Abstract
Desmoplakin (DSP), encoded by the DSP gene, is the main desmosome component and is abundant in the myocardial tissue. There are three DSP isoforms that assume the role of supporting structural stability through intercellular adhesion. It has been found that DSP regulates the transcription of adipogenic and fibrogenic genes, and maintains appropriate electrical conductivity by regulating gap junctions and ion channels. DSP is essential for normal myocardial development and the maintenance of its structural functions. Studies have suggested that DSP gene mutations are associated with a variety of hereditary cardiomyopathy, such as arrhythmia cardiomyopathy, dilated cardiomyopathy (DCM), left ventricular noncompaction, and is also closely associated with the Carvajal syndrome, Naxos disease, and erythro-keratodermia-cardiomyopathy syndrome with skin and heart damage. The structure and function of DSP, as well as the clinical manifestations of DSP-related cardiomyopathy were reviewed in this article.
Keywords: Desmoplakin, Desmosome, Mutation, Wnt Signaling Pathway, Cardiomyopathy
Desmoplakin (DSP) cardiomyopathy was classified in arrhythmogenic right ventricular cardiomyopathy (ARVC) before 2019. However, it has now been found that the disease is conceptually parallel to ARVC and left ventricular noncompaction (LVNC), both of which belong to arrhythmogenic cardiomyopathy (ACM). This review aims to provide current knowledge regarding the possible mechanisms and clinical features of DSP cardiomyopathy with additional attention to several cardiac diseases with similar symptoms to avoid misdiagnosis as far as possible.
DSP Structure and Distribution
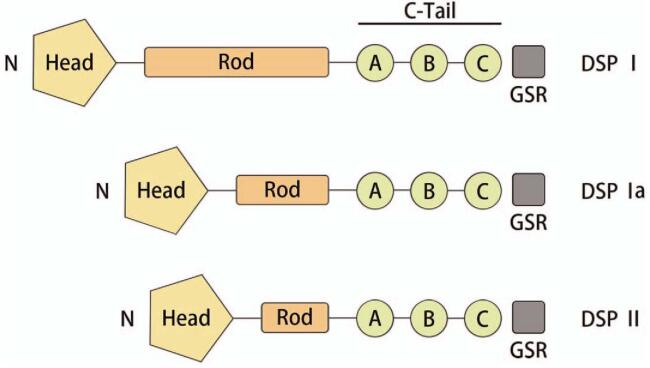
DSP is the most abundant component of the desmosome. Just like the other plakin superfamily members, it has a tripartite structure that includes a globular N-terminal plakin domain, a central alpha-helical rod domain, and a C-terminal tail domain. The carboxy-terminal tail (C-Tail) contains three plakin repeat domains (PRDs A, B, C), with a conserved linker joining PRDs B and C. A short glycine–serine–arginine rich (GSR) domain is found at the extreme C-terminal end of the protein.[1] The DSP gene, located on chromosome 6p24.3, undergoes alternative splicing to produce three isoforms: a long (DSP-I), an intermediate (DSP-Ia), and a short (DSP-II) isoform. DSP-I is identical to DSP-Ia and DSP-II in both the N and C terminal portions, with DSP-Ia containing only half of the rod domain and DSP-II missing approximately two-thirds of the rod domain [Figure 1].[2]
Figure 1.
The structure of DSP. The three isoforms differ in the length of the central rod domain (Rod). The C-Tail contains three PRDs (A–C) and a GSR domain that is thought to regulate DSP binding to IFs. The amino-terminal globular head domain (Head) mediates protein–protein interactions. C-Tail: Carboxy-terminal tail; GSR: Glycine–serine–arginine rich; DSP: Desmoplakin; IF: Intermediate filaments; PRDs: Plakin repeat domains.
DSP-I is the predominant cardiac isoform; however, it is also present in the skin. DSP-II was thought to be restricted to the skin; however, DSP-II transcripts have been found in the left atrium and ventricle, interventricular septum, sinistra auricle, and apex of the heart, but at a much lower expression level than that of DSP-I.[3] The expression levels of DSP-Ia in the epidermal keratinocytes, as well as cardiac tissues, have been reported to be low, and DSP-Ia is the only isoform in the aorta.[2]
DSP Functions
Cellular adhesion
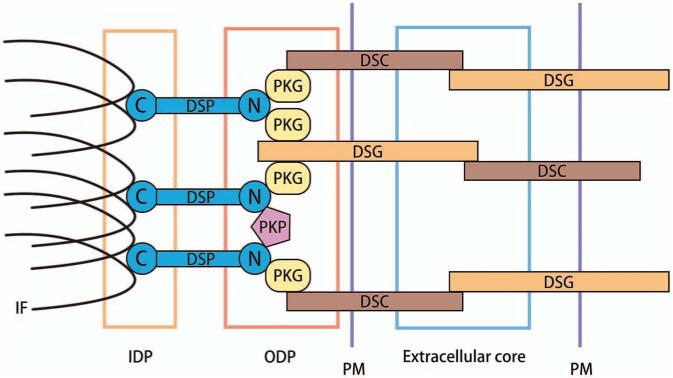
The cardiac intercalated disk (ID) connects adjacent cardiomyocytes and is classically comprised of three main structures: gap junctions, adherens junctions, and desmosomes [Figure 2].[4] DSP, together with desmoglein (DSG), desmocollin (DSC), plakoglobin (PKG), and plakophilin (PKP) constitute the desmosome in cardiomyocytes.[5] Desmosomal cadherins (DSGs and DSCs) extend into the extracellular core and outer dense plaque (ODP) to form dimers through heterophilic interactions in a Ca2+ dependent manner.[6] Extracellular calcium supports cadherin-mediated adhesion by allowing the cadherin extracellular domain to assume a rigid and functional conformation.[7] A member of the armadillo family, PKG, directly binds the cadherin cytoplasmic tails, and also interacts with DSP. Another armadillo family protein subgroup, PKP, connects the N-terminals of DSP with each other.[8] Intracellularlly, DSP and proteins of the armadillo/catenin (PKG and PKP) stabilize the desmosomal structure by linking membrane components with intermediate filaments (IFs).[9]
Figure 2.
