Abstract
Background
Host gene expression has emerged as a complementary strategy to pathogen detection tests for the discrimination of bacterial and viral infection. The impact of immunocompromise on host-response tests remains unknown. We evaluated a host-response test discriminating bacterial, viral, and noninfectious conditions in immunocompromised subjects.
Methods
An 81-gene signature was measured using real-time–polymerase chain reaction in subjects with immunocompromise (chemotherapy, solid-organ transplant, immunomodulatory agents, AIDS) with bacterial infection, viral infection, or noninfectious illness. A regularized logistic regression model trained in immunocompetent subjects was used to estimate the likelihood of each class in immunocompromised subjects.
Results
Accuracy in the 136-subject immunocompetent training cohort was 84.6% for bacterial versus nonbacterial discrimination and 80.8% for viral versus nonviral discrimination. Model validation in 134 immunocompromised subjects showed overall accuracy of 73.9% for bacterial infection (P = .04 relative to immunocompetent subjects) and 75.4% for viral infection (P = .30). A scheme reporting results by quartile improved test utility. The highest probability quartile ruled-in bacterial and viral infection with 91.4% and 84.0% specificity, respectively. The lowest probability quartile ruled-out infection with 90.1% and 96.4% sensitivity for bacterial and viral infection, respectively. Performance was independent of the type or number of immunocompromising conditions.
Conclusions
A host gene expression test discriminated bacterial, viral, and noninfectious etiologies at a lower overall accuracy in immunocompromised patients compared with immunocompetent patients, although this difference was only significant for bacterial infection classification. With modified interpretive criteria, a host-response strategy may offer clinically useful diagnostic information for patients with immunocompromise.
Keywords: host-pathogen interactions, gene expression profiling, immunocompromised host, molecular diagnostic techniques
This study evaluated the performance of a host gene expression test to discriminate bacterial, viral, and noninfectious illness in immunocompromised patients. Although test performance was marginally lower than in immunocompetent patients, this approach may provide useful diagnostic information in this high-risk population.
Difficulty distinguishing bacterial, viral, and noninfectious etiologies contributes to the excessive use of antibacterials, which drives antimicrobial resistance. Traditional pathogen-based diagnostic tests have low sensitivity, long time-to-result, require a priori suspicion of a pathogen, or fail to differentiate infection from colonization. The host response offers an alternative, unbiased diagnostic strategy that overcomes the limitations of pathogen-based strategies. Advances in transcriptomic technologies and machine learning enabled the identification of a number of host-based gene expression signatures that differentiate bacterial from viral infection [1–10]. However, the cohorts included insufficient numbers of immunocompromised subjects to confidently draw conclusions about utility in this patient population. Only 1 study reported sufficient numbers of immunocompromised patients, children with febrile neutropenia, but failed to identify a discriminating host gene expression signature [11]. How these findings relate to adults and to different types of immunocompromise remains unknown.
Identifying the presence and cause of infection is more challenging in immunocompromised patients. They are susceptible to a wider array of potential pathogens, progress rapidly clinically, often require invasive procedures for early and accurate diagnosis, and are frequently colonized with potentially pathogenic organisms, making diagnosis even more difficult [12–14].
Given these considerations, it is important to understand what role host-based infectious disease diagnostics can have in immunocompromised patients. Host gene expression signatures rely on the host’s ability to mount an immune response, which may be muted or altered in immunocompromised patients in ways that affect signature performance. However, host gene expression diagnostic tests have been used successfully in immunocompromised subjects for noninfectious diseases such as allograft rejection [15] and in an immunocompromised murine model for the diagnosis of fungal infection [16], suggesting this strategy could complement traditional infectious disease diagnostics.
We previously published a microarray-derived transcriptomic signature for discriminating bacterial, viral, and noninfectious causes of illness with an overall accuracy of 87% [5]. These signatures were translated into a real-time polymerase chain reaction (PCR) test [17]. In this study, we validated the test’s ability to distinguish bacterial, viral, and noninfectious causes of illness in immunocompromised patients. We aimed to understand the effect of an immunocompromised state on the performance of a host-response test for infection and the possible role such a test might play in this patient population.
METHODS
Subject Enrollment
Subjects were enrolled prospectively for sample collection in the emergency departments (EDs) of 4 hospitals from 2006 through 2016. Additional details regarding subject enrollment can be found in the Supplementary Methods.
Clinical Adjudication and Subject Selection
Clinical adjudication served as the reference standard, which was performed after enrollment but prior to gene expression measurements. Adjudications were conducted by a team of multidisciplinary physicians who had full access to the medical record, as previously described [5, 17–19].
Immunocompromised subjects included those with human immunodeficiency virus (HIV) with a CD4 count less than 200 cells/µL, solid-organ transplant recipients, those actively receiving cancer chemotherapy, and those receiving immunomodulatory drugs (disease-modifying antirheumatic drugs, chronic steroids [10 mg of prednisone equivalent daily for >30 days], or acute high dose steroids [60 mg of prednisone equivalent for ≥3 days]). Only subjects adjudicated as having a microbiologically confirmed bacterial infection, microbiologically confirmed viral infection, or confirmed noninfectious illness (NI) were included in this study. More information regarding subject selection can be found in the Supplementary Methods.
Host Gene Expression Measurement
Host gene expression was measured as previously described [17, 20]. In brief, peripheral whole blood was collected in PAXgene Blood RNA tubes (Qiagen) at the time of enrollment. Total RNA was extracted followed by generation of a complementary DNA library. Semiquantitative, real-time PCR (RT-PCR) was performed on custom TaqMan low-density arrays (TLDAs) (Applied Biosystems). The TLDA cards were customized to quantify 81 gene targets (Supplementary Table 1) that were originally selected via microarray technologies to maximize performance accuracy as previously described [17].
