Abstract
While diagnosis of COVID-19 relies on qualitative molecular testing for the absence or presence of SARS-CoV-2 RNA, quantitative viral load determination for SARS-CoV-2 has many potential applications in antiviral therapy and vaccine trials as well as implications for public health and quarantine guidance. To date, no quantitative SARS-CoV-2 viral load tests have been authorized for clinical use by the FDA. In this study, we modified the FDA emergency use authorized qualitative RealTime SARS-CoV-2 assay into a quantitative SARS-CoV-2 Laboratory Developed Test (LDT) using newly developed Abbott SARS-CoV-2 calibration standards. Both analytical and clinical performance of this SARS-CoV-2 quantitative LDT was evaluated using nasopharyngeal swabs (NPS). We further assessed the correlation between Ct and the ability to culture virus on Vero CCL81 cells. The SARS-CoV-2 quantitative LDT demonstrated high linearity with R2 value of 0.992, high inter- and intra-assay reproducibility across the dynamic range (SDs ± 0.08–0.14 log10 copies/mL for inter-assay reproducibility and ± 0.09 to 0.19 log10 copies/mL for intra-assay reproducibility). Lower limit of detection was determined as 1.90 log10 copies/mL. The highest Ct at which CPE was detected ranged between 28.21–28.49, corresponding to approximately 4.2 log10 copies/mL. Quantitative tests, validated against viral culture capacity, may allow more accurate identification of individuals with and without infectious viral shedding from the respiratory tract.
1. Introduction
The diagnosis of COVID-19 relies primarily on nucleic acid amplification tests for SARS-CoV-2 RNA targets. To date, these tests have been authorized by the FDA as qualitative tests, reporting the presence or absence of viral RNA in a clinical specimen. Quantitative assays of SARS-CoV-2 viral load (RNA copies/mL) have clear applications in preclinical and clinical trials of antiviral therapies and may have public health implications for contact tracing and quarantine guidance for individual patients. However, while numerous studies have reported the dynamics of SARS-CoV-2 Ct following infection using PCR-based assays, the relationship between viral load and infectivity, symptom severity and mortality remains unclear [1,2].
For PCR-based tests, the cycle threshold (Ct), defined as the number of PCR cycles needed to amplify the target viral RNA so that it can be detected over background, is inversely correlated with viral load. Due to different assay design strategies used by manufacturers, there are concerns about accurately comparing Ct values across qualitative assays [3], [4], [5]. Once there is a universal standard for SARS-CoV-2 such as a World Health Organization International Standard, viral load could be estimated based on the Ct value from the FDA Emergency Use Authorization (EUA) SARS-CoV-2 assays. Recently, several studies have shown an inverse correlation between the Ct and the ability to culture SARS-CoV-2 in vitro as a measure of infectivity [6], [7], [8].
Here, we describe the modification of the EUA-approved qualitative RealTime SARS-CoV-2 assay (Abbott Molecular, Des Plaines, IL) [9,10] into a SARS-CoV-2 quantitative LDT using Abbott SARS-CoV-2 calibration standards that correlate Ct and viral load. The analytical performance of the SARS-CoV-2 quantitative LDT was evaluated using commercially available SARS-CoV-2 material or using the Abbott SARS-CoV-2 material. Analytical analysis consisted of linearity, limit of detection, the inter-run and intra-run reproducibility. Using nasopharyngeal swab samples, the clinical performance was compared to three EUA SARS-CoV-2 qualitative assays which also included inter-laboratory agreement of the SARS-CoV-2 quantitative LDT. Using measurements from the SARS-CoV-2 quantitative LDT, we also assessed the correlation between Ct (viral load) and infectivity in clinical samples based on cytopathic effects (CPE) observed in Vero cells grown in culture.
2. Materials and methods
2.1. Development of the RealTime SARS-CoV-2 quantitative LDT
Open mode functionality on m2000sp/rt system was utilized to develop this SARS-CoV-2 quantitative LDT by using EUA Abbott SARS-CoV-2 qualitative reagents. Identical extraction and amplification/detection protocols developed for the RealTime SARS-CoV-2 qualitative EUA assay were also used for the development of the RealTime SARS-CoV-2 quantitative LDT. Specifically, 500 µl of sample was used for sample extraction, viral nucleic acid bound to the microparticles was eluted with 90 µl of elution buffer and 40 µl of the eluate was used for the amplification and detection reaction. The RealTime SARS-CoV-2 qualitative EUA assay and the RealTime SARS-CoV-2 quantitative LDT both utilize 10 unread cycles as part of their amplification and detection. In this assay, two calibrator levels (3 log10 RNA copies/mL and 6 log10 RNA copies/mL) tested in triplicate were used to generate a calibration curve and three control levels (negative, low positive at 3 log10 RNA copies/mL and high positive at 5 log10 RNA copies/mL) were included in each run for quality management.
2.2. Generation of Abbott SARS-CoV-2 calibration standard
A calibrator used for SARS-CoV-2 quantitative LDT was derived from virus propagated in culture on Vero cells (ATCC CCL-81). Briefly, nasopharyngeal (NP) swabs (New York Biologics, NY) containing viable SARS-CoV-2 were eluted into viral transport media (VTM, Becton Dickenson) and three specimens with the lowest Ct values were selected for lot production. Initially, 100 µL of VTM diluted in 1.9 mL of fresh MEM media (10–009-CV) without FBS was overlaid on a 10 cm plate with 2 × 106 cells and incubated for 2 h with rocking every 30 min to keep the monolayer from drying. The inoculum was removed by aspiration, monolayers were washed with 5 mL of 1X PBS, and 10 mL of fresh complete media containing 10% FBS was added. Cytopathic effects (CPE) developed within 4–5 days and both cells and supernatant were harvested. Primary lysates were freeze/thawed once and used to repeat infections on ten 10 cm plates, expanding each sample to 100 mL of virus lysate.
