Abstract
Light-field fluorescence microscopy can record large-scale population activity of neurons expressing genetically-encoded fluorescent indicators within volumes of tissue. Conventional light-field microscopy (LFM) suffers from poor lateral resolution when using wide-field illumination. Here, we demonstrate a structured-illumination light-field microscopy (SI-LFM) modality that enhances spatial resolution over the imaging volume. This modality increases resolution by illuminating sample volume with grating patterns that are invariant over the axial direction. The size of the SI-LFM point-spread-function (PSF) was approximately half the size of the conventional LFM PSF when imaging fluorescent beads. SI-LFM also resolved fine spatial features in lens tissue samples and fixed mouse retina samples. Finally, SI-LFM reported neural activity with approximately three times the signal-to-noise ratio of conventional LFM when imaging live zebrafish expressing a genetically encoded calcium sensor.
1. Introduction
Dissecting the complex dynamics of brain activity in model organisms requires large-scale and high-resolution volumetric imaging techniques. The combination of fluorescence microscopy and genetically encoded fluorescent indicators enable a variety of functional imaging experiments that record neural activity at various resolutions, imaging speeds, and imaging fields-of-view (FOVs) [1–3]. Light-field microscopy (LFM) is a useful volumetric imaging modality in the study of biological samples. LFM reconstructs three-dimensional (3D) volumes at the camera frame rate by capturing both two-dimensional (2D) spatial and 2D angular information without mechanical scanning of either the sample or the excitation source. LFM captures light-field information by imaging the conjugate sample plane after the tube lens on a microlens array (MLA), and then relaying the information from the focal plane of the MLA to the camera sensor. By illuminating the fluorophores at the imaging volume with the minimal instantaneous peak power, LFM has demonstrated various fast speed volumetric imaging, but at the cost of sacrificing the lateral resolution for the axial extent. To date, LFM was able to image the neural activity from worm and zebrafish [4]. However, LFM’s spatial resolution was comparable to the zebrafish neuron size, and additional computational methods were needed to unmix activity between neighboring neurons. Multiple advances have tried to improve the resolution of LFM. Compressive LFM [5] and sparse decomposition LFM [6] improved the resolution of conventional LFM by relying on the sparsity of temporal signal. These methods worked well to localize active neurons, but would be insufficient for structural imaging without dynamic information. Recently developed defocused LFM moderately improved the lateral resolution and extended the depth-of-field by placing the microlens array in a position after the focus of the tube lens [7]. LFMs using customized MLAs, such as eXtended LFM (XLFM) [8] and confocal LFM (CLFM) [9], have demonstrated whole brain imaging of freely moving zebrafish, but are not widely disseminated because of the lens customization. By manipulating the illumination, LFM with selective illumination [10, 11] and speckle illumination [12] has also shown better lateral resolution over wide-field illumination LFM. However, a key limitation for all these methods is that their improvement in lateral resolution cannot exceed ∼0.7 the size of the conventional LFM’s PSF, due to the diffraction limit.
Structured illumination microscopy (SIM) is a super-resolution imaging technique that can achieve sub-diffraction limit resolution. SIM explores the spatial frequency space beyond the cutoff frequency of the optical system by sequentially imaging the sample with high spatial frequency grating illumination patterns with different orientations and phases [13]; these patterns demodulate high spatial frequency information from the sample into the captured frequencies of the optical system. Image reconstruction combines the high spatial frequency information from all images to generate a high-resolution image. SIM can theoretically capture spatial information well beyond the optical system’s cutoff frequency. Compared with deconvolution techniques, SIM can extract high spatial frequency features without making assumptions about the structure of the sample or microscope [14]. Structured illumination has combined with wide-field microscopy [13] and light-sheet microscopy [15] to improve resolution in a variety of live-cell imaging experiments [16]. These microscopes have thus far volumetrically imaged samples by sequentially scanning the various depths of the sample one focal plane at a time, or acquired multiplane images with an image-splitting prism [17].
In this work, we integrated SIM with LFM to establish a new imaging scheme called structured-illumination LFM (SI-LFM). SI-LFM combined the advantages of SIM and LFM to image volumes of samples with high spatial resolution and contrast without scanning. While similar approaches such as hybrid light-sheet LFM can achieve similar gains, SI-LFM simplified the system and employed only one objective shared between the excitation and imaging paths. We instantiated volumetric structured illumination by using the super-pixel method to produce an illumination pattern that is invariant over depth [18, 19]. SI-LFM had more refined lateral resolution than conventional LFM when imaging calibration samples, lens tissue, and fixed mouse retina. We also validated SI-LFM by imaging live zebrafish, a model organism for optical fluorescence imaging and neuroscientific studies. The model’s nearly transparent body and available toolkit for genetic expression of fluorescent protein sensors [20] support volumetric optical interrogation of the zebrafish brain. SI-LFM demonstrated high-resolution large-scale and simultaneous recording of neural activity from hundreds of neurons in the zebrafish brain reported by the green-fluorescent calmodulin protein (GCaMP6s) sensor [21].
2. Methods
2.1. DMD pattern generation and characterization of volumetric grating illumination
The intensity profile of the 3D structured illumination was
| (1) |
where φ was the phase shift, and and were spatial frequencies in the x and y directions, respectively. This manifested as an interference pattern between two symmetrical plane waves with opposing propagation in the lateral direction; such an illumination pattern in the sample space corresponded to two spots in the Fourier plane. We generated this pattern using a DMD (DLP LightCrafter 4500, Texas Instruments Inc.). The DMD had a resolution of 1024 × 912 pixels, and a 4225 Hz refresh rate when preloaded with 1-bit patterns. We designed the holographic pattern for the DMD using the super-pixel encoding method [18, 19]. This method groups blocks of 4 × 4 pixels within the DMD into super-pixels for complex-field modulation of the first-order diffraction beam. We validated the generated DMD pattern by calculating the Fourier transform of the DMD pattern, which was comparable to the targeted amplitude and phase. We generated ten patterns in total: one pattern for WF-LFM imaging and nine patterns for SI-LFM (three directions: 0°, 60° and 120°; three phases in each direction: 0°, 120° and 240°). We experimentally tested the invariant range of super-pixel pattern, which is related to the interference angle (α) between the two plane waves. The theoretical invariant range of the pattern () was:
| (2) |
where w was the dimension of pattern’s size along the interference direction (295 μm in our design), g was the distance between these two beams at the back aperture of the objective, f was the focal length of the objective, λ was the excitation wavelength, and was the spatial frequency of the pattern.
We experimentally tested the intensity distribution of the pattern by imaging a fluorescent sample with conventional microscopy. We moved the target along the axial direction from −150 µm to 200 µm with a 1 µm step size using a motorized stage, and acquired an image at each position. We focused at each position by fine-tuning the focal distance of the macro lens. We defined the experimental invariant axial range as the range of depths that had a sinusoidal amplitude larger than 70% of amplitude at z = 0 µm.