Gene Expression–Based Classification Model
Normalized data from a training cohort of 136 immunocompetent subjects with confirmed bacterial infection, viral infection, or NI and no known immunocompromising condition were used to fit a regularized logistic regression model (lasso). This fixed-weight logistic regression model was then validated in a cohort of 134 subjects with at least 1 immunocompromising condition. More information regarding the gene expression–based classification model can be found in the Supplementary Methods.
Test Characteristics
Test sensitivity and specificity were assessed using a winner-take-all approach, where the highest independent probability determined the subject’s diagnosis. In this scenario, 3 probabilities were generated: probability of bacterial infection, probability of viral infection, and probability of NI. We also reported performance using thresholds defined by quartiles. The sensitivity and specificity for each quartile were calculated by omitting the number of subjects in that quartile from the numerator and dividing by all subjects. Higher quartiles have high specificities to rule in disease while lower quartiles have high sensitivities to rule out disease.
Statistical Analysis
The test was evaluated for its ability to distinguish between bacterial versus nonbacterial, viral versus nonviral, and noninfectious versus infectious illnesses. Test performance was evaluated by comparing area under the receiver operating characteristic (ROC) curve (AUC). The DeLong test was used to compare AUCs between the training (immunocompetent) and validation (immunocompromised) cohorts [21]. Kruskal-Wallis test was used to compare predicted probabilities based on type of immunocompromising condition. Mann-Whitney U test was used to compare test probabilities based on the number of immunocompromising conditions (1 vs >1). A Fisher’s exact test was used to compare overall test accuracy and test accuracy based on type and number (1 vs >1) across immunocompromising condition. P values < .05 were considered statistically significant. All analyses were performed in the R Environment for Statistical Computing version 3.5.0.
RESULTS
Clinical Cohorts
The training (immunocompetent) cohort included 136 subjects: 43 bacterial (32%), 52 viral (30%), and 41 NI (38%) (Table 1). The independent validation cohort included 134 immunocompromised subjects: 64 bacterial (48%), 28 viral (21%), and 42 NI (31%) (Supplementary Table 2). Both the immunocompetent and immunocompromised cohorts were demographically heterogenous, encompassing a wide age range and racial diversity (Table 2). The mean white blood cell (WBC) count in the immunocompromised cohort was 11.2 × 109 cells/L (SD, 7.4) compared with 10.8 × 109 cells/L (SD, 7.4) in the immunocompetent cohort (P = .62). Four subjects had absolute neutrophil counts <1.5 × 109 cells/L.
Table 1.
Subject Demographic Characteristics
| No. of Subjects | Gender (M/F), n/n | Mean (Range) Age, Years | Race (B/W/O), n/n/n | |
|---|---|---|---|---|
| Immunocompetent cohort | 136 | 66/68 | 50 (14–94) | 69/58/9 |
| Bacterial | 43 | 23/20 | 50 (16–94) | 22/19/2 |
| Viral | 41 | 17/34 | 49 (14–88) | 28/19/5 |
| Noninfectious illness | 52 | 26/14 | 50 (21–87) | 19/20/2 |
| Immunocompromised cohort | 134 | 68/66 | 53 (19–80) | 50/81/3 |
| Bacterial | 64 | 31/33 | 53 (19–79) | 28/34/2 |
| Viral | 28 | 12/16 | 53 (25–80) | 9/18/1 |
| Noninfectious illness | 42 | 25/17 | 53 (26–74) | 13/29/0 |
Abbreviations: B, Black/African American; F, female; M, male; O, other or unknown; W, White/Caucasian.
Table 2.
Number of Subjects per Immunocompromising Condition
| Bacterial, n | Viral, n | NI, n | Total, n (%) | |
|---|---|---|---|---|
| Immunomodulatory agent | 21 | 16 | 22 | 59 (44.0) |
| Solid-organ transplant | 25 | 5 | 8 | 38 (28.4) |
| Chemotherapy | 14 | 5 | 9 | 28 (20.9) |
| HIV/AIDS | 4 | 2 | 3 | 9 (6.7) |
| Total | 64 | 28 | 42 | 134 |
| Proportion, % | 47.8 | 20.8 | 31.3 | … |
All patients with HIV/AIDS had a CD4 count <200 cells/mm3. Immunomodulatory agents included disease modifying antirheumatic drugs, chronic steroids (>10 mg of prednisone equivalent daily for >30 days), and acute high-dose steroids (60 mg prednisone equivalent for ≥3 days).
Abbreviations: HIV, human immunodeficiency virus; NI, noninfectious illness.
Assessing Technical Variation
Data for the immunocompetent and immunocompromised cohorts were generated in 2 separate batches, potentially leading to batch effect. We therefore included 15 technical replicates from the training batch in the validation batch. Correlation was high for the 81 gene targets as measured in these 15 replicates (median Pearson correlation coefficient of 0.96). Normalized expression for each gene was similar in the immunocompetent and immunocompromised groups (Supplementary Figure 1).
Differentially Expressed Genes
In the bacterial versus nonbacterial model, the immunocompetent and immunocompromised cohorts demonstrated 57 and 19 differentially expressed genes (DEGs), respectively (Figure 1). Three genes (TNFAIP6, CCDC19, CHI3L1) were differentially expressed in the immunocompromised cohort but not in the immunocompetent cohort. In the viral model, there were 65 DEGs in the immunocompetent training data, which included all 26 DEGs in the immunocompromised cohort (Figure 1). Although there were fewer DEGs in the immunocompromised cohort relative to the immunocompetent cohort, the specific DEGs were conserved. Differential expression of each target gene stratified by adjudicated phenotype demonstrated phenotype-specific differences for many genes (Figure 2), particularly interferon-stimulated genes among subjects with viral infection.
