Abstract
Pathogen populations in estuarine areas are dynamic, as they are subject to multiple natural and anthropogenic challenges. Heavy rainfall events bring instability to the aquatic environment in estuaries, causing changes in pathogen populations and increased environmental sanitation and public health concerns. In this study, we investigated the effects of heavy precipitation on the occurrence of pathogens in the Puzi River estuary, which is adjacent to the largest inshore oyster farming area in Taiwan. Our results indicated that Vibrio parahaemolyticus and adenovirus were the most frequently detected pathogens in the area. There was a significant difference (Mann-Whitney U test, p < 0.01) in water quality parameters, including total coliform, Escherichia coli, water temperature, turbidity, salinity, and dissolved oxygen, between groups with and without V. parahaemolyticus. In addition, the detection rate was negatively correlated with the average daily rainfall (r2 > 0.8). There was no significant difference between water quality parameters and the presence/absence of adenovirus, but a positive correlation was observed between the average daily rainfall and the detection rate of adenovirus (r2 ≥ 0.75). We conclude that heavy precipitation changes estuarine water quality, causing variations in microbial composition, including pathogens. As extreme weather events become more frequent due to climate change, the potential impacts of severe weather events on estuarine environments require further investigation.
Introduction
In estuaries, salinity levels change remarkably because of the mixing of tidal flows and the seasonal variability in the amount of freshwater carried by. Steep salinity gradients in estuaries have been shown to result in large changes in bacterial diversity, community structure, and potential growth rates [1–3]. Other factors in estuaries, such as temperature, nutrients, turbidity, and many physical determinants, are known to affect microbial populations with respect to population size, community structure, and the predominant species [1, 3–10]. Many pathogens, including Vibrio spp., Salmonella spp., Aeromonas spp., Streptococci, Bacillus cereus, Clostridium difficile, hepatitis A virus, norovirus, and adenovirus, have been frequently found in estuarine waters [7–9, 11–16]. These pathogens may reach estuaries through the discharge of untreated sewage, wildlife waste, animal farming wastewater, or surface runoff, which could be further affected by the estuarine environment [17–19].
Estuarine waters are rich in organic matter, which makes them an excellent habitat for aquaculture. Some of the most common aquatic species in these environments are euryhaline fish, crustaceans, and mollusks. Aquatic pathogens can cause economic losses due to an increase in disease and mortality in aquaculture species [20–24]. Some aquaculture species are filter feeders, which can cause transmission of pathogens to humans through foodborne infections [25]. In some extreme cases, Vibrio organisms such as Vibrio vulnificus and Vibrio cholerae have been reported to cause illness or lethal septicemia in humans after exposure to pathogen-containing seawater [3, 7, 15, 26]. Therefore, to maintain public health, it is urgent to survey pathogens in estuarine environments.
Seasonal changes affect microbial communities in many aquatic habitats. In estuaries, the communities of bacterioplankton show predictable seasonal patterns that might be associated with salinity [2]. Recently, our group has demonstrated that seasonal changes affect the detection rates and concentrations of human adenovirus in aquatic environments, which is associated with human gastroenteritis. Evidence from previous studies also suggest that the physical determinants and chemical compositions of water bodies might vary seasonally and, consequently, affect microbial communities [27, 28]. Climate change is an important global issue. Due to climate change, the frequency of extreme weather events has increased in recent years. This has caused fluctuations in some physical determinants, including ambient temperature and precipitation, which could interfere with seasonal rhythms in aquatic environments [1, 8]. As the occurrence of extreme weather events becomes more frequent [17, 29, 30], the seasonal pattern of climate determinants has become less predictable, which in turn may affect the overall water quality, biochemical cycles, organism life cycles, and aquatic ecosystems [17, 31, 32].
The effects of climate change on pathogens require further investigation. Research on the impact of climate change on public health has emerged in recent years. For example, various studies have explored potential associations between climate change and waterborne disease infection [26, 32]. Accumulated data have shown that climate change is an important factor associated with the occurrence of certain diseases [33–36]. Nonetheless, most studies have focused on the connection between climate change and pathogenic outbreaks, while the effect of climate on the occurrence of pathogens in estuarine environments is still unclear. According to the 2015 Annual Report by the Taiwanese Council of Agriculture, oyster farming near the estuary of the Puzi River accounts for 43% of the entire oyster production in Taiwan. Precipitation patterns have changed substantially over the past few decades owing to climate change in Taiwan [35, 37]. Whether heavy precipitation affects the occurrence of pathogens in the Puzi River estuary, however, remains unclear. Increased pathogen load is a potential risk to human health from water-transmitted pathogens. For these reasons, we investigated the effects of precipitation events on the occurrence of pathogens in the water bodies of the Puzi River estuary. Water samples around the Puzi River estuary were collected quarterly to monitor seasonal changes. To screen for dynamic changes in precipitation events, water samples were taken at the same locations on the first, third, eighth, and twelfth days after the end of heavy precipitation events. Commonly found pathogens included Vibrio spp., C. difficile, B. cereus, norovirus (NoV), hepatitis A virus (HAV), and adenovirus (AdV). These were examined, and the potential effects of intensive precipitation events on microbial water quality in the estuary were also investigated.
Materials and methods
Collection of water samples
Water samples were collected from downstream of the Puzi River, at the Dongshing Fishing Port, and from the inshore waters adjacent to the oyster farms. All collections are the publicly accessible sites, which specific permission for collecting water samples was obtained from the central government. The ethics statement of this study had been proved by Centers for Disease Control, Ministry of Health and Welfare of Taiwan central government (MOHW105-CDC-C-114-112601 and 106-CDC-C-114-122601). There is not any endangered/protected land or species were involved in this study. A total of eight sites were investigated during periods of precipitation events; however, only five samples were collected during rain events. The collection of three offshore samples were banned by the Taiwan Coast Guard Administration for safety reasons. Moreover, sample collection was also carried out quarterly once per season from winter (December to February was defined as the winter season) in 2016 to winter in 2017 to monitor seasonal changes in water quality background. The longitude and latitude for each sampling site are illustrated in Fig 1.
Fig 1. Sampling locations around the estuarine areas of the Puzi River in Taiwan.

