Abstract
We recently characterized a 3-O-α-D-galactosyl-α-L-arabinofuranosidase (GAfase) for the release of α-D-Gal-(1→3)-L-Ara from gum arabic arabinogalactan protein (AGP) in Bifidobacterium longum subsp. longum JCM7052. In the present study, we cloned and characterized a neighboring α-galactosidase gene (BLGA_00330; blAga3). It contained an Open Reading Frame of 2151-bp nucleotides encoding 716 amino acids with an estimated molecular mass of 79,587 Da. Recombinant BlAga3 released galactose from α-D-Gal-(1→3)-L-Ara, but not from intact gum arabic AGP, and a little from the related oligosaccharides. The enzyme also showed the activity toward blood group B liner trisaccharide. The specific activity for α-D-Gal-(1→3)-L-Ara was 4.27- and 2.10-fold higher than those for melibiose and raffinose, respectively. The optimal pH and temperature were 6.0 and 50 °C, respectively. BlAga3 is an intracellular α-galactosidase that cleaves α-D-Gal-(1→3)-L-Ara produced by GAfase; it is also responsible for a series of gum arabic AGP degradation in B. longum JCM7052.
Keywords: α-D-galactosidase, glycoside hydrolase family 36, gum arabic AGP, Bifidobacterium longum
Abbreviations
AGP, arabinogalactan protein; Arap, L-arabinopyranose; Araf, L-arabinofuranose; pNP, p-nitrophenyl; GH, glycoside hydrolase family; HPAEC-PAD, high-performance anion-exchange chromatography with pulsed amperometric detection; TLC, thin-layer chromatography.
Galactosyl saccharides with α-linkage are present in natural oligosaccharides, polysaccharides and glycoconjugates, including raffinose, melibiose, stachyose, loliose, galactomannan, arabinogalactan, and blood group B antigens. α-D-Galactosidase (EC 3.2.1.22), which catalyzes terminal α-galactose hydrolysis, has been classified into six glycosyl hydrolase (GH) families based on amino acid sequence similarity: GH4, GH27, GH36, GH57, GH97, and GH110. In particular, GH36 α-galactosidases are widely distributed in archaea,1) bacteria,2),3),4) fungi,5),6),7),8) and plant.9),10),11) To date, several bifidobacterial α-galactosidases belonging to GH36 were cloned and characterized from Bifidobacterium longum subsp. longum NCC2705,12) B. longum DJO10A,13) B. bifidum NCIMB41171,3) B. adolescentis DSM20083,14),15) and B. breve 203.4) These enzymes are possibly required for the assimilation of α-galactosyl oligosaccharides in the human intestine. Gum arabic is a kind of arabinogalactan protein (AGP) used for food additive and has been reported to increase bifidobacteria, especially in B. longum.16),17),18),19) Gum arabic AGP is composed of type II arabinogalactan chains with a β1,3-galactan backbone and β1,6-galactan side chains and are modified by α-D-Gal-(1→3)-α-L-Araf-(1→3)- and β-L-Arap-(1→3)-α-L-Araf-(1→3)- in terminus.20),21),22) Previously, Saisin et al. purified an intracellular α-galactosidase from B. longum JCM7052 induced by gum arabic AGP.23) However, the role of α-galactosidase in the gum arabic AGP assimilation remained unclear. We recently characterized a key enzyme, 3-O-α-D-galactosyl-α-L-arabinofuranosidase (BLGA_00340; GAfase), that releases α-D-Gal-(1→3)-L-Ara from gum arabic AGP in B. longum JCM7052.24) The α-galactosidase gene (BLGA_00330; blAga3) was flanked by GAfase gene. In this study, we characterized BlAga3 as an intracellular enzyme for the degradation of α-D-Gal-(1→3)-L-Ara released by GAfase.
