Abstract
Background
Provider-collected nasopharyngeal specimens for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) molecular testing are the standard of care in many clinical settings, but patient-collected saliva and anterior nares specimens are less invasive and more flexible alternatives. Prior studies comparing specimen types for SARS-CoV-2 molecular testing have been limited by small sample sizes and low pretest probability. We conducted a large observational study among symptomatic adults at 7 emergency departments of Kaiser Permanente Southern California to examine sensitivity of SARS-CoV-2 molecular tests by specimen type and patient characteristics.
Methods
Provider-collected nasopharyngeal/oropharyngeal (NP/OP) specimens and patient-collected saliva and anterior nares specimens were collected at the same visit and analyzed with the Roche cobas® SARS-CoV-2 assay. Patients were considered truly positive for SARS-CoV-2 if any of the three specimens was positive and negative if all three specimens were negative. Factors associated with discordant and missed positive results were examined with multivariable logistic regression.
Results
Of 2112 patients, 350 (16.6%) were positive for SARS-CoV-2. Sensitivity of NP/OP was 93.7% (95% confidence interval [CI] 90.6%–96.0%), sensitivity of saliva was 87.7% (83.8%–91.0%), and sensitivity of anterior nares was 85.4% (81.3%–89.0%). Patients ages 18–39 years versus ≥40 years were more likely to have discordant results [adjusted odds ratio (aOR) 1.97 (1.12–3.45)], as were patients with <4 symptoms versus ≥4 [aOR 2.43 (1.39–4.25)]. Cycle threshold values were higher for saliva and anterior nares than NP/OP specimens, as well as for specimens in discordant versus concordant sets and patients with fewer symptoms.
Conclusion
This study provides robust evidence that patient-collected saliva and anterior nares are sensitive for SARS-CoV-2 molecular testing in emergency department settings, particularly among adults ages ≥40 years and those with multiple symptoms. Higher sensitivity of provider-collected NP/OP specimens must be weighed against the benefits of patient-collected specimens in tailored strategies for SARS-CoV-2 testing.
Keywords: SARS-CoV-2, COVID-19, Molecular diagnostic test, Saliva, Anterior nares, Self-collection
List of abbreviations
| aOR | adjusted odds ratio |
| CI | confidence interval |
| COVID-19 | coronavirus disease 2019 |
| Ct | cycle threshold |
| ED | emergency department |
| EHR | electronic health record |
| KPSC | Kaiser Permanente Southern California |
| NP | nasopharyngeal |
| NP/OP | nasopharyngeal/oropharyngeal |
| RT-PCR | reverse transcription polymerase chain reaction |
| SARS-CoV-2 | severe acute respiratory syndrome coronavirus 2 |
| T1 | target gene 1 (ORF1 a/b) |
| T2 | target gene 2 (E gene) |
1. Introduction
Widespread testing for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continues to be vital to the coronavirus disease 2019 (COVID-19) pandemic response. Individuals who test positive can be isolated, and their contacts can also be tested and take other protective measures. Despite repeated calls from healthcare professionals and community advocates for greater access to SARS-CoV-2 testing, the United States and other countries have struggled throughout the pandemic to provide adequate testing [1,2].
One consideration in determining testing strategies has been the sensitivity of SARS-CoV-2 tests. Sensitivity of real-time reverse transcription polymerase chain reaction (RT-PCR) tests has generally been higher than rapid molecular or antigen tests [3], though sensitivity may depend on specimen type and quality of collection [4]. For surveillance purposes, test sensitivity is arguably less critical if individuals are frequently tested [5]. In contrast, for healthcare organizations mandated to deliver high quality care, test sensitivity is paramount to avoid missing infections and putting patients and their contacts at risk.
The standard of care for SARS-CoV-2 testing in many healthcare settings has been nasopharyngeal (NP) swabs collected in transport medium and tested by RT-PCR. However, invasive NP swabs can be uncomfortable for patients and may elicit coughing or gagging, increasing potential exposure for the healthcare provider collecting the specimen [6]. Furthermore, the long, thin swabs required to reach the nasopharynx have periodically been unavailable due to high global demand. Relying on NP swabs alone to ramp up SARS-CoV-2 testing has proven infeasible.
Despite prior studies suggesting that saliva or anterior nares swabs are viable alternatives to NP swabs for SARS-CoV-2 RT-PCR testing [[7], [8], [9], [10]], evidence remains limited in clinical scenarios where testing is needed most – among symptomatic emergency department (ED) and ambulatory patients. Most prior studies of patient-collected specimens have been conducted among patients hospitalized with COVID-19 with high pretest probability or among asymptomatic groups in which very few individuals tested positive [[10], [11], [12], [13], [14]]. There is still a critical need for adequately powered studies comparing sensitivity by patient demographic and clinical characteristics to inform tailored strategies for SARS-CoV-2 testing.
