Abstract
Background:
Prenatal evaluation of fetal lung maturity (FLM) is a challenge, and an effective non-invasive method for prenatal assessment of FLM is needed. The study aimed to establish a normal fetal lung gestational age (GA) grading model based on deep learning (DL) algorithms, validate the effectiveness of the model, and explore the potential value of DL algorithms in assessing FLM.
Methods:
A total of 7013 ultrasound images obtained from 1023 normal pregnancies between 20 and 41 + 6 weeks were analyzed in this study. There were no pregnancy-related complications that affected fetal lung development, and all infants were born without neonatal respiratory diseases. The images were divided into three classes based on the gestational week: class I: 20 to 29 + 6 weeks, class II: 30 to 36 + 6 weeks, and class III: 37 to 41 + 6 weeks. There were 3323, 2142, and 1548 images in each class, respectively. First, we performed a pre-processing algorithm to remove irrelevant information from each image. Then, a convolutional neural network was designed to identify different categories of fetal lung ultrasound images. Finally, we used ten-fold cross-validation to validate the performance of our model. This new machine learning algorithm automatically extracted and classified lung ultrasound image information related to GA. This was used to establish a grading model. The performance of the grading model was assessed using accuracy, sensitivity, specificity, and receiver operating characteristic curves.
Results:
A normal fetal lung GA grading model was established and validated. The sensitivity of each class in the independent test set was 91.7%, 69.8%, and 86.4%, respectively. The specificity of each class in the independent test set was 76.8%, 90.0%, and 83.1%, respectively. The total accuracy was 83.8%. The area under the curve (AUC) of each class was 0.982, 0.907, and 0.960, respectively. The micro-average AUC was 0.957, and the macro-average AUC was 0.949.
Conclusions:
The normal fetal lung GA grading model could accurately identify ultrasound images of the fetal lung at different GAs, which can be used to identify cases of abnormal lung development due to gestational diseases and evaluate lung maturity after antenatal corticosteroid therapy. The results indicate that DL algorithms can be used as a non-invasive method to predict FLM.
Keywords: Convolutional neural network, Deep learning algorithms, Grading model, Normal fetal lung, Fetal lung maturity, Gestational age, Artificial intelligence
Introduction
The leading cause of neonatal morbidity and mortality in preterm and term fetuses is lung immaturity.[1] Lung immaturity leading to surfactant deficiency is related to neonatal respiratory morbidity (NRM); despite advances in the treatment of NRM, it still represents the most common complication in infants born preterm and even early term (<39 weeks).[2,3] In addition, some gestational diseases, such as gestational diabetes mellitus (GDM),[4] pre-eclampsia (PE),[5] oligohydramnios,[6] and fetal intrauterine growth restriction,[7] can also affect the maturation of the fetal lungs. It is important for clinicians to evaluate the fetal lung maturity (FLM) in the third trimester, particularly after 34 weeks of gestation when the risk of NRM ranges from 5% to 20%, and determine the use of antenatal corticosteroid (ACS) therapy or plan the place and time of elective delivery in the presence of late pregnancy complications.[8,9] Prenatal evaluation of FLM is challenging; however, it is important to evaluate FLM a few weeks before delivery, especially in cases of planned cesarean sections, to avoid iatrogenic prematurity.
In current clinical practice, evaluation of FLM relies on testing different components of the amniotic fluid, which requires amniocentesis.[10,11] Amniocentesis is an invasive procedure and is associated with preterm labor, premature rupture of membranes, fetomaternal hemorrhage, fetal injury, placental abruption, and (rarely) fetal or maternal death.[12,13] The American College of Obstetricians and Gynecologists (ACOG) suggests that FLM should be confirmed in a low-risk singleton pregnancy if elective delivery is considered before 39 weeks of gestation.[11] The use of a non-invasive method would be ideal to determine FLM.
For decades, the prediction of lung maturity by non-invasive methods has been extensively explored. Several approaches have been attempted, including comparison of fetal lung echogenicity with the placenta,[14] fetal gut,[15] liver,[16] gray-scale measurements,[17,18] fetal pulmonary artery Doppler velocimetry,[19] and lung tissue motion.[20] In recent years, quantitative texture analysis, a powerful technique for extracting information from ultrasound images and quantifying tissue changes, has been used to predict FLM.[21,22] These studies generally showed a good correlation with FLM, but the diagnostic accuracy was inadequate for clinical use.
Over the years, powerful artificial intelligence (AI) techniques have been developed for the medical profession especially in fields that require imaging data analysis, such as radiology and ultrasound, due to the advancement in computer technology and image resolution.[23,24] AI techniques, particularly deep learning (DL) algorithms, are garnering increased attention in research due to their outstanding performance in image recognition tasks.[25] DL algorithms can automatically make a quantitative assessment of complex medical image characteristics and extract subtle changes in the aspect of texture information that are invisible to the human eye.[23,25] The use of DL algorithms in ultrasound has previously been investigated for medical diagnostic applications, including in breast cancer, liver disease, and other diseases.[25,26] In recent years, some studies have demonstrated that an automatic quantitative ultrasound texture analysis algorithm based on AI can extract features from fetal lung ultrasound images, showing a strong correlation with both gestational age (GA) and the results of FLM testing of the amniotic fluid.[1,22] However, the same problems identified in other quantitative imaging methods using traditional machine algorithms persist, such as the lack of robustness of blind detection due to changes in acquisition conditions.[23] Nevertheless, a new method using DL algorithms may help to overcome these problems.[25] To date, the value of using DL algorithms by analyzing fetal lung ultrasound images to predict FLM has not been widely demonstrated.
In this study, we have established a normal fetal lung GA grading model based on DL algorithms by extracting the ultrasound image information of normal fetal lungs at different GAs. We evaluated the accuracy of the model in identifying the lung images at different GAs and the ability of the features extracted by DL algorithms to provide information related to GA. We expect that this model, which is based on normal fetal lung data, may help to identify abnormal lung development caused by gestational diseases, such as GDM, PE, or oligohydramnios, and assess lung maturity after ACS therapy. Additionally, we hope that this study will lay the foundation for DL algorithms as a non-invasive method for assessing FLM.
Methods
Ethical approval
The study was in compliance with the relevant ethical laws and approved by the Ethics Committee of Beijing Obstetrics and Gynecology Hospital, Capital Medical University (No. 2018-KY-003-03).
