Abstract
Bifidobacterium longum subsp. infantis ATCC 15697 possesses five α-L-fucosidases, which have been previously characterized toward fucosylated human milk oligosaccharides containing α1,2/3/4-linked fucose [Sela et al.: Appl. Environ. Microbiol., 78, 795-803 (2012)]. In this study, two glycoside hydrolase family 29 α-L-fucosidases out of five (Blon_0426 and Blon_0248) were found to be 1,6-α-L-fucosidases acting on core α1,6-fucose on the N-glycan of glycoproteins. These enzymes readily hydrolyzed p-nitrophenyl-α-L-fucoside and Fucα1-6GlcNAc, but hardly hydrolyzed Fucα1-6(GlcNAcβ1-4)GlcNAc, suggesting that they de-fucosylate Fucα1-6GlcNAcβ1-Asn-peptides/proteins generated by the action of endo-β- N-acetylglucosaminidase. We demonstrated that Blon_0426 can de-fucosylate Fucα1-6GlcNAc-IgG prepared from Rituximab using Endo-CoM from Cordyceps militaris. To generate homogenous non-fucosylated N-glycan-containing IgG with high antibody-dependent cellular cytotoxicity (ADCC) activity, the resulting GlcNAc-IgG has a potential to be a good acceptor substrate for the glycosynthase mutant of Endo-M from Mucor hiemalis. Collectively, our results strongly suggest that Blon_0426 and Blon_0248 are useful for glycoprotein glycan remodeling.
Keywords: core fucose, fucosidase, GH29, gut bacteria, probiotics
Abbreviations
ADCC, antibody-dependent cellular cytotoxicity; FL, fucosyllactose; GH, glycoside hydrolase; HMO, human milk oligosaccharide; LNFP, lacto-N-fucopentaose; pNP, para-nitrophenol.
INTRODUCTION
Bifidobacterium bifidum and B. longum subsp. infantis are the predominant commensals found in the gut of breast-fed infants. The ability to utilize human milk oligosaccharides (HMOs) is believed to be a factor in their predominance. It has been reported that these two bifidobacteria assimilate HMOs in distinct ways. 1) 2) 3) We assume that B. bifidum degrades HMOs into monosaccharides and disaccharides on the outer surface of the cells, since α-sialidases, 4) 5) α-L-fucosidases, 6) 7) 8) β- N -acetylhexosaminidases, 9) β-galactosidases, 9) and lacto- N -biosidase 10) 11) from B. bifidum JCM 1254 are predicted to be C-terminally membrane-anchored extracellular enzymes. Lacto- N -biose I (Galβ1-3GlcNAc), released from a major core tetrasaccharide lacto- N -tetraose (Galβ1-3GlcNAcβ1-3Galβ1-4Glc) by lacto- N -biosidase, is then incorporated into the cytosol by an ABC-type transporter 12) and degraded by intracellular lacto- N -biose I phosphorylase. 13) On the other hand, B. longum subsp. infantis is believed to incorporate intact HMOs and degrade them in the cytosol, since B. longum subsp. infantis ATCC 15697 possesses several sugar transporters with different specificities for various oligosaccharides 14) 15) and intracellular glycosidases, such as α-sialidases 16) and α-L-fucosidases 17) involved in the release of terminal sugars, in addition to β- N -acetylhexosaminidases and β-galactosidases involved in the degradation of core structures. 18)
The genome of B. longum subsp. infantis ATCC 15697 contains five α-L-fucosidase genes, which have been previously cloned and characterized. 17) All α-L-fucosidases are predicted to be intracellular enzymes. Blon_2335, belonging to glycoside hydrolase family 95 (GH95), prefers α1,2-L-fucosidic linkage, whereas Blon_2336, belonging to GH29 subfamily B (GH29_B), shows specificity for α1,3/4-L-fucosidic linkages likely to other GH29_B enzymes, such as AfcB from B. bifidum 8) and BT_2192 from Bacteroides thetaiotaomicron . 19) Blon_2336 may recognize branched structures [Galβ1-3/4(Fucα1-4/3)GlcNAc], as previously reported in AfcB. 20) Blon_0346 was initially classified into GH29, however, it later separated into a new family, GH151, which hydrolyzes p-nitrophenyl-α-L-fucoside (pNP-α-Fuc) and Fucα1-2Gal, but not 2´-fucosyllactose (2´-FL, Fucα1-2Galβ1-4Glc). Blon_0248 and Blon_0426, belonging to GH29_A, were reported to act on neither di- nor trisaccharides, such as 2´-FL, 3-fucosyllactose [3-FL, Galβ1-4(Fucα1-3)Glc] and Fucα1-2Gal, however, they were found to slowly hydrolyze the α1,2-L-fucosidic linkage in lacto- N -fucopentaose I (LNFP I, Fucα1-2Galβ1-3GlcNAcβ1-3Galβ1-4Glc) and the α1,3-L-fucosidic linkage in lacto- N -fucopentaose III [LNFP III, Galβ1-4(Fucα1-3)GlcNAcβ1-3Galβ1-4Glc].