The relative position of DSP in desmosome. The DSGs and DSCs extend into the extracellular core and ODP to establish, contact, and adhere to neighboring cells. Cadherin cytoplasmic tails associate with linker proteins, PKG, PKP, and DSP. DSP binds IF within the IDP, serving to tether the IF to the PM. DSC: Desmocollin; DSG: Desmoglein; DSP: Desmoplakin; IDP: Inner dense plaque; IF: Intermediate filaments; ODP: Outer dense plaque; PKG: Plakoglobin; PKP: Plakophilin; PM: Plasma membrane.
Cytoskeletal dynamics
DSP binds IFs within the inner dense plaque (IDP), thereby tethering the IFs to the plasma membrane (PM).[6] The main IF protein in mature striated myocytes is desmin.[10] The association of DSP with desmin is dependent on sequences within the linker region and C-terminal extremity of DSP, where the B and C subdomains contribute to efficient binding. A potentially phosphorylatable serine residue in the C-terminal extremity of DSP affects its association with desmin.[11]
Three juxtaposed PRDs of DSP exhibit a diverse range of IF binding surfaces.[12] Small angle X-ray scattering analysis revealed that the three DSP PRDs and linker form an elongated “beads on a string” structure.[13] The DSP linker exhibits a degree of flexibility, implying that it may facilitate domain motions to provide appropriate geometric positioning of flanking PRDs and allow dynamic recognition of targets.[12]
Conduction–repolarization kinetics
Two different types of electrical coupling allow for a mixed-mode of intercellular signaling across neighboring cardiomyocytes at the ID, namely (i) direct coupling through gap junctions and (ii) ephaptic coupling involving ion channel complexes.[9] The former is described in Sub-section Cx43, while this section focuses on the latter.
Voltage-gated sodium channels (Nav1.5) in the ID support cardiac muscle contraction. DSP silencing decreases expression level and abnormal distributions of connexin 43 (Cx43) and Nav1.5. Moreover, DSP suppression was shown to decrease sodium current in cultured cells which slowed conduction velocity.[14] In DSP-H1684R-carrying induced pluripotent stem cell (iPSC) cardiomyocytes, lower amplitudes of currents through Nav1.5 as well as L-type calcium channels, shortening of action-potential, and elevated amplitudes of current through transient-outward potassium channels were observed.[15] For the DSP genetic variants, besides the slowing of conduction attributed to structural heart remodeling, they also affect multiple ion channel activities, thereby aggravating arrhythmic manifestation.
Embryonic development
Homozygous deletion of DSP in mice resulted in embryonic lethality, with evidence of growth arrest before the embryonic day of development 6.5 (E6.5) and cardiac abnormalities. The most likely cause of death was a defect in cell adhesion during egg cylinder elongation, but it is noteworthy to observe that cell proliferation was also impaired.[5] In frog embryos, DSP is required during the process of radial intercalation, where basally located cells move into the outer epidermal layer. Suppressed DSP levels were shown to result in the failure of radially intercalating cells to expand their apical surface, thereby reducing the number of differentiated multiciliated and secretory cells.[16]
Underlying Mechanisms of DSP Cardiomyopathy
Wnt/β-catenin pathway
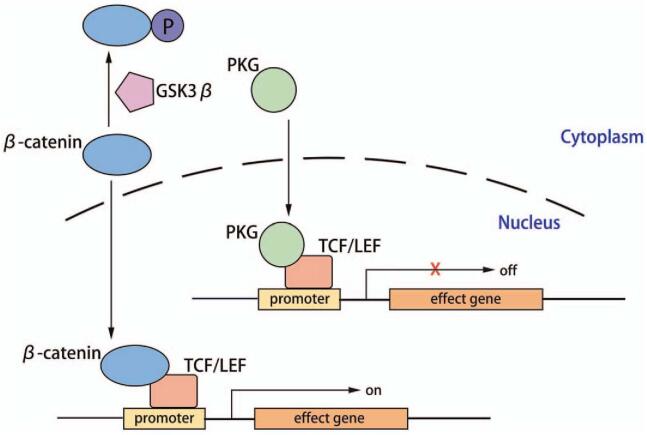
The Wnt/β-catenin signaling is a key regulator of myogenesis vs. adipogenesis. In the absence of Wnt stimulus, cytoplasmic β-catenin forms a “destruction complex” with APC/Axin/CK-1a/GSK-3β and is then phosphorylated and ubiquitinated.[17] Upon binding of the Wnt family protein to the receptor Frizzle protein, as well as to the co-receptors (lipoprotein receptor-related proteins 5 and 6 [LRP5 and LRP6]) on the PM, the intracellular disheveled (Dvl) phosphoprotein is activated, leading to the inactivation of the degradation complex and cytoplasmic accumulation of β-catenin.[18] Then, β-catenin is translocated to the nucleus, where it converts the T-cell factor/lymphoid enhancer factor (TCF/LEF) family DNA binding proteins into transcriptional activators. Transcription factor 7-like 2 (TCF7L2; previously referred to as TCF4), a member of the TCF/LEF family,[19] has two different sites for the binding of β-catenin and PKG. Only when PKG is not sequestered in the PM as part of desmosomes, is it able to participate in cell signaling.[20] PKG and β-catenin are ∼85% similar in their primary structure; however, they differ in their propensities to bind TCF7L2. Accordingly, β-catenin initiates gene expression (such as c-Myc, cyclin D1, etc.), whereas PKG exhibits a weak transcriptional activity [Figure 3].[21]
Figure 3.
The Wnt/β-catenin pathway. When PKG translocates from desmosomes to the nucleus, it competes with β-catenin for binding to the TCF7L2/LEF1 transcription factors, leading to suppression of the Wnt pathway, which means an increase in adipogenesis and fibrosis and adipogenesis and a decrease in myogenesis. GSK3β: Glycogen synthase kinase-3 beta; PKG: Plakoglobin; TCF/LEF: T-cell factor/lymphoid enhancer factor.
