blockade. Cancer cell 32, 253–267 e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibue T, and Weinberg RA (2017). EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol 14, 611–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smigiel JM, Parameswaran N, and Jackson MW (2017). Potent EMT and CSC Phenotypes Are Induced By Oncostatin-M in Pancreatic Cancer. Mol Cancer Res 15, 478–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, and Tavare S (2013). Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proceedings of the National Academy of Sciences of the United States of America 110, 4009–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. (2005). Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. The New England journal of medicine 352, 987–996. [DOI] [PubMed] [Google Scholar]
- Tanay A, and Regev A (2017). Scaling single-cell genomics from phenomenology to mechanism. Nature 541, 331–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry S, Savagner P, Ortiz-Cuaran S, Mahjoubi L, Saintigny P, Thiery JP, and Chouaib S (2017). New insights into the role of EMT in tumor immune escape. Mol Oncol 11, 824–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, and Suva ML (2019). Deciphering Human Tumor Biology by Single-Cell Expression Profiling. Annual Review of Cancer Biology Volume 3. [Google Scholar]
- Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak K, Fisher JM, Rodman C, Mount C, Filbin MG, et al. (2016). Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. In Nature, (England), pp. 309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher AS, Tirosh I, Hebert C, Yizhak K, Neftel C, Filbin MG, Hovestadt V, Escalante LE, Shaw ML, Rodman C, et al. (2017). Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. (2010). Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer cell 17, 98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakimoto H, Mohapatra G, Kanai R, Curry WT Jr., Yip S, Nitta M, Patel AP, Barnard ZR, Stemmer-Rachamimov AO, Louis DN, et al. (2012). Maintenance of primary tumor phenotype and genotype in glioblastoma stem cells. Neuro-oncology 14, 132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Hu B, Hu X, Kim H, Squatrito M, Scarpace L, deCarvalho AC, Lyu S, Li P, Li Y, et al. (2017). Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer cell 32, 42–56 e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen PY, Weller M, Lee EQ, Alexander BA, Barnholtz-Sloan JS, Barthel FP, Batchelor TT, Bindra RS, Chang SM, Chiocca EA, et al. (2020). Glioblastoma in Adults: A Society for Neuro-Oncology (SNO) and European Society of Neuro-Oncology (EANO) Consensus Review on Current Management and Future Directions. Neuro-oncology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West NR, Hegazy AN, Owens BMJ, Bullers SJ, Linggi B, Buonocore S, Coccia M, Gortz D, This S, Stockenhuber K, et al. (2017). Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat Med 23, 579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia C, Fan J, Emanuel G, Hao J, and Zhuang X (2019). Spatial transcriptome profiling by MERFISH reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression. Proceedings of the National Academy of Sciences of the United States of America 116, 19490–19499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Chen AX, Gartrell RD, Silverman AM, Aparicio L, Chu T, Bordbar D, Shan D, Samanamud J, Mahajan A, et al. (2019). Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat Med 25, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Zhang S, Fan X, Wu Q, Yan L, Dong J, Zhang H, Li L, Sun L, Pan N, et al. (2018). A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 555, 524–528. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Mouse model NMF programs, related to Figure 1.
Table S2: MGG23 NMF programs, related to Figure 3.
Table S3: PC1 and PC2 associated genes, related to Figure 5.
Table S4: Correlation of each T cell specific gene expression to total T cell signal or MES-like score, related to Figure 6.
Data Availability Statement
Data generated for this study are available through the Gene Expression Omnibus (GEO: GSE168004). The Code supporting the current study is available from the corresponding authors on request. Additional datasets used for this study were obtained from the following studies: GSE GSE131928 (Neftel et al., 2019), GSE GSE134707 (Geirsdottir et al., 2019), GSE GSE124335 (Masuda et al., 2019), GSE GSE104276 (Zhong et al., 2018), GSE GSE89567 (Venteicher et al., 2017), GSE GSE70630 (Tirosh et al., 2016), GSE GSE102130 (Filbin et al., 2018), SRA PRJNA482620 (Zhao et al., 2019) and (Wang et al., 2017).