Abstract
Background
Tumor microenvironment (TME) plays important roles in different cancers. Our study aimed to identify molecules with significant prognostic values and construct a relevant Nomogram, immune model, competing endogenous RNA (ceRNA) in lung adenocarcinoma (LUAD).
Methods
“GEO2R,” “limma” R packages were used to identify all differentially expressed mRNAs from Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) databases. Genes with P-value <0.01, LogFC>2 or <-2 were included for further analyses. The function analysis of 250 overlapping mRNAs was shown by DAVID and Metascape software. By UALCAN, Oncomine and R packages, we explored the expression levels, survival analyses of CDK2 in 33 cancers. “Survival,” “survminer,” “rms” R packages were used to construct a Nomogram model of age, gender, stage, T, M, N. Univariate and multivariate Cox regression were used to establish prognosis-related immune forecast model in LUAD. CeRNA network was constructed by various online databases. The Genomics of Drug Sensitivity in Cancer (GDSC) database was used to explore correlations between CDK2 expression and IC50 of anti-tumor drugs.
Results
A total of 250 differentially expressed genes (DEGs) were identified to participate in many cancer-related pathways, such as activation of immune response, cell adhesion, migration, P13K-AKT signaling pathway. The target molecule CDK2 had prognostic value for the survival of patients in LUAD (P = 5.8e-15). Through Oncomine, TIMER, UALCAN, PrognoScan databases, the expression level of CDK2 in LUAD was higher than normal tissues. Pan-cancer analysis revealed that the expression, stage and survival of CDK2 in 33 cancers, which were statistically significant. Through TISIDB database, we selected 13 immunodepressants, 21 immunostimulants associated with CDK2 and explored 48 genes related to these 34 immunomodulators in cBioProtal database (P < 0.05). Gene Set Enrichment Analysis (GSEA) and Metascape indicated that 49 mRNAs were involved in PUJANA ATM PCC NETWORK (ES = 0.557, P = 0, FDR = 0), SIGNAL TRANSDUCTION (ES = –0.459, P = 0, FDR = 0), immune system process, cell proliferation. Forest map and Nomogram model showed the prognosis of patients with LUAD (Log-Rank = 1.399e-08, Concordance Index = 0.7). Cox regression showed that four mRNAs (SIT1, SNAI3, ASB2, and CDK2) were used to construct the forecast model to predict the prognosis of patients (P < 0.05). LUAD patients were divided into two different risk groups (low and high) had a statistical significance (P = 6.223e-04). By “survival ROC” R package, the total risk score of this prognostic model was AUC = 0.729 (SIT1 = 0.484, SNAI3 = 0.485, ASB2 = 0.267, CDK2 = 0.579). CytoHubba selected ceRNA mechanism medicated by potential biomarkers, 6 lncRNAs-7miRNAs-CDK2. The expression of CDK2 was associated with IC50 of 89 antitumor drugs, and we showed the top 20 drugs with P < 0.05.
Conclusion
In conclusion, our study identified CDK2 related immune forecast model, Nomogram model, forest map, ceRNA network, IC50 of anti-tumor drugs, to predict the prognosis and guide targeted therapy for LUAD patients.
Keywords: tumor microenvironment, CDK2, pan-cancer analysis, nomogram model, prognostic model, ceRNA
Introduction
Lung cancer is the third leading cause of death in the world (Siegel et al., 2020; Kara et al., 2021), which is classified into small cell lung cancer (SCLC), lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD) (Wu Y. et al., 2021). Non-small cell lung cancer (NSCLC) accounted for 85% of lung cancer and the 5-year survival rate of the patients is less than 20% (Bade and Dela Cruz, 2020). LUAD is the most important type of NSCLC (Chen and Zhou, 2021; Hou and Yao, 2021; Zhou C. et al., 2021). In recent years, although the treatment of lung cancer is diversified, such as chemotherapy, immunotherapy, targeted therapy, the prognosis of patients with advanced lung cancer is still poor (Noreldeen et al., 2020; Wang et al., 2020). More than 60% of LUAD patients missed the best targeted treatment time due to the difficulty of diagnosis, which can reduce the survival rate (Kris et al., 2014; Chen et al., 2016). In recent years, the resistance of the majority of LUAD patients to multiple antitumor drugs has led to a decrease in the cure rate of LUAD. Therefore, it is very necessary to explore the early gene markers and treatment targets for the prognosis of patients (He et al., 2021).
The immune system is currently recognized as a determinant of cancer (Janssen et al., 2017). With the development of modern technology, new immunotherapy drugs have made remarkable achievements and improved the prognosis of patients (Yang et al., 2021). Combined with various clinical studies, immunotherapy may replace the traditional treatment (Zhai et al., 2021). Understanding TME and recognizing genes related to TME can provide new ideas for immunotherapy. In the TME, many cancer cells and immune cells mediate signaling pathways (Marwitz et al., 2021), involving in tumor progression and drug resistance (Santaniello et al., 2019; Ling et al., 2020). In view of the low treatment rate of LUAD patients with high pathogenicity, it is imperative to find new gene markers (Zhou C. S. et al., 2021).
The mechanism of immune infiltration plays an irreplaceable role in the progress of various cancers (Esenboga et al., 2021; Huang H. et al., 2021; Jomrich et al., 2021; Mungan et al., 2021). The novel systemic immune-inflammation index (SII) is a new kind of marker, including peripheral lymphocytes, neutrophils, and platelets (Ju et al., 2021). SII features play important roles in various cancers, such as Esophageal Squamous Cell Carcinoma (Geng et al., 2016), hepatocellular carcinoma (Hu et al., 2014), and prostate cancer (Lolli et al., 2016).
As for a non-coding RNA, lncRNA was limited to code protein (Chen and Zhang, 2021). In recent years, there has been increasing evidence that the expression of lncRNA involves in cancer progression, such as cancer metastasis (Li et al., 2016; Kim et al., 2018), drug resistance (Han et al., 2017), and apoptosis (Zhao et al., 2017). ceRNA network was composed of mRNAs, lncRNAs, and miRNAs (Chen Y. et al., 2021). lncRNA MALAT1 modulates cell migration, proliferation by sponging miRNA146a to regulate CXCR4 in acute myeloid leukemia (Sheng X. F. et al., 2021). ceRNA network represents a novel layer of gene regulation that controls both physiological and pathological processes (Zhang et al., 2021).
Our study aims to explore immune-related genes and construct forecast model for clinical guidance and prognosis analysis in LUAD using TCGA and GEO common databases. The relationship between target genes and immune cells is studied through various authoritative databases. We built the lncRNAs-miRNAs-CDK2 ceRNA network innovatively, found potential prognostic markers with LUAD. Our study may provide new molecular and therapeutic strategies for the treatment and prognosis of LUAD patients.
Materials and Methods
Gene Expression mRNA Seq, Clinical Data Collection
We downloaded the gene expression, clinical data, Pan-Cancer Atlas Hub from GEO database1 (Chen C. et al., 2021), UCSC Xena2 (Li F. et al., 2021), GDSC3.
mRNA-Based Survival Prediction in LUAD
Four hundred and thirty three LUAD tissues and 19 normal tissues were selected from the GSE68465 (GPL96). By GEO2R (Liu et al., 2019), P < 0.01, Log FC > 2 or Log FC < -2 were defined as screening criteria. Next, Metascape4 (Liu et al., 2020), KEGG5 (Zheng et al., 2021), and DAVID6 (Li J. et al., 2021) databases were used to analyze biological process (BP), cellular component (CC), molecular function (MF) and related pathways of differentially expressed mRNAs (DEmRNAs).
Validation of CDK2 in Different Databases
UALCAN7 (Liu H. et al., 2021) is a comprehensive, user-friendly, and interactive web resource for analyzing cancer OMICS data. Combing with clinical data, we explored the correlation between CDK2 expression and clinical indicators, such as age, grade, sex, smoking habits, stage, TP-53 mutation, weight. Heat maps were showed the positive and negative related genes with CDK2. We analyzed the overall survival (OS) and Relapse-Free Survival (RFS) about CDK2 for LUAD patients by PrognoScan8 (Liu X. et al., 2021).
Further Study About CDK2 Expression in 33-Cancers: Pan-Cancer Analysis
Compared with normal lung tissues, CDK2 expression was highly expressed (P < 0.05) in LUAD by Oncomine9 (Dai et al., 2021) and TIMER10 (Wu R. et al., 2021) databases. As for 33-cancers, we studied the relationships between the expression of CDK2 and clinical stage, survival conditions through different “R” packages.
Batch Related Genes, ROC Analysis, Functional Enrichment, GSEA Analysis in LUAD
To research the major molecule CDK2, we showed 50 positive and negative genes in LUAD by heat maps (Person test method). The 1, 3, 5, and 8 years’ ROC curves were constructed by ‘‘ROC’’ R package. The abscissa of the prediction model was False Positive (FP) and the ordinate was True Positive (TP). Next, we screened out the top 300 genes with the most significant positive correlation with CDK2 for enrichment analysis. Bar and bubble charts showed the classical functions of 300 genes by ‘‘cluster-profiler’’ R package. We downloaded the GSEA4.1.011 (Cao et al., 2021a) to investigate Kyoto Encyclopedia of Genes and Genomes (KEGG) of CDK2-related 300 genes. The top of 50 terms were sorted by P-value in circle graph.
Immune Infiltration of CDK2 in LUAD
CIBERSORT12 (Tan et al., 2021) is an analysis tool to provide an estimation of the abundances of member cell types in a mixed cell population using gene expression data. We selected immune cells associated with CDK2 (Pearson correlation coefficient r > 0.15). Four CDK2-related immune cells were shown in circle graph.
Correlation Analysis of Immune Cells and CDK2 Expression
R package was used to analyze the relation of CDK2 and immune cell infiltration score (P ≤ 0.05). A total of 33-cancers, according to the median of CDK2 gene expression, the samples were divided into high expression groups and low expression groups. Then, we discussed the different expression of immune cells (P < 0.05). The above results were shown by scatter plots and box plots.
Exploring CDK2-Related Immune Genes and GSEA Analysis
TISIDB13 (Huang X. Y. et al., 2021) is a web portal for tumor and immune system interaction, which integrates multiple heterogeneous data types. First, we found CDK2-related Immunomodulators with P-value < 0.05. Next, cBioPortal14 (Lu et al., 2021) was used to explore the genes related to Immunomodulators. DAVID and STRING15 (Zhuang et al., 2021) were used to analyze the Gene Ontology (GO), protein interaction of 49 genes. GSEA 4.1.0 further analyzed the NOM p-val and FDR q-val of these immune related genes.
Combining Clinical Data and Constructing Nomogram Model
Through LUAD clinical data, “survival,” “survminer” R packages were used to construct the COX regression model of age, gender, and stage. Forest plot (Xiang et al., 2021) was shown related Hazard ratio (HR), Log-Rank, Concordance index of LUAD. “rms” R package was used to structure the Nomogram model (Liu et al., 1976) of age, gender, stage, T, N, M.
Univariate, Multivariate Cox Regression and Time ROC Analysis
Using “Survival” R package, we explored the univariate Cox analysis of 49 genes (P < 0.05). Different P-value and HR of statistically significant genes were shown in forest map. According to the results of univariate regression analysis, the significant genes were further analyzed by multivariate analysis. According to the risk score of genes selected by multiple factors, the LUAD samples were divided into lower-risk and higher-risk groups. The risk score was as follows: Risk score = Expgene1 × Coefgene1+Expgene2 × Coefgene2+ Expgene3 × Coefgene3+ Expgene4 × Coefgene4. “survival,” “survminer,” “survival ROC” R packages were performed survival probability curve and ROC curve. Combining the patient’s high and low risk situation and the state of life, death, we used “pheatmap” R package to show the heat map.
The CDK2 Expression Was Further Verified by Histochemistry
We use Human Protein Atlas (HPA) database to verify CDK2 expression in LUAD tissues and normal lung tissues.
Identification of CDK2 Related ceRNA Network
We explored CDK2 related miRNAs from Targetscan16 (Barnett-Itzhaki et al., 2021), miRWalk17 (Sheng L. P. et al., 2021), mirDB18 (Ren et al., 2021), Starbase19 (Li et al., 2014). All miRNAs from four databases were analyzed by Venn map20. Kaplan–Meier Plotter software was used to explore the prognostic value of miRNAs with LUAD. The lncRNAs that regulate common miRNAs were screened by Starbase database (Li et al., 2014). The Cytoscape (Han et al., 2021) was used to build ceRNA network and Cytohubba selected the key node genes according to the degree in the network.
The Expression of CDK2 Was Verified by Polymerase Chain Reaction (PCR)
We did the basic experimental verification of PCR about CDK2 in LUAD cell lines (A549, H1299, H1975) and normal bronchial epithelial cell line (BEAS-2B). GraphPad Prism721 software was used to count the differences between cancer cell lines and normal cell line.
Correlation Between CDK2 and IC50 of Anti-tumor Drugs
We downloaded the response data of 192 anti-tumor drugs in more than 1000 cancer cell lines. “Ggplot2” R package was used to explore the correlation between the CDK2 expression and IC50 of 192 anti-tumor drugs by box diagrams and point diagrams.