Compared to the wild type, the frame-shift variant DSP c.832delG in HEK293T cells was shown to upregulate the nuclear junction plakoglobin (JUP) while downregulating β-catenin.[22] Suppression of DSP expression in cardiac myocytes led to PKG release from desmosomes, their translocation to the nucleus, and a 2-fold reduction in canonical Wnt/β-catenin signaling by binding the TCF7L2/LEF1 transcription factors. This led to elevated expression of adipogenic and fibrogenic genes in vitro and abnormal cardiac adipose tissue and fibrosis in vivo.[23] A subset of cardiac fibro-adipogenic progenitors differentiate to adipocytes through a Wnt-dependent mechanism. Activation of the canonical Wnt signaling was shown to rescue adipogenesis in a dose-dependent manner.[24]
Hippo/yes-associated protein pathway
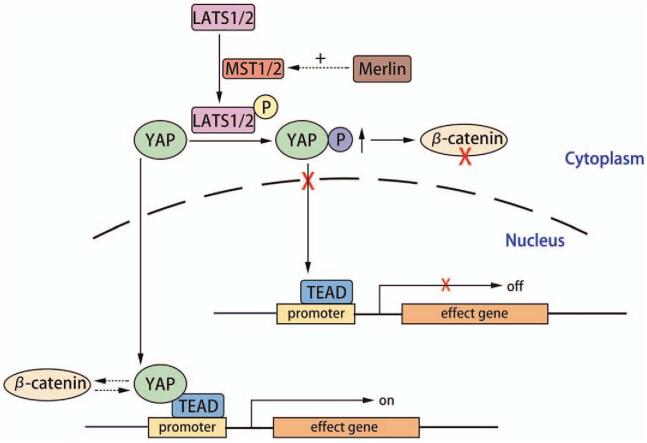
When the Hippo pathway is inactive, yes-associated protein (YAP) and tafazzin (TAZ) (transcriptional coactivators) are unphosphorylated and localized in the nucleus for transcriptional enhancer factor domain (TEAD) binding and activation of gene transcription.[25] YAP activation stimulates cardiomyocyte proliferation, cardiac morphogenesis, and myocardial trabeculation.[26] The Hippo kinase cascade, comprising mammalian STE20-like protein kinases 1/2 (MST1/2) and large tumor suppressor kinases 1/2 (LATS1/2), forms the core of the Hippo pathway. MST1/2 phosphorylates and activates LATS1/2, which in turn phosphorylates YAP or TAZ. Phosphorylation inhibits YAP and TAZ transcriptional activity by promoting their nuclear export and cytoplasmic sequestration.[27] Molecular remodeling of the IDs, including DSP, in human hearts using arrhythmogenic cardiomyopathy (ACM) activates the neurofibromin (NF) 2 gene (that encodes merlin, a tumor suppressor), resulting in cascade phosphorylation of the Hippo kinases, inactivation of YAP, and enhanced adipogenesis [Figure 4].[28]
Figure 4.
The Hippo/YAP pathway. Activated Hippo pathways and suppressed YAP signaling leads to an increase in apoptosis as well as Wnt/β-catenin pathway suppression through interactions between YAP and β-catenin in the nucleus, and pYAP-mediated sequestration of β-catenin in the cytosol. LATS1/2: Large tumor suppressor kinases 1/2; MST1/2: Mammalian STE20-like protein kinases 1/2; TEAD: Transcriptional enhancer factor domain; YAP: Yes-associated protein.
Hippo signaling inactivates Wnt through the interactions of YAP and β-catenin on Sox2 and Snai2 genes. This was shown by the close proximity of the conserved TCF/LEF binding elements (CTTTG) to the Sox2 and Snai2 downstream candidate YAP/TEAD sites.[29] The transcriptionally inactive effector of the Hippo pathway, phosphorylated YAP (pYAP), leads to sequestration of β-catenin in the cytosol.[28]
Cx43
DSP prevents lysosomal-mediated degradation of Cx43. DSP loss initiates the activation of extracellular signal-regulated kinase 1/2–mitogen-activated protein kinase (ERK1/2–MAPK) and elevates the phosphorylation of S279/282 of Cx43, which signals clathrin-mediated internalization and subsequent lysosomal degradation of Cx43.[30] DSP loss is also associated with a primary reduction in Cx43 levels and molecular dissociation of the mechanical junction complex in neonatal ventricular cardiomyocytes, suggesting that it is a primary stabilizer of connexin.[31]
DSP promotes membrane localization of Cx43. Cx43 delivery to the membrane is facilitated by microtubules and is dependent on the plus-end microtubule-binding protein end-binding 1 (EB1) and its interactions with DSP.[32] The microtubule-binding protein EB1 interacts with a region of the DSP N-terminus, which enables DSP to modify microtubule organization and dynamics near cell–cell contact sites. Mutations in the DSP N-termini impair Cx43 membrane localization and gap junction function by either directly disrupting the association with EB1 (N458Y, I533T) or by causing DSP mislocalization (N287K), and thus a corresponding mislocalization of EB1.[33] Haploinsufficiency of DSP is sufficient to cause significant Cx43 mislocalization.[34]
Inflammation
In cardiac myocytes of ACM, nuclear factor-κB signaling is activated and large amounts of inflammatory cytokines and chemotactic molecules are expressed.[35] Multifocal cardiomyocyte necrosis initiates a neutrophil-dominated inflammatory response, which also involves macrophages and T cells. During chronic disease progression, macrophages and T cells persist within mature scars and are present in expanding interstitial fibrosis.[36]18F-fluorodeoxyglucose positron emission tomography (FDG PET) scans revealed typical myocardial inflammation in the left ventricle (LV). Histological analyses of samples from DSP patients revealed inflammatory infiltrates and scarring in the left ventricular myocardium.[37]
DSP Cardiomyopathy Clinical Manifestations
Left-dominant ACM
ACM is a group of myocardial diseases that cannot be explained by ischemia, hypertension, valvular disease, etc., and can be caused by systemic diseases (eg, sarcoidosis, amyloidosis), infection (eg, Chagas disease), genetic factors (eg, ARVC, arrhythmogenic left ventricular cardiomyopathy (ALVC), lamin A/C (LMNA) gene-associated cardiomyopathy), and by ion channel diseases. It manifests with cardiac arrhythmias, syncope, sudden cardiac death (SCD), and cardiac failure in advanced stages.[38] DSP cardiomyopathy is a distinct form of ACM that is characterized by episodic myocardial injury, left ventricular fibrosis that precedes systolic dysfunction, and a high incidence of ventricular arrhythmias.[37]
DSP mutation is associated with LV involvement and may be indicative of worse prognosis and elevated SCD risk.[39] Docekal et al[40] identified a novel DSP gene mutation (c.3735_3741dupAAATCGA) in a 54-year-old male patient who suffered unheralded syncope and SCD while running. After coronary angiography had excluded any epicardial coronary artery disease, cardiac magnetic resonance (CMR) revealed severe LV global hypokinesis, LV dilation, and mid-myocardial delayed enhancement in the basal to apical inferoseptal wall. Chmielewski et al[41] reported that a c.3737dupA variant in the DSP gene impacted the rod domain of all three isoforms, causing a mildly dilated LV with lateral wall and septum hypokinesis in a teenage girl. LV dysfunction and structural LV involvement by CMR were found to be significantly more prevalent among patients with nonmissense mutations than among patients with DSP missense mutations.[42] An equally high arrhythmic potential was revealed for both missense and truncating mutations of DSP, while 41% of probands had an SCD victim among their relatives.[43]
Clinically, DSP cardiomyopathy may overlap ARVC, LVNC, dilated cardiomyopathy (DCM), and acute myocardial inflammatory syndromes, which may lead to a misdiagnosis.