Sites A and B were downstream of the Puzi River, sites C to E were fishing ports, and sites F to H were offshore oyster farms. The geographical coordinates (latitude, longitude) are as follows: A (23.458433, 120.177433), B (23.445306, 120.165667), C (23.452656, 120.138769), D (23.450560, 120.139850), E (23.451032, 120.137538), F (23.473775, 120.120292), G (23.444178, 120.132592), and H (23.410706, 120.120975).
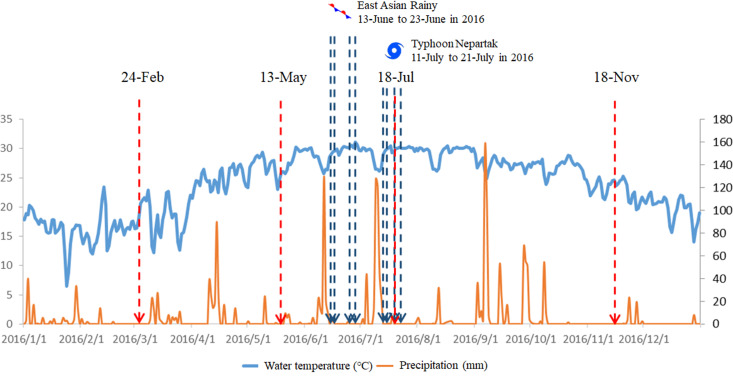
To evaluate the impact of heavy precipitation on water quality around the estuary in this study, water samples were collected from the same areas after heavy precipitation events. The precipitation intensity classification is based on the definitions of the Central Weather Bureau in Taiwan: 1) heavy rain, more than 80 mm within 24 h; 2) extremely heavy rain, more than 200 mm within 24 h, or more than 100 mm within 3 h. The dynamic changes in water quality parameters were evaluated on the first, third, eighth, and twelfth days after the events were established. The rainfall data were obtained through the website of the Taiwan Central Weather Bureau Observation Data Inquire System (http://e-service.cwb.gov.tw/HistoryDataQuery/index.jsp). The weather parameters of the two events were documented in Fig 2. One was classified as heavy rain, which occurred in the East Asian Rainy season on June 12. The other rainfall event was classified as extremely heavy rain, which appeared after Typhoon Nepartak on July 10 (see Table 1). In both events, the precipitation tapered off after the first couple of days. Although some rainfall occurred afterwards, the precipitation was no more than 5% of the first three days.
Fig 2. Timeline of weather parameters around estuarine areas of the Puzi River and the sampling dates.
The dashed red arrow bar indicates the date for seasonal sampling. The dashed blue arrow bar indicates the sampling dates during rainfall events.
Table 1. Seasonal water quality parameters around the estuarine areas of the Puzi River.
| Water quality parameters | Sampling dates | ||||
|---|---|---|---|---|---|
| 2016/2/24 (n = 8) | 2016/5/13 (n = 8) | 2016/7/18 (n = 8) | 2016/11/18 (n = 8) | 2017/2/8 (n = 8) | |
| Heterotrophic plate count (CFU/mL) | 354±1,387 | 12,366±5,218 | 38,912±11,310 | 3,566±1,623 | 335±1421 |
| Total Coliform (CFU/100mL) | 254±511 | 409±502 | 631±2,023 | 280±2662 | 224±483 |
| Escherichia coli (CFU/100mL) | 23±79 | 48±40 | 130±255 | 20±22 | 22±83 |
| pH | 8.6±0.6 | 8.0±0.2 | 7.9±0.1 | 7.6±0.2 | 8.4±0.3 |
| Turbidity (NTU) | 15.2±20.8 | 11.9±28.2 | 39.2±34.6 | 27.3±27.1 | 16.7±19.8 |
| Salinity (%) | 29.9±1.5 | 27.5±1.9 | 9.3±6.8 | 21.6±12.2 | 20.7±5.3 |
| Dissolved oxygen (mg/L) | 7.0±0.4 | 4.8±0.4 | 4.5±0.9 | 5.7±0.8 | 6.9±0.3 |
| Average water temperature (°C) | 18.2±0.9 | 29.9±0.7 | 28.9±0.4 | 27.0±1.2 | 20.2±1.2 |
The Figs of this study have been placed in website Figshare (https://doi.org/10.6084/m9.figshare.14502780).
Water quality examination
About 3-L of water sample was collected for testing during each sampling. The samples were stored at 4°C and transported to the laboratory within 6 h. Water samples were subjected to a spectrum of quality tests, including temperature, pH, dissolved oxygen, turbidity, and microbial analyses. The water temperature, pH, and dissolved oxygen were determined using a portable multi-parameter water quality meter (HI9828, Hanna Instruments, USA) immediately after sample collection. Turbidity was determined using a ratio turbidimeter (Waterproof Portable TN100, Eutech Instruments, Singapore). For microbial analysis, heterotrophic plate count was performed using the spread plate method with m-HPC agar according to the standard protocol (Methods 9215C) [38]. Total coliform was measured by membrane filtration and differential medium as described in the standard method for water and wastewater examination (Methods 9222 B) [38]. The quality control and quality assurance methods in this study were carried out as described in our previous report [27].
Sample pretreatment for microbial analyses
For detection of specific microbial pathogens, a water sample (300 mL) was filtered through a 47 mm GN-6 membrane (Pall, Mexico City, Mexico) with a pore size of 0.45 μm, which was held by a stainless filter holder. Subsequently, the membranes were used for the enrichment of microbes, as described in the next section. For the detection of the virus, the virus was recovered by the method described in our earlier studies [27, 39, 40]. In short, each water sample (1 L) was filtered through 47 mm GN-6 membranes (Pall, Mexico City, Mexico) with a pore size of 0.45 μm. Viruses attached to the filter membrane were scraped manually into 50 mL of sterilized phosphate-buffered saline (PBS). Subsequently, the eluent was transferred into two conical centrifuge tubes (50 mL each) and centrifuged at 2600×g for 30 min (KUBOTA Model 2420 Compact Tabletop Centrifuge, Japan). The pellet was resuspended in PBS (5 mL) for total viral DNA extraction at 4°C.