Study design
This was a retrospective study, and cases of this study were collected at the Department of Ultrasound at the Beijing Obstetrics and Gynecology Hospital, Maternal and Child Health Centre, Capital Medical University, Beijing, China, between January 2015 and March 2018. This study population included only singleton, non-anomalous births at 20 to 41 + 6 weeks with no neonatal respiratory diseases and the Apgar scores of newborns were ≥8 at 1, 5, and 10 min after birth. GA was determined by the last menstrual period and verified using first-trimester dating ultrasound (crown-rump length). Gestational hypertension, GDM, fetal growth restriction, ACS therapy, intrauterine infection, oligohydramnios, and other conditions that may affect fetal lung development, multiple pregnancies, fetal structure, and chromosomal abnormalities were excluded from the study.
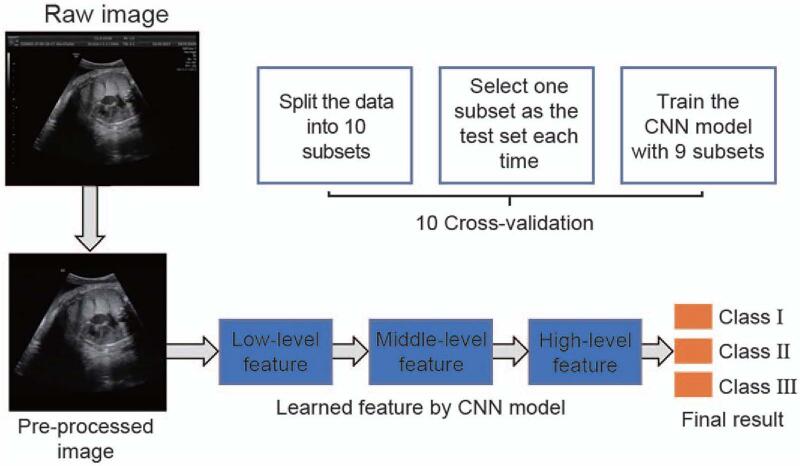
A total of 1023 cases were included in this study. We collected 7013 images of the axial cross-section of the fetal chest at the level of the four-chamber view of the fetal heart during routine ultrasound examination at 20 to 41 + 6 weeks of pregnancy. The standard images are shown in Figure 1 (raw image). Images were discarded if the lung area contained color Doppler, measurement caliper overlays, or obvious acoustic shadows created by bony structures. The quality of all images was inspected by two sonographers. To explore whether DL algorithms could be used to recognize fetal lung ultrasound images at different GAs, we divided the images into three categories based on GA: class I: 20 to 29 + 6 weeks; class II: 30 to 36 + 6 weeks; class III: 37 to 41 + 6 weeks. While selecting 30 and 37 weeks as the cutoff points, the distribution of ultrasound images at each class can be relatively balanced; also, preterm birth is defined as birth before 37 weeks of gestation. The number of ultrasound images in each class was 3323, 2142, and 1548, respectively.
Figure 1.
Process of DL algorithm to establish a normal fetal lung GA grading model. CNN: Convolutional neural network; DL: Deep learning; GA: Gestational age.
All images were collected by sonographers in obstetrics and gynecology who had >2 years of work experience. Eight different ultrasound machines provided by six different manufacturers: GE Voluson E8/E10 (GE Healthcare Austria GmbH & Co OG, Zipf, Austria), HI VISION Preirus (Hitachi Aloka Medical, Ltd., Tokyo, Japan), SIEMENS Acuson S2000 (Siemens Medical Systems, Mountain View, CA, USA), TOSHIBA Aplio500SMI (Toshiba Medical System Corporation, Tokyo, Japan), SAMSUNG WS80A (Samsung Medison Corporation, Seoul, Korea), and PHILIPS EPIQ7/EPIQ7C (Philips Healthcare, Bothell, WA, USA) were used for data acquisition. To obtain high-quality images, sonographers would adjust the machine settings, such as depth, gain, magnification, frequency, and time gain compensation, based on the specific conditions used during the examination.
Algorithm selection and machine learning
In our study, we applied a DL algorithm. The framework of our research process is shown in Figure 1, which includes the following three steps.
Step 1: Ultrasound image pre-processing
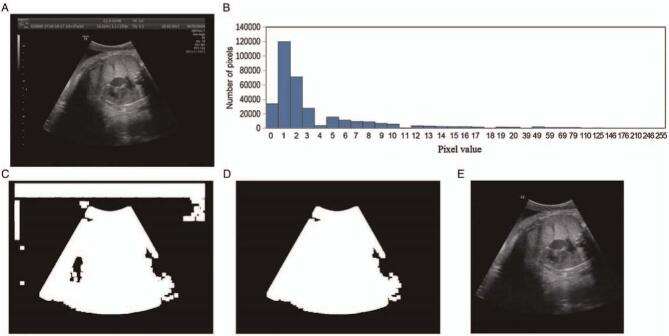
The ultrasound image directly exported from the ultrasound workstations is generally marked with some machine parameters in the upper left or right corner of the image. To remove irrelevant information from the image, we performed a pre-processing algorithm on each image as shown in Figure 2. We observed that there is an intensity difference between the region of interest (ROI) and the surrounding area in the ultrasound image; and we used the threshold method to segment the ROI. First, for an ultrasound image, we calculated the overall intensity distribution of the entire image as shown in Figure 2B. Then, we identified an appropriate threshold by the intensity distribution and binarized the image based on the threshold. Finally, we selected the largest area in the binarized image and removed the ROI based on this area.
Figure 2.
Pre-processing of image. (A) Raw ultrasound image; (B) intensity histogram of the image; (C) select an appropriate threshold based on the intensity histogram to binarize the image; (D) select the largest connected area based on the binary image; (E) cropped image according to the largest connected area.
Step 2: Build a classification network
We designed a convolutional neural network to identify different categories of fetal lung ultrasound images. The network automatically learns appropriate features from the training data and inputs the features into the classifier so that the feature extractor and classifier can be learned together. The network mainly contains convolution, pooling, non-linear, and fully connected operations. A convolutional layer extracts different features from the output feature maps of the previous layer based on a set of learnable filters. A pooling layer replaces the output at a specific location with a summary statistic of the nearby outputs and reduces the spatial size of the output feature maps. Non-linear operation increases the non-linear properties of the overall network. Several fully connected layers were used in the neural network to model high-level reasoning. Each unit of a fully connected layer has connections to all units in the previous layer.