Masquerading as ARVC
ARVC is a primary myocardial disorder that is usually expressed with severe ventricular arrhythmias accompanied by structural and functional alterations of the right ventricle (RV).[42] It is associated with mutations in desmosomal genes, specifically DSP (10%–15%), PKP2 (30%–35%), DSG2 (10%), and DSC (2%–5%), in addition to nondesmosomal genes (<1%) encoding for transforming growth factor (TGF) β3, human ryanodine receptor (RyR) 2 and the transmembrane protein (TMEM) 43.[44,45] Autosomal dominant mutations in the DSP gene can give rise to ARVC without cutaneous involvement, which is referred to as ARVC8.[46] In ARVC, the pathological process of myocyte degeneration and subsequent fibrous, as well as fatty substitution, is regional. It starts from the sub-epicardial layers; therefore, functional alterations in the multi-layered left ventricular myocardium are delayed to become apparent on two-dimensional echo as opposed to those of the thinner right ventricular wall.[47] Diagnosis is based on “Task Force Criteria,” a scoring system that takes into account structural and electrical abnormalities and family history, as well as genetic findings. This criterion is divided into major and minor criteria.[48] Although DSP cardiomyopathy has a similar desmosomal molecular basis to PKP2-associated ARVC, diagnostic and risk stratification criteria that work well for PKP2-associated ARVC are poor for DSP cardiomyopathy.[37]
Masquerading as LVNC
LVNC is a heterogeneous myocardial disorder that is characterized by prominent trabeculae, intratrabecular recesses, and a left ventricular myocardium with two distinct layers: compacted and noncompacted.[49] Williams et al[50] reported a premature truncation of the DSP-1 C-terminus, associated with a homozygous 2 bp deletion (5208_5209delAG), which led to Carvajal syndrome (CS) (see Sub-section Naxos/CS) comprising severe early-onset heart failure with features of noncompaction cardiomyopathy. DSP c.1339C>T is associated with an aggressive clinical phenotype of left-dominant ACM and LVNC.[51]
Masquerading as DCM
Mutations affecting the desmosome ODP (DSG2, PKG, PKP2, and the N-terminal of DSP) result in ARVC with an ordinary phenotype. However, mutations at the IDP, particularly those affecting the desmin-binding site of DSP, result in ARVC with predominant LV involvement and clinical overlaps with DCM.[52] Through autosomal recessive transmission, compound heterozygotic DSP variants are associated with an early-onset of nonsyndromic DCM.[53] Heliö et al[54] reported a DSP c.6310delA (p.Thr2104Glnfs∗12) variant in 17 individuals, 11 (65%) of whom fulfilled the DCM diagnostic criteria. Episodic myocardial injury in DSP cardiomyopathy contributes to progressive fibrosis that precedes the development of LV systolic dysfunction, an important difference when compared to typical DCM.[37]
Masquerading as acute myocardial inflammatory syndromes
In a case of male monozygotic twins who presented with symptoms of myocarditis and CMR, Kissopoulou et al[55] reported a nonsense heterozygous variant in the DSP gene (c.2521_2522del, p.Gln841Aspfs∗9), demonstrating an affected LV with comprehensive inflammatory, subepicardial changes consistent with myocarditis. Familial screening revealed that 39% of 28 DSP variant carriers with a documented episode of acute myocarditis had an ALVC phenotype.[56] In two young brothers with recurrent myocarditis triggered by physical exercise, screening of 218 cardiomyopathy-related genes revealed the presence of the heterozygous truncating variant p.Arg1458Ter in DSP.[57] Reichl et al[58] reported a case of a young male with acute myocarditis as the first presentation of DSP variant-associated ACM. FDG PET scans in four DSP cases revealed that acute LV myocardial injury is associated with myocardial inflammation, initially misdiagnosed as cardiac sarcoidosis or myocarditis.[37] Recurrent myoinflammatory episodes and signs of fatty replacement on CMR (regions with chemical shift artifacts on cinematic sequences, late gadolinium enhancement, elevated T2 values, and low or normal T1 values) may help identify such patients.[59]
Cardio-cutaneous disorder
DSP mutation carriers commonly complain of the painful, rough, hard skin on their feet and the need to regularly file or cut or shave this off using a razor. ACM patients with palmoplantar keratoderma (PPK) and curly hair should consider genetic screening programmes to identify high risk family members.[60]
Naxos/CS
Naxos disease is a recessive association of ARVC with wooly hair and diffuse PPK.[61] Mutations in genes encoding PKG and DSP have been identified as the main causes of Naxos disease.[62] In the Naxos disease variety described in families from Ecuador and Israel (Arab families), two different mutations of the DSP gene (Dsp7901del1G and DspG2375R) were shown to truncate the proteins at the C-terminal domains. Defects in the linking sites of these proteins can interrupt the contiguous chain of cell adhesion, particularly under conditions of increased mechanical stress or stretch, leading to cell death, progressive loss of the myocardium and fibro-fatty replacement.[62] Cardiac anomalies that are characterized by ventricular arrhythmias with ventricular extrasystoles and tachycardia as well as histologic features of the myocardium are consistent with ARVC, but in a more severe form of dysplasia with major dilatation of the RV.[63] An ARVC usually manifests by the adolescence period with syncope, ventricular tachycardia or SCD, with almost 100% penetrance. Cardiac failure symptoms usually appear in the final stages.[64]
