Abstract
The assumption of near-universal bacterial detection by pattern recognition receptors is a foundation of immunology. The limits of this pattern recognition concept, however, remain undefined. As a test of this hypothesis, we determined if mammalian cells can recognize bacteria that they have never had the natural opportunity to encounter. These bacteria were cultivated from the deep Pacific Ocean, where the genus Moritella was identified as a common constituent of the culturable microbiota. Most deep-sea bacteria contained cell wall lipopolysaccharide (LPS) structures that were expected to be immuno-stimulatory, and some deep-sea bacteria activated inflammatory responses from mammalian LPS receptors. However, LPS receptors were unable to detect 80% of deep-sea bacteria examined, with LPS acyl chain length being identified as a potential determinant of immuno-silence. The inability of immune receptors to detect most bacteria from a different ecosystem suggests that pattern recognition strategies may be defined locally, not globally.
One sentence summary
LPS immune detection strategies are tailored for host-microbe interactions that are possible in nature.
Introduction
The concept of pattern recognition, initially introduced by Janeway, posits that multicellular eukaryotes should have the ability to detect all microbes in the environment (1). This assumption of near-universal microbial detection is a foundation of modern immunology, and relies on the ability of multicellular organisms to detect infections through the actions of a set of cellular proteins known as pattern recognition receptors (PRRs) (2). PRRs recognize potentially infectious agents by detecting specific molecules that are common to large classes of microbes. These molecules, commonly microbial cell wall components or nucleic acids, are known as pathogen associated molecular patterns (PAMPs). Well-characterized examples of PAMPs include the lipid A region of bacterial lipopolysaccharides (LPS), the flagellin subunit of bacterial flagella and double stranded DNA. Each of these molecules is important for the viability or fitness of the organism that produces them. As such, they are highly stable and prevalent in the microbial world (3).
Based on the above-described scenario, PRRs should have the capacity to detect all members of a given class of microbe. The only exceptions to this statement should be host-adapted microbes that evolved strategies to alter their PAMPs to prevent PRR detection. For example, many commensal bacteria co-exist with mammalian hosts and avoid immune detection, whereas pathogens avoid detection to exploit the host (4–6). These exceptions represent the result of co-evolution between host and microbe. For bacteria not closely associated with a host, there are adaptive tradeoffs to altering PAMP structure that keep these strategies the rarity, rather than the norm (7–9). As such, outside of the intense host-microbial interface, mammalian PRRs are presumed to detect all bacteria they encounter.
However, virtually all knowledge of microbial detection has been derived from studies of bacteria that overlap ecologically with mammals, either in the same habitat or within mammalian hosts (10). With few exceptions (11–13), these studies focused on bacteria that inhabit terrestrial or shallow-depth aquatic environments (14), where mammals abound (15). But what of the bacteria that occupy different ecological niches? Is the selective advantage of PAMP detection exclusive to hosts and microbes from sympatric environments? More simply stated, does the pattern recognition concept have limits?
In this study, we sought to test assumptions of the pattern recognition concept by examining the ability of human and murine cells to recognize bacteria that they would have not had the natural opportunity to encounter. These bacteria were collected and cultivated during an expedition to the remote and deep Pacific Ocean, where samples were collected from deep-sea seamounts with abundant and diverse invertebrate life, and that are unoccupied by any resident mammals (tables S1, S2).
Results
We spent 20 days onboard the Schmidt Ocean Institute’s Research Vessel (R/V) Falkor within the boundaries of Kiribati’s Phoenix Islands Protected Area (PIPA), which is a no-take marine protected area and the largest and deepest UNESCO World Heritage site (16). We leveraged the opportunity to collect bacteria from the deep sea (>200m); a distinct ecological niche that is inhospitable to terrestrial life as it is principally devoid of light, under high pressure (>20 atmospheres), and cold (2–10°C) (17). In addition, the deep-sea lacks residential mammalian life (15). Although some marine mammals access the deep sea through intermittent diving, the primary habitat of marine mammals is the photic zone of the ocean (table S1; (15)). Bacteria from these remote, deep-sea ecosystems were therefore of high interest, as mammals would not have had the natural opportunity to interact extensively with bacteria from this ecosystem.
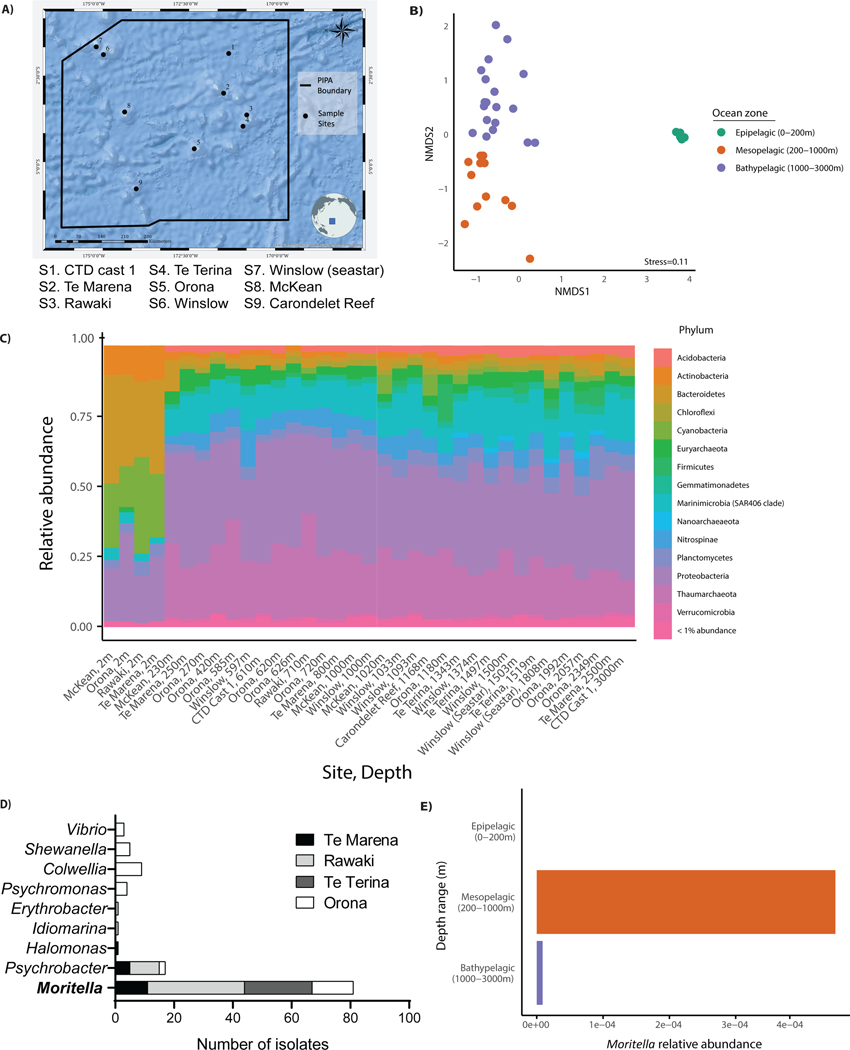
At nine sites within the boundaries of PIPA (Figure 1A), we collected seawater at stratified depths to assess bacterial community composition by amplicon sequencing of 16S ribosomal RNA (rRNA) genes. There was an observable shift in bacterial community composition between shallow (2m) and deep (>200m) seawater samples (Figure 1B and 1C). Thus, distinct microbial communities exist in deep-sea regions where mammalian populations are minimal. While the amplicon sequencing data obtained provided insights into the bacterial community composition of PIPA, our main goal was to obtain culturable bacteria for experimental analysis.
Figure 1.
Bacterial sample collection during the R/V Falkor’s 2017 expedition to the Phoenix Islands Protected Area, Kiribati. (A) Site map showing nine sites sampled. (B) Bray-Curtis non-metric multidimensional scaling ordination of total microbial community composition, based on 16S amplicon data from all stations. (C) Overview of the microbial community composition at stations S1-S9 as determined by 16S rRNA amplicon analysis. (D) Streak-purified bacteria strains isolated and sequenced from seawater and substrates collected at stations S2-S5. (E) Moritella relative 16S amplicon abundance in seawater samples collected from different depths at stations S2-S5.
To identify culturable bacteria under laboratory conditions, we sampled four sites (S2-S5) from 200–3000m depth for seawater, sediment, coral tissue, sponge tissue, and the gut contents of corallivorous sea stars. After sample collection at sea, bacterial colonies were grown from deep-sea substrates and seawater to build our ‘experimental toolbox’ of bacterial strains. A total of 117 bacterial colonies were isolated and streak purified from S2-S5. 16S ribosomal sequencing of streak purified strains identified all culturable bacteria to be of the class Gammaproteobacteria, and that 70% (82 of 117) of strains were of the genus Moritella (Figure 1D). Notably, Moritella were not detected in surface seawater samples (collected from 2m) (Figure 1E). Therefore, the Moritella genus is a common culturable constituent among our samples, exclusively found in the deeper waters of PIPA. A subset of these Moritella strains served as the foundation for our experimental analyses.
Moritella are Gram negative, which enabled us to determine the ability of mammalian LPS receptors to detect these bacteria. Mouse macrophages display monomers of the LPS receptors CD14 and TLR4 (associated with MD-2) at their plasma membrane, where they survey the extracellular space for this PAMP. When CD14 and TLR4 bind LPS, they are lost from the cell surface. CD14 is immediately internalized into endosomes whereas TLR4 must first dimerize before also being endocytosed (18–20). The LPS-induced loss of CD14 and TLR4 monomers from the plasma membrane can be monitored by flow cytometry, which represents a rapid assay that enables LPS-PRR interactions to be tracked for endogenous receptors in their natural setting (on the macrophage surface) (18–20).
Using this flow cytometry-based assay, we exposed murine immortalized bone marrow-derived macrophages (iBMDMs) to live bacteria for 20 minutes and measured the loss of receptors from the cell surface. E. coli LPS interacts efficiently with both receptors (19). Accordingly, live E. coli was used as a benchmark to delineate whether each Moritella strain engaged CD14 and TLR4. We limited the bacterial strains tested to those that were streak purified while onboard the R/V Falkor (n=50). Of the strains examined, 88% were of the genus Moritella (n=44), all isolated from >550m depth.