Bifidobacterium longum JCM7052 encoded three GH36 α-galactosidase candidate genes: BLGA_00330 (blAga3), BLGA_18610 (blAga2), and BLGA_18750 (blAga1).25) Based on previous study, BlAga1 is likely for the assimilation of raffinose, melibiose, and stachyose having α-(1→6)-galactosyl linkages.26),27),28) BlAga2 is likely for the degradation of α-galactobioses linked through α-(1→4)- and/or α-(1→3)-glycosidic bonds, based on the previous study for MelE (99 % identity with BlAga2) of B. breve UCC2003.27) BlAga3 exhibited 39.9 % sequence identities with BlAga1 at 83 % coverage; however, it is not significantly similar with BlAga2. BlAga3 exhibited 47.0 % sequence identity at 84 % coverage with a previously characterized GH36 α-galactosidase from Streptomyces sp. S27 ACCC41168 (GenBank ID: ACN78885.1).29) According to the signalP 4.1 and InterPro servers, the three α-galactosidase candidates lack the putative signal peptide and terminal transmembrane domain, suggesting that they are intracellular enzymes. According to previous research that proposed the classification of subfamily in GH36, BlAga1 and BlAga2 were classified into GH36 subfamilies I and II, respectively (Fig. 1).30) Except for Aga from B. breve 203 and MelE from B. breve UCC2003, almost all characterized bifidobacterial α-galactosidases were classified into subfamily I. The alignment of these enzymes exhibited that the catalytic nucleophile (D452), acid/base catalyst (D519), and residues involved in substrate binding (D340 and D341) were conserved in BlAga3, as well as other characterized GH36s15) (Fig. S1 ; see J. Appl. Glycosci. Web site). Furthermore, BlAga3 conserved C-x-x-G-x-x-R motif in the catalytic domain that was characteristic of subfamily I α-galactosidase.30) Because BlAga3 is somewhat differentiated from previously characterized bifidobacterial α-galactosidases classified into GH36 subfamily Ⅰ, BlAga3 was further divided into subfamily Ib.
Fig. 1. The phylogenetic relationships of bifidobacterial GH36 α-galactosidases.
The phylogenetic tree of BlAga3 with homologous proteins from bifidobacteria was constructed by the neighbor-joining method using the aligned sequences; for the construction, the program Clustal W was implemented in the MEGA7 software. The protein names or locus tags are shown alongside Bifidobacterium strains as follows: B. adolescentis DSM 20083 Aga (GenBank ID: AAD30994.2), B. bifidum NCIMB 41171 MelA (ABD96085.1), B. breve 203 Aga (AAK96217.2), B. breve 203 Aga2 (ABB76662.1), B. longum DJO10A GalA1 (ACD98928.1), B. longum NCC2705 AgA (AAN25312.1), B. longum VMKB44 AglL (AAG02023.1), B. breve UCC2003 RafA (ABE96531.1), B. breve UCC2003 MelE (ABE96518.1), B. longum JCM1217 BLLJ_1872 (BAJ67536.1), B. longum JCM1217 BLLJ_1885 (BAJ67549.1), B. longum JCM7052 BlAga3 (BBV22622.1), B. longum JCM7052 BlAga1 (BBV24464.1), and B. longum JCM7052 BlAga2 (BBV24450.1). BlAga3 characterized in this study is enclosed in the dashed-line box.
The genomic DNA of B. longum JCM7052 (GenBank Accession No. AP022379) was extracted using a Fast Pure DNA Kit (Takara Bio Inc., Otsu, Japan). The blAga3 gene was amplified by PCR using genomic DNA as a template. The forward (5′-AGGAGATATACCATGTTTCCTTCCGTATCGGT-3′) and reverse (5′-GTGGTGGTGCTCGAGGACTTTGACGATTTCAA-3′) primers were designed from nucleotides 7–23 and 2132–2148, respectively. The underlined letters represent the nucleotides complementary to the template. Subsequently, the amplicon was cloned into the pET-23d vector (Novagen, Inc., Madison, WI, USA) using an In-Fusion HD Cloning Kit (Clontech Laboratories Inc., Palo Alto, CA, USA). The plasmid was transformed into Escherichia coli BL21 (λDE3) cells (Genlantis, Inc., San Diego, CA, USA) and then grown at 37 °C using the Overnight Express Autoinduction System (Novagen). The cell culture was subsequently centrifuged, and the resultant pellet was then resuspended in the BugBuster Protein Extraction Reagent (Novagen). The His-tagged BlAga3 protein was purified with a column containing the TALON metal affinity resin (Clontech). The 50 mM imidazole fraction containing the eluted protein was desalted and concentrated using an ultrafiltration membrane with a 10-kDa cutoff (Millipore Co., Billerica, MA, USA). The purified recombinant BlAga3 migrated as a single band with an apparent molecular mass of 80 kDa on SDS-PAGE, which corresponded to its calculated molecular mass of 80,490 Da (Fig. S2 ; see J. Appl. Glycosci. Web site).
