Abstract
Patient-derived cancer organoids (PCOs) are in vitro organotypic models that reflect in vivo drug response, thus PCOs are an accessible model for cancer drug screening in a clinically relevant timeframe. However, current methods to assess the response of PCOs are limited. Here, a custom swept-source optical coherence tomography (OCT) system was used to rapidly evaluate volumetric growth and drug response in PCOs. This system was optimized for an inverted imaging geometry to enable high-throughput imaging of PCOs. An automated image analysis framework was developed to perform 3D single-organoid tracking of PCOs across multiple time points over 48 hours. Metabolic inhibitors and cancer therapies decreased PCOs volumetric growth rate compared to control PCOs. Single-organoid tracking improved sensitivity to drug treatment compared to a pooled analysis of changes in organoid volume. OCT provided a more accurate assessment of organoid volume compared to a volume estimation method based on 2D projections. Single-organoid tracking with OCT also identified heterogeneity in drug response between solid and hollow PCOs. This work demonstrates that OCT and 3D single-organoid tracking are attractive tools to monitor volumetric growth and drug response in PCOs, providing rapid, non-destructive methods to quantify heterogeneity in PCOs.
1. Introduction
Patient-derived cancer organoids (PCOs) are three-dimensional (3D) in vitro cultures isolated from tumor biopsies that recapitulate features of the in vivo tumor, including gene expression, cell morphology, and cell phenotype [1–3]. PCOs reflect in vivo drug sensitivity, offering a platform to perform high-throughput drug screens. Drug screens using PCOs have the potential to optimize treatment selection, improve treatment efficacy, and reduce side effects and toxicity [4,5]. Subclones captured in PCOs have also been shown to mirror the heterogeneity found in the original tumors, which often leads to treatment resistance [1,6,7]. This heterogeneity must be considered for robust predictions of patient drug sensitivity [7,8]. Heterogeneity manifests itself in PCOs both within (cell-level) and between (organoid-level) organoids, which motivates the assessment of large populations of PCOs to determine drug sensitivity. However, current methods to assess in vitro drug response in PCOs are limited because they are generally destructive, prohibiting time-course measurements (e.g., immunohistochemistry, single-cell sequencing/imaging, flow cytometry) or lack the ability to volumetrically image individual PCO size and morphology (e.g., brightfield microscopy, plate reader assays). There is a need to quantify volumetric growth and drug response in PCOs using non-destructive tools that are sensitive to organoid-level heterogeneity.
Optical coherence tomography (OCT) is a label-free imaging technique that detects light backscattered from a sample to reconstruct depth-resolved structure. OCT is based on low-coherence interferometry and provides rapid acquisition times of large 3D volumes (hundreds of mm3). Therefore, OCT can capture micron-scale volumetric information from large populations of PCOs. Additionally, OCT can track the volumetric growth of individual PCOs over time without destructive processing or exogenous labels. Previous studies have used OCT to monitor drug response in PCOs [9–15]. However, these previous studies have considered only a single PCO or pooled response across all PCOs in a well over time, and have not used single-organoid tracking to monitor heterogeneity in response on the individual PCO level.
Heterogeneity at the organoid-level results in sub-populations of organoids with differing treatment sensitivities [8,16]. Therefore, accurate OCT measurements of drug response requires imaging many PCOs over time to capture the response of these sub-populations. To compensate for organoid-level variability, the relative volumetric change from baseline for each PCO must also be captured. Accordingly, quantitative image analysis methods to automatically quantify and track organoids over time are needed for robust OCT-based high-throughput drug screens.
Here, we provide the first demonstration of 3D single-organoid tracking of PCOs using OCT. An image analysis framework was developed to automatically quantify organoid-level changes in volumetric growth. We measured the response to two chemotherapies (cisplatin, paclitaxel) and two metabolic inhibitors (sodium cyanide, 2-deoxy-glucose) across two patient-derived colorectal PCO lines. A quantitative image analysis pipeline was developed to track volumetric growth and drug response in each PCO before treatment and over multiple time points post-treatment up to 48 hours. Linear mixed-effect models were then applied to these organoid-level volumes over time, which provided improved sensitivity to drug response compared to pooling organoid volume over time (i.e., averaging all PCO volumes in a well, treatment group, or time point). Direct volume measurement using OCT provided more accurate quantification of PCO volume compared to a volume estimation technique based on 2D projections, similar to volume estimates commonly used with brightfield microscopy. This 2D volume estimation error was minimized in PCOs that were highly spherical. Lastly, volumetric growth rate was found to be heterogeneous within one patient, and this heterogeneity was attributed to PCO morphological sub-type (hollow or solid). This work demonstrates that OCT and 3D single-organoid tracking are attractive tools to monitor volumetric growth and drug response in PCOs, and that 3D single-organoid tracking provides improved sensitivity to PCO drug response and heterogeneity.
2. Methods
2.1. Patient-derived cancer organoid culture
PCOs were generated from two patient-derived metastatic colorectal cancer lines (P1 and P2), according to a previously published protocol [17]. The base culture medium used was DMEM/F12 (ThermoFisher) supplemented with 10% FBS (Sigma), and 1% Penicillin-Streptomycin (Sigma). Briefly, previously grown PCOs were singularized with 0.25% trypsin, resuspended in base medium, and mixed with Matrigel (Corning # 354234) at a 1:1 ratio. The cell-Matrigel mixture was pipetted onto the glass surface of each well in a 24-well glass bottom plate (black frame, #0 cover glass, Cellvis, P24-0-N). The plate was incubated at 37°C for 2-3 minutes to allow the mixture to solidify and then inverted to ensure PCOs were suspended in the Matrigel. The mixture was left to solidify for at least 20 minutes, then base medium supplemented with Wnt3a-conditioned medium (1:1 ratio) was added to each well. Medium was refreshed every two days. Aberrant Wnt/β-catenin signaling is a hallmark of colorectal cancer, and previous studies show that medium conditioned with Wnt3a supports colorectal PCO growth [18]. Wnt3a-conditioned medium is generated by harvesting medium in which murine L Wnt3a cells (ATCC, CRL-2647) have been cultured.
2.2. Treatment protocol
Five wells for each PCO line were grown for 14 days before treatment. At day 14, media in the wells were replaced with media containing sodium cyanide, 2-deoxy-glucose (2DG), cisplatin, or paclitaxel (Fig. 1(B), Drug Treatment). Media in the control wells were refreshed with unaltered media (no treatments). Specifics of each treatment group, including the mechanism of action, concentration used, and number of PCOs per group, are listed in Table 1.
Fig. 1.
Optical Coherence Tomography and 3D Single-Organoid Tracking of Patient-Derived Cancer Organoids. An overview of the custom OCT instrument, experimental protocol, and quantitative image analysis pipeline. (A) A diagram of the custom swept-source OCT instrument used in this study. The 100kHz swept-source laser used for this study had a center wavelength of 1040nm and a bandwidth of 109nm. The off-axis inverted geometry enabled imaging of patient-derived cancer organoids in a standard glass-bottom multiwell plate. BD: Balanced detector, GP: XY galvanometer pair, FBG: fiber Bragg grating, Circ.: Fiber circulator, DC: Dispersion compensation, M: mirror. AI/AO: analog in/analog out. (B) Experimental protocol illustration including drug treatment and imaging time points. (C) Graphical representation of the image analysis pipeline, which includes registration of all frames in each image time series, 3D organoid segmentation, single-organoid tracking, and quantification of organoid volume. Scale bar: 500µm.