In this study, we demonstrate that Blon_0248 and Blon_0426 are 1,6-α-L-fucosidases. Although an α1,6-fucosidic linkage may not exist in HMOs, it is found in the core-fucosylated N -glycans [R-GlcNAcβ1-4(Fucα1-6)GlcNAcβ1-Asn] in animal glycoproteins. Although Blon_0248 and Blon_0426 did not act on the intact core-fucosylated N -glycoprotein, they removed fucose from Fucα1-6GlcNAcβ1-Asn-protein, which could be generated by the action of endo-β- N -acetylglucosaminidase. Since B. longum subsp. infantis possesses an extracellular membrane-anchored endo-β- N -acetylglucosaminidase, 21) Blon_0248 and Blon_0426 may be involved in the degradation of fucose-containing N -glycopeptides.
MATERIALS AND METHODS
Cloning and expression of Blon_0248 and Blon_0426.
Full length DNA fragments of Blon_0248 and Blon_0426 were amplified by high-fidelity PCR using genomic DNA from B. longum subsp. longum ATCC 15697 (= JCM 1222) as a template with the following primer pairs: common forward primer: AAACATATGGTGTTGTTCATGGCCAA; reverse primers: TTTGTCGACGCGACGGACGAAGTGCA for Blon_0248 and TTTGTCGACGTGTCGAGCAAAACGCA for Blon_0426. The amplified fragments were treated with NdeI and SalI and ligated into the corresponding sites of pET-23b. The nucleotide sequence was confirmed by sequencing. Escherichia coli Rosetta (DE3) pLacI (Novagen, Germany) was transformed with pET-23b/Blon_0248 and pET-23b/Blon_0426. Both transformants were cultured in Luria-Bertani (LB) liquid medium containing 100 µg/mL ampicillin at 25 ºC until an optical density at 600 nm reached to 0.5 (ca. 14 h). IPTG was then added to the culture at a final concentration of 0.5 mM. The cells were cultured for an additional 26 h at 16 ºC to express Blon_0248, and for an additional 7 h at 25 ºC to express Blon_0426. The cells were harvested and lysed using BugBuster Protein Extraction Reagent (Novagen, Germany). After centrifugation, the supernatant was applied to a HisTrap HP column (1 mL, GE Healthcare, UK). The column was then washed with 5 mM imidazole in 50 mM sodium phosphate buffer (pH 7.0) containing 250 mM NaCl, followed by the elution of the adsorbed proteins by adding 250 mM imidazole to the same buffer. The active fraction was applied to gel filtration using an ÄKTA explorer equipped with a Superdex 200 10/300 GL column (GE Healthcare) using 50 mM sodium phosphate buffer (pH 7.0) containing 150 mM NaCl. The active fraction was concentrated with Microcon (Millipore) and dialyzed against 50 mM sodium acetate buffer (pH 6.0). The protein concentration was determined by Protein Assay Reagent (Bio-Rad, CA, USA) using bovine serum albumin (BSA) as a standard. Purified enzyme was analyzed by SDS-PAGE using a 10 % polyacrylamide gel under reducing conditions. Proteins were stained with CBB Stain One (Nacalai Tesque, Japan). Precision Plus Protein Dual Color Standards (Bio-Rad) were used as markers.
Enzyme assay.
Substrates were incubated with the enzymes at 37 ºC for the indicated time in 50 mM sodium acetate buffer (pH 6.0). For pNP-glycosides (Sigma-Aldrich), the reaction was stopped by the addition of 1.5 times volume of 1.0 M Na 2 CO 3 (pH 10.9). The amount of pNP released was determined by measuring the absorbance at 405 nm. For oligosaccharides, each substrate (approximately 10 µg) was incubated with 0.05 µg of enzyme for 2 h in 30 µL of the buffer. The reaction mixture was analyzed by silica-gel TLC (Merck 5553, Germany) with 1-butanol:acetic acid:water (2:1:1, by volume) as a developing solvent and visualized with spraying diphenylamine-aniline-phosphoric acid reagent (0.1 g diphenylamine, 0.1 mL aniline, and 1.0 mL phosphoric acid dissolved in 10 mL acetone) followed by heating at 140 ºC for 15 min. To determine the concentration of the released fucose, fucose dehydrogenase was used. Formed NADPH reduces Cu 2+ to Cu + , where the latter interacts with neocuproine to yield a complex with a maximal absorbance at 455 nm. 22)
Preparation of recombinant Endo-CoM and Fuca1-6GlcNAc-IgG.