CS may be considered a variant of Naxos syndrome, as it is also an autosomal recessive genetic disorder that is caused by mutations in the DSP gene. CS is characterized by a triad of left ventricular dilated ACM, striated and focal PPK and woolly hair.[65] Cutaneous findings of the Naxos/CS vary with age; woolly hair appears since birth, whereas PPK develops during the first year of life as infants begin using their hands and feet.[66] Truncating mutations in most patients developing cardiac disease are located in exons 23 and 24.[67] Guerra et al[68] reported that two mutations (c.4788delA and c.6091_6092delTT) in the DSP gene truncate the C-terminal plakin-repeat subdomains from DSP-I, resulting in protein loss-of-function. An unfavorable prognosis of the CS is associated with an early onset of the disease, high risk of SCD and a rapidly developing cardiac failure, consequent to the progressive dilatation of the cardiac chambers.[65]
Dominant heterozygous mutations in DSP have also been reported, and found to be associated with hypo/oligodontia in addition to the common CS triad. A de novo heterozygous missense mutation (c.1790 C>T, p.Ser597Leu) in exon 14 of the DSP gene was shown to lead to tooth agenesis ranging from the absence of the lower left second molar to 15 missing teeth (the typical pattern of oligodontia being absent in second premolars and absent in second and third molars) in affected family members.[69] These mutations are located in the N-terminal domain of DSP, which probably disrupts desmosome scaffold building by integrating abnormal DSP molecules through a dominant-negative mechanism.[70]
Erythro-keratodermia-cardiomyopathy syndrome
The Erythro-keratodermia-cardiomyopathy (EKC) syndrome was first described by Boyden et al[71]De novo heterozygous missense mutations clustered around the fourth spectrin domain (Spectrin Repeat 6) of DSP manifests a generalized erythro-keratodermia, ichthyosis, and progressive cardiomyopathy, alongside features such as PPK, enamel defects, nail onychodystrophy, and hair abnormalities. Missense mutations in EKC syndromes cause transitions of the native amino acid to proline.[72] DSP mutations in EKC syndrome patients affect desmosomal proteins and Cx43 localization in the skin, which result in desmosome aggregation, widening of intercellular spaces, and lipid secretory defects.[71]
Conclusions and Perspectives
DSP plays an important role in myocardium development as well as in the maintenance of normal structural functions. Regarding the functions of DSP, studies should aim at evaluating the mechanisms involved, including expression regulation and functional differences of different isoforms, and the influence of DSP on the electrical activity of myocytes, and the stability of intercellular connections. DSP cardiomyopathy is a unique ACM that is characterized by intermittent myocardial injury, left ventricular fibrosis prior to systolic dysfunction, and a high incidence of ventricular arrhythmia. Genotype-specific methods should be used for diagnosis and risk stratification. However, based on gene sequencing of normal populations and patients with cardiomyopathy, DSP genes have been shown to exhibit complex polymorphisms, and further studies are needed to clarify the relationships between these DSP gene variants and cardiomyopathy, evaluate the possible pathogenic mechanisms, and inform drug as well as gene therapy targets.
Funding
This study was supported by a grant from the Capital's Funds for HealthImprovement and Research (No. CRF 2020-2-2062).
Conflicts of interest
None.
Footnotes
How to cite this article: Yuan ZY, Cheng LT, Wang ZF, Wu YQ. Desmoplakin and clinical manifestations of desmoplakin cardiomyopathy. Chin Med J 2021;134:1771–1779. doi: 10.1097/CM9.0000000000001581
References
- 1.Al-Jassar C, Bikker H, Overduin M, Chidgey M. Mechanistic basis of desmosome-targeted diseases. J Mol Biol 2013; 425:4006–4022. doi: 10.1016/j.jmb.2013.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cabral RM, Wan H, Cole CL, Abrams DJ, Kelsell DP, South AP. Identification and characterization of DSPIa, a novel isoform of human desmoplakin. Cell Tissue Res 2010; 341:121–129. doi: 10.1007/s00441-010-0989-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uzumcu A, Norgett EE, Dindar A, Uyguner O, Nisli K, Kayserili H, et al. Loss of desmoplakin isoform I causes early onset cardiomyopathy and heart failure in a Naxos-like syndrome. J Med Genet 2006; 43:e5.doi: 10.1136/jmg.2005.032904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Towbin JA, McKenna WJ, Abrams DJ, Ackerman MJ, Calkins H, Darrieux FCC, et al. 2019 HRS expert consensus statement on evaluation, risk stratification, and management of arrhythmogenic cardiomyopathy. Heart Rhythm 2019; 16:e301–e372. doi: 10.1016/j.hrthm.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Garrod D, Chidgey M. Desmosome structure, composition and function. Biochim Biophys Acta 2008; 1778:572–587. doi: 10.1016/j.bbamem.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 6.Kottke MD, Delva E, Kowalczyk AP. The desmosome: cell science lessons from human diseases. J Cell Sci 2006; 119:797–806. doi: 10.1242/jcs.02888. [DOI] [PubMed] [Google Scholar]
- 7.Pokutta S, Weis WI. Structure and mechanism of cadherins and catenins in cell-cell contacts. Annu Rev Cell Dev Biol 2007; 23:237–261. doi: 10.1146/annurev.cellbio.22.010305.104241. [DOI] [PubMed] [Google Scholar]
