Abstract
Background and study aims Small bowel ulcerations are efficiently detected with deep learning techniques, whereas the ability to diagnose Crohnʼs disease (CD) in the colon with it is unknown. This study examined the ability of a deep learning framework to detect CD lesions with pan-enteric capsule endoscopy (CE) and classify lesions of different severity.
Patients and methods CEs from patients with suspected or known CD were included in the analysis. Two experienced gastroenterologists classified anonymized images into normal mucosa, non-ulcerated inflammation, aphthous ulceration, ulcer, or fissure/extensive ulceration. An automated framework incorporating multiple ResNet-50 architectures was trained. To improve its robustness and ability to characterize lesions, image processing methods focused on texture enhancement were employed.
Results A total of 7744 images from 38 patients with CD were collected (small bowel 4972, colon 2772) of which 2748 contained at least one ulceration (small bowel 1857, colon 891). With a patient-dependent split of images for training, validation, and testing, ulcerations were diagnosed with a sensitivity, specificity, and diagnostic accuracy of 95.7 % (CI 93.4–97.4), 99.8 % (CI 99.2–100), and 98.4 % (CI 97.6–99.0), respectively. The diagnostic accuracy was 98.5 % (CI 97.5–99.2) for the small bowel and 98.1 % (CI 96.3–99.2) for the colon. Ulcerations of different severities were classified with substantial agreement (κ = 0.72).
Conclusions Our proposed framework is in excellent agreement with the clinical standard, and diagnostic accuracies are equally high for the small bowel and colon. Deep learning approaches have a great potential to help clinicians detect, localize, and determine the severity of CD with pan-enteric CE.
Introduction
Crohn's disease (CD) belongs to the group of chronic inflammatory bowel diseases 1 . Cardinal lesions are mucosal ulcerations ranging from small aphthous ulcerations to large ulcers and fissures. Typically, CD has a segmental distribution, and the entire gastrointestinal tract may be involved, although the disease is most often located in the terminal ileum and right colon (ileocecal CD) 2 .
In recent years, technological advances have improved modalities for diagnosing and monitoring CD. Capsule endoscopy (CE) is non-invasive, patient-friendly, and highly sensitive for the earliest lesions of CD 3 4 . In patients with suspected CD and a normal ileocolonoscopy, the European Society of Gastrointestinal Endoscopy (ESGE) and the European Crohnʼs and Colitis Organization (ECCO) recommends CE as first line modality for investigating the small bowel in patients without obstructive symptoms 5 6 . Colon CE was introduced in 2006, and pan-enteric CE is now available allowing a direct and detailed evaluation of the entire gastrointestinal mucosa. However, the role of pan-enteric CE in patients with suspected or known CD remains to be established.
The camera pill captures more than 50,000 images of the gastrointestinal tract, and a significant limitation with CE is the time-consuming manual video analysis. In previous series, reading times above 40 to 50 minutes were reported for small bowel CE 3 , and pan-enteric CE takes more than 60 minutes to interpret for CD. Hence, lesions may be missed due to readerʼs fatigue or distraction. Better ways to optimize the work of the GI specialist without affecting the diagnostic accuracy of CE would be helpful in clinical practice.
Utilizing artificial intelligence (AI) – especially deep learning techniques–has received great attention in recent years. Multiple clinical settings have been studied including the ability to analyze endoscopy images and aid clinical decision-making 7 8 9 . A recent meta-analysis showed a high sensitivity and specificity of deep learning techniques for ulcer detection in the small bowel 8 , whereas the ability to diagnose CD in the colon is unknown. Results are promising, and AI could have a pivotal role in the future of non-invasive diagnosis of CD with pan-enteric CE. The aim of this study was to examine the ability of a deep learning framework to detect CD lesions in single images of the small bowel or colon captured with pan-enteric CE, determine the localization of lesions, and the ability to characterize lesions of different severity.
Patients and methods
Study design
Patients with suspected or known CD were recruited from three centers in the Region of Southern Denmark managing adult patients with inflammatory bowel diseases. All patients were prospectively enrolled in a clinical trial examining non-invasive modalities for diagnosing suspected CD (http://ClinicalTrials.gov Identifier NCT03134586) or assessing treatment response in patients with known CD ( http://ClinicalTrials.gov Identifier NCT03435016).
CD was clinically suspected in patients with diarrhea and/or abdominal pain for more than 1 month (or repeated episodes of diarrhea and/or abdominal pain) associated with a fecal calprotectin > 50 mg/kg and at least one additional finding suggesting CD: elevated inflammatory markers, anemia, fever, weight loss, perianal abscess/fistula, a family history of inflammatory bowel disease, or suspicion of CD after sigmoidoscopy.
Patients with an established diagnosis of CD based on ECCO criteria 10 were included if they had clinical disease activity (Harvey-Bradshaw Index ≥ 5 or Crohnʼs Disease Activity Index ≥ 150), endoscopic activity (Simple Endoscopic Score for Crohnʼs disease ≥ 3), and a clinical indication for medical treatment with corticosteroids or biological therapy.