The network architecture is designed based on the structure of DenseNet as shown in Figure 3. It consists of a convolution layer, a pooling layer, four dense blocks, three transition layers, a global pooling layer, and two fully connected layers. The first convolutional layer uses a 7 × 7 convolution kernel and the stride is set to 2 × 2. The transition layer is composed of a convolution layer and a pooling layer. The convolutional layer uses a 1 × 1 convolution and halves the number of channels in the feature map. Each pooling layer is a 2 × 2 average pooling layer with a stride of 2 × 2. The dense block consists of a series of convolution operations and concatenation operations, the structure of which is shown in Figure 4. The numbers of layers of the four dense blocks are six, four, four, and four, respectively. The global pooling layer averages the feature maps of each channel and outputs the same size output for inputs of different spatial sizes. In this architecture, all convolutional layers are followed by batch normalization and rectified linear units (ReLUs). The first fully connected layer is followed by the ReLUs, and the last fully connected layer is followed by soft-max units to output the probabilities. These are the main components of our network, which completed the construction of the model.
Figure 3.
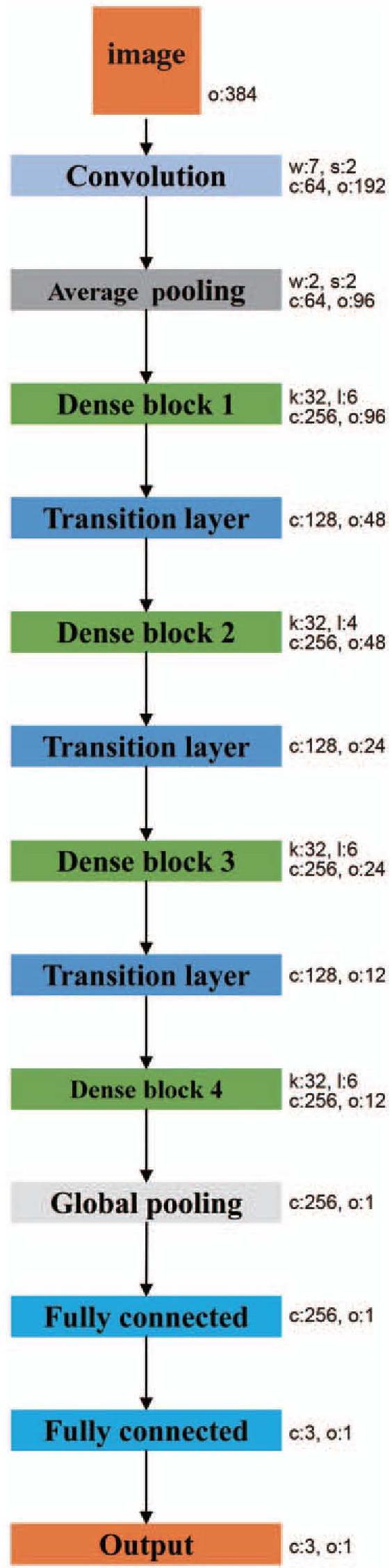
The architecture of the classification network. The network consists of a convolutional layer, an average pooling layer, four dense blocks, three transition layers, a global average pooling layer, and two fully connected layers. The parameter “w” indicates the kernel size, “s” indicates the stride, “c” indicates the output channels, “o” indicates the spatial size of the feature, “k” indicates the growth rate, and “l” indicates the number of layers of each dense block.
Figure 4.
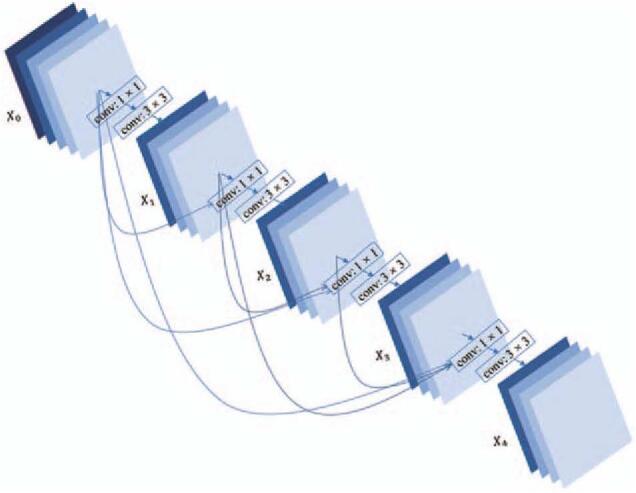
The architecture of a four-layer dense block. AUC: Area under the curve; ROC: Receiver operating characteristic.
After the design of the network architecture, the network produces the probabilities of each image from one of our three classes. To train the network, we used cross-entropy as the loss function, which is defined as:
where (x, y) is a sample from the dataset, yi indicates the ground-truth label of x, and pi(x) is the output probability of x belonging to the class i. By minimizing the cross-entropy loss, the network is trained to fit the images and provide the correct classifications.
Step 3: Build the training dataset and independent test dataset
To validate the performance of our model, ten-fold cross-validation was performed. We randomly split our data into ten subsets. In each fold, one subset was used as the independent test set, and the remaining subsets were used as the training set. Then, one subset was used for validation, and eight subsets were used to train the parameters of the network. During the training stage, all hyperparameters were determined based on the validation set. We oversampled the samples of minority classes, such that the numbers of the three classes remain similar due to the different number of training samples in each class.
Experimental setting
Training the network with only these samples can result in over-fitting due to the small size of the training set t. To avoid over-fitting, data augmentation was performed in our experiments. Random rotation, crop, and flipping were used for all the training samples. First, we randomly cropped a region from the image using the algorithm used by AlexNet[27] and then resized the region to 300 × 300 pixels. Second, each training image was randomly rotated within the range of (−30, 30). Finally, we randomly flipped each image with respect to the x- and y-axes.
Before training, the weights of the network were initialized using the method proposed by He et al.[28] The network was trained using stochastic gradient descent with a momentum of 0.9 and a weight decay of 0.0001. The batch size was set to 16. The initial learning rate was set to 0.01 and decreased to 0.0001 by a “cosine” learning rate policy.[29] The dropout (rate = 0.2) strategy was used in the global pooling layer to improve the generalization capability of the model. The network was implemented in Python based on the DL library of Keras. It took about 4 h to train the network using a graphics processing unit (GPU) of NVIDIA GeForce GTX 1080 Ti (NVIDIA, Santa Clara, CA, USA).