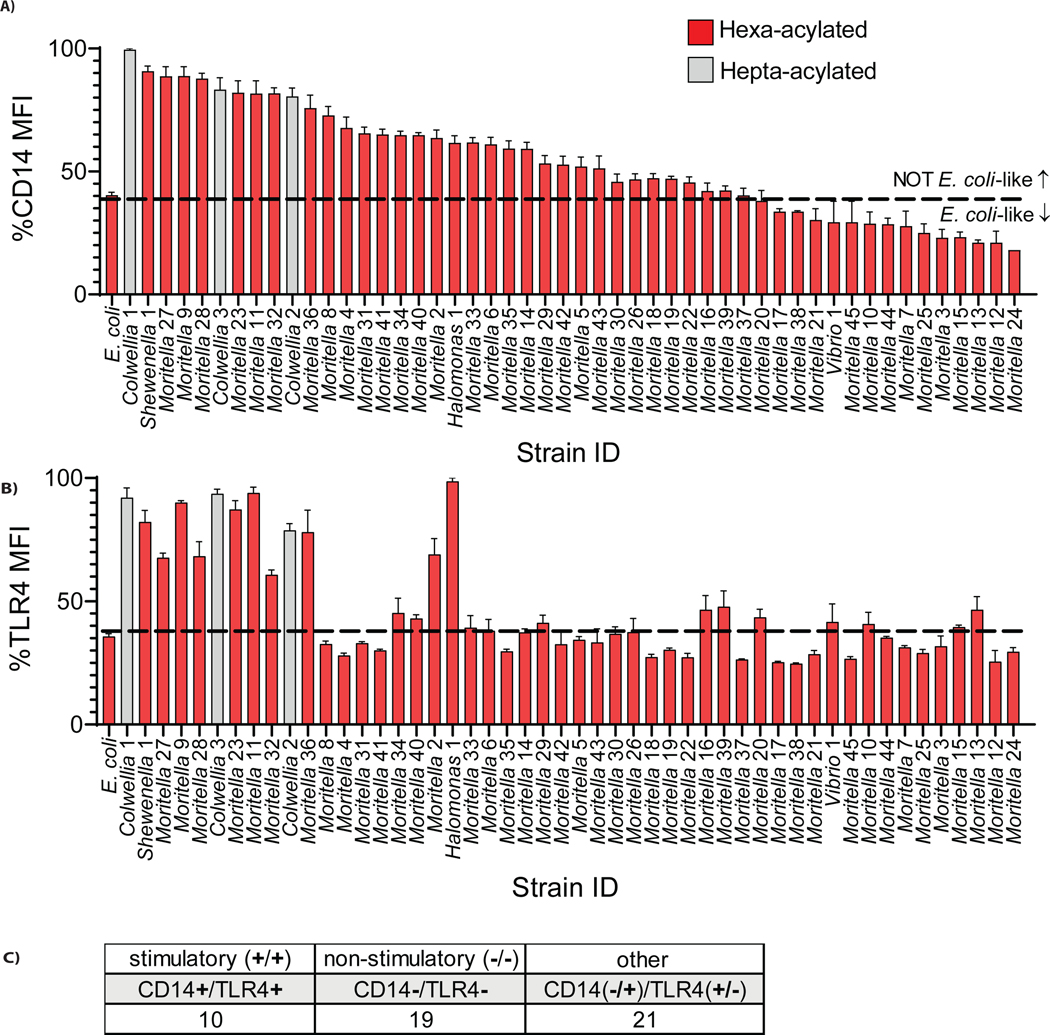
Compared to E. coli, which stimulated CD14 and TLR4 loss from the cell surface, specific marine bacterial strains were grouped into three categories (Figure 2A–C). Category 1 strains behaved similarly to E. coli, in that loss of both PRRs occurred. Category 2 strains were unable to engage either PRR. Category 3 strains engaged only one receptor. Overall, 10 strains engaged both receptors, 19 engaged neither, 21 engaged one receptor but not the other (Figure 2C). These findings were notable for two reasons. First, our finding that any strains of bacteria stimulated CD14 and TLR4 provides direct support for a central tenet of the concept of Pattern Recognition—that PRRs have the capacity to detect previously unencountered bacteria. While these findings are consistent with current dogma, it was unexpected that 80% (40/50) of bacteria displayed evidence of immune evasiveness, as defined by an inability to stimulate one or both of CD14 and TLR4. Deep-sea microbial species may therefore represent a reservoir of molecules with unpredictable inflammatory activities.
Figure 2.
Grouping of marine bacterial strains into three categories based on engagement with CD14 and TLR4 on mouse macrophages. (A,B) Surface expression of CD14 (A) and TLR4 (B) as measured by mean fluorescence intensity (MFI) on iBMDMs exposed to live deep-sea bacteria strains was compared to live E. coli (MOI=50). The color of columns represents the predicted acyl chain number for the LPS lipid A expressed, as described in Table 1. Dashed lines are used to delineate whether bacterial strains are stimulatory or silent to CD14 or TLR4, as compared to E. coli. (C) Summary of murine CD14 and TLR4 engagement by strains tested.
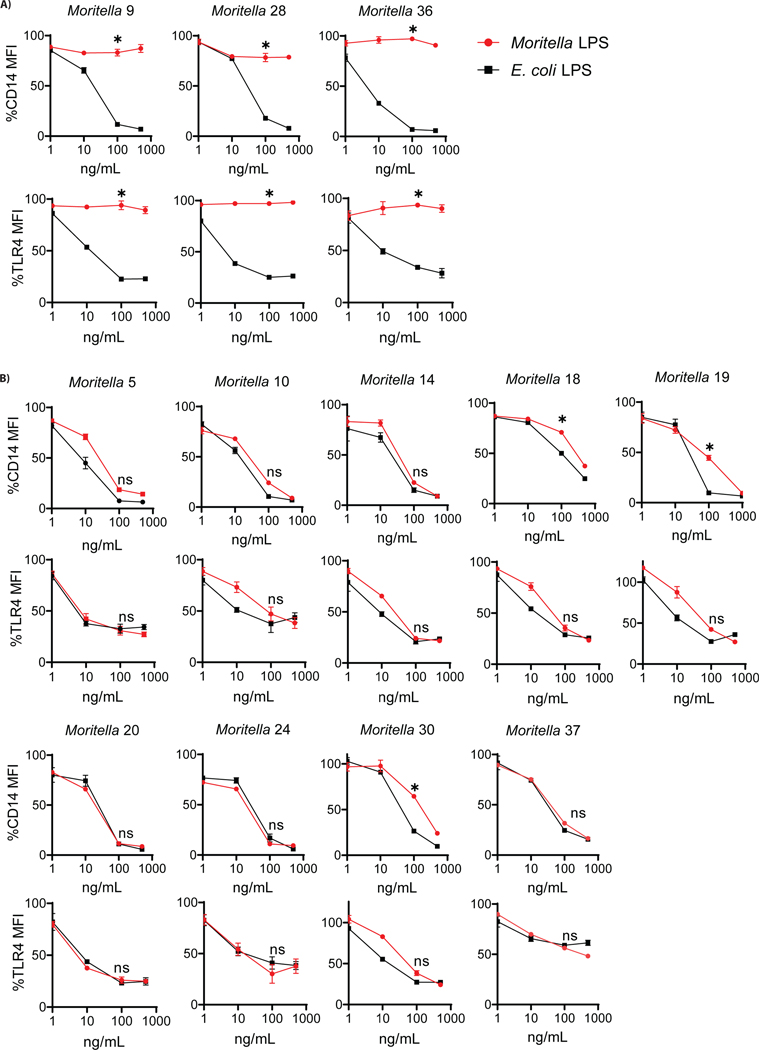
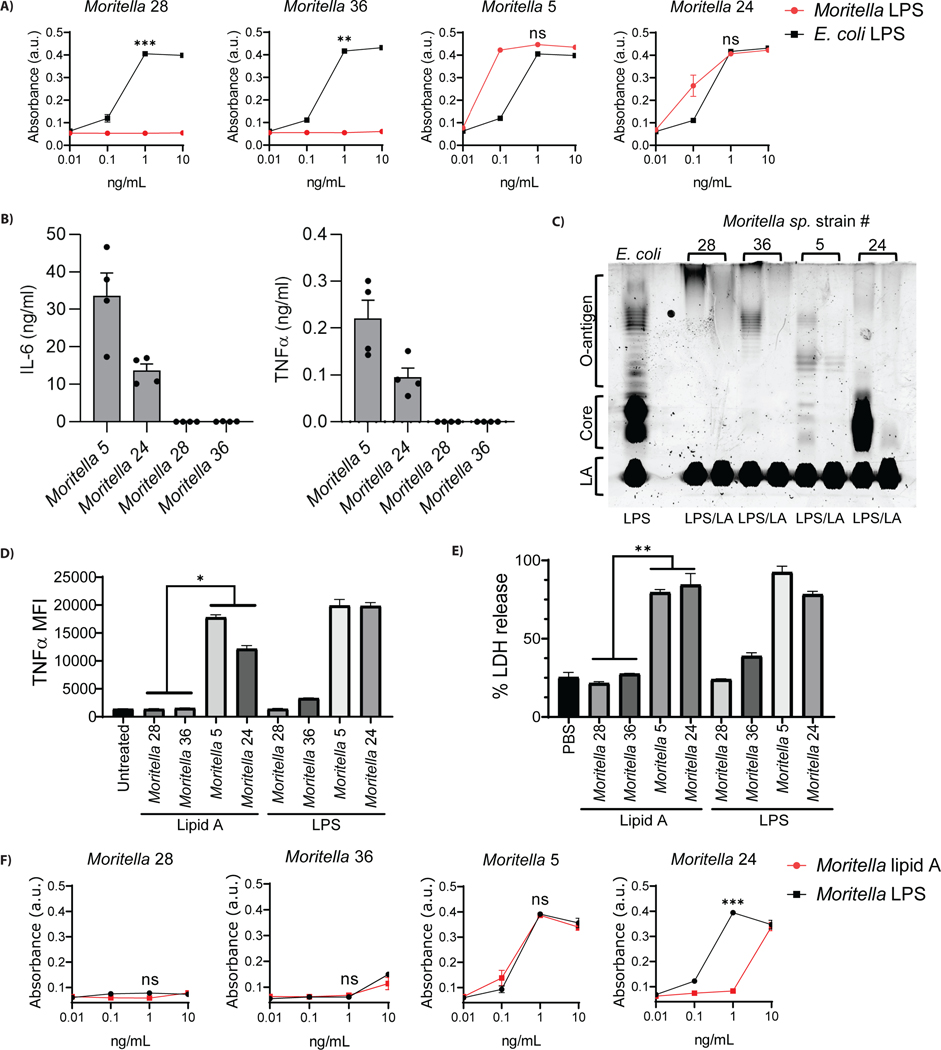
A possible explanation for the inability of deep-sea bacteria to stimulate CD14 or TLR4 is that the cell wall may contain features that prevent access of PRRs to immuno-stimulatory LPS. To address this possibility, we isolated LPS from select Moritella strains to evaluate if purified LPS behaved similarly to whole bacteria. Twelve Moritella strains over the range of CD14-TLR4 engagement demonstrated in Figure 2 were selected for this analysis. Specifically, three strains were selected that engaged neither PRR, and nine strains were selected that displayed a range of PRR engagement. In all analyses, purified LPS from each Moritella strain was compared to E. coli LPS (Figure 3). Dose response curves demonstrated that bacterial cells that were unable to engage CD14 and TLR4 yielded LPS that did not promote PRR loss from the cell surface, as compared to E. coli LPS (Figure 3A). Interestingly, all strains that had a partial phenotype when bacterial cells were used as stimuli (e.g. those that stimulated TLR4, but not CD14) were fully stimulatory for both PRRs when purified LPS was used (Figure 3B). This analysis therefore allowed us to classify LPS from Moritella in a binary fashion—either immuno-stimulatory or immuno-silent. Note, we chose the term “silent” rather than “evasive”, as the latter term suggests intent. The immuno-silent LPS preparations were isolated from Moritella 9, 28 and 36, the latter two of which were selected for further analyses.