All patients had a standardized work-up including medical history, physical examination, blood and stool samples, ileocolonoscopy, pan-enteric CE, magnetic resonance imaging enterocolonography, and bowel ultrasound.
Capsule endoscopy procedure
Pan-enteric CE was performed with the PillCam Crohn's capsule (Medtronic, Dublin, Ireland) after overnight fasting and bowel preparation with 2 + 2 L of polyethylene glycol (Moviprep) as previously described by ESGE 11 . Videos were analyzed with the PillCam Software v9.
Image selection and classification
Images with a normal mucosa or CD lesions located in the small bowel or colon were manually searched and randomly collected by three gastroenterologists with experience in CE and inflammatory bowel diseases (M.D.J., J.B.B. and M.L.H.). Images were anonymized and assigned into one of the following 13 categories by authors M.D.J. and J.B.B.:
Small bowel: normal mucosa, normal mucosa with lymphoid hyperplasia, normal mucosa with bubbles and/or debris, non-ulcerated inflammation, aphthous ulceration, ulcer, fissure / large ulcer
Colon: normal mucosa, normal mucosa with bubbles and/or debris, non-ulcerated inflammation, aphthous ulceration, ulcer, fissure/large ulcer
The following definitions were used for image classification:
Normal: No or minimal luminal content (estimated < 1 mm in size) and mucosa without erythema, edema or mucosal breaks
Debris: Dark fluid or solid luminal content without surrounding erythema or mucosal break
Bubbles: Luminal pocket of air reflecting the flashlight
Aphthous ulceration: Small superficial mucosal break with surrounding erythema (estimated < 5 mm in size)
Ulcer: Mucosal break with loss of substance and fibrin
Fissure: Longitudinal ulcer
Large ulcer: Ulcer involving > 50 % of the lumen
Non-ulcerated inflammation: Erythema and edema without mucosal break
In case of disagreement, a consensus decision was reached. If more lesions were seen in the same image, the most severe lesion determined the overall classification. The visual illustration of image classification is in Fig.1 .
Fig. 1.
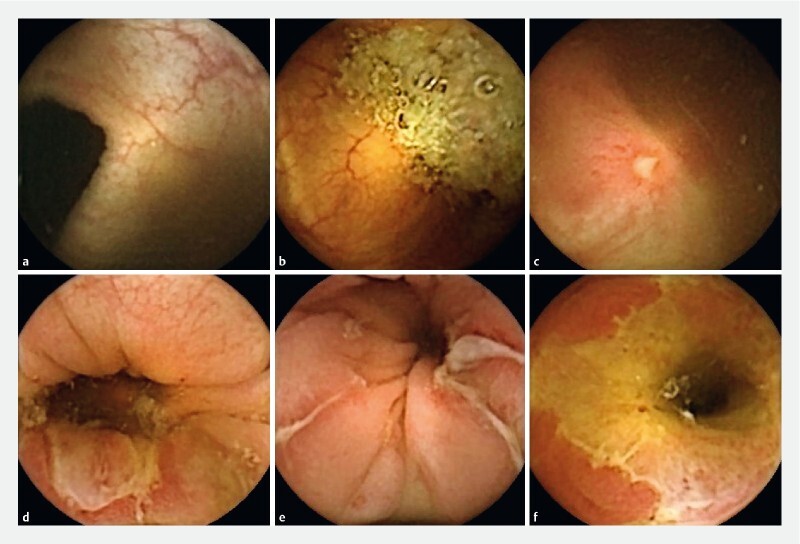
Examples of image classification. a Normal colon. b Normal colon with debris. c Aphthous ulceration in the colon; d Ulcer in the colon. e Fissures in the colon; f Large ulcer in the terminal ileum.
Image processing
After manual classification of all collected images, the images needed to be preprocessed in order to effectively train the deep learning algorithms. Since the original images contain text information near the corners, the Chan-Vese segmentation via graph cuts was used to extract the binary mask and to segment the relevant information from the image 12 .
Four different algorithms were employed for textural improvements:
Contrast increase: The original image is split into three individual intensity channels (red, green, and blue channels, RGB image). In each channel, the adjustment was applied, where the bottom 1 % and the top 1 % of all pixel values were saturated. At the end, grayscale channels were merged back to form an enhanced RGB image.
Histogram equalization: The input RGB image was converted to a different color space that describes colors similarly to how the human eye tends to perceive them. In this color space, hue (H) channel specifies the base color, saturation (S) channel represents the vibrancy of the color and captures the amount of gray in a particular color, and value (V) channel in conjunction with S channel describes the intensity or brightness of the color. In the next step, a contrast-limited adaptive histogram equalization (CLAHE) was applied on the V channel 13 . This non-parametric equalization operates on small regions in the image and computes histograms corresponding to distinct sections of the image. Subsequently, it uses them to redistribute the lightness values of the image. In the last step, the enhanced HSV image was converted back to RGB color space.
Gradient X: The original RGB image was converted to a single grayscale image by forming a weighted sum of the red, green, and blue component. In the next step, the X-directional gradient of the grayscale image was extracted. The resulting image was then copied into three color channels of RGB to form the output image.
