Abstract
Since the first described human infection with SARS-CoV-2 in December of 2019 many subunit protein vaccines have been proposed for use in humans. Subunit vaccines use one or more antigens suitable for eliciting a robust immune response. However, the major concern is the efficacy of subunit vaccines and elicited antibodies to neutralize the variants of SARS-CoV-2 like B.1.1.7 (Alpha), B.1.351 (Beta) and P1 (Gamma), B.1.617 (Delta) and C.37 (Lambda). The Spike protein (S) is a potential fragment for use as an antigen in vaccine development. This protein plays a crucial role in the first step of the infection process, as it binds to Angiotensin-Converting Enzyme 2 (ACE2) receptor and enters the host cell after binding. Immunization-induced specific antibodies against the receptor binding domain (RBD) may block and effectively prevent virus invasion. The focus of this review is the impact of spike mutated variants of SARS-CoV2 (Alpha, Beta, Gamma, Delta, and Lambda) on the efficacy of subunit recombinant vaccines. To date, a low or no significant impact on vaccine efficacy against Alpha and Delta variants has been reported. Such an impact on vaccine efficacy for Beta, Delta, Gamma, and Lambda variants may be even greater compared to the Alpha variant. Nonetheless, more comprehensive analyses are needed to assess the real impact on vaccine efficacy brought about by SARS-CoV-2 variants.
Keywords: COVID-19, SARS-CoV2, Variant, Mutation, Vaccine, Spike protein, Subunit vaccine
Introduction
The routine living status in the world stopped by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic. It is causing significant health, social and economic global burden.1 The SARS-CoV-2 is a positive-sense, single-stranded RNA virus that is rapidly evolving and continually accrues genomic mutations as it continues to be transmitted.2,3 Since the first described human infection with SARS-CoV-2 in Wuhan in December of 2019,4,5 many subunit protein vaccines have been presented for use in humans.6 Still, viral genomic mutations leading to new variants is a real challenge in tackling this pandemic worldwide.7 A primary focus of this review is to assess the impact of spike mutated variants of SARS-CoV2 (B.1.1.7, B.1.351, and B.1.1.28.1) on the efficacy of subunit recombinant vaccines designed to elicit an immune response against the spike protein of SARS-CoV2.
Methods
Independent searches in PubMed, Web of Science, and Global Index Medicus were conducted by two researchers in June 2021. The search strategy consisted of a word combination covering the following areas (Covid-19 OR SARS-CoV-2) AND Vaccine AND B.1.1.7 Variant OR B.1.351 variant OR B.1.1.28.1 Variant OR B.1.617.2 OR C.37 variant. The search had no geographic or language restrictions and included all studies reporting on efficacy of SARS-CoV-2 vaccines against variants Alpha, Beta, Gamma, Delta, and Lambda. Herein we narratively describe the main findings of the included studies.
The SARS-CoV-2 genomic features
The SARS-CoV-2 is a spherical-shaped virion with a positive-strand RNA virus, with a genome length of approximately 29,700 nucleotides.8 The 5ˊ end includes more than two-thirds of the genome and comprises a long ORF1ab poly protein, which encodes 16 non-structural proteins.8,9 The 3ˊ end encodes four major structural proteins, namely the spike (S) protein, nucleocapsid (N) protein, membrane (M) protein, and envelope (E) protein.8 Additionally, the virus contains six accessory proteins encoded by ORF3a, ORF6, ORF7a, ORF7b, ORF8, and ORF9b genes.8,10 ORF3a is a trans membrane protein that forms ion channels in the host membrane and ORF7a encodes a type I trans membrane protein(10). ORF8 is a protein with an N-terminal signal sequence for transport to the endoplasmic reticulum and ORF9b suppresses IFN-I response(9).
Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein
The S protein, a glycoprotein, is a homotrimer that plays a crucial role in the first step of the infection.11,12 The monomer of S protein contains 1273 amino acids and a molecular weight of about 140 kDa.13,14 The primary function of S protein is to bind to the Angiotensin-Converting Enzyme 2 (ACE2) receptor entering the host cell after binding.14, 15, 16 ACE2 is a type I integral membrane protein that functions as a carboxy peptidase, cleaving angiotensin II to angiotensin regulating blood pressure.12,17 The virus binds to ACE2 on the host cell (lungs, heart, etc.) for virus entry and subsequent pathogenesis, resulting in severe respiratory infection.12,15, 16, 17 Therefore, the S protein-ACE2 interaction is an easy target for vaccines.15,18 First, it is exposed at the surface and can be recognized directly by the host immune system.15,18,19 Second, it mediates the interaction with the host cell binding to the ACE2 receptor, which is essential for subsequent virus entry into target cells causing subsequent pathogenicity.15,18,19 Finally, the homolog proteins had been already used for vaccine development against SARS-CoV and MERS-CoV and proved effective.1,18,20,21
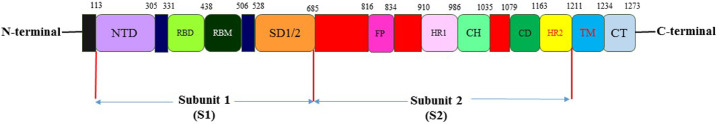
The S protein comprises two functional subunits that include an N-terminal S1 subunit (Val16–Gln690). It consists of S1A, S1B, S1C, and S1D domains. The latter is responsible for binding to the host cell receptor. A C-terminal membrane-proximal S2 subunit contains the essential elements and is responsible for fusion of the viral and cellular membranes.11,14, 15, 16,21, 22, 23, 24 This subunit has four parts including an internal membrane fusion peptide (FP), two 7-peptide repeats, a membrane-proximal external region, and a trans membrane domain (TM).15,21,23 The S1A domain, alluded to as the N-terminal domain (NTD), recognizes carbohydrates, such as sialic acid, required for virus attachment to host cell surface.15,21,23 The S1B domain (Arg319–Phe541), referred to as the receptor-binding domain (RBD) of the SARS-CoV-2 spike protein, interacts with the human ACE2 receptor.15,21,23 The S1 subunit can be defined further in two domains termed the N-terminal domain (NTD) and the C-terminal domain (CTD).15 The RBD is located in the CTD.13,15 The structural parts within the S2 subunit include three long α-helices, multiple α-helical segments, extended twisted β-sheets, membrane-spanning α-helix, and an intracellular cysteine-rich segment.11,15,22,25 The PRRA sequence motif located between the S1 and S2 subunits in SARS-CoV-2 presents a furin-cleavage site. In the S2 subunit, there is a second proteolytic cleavage site S20, upstream of the fusion peptide. Both these cleavage sites participate in viral entry into host cells 11,15,22 (Fig. 1).
Fig. 1.
The representative scheme of functional domains in S protein of SARS-CoV-2.
The S protein is a potential fragment to be used as antigens in vaccine designing including various forms of the full-length S protein, RBD domain, S1 subunit, NTD, and FP.11,15,22 The RBD of S protein interacts with the ACE2 receptor on host cells directly. The RBD immunization-induced specific antibodies may block this recognition and effectively prevent the viral invasion.15,17 Most SARS-CoV-2 subunit vaccines currently under development use RBD as the antigen.19, 20, 21 Moreover, the RBD domain was also used to develop SARS-CoV and MERS-CoV vaccines.19, 20, 21,26
Subunit vaccines against SARS-CoV-2
Subunit vaccines use one or more antigens suitable for eliciting a robust immune response.18 In theory, the subunit vaccine is very easy and safe, but in practice, it requires a suitable adjuvant to stimulate the host immune response.13,26,27 Several previous attempts were partially successful with SARS-CoV.18,28 Immunization of animals with the S1 RBD domain fused with the IgG1 FC portion (RBD-FC) induced highly potent antibodies, which could bind with the RBD domain of the S1 domain, completely neutralized SARS-CoV, and inhibited SARS-CoV entry into Vero E6 cells.18,26,29,30
So far, several companies and academics have started plans on the SARS-CoV-2 subunit vaccine (Table 1). Almost all of them use the S protein as an antigenic target.1,6,18,28,31 For example, Novavax, Inc. announced to have produced multiple nanoparticle vaccine candidates based on S protein.32,33 Besides, the Pasteur Institute of Iran and Finley institute of Cuba also prepared subunit vaccines against SARS-CoV-2.34 To facilitate the development of a SARS-CoV-2 vaccine, the preferred adjuvant varied widely, including the classic aluminum adjuvant that enhances the immune response by facilitating phagocytosis and slowing the diffusion of antigens from the injection site.6,18,27,35, 36, 37 It can efficiently stimulate the Th2 immune response upon injection. The F59, MF59 have already been used in flu vaccines in Europe and the United States.31,37, 38, 39, 40, 41, 42 The mechanism of MF59 is to recruit immune cells to induce antigen-specific immune responses. The adjuvant system (AS) comprises a series of adjuvants improved by GlaxoSmithKline (GSK), including AS01, AS02, AS03, and AS04.30,35,37,41, 42, 43 Among them, AS01 is a liposome adjuvant.37,44
Table 1.