2.2. Imaging setup
SI-LFM utilizes an epifluorescence microscope configuration, where the excitation path and the imaging path pass through a common objective [11]. We used either an air-coupled objective (20×/0.75NA, Olympus) for beads, tissue and retina imaging, or a water-coupled objective (XLUMPlanFL, 20×/1.0 NA, Olympus) for zebrafish imaging. We used a 505 nm laser (OBIS, Coherent) as the excitation light source. The laser first passed through a 504/12 nm bandpass filter (F01-504/12-25, Semrock) and an 8× beam expander to match the size of the DMD surface. The beam then reflected off of the DMD and subsequently passed through a Fourier lens. A spatial filter at the focal plane of the Fourier lens filtered for the first-order diffraction pattern, which represented the two eventual plane waves in the sample. The diffraction pattern then passed through a second relay lens pair, reflected off of a dichroic mirror (69008bs, Chroma), and entered the objective. The designed holographic pattern generated an interference pattern that was invariant along the axial direction around the focal plane of the objective in the sample space.
We imaged the structurally illuminated samples using a conventional LFM [11]. Sample emission after the objective and dichroic mirror initially passed through a 534/20 nm bandpass filter (FF01-534/20-25, Semrock) in the imaging path. We then used a 175 mm tube lens, producing a magnification of 19.4×. A microlens array (RPC Photonics, MLA-S100-f10) at the conjugate sample plane generated the light-field. The microlens array had an f-number of f/10, matching the f-number in the used portion of the tube lens (f/13). A five-axis kinematic mount (Thorlabs, K5X1) provided fine alignment of the MLA to the optical axis. A 1:1 relay macro lens (Tamron, 70-300 mm) then imaged the focal plane of the microlens array onto a 4.2-megapixel scientific complementary metal-oxide-semiconductor (sCMOS) camera (Flash4v3, Hamamatsu). We controlled the DMD with the DLPLCR4500 GUI provided by Texas Instruments. We generated a 24-bit image that contained the 10 DMD patterns and stored the image into the flash memory of the DMD. We configured the sequence mode of the software to sequentially display the patterns with equal exposure times in a continuous loop. The DMD provided a falling-edge trigger at the onset of each displayed pattern. This trigger synchronized each displayed pattern with one exposure of the camera.
2.3. Image reconstruction
We reconstructed the high-resolution light-field image in two steps, a light-field reconstruction and a SIM reconstruction. For light-field reconstruction, we first obtained the lenslet parameters of the image using the guides within the Light Field Display software (Version 2010), an open-source, cross-platform, GPU-accelerated software package for real-time viewing of microscope light fields provided by the Stanford Computer Graphics Laboratory [22]. We then employed these parameters within the shift-and-add algorithm to refocus each of the 9 patterned-illumination raw light-field images over depth [23, 24]. Each group of 9 volumetric reconstructions created one high-resolution SIM reconstruction.
We built a custom SIM reconstruction algorithm based on a previously published two-step strategy [25]. The first step preprocessed the 9 volumetric light-field reconstructions obtained with patterned illumination on a layer-by-layer basis; at each depth, the algorithm normalized the intensity and subtracted the background from the group of nine images. The second step reconstructed the high-resolution SIM image by integrating the information from the nine pre-processed images [25]. For each image, the algorithms extracted the spatial components at the three primary frequencies and deconvolved each component using a Wiener filter with a FWHM of 4.4 µm by following a published method [25]. The algorithm then shifted the three components to their true position in frequency space based on the orientation and lateral spatial frequency of the grating. The combination of all nine frequency components (3 components in each of the 3 directions) in this representation formed the complete spatial frequency representation. The inverse Fourier transform of this representation generated the complete SIM reconstruction with high resolution.
2.4. Bead preparation and imaging
The 2D bead samples consisted of 0.5 µm diameter fluorescent beads solution (L3280-1ML, Sigma) diluted at 1:1000 in deionized water. We added 50 µL of the diluted solution on a microscope slide covered with a thin glass cover.
The 3D bead samples consisted of the same sub-micron beads diluted at 1:10000 in a solution of 1.5% agarose by weight and deionized water. We placed 100 µL of the mixed solution on top of a thick microscope slide, and then placed a thin cover glass on top of the solution after the solution solidified.
We put the samples on a holder supported by a motorized x-y-z stage. For 2D sample imaging, we moved the sample over the axial direction in 1 µm steps. For 3D sample imaging, we positioned the sample at the principal focal plane.
2.5. Tissue preparation
We painted a piece of Thorlabs lens tissue with a green office highlighter. We placed the tissue on top of a glass slide for imaging after the tissue dried, and covered the tissue with a cover slip for imaging.
2.6. Retina sample preparation
We used a Ai148:Pcp2-Cre mouse strain for retina immunohistology and imaging. We generated this mouse strain by breeding Ai148 (TIT2L-GC6f-ICL-tTA2)-D mice [26] with Pcp2-Cre mice [27]. We genotyped the resulting litter to ensure Cre-dependent GCaMP6f expression in retinal bipolar cells [27, 28]. We housed mice in a pathogen free facility with a 12 hr alternate day-night light cycle, and with access to food and water. We used mice between the age of 1 month and 1 year for imaging. No sex specific differences were found in the immunohistology results (n = 3 mice). We euthanized the animals under room light through decapitation. We enucleated the eyes from these animals, and fixed the eye in a solution of paraformaldehyde (PFA, 4% in PBS) for 30 minutes at room temperature. We rinsed the sample with phosphate buffered saline (PBS, pH 7.4) three times. Under a stereo microscope we hemisected each eye along the ora serrata, and then removed the vitreous attachment. We finally stored the eyes at 4°C in PBS with sodium azide for antibody treatment.
Prior to treating the retina in primary antibody, we blocked the retina sample with 3% normal donkey serum, 0.3% Triton-X100, and 0.02% sodium azide for 1-3 hr in PBS. We then used chicken anti-GFP (1:1000, A10262, Invitrogen) to stain cells expressing GCaMP6f and mouse anti-Pcp2 (1:500, sc-137064, Santa Cruz Biotechnology) to counter-stain for Pcp2 expressing bipolar cells at 4°C for 5-7 days. We then washed the retina sample 2-3 times in PBS for a total of 3 hours at room temperature. We finally incubated the samples overnight in a light isolating container at 4°C with secondary antibodies conjugated to Alexa Fluor 488 for GFP (1:500, A-11039, Invitrogen) and Alexa Fluor 555 for Pcp2 (1:500, A31570, Invitrogen) at 1:500. Following the secondary incubation, we washed the retina samples again in PBS, removed the sclera and mounted the samples on slides using Aqua-Poly/Mount (Polysciences). We stored the samples in a light isolating container at 4°C for subsequent imaging.
2.7. Zebrafish preparation and neuron detection
We imaged albino zebrafish expressing GCaMP6s under the huc promoter at 5-7 days post fertilization using the Tol2kit system [20]. Before imaging, we embedded the fish within low melting point agarose gel (1.5% by weight in egg water) and then drew the fish and agarose inside a glass capillary with an inner diameter of 1.0 mm. After the agarose gel solidified, we extruded two-thirds of the larvae body from the capillary. We removed the agarose in the front of the fish head to allow for breathing. We then mounted the capillary horizontally inside a 3D printed water tank filled with egg water. For all imaging experiments, we used an illumination intensity of 0.5 mW/mm2. We imaged all specimens at room temperature. After 3 min of the control recording, we gently injected 1 mL of 200 mM PTZ (P6500, Sigma-Aldrich) into the water tank to obtain a final PTZ concentration of 15 mM. We captured calcium activity with WF-LFM and SI-LFM during the same trial with interleaved blocks of 9 frames.