Figure 3.
Dose-response curves testing ability of purified LPS from Moritella strains to induce PRR loss from the surface of mouse macrophages in comparison to E. coli LPS. (A) Purified LPS from three Moritella strains predicted to be hexa-acylated did not behave similarly to purified E. coli LPS in flow cytometry assays measuring engagement of CD14 and TLR4 on murine iBMDMs. (B) Purified LPS from nine other Moritella strains predicted to be hexa-acylated behaved similarly to E. coli LPS. Red circles (●) indicate Moritella LPS and black squares (■) indicate E. coli LPS. Statistical analysis based on comparison of 100 ng/mL dose from Moritella LPS with 100 ng/mL dose of E. coli LPS. (*p ≤ 0.01, ns = not significant).
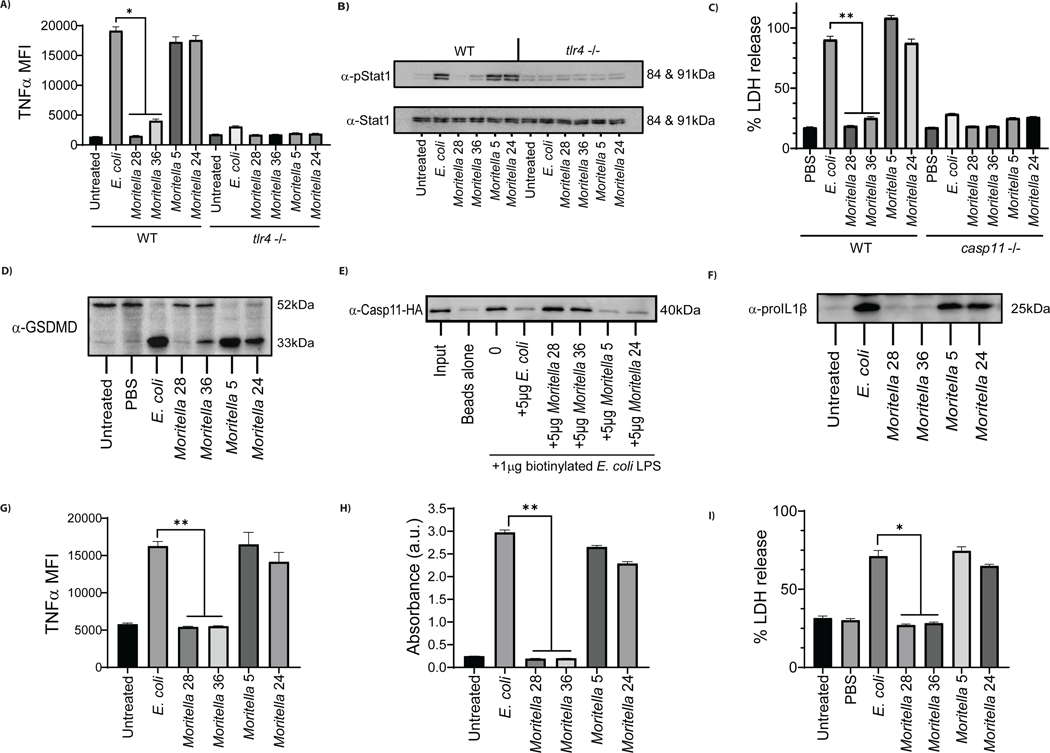
We reasoned that if LPS from Moritella 28 and 36 are incapable of stimulating CD14 and TLR4, then these LPS preparations should be unable to stimulate TLR4-dependent inflammatory responses. Consistent with our findings when assessing CD14 or TLR4 engagement, LPS from Moritella 5 and 24 induced TNFα production and STAT1 phosphorylation (Figure 4A, B). LPS from strains that did not stimulate CD14 or TLR4 were unable to induce these inflammatory responses (Figure 4A, B). All TNFα and STAT1 responses observed upon LPS stimulations were abolished when assays were performed on TLR4-deficient cells, as expected (21).
Figure 4.
Functional analysis of purified LPS from silent (#28, 36) or stimulatory (#5, 24) Moritella strains in murine and human macrophages. (A,B) Accumulation of TNFα after 3.5 hrs (A) and phosphorylation of STAT1 after 2.5 hrs (B) measured in wild type and Tlr4−/− iBMDMs incubated with 100 ng/mL LPS from Moritella strains or E. coli. (C) The release of LDH, 3 hours post-electroporation of wild type and Casp11−/− iBMDMs with 1 μg of LPS from Moritella strains and E. coli. (D) Cleavage of GSDMD, 3 hours post-electroporation of wild type iBMDMs with 1 μg of LPS from Moritella strains and E. coli. (E) Binding of HA-tagged caspase-11 to LPS supplied in excess (5 μg) from Moritella strains or E. coli, as measured by the ability of LPS to compete off biotinylated E. coli LPS (1 μg). (F) The production of pro-IL-1β, 2.5 hours post-treatment of human THP1 cells incubated with 50 ng/mL of LPS from Moritella strains compared to E. coli LPS. (G) Accumulation of TNFα, 4 hours post-treatment of human THP1 cells incubated with 100 ng/mL of LPS from Moritella strains compared to E. coli LPS. (H) Engagement of human TLR4 in human TLR4/NF-κB/SEAP reporter HEK293 cells by LPS from Moritella strains compared to E. coli LPS. (I) The release of LDH, 2.5 hours post-electroporation of human THP1 cells with 1 μg of LPS from Moritella strains compared to E. coli LPS. (*p ≤ 0.01 and ** p ≤ 0.001)
In addition to CD14 and TLR4, caspase-11 recognizes LPS in the cytosol of macrophages. Engagement of caspase-11 by E. coli LPS results in a lytic form of cell death called pyroptosis (22–24). Pyroptosis can be assessed by monitoring the release of the cytosolic enzyme lactate dehydrogenase (LDH) into the extracellular space (25) along with the cleavage of the pore forming protein, gasdermin D (GSDMD) (26). As expected (27, 28), electroporation of iBMDMs with E. coli LPS stimulated pyroptosis, as assessed by LDH release and GSDMD cleavage (Figure 4C,D). LPS from Moritella 5 and 24 also stimulated robust LDH released after electroporation. Notably, LPS from Moritella strains 28 and 36, which did not stimulate CD14 and TLR4, induced no LDH release from iBMDMs (Figure 4C). All LDH release observed was abolished when experiments were performed in caspase-11 deficient cells, as expected (22–24). Consistent with these results, partial or no cleavage of GSDMD was observed in iBMDMs electroporated with LPS from Moritella strains 28 and 36 (Figure 4D). Conversely, full cleavage of GSDMD was induced by LPS from E. coli, Moritella 5 and Moritella 24 (Figure 4D). To determine if differences in pyroptosis induction related to an ability to interact with caspase-11, in vitro experiments with purified LPS were performed. Using a procedure to assess PRR interactions with E. coli LPS or oxidized lipids (29), we assessed the ability of Moritella LPS to compete with biotinylated E. coli LPS for interactions with caspase-11. Biotinylated E. coli LPS formed a complex with caspase-11 in vitro (Figure 4E). This interaction was prevented when reactions were performed in the presence of excess non-biotinylated E. coli LPS, or in the presence of immuno-stimulatory LPS from Moritella 5 or Moritella 24 (Figure 4E). In contrast, LPS from the immuno-silent Moritella strains 28 and 36 did not compete with biotinylated E. coli LPS for binding to caspase-11 (Figure 4E). These results suggest that only immuno-stimulatory LPS can interact with caspase-11, a finding that likely explains the pyroptosis-inducing activity of the Moritella strains examined.
To establish whether the phenotypes observed in murine cells extend to other species, studies were performed in human THP1 monocytes and the LPS detection system from horseshoe crab crustaceans. Akin to the observations made in murine systems, LPS from Moritella 28 and 36 did not stimulate the production of pro-IL1β or TNFα in THP1 monocytes, as compared to LPS from E. coli, Moritella 5 or 24 (Figures 4F, G). To validate these findings, a HEK293-BLUE reporter system was used to assess direct engagement of human CD14, TLR4 and MD-2. In these cells, LPS interactions with these PRRs stimulates the secretion of alkaline phosphatase (SEAP). We found that only immuno-stimulatory LPS induced SEAP production by HEK293-BLUE cells (Figure 4H), thereby suggesting that silent Moritella LPS from strains 28 and 36 do not engage PRRs in the TLR4 pathway. Finally, electroporated LPS from Moritella 5 and 24 induced THP1 monocyte pyroptosis to an extent comparable to that elicited by E. coli LPS (Figure 4I). LPS from Moritella 28 and 36 induced minimal LDH release from electroporated THP1 monocytes (Figure 4I).
The horseshoe crab Limulus polyphemus contains a distinct LPS detection system than terrestrial mammals (30). Upon binding of LPS to the protein Factor C from L. polyphemus, a complement-like protease cascade is stimulated that results in a defensive coagulation response. Although it is an aquatic animal, L. polyphemus occupies shallow marine environments (31). Interestingly, LPS preparations that could stimulate detection (or not) by mammalian LPS receptors exhibited the identical pattern of activity of engagement with Factor C. LPS from Moritella 28 and 36 did not engage Factor C, whereas LPS from Moritella 5 and 24 did so, similarly to E. coli LPS (Figure 5A).
Figure 5.
