Abstract
Mitochondria play a critical role in numerous cell types and diseases, and structure and function of mitochondria can vary greatly among cells or within different regions of the same cell. However, there are currently limited methodologies that provide direct assessments of mitochondrial function in vivo, and contemporary measures of mitochondrial energy conversion lack the spatial resolution necessary to address cellular and subcellular heterogeneity. Here, we describe a novel metabolic imaging approach that provides direct measures of mitochondrial energy conversion with single-cell and subcellular resolution by evaluating NADH autofluorescence kinetics during the mitochondrial redox after cyanide experiment (mitoRACE). mitoRACE measures the rate of NADH flux through the steady-state mitochondrial NADH pool by rapidly inhibiting mitochondrial energetic flux, resulting in an immediate, linear increase in NADH fluorescence proportional to the steady-state NADH flux rate, thereby providing a direct measure of mitochondrial NADH flux. The experiments presented here demonstrate the sensitivity of this technique to detect physiological and pharmacological changes in mitochondrial flux within tissues of living animals and reveal the unique capability of this technique to evaluate mitochondrial function with single-cell and subcellular resolution in different cell types in vivo and in cell culture. Furthermore, we highlight the potential applications of mitoRACE by showing that within single neurons, mitochondria in neurites have higher energetic flux rates than mitochondria in the cell body. Metabolic imaging with mitoRACE provides a highly adaptable platform for evaluating mitochondrial function in vivo and in single cells with potential for broad applications in the study of energy metabolism.
Keywords: Metabolism, Oxidative Phophorylation, Skeletal Muscle
Introduction
Mitochondria serve a fundamental role in cellular energy conversion and are therefore critical for many cellular processes across a wide range of cell types. Not surprisingly, deficits in mitochondria have been implicated in numerous pathologies(Suomalainen & Battersby, 2018) and chronic reductions in mitochondrial function are associated with increases in cardiometabolic risk. As a result, assessment of mitochondrial function has been a major area of interest across much of biomedical research for decades(Picard, Wallace, & Burelle, 2016). Classical in vitro measures of oxygen consumption rates in mitochondria removed from the cellular environment or cells in culture have provided a wide variety of metabolic insights(Hebert-Chatelain et al., 2016; Pesta & Gnaiger, 2012; Weinberg et al., 2019), and in vivo assessments of high energy phosphates(Younkin et al., 1984), oxygen saturation(Grassi, Gladden, Samaja, Stary, & Hogan, 1998; Ryan, Southern, Reynolds, & McCully, 2013), and carbon labeling(Befroy et al., 2007) have provided integrative, non-invasive measure of oxidative metabolic rates in humans. However, each of the above techniques only provide a single bulk assessment of generally many thousands of cells. Since mitochondrial structure, composition, and function are tuned to match that of the cells in which they reside(Johnson, Harris, Blair, & Balaban, 2007), and different mitochondrial subpopulations within the same cell are now being reported in an increasing number of cell types(C. K. E. Bleck, Y. Kim, T. B. Willingham, & B. Glancy, 2018; Cheng, Hou, & Mattson, 2010), the lack of cellular or subcellular resolution in current methodologies provides a major limitation to the assessment of mitochondrial function in tissues with mixed cell types, scarce human biopsy tissue, primary cell cultures with multiple cell types, cells with varied gene transfection efficiencies, and cells with heterogeneous mitochondrial populations such as neurons, salivary glands, brown fat, cardiac, and skeletal muscle, among others. To address the spatial limitations of contemporary mitochondrial function techniques, we developed a novel metabolic imaging approach to provide direct measures of the rate of mitochondrial energy conversion with single-cell and subcellular resolution by using multiphoton microscopy to assess reduced nicotinamide adenine dinucleotide (NADH) autofluorescence kinetics during the mitochondrial redox after cyanide experiment (mitoRACE).
NADH is the primary electron donor for mitochondrial oxidative phosphorylation, and the subsequent transport of electrons down the electron transport chain (ETC) is coupled to the translocation of protons across the inner mitochondrial membrane, generating the electrochemical gradient used to produce ATP. Because NADH is inherently fluorescent and the oxidized NAD+ molecule is not, NADH fluorescence has long provided an organic, well-adopted marker of mitochondrial redox activity(Blinova et al., 2005; Chance & Williams, 1955; Mayevsky & Rogatsky, 2007). More recently, advancements in multiphoton microscopy have expanded the applications of NADH autofluorescence to the investigation of mitochondrial metabolism in live cells and intact tissues(Bakalar et al., 2012; Schroeder et al., 2010). In living cells, the NADH signal is derived from the steady-state mitochondrial NADH pool(Blinova et al., 2005; Chance, Cohen, Jobsis, & Schoener, 1962; Chance & Williams, 1955; Mayevsky & Rogatsky, 2007), representing a balance between production by fuel dehydrogenases and utilization by Complex I of the ETC. Therefore, changes to either the rate of production or utilization may alter the steady-state NADH concentration, and complete inhibition of the ETC has been used previously to fully reduce the NADH pool and quantify the mitochondrial redox status prior to inhibition(Bakalar et al., 2012; Schroeder et al., 2010). However, determining mitochondrial redox status alone does not provide specific information on the rate of mitochondrial energy conversion which has been the classical marker of mitochondrial function for the last 50+ years. Thus, there remains a great need for a spatially resolved measure of mitochondrial energetic flux.
The experiments presented here demonstrate the utility of mitoRACE to evaluate mitochondrial flux with spatially and temporally resolved measures of NADH autofluorescence. Similar to optical techniques using rapid onset ischemia to evaluate oxidative metabolism(Ryan et al., 2013), mitoRACE measures the rate of NADH flux through the steady-state mitochondrial NADH pool by rapidly inhibiting mitochondrial energetic flux via superfusion of cyanide, an inhibitor of the ETC (Complex IV). Rapid inhibition of flux through the ETC results in an immediate, linear increase in NADH fluorescence proportional to the steady-state NADH flux rate, thereby providing a direct measure of mitochondrial NADH flux. To demonstrate the sensitivity of mitoRACE in assessing changes in metabolism associated with physiological and pharmacological metabolic perturbations, we report mitochondrial NADH flux rates at rest and in the post-electrically stimulated, uncoupled, and fasted metabolic states in skeletal muscle of live mice with single-cell and subcellular resolution. Furthermore, we reveal the unique capability of this technique to evaluate mitochondrial function in different cell types within intact tissues in vivo and in heterogenous cell culture populations. Moreover, by enabling simultaneous assessment of mitochondria in different regions of the same cell, we show that the rate of mitochondrial energy conversion differs between the neurites and the cell body in primary neurons. The multiscale applicability of mitoRACE from in vivo tissues to within individual cells in culture provides a valuable tool to uniquely assess metabolic relationships across a wide variety of models in both health and disease.
Methods
Ethical Approval
All mouse procedures were approved by the of the National Heart, Lung, and Blood Institute Animal Care and Use Committee and performed in compliance with the guidelines of the Animal Care and Welfare Act.
Animals
All mice were housed indoors with food and water provided ad libitum. For 3D motion tracking under the microscope, mice with endogenous mCherry fluorescence within the vasculature were used in all in vivo experiments except for uncoupling experiments using TMRM. To generate a knock-in mouse line with mCherry-tagged to the N-terminus of endogenous non-muscle myosin 2A, the mCherry sequence was inserted at the 5′ of the initiating ATG codon in exon 2. The 4-kb 5’ arm immediately upstream of the ATG is followed by the mCherry-tagged exon2 plus 160-bp DNA sequence immediately 3’ of exon2, a loxP-flanked PGK-Neor cassette and the 2-kb 3’ arm. Nucleotide sequences of the cloned DNA fragments were confirmed by sequencing. Male and Female Mice 2–3 months of age were used for flux experiments. For uncoupling experiments, wild-type C57BL/6 mice (Taconic Biosciences) were used to allow for multiplex imaging with TMRM. Mice were euthanized via anoxia (nitrogen exposure).
Multiphoton laser-scanning fluorescence microscopy
Mice were anaesthetized continuously with 3% isoflurane by nose cone and secured to a custom, 3D-printed acrylic water-heated perfusion platform using medical adhesive tape. The tibialis anterior muscle fibers were exposed by surgically removing skin and fascia. Using a water-heated reservoir suspended ~70cm above the specimen, the exposed muscle fibers were continuously perfused with buffer flowing from the reservoir through a water-jacketed perfusate line to the objective. Buffer flowing from the objective head created a ~7ml perfusate pool surrounding the muscle and allowed for optical coupling of the objective and sample. For in vivo muscle experiments, Tyrodes buffer (pH = 7.1) containing 10mM HEPES, 137mM sodium chloride (NaCl), 4.5mM potassium chloride (KCl), 0.5mM magnesium sulfate (MgSO4), 0.5mM potassium phosphate (KPO4), 10mM glucose, and 1.8mM calcium chloride (CaCl2) was maintained at 37°C and continuously bubbled with 100% O2 in the reservoir. Cell culture NADH flux experiments were performed using a programmable microperfusion system (MultiChannel Systems) and Quick Change (Warner Instruments) imaging system with heated base (37°C ) to produce rapid, laminar flow of perfusate across cells. NADH autofluorescence was imaged using an upright Leica SP8 multiphoton microscope with resonant scanning and a Nikon 25x (1.1 NA) water-immersion objective. NADH fluorescence was excited using a Ti:Sapphire laser tuned to 710 nm or 750 nm and emission detected by a non-descanned Leica HyD detector in the 414–538 nm range. In the longer, resting (and fasted) metabolic experiments, field of view was maintained by rapidly imaging 3D volumes (32 μm in depth) of the discrete mCherry signal (vasculature) and employing a custom 3D-tracking software(Bakalar et al., 2012) to automatically manipulate the objective and stage in real-time to correct for tissue movement. Additional emission from 538–605 nm was acquired during experiments using mCherry 3D tracking and TMRM to enable multiplex detection of these fluorophores. Optical calibration experiments evaluating the relationship between NADH fluorescence intensity and NADH concentration were performed in the absence of cells or tissue by dissolving NADH (Sigma-Aldrich) in Tyrodes solution at incremental concentrations (0.5–3.0mM) and complete oxidation of NADH in solution was achieved by addition of 10mM sodium pyruvate (Sigma-Aldrich) and excess lactate dehydrogenase (Sigma-Aldrich).
Mitochondrial Redox After Cyanide Experiment (mitoRACE)
To explore the utility of mitoRACE as an assessment of mitochondrial energetic flux, NADH autofluorescence and redox kinetics were first evaluated in the intact, murine tibialis anterior muscle in vivo using two-photon excitation. For each mitoRACE, basal NADH autofluorescence was first evaluated at rest for ~10–15sec to establish baseline signal intensity. Next, steady-state mitochondrial NADH utilization was rapidly inhibited by the addition of 0.25% vol/vol of 2M stock NaCN (Sigma-Aldrich) to the perfusate, resulting in a final NaCN concentration of 5mM and complete inhibition of oxidative phosphorylation. The fully reduced NADH signal was quantified as the average fluorescence intensity of the new steady-state after superfusion of NaCN. Analysis of mitochondrial NADH redox status and kinetics were performed using ImageJ (NIH). Mitochondrial redox status was calculated as the baseline NADH fluorescence intensity relative to the fully reduced signal (% NADH reduced). Mitochondrial NADH flux was quantified as the rate of increase relative to the fully reduced NADH signal during the initial 30% of the change in NADH fluorescence observed after superfusion of NaCN. All basal redox status and mitochondrial NADH flux analyses were performed on raw images, and a single pixel mean filter was used to identify subcellular regions of interests for the subcellular flux analyses in oxidative muscle fibers and cultured neurons.