To address this need, we conducted a large observational study among a diverse population of symptomatic ED patients to compare provider-collected nasopharyngeal/oropharyngeal (NP/OP) specimens and patient-collected saliva and anterior nares specimens for SARS-CoV-2 RT-PCR testing.
2. Materials and methods
2.1. Study setting
This cross-sectional study was conducted at 7 EDs at Kaiser Permanente Southern California (KPSC) from 18 August to 2 November 2020. KPSC is an integrated healthcare system serving over 4.6 million members with diverse racial/ethnic and socioeconomic backgrounds similar to the Southern California population [15]. A comprehensive electronic health record (EHR) captures all aspects of inpatient and outpatient care, including diagnoses and laboratory tests.
2.2. Patient consent
As the study was conducted as part of real-world care delivery, research staff were not present at study sites. Information sheets in English or Spanish were distributed by providers to patients with COVID-19 symptoms. Patients who decided to participate in the study provided verbal agreement prior to testing. The KPSC Institutional Review Board approved the study waiving the requirement for written consent due to minimal risk to study participants.
2.3. Study design
Adult patients presenting to EDs with acute symptoms concerning for COVID-19 were eligible to participate in the study if their attending physician placed an order for a standard NP/OP SARS-CoV-2 test. This test order triggered an alert in the EHR, prompting test orders for patient collection of saliva and anterior nares specimens.
During the study period, NP/OP combination specimens were the standard of care for SARS-CoV-2 RT-PCR testing at KPSC. Per KPSC guidelines, NP/OP specimens were collected by nurses using a long, thin swab to swab the back of the throat, avoiding the tongue, followed by inserting the same swab into one nostril parallel to the palate for a few seconds to absorb nasopharyngeal secretions, and then placing the swab in a tube with viral transport media. After collection of the NP/OP swab, nurses observed and instructed patients to self-collect saliva and anterior nares swabs. Saliva was collected ≥30 min after eating, drinking, or chewing gum using a SpectrumDNA™ saliva collection device (Spectrum Solutions, Draper, UT). This kit was selected based on suitability for potential future home self-collection and mailing of saliva specimens for SARS-CoV-2 testing. Per manufacturer instructions, patients were asked to 1) provide approximately 2 mL of spit into the collection tube measured by a standardized wavy black line, 2) screw on a separate cap to release a stabilizing solution, and 3) shake for 5 s. Anterior nares swabs were collected by asking the patient to insert a shorter, thicker swab less than 1 in. into the nostril, rotate the swab against the nostril wall several times, repeat with the other nostril, and place in a tube with viral transport media. Specimens were tested within approximately 26 h using the Roche cobas® SARS-CoV-2 assay on the Roche cobas® 8800 System.
Patient demographic characteristics were ascertained from EHR data. To identify symptoms at time of testing and time since symptom onset, trained research associates reviewed medical charts of all patients with positive results, using a chart abstraction form to record presence of symptoms including cough, chest pain, dyspnea, chills, fever, headache, congestion, rhinorrhea, sore throat, anosmia, fatigue, myalgia, abdominal pain, vomiting, and nausea. A physician investigator was consulted when symptom descriptions were unclear. Cycle threshold (Ct) values for both targets 1 (T1: ORF1 a/b) and 2 (T2: E gene) were extracted for all positive samples in the study from the Roche cobas® 8800 System. A Ct value represents the number of cycles needed to amplify the SARS-CoV-2 RNA target to a detectable level, with values generally inversely proportional to the concentration of the viral target inoculum in the sample. The Roche cobas® 8800 System uses a complex kinetic algorithm rather than a Ct value cutoff to define results.
2.4. Sample size
The study was powered to assess sensitivity of each of specimen type (NP/OP, saliva, and anterior nares) compared to a composite measure in which the result was considered truly positive if any of the three specimens were positive and negative if all three of the specimens were negative. To assess a difference of 5 percentage points between each of the three specimen types and the composite measure, assuming 80% power and alpha of 0.05, 341 patients with a positive result were required. We expected between 15%–20% of patients to test positive based on trends among ED patients at KPSC, requiring approximately 1705 to 2274 total patients.
2.5. Analyses
We included patients with all three specimens collected in primary analyses. We compared demographic characteristics of patients who had all negative specimens and those with at least one positive using chi-square tests. We assessed sensitivity in comparison to the composite measure, stratifying by sex, age, days of symptoms prior to testing, and number of symptoms at testing. For patients with at least one positive result, we compared characteristics for those with concordant results (all three specimens positive) and discordant results (one or two specimens positive) using chi-square tests.