The optimal pH for enzyme activity was determined using 1 mM p-nitrophenyl (pNP)-α-Gal as a substrate between pH 5.0 and 8.0 using the following buffers: 50 mM sodium acetate (pH 5.0–6.0), sodium phosphate (pH 6.0–7.0), and Tris-HCl (pH 7.0–8.0) at 40 °C. The optimal temperature of enzyme activity was determined using 50 mM sodium acetate buffer (pH 6.0) at 30–55 °C. In both cases, samples were preincubated at each temperature for 5 min before adding enzyme, then incubated with enzyme for 20 min at each temperature. The optimal pH and temperature for pNP-α-Gal were 6.0 and 50 °C, respectively (Fig. 2).
Fig. 2. Optimal pH and temperature of BlAga3.
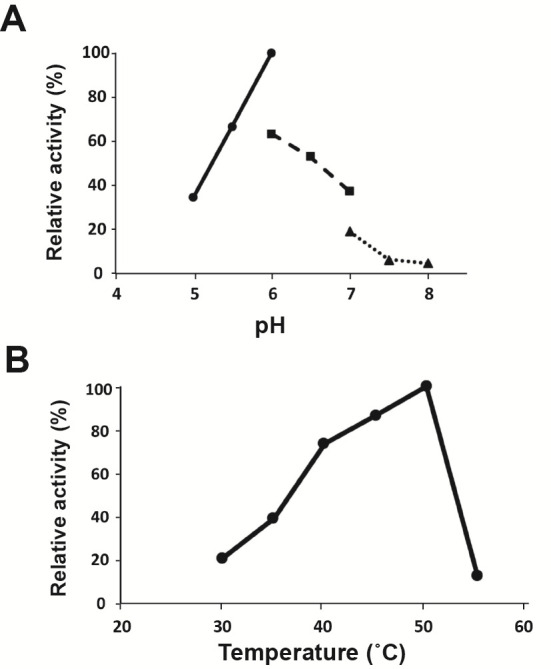
A, pH dependence of BlAga3 activity in various buffers at 40 °C for 20 min. Sodium acetate buffer (closed circle and solid line), sodium phosphate buffer (closed square and dashed line), and Tris-HCl buffer (closed triangle and dotted line) were used. Enzyme activities are expressed as a percentage of the activity in sodium acetate buffer at pH 6.0. B, the temperature dependence of BlAga3 activity at pH 6.0 for 20 min. The enzymatic activities are expressed as the percentage of the activity at 50 °C.
Substrate specificity toward pNP substrates was assayed by incubating 5 mM substrates with recombinant BlAga3 (2.0 µg/mL at final concentration) in 40 μL of 50 mM sodium acetate buffer (pH 6.0) at 37 °C for 19 h. Almost all pNP substrates were obtained from Sigma-Aldrich Corp. (St. Louis, MO, USA). pNP-β-Araf was synthesized as previously described.31) Reaction products were analyzed by thin-layer chromatography (TLC) using silica gel 60 aluminum plates (Merck KGaA, Darmstadt, Germany) with a 7:1:2 (v/v/v) 1-propanol/EtOH/water solvent mixture, and the separated sugars were visualized by spraying orcinol-sulfate reagent on the plates.32) Thus, BlAga3 exhibited activity to pNP-α-Gal and weak activity to pNP-β-Arap, but not to other pNP substrates tested (Fig. 3).