Dehazing: First, the complement image of the original RGB input was computed and dehazing algorithm that relies on a dark channel prior was used. The algorithm was originally designed to reduce the atmospheric haze, and it is based on the observation that unhazy images contain pixels that have low signal in color channels, which is also our case in CE images. At the end, the complement image was derived again and used as an enhanced output image.
For better illustration of all four described methods, their visual outcomes on three random samples are provided in Fig.2 . All possible subsets of these image transformations were evaluated, but the highest performance was achieved when all variants (original image plus four new variants) were considered together. It demonstrates that every single transformation adds to the robustness of our proposed system.
Fig. 2.
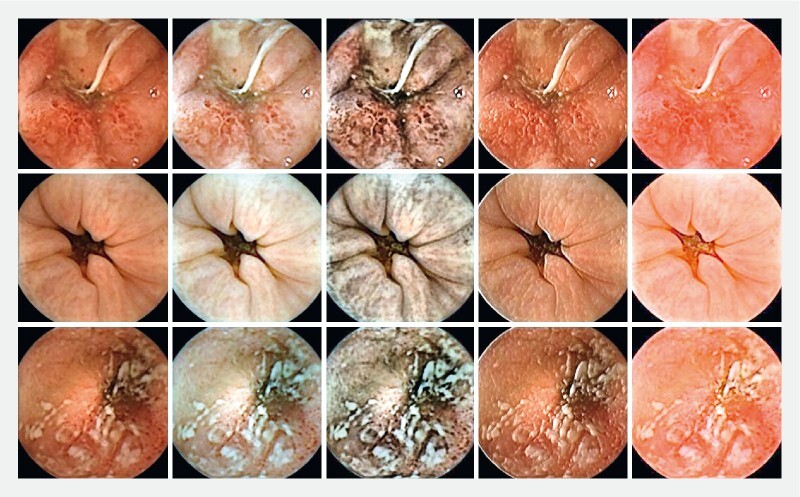
Illustration of applied texture enhancement methods on three random samples from our collected data. The five columns correspond to original images, contrast increase images, histogram equalization images, gradient x images, and dehazing images, respectively.
Using the image processing steps previously described, five separate datasets were created; one for original images and the remaining four for enhanced images. For each dataset, a separate deep learning model was trained and at the end, the results were merged to a single classification output. Configuration of all five models, however, was the same. The only difference between them was that they were trained either on the original set of images or on a set of images with a specific texture enhancement method.
Splits for training and validation
Two separate data splits were performed to evaluate our automated framework:
Random split: In this split, 70 % of all input images from each category were randomly chosen for training, 10 % for validation, and remaining 20 % were used as an independent test set.
Patient split: In this split, all images from a single patient were used either for training or for testing. The ratio between training, validation, and testing samples was as close as possible to the previous split.
Because the size of the training dataset was not sufficient for a deep learning algorithm, a data augmentation was employed. In the training part of the dataset, each image was rotated by 90°, 180°, and 270°. Together with the mirroring operation applied on original and rotated samples, the augmentation step resulted in seven new training samples that were derived from each original image.
The training process was done using a fine-tuning approach that is known as transfer learning 14 . The stochastic gradient descent was utilized with the momentum optimizer and the initial learning rate of 0.001. The mini-batch size of 8 images was used. Each model was trained for 30 epochs. In this work, only results obtained on the independent test set that was not used during the training process are reported.
ResNet-50 architecture pre-trained on ImageNet was employed 15 . The last three layers were removed and replaced with new fully-connected layer, softmax layer, and classification layer. The last layer was set to classify images directly to our 13 desired categories. All images were resized to an appropriate input size using the bicubic interpolation, and all tests were performed using MATLAB R2019b.
After training, all five models were evaluated on test images. For each test image variant, softmax probabilities for each output class were extracted. At the end, corresponding probabilities were multiplied (five values for each output class, one from each model) and assigned the test image to the output class with the highest value. Illustration of this approach is provided in Fig.3 .
Fig. 3.
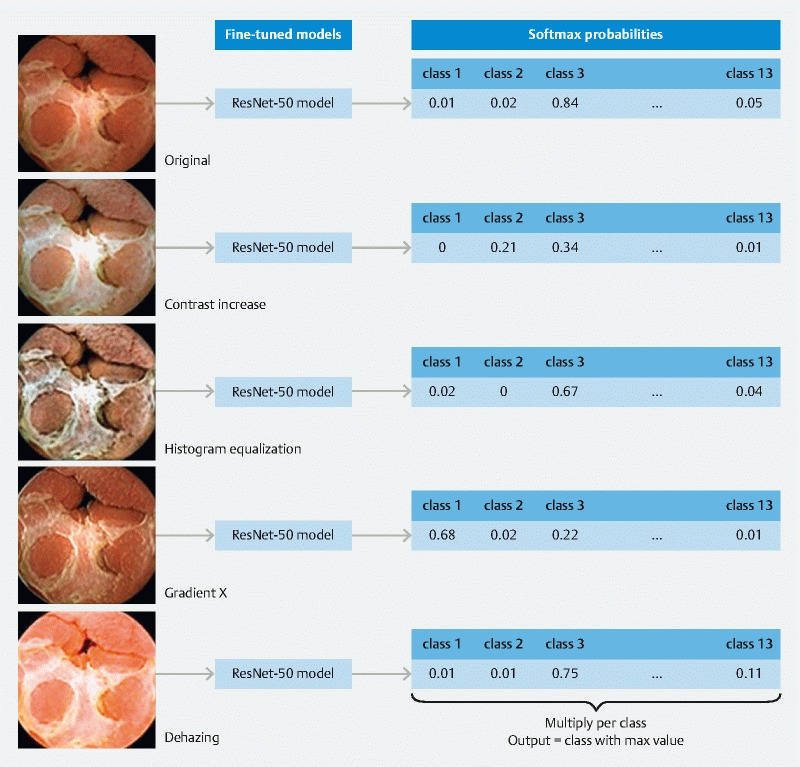
Illustration of the classification process.