All Escherichia coli strains and broth for recombinant proteins production were utilized as described previously. 23) E. coli expressing His 6 -tagged Endo-CoM from Cordyceps militaris was precultured in the liquid medium containing 1.25 % triptone, 2.5 % yeast extract, 0.85 % NaCl, 0.4 % glycerol, 20 mM Tris/HCl (pH 7.2), and 30 mg/L ampicillin at 37 ºC overnight. Thereafter, the preculture was inoculated into 250 mL of LB medium containing 30 mg/L ampicillin. Upon reaching an OD 600 of 0.8–1.4, the cells were cultured with 0.4 mM IPTG at 15 ºC overnight, then were lysed using an ultrasonic disruptor on ice before centrifuging. His 6 -tagged Endo-CoM was purified from the clarified supernatant using a HisTrap FF column (1 mL, GE Healthcare) according to the protocol provided by the manufacturer. Rituximab (Rituxan ® ), an anti-CD20 monoclonal antibody, was obtained from Zenyaku Kogyo Co., Ltd. (Tokyo, Japan). Fucα1-6GlcNAc-Rituximab was prepared as follows: 195 μg of Endo-CoM was incubated with 16.6 mg of Rituximab in 100 µL of sodium acetate buffer (pH 5.0) at 30 ºC for 6 h. The reaction mixture was then subjected to a HiTrap™ Protein A HP column (1 mL, GE Healthcare) to obtain a 15 μg/μL solution of Fucα1-6GlcNAc-Rituximab. The protein concentration was measured by NanoDrop Protein A280 analysis.
LC-MS/MS analysis.
The LC-MS/MS system was comprised of a Waters Acquity H-Class Bio UHPLC System with an MS/MS detector. A Waters Vion IMS Qtof instrument was operated in positive ion/sensitivity mode at an m/z range of 400–4,000. The capillary voltage was set at 2.75 kV and the cone voltage at 140 V with a source temperature of 150 ºC and a desolvation temperature of 600 ºC. Instrument control, data processing, and deconvolution were performed using the Waters UNIFI software v. 1.8.2. with an advanced maximum entropy (MaxEnt) based procedure. The samples were analyzed on a Waters ACQUITY UPLC BEH C4 column (300 Å, 1.7 μm, 2.1 × 50 mm) at 80 ºC with a gradient of 0.1 % formic acid in water and 0.1 % formic acid in acetonitrile (5–50 %, 0.1 % formic acid in acetonitrile for 5 min).
RESULTS
Blon_0248 and Blon_0426 are 1,6-α-L-fucosidases.
Five α-L-fucosidases from B. longum subsp. infantis ATCC 15697 have been previously characterized. 17) Among these, Blon_0248 and Blon_0426 were reported to hydrolyze very slowly the α1,2/3-L-fucosidic linkages in LNFP I and LNFP III; however, neither were found to act on di- nor trisaccharides, including Fucα1-2Gal, 2´-FL, and 3-FL. In this study, we found that Blon_0248 and Blon_0426 are 1,6-α-L-fucosidases acting on Fucα1-6GlcNAc.
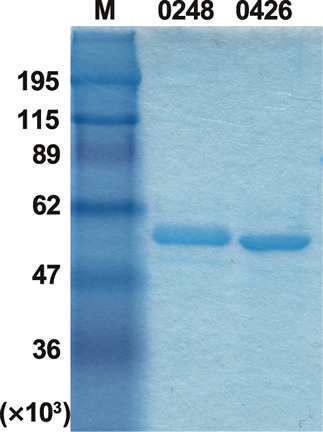
Blon_0248 and Blon_0426 have very similar amino acid sequences. Namely, they are the same length (449-aa polypeptide) with an identical N-terminal 402-aa polypeptide and variable C-terminal 47-aa polypeptide in which 19 residues were replaced. We expressed C-terminally His 6 -tagged recombinant Blon_0248 and Blon_0426 using E. coli Rosetta (DE3) pLacI. Blon_0426 was successfully expressed at 25 ºC in the presence of 0.5 mM IPTG, whereas Blon_0248 required a temperature of 16 ºC during induction. The recombinant enzymes were purified from cell-free extract by His-affinity column chromatography, followed by gel filtration. The purity and molecular size of the proteins was determined by SDS-PAGE. Blon_0426 was found to migrate slightly faster than Blon_0248, although the calculated mass was closely similar (50,266 and 50,122, respectively) ( Fig. 1 ). Both proteins specifically hydrolyzed pNP-α-Fuc, but not any of the other pNP-glycosides tested (pNP-α/β-Glc, pNP-α/β-Gal, pNP-α/β-Man, pNP-α/β-Xyl, pNP-α/β-Ara f , pNP-α/β-GlcNAc, pNP-α/β-GalNAc, and pNP-β-Fuc) (data not shown). The general properties were determined using pNP-α-Fuc as a substrate. The optimum pH for Blon_0248 and Blon_0426 was in a range of 6.0–6.5, which is consistent with the previous report. 17) Blon_0248 was found to be stable between the narrow range around pH 6.0, whereas Blon_0426 was stable between pH 5.0–7.5. Although Blon_0426 was stable at 37 ºC for 1.0 h incubation, Blon_0248 was gradually inactivated, even after incubation at 30 ºC in the optimal pH buffer (50 mM sodium acetate, pH 6.0). Considering the result of SDS-PAGE, Blon_0248 may be partially unfolded, which suggest it has relatively unstable properties. Therefore, Blon_0426 was analyzed further in the subsequent experiments.
Fig. 1. SDS-PAGE of purified Blon_0248 and Blon_0426. Proteins were analyzed by SDS-PAGE under reducing conditions. M, markers.
Substrate specificity of Blon_0426.
