Abstract
Feeding is essential for animal survival and reproduction and is regulated by both internal states and external stimuli. However, little is known about how internal states influence the perception of external sensory cues that regulate feeding behavior. Here, we investigated the neuronal and molecular mechanisms behind nutritional state-mediated regulation of gustatory perception in control of feeding behavior in the brown planthopper and Drosophila. We found that feeding increases the expression of the cholecystokinin-like peptide, sulfakinin (SK), and the activity of a set of SK-expressing neurons. Starvation elevates the transcription of the sugar receptor Gr64f and SK negatively regulates the expression of Gr64f in both insects. Interestingly, we found that one of the two known SK receptors, CCKLR-17D3, is expressed by some of Gr64f-expressing neurons in the proboscis and proleg tarsi. Thus, we have identified SK as a neuropeptide signal in a neuronal circuitry that responds to food intake, and regulates feeding behavior by diminishing gustatory receptor gene expression and activity of sweet sensing GRNs. Our findings demonstrate one nutritional state-dependent pathway that modulates sweet perception and thereby feeding behavior, but our experiments cannot exclude further parallel pathways. Importantly, we show that the underlying mechanisms are conserved in the two distantly related insect species.
Author summary
Food intake is critical for animal survival and reproduction and is regulated both by internal states that signal appetite or satiety, and by external sensory stimuli. It is well known that the internal nutritional state influences the strength of the chemosensory perception of food signals. Thus, both gustatory and olfactory signals of preferred food are strengthened in hungry animals. However, the molecular mechanisms behind satiety-mediated modulation of taste are still not known. We show here that cholecystokinin-like (SK) peptide in brown planthopper and Drosophila signals satiety and inhibits sugar attraction by lowering the activity of sweet-sensing gustatory neurons and transcription of a sugar receptor gene, Gr64f. We show that SK peptide signaling reflects the nutritional state and inhibits feeding behavior. Re-feeding after starvation increases SK peptide expression and spontaneous activity of SK producing neurons. Interestingly, we found that SK peptide negatively regulates the expression of the sweet gustatory receptor and that activation of SK producing neurons inhibits the activity of sweet-sensing gustatory neurons (GRNs). Furthermore, we found that one of the two known SK peptide receptors is expressed in some sweet-sensing GRNs in the proboscis and proleg tarsi. In summary, our findings provide a mechanism that is conserved in distantly related insects and which explains how feeding state modulates sweet perception to regulate feeding behavior. Thus, we have identified a neuropeptide signal and its neuronal circuitry that respond to satiety, and that regulate feeding behavior by inhibiting gustatory receptor gene expression and activity of sweet sensing GRNs.
Introduction
Neuronal control of feeding is interesting for at least two reasons: in the human population there is a growing problem with excess food consumption causing obesity and associated severe health problems, and secondly, pest insects consume large amounts of our crops worldwide. Both problems are very costly to society. Therefore, understanding mechanisms behind regulation of food search, feeding and satiety is of great interest. In general, animal behavior is guided by internal states and external stimuli [1–5]. Hence, behavioral decisions depend on the integration of signals from the internal and external environment by circuits of the brain. For instance, feeding behavior, which is essential for the survival and reproduction of animals, is regulated by the nutritional state of the organism and depends on the efficacy of chemosensory organs in localizing food in the environment [3,6–12]. Thus, internal nutrient sensors monitor the energy homeostasis and signal hunger or satiety to the nervous system. Hunger signals received by brain circuits are relayed to sense organs to increase their sensitivity. Hence, in a hungry animal the sensory threshold is lowered in olfactory and gustatory receptors that respond to food cues and thereby increases appetitive behavior and food seeking [6,10–13]. Concomitantly, the hunger signals increase the detection threshold for aversive stimuli, such as bitter tastants [11,14]. After food ingestion, satiety signals lower the attractive sensory thresholds and also act on neuronal circuits that regulate feeding behavior, thereby stopping further food intake [12,15].
In Drosophila and other insects, the modulation of sensory gain in appetitive behavior is largely dependent on neuropeptides and peptide hormones [4,12,15–17]. These signaling systems also orchestrate animal behavior and link internal and external sensory cues [see [17–19]]. Thus, several neuropeptides are known to trigger appetitive behavior, foraging, and mobilize energy stores, at the same time as they suppress other conflicting behaviors such as sleep and reproductive behavior [see [17–20]]. At the sensory level neuropeptides such as short neuropeptide F (sNPF), myoinhibitory peptide (MIP), CCH2amide and tachykinin (TK) regulate the sensitivity of subpopulations of olfactory receptor neurons (ORNs) to promote food seeking in hungry flies [6,10,21–24]. Also gustatory neurons in Drosophila are modulated by neuropeptides to regulate sugar and bitter sensitivities [11]. Hence, in hungry flies neuropeptide F (NPF), via dopamine cells, increases sweet sensitivity in Gr5a expressing cells and sNPF decreases bitter sensitivity [11]. NPF and sNPF are also known to regulate aspects of feeding, metabolism and sleep [25–32], suggesting action at several levels of the organism. Another neuropeptide known to regulate food intake in Drosophila is drosulfakinin, DSK, related to the mammalian peptide cholecystokinin (CCK) [33]. CCK in mammals and sulfakinins (SKs) in insects, including Drosophila, signal satiety and decrease food ingestion [34–38]. DSK has furthermore been reported to modulate aggression and courtship behavior in Drosophila [39–41]. The first insect SK was identified from head extracts of the cockroach, Rhyparobia maderae [42]. Characteristic of SKs is that it contains a sulfated tyrosine residue (DY(SO3)GHM/LRFamide) [43]. In a variety of insects, such as the desert locust, the German cockroach, and the red flour beetle, SK significantly reduced food intake [44–46]. SK is also known to inhibit the activity of digestive enzymes in the migratory locust and brown planthopper [35,47].
In Drosophila, upon feeding, DSK is released and the meal is terminated [12,15,33]. However, it is not known how DSK acts to decrease feeding. The mechanisms controlling sweet gustation and feeding in insects are remarkably similar to those in vertebrates. Sweet-sensing neurons are housed in taste sensilla in the labellum on the proboscis (the insect equivalent of the vertebrate tongue), the tarsal segments of the legs, and the pharynx [48–50]. In Drosophila, nine types of gustatory receptors (Gr) participate in sweet sensation. One of them, Gr64f, is a co-receptor that is required in combination with other gustatory receptors for sugar detection in Drosophila [49–51]. However, the mechanisms behind how the feeding state modulates sugar receptor gene expression and sweet-sensing neurons activity are poorly known.
We chose to study SK signaling in regulation of gustatory input and feeding behavior in two insect species, the brown planthopper Nilaparvata lugens and the genetic model insect Drosophila melanogaster. The brown planthopper is a serious pest on rice in Asia and causes damage costing more than 300 million US dollars annually [52]. N. lugens is a monophagous pest that pierce the rice stem and sucks sap, thereby transferring a virus [53] that destroys the plant [54]. With the magnificent genetic toolbox available, Drosophila has been extensively used as a model to study regulation of feeding, metabolism and sensory mechanisms underlying food seeking [see [4,12,13,15,17,55]]. Thus, we combine two insect species in our quest to understand how a neuropeptide regulates gustatory perception of food-related taste and ensuing initiation of food ingestion.
Using N. lugens in an RNA-seq transcriptome screen for altered gene expression after downregulation of SK, we found among others an upregulation of sweet-sensing gustatory receptors and the takeout (to) gene. Analysis of manipulations of SK and its receptor SKR, as well as gustatory receptors and to, suggests that feeding-induced SK signaling downregulates sweet sensing receptors and that to is a mediator of SK signaling in the planthopper. Further experiments in Drosophila unravel mechanisms behind the satiety signaling that modulates gustatory neurons. Feeding upregulates Dsk transcription and increases spontaneous activity and calcium signaling in DSK expressing median protocerebrum (MP) neurons. Furthermore, feeding downregulates Gr64f expression and starvation increases Gr64f. Optogenetic activation of Gr64f neurons increases the motivation to feed. In addition to this, knockdown of dsk leads to an upregulation of Gr64f transcription and activation DSK expressing MP neurons inhibit the sensitivity of gustatory neurons. Remarkably, we found expression of the DSK receptor CCKLR-17D3 in a subpopulation of the Gr64f GRNs in proleg tarsi, proboscis and maxillary palps and this receptor is downregulated in the appendages after feeding. Finally, knockdown of 17D3 in sweet sensing GRNs decreases the PER. Thus, in summary DSK signaling modulates the sensitivity of sweet-sensing GRNs in a nutrient-dependent fashion in both the planthopper and Drosophila, suggesting a conserved peptidergic signal pathway in these distantly related insects. It should be emphasized that our experiments cannot exclude further components in the state-dependent regulation of gustatory sensitivity, including contributions of other neuropeptides or neuromodulators, such as those indicated earlier in this introduction.
Results
Sulfakinin (SK) reduces food intake in N. lugens
In several insect species, SK is known to inhibit feeding and act as a satiety factor [33–35,44–46,56,57]. To test whether SK induces satiety also in N. lugens, we injected 4th instar nymph with 20 pmol of either of four types of N. lugens sulfakinins (NlSKs), namely nsNlSK1 (non-sulfated SK1), sNlSK1 (sulfated SK1), nsNlSK2 (non-sulfated SK2) and sNlSK2 (sulfated SK2), dissolved in PBS. After 24h we compared the food uptake to a control group, which was injected with PBS. N. lugens injected with sNlSK1 or sNlSK2 consumed 50%-70% less food than the PBS-injected control. However, non-sulfated SKs (nsNlSK1 and nsNlSK2) had no impact on the feeding behavior of N. lugens (Fig 1A).
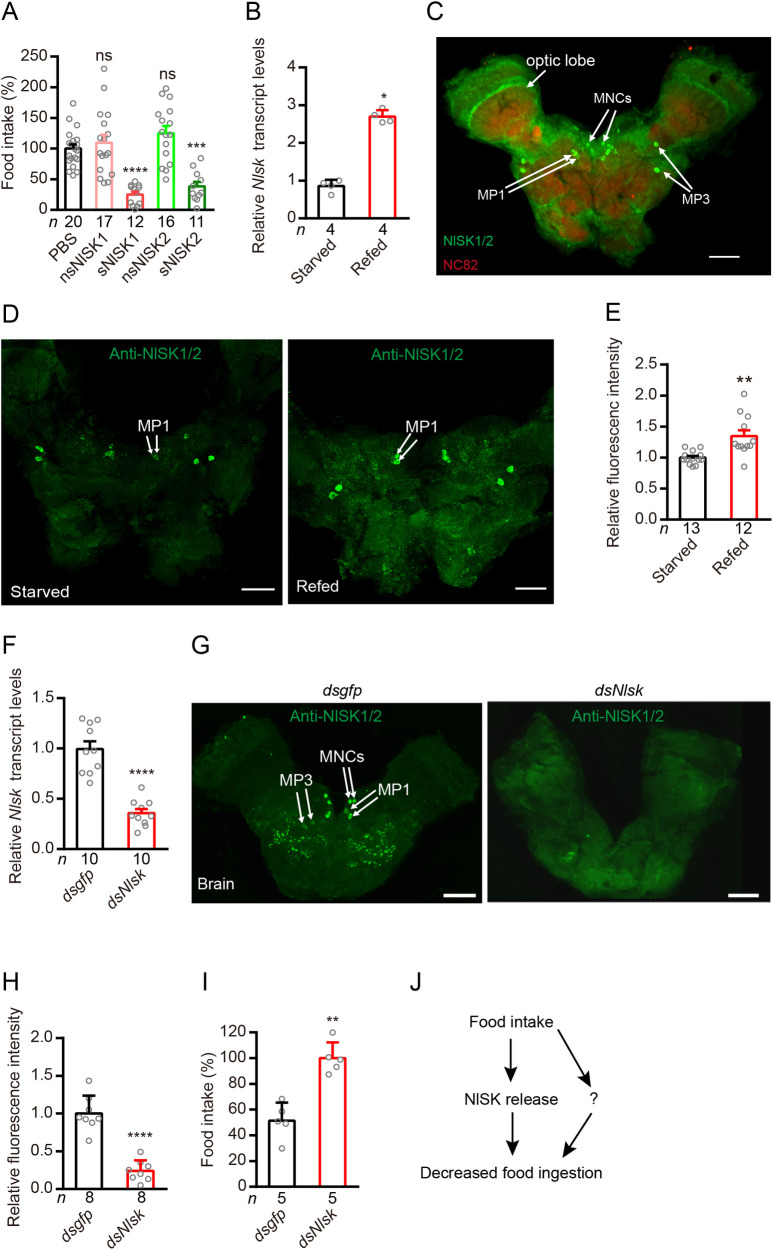
Fig 1. Sulfakinin (NlSK) and its receptor (SKR) signal satiety and inhibit feeding in the rice planthopper.
(A) Injection of N. lugens sulfated sulfakinins inhibits food intake in the brown planthopper. Fifty nanoliter of PBS (as control) and four sulfakinins (20 pmol/insect) were injected into 4rd instar nymph of brown planthopper. Non-sulfated Sulfakinins (nsNlSK1 and 2) have no effect. All data are presented as means ± s.e.m in this manuscript. ns: not significant, ***p < 0.001, ****p < 0.0001; One-way ANOVA followed by Tukey’s multiple comparisons test. (B) Refeeding for 5 hr after 24 hr starvation increases Nlsk mRNA transcript. *p < 0.05; Mann–Whitney test. (C) NlSK expression in the brain is revealed by anti-NlSK1/2 (green) and anti-nc82 (red). Two pairs of MP1 (medial protocerebrum), two pairs of MP3 neurons, and median neurosecretory cells (MNCs) are indicated. Bar: 50 μm. (D and E) Refeeding after 24h starvation increases NlSK peptide expression as revealed by anti-NlSK1/2 immunolabeling. Scale bars, 50 μm. **p < 0.01; Student’s t test. Note that we measure SK levels in neuronal MP1 cell bodies where production occurs. (F) Downregulation of Nlsk gene using Nlsk-RNAi leads to a reduction in mRNA expression level. ****p < 0.0001; Student’s t test. (G and H) Downregulation of Nlsk gene using Nlsk-RNAi (dsNlsk) leads to a reduction in NlSK1/2 immunoreactivity. In (G) we show NlSK1/2 immunoreactivity in brains of dsgfp- or dsNlsk-injected planthoppers. Scale bar: 50 μm. (H) Intensity of anti-NlSK1/2 immunoreactivity in brain. All data are presented as means ± s.e.m. ****p < 0.0001; Student’s t test. (I) Downregulation of Nlsk gene using Nlsk-RNAi (dsNlsk) increases the food intake. **p < 0.01; Mann–Whitney test. (J) Simplified model showing that NlSK inhibits feeding. Note that this model and those in subsequent figures of planthopper data are highly simplified and serve to summarize our data, rather that presenting accurate signaling pathways. The question mark indicates one or more possibly additional signals.