Statistics
Manual classification of images served as reference standard. The sensitivity, specificity, and diagnostic accuracy of our automated framework for detection of CD lesions was calculated from 2 × 2 contingency tables with 95 % confidence intervals (CI). For the overall evaluation of sensitivity and specificity, a lesion was considered true positive if it was detected in accordance with manual reading irrespective of the localization (i. e. an ulceration in the small bowel classified as an ulceration in the colon is a true positive for detection of ulceration overall). Agreement between the gold standard and our automated framework for lesion classification was assessed with kappa statistics. Kappa values were interpreted the following way: absence of agreement 0, slight agreement < 0.20, fair agreement 0.21 to 0.40, moderate agreement 0.41 to 0.60, substantial agreement 0.61 to 0.80, and almost perfect agreement > 0.81 as proposed by Landis and Koch 16 .
Ethics
The above-mentioned studies were approved by the Local Ethics Committee of Southern Denmark (S-20150189 and S-20170188) and the Danish Data Protection Agency (journal number 16/10457 and 18/11210). All patients gave informed consent before participation including permission to use anonymized CE videos for additional analysis.
Results
A total of 38 patients were included in the study of which 33 patients were examined for clinically suspected CD and five patients had an established diagnosis of CD. After ileocolonoscopy with biopsies, pan-enteric CE and MR-enterocolonography, 31 patients were diagnosed with active CD. Ulcerations were located in the small bowel, colon, and small bowel plus colon in 12, 10 and 9 patients, respectively.
Overall, 7744 anonymized image frames (small bowel 4972, colon 2772) were manually collected and annotated. 2748 of them contained at least one ulceration (small bowel 1857, colon 891). A total of 408 images showed non-ulcerative inflammation in patients with concomitant lesions consistent with CD or an established diagnosis of CD. The number of images and specific lesions used for training, validation, and testing in both splits are shown in Table 1 .
Table 1. Number of images used for training, validation, and testing in both considered splits.
| Random split | Patient split | Total | ||||||
| Training (after augmentation) | Validation | Testing | Training (after augmentation.) | Validation | Testing | |||
| Small bowel | Normal | 712 (5,696) |
101 | 204 | 714 (5,712) |
101 | 202 | 1017 |
| Normal with bubbles/debris | 1415 (11,320) |
202 | 406 | 1,429 (11,432) |
202 | 392 | 2023 | |
| Lymphoid hyperplasia | 32 (256) |
4 | 10 | 32 (256) |
4 | 10 | 46 | |
| Non-ulcerated inflammation | 21 (168) |
2 | 6 | 22 (176) |
2 | 5 | 29 | |
| Aphthous ulceration | 514 (4,112) |
73 | 148 | 538 (4,304) |
73 | 124 | 735 | |
| Ulcer | 504 (4,032) |
72 | 144 | 520 (4,160) |
72 | 128 | 720 | |
| Fissure/large ulcer | 280 (2,240) |
40 | 82 | 285 (2,280) |
40 | 77 | 402 | |
| Colon | Normal | 150 (1,200) |
21 | 44 | 154 (1,232) |
21 | 40 | 215 |
| Normal with bubbles/debris | 901 (7,208) |
128 | 258 | 916 (7,328) |
128 | 243 | 1287 | |
| Non-ulcerated inflammation | 266 (2,128) |
37 | 76 | 270 (2,160) |
37 | 72 | 379 | |
| Aphthous ulceration | 184 (1,472) |
26 | 54 | 193 (1,544) |
26 | 45 | 264 | |
| Ulcer | 237 (1,896) |
33 | 68 | 254 (2,032) |
33 | 51 | 338 | |
| Fissure/large ulcer | 203 (1,624) |
28 | 58 | 219 (1,752) |
28 | 42 | 289 | |
| Total |
5,419
(43,352) |
767 | 1,558 |
5,546
(44,368) |
767 | 1,431 | 7744 | |
Lesion classification
Our automated framework was evaluated on three different levels. The first level is a multiclass classification, where 13 classes used in the training process were considered. For the patient split, the algorithm was tested on 1431 image frames ( Table 2 ). The agreement between the automated framework and manual reading was substantial (κ = 0.74). Using a random split of patients for training, validation, and testing, an almost perfect agreement was achieved on 1558 images (κ = 0.89).