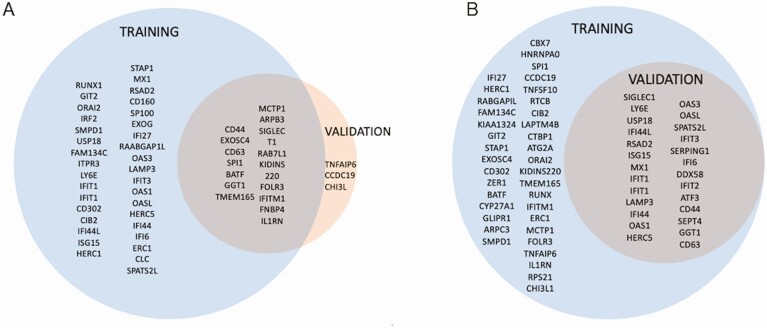
Figure 1.
Differentially expressed genes in bacterial (A) and viral (B) models. A, Of the 81 gene targets measured, there were 57 and 19 differentially expressed genes in the bacterial model for the immunocompetent and immunocompromised cohorts, respectively. B, There were 63 and 26 differentially expressed genes in the viral model for the immunocompetent and immunocompromised cohorts, respectively.
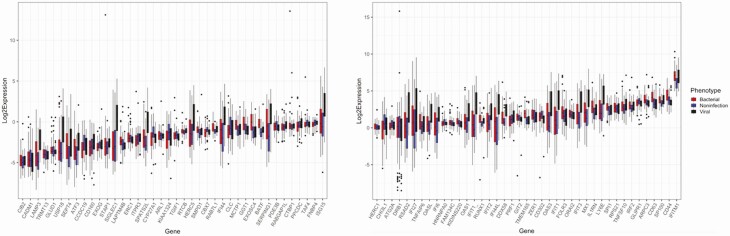
Figure 2.
Differential expression stratified by adjudicated phenotype. Post-normalization differential expression of each gene target in the signature stratified by adjudicated phenotype shows phenotype-specific differences for some genes. Gene targets were split across 2 graphs ordered by mean normalized expression values across all 3 phenotypes to facilitate visualization of phenotype-specific differences.
Training in Immunocompetent Subjects
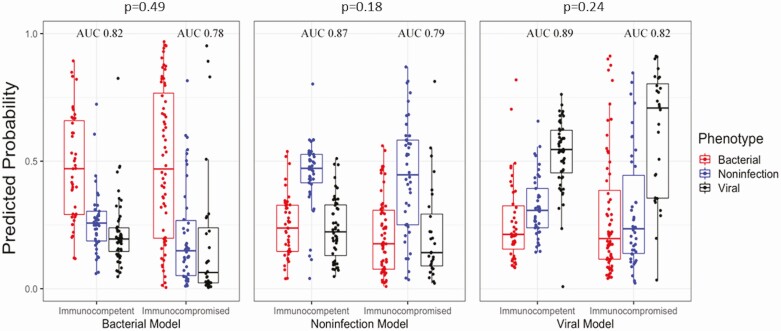
We first trained and evaluated performance of a logistic regression model in the immunocompetent training cohort. Performance in this training cohort was previously reported [17]. However, 15 subjects in this previously published training cohort had immunocompromising conditions and were therefore removed from model training. They were instead included in the immunocompromised validation cohort. Due to this change in the training cohort composition, we trained a new model and assessed performance using nested leave-one-out cross-validation (LOOCV). Each of the 3 models distinguished the specified group from the 2 alternative etiologies (Figure 3). The AUCs, sensitivities, and specificities using a winner-takes-all approach were .82 (95% confidence interval [CI], .73–.90), 65.1% (95% CI, 50.9–79.4%), and 93.5% (95% CI, 88.6–98.5%) for bacterial infection; .89 (95% CI, .83–.95), 84.6% (95% CI, 74.8–94.4%), and 78.6% (95% CI, 69.8–87.3%) for viral infection; and .87 (95% CI, .79–.95), 73.1% (95% CI, 59.6–86.7%), and 89.5% (95% CI, 83.3–95.6) for NI, respectively (Table 3).
Figure 3.
Illness etiology probabilities. These boxplots report predicted probabilities based on a fixed-weight model trained on the immunocompetent cohort. The distribution of probabilities for 3 models is shown: bacterial vs nonbacterial (left), noninfectious vs others (center), and viral vs nonviral (right). For each model, subjects adjudicated as having bacterial infection (red), noninfectious illness (blue), or viral infection (black) are displayed. All subjects (136 in the immunocompetent cohort, 134 in the immunocompromised cohort) are represented in each of the 3 models. P values are reported for the comparison of overall accuracy between immunocompetent and immunocompromised subjects. Abbreviation: AUC, area under the curve.
Table 3.