Statistical analysis
Different metrics were used to evaluate our method, including accuracy, sensitivity, specificity, macroF1 score, microF1 score, and confusion matrix. All metrics were performed using Python. In the n-class classification problem, a confusion matrix has n rows and n columns, and each row of the matrix represents the instances in a predicted class, whereas each column represents the instances in an actual class. The accuracy, sensitivity, and specificity of class i are defined as
where tpi is the number of correctly classified samples of class i, fpi is the number of negatives falsely classified as class i, tni is the number of correctly classified negatives, and fni is the number of samples of class i falsely classified as negative. The accuracy of each class is calculated by treating the classifier as multiple binary classifiers, using the samples of class i as positive, and the remaining samples as negatives. The accuracy of class i is the proportion of samples that were correctly classified. Sensitivity is the proportion of positive samples that were correctly classified, whereas specificity is the proportion of the negative samples that were correctly classified. The total accuracy is also used and is defined as
The macroF1 score and microF1 score are defined as
The scores of macroF1 and microF1 are used to assess the quality of problems with multiple classes. When the score of macroF1 or microF1 equals 1, the classifier is the best. When the score equals 0, the classifier is the worst. We also used receiver operating characteristic (ROC) curves and area under the curve (AUC) to evaluate our method. The ROC curve is created by plotting the true positive rate against the false positive rate at various threshold settings. The AUC metric computes the area under a discretized curve of true-positive vs. false-positive rates. AUC around 0.5 is the same thing as a random guess. The further away the AUC is from 0.5, the better. Also, the micro-average AUC (mAUC) and macro-average AUC (MAUC) are used to assess the quality of problems with multiple classes. The Delong test was used to compare ROC curves.
Results
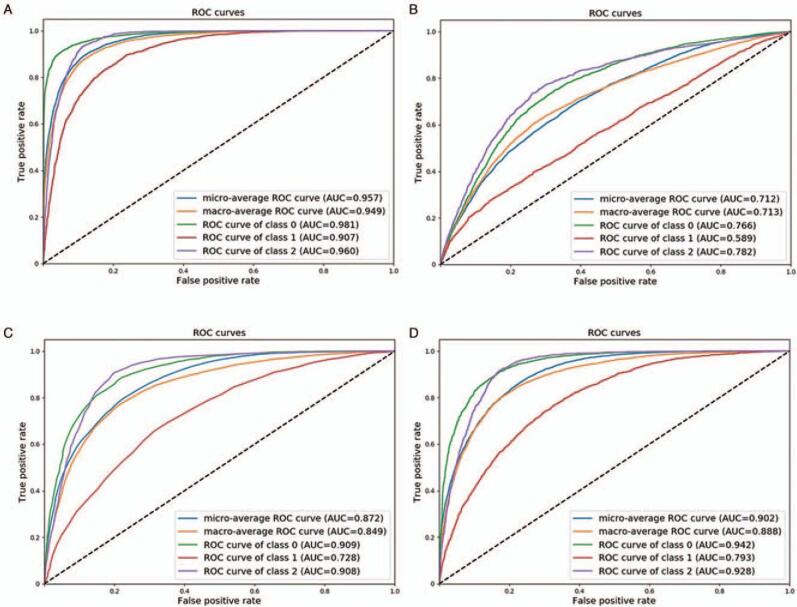
A total of 7013 ultrasound images obtained from 1023 pregnancies were analyzed in this study. The clinical characteristics and neonatal outcomes of the study population are displayed in Table 1. The composition of the gestational weeks of each class and the number of images are displayed in Table 2. The detailed results of the ten-fold cross-validation are described in Table 3. The sensitivities of the three classes in the independent test set were 91.7%, 69.8%, and 86.4%, respectively. The specificities of the three classes in the independent test set were 76.8%, 90.0%, and 83.1%, respectively. The total accuracy was 83.8%. The confusion matrix of the proposed method is shown in Table 4. Figure 5A shows the ROC curves, including the ROC curves of each class, the micro-average ROC curve, and the macro-average ROC curve. The AUC of each class was 0.982, 0.907, and 0.960, respectively. The mAUC was 0.957, and the MAUC was 0.949.
Table 1.
Clinical characteristics and neonatal outcomes of the 1023 cases of study population.
| Variable | Mean ± SD or n (%) |
| Maternal age, years | 27.1 ± 2.4 |
| GA at delivery, weeks | 39.2 ± 1.0 |
| Cesarean delivery | 138 (13.5) |
| Birth weight, g | 3358.8 ± 306.4 |
| Birth height, cm | 50.1 ± 0.8 |
GA: Gestational age; SD: standard deviation.
Table 2.
The composition gestational weeks of each class and the number of images.
| Class | Gestational week | Number of images (%) |
| Class I (n = 3323) | 20 | 11 (0.3) |
| 21 | 97 (2.9) | |
| 22 | 712 (21.4) | |
| 23 | 1625 (48.9) | |
| 24 | 175 (5.3) | |
| 25 | 120 (3.6) | |
| 26 | 161 (4.9) | |
| 27 | 82 (2.5) | |
| 28 | 90 (2.7) | |
| 29 | 250 (7.5) | |
| Class II (n = 2142) | 30 | 549 (25.6) |
| 31 | 487 (22.7) | |
| 32 | 182 (8.5) | |
| 33 | 116 (5.4) | |
| 34 | 238 (11.1) | |
| 35 | 252 (11.8) | |
| 36 | 318 (14.9) | |
| Class III (n = 1548) | 37 | 365 (23.6) |
| 38 | 395 (25.5) | |
| 39 | 404 (26.1) | |
| 40 | 336 (21.7) | |
| 41 | 48 (3.1) |
Table 3.