Modulation of in vivo metabolism
Muscle metabolism was physiologically and pharmacologically modulated in vivo using muscle contraction, fasting, and uncoupling of the mitochondrial inner membrane. To induce contraction of the TA muscle in vivo, electrodes were sutured to the sciatic nerve proximal to the peroneal and tibial branches and 8V electrical stimulation (Grass stimulator) was applied at 10Hz for 2 minutes. In fasting experiments, metabolism was modulated using 24 hours of nutrient deprivation to induce a state of torpor. In uncoupling experiments, 0.05% vol/vol of 10mM stock TMRM was first added to perfusate to enable multiplex metabolic imaging of mitochondrial membrane potential and NADH. After collection of baseline NADH and TMRM baseline signals, 0.1% vol/vol of 10mM stock FCCP (Sigma-Aldrich) in DMSO was added to the perfusate for a final FCCP concentration of 10μM. Skeletal muscle was incubated in 10μM FCCP for 30seconds to allow for complete dissipation of mitochondrial membrane potential before the addition of cyanide.
Muscle fibers isolation and culture
Flexor digitorum brevis (FDB) myofibers were dissected in vivo from mice anaesthetized with 3% isoflurane. Skin and fascia were surgically removed from the plantar surface of the foot and the FDB muscle was detached from the bone at the distal and proximal tendons. Muscles fiber for isolation were digested with 3 mg ml−1 collagenase D (Roche) in 10mL Tyrodes buffer (pH = 7.1) containing 10mM HEPES, 137mM sodium chloride (NaCl), 4.5mM potassium chloride (KCl), 0.5mM magnesium sulfate (MgSO4), 0.5mM potassium phosphate (KPO4), 10mM glucose, and 1.8mM calcium chloride (CaCl2) at 37°C in agitating bath for 60 minutes, and muscles for culture were digested with 3 mg ml−1 collagenase D (Roche) in 10mL of Dulbecco’s Modified Eagle’s Medium (DMEM)/F12 supplemented with 20%FBS, 2% Serum Replacement 2 (Sigma Aldrich), Glutamax, and penicillin/streptomycin (1:100) at 37 °C without agitation for 120 minutes. After digestion, all fibers were transferred to a gently agitated and suspended in 2mL microcentrifuge tube using a transfer pipet and immediately plated onto 35mm petri dishes with Matrigel coating (Corning) containing 5mL of the respective media. mitoRACE experiments in isolated fibers were performed 15–30 minutes after plating cells in the dish. Cultured muscle fibers were incubated at 37 °C in 5mL DMEM containing 2%FBS and penicillin/streptomycin (1:100) for 72 hours to allow for proliferation of satellite cells and growth of myoblasts. For mitoRACE experiments, Tyrode’s and DMEM were used for perfusate in isolated and cultured muscle fibers, respectively.
Hippocampal neuron isolation and culture
The hippocampus was dissected out from P1-P5 mouse brain, gently minced, and enzymatically digested with 0.025% Trypsin in Hank’s Balance Salt Solution (HBSS) solution at 37°C for 15 minutes. The digested tissues were then washed with 5ml of HBSS for 3 times, and mechanically dissociated with trituration. The dissociated cells were plated in 35mm Petri dishes coated with poly-D-lysine in Neurobasal Media containing B27 supplement, 0.5mM Glutamine, 1mM HEPES, and Penicillin/Streptomycin (1:200) and incubated for 120 hours at 37 °C. mitoRACE experiments were performed in cultured hippocampal neurons and glial cells using HBSS as perfusate.
NADH fluorescence lifetime measurements
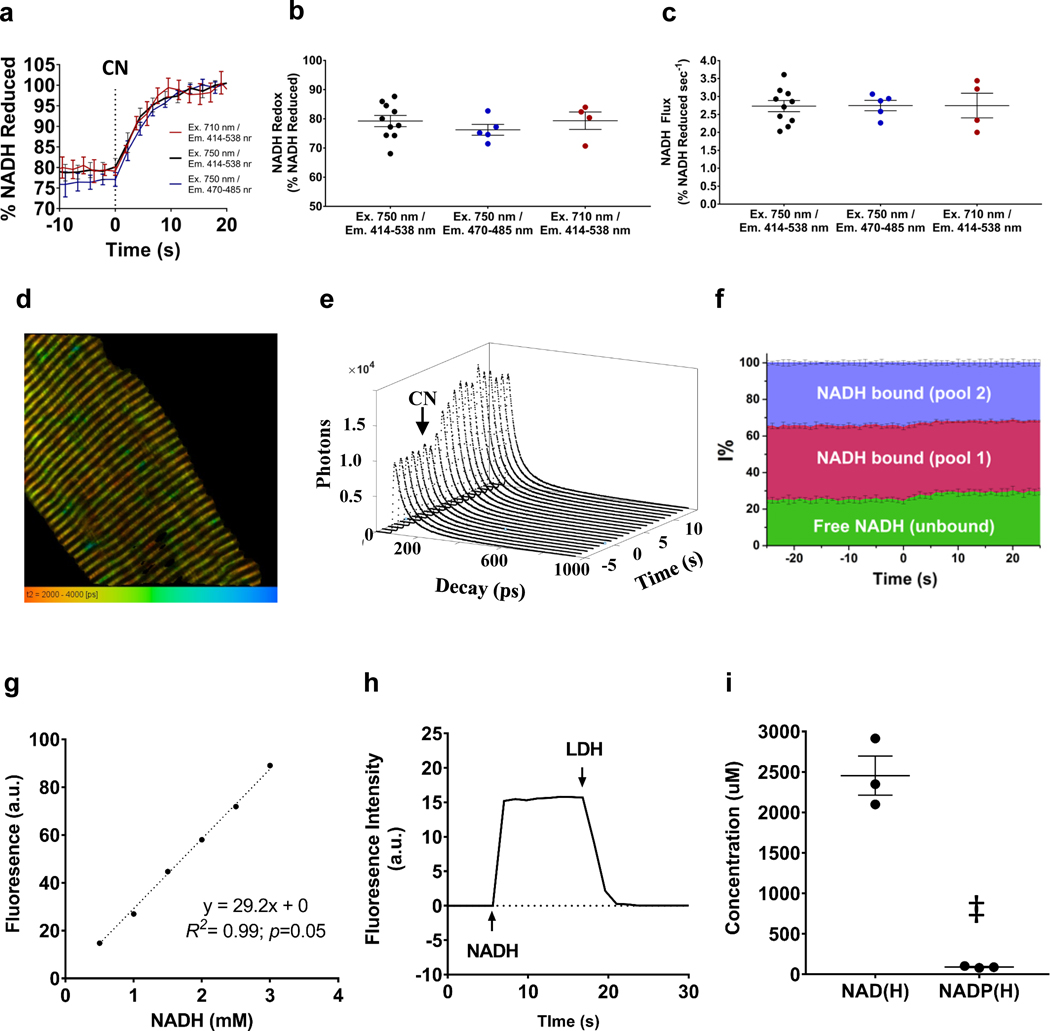
To determine whether changes to the proportion of bound or free species were contributing to the increase in NADH signal observed upon cyanide inhibition, using the same multiphoton system, we integrated real-time, in vivo NADH fluorescence lifetime measurements (FLIM) by synchronizing the 80 MHz synchronization signal from the Ti:Sapphire laser and the output from the HyD detector using a single-photon counting card (SPC150, Becker&Hickl). Data were collected using the SPCM64 software (Becker&Hickl). The observation window of 12.5 ns is set by the repetition rate of the laser, Time-Correlated Single Photon Counting (TCSPC) histograms with 1024 time channels (12 ps per channel) were integrated from a whole field of view at intervals of 1 s, yielding ~104 photons in the peak channel, for a total recording time of 60 – 80 s. Analysis was performed with the SPCImage software (Becker&Hickl), decays were fit with a three-component exponential function: where Ai and τi are the amplitude and lifetime of the i-th species of NADH in the sample. This model accounts for the lifetime values associated with the different species of NADH in mitochondria as previously described(Blinova et al., 2005; Blinova et al., 2008): free, bound pool 1 (putatively to dehydrogenase enzymes), and bound pool 2 (putatively associated to Complex I). The instrument response function (IRF) was collected using second harmonic generation from urea crystals and used for convolution during data fitting. Fitting of the data with this procedure yielded reduced χ2 values <1.2, indicating suitable goodness of fit for the model used. The amplitudes (Ai) and lifetimes (τi) thus obtained were used to compute the intensity contribution of each NADH species according to . The evolution of these contributions with time were analyzed for basal conditions, and after addition of NaCN and are reported here as percentage of the overall observed intensity. Data presented in Figure 3f are the average (±SD) of 3 biological replicates, each comprised of 2 technical replicates (contralateral limbs).
Figure 3. Calibration of NADH autofluorescence.
a. NADH autofluorescence measures during mitoRACE at excitation (Ex.) and emission detection (Em.) wavelengths of Ex710 / Em414 – 538nm (n = 4 mice), ex750 / em414 – 538nm (n = 10 mice), and ex750 / em470 – 485 nm (n = 5 mice). b-c. Redox status (b) and mitochondrial flux rates (c) measured with different excitation and emission strategies. d. Representative fluorescent lifetime imaging (FLIM) of isolated muscle fiber. e. NADH TCSPC decays during mitoRACE. f. Contribution of NADH lifetime species to overall fluorescence intensity signal during mitoRACE calculated from lifetime data (n = 6 muscles from 3 mice). Cyanide addition at t = 0. g. Representative data of linear correlation between NADH fluorescence intensity and NADH concentration in solution. h. Representative data of full oxidation of NADH in solution by 10 mM sodium pyruvate and excess lactate dehydrogenase. i. Nucleotide concentrations as measured by high pressure liquid chromatography (HPLC) in mitochondria isolated from lower limb murine skeletal muscle (n = 3 replicates from 21 mice). ‡ indicates significantly different from NAD(H) (p < 0.05).