Results
Differentially Expressed mRNAs for GSE68465
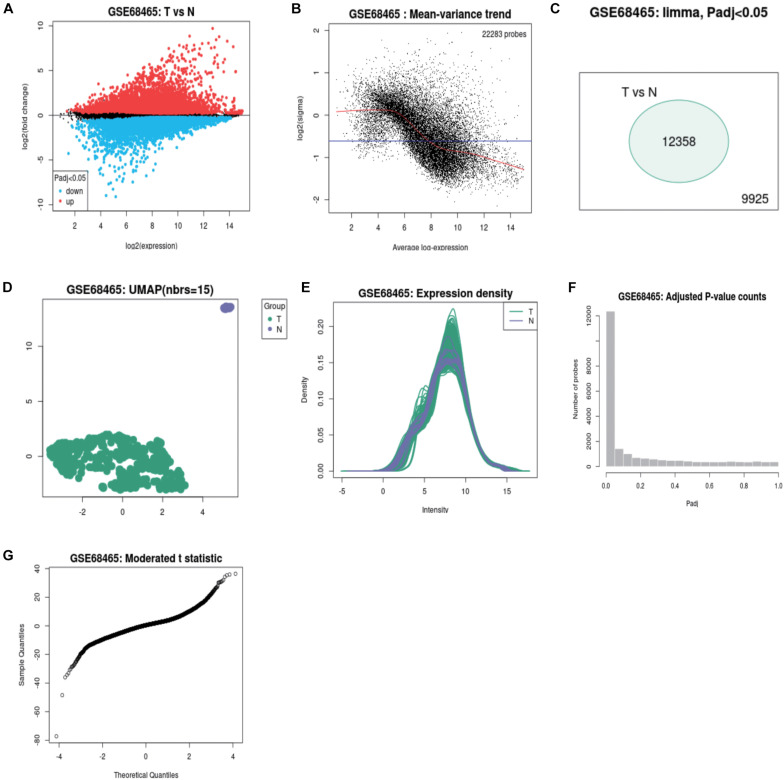
A total of 250 genes were chosen by GEO2R according to the P-value and LogFC. 161 mRNAs were highly expressed and 78 mRNAs were expressed at lower levels in 19 adjacent non-LUAD tissues and 433 LUAD tissues. The study design of this project was shown in Figure 1. The volcano map, scatter plots, peak plot of GSE68465 samples were displayed in Figure 2. A total of 15 highly expressed genes and 15 low expressing genes were shown in Table 1.
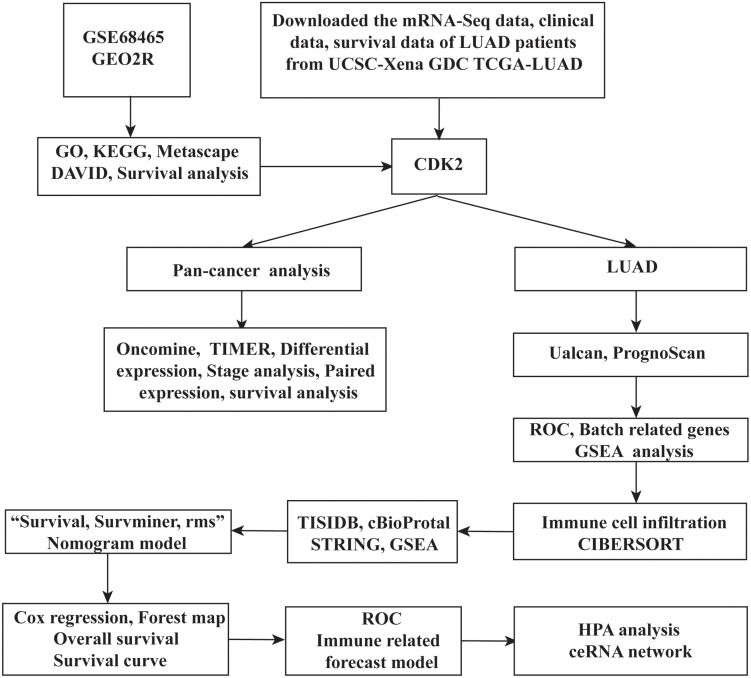
FIGURE 1.
The overall research ideas of this paper were shown as follows.
FIGURE 2.
The volcano map, scatter plot, peak plot of GSE68465 samples (A–G).
TABLE 1.
The top of 15 higher expressed and 15 lower expressed genes.
| Gene.symbol | logFC | adj.P.Val | P.Value |
| HLA-DRA | 9.71 | 4.20E-134 | 5.65E-138 |
| COL3A1 | 7.43 | 2.84E-132 | 6.37E-136 |
| VWF | 5.56 | 5.15E-130 | 1.39E-133 |
| IGHG1 | 7.66 | 1.34E-125 | 4.79E-129 |
| IGHA2 | 8.28 | 4.27E-116 | 1.92E-119 |
| RGS1 | 8.84 | 1.47E-91 | 1.91E-94 |
| IGK | 4.9 | 4.78E-111 | 2.79E-114 |
| COL3A1 | 6.72 | 2.56E-110 | 1.61E-113 |
| HLA-DRB1 | 6.25 | 5.15E-109 | 3.46E-112 |
| IGKC | 4.86 | 1.02E-108 | 7.33E-112 |
| CDH5 | 5.3 | 3.64E-108 | 2.78E-111 |
| COL3A1 | 6.44 | 1.42E-98 | 1.40E-101 |
| HLA-DRB5 | 5.9 | 1.88E-94 | 2.11E-97 |
| C1QA | 6.8 | 2.89E-94 | 3.37E-97 |
| HLA-DPA1 | 5.79 | 6.27E-93 | 7.88E-96 |
| HBZ | –9.92 | 1.40E-262 | 6.29E-267 |
| DCT | –4.69 | 1.02E-179 | 9.14E-184 |
| HBG2 | –6.77 | 9.77E-133 | 1.75E-136 |
| ALAS2 | –4.84 | 8.02E-126 | 2.52E-129 |
| HBG2 | –9.11 | 1.26E-120 | 5.10E-124 |
| HBG1 | –6.15 | 3.06E-111 | 1.65E-114 |
| HBE1 | –7.47 | 4.18E-108 | 3.38E-111 |
| AFP | –6.52 | 1.69E-101 | 1.44E-104 |
| DCT | –4.83 | 1.76E-101 | 1.58E-104 |
| AHSG | –8.98 | 5.18E-99 | 4.88E-102 |
| SOX10 | –4.59 | 6.18E-98 | 6.38E-101 |
| DCT | –7.69 | 7.40E-95 | 7.97E-98 |
| PMEL | –5.2 | 1.66E-93 | 2.01E-96 |
| AHSG | –7.56 | 2.57E-89 | 3.58E-92 |
| NANOG | –6.52 | 4.60E-86 | 6.81E-89 |
GO, KEGG Analysis of 250 DEmRNAs
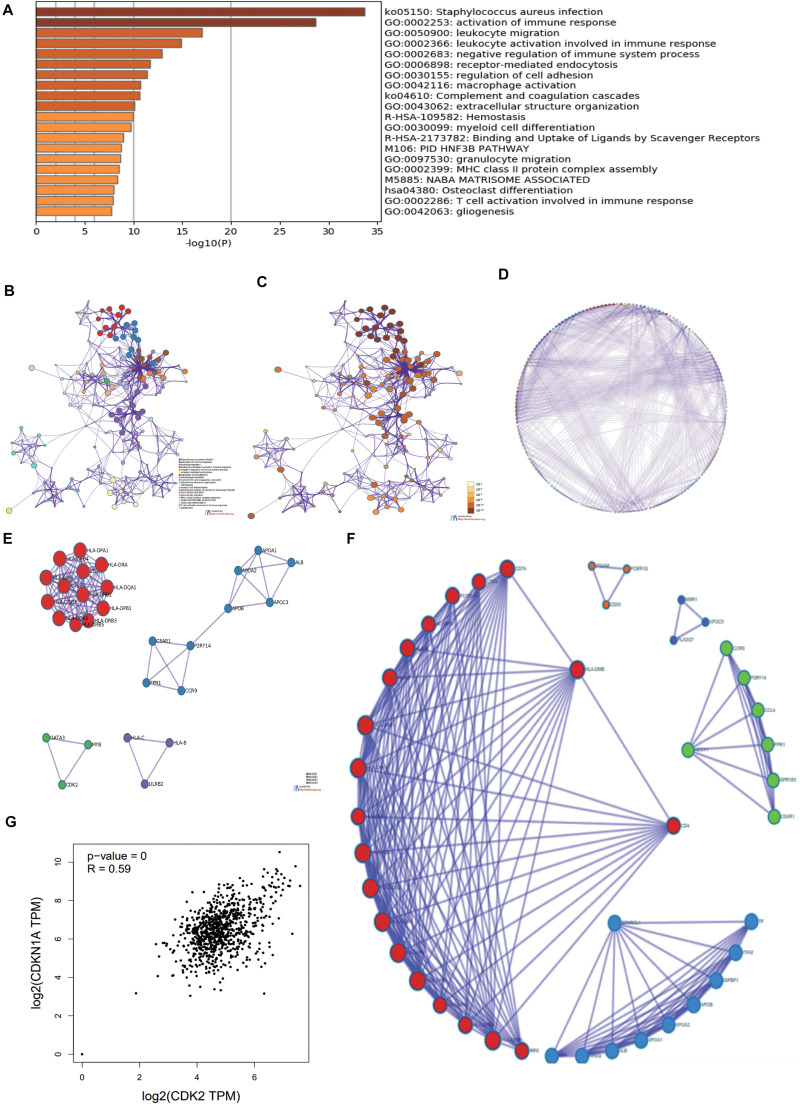
Through Metascape software, we identified a number of pathways significantly enriched, including activation of immune response, regulation of cell adhesion, T cell activation involved in immune response, regulation of cell adhesion, PID-HNF3B PATHWAY (Figure 3A). The interactions between different pathways were shown in Figure 3B. Figures 3C,D drew the P-value of different pathways. According to the relationship of DE mRNAs, several protein analyses were performed in the Figure 3E. CDK2, MYB, GATA3 related to each other in some tumor pathway and the results were listed in circle graph (Figure 3F). DAVID was used to analyze the KEGG results of genes and we found 9 mRNAs participated in the classic P13-AKT signaling pathway, such as MYB, COL3A1, COL4A1, CSF1R, CDK2, ITGB7, LAMA2, TLR2, and VWF. Combining with the Overall Survival (OS) of LUAD patients, we chose CDK2 as targeted molecule (P < 0.05). KEGG software further was used to identify the upstream molecules of CDK2 in P13-AKT signaling pathway. Finally, the CDKN1A might regulate downstream molecule CDK2 to influence cell cycle progression in LUAD. Through the scatter plot, there was a positive correlation between CDK2 and CDKN1A using GEPIA22 (P = 0, R = 0.59) (Figure 3G).
FIGURE 3.
The GO, KEGG analysis of 250 DEmRNAs. (A) Through Metascape software, we found most enriched items. (B) The related paths were shown in the network diagram. (C,D) the P-value of different pathways. (E) According to the relationship of DE mRNAs, several protein analyses were performed. (F) CDK2, MYB, GATA3 were connected each other in some tumor pathway from the circle graph. (G) There was a positive correlation between CDK2 and CDKN1A using GEPIA.
Further Study of CDK2 and Its Prognostic Values in LUAD
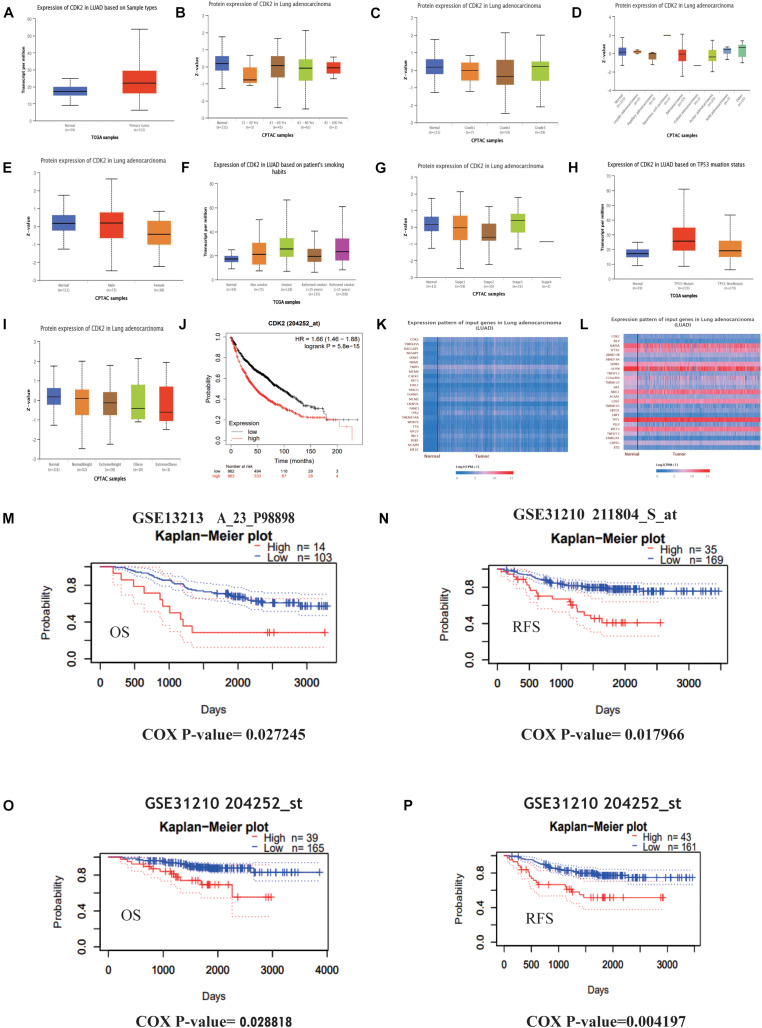
Through the Ualcan database, we studied the expression of CDK2 in LUAD tissues and normal tissues. CDK2 expression was high in 515 LUAD tissues in comparison with 59 normal tissues (P = 1.624E-12) (Figure 4A). Different ages had differential expression levels of CDK2 (Normal-vs.-Age 61–80, P = 1.044E-2) (Figure 4B). The expression of CDK2 was associated with various clinical features, such as Grade (Normal-vs.-Grade2, P = 5.088808E-03) (Figure 4C), the types of adenocarcinoma (Normal-vs.- Adenocarcinoma, P = 2.195E-02) (Figure 4D), gender (Male-vs.-Female, P = 1.338E-03) (Figure 4E), smoking habits (Normal-vs.-Smoker, P = 1.319E-11) (Figure 4F), stage (Normal-vs.-stage1, P = 3.458E-02) (Figure 4G), TP-53 mutation state (Normal-vs.-TP53 mutation, P = 1.624E-12) (Figure 4H), Weight (Normal-vs.-weight, P = 2.978E-2) (Figure 4I). The expression of CDK2 was related to the survival and prognosis of patients with LUAD (HR = 1.66, P = 5.8e-15) (Figure 4J). The higher the expression of CDK2, the shorter the survival time. Through this database, we discovered the CDK2-related genes and the heat maps were shown in Figures 4K,L. Many molecules involved in tumor classical signaling pathways were related to CDK2 expression. PrognoScan database: A new database for meta-analysis of the prognostic value of genes. We further found the expression of CDK2 influenced the OS and RFS of LUAD patients in different GSE datasets (GSE13213: COX P-value = 0.027245; GSE31210: COX P-value = 0.017966; GSE31210: COX P-value = 0.028818; GSE31210: COX P-value = 0.004197) (Figures 4M–P).