Performance Characteristics in the Training (Immunocompetent) and Validation (Immunocompromised) Cohorts
| Test Group | AUC (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | Overall Accuracy, % | LR+ (95% CI) | LR− (95% CI) |
|---|---|---|---|---|---|---|
| Immunocompetent cohort | ||||||
| Bacterial infection | .82 (.73–.90) | 65.1 (50.9–79.4) | 93.5 (88.6–98.5) | 84.5 | 10.09 (4.52–22.56) | .37 (.25–.56) |
| Viral infection | .89 (.83–.95) | 84.6 (74.8–94.4) | 78.6 (69.8–87.3) | 80.9 | 3.95 (2.58–6.04) | .20 (.10–.37) |
| Immunocompromised cohort | ||||||
| Bacterial infection | .78 (.71–.86) | 62.5 (50.6–74.4) | 84.3 (75.8–92.8) | 73.9 | 3.98 (2.24–7.07) | .44 (.32–.62) |
| Viral infection | .82 (.72–.90) | 78.6 (63.4–93.8) | 74.5 (66.2–82.8) | 75.4 | 3.08 (2.11–4.51) | .29 (.14–.59) |
| Combined cohort | ||||||
| Bacterial infection | .82 (.77–0.87) | 80.4 (72.3–87.9) | 80.4 (74.3–86.4) | 80.4 | 4.10 (2.96–5.66) | .24 (.17–.36) |
| Viral infection | .85 (.80–.90) | 66.3 (55.9–76.6) | 91.1 (87.0–95.1) | 83.4 | 7.45 (4.58–11.96) | .37 (.27–.51) |
| Immunocompetent subgroup | ||||||
| Bacterial infection | … | 86.0 (75.7–96.4) | 83.9 (76.4–91.3) | 84.6 | 5.34 (3.31–8.61) | .17 (.08–.35) |
| Viral infection | … | 69.2 (56.7–81.8) | 95.2 (90.7–99.8) | 85.3 | 14.42 (5.49–38.48) | .32 (.21–.49) |
| Immunocompromised subgroup | ||||||
| Bacterial infection | … | 76.6 (66.2–86.9) | 75.7 (65.7–85.8) | 76.1 | 3.2 (2.04–4.87) | .31 (.19–.49) |
| Viral infection | … | 60.7 (42.6–78.8) | 87.8 (81.5–94.0) | 81.3 | 4.95 (2.74–8.93) | .44 (.28–.71) |
Performance metrics for the immunocompetent cohort and combined cohort were obtained using LOOCV. Values for the immunocompromised cohort were based on parameters and thresholds set in the immunocompetent cohort.
Abbreviations: AUC, area under the curve; CI, confidence interval; LOOCV, leave-one-out cross-validation; LR+, likelihood ratio positive; LR−, likelihood ratio negative.
Validation in Immunocompromised Subjects
This model, trained on the immunocompetent subjects, was then applied to the 134-subject immunocompromised validation cohort. The AUCs, sensitivities, and specificities were .78 (95% CI, .71–.86), 62.5% (95% CI, 50.6–74.4%), and 84.3% (95% CI, 75.8–92.8%) for bacterial infection; .82 (95% CI, .72–.90), 78.6% (95% CI, 63.4–93.8%), and 74.5% (95% CI, 66.2–82.8%) for viral infection; and .79 (95% CI, .71–.88), 57.1% (95% CI, 42.2–72.1%), and 89.1% (95% CI, 82.3–95.5%) for NI, respectively (Table 3, Figure 3). Although the immunocompromised cohort demonstrated lower AUCs compared with the immunocompetent cohort, AUCs were not significantly different (P = .49 for bacterial infection, P = .24 for viral infection, P = .18 for NI). However, the lower overall accuracy for bacterial infection in immunocompromised versus immunocompetent subjects was statistically significant (73.9% vs 84.5%, P = .04). Differences in accuracy between the immunocompetent and immunocompromised groups were not significant for viral infection (80.9% vs 75.4%, P = .30) and NI (84.5% vs 79.1%, P = .27) (Table 3).
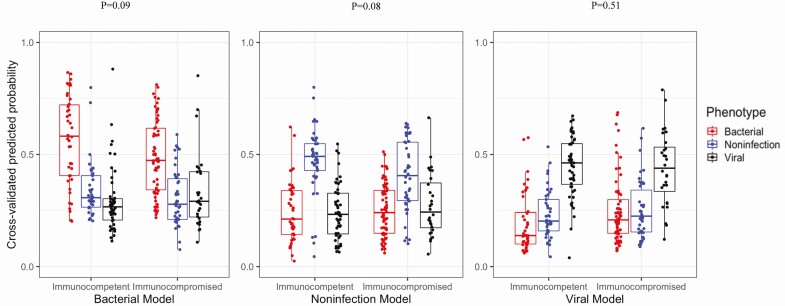
This lower performance in subjects with immunocompromising conditions could have been due to technical variability between batches. To assess this possibility, we combined the immunocompetent and immunocompromised subjects into a single cohort and classified subjects using LOOCV, which was then stratified by immune status. As shown in Figure 4, the host gene expression test discriminated bacterial, viral, and NI etiologies with AUC values of .82 (95% CI, .77–.87), .85 (95% CI, .80–.90), and .84 (95% CI, .78–.90), respectively (Table 3). Performance was lower in the immunocompromised cohort and neared statistical significance for bacterial infection (84.6% vs 76.1%, P = .09) and for the NI group (86.0% vs 77.6%, P = .08).
Figure 4.
LOOCV on combined immunocompromised and immunocompetent cohort. These boxplots report predicted probabilities from LOOCV on a single cohort of 136 immunocompetent and 134 immunocompromised subjects. The distribution of probabilities for 3 models is shown: bacterial vs nonbacterial (left), noninfectious vs others (center), and viral vs nonviral (right). For each model, subjects adjudicated as having bacterial infection (red), noninfectious illness (blue), or viral infection (black) are displayed. Abbreviation: LOOCV, leave-one-out cross-validation.