We incubated Blon_0426 with various fucose-containing oligosaccharides and analyzed the reaction mixture using TLC ( Figs. 2A and 2B ). Blon_0426 was found to release a small amount of fucose from Fucα1-2Gal and 2´-FL, which was in contrast to the results reported by the previous study. 17) Interestingly, in this condition, Fucα1-6GlcNAc was almost completely hydrolyzed. However, only a trace amount of fucose was released from Fucα1-6(GlcNAcβ1-4)GlcNAc. Due to the contamination of free fucose in the substrate, we were unable to detect the release of fucose from LNFP III, however, the substrate largely remained after incubation. LNFP II, Lewis a/x trisaccharides, Lewis y tetrasaccharide, sialyl Lewis a/x tetrasaccharides, and fucoidan were completely resistant. The substrate specificity of Blon_0248 was very similar to that of Blon_0426; namely Blon_0248 readily hydrolyzed Fucα1-6GlcNAc and partially hydrolyzed Fucα1-2Gal, but hardly hydrolyzed Fucα1-6(GlcNAcβ1-4)GlcNAc ( Fig. 2C ). Other substrates except for 2’-FL were resistant to Blon_0248 (data not shown).
Fig. 2. Substrate specificity of Blon_0426 and Blon_0248.
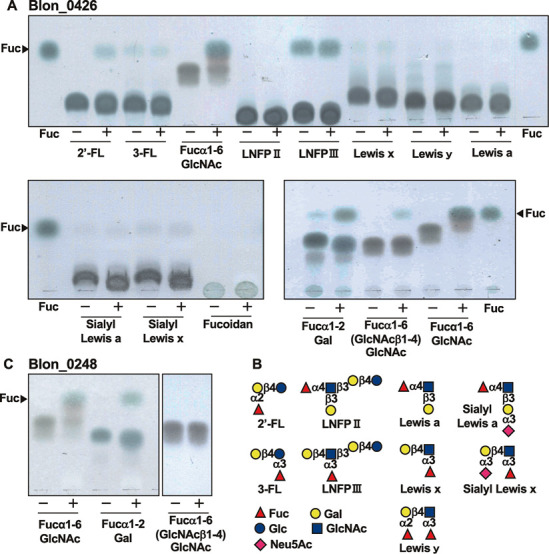
A, Blon_0426 was incubated with fucosylated oligosaccharides and analyzed by TLC. B, Structures of oligosaccharides tested. C, Blon_0248 was incubated with Fucα1-6GlcNAc, Fucα1-2Gal and Fucα1-6(GlcNAcβ1-4)GlcNAc and analyzed by TLC.
The Km and kcat values of Blon_0426 for pNP-α-Fuc, Fucα1-2Gal, 2’-FL, and Fucα1-6GlcNAc were determined ( Table 1 ). The kcat/Km value was approximately 10-fold higher for Fucα1-6GlcNAc than for Fucα1-2Gal, suggesting that Blon_0426 is a 1,6-α-fucosidase.
Table 1.
Kinetic parameters of Blon_0426 for various substrates.
| Km (mM) | kcat (s –1 ) | kcat / K m (s –1 mM –1 ) | |
|---|---|---|---|
| pNP-α-Fuc | 0.44 | 70.8 | 161 |
| Fucα1-2Gal | 1.94 | 12.6 | 6.5 |
| 2´-FL | 2.78 | 2.4 | 0.9 |
| Fucα1-6GlcNAc | 0.48 | 29.1 | 61 |
Action of Blon_0426 on Fucα1-6GlcNAc-carrying immunoglobulin.
The Fucα1-6GlcNAc structure is frequently found on the di- N , N ’-acetylchitobiose core in the N -glycan of animal glycoproteins. Since Blon_0426 preferably hydrolyzed Fucα1-6GlcNAc rather than Fucα1-6(GlcNAcβ1-4)GlcNAc, we speculated that this enzyme is involved in the degradation of Fucα1-6GlcNAc-carrying glycoproteins or glycopeptides generated from core-fucosylated N -glycoproteins or N -glycopeptides by the action of endo-β- N -acetylglucosaminidase. To confirm this hypothesis, we used Rituximab, an IgG monoclonal antibody, as a substrate, which has a heterogenous core-fucosylated N -glycan attached to Asn-297 in the constant region of a heavy chain. First, we prepared Fucα1-6GlcNAc-IgG by using Endo-CoM from Cordyceps militaris . 23) Then, Fucα1-6GlcNAc-IgG was incubated with Blon_0426 and analyzed by LC-MS/MS ( Fig. 3 ). When 10 % weight of enzyme was used for degradation, fucose was partially removed, namely the peak of the doubly fucosylated substrate (Fuc-GlcNAc/Fuc-GlcNAc, MS = 144,885 Da) was reduced, while those of the singly fucosylated form (GlcNAc/Fuc-GlcNAc, MS = 144,738 Da, theoretically 144,739 Da) and de-fucosylated form (GlcNAc/GlcNAc, MS= 144,592 Da, theoretically 144,593 Da) were generated ( Fig. 3B ). By increasing the amount of enzyme, almost all fucose was successfully removed ( Fig. 3C ).