- 8.Delva E, Tucker DK, Kowalczyk AP. The desmosome. Cold Spring Harb Perspect Biol 2009; 1:a002543.doi: 10.1101/cshperspect.a002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manring HR, Dorn LE, Ex-Willey A, Accornero F, Ackermann MA. At the heart of inter- and intracellular signaling: the intercalated disc. Biophys Rev 2018; 10:961–971. doi: 10.1007/s12551-018-0430-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brodehl A, Pour Hakimi SA, Stanasiuk C, Ratnavadivel S, Hendig D, Gaertner A, et al. Restrictive cardiomyopathy is caused by a novel homozygous desmin (DES) mutation p.Y122H leading to a severe filament assembly defect. Genes (Basel) 2019; 10:918.doi: 10.3390/genes10110918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lapouge K, Fontao L, Champliaud MF, Jaunin F, Frias MA, Favre B, et al. New insights into the molecular basis of desmoplakin- and desmin-related cardiomyopathies. J Cell Sci 2006; 119(Pt 23):4974–4985. doi: 10.1242/jcs.03255. [DOI] [PubMed] [Google Scholar]
- 12.Mohammed F, Trieber C, Overduin M, Chidgey M. Molecular mechanism of intermediate filament recognition by plakin proteins. Biochim Biophys Acta Mol Cell Res 2020; 1867:118801.doi: 10.1016/j.bbamcr.2020.118801. [DOI] [PubMed] [Google Scholar]
- 13.Kang H, Weiss TM, Bang I, Weis WI, Choi HJ. Structure of the intermediate filament-binding region of desmoplakin. PLoS One 2016; 11:e0147641.doi: 10.1371/journal.pone.0147641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Q, Deng C, Rao F, Modi RM, Zhu J, Liu X, et al. Silencing of desmoplakin decreases connexin43/Nav1.5 expression and sodium current in HL-1 cardiomyocytes. Mol Med Rep 2013; 8:780–786. doi: 10.3892/mmr.2013.1594. [DOI] [PubMed] [Google Scholar]
- 15.Gusev K, Khudiakov A, Zaytseva A, Perepelina K, Makeenok S, Kaznacheyeva E, et al. Impact of the DSP-H1684R genetic variant on ion channels activity in iPSC-Derived cardiomyocytes. Cell Physiol Biochem 2020; 54:696–706. doi: 10.33594/000000249. [DOI] [PubMed] [Google Scholar]
- 16.Bharathan NK, Dickinson AJG. Desmoplakin is required for epidermal integrity and morphogenesis in the Xenopus laevis embryo. Dev Biol 2019; 450:115–131. doi: 10.1016/j.ydbio.2019.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rao TP, Kuhl M. An updated overview on Wnt signaling pathways: a prelude for more. Circ Res 2010; 106:1798–1806. doi: 10.1161/CIRCRESAHA.110.219840. [DOI] [PubMed] [Google Scholar]
- 18.Wang Q, Lv Q, Bian H, Yang L, Guo KL, Ye SS, et al. A novel tumor suppressor SPINK5 targets Wnt/beta-catenin signaling pathway in esophageal cancer. Cancer Med 2019; 8:2360–2371. doi: 10.1002/cam4.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui J, Duan J, Chu J, Guo C, Xi M, Li Y, et al. Chikusetsu saponin IVa protects pancreatic β cell against intermittent high glucose-induced injury by activating Wnt/β-cateninTCF7L2 pathway. Aging 2020; 12:1591–1609. doi: 10.18632/aging.102702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aktary Z, Pasdar M. Plakoglobin: role in tumorigenesis and metastasis. Int J Cell Biol 2012; 2012:189521.doi: 10.1155/2012/189521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lombardi R, da Graca Cabreira-Hansen M, Bell A, Fromm RR, Willerson JT, Marian AJ. Nuclear plakoglobin is essential for differentiation of cardiac progenitor cells to adipocytes in arrhythmogenic right ventricular cardiomyopathy. Circ Res 2011; 109:1342–1353. doi: 10.1161/CIRCRESAHA.111.255075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin X, Ma Y, Cai Z, Wang Q, Wang L, Huo Z, et al. Next-generation sequencing identified novel Desmoplakin frame-shift variant in patients with Arrhythmogenic cardiomyopathy. BMC Cardiovasc Disord 2020; 20:74.doi: 10.1186/s12872-020-01369-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Gras E, Lombardi R, Giocondo MJ, Willerson JT, Schneider MD, Khoury DS, et al. Suppression of canonical Wnt/beta-catenin signaling by nuclear plakoglobin recapitulates phenotype of arrhythmogenic right ventricular cardiomyopathy. J Clin Invest 2006; 116:2012–2021. doi: 10.1172/JCI27751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lombardi R, Chen SN, Ruggiero A, Gurha P, Czernuszewicz GZ, Willerson JT, et al. Cardiac fibro-adipocyte progenitors express desmosome proteins and preferentially differentiate to adipocytes upon deletion of the desmoplakin gene. Circ Res 2016; 119:41–54. doi: 10.1161/CIRCRESAHA.115.308136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev 2016; 30:1–17. doi: 10.1101/gad.274027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Gise A, Lin Z, Schlegelmilch K, Honor LB, Pan GM, Buck JN, et al. YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proc Natl Acad Sci U S A 2012; 109:2394–2399. doi: 10.1073/pnas.1116136109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao G, Qiu Y, Zhang HM, Yang D. Intercalated discs: Cellular adhesion and signaling in heart health and diseases. Heart Fail Rev 2019; 24:115–132. doi: 10.1007/s10741-018-9743-7. [DOI] [PubMed] [Google Scholar]