Enrichment of bacterial isolates
For the detection of various pathogens, water samples were enriched according to their optimal growth conditions. For Vibrio spp., the water samples were enriched with alkaline peptone water (APW) and selectively cultured on CHROMagar™ Vibrio and thiosulfate-citrate-bile salts-sucrose agar (TCBSA). For V. parahaemolyticus and V. cholerae, samples were incubated at 37°C for 24 h. For V. vulnificus, samples were incubated at 30°C for 24 h. For Clostridium difficile, samples were enriched with cycloserine-cefoxitin fructose broth (CCFB) and selectively cultured on CHROMagar™ Clostridium difficile and cycloserine-cefoxitin fructose agar (CCFA), and incubated at 37°C for 24 h to 48 h under anaerobic conditions. For B. cereus, samples were enriched with trypticase soy polymyxin broth (TSPB), selectively cultured on CHROMagar™ Bacillus cereus and mannitol egg yolk polymyxin agar (MEYP), and incubated at 37°C for 24 h to 48 h.
Microbial DNA extraction and identification
For bacterial detection, DNA was extracted from the second enrichment broth (1 mL) using the MagPurix bacteria DNA Extraction Kit ZP02006. For virus detection, DNA was extracted from the pellet as described in the previous section. Total viral DNA/RNA was extracted using the Viral Nucleic Acid Extraction Kit ZP02003. MagPurix 12s Automated Nucleic Acid Purification System (Zinexts Life Science Corp., Taiwan) was employed for DNA extraction. PCR was carried out on the final DNA extraction product (100 μL) to confirm the species of pathogens. The experimental conditions of primer sequences, positive controls of pathogens, and their references are summarized in Table 2.
Table 2. The primer sequences and positive controls of pathogens.
| Pathogens | Target gene | Size | Sequence (5′ to 3′) | Positive control | References for reaction materials and PCR condition |
|---|---|---|---|---|---|
| Bacillus cereus | Bal | 533 | BalF: 5′-TGCAACTGTATTAGCACAAGCT-3′ | B. cereus, ATCC 11778 | [41] |
| BalR: 5′-TACCACGAAGTTTGTTCACTACT-3′ | |||||
| Clostridium difficile | tcdB | 632 | tcdA-F 5′-GTATGGATAGGTGGAGAAGTCAGTG-3′ | C. difficile, ATCC 9689 | [42] |
| tcdA-R 5′-CGGTCTAGTCCAATAGAGCTAGGTC-3′ | |||||
| tcdA | 441 | tcdB-F 5′-GAAGATTTAGGAAATGAAGAAGGTGA-3′ | |||
| tcdB-R 5′-AACCACTATATTCAACTGCTTGTCC-3′ | |||||
| cdtA | 260 | cdtA-F 5′-ATGCACAAGACTTACAAAGCTATAGTG-3′ | |||
| cdtA-R 5′-CGAGAATTTGCTTCTATTTGATAATC-3′ | |||||
| cdtB | 179 | cdtB-F 5′-ATTGGCAATAATCTATCTCCTGGA-3′ | |||
| cdtB-R 5′-CCAAAATTTCCACTTACTTGTGTTG-3′ | |||||
| Vibrio cholera | ompW | 427 | VCompW-F: 5′-CACCAAGAAGGTGACTTTATTGTG-3′ | V. cholera, ATCC 14035 | [43] |
| VCompW-R: 5′-CGTTAGCAGCAAGTCCCCAT-3′ | |||||
| V. parahaemolyticus | Collagenase | 271 | VPC-F: 5′-GAAAGTTGAACATCATCAGCACGA-3′ | V. parahaemolyticus, ATCC 17802 | [44] |
| VPC-R: 5′-GGTCAGAATCAAACGCCG-3′ | |||||
| V. vulnificus | vvhA | 505 | FDAvvhA-F: 5′ -CCGCGGTACAGGTTGGCGCA-3′ | V. vulnificus, ATCC 27562 | [43] |
| FDAvvhA-R: 5′-CGCCACCCACTTTCGGGCC-3′ | |||||
| Hepatitis A virus | 3D | 412 | HAV1: 5′-TTTGGTTGGATGAAAATGGTT-3′ | Internal controls: | [45] |
| HAV4: 5′-ATTCTACCTGCTTCTCTAATC-3′ | |||||
| 233 | HAV2: 5′-CAACCTGTCCAAAAGATGAAT-3′ | ||||
| HAV3: 5′-ACCAACATCTCCGAATCTTA-3′ | |||||
| Norovirus | ORF2 | Group I, GI: | [46] | ||
| COG1F: 5′-CGYTGGATGCGNTTYCATGA-3′ | |||||
| 377 | G1-SKF: 5′-CTGCCCGAATTYGTAAATGA-3′ | ||||
| 330 | G1-SKR: 5′-CCAACCCARCCATTRTACA-3′ | ||||
| 386 | Group II GII: | Bacteria universal 16S rDNA PCR for avoiding false negative results | |||
| 344 | COG2F: 5′- CARGARBCNATGTTYAGRTGGATGAG-3′ | ||||
| G2-SKF: 5′- CNTGGGAGGGCGATCGCAA -3′ | |||||
| G2-SKR: 5′- CCRCCNGCATRHCCRTTRTACAT -3′ | |||||
| Adenovirus | Hexon neHexon | 301 | Hex: | [47] | |
| Hex1 deg F: 5′-GCCSCARTGGKCWTACATGCACATC-3′ | |||||
| Hex2 deg R:5′-CAGCACSCCICGRATGTCAAA-3′ | |||||
| 171 | neHex: | ||||
| neHex 3deg F: 5′-GCCCGYGCMACIGAIACSTACTTC-3′ | |||||
| neHex 4deg R: 5′-CCYACRGCCAGIGTRWAICGMRCYTTGTA-3′ |
Statistical analysis
The Mann-Whitney U test was performed to examine the correlation between physical and biological water quality parameters and the occurrence of pathogens after a precipitation event, and the correlation between cumulative rainfall of a precipitation event and the occurrence of pathogens. The average daily precipitation is calculated as follows:
RE: Cumulative precipitation depth (mm/24 h) during the event;
RN: Cumulative precipitation depth (mm/24 h) during the sampling period (days 1 to N);
RED: Duration of the precipitation event in days;
RND: Cumulative days counted from the first day of the precipitation event to the sampling day.