To locate putative neurons from the imaging movies, we first registered the reconstructed videos with a subpixel image registration algorithm [29]. We then spatially filtered the imaged planes with a Gaussian spatial filter with low and high spatial cutoff frequencies at 33 cycles/mm and 200 cycles/mm, respectively. Lastly, we extracted active neurons and calcium transients with the CaImAn neuron analysis method [30], and displayed the neurons with a depth-encoded color map. For active neurons, we quantified the SNR, defined as the ratio of peak ΔF/F to the baseline noise.
2.8. Ethics statement
The Duke Institutional Animal Care and Use Committee (IACUC) approved all animal experiments.
3. Results
3.1. SI-LFM illuminates samples with a high contrast volumetric grating pattern
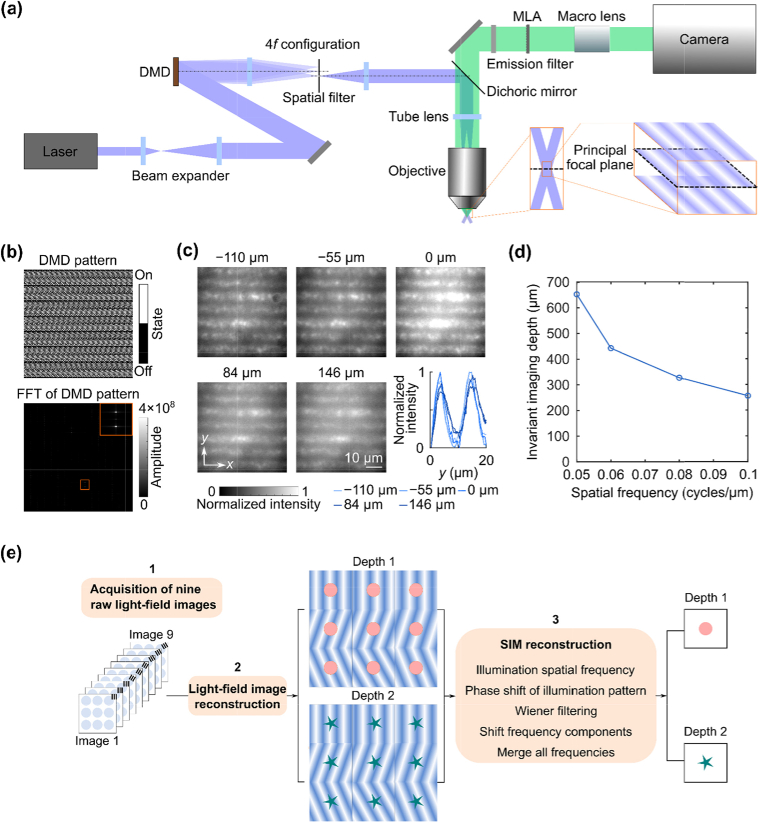
SI-LFM is a hybrid imaging modality that combines light-field detection with volumetric structured illumination. SI-LFM illuminated the sample volume with a 3D grating pattern that is invariant along the axial direction. We generated this pattern using a digital micromirror device (DMD) in the illumination path; the DMD modulated the phase and amplitude of the first-order diffraction pattern using the super-pixel holography method (Fig. 1(a)) [18]. We designed the holographic illumination pattern (Fig. 1(b), top; see Supplement 1 (4.5MB, pdf) , Fig. S1; Methods) to produce two symmetric plane waves with opposing lateral propagation (Fig. 1(b)). The interference between these plane waves within the volume around the principal focal plane of the objective generated the axially-invariant 3D sinusoidal grating illumination patterns [19].
Fig. 1.
– SI-LFM illuminates samples with an axially invariant pattern generated by super-pixel diffraction. (a) The schematic drawing shows the imaging setup of SI-LFM. A DMD in the imaging path formed the grating excitation pattern, and a conventional light-field imaging path collected the sample emission. (b) The DMD pattern (top) produced a first-order diffraction pattern in Fourier space (bottom); the top-right inset of the bottom panel is a zoomed-in view of the two peaks corresponding to two symmetric plane waves. (c) The interference between these two plane waves generated a grating pattern that was invariant at different axial positions. Bottom inset: The averaged intensity profile (averaged along the x-direction, n = 50 grating cycles) of the volumetric grating pattern with lateral spatial frequency 0.1 cycle/µm retained contrast over 250 µm of depth. The non-uniformity of the tape sample induced small, but repeatable, local intensity variations. (d) The invariant imaging depth decreased as the spatial frequency of the illumination pattern increased. (e) The image reconstruction includes a light-field reconstruction step and a SIM reconstruction step.
To experimentally characterize the grating quality along the axial direction, we illuminated the grating pattern on a fluorescent tape sample that could be moved axially and acquired the resulting fluorescence with conventional wide-field microscopy (Methods). When the lateral spatial frequency was 0.1 cycle/µm at the imaging plane, we consistently observed grating patterns with high contrast over an axial range of 256 µm (Fig. 1(c)); the modulation depth of the gratings at each depth throughout this invariant axial range was greater than 70% of the modulation depth of the grating at the principal focal plane (Fig. 1(c), bottom inset). The invariant axial range of volumetric grating illumination patterns decreased as lateral spatial frequency increased from 0.05 to 0.1 cycle/µm (Fig. 1(d)). This relationship occurred because high spatial frequency inputs require large relative angles between the two incident plane waves, resulting in a smaller interference range in the axial direction (see Supplement 1 (4.5MB, pdf) , Fig. S2). Because we targeted our illumination strategy for samples with an axial extent of ∼80 µm, we used 0.1 cycle/µm for all imaging experiments, which corresponded to 70% of the measured cutoff frequency of the conventional LFM system.
SI-LFM imaged the illuminated sample using conventional LFM (Fig. 1(e), Methods). We acquired nine volumetric light-field images, using illumination gratings with three different orientations and three phases at each orientation. We then reconstructed the high-resolution volume from these nine images through light-field reconstruction and subsequent SIM reconstruction at each layer of the volume. We also acquired images with non-patterned illumination for comparison.