In order to assess viral stock concentration, heat-inactivated viral stock was tested in triplicate with the RealTime SARS CoV-2 qualitative assay. Quantitation was determined by plotting the sample Ct vs the expected SARS CoV-2 concentration obtained from the SeraCare AccuPlex SARS-CoV-2 Verification Panel (Material Number 0505–0129, SeraCare, Milford, MA). The verification material contained selected sequences from ORF1a, RdRp, N, and E with concentrations ranging from 103 to 105 copies/ml. The slope/intercept derived from the linear equation (y = mx + b) of the Accuplex SARS-CoV-2 data was used to obtain quantitation of the viral stock. Due to the high concentration of the viral stock, an intermediate material targeting 8 log10 RNA copies/mL was generated using the RNA Storage solution (Catalog Number AM7001, ThermoFisher, Waltham, MA). This intermediate, Abbott SARS-CoV-2 material was used to generate two calibrators with target concentrations of 3 and 6 log10 RNA copies/mL. Final calibrator concentration was verified against SeraCare AccuPlex SARS-CoV-2 Verification Panel. The above dilution process was also followed for the preparation of SARS-CoV-2 high positive control.
2.3. Evaluation of SARS-CoV-2 quantitative LDT analytical performance
Linearity of the RealTime SARS-CoV-2 quantitative LDT was evaluated with the BEI SARS-CoV-2 viral stock (NR-52281; USA-WA1/2020), SeraCare AccuPlex SARS-CoV-2 Verification panel, and the Abbott SARS-CoV-2 material. The copy number of the BEI stock (lot #70033175) was 2.07 × 109 genome equivalents/mL determined by the BioRad QX200 Droplet Digital PCR (ddPCR™) System. For the BEI and the Abbott SARS-CoV-2 material, an intermediate concentration targeting 8 log10 copies/mL was diluted using RNA Storage solution. This material was then serially diluted to 1.7 log10 copies/mL and tested in 2 to 6 replicates at each dilution on different days. SeraCare verification panel was tested neat along with one additional dilution level at 2 log10 RNA copies/mL over three days.
The limit of detection (LOD) was determined by testing serial dilutions of the Abbott SARS-CoV-2 material on different days. Inactivated SARS-CoV-2 whole genome virus was diluted in the RNA Storage solution and used to prepare serial dilutions in Log10 RNA copies/mL: 5.00, 4.00, 3.00, 2.70, 2.40, 2.00, and 1.70 and replicates ranging from 6 to 22 at each dilution. LOD was defined as the concentration of the lowest dilution that could be detected with > 95% probability.
Inter- and intra-run reproducibility was assessed with the Abbott SARS-CoV-2 material diluted to two different viral load concentrations near the LOD (2.40 and 2.70 log10 copies/mL) and another at a higher viral load (5.0 Log10 copies/mL). The prepared dilutions were tested with a minimum of three replicates across three different days.
2.4. Clinical performance of the RealTime SARS-CoV-2 quantitative LDT
Ct correlation between the RealTime SARS-CoV-2 quantitative LDT and three other EUA SARS-CoV-2 qualitative assays was assessed. The University of Washington comparison was carried out using the EUA CDC 2019-nCoV Real-Time PCR Diagnostic Panel (30 positive and 30 negative specimens) while the Northwell Health Laboratories comparison included Roche cobas SARS-CoV-2 assay (30 positive) and Abbott Alinity m SARS-CoV-2 assay (10 positive and 30 negative specimens) [11]. Inter-laboratory comparison of the RealTime SARS-CoV-2 quantitative LDT was evaluated using 89 remnant clinical specimens (26 quantifiable specimens, 3 specimens <2.0 log10 copies/mL, 35 negative specimens and 25 specimens which were greater than the upper limit of quantitation (7.0 log10 copies/mL)) collected after standard of care SARS-CoV-2 testing on the above platforms. All samples from the University of Washington were de-identified, excess clinical material and were deemed to be non-human subjects by the respective Institutional Review Board. Northwell Health Laboratories study was approved by the respective Institutional Review Board (IRB number 21–0284).
2.5. Correlation of Ct/viral load with in vitro culture
De-identified frozen NP swabs (n = 459) in VTM were sourced from multiple hospital systems across the United States (Supplementary Table S1). A 1 mL aliquot of VTM was heat-inactivated at 65 °C for 30 min and then processed with the RealTime SARS-CoV-2 quantitative LDT. The first of 5 consecutive runs included Abbott SARS-CoV-2 standards in triplicate. Non-inactivated specimens were kept at 4 °C to avoid a freeze/thaw cycle until culturing of selected positive samples was performed later the same day. Duplicate wells of Vero cells (3 × 105 cells/100 µl) were co-plated with 100 µl of patient VTM overnight and cultured for ≥96 h. CPE was determined by microscopy and a corresponding decrease in Viral Tox Glo luminescence (Cat# G8941, Promega, Madison WI).
2.6. Statistical analysis
Linear regression was used to evaluate assay's linearity, Ct method and inter-lab assay comparison. LOD and reproducibility was assessed by evaluating mean and standard deviation. (Microsoft Office Excel 365 software, Microsoft, Redmond, WA).