FIGURE 4.
Further study of CDK2 and prognosis analysis in LUAD. (A) The expression of CDK2 was highly in 515 LUAD tissues than 59 normal tissues. (B) Different ages might have differential expression levels of CDK2. (C) The expression of CDK2 was associated with Grade. (D) The types of adenocarcinoma. (E) Gender. (F) Smoking habits. (G) Stage. (H) TP-53 mutation state. (I) Weight. (J) The expression of CDK2 was related to the survival and prognosis of patients with LUAD. (K,L) CDK2-related genes and heat maps. (M–P) The expression of CDK2 influenced the OS and RFS of LUAD patients.
The Expression of CDK2 in Pan-Cancer Analysis
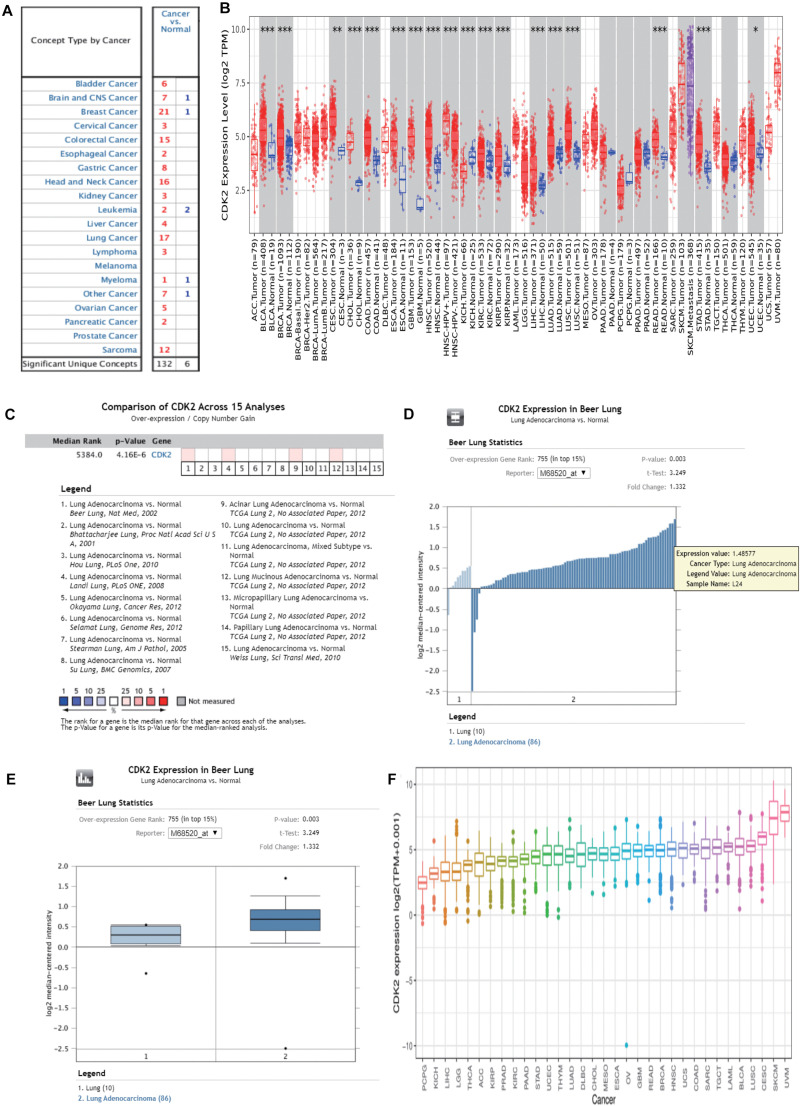
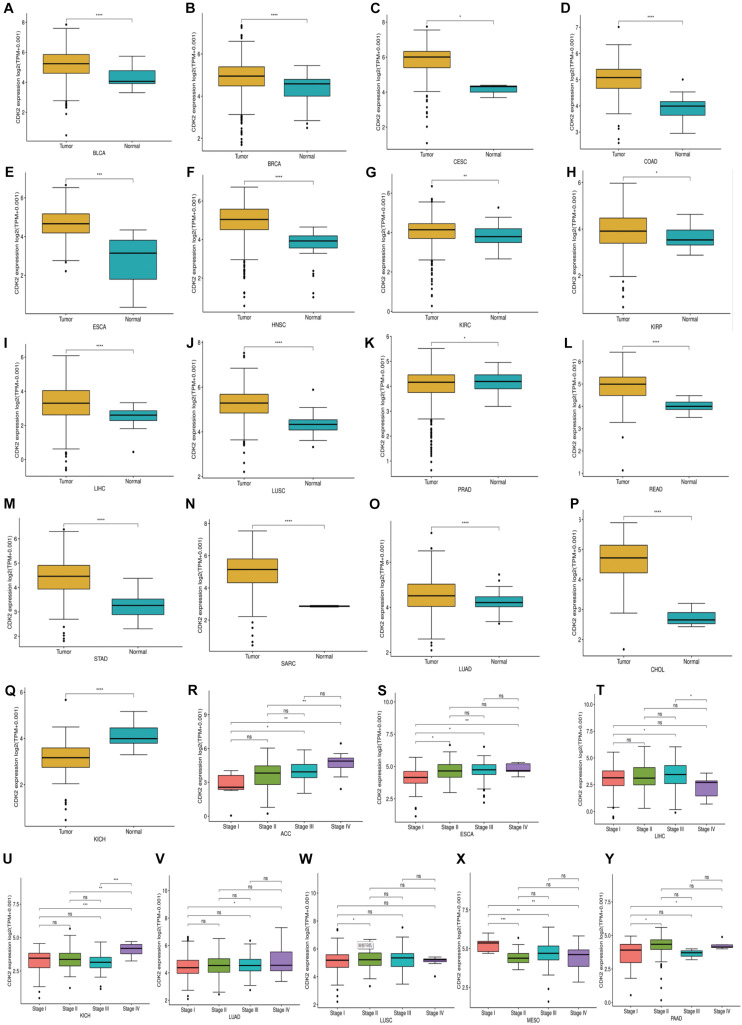
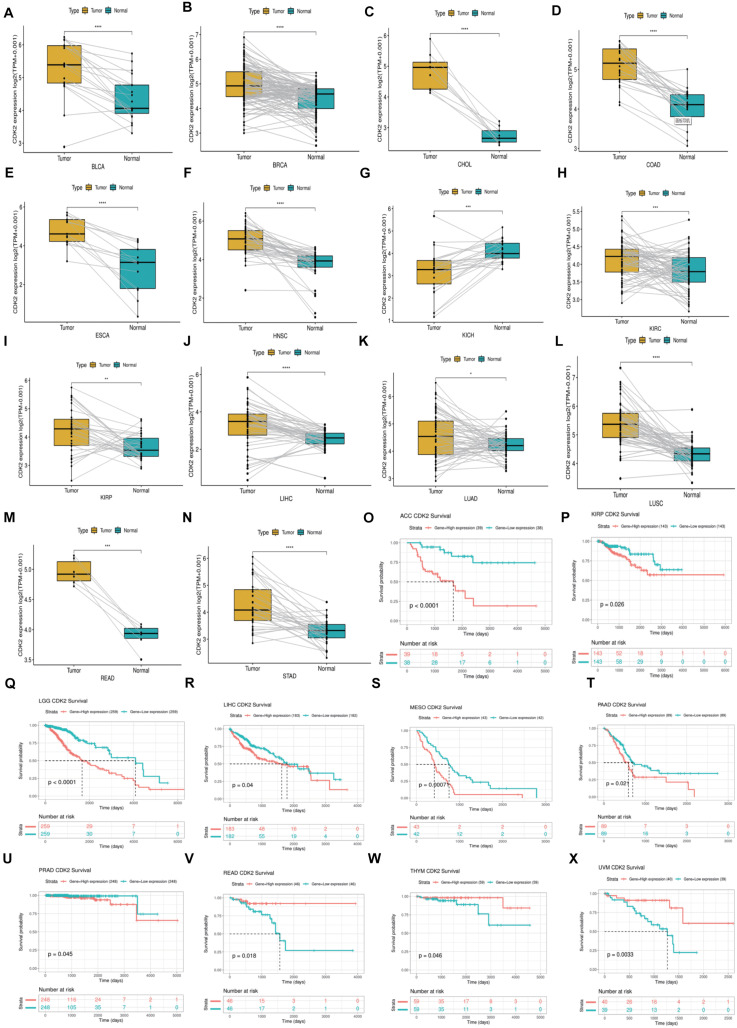
Through the Oncomine database, CDK2 expression was higher in 15 cancer tissues in comparison with normal tissues (Figure 5A). There was a great difference between cancer tissue and normal tissue. The statistical significance between normal and tumor tissues was further found in the TIMER database (the more “∗” symbol, the greater the difference) (Figure 5B). Mata analysis of 15 published studies on LUAD showed that expression of CDK2 was higher in LUAD (Median Rank = 5384.0, P = 4.16E-6) (Figure 5C). By t-test, box plot and peak plot of CDK2 in LUAD were shown in Figures 5D,E (t-Test = 3.249, Fold Change = 1.332, P = 0.003). Using the R package, we ranked CDK2 by its expression in 33 cancers (Figure 5F). Through different R packages, the expression of CDK2 was relatively higher in 17 cancers (P < 0.05) (Figures 6A–Q). The different expression levels of CDK2 had statistical significance on the stage of patients (P < 0.05) (Figures 6R–Y). There was significant difference in the expression of CDK2 between stage I and stage III, I and IV, II, and IV LUAD (∗/∗∗). For different types of cancer, we conducted paired differential expression of CDK2. Simultaneously, we found that there was statistical significance in 14 cancer types (P < 0.05) (Figures 7A–N). In 10 cancer types, high CDK2 expression was associated with poor prognosis of the patients (P < 0.05) (Figures 7O–X).
FIGURE 5.
The expression of CDK2 in pan-cancer analysis. (A) Through the Oncomine database, the expression of CDK2 was higher in 15 cancer tissues than normal tissues. (B) The expression of CDK2 in TIMER database. (C) Mata analysis of 15 published studies on LUAD showed that the expression of CDK2 was high in LUAD. (D,E) By t-test, Box plot and peak plot of CDK2 in LUAD. (F) The expression in 33 cancer, a pan-cancer analysis.
FIGURE 6.
The expression and stage of different cancer. (A–Q) The expression of CDK2 was relatively high in 17 cancers. (R–Y) The different expression levels of CDK2 had statistical significance on the stage of patients. (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
FIGURE 7.