- 28.Chen SN, Gurha P, Lombardi R, Ruggiero A, Willerson JT, Marian AJ. The hippo pathway is activated and is a causal mechanism for adipogenesis in arrhythmogenic cardiomyopathy. Circ Res 2014; 114:454–468. doi: 10.1161/CIRCRESAHA.114.302810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, et al. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science 2011; 332:458–461. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kam CY, Dubash AD, Magistrati E, Polo S, Satchell KJF, Sheikh F, et al. Desmoplakin maintains gap junctions by inhibiting Ras/MAPK and lysosomal degradation of connexin-43. J Cell Biol 2018; 217:3219–3235. doi: 10.1083/jcb.201710161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyon RC, Mezzano V, Wright AT, Pfeiffer E, Chuang J, Banares K, et al. Connexin defects underlie arrhythmogenic right ventricular cardiomyopathy in a novel mouse model. Hum Mol Genet 2014; 23:1134–1150. doi: 10.1093/hmg/ddt508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaw RM, Fay AJ, Puthenveedu MA, von Zastrow M, Jan YN, Jan LY. Microtubule plus-end-tracking proteins target gap junctions directly from the cell interior to adherens junctions. Cell 2007; 128:547–560. doi: 10.1016/j.cell.2006.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel DM, Dubash AD, Kreitzer G, Green KJ. Disease mutations in desmoplakin inhibit Cx43 membrane targeting mediated by desmoplakin-EB1 interactions. J Cell Biol 2014; 206:779–797. doi: 10.1083/jcb.201312110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gomes J, Finlay M, Ahmed AK, Ciaccio EJ, Asimaki A, Saffitz JE, et al. Electrophysiological abnormalities precede overt structural changes in arrhythmogenic right ventricular cardiomyopathy due to mutations in desmoplakin – A combined murine and human study. Eur Heart J 2012; 33:1942–1953. doi: 10.1093/eurheartj/ehr472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chelko SP, Asimaki A, Lowenthal J, Bueno-Beti C, Bedja D, Scalco A, et al. Therapeutic modulation of the immune response in arrhythmogenic cardiomyopathy. Circulation 2019; 140:1491–1505. doi: 10.1161/CIRCULATIONAHA.119.040676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lubos N, van der Gaag S, Gercek M, Kant S, Leube RE, Krusche CA. Inflammation shapes pathogenesis of murine arrhythmogenic cardiomyopathy. Basic Res Cardiol 2020; 115:42.doi: 10.1007/s00395-020-0803-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith ED, Lakdawala NK, Papoutsidakis N, Aubert G, Mazzanti A, McCanta AC, et al. Desmoplakin cardiomyopathy, a fibrotic and inflammatory form of cardiomyopathy distinct from typical dilated or arrhythmogenic right ventricular cardiomyopathy. Circulation 2020; 141:1872–1884. doi: 10.1161/CIRCULATIONAHA.119.044934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao S, Puthenvedu D, Lombardi R, Chen SN. Established and emerging mechanisms in the pathogenesis of arrhythmogenic cardiomyopathy: a multifaceted disease. Int J Mol Sci 2020; 21:6320.doi: 10.3390/ijms21176320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blusztein DI, Zentner D, Thompson T, Jayadeva P, Liang D, Wang R, et al. Arrhythmogenic right ventricular cardiomyopathy: areview of living and deceased probands. Heart Lung Circ 2019; 28:1034–1041. doi: 10.1016/j.hlc.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 40.Docekal JW, Lee JC. Novel gene mutation identified in a patient with arrhythmogenic ventricular cardiomyopathy. HeartRhythm Case Rep 2017; 3:459–463. doi: 10.1016/j.hrcr.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chmielewski P, Truszkowska GT, Kukla P, Zakrzewska-Koperska J, Spiewak M, Stepien-Wojno M, et al. A novel DSP truncating variant in a family with episodic myocardial injury in the course of arrhythmogenic cardiomyopathy – A possible role of a low penetrance NLRP3 variant. Diagnostics (Basel) 2020; 10:955.doi: 10.3390/diagnostics10110955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castelletti S, Vischer AS, Syrris P, Crotti L, Spazzolini C, Ghidoni A, et al. Desmoplakin missense and non-missense mutations in arrhythmogenic right ventricular cardiomyopathy: Genotype-phenotype correlation. Int J Cardiol 2017; 249:268–273. doi: 10.1016/j.ijcard.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 43.Protonotarios A, Anastasakis A, Panagiotakos DB, Antoniades L, Syrris P, Vouliotis A, et al. Arrhythmic risk assessment in genotyped families with arrhythmogenic right ventricular cardiomyopathy. Europace 2016; 18:610–616. doi: 10.1093/europace/euv061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herren T, Gerber PA, Duru F. Arrhythmogenic right ventricular cardiomyopathy/dysplasia: a not so rare “disease of the desmosome” with multiple clinical presentations. Clin Res Cardiol 2009; 98:141–158. doi: 10.1007/s00392-009-0751-4. [DOI] [PubMed] [Google Scholar]
- 45.Li J, Radice GL. A new perspective on intercalated disc organization: Implications for heart disease. Dermatol Res Pract 2010; 2010:207835.doi: 10.1155/2010/207835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rampazzo A. Genetic bases of arrhythmogenic right ventricular cardiomyopathy. Heart Int 2006; 2:17.doi: 10.4081/hi.2006.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sen-Chowdhry S, Syrris P, Prasad SK, Hughes SE, Merrifield R, Ward D, et al. Left-dominant arrhythmogenic cardiomyopathy: An under-recognized clinical entity. J Am Coll Cardiol 2008; 52:2175–2187. doi: 10.1016/j.jacc.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 48.Marcus FI, McKenna WJ, Sherrill D, Basso C, Bauce B, Bluemke DA, et al. Diagnosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia: Proposed modification of the Task Force Criteria. Eur Heart J 2010; 31:806–814. doi: 10.1093/eurheartj/ehq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Towbin JA, Lorts A, Jefferies JL. Left ventricular non-compaction cardiomyopathy. Lancet 2015; 386:813–825. doi: 10.1016/s0140-6736(14)61282-4. [DOI] [PubMed] [Google Scholar]
- 50.Williams T, Machann W, Kuhler L, Hamm H, Muller-Hocker J, Zimmer M, et al. Novel desmoplakin mutation: juvenile biventricular cardiomyopathy with left ventricular non-compaction and acantholytic palmoplantar keratoderma. Clin Res Cardiol 2011; 100:1087–1093. doi: 10.1007/s00392-011-0345-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lopez-Ayala JM, Gomez-Milanes I, Sanchez Munoz JJ, Ruiz-Espejo F, Ortiz M, Gonzalez-Carrillo J, et al. Desmoplakin truncations and arrhythmogenic left ventricular cardiomyopathy: characterizing a phenotype. Europace 2014; 16:1838–1846. doi: 10.1093/europace/euu128. [DOI] [PubMed] [Google Scholar]