3. Results
3.1. Modification of RealTime SARS-CoV-2 EUA assay
LDT functionality (open mode on m2000sp/rt) was utilized to develop the modified RealTime SARS-CoV-2 quantitative LDT. This modified LDT is an automated and quantitative real-time PCR assay that uses extraction and amplification/detection protocols developed for the RealTime SARS-CoV-2 qualitative EUA assay [9,10]. Data reduction parameters were updated to include three control levels (one high positive SARS-CoV-2 control, one low positive SARS-CoV-2 control and one negative control) that were used for quality management of the assay, and two calibration levels (3.00 log10 copies/ml and 6.00 log10 copies/ml) that were used to generate calibration curves for the quantitative assay.
3.2. Analytical performance of the RealTime SARS-CoV-2 quantitative LDT
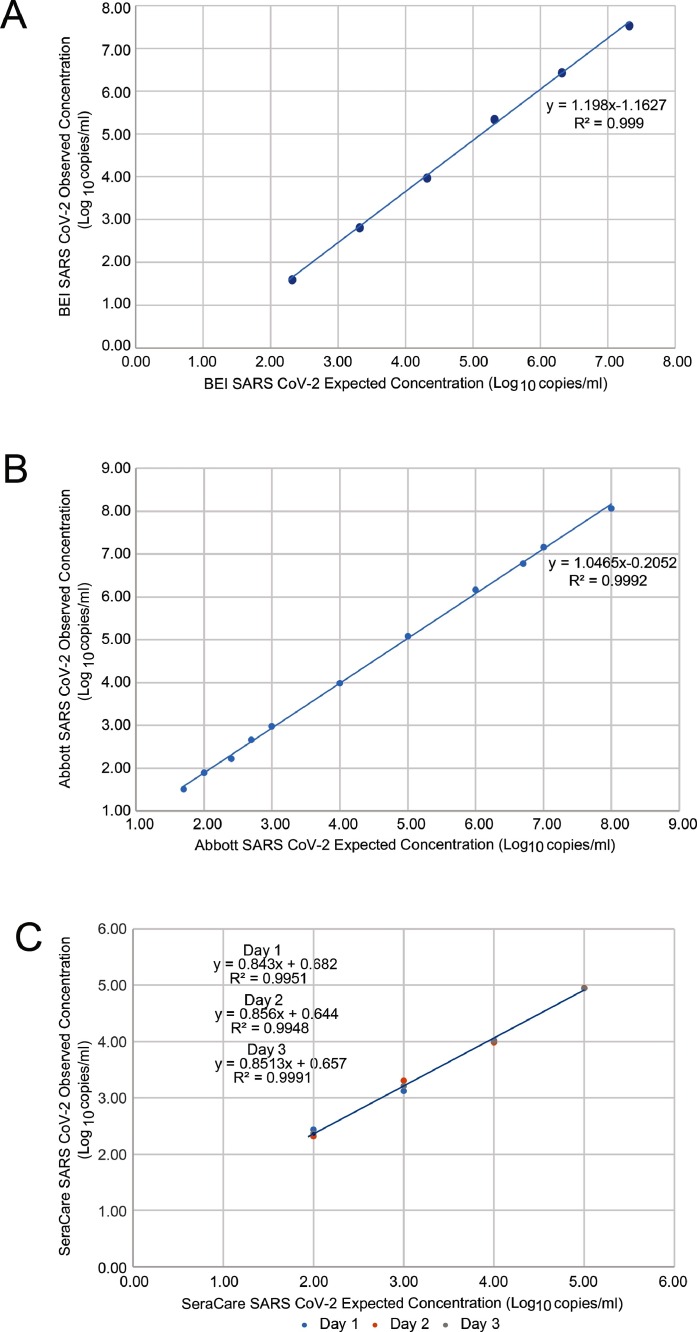
Excellent assay linearity was demonstrated across BEI SARS-CoV-2 viral stock (R2 = 0.9985; Fig. 1 A), Abbott SARS-CoV-2 material (R2 = 0.9992; Fig. 1 B) and SeraCare Verification Panel (R2 > 0.99; Fig. 1 C). These results demonstrated that the RealTime SARS-CoV-2 quantitative LDT can produce an accurate quantification value across the range of 1.7 to 8.0 log10 copies/ml. Using the dilution series of the Abbott SARS-CoV-2 material, the LOD was determined to be 1.90 log10 copies/mL (79 copies/mL in NPS) having a 100% detection rate with a SD ± 0.23 (Table 1 ). High inter- and intra-assay reproducibility was seen across the dynamic range (Table 2 ) with the SD of 0.14 at the lowest concentration tested (2.4 log10 copies/mL) and 0.08 at the highest concentration tested (5.0 log10 copies/mL) in the inter-run analysis. We observed an SD of 0.19 and 0.09 at the same concentrations in the intra-run analysis.
Fig. 1.
RealTime SARS-CoV-2 quantitative LDT linearity assessed with (A) BEI SARS-CoV-2 viral stock (B) Abbott SARS-CoV-2 viral stock and (C) SeraCare Accuplex SARS-CoV-2 verification panel.
Table 1.
Limit of detection of the RealTime SARS-CoV-2 quantitative LDT.
| Target Concentration (log10 copies/mL) | Observed Mean Concentration (log10 copies/mL ± SD) | SARS-CoV-2Mean Ct ± SD ( + 10 unread cycles) | %Detection (n = replicates detected/tested) |
|---|---|---|---|
| 3.00 | 2.98 ± 0.12 | 23.09 (33.09) ± 0.37 | 100 (6/6) |
| 2.70 | 2.66 ± 0.14 | 24.15 (34.15) ± 0.33 | 100 (9/9) |
| 2.40 | 2.22 ± 0.10 | 25.40 (35.40) ± 0.30 | 100 (9/9) |
| 2.00 | 1.90 ± 0.23 | 27.06 (37.06) ± 0.88 | 100 (22/22) |
| 1.70 | 1.51 ± 0.32 | 28.40 (38.40) ± 1.13 | 91 (20/22) |
| Neg | Not detected | N/A | 0 (0/30) |
Table 2.