Effects of Nlsk gene silencing on food intake
Next, we asked whether the gene expression level of Nlsk is affected by the satiety state. Indeed, we found that the Nlsk mRNA level is significantly higher in brown planthoppers that had been refed after 24 h starvation, compared to starved ones (Fig 1B). We furthermore examined the expression pattern of the NlSK peptides in the brain using anti-NlSK1/2 (Fig 1C). NlSK1/2 immunoreactivity was observed in the median neurosecretory cells (MNCs) as previously reported in Drosophila [33], as well as in two pairs of cells in the median protocerebrum (MP1 and MP3) also shown earlier in Drosophila [41,58]. We then asked whether refeeding leads to more NISK release from neurons. Our data showed that refeeding increases the immunolabeling intensity in the cell bodies of NlSK MP1 neurons compared with starved animals (Fig 1D and 1E). This might be explained by increased SK production accompanying the release at axon terminations. To determine the role of the Nlsk gene in feeding behavior, we performed RNAi injection experiments. The efficacy of the RNAi was tested by qPCR of whole animals and we found that the expression of Nlsk was significantly reduced (Fig 1F). The NlSK1/2 immunoreactivity in the brain was also significantly reduced by the RNAi (Fig 1G and 1H). Feeding experiments showed that the dsNlsk-injected planthoppers consumed approximately 2-fold more food than the dsgfp-injected controls (Fig 1I). Taken together, these results show that NlSK signaling reflects the nutritional state of the animal and inhibits feeding behavior (Fig 1J). However, we cannot rule out there are other neuromodulators that also play important roles in the satiety-induced regulation of food ingestion.
Global gene expression profiling of N. lugens in response to Nlsk gene-silencing
To unravel the molecular mechanisms underlying control of feeding behavior by Nlsk signaling, we performed transcriptome expression profiling (RNA-seq quantification) of N. lugens after Nlsk knockdown by dsNlsk injection. Illumina sequencing libraries were constructed by using mRNA from the dsNlsk- or dsgfp (control)-injected 4th instar brown planthoppers. We obtained 49,055,819.5 and 47,254,495 clean reads on average from samples of Nlsk–depleted and control groups, respectively (S2 Table). After removing low-quality regions, adapters, and possible contamination, we obtained more than 6 giga base clean bases with a Q20 percentage over 96%, Q30 percentage over 92%, and a GC percentage between 49.12 and 51.53% (S2 Table). After alignment by Bowtie, 61.01–65.66% and 61.97–66.21% unique reads were mapped into the reference genome of N. lugens (S3 Table). For non-model insects, this level of unique reads coverage (over 60%) is acceptable for further analysis. All of the RNA sequence data in this article have been deposited in the NCBI SRA database and are accessible in PRJNA657327 (SRR12460889-96).
The mapped reads were used to quantify the expression profiling by FPKM (fragments per kilobase of transcript per million mapped fragments) method (S4 Table). The correlation among four biological replicates in each treatment was then assessed, in terms of the whole genomic FPKM value by using the Pearson method [59]. The heatmap reveals that the square of correlation value ranged from 0.886 to 0.925 and from 0.861 to 0.916 in the dsNlsk–injected and dsgfp-injected control groups, respectively (Fig 2A). To identify differentially expressed genes (DEGs) in response to Nlsk silencing, the DEGs were screened according to the Noiseq method, which is effective in controlling the rate of false positives [60]. With a cutoff of |Log2 fold change| > 1 and p < 0.05, a total of 190 DEGs were identified in Nlsk–depleted brown planthoppers, including 111 up-regulated and 79 down-regulated DEGs (Fig 2B and S5 Table).
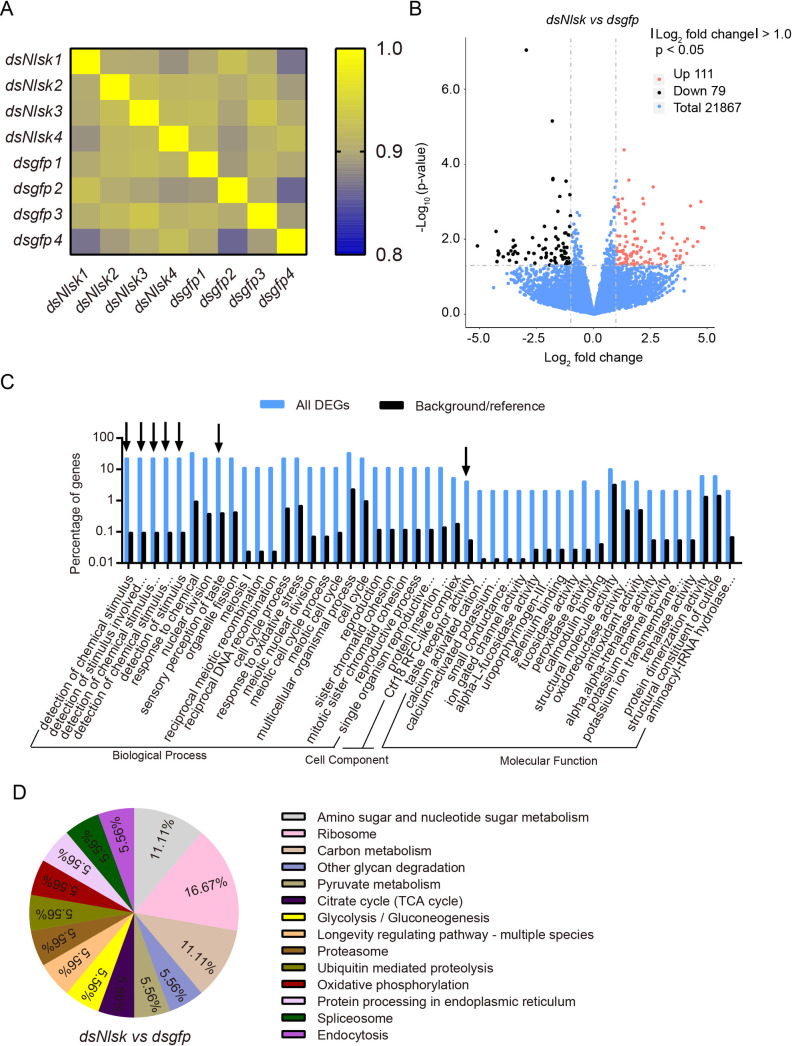
Fig 2. Global gene expression profile in the rice planthopper after Nlsk gene knockdown.
(A) Heatmap showing the square of correlation value from four biological replicates of dsNlsk and dsgfp groups analyzed by RNA-seq. The square of correlation value was assessed by using the Pearson correlation. (B) Volcano plot of differentially expressed genes comparing the dsgfp and dsNlsk treated brown planthoppers. The red dots indicate significantly (p < 0.05 and > 2-fold) upregulated genes. The black dots indicate significantly (p < 0.05 and > 2-fold) downregulated genes. (C) WEGO (Web Gene Ontology Annotation Plotting) output for Nlsk regulated genes. The histogram shows the percent of genes with GO terms enriched in each category. Arrows point to interesting GO categories affected by Nlsk knockdown. Hypergeometric test (FDR-adjusted): black arrow indicated p < 0.05. (D) The pie chart shows the percentage of regulated transcripts distribution in different pathways/processes identified by KEGG pathway analysis.
To obtain a functional classification, these DEGs were assigned to three main GO (Gene Ontology) categories: biological processes (37, 71.42%), cellular components (4, 14.29%), and molecular functions (5, 14.29%) which were further organized into 47 subcategories (Fig 2C and S5 and S6 Tables). Furthermore, GO analysis revealed an enrichment of GO terms related to detection of chemical stimulus, detection of stimulus involved in sensory perception, detection of chemical stimulus involved in sensory perception of taste, detection of stimulus and taste receptor activity (Fig 2C and S6 Table) suggesting that genes involved in these processes are regulated by Nlsk. For further categorization, the annotated genes were also mapped to KO (KEGG orthologous) terms in the KEGG database. In total, 50 genes (26.32%) could be accessed with a KO number (S5 Table) and were significantly enriched in several vital physiological processes associated with sugar and carbon metabolism (Fig 2D). For example, we identified upregulation of a gene encoding the putative juvenile hormone (JH) binding protein takeout (Nlto, Gene ID: 111048647, 111048649, and 111045963), the ortholog of which, in Drosophila, plays an important role in circadian activity and feeding behavior [61,62]. Other upregulated genes of interest are associated with sensory perception of sweet taste, including gustatory receptors (Gr) for sugar taste Gr64f-like (NlGr64f, 111045484) and Gr43a-like (NlGr43a, 111056166) (S4 and S5 Tables) [51,63,64]. Furthermore, genes coding for NADH-cytochrome b5 reductase 2-like (CYB5R2, 111046783), and NADH-cytochrome b5 reductase 4-like (CYB5R4, 111052735), the orthologs of which, in mice, play an important role in sugar metabolism [65], were down-regulated. Meanwhile, the Nlsk gene (Nlsk, 111047610 and 111060391) as positive indicator of our transcriptome data, was significantly repressed after Nlsk gene silencing (S4 and S5 Tables). To validate our RNA-seq data, we performed qRT-PCR on six selected genes, three up-regulated and three down-regulated genes in the dsNlsk-injection. The qPCR results are consistent with the RNA-seq data (S1 Fig and S4 Table).
Nlsk positively regulates takeout gene expression that inhibits feeding behavior of brown planthopper
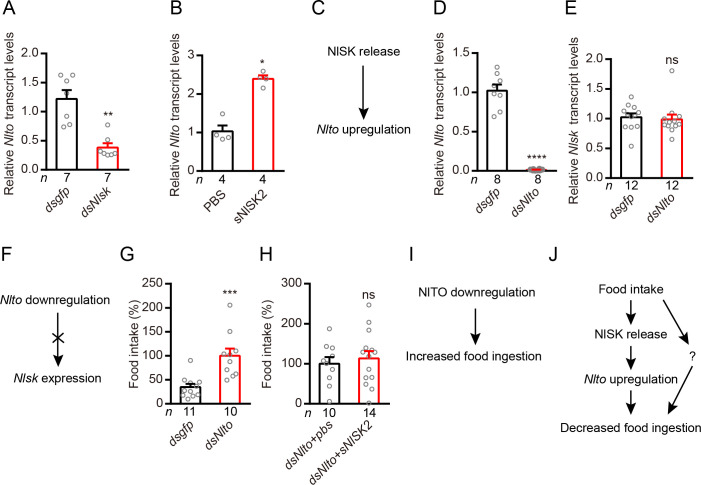
Takeout (To) has been shown to play an important role in the feeding behavior of Drosophila [61,62]. To confirm whether Nlsk knockdown impairs the expression of takeout gene (Nlto), we performed qRT-PCR to quantify the expression level of Nlto gene from the dsNlsk-injected and dsgfp-injected brown planthopper. Our results show that knockdown of Nlsk expression significantly down-regulates Nlto expression (Fig 3A). Furthermore, we found that injection of sNlSK2 significantly induces expression of Nlto gene (Fig 3B). These results indicate that NlSK signaling positively regulates the expression of Nlto gene (Fig 3C). Next, we asked whether down-regulation of Nlto gene could affect expression of Nlsk. After silencing of Nlto gene, we did not observe any change in the expression of the Nlsk gene (Fig 3D and 3E). These results indicate that Nlto has little impact on expression of Nlsk (Fig 3F). Since to regulates feeding behavior in Drosophila [61,62], we were interested to test its role in the planthopper. Indeed, we observed that brown planthoppers with silenced Nlto gene consume more food than the dsgfp-injected controls (Fig 3G). Interestingly, sNISK2 injection does not inhibit feeding in animals with simultaneous Nlto knockdown (Fig 3H). Hence, NlTO inhibits food ingestion (Fig 3I). Together with previous findings, we propose that feeding induces secretion of NlSK, which promotes release of Takeout and inhibition of feeding behavior in the brown planthopper (Fig 3J). Again, we should point out that it is likely that there are further molecular components involved in regulation of feeding state-dependent food ingestion (Fig 3J).
Fig 3. Takeout (to) is a downstream signal of SK that inhibits food intake in the rice planthopper.
(A) Knockdown of Nlsk results in downregulation of Nlto gene in the brown planthopper. All data are presented as means ± s.e.m. **p < 0.01; Mann–Whitney test. (B) Injection of sNlSK2 increases gene expression level of Nlto. *p < 0.05; Mann–Whitney test. (C) Model of the NlSK promotes Nlto expression. (D) Downregulation of Nlto gene using Nlto-RNAi leads to a reduction in mRNA expression level. ****p < 0.0001; Student’s t test. (E) Knockdown of Nlto have no impacts on expression of Nlsk gene in brown planthopper. All data are presented as means ± s.e.m. ns, no significant difference; Mann–Whitney test. (F) Model showing that Nlto has no effects on Nlsk expression. (G) Food intake after silencing Nlto gene. The dsNlto-injected nymphs eat three times more food than dsgfp-injected nymphs in the normal conditions. All data are presented as means ± s.e.m. ***p < 0.001; Mann–Whitney test. (H) Food intake after silencing the Nlto gene with injection of pbs or sNlSK2. No difference was observed between these two conditions. All data are presented as means ± s.e.m. ns, no significant difference; Student’s t test. (I) Model of the Takeout in the food inhibition. (J) Simplified model of the Takeout as a downstream signal of NlSK involved in the feeding inhibition. The question mark indicates one or more possibly additional signals.
Nlsk negatively regulates gustatory sugar receptor Gr64f expression and thereby reduces feeding in the brown planthopper
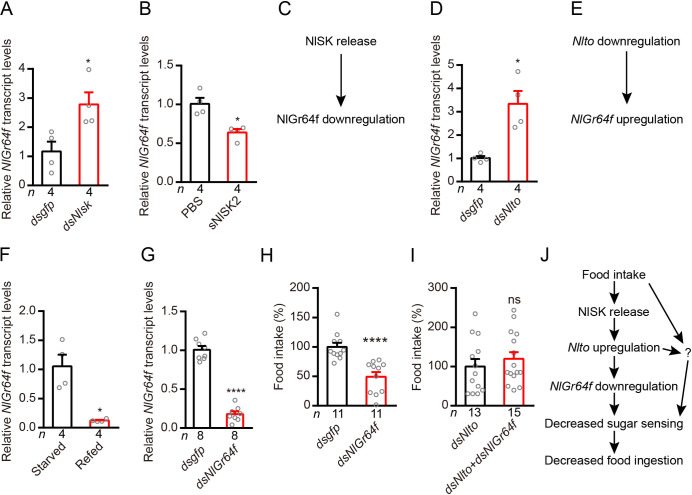
Interestingly, we found that silencing Nlsk gene leads to an up-regulation of gene transcription of the sugar-sensing gustatory receptor Gr64f (Fig 4A). Furthermore, injection of sNlSK2 down-regulates Gr64f gene expression (Fig 4B). These results indicate that NlSK signaling negatively regulates expression of NlGr64f (Fig 4C). Next, we showed that silencing of Nlto also increased the expression of NlGr64f (Fig 4D) indicating that Nlto inhibits expression of NlGr64f (Fig 4E). We found that the expression of NlGr64f was also influenced by feeding state and displays a phenotype opposite to that of the Nlsk gene (Fig 1B). Thus, refeeding after starvation inhibits the expression of NlGr64f (Fig 4F). Then, we asked whether silencing of the NlGr64f gene impairs feeding behavior. Down-regulation of the NlGr64f gene indeed diminishes food intake compared to control animals (Fig 4G and 4H). Next, we tested whether NlGr64f is mediating the effects of NIto. The double knockdown of Nlto and NlGr64f has no significant difference with Nlto knockdown alone in the food consumption (Fig 4I). These results suggest that food intake promotes NlSK production and release, which in turn induces Nlto expression and inhibits NlGr64f expression and leads to decreased food consumption. However, our findings indicate that food intake and Nlto might target also other factors, which affect food consumption (Fig 4J). We also found that Nlsk manipulations alter the expression of another sweet-sensing gustatory receptor, NlGr43a. Injection of dsNlsk increased NlGr43a expression, whereas injection of sNlSK2 decreased its expression (S2A and S2B Fig). However, we focused on Gr64f in the following analysis of Drosophila.