The type of protein subunit vaccines against SARS-CoV-2 registered and in clinical phase trials in various countries.
| Number | Type of subunit vaccine | Company | Current clinical phase trial number |
|---|---|---|---|
| 1 | SARS-CoV-2 rS/Matrix M1-Adjuvant (Full length recombinant SARS-CoV-2 glycoprotein nano particle vaccine adjuvant with Matrix M) | Novavax |
NCT04611802 EUCTR2020-004123-16-GB NCT04583995 |
| 2 | Recombinant SARS-CoV-2 vaccine (CHO Cell)This is not a pattern of 'ctgov' external object linking. | Anhui Zhifei Longcom Biopharmaceutical + Institute of Microbiology, Chinese Academy of Sciences |
NCT04646590 |
| 3 | KBP-COVID-19 (RBD-based) | Kentucky Bioprocessing Inc. | NCT04473690 |
| 4 | VAT00002: SARS-CoV-2 S protein with adjuvant | Sanofi Pasteur + GSK | PACTR202011523101903 |
| 5 | SCB-2019 + AS03 or CpG 1018 adjuvant plus Alum adjuvant (Native like Trimeric subunit Spike Protein vaccine) | Clover Biopharmaceuticals Inc./GSK/Dynavax | NCT04672395 |
| 6 | COVAX-19® Recombinant spike protein + adjuvant | Vaxine Pty Ltd. | NCT04453852 |
| 7 | MVC-COV1901 (S-2P protein + CpG 1018) | Medigen Vaccine Biologics + Dynavax + National Institute of Allergy and Infectious Diseases (NIAID) | NCT04695652 |
| 8 | FINLAY-FR1 anti-SARS-CoV-2 Vaccine (RBD + adjuvant) | Instituto Finlay de Vacunas | RPCEC00000332 |
| 9 | FINLAY-FR-2 anti-SARS-CoV-2 Vaccine (RBD chemically conjugated to tetanus toxoid plus adjuvant) | Instituto Finlay de Vacunas | RPCEC00000354 |
| 10 | EpiVacCorona (EpiVacCorona vaccine based on peptide antigens for the prevention of COVID-19) | Federal Budgetary Research Institution State Research Center of Virology and Biotechnology “Vector” | NCT04780035 |
| 11 | RBD (baculovirus production expressed in Sf9 cells) Recombinant SARS-CoV-2 vaccine (Sf9 Cell) | West China Hospital + Sichuan University | NCT04718467 |
| 12 | IMP CoVac-1 (SARS-CoV-2 HLA-DR peptides) | University Hospital Tuebingen | NCT04546841 |
| 13 | UB-612 (Multitope peptide based S1-RBD-protein based vaccine) | COVAXX + United Biomedical Inc | NCT04683224 |
| 14 | AdimrSC-2f (Recombinant RBD +/− Aluminium) | Adimmune Corporation | NCT04522089 |
| 15 | CIGB-669 (RBD + AgnHB) | Center for Genetic Engineering and Biotechnology (CIGB) | RPCEC00000345 |
| 16 | CIGB-66 (RBD + aluminium hydroxide) | Center for Genetic Engineering and Biotechnology (CIGB) | RPCEC00000359 |
| 17 | Recombinant Sars-CoV-2 Spike protein, Aluminum adjuvanted | Nanogen Pharmaceutical Biotechnology |
NCT04683484 |
| 18 | Recombinant protein vaccine S-268019 (using Baculovirus expression vector system) | Shionogi | jRCT2051200092 |
| 19 | SARS-CoV-2-RBD-Fc fusion protein | University Medical Center Groningen + Akston Biosciences Inc. | NCT04681092 |
| 20 | COVAC-1 and COVAC-2 sub-unit vaccine (spike protein) + SWE adjuvant | University of Saskatchewan | NCT04702178 |
| 21 | GBP510, a recombinant surface protein vaccine with adjuvant AS03 (aluminium hydroxide) | SK Biosciences Co. Ltd. And CEPI | NCT04750343 |