Table 2. Algorithm testing on 1431 images with a patient split used for training.
| Deep learning framework | |||||||||||||||
| Colon_aphtae | _debris | _fissure | _non-ulc. Inflamm. | _normal | _ulcer | SB_aphtae | _fissure | _non-ulc. Inflamm. | _normal | _debris | _lymph. hyp. | _ulcer | Total | ||
| Gold standard | Colon_aphtae | 34 | 2 | 0 | 3 | 0 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 45 |
| _debris | 0 | 223 | 0 | 0 | 15 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 243 | |
| _fissure | 6 | 1 | 0 | 6 | 0 | 20 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | 42 | |
| _non-ulc. Inflamm. | 21 | 2 | 1 | 39 | 1 | 2 | 1 | 0 | 4 | 0 | 1 | 0 | 0 | 72 | |
| _normal | 0 | 6 | 0 | 0 | 34 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 40 | |
| _ulcer | 9 | 1 | 0 | 8 | 0 | 27 | 0 | 5 | 0 | 0 | 0 | 0 | 1 | 51 | |
| SB_aphtae | 0 | 0 | 0 | 0 | 0 | 0 | 109 | 0 | 4 | 0 | 0 | 0 | 11 | 124 | |
| _fissure | 0 | 0 | 1 | 0 | 0 | 5 | 3 | 50 | 1 | 0 | 3 | 0 | 14 | 77 | |
| _non-ulc. Inflamm. | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 5 | |
| _normal | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 186 | 16 | 0 | 0 | 202 | |
| _debris | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 17 | 374 | 0 | 1 | 392 | |
| _lymph. hyp. | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 8 | 0 | 10 | |
| _ulcer | 1 | 0 | 0 | 1 | 0 | 8 | 46 | 39 | 0 | 0 | 4 | 0 | 29 | 128 | |
| Total | 71 | 236 | 2 | 58 | 50 | 65 | 164 | 105 | 9 | 203 | 404 | 8 | 56 | 1431 | |
The matrix displays the number of images according to their classification with the gold standard and deep learning framework depending on the location. Lesions were assigned to one of 13 predefined categories. The inter-modality agreement was substantial (κ = 0.74).
The framework was able to firmly distinguish between the small bowel and colon. For the random split, only four of 558 images of the colon were misclassified as the small bowel (29 of 493 for the patient split), and only seven of 1000 images of the small bowel were misclassified as the colon (18 of 938 for the patient split).
Diagnostic accuracy
The second set of tests was focused on the diagnostic accuracy for the detection of CD. For the patient split, the automated framework detected ulcerations consistent with CD with a sensitivity, specificity, and diagnostic accuracy of 95.7 % (CI 93.4–97.4), 99.8 % (CI 99.2–100), and 98.4 % (CI 97.6–99.0), respectively ( Table 3 ). The diagnostic accuracy was similar for lesions located in the small bowel and colon – 98.5 % (CI 97.5–99.2) and 98.1 % (CI 96.3–99.2), respectively. For detection of CD including non-ulcerated inflammation, the sensitivity, specificity, and diagnostic accuracy was 96.1 % (CI 94.2–97.6), 99.9 % (CI 99.4–100), and 98.5 % (CI 97.7–99.0), respectively.
Table 3. Diagnostic accuracy, sensitivity and specificity for detection of ulcerations in the small bowel and colon in patients with suspected or known Crohnʼs disease.
| TP | TN | FP | FN | Accuracy (%) | 95 %CI | Sensitivity (%) | 95 %CI | Specificity (%) | 95 %CI | |
| Patient split | ||||||||||
|
317 | 602 | 2 | 12 | 98.50 | 97.50–99.18 | 96.35 | 93.72–98.10 | 99.67 | 98.81–99.96 |
|
130 | 283 | 0 | 8 | 98.10 | 96.29–99.18 | 94.20 | 88.90–97.46 | 100 | 98.70–100 |
|
447 | 885 | 2 | 20 | 98.38 | 97.55–98.98 | 95.72 | 93.46–97.36 | 99.77 | 99.19–99.97 |
| Random split | ||||||||||
|
359 | 620 | 0 | 15 | 98.49 | 97.52–99.15 | 95.99 | 93.47–97.74 | 100 | 99.41–100 |
|
174 | 302 | 0 | 6 | 98.76 | 97.31–99.54 | 96.67 | 92.89–98.77 | 100 | 98.79–100 |
|
533 | 922 | 0 | 21 | 98.58 | 97.83–99.12 | 96.21 | 94.26–97.64 | 100 | 99.60–100 |
Data are shown for two different split of images used training, validation and testing of the deep learning framework.
TP, true positive; TN, true negative; FP, false positive; FN, false negative.
For the random split, the sensitivity, specificity, and diagnostic accuracy was 96.2 % (CI 94.3–97.6), 100 % (CI 99.6–100), and 98.6 % (CI 97.8–99.1), respectively, with similar results for lesions located in the small bowel and colon ( Table 3 ). Ulcerations plus non-ulcerated inflammation was detected with a sensitivity 97.2 % (CI 96.6–98.3), specificity 100 % (CI 99.6–100) and diagnostic accuracy 98.8 % (CI 98.2–99.3).