Table 1. Treatment groups, drug concentrations, and number of PCOs (n) assessed for P1 and P2 lines.
| Treatment Group | Drug Class | Mechanism of Action | Concentration | n (P1) | n (P2) |
|---|---|---|---|---|---|
| Sodium Cyanide [19] | Metabolic Inhibitor | Mitochondrial Complex IV Inhibition | 4mmol/L | 28 | 23 |
| 2DG [20] | Metabolic Inhibitor | Glucose-6-Phosphate Production Inhibitor | 10mmol/L | 24 | 26 |
| Cisplatin [21] | Chemotherapy | DNA Replication Inhibition | 33µmol/L | 21 | 14 |
| Paclitaxel [22] | Chemotherapy | Microtubule Stabilization | 0.5µmol/L | 16 | 17 |
| Control (No Treatment) | n/a | n/a | n/a | 35 | 18 |
|
| |||||
| Total | 124 | 98 | |||
2.3. Optical coherence tomography imaging
All OCT measurements were performed on a custom inverted swept-source OCT system (Fig. 1(A)) [23]. The swept-wavelength source (Axsun Technologies) had a center wavelength of 1040nm, bandwidth of 109nm, and a line rate of 100kHz. The calculated axial resolution was 6.5µm in culture medium (n = 1.33 at 1040nm). The lateral resolution was 16µm (Thorlabs, LSM03-BB, NA 0.55). This low numerical aperture scan lens was chosen to ensure that the PCOs in culture were within the depth-of-focus of the lens. A line clock was generated using a fiber Bragg grating with narrowband reflectivity at 990nm (AC Photonics) and a fast photodetector (Newport, 1811-AC-FC, 125 MHz bandwidth). The signal from the balanced detector (Newport, 1617-AC-FC, 800MHz bandwidth) and line clock photodetector were digitized using a high-speed digitizer (AlazarTech, ATS9350, 12 bit, 500 MS/s). The 90:10 and 50:50 fiber-coupled splitters were wideband (+100nm) with a center wavelength of 1064nm (Thorlabs, 90:10 TW1064R2A2A, 50:50 TW1064R5A2A). The circulators were wideband and polarization insensitive (AC Photonics). The beam was scanned using an XY galvanometer pair (Cambridge, 6210H). The scan waveform and frame trigger was output using an analog in/analog out data acquisition (DAQ) board (National Instruments, BNC-2110 and PCIe-6353). The computer used for acquisition was custom built with an Intel i7 quad-core processor, 32GB DDR3 RAM, GTX 550 Ti video card, and 240GB SSHDD. To eliminate specular reflection from the air-glass interface of the glass-bottom multiwell plate, the scan lens was mounted on a custom rotational mount to tilt the lens and XY galvanometers off-axis by 9° in the X direction.
Each well within a 24-well plate was imaged with a 6mm x 6mm lateral field-of-view (XY pixel size: 6µm). The volumes contain 1000 A-scans per B-scan, 1000 B-scans per volume, and 4 repeated B-scans per position. The A-scan length was 1337 pixels. Standard OCT processing was used to calculate intensity images [24] and the repeated B-scans were averaged for each position. Data was acquired pre-treatment, immediately post-treatment and at 12 hours, 24 hours, 36 hours, and 48 hours post-treatment.
The XYZ stage repositioning was automated with µManager [25]. OCT acquisition was performed using a custom LabView script. For each of the 2 plates, a total of five wells were imaged. Total imaging time per plate was less than 5 minutes, capturing 124 PCOs across five wells for P1 and 98 PCOs across five wells for P2 (Table 1). Note that the first well of the 24-well plate was used to assist in positioning the plate before imaging, which enabled reproducible measurements of the same field-of-view over multiple time-points (Fig. 1(B)).
2.4. Quantitative image analysis
A framework for automated volumetric image analysis was developed to process OCT volumes and track each PCO in 3D across multiple time points (Fig. 1(C)). This extends previous work that developed 2D single-organoid tracking for autofluorescence images [26]. Linkage of each organoid over time was performed via a 3D centroid matching-based approach with single-particle tracking. This pipeline was implemented using MATLAB (2019b, Mathworks), except when specifically noted.
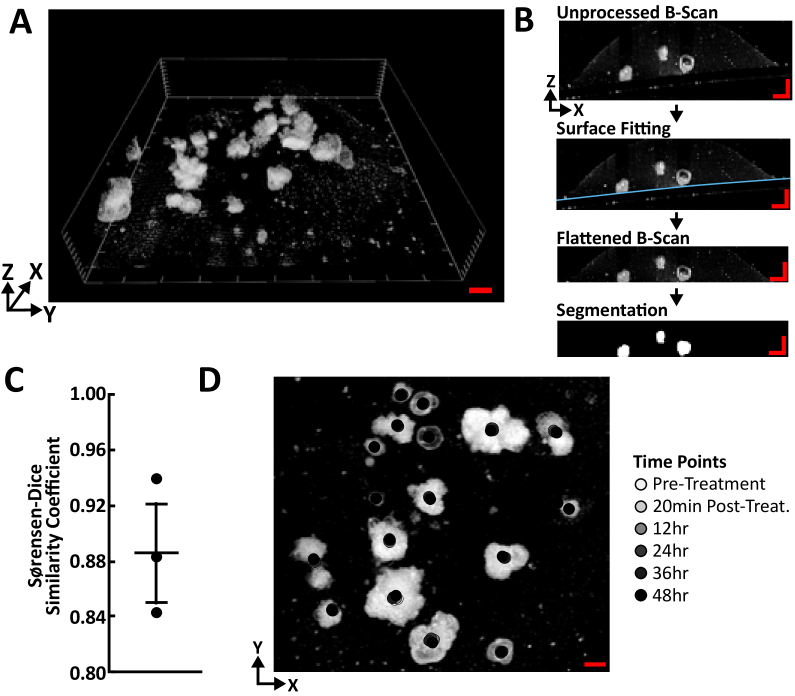
Preprocessing of each OCT volume was required before reliable segmentation and single-organoid tracking could be achieved (Fig. 1(C)). The goal here was to take OCT volumes that were captured off-axis and included the glass bottom of the multiwell plate and provide a flattened OCT volume that contains only organoids (Fig. 2(A),(B)). Figure 2(B) shows the image processing workflow for OCT volume image processing. First, the glass bottom was initially detected by calculating the Z gradient of the OCT volume and recording the coordinate of the maximum Z gradient value, which corresponded to the location of the air-glass interface. This provided a two-dimensional (2D) matrix of the location of the air-glass interface in Z for each XY location. A 3D second-order polynomial surface was then fit to the air-glass interface using least absolute residual (LAR) fitting (curvefit, MATLAB). A constant value based on the thickness of a #0 coverslip (n = 1.50 at 1040 nm) was added to the fitted coordinate values, which moved the coordinate values to the location of the glass-medium interface. These coordinate values were then used to shift each A-scan in the Z direction to yield a flattened volume that contained only organoids in the volumetric field-of-view. There was typically a large region in the volume in the Z direction that did not contain organoids, therefore each volume was cropped to contain only 200 pixels in Z. The final volume size was 1000 pixels by 1000 pixels by 200 pixels.
Fig. 2.
Quantitative Image Analysis of OCT Volumes A quantitative image analysis workflow was developed that enabled volumetric growth tracking of PCOs using OCT. (A) Representative OCT volume of PCOs pre-treatment. (B) Tracking of volumetric growth of organoids was achieved using an image processing workflow shown here with OCT B-scans for clarity. The goal was to correct for the off-axis imaging geometry and provide a flattened OCT volume that contains only organoids. The air-media interface was detected in 3D and used to shift each A-scan in the Z direction to yield a flattened volume that contained only organoids in the volumetric field-of-view. A segmentation algorithm based on OCT intensity was then used to segment PCOs. (C) The automated segmentation algorithm was compared against manual segmentation. The Sørenson-Dice similarity coefficients for n = 3 pairs of automatically and manually segmented OCT volumes are plotted here as individual data points along with the mean and standard error. (D) Representative 3D single-organoid tracking results for an OCT time course. Each organoid was tracked over multiple time points, and the tracked centroid is displayed as an overlay to a 2D maximum intensity projection of pre-treatment OCT volume. Scale bar: 500µm.
Registration of each OCT volume time series was achieved using both a registration mark and rigid image registration (Fig. 1(B)). A registration mark was etched on the glass bottom of the first well of each plate using a diamond-tip scribe and was referenced at every time point to precisely position the plate before imaging to minimize XYZ drift. After acquisition, XY drift between frames was corrected through rigid image registration, which registers two frames by maximizing the cross-correlation of pixel values (dftregistration, MATLAB Central File Exchange) [27,28]. The maximum intensity projection (MIP) of the preprocessed OCT volumes at each time point was used to calculate the XY shift values needed to align the volumes.
PCO segmentation was performed on each OCT volume independently to capture changes in organoid volume (Fig. 1(C)). A segmentation algorithm based on OCT intensity segmented PCOs after OCT volume preprocessing. First, the PCOs were initially segmented using two-class k-means clustering (kmeans, MATLAB). Postprocessing of the segmented regions involved filling holes in the volumes (imfill, MATLAB). Then any segmented region smaller than 5 pixels by 5 pixels by 5 pixels was removed (bwareaopen, MATLAB) to set a lower bound on the organoid size of ∼1.5 × 10−5 mm3 (equivalent organoid diameter of 32 µm). Filling holes in the PCO volume was performed for PCOs with either a solid morphology (cellular core) or hollow morphology (non-cellular core) [26,29,30]. The volume and 3D centroid of each segmented PCO was measured for each time point (regionprops3, MATLAB). The voxel size (6 µm x 6 µm x 3.5 µm) was used to scale the volume measurements from pixels to mm3. The performance of the automated segmentation algorithm was compared against manual segmentation. OCT volumes were manually segmented and the Sørenson-Dice similarity coefficient was calculated for pairs of automatically segmented and manual segmented volumes (Fig. 2(C), n = 3, mean ± SEM = 0.89 ± 0.03).