Results of ten-fold cross-validation for fetal lung gestational age grading model, (%).
| Class I | Class II | Class III | ||||||||
| Validation sequence | Specificity | Sensitivity | Accuracy | Specificity | Sensitivity | Accuracy | Specificity | Sensitivity | Accuracy | Average accuracy |
| 1 | 78.9 | 90.5 | 92.9 | 88.9 | 73.9 | 84.8 | 84.2 | 81.2 | 91.3 | 82.9 |
| 2 | 76.4 | 90.3 | 93.7 | 91.1 | 74.6 | 84.2 | 82.2 | 83.4 | 89.5 | 84.0 |
| 3 | 77.0 | 92.3 | 93.4 | 87.2 | 67.8 | 83.0 | 83.5 | 88.7 | 89.3 | 83.7 |
| 4 | 78.2 | 90.5 | 92.1 | 88.2 | 73.9 | 84.4 | 84.1 | 85.5 | 91.4 | 84.5 |
| 5 | 78.9 | 92.6 | 93.7 | 91.2 | 72.2 | 85.2 | 84.3 | 88.3 | 91.5 | 85.2 |
| 6 | 75.9 | 93.2 | 91.8 | 93.7 | 63.3 | 83.0 | 81.6 | 94.7 | 91.5 | 84.3 |
| 7 | 74.2 | 91.4 | 93.3 | 88.4 | 68.1 | 84.1 | 82.5 | 82.2 | 88.6 | 82.4 |
| 8 | 73.1 | 96.0 | 92.8 | 91.6 | 67.0 | 83.3 | 84.7 | 81.8 | 91.2 | 84.1 |
| 9 | 78.2 | 87.8 | 91.9 | 87.7 | 71.6 | 83.3 | 82.1 | 87.5 | 91.1 | 83.2 |
| 10 | 76.6 | 92.7 | 93.5 | 92.0 | 65.9 | 84.3 | 82.0 | 90.6 | 90.2 | 84.0 |
| Average | 76.8 | 91.7 | 92.9 | 90.0 | 69.8 | 84.2 | 83.1 | 86.4 | 90.6 | 83.8 |
Table 4.
Confusion matrix of our method of fetal lung gestational age grading model.
| Predicted class | |||
| True class | Class I | Class II | Class III |
| Class I | 3046 | 258 | 19 |
| Class II | 215 | 1495 | 432 |
| Class III | 5 | 206 | 1337 |
Figure 5.
(A) The ROC curves of our model; (B) the ROC curves of NB; (C) the ROC curves of RF; (D) the ROC curves of SVM. NB: Naïve Bayes; RF: Random forests; ROC: Receiver operating characteristic; SVM: Support vector machine.
We also compared our model to other methods, such as random forests (RF), support vector machine (SVM), and naïve Bayes (NB).[30] To train these classifiers, 481 features were extracted, including an intensity histogram, histogram of oriented gradient, and gray-level co-occurrence matrix.[31] Principle component analysis was also performed to further reduce the dimensionality of features, resulting in 128 features. Finally, the three models were trained based on these features. Table 5 presents a comparison of the different models, as well as the results of the validation set. Indeed, our method achieved the best performance and the over-fitting problem is negligible. Figures 5B to 5D show the ROC curves of NB, RF, and SVM, respectively. Our model shows impressive improvements when compared with NB, RF, and SVM. The macroF1/microF1 score of our method was 81.8%/83.8%. The microF1/macroF1 scores of NB, RF, and SVM were 52.9%/51.4%, 69.3%/65.8%, and 73.2%/71.1%, respectively. Our DL-based model was compared with other NB and showed a higher AUC in three classes (class I: AUC: 0.981, 95% confidence interval [CI]: 0.979–0.984 vs. AUC: 0.766, 95% CI: 0.755–0.777, P value < 0.001; class II: AUC: 0.907, 95% CI: 0.900–0.914 vs. AUC: 0.593, 95% CI: 0.575–0.604, P value < 0.001; class III: AUC: 0.960, 95% CI: 0.956–0.964 vs. AUC: 0.782, 95% CI: 0.769–0.795, P value < 0.001). Our DL-based model was also compared with other RF and showed a higher AUC in three classes (class I: AUC: 0.981, 95% CI: 0.979–0.984 vs. AUC: 0.911, 95% CI: 0.903–0.916, P value < 0.001; class II: AUC: 0.907, 95% CI: 0.900–0.914 vs. AUC: 0.724, 95% CI: 0.716–0.741, P value < 0.001; class III: AUC: 0.960, 95% CI: 0.956–0.964 vs. AUC: 0.908, 95% CI: 0.901–0.916, P value < 0.001). Our DL-based model was also compared with other SVM and showed a higher AUC on three classes (class I: AUC: 0.981, 95% CI: 0.979–0.984 vs. AUC: 0.942, 95% CI: 0.937–0.947, P value < 0.001; class II: AUC: 0.907, 95% CI: 0.900–0.914 vs. AUC: 0.795, 95% CI: 0.782–0.804, P value < 0.001; class III: AUC: 0.960, 95% CI: 0.956–0.964 vs. AUC: 0.928, 95% CI: 0.922–0.934, P value < 0.001).
Table 5.
Comparison of diagnostic efficacy among different models, (%).
| Models | microF1 | macroF1 | mAUC | MAUC |
| Naïve Bayes | 52.9 | 51.4 | 71.3 | 71.4 |
| Random forests | 69.3 | 65.8 | 87.3 | 74.7 |
| Support vector machine | 73.2 | 71.1 | 90.2 | 88.8 |
| Model in current study | 83.8 | 81.8 | 95.8 | 94.9 |
| Model by validation set | 84.1 | 82.2 | 95.9 | 95.1 |
mAUC: Micro-average area under the curve; MAUC: Macro-average area under the curve.
Discussion
In the current clinical practice, the evaluation of FLM relies on amniocentesis, an invasive procedure that is used to analyze the different components of the amniotic fluid.[1,2] The latest guidelines of ACOG and the Society for Maternal Fetal Medicine emphasized that the role of amniotic fluid testing is becoming increasingly limited and no longer has clinical utility. Therefore, this method should not be used to assess FLM.[32] Meanwhile, the use of non-invasive methods to predict lung maturity has been extensively explored with results showing a good correlation with FLM, but the diagnostic accuracy was inadequate for clinical use. In recent years, despite all the advances in the treatment of NRM, it remains a leading cause of neonatal morbidity and mortality in infants born late preterm (28–36 + 6 weeks’ gestation) and even in early term (37–38 + 6 weeks).[2,3] Therefore, it is necessary to explore effective and non-invasive methods for prenatal assessment of FLM. Compared with traditional algorithms, DL algorithms do not require manually designed features or learning a classifier based on the features to obtain classification results. Although traditional machine learning algorithms have achieved relatively good results in medical image processing, manual design features require a deep understanding of the image and the problem. Ultrasound imaging has the disadvantages of large noise, irregularity, and blur; making it difficult to design appropriate features to describe the information of interest. Therefore, it is difficult for traditional algorithms to deal with the classification of fetal lungs based on ultrasound images. In contrast, DL algorithms can automatically extract task-related features from the training data and learn a classifier, which is an end-to-end method.