Reproducibility of the RealTime SARS-CoV-2 quantitative LDT across the dynamic range.
| Nominal SARS-CoV-2log10 copies/mL | No. samples tested | Observed Mean(log10 copies/mL ± SD) | |
|---|---|---|---|
| Inter-Assay Reproducibility | 5.00 | 11 | 5.08 ± 0.08 |
| 2.70 | 9 | 2.66 ± 0.14 | |
| 2.40 | 9 | 2.22 ± 0.10 | |
| Intra-Assay Reproducibility | 5.00 | 4 | 5.03 ± 0.09 |
| 2.70 | 3 | 2.73 ± 0.19 | |
| 2.40 | 3 | 2.26 ± 0.11 |
3.3. Clinical performance
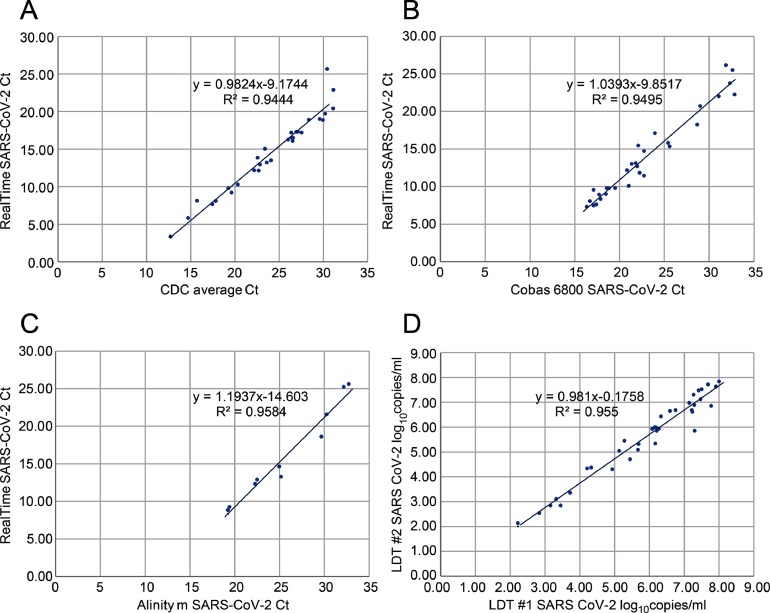
Ct's obtained with the RealTime SARS-CoV-2 quantitative LDT correlated well with those obtained by the EUA CDC 2019-nCoV assay (R2 = 0.9444; Fig. 2 a), cobas SARS-CoV-2 assay (R2 = 0.9495; Fig. 2 b) and Alinity m SARS-CoV-2 assay (R2 = 0.9584; Fig. 2 c). Inter-lab comparison of the RealTime SARS-CoV-2 quantitative LDT demonstrated excellent correlation (R2 = 0.955; Fig. 2 d) with a mean bias of 0.29 log10 RNA copies/mL (data not shown).
Fig. 2.
(A) Ct correlation between the EUA CDC 2019-nCoV assay and the RealTime SARS-CoV-2 quantitative LDT; (B) Ct correlation between the cobas SARS-CoV-2 EUA assay and the RealTime SARS-CoV-2 quantitative LDT; (C) Ct correlation between the Alinity m SARS-CoV-2 EUA assay and the RealTime SARS-CoV-2 quantitative LDT (D) Inter-laboratory comparison of RealTime SARS-CoV-2 quantitative LDT.
3.4. Correlation of CT/viral titer with culturing in clinical samples
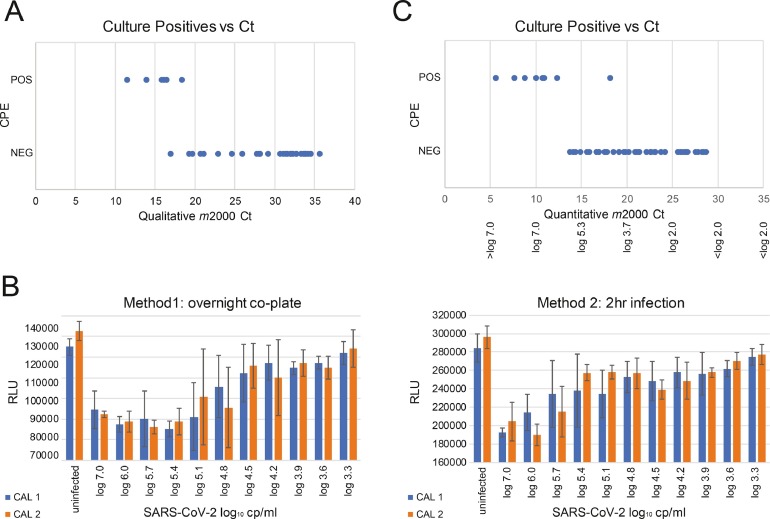
Numerous groups have sought to determine the Ct or viral load at which SARS-CoV-2 is transmissible [[6], [7], [8], [12], [13], [14], [15], [16], [17]]. One approximation is the ability to culture virus in vitro. A pilot experiment was performed with Vero CCL81 cells in 12 well plates using 36 nasopharyngeal swabs in VTM spanning a range of Cts (7.5–32) previously determined by the m2000 RealTime SARS-CoV-2 Qualitative assay [9,10]. CPE readily developed within 2–4 days as assessed by microscopy, however, only in those specimens with the lowest Cts (Fig. 3 a). Non-quantitative results indicated a cutoff of 18.27, or 28.27 when adjusting for the 10 unread cycles and qPCR/culture input volumes. SARS-CoV-2 culture was then adapted to a 96-well format to enable a quantitative measure of CPE using the Viral Tox Glo system (Fig. 3 b). We validated the assay by plating serial dilutions of the high titer calibrators in quadruplicate. After 96 h in culture, CPE was evaluated by both microscopy and luminescence, with a decrease in RLUs corresponding to the cell death observed visually. Co-plating of trypsinized cells with VTM at 1:1 v/v (Method 1) resulted in greater reproducibility and sensitivity compared to 2 h infections (Method 2) with cells seeded the night before. For Method 1, an apparent cut-off of 62,500 copies/mL (4.8 log10 copies/mL) was determined, wherein 2/4 replicates were infected for both calibrators, although CPE could still be detected in 1 of 4 replicates at 15,625 copies/mL (4.2 log10 copies/mL), which equates to a Ct of 18.5 (28.5 without unread cycles; Fig. 3 b, left panel). By contrast, a cut-off of 125,000 copies/ml (5.1 log10 copies/ml) at Ct = 15.65 (25.65) was observed for Method 2, with no dilutions lower than this showing evidence of successful infection (Fig. 3 b, right panel). Note that large error bars near these cutoffs indicate variability in replicates, wherein high RLU values (uninfected) are averaged with low values (infected).