Fig. 3. LC-MS/MS analysis of Fucα1-6GlcNAc-IgG treated with Blon_0426.
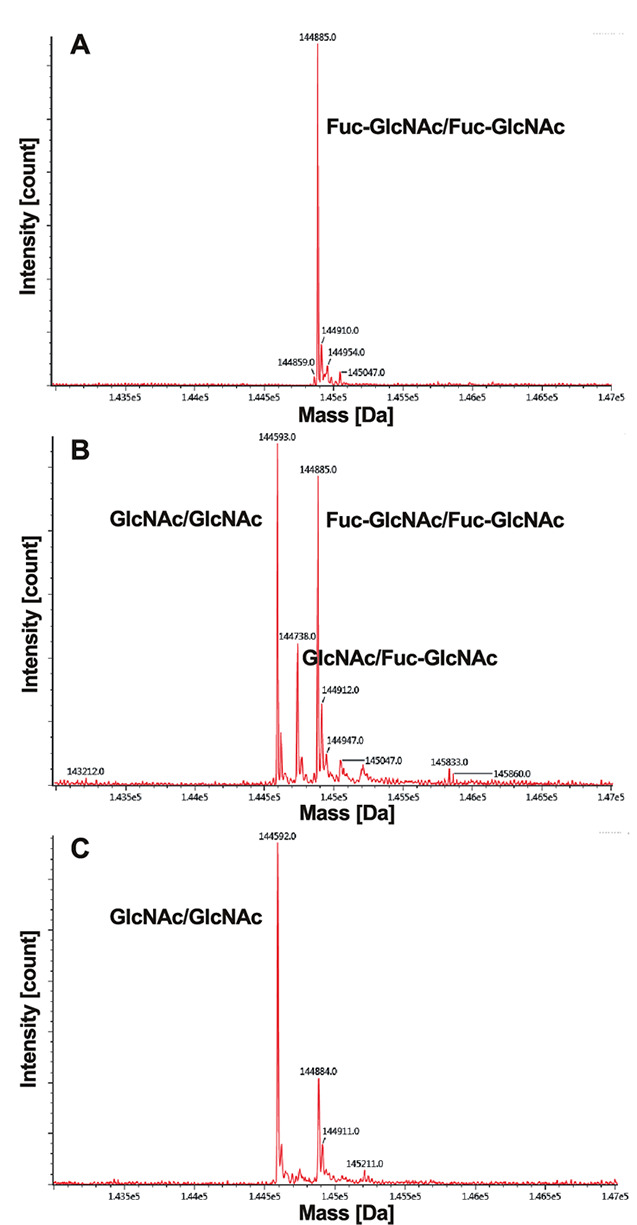
A, Fucα1-6GlcNAc-IgG; B, Fucα1-6GlcNAc-IgG treated with 10 % weight of Blon_0426; C, Fucα1-6GlcNAc-IgG treated with 50 % weight of Blon_0426.
DISCUSSION
α-L-Fucosidases are widely distributed in various organisms, including animals, plants, bacteria, and archaea. The majority of the animal α-L-fucosidases that have been characterized are lysosomal enzymes that show a broad substrate specificity for α-1,2/3/4/6-fucosidic linkages. On the other hand, bacterial α-L-fucosidases generally show a relatively narrow specificity. In fact, B. longum subsp. infantis have five α-L-fucosidases with different substrate specificities. 17) One of the major nutritional sources of B. longum subsp. infantis in the gut of breast-fed infant is HMOs, which are not digested or adsorbed by the host and are thus assimilated by the gut microbiota. In HMOs, there are four types of fucosidic linkages: Fucα1-2Gal, Fucα1-3Glc, Fucα1-3GlcNAc, and Fucα1-4GlcNAc. GH95 Blon_2335 and GH29 Blon_2336, which are encoded by tandemly arranged genes within HMO gene cluster, 18) are able to cleave all types of fucosidic linkages in HMOs. 17)
In other glycans in humans, three linkages, Fucα1-2Gal, Fucα1-3GlcNAc, and Fucα1-4GlcNAc, are found in the ABO and Lewis blood groups in the termini of the glycans of glycoproteins and glycolipids. These fucosidic linkages may also be cleavable by Blon_2335 and Blon_2336. In addition to these three linkages, Fucα1-6GlcNAc is frequently found in the core of N -glycans in glycoproteins. The core α1,6-fucose is attached by Fut8 fucosyltransferase, and this modification on N -glycoproteins plays many important biological roles. 24) In the intestines, the core-fucosylated glycoproteins are secreted from the host and also present as various foods originated from animals. It is well-known that the core fucose is highly abundant in human milk glycoproteins, such as lactoferrin 25) and secretory IgA. 26) The majority of milk proteins are digested by proteases and peptidases in the digestive tract, resulting in short peptides and amino acids being adsorbed from the small intestine. However, bulky N -glycopeptides may not be adsorbed and reach the lower intestine. B. longum subsp. infantis has extracellular GH18 endo-β- N -acetylglucosaminidase (EndoBI-1) that acts on N , N ´-diacetylchitobiose in N -glycans with both fucosylation and non-fucosylation. 21) Released distal oligosaccharides have been reported to be selective growth substrates for B. longum subsp. infantis . 27) Since Blon_0426 and Blon_0248, which are able to cleave Fucα1-6GlcNAc-protein, are localized in the cytosol, Fucα1-6GlcNAc-peptides derived from lactoferrin may be incorporated into the cytosol through an unknown transporter and assimilated there ( Fig. 4A ).