Logistic regression analyses were performed to estimate adjusted odds ratios (aORs) for the association between patient characteristics and discordant results overall, and with specimen type resulting in a missed positive. Factors considered in the logistic regression models included age, sex, and number and duration of symptoms at testing. For parsimony and due to the small number of discordant results, factors with weak (non-significant) associations were not included in the final models.
Median Ct values and interquartile ranges for T1 and T2 were described for all three specimens for sets with concordant and discordant results. The distribution of the means of T1 and T2 values were compared using the paired t-test and examined by days of symptoms prior to testing and number of symptoms at testing.
We conducted several additional analyses to supplement results. First, we conducted a sensitivity analysis excluding a small proportion of specimens (5.8%) that were inadvertently tested using the Aptima® SARS-CoV-2 assay (Hologic Panther® System). In a second sensitivity analysis, we included individuals who had NP/OP specimens but only one of saliva or anterior nares specimens. Third, to facilitate comparison with other studies, we conducted a secondary analysis using NP/OP specimens as a gold standard and assessed sensitivity and specificity of saliva and anterior nares swabs. Specificity was not assessed in primary analyses, as specificity would be 100% compared to the composite measure. Although false positives are possible, usually due to contamination, specificity of SARS-CoV-2 RT-PCR assays is very high [16].
3. Results
3.1. Patient characteristics
The study included 2112 patients with all three specimens, of whom 1160 patients (54.9%) were female, 864 (40.9%) were ages 18–39 years, and 1103 (52.2%) were Hispanic (Table 1 ). There were 350 patients (16.6%) with a positive result, most of whom were male (191 [54.6%]), ages 40–59 years (145 [41.4%]), and Hispanic (236 [67.4%]). The majority [279 (79.7%)] were positive by all three specimens, and 26 (7.4%) were positive by two of the three specimens, 25 (7.1%) were positive by NP/OP only, 14 (4.0%) were positive by saliva only, and 6 (1.7%) were positive by anterior nares only (Table S1).
Table 1.
Characteristics of emergency department patients with three specimens tested for SARS-CoV-2.
| All specimens negative |
Any specimen positive |
Total |
P-value | |
|---|---|---|---|---|
| n (%) | n (%) | n (%) | ||
| Sex | <0.001 | |||
| Female | 1001 (56.8) | 159 (45.4) | 1160 (54.9) | |
| Male | 761 (43.2) | 191 (54.6) | 952 (45.1) | |
| Age at test (years) | <0.001 | |||
| 18–39 | 751 (42.6) | 113 (32.3) | 864 (40.9) | |
| 40–59 | 502 (28.5) | 145 (41.4) | 647 (30.6) | |
| ≥60 | 509 (28.9) | 92 (26.3) | 601 (28.5) | |
| Race/ethnicity | <0.001 | |||
| Asian | 114 (6.5) | 10 (2.9) | 124 (5.9) | |
| Black | 174 (9.9) | 14 (4.0) | 188 (8.9) | |
| Hispanic | 867 (49.2) | 236 (67.4) | 1103 (52.2) | |
| Other/Unknown | 209 (11.9) | 47 (13.4) | 256 (12.1) | |
| Pacific Islander | 16 (0.9) | 4 (1.1) | 20 (0.9) | |
| White | 382 (21.7) | 39 (11.1) | 421 (19.9) | |
| Total | 1762 (83.4) | 350 (16.6) | 2112 (100) |
Abbreviations: RT-PCR, reverse transcription polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Patients were included in the primary analysis if they had a provider-collected nasopharyngeal/oropharyngeal specimen and patient-collected saliva and anterior nares specimens tested with RT-PCR for SARS-CoV-2.
3.2. Sensitivity by specimen type
Compared to the composite measure, sensitivity of NP/OP was 93.7% (95% confidence interval 90.6%–96.0%), sensitivity of saliva was 87.7% (83.8%–91.0%), and sensitivity of anterior nares was 85.4% (81.3%–89.0%) (Table 2 ). The difference in sensitivity of NP/OP versus saliva was 6.0% (2.0%–10.0%), while the difference in sensitivity between NP/OP and anterior nares was 8.3% (4.6%–11.9%). The difference between saliva and anterior nares was 2.3% (−1.4%–6.0%).
Table 2.