- 52.Tsatsopoulou AA, Protonotarios NI, McKenna WJ. Arrhythmogenic right ventricular dysplasia, a cell adhesion cardiomyopathy: Insights into disease pathogenesis from preliminary genotype – Phenotype assessment. Heart 2006; 92:1720–1723. doi: 10.1136/hrt.2005.081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Surmacz R, Franaszczyk M, Pyda M, Płoski R, Bilińska ZT, Bobkowski W. Autosomal recessive transmission of familial nonsyndromic dilated cardiomyopathy due to compound desmoplakin gene mutations. Pol Arch Intern Med 2018; 128:785–787. [DOI] [PubMed] [Google Scholar]
- 54.Heliö K, Kangas-Kontio T, Weckstrom S, Vanninen SUM, Aalto-Setala K, Alastalo TP, et al. DSP p.(Thr2104Glnfs∗12) variant presents variably with early onset severe arrhythmias and left ventricular cardiomyopathy. BMC Med Genet 2020; 21(19): doi: 10.1186/s12881-020-0955-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kissopoulou A, Fernlund E, Holmgren C, Isaksson E, Karlsson JE, Green H, et al. Monozygotic twins with myocarditis and a novel likely pathogenic desmoplakin gene variant. ESC Heart Fail 2020; 7:1210–1216. doi: 10.1002/ehf2.12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Piriou N, Marteau L, Kyndt F, Serfaty JM, Toquet C, Le Gloan L, et al. Familial screening in case of acute myocarditis reveals inherited arrhythmogenic left ventricular cardiomyopathies. ESC Heart Fail 2020; 7:1520–1533. doi: 10.1002/ehf2.12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poller W, Haas J, Klingel K, Kuhnisch J, Gast M, Kaya Z, et al. Familial recurrent myocarditis triggered by exercise in patients with a truncating variant of the desmoplakin gene. J Am Heart Assoc 2020; 9:e015289.doi: 10.1161/JAHA.119.015289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reichl K, Kreykes SE, Martin CM, Shenoy C. Desmoplakin variant-associated arrhythmogenic cardiomyopathy presenting as acute myocarditis. Circ Genom Precis Med 2018; 11:e002373.doi: 10.1161/CIRCGEN.118.002373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alley R, Grizzard JD, Rao K, Markley R, Trankle CR. Inflammatory episodes of desmoplakin cardiomyopathy masquerading as myocarditis: unique features on cardiac magnetic resonance imaging. JACC Cardiovasc Imaging 2020; S1936-878X(20)30711-7. doi: 10.1016/j.jcmg.2020.07.028. [DOI] [PubMed] [Google Scholar]
- 60.Maruthappu T, Posafalvi A, Castelletti S, Delaney PJ, Syrris P, O’Toole EA, et al. Loss-of-function desmoplakin I and II mutations underlie dominant arrhythmogenic cardiomyopathy with a hair and skin phenotype. Br J Dermatol 2019; 180:1114–1122. doi: 10.1111/bjd.17388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Islam AM, Rahman MT, Chowdhury AH. Cardiocutaneous syndrome (Naxos disease) in a Bangladeshi boy. Cardiovasc Diagn Ther 2016; 6:462–465. doi: 10.21037/cdt.2016.03.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Protonotarios N, Tsatsopoulou A. Naxos disease: cardiocutaneous syndrome due to cell adhesion defect. Orphanet J Rare Dis 2006; 1:4.doi: 10.1186/1750-1172-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li GL, Saguner AM, Fontaine GH. Naxos disease: from the origin to today. Orphanet J Rare Dis 2018; 13:74.doi: 10.1186/s13023-018-0814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leopoulou M, Mattsson G, LeQuang J, Pergolizzi JV, Varrassi G, Wallhagen M, et al. Naxos disease – A narrative review. Expert Rev Cardiovasc Ther 2020; 18:801–808. doi: 10.1080/14779072.2020.1828064. [DOI] [PubMed] [Google Scholar]
- 65.Yermakovich D, Sivitskaya L, Vaikhanskaya T, Danilenko N, Motuk I. Novel desmoplakin mutations in familial Carvajal syndrome. Acta Myol 2018; 37:263–266. [PMC free article] [PubMed] [Google Scholar]
- 66.Bardawil T, Khalil S, Bergqvist C, Abbas O, Kibbi AG, Bitar F, et al. Genetics of inherited cardiocutaneous syndromes: a review. Open Heart 2016; 3:e000442.doi: 10.1136/openhrt-2016-000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Polivka L, Bodemer C, Hadj-Rabia S. Combination of palmoplantar keratoderma and hair shaft anomalies, the warning signal of severe arrhythmogenic cardiomyopathy: a systematic review on genetic desmosomal diseases. J Med Genet 2016; 53:289–295. doi: 10.1136/jmedgenet-2015-103403. [DOI] [PubMed] [Google Scholar]
- 68.Guerra L, Magliozzi M, Baban A, Di Mambro C, Di Zenzo G, Novelli A, et al. Palmoplantar keratoderma and woolly hair revealing asymptomatic arrhythmogenic cardiomyopathy. Acta Derm Venereol 2019; 99:831–832. doi: 10.2340/00015555-3216. [DOI] [PubMed] [Google Scholar]
- 69.Chalabreysse L, Senni F, Bruyere P, Aime B, Ollagnier C, Bozio A, et al. A new hypo/oligodontia syndrome: carvajal/Naxos syndrome secondary to desmoplakin-dominant mutations. J Dent Res 2011; 90:58–64. doi: 10.1177/0022034510383984. [DOI] [PubMed] [Google Scholar]
- 70.Samuelov L, Sprecher E. Inherited desmosomal disorders. Cell Tissue Res 2015; 360:457–475. doi: 10.1007/s00441-014-2062-y. [DOI] [PubMed] [Google Scholar]
- 71.Boyden LM, Kam CY, Hernandez-Martin A, Zhou J, Craiglow BG, Sidbury R, et al. Dominant de novo DSP mutations cause erythrokeratodermia-cardiomyopathy syndrome. Hum Mol Genet 2016; 25:348–357. doi: 10.1093/hmg/ddv481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee JYW, McGrath JA. Mutations in genes encoding desmosomal proteins: spectrum of cutaneous and extracutaneous abnormalities. Br J Dermatol 2021; 184:596–605. doi: 10.1111/bjd.19342. [DOI] [PubMed] [Google Scholar]