Quantification of mitochondrial NAD(H) and NADP(H) concentration using high pressure liquid chromatography (HPLC)
Mitochondria were isolated from murine, lower limb skeletal muscle as previously described(Glancy & Balaban, 2011) and further purified by centrifugation for 40 min at 68,000G with Percoll Solution (30% vol/vol Percoll, 250 mM sucrose, 10 mM HEPES, 0.1% wt/vol fatty acid-free BSA, pH 7.4). Purified skeletal muscle mitochondria were dissolved with 50 μl of KPBS (136 mM KCl, 10 mM KH2PO4 pH7.25, mixed with cold 80% final methanol (HPLC grade, J.T. Baker, USA) (Chen, Freinkman, Wang, Birsoy, & Sabatini, 2016), vortexed vigorously for 3 min, and sonicated in a Branson Ultrasonic bath (Fisher Scientific, USA) for 5 min. The lysate was centrifuged at 16 x g for 15 min at 4 °C. The supernatant was transferred to a fresh tube and dried by a Savant SpeedVac equipped with a cold trap (Thermo Fisher Scientific, USA). The pellet was reconstituted with 50 μl of 10 mM KH2PO4, 1 % methanol and centrifuged at 16 x g for 5 min to remove any insoluble particles. The supernatant was transferred to a HPLC sample vial (Thermo Fisher Scientific, USA). Target metabolites were analyzed by a modified HPLC method as described previously(Lazzarino, Di Pierro, Tavazzi, Cerroni, & Giardina, 1991). The 15 μl samples were analyzed in 0.7 ml/min flow for 53 minutes using the Agilent 1100 HPLC (Agilent Technologies, USA) has equipped with a reverse phase column, SUPERCOSIL LC-18-T, 150 × 3 mm, 3 μm (SUPELCO Analytical, USA) with two buffers, Buffer A, 10 mM KH2PO4 pH6.98, 10 mM tetrabutylammonium hydroxide, 1 % methanol; Buffer B, 100 mM KH2PO4 pH5.47, 2.8 mM tetrabutylammonium hydroxide, 30 % methanol. The gradient was starting with 0 % of buffer B and changed % of B as follows: 6 min as 0 %, 8 min at up to 40 %, 11 min at up to 44, 10 min at up to 100 %. 100% B was held for 5 min and then back to the initial setting within 1 min and re-equilibrated the column for 12 min at 0 %. The method was verified and calibrated with various concentrations (200 ~ 800 pmol) of NAD(H) and NADP(H) purchased from Sigma. Those metabolites were detected at 260 nm and all data were analyzed using Agilent software, ChemStation B. 04.03 (Agilent Technologies, USA).
Statistical analysis
All values are reported as mean±SE. Mitochondrial NADH flux rates were calculated by performing bivariate correlation analysis (%NADH/time) in Microsoft Excel using time course measurements of raw NADH fluorescence (all R2 ≥ 0.9). Mean redox status and mean NADH flux rates were compared between groups using two-tailed, paired T-Test to compare means. Significance for all comparisons was accepted at p < 0.05.
Results
mitoRACE
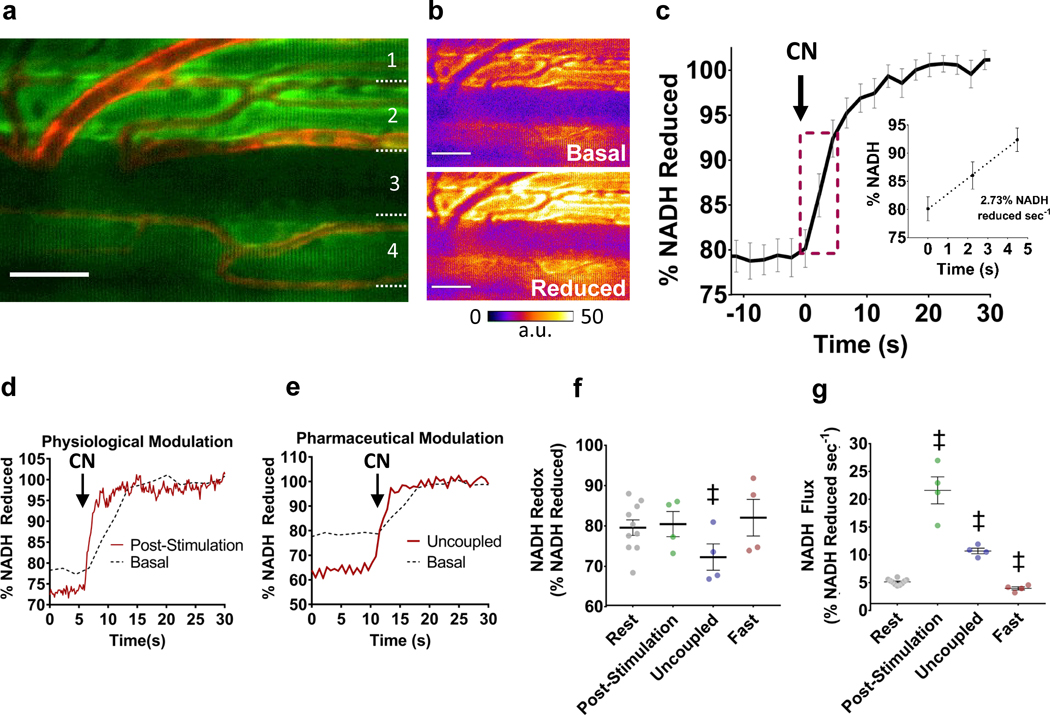
To explore the utility of mitoRACE as an assessment of mitochondrial energetic flux, NADH autofluorescence and redox kinetics were first evaluated in the intact, murine tibialis anterior muscle using two-photon excitation (Fig. 1a), and in vivo NADH autofluorescence before and after inhibition of oxidative phosphorylation via cyanide is shown in Figure 1b. Inhibition of mitochondrial ETC flux in living skeletal muscle by superfusion of sodium cyanide (5mM) resulted in an instantaneous, linear increase in NADH fluorescence at a rate of 2.7% of the total NADH pool per second (%NADH·sec−1) (Fig. 1b,c; Supplementary Video 1). We next evaluated the responsiveness of this technique to physiological and pharmacological modulation of metabolism in skeletal muscle in vivo using electrically-stimulated contractions, fasting, and uncoupling of the mitochondrial membrane potential (Fig. 1d, e, f, g). Examples of NADH redox kinetics following physiological and pharmacological modulation of mitochondrial flux are shown in Figure 1. Acute physiological modulation of muscle metabolism by electrically stimulated muscle contraction resulted in a ~7-fold increase (p<0.05) in mitochondrial NADH flux (19.2±2.8% NADH sec−1) immediately after cessation of contractions compared to basal flux rates (Fig. 1d, f, g). Alternatively, a longer-term physiological modulation of mitochondrial flux due to fasting was associated with a 43.4% decline in basal flux (1.6±0.8% NADH sec−1) (Fig. 1f, g). No changes in mitochondrial redox state were observed in the post-stimulation and fasted states (Fig. 1f, g). Uncoupling of mitochondria by FCCP resulted in a ~3-fold increase (p<0.05) in mitochondrial flux (8.5±0.5% NADH sec−1) (Fig. 1e, g), and reduced redox status by ~8% (p<0.05) compared to basal (Fig. 1e, f).
Figure 1. Mitochondrial Redox After Cyanide Experiment (mitoRACE).
a. Average in vivo NADH autofluorescence in murine TA muscle. b. Representative NADH autofluorescence before and after inhibition of oxidative phosphorylation via superfusion of 5mM cyanide. c. Average mitochondrial NADH fluorescence response of whole muscle tissues (TA) during mitoRACE and rate of increase in NADH fluorescence following inhibition of oxidative phosphorylation (n = 10 mice). d-e. Representative data from post-stimulation (d) and uncoupled (e) mitoRACE compared to rest. f-g. Redox status (f) and mitochondrial flux rates (g) at rest (n = 10 mice) and during the post-stimulation (n = 4 mice), uncoupled (n = 4 mice), and fasted states (n = 4 mice). ‡ indicates significantly different from rest (p < 0.05).
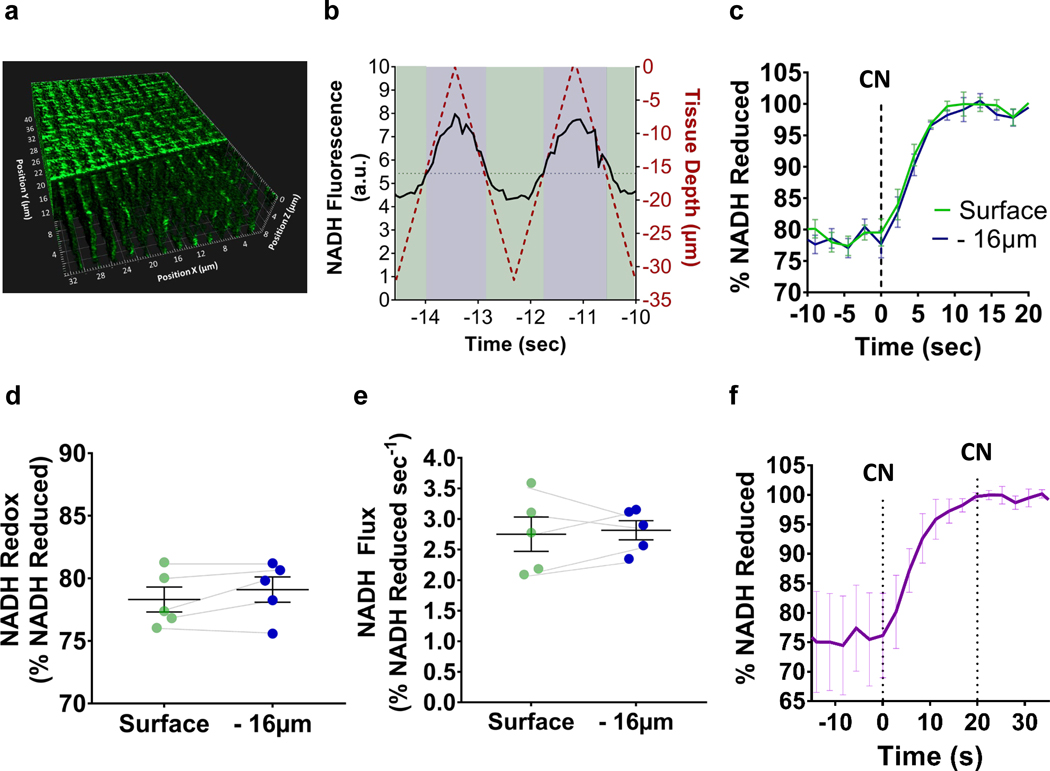
We next evaluated the potential influence of a cyanide diffusion gradient on measures of NADH flux within the 3D volume of the muscle fibers (Fig. 2a). Using rapid 3D time course imaging during mitoRACE (Fig. 2b), we confirmed that NADH flux and NADH redox status were the same between the surface and 16 μm into the densely packed muscle fibers, indicating that due to its high water solubility, cyanide was immediately taken up uniformly throughout the muscle fibers, and (Fig. 2c, d and e). Full reduction of mitochondrial NADH pool was confirmed by the addition of cyanide after the initial reduction event, as titration of cyanide to 10 mM did not result in any further increase in NADH autofluorescence, indicating complete and uniform inhibition of oxidative phosphorylation (Fig. 2f) during mitoRACE.
Figure 2. Rapid 3D time course imaging evaluates in vivo metabolism across tissue depths.
a. 3D rendering of skeletal muscle NADH autofluorescence (TA) from surface to 16 μm tissue depth. b. Rapid 3D time course imaging simultaneously evaluates NADH autofluorescence kinetics across tissues depths. c. mitoRACE at different tissue depths (n = 4 mice). d-e. Redox status (d) and mitochondrial flux rates (e) at different tissue depths (n = 4 mice). f. Average NADH fluorescence response to the titration of cyanide to 10 mM after initial reduction by 5 mM cyanide (n = 3 mice).