Tracking of each PCO over time was achieved using an open-source single-particle tracking tool (Fig. 1(C), TrackMate, ImageJ/FIJI) [31]. TrackMate achieves single-particle tracking by detecting particles in each frame, linking detected particles from frame-to-frame, and combining all links into the most likely tracks [32]. Volume time series were generated using MATLAB with 3D Gaussian particles (kernel: 10 × 10 × 10 pixels) at the 3D centroid of each segmented PCO. These centroids were then identified in each frame using the difference-of-Gaussian blob detector (options: 10 pixel diameter, no spot filtering). The linear assignment problem (LAP) algorithm was chosen to perform particle tracking because it is sensitive to events such as particle drift, particle merging, or over-/under-segmentation (options: 200 pixel maximum linking distance, 200 pixel max gap-closing distance, 2 frame max gap-closing distance). All key parameters were initially chosen by visual inspection of the tracking results for the P1 control time series, and then applied to all other image time series. Once completed for an OCT volume time series, the single-organoid tracking results were used to link the organoid volume measurements over time.
2.5. Organoid volume estimation and comparison
One approach for measuring organoid volume involves diameter measurements from 2D images of organoids (e.g., brightfield microscopy) that are used in an equation to estimate the organoid volume [10,33,34]. This approach assumes that each PCO is spherical (Eq. (1)).
| (1) |
where d is the diameter of the 2D projection of each PCO. To create 2D images for organoid diameter measurements, first MIPs were calculated for each pre-treatment OCT volume. These provide similar information to brightfield microscopy images as both OCT and brightfield use light scattering as contrast. To automatically measure the diameter of each organoid, 2D PCO segmentation was performed corresponding to the 3D case described above. 2D segmentation was achieved using two-class k-means clustering (kmeans, MATLAB). Postprocessing of the segmented regions involved filling holes (imfill, MATLAB). Then any segmented region smaller than 5 pixels by 5 pixels was removed (bwareaopen, MATLAB) to set a lower bound on the organoid size of approximately 32 µm diameter. The diameter was then quantified for each 2D segmented organoid (regionprops, MATLAB), and these diameters were used to calculate the estimated organoid volume (Eq. (1)). The estimated organoid volume (Eq. (1), “2D Est.”) was then compared to the measured organoid volume (regionprops3, MATLAB, “OCT”) for each organoid at pre-treatment, and the volume estimation error between the two values was quantified by calculating the percent error ().
Sphericity is a measure of how close a 3D volume is to a sphere and is based on variables (volume and surface area) that are easily measured for each segmented PCO [35]. Measuring organoid sphericity for each PCO enables a quantitative investigation of the error between direct volume measurement using OCT and volume estimation from 2D diameter measurements for organoids with different shapes.
| (2) |
where V and SA are the organoid volume and surface area measured using 3D segmentation of each PCO from OCT volumes (regionprops3, MATLAB).
2.6. Statistics
All statistical analyses were performed using MATLAB or R. Open-source MATLAB toolboxes used were Gramm, a data visualization and statistics package, and drtoolbox, a dimensionality reduction package [36,37]. Open-source R toolboxes included nlme, a mixed effect modeling package, and lsmeans, a least-squares means estimation package [38,39].
Organoid-level-normalized time series were calculated for each organoid as Xpost-treatment/Xpre-treatment_organoid, where X is the organoid volume. Linear mixed-effect models were used to analyze these organoid-level-normalized time series for each treatment and PCO line, specifically a longitudinal random-intercept model was used (nlme, R) [38]. Random-intercept models compensate for organoid variability at pre-treatment, which enables a more accurate assessment of individual PCO drug response [40,41]. A random-intercept model was specified as , where Y is the pre-treatment-normalized organoid volume, Time is a categorical variable encoding time, Treatment is a categorical variable encoding the five treatment groups, Organoid is the organoid-level-normalized value at 20 minutes post-treatment, and Error is the random error in the model. Time is defined as a categorical variable to avoid the assumption of linearity between time and organoid volume. Organoid here is encoded as a random effect to account for variability at the organoid-level. Pre-treatment values were excluded from the analysis because all time points were normalized to pre-treatment values (i.e., pre-treatment variance is one). A first-order autoregressive (i.e., AR(1)) covariance structure was used to account for the correlations found in these time-course data, where the correlation between time points decreases with increasing separation in time. Pairwise comparisons were performed between all treatment groups at each time point using least-squares means (lsmeans, R) [39]. Tukey’s Honest Significant Difference method was used to account for multiple comparisons.
Comparison between single-organoid tracking and pooled analysis was performed. The pooled analysis was the same as the single-organoid tracking analysis, except for the normalization approach and statistical model. To mimic pooled organoid data without organoid tracking, data for each organoid was normalized to the well-level pre-treatment mean as Xpost-treatment/Xpre-treatment_well, where X is the organoid volume. A fixed effects model was specified as , where Y is the well-level-normalized organoid volume, Time is a categorical variable encoding time, Treatment is a categorical variable encoding the five treatment groups, and Error is the random error in the model. Pre-treatment values were also not included in the analysis. All other parts of the pooled analysis were the same as the organoid-level analysis including the AR(1) covariance structure, least-squares mean for pairwise comparisons, and correction for multiple comparisons.
The difference between OCT-measured organoid volume and 2D-estimated organoid volume was tested using the non-parametric Wilcoxon Signed Rank test. The correlation between volume estimation error and OCT-measured organoid volume was quantified using Spearman’s correlation coefficient. The correlation between volume estimation error and organoid sphericity was quantified using Spearman’s correlation coefficient.
For the growth rate heterogeneity analysis, single-component Gaussian distributions were fit to the pre-treatment normalized organoid volume at 48 hours post-treatment for each of the treatment groups. Single-component Gaussian were chosen from visual inspection of the data, which was unimodal and normally distributed for each treatment group. Then, PCOs with hollow morphologies were identified via visual inspection. The Z-score for all PCOs were calculated and stratified into hollow or solid morphology groups, where X is the volume of an organoid in a treatment group, µTreatment is the mean volume of all organoids within that treatment group, and σTreatment is the variance of all organoids within that treatment group. The Z-score enables comparison between treatment groups. The non-parametric Mann-Whitney U test was used to test the difference between the hollow and solid morphology groups.
3. Results
3.1. Drug response reduces organoid volumetric growth rate
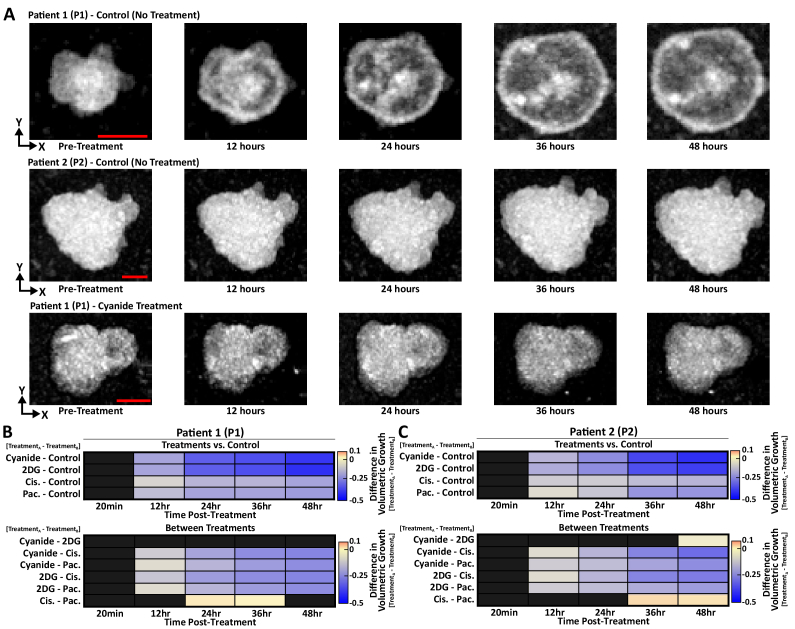
Drug response changes PCO volume or volumetric growth rate. Figure 3(A) shows representative MIP of individual PCOs imaged over time, including one each from the P1 and P2 control (no treatment) groups and one from the P1 cyanide treated group. Single-organoid tracking was used to track the volumetric growth of PCOs in response to treatments, which were compared against PCOs grown in a control well. In addition, heterogeneity exists in both organoid morphology and size. Again, singe-organoid tracking enabled normalization of each PCO volume to its value pre-treatment, so that relative changes in organoid volume can be compared across organoids of different sizes. Linear mixed models were fit to the pre-treatment-normalized organoid volume time-series of all organoids from a PCO line (P1 or P2). The change in organoid volume caused by drug treatment was quantified for all five treatment groups. Least-squares means was used to calculate the pairwise differences between each treatment group and control for each PCO line (Fig. 3). Figure 3 also shows the pairwise differences between all treatment groups for P1 and P2.