Fig. 3. TLC analysis of BlAga3 reactions to pNP substrates.
The pNP substrates were incubated in the absence (lane a) or presence (lane b) of the recombinant BlAga3 at 37 °C for 16 h. Lane 1, galactose standard; lane 2, L-arabinose standard. pNP-α-Gal (lane 3), pNP-β-Gal (lane 4), pNP-α-Arap (lane 5), pNP-β-Arap (lane 6), pNP-α-Xyl (lane 7), pNP-β-Xyl (lane 8), pNP-α-Glc (lane 9), and pNP-β-Glc (lane 10) were used as substrates.
To determine the substrate specificity of BlAga3, we used several oligosaccharides containing α1,3 galactosyl, α1,6 galactosyl, and β1,3 arabinopyranosyl linkages, as shown in Table 1. The blood group B-related oligosaccharides (liner B-2 trisaccharide, blood group B trisaccharide, and blood group B pentasaccharide) were obtained from Dextra Laboratories Ltd. (Reading, UK) and gum arabic AGP-related oligosaccharides (S3GA and S5GA) were prepared using previously described method.24) The reaction mixture consists of 50 mM sodium acetate buffer (pH 6.0), and substrates were incubated with BlAga3 at 45 °C for a suitable time, as shown in the legend of Table 1. The reaction was terminated by adding 5 % trichloroacetic acid (TCA) to one-fifth of the reaction mixture, and the diluted products were analyzed by high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) using a CarboPac PA-1 column (φ 4 × 250 mm; Dionex Corp., Sunnyvale, CA, USA) that was eluted at a flow rate of 1.0 mL/min using the following gradient: 0‒5 min, 100 % eluent A (0.1 M NaOH); 5‒30 min, 0‒100 % eluent B (0.5 M sodium acetate in 0.1 M NaOH); and 30‒35 min, 100 % eluent B. One unit of enzyme activity was defined as the amount of enzyme required to produce 1 μmol of galactose or arabinose per minute. BlAga3 hydrolyzed α-D-Gal-(1→3)-L-Ara in reducing form as the most suitable substrate in naturally occurring oligosaccharides (Table 1). Although BlAga3 acted to α-D-Gal-(1→3)-α-L-Araf-OMe in non-reducing form, the activity was 4.48-fold lower than that for α-D-Gal-(1→3)-L-Ara. It indicated that the structural degree of freedom of Ara at subsite +1 was important for the enzymatic activity. Furthermore, BlAga3 exhibited a little activity to S3GA and S5GA, and not to gum arabic AGP or larch AGP (Fig. S3; see J. Appl. Glycosci. Web site). The enzyme also reacted to blood group B liner trisaccharide with 5.85-fold lower activity than α-D-Gal-(1→3)-L-Ara but showed little reaction to blood group B branched trisaccharide, indicating that a branched form hindered the access of the enzyme to the terminal Gal. The enzyme also exhibited a little activity for β1,3-L-arabinopyranosyl linkages. However, the enzymatic activity for β-L-Arap-(1→3)-L-Ara was 156-fold lower than α-D-Gal-(1→3)-L-Ara. The bifunctional property was due to structural similarity between D-Gal and L-Arap as known in GH36 and GH27 enzymes.33),34) BlAga3 is an α-galactosidase that mainly cleaves α-D-Gal-(1→3)-L-Ara.
Table 1.