Sensitivity of specimen types for SARS-CoV-2 testing by patient characteristics.a
| Total positive patients | NP/OP |
Saliva |
Anterior nares |
||||
|---|---|---|---|---|---|---|---|
| Positive by test | Sensitivity % (95% CI) |
Positive by test | Sensitivity % (95% CI) |
Positive by test | Sensitivity % (95% CI) |
||
| Total | 350 | 328 | 93.7 (90.6–96.0) | 307 | 87.7 (83.8–91.0) | 299 | 85.4 (81.3–89.0) |
| Sex | |||||||
| Female | 159 | 152 | 95.6 (91.1–98.2) | 143 | 89.9 (84.2–94.1) | 135 | 84.9 (78.4–90.1) |
| Male | 191 | 176 | 92.1 (87.4–95.5) | 164 | 85.9 (80.1–90.5) | 164 | 85.9 (80.1–90.5) |
| Age (years) | |||||||
| 18–39 | 113 | 103 | 91.2 (84.3–95.7) | 96 | 85.0 (77.0–91.0) | 90 | 79.6 (71.0–86.6) |
| 40–59 | 145 | 138 | 95.2 (90.3–98.0) | 128 | 88.3 (81.9–93.0) | 130 | 89.7 (83.5–94.1) |
| ≥60 | 92 | 87 | 94.6 (87.8–98.2) | 83 | 90.2 (82.2–95.4) | 79 | 85.9 (77.0–92.3) |
| Days of symptoms prior to test | |||||||
| 0–2 | 100 | 92 | 92.0 (84.8–96.5) | 87 | 87.0 (78.8–92.9) | 84 | 84.0 (75.3–90.6) |
| 3–6 | 153 | 143 | 93.5 (88.3–96.8) | 137 | 89.5 (83.6–93.9) | 136 | 88.9 (82.8–93.4) |
| ≥7 | 97 | 93 | 95.9 (89.8–98.9) | 83 | 85.6 (77.0–91.9) | 79 | 81.4 (72.3–88.6) |
| Number of symptoms | |||||||
| 1–3 | 100 | 90 | 90.0 (82.4–95.1) | 81 | 81.0 (71.9–88.2) | 77 | 77.0 (67.5–84.8) |
| 4–5 | 112 | 104 | 92.9 (86.4–96.9) | 100 | 89.3 (82.0–94.3) | 94 | 83.9 (75.8–90.2) |
| 6–15 | 138 | 134 | 97.1 (92.7–99.2) | 126 | 91.3 (85.3–95.4) | 128 | 92.8 (87.1–96.5) |
Abbreviations: CI, confidence interval; NP/OP, nasopharyngeal/oropharyngeal; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Sensitivity as assessed in comparison to a composite measure, in which patients were considered truly positive if any of the three specimens was positive and truly negative if all three specimens were negative.
In stratified analyses, test sensitivity varied by patient characteristics (Table 2 and Fig. 1 ). Sensitivity was higher among women than men for NP/OP [95.6% (91.1%–98.2%)] and saliva [89.9% (84.2% -94.1%)], but similar among women and men for anterior nares. Sensitivity of all three specimens was lower among individuals ages 18–39 years compared to those ages ≥40 years; sensitivity was highest among those ages 40–59 years for NP/OP [95.2% (90.3%–98.0%)] and anterior nares [89.7% (83.5%–94.1%)] and those ages ≥60 years for saliva [90.2% (82.2%–95.4%)]. Sensitivity peaked 3–6 days after symptom onset for saliva [89.5% (83.6%–93.9%)] and anterior nares [88.9% (82.8%–93.4%)], but for NP/OP was higher ≥7 days after symptom onset [89.5% (83.6%–93.9%)]. For all specimen types, sensitivity increased with number of symptoms.
Fig. 1.
Sensitivity of specimen types for SARS-CoV-2 RT-PCR Testing by days of symptoms prior to test and number of symptoms.
Abbreviations: NP/OP, nasopharyngeal/oropharyngeal; RT-PCR, reverse-transcriptase polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Sensitivity of NP/OP, anterior nares, and saliva specimens for SARS-CoV-2 testing by (Fig. 1A) days of symptoms prior to test and (Fig. 1B) number of symptoms.
3.3. Factors associated with discordant results
In multivariable analyses, patients ages 18–39 years were twice as likely to have a discordant result [aOR 1.97 (1.12–3.45)] and have positive results missed by each of the specimen types (aOR range 1.55 to 2.14) (Table 3 ). Patients with <4 symptoms versus ≥4 symptoms were more than twice as likely to have discordant results [aOR 2.43 (1.39–4.25)] and have a positive missed by all three specimens (aOR range 2.21 to 2.63). Days of symptoms prior to testing was not significantly associated with discordant results.