Fig. 3.
Treatment-induced Changes in Patient-Derived Cancer Organoid Volume. Single-organoid tracking quantified changes in organoid volume for each treatment group of PCOs over time. (A) Representative MIP of individual PCOs imaged over 48 hours, including one each from the P1 and P2 control groups and one from the P1 cyanide treated group. The 20-minute post-treatment time point was omitted for clarity as there was no change in PCO volume. (B) Heatmaps show the pairwise differences between pre-treatment-normalized organoid volume for all treatment groups for P1. Top: Pairwise differences between each treatment and control for P1. Bottom: Pairwise differences between drug treatments (excluding control) for P1. (C) Heatmaps show the pairwise differences between pre-treatment-normalized organoid volume for all treatment groups for P2. Top: Pairwise differences between each treatment and control for P2. Bottom: Pairwise differences between drug treatments for P2. All pairwise differences were calculated from the linear mixed-effect models via least-squares means and are significant at the p < 0.05 level. Black boxes indicate that these differences are not significant (p > 0.05, black boxes). 2DG: 2-deoxyglucose, Pac: paclitaxel, Cis: cisplatin. Table 1 shows the number of PCOs in each treatment group for each patient. Scale bar: 50µm.
PCOs from P1 and P2 had similar drug response patterns. For P1 groups treated with metabolic inhibitors (cyanide, 2-deoxyglucose), lower organoid volumes compared to control were observed starting at 12 hours for the cyanide (p < 0.001) and 2DG (p < 0.001) groups (Fig. 3(B)). P1 PCOs treated with cisplatin or paclitaxel were significantly different from control beginning at 12 hours (p < 0.01, Fig. 3(B)). For P2 PCOs treated with metabolic inhibitors, lower organoid volumes compared to control were observed starting at 12 hours for the cyanide (p < 0.001) and 2DG (p < 0.001) groups (Fig. 3(C)). For P2 PCOs treated with cisplatin or paclitaxel, organoid volume was lower than control starting at 12 hours (p < 0.01, Fig. 3(C)). Heatmaps (Fig. 3(B),(C)) also show the pairwise differences between treatment groups.
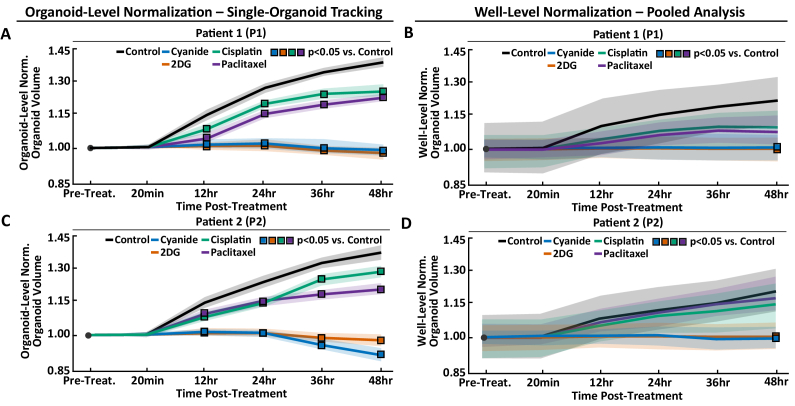
In both P1 and P2 control groups analyzed using single-organoid tracking, PCOs continue to grow over 48 hours, with a statistically significant increase in organoid volume of 32% and 30%, respectively, at 48 hours compared to pre-treatment (Fig. 4(A),(C) p < 0.001 pre-treatment vs. each time-point post-treatment using organoid-level normalization).
Fig. 4.
Comparison of Single-Organoid Tracking and Pooled Analysis. Comparison of single-organoid tracking and pooled analysis using time series data from Patient 1 (P1) and Patient 2 (P2) PCOs. Normalization refers to dividing the value of each organoid at each time-point by either its own pre-treatment value (organoid-level normalization) or the well-level mean (well-level normalization). (A,B) Pre-treatment normalized organoid volume for P1 using organoid-level normalization (A) and well-level normalization (B) for each treatment group. (C,D) Pre-treatment normalized organoid volume for P2 using organoid-level normalization (C) and well-level normalization (D) for each treatment group. Data in A,C were analyzed using a linear mixed-effect model that uses single-organoid tracking to account for organoid-level variability. Data in B,D were analyzed using a linear mixed-effect model that does not account for organoid-level variability due to the lack of single-organoid tracking. Data plotted is mean (lines) ± standard error of the mean (shaded area). Significant differences between a treatment and control (p < 0.05) are indicated with a square color-coded for the treatment group (see legend). Table 1 shows the number of PCOs in each group and for each patient.
3.2. Single-organoid tracking increases sensitivity to drug response
The volumetric growth of each PCO was quantified using single-organoid tracking. Pre-treatment normalization of each organoid to its initial volume accounts for heterogeneity before treatment, and thereby provides more robust analysis of treatment effects. A comparison between single-organoid tracking and a pooled analysis was performed. For the single-organoid tracking, the volume for each PCO was normalized to the pre-treatment volume of the same PCO. For the pooled analysis, each PCO was normalized to the well-level mean of all PCOs in that well. Figure 3 shows organoid volume for P1 and P2 analyzed using single-organoid tracking (organoid-level normalization) or pooled analysis (well-level normalization). Single-organoid tracking (Fig. 4(A),(C)) yields lower variability compared to pooled analysis (Fig. 4(B),(D)), as the well-level mean does not account for organoid-level variability. Importantly, the statistical model that can be applied to organoid-level time series provided by single-organoid tracking increases statistically significant differences between treatment. Overall, single-organoid tracking improves sensitivity to drug response compared to pooled analysis.
3.3. Optical coherence tomography directly quantifies organoid volume
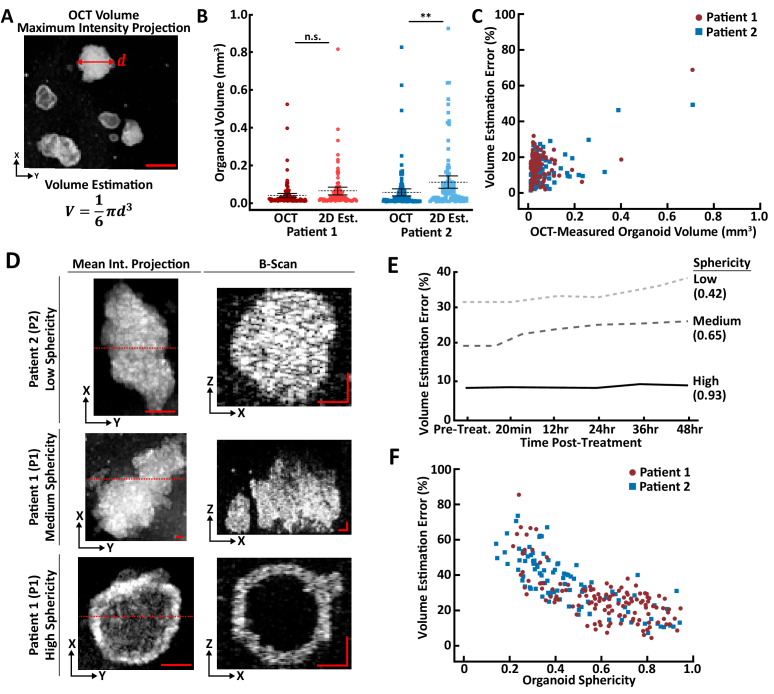
The PCO diameter in 2D images, typically brightfield microscopy images, are often used to estimate changes in PCO volume. This estimation procedure assumes that each PCO is spherical, and therefore large errors can occur with irregular shaped organoids. OCT enables direct measurement of PCO volume, which eliminates the need for volume estimation. Figure 4 compares 3D measurements of PCO volumes using OCT to 2D volume estimates using OCT 2D maximum intensity projections (MIPs) that mimic brightfield measurements. A graphical illustration and equation for volume estimation based on 2D diameter measurements in OCT MIPs is shown (Fig. 5(A)). Dot plots of the OCT measured and 2D estimated PCO volumes are shown for both P1 and P2 (Fig. 5(B)). The distributions of OCT volumes are log-normal, where there are more small organoids than large organoids, which agrees with qualitative observations of the sizes of PCOs present in the culture. Note that organoids smaller than 32 µm diameter are excluded from this analysis.