Impact of Immunocompromising Condition
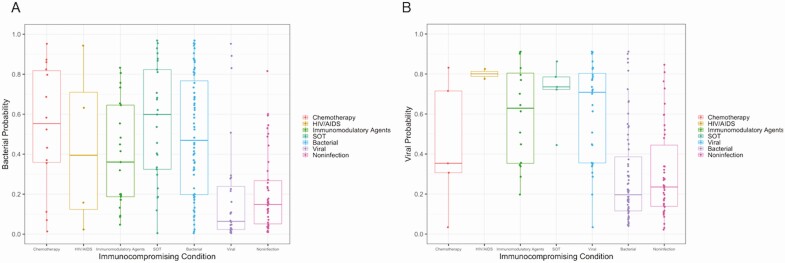
To assess if test performance varied by the type or number of immunocompromising conditions, we stratified predictions by these parameters. As shown in Figure 5, there was a large spread of predicted probabilities within each category of immunocompromising condition, such that no statistically significant differences were observed. Samples were unavailable to functionally measure immune response. We, therefore, used the number of immunocompromising conditions as a proxy for the severity of immunocompromise. However, there were no differences in bacterial or viral diagnostic accuracy based on the number of immunocompromising conditions (bacterial: 75.2% for 1 condition vs 69.0% for multiple immunocompromising conditions, P = .48; viral: 74.3% for 1 condition vs 79.3% for multiple immunocompromising conditions, P = .64).
Figure 5.
Bacterial (A) and viral (B) probabilities by type of immunocompromising condition. Predicted probabilities for clinical subgroups in the immunocompromised validation cohort. The boxplots indicate median (horizontal line), interquartile ranges (top–bottom box borders), and the whiskers extend to the highest and lowest predictions. In panel A, all bacterial subjects are plotted by type of immunocompromising condition while all subjects adjudicated as viral or noninfectious are listed in the respective columns on the right side of the graph. In panel B, all viral subjects are plotted by type of immunocompromising condition while all subjects adjudicated as bacterial or noninfectious are listed in the respective columns on the right side of the graph. Abbreviations: HIV, human immunodeficiency virus; SOT, solid-organ transplant.
Performance by Quartile
A winner-take-all scheme uses the highest predicted probability to assign class. Since we report results for 3 models, the opportunities for errors increase, especially in comparison to schemes that only have 2 classes. Therefore, we re-calculated performance characteristics using test quartiles. Specifically, this scheme provided greater ability to rule in or rule out bacterial and viral infection for subjects in the highest and lowest quartiles, respectively. The lowest quartile for bacterial infection had sensitivities of 95.3% and 90.1% in the immunocompetent and immunocompromised cohorts, respectively (Table 4). The highest quartile for bacterial infection had specificities of 91.4% (positive likelihood ratio [+LR], 4.87) and 91.4% (+LR, 6.51) in the immunocompetent and immunocompromised cohorts, respectively. Performance was similarly high in cases of viral infection. Quartile 1 for viral infection had sensitivities of 98.1% (negative LR [−LR], 0.03) and 96.4% (−LR, 0.05) in the immunocompetent and immunocompromised cohorts, respectively. Quartile 4 for viral infection had specificities of 95.2% (+LR, 8.81) and 84.0% (+LR, 2.45) in the immunocompetent and immunocompromised cohorts, respectively.
Table 4.
Test Statistics per Quartile
| Quartile | Bacterial | Non-bacterial | Likelihood Ratio | Test Purpose | Sensitivity in Quartile | Specificity in Quartile |
|---|---|---|---|---|---|---|
| A: Immunocompetent cohort, bacterial vs nonbacterial | ||||||
| Quartile 1 (lowest) | 2 | 32 | 0.07 | Rule out | 95.3 | |
| Quartile 2 | 4 | 30 | 0.15 | Rule out | 90.1 | |
| Quartile 3 | 12 | 23 | 2.92 | Rule in | 75.3 | |
| Quartile 4 (highest) | 25 | 8 | 4.87 | Rule in | 91.4 | |
| B: Immunocompromised cohort, bacterial vs nonbacterial | ||||||
| Quartile | Bacterial | Non-bacterial | Likelihood Ratio | Test Purpose | Sensitivity in Quartile | Specificity in Quartile |
| Quartile 1 (lowest) | 6 | 28 | 0.17 | Rule out | 90.1 | |
| Quartile 2 | 12 | 21 | 0.27 | Rule out | 81.3 | |
| Quartile 3 | 18 | 15 | 3.33 | Rule in | 78.6 | |
| Quartile 4 (highest) | 28 | 6 | 6.51 | Rule in | 91.4 | |
| C: Immunocompetent cohort, viral vs nonviral | ||||||
| Quartile | Viral | Nonviral | Likelihood Ratio | Test Purpose | Sensitivity in Quartile | Specificity in Quartile |
| Quartile 1 (lowest) | 1 | 33 | 0.03 | Rule out | 98.1 | |
| Quartile 2 | 4 | 30 | 0.12 | Rule out | 92.3 | |
| Quartile 3 | 17 | 17 | 3.32 | Rule in | 79.7 | |
| Quartile 4 (highest) | 30 | 4 | 8.81 | Rule in | 95.2 | |
| D: Immunocompromised cohort, viral vs nonviral | ||||||
| Quartile | Bacterial | Non-bacterial | Likelihood Ratio | Test Purpose | Sensitivity in Quartile | Specificity in Quartile |
| Quartile 1 (lowest) | 1 | 33 | 0.05 | Rule out | 96.4 | |
| Quartile 2 | 1 | 33 | 0.05 | Rule out | 96.4 | |
| Quartile 3 | 9 | 23 | 3.08 | Rule in | 78.3 | |
| Quartile 4 (highest) | 17 | 17 | 2.45 | Rule in | 84.0 | |
Results are presented for the bacterial vs nonbacterial model in the immunocompetent cohort (A) and immunocompromised cohort (B). Results for the viral vs nonviral model are presented for the immunocompetent (C) and immunocompromised (D) cohorts. Since the purpose of the lower quartiles (1 and 2) is to “rule out” the specified condition, only sensitivity is reported. The sensitivity and specificity for each quartile were calculated by omitting the number of subjects in that quartile from the numerator and dividing by all subjects. Higher bands have high specificities to rule in disease while lower bands have high sensitivities to rule out disease. For example, the sensitivity in quartile 2 of Table 8a is calculated by (2 + 12 + 25)/(2 + 4 + 12 + 25) and the specificity in quartile 3 of Table 8a is calculated by (32 + 30 + 8)/(32 + 30 + 23 + 8).