Severity of Crohnʼs lesions
Images were grouped according to the type of lesion irrespective of their location in the small bowel or colon, and the ability of the automated framework to determine the severity of lesions was compared with manual reading. For the patient split, normal mucosa, aphthous ulcerations, ulcers and fissures/ large ulcers were classified with substantial agreement (κ = 0.72, Table 4 ). Using a random split for training and testing, the agreement was almost perfect (κ = 0.90).
Table 4. Classification of images with the gold standard and deep learning framework according to the severity of ulcerations regardless their localization.
| Deep learning framework | ||||||
| Normal | Aphthae | Ulcer | Fissure/large ulcer | Total | ||
| Gold standard | Normal | 885 | 0 | 1 | 1 | 887 |
| Aphtae | 9 | 146 | 14 | 0 | 169 | |
| Ulcer | 14 | 56 | 65 | 44 | 179 | |
| Fissure/large ulcer | 11 | 9 | 39 | 60 | 119 | |
| Total | 919 | 211 | 119 | 105 | 1354 | |
Data are shown for the patient split used for training. The inter-modality agreement for severity of lesions is substantial (κ = 0.72).
Discussion
CE is patient-friendly and non-invasive, and, compared to cross-sectional imaging, highly sensitive for the earliest lesions of CD 4 17 . Additional information obtained with CE about the proximal distribution of CD affects the prognosis and medical treatment 1 18 19 . Hence, CE is the preferred method for examining the small intestine in patients with suspected CD without obstructive symptoms 5 6 . With the Crohnʼs capsule, pan-enteric evaluation in one procedure is now feasible. Although the role of pan-enteric CE in CD is not yet established, it could play a major role in a future algorithm for noninvasive diagnosis and monitoring of CD.
The risk of capsule retention, required bowel preparation, and time consumption used for video analysis are important limitations for the clinical use of pan-enteric CE. Our study addresses the use of deep learning algorithms for optimizing the video analysis. At present time, there are no evidence-based recommendations regarding the optimal reading protocol for analyzing CE recordings 20 . With the existing software, reading times can be reduced by increasing the frame rate or the number of images seen simultaneously, or by using a quick view function (i. e. only a fraction of images is shown). Increasing the speed, however, results in lower detection rates 21 . Although missed lesions is undesirable, these techniques may be justified in patients with diffuse involvement of the gastrointestinal tract, e. g. CD. Deep learning algorithms are attractive because of their potential for fast video analysis while maintaining a high diagnostic accuracy.
Previous studies in this field were focused on the small bowel. In a retrospective study by Aoki et al. including 5800 images of erosions and ulcers, and 10,000 normal images, lesions were detected with a 90.8 % diagnostic accuracy and an AUC of 0.958 22 . Interestingly, the degree of obscuration due to bubbles, debris, and bile reduced the sensitivity, regardless of the lesion size. The false negative rate was 19.4 % and 8.5 % in patients with major and minor obscuration, respectively ( P = 0.001). Klang et al. developed a deep learning algorithm for the automated detection of small bowel ulcers in patients with CD 23 . With 7391 images of ulcerations and 10,249 images of normal mucosa, a diagnostic accuracy of 96.7 % was achieved. The algorithm required a median of less than 3.5 minutes to analyze a complete small bowel CE. A recent meta-analysis on this topic showed sensitivity and specificity of 95 % (CI 89–98) and 94 % (CI 90–96), respectively for ulcer detection in the small bowel with deep learning algorithms 8 .
The largest retrospective study performed so far was not included in the meta-analysis, however. Ding et al. collected 113,426,569 images from 6970 patients examined with small bowel CE performed on various indications 24 . In this extensive multi-center analysis, automated CE analysis achieved a per lesion sensitivity of 98.1 % (CI 96.0–99.2) and a specificity of 100 % (99.9–100) for detection of ulcers. Inflammation was diagnosed with a 93,9 % sensitivity (CI 92.6–94.9). The deep learning algorithm identified abnormalities with a higher sensitivity and significantly shorter reading times compared to manual analysis (5.9 ± 2.2 minutes vs. 96.6 ± 22.5 minutes, P < 0.001).
To the best of our knowledge, this is the first study to examine the use of deep learning for detection of CD in the both the small bowel and colon. It is also the first study to apply texture enhancement methods for capsule endoscopy images. In 7744 images collected from patients with clinically suspected or known CD, our automated framework diagnosed ulcerations with an almost perfect sensitivity and specificity (> 95 %) compared to manual analysis by two gastrointestinal experts. In our test set of 1558 images, only four colon images (about 0.26 % of all test samples) and seven small bowel images (about 0.45 % of all test samples) were misclassified. Typical reasons include abnormalities in inputs caused by some rare artifacts.
It should be emphasized, that we did not use a grading scale to evaluate the bowel cleansing and image quality in each frame although non-diagnostic CEs were excluded from the analysis (i. e. large amount of debris precluding a complete examination). Instead, we randomly selected images from patients examined for CD and classified them according to the type of lesion and presence of debris or bubbles. Our aim was create an algorithm that could discriminate a normal mucosa with debris or bubbles from CD lesions. We achieved a similar high diagnostic accuracy for detection of ulcerations in the small bowel and colon. Although the image quality was not included in our analysis, the impact of obscuration found by Aoki et al. 22 did not result in a lower sensitivity for detection of CD in the colon.