Fig. 5.
Quantification of Volume Estimation Error. Percent error between direct volume measurement using OCT and volume estimation from diameter measurements of 2D maximum intensity projections (MIPs) of OCT volumes that mimic brightfield measurements. (A) Illustration of the estimated organoid volume (V) based on a diameter measurement (d) in a 2D MIP of an OCT volume. (B) Dot plots of organoid volumes either directly measured from 3D OCT (“OCT”) or estimated from 2D MIPs of OCT volumes (“2D Est.”, as shown in (A)). Statistical significance was assessed with the non-parametric Wilcoxon Signed Rank test (** for p < 0.01; n.s., not significant). Mean (dotted line) and 95% confidence interval (solid error bars) are shown. (C) Plot of the volume estimation error as a function of OCT-measured organoid volume for all organoids pre-treatment. Volume estimation error is the percent error () between the OCT-measured organoid volume and 2D-estimated organoid volume (as shown in (A)). (Spearman’s ρ = 0.37, p < 0.01) (D) The mean intensity projection (top-down view) of each representative PCO and a B-scan chosen from the center (denoted with a red dashed line) of each organoid. These PCOs were chosen to be representative of low (0.42), medium (0.65), and high sphericity (0.93), see Eq. (2). Scale bar: 50 µm. (E) Plot of the volume estimation error for the three representative PCOs (shown in d) at pre-treatment and multiple time points post-treatment. The high sphericity organoid had the lowest error (9% at 48 hours) and low sphericity organoid had the highest error (38% at 48 hours post-treatment), with the medium sphericity organoid in between (25% at 48 hours post-treatment). (F) Plot of the volume estimation error as a function of organoid sphericity for all PCOs at 48 hours post-treatment (Spearman’s ρ = -0.85, p < 0.01). Table 1 shows the number of PCOs for each patient.
Figure 5(C) quantifies the volume estimation error (percent error) between the OCT-measured volume and 2D-estimated volume for every PCO pre-treatment (n = 124 PCOs for P1, and n = 98 PCOs for P2). A linear model was fit to the volume estimation error as a function of measured PCO volume. There was a moderate positive correlation (Spearman’s ρ = 0.37, p < 0.01) between the volume estimation error and the measured PCO volume, which shows that estimating PCO volume from 2D diameter measurements becomes increasingly inaccurate with larger PCOs.
Next, the relationship between PCO morphology (sphericity, see Eq. (2)) and the volume estimation error (percent error) as a function of time after treatment was examined. Figure 5(D),(E) shows three representative PCOs and their volume estimation error as a function of time. In addition, Fig. 5(F) shows the volume estimation error at 48 hours as a function of sphericity. Three representative PCOs were chosen to be representative of low, medium, and high sphericity (Fig. 5(D)). Figure 5(D) shows the mean intensity projection (top-down view) of the entire PCO and a representative B-scan chosen from the center of the PCO (denoted with a red dashed line) for each PCO. Figure 5(E) shows the volume estimation error for each of these representative PCOs at pre-treatment and at multiple time points post-treatment. As expected, the high sphericity PCO (sphericity = 0.93) has the lowest volume estimation error (9% at 48 hours post-treatment), whereas the PCO with the lowest sphericity (sphericity = 0.42) has the highest error (38% at 48 hours post-treatment). The medium sphericity PCO (sphericity = 0.65) had a volume estimation error (25% at 48 hours post-treatment) that fell between the other two PCOs. To see if this observation held for all PCOs used in the study, Fig. 5(F) shows the volume estimation error at 48 hours post-treatment as a function of organoid sphericity for all PCOs from both P1 and P2. There is an inverse relationship between volume estimation error and organoid sphericity (Spearman’s ρ = -0.85, p < 0.01), with higher organoid sphericity resulting in lower volume estimation errors.
3.4. Patient 1 organoids exhibit growth rate heterogeneity
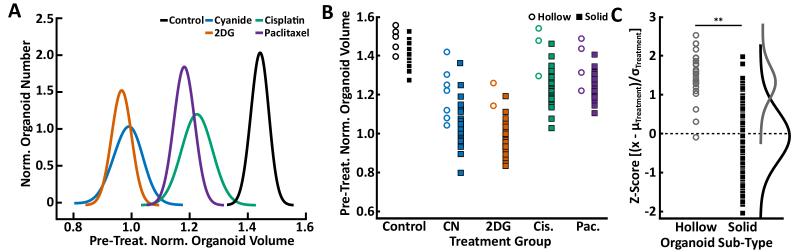
Additional analysis was performed to explore heterogeneity in PCO growth rate. Patient 1 had PCOs with solid or hollow morphologies (Fig. 5(D), middle and bottom panels, respectively). Patient 1 had 103 PCOs with a solid morphology and 21 with a hollow morphology. Patient 2 only had PCOs with solid morphologies (Fig. 5(D), top panel). Figure 6(A) shows single-component Gaussian distribution fits to the pre-treatment normalized PCO volumes for each of the Patient 1 treatment groups at 48 hours. Stratifying the Patient 1 PCOs into hollow and solid sub-types qualitatively showed that the hollow PCOs in each treatment group had higher growth rates relative to the treatment group mean (Fig. 6(B)). Further quantitative analysis was performed by calculating the Z-score for each PCO relative to all other PCOs in that treatment group (Fig. 6(C)). PCOs with positive Z-scores had growth higher than the mean of all other PCOs in that treatment group, while PCOs with negative Z-scores had growth lower than the mean of all other PCOs in that treatment group. This analysis quantitatively shows that hollow PCOs had higher pre-treatment normalized organoid volume at 48 hours compared to solid PCOs (Fig. 6(C), p < 0.01).
Fig. 6.
Volumetric Growth Rate Heterogeneity. Heterogeneity analysis of pre-treatment normalized organoid volume at 48 hours for Patient 1 (P1) PDCOs. (A) Single-component Gaussian distributions were separately fit to the pre-treatment normalized PCO volumes for each of the P1 treatment groups at 48 hours. (B) Dot plots of the pre-treatment normalized organoid volume at 48 hours for PCOs in each of the P1 treatment groups. PCOs were stratified into hollow and solid morphology sub-types based on visual inspection of the images. Hollow organoids for each group are plotted using open circle markers and solid organoids are plotted using solid square markers. CN: cyanide, Cis.: cisplatin, Pac.: paclitaxel. (C) The Z-score for all pre-treatment normalized P1 PCOs at 48 hours were calculated and stratified into hollow or solid morphological sub-types. The Z-score enables comparison between treatment groups. Statistical significance was assessed using the non-parametric Mann-Whitney U test (** for p < 0.01). Distributions of the Z-scores were plotted from the dot plot data (gramm Package, MATLAB). Table 1 shows the number of PCOs for Patient 1.
4. Discussion
Clinicians base the selection of a treatment for an individual cancer patient on biomarkers (e.g., mutational profile, medical imaging, histology, and stage) with limited ability to predict response from heterogeneous populations of tumor cells. Drug screening with PCOs is a promising approach to optimize treatment decisions for individual cancer patients. However, non-destructive techniques to quantify drug response on an individual PCO level are needed to capture intra-patient heterogeneity. Here, we provide the first demonstration of 3D single-organoid tracking using OCT to assess volumetric growth and drug response in PCOs. Single-organoid tracking enables organoid-level measurements of drug response over time that improves sensitivity over bulk measurements across an entire well. OCT is inherently non-destructive, rapid, and label-free, which is attractive for capturing heterogeneity in drug response across numerous PCOs from the same patient.