The statistical analyses were performed using Paleontological Statistics Version 3.14 (PAST, University of Oslo, Norway) and SigmaPlot V.10.0. software (Systat Software Inc., US). Differences were considered significant when the p value was ≤ 0.05.
The Figs, Raw data and Tables of this study have been placed in website Figshare (https://doi.org/10.6084/m9.figshare.14502780).
Results
Pathogen detection rates of seasonal levels and after rainfall events
Of all the pathogens examined in this study, V. parahaemolyticus and AdVs were the most frequently detected bacteria and viruses, with detection rates of 90% (29/32) and 25% (8/32), respectively (Table 3). Since there was no statistically significant difference in detection rates of V. parahaemolyticus and AdVs according to seasonal changes, the data from 4 seasons were combined to compare to the detection rates after precipitation events. After heavy rainfall, the detection rate of V. parahaemolyticus immediately dropped to 40% and then gradually increased to the original high level. On the other hand, the detection rate of AdVs reached 80% and decreased to 12.5% after 12 days. As for the other pathogens tested in this study, i.e., V. cholerae, V. vulnificus, B. cereus, C. difficile, NoV, and HAV, they were detected occasionally during quarterly tests and not affected by precipitation events.
Table 3. The presence of pathogens in estuarine areas of the Puzi River after rainfall.
| Days after heavy precipitation (by East Asian Rainy) | Days after extremely heavy precipitation (by Typhoon Nepartak) | Seasonal background | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Day 1 | Day 3 | Day 8 | Day 12 | Day 1 | Day 3 | Day 8 | Day 12 | n = 32 | |
| (n = 5) | (n = 8) | (n = 8) | (n = 8) | (n = 8) | (n = 8) | (n = 8) | (n = 8) | ||
| Vibrio parahaemolyticus * | 2 | 6 | 6 | 8 | 6 | 7 | 7 | 8 | 29 |
| Vibrio cholera | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 4 |
| Vibrio vulnificus | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 |
| Bacillus cereus | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 1 |
| Clostridium difficile | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 |
| Adenovirus* | 4 | 5 | 5 | 3 | 5 | 5 | 3 | 1 | 8 |
| Norovirus (G1/G2) | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 2 |
| HAV (hepatitis A virus) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
*indicates dominant pathogen.
Correlation between precipitation and the occurrence of pathogens
Fig 3 shows the correlation between the average daily precipitation and detection rates of V. parahaemolyticus and AdVs. In both cases, the average daily precipitation decreased with time from the first day of the precipitation event to the twelfth sampling day, while the detection rates for V. parahaemolyticus increased in the same period (Fig 3A and 3B). There was a negative correlation between precipitation and detection rates for V. parahaemolyticus (r2 > 0.8). The extremely heavy rain did not increase the slope value of the correlation curve between the detection rate of V. parahaemolyticus and daily precipitation. The detection rate of V. parahaemolyticus in quarterly tests was about 90% to 100% in the estuary of the Puzi River. In contrast, a positive correlation was found between the detection rates of AdVs and precipitation (Fig 3C and 3D, r2 ≥ 0.75). The quarterly detection rate of AdVs was approximately 25% in estuary areas. After both precipitation events, the detection rates of AdVs rose to 60%–80% and returned to the original levels within 12 days after the events. A higher precipitation level did not further enhance the detection rate of AdVs.
Fig 3. Correlation between the detection rates of pathogens and the averages of daily precipitation.
Differences in water quality parameters and the occurrence of pathogens
Table 3 presents the data on water quality parameters, including temperature, pH, salinity, dissolved oxygen, turbidity, and microbial indicators, from quarterly samples. Seasonal changes were observed in environmental parameters, such as temperature, dissolved oxygen, and several biological parameters. It should be pointed out that the third quarterly samples were taken on the eighth day after extremely heavy precipitation events, which could have interfered with the regular pattern.
The effects of precipitation events on environmental parameters are summarized in Tables 4 and 5. The parameters of heterotrophic plate count (HPC), total coliform, and E. coli increased significantly immediately after the occurrence of heavy rainfall and gradually decreased. In addition, rainfall causes an increase in turbidity and dissolved oxygen in water, which decreases over time afterwards. Rainfall also dropped the water temperature by 3 degrees and decreased salinity drastically. Rainfall has little effect on the changes in pH levels in estuarine areas. The average water quality parameters variation by different sampling sites was shown on S1–S3 Tables.
Table 4. Water quality parameters around the estuarine areas of the Puzi River after heavy precipitation.
| Sampling events | Days after heavy precipitation (by East Asian Rainy) | |||
|---|---|---|---|---|
| Water quality parameters | Day 1 (n = 5) | Day 3 (n = 8) | Day 8 (n = 8) | Day 12 (n = 8) |
| Heterotrophic plate count (CFU/mL) | 86,093±20,618 | 63,142±12,910 | 16,366±7,819 | 3,378±1,720 |
| Total Coliform (CFU/100mL) | 9,700±2,953 | 5,351±1,507 | 941±645 | 78±75 |
| Escherichia coli (CFU/100mL) | 557±222 | 253±106 | 188±19 | 1±1 |
| pH | 7.6±0.1 | 7.6±0 | 7.7±0.1 | 7.9±0 |
| Turbidity (NTU) | 78.5±32.4 | 25.8±6.7 | 20.5±11.6 | 1.8±0.4 |
| Salinity (PSU) | 2.9±1.1 | 8.5±2.1 | 11.8±2.7 | 15.7±2.2 |
| Dissolved oxygen (mg/L) | 5.3±0.1 | 4.7±0.1 | 4.2±0.1 | 3.8±0.2 |
| Average water temperature (°C) | 26.4±0.1 | 28.0±0.2 | 28.9±0.3 | 29.3±3.1 |
| Cumulative precipitation (mm) | 161 | 165 | 167 | 169 |
| Average daily precipitation (mm) | 53.7 | 33.0 | 16.7 | 12.1 |
Table 5. Water quality parameters around the estuarine areas of the Puzi River after extreme heavy precipitation.