Endoscopic disease severity is currently based on validated scores with ileocolonoscopy or CE: Crohn's Disease Endoscopic Index of Severity (CDEIS), Simple Endoscopic Score for Crohn's Disease (SES-CD), Lewis Score or Capsule Endoscopy Crohn's Disease Activity Index (CECDAI) 17 . Common denominators in these scores are ulcer size and the affected surface. No previous study of deep learning algorithms included lesion characterization, which is fundamental for determining the disease severity. In this study, ulcerations were classified as aphthous ulcerations, ulcers, or fissures/large ulcers with a substantial to almost perfect agreement compared to manual reading. In a recent study, Barash et al. found an agreement between manual reading and deep learning of 67 % for discriminating ulcers of different severity with small bowel CE (grades 1–3 from mild to severe) 25 . There was excellent accuracy when comparing grade 1 ulcerations with grade 3 ulcerations (specificity and sensitivity of 0.91 and 0.91, respectively). These results encourage a future role of deep learning algorithms for autonomous assessment of the disease severity in CD.
There are some limitations to this study. First, two different splits for training and testing were applied. With the random split, there is a risk of bias in favor of the automated classification because images from the same patient are included for training and testing. Hence, the algorithm may recognize lesions with similar appearance from the same patient, which tends to increase the diagnostic accuracy. With the patient split, images from the same patient were used either for training or for testing. This, however, tends to lower the diagnostic accuracy because of variance in visual appearance between patients (color, lighting, debris, bubbles, lesions types, localization, etc.). Validation of our results in a larger cohort will overcome this issue. Second, the number of patients was limited and the analysis was retrospective, although patients were recruited from two ongoing prospective studies of patients with suspected or known CD based on accepted clinical criteria. Third, this study – similar to previous studies – included static images of normal mucosa and CD lesions, and results cannot be generalized to full-length CEs. Our results need validation on full length video sequences. This step is pivotal before clinical implementation of the framework. Fourth, the algorithm performed equally well in the small bowel and colon. However, we did not include a grading scale to evaluate the bowel cleansing and image quality in each image frame. Finally, data augmentation was used in the analysis to increase the number of samples. This is very common in studies employing deep learning techniques. It should be emphasized that this process only applies for training the algorithm.
Conclusions
In conclusion, we built a robust and efficient framework for automated recognition of CD lesions with various severities located in the small bowel and colon. The technical solution relies on combined multiple pre-trained deep learning models and a unique image preprocessing step. The framework was extensively evaluated using different testing scenarios, and we report results with almost perfect agreement with the clinical standard. These results are promising for future automated diagnosis of CD. Deep learning approaches have great potential to help clinicians detect, localize, and determine the severity of CD with pan-enteric CE.
Footnotes
Competing interests The authors declare that they have no conflict of interest.
References
- 1.Torres J, Mehandru S, Colombel J F et al. Crohn's disease. Lancet. 2017;389:1741–1755. doi: 10.1016/S0140-6736(16)31711-1. [DOI] [PubMed] [Google Scholar]
- 2.Burisch J, Kiudelis G, Kupcinskas L et al. Natural disease course of Crohn's disease during the first 5 years after diagnosis in a European population-based inception cohort: an Epi-IBD study. Gut. 2019;68:423–433. doi: 10.1136/gutjnl-2017-315568. [DOI] [PubMed] [Google Scholar]
- 3.Koulaouzidis A, Iakovidis D K, Karargyris A et al. Optimizing lesion detection in small-bowel capsule endoscopy: from present problems to future solutions. Expert Rev Gastroenterol Hepatol. 2015;9:217–235. doi: 10.1586/17474124.2014.952281. [DOI] [PubMed] [Google Scholar]
- 4.Jensen M D, Nathan T, Rafaelsen S R et al. Diagnostic accuracy of capsule endoscopy for small bowel Crohn's disease is superior to that of MR enterography or CT enterography. Clin Gastroenterol Hepatol. 2011;9:124–129. doi: 10.1016/j.cgh.2010.10.019. [DOI] [PubMed] [Google Scholar]