3.2. SI-LFM provides superior resolution in imaging fluorescent beads samples
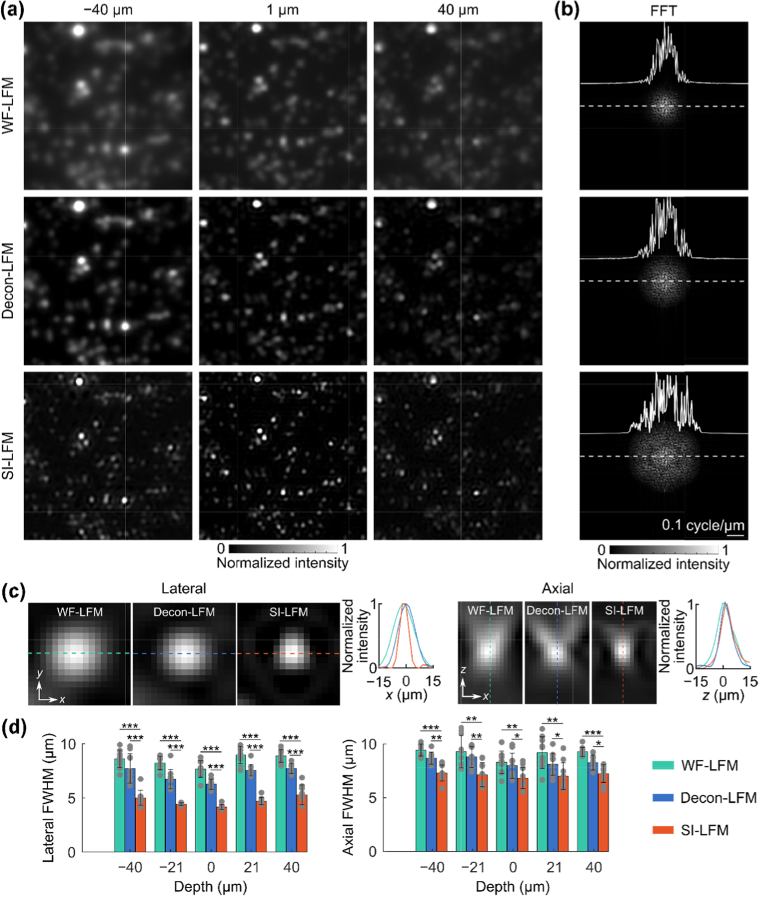
We validated the SI-LFM’s potential for high-resolution imaging by initially imaging 0.5 µm fluorescent beads. We first imaged a 2D sample using SI-LFM, conventional wide-field light-field microscopy (WF-LFM), and deconvolved light-field microscopy (Decon-LFM). We compared the images acquired by the three modalities at the depths of −40 μm, 0 μm, and 40 μm relative to the principal focal plane. At each depth, beads imaged by WF-LFM were blurry and had large spot sizes (Fig. 2(a), upper). The spot size of beads imaged by Decon-LFM was smaller than the spot size of beads imaged by WF-LFM, demonstrating the limited effectiveness of deconvolution (Fig. 2(a), middle). Beads imaged by SI-LFM were brighter and even more localized than beads imaged by Decon-LFM and WF-LFM (Fig. 2(a), bottom).
Fig. 2.
– SI-LFM improved resolution at multiple depths when imaging a two-dimensional fluorescent beads sample. (a) Images of fluorescent beads obtained through WF-LFM (upper), Decon-LFM (middle), and SI-LFM (bottom) showed that deconvolution and structured illumination improved the spot size. (b) Frequency analysis of the bead images demonstrated that the range of spatial frequency components in SI-LFM images was larger than the range of components in the images produced by the other two imaging modalities. We subtracted mean from the images to remove the constant component of the frequency representation at the origin. The profiles show the 1D frequency spectra along the dashed lines. (c) Zoomed-in images of beads along the lateral and axial directions show that the PSF obtained by SI-LFM was smaller than the PSF obtained by other light-field modalities. (d) Quantitative comparison between the FWHMs of the beads obtained by the three imaging modalities showed that the lateral and axial spot sizes of beads imaged by SI-LFM were significantly smaller than the lateral and axial spot sizes of beads imaged by the other two modalities (dots are individual data values; error bars are standard deviations; n = 10; ***p < 0.001, **p < 0.01, *p < 0.05, two-sided Wilcoxon rank-sum test).
To visualize these three modalities’ ability to capture high spatial frequencies, we calculated the Fourier transform of the images at z = 0 µm (Fig. 2(b)). The spatial frequency components produced by SI-LFM extended beyond the range of spatial frequencies produced by WF-LFM and Decon-LFM. We also quantified the spot size of the imaging modalities at each depth by fitting the intensity profiles of the beads to Gaussian profiles and computing the full-widths at half-maximum (FWHMs) along both lateral and axial directions (Fig. 2(c)). Beads imaged by SI-LFM had significantly smaller lateral and axial spot sizes than beads imaged by Decon-LFM and WF-LFM at nearly all depths (Fig. 2(d)). The lateral and axial spot sizes of the WF-LFM and Decon-LFM grew as the distance from the principal focal plane increased from 0 µm to 40 µm, but the spot sizes of SI-LFM were similar across multiple depths.
We next quantified the performance of WF-LFM, Decon-LFM, and SI-LFM through imaging of beads distributed over various depths. We plotted all the detected beads in a maximum intensity projection image with depth-encoded color, where beads close to the principal focal plane appear blue and beads far away from the principal plane appear red. When imaging the same beads, SI-LFM produced the smallest bead spots among all LFM imaging modalities (Fig. S3a-c). At the principal focal plane (z = 0 μm), the lateral spot size of beads imaged by SI-LFM was 0.65 of the lateral spot size of beads imaged by Decon-LFM, and 0.56 of the size of beads imaged by WF-LFM. In addition, both SI-LFM and Decon-LFM suppressed the background that was apparent in the WF-LFM image. The FWHMs in both the lateral and axial directions of beads over different depths were the smallest when imaged by SI-LFM among the three imaging modalities (Fig. S3d).
3.3. SI-LFM resolved fine fiber structures over multiple depths when imaging lens tissue
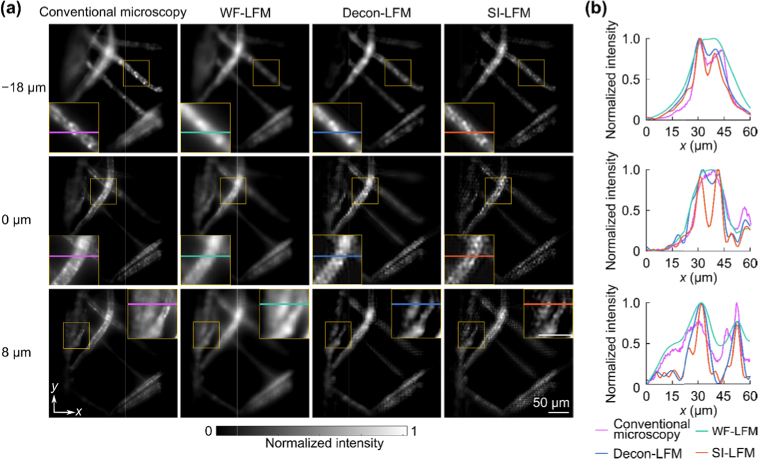
We next evaluated the resolution improvement of SI-LFM by imaging lens tissue marked with a green fluorescent highlighter (Methods). Analogous to our beads experiment, we performed volumetric imaging with WF-LFM, Decon-LFM, and SI-LFM. We also used conventional microscopy to sequentially scan all depths and produce a volumetric ground truth with high lateral resolution. Again, WF-LFM produced blurry images of tissue structure at all the three representative depths (Fig. 3(a)), while Decon-LFM resolved small features more clearly than WF-LFM. SI-LFM resolved the smallest features among the tissue fibers. The line intensity profiles across narrow lens tissue fibers also demonstrated the superior resolving power of SI-LFM (Fig. 3(b)).
Fig. 3.
– SI-LFM resolved tissue fibers with more details than WF-LFM. (a) At depths of −18 µm, 0 µm, and 8 µm, SI-LFM revealed fine structure within the fibers of tissue paper that were not visible within the images obtained with WF-LFM and Decon-LFM. A volumetric image stack acquired with conventional microscopy served as the ground truth. (b) At multiple depths, the intensity profiles of the features labeled with color lines also show that SI-LFM revealed structures with smaller widths, similar to the structural details captured by conventional microscopy.