Cross-species evasion of LPS receptor activity by deep-sea Moritella strains. (A) Engagement of Limulus polyphemus factor C by LPS from Moritella strains compared to E. coli LPS. (B) Inflammatory response to LPS from Moritella strains in vivo. IL-6 and TNFα plasma levels were measured 4 hours post intraperitoneal injection of mice with LPS from stimulatory (#5, 24) or silent (#28, 36) Moritella strains. (C) 10 μg of E. coli LPS was compared to 10 μg of LPS or lipid A (LA) from Moritella strains visualized on a polyacrylamide gel stained with ProQ Emerald LPS staining solution. (D) Accumulation of TNFα after 3.5 hr stimulations of iBMDMs with 100 ng/mL lipid A compared to LPS from Moritella strains. (E) The release of LDH 24 hours post-electroporation of iBMDMs with 1μg of lipid A or LPS from Moritella strains. (F) Engagement of Limulus polyphemus factor C by LPS and lipid A from Moritella strains. (*p ≤ 0.01, ** p ≤ 0.001 and ***<0.0001)
To determine if the immuno-silence of select Moritella LPS extended to an in vivo setting, inflammatory responses in mice were examined. Intraperitoneal injections of LPS from Moritella 5 and 24 induced the rapid accumulation of cytokines TNFα and IL-6 in the plasma (Figure 5B). In contrast, these cytokines were not detected in the plasma of mice injected with Moritella 28 and 36 (Figure 5B). Overall, symmetrical observations were made in mice, in human and murine cells, and in the in vitro system offered by the horseshoe crab. All these systems revealed immuno-silent LPS from select Moritella.
The LPS molecule of Gram-negative bacteria is composed of three regions. The hydrophobic lipid A region anchors LPS to the bacterial outer membrane, whereas the water-soluble core oligosaccharide and O-antigen extend from the lipid A anchor into the aqueous extracellular space (32). It was possible that the silent activity of select Moritella strains was due to specific features of the O-antigen region that prevent PRR detection of lipid A. If this possibility was correct, then removal of the O-antigen should render immuno-silent molecules stimulatory. Thus, we purified lipid A after hydrolysis of the O-antigen from the LPS preparations of interest. This procedure resulted in the near-complete disappearance of the core and/or O-antigen regions, as assessed by silver staining (Figure 5C). Removal of O-antigen did not increase the stimulatory activity of the immuno-silent lipid A preparations. Lipid A from immuno-stimulatory Moritella strains remained capable of inducing TNFα production, whereas lipid A from silent Moritella strains 28 and 36 remained incapable of inducing this inflammatory response (Figure 5D). The same pattern of engagement for Moritella lipid A was observed for murine caspase-11 engagement, as indicated by pyroptosis induction after electroporation (Figure 5E). Finally, neither LPS nor lipid A from Moritella 28 and 36 was able to stimulate Factor C (Figure 5F). These data indicate that lipid A was not being hidden from LPS receptors by other structural regions of the LPS molecule. We therefore conclude that the silence of some deep-sea bacteria is intrinsic to the lipid A structure of LPS and extends across the evolutionary tree of PRRs.
The features of lipid A that are associated with immuno-stimulatory activity of E. coli include a hexa-acylated and bis-phosphorylated di-glucosamine backbone (32). LPS structures that contain more (or less) than six acyl chains are weakly stimulatory to PRRs, and LPS structures containing mono-phosphorylated LPS are similarly immune-evasive. Additionally, specific modifications to the lipid A, such as aminoarabinose, phosphoethanolamine (PEtn), or the presence of cardiolipin in LPS preparations are associated with non-stimulatory activity (10, 33). To determine if deep-sea LPS display any of these immuno-evasive lipid A features, structural composition analysis was performed. We first performed a large-scale analysis of lipid A structures isolated from all 50 deep-sea bacteria we examined at the start of this study (Table 1). We extracted lipid A and predicted structural composition by MALDI-TOF mass spectrometry (MS) in the negative ion mode (34). Mass spectral data were used to predict the following within lipid A: (1) the number of acyl chains likely to be present, (2) the loss of a phosphate from the di-glucosamine sugar backbone (mono-phosphorylation), (3) the addition of aminoarabinose or PEtn to the di-glucosamine sugar backbone and 4) the presence of cardiolipin in the lipid A extractions.
Table 1.
MALDI-TOF MS structural data and endogenous murine CD14-TLR4 engagement by bacterial strain. Structural data reported for each strain: 1) The primary m/z peak value, 2) the number of acyl chains predicted for the m/z peak value reported, 3) mono-phosphorylation of the lipid A di-glucosamine backbone, and 4) the addition of phosphoethanolamine (PEtn) to the lipid A di-glucosamine backbone. CD14-TLR4 engagement is reported as Yes/No compared to E. coli.
| Strain Identification | 1° ionizable m/z peak value | Predicted # acyl chains | Minor mono- (Yes/No) | PEtn addition (Yes/No) | Cardiolipin (Yes/No) | CD14 internalization | TLR4/MD2 dimerization |
|---|---|---|---|---|---|---|---|
| E. coli | 1798 | 6 | No | No | No | Yes | Yes |
| Colwellia 1 | 2048 | 7 | Yes | No | No | No | No |
| Shewenella 1 | 1727 | 6 | Yes | No | No | No | No |
| Moritella 27 | 1792 | 6 | Yes | No | No | No | No |
| Moritella 9 | 1755 | 6 | Yes | Yes | No | No | No |
| Moritella 28 | 1739 | 6 | No | No | No | No | No |
| Colwellia 3 | 2047 | 7 | Yes | Yes | No | No | No |
| Moritella 23 | 1739 | 6 | No | No | No | No | No |
| Moritella 11 | 1798 | 6 | Yes | Yes | No | No | No |
| Moritella 32 | 1768 | 6 | No | No | No | No | No |
| Colwellia 2 | 2020 | 7 | Yes | No | No | No | No |
| Moritella 36 | 1739 | 6 | No | No | No | No | No |
| Moritella 8 | 1658 | 6 | No | No | No | No | Yes |
| Moritella 4 | 1766 | 6 | Yes | Yes | No | No | Yes |
| Moritella 31 | 1740 | 6 | Yes | No | No | No | Yes |
| Moritella 41 | 1740 | 6 | No | No | No | No | Yes |
| Moritella 34 | 1739 | 6 | Yes | No | No | No | No |
| Moritella 40 | 1739 | 6 | Yes | No | No | No | No |
| Moritella 2 | 1740 | 6 | Yes | No | No | No | No |
| Halomonas 1 | 1739 | 6 | Yes | No | No | No | No |
| Moritella 33 | 1739 | 6 | Yes | No | No | No | No |
| Moritella 6 | 1740 | 6 | Yes | No | No | No | No |
| Moritella 35 | 1766 | 6 | Yes | No | No | No | Yes |
| Moritella 14 | 1790 | 6 | Yes | No | No | No | No |
| Moritella 29 | 1740 | 6 | Yes | No | Yes | No | No |
| Moritella 42 | 1739 | 6 | No | No | No | No | Yes |
| Moritella 5 | 1739 | 6 | No | No | No | No | No |
| Moritella 43 | 1769 | 6 | Yes | No | No | No | Yes |
| Moritella 30 | 1764 | 6 | No | No | No | No | No |
| Moritella 26 | 1766 | 6 | Yes | No | No | No | No |
| Moritella 18 | 1767 | 6 | Yes | No | No | No | Yes |
| Moritella 19 | 1766 | 6 | Yes | No | No | No | Yes |
| Moritella 22 | 1739 | 6 | Yes | No | No | No | Yes |
| Moritella 16 | 1764 | 6 | Yes | No | No | No | No |
| Moritella 39 | 1766 | 6 | No | No | No | No | No |
| Moritella 37 | 1766 | 6 | No | No | No | No | Yes |
| Moritella 20 | 1767 | 6 | Yes | No | No | No | No |
| Moritella 17 | 1790 | 6 | No | No | Yes | Yes | Yes |
| Moritella 38 | 1768 | 6 | Yes | No | No | Yes | Yes |
| Moritella 21 | 1659 | 6 | No | No | Yes | Yes | Yes |
| Vibrio 1 | 1754 | 6 | No | No | Yes | No | Yes |
| Moritella 45 | 1807 | 6 | No | No | No | Yes | Yes |
| Moritella 10 | 1767 | 6 | Yes | No | No | No | Yes |
| Moritella 44 | 1807 | 6 | No | No | Yes | Yes | Yes |
| Moritella 7 | 1766 | 6 | Yes | No | No | Yes | Yes |
| Moritella 25 | 1655 | 6 | No | No | Yes | Yes | Yes |
| Moritella 3 | 1766 | 6 | Yes | No | No | Yes | Yes |
| Moritella 15 | 1766 | 6 | Yes | No | No | No | Yes |
| Moritella 13 | 1764 | 6 | Yes | No | No | No | Yes |
| Moritella 12 | 1791 | 6 | No | No | No | Yes | Yes |
| Moritella 24 | 1784 | 6 | No | No | No | Yes | Yes |
These structural analyses revealed that several bacteria contain immuno-silent modifications to their lipid A. Specifically, the Colwellia strains examined contained a primary ionizable m/z peak >2000 atomic mass units (amu), which is predicted to be hepta-acylated (Table 1). Additionally, we observed PEtn additions (Δm/z of 123 amu) to the lipid A backbone of select strains, as well as the presence of cardiolipin (repeating Δm/z of 12 amu) in others (Table 1). These bacteria were identified in Figure 2 as containing LPS that prevents detection by CD14 or TLR4, an observation that is likely explained by the structural changes to the lipid A we identified.
In contrast to the examples offered above, which are consistent with current views of LPS-PRR interactions, most strains examined (including all 44 Moritella strains) contained a primary ionizable m/z peak ranging from 1650–1800 amu (Table 1). These spectra are consistent with hexa-acylated lipid A being present in all Moritella examined. Similar findings were observed for lipid A structures from Halomonas, Shewanella, and Vibrio strains (Table 1). All Moritella strains were also predicted to contain bis-phosphorylated lipid A, although some strains had minor populations of mono-phosphorylation (Δm/z of 80) (Table 1). These features of lipid A—hexa-acylated and bis-phosphorylated—are commonly associated with immuno-stimulatory LPS, such as that from E. coli. Yet, these E. coli-like lipid A structures from deep-sea bacteria were either immuno-stimulatory or immuno-silent. Indeed, of the Moritella examined structurally, 77.3% (34/44) displayed evidence of immuno-silence when bacteria were used to stimulate murine cells (Figure 2). The immuno-silence of Moritella 9 may be explained by the presence of a PEtn on its lipid A di-glucosamine backbone (Table 1) (35). Moritella 28 and 36, in contrast, did not have any observable backbone modifications to explain why their LPS was immuno-silent across multiple experimental systems.