The Paired expression and survival analysis of CDK2. (A–N) For different types of cancer, we carried out paired differential expression of CDK2. (O–X) In 10 kinds of cancers, the expression of CDK2 was related to the prognosis of patients. (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Functional Enrichment Analysis of CDK2
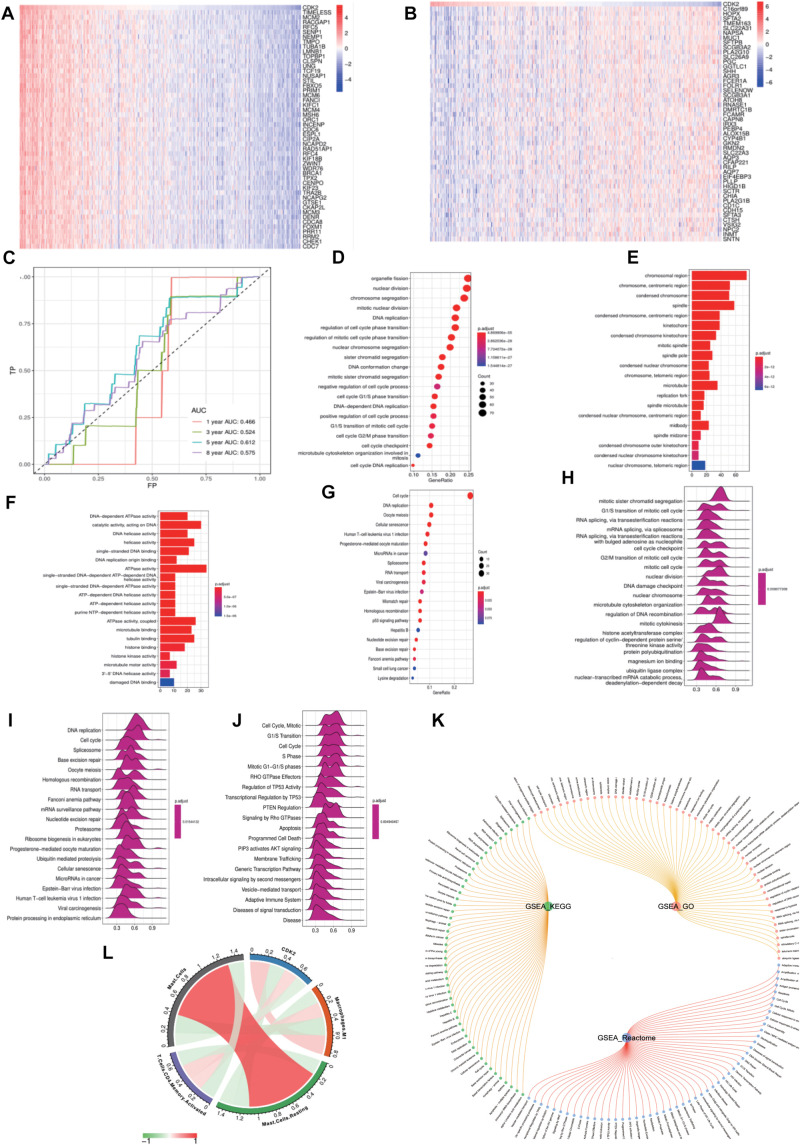
We compared CDK2 high expression groups with low expression groups in LUAD, and Figures 8A,B showed the top differentially expressed genes in two groups of cancer specimens. We took FP as the abscissa and TP as the ordinate and showed the AUC curve of 1, 3, 5, and 8 years to forecast the survival of patients. The areas under the curve were 0.465, 0.524, 0.612, and 0.575 (Figure 8C). “Clusterprofiler” R package was used to show the function analysis. And CDK2 was related to the DNA replication, regulation of cell cycle, cell cycle checkpoint, P53 signaling pathway. The bubble and bar charts were shown in Figures 8D–G. Wave charts about GO, KEGG, Reactome showed that CDK2 involved in classic tumor signaling pathways, such as regulation of TP53 activity, PTEN regulation, apoptosis, P13K-AKT signaling pathway (Figures 8H–J). The circle graph was shown in Figure 8K. For different immune cells, CDK2 was positively correlated with CD4 T cells (Cor = 0.1679, P = 0.0003), macrophage M1 (Cor = 0.2437, P = 1.40E-07) and negatively correlated with Mast cells (Cor = –0.1545, P = 0.0009) by CIBERSOPT algorithm (Figure 8L).
FIGURE 8.
Functional enrichment analysis of CDK2. (A,B) The top 50 heat maps of CDK2 positive and negative genes were shown. (C) The AUC curve of 1, 3, 5, and 8 years to forecast the survival of patients. (D–G) CDK2 was related to the DNA replication, regulation of cell cycle, cell cycle checkpoint, P53 signaling pathway. (H–J) Wave charts about GO, KEGG, Reactome showed that CDK2 involved in classic tumor signaling pathways, such as regulation of TP53 activity, PTEN regulation, apoptosis, P13K-AKT signaling pathway. (K) The circle graph of pathways. (L) CDK2 was positively correlated with CD4 T cells (Cor = 0.1679, P = 0.0003), macrophage M1 (Cor = 0.2437, P = 1.40E-07) and negatively correlated with Mast cells (Cor = –0.1545, P = 0.0009) by CIBERSOPT algorithm.
Characteristics of CDK2 Immune Cells Infiltration
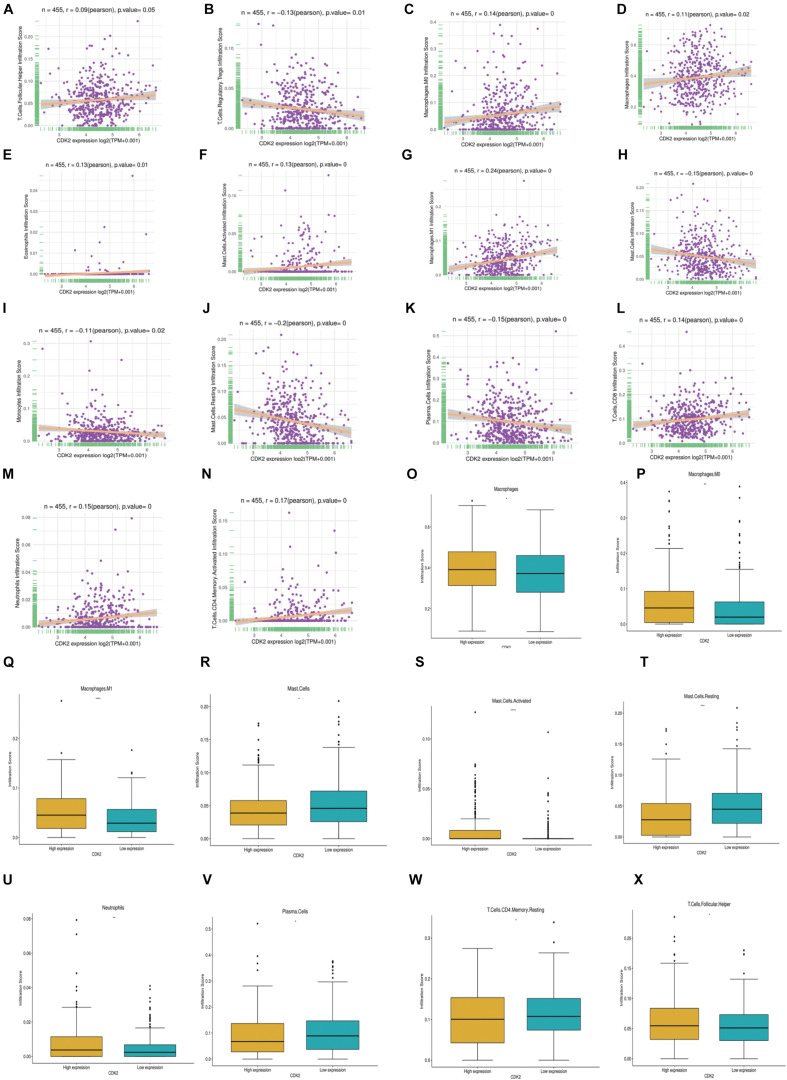
In LUAD, we further investigated different expression of CDK2 in different immune cell types. We found that 14 immune cells were closely related to CDK2 expression, such as T cells Follicular (P = 0.05, r = 0.09), T cells Regulatory Tregs (P = 0.01, r = –0.13), Macrophages.M0 (P = 0, r = 0.14), Macrophages (P = 0.02, r = 0.11), Eosinophils (P = 0.01, r = 0.13), Mast cells activated (P = 0, r = 0.13), Macrophages M1 (P = 0, r = 0.24), Mast cells (P = 0, r = –0.15), Monocytes (P = 0.02, r = –0.11), Mast cells Resting (P = 0, r = –0.2), Plasma cells (P = 0, r = –0.15), Tcells CD8 (P = 0, r = 0.14), Neutrophils (P = 0, r = 0.15), T cells CD4 (P = 0, r = 0.17) (Figures 9A–N). According to the median value of CDK2 expression, patients with LUAD were divided into the high expression groups and low expression groups. In different groups, the expression of 10 immune cells had systematic differences (P < 0.05) (Figures 9O–X).
FIGURE 9.
Characteristics of CDK2 immune cells infiltration. (A–N) 14 immune cells (T cells Follicular, T cells Regulatory Tregs, Macrophages.M0, Macrophages, Eosinophils, Mast cells activated, Macrophages M1, Mast cells, Monocytes, Mast cells Resting, Plasma cells, T cells CD8, Neutrophils, T cells CD4) were closely related to the expression of CDK2 (P ≤ 0.05). (O–X) In different CDK2 expression groups, the expression of immune cells had systematic differences (P < 0.05).
Construction of Immune Related Forecasting Model
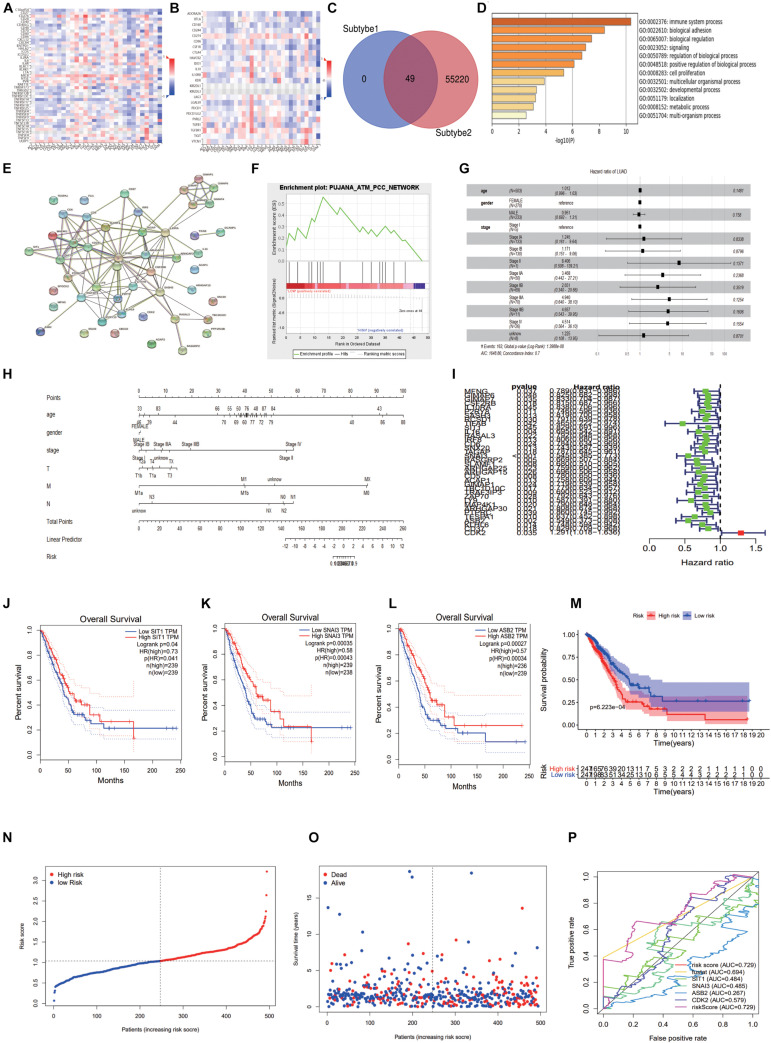
Through the TISIDB database, we found CDK2-related immunomodulators and the heat maps of immunopotentiators and immunosuppressants were shown in Figures 10A,B. By sorting the P-value (P < 0.05), we identified 13 immunosuppressants (ADORA2A, BTLA, CD274, CSF1R, IL10, KDR, LAG3, LGALS9, PDCD1, PDCD1LG2, TGFB1, TIGIT, VTCN1) and 21 immunopotentiators (CD27, CD276, CD28, CD40LG, CD48, CD70, CD80, CXCL12, CXCR4, ENTPD1, HHLA2, ICOSLG, IL2RA, IL6, IL6R, KLRC1, MICB, PVR, TMEM173, TNFRSF13B, ULBP1) with a high correlation of CDK2. In the cBioProtal, we explored forty-nine genes associated with 34 immunomodulators. Clinical data and gene expression data were downloaded from TCGA database. Forty-nine genes were mixed from the TCGA using “perl” and “R” package (Figure 10C). Through the Metascape database, the most abundant pathway of 49 genes was the immune system process, other function terms were biological adhesion, biological regulation, cell proliferation (Figure 10D). The BP, CC, MF was showed specifically in Table 2. The protein network interaction (PPI) of these 49 immune related genes were shown in the Figure 10E (P < 1.0e-16). GSEA analyzed the function, ES, NES, NOM p-val, FDR q-val of 49 genes. The statistically significant items were PUJANA_ATM_PCC_NETWORK and INTRACELLULAR_SIGNAL_TRANSDUCTION (P < 0.01) (Figure 10F and Table 3). Combing with clinical data and expression matrix, univariate and multivariate regression analyses were performed on age, gender and tumor stage (Total P = 1.399e-08) (Table 4). The forest map showed HR and concordance index (0.7) of LUAD. As predicted, the tumor stage was an independent risk factor for the prognosis of patients with LUAD (Figure 10G). Nomogram model combined several clinical factors, such as age, gender, stage, T, N, M to intuitively analyze the prognosis of LUAD (Figure 10H). Each patient can be assessed by nomogram model based on baseline clinical data. Univariate regression analysis showed that 36 genes were associated with the prognosis of LUAD (P < 0.05). We showed the HR, HR95L, HR95H, P-value of these 36 significant genes. CDK2 was an independent prognostic gene in LUAD (HR > 1) (Table 5 and Figure 10I). Multivariate regression analysis was used to analyze the 36 genes and only four genes (SIT1, SNAI3, ASB2, and CDK2) were included in the prediction model (Table 6). Through the GEPIA database, SIT1 (P = 0.04), SNAI3 (P = 0.00035), ASB2 (P = 0.00027) were lower expressed in LUAD (P < 0.05) (Figures 10J–L). To verify the prognostic model, the risk curve showed that the high-risk group was more lethal to lung cancer patients (P = 6.223E-04) (Figure 10M). Based on different risk scores, patients were divided into high and low risk groups using “R” package (Figures 10N,O). The ROC curve showed the different AUC of 4 interesting genes and CDK2 had more predictive value in the prognostic model (AUC = 0.579) (Figure 10P).