DISCUSSION
Difficulty differentiating bacterial, viral, and noninfectious disease leads to diagnostic uncertainty and antibiotic overuse. Beyond pathogen-focused diagnostics, the host response offers a complementary strategy to identify the presence of infection and its underlying cause. Numerous host transcriptomic signatures for bacterial, viral, fungal, mycobacterial, and parasitic infections have been defined [1–10, 22], but none of these studies included significant numbers of immunocompromised subjects. In our prior research, we developed a host-response signature that discriminates bacterial and viral infection with an overall accuracy of 87%. In this study, we applied our previously developed host-response test to a heterogenous cohort of immunocompromised subjects and classified them by their underlying bacterial, viral, or noninfectious gene expression patterns. We compared these results with clinically adjudicated phenotypes. The results showed that host gene expression accurately discriminated bacterial, viral, and noninfectious etiologies of illness in the immunocompromised host, but at a lower accuracy than in immunocompetent patients. These differences could be overcome by alternative reporting schemes such as the use of quartiles. These results suggest that a host-response strategy may offer clinically useful and complementary diagnostic information for immunocompromised patients.
Prior studies showed that host-based diagnostics are applicable to immunocompromised populations and can guide antimicrobial stewardship in this patient population [23]. Procalcitonin, a widely used biomarker to differentiate bacterial and nonbacterial etiologies of illness, can be useful for infectious disease diagnosis in immunocompromised patients [24]. Pro-adrenomedullin (pro-ADM) and urinary chemokines may also be useful diagnostic biomarkers [24, 25]. A recent bacterial versus viral mRNA signature was evaluated in a cohort that included a small number of immunocompromised subjects (n = 31), but observed no difference compared with immunocompetent subjects [26]. Other host gene expression tests—for example, Allomap—have been successfully developed and implemented for noninfectious conditions such as acute cellular rejection in cardiac transplant patients [27]. We could only identify 1 prior study focused specifically on host gene expression in immunocompromised patients with infections. Wahlund et al [11] studied 63 children with febrile neutropenia but was unable to identify a discriminating signature. Several reasons could explain this, including their focus on neutropenia, which may have resulted in too severe a derangement to support a host gene expression approach; a comparatively small sample size; or sample collection after a diagnosis was made (after treatment had already begun).
In this study, the only statistically significant difference due to immune status that we observed in this test was in the diagnosis of bacterial infection. However, performance was generally lower in the immunocompromised cohort and likely failed to reach statistical significance due to sample size. Reasons for this difference may be technical in nature. For example, test performance is typically lower in independent validation than in the training cohort. Training a model on all subjects together should overcome this issue, yet it did not resolve the lower performance seen in immunocompromised subjects, although the difference was not statistically significant. It is therefore more likely that biological differences between the immunocompromised and immunocompetent groups explain this lower performance. Since the source of the host response measured in this test is circulating WBCs, it is plausible that this response may be different in those with immunocompromise. Several genes, including some evaluated here, have been shown to be altered in immunocompromised patients [28–32]. For example, expression of MX1 was decreased in patients with systemic lupus erythematous after initiation of immunosuppressant therapy. Bergallo et al [32] identified a 6-gene host signature (all of which were represented in this study) that remained useful in immunocompromised subjects but only if the weight assigned to 3 particular genes (IFI27, IFTI1, ISG15) was limited. We also investigated whether there were any common features among subjects who tended to have incorrect or low predicted probabilities, although no pattern could be identified.
The lower overall performance in the immunocompromised population can be largely mitigated through alternative reporting schemes. Rather than reporting results as positive or negative for every subject, indeterminate zones could be developed such that results in the lowest and highest quartiles offer the greatest diagnostic confidence. Results in the middle quartiles might still be useful but would be associated with greater uncertainty. There is precedent for this type of stratified reporting for other host-response signatures [26, 33–36].
One limitation of this study is that it did not assess host gene expression response to fungal infections, which is an important pathogen class in immunocompromised patients [37]. Additionally, the gene expression measurements were limited to the 81 genes included on a custom TLDA assay. This limits our ability to identify additional DEGs that might be unique to this population and would subsequently offer a better test. Even if an optimized signature could be found in this population, it becomes impractical to develop and use different host-response tests for different populations. Ideally, a single signature would be broadly applicable to both immunocompromised and immunocompetent patients. Given this desired application, we originally identified this signature in a biologically heterogeneous discovery cohort that included immunocompromised subjects [5, 17]. In so doing, the effects of immunocompromise were accounted for by the model at the time of discovery. Another limitation is that we were unable to functionally assess immune status and relied on a compatible medical diagnosis or treatment to define immunocompromise. Despite this limitation, this mirrors actual clinical practice where a diagnosis of immunocompromise is based on underlying conditions or their treatments. Additionally, while a large cohort of immunocompromised subjects was studied here, subgroups were small, limiting the power to detect differences due to the specific types of immunocompromise, which are expected to have a highly variable impact on the immune system. Last, clinical adjudication is an imperfect reference standard, which explains at least some discrepancies with the host-response test.