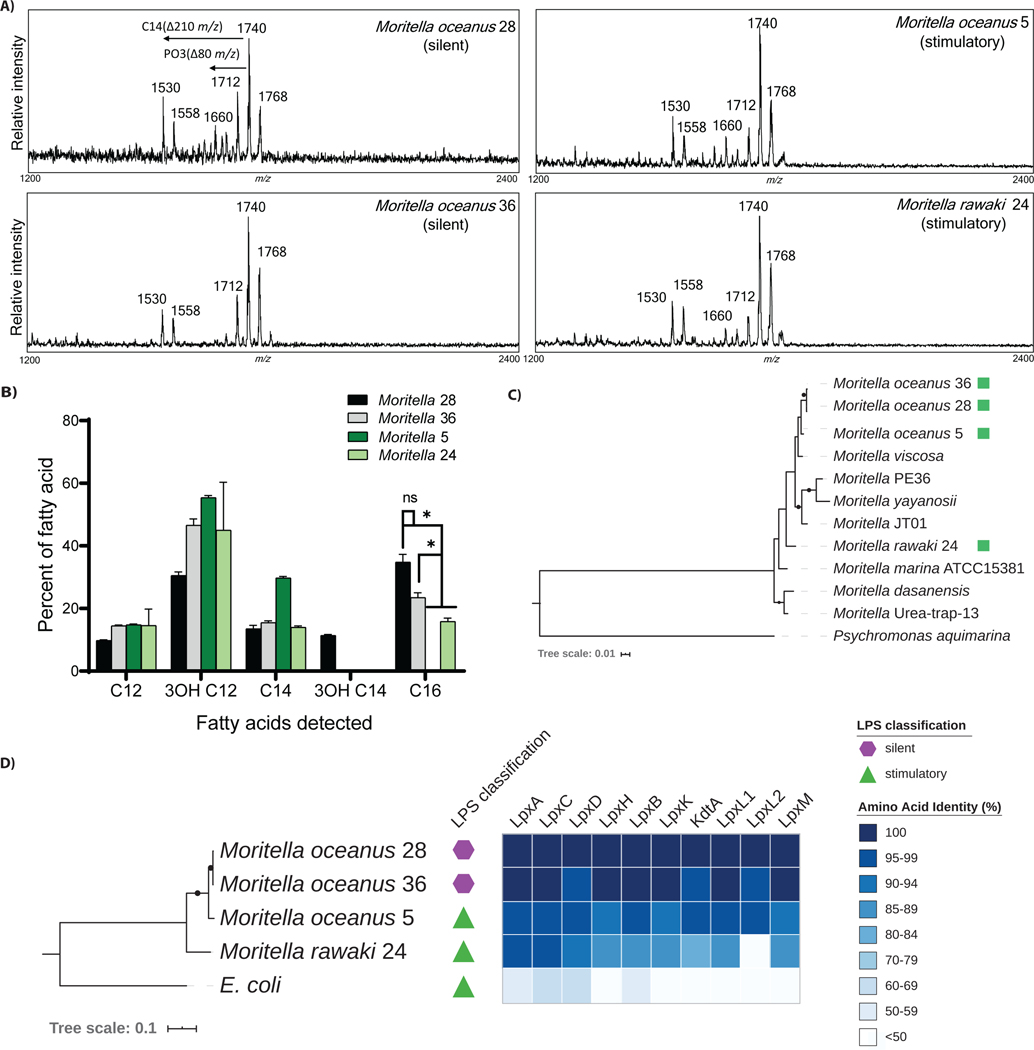
To confirm the MALDI-TOF MS analyses, we employed a newly described gentle extraction method, known as Fast Lipid Analysis Technique (FLAT) (36). FLAT enables the detection of very small quantities of lipid A within a bacterial colony, without the need for centrifugation or lyophilization. The mass spectra obtained by this distinct method were consistent with those we obtained using other methods (Table 1), in that all four Moritella strains of interest (2 immuno-stimulatory and 2 immuno-silent) were predicted to be hexa-acylated and bis-phosphorylated (Figure 6A).
Figure 6.
Further characterization of immuno-silent and immuno-stimulatory Moritella strains. (A) MALDI-TOF MS spectra of Moritella lipid A generated using the FLAT technique. (B) Relative fatty acid content in lipid A derived from silent and stimulatory Moritella as determined by gas chromatography–mass spectrometry (GC-MS) (*p ≤ 0.01). (C) Whole genome phylogeny of Moritella strains. The amino acid phylogeny was inferred using maximum likelihood from a concatenated alignment of 120 single copy genes (56) generated from the four newly-sequenced Moritella genomes (green squares) and other publicly available assemblies. Black dots on branches indicate bootstrap support >75%. (D) Degree of sequence conservation for enzymes in the lipid A biosynthesis pathway. The maximum likelihood phylogeny at left is based on a concatenated amino acid alignment of the 10 indicated lipid A biosynthesis enzymes from each genome. The heatmap depicts the % amino acid identity for each individual enzyme in the pathway, as compared to Moritella oceanus 28. Black dots on tree branches indicate bootstrap support > 75%.
To further assess the structural features of Moritella lipid A, gas chromatography-mass spectrometry (GC-MS) was performed. This analysis enables the determination of the composition of acyl chains present on lipid A (37). We found that immuno-silent strains (Moritella 28 and 36) contained lipid A with the highest amount of C16 acyl chains (Figure 6B). Conversely, the stimulatory strains contained lipid A with zero C16 chains (Moritella 5) or low amounts of C16 chains (Moritella 24). Instead, these stimulatory strains had mainly C12 and C14 length acyl chains (Figure 6B). These findings were notable, as C14 and C12 length acyl chains are the optimal lengths for robust activation of TLR4 signaling (38, 39). C16 chains, in contrast, are recognized as being non-optimal lengths for productive interactions with MD-2 (40). Indeed, in the context of synthetic lipid A mimics, those with C16 chains cannot bind MD-2 at all—and are consequently immuno-silent (40). Consistent with this idea, the highest amount of C16 acyl chains were found in the immuno-silent Moritella strains 28 and 36. Other than these C16 differences, the GC analysis revealed similar patterns of fatty acid composition in the strains examined, with no strain-specific differences in short or odd-length fatty acids detected. Thus, we propose that immuno-silence is not the result of an unusual addition to the lipid A molecules found in the deep sea, but rather results from the existence of abundant C16 acyl chains found in select strains, which determines PRR interactions.
We sought to determine how each Moritella strain related to each other and to previously identified Moritella species. A combination of Nanopore and Illumina sequencing was used to determine the genome sequence of the two silent and the two stimulatory Moritella strains (table S3). We determined the phylogenetic relationship between these strains and other sequenced Moritella genomes (Figure 6C). This analysis indicated that our four Moritella strains are evolutionarily distinct from known Moritella. Average nucleotide identity analysis (41, 42) indicated that Moritella 24 represents a distinct species, whereas Moritella 5, 28, and 36 represent substrains of a second new species (table S4). We tentatively propose these species be designated Moritella oceanus (formerly Moritella 5, 28, and 36) and Moritella rawaki (formerly Moritella 24).
Pangenome analysis revealed a set of protein-coding sequences that distinguished the two immuno-silent from the two immuno-stimulatory strains. Specifically, both immuno-silent Moritella strains encoded a common set of 192 protein-coding sequences (table S5) not found in either immuno-stimulatory strain. 55 protein-coding genes were found exclusively in both stimulatory strains (table S6). Most of these genes lack an annotated function. Whether any of these 55 genes affect immuno-silence or detection is unknown, as is the degree of sequence conservation across other (un-sequenced, but behaviorally similar (as in Figure 2)) Moritella strains.
We identified the lipid A biosynthesis enzymes and their protein coding sequences (32) within the genomes of our four Moritella strains (fig. S1). We identified one copy of each enzyme, with the exception of LpxL, which had two protein coding sequences present in each genome. We determined the degree of sequence conservation for enzymes in the lipid A biosynthesis pathway between these four Moritella strains and E. coli (Figure 6D). While the E. coli lipid A enzymes were distinct from those present in any Moritella strain, the most notable distinctions came from comparisons within Moritella. The enzymes that build lipid A in immuno-silent M. oceanus 28 and M. oceanus 36 are (on a network scale) highly similar to each other. The analogous enzymes in immuno-stimulatory M. oceanus 5 or M. rawaki 24 are also highly similar to each other, but differed from the corresponding enzymes in immuno-silent M. oceanus strains. Based on this analysis, the entire lipid A biosynthetic pathway may contribute to the distinct inflammatory activities of select deep-sea Moritella LPS structures.
Discussion
In this study, we tested the limits of the pattern recognition hypothesis by asking if mammalian PRRs could detect bacteria from a different ecosystem. Two major conclusions were drawn. First, some deep-sea bacteria can be detected by mammalian LPS receptors, although immuno-stimulation of both CD14 and TLR4 represented a minor population of all bacteria examined (20% (10/50)). Nonetheless, the identification of even a single bacterium with immuno-stimulatory activity validates a central principle of Pattern Recognition—that our innate immune system can detect previously unencountered bacteria. Secondly, we found that mammalian PRRs were unable to detect the majority of LPS displayed on live, cultured bacterial strains from the deep sea (Figure 2), revealing that the rules of innate immune detection may be more limited than previously appreciated. Thus, we posit a revision to the pattern recognition concept—PRRs should have the capacity to detect all bacteria that exist in the same general habitat as the host. In other words, innate immunity follows local (not global) rules of engagement (fig. S2).
It is unlikely that deep-sea bacteria experience any fitness benefit from evading detection of mammalian PRRs, even those expressed by mammals that occasionally dive to the depths that these microbes inhabit (table S1). Inhabiting an environment devoid (or nearly devoid) of mammals likely creates a scenario where there is no selective advantage to bacterial LPS being immune-evasive, immuno-silent, or immuno-stimulatory. Bacteria from extreme, non-mammalian environments may have cell wall structures that are uniquely optimized to their environment, and when habitats are artificially crossed, bacteria may encounter a mammalian host in which they are accidentally immune-evasive. The consequences of such interactions are difficult to predict, but are important to consider in this age of increasing globalization and exploitation of deep-sea resources (43).