To evaluate the presence of non-NADH autofluorescence(Huang, Heikal, & Webb, 2002), mitochondrial NADH kinetics were measured at excitation wavelengths of 710 nm and 750 nm as well as emission detections of 414 – 538 nm and 470 – 485 nm, and measures of redox status and flux rates were consistent across all imaging strategies (Fig. 3a,b,c). In addition, the potential effect of increased binding of NADH on the changes in NADH fluorescence intensity observed during mitoRACE were evaluated using time-resolved in vivo NADH lifetime measurements(Fig. 3d), and while the absolute intensity of each pool increased following the addition of cyanide(Fig. 3e), the contribution of bound NADH lifetime species to the overall fluorescence signal actually decreased 2.1% (p<0.05) (Fig. 3f). The linear relationship between physiological concentrations of NADH and fluorescence intensity was established by titration of NADH in solution (Fig. 3g), and complete conversion of NADH to NAD in solution demonstrated that a fully oxidized NAD pool resulted in negligible fluorescence (<0.05% of fully reduced signal) in our optical system (Fig. 3h). To quantify NADH kinetics, mitochondrial nucleotide concentrations were measured using (Fig. 3i), and the fully reduced NADH fluorescence signal (100% NADH) was normalized to the measured 2.45±0.24mM mitochondrial NADH concentration.
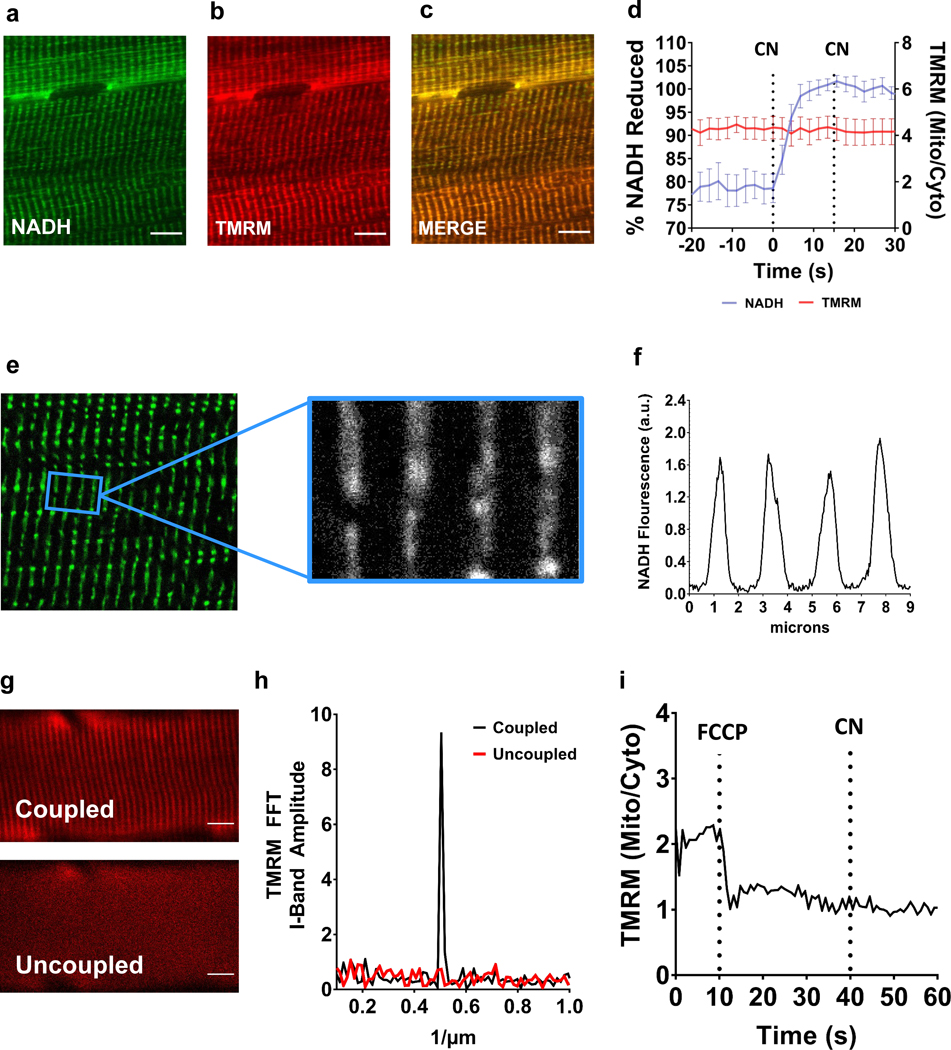
Multiplex imaging of NADH (Fig. 4a) and a mitochondrial membrane potential fluorophore, tetramethylrhodamine (TMRM) (Fig. 4b), confirmed the mitochondrial localization of the NADH signal in muscle (Fig. 4c) and allowed for simultaneous optical measurements of mitochondrial electron flux (NADH) and the proton electrochemical gradient (Ψ) across the inner mitochondrial membrane during mitoRACE (Fig. 4d). In addition to having strong colocalization with the mitochondrial TMRM signal, there was very little cytosolic signal in the muscle and the fluorescence signal from the spaces in between the mitochondrial network was only ~3% of the intramuscular NADH signal (Fig. 4e,f). Multiplex metabolic imaging with mitoRACE was also used to confirm a loss of Ψ across the mitochondrial inner membrane during uncoupling experiments, and indeed, uncoupling of mitochondria by FCCP resulted in complete dissipation of mitochondrial membrane potential (Ψ) (Fig. 4g,h,i),
Figure 4. Multiphoton mitochondrial NADH autofluorescence.
a-c.in vivo multiphoton NADH autofluorescence (a) and mitochondrial membrane potential as indicated with TMRM (b) are colocalized (c) in murine skeletal muscle. d. Average in vivo multiphoton NADH autofluorescence and TMRM signal during mitoRACE (n = 4 mice). e-f. Intermyofibrillar NADH autofluorescence in skeletal muscle is localized to I-Band mitochondria. g. Mitochondrial membrane potential (Ψ) as indicated by TMRM fluorescence at rest (top) and following uncoupling (bottom) by 10 μM FCCP. h. Representative Fast Fourier Transform (FFT) of TMRM signal at rest and following uncoupling. i. Representative mitochondrial/cytoplasmic TMRM signal ratio during mitoRACE in the uncoupled state (10 μM FCCP).
Simultaneous Assessment of Heterogeneous Cell Types
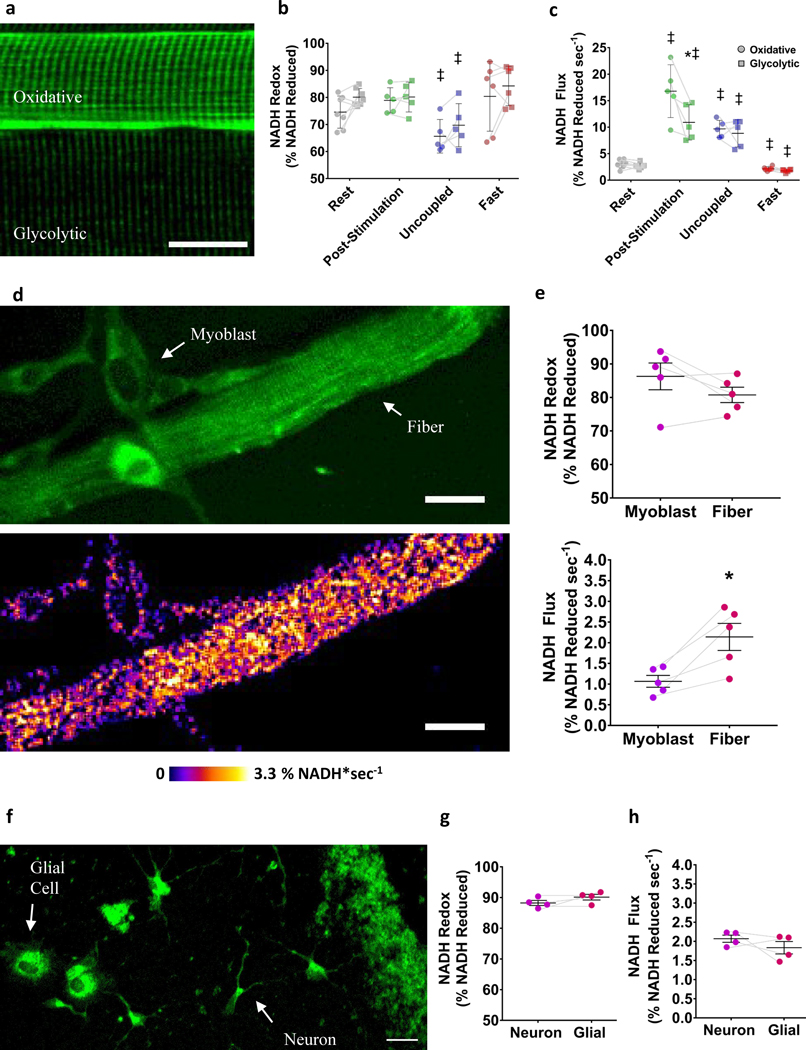
To demonstrate the capacity of mitoRACE to simultaneously evaluate different cell types within living tissues, we integrated the morphological and metabolic information from mitoRACE performed in skeletal muscle to differentially assess metabolism in different muscle cell types. In living skeletal muscle tissues, oxidative fibers were readily identified by increased mitochondrial density (Fig. 5a), a grid-like mitochondrial network structure (Fig. 2a), the presence of PVM pools, and high capillarization (Fig. 1a); whereas glycolytic fibers have lower mitochondrial content (Fig. 5a), a more perpendicular mitochondrial network structure colocalized to the I-band (Fig. 2a), and a lack of capillarization (Fig. 1a). Oxidative and glycolytic muscle fibers (n=7) within the same muscle were compared across all conditions. At rest, basal redox status and mitochondrial flux rates were not different between oxidative and glycolytic muscle fibers (Fig. 5b,c). Moreover, no differences in redox status were found between oxidative and glycolytic fibers in the post-stimulation, uncoupled, or fasted states (Fig. 5b). While mitochondrial flux was similar in oxidative and glycolytic muscle fibers at rest (Fig. 5c), flux rates following muscle contractions were significantly higher (p<0.05) in oxidative fibers (16.8±1.5% NADH sec−1) compared to glycolytic fibers (10.9±2.2% NADH sec−1) (Fig. 5c). Conversely, the increases in mitochondrial flux rates observed with uncoupling were similar in magnitude among oxidative and glycolytic fibers (Fig.5c). Flux rates measured in the fasted state were also similar between oxidative and glycolytic fibers (Fig. 5c).
Figure 5. Simultaneous assessment of heterogeneous cell types.
a. Representative averaged image of oxidative and glycolytic fibers. b-c. Redox status (b) and mitochondrial flux rates (c) of oxidative and glycolytic fibers in the basal (n = 7 fibers from 5 mice), post-stimulated (n = 5 fibers from 4 mice), uncoupled (n = 5 fibers from 4 mice), and fasted (n = 6 fibers from 4 mice) states in vivo. ‡ indicates significantly different from rest (p < 0.05). * indicates statistical difference between fiber types within the same muscle (p < 0.05). d. Representative averaged image of cultured FDB fiber and myoblast (top) and mitochondrial NADH flux rate map (bottom). e. Redox status (top) and mitochondrial flux rates (bottom) of fiber and myoblast (n = 5 cells from 3 biological samples). * indicates statistical difference between cell types (p < 0.05). f. NADH autofluorescence in cultured neurons and glial cells from murine hippocampal tissues. g-h. Redox status (g) and mitochondrial flux rates (h) of neuron and glial cells (n = 4 cells from 3 biological samples).