PCOs typically vary in size and morphology due to the 3D culturing technique, so single-organoid tracking is needed to account for this baseline heterogeneity [42,43]. OCT has been used to image drug response in organoids by tracking volumetric growth, measuring the amount of cell death, or quantifying changes in cellular-level dynamics [10–12,14]. None of these previous OCT studies tracked each organoid over time, but instead measured a single time point or calculated averages over all organoids in a well. Here, we demonstrate a framework to link the same PCO across multiple OCT imaging time points. These methods can be used to measure drug response in PCOs that accounts for baseline PCO variability. In addition, statistical techniques such as mixed-effect models can use these time-course data to account for organoid-level heterogeneity and provide improved sensitivity to drug response [40,44]. Our results show that single-organoid tracking of PCOs reduces variability and increases statistically significant differences between the treatment groups.
To demonstrate the benefit of OCT for drug response measurements in PCOs, we compared OCT direct volume measurements to 2D volume estimation that mimics standard brightfield measurements. Interestingly, while there was error for organoids from P1 and P2, the distribution of actual and estimated PCO volumes were only significantly different for PCOs from P2. Further examination of volume estimation error as a function of sphericity showed that error decreased as sphericity increased. PCOs from P2 had lower sphericities, which may explain the difference in the volume distributions between OCT-measured volume and 2D-esimtated volume. Our results show that assessing the volumetric growth rate of PCOs offers improved accuracy compared to traditional 2D estimates similar to brightfield diameter measurements.
Lastly, P1 PCOs exhibited two morphological sub-types, solid and hollow, which were identified by visual inspection of the OCT images. The hollow morphology is consistent with cystic morphologies observed in previous studies of colorectal PCOs, and is thought to reflect the genetic and phenotypic in vivo heterogeneity of the patient’s tumor [30]. Hollow PCOs grew more over 48 hours compared to solid PCOs, which may be a result of differential gene expression between the two morphologies or the lack of an apoptotic core [29,30]. Note that differences in the density of hollow compared to solid organoids were not taken into consideration in the volume estimation, which assumes that all organoids are dense and non-cystic. Future studies could set a threshold on the OCT signal to only quantify the cellular regions of cystic organoids in the volume calculations. Our results show that 3D single-organoid tracking and OCT can be used to identify growth rate heterogeneity, and differences in growth rate between hollow and solid PCOs.
This study illustrates a framework for robust drug response measurements of PCOs using OCT and 3D single-organoid tracking that accounts for pre-treatment heterogeneity in PCO volume. OCT can provide rapid volumetric images of organoids, which could be used to characterize diverse drug response across numerous PCOs derived from the same patient. The framework developed in this manuscript can be extended to evaluate other aspects of the PCO system, including basic scientific questions about the growth dynamics and heterogeneity of PCOs.
In addition, 3D single-organoid tracking could be further developed to enable multimodal optical imaging of the same PCOs over time. We envision that multiple optical modalities, such as autofluorescence imaging [26,29,45–50] and functional extensions of OCT [11–13,51–55], could be used with 3D single-organoid tracking to monitor each PCO over time. Autofluorescence imaging of the metabolic coenzymes NAD(P)H and FAD could provide label-free metabolic information of each organoid and has already been demonstrated to be a powerful tool for assessing PCOs [26,29,45–50]. Fluorescence lifetime of NAD(P)H and FAD reports on the binding activity of these enzymes, and can provide additional information on the specific metabolic program of cells [47,50,56–59]. Images taken using other modalities at higher magnification [60–62] could provide cellular and sub-cellular information for each PCO, which could then be integrated with OCT to provide a comprehensive readout of organoid health and drug response. Images at multiple scales could also provide insight into the relationship between the OCT signal in PCOs and its corresponding cellular and sub-cellular organization. Lastly, because OCT is non-destructive, methods such as genomic sequencing, transcriptomics, proteomics, and metabolomics could be used in tandem to provide complementary information on drug response [2,8,63,64].
We believe that OCT coupled with 3D single-organoid tracking can be applied to improve clinical treatment selection, basic science, and drug development.
Acknowledgements
The authors thank Tongcheng Qian for his useful comments on the manuscript.
Funding
Stand Up To Cancer10.13039/100009730 (SU2C-AACR-IG-08-16); National Science Foundation10.13039/100000001 (CBET-1642287); National Institutes of Health10.13039/100000002 (P30 CA014520, R01 CA185747, R37 CA226526).
Disclosures
Authors have no relevant financial interests in the manuscript and no other potential conflicts of interest to disclose.
Data availability
Data underlying the results presented in this paper are not publicly available at this time but may be obtained from the authors upon reasonable request.
References
- 1.Sachs N., de Ligt J., Kopper O., Gogola E., Bounova G., Weeber F., Balgobind A. V., Wind K., Gracanin A., Begthel H., Korving J., van Boxtel R., Duarte A. A., Lelieveld D., van Hoeck A., Ernst R. F., Blokzijl F., Nijman I. J., Hoogstraat M., van de Ven M., Egan D. A., Zinzalla V., Moll J., Boj S. F., Voest E. E., Wessels L., van Diest P. J., Rottenberg S., Vries R. G. J., Cuppen E., Clevers H., “A living biobank of breast cancer organoids captures disease heterogeneity,” Cell 172(1-2), 373–386.e10 (2018). 10.1016/j.cell.2017.11.010 [DOI] [PubMed] [Google Scholar]
- 2.Vlachogiannis G., Hedayat S., Vatsiou A., Jamin Y., Fernandez-Mateos J., Khan K., Lampis A., Eason K., Huntingford I., Burke R., Rata M., Koh D. M., Tunariu N., Collins D., Hulkki-Wilson S., Ragulan C., Spiteri I., Moorcraft S. Y., Chau I., Rao S., Watkins D., Fotiadis N., Bali M., Darvish-Damavandi M., Lote H., Eltahir Z., Smyth E. C., Begum R., Clarke P. A., Hahne J. C., Dowsett M., de Bono J., Workman P., Sadanandam A., Fassan M., Sansom O. J., Eccles S., Starling N., Braconi C., Sottoriva A., Robinson S. P., Cunningham D., Valeri N., “Patient-derived organoids model treatment response of metastatic gastrointestinal cancers,” Science 359(6378), 920–926 (2018). 10.1126/science.aao2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saito Y., Muramatsu T., Kanai Y., Ojima H., Sukeda A., Hiraoka N., Arai E., Sugiyama Y., Matsuzaki J., Uchida R., Yoshikawa N., Furukawa R., Saito H., “Establishment of patient-derived organoids and drug screening for biliary tract carcinoma,” Cell Rep. 27(4), 1265–1276.e4 (2019). 10.1016/j.celrep.2019.03.088 [DOI] [PubMed] [Google Scholar]
- 4.Xu H., Lyu X., Yi M., Zhao W., Song Y., Wu K., “Organoid technology and applications in cancer research,” J. Hematol. Oncol. 11(1), 116 (2018). 10.1186/s13045-018-0662-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Granat L. M., Kambhampati O., Klosek S., Niedzwecki B., Parsa K., Zhang D., “The promises and challenges of patient-derived tumor organoids in drug development and precision oncology,” Anim. Models Exp. Med. 2(3), 150–161 (2019). 10.1002/ame2.12077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kopper O., de Witte C. J., Lõhmussaar K., Valle-Inclan J. E., Hami N., Kester L., Balgobind A. V., Korving J., Proost N., Begthel H., van Wijk L. M., Revilla S. A., Theeuwsen R., van de Ven M., van Roosmalen M. J., Ponsioen B., Ho V. W. H., Neel B. G., Bosse T., Gaarenstroom K. N., Vrieling H., Vreeswijk M. P. G., van Diest P. J., Witteveen P. O., Jonges T., Bos J. L., van Oudenaarden A., Zweemer R. P., Snippert H. J. G., Kloosterman W. P., Clevers H., “An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity,” Nat. Med. 25(5), 838–849 (2019). 10.1038/s41591-019-0422-6 [DOI] [PubMed] [Google Scholar]