Finally, it is worth considering whether deep-sea invertebrates may have evolved PRRs to detect LPS structures that are common to bacteria found in these habitats. This consideration is notable, as the functions of LPS receptors have been explored almost exclusively in mammals. In fact, genomic analysis of numerous organisms has demonstrated a notable lack of the CD14-MD-2-TLR4 system in marine fish and invertebrates. While proteins that display homology to TLR4 can sometimes be identified in fish or invertebrates, the significance of this homology in unclear, as CD14 and MD-2 can almost be considered mammal-specific factors. Identification of invertebrate taxa in the deep sea is ongoing, and many species are yet unknown, however deep-sea corals and sponges are among the most abundant foundational macrofauna (44, 45). Shallow-water cnidarians have an abundance of genes that have been bioinformatically annotated as PRRs (46); however, whether these PRRs also exist abundantly in deep-sea taxa is unknown. One study has examined flagellin signaling in cnidarians (a shallow-water anemone) (47), but nothing is yet known about LPS detection events in any marine invertebrate. As such, it is unclear whether LPS detection systems are common outside of the mammalian lineage. Indeed, even the plant Arabidopsis thaliana, which was once thought to use the protein LORE as an LPS receptor, is now recognized to not detect LPS (48). While these evolution-function considerations remain to be addressed, our data suggest that the rules of innate immune engagement are defined locally (not globally), and thus provide a mandate for further exploration of host-microbe interactions in diverse ecosystems.
Materials and Methods
Study Design
We designed this study to evaluate the ability of mammalian PRRs to detect deep-sea bacteria via the PAMP, LPS. Therefore, we cultured bacterial strains from the deep-sea of PIPA to screen in a flow cytometry-based assay for their ability to interact with endogenous murine PRRs, CD14 and TLR4. Our results indicated that 80% of live bacteria strains did not engage with murine CD14 and/or TLR4 and were categorized as “silent” to these PRRs. To confirm that LPS from deep-sea bacteria was silent, we purified LPS from stimulatory and silent Moritella strains to characterize the downstream innate immune response to extracellular and intracellular LPS in murine and human cells. LPS from Moritella strains that were silent to murine and human PRRs did not initiate any downstream innate immune response. LPS from silent Moritella strains also did not engage with murine PRRs in vivo or in Limulus polyphemus. We then hydrolyzed and purified lipid A from Moritella LPS and confirmed that the activity of silent LPS was a direct result of the structure of the lipid A moiety. To delineate the differences between stimulatory and silent Moritella lipid A, we characterized the structure of lipid A and sequenced the genomes of each strain of interest.
Deep-sea sampling
In October 2017, the R/V Falkor, operated by the Schmidt Ocean Institute, conducted 17 remotely operated vehicle (ROV) surveys in the Phoenix Islands Protected Area (cruise FK171005) (detailed in (45)) using the 4,500-m rated ROV SuBastian. On the PIPA expedition, ROV SuBastian was equipped with an Insite Pacific Mini Zeus HD 1080i CMOS camera for situational awareness and an Insite Zeus Plus or SULIS 4K 12× Zoom camera for science surveys (5.1–51 mm wide angle zoom lens). In addition, the vehicle hosted a Seabird FastCAT CTD Sensor (SBE49) and a Paroscientific 8000 Series Submersible Depth Sensor for measuring depth of observations (as in (49)). In addition to 17 ROV dives, 7 CTD casts were deployed and retrieved in PIPA, from which water was filtered for microbial extraction or culturing, as below.
Isolation of culturable bacteria strains
At five atolls within the boundaries of PIPA (depths 270–2500m), seawater, soft coral tissue, glass sponge tissue, and sea star gut content were collected to use as substrate to grow deep-sea bacteria. Tissue and gut content were homogenized in 5mL of PBS pH 7.4 in sterile 15 mL falcon tubes. 200 μl of substrate was loaded onto individual Difco marine agar 2216 (Becton Dickenson, 212185) plates without antibiotics in a sterile fume hood aboard the R/V Falkor. Plates were incubated at 4°C for 5–14 days, protected from light, until colonies formed. Following bacteria colony formation, strains were streak purified and glycerol stocks (50% marine media, 25% water, 25% glycerol) were stored at −80°C in 2.0 mL cryovials. Upon expedition completion, glycerol stocks were transported on dry ice by airplane to be stored at −80°C in Boston, MA.
Amplicon sequencing
Amplicon sequencing is described in the Supplementary Methods.
Sequencing bacterial strains from liquid culture
Bacterial gene sequencing is described in the Supplementary Methods.
Moritella genome sequencing and analysis
Genome sequencing and analysis are described in the Supplementary Methods.
Cell lines
Wild type, Tlr4−/−, and Casp11−/− immortalized bone marrow derived macrophages (iBMDMs) were cultured in DMEM with 10% FBS, 1% penicillin and streptomycin, and 1% supplementation with L-glutamine and sodium pyruvate. This media is referred to as complete DMEM. Prior to experimentation, cells were washed with PBS pH 7.4 and lifted with PBS pH 7.4 containing 2 mM EDTA. Otherwise, cells were passaged 1:10 every 2–3 days. During routine passage, cells were washed with PBS pH 7.4 and detached with 0.25% trypsin. Trypsin was deactivated by adding serum containing media; after which cells were spun down and re-suspended in fresh media.
LPS and lipid A preparations
LPS and lipid A preparations are described in the Supplementary Methods.
Lipid A extraction for MALDI-TOF MS
Bacteria strains (n=50) were grown for 48–72 hours to OD600=1 in 5 mL marine media at 14.5°C in a shaking incubator (180rpm). Bacteria were pelleted and frozen for future extraction of lipid A. Frozen bacterial pellets were thawed on ice and lipid A was extracted from pellets using the isobutyric acid method (50). Briefly, in screw-cap tubes, pellets were resuspended in 70% isobutyric acid and 1 M ammonium hydroxide (5:3 [vol/vol]; 400 μL total). Tubes were incubated for 60 minutes at 100°C with occasional vortexing to liberate the lipid A. Products were cooled on ice, centrifuged for 5min at 8,000 x g prior to harvesting the supernatant into new 1.5 mL tubes containing an equal volume (400 μl) of endotoxin-free ddH2O. Samples were frozen on dry ice for 30 min and lyophilized. Lyophilized samples were washed with methanol and lipid A was extracted from the remaining pellet with 50 μL of chloroform:methanol:water (3:1.5:0.25 [vol/vol]). The final lipid A product was mixed with Dowex beads and centrifuged for 5 min at 5,000 x g. 1 μl of the lipid A product (supernatant) was resuspended with 1 μl of 10 mg/mL Norharmane matrix (Sigma, N6252, (51)) resuspended in chloroform: methanol (2:1 [vol/vol]) at a ratio of 1:1 and spotted on a steel re-usable MALDI plate. Spots were air dried and samples were analyzed on a Bruker Microflex mass spectrometer (Bruker Daltonics, Billerica, MA) in the negative-ion mode with reflectron mode. The spectrometer was calibrated with an electron spray tuning mix prior to every run (Agilent, Palo Alto, CA).
FLAT technique lipid A extraction for MALDI-TOF MS
Microbial colony smears or liquid samples were applied to a target location on a stainless steel MALDI plate. The target plate was incubated in a 70% citric acid buffer in a humidified chamber for 30 minutes at 110ºC. The MALDI plate was washed with deionized water from a squeeze bottle, allowed to air dry, then 1 μL of Norharmane matrix solution was applied (10 mg/mL in 12:6:1, v/v/v chloroform/methanol/water) to each target location (36). Following the method of Leung et al. (52), spectra were acquired from target locations in negative ion mode using a Microflex LRF MALDI-TOF MS (Bruker, Billerica MA) in reflectron mode with a limited mass range of 1,000 – 2,400 m/z. Typically, 300 laser shots were summed to acquire each mass spectrum.
MALDI-TOF MS Data Analysis
Replicate mass-to-charge (m/z) spectra data was generated for each lipid A. Bruker Daltonics flexAnalysis software was used to analyze all spectral data generated. Lipid A was classified as hexa- or hepta-acylated based on the m/z ratio of its primary spectral peak. Based on previous MS-based studies on LPS lipid A (35, 53–55), peaks with m/z ratios between 1650–1800 atomic mass units (amu) are predicted to by hexa-acylated, while peaks above 2000 amu are predicted to be hepta-acylated. Using these parameters, the acyl chain number present in the lipid A of each strain was predicted. In addition, the loss or addition of a phosphate to the di-glucosamine backbone was indicated by a m/z change of 80 amu, and the addition of PEtn was indicated by a m/z change 123amu. The presence of the outer membrane molecule, cardiolipin, was indicated by a m/z ratio value of 1456 amu and single carbon loss/additions.
Gas Chromatography-Mass Spectrometry (GC-MS) analysis
GC-MS is described in is described in the Supplementary Methods.
Murine macrophage stimulations and analysis
Macrophages stimulations and analysis are described in the Supplementary Methods.
In vivo LPS challenge
10-week old mice were injected intraperitoneally with 1 mg/kg of LPS purified from Moritella sp. strains 5, 24, 28 or 36. Blood samples were collected 4 hours post injection and plasma were obtained by centrifugation at 2000g for 10 min. IL-6 and TNFα plasma concentrations were measured by ELISA (Biolegend). All animal procedures were approved by the Institutional Animal Care and Use Committee.
Human THP1 cell stimulations and analysis
THP1 stimulations are described in the Supplementary Methods.
HEK-Blue 293T reporter cell line (InvivoGen, hkb-htlr4)
In a 96-well plate, 0.5 × 105 cells/well were plated in culture media for real-time detection of secreted alkaline phosphatase (InvivoGen, hb-det2). In triplicate, 100 ng/ml of Moritella LPS or E. coli O111:B4 LPS was added to each well. Plates were incubated for 24 hours at 37°C, 5% CO2. Secreted alkaline phosphatase was determined by reading the absorbance at 635 nm (Tecan, model Spark 10M). The statistical analysis was done using Prism. One-way analysis of variance (ANOVA) was performed assuming Gaussian distribution, no pairing, and multiple comparisons to E. coli O111:B4 LPS. Equal standard deviation was not assumed, and Brown-Forsythe and Welch ANOVA tests were performed to determine if there were significant differences in absorbance between 293T cells incubated with Moritella LPS and E. coli LPS. Significance was reported to a 99.9% confidence interval (p ≤ 0.001).