FIGURE 10.
Construction of immune related forecasting model. (A,B) The heat maps of immunopotentiators and immunosuppressants about CDK2. (C) 49 genes were mixed from the TCGA using “perl” and “R” package. (D) The most abundant pathway of 49 genes was immune system process. (E) The protein network interaction of these 49 immune related genes. (F) GSEA analyzed the function, ES, NES, NOM p-val, FDR q-val of 49 genes. (G) The forest map showed Hazard ratio and concordance index (0.7) of LUAD. (H) Nomogram model combined several clinical factors, such as age, gender, stage, T, N, M. (I) We showed the Hazard ratio (HR), HR95L, HR95H, p-value of these 36 significant genes. (J–L) Through the GEPIA database, SIT1, SNAI3, ASB2 were lower expressed in LUAD. (M) The risk curve showed that the high-risk group was more lethal to lung cancer patients. (N,O) The high and low risk maps. (P) The ROC curve showed the different AUC of four interesting genes.
TABLE 2.
The BP, CC, and MF were showed specifically of 49 immune related genes.
| GO term | Subgroup | Enrichment score |
| Positive regulation of GTPase activity | BP | 6.911710228 |
| B cell receptor signaling pathway | BP | 28.92678725 |
| Signal transduction | BP | 3.699937904 |
| T cell differentiation | BP | 39.05116279 |
| Regulation of small GTPase mediated signal transduction | BP | 11.65706352 |
| T cell receptor signaling pathway | BP | 10.55436832 |
| Positive regulation of T cell proliferation | BP | 19.5255814 |
| T cell costimulation | BP | 15.019678 |
| Positive regulation of T cell receptor signaling pathway | BP | 97.62790698 |
| Innate immune response | BP | 4.540832883 |
| G-protein coupled purinergic nucleotide receptor signaling pathway | BP | 55.78737542 |
| Adaptive immune response | BP | 7.915776241 |
| Positive regulation of Rho protein signal transduction | BP | 30.03935599 |
| Response to lipopolysaccharide | BP | 7.143505389 |
| Positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway | BP | 27.89368771 |
| Regulation of defense response to virus by virus | BP | 27.89368771 |
| Positive regulation of B cell proliferation | BP | 20.02623733 |
| Peptidyl-tyrosine autophosphorylation | BP | 19.5255814 |
| Release of sequestered calcium ion into cytosol | BP | 19.0493477 |
| Immunological synapse | CC | 44.66666667 |
| T cell receptor complex | CC | 63.27777778 |
| Membrane | CC | 2.070909091 |
| Plasma membrane | CC | 1.658335355 |
| Cytosol | CC | 1.717948718 |
| Integral component of plasma membrane | CC | 2.146525324 |
| Membrane raft | CC | 5.529126214 |
| GTPase activator activity | MF | 10.75651135 |
| GTP binding | MF | 5.861458333 |
| Receptor activity | MF | 6.914900154 |
| G-protein coupled purinergic nucleotide receptor activity | MF | 53.59047619 |
| Protein tyrosine kinase activity | MF | 8.461654135 |
| SH2 domain binding | MF | 25.87126437 |
TABLE 3.
The GSEA analysis of 49 mRNAs.
| Name | Size | ES | NES | NOM p-val | FDR q-val | FWER p-val | Rank at max | Leading edge |
| PUJANA_ATM_PCC_NETWORK | 16 | 0.5574023 | 1.7549632 | 0 | 0 | 0 | 13 | Tags = 50%, list = 27%, signal = 46% |
| LEE_DIFFERENTIATING_T_LYMPHOCYTE | 16 | 0.5064702 | 1.1931181 | 0.33333334 | 0.6333333 | 0.8 | 22 | Tags = 69%, list = 45%, signal = 84% |
| SMID_BREAST_CANCER_NORMAL_LIKE_UP | 22 | 0.45970738 | 0.9803642 | 0.4 | 0.4888889 | 0.8 | 12 | Tags = 36%, list = 24%, signal = 27% |
| SMID_BREAST_CANCER_LUMINAL_B_DN | 15 | 0.344885 | 0.68991035 | 1 | 0.8833334 | 1 | 1 | Tags = 13%, list = 2%, signal = 9% |
| GO_REGULATION_OF_INTRACELLULAR_SIGNAL_ TRANSDUCTION | 16 | -0.45997676 | -1.2494261 | 0 | 0 | 0 | 2 | Tags = 6%, list = 4%, signal = 4% |
TABLE 4.
Univariate and multivariate regression analysis of age, gender, and tumor stage.
| coef | exp(coef) | se(coef) | z | p | |
| Age | 0.011894 | 1.011965 | 0.008224 | 1.446 | 0.148 |
| genderMALE | –0.050014 | 0.951216 | 0.162323 | –0.308 | 0.758 |
| stageStage IA | 0.219186 | 1.245063 | 1.044445 | 0.21 | 0.834 |
| stageStage IB | 0.158106 | 1.17129 | 1.043788 | 0.151 | 0.88 |
| stageStage II | 2.128962 | 8.406135 | 1.432179 | 1.487 | 0.137 |
| stageStage IIA | 1.243594 | 3.468057 | 1.051101 | 1.183 | 0.237 |
| stageStage IIB | 0.9751 | 2.651431 | 1.047535 | 0.931 | 0.352 |
| stageStage IIIA | 1.597291 | 4.939634 | 1.042318 | 1.532 | 0.125 |
| stageStage IIIB | 1.538441 | 4.657324 | 1.096495 | 1.403 | 0.161 |
| stageStage IV | 1.50713 | 4.51376 | 1.060871 | 1.421 | 0.155 |
| P = 1.399e-08 | |||||
TABLE 5.
The analysis of Univariate regression about 36 genes.
| Id | HR | HR.95L | HR.95H | P-value |
| MFNG | 0.788791335 | 0.631264471 | 0.985627735 | 0.036858598 |
| GIMAP6 | 0.825241244 | 0.68227954 | 0.998158484 | 0.04782116 |
| GIMAP7 | 0.833265382 | 0.7035123 | 0.98694962 | 0.034680373 |
| CSF2RB | 0.814678719 | 0.687175564 | 0.965839662 | 0.018264635 |
| IL10RA | 0.838397617 | 0.705873794 | 0.995802041 | 0.044655471 |
| P2RY8 | 0.746446286 | 0.595538414 | 0.935593818 | 0.011158897 |
| SASH3 | 0.819264713 | 0.700490753 | 0.958177773 | 0.01261093 |
| RCSD1 | 0.790672034 | 0.639423589 | 0.977696595 | 0.030144597 |
| TIFAB | 0.465083695 | 0.222080992 | 0.973981795 | 0.042370157 |
| SIT1 | 0.829408759 | 0.690741005 | 0.995914365 | 0.045088923 |
| IL16 | 0.694821369 | 0.541782159 | 0.89109013 | 0.004125937 |
| RASAL3 | 0.791834804 | 0.648056113 | 0.967512448 | 0.022429427 |
| IRF8 | 0.805982708 | 0.679749247 | 0.955658471 | 0.013071014 |
| CD6 | 0.783523435 | 0.633680321 | 0.968799176 | 0.024280529 |
| SNX20 | 0.742751039 | 0.587258851 | 0.939413865 | 0.013084672 |
| TAGAP | 0.786867105 | 0.644621205 | 0.960501822 | 0.018468635 |
| SNAI3 | 0.545360728 | 0.384584195 | 0.773350355 | 0.000668422 |
| RASGRP2 | 0.669309183 | 0.506911612 | 0.883733519 | 0.004630788 |
| SLAMF1 | 0.679757859 | 0.510377564 | 0.905350821 | 0.008290951 |
| ARHGAP25 | 0.759490747 | 0.599527918 | 0.962134 | 0.022615883 |
| ARHGAP15 | 0.696431097 | 0.506489419 | 0.957603961 | 0.025975442 |
| CD5 | 0.779912525 | 0.649789891 | 0.936092658 | 0.007605839 |
| ACAP1 | 0.758479625 | 0.609407945 | 0.944016806 | 0.013287558 |
| GIMAP1 | 0.718737917 | 0.539208263 | 0.958042056 | 0.024304367 |
| TBC1D10C | 0.778901559 | 0.634277612 | 0.956501738 | 0.01711004 |
| TRAF3IP3 | 0.69040004 | 0.522777097 | 0.911769505 | 0.009030209 |
| ZAP70 | 0.792228171 | 0.643311406 | 0.975616893 | 0.028356063 |
| LY9 | 0.586557529 | 0.391159073 | 0.879564757 | 0.009858385 |
| MAP4K1 | 0.790353869 | 0.64826384 | 0.963587971 | 0.019976631 |
| ARHGAP30 | 0.808036247 | 0.674219373 | 0.968412661 | 0.021030946 |
| PTPRC | 0.859646397 | 0.744782085 | 0.992225703 | 0.038770457 |
| TESPA1 | 0.637294984 | 0.452324142 | 0.897906742 | 0.010006127 |
| ASB2 | 0.549252388 | 0.373292675 | 0.808154581 | 0.002358241 |
| KLHL6 | 0.747994992 | 0.593856228 | 0.942141348 | 0.013656291 |
| CD37 | 0.828826076 | 0.709387822 | 0.968373918 | 0.018042429 |
| CDK2 | 1.290598521 | 1.017922999 | 1.636316836 | 0.035146972 |
TABLE 6.
The analysis of multivariate regression about four genes.
| id | coef | HR | HR.95L | HR.95H | P-value |
| SIT1 | 0.255264409 | 1.290802876 | 0.925388716 | 1.800510461 | 0.132760088 |
| SNAI3 | –0.400450665 | 0.670018024 | 0.422135602 | 1.063459586 | 0.089332143 |
| ASB2 | –0.799324198 | 0.449632724 | 0.219800019 | 0.919788756 | 0.028601817 |
| CDK2 | 0.203710988 | 1.22594379 | 0.967951804 | 1.552699391 | 0.09107145 |
HPA Analysis and Construction of Predictive ceRNA Network in LUAD
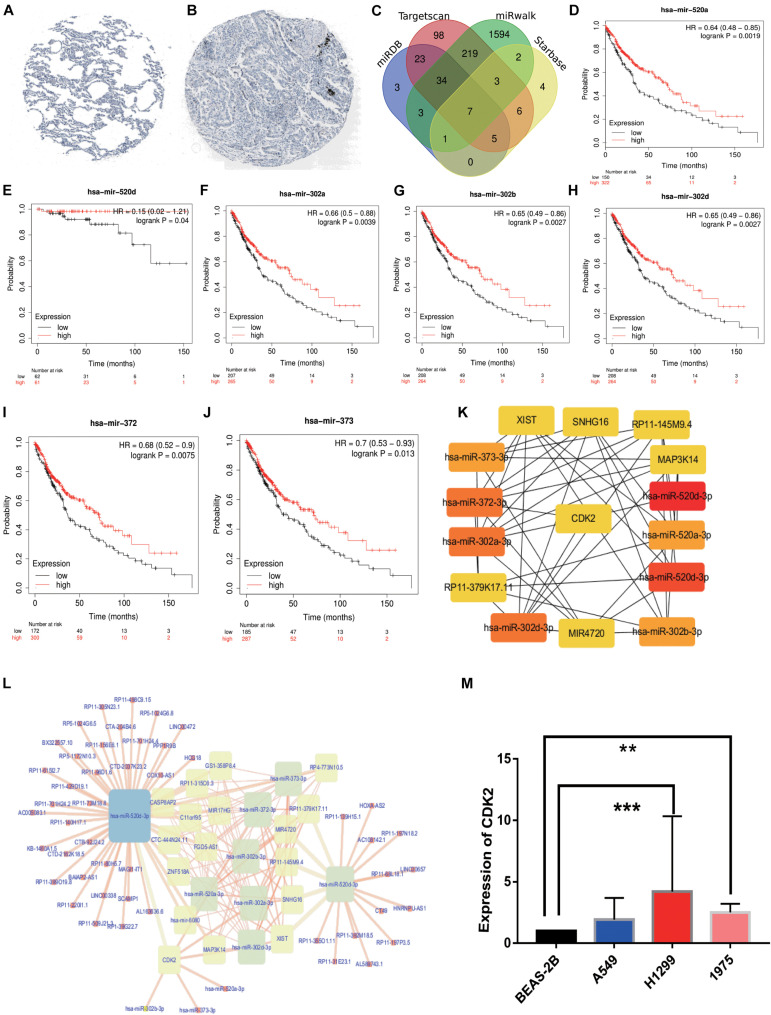
Immunohistochemical result showed that the CDK2 expression in LUAD tissues was significantly higher than that in normal lung tissues (Figures 11A,B). We selected 455 miRNAs related to CDK2 from TargetScan database. 10,018 miRNAs were found in miRWalk database. And we explored 76 miRNAs from mirDB and 28 miRNAs from Starbase. Combining with differentially expressed miRNAs in LUAD, 7 miRNAs were joined in the network using Venn map, such as hsa-miR-302b-3p, hsa-miR-372-3p, hsa-miR-302a-3p, hsa-miR-373-3p, hsa-miR-520a-3p, hsa-miR-520d-3p, and hsa-miR-302d-3p (Figure 11C). These miRNAs had obvious influence on the prognosis of LUAD patients (P < 0.05) (Figures 11D–J). We used the Starbase database to find the potential lncRNAs that regulate seven miRNAs. These coding genes and non-coding genes were interacted with each other by using Cytoscape software (Figure 11L). According to the degree in the network, we screened out the top 15 genes using CytoHubba (six lncRNAs: XIST, SNHG16, RP11-145M9.4, MAP3K14, MIR4720, and RP11-379K17.11) (Figure 11K).