Substrate specificities of BlAga3.
| Substrate | Structure | Conc. (mM) |
Specific activity (unit/mg) |
Relative activityg (%) |
|---|---|---|---|---|
| GAa | αGal-(1→3)-L-Ara* | 2.0 | 101 | 100 |
| pNP-α-Gala | αGal-pNP | 5.0 | 337 | 335 |
| Melibioseb | αGal-(1→6)-Glc* | 5.0 | 23.6 | 23.4 |
| Raffinoseb | αGal-(1→6)-αGlc-(1↔2)-βFruf | 5.0 | 48.0 | 47.7 |
| Stachyoseb | αGal-(1→6)-αGal-(1→6)-αGlc-(1↔2)-βFruf | 5.0 | 3.38 | 3.36 |
| GA-Meb | αGal-(1→3)-α-L-Araf-OMe | 2.0 | 22.4 | 22.3 |
| AAc, f | β-L-Arap-(1→3)-L-Ara* | 2.0 | 0.647 | 0.643 |
| AA-Mec | β-L-Arap-(1→3)-α-L-Araf-OMe | 2.0 | Trace | Trace |
| pNP-β-Arapc | β-L-Arap-pNP | 5.0 | 1.06 | 1.06 |
| Liner B-2 trisaccharided | αGal-(1→3)-βGal-(1→4)-GlcNAc* | 0.5 | 17.2 | 17.1 |
| Blood group B trisaccharidee | αGal-(1→3)-[αFuc-(1→2)]-Gal* | 0.5 | 0.610 | 0.607 |
| Blood group B pentasaccharidee | αGal-(1→3)- [αFuc-(1→2)]-βGal-(1→4)- [αFuc-(1→3)]-Glc* | 0.05 | Trace | Trace |
| S3GAe | αGal-(1→3)-α-L-Araf-(1→3)- [α-L-Araf-(1→4)]-βGal-(1→6)-Gal* | 0.5 | 0.643 | 0.639 |
| S5GAe | αGal-(1→3)-α-L-Araf-(1→3)-[αRha-(1→4)-βGlcA-(1→6)]-[α-L-Araf-(1→4)]-βGal-(1→6)-Gal* | 0.5 | 0.514 | 0.511 |
*Represents reducing end of the oligosaccharide. a 0.099 μg/mL BlAga3 was incubated for 20 min. b 0.099 μg/mL BlAga3 was incubated for 2 h. c 0.099 μg/mL BlAga3 was incubated for 6 h. d 0.020 μg/mL BlAga3 was incubated for 1 h. e 0.020 μg/mL BlAga3 was incubated for 5 h. f The specific activity was calculated on the value for released L-arabinose by one-half. g Relative activity is expressed as the percentage of the activity toward α-D-Gal-(1→3)-L-Ara.
Although BlAga3 also exhibited the activity to melibiose, raffinose, and stachyose, as is the case for most other bifidobacterial GH36 α-galactosidases, the specific activities were 4.27-fold, 2.10-fold, and 29.8-fold lower than that for α-D-Gal-(1→3)-L-Ara, respectively (Table 1). The kinetic parameters of BlAga3 were determined using 0.1–10 mM α-D-Gal-(1→3)-L-Ara and 1.0-50 mM raffinose as substrates. The reaction mixture of 40 µL containing 50 mM sodium acetate buffer (pH 6.0) and 0.20 or 0.79 μg/mL BlAga3 was incubated at 45 °C for 20 min in the case of α-D-Gal-(1→3)-L-Ara and raffinose, respectively. The reactions were terminated by adding 10 μL of 5 % TCA, and the products were analyzed using HPAEC-PAD. The comparison of kinetic parameter study showed that Km of BlAga3 for α-D-Gal-(1→3)-L-Ara was 38.2-fold lower than that for raffinose, and kcat/Km value for α-D-Gal-(1→3)-L-Ara was 15.0-fold higher than that for raffinose (Table 2). It suggested that the difference of substrate affinity between α-D-Gal-(1→3)-L-Ara and raffinose contributed to the activities.
Table 2.