In this study, we developed and validated a normal fetal lung GA grading model that could accurately identify fetal lung ultrasound images of different GAs. The sensitivity of each class in the independent test set was 91.7%, 69.8%, and 86.4%, respectively, and the specificity of each class in the independent test set was 76.8%, 90.0%, and 83.1%, respectively. The total accuracy reached 83.8%, and the AUC of each class was 0.982, 0.907, and 0.960, respectively. The grading model had good stability and repeatability and was less disturbed by external conditions. Since GA is strongly associated with FLM and maybe the best estimator of risk for respiratory distress syndrome,[33] the findings of this study confirmed that DL algorithms have great potential and research value in evaluating FLM. For any imported fetal lung ultrasound images of 20 to 41 + 6 weeks, the grading model automatically outputs the classification of gestational weeks of the images by extracting subtle image information that is not visible to the naked eye. The results of our study provide preliminary evidence for the use of AI approaches for the prenatal prediction of FLM and may support future research evaluating the relationship between DL algorithms and lung maturity.
Animal experiments and laboratory tests have shown that gestational diseases have an impact on fetal lung development. Winn et al[5] reported that pregnancies complicated by PE were associated with delayed fetal lung maturation biochemical profile, as shown by both the lecithin to sphingomyelin ratio and TDx-fetal lung maturity (TDx-FLM II, Abbott Laboratories, Abbott Park, IL, USA) assay values at GAs between 33 and 36 weeks. Najrana et al[6] suggested that increased external compression secondary to severe oligohydramnios can compromise lung cell size and interfere with epithelial and endothelial development. Baack et al[33] used a rat model and found that late-gestational diabetes affected the maturation rate of the fetal lung by reducing pulmonary angiogenesis. The results of these studies indicate that some gestational diseases can affect FLM. The latest research by Du et al[34] found that GDM, PE, and normal fetal lungs can be quickly and accurately classified by ultrasound-based radiomics techniques, indicating that differences in fetal lung development between gestational diseases (GDM and PE), and normal pregnancies were highly significant. Our study used DL algorithms to extract information from numerous normal fetal lung ultrasound images to establish a normal fetal lung grading model that can help to identify abnormal lung development that can result from some gestational diseases. The abnormal lungs’ grading results often do not match the actual GA. In clinical practice, women at risk of preterm birth between 24 and 34 + 6 weeks of gestation require ACS therapy. After ACS therapy, we can consider that the risk of delivery is reduced if the grading model evaluates the fetal lung as class III, which is equivalent to the 37 and 41 + 6 weeks’ gestation level of normal lungs. Our grading model can be useful for clinical decision-making.
Previous studies have explored various non-invasive methods for evaluating FLM by analyzing ultrasound images. In a study by Maeda et al,[18] fetal lung ultrasonic gray-level histogram width (GLHW) increased with fetal lung development; however, there was no change in liver GLHW. The lung/liver GLHW ratios were <1 at 24 to 29 weeks; however, they were ≥1 at 30 to 35 weeks. The authors of this study believed that GLHW was reliable, since it is reproducible in various gain settings with different ultrasonic imaging devices. Tekesin et al[35] evaluated the mean gray value of fetal lungs (MGL), showing a changing pattern with fetal lung development. However, no significant differences were observed after 32 weeks of gestation. A study by Maeda et al[18] showed that the MGL was less reliable, since the gray level varied according to the gain changes among various machines. Serizawa and Maeda[17] applied ultrasonic GLHW to predict fetal lung immaturity. The results of the study showed that fetal lung-to-liver GLHW ratios combined with GA predicted respiratory distress syndrome, with a sensitivity of 0.96 and a specificity of 0.72, which was comparable to invasive amniotic fluid tests. These studies demonstrated a correlation between quantitative image analysis and FLM. However, these methods can be affected by ultrasonic imaging equipment and operators, and the diagnostic accuracy is reduced, which hinders the clinical application of quantitative image analysis and prediction of FLM. Our DL method was not affected by the sonographers or ultrasonic parameters, such as the type of ultrasonic equipment or transducer.
In recent years, powerful quantitative techniques for ultrasound image analysis have been developed due to improvements in computer technology and image resolution.[36] Palacio et al[22] used an automatic quantitative ultrasound analysis (AQUA) texture extractor to extract descriptors most relevant to FLM, using the TDx FLM assay II test (TDx-FLM) as a reference. The results of this study showed that the imaging biomarker based on AQUA predicted FLM with a sensitivity of 95.1%, a specificity of 85.7%, and an accuracy of 90.3%. Palacio et al[21] and Bonet-Carne et al[23] applied quantitative ultrasound texture analysis of the fetal lung (quantusFLM; www.quantusflm.com; Transmural Biotech, Barcelona, Spain) that combined texture extraction with machine learning methods to predict neonatal respiratory diseases. The quantusFLM predicted NRM with a sensitivity, specificity, and accuracy of 74.3% to 86%, 87.0% to 88.6%, and 86.5%, respectively. This study provided evidence that quantitative texture analysis of lung ultrasound images could predict neonatal respiratory diseases with similar accuracy to current tests using amniotic fluid.[23] However, the common problems of quantitative imaging methods using traditional machine algorithms still exist, such as the lack of robustness of blind detection due to changes in acquisition conditions.[23] Our study explored a new AI method based on DL algorithms that can help to improve these problems.[25]
Our method is a new AI method based on DL algorithms, which recognizes the specific characteristics of ultrasound images related to the GA and consequently establishes a classification algorithm. This method is reliable and robust to small variations in the conditions of image acquisition, including depth and changes in the gain of the image, and does not need to be compared with other tissues (fetal liver and placenta). Compared with traditional machine learning algorithms, DL algorithms have the advantage of automatically extracting features related to the maturity of fetal lungs from ultrasound images, which is different from traditional machine algorithms that require the manual design of features. Through the end-to-end algorithm implementation, the model can optimize the feature extraction and classification module for increased accuracy of the classification. To reflect the superiority of our algorithm, we compared the performance of traditional machine learning algorithms and DL algorithms in recognizing fetal lung ultrasound images at different GAs. The comparison results are shown in Table 5. We concluded that the performance of DL algorithms was superior to that of traditional machine learning algorithms. In addition, the entire algorithm can automatically process images without additional operations. Furthermore, DL algorithms use convolutional neural networks to classify images and GPUs to accelerate model calculations. Generally, dozens of images can be processed in 1 s.