To establish the applicability of mitoRACE in evaluating heterogenous populations of cells in culture, we measured mitochondrial NADH flux rates in cultured, murine flexor digitorum brevis (FDB) myofibers and the myoblasts derived from the myofiber-associated satellite cells. After dissection and digestion of the FDB, the natural process of muscle regeneration results in the activation of satellite cells and subsequent formation of new myoblasts, offering a relevant model of the cellular heterogeneity often found in cell cultures (Fig. 5d). After 72 hours, we observed multiple myoblasts surrounding the parent FDB myofibers (Fig. 5d), and while redox status was not significantly different between cell types (Fig. 5e), mitochondrial NADH flux rates as measured by mitoRACE were ~56% slower in myoblasts (1.1±0.2 %NADH·sec−1) compared to their parent myofiber (2.0±0.3 %NADH·sec−1(Fig. 5d, e; Supplementary Video 2). To demonstrate the utility of mitoRACE in a non-muscle model, we cultured primary hippocampal neurons from neonatal mice (Fig. 5f). A challenge to evaluating mitochondrial function in primary cultured neurons is the contamination of the cell population with glial cells within the dish(Seibenhener & Wooten, 2012). However, the spatial information from mitoRACE allowed for glial cells to be readily identified based on morphology (Fig. 5f), and single-cell evaluation of neurons and glial cells did not reveal any differences between these cell types (Fig. 5g,h).
Mitochondrial Network Regional Flux Analysis
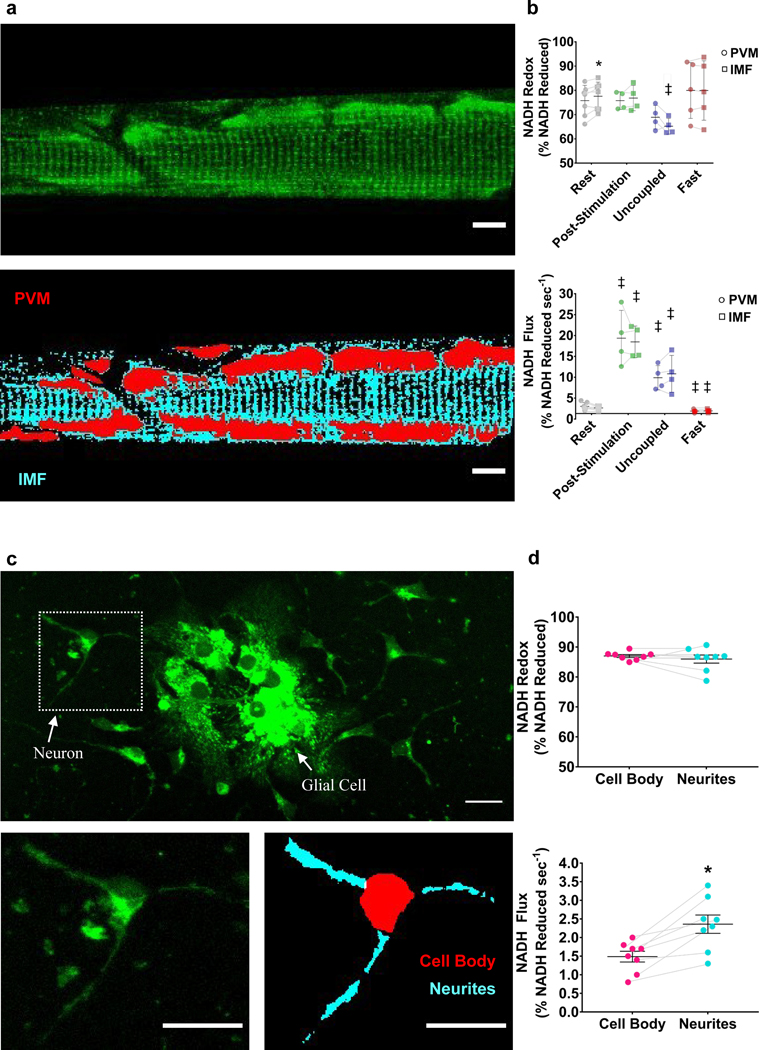
We next evaluated the capability of mitoRACE to evaluate metabolic flux in different subcellular regions of the mitochondrial network within a single cell. In skeletal muscle, subcellular spatial resolution allowed for distinction between the high density paravascular mitochondria (PVM) near the capillaries along the edge of the cell and the intramyofibrillar (IMF) mitochondrial subfractions of network located in the interior of the cell (Fig. 6a). Using this spatial information, we were able to evaluate different subcellular mitochondrial pools (PVM and IMF) within the same oxidative muscle fibers across all conditions (Fig. 6b). At rest, PVM (75.8±2.4% reduced) were slightly, but significantly (p<0.05) more oxidized than IMF mitochondrial pools (77.6±2.2% reduced) (Fig. 6b). However, no differences in basal redox status were found between PVM and IMF mitochondrial pools following muscle post-stimulation or in the fasted state (Fig. 6b). Uncoupled IMF mitochondrial pools (65.21±1.6%) were significantly more reduced (~12%) compared to resting IMF redox status (p<0.05), but no differences were observed between PVM and IMF mitochondrial pools within the same fiber in the uncoupled state (Fig. 6b). No differences in mitochondrial flux rates were found between PVM and IMF mitochondrial pools in the post-stimulation, fasted, or uncoupled state (Fig. 6b). To demonstrate the utility of mitoRACE in identifying differences in mitochondrial function among subcellular compartments not connected by mitochondrial subnetworks, we also evaluated mitochondrial NADH flux in the neurites and cell body of cultured hippocampal neurons (Fig. 6c), and these experiments revealed a 55% higher mitochondrial NADH flux rate in neurites (2.5±0.4 %NADH·sec−1) relative to the cell body (1.6±0.2 %NADH·sec−1) (Fig. 6d).
Figure 6. Mitochondrial network regional flux analysis.
a. Representative average image (top) and analysis of PVM and IMF mitochondrial subfractions (bottom) in an oxidative muscle fiber. b. Redox status (top) and mitochondrial flux rates (bottom) of PVM and IMF mitochondria in the basal (n = 7 fibers from 5 mice), post-stimulated (n = 4 fibers from 4 mice), uncoupled (n = 4 fibers from 4 mice), and fasted (n = 5 fibers from 4 mice) states in vivo. ‡ indicates significantly different from rest (p < 0.05). * indicates statistical difference between mitochondrial subfractions within the same cell (p < 0.05). c. Representative average image of primary cultured neurons and glial cells from hippocampal tissues (top) and analysis of cell body and dendrite mitochondrial NADH flux in single neuron (bottom). d. Redox status (top) and mitochondrial flux rates (bottom) of cell body and dendrite mitochondria (n = 8 cells from 3 biological samples). * indicates statistical difference between the cell body and neurite subcellular regions (p < 0.05).
Discussion
Multiphoton metabolic imaging with mitoRACE provides spatially-resolved measures of mitochondrial energy conversion that are responsive to acute and chronic alterations in metabolic status. The experiments presented here demonstrate the utility of mitoRACE in evaluating energy metabolism with single-cell and subcellular resolution in intact tissues and cell culture. Indeed, there is very little information available related to single-cell and subcellular mitochondrial responses to metabolic perturbations in intact cells and tissues, and future applications of mitoRACE using NADH autofluorescence could greatly expand the physiological and biochemical investigation of mitochondrial function in living cells.
Since first described by Chance and Williams(Chance & Williams, 1955), optical techniques using NADH autofluorescence have become increasingly useful in the evaluation of mitochondrial energy metabolism. However, the potential to evaluate mitochondrial energetic flux using NADH redox kinetics during the transition from resting steady-state to fully reduced has not yet been explored. Here, we demonstrate that, in addition to evaluating mitochondrial redox status, intravital multiphoton excitation of NADH autofluorescence can provide real-time measures of mitochondrial energetic flux in living tissue. Due to the spatially resolved nature of this technique, the calculated NADH flux rates are inherently normalized to mitochondrial content, the observed rate of increase in %NADH during mitoRACE (2.73% NADH·sec−1) is equivalent to 68.3 μM NADH·sec−1 when quantified using the measured 2.45±0.24mM mitochondrial NADH concentration. Thus, assuming a ~10% mitochondrial content per muscle volume(C. K. E. Bleck et al., 2018; Larsen et al., 2012) and the 2:1 stoichiometry of NADH:oxygen in oxidative phosphorylation, our measured NADH flux in resting skeletal muscle translates to 205 nmol O2·g muscle−1·min−1, consistent with previously reported bulk tissue O2 flux rates between 80–412 nmol O2·g−1·min−1 in resting skeletal muscle using several different methods(Grassi, Gladden, Stary, Wagner, & Hogan, 1998; Hood, Gorski, & Terjung, 1986; Kushmerick, Meyer, & Brown, 1992; Marcinek, Ciesielski, Conley, & Schenkman, 2003; Wust et al., 2011). While there are certainly sources of electrons that feed oxidative phophorlyation other than NADH:Complex I, such as FAD:Complex II, NADH serves as the primary electron donor for oxidative phorphorylation in mammalian cells when using carbohydtrates, fats, and ketones as substrates, and our measures of flux are in agreement with intergrative oxygen consumption measures that include FAD:Complex II-mediated flux. Although the rate of mitochondrial NADH flux has not been specifically quantified in previous studies, the magnitude of increase in NADH fluorescence from baseline to fully reduced observed in our experiments (21%) is also comparable to previous studies (25%) evaluating changes in NADH fluorescence in living skeletal muscle following inhibition of oxidative phosphorylation via anoxia(Schroeder et al., 2010), suggesting that full reduction skeletal muscle mitochondria was achieved. Moreover, the magnitude and rate of NADH reduction were preserved across tissue depths and no further increase in NADH fluorescence was observed following the superfusion of additional cyanide, indicating that inhibition of oxidative phosphorylation was immediate and complete throughout the cross-sectional area of the superficial muscle fibers. We did attempt to inhibit the ETC directly at Complex I instead of Complex IV but found that ETC inhibitors with low water solubility such as rotenone and antimycin A are taken up too slowly into cells to be used for rapid, complete inhibition of NADH utilization. Nonetheless, inhibition of Complex IV would still be expected to fully block the tightly coupled electron flow and proton pumping across ETC, and the only potential leak from the ETC, reactive oxygen species (ROS), has been shown to be much less than 1% of total electron flux under normal conditions(Goncalves, Quinlan, Perevoshchikova, Hey-Mogensen, & Brand, 2015). Thus, the ROS contribution to the total NADH flux as measured by mitoRACE is negligible. Taken together, these experiments support the use of mitoRACE as an in vivo assessment of basal mitochondrial NADH flux in superficial skeletal muscles, organs, and other models compatible with multiphoton excitation of NADH autofluorescence.