3.4. SI-LFM reported sub-cellular neural structure when imaging scattering retina samples
Volumetric imaging can be used to characterize the structural profile and the laminar organization of genetically targeted cell types in the retina. Such deep, layer-by-layer imaging could be obscured by the retina’s high scattering coefficient (273 cm−1 at 514.5 nm) [31–33]. We examined the effect of scattering on the image quality of multiple light-field modalities. We imaged whole-mount samples of fixed retina from Ai148:Pcp2-Cre transgenic mice that expressed GCaMP6f in a subset of bipolar cells (BCs) [27, 34]. The BCs extend approximately 30 µm in the axial direction; the axons making synaptic contacts with retinal ganglion cells in the inner plexiform layer, while dendrites and somas occupy the outer plexiform layer and inner nuclear layer, respectively [35]. We first confirmed GCaMP6f expression in the soma and the axon by conventional epifluorescence imaging and confocal imaging (see Supplement 1 (4.5MB, pdf) , Fig. S4a-b).
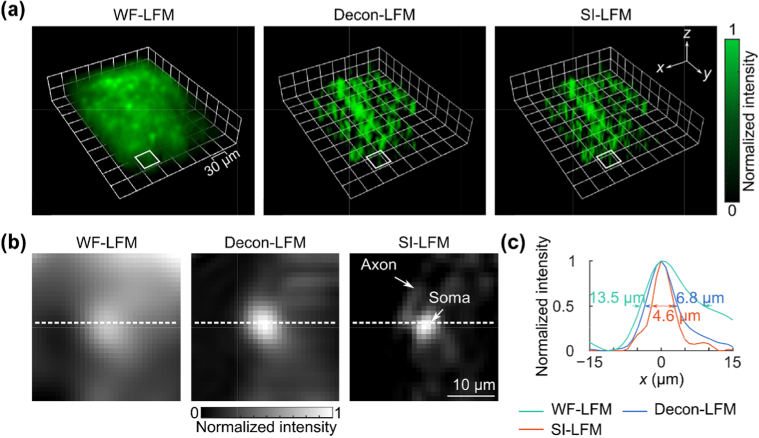
We then acquired volumetric images of the retina using the same three LFM modalities. The reconstructed 3D images from WF-LFM exhibited spatial blurring of the somatic and axonal structures (Fig. 4(a)); these images also displayed high background noise that progressively increased with depth (Fig. 4(a)). The reconstructed 3D images of the BCs from Decon-LFM resolved the locations of somas of individual BCs across the volume (Fig. 4(a)). The spatial resolution of BC structures in the images from SI-LFM was higher than the resolution from the same structures imaged by WF-LFM and Decon-LFM (Fig. 4(a)). SI-LFM produced the best images of BC sub-cellular structures, clearly showing both the BC soma and axons (Fig. 4(b)). The FWHMs of clearly resolved BC somas imaged by SI-LFM were smaller than the FWHMs of somas imaged by other imaging modalities (Fig. 4(c)). Although SI-LFM yielded increased clarity, we observed scattering induced deconvolution artifacts (see Supplement 1 (4.5MB, pdf) , Fig. S4c-e). Together, these experiments confirmed the superior resolution of SI-LFM among the three light-field imaging modalities when imaging in scattering tissue. Increased scattering with depth reduced the contrast of the excitation beam at depth; at z = –35 µm from the principal focal plane, the excitation grating pattern amplitude was only 5% of the amplitude of the pattern at 0 µm (see Supplement 1 (4.5MB, pdf) , Fig. S5a). This blurred excitation pattern limited the spatial resolution of deeper structures. For example, SI-LFM and Decon-LFM provided qualitatively similar images at z = −35 µm (see Supplement 1 (4.5MB, pdf) , Fig S5b and Fig S5c).
Fig. 4.
– SI-LFM resolved the sub-cellular structure of neurons labeled by GCaMP6f within a fixed mouse retina with more detail than WF-LFM. (a) Volumetric images showed that SI-LFM acquired sharp retina features missed by the other two imaging modalities. All three images are thresholded at a normalized intensity of 0.4 such that only values above this threshold are shown. Individual cells appear as refined footprints in the SI-LFM volumetric rendering, but lose resolution in the WF-LFM and Decon-LFM images. (b) The magnified views of the white boxes in Fig. 4(a) show the soma and axon of a neuron in the SI-LFM image, but not in images acquired by other modalities. (c) The intensity profile along the dashed line in panel (b) showed that among the three light-field imaging modalities, SI-LFM imaged the cell with the smallest FWHMs along the x-direction.
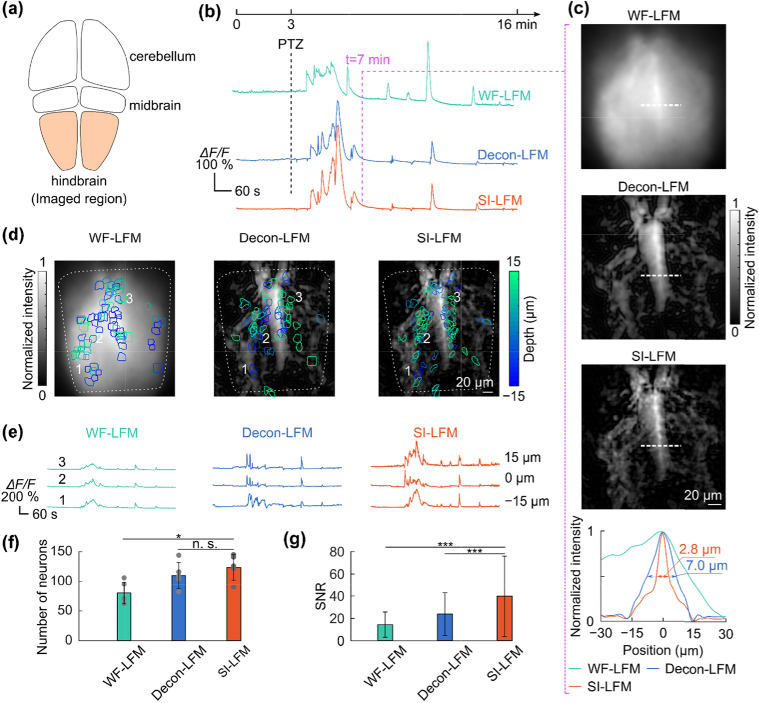
3.5. SI-LFM reported calcium activity in zebrafish larvae with high fidelity
We validated the performance of SI-LFM by recording the hindbrain activity (Fig. 5(a)) of GCaMP6s-expressing larval zebrafish. We imaged the same volume of zebrafish brain with WF-LFM, Decon-LFM, and SI-LFM, before and after the delivery of pentylenetetrazole (PTZ), a seizure-inducing chemical agent [36, 37] (Fig. 5(b)). To eliminate the motion induced by fish movement, we registered each individual frames to an average reference volume (see Supplement 1 (4.5MB, pdf) , Fig. S6; Methods). Before the delivery of PTZ, zebrafish hindbrain region showed only small calcium transients with amplitudes less than 10% ΔF/F. A few minutes after the delivery of PTZ, zebrafish showed large calcium transients with amplitudes greater than 200% ΔF/F (Fig. 5(b)). We assessed the image quality of the three light-field modalities by quantifying the intensity distribution through a midline feature of the hindbrain (Fig. 5(c); white dashed lines). SI-LFM resolved this feature with greater sharpness than the other two modalities (Fig. 5(c), bottom). The expression pattern in the dorsal region of the fish imaged by SI-LFM matched well to the light-sheet imaging results (Fig. S7). We also computationally detected the active neurons (Fig. 5(d); Methods) and calculated the calcium activity of each active neuron (Fig. 5(e); Methods). SI-LFM identified significantly more active neurons than Decon-LFM and WF-LFM (p = 0.03, two-sided Wilcoxon rank-sum test, n = 5 fish.). SI-LFM also provided significantly higher signal-to-noise-ratio (SNR) than Decon-LFM and WF-LFM from detected neurons (p < 10−40, two-sided Wilcoxon rank-sum test. n = 625, 555, and 405 neurons respectively from SI-LFM, Decon-LFM, and WF-LFM, from 5 fish).