Table 3.
Factors associated with discordant results between specimen types or, for each specimen type, missing a positive resulta
| Discordant OR (95% CI) | Missed by NP/OP OR (95% CI) | Missed by saliva OR (95% CI) | Missed by nares OR (95% CI) | |
|---|---|---|---|---|
| Age < 40 years versus ≥40 years | 1.97 (1.12–3.45) | 1.81 (0.74–4.43) | 1.55 (0.79–3.06) | 2.14 (1.14–4.03) |
| 3–6 of symptoms prior to testing versus 0–2 days | 0.95 (0.49–1.84) | 0.91 (0.34–2.45) | 0.87 (0.39–1.93) | 0.76 (0.36–1.62) |
| ≥7 of symptoms prior to testing versus 0–2 days | 1.82 (0.90–3.67) | 0.61 (0.17–2.16) | 1.38 (0.59–3.20) | 1.60 (0.73–3.49) |
| <4 symptoms present at testing versus ≥4 symptoms | 2.43 (1.39–4.25) | 2.21 (0.92–5.36) | 2.34 (1.20–4.53) | 2.63 (1.40–4.93) |
Abbreviations: CI, confidence interval; NP/OP, nasopharyngeal/oropharyngeal; OR, odds ratio.
All models adjusted for other variables shown in table.
Patients were considered truly positive if any of the three specimens were positive and truly negative if all three specimens were negative.
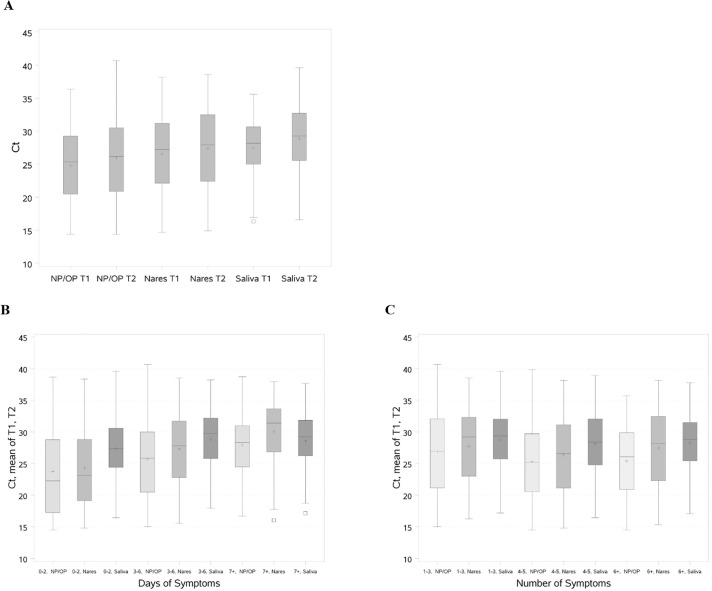
3.4. Ct values
Ct values varied by specimen type (Fig. 2A and Table S2). Median (interquartile range) values for T1 and T2 were significantly higher for saliva [T1, 28.2 (25.0–30.6); T2, 29.3 (25.6–32.7)] and anterior nares [T1, 27.2 (22.1–31.2); T2, 27.9 (22.4–32.5)] than for NP/OP specimens [T1, 25.3 (20.5–29.3); T2, 26.1 (20.9–30.5)]. For patients with discordant results, the Ct values for each of the three specimens were significantly higher than for patients who had concordant positive results (Table S2). Ct values were higher for all three specimens among patients tested after 3–6 days of symptoms compared to 0–2 days, and higher for NP/OP and anterior nares, but not saliva, for patients tested ≥7 days of symptoms (Fig. 2B). Ct values were also higher for all three specimens for patients with <4 symptoms compared to those with ≥4 symptoms (Fig. 2C).
Fig. 2.
Ct values for SARS-CoV-2 RT-PCR testing by specimen type, days of symptoms prior to test, and number of symptoms.
Abbreviations: Ct, cycle threshold; NP/OP, nasopharyngeal/oropharyngeal; RT-PCR, reverse-transcriptase polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; T1, Target 1 (ORF1 a/b); T2, Target 2 (E gene).
Fig. 2A shows the distribution of Ct values (median, interquartile range, maximum and minimum) in box plots for each specimen type and target. The distribution of the mean of T1 and T2 Ct values (median, interquartile range, maximum and minimum) are shown in box plots for each specimen type by (Fig. 2B) days of symptoms prior to test and (Fig. 2C) number of symptoms. The + indicates the mean value and □ indicates outliers.