Fig. 3.
A. COVID-19 positive nasopharyngeal swabs in VTM (n = 36) with Cts ranging from 7.5 to 32 determined by the non-quantitative assay were cultured on Vero cells. After adjusting for sample dilution and PCR input, the highest Ct where cytopathic effects (CPE) were detected was 18.27. B. SARS-CoV-2 calibrator stocks were cultured in 96-well format and cytopathic effects were quantified with the Viral Tox Glo system, comparing overnight co-plating (Method 1) or 2 h infection (Method 2). The highest Ct producing CPE was 18.49, corresponding to 4.2 log10 copies/ml. C. Clinical samples (n = 459) were screened with the RealTime SARS-CoV-2 quantitative LDT and identified 51 positives that were tested for their ability to infect in culture. A total of 9/51 (11.1%) demonstrated CPE, the majority having a Ct < 12 (> 1000,000 copies/mL). The lowest titer demonstrating CPE in culture had a Ct = 18.21 (28.21 w/o dark cycles), corresponding to 4.31 log10 copies/mL.
Using the in vitro culture method and NP clinical specimens (n = 459) sourced from multiple hospital systems across the United States (Supplementary Table S1), we explored the relationship between Ct, viral copy number/mL, and infectivity. An aliquot of each VTM was heat-inactivated, extracted, and measured using the newly developed SARS-CoV-2 RealTime quantitative LDT. Five successive experiments of 93 samples with calibrators and controls were run per day and positive specimens were infected in duplicate on Vero cells that evening. A total of 51 positive specimens were identified (11.1%) by qPCR, the majority (> 75%) having a Ct > 15 [25] (Supplementary Table S1). Only 9 specimens (17.6%) induced CPE: 8/9 had Cts ≤ 12.4 (22.4), corresponding to ≥ 6.2 log10 copies/mL or > 1,000,000 copies/mL, and 1/9 had a Ct = 18.21 (28.21), corresponding to 4.31 log10 copies/mL, or 20,417 copies/mL (Fig. 3 c). These results mirrored our prior experimental data and confirmed that a Ct ≤ 28.5 with a titer ≥ 4.2 log10 RNA copies/mL ( > 16,000 cp/ml) is required for successful Vero cell culture of SARS-CoV-2.
4. Discussion
Using Abbott SARS-CoV-2 calibrators to correlate viral load with Ct, we calibrated and modified the existing RealTime SARS-CoV-2 EUA assay into a quantitative measure of viral load. The SARS-CoV-2 quantitative LDT demonstrated broad assay linearity, reproducibility across the dynamic range, and an LOD of approximately 1.9 log10 copies/mL (79 copies/mL) at a Ct of 27 (37 without unread cycles). This study demonstrated that Ct values had a high degree of correlation (R2 > 0.94) between different SARS-CoV-2 EUA assays. Inter-lab comparison of the RealTime SARS-CoV-2 quantitative LDT demonstrated excellent correlation (R2=0.955) with a mean bias of 0.29 log10 RNA copies/mL. Additionally, quantitation of samples between 7 and 8 log10 RNA copies/mL (n = 13) demonstrated good correlation with a mean bias of 0.07 log10 RNA copies/mL, thus suggesting that the dynamic range could be expanded to 8 log10 RNA copies/mL. This change may have significant workflow and turnaround time impact for the laboratory as fewer dilutions and repeat testing would be required prior to result reporting. Viral culture was performed as a final step to correlate the viral load measured with the quantitative assay in clinical samples and potential infectivity, measured as cytopathic effect in culture. We conducted three separate culture studies that achieved consistent results.