Kinetic parameters of BlAga3.
|
Km (mM) |
kcat (s-1) |
kcat /Km (s-1・mM-1) |
|
|---|---|---|---|
| α-D-Gal-(1→3)-L-Ara | 0.774 ± 0.014 | 107 ± 21 | 138 |
| Raffinose | 29.5 ± 1.0 | 270 ± 6 | 9.17 |
The transglycosylation reactions were performed using pNP-α-Gal as the donor and 1-alkanols as acceptors (Fig. S4; see J. Appl. Glycosci. Web site). The 40 μL reaction mixture containing 12.5 mM pNP-α-Gal was incubated at 40 °C for 30 min or 2 h with 2.0 µg/mL of BlAga3 in 50 mM sodium acetate buffer (pH 6.0) with 10 % methanol, ethanol, or 1-propanol as the acceptor. TLC analysis of the reaction products revealed that BlAga3 exhibited transglycosylation activity similar to other characterized GH36 enzymes.
In conclusion, BlAga3 is an intracellular α-galactosidase that preferentially cleaves α-D-Gal-(1→3)-L-Ara rather than β-L-Arap-(1→3)-L-Ara in GAfase-releasing disaccharides. Therefore, it suggests that α-D-Gal-(1→3)-L-Ara is transported through ABC transporter modules (BLGA_00300, 00310, 00320, and 00350) encoded in the same gene cluster after acting of GAfase, but β-L-Arap-(1→3)-L-Ara may not be internalized based on our recent study that B. longum JCM7052 assimilated α-D-Gal-(1→3)-L-Ara but not β-L-Arap-(1→3)-L-Ara.24) Therefore, BlAga3 and GAfase mediate a series of gum arabic AGP degradation in B. longum JCM7052. Bifidobacterium longum JCM7052 may have three GH36 α-galactosidases with different roles. The difference in the substrate specificity of GH36 α-galactosidases in B. longum is considered to reflect the variety of α-galactose-containing carbohydrates that can be encountered by B. longum living in the human intestine.
CONFLICTS OF INTEREST
The authors declare no conflict of interests.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by JSPS KAKENHI Grant-in-Aid, Grant Numbers 19J20806 (to Y.S.) and 19K05816 (to K.F.).
REFERENCES
- 1).Brouns S.J., Smits N., Wu H., Snijders A.P., Wright P.C., de Vos W.M., and van der Oost J.: Identification of a novel α-galactosidase from the hyperthermophilic archaeon Sulfolobus solfataricus. J. Bacteriol., 188, 2392–2399 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Cao Y., Huang H., Meng K., Yang P., Shi P., Wang Y., Luo H., Zhang Z., Wu N., and Yao B.: Cloning and functional expression of an α-galactosidase from Yersinia pestis biovar Microtus str. 91001. Biosci. Biotechnol. Biochem., 72, 2203–2205 (2008). [DOI] [PubMed] [Google Scholar]
- 3).Goulas T., Goulas A., Tzortzis G., and Gibson G.R.: A novel α-galactosidase from Bifidobacterium bifidum with transgalactosylating properties: gene molecular cloning and heterologous expression Appl. Microbiol. Biotechnol., 82, 471–477 (2009). [DOI] [PubMed] [Google Scholar]
- 4).Zhao H., Lu L., Xiao M., Wang Q., Lu Y., Liu C., Wang P., Kumagai H., and Yamamoto K.: Cloning and characterization of a novel α-galactosidase from Bifidobacterium breve 203 capable of synthesizing Gal-α-1,4 linkage. FEMS Microbiol. Lett., 285, 278–283 (2008). [DOI] [PubMed] [Google Scholar]
- 5).Ademark P., de Vries R.P., Hägglund P., Stålbrand H., and Visser J.: Cloning and characterization of Aspergillus niger genes encoding an α-galactosidase and a β-mannosidase involved in galactomannan degradation. Eur. J. Biochem., 268, 2982–2990 (2001). [DOI] [PubMed] [Google Scholar]