Mitochondrial NADH flux measured by mitoRACE showed acute responsiveness to both physiological and pharmacological modulation, and the magnitude of changes in flux consequent to these perturbations were similar to values previously reported by in vivo studies evaluating changes in oxygen consumption associated with muscle contraction(Grassi, Gladden, Samaja, et al., 1998; Hood et al., 1986; Marcinek et al., 2003), dietary fasting(Marvyn, Bradley, Mardian, Marks, & Duncan, 2016; Webb, Jagot, & Jakobson, 1982), and mitochondrial uncoupling(Marcinek et al., 2003). In agreement with previous studies evaluating whole muscle oxygen uptake in skeletal muscle(Grassi, Gladden, Samaja, et al., 1998; Hood et al., 1986; Marcinek et al., 2003), we found that physiological modulation of metabolism by muscle contraction was associated with a significantly larger increase in mitochondrial flux (~7 fold) (Grassi, Gladden, Samaja, et al., 1998; Hood et al., 1986; Marcinek et al., 2003) compared to pharmacological modulation by uncoupling (~3 fold)(Marcinek et al., 2003), reflecting the different metabolic mechanisms involved in these states. Although the mechanisms by which energy homeostasis is maintained during muscle contraction are not completely understood, reductions in cellular free energy (ΔGATP)(Combs, Aletras, & Balaban, 1999; Glancy, Willis, Chess, & Balaban, 2013; Kushmerick et al., 1992; Ryschon, Fowler, Wysong, Anthony, & Balaban, 1997) and increases in cytoplasmic [Ca+](Glancy et al., 2013; Territo, Mootha, French, & Balaban, 2000) during muscle contractions contribute to increases in mitochondrial flux by altering the thermodynamic driving forces of oxidative phosphorylation and increasing the activation state of the ETC(Glancy et al., 2013; Willis, Jackman, Messer, Kuzmiak-Glancy, & Glancy, 2016). In contrast, uncoupling of the inner mitochondrial membrane with FCCP increases flux by shifting a single driving force, membrane potential (ΔΨ), without altering conductance of the ETC. Therefore, it would be expected that muscle contraction would produce a greater mitochondrial flux rate compared to uncoupling. These findings illustrate the unique ability of mitoRACE to evaluate metabolic responses to physiological and pharmacological perturbations in vivo and highlight the potential application of this technique to the study of transient activation states and pharmaceutical treatment conditions. Furthermore, the observed reduction in basal metabolic rate associated with fasting in the present study further demonstrates how this technique may be used to evaluate chronic changes in mitochondrial function associated with pathology, interventions, and/or knockout models.
Similar to other mammalian tissues, skeletal muscle is composed of multiple cell types, and it is therefore important to consider cellular heterogeneity when evaluating mitochondrial flux in skeletal muscle and other tissues. For example, it has been well established that striated muscle cells can present different phenotypes within a single tissue that favor either oxidative or glycolytic metabolism(Christopher K. E. Bleck, Yuho Kim, T. Bradley Willingham, & Brian Glancy, 2018; Kushmerick, Moerland, & Wiseman, 1993; Leary et al., 2003; Mogensen & Sahlin, 2005; Schiaffino & Reggiani, 2011). Certainly, whole muscle(Kushmerick et al., 1992; Meyer, Brown, & Kushmerick, 1985) and partial volume(Forbes, Paganini, Slade, Towse, & Meyer, 2009) assessments of skeletal muscle mitochondrial function in vivo have found lower mitochondrial capacities to be associated with more glycolytic muscle tissues, but these techniques lack the spatial resolution required for the differential evaluation of oxidative and glycolytic muscle fibers within a single tissue. By employing spatially-resolved multiphoton microscopy, we were able to distinguish oxidative and glycolytic fiber types within a single tissue and concurrently evaluate mitochondrial flux in each of the separate fibers. Consistent with previous studies in skeletal muscle(Kushmerick et al., 1992; Schroeder et al., 2010), we found no differences in basal mitochondrial flux or redox status between oxidative and glycolytic fiber types, suggesting that mitochondrial activity was not significantly different at rest. However, post-stimulation flux rates were ~54% greater on average in oxidative fibers compared to glycolytic fibers in our experiments. By additionally accounting for the at least three-fold higher mitochondrial content in oxidative compared to glycolytic muscles(C. K. E. Bleck et al., 2018), mitoRACE was able to detect a 4.6-fold difference in the rate of mitochondrial energy conversion within adjacent cells in vivo. Notably, post-stimulation flux measures were obtained immediately after muscle contraction, and the difference in flux rates found between fiber types in the present study is in agreement with previous studies reporting heterogeneity(Vandenborne, Walter, Leigh, & Goelman, 1993) and multicomponent(Forbes et al., 2009) recovery kinetics of whole muscle. Following serial muscle contractions, mitochondrial flux is increased in both oxidative and glycolytic fiber types(Kushmerick et al., 1992), but the metabolic response to contraction is not consistent across fiber types. For example, glycolytic fibers have a greater contribution of anaerobic metabolism during sustained exercise, and therefore experience greater declines in pH (muscle acidosis)(Kushmerick et al., 1992; Tesch, Sjodin, & Karlsson, 1978; Vandenborne et al., 1993) which may attenuate mitochondrial flux rates during recovery(Paganini, Foley, & Meyer, 1997; Walsh, Tiivel, Tonkonogi, & Sahlin, 2002). Moreover, some evidence suggests that glycolytic ATP production may contribute to recovery in glycolytic fiber types, and therefore, reduce the driving forces for mitochondrial oxidative phosphorylation immediately following exercise(Conley, Blei, Richards, Kushmerick, & Jubrias, 1997; Crowther, Carey, Kemper, & Conley, 2002). Oxidative fibers rely almost entirely on mitochondrial oxidative phosphorylation for ATP during exercise and recovery(Paganini et al., 1997). Thus, oxidative fibers are more resistant to muscle acidosis caused by repeated muscle contractions(Kushmerick et al., 1992) and may have higher demands on oxidative phosphorylation during recovery. The differential response to exercise among muscle cells observed in the present study showcases how the spatial acuity of this technique can identify cellular heterogeneity in mitochondrial function within a single tissue. In addition to characterizing the functional roles of different cell types within living tissues, mitoRACE may also provide a practical approach to evaluating mitochondrial function in more complex tissues and other models with diverse cell populations. Using primary cell cultures from muscle and brain tissues, we further demonstrated how single-cell applications of mitoRACE can provide a novel approach to evaluating metabolism in specific subgroups of cells within complex cell populations. The combination of single-cell functional measures such as mitoRACE with single-cell sequencing technologies(Tang et al., 2009) may prove particularly beneficial to the discovery of new mechanisms and the identiffication of metabolic heterogeneity associated with pathology(Hai et al., 2019)
While the structure-function relationships of mitochondrial networks are not completely understood, recent studies have found that subcellular differences in mitochondrial morphology(Glancy et al., 2015), connectivity(C. K. E. Bleck et al., 2018; Glancy et al., 2018; Glancy et al., 2015), and interactions with other organelles(Benador et al., 2018; C. K. E. Bleck et al., 2018) are associated with intracellular differences in mitochondrial function. Therefore, the development of new metabolic imaging techniques that provide spatial and temporal information at the scales of mitochondrial function (microns and seconds, respectively) is critical to understanding the relationships between cell topography, mitochondrial morphology, and the subcellular specialization of mitochondrial function. Here, we demonstrate how mitoRACE can provide a novel approach to evaluating mitochondrial flux in the intact mitochondrial reticulum with the capacity to evaluate subcellular dynamics. Consistent with previous studies(Schroeder et al., 2010), we found PVM to be slightly, but significantly more oxidized at rest compared to IMF mitochondria. However, we did not find any differences in mitochondrial flux between PVM and IMF mitochondrial pools, contradicting several previous in vitro studies evaluating oxygen consumption rates of isolated mitochondria(Cogswell, Stevens, & Hood, 1993; Ferreira et al., 2010). The discrepancy between our in vivo findings and studies using isolated mitochondria could be explained, in part, by the connectivity of mitochondrial networks(Christopher K. E. Bleck et al., 2018; Glancy et al., 2018; Glancy et al., 2015). If mechanisms such as the conduction of membrane potential can efficiently distribute the energy transduction efforts across the mitochondrial network(Christopher K. E. Bleck et al., 2018; Glancy et al., 2018; Glancy et al., 2015), it would be expected that flux would remain consistent throughout the network and between mitochondrial subfractions. We also found PVM and IMF mitochondria to have similar increases in flux rates following uncoupling, indicating that energy distribution across the network is sustained when faced with significant increases in energy demand. Additionally, mitochondrial flux in PVM and IMF subfractions remained consistent during muscle contraction. Despite the immense increase in energy demand created by the contraction of myofibrils localized to the interior region of the cell, mitochondrial flux increased equally across the interior (IMF) and peripheral (PVM) regions of the network. Furthermore, unlike resting conditions, there was no difference in redox status between PVM and IMF pools following muscle contractions and uncoupling, suggesting that redox status across the network was normalized by the presence of a metabolic challenge.
We further evaluated subcellular specialization of energy metabolism by comparing mitochondrial NADH flux rates between the neurites and the cell body of hippocampal neurons. In contrast to the relatively homogenous structure of the muscle cell, neuronal cells have specialized subcellular compartments with unique functionalities. For example, neurites form thin projections from the cell body that are responsible for receiving and transmitting messages, whereas the cell body performs signal processing and housekeeping(Cheng et al., 2010). Indeed, we found that mitchondrial NADH flux rates were signficiantly higher in the nuerites compared to the cell body, indicating that nuerites may experience higher energy demands from post-synaptic ion cycling and dendritic outgrowth. These experiments in living muscle tissue and cultured neurons demonstrate how in vivo and in vitro applications of this technique can be employed to identify differences in metabolic flux among subcellular regions. Such coordinated measurements of morphology and metabolism provide a unique opportunity to investigate structure-function mechanisms across cell types and models, and future applications of mitoRACE could substantially advance inquiry into subcellular energy metabolism and the physiological role of the mitochondrial connectome(C. K. E. Bleck et al., 2018).
Evaluating mitochondrial function in living tissues has been an ongoing challenge for researchers and clinicians for over a hundred years(Mayevsky & Rogatsky, 2007). While in vivo measurements have the obvious benefit of physiological relevance, intravital metabolic imaging can be limited by tissue viability, movement, and optical calibration practices. Therefore, it should be considered that anesthesia(Hanley, Ray, Brandt, & Daut, 2002) and surgical procedures could have altered the metabolic status of the muscle tissues evaluated in the present study. Specifically, isoflurane has been shown to reduce Complex I activity up to 20% in isolated mitochondria at physiological concentrations. However, the use of volatile anesthetics is required for in vivo experiments, and our measures of flux are similar measures of oxygen consumption in experiments using other anesthetics, including injectable agents(Grassi, Gladden, Stary, et al., 1998; Hanley et al., 2002; Hood et al., 1986; Kushmerick et al., 1992; Marcinek et al., 2003; Wust et al., 2011). Notwithstanding, we did find strong colocalization of TMRM and mitochondrial NADH fluorescence signals, indicating that mitochondrial function was sufficient to sustain membrane potential during anesthesia and all succeeding surgical procedures. Notably, tissue viability did decrease substantially following dissipation of membrane potential by uncoupler, resulting in severe tissue deformation within ~5 minutes. Tissue movement also limited our ability to maintain field of view during electrical stimulation, and for this reason, stimulated flux rates were evaluated immediately after (<5 sec) the 2 min electrical stimulation bout. Certainly, recovery kinetics of oxidative metabolism are exponential(Forbes et al., 2009), and measurements during or closer to the termination of exercise would have likely resulted in considerably greater flux rates.