Fig 4. Sulfakinin inhibits expression of the sweet gustatory receptor Gr64f, which promotes food ingestion in the rice planthopper.
(A) Downregulation of Nlsk gene using Nlsk-RNAi (dsNlsk) leads to up-regulation of transcript of sweet sensing NlGr64f. *p < 0.05; Mann–Whitney test. (B) Injection of sNlSK2 leads to down-regulation of NlGr64f gene. *p < 0.05; Mann–Whitney test. (C) Model showing that NlSK inhibits NlGr64f expression. (D) Downregulation of Nlto gene using Nlto-RNAi (dsNlto) leads to up-regulation of transcript of sweet sensing NlGr64f. *p < 0.05; Mann–Whitney test. (E) Model showing that Nlto inhibits NlGr64f expression. (F) Refeeding for 5 hr after 24 hr starvation decreases NlGr64f transcript. *p < 0.05; Mann–Whitney test. (G) Downregulation of NlGr64f gene using NlGr64f-RNAi (dsNlGr64f) leads to a reduction in mRNA expression level. ****p < 0.0001; Student’s t test. (H) Downregulation of NlGr64f gene decreases the food intake of brown planthopper. ****p < 0.0001; Mann–Whitney test. (I) Double knockdown of Nlto and NlGr64f does not rescue the increased food consumption seen with Nlto knockdown alone. All data are presented as means ± s.e.m. ns, no significant difference; Student’s t test. (J) Simplified model showing that the feeding state regulates SK signaling which in turn modulates Nlto and NlGr64f signaling (sweet sensing) and thereby feeding. The question mark indicates one or more possibly additional signals.
Drosulfakinins (DSKs) signal satiety in Drosophila
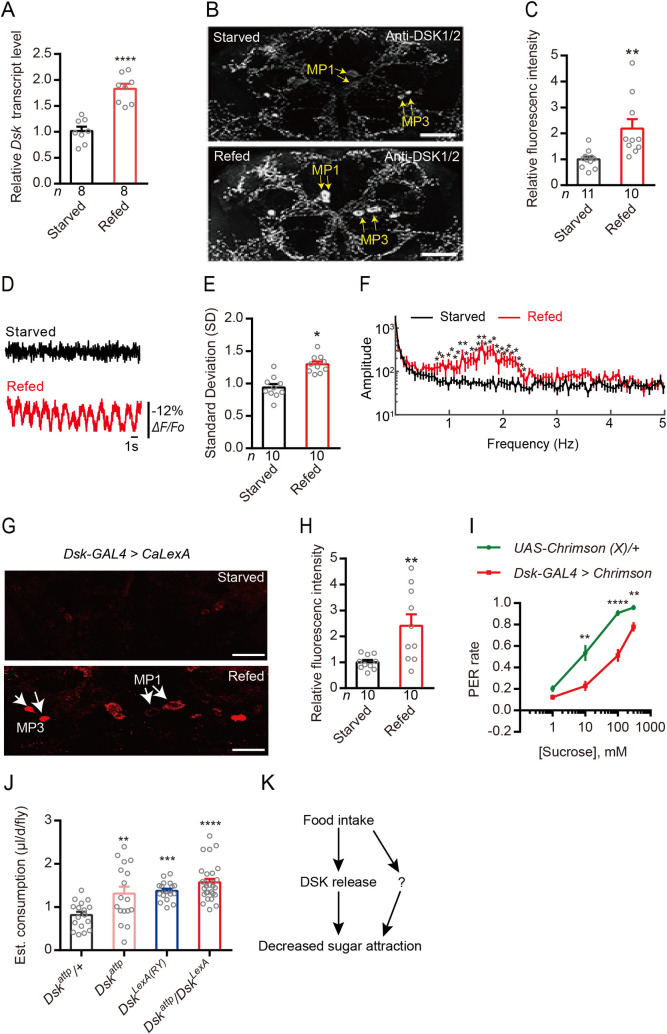
Next, we used the genetic model insect, Drosophila melanogaster, to further investigate mechanisms of DSK signaling in modulation of gustatory sugar reception and feeding. Earlier studies have implicated DSK signaling in regulation of food intake [33,37,66,67]. First, we found that the Dsk mRNA level is significantly higher in heads of flies refed after 24 h starvation, compared to those from starved flies (Fig 5A). Consistent with this, immunohistochemistry showed that the level of DSK peptides (anti-DSK1/2) in refed flies is higher than that in starved ones (Fig 5B and 5C). These results are similar to the findings in the brown planthopper (Fig 1D and 1E). To monitor activity in Dsk expressing neurons related to feeding, we next expressed the genetically encoded fluorescent voltage indicator ArcLight [68] in Dsk-GAL4 neurons. We observed a higher level of spontaneous activity in the Dsk-expressing medial protocerebrum (MP1) neurons [41] in refed flies compared to that in starved flies (Fig 5D–5F). We ruled out the possibility that this change in activity was induced by altered expression level of the GAL4 RNA that was under control by the Dsk gene promoter, since we found that the GAL4 expression in the Dsk-GAL4/+ fly was not influenced by feeding state (S3 Fig). Furthermore, we utilised an activity reporter system, calcium-dependent nuclear import of LexA (CaLexA) [69], to reveal whether increased activity of the Dsk-expressing MP1 neurons correlates with refeeding after starvation. Our analysis shows that flies refed after starvation exhibit significantly increased Ca2+ activity in the MP1 neurons, compared to starved flies (Fig 5G and 5H). To further confirm our findings, we employed a red-shifted channel rhodopsin, CsChrimson [70] as an optogenetic effector under control of the Dsk-GAL4. After light activation of Dsk-GAL4 expressing neurons, we found that the PER (proboscis extension response) of flies was significantly decreased compared with control flies (Fig 5I). However, thermal activation of DSK neurons expressing UAS-dTrpA1 under Dsk-GAL4 control did not inhibit feeding behavior (S4 Fig). One reason may be that the long-term activation ultimately reduces the signal from the DSK neurons and/or that the sustained activation desensitizes the response to DSK in target neurons. We also found that Dsk mutants display increased motivation to feed in the capillary feeding (CAFE) assay [71] (Fig 5J). Similar results were also previously reported [66]. However, silencing of the Dsk gene using Dsk-GAL4 has no significant effect on feeding behavior in Drosophila as shown in PER and CAFE experiments (S5A and S5B Fig). It might be that in our hand RNAi is not sufficient induce a behavior phenotype. Another possibility is that Dsk is not the only satiety signal which regulates sweet sensing or feed behavior. These results suggest that re-feeding after starvation increases DSK expression and activates Dsk-expressing MP neurons, which release DSK to inhibit feeding behavior (Fig 5K). Our data (S4 and S5 Figs) also suggest that food intake might decrease sugar attraction through additional pathways (see Fig 5K).
Fig 5. Drosulfakinin (DSK) signals satiety and inhibits feeding in Drosophila.
(A) Refeeding after 24h starvation increases Dsk transcript in the head of Drosophila. ****p < 0.0001; Student’s t test. (B and C) Refeeding after 24h starvation increases DSK peptide expression as revealed by anti-DSK1/2 immunolabeling. Yellow arrows indicated the MP1 and MP3 neurons. Scale bars, 50 μm. **p < 0.01; Student’s t test. (D) Representative optical recordings of spontaneous membrane activity in median protocerebrum (MP1) neurons (Somata) in the starved (0.5% agar for 24 hours) or in the refed (fed 1.5 h after 24 h starvation in 0.5% agar) of flies that Dsk-GAL4 drive the voltage indicator ArcLight maintained in 12 hr:12 hr light:dark conditions. (E) Standard deviations (SDs) over the recording trial were computed for each field. Refed SD is significantly greater than starved. *p < 0.05; Student’s t test. (F) Power spectrum was computed for each terminal field using fast Fourier transform with 0.05 Hz bin width. Re-fed power is significantly greater than starved power between 1 Hz to 2.5 Hz. n = 14 for starved and n = 30 for refed. *p < 0.05, **p < 0.01; Student’s t test. (G) Representative images showing CaLexA signals in DSK expressing MP1 neurons of flies exposed to starvation and refeeding. Starved: flies were raised in 0.5% agar for 24 hours; Refed: flies were raised in fly food 1.5 h after 24 h starvation in 0.5% agar. Red: maximal intensity of CaLexA signals. Scale bar, 50 μm. (H) Quantification of the signal intensity of CaLexA signals in DSK expressing MP neurons from flies treated with conditions shown in (G). **p < 0.01; Student’s t test. (I) Optogenetic activation (19 μm/mm2) of Dsk-GAL4 neurons expressing Chrimson is sufficient to inhibit proboscis responses. A fly immobilized in a pipet tip was stimulated on the labellum with different concentrations of sucrose in the presence of light stimulus. We used two values to score the PER assay. A score of 1.0 indicates a fly that extended its proboscis and ingested after being presented with the probe. The score was 0 if the fly failed to extend its proboscis. n = 10 trials. **p < 0.01, ****p < 0.0001; Mann–Whitney test. (J) Three Dsk mutants show increased food consumption compared to wildtype in the CAFE essay. **p<0.01, ***p<0.001, ****p<0.0001, Kruskal–Wallis test followed by Dunn’s multiple comparisons test. (K) Simplified model showing that DSK inhibits feeding. Note that this model and those in subsequent figures with Drosophila data are highly simplified and serve to summarize our data, rather that presenting complete and accurate signaling pathways. The question mark indicates one or more possibly additional signals.
The sugar receptor Gr64f promotes motivation to feed in Drosophila
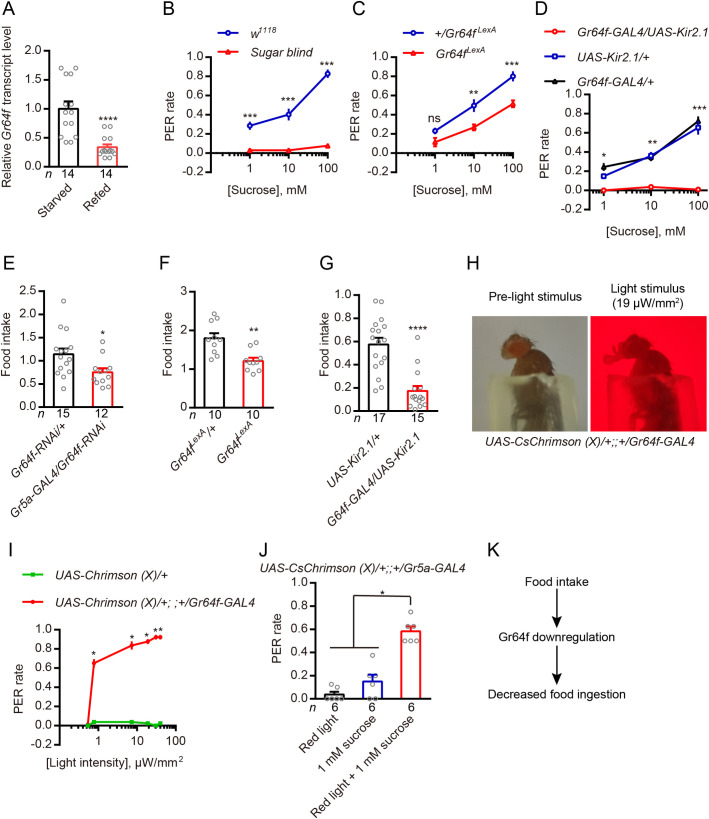
We next tested whether sugar-sensing GRNs promote feeding behavior in Drosophila. First, we found that the level of Gr64f transcription was down-regulated in the refed flies compared with starved ones (Fig 6A), which is opposite to the Dsk transcription (Fig 5A). Then, we utilized mutants that lacked all eight known sugar receptors (sugar blind) or the critical co-receptor Gr64f (Gr64fLexA) in tests of sugar sensing [72,73] by PER analysis. As expected, sugar blind or Gr64fLexA flies show almost no, or lower, responses to sucrose solutions (Fig 6B and 6C), similar to flies with their sweet GRNs selectively silenced (Fig 6D). We also find that Gr64f mutants or flies with Gr64f gene knockdown in the Gr5a-GAL4 expressing neurons consume less food than controls (Fig 6E and 6F). This is similar to flies with their sweet GRNs selectively silenced by expressing a hyperpolarizing potassium channel (Fig 6G). Next, we tested whether activation of sweet neurons is sufficient to induce feeding behavior. We expressed the optogenetic effector CsChrimson under control of the Gr64f-GAL4. As with the preceding results, control animals display a low rate of PERs upon stimulation with light stimuli. However, the UAS-CsChrimson (X)/+;;+/Gr64f-GAL4 animals show a prominent dose-dependent PER responses under light stimuli (Fig 6H and 6I and S1 Movie). Similar results were also reported in previous work [74,75]. Furthermore, we found that increasing the baseline activity of sugar GRNs by opto-genetical activation increases the probability that the fly shows PER upon sugar stimulation (Fig 6J). These results indicate that starvation promotes Gr64f expression and activates sugar-sensing GRNs, reflecting a starved state that promotes feeding behavior (Fig 6K).
Fig 6. In Drosophila, Gr64f promotes feeding.
(A) Refeeding after 24h starvation decreases Gr64f transcript. ****p < 0.0001; Student’s t test. (B) The sugar blind mutant (all eight known sugar receptors mutated) show almost no motivation to feed in proboscis extension reflex (PER). n = 10 trials. ***p < 0.001; Mann–Whitney test. (C) Gr64f mutant (Gr64fLexA) flies show less motivation to feed in PER. n = 10 trials. ***p < 0.001, **p < 0.01, ns, no significant; Mann–Whitney test. (D) Silencing Gr64f-GAL4 neurons by expressing the hyperpolarizing channel Kir2.1 also caused loss of motivation to feed in PER. n = 10 trials. ***p < 0.001, **p < 0.01, *p < 0.05; Kruskal–Wallis test followed by Dunn’s multiple comparisons test. (E) Silencing the Gr64f gene using the Gr5a-GAL4 driver leads to decreased food consumption compared to wildtype flies in the CAFE essay. *p < 0.05; Student’s t test. Food intake is the estimated food consumption (μl) of one female per day. (F) Gr64f mutant (Gr64fLexA) flies show decreased food consumption compared to wildtypes in the CAFE essay. **p < 0.01; Student’s t test. (G) Silencing Gr64f-GAL4 neurons by expressing the hyperpolarizing channel Kir2.1 also leads to decreased food consumption in the CAFE assay. ****p < 0.0001; Mann–Whitney test. (H) Dynamics of light-induced proboscis extension after photoactivation in a fly expressing Gr64f-Gal4, UAS-CsChrimson. (I) Relationship between the intensity of the stimulating light and PER rate using the indicated flies. A fly immobilized in a pipet tip was stimulated with red light at the indicated intensity and the light-induced proboscis extension of the fly was recorded. We used two values to score the PER assay. A score of 1.0 indicates a fly that extended its proboscis after being presented with the light. The score was 0 if the fly failed to extend its proboscis. n = 8 trials. **p < 0.01, *p < 0.05; Mann–Whitney test. (J) Increasing the baseline activity of sweet GRNs (UAS-Chrimson (X)/+;; +/Gr5a-GAL4) by photo-activation with 0.78 μW/mm2 of red light increases the probability that the fly shows PER upon sugar stimulation (1 mM sucrose) compared with either red light stimulation (0.78 μW/mm2) or 1 mM sucrose alone. *p < 0.05; Kruskal–Wallis test followed by Dunn’s multiple comparisons test. (K) Simplified model showing that Gr64f promotes feeding.