Accounting for unread cycles (referred to as dark cycles), the range of Ct at which CPE was detected was 28.21–28.49 corresponding to approximately 16,000 RNA copies/mL (4.2 log10 copies/mL). While a culture cutoff of Ct = 33 (500 copies/mL, 2.7 log10 copies/mL) has been reported by others, our data consistently shows that titers more than 30 times greater (4.5 Cts) than this are required for culturing SARS-CoV-2 [14]. Numerous reports in the literature corroborate our findings that viral replication is seen from samples with Ct ≤ 28 [18,19]. In most reports, no replication was observed from samples above Ct > 30 (range of Ct 24–34) [6,7,14,20,21]. Of note, Ct values can be highly platform- and assay-specific, which may account for the large ranges of values seen across studies. The number of freeze-thaw cycles, how samples were sourced, and differences in cell lines used can also play a role in recovery. Here, we infected Vero CCL81 cells, whereas Vero E6 or Vero E6 lines over-expressing TMPRSS2 or ACE2 are considered more sensitive [22,23]. Some studies have reported a small percentage of samples with Ct ≥ 30 are able to be cultured, though the probability drops with each additional Ct [[13], [14], [15],17]. Other studies show a complete inability to culture virus [12,24,25]. Some of the variability may be due to the ratio of genomic to sub-genomic RNA being measured [26].
Our finding that a large viral load is needed to culture SARS-CoV-2 suggests that the ability to culture virus may be predictive of transmissibility. RT-PCR is extremely sensitive and can detect low levels of RNA shed persistently during a period when individuals may no longer be infectious [27]. Studies have reported no symptomatic COVID-19 cases with viral loads below 4 log10 copies/mL [8,[28], [29], [30]]. Indeed, the presence of viral RNA by itself is not proof of infectivity, which can only be determined by clinical investigation outside the lab, but a Ct > 30 found in air and surface samples has been associated with non-infectious samples [25]. Thus, a qualitative approach to interpreting RT-PCR assays will likely report as positive individuals with low levels of virus who may no longer be infectious [31]. Quantitative assays, validated against viral culture capacity, may allow more accurate identification of individuals with and without infectious viremia. It is conceivable that mutations that increase transmissibility will influence this value [32], [33], [34]. Nevertheless, given the close correspondence of rapid antigen test limits of detection ( ∼ 40,000 copies/swab; Ct ∼ 30) with what can be successfully cultured, this would argue that a cheaper, less sensitive test may be acceptable from a public health perspective to increase testing and reduce transmission [35,36].
As SARS-CoV-2 infection typically resolves in weeks, the application of a quantitative assay is limited in terms of treatment decision-making, though it may have major public health implications for decisions about patient quarantine/isolation time. Quantification of SARS-CoV-2 may also be useful for therapeutic clinical trials and vaccine development, and in screening efforts to direct contact tracing. Future research is needed to determine whether viral load is a clinical measure of disease severity.
Declaration of Competing Interest
WZ, EJDA, FS, and GB do not have conflicts of interest to disclose.
Acknowledgements
We thank Erin Quaco and Christopher Lark of Abbott Laboratories for technical assistance and Meei-Li Huang PhD for providing de-identified clinical specimens for the University of Washington studies. RWC is financially supported by the following grants: UM1-AI-106701; UM1-AI-068618; UW CFAR P30-AI-027757. ALG reports contract funding from Abbott and Gilead for testing, research funding from Merck, all outside of the submitted work.
MGB, DL, MA, KF, AO, DT and GAC are all employees and shareholders of Abbott Laboratories. The funder provided support in the form of salaries for authors MGB, DL, MA, KF, AO, DT, and GAC but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jcv.2021.104945.
Appendix. Supplementary materials
References
- 1.Walsh K.A., Jordan K., Clyne B., Rohde D., Drummond L., Byrne P., Ahern S., Carty P.G., O'Brien K.K., O'Murchu E., O'Neill M., Smith S.M., Ryan M., Harrington P. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J. Infect. 2020;81:357–371. doi: 10.1016/j.jinf.2020.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryan A., Fink S.L., Gattuso M.A., Pepper G., Chaudhary A., Wener M.H., Morishima C., Jerome K.R., Mathias P.C., Greninger A.L. SARS-CoV-2 viral load on admission is associated with 30-day mortality. Open Forum Infect. Dis. 2020;7:ofaa535. doi: 10.1093/ofid/ofaa535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rhoads D., Peaper D.R., She R.C., Nolte F.S., Wojewoda C.M., Anderson N.W., Pritt B.S. College of American Pathologists (CAP) microbiology committee perspective: caution must be used in interpreting the cycle threshold (Ct) value. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1199. [DOI] [PubMed] [Google Scholar]
- 4.Lieberman J.A., Pepper G., Naccache S.N., Huang M.L., Jerome K.R., Greninger A.L. Comparison of commercially available and laboratory-developed assays for in vitro detection of SARS-CoV-2 in clinical laboratories. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00821-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nalla A.K., Casto A.M., Huang M.W., Perchetti G.A., Sampoleo R., Shrestha L., Wei Y., Zhu H., Jerome K.R., Greninger A.L. Comparative performance of SARS-CoV-2 detection assays using seven different primer-probe sets and one assay kit. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bullard J., Dust K., Funk D., Strong J.E., Alexander D., Garnett L., Boodman C., Bello A., Hedley A., Schiffman Z., Doan K., Bastien N., Li Y., Van Caeseele P.G., Poliquin G. Predicting infectious SARS-CoV-2 from diagnostic samples. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gniazdowski V., Morris C.P., Wohl S., Mehoke T., Ramakrishnan S., Thielen P., Powell H., Smith B., Armstrong D.T., Herrera M., Reifsnyder C., Sevdali M., Carroll K.C., Pekosz A., Mostafa H.H. Repeat COVID-19 molecular testing: correlation of SARS-CoV-2 culture with molecular assays and cycle thresholds. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang C.G., Lee K.M., Hsiao M.J., Yang S.L., Huang P.N., Gong Y.N., Hsieh T.H., Huang P.W., Lin Y.J., Liu Y.C., Tsao K.C., Shih S.R. Culture-based virus isolation to evaluate potential infectivity of clinical specimens tested for COVID-19. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.01068-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abbott RealTime SARS-CoV-2 Assay. EUA Package Insert. Abbott Molecular 2020.