| Sampling events | Days after extreme heavy precipitation (by Typhoon Nepartak) | |||
|---|---|---|---|---|
| Water quality parameters | Day 1 (n = 8) | Day 3 (n = 8) | Day 8 (n = 8) | Day 12 (n = 8) |
| Heterotrophic plate count (CFU/mL) | 314,381±180,326 | 716,950±471,663 | 38,912±41,879 | 27,383±22,439 |
| Total Coliform (CFU/100mL) | 2,390±1,639 | 1,149±823 | 631±708 | 52±87 |
| Escherichia coli (CFU/100mL) | 133±128 | 81±33 | 130±95 | 24±21 |
| pH | 8.1±0.1 | 7.7±0.1 | 7.9±0.1 | 7.8±0.1 |
| Turbidity (NTU) | 55.3±19.6 | 30.7±14.7 | 30.2±11.1 | 11.9±4.6 |
| Salinity (PSU) | 2.9±1.0 | 1.1±0.6 | 9.3±3.1 | 15.3±1.4 |
| Dissolved oxygen (mg/L) | 7.4±0.6 | 4.5±0.2 | 4.5±0.6 | 4.8±0.4 |
| Average water temperature (°C) | 26.1±0.1 | 27.2±0.2 | 28.9±0.3 | 29.2±0.3 |
| Cumulative precipitation (mm) | 294 | 317 | 317 | 324 |
| Average daily precipitation (mm) | 97.8 | 63.3 | 32.4 | 23.1 |
Table 6 summarizes the results of statistical significance in water quality parameters between the groups with or without the occurrence of V. parahaemolyticus using the Mann-Whitney U test. The occurrence of V. parahaemolyticus was associated with higher salinity, higher temperature, lower turbidity, and lower numbers of total coliform and E. coli. In contrast, the occurrence of AdVs in the water samples after precipitation events was not associated with any of the water quality parameters in this study (Table 7).
Table 6. Comparisons between water quality parameters and the presence of Vibrio parahaemolyticus.
| Water quality parameters | Mann-Whitney U test | VP- Positive (n = 50) | VP-Negative (n = 11) | ||||
|---|---|---|---|---|---|---|---|
| Median | Q1 | Q3 | Median | Q1 | Q3 | ||
| Heterotrophic plate count (CFU/mL) | p = 0.68 | 78,000 | 14,053 | 304,867 | 61,500 | 23,883 | 91,133 |
| Total Coliform (CFU/100mL)* | p = 0.003 | 347 | 16 | 3,309 | 4,964 | 2,591 | 11,512 |
| Escherichia coli(CFU/100mL)* | p = 0.002 | 32 | 0 | 125 | 415 | 166 | 700 |
| Water temperature (°C)* | p = 0.007 | 31.0 | 30.1 | 32.3 | 29.0 | 26.5 | 29.9 |
| pH | p = 0.13 | 7.8 | 7.6 | 8.0 | 7.6 | 7.5 | 7.8 |
| Turbidity (NTU)* | p = 0.001 | 14.0 | 3.5 | 20.5 | 69.1 | 33.5 | 131.2 |
| Salinity (PSU)* | p<0.001 | 11.7 | 3.3 | 16.7 | 0.7 | 0.1 | 0.9 |
| Dissolved oxygen (mg/L) | p = 0.08 | 4.6 | 4.0 | 5.2 | 5.2 | 4.7 | 6.0 |
*indicates p<0.05; VP for Vibrio parahaemolyticus.
Q1 for first quartile; Q3 for third quartile.
Table 7. Comparisons between water quality parameters and the presence of adenovirus.
| Water quality parameters | Mann-Whitney U test | AdVs- Positive (n = 31) | AdVs-Negative (n = 30) | ||||
|---|---|---|---|---|---|---|---|
| Median | Q1 | Q3 | Median | Q1 | Q3 | ||
| Heterotrophic plate count (CFU/mL) | p = 0.17 | 95,733 | 22,998 | 304,217 | 25,383 | 2,418 | 255,000 |
| Total Coliform (CFU/100mL) | p = 0.21 | 2,399 | 38 | 5,186 | 453 | 42 | 2,297 |
| Escherichia coli(CFU/100mL) | p = 0.25 | 86 | 0 | 299 | 34 | 0 | 96 |
| Water temperature (°C) | p = 0.81 | 30.8 | 29.8 | 32.2 | 30.6 | 29.9 | 31.9 |
| pH | p = 0.47 | 7.8 | 7.6 | 8.0 | 7.7 | 7.5 | 8.0 |
| Turbidity (NTU) | p = 0.70 | 18.6 | 5.8 | 33.1 | 15.2 | 3.7 | 32.0 |
| Salinity (PSU) | p = 0.94 | 4.8 | 1.3 | 14.2 | 9.1 | 0.9 | 15.3 |
| Dissolved oxygen (mg/L) | p = 0.11 | 4.8 | 4.4 | 5.5 | 4.5 | 4.0 | 5.1 |
*indicates p<0.05; AdVs for adenovirus.
Q1 for first quartile; Q3 for third quartile.
Discussion
As inshore farming of oysters around the Puzi River estuary is the primary source of oysters in Taiwan, and human activities and livestock farms are located around the Puzi River basin [48], the wastewater from the river is an important contributor to the water quality of the estuary. Therefore, it is necessary to monitor the dynamics of microbial communities in these areas. Microbial communities in estuarine environments are easily affected by weather, river flow, tidal cycles, sea temperature, and salinity [1, 3, 10]. As severe weather events are likely to affect pathogens due to changes in environmental factors, they pose potential threats to human health, though the extent of this threat has not been determined. Vibrio spp. and AdV were the most frequently detected pathogens in this study. Previous studies have shown that halophilic Vibrio species are a major group of pathogens in estuarine environments [7, 8, 22]. As confirmed by PCR and DNA sequencing, we demonstrated that V. parahaemolyticus and AdVs (human adenovirus 41 [HAdV-41] and porcine adenovirus 5 [PAdV-5]) were the predominant pathogens in the Puzi River estuary.