DSK and takeout negatively regulate sugar receptor Gr64f expression in Drosophila
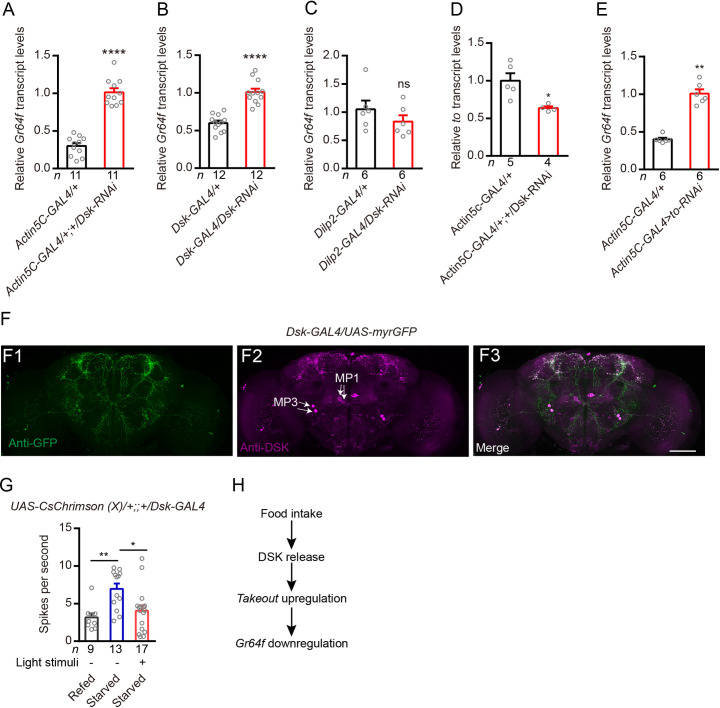
To test whether silencing Dsk gene expression affects the expression of Gr64f also in Drosophila, we used a UAS-Dsk-RNAi transgene targeted either globally, to Dsk-expressing neurons, or to insulin-producing cells (IPCs) [41]. Knockdown with the global GAL4 driver (Actin5C-GAL4), and the specific driver (Dsk-GAL4), resulted in up-regulation of the Gr64f gene (Fig 7A and 7B). However, diminishing of Dsk in the insulin producing cells (IPCs) using Dilp2-GAL4 (S6 Fig) has no significant effect on Gr64f gene expression (Fig 7C). Our previous studies showed that Dsk-GAL4 is expressed in two sets of MP neurons and a subset of the IPC neurons [33,41], and our data here indicate that DSK from MP neurons, but not from IPCs is responsible for affecting Gr64f expression. Similar to brown planthopper, in Drosophila, silencing of Dsk gene negatively regulates takeout (to) expression (Fig 7D). Thus, we asked whether to also negatively regulates Gr64f expression. Indeed, our results show that reducing to expression using an Actin5C-GAL4 driver upregulates Gr64f expression (Fig 7E).
Fig 7. In Drosophila DSK and TO inhibits Gr64f gene transcription and optogenetic activation of Dsk-GAL4 neurons decreases the sensitivity of gustatory neurons in starved flies.
(A and B) Relative expression levels of Gr64f transcripts in DSK deficient files. Depletion of Dsk, either globally in all cells (Actin5C-GAL4) or in the Dsk-GAL4-expressing cells increased Gr64f transcripts in starved flies. ****p < 0.0001; Student’s t test. (C) Silencing only Dsk in the IPCs with RNAi (Dilp2-GAL4/Dsk-RNAi; red bars) did not affect Gr64f transcript levels. ns: not significant; Mann–Whitney test. (D) Relative expression levels of to transcripts in Dsk deficient files. Depletion of Dsk in all cells (Actin5C-GAL4) decreased to transcripts in flies. *p < 0.05; Mann–Whitney test. (E) Relative expression levels of Gr64f transcripts in TO deficient files. Depletion of to in all cells (Actin5C-GAL4) increased Gr64f transcripts in flies. **p < 0.01; Mann–Whitney test. (F) Expression pattern of Dsk-GAL4 in the brain revealed by anti-GFP (F1) and anti-DSK (F2). Scale bars, 50 μm. (G) Electrophysiological recordings reveal that starved flies (24 h) show increased sensitivity of GRNs compared with refed flies (fed 1.5 h after starvation) and optogenetic activation of Dsk-GAL4 neurons (30 min) decreases the sensitivity of gustatory neurons in starved flies (24 h). **p < 0.001; *p < 0.05; Kruskal–Wallis test followed by Dunn’s multiple comparisons test. (H) Simplified model of the feeding state, DSK, TO and Gr64f interaction.
SK is not likely to be the only satiety signal that mediates nutritional state dependent reduction of sweet attraction
Next, we asked whether SK is the only satiety-induced neuropeptide that modulates sweet sensation. We found that starvation renders wild type flies more sensitive to sucrose than re-fed flies (S7A Fig). However, sugar blind, which is the sugar receptor mutant, displayed no difference of PER rate between starved and re-fed conditions (S7B Fig). Similar results were also observed when we silenced sweet GRNs by expressing the Kir2.1 channel under Gr64f-GAL4 control (S7C Fig). These findings are expected since we eliminated the sugar receptors or silenced the sweet-sensing GRNs. However, dsk mutant flies and flies with silencing of Dsk-GAL4 expressing neurons exhibited responses similar to wild type flies where starved flies are more sensitive than re-fed flies (S7D and S7E Fig). These results indicate that SK is not the only signal that contributes to a reduction of sweet attraction in re-fed flies.
Optogenetic stimulation of Dsk-GAL4 neurons decreases the sensitivity of gustatory neurons in starved flies
Next, we set out to identify the potential mechanism by which DSK neurons modulate Gr64f-expressing sugar-sensing GRNs. We found that sugar-sensing GRNs in the proboscis and Dsk-expressing neurons in the brain project their axons to overlapping areas in the subesophageal zone (SEZ) of the brain (S8A and S8B Fig). We did not detect Dsk-expressing neurons in the proboscis or proleg tarsi (S8C and S8D Fig). Our Dsk-GAL4 line labels eight MP neurons in the fly brain (Fig 7F), but, only MP1 neurons project to SEZ of the brain [41]. Next, we asked whether activation of Dsk-GAL4 neurons could impair the sensitivity of GRNs. As previous studies reported [62], we also observed that starved flies show increased sensitivity of GRNs compared with re-fed flies (Fig 7G). However, activation of Dsk-GAL4 neurons for 30 min decreases the sensitivity of GRNs in starved flies (Fig 7G). These results indicate that re-feeding induces release of DSK, which promotes TO and then inhibits sugar receptor expression and sugar-sensing neuron activity (Fig 7H).
Distribution of sulfakinin receptor (SKR) in the sweet-sensing gustatory neurons
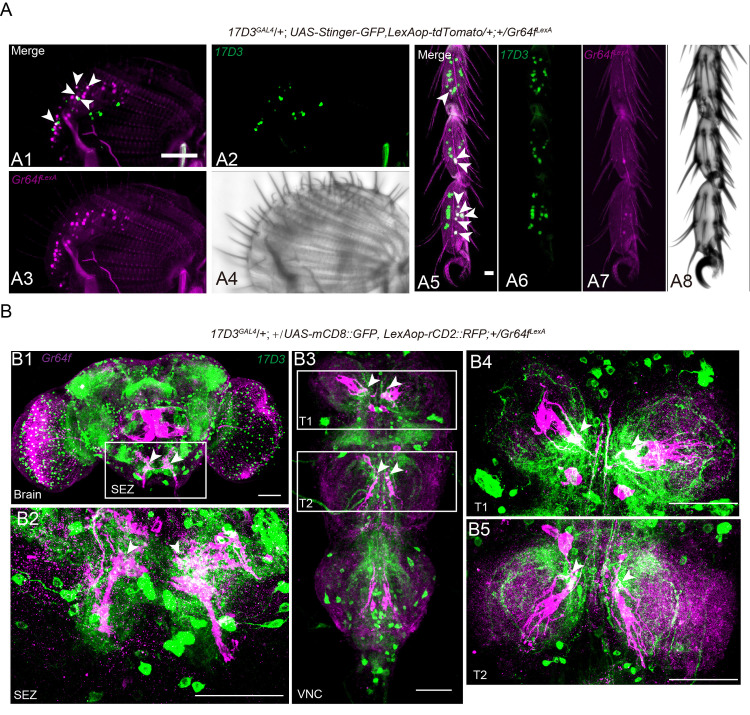
Earlier studies have shown that GRNs in Drosophila are modulated by neuropeptides to adjust sensitivities to sweet and bitter taste [11]. Hence, in hungry flies NPF, via dopaminergic cells, increases sweet sensitivity and sNPF decreases bitter sensitivity [11]. Thus, we asked whether DSK receptors are expressed in the sweet-sensing GRNs of the gustatory system. Two G-protein coupled receptors, the CCK-like receptors (CCKLR)-17D1 and CCKLR-17D3, have been identified as the receptors of DSK peptides in Drosophila [76–78]. As we failed to generate functional CCKLR-17D1 and CCKLR-17D3 antibodies, we produced a knock-in GAL4 into the start codon of each of CCKLR-17D1 and CCKLR-17D3 [41] for expression analysis (S9A Fig). We did not detect expression of 17D1GAL4 in cells of the leg tarsi, maxillary palps or proboscis (S9B Fig). However, we found that 17D3GAL4 drives expression broadly in cells of the proleg tarsi, proboscis and maxillary palps (S10 Fig). Next, we asked whether the CCKLR-17D3 receptor is co-expressed with sugar receptors in the gustatory receptor neurons (GRNs). Double labeling of Gr64fLexA and 17D3GAL4 expression (17D3GAL4; LexAop-RedStinger, UAS-stinger-GFP/+; +/Gr64fLexA) revealed that five pairs of neurons of the labellum and eight of each proleg tarsi are co-labeled by 17D3GAL4 and Gr64fLexA (Fig 8A). Thus, we have evidence that CCKLR-17D3, one of the two known sulfakinin receptors, is expressed in some of the Gr64f-expressing neurons in the proboscis and leg tarsi (Fig 8A). We also show here that Gr64fLexA and 17D3GAL4 labeled neurons of the proboscis extend their axons to overlapping areas in the SEZ of the brain (Fig 8B1 and 8B2). In addition, we also detected their axons in overlapping areas of the prothoracic neuromere of the ventral nerve cord (Fig 8B3–8B5).
Fig 8. In Drosophila the DSK receptor, CCKLR-17D3, is expressed in Gr64fLexA expressing neurons.
(A) 17D3GAL4, UAS-Stinger-GFP (green) superimposes with Gr64fLexA, LexAop > tdTomato (magenta) expressing cells in the labellum (A1-A4) and proleg tarsi (A5-A8) of flies. Overlap is in white. The white arrowheads (A1 and A5) indicate the positive neural cells co-labeled by 17D3GAL4 and Gr64LexA. Scale bar: 50 μm. (B) Double labeling of 17D3GAL4-expressing neurons and Gr64fLexA-expressing sweet neurons in the brain (B1), SEZ (B2), and VNC (B3-B5). SEZ: subesophageal zone; VNC: ventral nerve cord; T1-T3: thoracic ganglion 1–3. Scale bar: 50 μm.
CCKLR-17D3 signaling in GRNs mediates feeding-induced modulation of PER
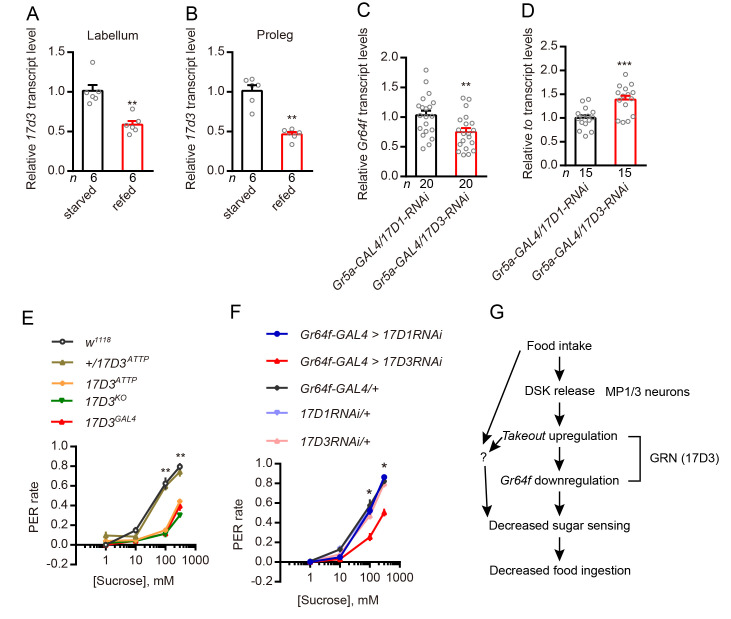
Next, we asked whether the inhibition of the PER by DSK that we observed is mediated by its receptor, CCLR-17D3, which is expressed by the sweet-sensing GRNs. To address this, we first found that the level of ccklr-17d3 transcription in the labellum and proleg is down-regulated in the refed flies compared to starved flies (Fig 9A and 9B), which is similar to the transcription of Gr64f (Fig 6A). Secondly, to exclude that DSK might have an indirect effect by acting on 17D3 receptors in other cells, we specifically silenced the two DSK receptors in Gr5a neurons. Only knockdown 17d3 gene expression in Gr5a neurons leads to the downregulation of Gr64f and an upregulation of to (Fig 9C and 9D). Thirdly, we found that 17D3 mutant flies exhibit decreased PER compared to control flies (Fig 9E). In contrast, 17D1 mutants do not display a significant difference to controls in our PER assay (S9C Fig). Furthermore, we knocked down either of the DSK receptors (CCKLR-17D1 and CCKLR-17D3) by crossing each of UAS-17D1-RNAi and UAS-17D3-RNAi with Gr64f-GAL4. We measured the PER in our behavioral assay and found that flies lacking 17D3 in sweet-sensing GRNs exhibit a significantly decreased PER, whereas 17D1 knockdown had no effect (Fig 9F). Thus, 17D3 expression in Gr64f expressing GRNs mediates the starvation-dependent enhancement of PER. Importantly, it should be noted that our data suggest that the 17D3 receptor mediates an inhibitory signal in the Gr64f expressing GRNs, and possibly other brain neurons. Thus, the DSK signal after food intake inhibits activity in Gr64f expressing GRNs, which reduces sweet sensing, and consequently knockdown of 17D3 leads to diminishment of this inhibition and thereby a decrease in PER. Other studies have also indicated inhibitory action of SK receptors in specific neurons. For instance, Wu et al. [41] found that 17D3 activation leads to an inhibition of activity in specific fruitless expressing neurons (P1) resulting in a suppression of sexual arousal and Wicher et al. [56] showed that SK, as a satiety signal, hyperpolarizes dorsal unpaired median neurons and thus affects general activity in a cockroach.
Fig 9. In Drosophila activity of the DSK receptor (CCKLR-17D3) is necessary for PER.