- 10.Degli-Angeli E., Dragavon J., Huang M.L., Lucic D., Cloherty G., KR Jerome, Greninger A.L., Coombs R.W. Validation and verification of the Abbott RealTime SARS-CoV-2 assay analytical and clinical performance. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anonymous. Centers for Disease Control and Prevention. CDC 2019–Novel coronavirus (2019-nCoV) Real-Time RT-PCR diagnostic panel (EUA) Instructions for use [package insert].
- 12.Andersson M., Arincibia-Carcamo C.V., Aukland K. SARS-CoV2 RNA detected in blood samples from patients with COVID-19 is not associated with infectious virus. medRxiv. 2020 doi: 10.1101/2020.05.21.20105486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basile K., McPhie K., Carter I., Alderson S., Rahman H., Donovan L., Kumar S., Tran T., Ko D., Sivaruban T., Ngo C., Toi C., O'Sullivan M.V., Sintchenko V., Chen S.C., Maddocks S., Dwyer D.E., Kok J. Cell-based culture of SARS-CoV-2 informs infectivity and safe de-isolation assessments during COVID-19. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.La Scola B., Le Bideau M., Andreani J., Hoang V.T., Grimaldier C., Colson P., Gautret P., Raoult D. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39:1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ladhani S.N., Chow J.Y., Atkin S., Brown K.E., Ramsay M.E., Randell P., Sanderson F., Junghans C., Sendall K., Downes R., Sharp D., Graham N., Wingfield D., Howard R., McLaren R., Lang N. Regular mass screening for SARS-CoV-2 infection in care homes already affected by COVID-19 outbreaks: implications of false positive test results. J. Infect. 2020 doi: 10.1016/j.jinf.2020.09.008:4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ladhani S.N., Chow J.Y., Janarthanan R., Fok J., Crawley-Boevey E., Vusirikala A., Fernandez E., Perez M.S., Tang S., Dun-Campbell K., Evans E.W., Bell A., Patel B., Amin-Chowdhury Z., Aiano F., Paranthaman K., Ma T., Saavedra-Campos M., Myers R., Ellis J., Lackenby A., Gopal R., Patel M., Brown C., Chand M., Brown K., Ramsay M.E., Hopkins S., Shetty N., Zambon M. Investigation of SARS-CoV-2 outbreaks in six care homes in London, April 2020. EClinicalMedicine. 2020;26 doi: 10.1016/j.eclinm.2020.100533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singanayagam A., Patel M., Charlett A., J Lopez Bernal, Saliba V., Ellis J., Ladhani S., Zambon M., Gopal R. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro. Surveill. 2020:25. doi: 10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.L'Huillier A.G., Torriani G., Pigny F., Kaiser L., Eckerle I. Culture-competent SARS-CoV-2 in nasopharynx of symptomatic neonates, children, and adolescents. Emerg. Infect. Dis. 2020;26:2494–2497. doi: 10.3201/eid2610.202403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perera R., Tso E., Tsang O.T.Y., Tsang D.N.C., Fung K., Leung Y.W.Y., Chin A.W.H., Chu D.K.W., Cheng S.M.S., Poon L.L.M., Chuang V.W.M., Peiris M. SARS-CoV-2 virus culture and subgenomic RNA for respiratory specimens from patients with mild coronavirus disease. Emerg. Infect. Dis. 2020;26:2701–2704. doi: 10.3201/eid2611.203219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brunink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 21.Young B.E., Ong S.W.X., Ng L.F.P., Anderson D.E., Chia W.N., Chia P.Y., Ang L.W., Mak T.M., Kalimuddin S., Chai L.Y.A., Pada S., Tan S.Y., Sun L., Parthasarathy P., Fong S.W., Chan Y.H., Tan C.W., Lee B., Rotzschke O., Ding Y., Tambyah P., Low J.G.H., Cui L., Barkham T., Lin R.T.P., Leo Y.S., Renia L., Wang L.F., Lye D.C., Singapore Novel Coronavirus Outbreak Research Team. Viral dynamics and immune correlates of COVID-19 disease severity. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harcourt J., Tamin A., Lu X., Kamili S., Sakthivel S.K., Murray J., Queen K., Tao Y., Paden C.R., Zhang J., Li Y., Uehara A., Wang H., Goldsmith C., Bullock H.A., Wang L., Whitaker B., Lynch B., Gautam R., Schindewolf C., Lokugamage K.G., Scharton D., Plante J.A., Mirchandani D., Widen S.G., Narayanan K., Makino S., Ksiazek T.G., Plante K.S., Weaver S.C., Lindstrom S., Tong S., Menachery V.D., Thornburg N.J. Severe acute respiratory syndrome coronavirus 2 from patient with coronavirus disease, United States. Emerg. Infect. Dis. 2020;26:1266–1273. doi: 10.3201/eid2606.200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., Nagata N., Sekizuka T., Katoh H., Kato F., Sakata M., Tahara M., Kutsuna S., Ohmagari N., Kuroda M., Suzuki T., Kageyama T., Takeda M. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. USA. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu J., Peng J., Xiong Q., Liu Z., Lin H., Tan X., Kang M., Yuan R., Zeng L., Zhou P., Liang C., Yi L., du Plessis L., Song T., Ma W., Sun J., Pybus O.G., Ke C. Clinical, immunological and virological characterization of COVID-19 patients that test re-positive for SARS-CoV-2 by RT-PCR. EBioMedicine. 