3.5. Additional analyses
Results of additional analyses were consistent with primary analyses. In a sensitivity analysis excluding 19 patients positive by the Aptima® SARS-CoV-2 assay instead of the Roche cobas® SARS-CoV-2 assay, results were similar to the primary analysis (Table S3). In a sensitivity analysis including 81 patients with NP/OP specimens and either saliva or anterior nares specimens, results were also similar to the primary analysis (Table S4). In analyses considering NP/OP as the gold standard, sensitivity was 88.7% (95% CI 84.8%–91.9%) for both saliva and anterior nares specimens, while specificity was 99.1% (98.5%–99.5%) for saliva and 99.6% (99.1%–99.8%) for anterior nares (Table S5).
4. Discussion
In this large study of symptomatic ED patients, sensitivity of SARS-CoV-2 RT-PCR testing was moderately high for all three specimen types, but higher for provider-collected NP/OP specimens (93.7%) than patient-collected saliva (87.7%) or anterior nares (85.4%) specimens. Positive results were missed by all three specimen types, but less frequently by NP/OP specimens. Our study suggests that patient-collected saliva and anterior nares are sensitive for SARS-CoV-2 molecular testing in ED settings, particularly among adults ages ≥40 years and those with multiple symptoms. However, NP/OP specimens may be preferred when the risks of missing positives outweigh the benefits of patient self-collection.
Our results are within range of prior studies comparing specimen types for SARS-CoV-2 RT-PCR testing. Many of these studies were conducted in populations other than ambulatory patients, such as hospitalized COVID-19 patients with high pretest probability [10,12,17,18], health care workers [10,19], or asymptomatic populations with few positives [20,21]. In ambulatory settings similar to our study, several studies reported high sensitivity of saliva (eg, >90%) [7,9,22], while other studies found moderate sensitivity of saliva (eg, 81.4%) [23] or anterior nares (e.g., 86.3%) [7], or low sensitivity of saliva (e.g., 68.6%) [24]. Differences in sensitivity between studies could be due to variation in specimen collection methods and quality of collection [25], SARS-CoV-2 assays and their molecular targets, or clinical characteristics of patients, all of which merit additional study.
Our study included 2112 patients, of whom 350 (16.6%) were positive by at least one of the three specimen types evaluated, allowing for more precise estimates than other studies and facilitating stratified and multivariable analyses. We found that younger adults and those with fewer symptoms were more likely to have discordant results and missed positives. Ct values for both targets were higher among discordant versus concordant specimen sets. Ct values were also higher among specimens from patients tested later after symptom onset (3–6 days versus 1–2 days) and those with fewer symptoms (<4 versus 4–15). These data are aligned with several other reports of lower test sensitivity or higher Ct values among asymptomatic versus symptomatic patients [23,[26], [27], [28]], but are in contrast to other studies reporting no significant differences [18,29].
These results have implications for prioritizing NP/OP, saliva, and anterior nares specimens in different clinical scenarios. NP/OP specimens had the highest sensitivity, suggesting that these specimens are preferred when possible, particularly for scenarios in which there may be severe consequences if positives are missed. However, saliva and anterior nares specimens detected some positives that were missed by NP/OP specimens. In addition, the slightly lower sensitivity of saliva and anterior nares specimens should be weighed against the benefits of patient self-collection, including lower risk to health care providers, potential for reducing testing burden in ambulatory settings, facilitating home self-collection testing programs, and less reliance on swab and transport media availability.
Our study had additional strengths and limitations. The large sample size across 7 EDs in Southern California enabled robust characterization of specimen sensitivity for SARS-CoV-2 RT-PCR testing by demographic and clinical characteristics. We used a composite measure as a gold standard so that sensitivity of the three specimens could be directly compared. However, there may have been variation in specimen collection practices across and within EDs, despite efforts to provide clear and standardized instructions. NP specimens were collected in combination with oropharyngeal specimens, which may increase sensitivity compared to NP specimens alone [30]. Saliva was collected using saliva collection kits with stabilizer, an approach that is more feasible for high throughput testing and home collection than plain saliva [31], but sensitivity could differ using plain saliva or other SARS-CoV-2 assays with different gene targets. In addition, symptoms and symptom onset were ascertained by chart review and may be subject to bias if provider notes were incomplete or inaccurate, or if patients did not accurately recall symptom onset.
In conclusion, in this large study among ED patients, sensitivity of SARS-CoV-2 RT-PCR testing was highest for provider-collected NP/OP specimens, followed by patient-collected saliva and anterior nares specimens. As pandemic dynamics continue to evolve, further work is needed to tailor SARS-CoV-2 testing strategies for different clinical and public health scenarios.