- 7.Sasaki N., Clevers H., “Studying cellular heterogeneity and drug sensitivity in colorectal cancer using organoid technology,” Curr. Opin. Genet. Dev. 52, 117–122 (2018). 10.1016/j.gde.2018.09.001 [DOI] [PubMed] [Google Scholar]
- 8.Schumacher D., Andrieux G., Boehnke K., Keil M., Silvestri A., Silvestrov M., Keilholz U., Haybaeck J., Erdmann G., Sachse C., Templin M., Hoffmann J., Boerries M., Schäfer R., Regenbrecht C. R. A., “Heterogeneous pathway activation and drug response modelled in colorectal-tumor-derived 3D cultures,” PLOS Genet. 15(3), e1008076 (2019). 10.1371/journal.pgen.1008076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jung Y., Klein O. J., Wang H., Evans C. L., “Longitudinal, label-free, quantitative tracking of cell death and viability in a 3D tumor model with OCT,” Sci. Rep. 6(1), 27017 (2016). 10.1038/srep27017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Y., Wang S., Guo Q., Kessel S., Rubinoff I., Chan L. L., Li P., Liu Y., Qiu J., Zhou C., “Optical coherence tomography detects necrotic regions and volumetrically quantifies multicellular tumor spheroids,” Cancer Res. 77(21), 6011–6020 (2017). 10.1158/0008-5472.CAN-17-0821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abd El-Sadek I., Miyazawa A., Tzu-Wei Shen L., Makita S., Fukuda S., Yamashita T., Oka Y., Mukherjee P., Matsusaka S., Oshika T., Kano H., Yasuno Y., “Optical coherence tomography-based tissue dynamics imaging for longitudinal and drug response evaluation of tumor spheroids,” Biomed. Opt. Express 11(11), 6231–6248 (2020). 10.1364/BOE.404336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oldenburg A. L., Yu X., Gilliss T., Alabi O., Taylor R. M., 2nd, Troester M. A., “Inverse-power-law behavior of cellular motility reveals stromal-epithelial cell interactions in 3D co-culture by OCT fluctuation spectroscopy,” Optica 2(10), 877–885 (2015). 10.1364/OPTICA.2.000877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu X., Fuller A. M., Blackmon R., Troester M. A., Oldenburg A. L., “Quantification of the effect of toxicants on the intracellular kinetic energy and cross-sectional area of mammary epithelial organoids by OCT fluctuation spectroscopy,” Toxicol Sci. 162(1), 234–240 (2018). 10.1093/toxsci/kfx245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang L., Yu X., Fuller A. M., Troester M. A., Oldenburg A. L., “Characterizing optical coherence tomography speckle fluctuation spectra of mammary organoids during suppression of intracellular motility,” Quant. Imaging Med. Surg. 10(1), 76–85 (2020). 10.21037/qims.2019.08.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberge C. L., Kingsley D. M., Faulkner D. E., Sloat C. J., Wang L., Barroso M., Intes X., Corr D. T., “Non-destructive tumor aggregate morphology and viability quantification at cellular resolution, during development and in response to drug,” Acta Biomater. 117, 322–334 (2020). 10.1016/j.actbio.2020.09.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phan N., Hong J. J., Tofig B., Mapua M., Elashoff D., Moatamed N. A., Huang J., Memarzadeh S., Damoiseaux R., Soragni A., “A simple high-throughput approach identifies actionable drug sensitivities in patient-derived tumor organoids,” Commun. Biol. 2(1), 78 (2019). 10.1038/s42003-019-0305-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fricke S. L., Payne S. N., Favreau P. F., Kratz J. D., Pasch C. A., Foley T. M., Yueh A. E., Van De Hey D. R., Depke M. G., Korkos D. P., Sha G. C., DeStefanis R. A., Clipson L., Burkard M. E., Lemmon K. K., Parsons B. M., Kenny P. A., Matkowskyj K. A., Newton M. A., Skala M. C., Deming D. A., “MTORC1/2 inhibition as a therapeutic strategy for PIK3CA mutant cancers,” Mol. Cancer Ther. 18(2), 346–355 (2019). 10.1158/1535-7163.MCT-18-0510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhan T., Rindtorff N., Boutros M., “WNT signaling in cancer,” Oncogene 36(11), 1461–1473 (2017). 10.1038/onc.2016.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Way J. L., “Cyanide intoxication and its mechanism of antagonism,” Annu. Rev. Pharmacol. Toxicol. 24(1), 451–481 (1984). 10.1146/annurev.pa.24.040184.002315 [DOI] [PubMed] [Google Scholar]
- 20.Zhang D., Li J., Wang F., Hu J., Wang S., Sun Y., “2-Deoxy-D-glucose targeting of glucose metabolism in cancer cells as a potential therapy,” Cancer Lett. 355(2), 176–183 (2014). 10.1016/j.canlet.2014.09.003 [DOI] [PubMed] [Google Scholar]
- 21.Dasari S., Tchounwou P. B., “Cisplatin in cancer therapy: molecular mechanisms of action,” Eur. J. Pharmacol. 740, 364–378 (2014). 10.1016/j.ejphar.2014.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weaver B. A., “How Taxol/paclitaxel kills cancer cells,” Mol. Biol. Cell 25(18), 2677–2681 (2014). 10.1091/mbc.e14-04-0916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kristin M. P., Chetan A. P., Christopher E. N., Devin R. M., Megan C. M., Craig L. D., Melissa C. S., “Longitudinal study of arteriogenesis with swept source optical coherence tomography and hyperspectral imaging,” Proc. SPIE 8934, 89341Z (2014). 10.1117/12.2042037 [DOI] [Google Scholar]
- 24.Drexler W., Fujimoto J. G., Optical Coherence Tomography, Biological and Medical Physics, Biomedical Engineering (Springer Berlin Heidelberg, 2008). [Google Scholar]
- 25.Edelstein A. D., Tsuchida M. A., Amodaj N., Pinkard H., Vale R. D., Stuurman N., “Advanced methods of microscope control using µManager software,” J. Biol. Methods 1(2), e10 (2014). 10.14440/jbm.2014.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gil D., Deming D., Skala M., “Patient-derived cancer organoid tracking with wide-field one-photon redox imaging to assess treatment response,” J. Biomed. Opt. 26(3), 036005 (2021). 10.1117/1.jbo.26.3.036005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guizar-Sicairos M., Thurman S. T., Fienup J. R., “Efficient subpixel image registration algorithms,” Opt. Lett. 33(2), 156–158 (2008). 10.1364/OL.33.000156 [DOI] [PubMed] [Google Scholar]
- 28.Guizar M., “Efficient subpixel image registration by cross-correlation,” (2020), retrieved January 14, 2020, https://www.mathworks.com/matlabcentral/fileexchange/18401-efficient-subpixel-image-registration-by-cross-correlation.
- 29.Favreau P. F., He J., Gil D. A., Deming D. A., Huisken J., Skala M. C., “Label-free redox imaging of patient-derived organoids using selective plane illumination microscopy,” Biomed. Opt. Express 11(5), 2591–2606 (2020). 10.1364/BOE.389164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kashfi S. M. H., Almozyan S., Jinks N., Koo B.-K., Nateri A. S., “Morphological alterations of cultured human colorectal matched tumour and healthy organoids,” Oncotarget 9(12), 10572–10584 (2018). 10.18632/oncotarget.24279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tinevez J. Y., Perry N., Schindelin J., Hoopes G. M., Reynolds G. D., Laplantine E., Bednarek S. Y., Shorte S. L., Eliceiri K. W., “TrackMate: An open and extensible platform for single-particle tracking,” Methods 115, 80–90 (2017). 10.1016/j.ymeth.2016.09.016 [DOI] [PubMed] [Google Scholar]
- 32.Jaqaman K., Loerke D., Mettlen M., Kuwata H., Grinstein S., Schmid S. L., Danuser G., “Robust single-particle tracking in live-cell time-lapse sequences,” Nat. Methods 5(8), 695–702 (2008). 10.1038/nmeth.1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McMurtrey R. J., “Analytic models of oxygen and nutrient diffusion, metabolism dynamics, and architecture optimization in three-dimensional tissue constructs with applications and insights in cerebral organoids,” Tissue Eng., Part C 22(3), 221–249 (2016). 10.1089/ten.tec.2015.0375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vergara M. N., Flores-Bellver M., Aparicio-Domingo S., McNally M., Wahlin K. J., Saxena M. T., Mumm J. S., Canto-Soler M. V., “Three-dimensional automated reporter quantification (3D-ARQ) technology enables quantitative screening in retinal organoids,” Development 144(20), 3698–3705 (2017). 10.1242/dev.146290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wadell H., “Volume, shape, and roundness of quartz particles,” J. Geol. 43(3), 250–280 (1935). 10.1086/624298 [DOI] [Google Scholar]
- 36.van der Maaten L., “drtoolbox: Matlab toolbox for dimensionality reduction,” (2019), retrieved December 12, 2019, https://lvdmaaten.github.io/drtoolbox/#usage.
- 37.Morel P., “Gramm: grammar of graphics plotting in Matlab,” The J. Open Source Soft. 3(23), 568 (2018). 10.21105/joss.00568 [DOI] [Google Scholar]
- 38.Bates D., DebRoy S., Sarkar D., Team R. C., “nlme: Linear and nonlinear mixed effects models,” (2018).