Similar to previous studies(Mayevsky & Rogatsky, 2007; Schroeder et al., 2010), we used the fully reduced mitochondrial NADH signal to calibrate our optical measurements (100% NADH). Although it was not possible to achieve complete oxidation of the mitochondrial NAD pool in vivo (0% NADH) due to tissue movement during prolonged FCCP exposure, full oxidation of NADH in solution established zero fluorescence (<0.05%) in our system, and in agreement with previous studies(Blinova et al., 2005), we found fluorescence intensity to be linearly associated with physiological NADH concentrations. When evaluating changes in NADH fluorescence intensity in living organisms, it is also important to consider the distribution of NADH species (bound/unbound) contributing the fluorescence signal. Specifically, studies have found that the binding of NADH can significantly increase fluorescence emission(Blinova et al., 2005), and the free NADH in buffer could have a ~10-fold less yield than bound NADH species found in mitochondria. Furthermore, a reorganization of NADH lifetime pools to favor a higher proportion of bound species could have contributed to the increase in NADH fluorescence observed during the in vivo experiments. Using real-time NADH lifetime measurements in vivo, we actually found a small decrease (~2%) in the contribution of bound NADH to the fluorescence signal, indicating that increases in fluorescence intensity following inhibition of oxidative phosphorylation were not influenced by changes in % bound NADH within the matrix. In addition to the binding status of mitochondrial NADH, non-NADH autofluorescence may also influence fluorescence measurements of NADH(Webb et al., 1982). Multiphoton NADH excitation can be attained at 700 – 800 nm (Expeak ~710 nm)(Huang et al., 2002), resulting in broad emission spectra between 400 – 600 nm (Empeak ~460 nm) (Huang et al., 2002; Piston, Masters, & Webb, 1995), and it has been demonstrated that other molecules in living cells have similar spectral properties in this range. Specifically, the emission/excitation properties of nucleotide nicotinamide adenine dinucleotide phosphate (NADPH) are identical to that of NADH(Huang et al., 2002). However, quantification of skeletal muscle mitochondrial nucleotide concentrations using high pressure liquid chromatography (HPLC) revealed that the NADP pool was ~30-fold lower in concentration than the mitochondrial NAD pool, indicating that the NADPH signal was negligible in skeletal muscle mitochondria. Moreover, NADPH does not experience pronounced enhancements in fluorescence emission within the mitochondrial matrix as seen with NADH (Blinova et al., 2005; Blinova et al., 2008; Estabrook, 1962) and increases in NADH fluorescence associated with anoxia are not accompanied by increases in NADP(Chance, 1970; Mayevsky & Rogatsky, 2007). To evaluate the potential influence of other sources of autofluorescence outside the peak excitation/emission of NADH(Huang et al., 2002), we measured mitochondrial redox status and flux rates at various excitation (710 nm and 750 nm) and emission (414 – 538 nm and 470 – 485 nm) wavelengths and found no differences in NADH kinetics across imaging strategies, further demonstrating that non-NADH autofluorescence has negligible influence on mitochondrial NADH flux rates during mitoRACE.
In summary, multiphoton metabolic imaging with mitoRACE provides direct, spatially-resolved measures of mitochondrial energy conversion, enabling single-cell and subcellular evaluation of energy metabolism from animals in vivo to individual cells in a dish. The versatility of this technique provides a highly adaptable platform with broad applications in the study of cellular energy metabolism, and future applications of this methodology may provide new insights into the dynamics of cellular energy transduction and the pathophysiology of mitochondrial and cellular dysfunction.
Supplementary Material
Key Points.
We developed a novel metabolic imaging approach that provides direct measures of the rate of mitochondrial energy conversion with single-cell and subcellular resolution by evaluating NADH autofluorescence kinetics during the mitochondrial redox after cyanide experiment (mitoRACE).
Measures of mitochondrial NADH flux by mitoRACE are sensitive to physiological and pharmacological perturbations in vivo.
Metabolic imaging with mitoRACE provides a highly adaptable platform for evaluating mitochondrial function in vivo and in single cells with potential for broad applications in the study of energy metabolism.
Acknowledgements
We want to thank Dr. Robert S. Adelstein for providing the mCherry non-muscle myosin IIA mouse, and NHLBI transgenic core for assistance in generating the mCherry non-muscle myosin IIA mouse line.
Funding
This work was funded by the Division of Intramural Research of the National Heart Lung, and Blood Institute (ZIAHL006221–02) and the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases (ZIA HL006221–02).
Footnotes
Competing Interests
The authors declare no competing interests.
References
- Bakalar M, Schroeder JL, Pursley R, Pohida TJ, Glancy B, Taylor J, . . . Balaban RS (2012). Three-dimensional motion tracking for high-resolution optical microscopy, in vivo. J Microsc, 246(3), 237–247. doi: 10.1111/j.1365-2818.2012.03613.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Befroy DE, Petersen KF, Dufour S, Mason GF, de Graaf RA, Rothman DL, & Shulman GI (2007). Impaired mitochondrial substrate oxidation in muscle of insulin-resistant offspring of type 2 diabetic patients. Diabetes, 56(5), 1376–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benador IY, Veliova M, Mahdaviani K, Petcherski A, Wikstrom JD, Assali EA, . . . Shirihai OS (2018). Mitochondria Bound to Lipid Droplets Have Unique Bioenergetics, Composition, and Dynamics that Support Lipid Droplet Expansion. Cell Metab, 27(4), 869–885 e866. doi: 10.1016/j.cmet.2018.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleck CKE, Kim Y, Willingham TB, & Glancy B. (2018). Subcellular connectomic analyses of energy networks in striated muscle. Nature Communications, 9(1), 5111. doi: 10.1038/s41467-018-07676-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleck CKE, Kim Y, Willingham TB, & Glancy B. (2018). Subcellular connectomic analyses of energy networks in striated muscle. Nat Commun, 9(1), 5111. doi: 10.1038/s41467-018-07676-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blinova K, Carroll S, Bose S, Smirnov AV, Harvey JJ, Knutson JR, & Balaban RS (2005). Distribution of mitochondrial NADH fluorescence lifetimes: steady-state kinetics of matrix NADH interactions. Biochemistry, 44(7), 2585–2594. doi: 10.1021/bi0485124 [DOI] [PubMed] [Google Scholar]
- Blinova K, Levine RL, Boja ES, Griffiths GL, Shi ZD, Ruddy B, & Balaban RS (2008). Mitochondrial NADH fluorescence is enhanced by complex I binding. Biochemistry, 47(36), 9636–9645. doi: 10.1021/bi800307y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chance B. (1970). The identification and control of metabolic states. Behav Sci, 15(1), 1–23. [DOI] [PubMed] [Google Scholar]
- Chance B, Cohen P, Jobsis F, & Schoener B. (1962). Intracellular oxidation-reduction states in vivo. Science, 137(3529), 499–508. [DOI] [PubMed] [Google Scholar]
- Chance B, & Williams GR (1955). A method for the localization of sites for oxidative phosphorylation. Nature, 176(4475), 250–254. [DOI] [PubMed] [Google Scholar]
- Chen WW, Freinkman E, Wang T, Birsoy K, & Sabatini DM (2016). Absolute Quantification of Matrix Metabolites Reveals the Dynamics of Mitochondrial Metabolism. Cell, 166(5), 1324–1337 e1311. doi: 10.1016/j.cell.2016.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A, Hou Y, & Mattson MP (2010). Mitochondria and neuroplasticity. ASN Neuro, 2(5), e00045. doi: 10.1042/AN20100019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogswell AM, Stevens RJ, & Hood DA (1993). Properties of skeletal muscle mitochondria isolated from subsarcolemmal and intermyofibrillar regions. Am J Physiol, 264(2 Pt 1), C383–389. doi: 10.1152/ajpcell.1993.264.2.C383 [DOI] [PubMed] [Google Scholar]
- Combs CA, Aletras AH, & Balaban RS (1999). Effect of muscle action and metabolic strain on oxidative metabolic responses in human skeletal muscle. J Appl Physiol (1985), 87(5), 1768–1775. doi: 10.1152/jappl.1999.87.5.1768 [DOI] [PubMed] [Google Scholar]
- Conley KE, Blei ML, Richards TL, Kushmerick MJ, & Jubrias SA (1997). Activation of glycolysis in human muscle in vivo. Am J Physiol, 273(1 Pt 1), C306–315. doi: 10.1152/ajpcell.1997.273.1.C306 [DOI] [PubMed] [Google Scholar]
- Crowther GJ, Carey MF, Kemper WF, & Conley KE (2002). Control of glycolysis in contracting skeletal muscle. I. Turning it on. Am J Physiol Endocrinol Metab, 282(1), E67–73. doi: 10.1152/ajpendo.2002.282.1.E67 [DOI] [PubMed] [Google Scholar]
- Estabrook RW (1962). Fluorometric measurement of reduced pyridine nucleotide in cellular and subcellular particles. Anal Biochem, 4, 231–245. [DOI] [PubMed] [Google Scholar]
- Ferreira R, Vitorino R, Alves RM, Appell HJ, Powers SK, Duarte JA, & Amado F. (2010). Subsarcolemmal and intermyofibrillar mitochondria proteome differences disclose functional specializations in skeletal muscle. Proteomics, 10(17), 3142–3154. doi: 10.1002/pmic.201000173 [DOI] [PubMed] [Google Scholar]
- Forbes SC, Paganini AT, Slade JM, Towse TF, & Meyer RA (2009). Phosphocreatine recovery kinetics following low- and high-intensity exercise in human triceps surae and rat posterior hindlimb muscles. Am J Physiol Regul Integr Comp Physiol, 296(1), R161–170. doi: 10.1152/ajpregu.90704.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glancy B, & Balaban RS (2011). Protein composition and function of red and white skeletal muscle mitochondria. Am J Physiol Cell Physiol, 300(6), C1280–1290. doi: 10.1152/ajpcell.00496.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glancy B, Hartnell LM, Combs CA, Femnou A, Sun J, Murphy E, . . . Balaban RS (2018). Power Grid Protection of the Muscle Mitochondrial Reticulum. Cell Rep, 23(9), 2832. doi: 10.1016/j.celrep.2018.05.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glancy B, Hartnell LM, Malide D, Yu ZX, Combs CA, Connelly PS, . . . Balaban RS (2015). Mitochondrial reticulum for cellular energy distribution in muscle. Nature, 523(7562), 617–620. doi: 10.1038/nature14614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glancy B, Willis WT, Chess DJ, & Balaban RS (2013). Effect of calcium on the oxidative phosphorylation cascade in skeletal muscle mitochondria. Biochemistry, 52(16), 2793–2809. doi: 10.1021/bi3015983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves RL, Quinlan CL, Perevoshchikova IV, Hey-Mogensen M, & Brand MD (2015). Sites of superoxide and hydrogen peroxide production by muscle mitochondria assessed ex vivo under conditions mimicking rest and exercise. J Biol Chem, 290(1), 209–227. doi: 10.1074/jbc.M114.619072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassi B, Gladden LB, Samaja M, Stary CM, & Hogan MC (1998). Faster adjustment of O2 delivery does not affect V(O2) on-kinetics in isolated in situ canine muscle. J Appl Physiol (1985), 85(4), 1394–1403. doi: 10.1152/jappl.1998.85.4.1394 [DOI] [PubMed] [Google Scholar]