Fig. 4. Function and application of Blon_0426 and Blon_0248.
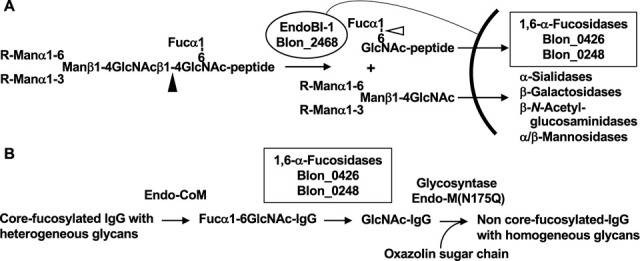
A, Proposal physiological function of Blon_0426 and Blon_0248 in B. longum subsp. infantis . The black arrowhead indicates the hydrolytic site of endo-β- N -acetylglucosaminidase EndoBl-1. The white arrowhead indicates the hydrolytic site of Blon_0426 and Blon_0248. B, Schematic of enzymatic N -glycan remodeling of pharmaceutical IgG.
The correct attachment of core α1,6-fucose by Fut8 is known to be important. 24) However, in anti-cancer antibody drugs, the presence of the core α1,6-fucose on N -glycan attached to the constant region of heavy chains was found to severely reduce the levels of ADCC (antibody-dependent cellular cytotoxicity) activity. 28) 29) Therefore, a method for the de-fucosylation of the antibody is required. Here, we showed the possibility that Blon_0426 can be used for the de-fucosylation of IgG in combination with endo-β- N -acetylglucosaminidase (Endo-CoM). By using the glycosynthase mutant of endo-β- N -acetylglucosaminidase (Endo-M N175Q) and chemically-activated oxazolin sugar, 30) 31) 32) uniform N -glycan could be added to GlcNAc-IgG ( Fig. 4B ). Bioactive glycoprotein with homogeneous N -glycan is needed for the proper evaluation of its biological activity. Several approaches have been already attempted by two groups, 33) 34) employing α-fucosidases from Lactobacillus casei (AlfC) 35) and Bacteroides fragilis NTNC 9343 (BfFucH, BF3242). 36) AlfC is a protein consisting of 344 amino acid residues with 24 % identity to Blon_0426, which shows rather strict specificity toward Fucα1-6GlcNAc. 35) By contrast, BfFucH is 434-amino-acid protein with 19 % identity to Blon_0426, which shows broad specificity toward α1-2/3/4/6-fucosidic linkages. 36) The enzyme studied here, Blon_0426 and Blon_0248, may also be useful for the glycan remodeling strategy.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
Acknowledgments
This work was supported by JSPS KAKENHI (grant numbers: 15K07448 and 18K05494) (to H.A.). We would like to thank the late Dr. Masashi Kiyohara for the contributions made during the initial stages of this study.
References
- 1).Sela D.A. and Mills D.A.: Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends. Microbiol., 18, 298–307 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Zivkovic A.M., German J.B., Lebrilla C.B., and Mills D.A.: Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc. Natl. Acad. Sci. USA, 108, Suppl. 1,4653–4658 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3).Asakuma S., Hatakeyama E., Urashima T., Yoshida E., Katayama T., Yamamoto K., Kumagai H., Ashida H., Hirose J., and Kitaoka M.: Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J. Biol. Chem., 286, 34583–34592 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Kiyohara M., Tanigawa K., Chaiwangsri T., Katayama T., Ashida H., and Yamamoto K.: An exo-α-sialidase from bifidobacteria involved in the degradation of sialyloligosaccharides in human milk and intestinal glycoconjugates. Glycobiology, 21, 437–447 (2011). [DOI] [PubMed] [Google Scholar]
- 5).Ashida H., Tanigawa K., Kiyohara M., Katoh T., Katayama T., and Yamamoto K.: Bifunctional properties and characterization of a novel sialidase with esterase activity from Bifidobacterium bifidum. Biosci. Biotechnol. Biochem., 82, 2030–2039 (2018). [DOI] [PubMed] [Google Scholar]
- 6).Katayama T., Sakuma A., Kimura T., Makimura Y., Hiratake J., Sakata K., Yamanoi T., Kumagai H., and Yamamoto K.: Molecular cloning and characterization of Bifidobacterium bifidum 1,2-α-L-fucosidase (AfcA), a novel inverting glycosidase (glycoside hydrolase family 95). J. Bacteriol., 186, 4885–4893 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Nagae M., Tsuchiya A., Katayama T., Yamamoto K., Wakatsuki S., and Kato R.: Structural basis of the catalytic reaction mechanism of novel 1,2-α-L-fucosidase from Bifidobacterium bifidum. J. Biol. Chem., 282, 18497–18509 (2007). [DOI] [PubMed] [Google Scholar]
- 8).Ashida H., Miyake A., Kiyohara M., Wada J., Yoshida E., Kumagai H., Katayama T., and Yamamoto K.: Two distinct α-L-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology, 19, 1010–1017 (2009). [DOI] [PubMed] [Google Scholar]