- 6).Nakai H., Baumann M.J., Petersen B.O., Westphal Y., Hachem M.A., Dilokpimol A., Duus J.O., Schols H.A., and Svensson B.: Aspergillus nidulans α-galactosidase of glycoside hydrolase family 36 catalyses the formation of α-galacto-oligosaccharides by transglycosylation. FEBS J., 277, 3538–3551 (2010). [DOI] [PubMed] [Google Scholar]
- 7).Mi S., Meng K., Wang Y., Bai Y., Yuan T., Luo H., and Yao B.: Molecular cloning and characterization of a novel α-galactosidase gene from Penicillium sp. F63 CGMCC 1669 and expression in Pichia pastoris. Enzyme Microb. Technol., 40, 1373–1380 (2007). [Google Scholar]
- 8).Margolles-Clark E., Tenkanen M., Luonteri E., and Penttilä M.: Three α-galactosidase genes of Trichoderma reesei cloned by expression in yeast. Eur. J. Biochem., 240, 104–111 (1996). [DOI] [PubMed] [Google Scholar]
- 9).Carmi N., Zhang G., Petreikov M., Gao Z., Eyal Y., Granot D., and Schaffer A.A.: Cloning and functional expression of alkaline α-galactosidase from melon fruit: similarity to plant SIP proteins uncovers a novel family of plant glycosyl hydrolases. Plant J., 33, 97–106 (2003). [DOI] [PubMed] [Google Scholar]
- 10).Lee R.-H., Lin M.-C., and Chen S.-C.: A novel alkaline α-galactosidase gene is involved in rice leaf senescence. Plant Mol. Biol., 55, 281–295 (2004). [DOI] [PubMed] [Google Scholar]
- 11).Lee R.H., Hsu J.H., Huang H.J., Lo S.F., and Chen S.C.: Alkaline α-galactosidase degrades thylakoid membranes in the chloroplast during leaf senescence in rice. New Phytol., 184, 596–606 (2009). [DOI] [PubMed] [Google Scholar]
- 12).Schell M.A., Karmirantzou M., Snel B., et al.: The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA, 99, 14422–14427 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13).Lee J.H., Karamychev V.N., Kozyavkin S.A., et al.: Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genom., 9, 247 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14).Leder S., Hartmeier W., and Marx S.P.: α-Galactosidase of Bifidobacterium adolescentis DSM 20083. Curr. Microbiol., 38, 101–106 (1999). [DOI] [PubMed] [Google Scholar]
- 15).Hinz S.W.A., Doeswijk-Voragen C.H.L., Schipperus R., van den Broek L.A.M., Vincken J.-P., and Voragen A.G.J.: Increasing the transglycosylation activity of α-galactosidase from Bifidobacterium adolescentis DSM 20083 by site-directed mutagenesis. Biotechnol. Bioeng., 93, 122–131 (2006). [DOI] [PubMed] [Google Scholar]
- 16).Patel S. and Goyal A.: Applications of natural polymer gum arabic: a review. Int. J. Food Prop., 18, 986–998 (2015). [Google Scholar]
- 17).Calame W., Weseler A.R., Viebke C., Flynn C., and Siemensma A.D.: Gum arabic establishes prebiotic functionality in healthy human volunteers in a dose-dependent manner. Br. J. Nutr., 100, 1269–1275 (2008). [DOI] [PubMed] [Google Scholar]
- 18).Terpend K., Possemiers S., Daguet D., and Marzorati M.: Arabinogalactan and fructo-oligosaccharides have a different fermentation profile in the Simulator of the Human Intestinal Microbial Ecosystem (SHIME®). Environ. Microbiol. Rep., 5, 595–603 (2013). [DOI] [PubMed] [Google Scholar]
- 19).Daguet D., Pinheiro I., Verhelst A., Possemiers S., and Marzorati M.: Arabinogalactan and fructooligosaccharides improve the gut barrier function in distinct areas of the colon in the Simulator of the Human Intestinal Microbial Ecosystem. J. Funct. Foods, 20, 369–379 (2016). [Google Scholar]
- 20).Street C.A. and Anderson D.M.: Refinement of structures previously proposed for gum arabic and other Acacia gum exudates. Talanta, 30, 887–893 (1983). [DOI] [PubMed] [Google Scholar]