| 22 | Razi Cov Pars, recombinant spike protein | Razi Vaccine and Serum Research Institute |
IRCT20201214049709N1 |
| 23 | MF59 adjuvanted SARS-CoV-2 Sclamp vaccine | The University of Queensland | NCT04495933 |
| 24 | SK SARS-CoV-2 recombinant surface antigen protein subunit (NBP2001) + adjuvanted with alum | SK Bioscience Co., Ltd. | NCT04760743 |
| 25 | SpFN (spike ferritin nanoparticle) uses spike proteins with a liposomal formulation QS21 (ALFQ) adjuvant. | Walter Reed Army Institute of Research (WRAIR) | NCT04784767 |
| 26 | EuCorVac-19; A spike protein using the recombinant protein technology and with an adjuvant. | POP Biotechnologies and EuBiologics Co.,Ltd |
NCT04783311 |
| 27 | ReCOV: Recombinant two-component spike and RBD protein COVID-19 vaccine (CHO cell). | Jiangsu Rec-Biotechnology | NCT04818801 |
Variant analysis of SARS-CoV-2 in spike protein mutations
Alpha (B.1.1.7 or VOC 202012/01 or 20B/501Y.V1) variant
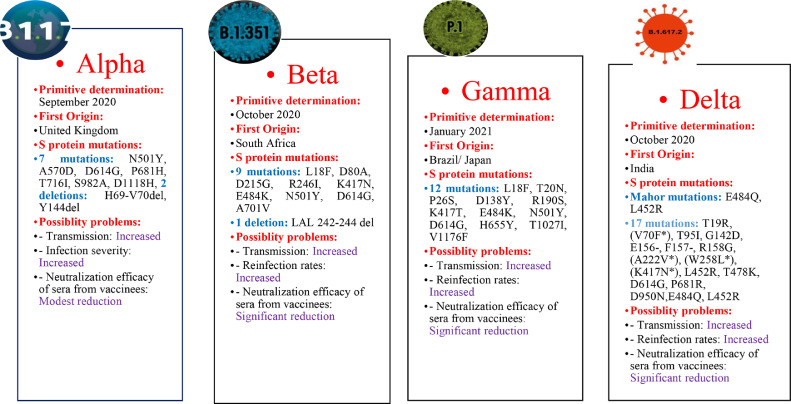
The B.1.1.7 (Alpha) variant was presented on December 14, 2020 by the United Kingdom (UK) authorities.45 The robust characterization of this variant which greater transmission, accompanying an increase in incidence, hospitalizations, and pressure on the health system.45, 46, 47 Epidemiological reports and modeling suggest that it spreads 56% faster than other lineages. Studies estimated a 35% (12–64%) increased risk of death associated with the Alpha variant. However, there were no scientific reports of more severe diseases in children and young people.46,48
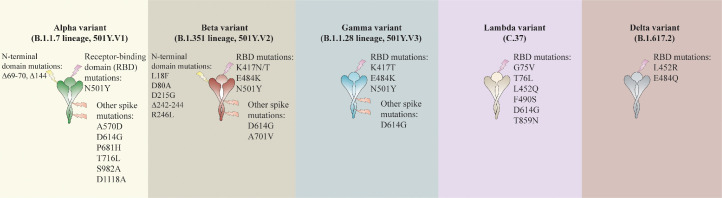
In comparison with ancestral viruses containing the D614G mutation, the Alpha variant has accumulated 23 mutations. Of these mutations, 14 are non-synonymous: [T1001I, A1708D, and I2230T] in the open reading frame ORF1ab; [N501Y, A570D, P681H, T716I, S982A, and D1118H] in the S protein; [Q27stop, R52I, and Y73C] in ORF8; and [D3L and S235F] in the nucleocapsid (N) protein]; six are synonymous: [C913T, C5986T, C14676T, C15279T, and T16176C] in ORF1ab; and [T26801C] in M (membrane) gene; and three are deletions: [SGF 3675-3677del] in ORF1ab; and [H69-V70del and Y144del] in S protein 45,48, 49, 50, 51, 52, 53, 54, 55, 56, 57 (Table 2).
Table 2.
The main characterizations of SARS-CoV-2 variants with mutated Spike protein.
Forty-seven percent of reported changes in the Alpha variant occur in the gene encoding for the S protein, including the RBD. These mutations can play a role in (i) changing the interaction with the human angiotensin-converting enzyme-2 (hACE2) receptor, increasing the infection rate; (ii) compromising efficacy of both neutralizing antibodies and specific T cells elicited either during natural infection or through vaccination; or (iii) altering sensitivity to neutralization by monoclonal antibodies or sera from convalescent patients, compromising the efficacy of treatments 45,48, 49, 50, 51, 52, 53, 54, 55, 56, 57 (Fig. 2).
Fig. 2.
Mutations in S protein of SARS-CoV-2 are generated different variants.