Previous studies have demonstrated that V. parahaemolyticus is a serious pathogen that causes foodborne illness in many outbreaks [24, 35, 41, 42]. This bacterium survives in high-temperature marine environments and is frequently isolated from a variety of seafood, including oysters, mollusks, and crustaceans. Vibrio species can also cause economic losses in aquaculture by interfering with the growth of mollusks and crustaceans [24]. After the events of heavy precipitation, Eichhornia crassipes, commonly known as water hyacinth, which grows in the Puzi River, washed down and piled up separately around the estuary. We found that the water temperature, turbidity, and salinity of the estuary areas were affected by heavy rainfall. Our data indicated that the occurrence of V. parahaemolyticus was significantly associated with several parameters such as temperature, turbidity, and salinity according to the results of the Mann-Whitney U test. Previous studies have shown that the occurrence and abundance of halophilic Vibrio are affected by changes in salinity, water temperature, and turbidity, which is consistent with the current study [8, 9, 35, 42, 43]. Interestingly, the heterotrophic plate count after the typhoon with extremely heavy rainfall was maintained at a high level for almost 2weeks, causing the issue of a warning to human health. It is important to survey whether there are unknown pathogens that have not yet been revealed in this study following events of extremely heavy precipitation.
AdVs are one of the most prevalent waterborne viruses, and they may be easily transmitted through water [14, 36, 44]. In this study, AdVs were found to be the most abundant DNA virus in water samples after heavy precipitation events. There was a positive correlation between the occurrence of adenovirus and the average daily precipitation; however, there was no significant difference between the occurrence of AdVs and any of the water quality parameters that were assayed in this study. It is plausible that the heavy precipitation carried the AdVs from polluted water upstream of the Puzi River and from contaminated adjacent urban areas to the estuary areas, and consequently elevated the AdVs detection rates. AdVs are highly contagious, and large numbers of AdVs can be excreted in human feces or urine from infected individuals [14, 28, 45, 46]. As one of the major origins of AdVs is human feces, AdVs have been proposed as an indicator of human fecal pollution [44, 46, 47]. In this study, the detection rate for AdVs increased significantly straightaway after rainfall events and returned to seasonal levels in 12 days. The pattern suggested that the AdVs gathered by heavy precipitation were run off continuously by water upstream. These data implied that the risk of AdVs infections increased in human and aquaculture species due to exposure to high levels of the virus in estuarine areas after heavy and extremely heavy precipitation events.
In this study, we revealed that the detection rates of two important pathogens were affected by rainfall events in opposite patterns. We proposed three possible underlying mechanisms that may influence the changes in organisms in estuarine areas after precipitation events. First, heavy and extremely heavy precipitation changed the environmental parameters, which may affect the advantages or disadvantages of the survival of predominant pathogens. In this study, we showed that the detection rates of V. parahaemolyticus decreased since the environmental conditions did not favor the growth of the organism. Second, precipitation events changed the detection rates of pathogens by carrying them into certain habitats and altering their occurrences. In this study, we found that the presence of adenovirus correlated only with the average daily precipitation, not the water quality parameters, suggesting that rainfall can bring adenovirus to the estuary. Third, heavy precipitation can promote the growth of microbes because heavy precipitation drastically elevates the heterotrophic plate counts. Moreover, the extent of microbial growth was enhanced even further in the extremely heavy precipitation event, suggesting that the intensity of precipitation played a role in changing the balance of microbial communities. A natural habitat may take a longer time to recover when it experiences extremely heavy events.
Current climate change models suggest that global warming may have altered the frequency and intensity of precipitation and extreme weather events [30, 48]. Changes in precipitation patterns threaten the ecosystem, aquaculture, and other forms of human activities [17, 26, 37]. Environmental changes from extreme weather events may also have direct or indirect impacts on marine microorganisms, especially in estuarine areas [1, 3, 7, 10, 13]. Historical climate data in Taiwan have suggested an increased incidence and intensity of extreme weather events over the past few years [37]. More studies on the changes in microbial communities are needed to reduce the threats of these events.
Conclusions
The correlation between the presence of commonly detected pathogens and the dynamic changes in water quality and microbial indices after heavy precipitation were investigated around the Puzi River estuary. We found that V. parahaemolyticus and AdVs were the most frequently detected pathogens in the estuarine environment after heavy precipitation. The detection rate of V. parahaemolyticus decreased immediately after heavy precipitation, followed by slowly returning to the original levels. On the other hand, the detection rates of AdVs surged to a high level after the rainfall events, followed by a gradual decrease to seasonal levels in 12 days. Microbial indices such as E. coli, total coliform, and heterotrophic plate count were elevated drastically by precipitation, suggesting that a favorable environment for the growth of microbes is created after rainfall events. As extreme weather events have become more frequent in recent years, more investigations are needed to survey the relative abundance of microbial communities after heavy precipitation events and to identify the pathogens of potential threats that have not yet emerged.
Supporting information
(DOCX)
(DOCX)
(DOCX)
Data Availability
The Figs, Raw data and Tables of this study are in the Supporting Information and on Figshare. (https://doi.org/10.6084/m9.figshare.14502780.v1).
Funding Statement
This research was supported by the Ministry of Science and Technology of Taiwan (MOST 105-2116-M-194-014), Centers for Disease Control, Taiwan, R.O.C. (MOHW105-CDC-C-114-112601 and 106-CDC-C-114-122601), Show Chwan Health Care System (RD108018), Wan Fang Hospital (108-wf-swf-10) and Ditmanson Medical Foundation Chiayi Christian Hospital-National Chung Cheng University Joint Research Program (R110-25). This research was also supported by the Center for Innovative Research on Aging Society (CIRAS) from the Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan.