The results of this study indicate that host gene expression offers a valuable, complementary diagnostic strategy for the immunocompromised population. Although the host-response test is far better than routinely ordered tests like WBC count or procalcitonin, it is not sufficiently accurate to stand independently of other diagnostic information. Used in conjunction with other clinical assessments, this host gene expression strategy could be a highly valuable diagnostic test. Further research with a larger sample size is required to understand what role host-based gene expression tests can play in various types of immunocompromise. Ideally, such studies would include functional immunological assessments and also evaluate the impact of fungal infection on the host response. Real-time determinations of clinical utility will be needed, which will be made more feasible with the development of point-of-need, host gene expression testing [38, 39].
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We are grateful for the contributions made by Christina Nix and Carolyne Whiting for their data-management support. We also acknowledge the contributions made by Anna Mazur and Pamela Isner in the laboratory.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. These funding sources had no role in the writing of the manuscript or the decision to submit it for publication. The corresponding authors had access to all the data in the study and bear final responsibility for the decision to submit for publication.
Financial support. This work was supported in part by the US Defense Advanced Research Projects. Agency (DARPA) (contract number N66001–09-C2082). This work was also supported in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number UM1AI104681. The work was also supported by the Eugene A. Stead Scholarship from Duke University School of Medicine and the Infectious Diseases Society of America Medical Scholars Program granted to R. E. M.
Potential conflict of interests. E. L. T., R. H., M. T. M., G. S. G., T. W. B., and C. W. W. have filed for a patent pertaining to the signatures discussed in this study (WO 2017/004390 A1). E. L. T., G. S. G., and C. W. W. are co-founders of Predigen, Inc. T. W. B. is a consultant for and holds equity in Predigen, Inc. E. L. T. reports patents pending for Biomarkers for the Molecular Classification of Bacterial Infection, Gene Expression Signatures Useful to Predict or Diagnose Sepsis and Methods of Using the Same, and Host-based Molecular Signatures of Human Infection with SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) (COVID-19 [coronavirus disease 2019]). G. S. G. reports patent US 14/214853 issued for Biomarkers for the Molecular Classification of Bacterial Infection. C. W. W. reports personal fees from bioMerieux and issued patents for Biomarkers for the Molecular Classification of Bacterial Infection and Methods of Identifying Infectious Disease and Assays for Identifying Infectious Disease. J. M. S. reports patents DU6795PROV and T-006820 pending for Transcriptional Signature of Aspergillus Infection and Transcriptional Signature of Candidemia Infection. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Bhattacharya S, Rosenberg AF, Peterson DR, et al. . Transcriptomic biomarkers to discriminate bacterial from nonbacterial infection in adults hospitalized with respiratory illness. Sci Rep 2017; 7:6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suarez NM, Bunsow E, Falsey AR, Walsh EE, Mejias A, Ramilo O. Superiority of transcriptional profiling over procalcitonin for distinguishing bacterial from viral lower respiratory tract infections in hospitalized adults. J Infect Dis 2015; 212:213–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramilo O, Allman W, Chung W, et al. . Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood 2007; 109:2066–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parnell GP, McLean AS, Booth DR, et al. . A distinct influenza infection signature in the blood transcriptome of patients with severe community-acquired pneumonia. Crit Care 2012; 16:R157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsalik EL, Henao R, Nichols M, et al. . Host gene expression classifiers diagnose acute respiratory illness etiology. Sci Transl Med 2016; 8:322ra11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herberg JA, Kaforou M, Wright VJ, et al. ; IRIS Consortium . Diagnostic test accuracy of a 2-transcript host RNA signature for discriminating bacterial vs viral infection in febrile children. JAMA 2016; 316:835–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sweeney TE, Perumal TM, Henao R, et al. . A community approach to mortality prediction in sepsis via gene expression analysis. Nat Commun 2018; 9:694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang BM, Shojaei M, Parnell GP, et al. . A novel immune biomarker IFI27 discriminates between influenza and bacteria in patients with suspected respiratory infection. Eur Respir J 2017; 49. [DOI] [PubMed] [Google Scholar]
- 9.Mejias A, Dimo B, Suarez NM, et al. . Whole blood gene expression profiles to assess pathogenesis and disease severity in infants with respiratory syncytial virus infection. PLoS Med 2013; 10:e1001549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heinonen S, Jartti T, Garcia C, et al. . Rhinovirus detection in symptomatic and asymptomatic children: value of host transcriptome analysis. Am J Respir Crit Care Med 2016; 193:772–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wahlund M, Sinha I, Broliden K, Saghafian-Hedengren S, Nilsson A, Berggren A.. The feasibility of host transcriptome profiling as a diagnostic tool for microbial etiology in childhood cancer patients with febrile neutropenia. Int J Mol Sci 2020; 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Timsit JF, Sonneville R, Kalil AC, et al. . Diagnostic and therapeutic approach to infectious diseases in solid organ transplant recipients. Intensive Care Med 2019; 45:573–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeNegre AA, Ndeffo Mbah ML, Myers K, Fefferman NH. Emergence of antibiotic resistance in immunocompromised host populations: a case study of emerging antibiotic resistant tuberculosis in AIDS patients. PLoS One 2019; 14:e0212969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Theodoropoulos N, Ison MG. Current issues in transplant infectious diseases. Curr Infect Dis Rep 2013; 15:453–4. [DOI] [PubMed] [Google Scholar]