The three mutations of the Alpha variant with the most significant potential to influence the biological characteristics of the virus are H69-V70del, N501Y, and P681H. How does Alpha distribution impact the efficacy of vaccines and treatments that are being administered globally? To date, the ALPHA variant appears to have low or no significant impact on vaccine effectiveness.32,45,48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59
Beta (B.1.351 or 20H/501Y.V2) variant
The B.1.351 60 variant, also known as 20H/501Y.V2, was first reported on December 18, 2020 by the Republic of South Africa.61 The significant feature of this variant is the higher transmission rate.49,62 The Beta variant has 12 non-synonymous mutations and one deletion compared to the Wuhan reference strain. Approximately 77% of these mutations are located in the S protein [L18F, D80A, D215G, LAL 242–244 del, R246I, K417N, E484K, N501Y, D614G, and A701V] while the remaining ones are located in ORF1a [K1655N], envelope (E) [P71L], and N [T205I] viral proteins.47,49,56,58,63, 64, 65 Most mutations are within two of the most immuno dominant regions, such as NTD and RBD domains. This issue suggests that Beta variant mutations could escape neutralizing antibodies and compromise vaccine efficacy 47,49,56,58,63, 64, 65 (Table 2).
Various studies determined that a combination of RBD and NTD mutations in the Beta spike protein considerably influences neutralization activity in vaccinated patients. Vaccine efficacy are more compromised against the Beta variant than against the varinat B.1.1.7 49,53,61, 62, 63,65, 66, 67, 68 (Fig. 2).
Gamma (P.1 or B.1.1.28.1) variant
The B.1.1.28.1. (Gamma or P.1) variant was first detected by Japan's National Institute of Infectious Diseases on January 6, 2021 and was isolated from four travelers who arrived in Tokyo coming from Amazonas, Brazil, on January 2, 2021 at airport control.47,61,64 The rapid increase in the number of hospital admissions was a significant problem with this variant. This variant contains 17 non-synonymous mutations: [L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, and V1176F] in S protein, [S1188L, K1795Q, and E5665D] in ORF1ab, [E92K] in ORF8, and [P80K] in N protein; 1 deletion: [SGF 3675-3677del] in ORF1ab; and four synonymous mutations. Gamma is the SARS-CoV-2 variant that accumulates the highest number of mutations in the S protein (12 mutations) 46,54,56,57,61,64,69,70 (Table 2).
The mutation N501Y is present in the three variants, while L18F, K417T, E484K, and D614G mutations are present in the Beta variants. This group of S mutations has important implications for the evasion of antibody-mediated immunity. The efficacy of serum neutralization against the virus holding the E484K mutation was decreased in both vaccination and convalescent sera 46,54,56,57,61,64,69 70 (Fig. 2).
Delta (B.1.617.2) variant
The SARS-CoV-2 B.1.617.2 (Delta) variant was first reported in the state of Maharashtra in October 2020 with subsequent dissemination throughout India. The virus sequences extracted in India had two critical amino acid substitutions (L452R and E484Q) in the RDB of the S glycoprotein. This “double mutant” has three sub-lineages (B.1.617.1, B.1.617.2, and B.1.617.3) characterized by the spike mutation L452R, whilst E484Q is present in B.1.617.1 and B.1.617.3 but not in B.1.617.2. The B.1.617.2 is now the major variant in the UK. The lineages have mutations in the gene encoding the SARS-CoV-2 S protein. The Delta genome has 17 mutation of which four are of particular concern. In the D614G, substitution at position 614, an aspartic acid-to-glycine substitution, is shared with other highly transmissible variants like Alpha, Beta, and Gamma. T478K is an exchange at position 478. The L452R is a substitution at position 452, confers a stronger affinity of the S protein for the ACE2 receptor and decreased recognition capability of the immune system. The P681R is a substitution at position 681, may boost cell-level infectivity of the variant.71, 72, 73, 74
Lambda (C.37) variant
The Lambda (C.37) variant was first reported in Peru in December of 2020. The Sequencing of the variant genome revealed a deletion (Δ3675- 3677) in open reading frame 1a (ORF1a) of wild type. The Lambda variant also has of a new deletion (Δ246-252) and multiple non-synonymous mutations (G75V, T76I, L452Q, F490S, D614G, and T859N) in S protein. The mutations L452Q and F490S are present in the spike RBD. The F490S mutation is associated with decreased susceptibility to antibody neutralization. This variant has 19 mutations that made it more transmissible or with increased resistance to antibodies induced by vaccination or by prior exposure to the virus.75, 76, 77
Other variants
The Eta (B.1.525) variant was first identified in Nigeria in December of 2020. Its spike mutations include E484K, D614G, Q677H which impact antibody neutralization and transmission. The Epsilon (B.1.427/B.1.429) variant was first identified in the USA in September of 2020 showing L452R, D614G as spike mutations, but their impact on antibody neutralization and transmission is unclear. The Theta (P.3) variant was first identified in the Philippines in January of 2021, with E484K, N501Y, D614G, P681H as spike mutations which impact antibody neutralization and transmission. The Kappa (B.1.617.1) variant was first identified in India in December of 2020, with L452R, E484Q, D614G, P681R as spike mutations which also impact antibody neutralization and transmission. The Iota (B.1.526) variant was first identified in the USA in December of 2020, with E484K, D614G, A701V as spike mutations which impact antibody neutralization. The Zeta (P.2) variant was first identified in Brazil in January of 2021, with E484K, D614G as spike mutations which impact antibody neutralization. All of the above variants have not been shown to cause more severe disease.