Limulus Amebocyte Lysate Pyrochrome assay
In triplicate, 20 ng/mL of Moritella LPS, Moritella lipid A, or E. coli O111:B4 LPS was sequentially diluted 10-fold from 20 ng/mL to 20 pg/mL in a 96-well plate. The total volume in each well was 50 μl/well. PBS was used as a negative control. Pyrochrome reagent was resuspended in Glucashield buffer according to the manufacturer’s protocol (Associates of Cape Cod, CG1500). 50 μl of Pyrochrome reagent (1:1 [vol/vol]) was added to each well for a final volume of 100μl/well. The final concentrations of LPS/lipid A measured in triplicate were: 10 ng/mL, 1 ng/mL, 100 pg/mL and 10 pg/mL. Absorbance was measured at 405 nm at 37°C for 60 minutes every 5 min by plate reader (Tecan, model Spark 10M). Data was analyzed when maximum absorbance was reached at 1 ng/mL. Data presentation and statistical analyses were performed using Prism. One-way analysis of variance (ANOVA) was performed assuming Gaussian distribution, no pairing, and multiple comparisons to either E. coli LPS or Moritella LPS. Equal standard deviation was not assumed, and Brown-Forsythe and Welch ANOVA tests were performed to determine if there were significant differences between conditions analyzed. Calculated p-values are as indicated in the figure captions.
Supplementary Material
Fig. S1. Lipid A pathway overview.
Fig. S2. Biological and ecological model of the discoveries made in this study.
Table S1. Reported ranges of Phoenix Islands Region marine mammals.
Table S2. Marine animals from different depths of the ocean.
Table S3. Moritella genomes sequenced in this study.
Table S4. Comparative analysis of all Moritella genomes.
Table S5. Genes found uniquely in immuno-silent Moritella.
Table S6. Genes found uniquely in immuno-stimulatory Moritella.
Table S7. Raw data file (Excel spreadsheet).
Acknowledgements
We thank members of the Kagan lab for helpful discussions on this project, and are grateful to have had the privilege to work in the PIPA, in the Republic of Kiribati. We wish to thank the Schmidt Ocean Institute and the Master and crew of the RV Falkor. We are grateful to the PIPA Implementation Office and Conservation Trust for support of this work. We thank the BU Marine Program and Cara Johnson (BU) for logistical support, and Brian R. C. Kennedy for mapping assistance. We thank Dr. Rosie Alegado (U. Hawai’i at Manoa C-MORE) for enabling preparatory materials prior to sail, and Dr. Koty Sharp (Roger Williams University) for helpful discussions.
Funding
J.C.K. is supported by NIH grants AI133524, AI093589, AI116550, and P30DK34854 and an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. Our work was conducted under PIPA Research Permit #4/17, funded by NOAA (#NA17OAR0110083 to R.R., E.C., and T.S.). D.R.G. thanks the International Centre for Cancer Vaccine Science project of the International Research Agendas program of the Foundation for Polish Science co-financed by the European Union under the European Regional Development Fund (MAB/2017/03) at the University of Gdansk. D.R.G. and R.K.E. thank the NIH for funding (AI123820 and AI147314).
Footnotes
Competing interests
Boston University and Boston Children’s Hospital have filed a patent application entitled “Immunomodulatory lipopolysaccharide compositions” with R.D.R, A.E.G., A.T. and J.C.K as inventors. J.C.K. holds equity and consults for IFM Therapeutics, Quench Bio and Corner Therapeutics. None of these relationships influenced the work performed in this study. The other authors declare that they have no competing interests.
Data and materials availability: Bacterial strains are available upon request from the authors; assistance will be provided in obtaining the required written permission from the Kiribati government. All bacterial genome sequences were deposited in NCBI under BioProject PRJNA639995. Genomic data for six Moritella isolates was deposited in GenBank under accession numbers CP056120-CP056125 (also see table S3). The 16S amplicon sequence data are available from the NCBI Sequence Read Archive (study SRP267655). All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.Janeway CA Jr., Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harbor symposia on quantitative biology 54 Pt 1, 1–13 (1989). [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R, Janeway CA, Innate immunity: impact on the adaptive immune response. Current Opinion in Immunology 9, 4–9 (1997). [DOI] [PubMed] [Google Scholar]
- 3.C. A. Janeway, Jr., R. Medzhitov, Innate immune recognition. Annual review of immunology 20, 197–216 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Vance RE, Isberg RR, Portnoy DA, Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell host & microbe 6, 10–21 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steimle A, Autenrieth IB, Frick J-S, Structure and function: Lipid A modifications in commensals and pathogens. International Journal of Medical Microbiology 306, 290–301 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Andersen-Nissen E, Smith KD, Strobe KL, Barrett SLR, Cookson BT, Logan SM, Aderem A, Evasion of Toll-like receptor 5 by flagellated bacteria. Proceedings of the National Academy of Sciences of the United States of America 102, 9247–9252 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Wang Z, Chen J, Ernst RK, Wang X, Influence of lipid A acylation pattern on membrane permeability and innate immune stimulation. Marine drugs 11, 3197–3208 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Da Silva GJ, Domingues S, Interplay between Colistin Resistance, Virulence and Fitness in Acinetobacter baumannii. Antibiotics (Basel, Switzerland) 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beceiro A, Tomás M, Bou G, Antimicrobial Resistance and Virulence: a Successful or Deleterious Association in the Bacterial World? Clinical Microbiology Reviews 26, 185–230 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan MM, Ernst O, Sun J, Fraser IDC, Ernst RK, Goodlett DR, Nita-Lazar A, Mass Spectrometry-based Structural Analysis and Systems Immunoproteomics Strategies for Deciphering the Host Response to Endotoxin. Journal of molecular biology 430, 2641–2660 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Kokoulin MS, Sokolova EV, Elkin YN, Romanenko LA, Mikhailov VV, Komandrova NA, Partial structure and immunological properties of lipopolysaccharide from marine-derived Pseudomonas stutzeri KMM 226. Antonie van Leeuwenhoek 110, 1569–1580 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Lorenzo FD, Palmigiano A, Paciello I, Pallach M, Garozzo D, Bernardini ML, Cono V, Yakimov MM, Molinaro A, Silipo A, The Deep-Sea Polyextremophile Halobacteroides lacunaris TB21 Rough-Type LPS: Structure and Inhibitory Activity towards Toxic LPS. Marine drugs 15, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorenzo FD, Palmigiano A, Albitar-Nehme S, Pallach M, Kokoulin M, Komandrova N, Romanenko L, Bernardini ML, Garozzo D, Molinaro A, Silipo A, Lipid A Structure and Immunoinhibitory Effect of the Marine Bacterium Cobetia pacifica KMM 3879T. European Journal of Organic Chemistry 2018, 2707–2716 (2018). [Google Scholar]
- 14.Scott AJ, Oyler BL, Goodlett DR, Ernst RK, Lipid A structural modifications in extreme conditions and identification of unique modifying enzymes to define the Toll-like receptor 4 structure-activity relationship. Biochimica et biophysica acta. Molecular and cell biology of lipids 1862, 1439–1450 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perrin WF, Würsig B, Thewissen JGM, Encyclopedia of marine mammals, (Elsevier Inc., Amsterdam, The Netherlands, 2009), vol. 2, pp. 1316. [Google Scholar]
- 16.Rotjan R, Jamieson R, Carr B, Kaufman L, Mangubhai S, Obura D, Pierce R, Rimon B, Ris B, Sandin S, Shelley P, Sumaila UR, Taei S, Tausig H, Teroroko T, Thorrold S, Wikgren B, Toatu T, Stone G, Establishment, management, and maintenance of the phoenix islands protected area. Advances in marine biology 69, 289–324 (2014). [DOI] [PubMed] [Google Scholar]
- 17.D’Amico S, Collins T, Marx J-C, Feller G, Gerday C, Psychrophilic microorganisms: challenges for life. EMBO Rep 7, 385–389 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zanoni I, Ostuni R, Marek LR, Barresi S, Barbalat R, Barton GM, Granucci F, Kagan JC, CD14 controls the LPS-induced endocytosis of Toll-like receptor 4. Cell 147, 868–880 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan Y, Zanoni I, Cullen TW, Goodman AL, Kagan JC, Mechanisms of Toll-like Receptor 4 Endocytosis Reveal a Common Immune-Evasion Strategy Used by Pathogenic and Commensal Bacteria. Immunity 43, 909–922 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akashi S, Saitoh S, Wakabayashi Y, Kikuchi T, Takamura N, Nagai Y, Kusumoto Y, Fukase K, Kusumoto S, Adachi Y, Kosugi A, Miyake K, Lipopolysaccharide interaction with cell surface Toll-like receptor 4-MD-2: higher affinity than that with MD-2 or CD14. The Journal of experimental medicine 198, 1035–1042 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B, Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science (New York, N.Y.) 282, 2085–2088 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, Hu L, Shao F, Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514, 187–192 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Kayagaki N, Wong MT, Stowe IB, Ramani SR, Gonzalez LC, Akashi-Takamura S, Miyake K, Zhang J, Lee WP, Muszyński A, Forsberg LS, Carlson RW, Dixit VM, Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science (New York, N.Y.) 341, 1246–1249 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Hagar JA, Powell DA, Aachoui Y, Ernst RK, Miao EA, Cytoplasmic LPS Activates Caspase-11: Implications in TLR4-Independent Endotoxic Shock. Science (New York, N.Y.) 341, 1250–1253 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rayamajhi M, Zhang Y, Miao EA, Detection of pyroptosis by measuring released lactate dehydrogenase activity. Methods Mol Biol 1040, 85–90 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi J, Gao W, Shao F, Pyroptosis: Gasdermin-Mediated Programmed Necrotic Cell Death. Trends in Biochemical Sciences 42, 245–254 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F, Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, Liu PS, Lill JR, Li H, Wu J, Kummerfeld S, Zhang J, Lee WP, Snipas SJ, Salvesen GS, Morris LX, Fitzgerald L, Zhang Y, Bertram EM, Goodnow CC, Dixit VM, Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Zanoni I, Tan Y, Di Gioia M, Springstead JR, Kagan JC, By Capturing Inflammatory Lipids Released from Dying Cells, the Receptor CD14 Induces Inflammasome-Dependent Phagocyte Hyperactivation. Immunity 47, 697–709.e693 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muta T, Miyata T, Misumi Y, Tokunaga F, Nakamura T, Toh Y, Ikehara Y, Iwanaga S, Limulus factor C. An endotoxin-sensitive serine protease zymogen with a mosaic structure of complement-like, epidermal growth factor-like, and lectin-like domains. The Journal of biological chemistry 266, 6554–6561 (1991). [PubMed] [Google Scholar]
- 31.Smith DR, Brockmann HJ, Beekey MA, King TL, Millard MJ, Zaldívar-Rae J, Conservation status of the American horseshoe crab, (Limulus polyphemus): a regional assessment. Reviews in Fish Biology and Fisheries 27, 135–175 (2017). [Google Scholar]