Funding
This work was supported by the Regional Research Committee of Kaiser Permanente Southern California [grant number KP-RRC-20200703] and Kaiser Permanente Southern California Internal Research Funds.
Declaration of Competing Interest
None.
Acknowledgments
We thank Nitin Dhamija, Ben Hsu, Robert McCormick, George Preciado, Michael Schwartzwald, Karen Shih, Anu Singh, and Ivan Wu for clinical leadership and facilitation of the study. We thank physicians, nurses, and laboratory and research colleagues for their assistance.
We also thank the patients of Kaiser Permanente and their partnership with us to improve their health. Their information, collected through our electronic health record systems, leads to findings that help us improve care for our members and that we can share with the larger community.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ajem.2021.08.034.
Appendix A. Supplementary data
Supplementary material
References
- 1.Schneider E.C. Failing the test - the tragic data gap undermining the U.S. pandemic response. N Engl J Med. 2020;383(4):299–302. doi: 10.1056/NEJMp2014836. [DOI] [PubMed] [Google Scholar]
- 2.The Lancet Respiratory M COVID-19 testing in the UK. Lancet Respir Med. 2020;8(11):1061. doi: 10.1016/S2213-2600(20)30445-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dinnes J., Deeks J.J., Adriano A., Berhane S., Davenport C., Dittrich S., et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev. 2020;8 doi: 10.1002/14651858.CD013705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green D.A., Zucker J., Westblade L.F., Whittier S., Rennert H., Velu P., et al. Clinical performance of SARS-CoV-2 molecular tests. J Clin Microbiol. 2020;58(8) doi: 10.1128/JCM.00995-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larremore D.B., Wilder B., Lester E., Shehata S., Burke J.M., Hay J.A., et al. Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Sci Adv. 2020 doi: 10.1126/sciadv.abd5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frazee B.W., Rodriguez-Hoces de la Guardia A., Alter H., Chen C.G., Fuentes E.L., Holzer A.K., et al. Accuracy and discomfort of different types of intranasal specimen collection methods for molecular influenza testing in emergency department patients. Ann Emerg Med. 2018;71(4):509–517. doi: 10.1016/j.annemergmed.2017.09.010. e1. [DOI] [PubMed] [Google Scholar]
- 7.Hanson K.E., Barker A.P., Hillyard D.R., Gilmore N., Barrett J.W., Orlandi R.R., et al. Self-collected anterior nasal and saliva specimens versus health care worker-collected nasopharyngeal swabs for the molecular detection of SARS-CoV-2. J Clin Microbiol. 2020;58(11) doi: 10.1128/JCM.01824-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kandel C., Zheng J., McCready J., Serbanescu M.A., Racher H., Desaulnier M., et al. Detection of SARS-CoV-2 from saliva as compared to nasopharyngeal swabs in outpatients. Viruses. 2020;12(11) doi: 10.3390/v12111314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCormick-Baw C., Morgan K., Gaffney D., Cazares Y., Jaworski K., Byrd A., et al. Saliva as an ALTERNATE SPECIMEN SOURCE FOR DETECtion of SARS-CoV-2 in symptomatic patients using cepheid xpert xpress SARS-CoV-2. J Clin Microbiol. 2020;58(8) doi: 10.1128/JCM.01109-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P., et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N Engl J Med. 2020;383(13):1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altamirano J., Govindarajan P., Blomkalns A.L., Kushner L.E., Stevens B.A., Pinsky B.A., et al. Assessment of sensitivity and specificity of patient-collected lower nasal specimens for sudden acute respiratory syndrome coronavirus 2 testing. JAMA Netw Open. 2020;3(6) doi: 10.1001/jamanetworkopen.2020.12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jamal A.J., Mozafarihashjin M., Coomes E., Powis J., Li A.X., Paterson A., et al. Sensitivity of nasopharyngeal swabs and saliva for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shakir S.M., Barker A.P., Hillyard D.R., Gilmore N., Barrett J.W., Orlandi R.R., et al. Combined self-collected anterior nasal and oropharyngeal specimens versus provider-collected nasopharyngeal swabs for the detection of SARS-CoV-2. J Clin Microbiol. 2020 doi: 10.1128/JCM.02291-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.To K.K., Tsang O.T., Yip C.C., Chan K.H., Wu T.C., Chan J.M., et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020;71(15):841–843. doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koebnick C., Langer-Gould A.M., Gould M.K., Chao C.R., Iyer R.L., Smith N., et al. Sociodemographic characteristics of members of a large, integrated health care system: comparison with US Census Bureau data. Perm J. 2012;16(3):37–41. doi: 10.7812/tpp/12-031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skittrall J.P., Wilson M., Smielewska A.A., Parmar S., Fortune M.D., Sparkes D., et al. Specificity and positive predictive value of SARS-CoV-2 nucleic acid amplification testing in a low-prevalence setting. Clin Microbiol Infect. 