- 5.Pennazio M, Spada C, Eliakim R et al. Small-bowel capsule endoscopy and device-assisted enteroscopy for diagnosis and treatment of small-bowel disorders: European Society of Gastrointestinal Endoscopy (ESGE) Clinical Guideline. Endoscopy. 2015;47:352–376. doi: 10.1055/s-0034-1391855. [DOI] [PubMed] [Google Scholar]
- 6.Maaser C, Sturm A, Vavricka S R et al. ECCO-ESGAR Guideline for Diagnostic Assessment in IBD Part 1: Initial diagnosis, monitoring of known IBD, detection of complications. J Crohns Colitis. 2019;13:144–164. doi: 10.1093/ecco-jcc/jjy113. [DOI] [PubMed] [Google Scholar]
- 7.Ahmad O F, Soares A S, Mazomenos E et al. Artificial intelligence and computer-aided diagnosis in colonoscopy: current evidence and future directions. Lancet Gastroenterol Hepatol. 2019;4:71–80. doi: 10.1016/S2468-1253(18)30282-6. [DOI] [PubMed] [Google Scholar]
- 8.Soffer S, Klang E, Shimon O et al. Deep learning for wireless capsule endoscopy: a systematic review and meta-analysis. Gastrointest Endosc. 2020;92:831–839 e838. doi: 10.1016/j.gie.2020.04.039. [DOI] [PubMed] [Google Scholar]
- 9.Nadimi E S, Buijs M M, Herp J et al. Application of deep learning for autonomous detection and localization of colorectal polyps in wireless colon capsule endoscopy. Comput Elect Eng. 2020:81.106531. [Google Scholar]
- 10.Gionchetti P, Dignass A, Danese S et al. 3rd European Evidence-based Consensus on the Diagnosis and Management of Crohn's Disease 2016: Part 2: Surgical Management and Special Situations. J Crohns Colitis. 2017;11:135–149. doi: 10.1093/ecco-jcc/jjw169. [DOI] [PubMed] [Google Scholar]
- 11.Spada C, Hassan C, Galmiche J P et al. Colon capsule endoscopy: European Society of Gastrointestinal Endoscopy (ESGE) Guideline. Endoscopy. 2012;44:527–536. doi: 10.1055/s-0031-1291717. [DOI] [PubMed] [Google Scholar]
- 12.Daněk O, Matula P, Maška M et al. Smooth Chan–Vese segmentation via graph cuts. Pattern Recognition Letters. 2012;33:1405–1410. [Google Scholar]
- 13.Zuiderveld K. Contrast Limited Adaptive Histogram Equalization. Graphics Gems IV Academic Press Professional. 1994:474–485. [Google Scholar]
- 14.Yosinski J, Clune J, Bengio Y et al. How transferable are features in deep neural networks? Adv Neural Informat Proc Syst. 2014:3320–3328. [Google Scholar]
- 15.He K, Zhang X, Ren S Deep residual learning for image recognition. IEEE Conference on Computer Vision and Pattern Recognition 2016. pp. 770–778.
- 16.Landis J R, Koch G G. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- 17.Annese V, Daperno M, Rutter M D et al. European evidence based consensus for endoscopy in inflammatory bowel disease. J Crohns Colitis. 2013;7:982–1018. doi: 10.1016/j.crohns.2013.09.016. [DOI] [PubMed] [Google Scholar]
- 18.Hansel S L, McCurdy J D, Barlow J M et al. Clinical Benefit of capsule endoscopy in crohn's disease: impact on patient management and prevalence of proximal small bowel involvement. Inflamm Bowel Dis. 2018;24:1582–1588. doi: 10.1093/ibd/izy050. [DOI] [PubMed] [Google Scholar]
- 19.Kopylov U, Nemeth A, Koulaouzidis A et al. Small bowel capsule endoscopy in the management of established Crohn's disease: clinical impact, safety, and correlation with inflammatory biomarkers. Inflamm Bowel Dis. 2015;21:93–100. doi: 10.1097/MIB.0000000000000255. [DOI] [PubMed] [Google Scholar]
- 20.Rondonotti E, Spada C, Adler S et al. Small-bowel capsule endoscopy and device-assisted enteroscopy for diagnosis and treatment of small-bowel disorders: European Society of Gastrointestinal Endoscopy (ESGE) Technical Review. Endoscopy. 2018;50:423–446. doi: 10.1055/a-0576-0566. [DOI] [PubMed] [Google Scholar]
- 21.Jensen M D, Brodersen J B, Kjeldsen J. Capsule endoscopy for the diagnosis and follow up of Crohn's disease: a comprehensive review of current status. Ann Gastroenterol. 2017;30:168–178. doi: 10.20524/aog.2016.0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aoki T, Yamada A, Aoyama K et al. Automatic detection of erosions and ulcerations in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest Endosc. 2019;89:357–363 e352. doi: 10.1016/j.gie.2018.10.027. [DOI] [PubMed] [Google Scholar]
- 23.Klang E, Barash Y, Margalit R Y et al. Deep learning algorithms for automated detection of Crohn's disease ulcers by video capsule endoscopy. Gastrointest Endosc. 2020;91:606–613 e602. doi: 10.1016/j.gie.2019.11.012. [DOI] [PubMed] [Google Scholar]
- 24.Ding Z, Shi H, Zhang H et al. Gastroenterologist-level identification of small-bowel diseases and normal variants by capsule endoscopy using a deep-learning model. Gastroenterology. 2019;157:1044–1054 e1045. doi: 10.1053/j.gastro.2019.06.025. [DOI] [PubMed] [Google Scholar]
- 25.Barash Y, Azaria L, Soffer S et al. Ulcer severity grading in video capsule images of patients with Crohn's disease: an ordinal neural network solution. Gastrointest Endosc. 2021;93:187–192. doi: 10.1016/j.gie.2020.05.066. [DOI] [PubMed] [Google Scholar]