(A and B) Refeeding after 24h starvation decreases 17d3 mRNA transcript in the labellum and proleg tarsi. All data are presented as means ± s.e.m. **p < 0.01; Mann–Whitney test. (C and D) Silencing of the 17d3 gene in sweet GRNs leads to downregulation of Gr64f and upregulation of the to gene. All data are presented as means ± s.e.m. **p < 0.01, ***p < 0.001; Student’s t test. (E) 17D3 mutants show decreased motivation to feed in PER. **p < 0.01; Kruskal–Wallis test followed by Dunn’s multiple comparisons test. (F) Silencing of 17D3 gene in Gr64f-GAL4-expressing neurons showed decreased motivation to feed in PER. *p < 0.05; Kruskal–Wallis test followed by Dunn’s multiple comparisons test. (G) Simplified model of SK as satiety signal that reflects internal states and inhibits sweet sensation (an external stimulus). Note that this model is simplified and does not exclude additional parallel signaling pathways that modulate gustation and feeding. The question mark indicates one or more possibly additional signals.
Discussion
Food seeking and feeding are under complex control by neuronal circuits as well as by neuropeptides and peptide hormones [3,12,15,40,55,79–81]. Thus, sensory systems, central circuits and interorgan communication, in a nutrient-dependent fashion, contribute to feeding decisions and regulation of food ingestion. In this study, we have analyzed mechanisms of nutrient state-dependent peptidergic regulation of gustatory inputs and feeding.
We show herein that in both the planthopper N. lugens and the fly Drosophila, SK signaling mediates satiety and decreases sensitivity of gustatory neurons (GRNs) expressing the gustatory sugar receptor Gr64f. Thus, SK release not only decreases food intake [33,34,82], but also downregulates attraction to sugar. We find that food ingestion diminishes and starvation increases Gr64f expression under control of SK signaling. In the brown planthopper, the gene takeout is upregulated by SK, and we showed that knockdown of takeout upregulates Gr64f and increases feeding in both insects.
We performed additional experiments in Drosophila to reveal further SK signaling mechanisms. Calcium and membrane activity in DSK expressing MP neurons in the brain increase after feeding, suggesting that these neurons receive nutrient signals. We could also show that optogenetic activation of DSK neurons in the brain decreases the PER, whereas activation of Gr64f expressing GRNs increases the PER. DSK neurons were found to make functional contacts with Gr64f expressing GRNs in the SEZ, and one of the two DSK receptors, CCKLR-17D3, is expressed in Gr64f expressing GRNs in proleg tarsi, labellum and maxillary palps. Furthermore, the ccklr-17D3 levels are downregulated in these appendages after feeding. Our data suggest that the CCKLR-17D3 inhibits activity in Gr64f expressing GRNs and hence knockdown of this receptor leads to a decrease in sugar sensitivity and reduced PER. In Drosophila, takeout knockdown not only increases feeding, but also expression of Gr64f, further suggesting a role of takeout in DSK-mediated satiety signaling. Hence, we find that food ingestion activates SK signaling in the two insects and that SK acts to decrease expression of a sweet receptor and thereby diminish food attraction and feeding (Fig 9G). However, our data also suggest that SK in not the only satiety-induced signal that contributes to nutritional state modulation of sugar gustation (see Fig 9G). As mentioned in the introduction, neuropeptides such as NPF and sNPF, as well as dopamine, are also known to regulate gustatory perception [11]. Furthermore, food search also depends on olfaction, and neuropeptides like MIP, CCH2amide and TK have been found to modulate the sensitivity of ORNs [6,10,21–24]. We can also not rule out the possibility that manipulating DSK signaling could affect food intake and dietary status and thus indirectly influence sweet sensing via other signaling pathways.
Invertebrate SKs and the vertebrate CCKs are known as satiety inducing peptides that regulate food ingestion [34,36,38,44–46,81,83,84]. However, these peptides also act in a multitude of other regulatory functions both centrally and in the periphery [37,38,85]. In insects SKs play additional roles in gut motility [42], digestive enzyme production and release [35,47,86], regulation of sexual arousal [41], as well as hyperactivity and aggression [39,67]. Similarly, in mammals CCK stimulates pancreatic enzyme secretion and release of insulin and glucagon, gallbladder contraction and gut motility, and is implicated in fear, anxiety, and aggression [see [36,38,85,87]]. In addition CCK has diverse roles in the brain as a neuromodulator regulating other neurotransmitter systems [see [85]]. Thus, in planthoppers and flies, one can expect that SK acts at several levels and that these actions are coordinated to generate a relevant behavioral and physiological outcome.
In Drosophila and planthopper there are cell bodies of SK-producing neurons only in the brain, and to our knowledge no SK peptide is produced in the intestine or other tissues [41,58]; see also FlyAtlas2 http://flyatlas.gla.ac.uk, [88]. We show here that four pairs of posterior DSK interneurons, MP1 and MP3, with wide arborizations in the brain are likely to underlie the regulation of Gr64f-expressing GRNs. These MP1 neurons display increased calcium and spontaneous electric activity after feeding, and we demonstrate here that the MP1 neurons have processes that superimpose GRN axon terminations in the SEZ [see also [41]]. Furthermore, optogenetic activation of DSK neurons rapidly inhibits the PER suggesting direct neuronal connections. A subset of the brain insulin-producing cells (IPCs) is also known to co-express DSK [33,39,41], but our experiments exclude these cells in modulation of GRNs. The IPCs may instead release DSK as a circulating hormone to act on the intestine in regulation of digestive enzymes as demonstrated in the planthopper and other insects [see [35,47,86]], but not yet shown in Drosophila.
We found that knockdown of takeout increases feeding, and also that this upregulates expression of Gr64f in planthopper and fly. Interestingly, it has been shown earlier that takeout is expressed in GRNs of the labellum in Drosophila and that mutant flies are deficient in sugar sensing and regulation of food ingestion [62]. Takeout mutants are also aberrant in their starvation-induced locomotor activity and display increased mortality during starvation [61,62]. Furthermore, the gene takeout is expressed also in the intestine and fat body, is under control by the circadian clock, and was proposed to link circadian rhythms and feeding behavior [61]. Since takeout encodes a putative juvenile hormone (JH) binding protein, it was suggested that it regulates levels of circulating JH and that this may impart the effects on locomotor activity and food intake, as well as effects on metabolism [62]. The role of takeout in the GRNs in modulation of sugar sensitivity by regulating Gr64f in the same cells requires further investigation.
As mentioned, SK acts at several levels of the CNS and periphery and modulates conflicting behaviors such as food search, feeding, sexual arousal and aggression [33,39,41,67]. These studies show that Dsk neurons increase aggression and decrease feeding and mating. The DSK-expressing MP neurons are central in suppression of both feeding (this study), mating [41] and aggression [66], but aggression was also found dependent on DSK in the IPCs [39,67]. Modulation of feeding and mating relies on different downstream circuits. For suppression of male sexual behavior DSK (from MP neurons) acts on male-specific fruitless-expressing P1 neurons that coordinate arousal-related behaviors such as sex, sleep and locomotion [41]. In parallel, as we show here, the same DSK neurons target GRNs to suppress sweet attraction and inhibit feeding. It is not clear at present how DSK released from IPCs suppresses feeding and what the hormonal DSK targets are [33].
Our data herein suggest that SK is not the only signal that modulates sweet attraction and feeding behavior in response to food ingestion. Other systems, such as the four widely arborizing SIFamide expressing neurons of the pars Intercerebralis, are also known to coordinate hunger and satiety signals to stimulate appetitive behavior and suppress mating behavior and sleep [18,89]. Also this SIFamide system acts at different levels, such as olfactory and gustatory circuits, as well as sleep and activity circuits and fruitless expressing neurons [18,90]. Several studies have also shown peptidergic modulation of ORNs and GRNs to promote state-dependent food seeking in Drosophila. Thus, nutrient dependent insulin signaling modulates sNPF and TK signaling to alter sensitivity of ORNs [6,10,21], whereas NPF and sNPF regulate sweet and bitter sensitivity, respectively [11]. NPF and sNPF are also known to act in other circuits to modulate feeding, metabolism and sleep [see e. g. [25–28,30–32]]. Previous studies found that short-term starvation changes dopamine signaling, which leads to sensitization of sweet sensing neurons [11]. However, in our experiments the starvation time is 24 hours, which is longer than in the earlier experiments. Thus, we assume that in our experiments the effect on the sugar response is not mediated by dopamine.
In summary, we show that DSK secreted from brain neurons regulates sugar sensitivity of DSK receptor-expressing GRNs in response to food ingestion and thereby diminishes food attraction (Fig 9G). Mechanistically this state-dependent desensitization of specific GRNs is by SK receptor mediated downregulation of Gr64f expression in GRNs, possibly involving action of takeout. Importantly, the mechanisms described herein are conserved in Drosophila and the brown planthopper, although these insects are only distantly related. It is likely that our findings have revealed only a portion of the mechanisms involved in mediating nutritional state dependent regulation of sensory perception and subsequent feeding behavior.
Materials and methods
Experimental insects and husbandry
The brown planthopper N. lugens was reared on ‘Taichung Native 1’ (TN1) rice (Oryza sativa L.) seedlings in the laboratory and maintained at 27 ± 1 ∘C, with 70 ± 10% relative humidity, under a 16 h: 8 h light dark photoperiod [91].
Flies were maintained on standard molasses/cornmeal/yeast/agar food at 25°C on a 12:12 LD cycle with humidity set to 60 ± 5% unless otherwise indicated. The following fly strains were ordered from Bloomington Stock Center: w1118 (Bloomington Stock number: #5905); Canton-S (#64349); Gr64f-GAL4 (#57668; [92]); Actin5c-GAL4 (#4414); UAS-ArcLight (#51056; [68]); UAS-RedStinger (#8547; [93]); UAS-CaLexA (#66542; [69]); UAS-Stinger-GFP (#84277); UAS-Chrimson, attp18 (#55134; [70]); UAS-Chrimson, attp40 (#55135; [70]); UAS-dTrpA1 [94]; UAS-NaChBac (#9467; [95]); LexAop-rCD2::RFP, UAS-mCD8::GFP (#67093); UAS-nSyb-spGFP1-10, LexAOP-CD4-spGFP11 (#64314; [96]); LexAOP-nSyb-spGFP1-10, UAS-CD4-spGFP11 (#64315; [96]); Δ17D1[Df(1)Exel9051] (#7762;17D1KO [78]). UAS-DskRNAi (THU2073), UAS-17D1RNAi (THU2656), UAS-17D3RNAi (THU2970), and UAS-Gr64fRNAi (TH04838.N) were purchased from Tsinghua Fly Center at the Tsinghua University [97,98]. UAS-Kir2.1 [99], LexAop2-IVS-nlstdTomato [93], Dsk-GAL4, Dsk-LexA, 17D3KO, 17D3GAL4, Dsk1, and UAS-mCD8::GFP have been described previously [41]. Dilp2-GAL4 (#37516; [100]) was kindly provided by Dr. Wei Song. Gr64fLexA and sugar blind flies (R1,Gr5aLexA;+; Δ61a, Δ64a-f) were a gift from Dr. Hubert Amrein [72,73]. DskattP, DskLexA(RY), 17D1attP, and 17D3attP were kind gifts from Dr. Yi Rao [101].
Peptide synthesis
Sulfakinin peptides were synthesized by Genscript (Nanjing, China) Co., Ltd. Peptides mass was confirmed by MS and the amount of peptide was quantified by amino acid analysis. The amino acid sequence of the peptides used in this study are: N. lugens sulfakinin 1: (NlSK1): SDDYGHMRFamide; sulfakinin 2: (NlSK2): GEADDKFDDYGHMRFamide; sulfated sulfakinin1 (sNlSK): SDDY(SO3H)GHMRFamide; sulfated sulfakinin2 (sNlSK2): GEADDKFDDY(SO3H)GHMRFamide.
Gene cloning and sequence analysis
We used the NCBI database with BLAST programs to carry out sequence alignment and analysis. Then we predicted Open Reading Frames (ORFs) with EditSeq. The primers were designed by Primer designing tool–NCBI. Total RNA Extraction was using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The cDNA template used for cloning was synthesized using the Biotech M-MLV reverse transcription kit and the synthesized cDNA template was stored at -20°C.
The transmembrane segments and topology of proteins were predicted by TMHMM v2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/) [102]. Multiple alignments of the complete amino acid sequences were performed with Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo). Phylogenetic tree was constructed using MEGA 7.0 software with the Maximum Likelihood Method and bootstrapped with 1000 replications [103].
Quantitative RT-PCR
The first-strand cDNA was synthesized with HiScript II Q RT SuperMix for qPCR (+gDNA wiper) kit (Vazyme, Nanjing, China) using an oligo(dT)18 primer and 500 ng total RNA template in a 10 μl reaction, following the instructions. Real-time qPCRs in the various samples used the UltraSYBR Mixture (with ROX) Kit (CWBIO, Beijing, China). The PCR was performed in 20 μl reaction including 4 μl of 10-fold diluted cDNA, 1μl of each primer (10 μM), 10 μl 2 × UltraSYBR Mixture, and 6 μl RNase-free water. The PCR conditions used were as follows: initial incubation at 95°C for 10 min, followed by 40 cycles of 95°C for 10 s and 60°C for 45 s. N. lugens 18S rRNA or Drosophila rp49 were used as an internal control (S1 Table). Relative quantification was performed via the comparative 2−△△CT method [104]. We used Drosophila heads to analyze the expression of Dsk and GAL4 in starved or refed condition.
RNA interference in N. lugens
For lab-synthesized dsRNA, gfp, Nlsk, Nlto and NlGr64f fragments were amplified by PCR using specific primers conjugated with the T7 RNA polymerase promoter (primers listed in S1 Table). dsRNA was synthesized by the MEGAscript T7 transcription kit (Ambion, Austin, TX, USA) according to the manufacturer’s instructions. Finally, the quality and size of the dsRNA products were verified by 1% agarose gel electrophoresis and the Nanodrop 1000 spectrophotometer and kept at -70°C until use.
The 4th instar nymph of brown planthoppers was used for injection of 60 nl of 5 μg/μl dsRNA per insect. Injection of an equal volume of dsgfp was used as negative control. RNAi efficiency was examined by qPCR using a pool of ten individuals on the 3rd day after dsRNA injections. The insects of the third day after dsRNA injection were used for feeding assay and gene relative expression analysis.
Feeding assay of N. lugens
The animals were food deprived for 5 h before onset of the experiment to ensure that all experimental animals were in the same nutritional state prior to the experiment. This was based on several periods of starvation (2, 5, 12, and 24 h) we tested, from which a starvation for 5 h gave the best results in terms of intraexperiment variation. The animals were starved in the morning and the experiments were done in the afternoon. The feeding assay method and artificial diet was adopted as previously reported with modification [105]. Briefly, the antifeedant potency of sulfakinins was measured in fourth instar nymphs of N. lugens. Prior to injection, the peptides were dissolved in PBS. Individual brown planthoppers were then injected with 40 nl of peptide solution (2.25 pmol/insect) or 40 nl of PBS in the lateral side of the abdomen using a FemtoJet system (Eppendorf-Nethler-Hinz, Hamburg, Germany). Immediately after injection, ten animals were placed in separate plastic containers (9 cm long and 2 cm in diameter), provided with 200 μl of artificial diet and allowed to feed ad libitum for 24 hours.