- 32.Raetz CR, Whitfield C, Lipopolysaccharide endotoxins. Annual review of biochemistry 71, 635–700 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopalco P, Stahl J, Annese C, Averhoff B, Corcelli A, Identification of unique cardiolipin and monolysocardiolipin species in Acinetobacter baumannii. Scientific Reports 7, 2972 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Therisod H, Labas V, Caroff M, Direct microextraction and analysis of rough-type lipopolysaccharides by combined thin-layer chromatography and MALDI mass spectrometry. Analytical chemistry 73, 3804–3807 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Pelletier MR, Casella LG, Jones JW, Adams MD, Zurawski DV, Hazlett KRO, Doi Y, Ernst RK, Unique structural modifications are present in the lipopolysaccharide from colistin-resistant strains of Acinetobacter baumannii. Antimicrob Agents Chemother 57, 4831–4840 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sorensen M, Chandler CE, Gardner FM, Ramadan S, Khot PD, Leung LM, Farrance CE, Goodlett DR, Ernst RK, Nilsson E, Rapid microbial identification and colistin resistance detection via MALDI-TOF MS using a novel on-target extraction of membrane lipids. Scientific Reports 10, 21536 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hittle LE, Powell DA, Jones JW, Tofigh M, Goodlett DR, Moskowitz SM, Ernst RK, Site-specific activity of the acyltransferases HtrB1 and HtrB2 in Pseudomonas aeruginosa lipid A biosynthesis. Pathogens and disease 73, ftv053–ftv053 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rietschel ET, Kirikae T, Schade FU, Mamat U, Schmidt G, Loppnow H, Ulmer AJ, Zähringer U, Seydel U, Di Padova F, Schreier M, Brade H, Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 8, 217–225 (1994). [DOI] [PubMed] [Google Scholar]
- 39.Park BS, Lee J-O, Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp Mol Med 45, e66–e66 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Facchini FA, Zaffaroni L, Minotti A, Rapisarda S, Calabrese V, Forcella M, Fusi P, Airoldi C, Ciaramelli C, Billod J-M, Schromm AB, Braun H, Palmer C, Beyaert R, Lapenta F, Jerala R, Pirianov G, Martin-Santamaria S, Peri F, Structure–Activity Relationship in Monosaccharide-Based Toll-Like Receptor 4 (TLR4) Antagonists. Journal of Medicinal Chemistry 61, 2895–2909 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Ciufo S, Kannan S, Sharma S, Badretdin A, Clark K, Turner S, Brover S, Schoch CL, Kimchi A, DiCuccio M, Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. International journal of systematic and evolutionary microbiology 68, 2386–2392 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S, High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nature communications 9, 5114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cordes EE, Levin LA, Exploration before exploitation. Science (New York, N.Y.) 359, 719 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Kennedy BRC, Cantwell K, Malik M, Kelley C, Potter J, Elliot K, Lobecker M, Sowers D, White M, France S, Mah C, Auscavitch S, Rotjan RD, The unknown and the unexplored: Quantifying what we now know, and still don’t know, about the Pacific deep-sea following the 3-year NOAA CAPSTONE Expeditions. . In review, Frontiers in Marine Science, (2019). [Google Scholar]
- 45.Auscavitch SR, Deere MC, Keller AG, Rotjan RD, Shank TM, Cordes EE, Oceanographic Drivers of Deep-Sea Coral Species Distribution and Community Assembly on Seamounts, Islands, Atolls, and Reefs Within the Phoenix Islands Protected Area. Frontiers in Marine Science 7, (2020). [Google Scholar]
- 46.Hamada M, Shoguchi E, Shinzato C, Kawashima T, Miller DJ, Satoh N, The complex NOD-like receptor repertoire of the coral Acropora digitifera includes novel domain combinations. Molecular biology and evolution 30, 167–176 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Brennan JJ, Messerschmidt JL, Williams LM, Matthews BJ, Reynoso M, Gilmore TD, Sea anemone model has a single Toll-like receptor that can function in pathogen detection, NF-κB signal transduction, and development. Proceedings of the National Academy of Sciences of the United States of America 114, E10122–e10131 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kutschera A, Dawid C, Gisch N, Schmid C, Raasch L, Gerster T, Schäffer M, Smakowska-Luzan E, Belkhadir Y, Vlot AC, Chandler CE, Schellenberger R, Schwudke D, Ernst RK, Dorey S, Hückelhoven R, Hofmann T, Ranf S, Bacterial medium-chain 3-hydroxy fatty acid metabolites trigger immunity in Arabidopsis plants. Science (New York, N.Y.) 364, 178–181 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Amon DJ, Kennedy BRC, Cantwell K, Suhre K, Glickson D, Shank TM, Rotjan RD, Deep-Sea Debris in the Central and Western Pacific Ocean. Frontiers in Marine Science 7, (2020). [Google Scholar]
- 50.El Hamidi A, Tirsoaga A, Novikov A, Hussein A, Caroff M, Microextraction of bacterial lipid A: easy and rapid method for mass spectrometric characterization. J Lipid Res 46, 1773–1778 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Scott AJ, Flinders B, Cappell J, Liang T, Pelc RS, Tran B, Kilgour DPA, Heeren RMA, Goodlett DR, Ernst RK, Norharmane matrix enhances detection of endotoxin by MALDI-MS for simultaneous profiling of pathogen, host and vector systems. Pathogens and Disease 74, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leung LM, Fondrie WE, Doi Y, Johnson JK, Strickland DK, Ernst RK, Goodlett DR, Identification of the ESKAPE pathogens by mass spectrometric analysis of microbial membrane glycolipids. Scientific Reports 7, 6403 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Y-Y, Chandler CE, Leung LM, McElheny CL, Mettus RT, Shanks RMQ, Liu J-H, Goodlett DR, Ernst RK, Doi Y, Structural Modification of Lipopolysaccharide Conferred by mcr-1 in Gram-Negative ESKAPE Pathogens. Antimicrob Agents Chemother 61, e00580–00517 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Apicella MA, Isolation and characterization of lipopolysaccharides. Methods Mol Biol 431, 3–13 (2008). [DOI] [PubMed] [Google Scholar]
- 55.Li Y, Powell DA, Shaffer SA, Rasko DA, Pelletier MR, Leszyk JD, Scott AJ, Masoudi A, Goodlett DR, Wang X, Raetz CR, Ernst RK, LPS remodeling is an evolved survival strategy for bacteria. Proceedings of the National Academy of Sciences of the United States of America 109, 8716–8721 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil P-A, Hugenholtz P, A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nature Biotechnology 36, 996–1004 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Obura D, Stone G, Mangubhai S, Bailey S, Yoshinaga A, Holloway C, Barrel R, Baseline Marine Biological Surveys of the Phoenix Islands, July 2000. Atoll Research Bulletin 589:1–61 (2011). [Google Scholar]
- 58.Kerr V, Wragg G, Southern Line Islands -Observations and Marine Survey Report 2008. (2008). [Google Scholar]
- 59.Kerr V, Wragg G, Phoenix Islands Conservation Survey 2006 Marine Survey Report. (2006). [Google Scholar]
- 60.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu Y-X, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG, Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology 37, 852–857 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP, DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods 13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO, The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic acids research 41, D590–D596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McMurdie PJ, Holmes S, phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PloS one 8, e61217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wickham H, ggplot2: Elegant Graphics for Data Analysis. (Springer-Verlag New York, 2016). [Google Scholar]
- 65.Team RC, R: A language and environment for statistical computing. (Foundation for Statistical Computing, Vienna, Austria, 2020). [Google Scholar]
- 66.Rudi K, Skulberg OM, Larsen F, Jakobsen KS, Strain characterization and classification of oxyphotobacteria in clone cultures on the basis of 16S rRNA sequences from the variable regions V6, V7, and V8. Appl Environ Microbiol 63, 2593–2599 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wick RR, Judd LM, Gorrie CL, Holt KE, Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Computational Biology 13, e1005595 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW, CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome research 25, 1043–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH, GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics (Oxford, England), (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stamatakis A, RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics (Oxford, England) 30, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Letunic I, Bork P, Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic acids research 47, W256–w259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li L, Stoeckert CJ Jr., Roos DS, OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome research 13, 2178–2189 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arkin AP, Cottingham RW, Henry CS, Harris NL, Stevens RL, Maslov S, Dehal P, Ware D, Perez F, Canon S, Sneddon MW, Henderson ML, Riehl WJ, Murphy-Olson D, Chan SY, Kamimura RT, Kumari S, Drake MM, Brettin TS, Glass EM, Chivian D, Gunter D, Weston DJ, Allen BH, Baumohl J, Best AA, Bowen B, Brenner SE, Bun CC, Chandonia J-M, Chia J-M, Colasanti R, Conrad N, Davis JJ, Davison BH, DeJongh M, Devoid S, Dietrich E, Dubchak I, Edirisinghe JN, Fang G, Faria JP, Frybarger PM, Gerlach W, Gerstein M, Greiner A, Gurtowski J, Haun HL, He F, Jain R, Joachimiak MP, Keegan KP, Kondo S, Kumar V, Land ML, Meyer F, Mills M, Novichkov PS, Oh T, Olsen GJ, Olson R, Parrello B, Pasternak S, Pearson E, Poon SS, Price GA, Ramakrishnan S, Ranjan P, Ronald PC, Schatz MC, Seaver SMD, Shukla M, Sutormin RA, Syed MH, Thomason J, Tintle NL, Wang D, Xia F, Yoo H, Yoo S, Yu D, KBase: The United States Department of Energy Systems Biology Knowledgebase. Nature Biotechnology 36, 566–569 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Katoh K, Standley DM, MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular biology and evolution 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJ, Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics (Oxford, England) 25, 1422–1423 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hirschfeld M, Ma Y, Weis JH, Vogel SN, Weis JJ, Cutting edge: repurification of lipopolysaccharide eliminates signaling through both human and murine toll-like receptor 2. Journal of immunology (Baltimore, Md. : 1950) 165, 618–622 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Lipid A pathway overview.
Fig. S2. Biological and ecological model of the discoveries made in this study.
Table S1. Reported ranges of Phoenix Islands Region marine mammals.
Table S2. Marine animals from different depths of the ocean.
Table S3. Moritella genomes sequenced in this study.
Table S4. Comparative analysis of all Moritella genomes.
Table S5. Genes found uniquely in immuno-silent Moritella.
Table S6. Genes found uniquely in immuno-stimulatory Moritella.
Table S7. Raw data file (Excel spreadsheet).