Fig. 5.
– SI-LFM detected active neurons in imaging living zebrafish larvae with high fidelity. (a) The schematic illustration shows sub-regions of the larval zebrafish brain, including the hindbrain target. (b) PTZ delivered at 3 min (top) induced seizures and large calcium transients in zebrafish (bottom). (c) At 7 min, the image acquired with SI-LFM at 2 µm depth resolved more brain structures than images acquired with the other two imaging modalities. The intensity distribution of the features labeled along white dash lines (bottom) was sharper. (d) The depth-coded colored masks labeled the active neurons imaged with different imaging modalities. (e) The calcium activity of representative neurons located at different depths indicated that SI-LFM recorded the same neural activity with the highest change in fluorescence. (f) SI-LFM identified significantly more active neurons than the WF-LFM (dots are individual data values; *p < 0.05, n.s. – not significant, two-sided Wilcoxon rank-sum test, n = 5 fish; error bars are standard deviations). (g) The calcium transients recorded by SI-LFM had significantly higher SNR than the transients recorded by other LFM techniques (***p < 0.001, *p < 0.05, n.s. – not significant, two-sided Wilcoxon rank-sum test; n = 625, 555, and 405 neurons respectively from SI-LFM, Decon-LFM, and WF-LFM, all from 5 fish; error bars are standard deviations).
4. Discussion and conclusion
Recording the detailed structure and physiology of individual cells within large populations is important for studying various biological systems. Here, we demonstrated that SI-LFM achieved high resolution within volumetric imaging by combining WF-LFM with a volumetric structured illumination that is invariant over the imaging depth. SI-LFM accessed spatial frequency ranges beyond the frequency range of conventional LFM. By acquiring volumetric information in a single frame, SI-LFM avoided the scan-induced artifacts of other scanner-based volumetric imaging modalities. We demonstrated the feasibility of SI-LFM through the imaging of fluorescent beads, lens tissue, fixed mouse retina expressing GCaMP6f, and live zebrafish expressing GCaMP6s. When imaging static fluorescent beads, SI-LFM produced smaller spot size than the existing LFM techniques. When imaging lens tissue and fixed retina sample, SI-LFM resolved more structural features within the specimen than existing LFM techniques. SI-LFM identified more neurons when imaging live zebrafish than existing LFM techniques; the calcium activity of individual zebrafish neurons imaged by SI-LFM displayed approximately three times the SNR of calcium activity of individual zebrafish neurons imaged by WF-LFM.
We can further optimize SI-LFM in terms of the FOV and imaging depth. There is a tradeoff between the lateral resolution and the FOV [19]. Our current lateral FOV, approximately 200 µm by 250 µm, is limited by the size of the DMD. Recently developed DMD models have substantially more pixels and surface area than the DMD used in our experiments. The surface of the DMD is conjugate to the sample, so larger DMDs could expand the FOV to cover areas larger than the zebrafish brain. Strong scattering of the excitation and emission light by biological tissue limits the imaging depth of SI-LFM. Recently developed red fluorescent calcium indicators [38] employ red-shifted excitation and emission light; these wavelengths would extend the imaging depth of SI-LFM by maintaining the grating intensity distribution deep into tissue and by retaining the light-field information as fluorescence exits the tissue. Chemical clearing methods [39, 40] could likewise reduce the scattering of fixed tissue and extend the depth of SI-LFM imaging.
SI-LFM can further studies of fast biological processes by improving data processing speeds and providing new forms of volumetric information. First, SI-LFM provided better resolution when compared with existing, augmented LFM modalities. The high-resolution of SI-LFM could improve the separation of neighboring neurons, and thus minimize the errors when segmenting neurons. Accurate neuron spatial masks could speed up the data processing by both reducing the workload of unmixing neural activity from background fluorescence and improve the fidelity of detecting transients within the resulting ΔF/F traces [41, 42]. Second, SI-LFM could image a variety of genetically targeted fluorescent sensors with high resolution over large volumetric FOVs; this ability could potentially capture diverse anatomical and functional features simultaneously in large volumes of biological samples.
Like other imaging modalities, SI-LFM trades off speed, spatial resolution, and field of view due to limitations in the camera frame rate or excitation patterning. However, SI-LFM’s moderate imaging resolution and speed can benefit several biological imaging applications that examine the fixed or dynamic spatial details within volumetric imaging. One such application of SI-LFM could be nanoscale 3D structural imaging of tissue when combined with expansion microscopy. Expansion microscopy uniformly increases the size of a biological sample such that features beyond the diffraction limit before expansion become larger and more readily imaged [43]. The combination of expansion microscopy and SIM has revealed the organization of the drosophila synaptonemal complex [44]. Combining the 3D imaging capability of SI-LFM with expansion microscopy could enable volumetric imaging of small, sub-diffraction features and identify regions of structural connectivity on larger scales than currently imaged. The rapid volumetric imaging of cleared, expanded tissue could help quickly capture the connectivity patterns over entire small model organisms [45]. Another potential application of SI-LFM could be long-term imaging of volumetric structural dynamics, such as the development of zebrafish embryogenesis. Another potential application of SI-LFM could be long-term imaging of volumetric structural dynamics, such as the development of zebrafish embryogenesis. Although light-sheet microscopy [46] and confocal microscopy [47] have monitored morphological changes in the embryo, they employ mechanical scanning of optics or the sample. SI-LFM bypasses such scanning and could simplify the reconstruction process and improve the accuracy of the registration between layers over long time series, while tracking cellular displacement with high accuracy. Altogether, our validation of SI-LFM’s imaging performance over multiple samples indicates that SI-LFM could help advance many imaging applications. By providing volumetric high-resolution imaging without scanning, SI-LFM will facilitate the evolution of volumetric biomedical imaging of relatively transparent samples at high volumetric acquisition rates.
Funding
National Science Foundation, Directorate for Engineering10.13039/100000084 (1847540); National Institute of Neurological Disorders and Stroke10.13039/100000065 (1UF1NS107678); Arnold and Mabel Beckman Foundation10.13039/100000997; Vallee Foundation10.13039/100007204; Alfred P. Sloan Foundation10.13039/100000879; Brain Research Foundation10.13039/100000882.