For studying the effect of gene silencing on the feeding behavior of N. lugens, the dsRNA-injected 4th instar nymphs were reared on rice seedlings in the laboratory and maintained at 27 ± 1 ∘C, with 70 ± 10% relative humidity, under a 16 h: 8 h light dark photoperiod to recover for 2 d. After 5 hours starvation, ten nymphs were transferred into separate plastic containers (9 cm long and 2 cm in diameter), provided with 200 μl of artificial diet and allowed to feed ad libitum for 24 hours as mentioned above. The feeding amount of brown planthopper in each feeding chamber was recorded after 24h. The experiment was repeated at least four times.
RNA-seq analysis
Total RNA of thirty 4th instar nymphs was isolated at day three after dsNlsk or dsgfp injections in the 4th instar nymphs using a TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. Library construction and sequencing was performed by Novogene with Illumina HiSeq2000 platform (Novogene Bioinformatics Technology Co.Ltd, Beijing, China). Raw sequence data were submitted to the Short Read Archive (SRA) database of NCBI under the accession numbers SRR12460889 (dsNlsk-1), SRR12460895 (dsNlsk-2), SRR12460894 (dsNlsk-3), SRR12460893 (dsNlsk-4) and SRR12460896 (dsgfp-1), SRR12460892 (dsgfp-2), SRR12460891 (dsgfp-3), and SRR12460890 (dsgfp4).
The raw data were analyzed after filtering the low-quality sequences. Sequences were aligned to the Nilaparvata lugens genome (https://www.ncbi.nlm.nih.gov/genome/?term=Nilaparvata+lugens) using Hisat2 v2.0.5. The expression level of genes from the RNA sequencing was normalized by the FPKM method (Fragments Per Kilobase of transcript sequence per Millions base pairs sequenced). This method considers the effect of sequencing depth and gene length for the reads count at the same time and is currently the most commonly used method for estimating gene expression levels. Differential expression analysis was performed using the DESeq2 R package (1.16.1). The clusterProfiler R package was used for Gene Ontology (GO) enrichment analysis and KEGG pathway analysis [106]. FDR-adjusted multiple tests were added to the hypergeometric test.
Generation of 17D1GAL4 knock-in line
To prepare the 17D1GAL4 line, we used CRISPR-HDR (clustered regularly interspaced short palindromic repeats–homology directed repair) method based on previous methods [41]. We chose the upstream and a downstream guide RNAs targeting the part of first exon using the CRISPR Optimal Target Finder: http://tools.flycrispr.molbio.wisc.edu/targetFinder/. In brief, the part of first CCKLR-17D1 coding exon was replaced by GAL4::p65 (S6A Fig). Firstly, two gRNAs (gRNA1: 5′-GATTTATAAACTCGGGTCGCA-3′; gRNA2: 5′-TCACCGACAGCGGAGATCTC-3′) against CCKLR-17D1 were inserted into pCFD4 as previously described [107]. We then fused GAL4::p65 into pHD-DsRed (Addgene #51434) between the EcoRI and the NdeI sites. Next, each homologous arm was subcloned into the pHD-DsRed vector too. We injected the modified pCFD4 and pHD-DsRed plasmids into the embryo of vas-Cas9 flies (# 51324). The correct insertion was confirmed by 3xP3-DsRed screening and recombination accuracy was confirmed by sequencing.
NSK/DSK antibody
Rabbit anti-DSK antibody was generated by using the peptide N′-FDDYGHMRFC-C′ that corresponds to the predicted DSK-1 and DSK-2 peptides as antigen as previously reported [41]. We used the DSK antibody to recognize both SK peptides of N. lugens and Drosophila since the antigen share the same sequence in two species.
Immunohistochemistry
Unless otherwise stated, fourth instar nymph of brown planthopper and 3-5-day old mated female flies were dissected under phosphate-buffered saline (PBS; pH 7.4) or Schneider’s insect medium (S2) as previously described [41,108]. The tissues were fixed in 4% paraformaldehyde in PBS for 30 min at room temperature. After extensive washing with PAT (0.5% Triton X-100, 0.5% bovine serum albumin in PBS), the tissues were incubated in primary antibody for 24 h at 4°C and in secondary antibody for 24 h at 4°C. Primary antibodies used: mouse anti-GFP (Sigma-Aldrich Cat# G6539, 1:1000), rabbit anti-RFP (Abcam Cat#ab62341, 1:1000), mouse anti-Bruchpilot (Developmental Studies Hybridoma Bank nc82, 1:30), rabbit anti-DSK (see antibody generation section, 1:100). Secondary antibodies used: donkey anti-mouse IgG conjugated to Alexa 488 (1:500) or Alexa 555 (1:500) and donkey anti-rabbit IgG conjugated to Alexa 488 (1:500) or Alexa 555 (1:500) (Molecular Probes). The samples were mounted in Vectorshield (Vector Laboratory). Images were acquired with Zeiss LSM 700 confocal microscopes, and were processed with Image J software [109].
To quantify NlSK level in the brain, we stained the brains with DSK antibody (Fig 1E and 1F). The dsgfp- or dsNlsk-injected samples were processed in parallel and using the same solution and imaged with the same laser power and scanning settings. With the imaged data, we got “Sum Slices” Z-projection of the sub-stacks encompassing whole brain to measure fluorescence intensity (Fdsgfp and FdsNlsk), then select a small region without signal as the background fluorescence (Bdsgfp and Bdsgfp) using ImageJ. Then we obtained the relative fluorescence of NlSK as the ratio of NlSK signal to the control signal ((FdsNlsk—BdsNlsk)/(Fdsgfp − Bdsgfp)).
Arclight imaging
Imaging of freshly dissected brain explants of starved (24 hours) or refed (1.5 hours refed after 24 hours starvation) was performed on a Zeiss 710 NLO Axio Examiner confocal microscope using a water immersion objective (Zeiss, Germany). ArcLight was excited with the 488 nm laser. The objective C-mount image was projected onto the 256 × 256 pixel chip controlled by Zen2010 software (Zeiss Germany). Images were recorded at a frame rate of roughly 80 Hz, and depicted optical traces were spatial averages of intensity of all pixels within the region of interest (ROI, in this study, cell bodies of MP1 neurons), with signals processed as previously reported [68]. Statistical analysis and plotting of the data were performed using Excel and Prism GraphPad.
CaLexA measurements
Calcium activity of DSK neurons following starvation and refed was measured using the calcium-dependent nuclear import of LexA (CaLexA) reporter system [69]. Female flies (4–7-days old) carrying Dsk-GAL4 and UAS-CaLexA system were collected. They were then divided in two groups: one group was starved for 24 hours in presence of water and another group was refed 1.5 hours after 24 hours starvation. Following this period, the fly brains were quickly dissected and were processed for immunohistochemistry. The GFP fluorescence were quantified as described above.
CsChrimson activation
For optogenetic stimulation, tester flies were collected within twelve hours after eclosion and transferred into a vial with regular food containing 200 μM all-trans retinal (116–31–4, Sigma-Aldrich). The vials were covered by aluminum foil to protect from light for 3–5 days before PER test. We immobilized flies on a glass slide with back down so that the proboscis was exposed to the upside and stimulated the animal with red (620 nm, 0.03 mW/mm2, Vanch Technology, Shanghai, China) light. Unless otherwise noted, light stimulation was presented continuously throughout the observation period. Light intensity was measured by placing an optical power meter (PS-310 V2, Gentec, Canada) nearby the location of glass slide.
dTrpA1 activation
Experimental flies were maintained at 22°C, cold anesthetized and loaded into CAFE vial, and allowed to recover for at least 30 min at 22°C. Vials containing flies were then placed at experimental temperatures (30°C) for 24 hours to recording the feeding amount.
Extracellular tip recording
7–10 days old females were used for electrophysiological recordings. All flies were kept in dark after eclosion and fed with 200 μM all-trans-retinal for 3 to 5 days. Before the assays, flies were transferred to a tube contained a filter paper with 2ml of all-trams-retinal solution (200 μM all-trans-retinal diluted in 2 ml ultrapure water) for 24 hours, and then some groups were refed for 1.5 hour. Electrophysiological recording from Drosophila labellar taste sensilla were implemented as previously reported [110]. A reference electrode was inserted into dorsal thorax of fly, and proboscis was fully extended. The recording electrode was approached to the tip of a single L-sensilla on labellum and covered ~ 50% of the total shaft length. Beadle-Ephrussi Ringer solution (B&E) was used as the reference electrode electrolyte, which contained 7.5 g NaCl, 0.35g KCl, and 0.279 g CaCl2∙2H2O in one liter of ultrapure water. Using 30 mM tricholine citrate solution (TCC) solved with 100 mM sucrose as the recording electrode electrolyte. Both reference and recording electrodes were capillary glass (borosilicate glass with filament, BF120-69-15; O.D.: 1.2mm, I.D.: 0.69mm) that was pulled on a P-97 puller (Sutter Instrument Corp). The recording electrode was connected to an amplifier (TastePROBE DTP-02, SYNTECH) and a data acquisition device (Axon Digidata 1550B, Molecular Device) under the control of Axon pCLAMP 10.6 software (Molecular Devices). Data was analyzed with Clampfit 10.7 software.
Proboscis extension response (PER) assays
We performed the PER assays at about 2:00–5:00 pm (ZT6–ZT10) at room temperature. 3-5-day-old females were used for the assays. Before the assays, flies were starved for 24 hours, and refed for 1.5 hour. We first anesthetized flies with carbon dioxide and stuck them on microscope slides. After one hour recovery, we tested flies with water. Proboscis were touched by a water drop, and if the fly did not extend its proboscis in three seconds, we performed the assays with different concentrations of sucrose. Two values were used in the PER assays. A score of 1 means a fly that extended its proboscis and ingested after being fed the sucrose water drop. If not, the score of that fly is 0. We averaged the scores of 5–10 flies as one replicate.
Capillary feeding (CAFE) assay
This method was modified from Ja et al. [71]. A vial (9 cm height × 2 cm diameter), filled with 5 ml of 1% agarose to provide water for the flies, was used for this assay. Capillaries (5 μl, VWR International) were exchanged daily with new ones containing fresh food solution (5% sucrose and 5% yeast extract dissolved in water). Typically, 24-h feeding of every fly is shown after 1 or 2 d of habituation in this assay. The amount of consumed food minus evaporation was quantified.
Statistics
We used the GraphPad Prism 7 software package to generate graphs and statistically analyze data. Data presented in this study were first verified for normal distribution by D’Agostino–Pearson normality test. If normally distributed, Student’s t test was used for pairwise comparisons, and one-way ANOVA was used for comparisons among multiple groups, followed by Tukey’s multiple comparisons. If not normally distributed, Mann–Whitney test was used for pairwise comparisons, and Kruskal–Wallis test was used for comparisons among multiple groups, followed by Dunn’s multiple comparisons. All data are presented as mean ± s.e.m. The sample sizes and statistical tests used for each experiment are stated in the figures or figure legends. The raw data of this project are shown in the S7 Table.
Supporting information
qRT-PCR analysis of six selected genes from RNA-seq data after Nlsk knockdown by RNAi in the brown planthopper (A-F). All data are presented as means ± s.e.m. ***p < 0.001, **p < 0.01, *p < 0.05; Mann–Whitney test.
(TIF)
(A) Downregulation of Nlsk gene using Nlsk-RNAi (dsNlsk) leads to up-regulation of transcript of sweet sensing NlGr43a. *p < 0.05; Mann–Whitney test. (B) Injection of sNlSK2 leads to down-regulation of NlGr43a gene. ***p < 0.001; Mann–Whitney test.
(TIF)
ns: not significant; Student’s t test.
(TIF)
All flies were kept in CAFE tubes for 24 hours at 30°C. ns: not significant; Kruskal–Wallis test followed by Dunn’s multiple comparisons test.
(TIF)
(A) Silencing the Dsk gene using the Dsk-GAL4 driver has no impact on feeding in the proboscis extension reflex (PER). n = 10 trials. ns: not significant; Mann–Whitney test. (B) Silencing the Dsk gene using the Dsk-GAL4 driver has no impact on feeding in the CAFE assay. n = 10 trials. ns: not significant; Mann–Whitney test.
(TIF)
Scale bars: 50 μm.
(TIF)
(A) Starved Canton-S showed more motivation to feed in the PER assay. n = 10 trials. *p < 0.05; Mann–Whitney test. (B and C) Sugar blind mutants and silencing sweet GRNs by expressing the Kir2.1 channel did not influence responses to starvation and re-feeding in the PER. n = 10 trials. ns, no significant; Mann–Whitney test. (D and E) Starved dsk mutants and flies with silencing of Dsk-GAL4 labeled neurons by expressing the Kir2.1 channel displayed more motivation to feed in PER. n = 10 trials. *p < 0.05; Mann–Whitney test.
(TIF)
Double labeling of Dsk-GAL4-expressing neurons and Gr64fLexA-expressing sweet gustatory neurons in the Drosophila brain (A: anterior and B: posterior), proleg (C) and proboscis (D). No Dsk signal was detected on the proleg and proboscis (C and D). Scale bar: 50 μm.
(TIF)
(A) Generation of knock-in of GAL4 into the CCKLR-17D1 locus. (B) No signal was detected when the 17D1GAL4 drives stinger and GFP in the proboscis and leg tarsi. Scale bar: 50 μm. (C) 17D1 mutants show no decreased motivation to feed in PER compared with control. **p < 0.01; Kruskal–Wallis test followed by Dunn’s multiple comparisons test.
(TIF)
Expression pattern of 17D3GAL4 visualized by UAS-mCD8::GFP in leg tarsi (A1-A3), proboscis and maxillary palps (B1-B3). Scale bar: 50 μm.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(MP4)
Acknowledgments
We thank Yi Rao (Peking University), Wei Zhang (Tsinghua University) and Wei Song (Wuhan University) for sharing fly strains. Other members of the Wu and Gao laboratory are thanked for helpful discussions.
Data Availability
Raw sequence Data of the RNA-seq analysis in this study are available in the Short Read Archive (SRA) database of NCBI with the accession numbers and NCBI URLs as below: dsNlsk_1, (SRR12460889, https://www.ncbi.nlm.nih.gov/sra/?term=SRR12460889), dsNlsk_2, (SRR12460895, https://www.ncbi.nlm.nih.gov/sra/?term=SRR12460895), dsNlsk_3, (SRR12460894, https://www.ncbi.nlm.nih.gov/sra/?term=SRR12460894), dsNlsk_4, (SRR12460893, https://www.ncbi.nlm.nih.gov/sra/?term=SRR12460893), dsgfp_1 (SRR12460896, https://www.ncbi.nlm.nih.gov/sra/?term=SRR12460896), dsgfp_2 (SRR12460892, https://www.ncbi.nlm.nih.gov/sra/?term=SRR12460892), dsgfp_3 (SRR12460891, https://www.ncbi.nlm.nih.gov/sra/?term=SRR12460891) and dsgfp_4 (SRR12460890, https://www.ncbi.nlm.nih.gov/sra/?term=SRR12460890).