The mutated spike variants of SARS-CoV-2 and vaccines efficacy reports
The main concern for healthcare systems related to the variants of SARS-CoV-2 are the potential compromise of antibody neutralization and subunit vaccines efficacy. Various reports demonstrate the interaction between SARS-CoV-2 and vaccine efficacy.46, 47, 48,57,64,70 In the present work, the main objective was to assess the impact of the variants that mutated in the target site of the vaccine (S protein) on subunit vaccines efficacy.
Yiska et al. demonstrated that functional SARS-CoV-2 S protein variants with mutations in RBD confer resistance to monoclonal antibodies or convalescent plasma and can be readily selected. Therefore, the virus S variants can resist commonly elicited neutralizing antibodies.78 Pengfei et al. showed that the B.1.351 variant is refractory to neutralization by most monoclonal antibodies against the N-terminal domain and multiple individual monoclonal antibodies against the receptor-binding motif of the RBD. Moreover, B.1.351 is markedly more resistant to neutralization by convalescent plasma and sera from individuals who have been vaccinated.49
Delphine et al. demonstrated that B1.351, but not B.1.1.7, may increase the risk of infection in immunized individuals.53 A description by Rees et al. shown that the neutralizing activity of some antibodies was dramatically reduced by Spike mutations (B.1.1.7).64 In Rita et al. reported the impact on antibody neutralization of a panel of authentic SARS-CoV-2 variants, including a B.1.1.7 isolate, chimeric strains with South African, Brazilian spike genes, and isogenic recombinant viral variants. They showed reduced inhibitory activity against viruses containing an E484K spike mutation as antibodies binding to spike RBD demonstrate diminished neutralization potency in vitro against some emerging variants.79
Xiu et al. revealed that UK and Brazil variants have no significantly decreased effect on vaccine impact and neutralization by sera from vaccinated individuals after two BNT162b2 (Pfizer) doses.80 Collier et al. did not observe a significant decrease in the ability of sera from vaccinated persons (Pfizer) to inhibit parental or mutant pseudo viruses, including only three (H69/V70del, N501Y, and A570D) S mutations.81 Wu et al. found no considerable impact in the neutralizing potency of sera from people who received the Moderna vaccine against the B.1.1.7 variant.82 Nevertheless, Wang et al. reported that B.1.351 variant was particularly more resistant to neutralization in people immunized with Pfizer or Moderna vaccines.49 On the other side, he reported considerable decrease in the neutralization of B.1.351 variants by sera from volunteers who were vaccinated with Moderna.49 Wilfredo et al. analyzed neutralization potency in individuals who received one or two doses of either BNT162b2 or mRNA-1273 vaccines, suggesting that a relatively small number of mutations, like B.1.351 can mediate potent escape from vaccine responses. Their results highlight the potential for variants to escape neutralizing humoral immunity and emphasize the need to develop broadly protective interventions against the evolving pandemic.83
In this review, we highlight the compromise of subunit vaccine efficacy against the variants. In the study by Arturo Chang et al. the vaccine based on recombinant dimeric receptor-binding domain (d-RBD, 50 g) on alum (FINLAY-FR-1A) in clinical development in Cuba, and they hypothesized that a single dose of that vaccine might be an effective booster for individuals with pre-existing immunity to SARS-CoV-2.34 The report by Shilong et al. focused on the ZF2001 vaccine (protein-based COVID-19). They indicated that the RBD-based protein subunit vaccine could be safe and immunogenic.30 Shinde et al. reported on a comprehensive analysis of the NVX-CoV2373 vaccine in 4387 participants. They revealed that efficacy against B.1.351 was 51.0%. The NVX-CoV2373 vaccine was efficacious in preventing Covid-19, which was predominantly mild to moderate and due to the B.1.351 variant.66
SARS-CoV-2 vaccine NVX-CoV2373, produced by Novavax is based on S protein derived from the SARS-CoV-2 Wuhan reference strain, has shown 85.6% efficacy against the B.1.1.7 variant (95.6% against the original strain) in phase 3 clinical trial.32 However, in a study in South Africa Callaway et al. reported a 49.4% efficacy against B.1.351 variant in the overall population.33 These interim data evidence a significant decrease in vaccine efficacy influenced by variants such as B.1.351.