FIGURE 11.
HPA analysis and ceRNA network. (A) Immunohistochemical results in normal lung tissues. (B) Immunohistochemical results in LUAD tissues. (C) The intersection results of the four databases were shown by Venn diagram. (D–J) The survival analysis of seven miRNAs. (K) 6lncRNAs-7miRNAs-CDK2 were considered as top 15 genes by CytoHubba. (L) ceRNA network was constructed by Cytoscape. (M) The PCR results about CDK2 expression in BEAS-2B, A549 (P = 0.0808), H1299 (P = 0.0006), H1975 (P = 0.0030). (**P < 0.01, ***P < 0.001).
The PCR Results of CDK2 Expression
Compared with the expression of CDK2 in BEAS-2B cell line, the expression of CDK2 in A549 cell line was increased, but there was no statistical difference (P = 0.0808). There were significant differences between H1299 cell line (P = 0.0006) and H1975 cell line (P = 0.0030) (Figure 11M).
Correlation Analysis of Drug Resistance
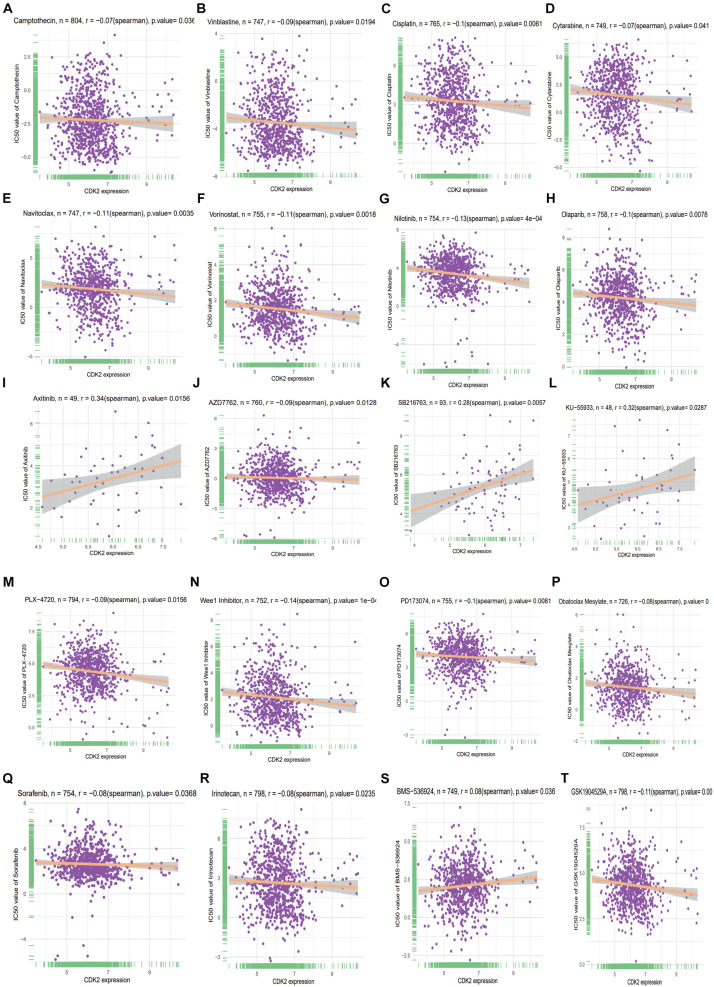
A total of 192 anti-tumor drugs were included in the study. The IC50 level of 89 anti-tumor drugs were related to the expression of CDK2. According to the size of P value (P < 0.05), we screened out the top 20 anti-tumor drugs with positive or negative correlation, such as Camptothecin (r = –0.074, P = 0.000018), Vinblastine (r = –0.085, P = 0.0000243), Cisplatin (r = –0.099, P = 0.0000843), Cytarabine (r = –0.0746, P = 0.0000975), Navitoclax (r = –0.106, P = 0.000158), Vorinostat (r = –0.113, P = 0.0002), Nilotinib (r = –0.127, P = 0.000258), Olaparib (r = –0.0966, P = 0.000302), Axitinib (r = 0.343, P = 0.000381), AZD7762 (r = –0.0902, P = 0.000382), SB216763 (r = 0.284, P = 0.000403), KU-55933 (r = 0.315, P = 0.000404), PLX-4720 (r = –0.0857, P = 0.000428), Wee1 Inhibitor (r = –0.141, P = 0.000780), PD173074 (r = –0.0963, P = 0.000794), Obatoclax Mesylate (r = –0.08009, P = 0.000872), Sorafenib (r = –0.076, P = 0.0009), Irinotecan (r = –0.0801, P = 0.00120), BMS-536924 (r = 0.0765, P = 0.0012), and GSK1904529A (r = –0.113, P = 0.0012) (Table 7 and Figures 12A–T).
TABLE 7.
Here are the top 20 anti-tumor drug resistance studies related to CDK2.
| Correlation | p value | Type | Label |
| –0.073624414 | 1.88E-05 | Camptothecin | Negative |
| –0.085496637 | 2.44E-05 | Vinblastine | Negative |
| –0.099132332 | 8.43E-05 | Cisplatin | Negative |
| –0.074684535 | 9.76E-05 | Cytarabine | Negative |
| –0.106844461 | 0.000158572 | Navitoclax | Negative |
| –0.11367658 | 0.000200639 | Vorinostat | Negative |
| –0.127941152 | 0.000258313 | Nilotinib | Negative |
| –0.096615555 | 0.000302571 | Olaparib | Negative |
| 0.343877551 | 0.000381858 | Axitinib | Positive |
| –0.090294733 | 0.000382049 | AZD7762 | Negative |
| 0.284802005 | 0.000403878 | SB216763 | Positive |
| 0.315892314 | 0.00040404 | KU-55933 | Positive |
| –0.085796235 | 0.00042863 | PLX-4720 | Negative |
| –0.141613081 | 0.000780249 | Wee1 Inhibitor | Negative |
| –0.09631745 | 0.000794901 | PD173074 | Negative |
| –0.080096541 | 0.000872875 | Obatoclax Mesylate | Negative |
| –0.076063235 | 0.000903871 | Sorafenib | Negative |
| –0.080170909 | 0.001206821 | Irinotecan | Negative |
| 0.076522222 | 0.00126849 | BMS-536924 | Positive |
| –0.113717172 | 0.001291814 | GSK1904529A | Negative |
FIGURE 12.
The top 20 anti-tumor drug resistance studies related to CDK2 (A–T).
Discussion
In recent years, the role of immune invasion in the progression of LUAD has been improved. Immunotherapy can resist tumor cells by activating the activity of immune molecules. It is necessary to find effective immune related-markers to predict prognosis and apply the individual therapy. And the study of TME is the key to overcome drug resistance (Li et al., 2020).
Our study first explored the prognostic molecule CDK2, which was differently expressed in LUAD tissues and adjacent non-LUAD tissues (P = 1.624E-12). High expression of CDK2 in LUAD has poor prognosis. In order to verify CDK2 in all kinds of cancers, we carried out a pan-cancer study. We further showed the expression, stage analysis, paired expression, survival condition in 33-cancer. The expression, stage and prognosis of CDK2 were obviously different in many cancers. According to published articles, a gene marker is no longer limited to one cancer, but is extended to various cancers, so the credibility has been improved than before. Based on CDK2, we searched for relevant immunomodulators and combined with the clinical expression data of LUAD to conduct univariate and multivariate regression models. Then the Nomogram forecast model of age, gender, stage, T, N, M provided the clinical significance and prognosis. The patient’s condition will be assessed according to the total risk score. Nomogram has a great effect on the prognosis of patients with LUAD. Finally, multivariate regression analysis showed that four mRNAs (SIT1, SNAI3, ASB2, and CDK2) were important for this immune model and significant for the prognosis of patients (P < 0.05). CDK2 was a risk molecule with independent prognosis (HR = 1.291, P = 0.035). This provides potential markers for targeted therapy with LUAD.
Cell-dependent kinases (CDKs) are involved in proliferation, DNA damage repair (Lim and Kaldis, 2013) and treatment of various tumors (Lin et al., 2021; Majumdar et al., 2021). A variety of mechanisms, including chaperone, positive phosphorylation and negative phosphorylation (Lee et al., 2021) regulate the activity of CDK family. Recently, CDK family molecules have been used as novel molecular markers for tumor-targeted therapy. As a star molecule, CDK2 (Kawakami et al., 2020) participates in classic pathways in various cancers, such as colorectal cancer (Somarelli et al., 2020), neuroblastoma (Poon et al., 2020), breast cancer (Hur et al., 2020), hepatocellular carcinoma (Hou et al., 2019), and prostate cancer (Washino et al., 2019). However, there are few studies on the mechanism of CDK2 in lung cancer. We studied the relationship between CDK2 and tumor immunity, providing a new idea for immunotherapy about LUAD.
In this study, we found that the expression of CDK2 was related to various immune cells including T cells (P = 0.05), Macrophages (P = 0), Eosinophils (P = 0.01), Mast cells (P = 0). Therefore, CDK2 can affect the tumorigenesis and proliferation by regulating the mechanism of immune inflammation. Combined with results of COX regression, target molecule CDK2 provides prognosis analysis and treatment strategy.
Cell cycle imbalance is common in different tumors (Chen T. et al., 2021). Cell-dependent kinase (CDK) is involved in many tumors related biological processes, such as cell cycle, immune checkpoint (Hume et al., 2020), RNA transcription (Al-Sanea et al., 2021), cell proliferation (Liu J. et al., 2021). Recently, there have been studies on CDK4 (Pandey et al., 2021) and CDK6 (Pandey et al., 2021), but the mechanism of CDK2 (El-Sattar et al., 2021) in cancer is relatively lacking. In our study, we deeply studied CDK2 and constructed the related immune prediction model. And CDK2 was associated in the P13K-AKT (Li Z. et al., 2021) signaling pathway in LUAD, inducing cell proliferation (Wu et al., 2020). Combing with clinical data, we constructed the forest map and Nomogram model (Cao et al., 2021b; Jang et al., 2021; Wang R. R. et al., 2021; Wang K. et al., 2021) of clinical characteristics. Through the features of patients, we can calculate the total score to evaluate the prognosis of patients. The prognosis of patients can be evaluated by a simple calculation method.
Compared with the current studies, the advantage is that we first established CDK2 related immune forecast model through the univariate, multivariate regression analysis and different “R” packages. This forecast was significant for LUAD patients. And the molecule CDK2 was related to various immune cells and may regulate mechanism in some way. The correlation between CDK2 and immune cells was listed by box diagram and point diagram. Compared with previous studies, they may research some markers in only signal cancer. But we explored detailed information of CDK2 in 33-cancer, a pan-cancer analysis was shown in our study. In various cancers, CDK2 has corresponding systematic significance with several clinical aspects (P < 0.05). CDK2 was associated with clinical stage, age, pathological type, TP-53 mutation, smoking with LUAD. According to the risk score, LUAD patients were divided into the high-risk group and low-risk group. There was a great difference between different risk curves (Yu et al., 2021). The survival time of the higher risk group was shorter than that of the lower risk group (P = 6.223E-04). This indicates that the immune related model is of great significance for the prognosis of patients with LUAD. All data were collected from reliable GEO database, TCGA, UCSC Xena and this increases the reliability of the data. In recent years, ceRNA network has certain significance in various cancers. The functions of many non-coding genes have been gradually explored (Yuan et al., 2014). Seven miRNAs (hsa-miR-302b-3p, hsa-miR-372-3p, hsa-miR-302a-3p, hsa-miR-373-3p, hsa-miR-520a-3p, hsa-miR-520d-3p, and hsa-miR-302d-3p), six lncRNAs (XIST, SNHG16, RP11-145M9.4, MAP3K14, MIR4720, and RP11-379K17.11) were considered as potential biomarkers in LUAD. Combined with the current literature, these non-coding genes have been studied in other cancers, but less in LUAD. This study provides potential therapeutic targets for LUAD and contributes to immunotherapy.