- 15.Abbo LM, Ariza-Heredia EJ. Antimicrobial stewardship in immunocompromised hosts. Infect Dis Clin North Am 2014; 28:263–79. [DOI] [PubMed] [Google Scholar]
- 16.Steinbrink JM, Zaas AK, Betancourt M, Modliszewski JL, Corcoran DL, McClain MT. A transcriptional signature accurately identifies Aspergillus infection across healthy and immunosuppressed states. Transl Res 2020; 219:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lydon EC, Henao R, Burke TW, et al. . Validation of a host response test to distinguish bacterial and viral respiratory infection. EBioMedicine 2019; 48:453–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lydon EC, Bullard C, Aydin M, et al. . A host gene expression approach for identifying triggers of asthma exacerbations. PLoS One 2019; 14:e0214871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langley RJ, Tsalik EL, van Velkinburgh JC, et al. . An integrated clinico-metabolomic model improves prediction of death in sepsis. Sci Transl Med 2013; 5:195ra95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lydon EC, Ko ER, Tsalik EL. The host response as a tool for infectious disease diagnosis and management. Expert Rev Mol Diagn 2018; 18:723–38. [DOI] [PubMed] [Google Scholar]
- 21.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988; 44:837–45. [PubMed] [Google Scholar]
- 22.Sweeney TE, Wong HR, Khatri P. Robust classification of bacterial and viral infections via integrated host gene expression diagnostics. Sci Transl Med 2016; 8:346ra91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robilotti E, Holubar M, Seo SK, Deresinski S. Feasibility and applicability of antimicrobial stewardship in immunocompromised patients. Curr Opin Infect Dis 2017; 30:346–53. [DOI] [PubMed] [Google Scholar]
- 24.El Haddad H, Chaftari AM, Hachem R, Chaftari P, Raad II. Biomarkers of sepsis and bloodstream infections: the role of procalcitonin and proadrenomedullin with emphasis in patients with cancer. Clin Infect Dis 2018; 67:971–7. [DOI] [PubMed] [Google Scholar]
- 25.Safa K, Magee CN, Azzi J. A critical review of biomarkers in kidney transplantation. Curr Opin Nephrol Hypertens 2017; 26:509–15. [DOI] [PubMed] [Google Scholar]
- 26.Mayhew MB, Buturovic L, Luethy R, et al. . A generalizable 29-mRNA neural-network classifier for acute bacterial and viral infections. Nat Commun 2020; 11:1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang KC. Clinical utilities of peripheral blood gene expression profiling in the management of cardiac transplant patients. J Immunotoxicol 2007; 4:209–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng X, Sun T, Yao P, et al. . Differential expression of innate immunity regulation genes in chronic HIV-1 infected adults. Cytokine 2020; 126:154871. [DOI] [PubMed] [Google Scholar]
- 29.Gebremicael G, Kassa D, Alemayehu Y, et al. . Gene expression profiles classifying clinical stages of tuberculosis and monitoring treatment responses in Ethiopian HIV-negative and HIV-positive cohorts. PLoS One 2019; 14:e0226137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimizu Y, Yasuda S, Kimura T, et al. . Interferon-inducible Mx1 protein is highly expressed in renal tissues from treatment-naïve lupus nephritis, but not in those under immunosuppressive treatment. Mod Rheumatol 2018; 28:661–9. [DOI] [PubMed] [Google Scholar]
- 31.Massanella M, Singhania A, Beliakova-Bethell N, et al. . Differential gene expression in HIV-infected individuals following ART. Antiviral Res 2013; 100:420–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bergallo M, Ferrari L, Faolotto G, et al. . Interferon signature in immunosuppressed patients with lower respiratory tract infections: dosage on bronchoalveolar lavage. Minerva Med 2020; 111:245–53. [DOI] [PubMed] [Google Scholar]
- 33.Sweeney TE, Khatri P. Benchmarking sepsis gene expression diagnostics using public data. Crit Care Med 2017; 45:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sweeney TE, Shidham A, Wong HR, Khatri P. A comprehensive time-course-based multicohort analysis of sepsis and sterile inflammation reveals a robust diagnostic gene set. Sci Transl Med 2015; 7:287ra71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McHugh L, Seldon TA, Brandon RA, et al. . A molecular host response assay to discriminate between sepsis and infection-negative systemic inflammation in critically ill patients: discovery and validation in independent cohorts. PLoS Med 2015; 12:e1001916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oved K, Cohen A, Boico O, et al. . A novel host-proteome signature for distinguishing between acute bacterial and viral infections. PLoS One 2015; 10:e0120012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lockhart SR, Guarner J. Emerging and reemerging fungal infections. Semin Diagn Pathol 2019; 36:177–81. [DOI] [PubMed] [Google Scholar]
- 38.Tsalik EL, Khine A, Talebpour A, et al. . Rapid, sample-to-answer host gene expression test to diagnose viral infection. Open Forum Infect Dis 2019; 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsalik EL, Henao R, Aydin M, et al. . FilmArray® m easurement of host response signatures rapidly discriminates viral, bacterial, and non-infectious etiologies of illness. Open Forum Infect Dis 2018; 5(Suppl 1):S586–6. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.