In the report of Xiaoying et al. B.1.1.7 remained sensitive to neutralization, albeit at moderately reduced levels, by serum samples from convalescent individuals and recipients of an mRNA vaccine (Moderna) and a protein nanoparticle vaccine (NVXCoV2373, Novavax).45
Mlcochova et al. showed evasion of the Delta variant from neutralizing antibodies present in convalescent patients, as well as in vaccinated individuals with two different vaccines in the UK (adenovirus vector (ChAdOx-1), and the other mRNA 19 (BNT162b2)). They demonstrated a reduced susceptibility of Delta to vaccine-elicited neutralization. In their data, the variant showed approximately 8 to 20-fold reduced sensitivity to vaccine-elicited antibodies. Serum neutralizing titers against the variant were significantly lower in vaccinated persons with ChadOx-1 as compared to BNT162b2. These data revealed that the Delta variant can evade from neutralizing antibodies in previously infected individuals, although severe disease in fully vaccinated was rare.84
In another report by Davis et al. reductions in the neutralization of B.1.617.1 and B.1.617.2 were 4.31- and 5.11-fold, respectively.71 In another report, Lustig and et al. demonstrated significant fold change reduction in neutralizing titers: Gamma (P.1) 2.3, Beta (B.1.351) 10.4, Delta 2.1, and 2.6. The fold reduction of the Alpha (B.1.1.7) variant was not significant.73
No published report demonstrated subunit vaccine efficacy on Delta variant, and all other reports were based on Pfizer and Oxford vaccines and their antibody neutralization. No published report demonstrated subunit vaccine efficacy against Delta variant.
Acevedo et al. observed greater infectivity mediated by the Lambda spike protein compared to D614G (lineage B) or Alpha and Gamma variants. In addition, neutralization was reduced by 3.05-fold for the Lambda variant, 2.33-fold for the Gamma variant and 2.03-fold for the Alpha variant.77 Tada et al. showed an average 2.3-3.3-fold reduction of antibody titers against Lambda variant.76
Conclusion
Subunit vaccines with strong immunogenic capacity can efficiently elicit host immune response. However, the major healthcare concern is a reduction of subunit vaccines efficacy in translated by lower antibody neutralization potency against SARS-CoV-2 Alpha, Beta, Gamma, Delta, and Lambda variants. To date, low or no significant impact on vaccine efficacy against Alpha variants has been reported. Concern about and Delta, Beta, Gamma, and Lambda mutations on vaccine efficacy and treatments is greater than for the Alpha variant. Further comprehensive analyses all over the world validated by clinical trials are clearly warranted.
Author contributions
The article was conceived and critically reviewed by Mehrdad Mohammadi and performed the literature search and drafted the work. Dr. Mohammadi Shayestepour approved the work and agreed to be accountable for the work. Dr. Hamed Mirzaei designed all of the figures.
Funding
None.
Conflicts of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Acknowledgments
We express our thanks to Sebastien Kenmoe, (MSc, Ph.D., Assistant Lecturer, Department of Microbiology and Parasitology University of Buea) for accompanying us in this research.