In recent years, nomogram model has been considered as a tool to predict tumor prognosis (Balachandran et al., 2015; Huang et al., 2016), such as colorectal cancer (Huang et al., 2016) and cervical cancer (Rose et al., 2015). Nomogram model meets our desire for biologically and clinically integrated models. It is not limited to one factor, but combines with various influencing factors to evaluate the prognosis of patients. We constructed nomogram model of age, gender, stage, T, N, M. According to the corresponding score of each influencing factor, the risk index of LUAD patients is estimated to evaluate the survival time.
According to our anti-tumor drug resistance research findings, the IC50 of four drugs (Axitinib, SB216763, KU-55933, BMS-536924) were positive with expression of CDK2. This indicates that cancer patients with high expression of CDK2 are prone to resistance to the above four drugs. Other 16 drugs were negative with the expression of CDK2. The patients with high expression of CDK2 had good response to the above 16 drugs and low resistance rate. Three drugs Cisplatin, Cytarabine, Nilotinib are considered as classic anti-cancer drugs. They are effective for patients with high expression of CDK2 cancer and it is not easy to cause drug resistance. Of course, the mechanism between CDK2 and drug resistance still needs to further be studied.
There are disadvantages in our study compared with the current articles. The target molecules lack of experimental verification and big data support. The regulatory mechanism of target genes is unclear.
In summary, immune-related forecast model was constructed by univariate regression and multivariate regression analysis to guide prognosis of LUAD. K–M curve verified the high-risk group had a poor prognosis (P < 0.01). Because of the correlation of CDK2 and various immune cells, CDK2 may be involved in tumor regulation of immune infiltration. CDK2 provides a new immunotherapy target for LUAD, which can improve the prognosis. The construction of ceRNA network provides a new way for exploring potential gene markers with LUAD. Drug resistance research can help patients choose a reasonable treatment plan, not blindly targeted therapy or immunosuppressive therapy.
Conclusion
In conclusion, we discovered a set of four genes, including expression of CDK2, has a significant prognostic value in LUAD. CDK2 expression is highly associated with immune responses in the cancer. We made some prediction to link CDK2 expression with drug responses and miRNA expression.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author Contributions
T-TL and RL conceived and designed the study. CH, J-PL, JY, and X-LJ collected the literature. T-TL drafted the manuscript. Y-QQ revised the manuscript. All the authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
- TME
tumor microenvironment
- ceRNA
competing endogenous RNA
- LUAD
lung adenocarcinoma
- GEO
Gene Expression Omnibus
- TCGA
The Cancer Genome Atlas
- GDSC
Genomics of Drug Sensitivity in Cancer
- DEG
differentially expressed genes
- GSEA
Gene Set Enrichment Analysis
- LUSC
lung squamous cell carcinoma
- NSCLC
non-small cell lung cancer
- SII
systemic immune-inflammation index
- CDK
cell-dependent kinases
- BP
biological process
- CC
cellular component
- MF
molecular function
- OS
The Overall Survival
- RFS
Relapse-Free Survival
- FP
false positive
- TP
true positive
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- AML
acute myeloid leukemia
- GO
Gene Ontology
- HPA
Human Protein Atlas
- PCR
polymerase chain reaction.
Funding. This work was supported by grants from the Major Scientific and Technological Innovation Project of Shandong Province (Grant No. 2018CXGC1212), the CSCO-Qilu Cancer Research Fund (Grant No. Y-Q201802-014), the Medical and Health Technology Innovation Plan of Jinan City (Grant No. 201805002), and Special fund for clinical research of Jinan City (201912011).
References
- Al-Sanea M. M., Obaidullah A. J., Shaker M. E., Chilingaryan G., Alanazi M. M., Alsaif N. A., et al. (2021). CDK2 Inhibitor with 3-hydrazonoindolin-2-one scaffold endowed with anti-breast cancer activity: design, synthesis, biological evaluation, and in silico insights. Molecules 26:412. 10.3390/molecules26020412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bade B. C., Dela Cruz C. S. (2020). Lung Cancer 2020: epidemiology, etiology, and prevention. Clin. Chest. Med. 41 1–24. 10.1016/j.ccm.2019.10.001 [DOI] [PubMed] [Google Scholar]
- Balachandran V. P., Gonen M., Smith J. J., DeMatteo R. P. (2015). Nomograms in oncology: more than meets the eye. Lancet Oncol. 16 e173–e180. 10.1016/S1470-2045(14)71116-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett-Itzhaki Z., Knapp S., Avraham C., Racowsky C., Hauser R., Bollati V., et al. (2021). Association between follicular fluid phthalate concentrations and extracellular vesicle microRNAs expression. Hum. Reprod. 36 1590–1599. 10.1093/humrep/deab063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Wu L., Lei X., Shi K., Shi L. (2021a). A signature of 13 autophagyrelated gene pairs predicts prognosis in hepatocellular carcinoma. Bioengineered 12 697–707. 10.1080/21655979.2021.1880084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Wu L., Lei X., Shi K., Shi L., Shi Y. (2021b). Long non-coding RNA-based signature for predicting prognosis of hepatocellular carcinoma. Bioengineered 12 673–681. 10.1080/21655979.2021.1878763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Chio C. L., Zeng H., Li Y. (2021). High expression of CD56 may be associated with favorable overall survival in intermediate-risk acute myeloid leukemia. Hematology 26 210–214. 10.1080/16078454.2021.1880734 [DOI] [PubMed] [Google Scholar]
- Chen J., Zhou R. (2021). Tumor microenvironment related novel signature predict lung adenocarcinoma survival. PeerJ 9:e10628. 10.7717/peerj.10628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Liu L., Zou Y., Hu X., Zhang W., Zhou T., et al. (2021). Nobiletin downregulates the SKP2-p21/p27-CDK2 axis to inhibit tumor progression and shows synergistic effects with palbociclib on renal cell carcinoma. Cancer Biol. Med. 18 227–244. 10.20892/j.issn.2095-3941.2020.0186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Zheng R., Baade P. D., Zhang S., Zeng H., Bray F., et al. (2016). Cancer statistics in China, 2015. CA Cancer J Clin 66 115–132. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- Chen Y., Zhang R. (2021). Long non-coding RNA AL139002.1 promotes gastric cancer development by sponging microRNA-490-3p to regulate hepatitis a virus cellular receptor 1 expression. Bioengineered 12 1927–1938. 10.1080/21655979.2021.1922329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zhou H., Wang Z., Huang Z., Wang J., Zheng M., et al. (2021). Integrated analysis of ceRNA network and tumor-infiltrating immune cells in esophageal cancer. Biosci. Rep. 41:BSR20203804. 10.1042/BSR20203804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W., Feng J., Hu X., Chen Y., Gu Q., Gong W., et al. (2021). SLC7A7 is a prognostic biomarker correlated with immune infiltrates in non-small cell lung cancer. Cancer Cell Int. 21:106. 10.1186/s12935-021-01781-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sattar N., Badawy E. H. K., AbdEl-Hady W. H., Abo-Alkasem M. I., Mandour A. A., Ismail N. S. M. (2021). Design and synthesis of new CDK2 inhibitors containing thiazolone and thiazolthione scafold with apoptotic activity. Chem. Pharm. Bull. (Tokyo) 69 106–117. 10.1248/cpb.c20-00714 [DOI] [PubMed] [Google Scholar]
- Esenboga K., Kurtul A., Yamanturk Y. Y., Tan T. S., Tutar D. E. (2021). Systemic immune-inflammation index predicts no-reflow phenomenon after primary percutaneous coronary intervention. Acta. Cardiol. 1–8. 10.1080/00015385.2021.1884786 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Geng Y., Shao Y., Zhu D., Zheng X., Zhou Q., Zhou W., et al. (2016). Systemic immune-inflammation index predicts prognosis of patients with esophageal squamous cell carcinoma: a propensity score-matched analysis. Sci Rep 6 39482. 10.1038/srep39482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P., Li J. W., Zhang B. M., Lv J. C., Li Y. M., Gu X. Y., et al. (2017). The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/beta-catenin signaling. Mol. Cancer 16:9. 10.1186/s12943-017-0583-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T., Zhou Y., Li D. (2021). Relationship between hepatocellular carcinoma and depression via online database analysis. Bioengineered 12 1689–1697. 10.1080/21655979.2021.1921552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M., Han Y., Cai C., Liu P., Chen Y., Shen H., et al. (2021). CLEC10A is a prognostic biomarker and correlated with clinical pathologic features and immune infiltrates in lung adenocarcinoma. J. Cell Mol. Med. 25 3391–3399. 10.1111/jcmm.16416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J., Yao C. (2021). Potential prognostic biomarkers of lung adenocarcinoma based on bioinformatic analysis. Biomed. Res. Int. 2021:8859996. 10.1155/2021/8859996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Wang Z., Huang S., Sun C., Zhao J., Shi J., et al. (2019). SKA3 Promotes tumor growth by regulating CDK2/P53 phosphorylation in hepatocellular carcinoma. Cell Death Dis. 10:929. 10.1038/s41419-019-2163-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Yang X. R., Xu Y., Sun Y. F., Sun C., Guo W., et al. (2014). Systemic immune-inflammation index predicts prognosis of patients after curative resection for hepatocellular carcinoma. Clin. Cancer Res. 20 6212–6222. 10.1158/1078-0432.CCR-14-0442 [DOI] [PubMed] [Google Scholar]
- Huang H., Kong L., Luan S., Qi C., Wu F. (2021). Ligustrazine suppresses platelet-derived growth factor-bb-induced pulmonary artery smooth muscle cell proliferation and inflammation by regulating the PI3K/AKT signaling pathway. Am. J. Chin. Med. 49 437–459. 10.1142/S0192415X21500208 [DOI] [PubMed] [Google Scholar]
- Huang X. Y., Liu J. J., Liu X., Wang Y. H., Xiang W. (2021). Bioinformatics analysis of the prognosis and biological significance of VCAN in gastric cancer. Immun. Inflamm. Dis. 9 547–559. 10.1002/iid3.414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. Q., Liang C. H., He L., Tian J., Liang C. S., Chen X., et al. (2016). Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J. Clin. Oncol. 34 2157–2164. 10.1200/JCO.2015.65.9128 [DOI] [PubMed] [Google Scholar]
- Hume S., Dianov G. L., Ramadan K. (2020). A unified model for the G1/S cell cycle transition. Nucleic Acids Res. 48 12483–12501. 10.1093/nar/gkaa1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur S., Kim J. H., Yun J., Ju Y. W., Han J. M., Heo W. (2020). Protein Phosphatase 1H, cyclin-dependent kinase inhibitor p27, and cyclin-dependent kinase 2 in paclitaxel resistance for triple negative breast cancers. J. Breast Cancer 23 162–170. 10.4048/jbc.2020.23.e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S. Y., Kim J. S., Baek S. Y., Lee H. A., Lee J. K. (2021). Proposed nomogram predicting neoplastic ampullary obstruction in patients with a suspected ampulla of Vater lesion on CT. Abdom. Radiol. (NY) 46 3128–3138. 10.1007/s00261-021-02975-3 [DOI] [PubMed] [Google Scholar]
- Janssen L. M. E., Ramsay E. E., Logsdon C. D., Overwijk W. W. (2017). The immune system in cancer metastasis: friend or foe? J. Immunother. Cancer 5:79. 10.1186/s40425-017-0283-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jomrich G., Paireder M., Kristo I., Baierl A., Ilhan-Mutlu A., Preusser M., et al. (2021). High systemic immune-inflammation index is an adverse prognostic factor for patients with gastroesophageal adenocarcinoma. Ann. Surg. 273 532–541. 10.1097/SLA.0000000000003370 [DOI] [PubMed] [Google Scholar]
- Ju Q., Huang T., Zhang Y., Wu L., Geng J., Mu X., et al. (2021). Systemic immune-inflammation index predicts prognosis in patients with different EGFR-mutant lung adenocarcinoma. Medicine (Baltimore) 100:e24640. 10.1097/MD.0000000000024640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kara A., Ozgur A., Tekin S., Tutar Y. (2021). Computational analysis of drug resistance network in lung adenocarcinoma. Anticancer Agents Med. Chem 10.2174/1871520621666210218175439 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Kawakami M., Mustachio L. M., Chen Y., Chen Z., Liu X., Wei C. H., et al. (2020). CDK2/9 Inhibitor CYC065 causes anaphase catastrophe and represses proliferation, tumorigenesis, and metastasis in aneuploid cancers. Mol. Cancer Ther. 20 477–489. 10.1158/1535-7163.MCT-19-0987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Piao H. L., Kim B. J., Yao F., Han Z., Wang Y., et al. (2018). Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet. 50 1705–1715. 10.1038/s41588-018-0252-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kris M. G., Johnson B. E., Berry L. D., Kwiatkowski D. J., Iafrate A. J., Wistuba I. I., et al. (2014). Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 311 1998–2006. 10.1001/jama.2014.3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. C., Hong K. H., Becker A., Tash J. S., Schonbrunn E., Georg G. I. (2021). Tetrahydroindazole inhibitors of CDK2/cyclin complexes. Eur. J. Med. Chem. 214:113232. 10.1016/j.ejmech.2021.113232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Jin Y., Pei X., Guo P., Dong K., Wang H., et al. (2021). Bioinformatics analysis and verification of gene targets for renal clear cell carcinoma. Comput. Biol. Chem. 92:107453. 10.1016/j.compbiolchem.2021.107453 [DOI] [PubMed] [Google Scholar]