Funding Statement
This work was supported by the National Natural Science Foundation of China to SFW (No. 32022011 & 31772205) (https://isisn.nsfc.gov.cn/egrantindex/funcindex/prjsearch-list?locale=zh_CN). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Davis WJ. Behavioural hierarchies. Trends Neurosci. 1979; 2:5–7. 10.1016/0166-2236(79)90003-1 [DOI] [Google Scholar]
- 2.Münch D, Ezra-Nevo G, Francisco AP, Tastekin I, Ribeiro C. Nutrient homeostasis—translating internal states to behavior. Curr Opin Neurobiol. 2020; 60:67–75. 10.1016/j.conb.2019.10.004 [DOI] [PubMed] [Google Scholar]
- 3.Sternson SM, Nicholas Betley J, Cao ZFH. Neural circuits and motivational processes for hunger. Curr Opin Neurobiol. 2013; 23(3):353–60. 10.1016/j.conb.2013.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim SM, Su CY, Wang JW. Neuromodulation of innate behaviors in Drosophila. Annu Rev Neurosci 2017; 40:327–48. 10.1146/annurev-neuro-072116-031558 [DOI] [PubMed] [Google Scholar]
- 5.Yapici N, Zimmer M, Domingos AI. Cellular and molecular basis of decision-making. EMBO rep. 2014; 15(10):1023–35. 10.15252/embr.201438993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Root Cory M, Ko Kang I, Jafari A, Wang Jing W. Presynaptic facilitation by neuropeptide signaling mediates odor-driven food search. Cell. 2011; 145(1):133–44. 10.1016/j.cell.2011.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inagaki Hidehiko K, Ben-Tabou de-Leon S, Wong AM, Jagadish S, Ishimoto H, et al. Visualizing neuromodulation In vivo: TANGO-mapping of dopamine signaling reveals appetite control of sugar sensing. Cell. 2012; 148(3):583–95. 10.1016/j.cell.2011.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yapici N, Cohn R, Schusterreiter C, Ruta V, Vosshall Leslie B. A taste circuit that regulates ingestion by integrating food and hunger signals. Cell. 2016; 165(3):715–29. 10.1016/j.cell.2016.02.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morton GJ, Cummings DE, Baskin DG, Barsh GS, Schwartz MW. Central nervous system control of food intake and body weight. Nature 2006; 443(7109):289–95. 10.1038/nature05026 [DOI] [PubMed] [Google Scholar]
- 10.Ko KI, Root CM, Lindsay SA, Zaninovich OA, Shepherd AK, et al. Starvation promotes concerted modulation of appetitive olfactory behavior via parallel neuromodulatory circuits. eLife. 2015; 4:e08298. 10.7554/eLife.08298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inagaki Hidehiko K, Panse Ketaki M, Anderson David J. Independent, reciprocal neuromodulatory control of sweet and bitter taste sensitivity during starvation in Drosophila. Neuron. 2014; 84(4):806–20. 10.1016/j.neuron.2014.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pool AH, Scott K. Feeding regulation in Drosophila. Curr Opin Neurobiol. 2014; 29:57–63. 10.1016/j.conb.2014.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grunwald Kadow IC. State-dependent plasticity of innate behavior in fruit flies. Curr Opin Neurobiol. 2019; 54:60–5. 10.1016/j.conb.2018.08.014 [DOI] [PubMed] [Google Scholar]
- 14.LeDue Emily E, Mann K, Koch E, Chu B, Dakin R, et al. Starvation-induced depotentiation of bitter taste in Drosophila. Curr Biol. 2016; 26(21):2854–61. 10.1016/j.cub.2016.08.028 [DOI] [PubMed] [Google Scholar]
- 15.Itskov PM, Ribeiro C. The dilemmas of the gourmet fly: The molecular and neuronal mechanism of feeding and nutrient decision making in Drosophila. Front Neurosci. 2013; 7:12. 10.3389/fnins.2013.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Audsley N, Weaver RJ. Neuropeptides associated with the regulation of feeding in insects. Gen Comp Endocrinol. 2009; 162(1):93–104. 10.1016/j.ygcen.2008.08.003 [DOI] [PubMed] [Google Scholar]
- 17.Nässel DR, Zandawala M. Recent advances in neuropeptide signaling in Drosophila, from genes to physiology and behavior. Prog Neurobiol. 2019; 179:101607. 10.1016/j.pneurobio.2019.02.003 [DOI] [PubMed] [Google Scholar]
- 18.Martelli C, Pech U, Kobbenbring S, Pauls D, Bahl B, et al. SIFamide translates hunger signals into appetitive and feeding behavior in Drosophila. Cell Rep. 2017; 20(2):464–78. 10.1016/j.celrep.2017.06.043 [DOI] [PubMed] [Google Scholar]
- 19.Nässel DR, Pauls D, Huetteroth W. Neuropeptides in modulation of Drosophila behavior: how to get a grip on their pleiotropic actions. Curr Opin Insect Sci. 2019; 36:1–8. 10.1016/j.cois.2019.03.002 [DOI] [PubMed] [Google Scholar]
- 20.Owusu-Ansah E, Perrimon N. Modeling metabolic homeostasis and nutrient sensing in Drosophila: implications for aging and metabolic diseases. Dis Model Mech. 2014; 7(3):343–50. 10.1242/dmm.012989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ignell R, Root CM, Birse RT, Wang JW, Nässel DR, et al. Presynaptic peptidergic modulation of olfactory receptor neurons in Drosophila. Proc Natl Acad Sci USA. 2009; 106(31):13070–5. 10.1073/pnas.0813004106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hussain A, Üçpunar HK, Zhang M, Loschek LF, Grunwald Kadow IC. Neuropeptides modulate female chemosensory processing upon mating in Drosophila. PLoS Biol. 2016; 14(5):e1002455. 10.1371/journal.pbio.1002455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farhan A, Gulati J, Groβe-Wilde E, Vogel H, Hansson BS, et al. The CCHamide 1 receptor modulates sensory perception and olfactory behavior in starved Drosophila. Sci Rep. 2013; 3:2765. 10.1038/srep02765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi W, Wang G, Wang L. A novel satiety sensor detects circulating glucose and suppresses food consumption via insulin-producing cells in Drosophila. Cell Res. 2021; 31(5):580–8. 10.1038/s41422-020-00449-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung BY, Ro J, Hutter SA, Miller KM, Guduguntla LS, et al. Drosophila neuropeptide F signaling independently regulates feeding and sleep-wake behavior. Cell Rep. 2017; 19(12):2441–50. 10.1016/j.celrep.2017.05.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee KS, You KH, Choo JK, Han YM, Yu K. Drosophila short neuropeptide F regulates food intake and body size. J Biol Chem. 2004; 279(49):50781–9. 10.1074/jbc.M407842200 [DOI] [PubMed] [Google Scholar]
- 27.Shao L, Saver M, Chung P, Ren Q, Lee T, et al. Dissection of the Drosophila neuropeptide F circuit using a high-throughput two-choice assay. Proc Natl Acad Sci U S A. 2017; 114(38):E8091–99. 10.1073/pnas.1710552114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nässel DR, Wegener C. A comparative review of short and long neuropeptide F signaling in invertebrates: Any similarities to vertebrate neuropeptide Y signaling? Peptides. 2011; 32(6):1335–55. 10.1016/j.peptides.2011.03.013 [DOI] [PubMed] [Google Scholar]
- 29.Lee S, Kim Y-J, Jones WD. Central peptidergic modulation of peripheral olfactory responses. BMC Biol. 2017; 15(1):35. 10.1186/s12915-017-0374-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pu Y, Zhang Y, Zhang Y, Shen P. Two Drosophila neuropeptide Y-like neurons define a reward module for transforming appetitive odor representations to motivation. Sci Rep. 2018; 8(1):11658. 10.1038/s41598-018-30113-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen P, Cai HN. Drosophila neuropeptide F mediates integration of chemosensory stimulation and conditioning of the nervous system by food. J Neurobiol. 2001; 47(1):16–25. 10.1002/neu.1012 [DOI] [PubMed] [Google Scholar]
- 32.Beshel J, Zhong Y. Graded encoding of food odor value in the Drosophila brain. J Neurosci 33(40):15693–704. 10.1523/JNEUROSCI.2605-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Söderberg JAE, Carlsson MA, Nässel DR. Insulin-producing cells in the Drosophila brain also express satiety-inducing cholecystokinin-like peptide, Drosulfakinin. Front Endocrinol. 2012; 3:109. 10.3389/fendo.2012.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu N, Smagghe G. Characterization of sulfakinin receptor 2 and its role in food intake in the red flour beetle, Tribolium castaneum. Peptides. 2014; 53:232–7. 10.1016/j.peptides.2013.12.011 [DOI] [PubMed] [Google Scholar]
- 35.Zels S, Dillen S, Crabbé K, Spit J, Nachman RJ, et al. Sulfakinin is an important regulator of digestive processes in the migratory locust, Locusta migratoria. Insect Biochem Mol Biol. 2015; 61:8–16. 10.1016/j.ibmb.2015.03.008 [DOI] [PubMed] [Google Scholar]
- 36.Dockray GJ. Cholecystokinin and gut–brain signalling. Regul Pept. 2009; 155(1–3):6–10. 10.1016/j.regpep.2009.03.015 [DOI] [PubMed] [Google Scholar]
- 37.Nässel DR, Williams MJ. Cholecystokinin-like peptide (DSK) in Drosophila, not only for satiety signaling. Front Endocrinol. 2014; 5:219. 10.3389/fendo.2014.00219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rehfeld JF. Cholecystokinin—From local gut hormone to ubiquitous messenger. Front Endocrinol. 2017; 8:47. 10.3389/fendo.2017.00047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agrawal P, Kao D, Chung P, Looger LL. The neuropeptide Drosulfakinin regulates social isolation-induced aggression in Drosophila. J Exp Biol. 2020; 223(Pt 2):jeb207407. 10.1242/jeb.207407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams MJ, Goergen P, Rajendran J, Klockars A, Kasagiannis A, et al. Regulation of aggression by obesity-linked genes TfAP-2 and Twz through octopamine signaling in Drosophila. Genetics. 2014; 196(1):349–62. 10.1534/genetics.113.158402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu SF, Guo C, Zhao H, Sun MS, Chen J, et al. Drosulfakinin signaling in fruitless circuitry antagonizes P1 neurons to regulate sexual arousal in Drosophila. Nat Commun. 2019; 10(1):4770. 10.1038/s41467-019-12758-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nachman R, Holman G, Haddon W, Ling N. Leucosulfakinin, a sulfated insect neuropeptide with homology to gastrin and cholecystokinin. Science. 1986; 234(4772):71–3. 10.1126/science.3749893 [DOI] [PubMed] [Google Scholar]
- 43.Nachman RJ, Holman GM, Cook BJ, Haddon WF, Ling N. Leucosulfakinin-II, a blocked sulfated insect neuropeptide with homology to cholecystokinin and gastrin. Biochem Biophys Res Commun. 1986; 140(1):357–64. 10.1016/0006-291x(86)91098-3 [DOI] [PubMed] [Google Scholar]
- 44.Wei Z, Baggerman G, J. Nachman R, Goldsworthy G, Verhaert P, et al. Sulfakinins reduce food intake in the desert locust, Schistocerca gregaria. J Insect Physiol. 2000; 46(9):1259–65. 10.1016/s0022-1910(00)00046-9 [DOI] [PubMed] [Google Scholar]
- 45.Maestro JL, Aguilar R, Pascual N, Valero M-L, Piulachs M-D, et al. Screening of antifeedant activity in brain extracts led to the identification of sulfakinin as a satiety promoter in the German cockroach. Eur J Biochem. 2001; 268(22):5824–30. 10.1046/j.0014-2956.2001.02527.x [DOI] [PubMed] [Google Scholar]
- 46.Yu N, Benzi V, Zotti MJ, Staljanssens D, Kaczmarek K, et al. Analogs of sulfakinin-related peptides demonstrate reduction in food intake in the red flour beetle, Tribolium castaneum, while putative antagonists increase consumption. Peptides. 2013; 41:107–12. 10.1016/j.peptides.2012.12.005 [DOI] [PubMed] [Google Scholar]
- 47.Guo D, Zhang S, Zhang YJ, Ma JY, Gao CF, Wu SF. Sulfakinin inhibits activity of digestive enzymes in the brown planthopper, Nilaparvata lugens. J Asia Pac Entomol. 2020; 23(4):1073–82. 10.1016/j.aspen.2020.09.004 PMID: [DOI] [Google Scholar]
- 48.Scott K. Gustatory processing in Drosophila melanogaster. Annu Rev Entomol. 2018; 63:15–30. 10.1146/annurev-ento-020117-043331 [DOI] [PubMed] [Google Scholar]
- 49.Dahanukar A, Lei Y-T, Kwon JY, Carlson JR. Two Gr genes underlie sugar reception in Drosophila. Neuron. 2007; 56:503–516. 10.1016/j.neuron.2007.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiao Y, Moon SJ, Montell C. A Drosophila gustatory receptor required for the responses to sucrose, glucose, and maltose identified by mRNA tagging. Proc Natl Acad Sci. 2007; 104(35):14110–5. 10.1073/pnas.0702421104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiao Y, Moon SJ, Wang X, Ren Q, Montell C. Gr64f is required in combination with other gustatory receptors for sugar detection in Drosophila. Curr Biol. 2008; 18(22):1797–801. 10.1016/j.cub.2008.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu SF, Zeng B, Zheng C, Mu XC, Zhang Y, Hu J, et al. The evolution of insecticide resistance in the brown planthopper (Nilaparvata lugens Stål) of China in the period 2012–2016. Sci Rep. 2018; 8(1):4586. 10.1038/s41598-018-22906-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang LX, Niu CD, Salgado VL, Lelito K, Stam L, Jia YL, et al. Pymetrozine activates TRPV channels of brown planthopper Nilaparvata lugens. Pestic Biochem Physiol. 2019; 153:77–86. 10.1016/j.pestbp.2018.11.005 [DOI] [PubMed] [Google Scholar]
- 54.Wang LX, Zhang YC, Tao S, Guo D, Zhang Y, Jia YL, et al. Pymetrozine inhibits reproductive behavior of brown planthopper Nilaparvata lugens and fruit fly Drosophila melanogaster. Pestic Biochem Physiol. 2020; 165:104548. 10.1016/j.pestbp.2020.02.014 [DOI] [PubMed] [Google Scholar]
- 55.Miroschnikow A, Schlegel P, Pankratz MJ. Making feeding decisions in the Drosophila nervous system. Curr Biol. 2020; 30(14):831–40. 10.1016/j.cub.2020.06.036 [DOI] [PubMed] [Google Scholar]
- 56.Wicher D, Derst C, Gautier H, Lapied B, Heinemann SH, Agricola HJ. The satiety signaling neuropeptide perisulfakinin inhibits the activity of central neurons promoting general activity. Front Cell Neurosci. 2007; 1:3. 10.3389/neuro.03.003.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Downer KE, Haselton AT, Nachman RJ, Stoffolano JG Jr. Insect satiety: Sulfakinin localization and the effect of drosulfakinin on protein and carbohydrate ingestion in the blow fly, Phormia regina (Diptera: Calliphoridae). J Insect Physiol. 2007; 53(1):106–12. 10.1016/j.jinsphys.2006.10.013 [DOI] [PubMed] [Google Scholar]
- 58.Nichols R, Lim IA. Spatial and temporal immunocytochemical analysis of drosulfakinin (Dsk) gene products in the Drosophila melanogaster central nervous system. Cell Tissue Res. 1996; 283(1):107–16. 10.1007/s004410050518 [DOI] [PubMed] [Google Scholar]