- Grassi B, Gladden LB, Stary CM, Wagner PD, & Hogan MC (1998). Peripheral O2 diffusion does not affect V(O2)on-kinetics in isolated insitu canine muscle. J Appl Physiol (1985), 85(4), 1404–1412. doi: 10.1152/jappl.1998.85.4.1404 [DOI] [PubMed] [Google Scholar]
- Hai P, Imai T, Xu S, Zhang R, Aft RL, Zou J, & Wang LV (2019). High-throughput, label-free, single-cell photoacoustic microscopy of intratumoral metabolic heterogeneity. Nat Biomed Eng, 3(5), 381–391. doi: 10.1038/s41551-019-0376-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley PJ, Ray J, Brandt U, & Daut J. (2002). Halothane, isoflurane and sevoflurane inhibit NADH:ubiquinone oxidoreductase (complex I) of cardiac mitochondria. J Physiol, 544(3), 687–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert-Chatelain E, Desprez T, Serrat R, Bellocchio L, Soria-Gomez E, Busquets-Garcia A, . . . Marsicano G. (2016). A cannabinoid link between mitochondria and memory. Nature, 539(7630), 555–559. doi: 10.1038/nature20127 [DOI] [PubMed] [Google Scholar]
- Hood DA, Gorski J, & Terjung RL (1986). Oxygen cost of twitch and tetanic isometric contractions of rat skeletal muscle. Am J Physiol, 250(4 Pt 1), E449–456. doi: 10.1152/ajpendo.1986.250.4.E449 [DOI] [PubMed] [Google Scholar]
- Huang S, Heikal AA, & Webb WW (2002). Two-photon fluorescence spectroscopy and microscopy of NAD(P)H and flavoprotein. Biophys J, 82(5), 2811–2825. doi: 10.1016/S0006-3495(02)75621-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DT, Harris RA, Blair PV, & Balaban RS (2007). Functional consequences of mitochondrial proteome heterogeneity. Am J Physiol Cell Physiol, 292(2), C698–707. [DOI] [PubMed] [Google Scholar]
- Kushmerick MJ, Meyer RA, & Brown TR (1992). Regulation of oxygen consumption in fast- and slow-twitch muscle. Am J Physiol, 263(3 Pt 1), C598–606. doi: 10.1152/ajpcell.1992.263.3.C598 [DOI] [PubMed] [Google Scholar]
- Kushmerick MJ, Moerland TS, & Wiseman RW (1993). Two classes of mammalian skeletal muscle fibers distinguished by metabolite content. Adv Exp Med Biol, 332, 749–760; discussion 760–741. [DOI] [PubMed] [Google Scholar]
- Larsen S, Nielsen J, Hansen CN, Nielsen LB, Wibrand F, Stride N, . . . Hey-Mogensen M. (2012). Biomarkers of mitochondrial content in skeletal muscle of healthy young human subjects. J Physiol, 590(14), 3349–3360. doi: 10.1113/jphysiol.2012.230185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzarino G, Di Pierro D, Tavazzi B, Cerroni L, & Giardina B. (1991). Simultaneous separation of malondialdehyde, ascorbic acid, and adenine nucleotide derivatives from biological samples by ion-pairing high-performance liquid chromatography. Anal Biochem, 197(1), 191–196. [DOI] [PubMed] [Google Scholar]
- Leary SC, Lyons CN, Rosenberger AG, Ballantyne JS, Stillman J, & Moyes CD (2003). Fiber-type differences in muscle mitochondrial profiles. Am J Physiol Regul Integr Comp Physiol, 285(4), R817–826. doi: 10.1152/ajpregu.00058.2003 [DOI] [PubMed] [Google Scholar]
- Marcinek DJ, Ciesielski WA, Conley KE, & Schenkman KA (2003). Oxygen regulation and limitation to cellular respiration in mouse skeletal muscle in vivo. Am J Physiol Heart Circ Physiol, 285(5), H1900–1908. doi: 10.1152/ajpheart.00192.200300192.2003 [pii] [DOI] [PubMed] [Google Scholar]
- Marvyn PM, Bradley RM, Mardian EB, Marks KA, & Duncan RE (2016). Data on oxygen consumption rate, respiratory exchange ratio, and movement in C57BL/6J female mice on the third day of consuming a high-fat diet. Data Brief, 7, 472–475. doi: 10.1016/j.dib.2016.02.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayevsky A, & Rogatsky GG (2007). Mitochondrial function in vivo evaluated by NADH fluorescence: from animal models to human studies. Am J Physiol Cell Physiol, 292(2), C615–640. doi: 10.1152/ajpcell.00249.2006 [DOI] [PubMed] [Google Scholar]
- Meyer RA, Brown TR, & Kushmerick MJ (1985). Phosphorus nuclear magnetic resonance of fast- and slow-twitch muscle. Am J Physiol, 248(3 Pt 1), C279–287. doi: 10.1152/ajpcell.1985.248.3.C279 [DOI] [PubMed] [Google Scholar]
- Mogensen M, & Sahlin K. (2005). Mitochondrial efficiency in rat skeletal muscle: influence of respiration rate, substrate and muscle type. Acta Physiol Scand, 185(3), 229–236. doi: 10.1111/j.1365-201X.2005.01488.x [DOI] [PubMed] [Google Scholar]
- Paganini AT, Foley JM, & Meyer RA (1997). Linear dependence of muscle phosphocreatine kinetics on oxidative capacity. Am J Physiol, 272(2 Pt 1), C501–510. doi: 10.1152/ajpcell.1997.272.2.C501 [DOI] [PubMed] [Google Scholar]
- Pesta D, & Gnaiger E. (2012). High-resolution respirometry: OXPHOS protocols for human cells and permeabilized fibers from small biopsies of human muscle. Methods Mol Biol, 810, 25–58. doi: 10.1007/978-1-61779-382-0_3 [DOI] [PubMed] [Google Scholar]
- Picard M, Wallace DC, & Burelle Y. (2016). The rise of mitochondria in medicine. Mitochondrion, 30, 105–116. doi: 10.1016/j.mito.2016.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piston DW, Masters BR, & Webb WW (1995). Three-dimensionally resolved NAD(P)H cellular metabolic redox imaging of the in situ cornea with two-photon excitation laser scanning microscopy. J Microsc, 178(Pt 1), 20–27. [DOI] [PubMed] [Google Scholar]
- Ryan TE, Southern WM, Reynolds MA, & McCully KK (2013). A cross-validation of near-infrared spectroscopy measurements of skeletal muscle oxidative capacity with phosphorus magnetic resonance spectroscopy. J Appl Physiol (1985), 115(12), 1757–1766. doi: 10.1152/japplphysiol.00835.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryschon TW, Fowler MD, Wysong RE, Anthony A, & Balaban RS (1997). Efficiency of human skeletal muscle in vivo: comparison of isometric, concentric, and eccentric muscle action. J Appl Physiol (1985), 83(3), 867–874. doi: 10.1152/jappl.1997.83.3.867 [DOI] [PubMed] [Google Scholar]
- Schiaffino S, & Reggiani C. (2011). Fiber types in mammalian skeletal muscles. Physiol Rev, 91(4), 1447–1531. doi: 10.1152/physrev.00031.2010 [DOI] [PubMed] [Google Scholar]
- Schroeder JL, Luger-Hamer M, Pursley R, Pohida T, Chefd’hotel C, Kellman P, & Balaban RS (2010). Short communication: Subcellular motion compensation for minimally invasive microscopy, in vivo: evidence for oxygen gradients in resting muscle. Circ Res, 106(6), 1129–1133. doi:CIRCRESAHA.109.211946 [pii] 10.1161/CIRCRESAHA.109.211946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibenhener ML, & Wooten MW (2012). Isolation and culture of hippocampal neurons from prenatal mice. J Vis Exp(65). doi: 10.3791/3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen A, & Battersby BJ (2018). Mitochondrial diseases: the contribution of organelle stress responses to pathology. Nat Rev Mol Cell Biol, 19(2), 77–92. doi: 10.1038/nrm.2017.66 [DOI] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, . . . Surani MA (2009). mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods, 6(5), 377–382. doi: 10.1038/nmeth.1315 [DOI] [PubMed] [Google Scholar]
- Territo PR, Mootha VK, French SA, & Balaban RS (2000). Ca(2+) activation of heart mitochondrial oxidative phosphorylation: role of the F(0)/F(1)-ATPase. Am J Physiol Cell Physiol, 278(2), C423–435. doi: 10.1152/ajpcell.2000.278.2.C423 [DOI] [PubMed] [Google Scholar]
- Tesch P, Sjodin B, & Karlsson J. (1978). Relationship between lactate accumulation, LDH activity, LDH isozyme and fibre type distribution in human skeletal muscle. Acta Physiol Scand, 103(1), 40–46. doi: 10.1111/j.1748-1716.1978.tb06188.x [DOI] [PubMed] [Google Scholar]
- Vandenborne K, Walter G, Leigh JS, & Goelman G. (1993). pH heterogeneity during exercise in localized spectra from single human muscles. Am J Physiol, 265(5 Pt 1), C1332–1339. doi: 10.1152/ajpcell.1993.265.5.C1332 [DOI] [PubMed] [Google Scholar]
- Walsh B, Tiivel T, Tonkonogi M, & Sahlin K. (2002). Increased concentrations of P(i) and lactic acid reduce creatine-stimulated respiration in muscle fibers. J Appl Physiol (1985), 92(6), 2273–2276. doi: 10.1152/japplphysiol.01132.2001 [DOI] [PubMed] [Google Scholar]
- Webb GP, Jagot SA, & Jakobson ME (1982). Fasting-induced torpor in Mus musculus and its implications in the use of murine models for human obesity studies. Comp Biochem Physiol A Comp Physiol, 72(1), 211–219. [DOI] [PubMed] [Google Scholar]
- Weinberg SE, Singer BD, Steinert EM, Martinez CA, Mehta MM, Martinez-Reyes I, . . . Chandel NS (2019). Mitochondrial complex III is essential for suppressive function of regulatory T cells. Nature, 565(7740), 495–499. doi: 10.1038/s41586-018-0846-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis WT, Jackman MR, Messer JI, Kuzmiak-Glancy S, & Glancy B. (2016). A Simple Hydraulic Analog Model of Oxidative Phosphorylation. Med Sci Sports Exerc, 48(6), 990–1000. doi: 10.1249/MSS.0000000000000884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wust RC, Grassi B, Hogan MC, Howlett RA, Gladden LB, & Rossiter HB (2011). Kinetic control of oxygen consumption during contractions in self-perfused skeletal muscle. J Physiol, 589(Pt 16), 3995–4009. doi: 10.1113/jphysiol.2010.203422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younkin DP, Delivoria-Papadopoulos M, Kelly R, Clancy R, Leigh JS, & Chance B. (1984). STUDIES OF OXIDATIVE METABOLITES USING 31-P NMR SPECTROSCOPY IN NEWBORN NEUROLOGIC DISORDERS. Pediatric Research, 18(4), 385–385. doi: 10.1203/00006450-198404001-01750 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.