- 9).Miwa M., Horimoto T., Kiyohara M., Katayama T., Kitaoka M., Ashida H., and Yamamoto K.: Cooperation of β-galactosidase and β-N-acetylhexosaminidase from bifidobacteria in assimilation of human milk oligosaccharides with type 2 structure. Glycobiology, 20, 1402–1409 (2010). [DOI] [PubMed] [Google Scholar]
- 10).Wada J., Ando T., Kiyohara M., Ashida H., Kitaoka M., Yamaguchi M., Kumagai H., Katayama T., and Yamamoto K.: Bifidobacterium bifidum lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure. Appl. Environ. Microbiol., 74, 3996–4004 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Ito T., Katayama T., Hattie M., Sakurama H., Wada J., Suzuki R., Ashida H., Wakagi T., Yamamoto K., Stubbs K.A., and Fushinobu S.: Crystal structures of a glycoside hydrolase family 20 lacto-N-biosidase from Bifidobacterium bifidum. J. Biol. Chem., 288, 11795–11806 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Suzuki R., Wada J., Katayama T., Fushinobu S., Wakagi T., Shoun H., Sugimoto H., Tanaka A., Kumagai H., Ashida H., Kitaoka M., and Yamamoto K.: Structural and thermodynamic analyses of solute-binding protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I. J. Biol. Chem., 283, 13165–13173 (2008). [DOI] [PubMed] [Google Scholar]
- 13).Kitaoka M., Tian J., and Nishimoto M.: Novel putative galactose operon involving lacto-N-biose phosphorylase in Bifidobacterium longum. Appl. Environ. Microbiol., 71, 3158–3162 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14).Garrido D., Kim J.H., German J.B., Raybould H.E., and Mills D.A.: Oligosaccharide binding proteins from Bifidobacterium longum subsp. infantis reveal a preference for host glycans. PLoS One, 6, e17315 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Sakanaka M., Hansen M.E., Gotoh A., Katoh T., Yoshida K., Odamaki T., Yachi H., Sugiyama Y., Kurihara S., Hirose J., Urashima T., Xiao J.Z., Kitaoka M., Fukiya S., Yokota A., Lo Leggio L., Abou Hachem M., and Katayama T.: Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis. Sci. Adv., 5, eaaw7696 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).Sela D.A., Li Y., Lerno L., Wu S., Marcobal A.M., German J.B., Chen X., Lebrilla C.B., and Mills D.A.: An infant-associated bacterial commensal utilizes breast milk sialyloligosaccharides. J. Biol. Chem., 286, 11909–11918 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17).Sela D.A., Garrido D., Lerno L., Wu S., Tan K., Eom H.J., Joachimiak A., Lebrilla C.B., and Mills D.A.: Bifidobacterium longum subsp. infantis ATCC 15697 α-fucosidases are active on fucosylated human milk oligosaccharides. Appl. Environ. Microbiol., 78, 795–803 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18).Sela D.A., Chapman J., Adeuya A., Kim J.H., Chen F., Whitehead T.R., Lapidus A., Rokhsar D.S., Lebrilla C.B., German J.B., Price N.P., Richardson P.M., and Mills D.A.: The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. USA, 105, 18964–18969 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).Sakurama H., Tsutsumi E., Ashida H., Katayama T., Yamamoto K., and Kumagai H.: Differences in the substrate specificities and active-site structures of two α-L-fucosidases (glycoside hydrolase family 29) from Bacteroides thetaiotaomicron. Biosci. Biotechnol. Biochem., 76, 1022–1024 (2012). [DOI] [PubMed] [Google Scholar]
- 20).Sakurama H., Fushinobu S., Hidaka M., Yoshida E., Honda Y., Ashida H., Kitaoka M., Kumagai H., Yamamoto K., and Katayama T.: 1,3-1,4-α-L-Fucosynthase that specifically introduces Lewis a/x antigens into type-1/2 chains. J. Biol. Chem., 287, 16709–16719 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Garrido D., Nwosu C., Ruiz-Moyano S., Aldredge D., German J.B., Lebrilla C.B., and Mills D.A.: Endo-β- N -acetylglucosaminidases from infant gut-associated bifidobacteria release complex N -glycans from human milk glycoproteins. Mol. Cell. Proteomics, 11, 775–785 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22).Cohenford M.A., Abraham A., Abraham J., and Dain J.A.: Colorimetric assay for free and bound L-fucose. Anal. Biochem., 177, 172–177 (1989). [DOI] [PubMed] [Google Scholar]