- Li J. H., Liu S., Zhou H., Qu L. H., Yang J. H. (2014). starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 42 D92–D97. 10.1093/nar/gkt1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Huang Y., Zhang Y., Wen J., Chen Y., Wang L., et al. (2021). Identification BCL6 and miR-30 family associating with Ibrutinib resistance in activated B-cell-like diffuse large B-cell lymphoma. Med. Oncol. 38:33. 10.1007/s12032-021-01470-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Meng H., Bai Y., Wang K. (2016). Regulation of lncRNA and its role in cancer metastasis. Oncol. Res. 23 205–217. 10.3727/096504016X14549667334007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Tao L., Cai W. (2020). Profiles of immune infiltration and prognostic immunoscore in lung adenocarcinoma. Biomed. Res. Int. 2020:5858092. 10.1155/2020/5858092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Chen C., Wang J., Wei M., Liu G., Qin Y., et al. (2021). Overexpressed PLAU and its potential prognostic value in head and neck squamous cell carcinoma. PeerJ 9:e10746. 10.7717/peerj.10746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S., Kaldis P. (2013). Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development 140 3079–3093. 10.1242/dev.091744 [DOI] [PubMed] [Google Scholar]
- Lin T., Li J., Liu L., Li Y., Jiang H., Chen K., et al. (2021). Design, synthesis, and biological evaluation of 4-benzoylamino-1H-pyrazole-3-carboxamide derivatives as potent CDK2 inhibitors. Eur. J. Med. Chem. 215:113281. 10.1016/j.ejmech.2021.113281 [DOI] [PubMed] [Google Scholar]
- Ling B., Huang Z., Huang S., Qian L., Li G., Tang Q. (2020). Microenvironment analysis of prognosis and molecular signature of immune-related genes in lung adenocarcinoma. Oncol. Res. 28 561–578. 10.3727/096504020X15907428281601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Song M., Sun X., Zhang X., Miao H., Wang Y. (2021). T-box transcription factor TBX1, targeted by microRNA-6727-5p, inhibits cell growth and enhances cisplatin chemosensitivity of cervical cancer cells through AKT and MAPK pathways. Bioengineered 12 565–577. 10.1080/21655979.2021.1880732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Zeng X., Han K., Jia X., Zhou M., Zhang Z., et al. (2021). The expression regulation of Cyclins and CDKs in ovary via miR-9c and miR-263a of scylla paramamosain. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 254:110567. 10.1016/j.cbpb.2021.110567 [DOI] [PubMed] [Google Scholar]
- Liu X. S., Gao Y., Liu C., Chen X. Q., Zhou L. M., Yang J. W., et al. (2020). Comprehensive analysis of prognostic and immune infiltrates for E2F transcription factors in human pancreatic adenocarcinoma. Front Oncol 10 10:606735. 10.3389/fonc.2020.606735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zhan Y., Xu W., Liu X., Geng Y., Liu L., et al. (2021). Prognostic and immunological role of Fam20C in pan-cancer. Biosci. Rep. 41:BSR20201920. 10.1042/BSR20201920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Chen G., Liu H., Li Z., Yang Q., Gu X., et al. (2019). Integrated bioinformatics analysis of miRNA expression in ewing sarcoma and potential regulatory effects of miR-21 via targeting ALCAM/CD166. Artif. Cells Nanomed. Biotechnol. 47 2114–2122. 10.1080/21691401.2019.1620760 [DOI] [PubMed] [Google Scholar]
- Liu Y., Li L., Jiang D., Yang M., Gao X., Lv K., et al. (1976). For survival prediction of patients with spinal metastasis from prostate cancer. Spine (Phila Pa 46 E364–E373. 10.1097/BRS.0000000000003888 [DOI] [PubMed] [Google Scholar]
- Lolli C., Caffo O., Scarpi E., Aieta M., Conteduca V., Maines F., et al. (2016). Systemic immune-inflammation index predicts the clinical outcome in patients with mCRPC treated with abiraterone. Front. Pharmacol. 7:376. 10.3389/fphar.2016.00376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H. P., Du X. F., Li J. D., Huang S. N., He R. Q., Wu H. Y., et al. (2021). Expression of cell division cycle protein 45 in tissue microarrays and the CDC45 gene by bioinformatics analysis in human hepatocellular carcinoma and patient outcomes. Med. Sci. Monit. 27:e928800. 10.12659/MSM.928800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar A., Burban D. J., Muretta J. M., Thompson A. R., Engel T. A., Rasmussen D. M. (2021). Allostery governs Cdk2 activation and differential recognition of CDK inhibitors. Nat. Chem. Biol. 17 456–464. 10.1038/s41589-020-00725-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marwitz S., Ballesteros-Merino C., Jensen S. M., Reck M., Kugler C., Perner S. (2021). Phosphorylation of SMAD3 in immune cells predicts survival of patients with early stage non-small cell lung cancer. J. Immunother. Cancer 9:e001469. 10.1136/jitc-2020-001469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungan I., Bostanci E. B., Turksal E., Tezcan B., Aktas M. N., Can M., et al. (2021). The predictive power of C-reactive protein- lymphocyte ratio for in-hospital mortality after colorectal cancer surgery. Cancer Rep. (Hoboken) 4:e1330. 10.1002/cnr2.1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noreldeen H. A. A., Liu X., Xu G. (2020). Metabolomics of lung cancer: Analytical platforms and their applications. J. Sep. Sci. 43 120–133. 10.1002/jssc.201900736 [DOI] [PubMed] [Google Scholar]
- Pandey K., Lee E., Park N., Hur J., Cho Y. B., Katuwal N. B., et al. (2021). Deregulated immune pathway associated with palbociclib resistance in preclinical breast cancer models: integrative genomics and transcriptomics. Genes (Basel) 12:159. 10.3390/genes12020159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon E., Liang T., Jamin Y., Walz S., Kwok C., Hakkert A. (2020). Orally bioavailable CDK9/2 inhibitor shows mechanism-based therapeutic potential in MYCN-driven neuroblastoma. J. Clin. Invest. 130 5875–5892. 10.1172/JCI134132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C., Li M., Zheng Y., Wu F., Du W., Quan R. (2021). Identification of diagnostic genes and vital microRNAs involved in rheumatoid arthritis: based on data mining and experimental verification. PeerJ 9:e11427. 10.7717/peerj.11427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose P. G., Java J., Whitney C. W., Stehman F. B., Lanciano R., Thomas G. M. (2015). Nomograms predicting progression-free survival, overall survival, and pelvic recurrence in locally advanced cervical cancer developed from an analysis of identifiable prognostic factors in patients from nrg oncology/gynecologic oncology group randomized trials of chemoradiotherapy. J. Clin. Oncol. 33 2136–2142. 10.1200/JCO.2014.57.7122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaniello A., Napolitano F., Servetto A., De Placido P., Silvestris N., Bianco C., et al. (2019). Tumour microenvironment and immune evasion in EGFR addicted NSCLC: hurdles and possibilities. Cancers (Basel) 11:1419. 10.3390/cancers11101419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng L. P., Han C. Q., Nie C., Xu T., Zhang K., Li X. J., et al. (2021). Identification of potential serum exosomal microRNAs involved in acinar-ductal metaplasia that is a precursor of pancreatic cancer associated with chronic pancreatitis. Medicine (Baltimore) 100:e25753. 10.1097/MD.0000000000025753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng X. F., Hong L. L., Li H., Huang F. Y., Wen Q., Zhuang H. F. (2021). Long non-coding RNA MALAT1 modulate cell migration, proliferation and apoptosis by sponging microRNA-146a to regulate CXCR4 expression in acute myeloid leukemia. Hematology 26 43–52. 10.1080/16078454.2020.1867781 [DOI] [PubMed] [Google Scholar]
- Siegel R. L., Miller K. D., Jemal A. (2020). Cancer statistics, 2020. CA Cancer J. Clin. 70 7–30. 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- Somarelli J. A., Roghani R. S., Moghaddam A. S., Thomas B. C., Rupprecht G., Ware K. E., et al. (2020). A Precision medicine drug discovery pipeline identifies combined CDK2 and 9 inhibition as a novel therapeutic strategy in colorectal cancer. Mol. Cancer Ther. 19 2516–2527. 10.1158/1535-7163.MCT-20-0454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Xu Q., Shi R., Zhang G. (2021). Bioinformatics analysis reveals the landscape of immune cell infiltration and immune-related pathways participating in the progression of carotid atherosclerotic plaques. Artif. Cells Nanomed. Biotechnol. 49 96–107. 10.1080/21691401.2021.1873798 [DOI] [PubMed] [Google Scholar]
- Wang C., Qiao W., Jiang Y., Zhu M., Shao J., Wang T., et al. (2020). The landscape of immune checkpoint inhibitor plus chemotherapy versus immunotherapy for advanced non-small-cell lung cancer: a systematic review and meta-analysis. J. Cell Physiol. 235 4913–4927. 10.1002/jcp.29371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Wu Z., Wang G., Shi H., Xie J., Yin L., et al. (2021). Survival nomogram for patients with bone metastatic renal cell carcinoma: a population-based study. Int. Braz. J. Urol. 47 333–349. 10.1590/S1677-5538.IBJU.2020.0195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. R., He M., Gui X., Kang Y. (2021). A nomogram based on serum cystatin C for predicting acute kidney injury in patients with traumatic brain injury. Ren Fail 43 206–215. 10.1080/0886022X.2021.1871919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washino S., Rider L. C., Romero L., Jillson L. K., Affandi T., Ohm A. M. (2019). Loss of MAP3K7 sensitizes prostate cancer cells to cdk1/2 inhibition and DNA damage by disrupting homologous recombination. Mol. Cancer Res. 17 1985–1998. 10.1158/1541-7786.MCR-18-1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Gao H., Ge W., He J. (2020). Over expression of PTEN induces apoptosis and prevents cell proliferation in breast cancer cells. Acta Biochim. Pol. 67 515–519. 10.18388/abp.2020_5371 [DOI] [PubMed] [Google Scholar]
- Wu R., Zhuang H., Mei Y. K., Sun J. Y., Dong T., Zhao L. L., et al. (2021). Systematic identification of key functional modules and genes in esophageal cancer. Cancer Cell Int. 21:134. 10.1186/s12935-021-01826-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Liu L., Shen X., Liu W., Ma R. (2021). Plakophilin-2 promotes lung adenocarcinoma development via enhancing focal adhesion and epithelial-mesenchymal transition. Cancer Manag. Res. 13 559–570. 10.2147/CMAR.S281663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M., Feng Y., Wang Y., Wang J., Zhang Z., Liang J., et al. (2021). Correlation between circulating interleukin-18 level and systemic lupus erythematosus: a meta-analysis. Sci. Rep. 11:4707. 10.1038/s41598-021-84170-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Yang L., Wang Y. (2021). [Immunotherapy for lung cancer: mechanisms of resistance and response strategy]. Zhongguo Fei Ai Za Zhi 24 112–123. 10.3779/j.issn.1009-3419.2021.101.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Chen X., Li Y., Wang Y., Cao X., Liu Z., et al. (2021). Pro-inflammatory cytokines as potential predictors for intradialytic hypotension. Ren Fail 43 198–205. 10.1080/0886022X.2021.1871921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J. H., Yang F., Wang F., Ma J. Z., Guo Y. J., Tao Q. F., et al. (2014). A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 25 666–681. 10.1016/j.ccr.2014.03.010 [DOI] [PubMed] [Google Scholar]
- Zhai Y., Zhao B., Wang Y., Li L., Li J., Li X., et al. (2021). Construction of the optimization prognostic model based on differentially expressed immune genes of lung adenocarcinoma. BMC Cancer 21:213. 10.1186/s12885-021-07911-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Jin X., Li J., Tian Y., Wang Q., Li X., et al. (2021). CeRNASeek: an R package for identification and analysis of ceRNA regulation. Brief Bioinform. 22:bbaa048. 10.1093/bib/bbaa048 [DOI] [PubMed] [Google Scholar]
- Zhao C., Wang Y., Jin H., Yu T. (2017). Knockdown of microRNA-203 alleviates LPS-induced injury by targeting MCL-1 in C28/I2 chondrocytes. Exp. Cell Res. 359 171–178. 10.1016/j.yexcr.2017.07.034 [DOI] [PubMed] [Google Scholar]
- Zheng S., Wang X., Fu Y., Li B., Xu J., Wang H., et al. (2021). Targeted next-generation sequencing for cancer-associated gene mutation and copy number detection in 206 patients with non-small-cell lung cancer. Bioengineered 12 791–802. 10.1080/21655979.2021.1890382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C. S., Feng M. T., Chen X., Gao Y., Chen L., Li L. D., et al. (2021). Exonuclease 1 (EXO1) is a potential prognostic biomarker and correlates with immune infiltrates in lung adenocarcinoma. Onco. Targets Ther. 14 1033–1048. 10.2147/OTT.S286274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Wang Y., Wang Y., Lei L., Ji M. H., Zhou G., et al. (2021). Predicting lung adenocarcinoma prognosis with a novel risk scoring based on platelet-related gene expression. Aging (Albany NY) 13 8706–8719. 10.18632/aging.202682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Z., Lin T., Luo L., Zhou W., Wen J., Huang H., et al. (2021). Exploring the mechanism of aidi injection for lung cancer by network pharmacology approach and molecular docking validation. Biosci. Rep. 41:BSR20204062. 10.1042/BSR20204062 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.