This study has some limitations that should be considered, including the fact that this was a retrospective study. The main limitation of this study was that the number of images was insufficient, especially images of classes II and III. Indeed, AI requires big data research, and more image data can improve the accuracy of the research. Furthermore, there was an obvious difference in the number of images collected in each gestational week. Class I images were mainly collected from 22 to 24 + 6 weeks, class II images were mainly collected from 30, 31, and 36 weeks, and class III images were rarely collected from 41 weeks. This may lead to inaccurate recognition of lung ultrasound images in some gestational weeks.
To conclude, this study demonstrated that the normal fetal lung GA grading model based on DL algorithms had good accuracy in identifying the ultrasound images of fetal lungs at different GAs and extracting information on fetal lung images related to GA. These results can support further research to establish the potential use of DL algorithms as a non-invasive predictive method of FLM. And the grading model can help to identify abnormal lung development caused by gestational diseases and assess lung maturity after ACS therapy.
Funding
This work was supported by a grant from the National Key Research and Development Program of China (No. 2016YFC1000104).
Conflicts of interest
None.
Footnotes
How to cite this article: Xia TH, Tan M, Li JH, Wang JJ, Wu QQ, Kong DX. Establish a normal fetal lung gestational age grading model and explore the potential value of deep learning algorithms in fetal lung maturity evaluation. Chin Med J 2021;134:1828–1837. doi: 10.1097/CM9.0000000000001547
Tai-Hui Xia and Man Tan contributed equally to this work.
References
- 1.Cobo T, Bonet-Carne E, Martínez-Terrón M, Perez-Moreno A, Elías N, Luque J, et al. Feasibility and reproducibility of fetal lung texture analysis by automatic quantitative ultrasound analysis and correlation with gestational age. Fetal Diagn Ther 2012; 31:230–236. doi: 10.1159/000335349. [DOI] [PubMed] [Google Scholar]
- 2.Teune MJ, Bakhuizen S, Gyamfi Bannerman C, Opmeer BC, van Kaam AH, van Wassenaer AG, et al. A systematic review of severe morbidity in infants born late preterm. Am J Obstet Gynecol 2011; 205:374.e1–374.e9. doi: 10.1016/j.ajog.2011.07.015. [DOI] [PubMed] [Google Scholar]
- 3.Hibbard JU, Wilkins I, Sun L, Gregory K, Haberman S, et al. Consortium on Safe Labor. Respiratory morbidity in late preterm births. JAMA 2010; 304:419–425. doi: 10.1001/jama.2010.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Luca AKC, Nakazawa CY, Azevedo BC, Rudge MVC, De Araújo Costa RA, Calderon IMP. Influence of glycemic control on fetal lung maturity in gestations affected by diabetes or mild hyperglycemia. Acta Obstet Gynecol Scand 2009; 88:1036–1040. doi: 10.1080/00016340903118018. [DOI] [PubMed] [Google Scholar]
- 5.Winn HN, Klosterman A, Amon E, Shumway JB, Artal R. Does preeclampsia influence fetal lung maturity? J Perinat Med 2000; 28:210–213. doi: 10.1515/JPM.2000.028. [DOI] [PubMed] [Google Scholar]
- 6.Najrana T, Ramos LM, Eid RA, Sanchez-Esteban J. Oligohydramnios compromises lung cells size and interferes with epithelial-endothelial development. Pediatr Pulmonol 2017; 52:746–756. doi: 10.1002/ppul.23662. [DOI] [PubMed] [Google Scholar]
- 7.Sasi A, Abraham V, Davies-Tuck M, Polglase GR, Jenkin G, Miller SL, et al. Impact of intrauterine growth restriction on preterm lung disease. Acta Paediatr 2015; 104:e552–e556. doi: 10.1111/apa.13220. [DOI] [PubMed] [Google Scholar]
- 8.Sengupta S, Carrion V, Shelton J, Wynn RJ, Ryan RM, Singhal K, et al. Adverse neonatal outcomes associated with early-term birth. JAMA Pediatr 2013; 16:1053–1059. doi: 10.1001/jamapediatrics.2013.2581. [DOI] [PubMed] [Google Scholar]
- 9.Gyamfi-Bannerman C, Thom EA, Blackwell SC, Tita ATN, Reddy UM, Saade GR, et al. Antenatal betamethasone for women at risk for late preterm delivery. N Engl J Med 2016; 374:1311–1320. doi: 10.1056/NEJMoa1516783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Besnard AE, Wirjosoekarto SAM, Broeze KA, Opmeer BC, Mol BWJ. Lecithin/sphingomyelin ratio and lamellar body count for fetal lung maturity: a meta-analysis. Eur J Obstet Gynecol Reprod Biol 2013; 169:177–183. doi: 10.1016/j.ejogrb.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 11.American College of Obstetricians and Gynecologists. ACOG practice bulletin no. 97: fetal lung maturity. Obstet Gynecol 2008; 112:717–726. doi: 10.1097/AOG.0b013e318188d1c2. [DOI] [PubMed] [Google Scholar]
- 12.Stark CM, Smith RS, Lagrandeur RM, Batton DG, Lorenz RP. Need for urgent delivery after third-trimester amniocentesis. Obstet Gynecol 2000; 95:48–50. doi: 10.1016/s0029-7844(99)00479-2. [DOI] [PubMed] [Google Scholar]
- 13.Gordon MC, Narula K, O'Shaughnessy R, Barth WH, Jr. Complications of third trimester amniocentesis using continuous ultrasound guidance. Obstet Gynecol 2002; 99:255–259. doi: 10.1016/s0029-7844(01)01715-x. [DOI] [PubMed] [Google Scholar]
- 14.Harman CR, Manning FA, Stearns E, Morrison I. The correlation of ultrasonic placental grading and fetal pulmonary maturation in five hundred sixty-three pregnancies. Am J Obstet Gynecol 1982; 143:941–943. doi: 10.1016/0002-9378(82)90478-1. [DOI] [PubMed] [Google Scholar]