- 59.Filloux C, Cédric M, Romain P, Lionel F, Christophe K, Dominique R, et al. An integrative method to normalize RNA-Seq data. BMC Bioinformatics. 2014; 15:188. 10.1186/1471-2105-15-188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tarazona S, García-Alcalde F, Dopazo J, Ferrer A, Conesa A. Differential expression in RNA-seq: A matter of depth. Genome Res. 2011; 21(12):2213–23. 10.1101/gr.124321.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sarov-Blat L, So WV, Liu L, Rosbash M. The Drosophila takeout gene is a novel molecular link between circadian rhythms and feeding behavior. Cell. 2000; 101(6):647–56. 10.1016/s0092-8674(00)80876-4 [DOI] [PubMed] [Google Scholar]
- 62.Meunier N, Belgacem YH, Martin J-R. Regulation of feeding behaviour and locomotor activity by takeout in Drosophila. J Exp Biol. 2007; 210(8):1424–34. 10.1242/jeb.02755 [DOI] [PubMed] [Google Scholar]
- 63.Miyamoto T, Slone J, Song X, Amrein H. A fructose receptor functions as a nutrient sensor in the Drosophila brain. Cell. 2012; 151(5):1113–25. 10.1016/j.cell.2012.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Z, Singhvi A, Kong P, Scott K. Taste representations in the Drosophila brain. Cell. 2004; 117(7):981–91. 10.1016/j.cell.2004.06.011 [DOI] [PubMed] [Google Scholar]
- 65.Xie J, Zhu H, Larade K, Ladoux A, Seguritan A, Chu M, et al. Absence of a reductase, NCB5OR, causes insulin-deficient diabetes. Proc Natl Acad Sci U S A. 2004; 101(29):10750–5. 10.1073/pnas.0404044101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu F, Deng B, Xiao N, Wang T, Li Y, Wang R, et al. A neuropeptide regulates fighting behavior in Drosophila melanogaster. eLife. 2020; 9:e54229. 10.7554/eLife.54229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Williams MJ, Goergen P, Rajendran J, Zheleznyakova G, Hägglund MG, Perland E, et al. Obesity-linked homologues TfAP-2 and Twz establish meal frequency in Drosophila melanogaster. PLoS Genet. 2014; 10(9):e1004499. 10.1371/journal.pgen.1004499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cao G, Platisa J, Pieribone Vincent A, Raccuglia D, Kunst M, Nitabach MN. Genetically targeted optical electrophysiology in intact neural circuits. Cell. 2013; 154(4):904–13. 10.1016/j.cell.2013.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Masuyama K, Zhang Y, Rao Y, Wang JW. Mapping neural circuits with activity-dependent nuclear import of a transcription factor. J Neurogenet. 2012; 26(1):89–102. 10.3109/01677063.2011.642910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Klapoetke NC, Murata Y, Kim SS, Pulver SR, Birdsey-Benson A, Cho YK, et al. Independent optical excitation of distinct neural populations. Nat Meth. 2014; 11(3):338–46. 10.1038/nmeth.2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ja WW, Carvalho GB, Mak EM, de la Rosa NN, Fang AY, Liong JC, et al. Prandiology of Drosophila and the CAFE assay. Proc Natl Acad Sci U S A. 2007; 104(20):8253–6. 10.1073/pnas.0702726104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fujii S, Yavuz A, Slone J, Jagge C, Song X, Amrein H. Drosophila sugar receptors in sweet taste perception, olfaction, and internal nutrient sensing. Curr Biol. 2015; 25(5):621–7. 10.1016/j.cub.2014.12.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yavuz A, Jagge C, Slone J, Amrein H. A genetic tool kit for cellular and behavioral analyses of insect sugar receptors. Fly. 2014; 8(4):189–96. 10.1080/19336934.2015.1050569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Inagaki HK, Jung Y, Hoopfer ED, Wong AM, Mishra N, Lin JY, et al. Optogenetic control of Drosophila using a red-shifted channelrhodopsin reveals experience-dependent influences on courtship. Nat Meth. 2014; 11(3):325–32. 10.1038/nmeth.2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moreira J-M, Itskov PM, Goldschmidt D, Baltazar C, Steck K, Tastekin I, et al. optoPAD, a closed-loop optogenetics system to study the circuit basis of feeding behaviors. eLife. 2019; 8:e43924. 10.7554/eLife.43924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kubiak TM, Larsen MJ, Burton KJ, Bannow CA, Martin RA, ZantelloMR, et al. Cloning and functional expression of the first Drosophila melanogaster sulfakinin receptor DSK-R1. Biochem Biophys Res Commun. 2002; 291(2):313–20. 10.1006/bbrc.2002.6459 [DOI] [PubMed] [Google Scholar]
- 77.Chen X, Peterson J, Nachman RJ, Ganetzky B. Drosulfakinin activates CCKLR-17D1 and promotes larval locomotion and escape response in Drosophila. Fly. 2012; 6(4):290–7. 10.4161/fly.21534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen X, Ganetzky B. A neuropeptide signaling pathway regulates synaptic growth in Drosophila. J Cell Biol. 2012; 196(4):529–43. 10.1083/jcb.201109044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jourjine N. Hunger and thirst interact to regulate ingestive behavior in flies and mammals. BioEssays. 2017; 39(5):1600261. 10.1002/bies.201600261 [DOI] [PubMed] [Google Scholar]
- 80.Nagata S, Zhou YJ. Chapter Four—Feeding-modulating neuropeptides and peptide hormones in insects. In: Jurenka R, editor. Advances in Insect Physiology: Academic Press. 2019; pp. 137–172. [Google Scholar]
- 81.Williams KW, Elmquist JK. From neuroanatomy to behavior: central integration of peripheral signals regulating feeding behavior. Nat Neurosci. 2012; 15(10):1350–5. 10.1038/nn.3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zels S, Verlinden H, Dillen S, Vleugels R, Nachman RJ, Broeck JV. Signaling properties and pharmacological analysis of two sulfakinin receptors from the red flour beetle, Tribolium castaneum. PLoS ONE. 2014; 9(4):e94502. 10.1371/journal.pone.0094502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Al-Alkawi H, Lange AB, Orchard I. Cloning, localization, and physiological effects of sulfakinin in the kissing bug, Rhodnius prolixus. Peptides. 2017; 98:15–22. 10.1016/j.peptides.2016.12.017 [DOI] [PubMed] [Google Scholar]
- 84.Oranth A, Schultheis C, Tolstenkov O, Erbguth K, Nagpal J, Hain D, et al. Food sensation modulates locomotion by dopamine and neuropeptide signaling in a distributed neuronal network. Neuron. 2018; 100(6):1414–1428.e1410. 10.1016/j.neuron.2018.10.024 [DOI] [PubMed] [Google Scholar]
- 85.Moran TH, Schwartz GJ. Neurobiology of cholecystokinin. Crit Rev Neurobiol. 1994; 9(1):1–28. [PubMed] [Google Scholar]
- 86.Harshini S, Nachman RJ, Sreekumar S. In vitro release of digestive enzymes by FMRF amide related neuropeptides and analogues in the lepidopteran insect Opisina arenosella (Walk.). Peptides. 2002; 23(10):1759–63. 10.1016/s0196-9781(02)00152-3 [DOI] [PubMed] [Google Scholar]
- 87.Zwanzger P, Domschke K, Bradwejn J. Neuronal network of panic disorder: The role of the neuropeptide cholecystokinin. Depress Anxiety. 2012; 29(9):762–74. 10.1002/da.21919 [DOI] [PubMed] [Google Scholar]
- 88.Chintapalli VR, Wang J, Dow JAT. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007; 39(6):715–20. 10.1038/ng2049 [DOI] [PubMed] [Google Scholar]
- 89.Terhzaz S, Rosay P, Goodwin SF, Veenstra JA. The neuropeptide SIFamide modulates sexual behavior in Drosophila. Biochem Biophys Res Commun. 2007; 352(2):305–10. 10.1016/j.bbrc.2006.11.030 [DOI] [PubMed] [Google Scholar]
- 90.Sellami A, Veenstra JA. SIFamide acts on fruitless neurons to modulate sexual behavior in Drosophila melanogaster. Peptides. 2015; 74:50–6. 10.1016/j.peptides.2015.10.003 [DOI] [PubMed] [Google Scholar]
- 91.Wu SF, Jv XM, Li J, Xu GJ, Cai XY, Gao CF. Pharmacological characterisation and functional roles for egg-laying of a β-adrenergic-like octopamine receptor in the brown planthopper Nilaparvata lugens. Insect Biochem Mol Biol. 2017; 87:55–64. 10.1016/j.ibmb.2017.06.008 [DOI] [PubMed] [Google Scholar]
- 92.Weiss LA, Dahanukar A, Kwon JY, Banerjee D, Carlson JR. The molecular and cellular basis of bitter taste in Drosophila. Neuron. 2011; 69(2):258–72. 10.1016/j.neuron.2011.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Meissner GW, Luo SD, Dias BG, Texada MJ, Baker BS. Sex-specific regulation of Lgr3 in Drosophila neurons. Proc Natl Acad Sci U S A. 2016; 113(9):E1256–65. 10.1073/pnas.1600241113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hamada FN, Rosenzweig M, Kang K, Pulver SR, Ghezzi A, Jegla, et al. An internal thermal sensor controlling temperature preference in Drosophila. Nature. 2008; 454(7201):217–20. 10.1038/nature07001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang L, Anderson DJ. Identification of an aggression-promoting pheromone and its receptor neurons in Drosophila. Nature. 2010; 463(7278):227–231. 10.1038/nature08678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Macpherson LJ, Zaharieva EE, Kearney PJ, Alpert MH, Lin T-Y, Turan Z, et al. Dynamic labelling of neural connections in multiple colours by trans-synaptic fluorescence complementation. Nat Commun. 2015; 6:10024. 10.1038/ncomms10024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Zhou DY, et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Meth. 2011; 8(5):405–7. 10.1038/nmeth.1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ni JQ, Liu LP, Binari R, Hardy R, Shim HS, CavallaroA, et al. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics. 2009; 182(4):1089–100. 10.1534/genetics.109.103630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Feng K, Palfreyman Mark T, Häsemeyer M, Talsma A, Dickson Barry J. Ascending SAG neurons control sexual receptivity of Drosophila females. Neuron. 2014; 83(1):135–48. 10.1016/j.neuron.2014.05.017 [DOI] [PubMed] [Google Scholar]
- 100.Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: Growth and diabetic phenotypes. Science, 2002; 296(5570):1118–20. 10.1126/science.1070058 [DOI] [PubMed] [Google Scholar]
- 101.Deng B, Li Q, Liu X, Cao Y, Li B, Qian Y, et al. Chemoconnectomics: Mapping chemical transmission in Drosophila. Neuron. 2019; 101(5):876–893.e4. https://10.1016/j.neuron.2019.01.045 [DOI] [PubMed] [Google Scholar]
- 102.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J Mol Biol. 305(5):567–80. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 103.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016; 33(7):1870–4. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001; 25(4):402–8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 105.Fu Q, Zhang Z, Hu C, Lai F, Sun Z. A chemically defined diet enables continuous rearing of the brown planthopper, Nilaparvata lugens (Stal) (Homoptera: Delphacidae). Appl Entomol Zool. 2001; 36(1):111–6. 10.1303/aez.2001.111 [DOI] [Google Scholar]
- 106.Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Research. 2007; 35:W193–200. 10.1093/nar/gkm226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Port F, Chen HM, Lee T, Bullock SL. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci U S A. 2014; 111(29):2967–76. 10.1073/pnas.1405500111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wu SF, Ja YL, Zhang YJ, Yang CH. Sweet neurons inhibit texture discrimination by signaling TMC-expressing mechanosensitive neurons in Drosophila. eLife. 2019; 8:e46165. 10.7554/eLife.46165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012; 9(7):671–5. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ling F, Dahanukar A, Weiss LA, Kwon JY, Carlson JR. The molecular and cellular basis of taste coding in the legs of Drosophila. J Neurosci 34(21):7148–64. 10.1523/JNEUROSCI.0649-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
qRT-PCR analysis of six selected genes from RNA-seq data after Nlsk knockdown by RNAi in the brown planthopper (A-F). All data are presented as means ± s.e.m. ***p < 0.001, **p < 0.01, *p < 0.05; Mann–Whitney test.
(TIF)
(A) Downregulation of Nlsk gene using Nlsk-RNAi (dsNlsk) leads to up-regulation of transcript of sweet sensing NlGr43a. *p < 0.05; Mann–Whitney test. (B) Injection of sNlSK2 leads to down-regulation of NlGr43a gene. ***p < 0.001; Mann–Whitney test.
(TIF)
ns: not significant; Student’s t test.
(TIF)
All flies were kept in CAFE tubes for 24 hours at 30°C. ns: not significant; Kruskal–Wallis test followed by Dunn’s multiple comparisons test.
(TIF)
(A) Silencing the Dsk gene using the Dsk-GAL4 driver has no impact on feeding in the proboscis extension reflex (PER). n = 10 trials. ns: not significant; Mann–Whitney test. (B) Silencing the Dsk gene using the Dsk-GAL4 driver has no impact on feeding in the CAFE assay. n = 10 trials. ns: not significant; Mann–Whitney test.
(TIF)
Scale bars: 50 μm.
(TIF)
(A) Starved Canton-S showed more motivation to feed in the PER assay. n = 10 trials. *p < 0.05; Mann–Whitney test. (B and C) Sugar blind mutants and silencing sweet GRNs by expressing the Kir2.1 channel did not influence responses to starvation and re-feeding in the PER. n = 10 trials. ns, no significant; Mann–Whitney test. (D and E) Starved dsk mutants and flies with silencing of Dsk-GAL4 labeled neurons by expressing the Kir2.1 channel displayed more motivation to feed in PER. n = 10 trials. *p < 0.05; Mann–Whitney test.
(TIF)
Double labeling of Dsk-GAL4-expressing neurons and Gr64fLexA-expressing sweet gustatory neurons in the Drosophila brain (A: anterior and B: posterior), proleg (C) and proboscis (D). No Dsk signal was detected on the proleg and proboscis (C and D). Scale bar: 50 μm.
(TIF)
(A) Generation of knock-in of GAL4 into the CCKLR-17D1 locus. (B) No signal was detected when the 17D1GAL4 drives stinger and GFP in the proboscis and leg tarsi. Scale bar: 50 μm. (C) 17D1 mutants show no decreased motivation to feed in PER compared with control. **p < 0.01; Kruskal–Wallis test followed by Dunn’s multiple comparisons test.
(TIF)
Expression pattern of 17D3GAL4 visualized by UAS-mCD8::GFP in leg tarsi (A1-A3), proboscis and maxillary palps (B1-B3). Scale bar: 50 μm.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(MP4)
Data Availability Statement
Raw sequence Data of the RNA-seq analysis in this study are available in the Short Read Archive (SRA) database of NCBI with the accession numbers and NCBI URLs as below: dsNlsk_1, (SRR12460889, https://www.ncbi.nlm.nih.gov/sra/?term=SRR12460889), dsNlsk_2, (SRR12460895, https://www.ncbi.nlm.nih.gov/sra/?term=SRR12460895), dsNlsk_3, (SRR12460894, https://www.ncbi.nlm.nih.gov/sra/?term=SRR12460894), dsNlsk_4, (SRR12460893, https://www.ncbi.nlm.nih.gov/sra/?term=SRR12460893), dsgfp_1 (SRR12460896, https://www.ncbi.nlm.nih.gov/sra/?term=SRR12460896), dsgfp_2 (SRR12460892, https://www.ncbi.nlm.nih.gov/sra/?term=SRR12460892), dsgfp_3 (SRR12460891, https://www.ncbi.nlm.nih.gov/sra/?term=SRR12460891) and dsgfp_4 (SRR12460890, https://www.ncbi.nlm.nih.gov/sra/?term=SRR12460890).