Disclosures
The authors declare no conflicts of interest.
Data availability
Data underlying the results presented in this paper are not publicly available at this time but may be obtained from the authors upon reasonable request.
Supplemental document
See Supplement 1 (4.5MB, pdf) for supporting content.
References
- 1.Dana H., Sun Y., Mohar B., Hulse B. K., Kerlin A. M., Hasseman J. P., Tsegaye G., Tsang A., Wong A., Patel R., “High-performance calcium sensors for imaging activity in neuronal populations and microcompartments,” Nat. Methods 16(7), 649–657 (2019). 10.1038/s41592-019-0435-6 [DOI] [PubMed] [Google Scholar]
- 2.Ahrens M. B., Orger M. B., Robson D. N., Li J. M., Keller P. J., “Whole-brain functional imaging at cellular resolution using light-sheet microscopy,” Nat. Methods 10(5), 413–420 (2013). 10.1038/nmeth.2434 [DOI] [PubMed] [Google Scholar]
- 3.Gong Y., Wagner M. J., Li J. Z., Schnitzer M. J., “Imaging neural spiking in brain tissue using FRET-opsin protein voltage sensors,” Nat. Commun. 5, 1–11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prevedel R., Yoon Y.-G., Hoffmann M., Pak N., Wetzstein G., Kato S., Schrödel T., Raskar R., Zimmer M., Boyden E. S., “Simultaneous whole-animal 3D imaging of neuronal activity using light-field microscopy,” Nat. Methods 11(7), 727–730 (2014). 10.1038/nmeth.2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pégard N. C., Liu H.-Y., Antipa N., Gerlock M., Adesnik H., Waller L., “Compressive light-field microscopy for 3D neural activity recording,” Optica 3(5), 517–524 (2016). 10.1364/OPTICA.3.000517 [DOI] [Google Scholar]
- 6.Yoon .Y.-G., Wang Z., Pak N., Park D., Dai P., Kang J. S., Suk H.-J., Symvoulidis P., Guner-Ataman B., Wang K., “S parse decomposition light-field microscopy for high speed imaging of neuronal activity,” Optica 7(10), 1457–1468 (2020). 10.1364/OPTICA.392805 [DOI] [Google Scholar]
- 7.Li H., Guo C., Kim-Holzapfel D., Li W., Altshuller Y., Schroeder B., Liu W., Meng Y., French J. B., Takamaru K.-I., “Fast, volumetric live-cell imaging using high-resolution light-field microscopy,” Biomed. Opt. Express 10(1), 29–49 (2019). 10.1364/BOE.10.000029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cong L., Wang Z., Chai Y., Hang W., Shang C., Yang W., Bai L., Du J., Wang K., Wen Q., “Rapid whole brain imaging of neural activity in freely behaving larval zebrafish (Danio rerio),” Elife 6, e28158 (2017). 10.7554/eLife.28158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rumyantsev O. I., Lecoq J. A., Hernandez O., Zhang Y., Savall J., Chrapkiewicz R., Li J., Zeng H., Ganguli S., Schnitzer M. J., “Fundamental bounds on the fidelity of sensory cortical coding,” Nature 580(7801), 100–105 (2020). 10.1038/s41586-020-2130-2 [DOI] [PubMed] [Google Scholar]
- 10.Truong T. V., Holland D. B., Madaan S., Andreev A., Keomanee-Dizon K., Troll J. V., Koo D. E., McFall-Ngai M. J., Fraser S. E., “High-contrast, synchronous volumetric imaging with selective volume illumination microscopy,” Commun. Biol. 3(1), 74 (2020). 10.1038/s42003-020-0787-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang D., Xu S., Pant P., Redington E., Soltanian-Zadeh S., Farsiu S., Gong Y., “Hybrid light-sheet and light-field microscope for high resolution and large volume neuroimaging,” Biomed. Opt. Express 10(12), 6595–6610 (2019). 10.1364/BOE.10.006595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor M. A., Nöbauer T., Pernia-Andrade A., Schlumm F., Vaziri A., “Brain-wide 3D light-field imaging of neuronal activity with speckle-enhanced resolution,” Optica 5(4), 345–353 (2018). 10.1364/OPTICA.5.000345 [DOI] [Google Scholar]
- 13.Turcotte R., Liang Y., Tanimoto M., Zhang Q., Li Z., Koyama M., Betzig E., Ji N., “Dynamic super-resolution structured illumination imaging in the living brain,” Proc. Natl. Acad. Sci. 116(19), 9586–9591 (2019). 10.1073/pnas.1819965116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gustafsson M. G., Shao L., Carlton P. M., Wang C. R., Golubovskaya I. N., Cande W. Z., Agard D. A., Sedat J. W., “Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination,” Biophys. J. 94(12), 4957–4970 (2008). 10.1529/biophysj.107.120345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang B.-J., Meza V. D. P., Stelzer E. H., “csiLSFM combines light-sheet fluorescence microscopy and coherent structured illumination for a lateral resolution below 100 nm,” Proc. Natl. Acad. Sci. 114(19), 4869–4874 (2017). 10.1073/pnas.1609278114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heintzmann R., Huser T., “Super-resolution structured illumination microscopy,” Chem. Rev. 117(23), 13890–13908 (2017). 10.1021/acs.chemrev.7b00218 [DOI] [PubMed] [Google Scholar]
- 17.Descloux A., Müller M., Navikas V., Markwirth A., Van den Eynde R., Lukes T., Hübner W., Lasser T., Radenovic A., Dedecker P., “High-speed multiplane structured illumination microscopy of living cells using an image-splitting prism,” Nanophotonics 9(1), 143–148 (2019). 10.1515/nanoph-2019-0346 [DOI] [Google Scholar]
- 18.Goorden S. A., Bertolotti J., Mosk A. P., “Superpixel-based spatial amplitude and phase modulation using a digital micromirror device,” Opt. Express 22(15), 17999–18009 (2014). 10.1364/OE.22.017999 [DOI] [PubMed] [Google Scholar]
- 19.Yang J., Gong L., Xu X., Hai P., Shen Y., Suzuki Y., Wang L. V., “Motionless volumetric photoacoustic microscopy with spatially invariant resolution,” Nat. Commun. 8(1), 1–7 (2017). 10.1038/s41467-016-0009-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwan K. M., Fujimoto E., Grabher C., Mangum B. D., Hardy M. E., Campbell D. S., Parant J. M., Yost H. J., Kanki J. P., Chien C. B., “The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs,” Dev. Dyn. 236(11), 3088–3099 (2007). 10.1002/dvdy.21343 [DOI] [PubMed] [Google Scholar]
- 21.Chen T.-W., Wardill T. J., Sun Y., Pulver S. R., Renninger S. L., Baohan A., Schreiter E. R., Kerr R. A., Orger M. B., Jayaraman V., “Ultrasensitive fluorescent proteins for imaging neuronal activity,” Nature 499(7458), 295–300 (2013). 10.1038/nature12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng R., Levoy M., Brédif M., Duval G., Horowitz M., Hanrahan P., “Light field photography with a hand-held plenoptic camera,” Phys. Rev. X 2, 1–11 (2005). 10.1103/PhysRevX.4.011031 [DOI] [Google Scholar]
- 23.Wilburn B., Joshi N., Vaish V., Talvala E.-V., Antunez E., Barth A., Adams A., Horowitz M., Levoy M., “High performance imaging using large camera arrays,” in ACM Transactions on Graphics (TOG), 24(3)(ACM, 2005), 765–776. 10.1145/1073204.1073259 [DOI] [Google Scholar]