- 21).Aspinall G.O. and Knebl M.C.: The location of α-D-galactopyranose residues in gum arabic. Carbohydr. Res., 157, 257–260 (1986). [Google Scholar]
- 22).Goodrum L.J., Patel A., Leykam J.F., and Kieliszewski M.J.: Gum arabic glycoprotein contains glycomodules of both extensin and arabinogalactan-glycoproteins. Phytochemistry, 54, 99–106 (2000). [DOI] [PubMed] [Google Scholar]
- 23).Saishin N. and Yamamoto I: α-Galactosidase purified from Bifidobacterium longum JCM 7052 grown on gum arabic. J. Biol. Macromol., 9, 71–80 (2009). [Google Scholar]
- 24).Sasaki Y., Horigome A., Odamaki T., Xiao J.-Z., Ishiwata A., Ito Y., Kitahara K., and Fujita K.: Novel 3-O-α-D-galactosyl-α-L-arabinofuranosidase for the assimilation of gum arabic arabinogalactan protein in Bifidobacterium longumsubsp. longum. Appl. Environ. Microbiol., 87, e02690–02620 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25).Yamamoto I., Ueno Y., Geshi M., Inagaki Y., Odamaki T., and Fujita K.: Complete genome sequence of Bifidobacterium longum subsp. longum JCM7052. Microbiol. Resour. Announc., 10, 01193–01120 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).Saishin N., Ueta M., Wada A., and Yamamoto I.: Purification and characterization of α-galactosidase I from Bifidobacterium longum subsp.longum JCM 7052. J. Biol. Macromol., 10, 13–22 (2010). [Google Scholar]
- 27).O’Connell K.J., Motherway M.O.C., O'Callaghan J., Fitzgerald G.F., Ross R.P., Ventura M., Stanton C., and van Sinderen D.: Metabolism of four α-glycosidic linkage-containing oligosaccharides by Bifidobacterium breve UCC2003. Appl. Environ. Microbiol., 79, 6280–6292 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Ejby M., Fredslund F., Andersen J.M., Žagar A.V., Henriksen J.R., Andersen T.L., Svensson B., Slotboom D.J., and Abou Hachem M.: An ATP binding cassette transporter mediates the uptake of α-(1,6)-linked dietary oligosaccharides in Bifidobacterium and correlates with competitive growth on these substrates. J. Biol. Chem., 291, 20220–20231 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Cao Y., Yuan T., Shi P., et al.: Properties of a novel α-galactosidase from Streptomyces sp. S27 and its potential for soybean processing. Enzyme Microb. Technol., 47, 305–312 (2010). [Google Scholar]
- 30).Fredslund F., Hachem M.A., Larsen R.J., Sorensen P.G., Coutinho P.M., Lo Leggio L., and Svensson B.: Crystal structure of α-galactosidase from Lactobacillus acidophilus NCFM: insight into tetramer formation and substrate binding. J. Mol. Biol., 412, 466–480 (2011). [DOI] [PubMed] [Google Scholar]
- 31).Kaeothip S., Ishiwata A.,Ito T., Fushinobu S., Fujita K., and Ito Y.: Preparation of p-nitrophenyl β-L-arabinofuranoside as a substrate of β-L-arabinofuranosidase. Carbohydr. Res., 382, 95–100 (2013). [DOI] [PubMed] [Google Scholar]
- 32).Holmes E.W. and O’Brien J.S.: Separation of glycoprotein-derived oligosaccharides by thin-layer chromatography. Anal. Biochem., 93, 167–170 (1979). [PubMed] [Google Scholar]
- 33).Sasaki Y., Togo N., Kitahara K., and Fujita K.: Characterization of a GH36 β-L-arabinopyranosidase in Bifidobacterium adolescentis. J. Appl. Glycosci., 65, 23–30 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34).Ichinose H., Fujimoto Z., Honda M., Harazono K., Nishimoto Y., Uzura A., and Kaneko S.: A β-L-arabinopyranosidase from Streptomyces avermitilis is a novel member of glycoside hydrolase family 27. J. Biol. Chem., 284, 25097–25106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