- 23).Seki H., Huang Y., Arakawa T., Yamada C., Kinoshita T., Iwamoto S., Higuchi Y., Takegawa K., and Fushinobu S.: Structural basis for the specific cleavage of core-fucosylated N-glycans by endo-β-N-acetylglucosaminidase from the fungus Cordyceps militaris. J. Biol. Chem., 294, 17143–17154 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).Takahashi M., Kuroki Y., Ohtsubo K., and Taniguchi N.: Core fucose and bisecting GlcNAc, the direct modifiers of the N-glycan core: their functions and target proteins. Carbohydr. Res., 344, 1387–1390 (2009). [DOI] [PubMed] [Google Scholar]
- 25).Yu T., Guo C., Wang J., Hao P., Sui S., Chen X., Zhang R., Wang P., Yu G., Zhang L., Dai Y., and Li N.: Comprehensive characterization of the site-specific N-glycosylation of wild-type and recombinant human lactoferrin expressed in the milk of transgenic cloned cattle. Glycobiology, 21, 206–224 (2011). [DOI] [PubMed] [Google Scholar]
- 26).Huang J., Guerrero A., Parker E., Strum J.S., Smilowitz J.T., German J.B., and Lebrilla C.B.: Site-specific glycosylation of secretory immunoglobulin A from human colostrum. J. Proteome Res., 14, 1335–1349 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Karav S., Le Parc A., Leite Nobrega de Moura Bell J.M., Frese S.A., Kirmiz N., Block D.E., Barile D., and Mills D.A.: Oligosaccharides released from milk glycoproteins are selective growth substrates for infant-associated bifidobacteria. Appl. Environ. Microbiol., 82, 3622–3630 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Shields R.L., Lai J., Keck R., O’Connell L.Y., Hong K., Meng Y.G, Weikert S.H.A., and Presta L.G.: Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human FcγRIII and antibody-dependent cellular toxicity. J. Biol. Chem., 277, 26733–26740 (2002). [DOI] [PubMed] [Google Scholar]
- 29).Shinkawa T., Nakamura K., Yamane N., Shoji-Hosaka E., Kanda Y., Sakurada M., Uchida K., Anazawa H., Satoh M., Yamasaki M., Hanai N., and Shitara K.: The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J. Biol. Chem., 278, 3466–3473 (2003). [DOI] [PubMed] [Google Scholar]
- 30).Umekawa M., Huang W., Li B., Fujita K., Ashida H., Wang L.X., and Yamamoto K.: Mutants of Mucor hiemalis endo-β-NJ. Biol. Chem.-acetylglucosaminidase show enhanced transglycosylation and glycosynthase-like activities. J. Biol. Chem., 283, 4469–4479 (2008). [DOI] [PubMed] [Google Scholar]
- 31).Umekawa M., Li C., Higashiyama T., Huang W., Ashida H., Yamamoto K., and Wang L.X.: Efficient glycosynthase mutant derived from Mucor hiemalis endo-β-N-acetylglucosaminidase capable of transferring oligosaccharide from both sugar oxazoline and natural N-glycan. J. Biol. Chem., 285, 511–521 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32).Umekawa M., Higashiyama T., Koga Y., Tanaka T., Noguchi M., Kobayashi A., Shoda S., Huang W., Wang L.X., Ashida H., and Yamamoto K.: Efficient transfer of sialo-oligosaccharide onto proteins by combined use of a glycosynthase-like mutant of Mucor hiemalis endoglycosidase and synthetic sialo-complex-type sugar oxazoline. Biochim. Biophys. Acta., 1800, 1203–1209 (2010). [DOI] [PubMed] [Google Scholar]
- 33).Lin C.W., Tsai M.H., Li S.T., Tsai T.I., Chu K.C., Liu Y.C., Lai M.Y., Wu C.Y., Tseng Y.C., Shivatare S.S., Wang C.H., Chao P., Wang S.Y., Shih H.W., Zeng Y.F., You T.H., Liao J.Y., Tu Y.C., Lin Y.S., Chuang H.Y., Chen C.L., Tsai C.S., Huang C.C., Lin N.H., Ma C., Wu C.Y., and Wong C.H.: A common glycan structure on immunoglobulin G for enhancement of effector functions. Proc. Natl. Acad. Sci. USA, 112, 10611–10616 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34).Giddens J.P., Lomino J.V., DiLillo D.J., Ravetch J.V., and Wang L.X.: Site-selective chemoenzymatic glycoengineering of Fab and Fc glycans of a therapeutic antibody. Proc. Natl. Acad. Sci. USA, 115, 12023–12027 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35).Rodríguez-Díaz J., Monedero V., and Yebra M.J.: Utilization of natural fucosylated oligosaccharides by three novel α-L-fucosidases from a probiotic Lactobacillus casei strain. Appl. Environ. Microbiol., 77, 703–705 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).Tsai T.I., Li S.T., Liu C.P., Chen K.Y., Shivatare S.S., Lin C.W., Liao S.F., Lin C.W., Hsu T.L., Wu Y.T., Tsai M.H., Lai M.Y., Lin N.H., Wu C.Y. and Wong C.H.: An effective bacterial fucosidase for glycoprotein remodeling. ACS Chem. Biol., 12, 63–72 (2017). [DOI] [PubMed] [Google Scholar]