- 24.Tian L., Wang J., Waller L., “3D differential phase-contrast microscopy with computational illumination using an LED array,” Opt. Lett. 39(5), 1326–1329 (2014). 10.1364/OL.39.001326 [DOI] [PubMed] [Google Scholar]
- 25.Lal A., Shan C., Xi P., “Structured illumination microscopy image reconstruction algorithm,” IEEE J. Sel. Top. Quantum Electron. 22(4), 50–63 (2016). 10.1109/JSTQE.2016.2521542 [DOI] [Google Scholar]
- 26.Daigle T. L., Madisen L., Hage T. A., Valley M. T., Knoblich U., Larsen R. S., Takeno M. M., Huang L., Gu H., Larsen R., “A suite of transgenic driver and reporter mouse lines with enhanced brain-cell-type targeting and functionality,” Cell 174(2), 465–480.e22 (2018). 10.1016/j.cell.2018.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Q., Ivanova E., Ganjawala T. H., Pan Z.-H., “Cre-mediated recombination efficiency and transgene expression patterns of three retinal bipolar cell-expressing Cre transgenic mouse lines,” Invest. Ophthalmol. Vis. Sci. 19, 1310 (2013). 10.1167/iovs.04-1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oberdick J., Smeyne R. J., Mann J. R., Zackson S., Morgan J. I., “A promoter that drives transgene expression in cerebellar Purkinje and retinal bipolar neurons,” Science 248(4952), 223–226 (1990). 10.1126/science.2109351 [DOI] [PubMed] [Google Scholar]
- 29.Guizar-Sicairos M., Thurman S. T., Fienup J. R., “Efficient subpixel image registration algorithms,” Opt. Lett. 33(2), 156–158 (2008). 10.1364/OL.33.000156 [DOI] [PubMed] [Google Scholar]
- 30.Giovannucci A., Friedrich J., Gunn P., Kalfon J., Brown B. L., Koay S. A., Taxidis J., Najafi F., Gauthier J. L., Zhou P., “CaImAn an open source tool for scalable calcium imaging data analysis,” Elife 8 8, e38173 (2019). 10.7554/eLife.38173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sardar D. K., Yust B. G., Barrera F. J., Mimun L. C., Tsin A. T., “Optical absorption and scattering of bovine cornea, lens and retina in the visible region,” Lasers Med. Sci. 24(6), 839–847 (2009). 10.1007/s10103-009-0677-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonzalez-Andrades M., de la Cruz Cardona J., Ionescu A. M., Mosse C. A., Brown R. A., “Photographic-based optical evaluation of tissues and biomaterials used for corneal surface repair: A new easy-applied method,” PLoS One 10, e0142099 (2015). 10.1371/journal.pone.0142099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacques S. L., “Optical properties of biological tissues: a review,” Phys. Med. Biol. 58(11), R37–R61 (2013). 10.1088/0031-9155/58/11/R37 [DOI] [PubMed] [Google Scholar]
- 34.Dhingra A., Sulaiman P., Xu Y., Fina M. E., Veh R. W., Vardi N., “Probing neurochemical structure and function of retinal ON bipolar cells with a transgenic mouse,” J. Comp. Neurol. 510(5), 484–496 (2008). 10.1002/cne.21807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolb H., Fernandez E., Nelson R., “Simple anatomy of the retina–webvision: the organization of the retina and visual system,” (1995). [PubMed]
- 36.Turrini L., Fornetto C., Marchetto G., Müllenbroich M., Tiso N., Vettori A., Resta F., Masi A., Mannaioni G., Pavone F., “Optical mapping of neuronal activity during seizures in zebrafish,” Sci. Rep. 7(1), 3025 (2017). 10.1038/s41598-017-03087-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J., Baraban S. C., “Network properties revealed during multi-scale calcium imaging of seizure activity in zebrafish,” Eneuro 6 (2019). doi: 10.1523/ENEURO.0041-19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohr M. A., Bushey D., Aggarwal A., Marvin J. S., Kim J. J., Marquez E. J., Liang Y., Patel R., Macklin J. J., Lee C.-Y., “jYCaMP: An optimized calcium indicator for two-photon imaging at fiber laser wavelengths,” Nat. Methods 8, 1–4 (2020). 10.1101/816660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hama H., Kurokawa H., Kawano H., Ando R., Shimogori T., Noda H., Fukami K., Sakaue-Sawano A., Miyawaki A., “Sca le: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain,” Nat. Neurosci. 14(11), 1481–1488 (2011). 10.1038/nn.2928 [DOI] [PubMed] [Google Scholar]
- 40.Chung ., Wallace J., Kim S.-Y., Kalyanasundaram S., Andalman A. S., Davidson T. J., Mirzabekov J. J., Zalocusky K. A., Mattis J., Denisin A. K., “Structural and molecular interrogation of intact biological systems,” Nature 497(7449), 332–337 (2013). 10.1038/nature12107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Theis L., Berens P., Froudarakis E., Reimer J., Rosón M. R., Baden T., Euler T., Tolias A. S., Bethge M., “Benchmarking spike rate inference in population calcium imaging,” Neuron 90(3), 471–482 (2016). 10.1016/j.neuron.2016.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soltanian-Zadeh S., Sahingur K., Blau S., Gong Y., Farsiu S., “Fast and robust active neuron segmentation in two-photon calcium imaging using spatiotemporal deep learning,” Proc. Natl. Acad. Sci. 116(17), 201812995 (2019). 10.1073/pnas.1812995116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wassie A. T., Zhao Y., Boyden E. S., “Expansion microscopy: principles and uses in biological research,” Nat. Methods 16(1), 33–41 (2019). 10.1038/s41592-018-0219-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cahoon C. K., Yu Z., Wang Y., Guo F., Unruh J. R., Slaughter B. D., Hawley R. S., “Superresolution expansion microscopy reveals the three-dimensional organization of the Drosophila synaptonemal complex,” Proc. Natl. Acad. Sci. 114(33), E6857–E6866 (2017). 10.1073/pnas.1705623114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGurk L., Morrison H., Keegan L. P., Sharpe J., O’Connell M. A., “Three-dimensional imaging of Drosophila melanogaster,” PLoS One 2, e834 (2007). 10.1523/ENEURO.0041-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keller P. J., “In vivo imaging of zebrafish embryogenesis,” Methods 62(3), 268–278 (2013). 10.1016/j.ymeth.2013.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugiyama M., Sakaue-Sawano A., Iimura T., Fukami K., Kitaguchi T., Kawakami K., Okamoto H., Higashijima S.-i., Miyawaki A., “Illuminating cell-cycle progression in the developing zebrafish embryo,” Proceedings of the National Academy of Sciences 106(49), 20812–20817 (2009). 10.1073/pnas.0906464106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data underlying the results presented in this paper are not publicly available at this time but may be obtained from the authors upon reasonable request.