2021;27(3):469. doi: 10.1016/j.cmi.2020.10.003. e9- e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagura-Ikeda M., Imai K., Tabata S., Miyoshi K., Murahara N., Mizuno T., et al. Clinical evaluation of self-collected saliva by quantitative reverse transcription-PCR (RT-qPCR), direct RT-qPCR, reverse transcription-loop-mediated isothermal amplification, and a rapid antigen test to diagnose COVID-19. J Clin Microbiol. 2020;58(9) doi: 10.1128/JCM.01438-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Azzi L., Carcano G., Gianfagna F., Grossi P., Gasperina D.D., Genoni A., et al. Saliva is a reliable tool to detect SARS-CoV-2. J Infect. 2020;81(1):e45–e50. doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tu Y.P., Jennings R., Hart B., Cangelosi G.A., Wood R.C., Wehber K., et al. Swabs collected by patients or health care workers for SARS-CoV-2 testing. N Engl J Med. 2020;383(5):494–496. doi: 10.1056/NEJMc2016321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yokota I., Shane P.Y., Okada K., Unoki Y., Yang Y., Inao T., et al. Mass screening of asymptomatic persons for SARS-CoV-2 using saliva. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Senok A., Alsuwaidi H., Atrah Y., Al Ayedi O., Al Zahid J., Han A., et al. Saliva as an alternative specimen for molecular COVID-19 testing in community settings and population-based screening. Infect Drug Resist. 2020;13:3393–3399. doi: 10.2147/IDR.S275152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams E., Bond K., Zhang B., Putland M., Williamson D.A. Saliva as a noninvasive specimen for detection of SARS-CoV-2. J Clin Microbiol. 2020;58(8) doi: 10.1128/JCM.00776-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yee R., Truong T., Pannaraj P.S., Eubanks N., Gai E., Jumarang J., et al. Saliva is a promising alternative specimen for the detection of SARS-CoV-2 in children and adults. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.02686-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caulley L., Corsten M., Eapen L., Whelan J., Angel J.B., Antonation K., et al. Salivary detection of COVID-19. Ann. Intern. Med. 2020 doi: 10.7326/M20-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kinloch N.N., Ritchie G., Brumme C.J., Dong W., Dong W., Lawson T., et al. Suboptimal biological sampling as a probable cause of false-negative COVID-19 diagnostic test results. J Infect Dis. 2020;222(6):899–902. doi: 10.1093/infdis/jiaa370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kociolek L.K., Muller W.J., Yee R., Dien Bard J., Brown C.A., Revell P., et al. Comparison of upper respiratory viral load distributions in asymptomatic and symptomatic children diagnosed with SARS-CoV-2 infection in pediatric hospital testing programs. J Clin Microbiol. 2020 doi: 10.1128/JCM.02593-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Migueres M., Mengelle C., Dimeglio C., Didier A., Alvarez M., Delobel P., et al. Saliva sampling for diagnosing SARS-CoV-2 infections in symptomatic patients and asymptomatic carriers. J Clin Virol. 2020;130:104580. doi: 10.1016/j.jcv.2020.104580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salvatore P.P., Dawson P., Wadhwa A., Rabold E.M., Buono S., Dietrich E.A., et al. Epidemiological correlates of PCR cycle threshold values in the detection of SARS-CoV-2. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee S., Kim T., Lee E., Lee C., Kim H., Rhee H., et al. Clinical course and molecular viral shedding among asymptomatic and symptomatic patients with SARS-CoV-2 infection in a community treatment center in the Republic of Korea. JAMA Intern Med. 2020 doi: 10.1001/jamainternmed.2020.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ek P., Bottiger B., Dahlman D., Hansen K.B., Nyman M., Nilsson A.C. A combination of naso- and oropharyngeal swabs improves the diagnostic yield of respiratory viruses in adult emergency department patients. Infect Dis (Lond) 2019;51(4):241–248. doi: 10.1080/23744235.2018.1546055. [DOI] [PubMed] [Google Scholar]
- 31.Landry M.L., Criscuolo J., Peaper D.R. Challenges in use of saliva for detection of SARS CoV-2 RNA in symptomatic outpatients. J Clin Virol. 2020;130:104567. doi: 10.1016/j.jcv.2020.104567. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material