References
- 1.Campbell BJ, Kirchman DL. Bacterial diversity, community structure and potential growth rates along an estuarine salinity gradient. The ISME journal. 2013;7(1):210–20. doi: 10.1038/ismej.2012.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fortunato CS, Herfort L, Zuber P, Baptista AM, Crump BC. Spatial variability overwhelms seasonal patterns in bacterioplankton communities across a river to ocean gradient. The ISME journal. 2012;6(3):554–63. doi: 10.1038/ismej.2011.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crump BC, Hopkinson CS, Sogin ML, Hobbie JE. Microbial biogeography along an estuarine salinity gradient: combined influences of bacterial growth and residence time. Applied and environmental microbiology. 2004;70(3):1494–505. doi: 10.1128/AEM.70.3.1494-1505.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ducklow H. Bacterial production and biomass in the oceans. Microbial ecology of the oceans. 2000;1:85–120. [Google Scholar]
- 5.Morris RM, Rappé MS, Connon SA, Vergin KL, Siebold WA, Carlson CA, et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature. 2002;420(6917):806–10. doi: 10.1038/nature01240 [DOI] [PubMed] [Google Scholar]
- 6.Austin B, Allen D, Mills A, Colwell R. Numerical taxonomy of heavy metal-tolerant bacteria isolated from an estuary. Canadian Journal of Microbiology. 1977;23(10):1433–47. doi: 10.1139/m77-212 [DOI] [PubMed] [Google Scholar]
- 7.Pruzzo C, Huq A, Colwell RR, Donelli G. Pathogenic Vibrio species in the marine and estuarine environment. Oceans and health: pathogens in the marine environment: Springer; 2005. p. 217–52. [Google Scholar]
- 8.Johnson CN. Influence of Environmental Factors on Vibrio spp. in Coastal Ecosystems. Microbiology spectrum. 2015;3(3). doi: 10.1128/microbiolspec.VE-0008-2014 [DOI] [PubMed] [Google Scholar]
- 9.Sayler G, Nelson J, Justice A, Colwell R. Incidence of Salmonella spp., Clostridium botulinum, and Vibrio parahaemolyticus in an estuary. Applied and environmental microbiology. 1976;31(5):723–30. doi: 10.1128/aem.31.5.723-730.1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flowers E. Microbial community structure & function in estuarine sediments: Cardiff University; 2013. [Google Scholar]
- 11.Rhodes MW, Kator H. Survival of Escherichia coli and Salmonella spp. in estuarine environments. Applied and Environmental Microbiology. 1988;54(12):2902–7. doi: 10.1128/aem.54.12.2902-2907.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaper J, Lockman H, Colwell R, Joseph S. Aeromonas hydrophila: ecology and toxigenicity of isolates from an estuary. Journal of Applied Bacteriology. 1981;50(2):359–77. doi: 10.1111/j.1365-2672.1981.tb00900.x [DOI] [PubMed] [Google Scholar]
- 13.Ferguson CM, Coote BG, Ashbolt NJ, Stevenson IM. Relationships between indicators, pathogens and water quality in an estuarine system. Water Research. 1996;30(9):2045–54. [Google Scholar]
- 14.Castignolles N, Petit F, Mendel I, Simon L, Cattolico L, Buffet-Janvresse C. Detection of adenovirus in the waters of the Seine River estuary by nested-PCR. Molecular and cellular probes. 1998;12(3):175–80. doi: 10.1006/mcpr.1998.0166 [DOI] [PubMed] [Google Scholar]
- 15.Gentry J, Vinjé J, Guadagnoli D, Lipp EK. Norovirus distribution within an estuarine environment. Applied and environmental microbiology. 2009;75(17):5474–80. doi: 10.1128/AEM.00111-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernandez-Morga J, Leon-Felix J, Peraza-Garay F, Gil-Salas B, Chaidez C. Detection and characterization of hepatitis A virus and Norovirus in estuarine water samples using ultrafiltration–RT-PCR integrated methods. Journal of applied microbiology. 2009;106(5):1579–90. doi: 10.1111/j.1365-2672.2008.04125.x [DOI] [PubMed] [Google Scholar]
- 17.Sterk A, Schijven J, de Nijs T, de Roda Husman AM. Direct and indirect effects of climate change on the risk of infection by water-transmitted pathogens. Environmental science & technology. 2013;47(22):12648–60. doi: 10.1021/es403549s [DOI] [PubMed] [Google Scholar]
- 18.Pandey PK, Kass PH, Soupir ML, Biswas S, Singh VP. Contamination of water resources by pathogenic bacteria. Amb Express. 2014;4(1):51. doi: 10.1186/s13568-014-0051-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Viau EJ, Lee D, Boehm AB. Swimmer risk of gastrointestinal illness from exposure to tropical coastal waters impacted by terrestrial dry-weather runoff. Environmental science & technology. 2011;45(17):7158–65. doi: 10.1021/es200984b [DOI] [PubMed] [Google Scholar]
- 20.Bellou M, Kokkinos P, Vantarakis A. Shellfish-borne viral outbreaks: a systematic review. Food and Environmental Virology. 2013;5(1):13–23. doi: 10.1007/s12560-012-9097-6 [DOI] [PubMed] [Google Scholar]
- 21.Lee R, Rangdale R, Croci L, Hervio Heath D, Lozach S. Bacterial pathogens in seafood. 2008. [Google Scholar]
- 22.Cañigral I, Moreno Y, Alonso JL, González A, Ferrús MA. Detection of Vibrio vulnificus in seafood, seawater and wastewater samples from a Mediterranean coastal area. Microbiological research. 2010;165(8):657–64. doi: 10.1016/j.micres.2009.11.012 [DOI] [PubMed] [Google Scholar]
- 23.Blackstone GM, Nordstrom JL, Vickery MC, Bowen MD, Meyer RF, DePaola A. Detection of pathogenic Vibrio parahaemolyticus in oyster enrichments by real time PCR. Journal of Microbiological Methods. 2003;53(2):149–55. doi: 10.1016/s0167-7012(03)00020-4 [DOI] [PubMed] [Google Scholar]
- 24.Joshi J, Srisala J, Truong VH, Chen I-T, Nuangsaeng B, Suthienkul O, et al. Variation in Vibrio parahaemolyticus isolates from a single Thai shrimp farm experiencing an outbreak of acute hepatopancreatic necrosis disease (AHPND). Aquaculture. 2014;428:297–302. [Google Scholar]