- 39.Lenth R. V., “Least-squares means: The R Package lsmeans,” J. Stat. Soft. 69(1), 1–33 (2016). 10.18637/jss.v069.i01 [DOI] [Google Scholar]
- 40.Gibbons R. D., Hedeker D., DuToit S., “Advances in analysis of longitudinal data,” Annu. Rev. Clin. Psychol. 6(1), 79–107 (2010). 10.1146/annurev.clinpsy.032408.153550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harrison X. A., Donaldson L., Correa-Cano M. E., Evans J., Fisher D. N., Goodwin C. E. D., Robinson B. S., Hodgson D. J., Inger R., “A brief introduction to mixed effects modelling and multi-model inference in ecology,” PeerJ 6, e4794 (2018). 10.7717/peerj.4794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borten M. A., Bajikar S. S., Sasaki N., Clevers H., Janes K. A., “Automated brightfield morphometry of 3D organoid populations by OrganoSeg,” Sci. Rep. 8(1), 5319 (2018). 10.1038/s41598-017-18815-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tirier S. M., Park J., Preußer F., Amrhein L., Gu Z., Steiger S., Mallm J.-P., Krieger T., Waschow M., Eismann B., Gut M., Gut I. G., Rippe K., Schlesner M., Theis F., Fuchs C., Ball C. R., Glimm H., Eils R., Conrad C., “Pheno-seq – linking visual features and gene expression in 3D cell culture systems,” Sci. Rep. 9(1), 12367 (2019). 10.1038/s41598-019-48771-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verbeke G., Fieuws S., Molenberghs G., Davidian M., “The analysis of multivariate longitudinal data: a review,” Stat. Methods Med. Res. 23(1), 42–59 (2014). 10.1177/0962280212445834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cannon T. M., Shah A. T., Walsh A. J., Skala M. C., “High-throughput measurements of the optical redox ratio using a commercial microplate reader,” J. Biomed. Opt. 20(1), 010503 (2015). 10.1117/1.JBO.20.1.010503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walsh A. J., Cook R. S., Sanders M. E., Aurisicchio L., Ciliberto G., Arteaga C. L., Skala M. C., “Quantitative optical imaging of primary tumor organoid metabolism predicts drug response in breast cancer,” Cancer Res. 74(18), 5184–5194 (2014). 10.1158/0008-5472.CAN-14-0663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kolenc O. I., Quinn K. P., “Evaluating cell metabolism through autofluorescence imaging of NAD(P)H and FAD,” Antioxid. Redox Signaling 30(6), 875–889 (2019). 10.1089/ars.2017.7451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hogan M. C., Stary C. M., Balaban R. S., Combs C. A., “NAD(P)H fluorescence imaging of mitochondrial metabolism in contracting Xenopus skeletal muscle fibers: effect of oxygen availability,” J. Appl. Physiol. 98(4), 1420–1426 (2005). 10.1152/japplphysiol.00849.2004 [DOI] [PubMed] [Google Scholar]
- 49.Palmer S., Litvinova K., Dunaev A., Fleming S., McGloin D., Nabi G., “Changes in autofluorescence based organoid model of muscle invasive urinary bladder cancer,” Biomed. Opt. Express 7(4), 1193–1200 (2016). 10.1364/BOE.7.001193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sharick J. T., Favreau P. F., Gillette A. A., Sdao S. M., Merrins M. J., Skala M. C., “Protein-bound NAD(P)H lifetime is sensitive to multiple fates of glucose carbon,” Sci. Rep. 8(1), 5456 (2018). 10.1038/s41598-018-23691-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chhetri R. K., Phillips Z. F., Troester M. A., Oldenburg A. L., “Longitudinal study of mammary epithelial and fibroblast co-cultures using optical coherence tomography reveals morphological hallmarks of pre-malignancy,” PLoS One 7(11), e49148 (2012). 10.1371/journal.pone.0049148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thouvenin O., Fink M., Boccara A. C., “Dynamic multimodal full-field optical coherence tomography and fluorescence structured illumination microscopy,” J. Biomed. Opt. 22(02), 1 (2017). 10.1117/1.JBO.22.2.026004 [DOI] [PubMed] [Google Scholar]
- 53.An R., Merrill D., Avramova L., Sturgis J., Tsiper M., Robinson J. P., Turek J., Nolte D. D., “Phenotypic profiling of raf inhibitors and mitochondrial toxicity in 3D tissue using biodynamic imaging,” J. Biomol. Screening 19(4), 526–537 (2014). 10.1177/1087057113516674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hao S., Daniel M., Ran A., John T., Daniela M., David D. N., “Biodynamic imaging for phenotypic profiling of three-dimensional tissue culture,” J. Biomed. Opt. 22(1), 016007 (2017). 10.1117/1.JBO.22.1.016007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Z., An R., Swetzig W. M., Kanis M., Nwani N., Turek J., Matei D., Nolte D., “Intracellular optical doppler phenotypes of chemosensitivity in human epithelial ovarian cancer,” Sci. Rep. 10(1), 17354 (2020). 10.1038/s41598-020-74336-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Okkelman I. A., Neto N., Papkovsky D. B., Monaghan M. G., Dmitriev R. I., “A deeper understanding of intestinal organoid metabolism revealed by combining fluorescence lifetime imaging microscopy (FLIM) and extracellular flux analyses,” Redox Biol. 30, 101420 (2020). 10.1016/j.redox.2019.101420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Datta R., Heaster T., Sharick J., Gillette A., Skala M., “Fluorescence lifetime imaging microscopy: fundamentals and advances in instrumentation, analysis, and applications,” J. Biomed. Opt. 25(07), 1 (2020). 10.1117/1.JBO.25.7.071203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blacker T. S., Mann Z. F., Gale J. E., Ziegler M., Bain A. J., Szabadkai G., Duchen M. R., “Separating NADH and NADPH fluorescence in live cells and tissues using FLIM,” Nat. Commun. 5(1), 3936 (2014). 10.1038/ncomms4936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Skala M. C., Riching K. M., Gendron-Fitzpatrick A., Eickhoff J., Eliceiri K. W., White J. G., Ramanujam N., “In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia,” Proc. Natl. Acad. Sci. U. S. A. 104(49), 19494–19499 (2007). 10.1073/pnas.0708425104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bode K. J., Mueller S., Schweinlin M., Metzger M., Brunner T., “A fast and simple fluorometric method to detect cell death in 3D intestinal organoids,” BioTechniques 67(1), 23–28 (2019). 10.2144/btn-2019-0023 [DOI] [PubMed] [Google Scholar]
- 61.Au - van Ineveld R. L., Au - Ariese H. C. R., Au - Wehrens E. J., Au - Dekkers J. F., Au - Rios A. C., “Single-cell resolution three-dimensional imaging of intact organoids,” JoVE 160, e60709 (2020). 10.3791/60709 [DOI] [PubMed] [Google Scholar]
- 62.Dekkers J. F., Alieva M., Wellens L. M., Ariese H. C. R., Jamieson P. R., Vonk A. M., Amatngalim G. D., Hu H., Oost K. C., Snippert H. J. G., Beekman J. M., Wehrens E. J., Visvader J. E., Clevers H., Rios A. C., “High-resolution 3D imaging of fixed and cleared organoids,” Nat. Protoc. 14(6), 1756–1771 (2019). 10.1038/s41596-019-0160-8 [DOI] [PubMed] [Google Scholar]
- 63.Rodon J., Soria J. C., Berger R., Miller W. H., Rubin E., Kugel A., Tsimberidou A., Saintigny P., Ackerstein A., Braña I., Loriot Y., Afshar M., Miller V., Wunder F., Bresson C., Martini J. F., Raynaud J., Mendelsohn J., Batist G., Onn A., Tabernero J., Schilsky R. L., Lazar V., Lee J. J., Kurzrock R., “Genomic and transcriptomic profiling expands precision cancer medicine: the WINTHER trial,” Nat. Med. 25(5), 751–758 (2019). 10.1038/s41591-019-0424-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen K. Y., Srinivasan T., Lin C., Tung K. L., Gao Z., Hsu D. S., Lipkin S. M., Shen X., “Single-cell transcriptomics reveals heterogeneity and drug response of human colorectal cancer organoids,” Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2018, 2378–2381 (2018). 10.1109/EMBC.2018.8512784 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data underlying the results presented in this paper are not publicly available at this time but may be obtained from the authors upon reasonable request.