2020;59 doi: 10.1016/j.ebiom.2020.102960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou J., Otter J.A., Price J.R., Cimpeanu C., Garcia D.M., Kinross J., Boshier P.R., Mason S., Bolt F., Holmes A.H., Barclay W.S. Investigating SARS-CoV-2 surface and air contamination in an acute healthcare setting during the peak of the COVID-19 pandemic in London. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alexandersen S., Chamings A., Bhatta T.R. SARS-CoV-2 genomic and subgenomic RNAs in diagnostic samples are not an indicator of active replication. Nat. Commun. 2020;11:6059. doi: 10.1038/s41467-020-19883-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corcorran M.A., Olin S., Rani G., Nasenbeny K., Constantino-Shor C., Holmes C., Quinnan-Hostein L., Solan W., Snoeyenbos Newman G., Roxby A.C., Greninger A.L., Jerome K.R., Neme S., Lynch J.B., Dellit T.H., Cohen S.A. Prolonged persistence of PCR-detectable virus during an outbreak of SARS-CoV-2 in an inpatient geriatric psychiatry unit in king county, Washington. Am. J. Infect. Control. 2021;49:293–298. doi: 10.1016/j.ajic.2020.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arons M.M., Hatfield K.M., Reddy S.C., Kimball A., James A., Jacobs J.R., Taylor J., Spicer K., Bardossy A.C., Oakley L.P., Tanwar S., Dyal J.W., Harney J., Chisty Z., Bell J.M., Methner M., Paul P., Carlson C.M., McLaughlin H.P., Thornburg N., Tong S., Tamin A., Tao Y., Uehara A., Harcourt J., Clark S., Brostrom-Smith C., Page L.C., Kay M., Lewis J., Montgomery P., Stone N.D., Clark T.A., Honein M.A., Duchin J.S., Jernigan J.A., Public H.-.S., King C., Team CC-I Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N. Engl. J. Med. 2020;382:2081–2090. doi: 10.1056/NEJMoa2008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saviddes C., Siegel R. Asymptomatic and pre-symptomatic transmission of SARS-CoV-2: a systematic review. medRxiv. 2020 doi: 10.1101/2020.06.11.20129072. [DOI] [Google Scholar]
- 30.He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., Lau Y.C., Wong J.Y., Guan Y., Tan X., Mo X., Chen Y., Liao B., Chen W., Hu F., Zhang Q., Zhong M., Wu Y., Zhao L., Zhang F., Cowling B.J., Li F., Leung G.M. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 31.Jefferson T., Spencer E., Brassey J., Heneghan C. Viral cultures for COVID-19 infectivity assessment–A systematic review. medRxiv. 2020 doi: 10.1101/2020.08.04.20167932:2020.08.04.20167932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sabino E.C., Buss L.F., Carvalho M.P.S., Prete C.A., Jr., Crispim M.A.E., Fraiji N.A., Pereira R.H.M., Parag K.V., da Silva Peixoto P., Kraemer M.U.G., Oikawa M.K., Salomon T., Cucunuba Z.M., Castro M.C., de Souza Santos A.A., Nascimento V.H., Pereira H.S., Ferguson N.M., Pybus O.G., Kucharski A., Busch M.P., Dye C., Faria N.R. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet. 2021;397:452–455. doi: 10.1016/S0140-6736(21)00183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., Mlisana K., von Gottberg A., Walaza S., Allam M., Ismail A., Mohale T., Glass A.J., Engelbrecht S., Van Zyl G., Preiser W., Petruccione F., Sigal A., Hardie D., Marais G., Hsiao M., Korsman S., Davies M.-.A., Tyers L., Mudau I., York D., Maslo C., Goedhals D., Abrahams S., Laguda-Akingba O., Alisoltani-Dehkordi A., Godzik A., Wibmer C.K., Sewell B.T., Lourenço J., Alcantara L.C.J., Pond S.L.K., Weaver S., Martin D., Lessells R.J., Bhiman J.N., Williamson C., de Oliveira T. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020 doi: 10.1101/2020.12.21.20248640:2020.12.21.20248640. [DOI] [Google Scholar]
- 34.Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., Hinsley W.R., Laydon D.J., Dabrera G., Á O'Toole, Amato R., Ragonnet-Cronin M., Harrison I., Jackson B., Ariani C.V., Boyd O., Loman N.J., McCrone J.T., Gonçalves S., Jorgensen D., Myers R., Hill V., Jackson D.K., Gaythorpe K., Groves N., Sillitoe J., Kwiatkowski D.P., Flaxman S., Ratmann O., Bhatt S., Hopkins S., Gandy A., Rambaut A., Ferguson N.M. Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: insights from linking epidemiological and genetic data. medRxiv. 2021 doi: 10.1101/2020.12.30.20249034:2020.12.30.20249034. [DOI] [Google Scholar]
- 35.Pekosz A., Cooper C.K., Parvu V., Li M., Andrews J.C., Manabe Y.C., Kodsi S., Leitch J., Gary D.S., Roger-Dalbert C. Antigen-based testing but not real-time PCR correlates with SARS-CoV-2 virus culture. medRxiv. 2020 doi: 10.1101/2020.10.02.20205708:2020.10.02.20205708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perchetti G.A., Huang M.-.L., Mills M.G., Jerome K.R., Greninger A.L. Analytical sensitivity of the Abbott BinaxNOW COVID-19 Ag Card. J. Clin. Microbiol. 2021;59 doi: 10.1128/JCM.02880-20. e02880–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.