- 15.Zilianti M, Fernandez S. Correlation of ultrasonic images of fetal intestine with gestational age and fetal maturity. Obstet Gynecol 1983; 62:569–573. doi: 10.1016/0378-5122(83)90009-9. [PubMed] [Google Scholar]
- 16.Feingold M, Scollins J, Cetrulo CL, Koza D. Fetal lung to liver reflectivity ratio and lung maturity. J Clin Ultrasound 1987; 15:384–387. doi: 10.1002/jcu.1870150605. [DOI] [PubMed] [Google Scholar]
- 17.Serizawa M, Maeda K. Noninvasive fetal lung maturity prediction based on ultrasonic gray level histogram width. Ultrasound Med Biol 2010; 36:1998–2003. doi: 10.1016/j.ultrasmedbio.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 18.Maeda K, Utsu M, Yamamoto N, Serizawa M. Echogenicity of fetal lung and liver quantified by the grey-level histogram width. Ultrasound Med Biol 1999; 25:201–208. doi: 10.1016/s0301-5629(98)00160-4. [DOI] [PubMed] [Google Scholar]
- 19.Azpurua H, Norwitz ER, Campbell KH, Funai EF, Pettker CM, Kleine M, et al. Acceleration/ejection time ratio in the fetal pulmonary artery predicts fetal lung maturity. Am J Obstet Gynecol 2010; 203:40.e1–40.e8. doi: 10.1016/j.ajog.2010.01.075. [DOI] [PubMed] [Google Scholar]
- 20.La Torre R, Cosmi E, Anceschi MH, Piazze JJ, Piga MD, Cosmi EV. Preliminary report on a new and noninvasive method for the assessment of fetal lung maturity. J Perinat Med 2003; 31:431–434. doi: 10.1515/JPM.2003.067. [DOI] [PubMed] [Google Scholar]
- 21.Palacio M, Bonet-Carne E, Cobo T, Perez-Moreno A, Sabrià J, Richter J, et al. Prediction of neonatal respiratory morbidity by quantitative ultrasound lung texture analysis: a multicenter study. Am J Obstet Gynecol 2017; 217:196.e1–196.e14. doi: 10.1016/j.ajog.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palacio M, Cobo T, Martínez-Terrón M, Rattá GA, Bonet-Carné E, Amat-Roldán I, et al. Performance of an automatic quantitative ultrasound analysis of the fetal lung to predict fetal lung maturity. Am J Obstet Gynecol 2012; 207:504.e1–504.e5. doi: 10.1016/j.ajog.2012.09.027. [DOI] [PubMed] [Google Scholar]
- 23.Bonet-Carne E, Palacio M, Cobo T, Perez-Moreno A, Lopez M, Piraquive JP, et al. Quantitative ultrasound texture analysis of fetal lungs to predict neonatal respiratory morbidity. Ultrasound Obstet Gynecol 2015; 45:427–433. doi: 10.1002/uog.13441. [DOI] [PubMed] [Google Scholar]
- 24.Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL. Artificial intelligence in radiology. Nat Rev Cancer 2018; 18:500–510. doi: 10.1038/s41568-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou LQ, Wang JY, Yu SY, Wu GG, Wei Q, Deng YB, et al. Artificial intelligence in medical imaging of the liver. World J Gastroenterol 2019; 25:672–682. doi: 10.3748/wjg.v25.i6.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park HJ, Kim SM, La Yun B, Jang M, Kim B, Jang JY, et al. A computer-aided diagnosis system using artificial intelligence for the diagnosis and characterization of breast masses on ultrasound: added value for the inexperienced breast radiologist. Medicine (Baltimore) 2019; 98:e14146.doi: 10.1097/MD.0000000000014146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Adv Neural Inform Process Syst 2012; 25:1097–1105. doi: 10.1145/3065386. [Google Scholar]
- 28.He K, Zhang X, Ren S, Sun J. Delving Deep Into Rectifiers: Surpassing Human-level Performance on Imagenet Classification. In: Proceedings of the IEEE International Conference on Computer Vision; 2015:1026–1034. doi: 10.1109/ICCV.2015.123. [Google Scholar]
- 29.Hinton GE, Osindero S, Teh YW. A fast-learning algorithm for deep belief nets. Neural Comput 2006; 18:1527–1554. doi: 10.1162/neco.2006.18.7.1527. [DOI] [PubMed] [Google Scholar]
- 30.Cortes C, Vapnik V. Support-vector networks. Mach Learn 1995; 20:273–297. doi: 10.1023/A:1022627411411. [Google Scholar]
- 31.Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans Syst Man Cybern 1973; SMC-3:610–621. doi: 10.1109/TSMC.1973.4309314. [Google Scholar]
- 32.Johnson LM, Johnson C, Karger AB. End of the line for fetal lung maturity testing. Clin Biochem 2019; 71:74–76. doi: 10.1016/j.clinbiochem.2019.07.003. [DOI] [PubMed] [Google Scholar]
- 33.Baack ML, Forred BJ, Larsen TD, Jensen DN, Wachal AL, Khan MA, et al. Consequences of a maternal high-fat diet and late gestation diabetes on the developing rat lung. PLoS One 2016; 11:e0160818.doi: 10.1371/journal.pone.0160818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du Y, Fang Z, Jiao BDJ, Xi G, Zhu C, Ren Y, et al. Application of ultrasound-based radiomics technology in fetal lung texture analysis in pregnancies complicated by gestational diabetes or pre-eclampsia. Ultrasound Obstet Gynecol 2021; 57:804–810. doi: 10.1002/uog.22037. [DOI] [PubMed] [Google Scholar]
- 35.Tekesin I, Anderer G, Hellmeyer L, Stein W, Kühnert M, Schmidt S. Assessment of fetal lung development by quantitative ultrasonic tissue characterization: a methodical study. Prenat Diagn 2004; 24:671–676. doi: 10.1002/pd.951. [DOI] [PubMed] [Google Scholar]
- 36.Insana MF, Garra BS, Rosenthal SJ, Hall TJ. Quantitative ultrasonography. Med Prog Technol 1989; 15:141–153. doi: 10.1007/BF02442178. [PubMed] [Google Scholar]