- 25.Polo D, Varela MF, Romalde JL. Detection and quantification of hepatitis A virus and norovirus in Spanish authorized shellfish harvesting areas. International journal of food microbiology. 2015;193:43–50. doi: 10.1016/j.ijfoodmicro.2014.10.007 [DOI] [PubMed] [Google Scholar]
- 26.Baker-Austin C, Trinanes JA, Taylor NG, Hartnell R, Siitonen A, Martinez-Urtaza J. Emerging Vibrio risk at high latitudes in response to ocean warming. Nature Climate Change. 2013;3(1):73–7. [Google Scholar]
- 27.Tao C-W, Hsu B-M, Kao P-M, Huang W-C, Hsu T-K, Ho Y-N, et al. Seasonal difference of human adenoviruses in a subtropical river basin based on 1-year monthly survey. Environmental Science and Pollution Research. 2016;23(3):2928–36. doi: 10.1007/s11356-015-5501-8 [DOI] [PubMed] [Google Scholar]
- 28.Huang Z-M, Hsu B-M, Kao P-M, Chang T-Y, Hsu T-K, Ho Y-N, et al. Prevalence, quantification, and typing of human adenoviruses detected in river water in Taiwan. Environmental Science and Pollution Research. 2015;22(11):8359–66. doi: 10.1007/s11356-014-4000-7 [DOI] [PubMed] [Google Scholar]
- 29.Bates B, Kundzewicz ZW, Wu S, Palutikof J. Climate change and water. Technical paper of the intergovernmental panel on climate change, IPCC Secretariat, Geneva, 210 pp. ISBN 978-92-9169-123-4; 2008. [Google Scholar]
- 30.Dore MH. Climate change and changes in global precipitation patterns: what do we know? Environment international. 2005;31(8):1167–81. doi: 10.1016/j.envint.2005.03.004 [DOI] [PubMed] [Google Scholar]
- 31.Lindgren E, Jaenson TG. Lyme borreliosis in Europe: influences of climate and climate change, epidemiology, ecology and adaptation measures. WHO Regional Office for Europe Copenhagen; 2006. [Google Scholar]
- 32.Turner LM, Alsterberg C, Turner AD, Girisha S, Rai A, Havenhand JN, et al. Pathogenic marine microbes influence the effects of climate change on a commercially important tropical bivalve. Scientific Reports. 2016;6. doi: 10.1038/s41598-016-0015-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen M-J, Lin C-Y, Wu Y-T, Wu P-C, Lung S-C, Su H-J. Effects of extreme precipitation to the distribution of infectious diseases in Taiwan, 1994–2008. PLoS One. 2012;7(6):e34651. doi: 10.1371/journal.pone.0034651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Curriero FC, Patz JA, Rose JB, Lele S. The association between extreme precipitation and waterborne disease outbreaks in the United States, 1948–1994. American journal of public health. 2001;91(8):1194–9. doi: 10.2105/ajph.91.8.1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsiao H-I, Jan M-S, Chi H-J. Impacts of Climatic Variability on Vibrio parahaemolyticus Outbreaks in Taiwan. International journal of environmental research and public health. 2016;13(2):188. doi: 10.3390/ijerph13020188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sinclair R, Jones E, Gerba CP. Viruses in recreational water-borne disease outbreaks: A review. Journal of Applied Microbiology. 2009;107(6):1769–80. doi: 10.1111/j.1365-2672.2009.04367.x [DOI] [PubMed] [Google Scholar]
- 37.Chang Y, Lee M-A, Lee K-T, Shao K-T. Adaptation of fisheries and mariculture management to extreme oceanic environmental changes and climate variability in Taiwan. Marine Policy. 2013;38:476–82. [Google Scholar]
- 38.APHA. Standard Methods for the Examination of Water and Wastewater, 22nd edn. Association APH, editor. Washington, DC: American Public Health Association; 2012. [Google Scholar]
- 39.Hsu B-M, Chen C-H, Kung C-M, Wan M-T, Shen S-M. Evaluation of enterovirus recovery in surface water by different adsorption and elution procedures. Chemosphere. 2007;66(5):964–9. doi: 10.1016/j.chemosphere.2006.06.054 [DOI] [PubMed] [Google Scholar]
- 40.Huang W-C, Chou Y-P, Kao P-M, Hsu T-K, Su H-C, Ho Y-N, et al. Nested-PCR and TaqMan real-time quantitative PCR assays for human adenoviruses in environmental waters. Water Science and Technology. 2016;73(8):1832–41. doi: 10.2166/wst.2016.004 [DOI] [PubMed] [Google Scholar]
- 41.Daniels NA, MacKinnon L, Bishop R, Altekruse S, Ray B, Hammond RM, et al. Vibrio parahaemolyticus infections in the United States, 1973–1998. Journal of Infectious Diseases. 2000;181(5):1661–6. doi: 10.1086/315459 [DOI] [PubMed] [Google Scholar]
- 42.Su Y-C, Liu C. Vibrio parahaemolyticus: a concern of seafood safety. Food microbiology. 2007;24(6):549–58. doi: 10.1016/j.fm.2007.01.005 [DOI] [PubMed] [Google Scholar]
- 43.Vezzulli L, Pezzati E, Brettar I, Höfle M, Pruzzo C. Effects of Global Warming on Vibrio Ecology. Microbiology spectrum. 2015. doi: 10.1128/microbiolspec.VE-0004-2014 [DOI] [PubMed] [Google Scholar]
- 44.Jiang SC. Human Adenoviruses in Water: Occurrence and Health Implications: A Critical Review. Environmental Science & Technology. 2006;40(23):7132–40. doi: 10.1021/es060892o [DOI] [PubMed] [Google Scholar]
- 45.Wold W, Horwitz M. Adenoviruses. Fields virology. 2007;2:2395–436. doi: 10.1385/1-59745-166-5:157 [DOI] [PubMed] [Google Scholar]
- 46.Albinana-Gimenez N, Miagostovich MP, Calgua B, Huguet JM, Matia L, Girones R. Analysis of adenoviruses and polyomaviruses quantified by qPCR as indicators of water quality in source and drinking-water treatment plants. water research. 2009;43(7):2011–9. doi: 10.1016/j.watres.2009.01.025 [DOI] [PubMed] [Google Scholar]
- 47.Bofill-Mas S, Calgua B, Clemente-Casares P, La Rosa G, Iaconelli M, Muscillo M, et al. Quantification of human adenoviruses in European recreational waters. Food and Environmental Virology. 2010;2(2):101–9. [Google Scholar]
- 48.IPCC. Intergovernmental Panel on Climate Change 5th Assessment Report. Cambridge and New York: IPCC, 2014. [Google Scholar]