References
- 1.Kyriakidis NC, López-Cortés A, González EV, Grimaldos AB, Prado EO. SARS-CoV-2 vaccines strategies: a comprehensive review of phase 3 candidates. NPJ Vaccines. 2021;6(1):28. doi: 10.1038/s41541-021-00292-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mohammadi M, Meskini M, do Nascimento, Pinto AL. Novel coronavirus (COVID-19) overview. J Public Health. 2020:1–9. doi: 10.1007/s10389-020-01258-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zamani B, Moeini Taba SM, Shayestehpour M. Systemic lupus erythematosus manifestation following COVID-19: a case report. J Med Case Rep. 2021;15(1):29. doi: 10.1186/s13256-020-02582-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mercatelli D, Giorgi FM. Geographic and Genomic Distribution of SARS-CoV-2 Mutations. Front Microbiol. 2020;11:1–12. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kouhsari E, Azizian K, Sholeh M, et al. Clinical, epidemiological, laboratory, and radiological characteristics of novel Coronavirus (2019-nCoV) in retrospective studies: a systemic review and meta-analysis. Indian J Med Microbiol. 2021;39(1):104–115. doi: 10.1016/j.ijmmb.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heinz FX, Stiasny KJWKW. Profiles of current COVID-19 vaccines. 2021:1–13. [DOI] [PMC free article] [PubMed]
- 7.Guruprasad LJPS, Function Bioinformatics. Human SARS CoV‐2 spike protein mutations. Proteins. 2021;89(5):569–576. doi: 10.1002/prot.26042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khailany RA, Safdar M, Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020;19 doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finkel Y, Mizrahi O, Nachshon A, et al. The coding capacity of SARS-CoV-2. Nature. 2021;589(7840):125–130. doi: 10.1038/s41586-020-2739-1. [DOI] [PubMed] [Google Scholar]
- 10.Arya R, Kumari S, Pandey B, et al. Structural insights into SARS-CoV-2 proteins. J Mol Biol. 2021;433(2) doi: 10.1016/j.jmb.2020.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yurkovetskiy L, Wang X, Pascal KE, et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183(3):739–751. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie Y, Karki CB, Du D, et al. Spike proteins of SARS-CoV and SARS-CoV-2 utilize different mechanisms to bind with human ACE2. Front Mol Biosci. 2020;7:392. doi: 10.3389/fmolb.2020.591873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Y, Yang C, Xu X-f, Xu W, Liu S-w. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3(1):237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai Y, Zhang J, Xiao T, et al. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369(6511):1586. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hussain M, Jabeen N, Raza F, et al. Structural variations in human ACE2 may influence its binding with SARS-CoV-2 spike protein. J Med Virol. 2020;92(9):1580–1586. doi: 10.1002/jmv.25832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Creech CB, Walker SC, Samuels RJ. SARS-CoV-2 Vaccines. JAMA. 2021;325(13):1318–1320. doi: 10.1001/jama.2021.3199. [DOI] [PubMed] [Google Scholar]
- 19.Salvatori G, Luberto L, Maffei M, et al. SARS-CoV-2 SPIKE PROTEIN: an optimal immunological target for vaccines. J Transl Med. 2020;18:1–3. doi: 10.1186/s12967-020-02392-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samrat SK, Tharappel AM, Li Z. Li HJVr. Prospect of SARS-CoV-2 spike protein: potential role in vaccine and therapeutic development. Virus Res. 2020:198141. doi: 10.1016/j.virusres.2020.198141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duan L, Zheng Q, Zhang H, Niu Y, Lou Y, Wang H. The SARS-CoV-2 spike glycoprotein biosynthesis, structure, function, and antigenicity: implications for the design of spike-based vaccine immunogens. Front Immunol. 2020;11:2593. doi: 10.3389/fimmu.2020.576622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki YJ, Gychka SG. SARS-CoV-2 spike protein elicits cell signaling in human host cells: implications for possible consequences of COVID-19 vaccines. Vaccines. 2021;9(1):36. doi: 10.3390/vaccines9010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berger I. Schaffitzel CJCr. The SARS-CoV-2 spike protein: balancing stability and infectivity. Cell Res. 2020;30(12):1059–1060. doi: 10.1038/s41422-020-00430-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guruprasad L. Evolutionary relationships and sequence-structure determinants in human SARS coronavirus-2 spike proteins for host receptor recognition. Proteins. 2020;88(11):1387–1393. doi: 10.1002/prot.25967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson CB, Zhang L, Farzan M, Choe H. Functional importance of the D614G mutation in the SARS-CoV-2 spike protein. Biochem Biophys Res Commun. 2021;538:108–115. doi: 10.1016/j.bbrc.2020.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Wang L, Cao H, CJJomv Liu. SARS-CoV-2 S1 is superior to the RBD as a COVID-19 subunit vaccine antigen. J Med Virol. 2021;93(2):892–898. doi: 10.1002/jmv.26320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khalaj-Hedayati A. Protective immunity against SARS subunit vaccine candidates based on spike protein: lessons for Coronavirus vaccine development. J Immunol Res. 2020;2020 doi: 10.1155/2020/7201752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malik JA, Mulla AH, Farooqi T, Pottoo FH, Anwar S, Rengasamy KRJB, et al. Targets and strategies for vaccine development against SARS-CoV-2. Biomed Pharmacother. 2021;137:111254. doi: 10.1016/j.biopha.2021.111254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dar HA, Waheed Y, Najmi MH, Ismail S, Hetta HF, Ali A, et al. Multiepitope Subunit Vaccine Design against COVID-19 Based on the Spike Protein of SARS-CoV-2: An In Silico Analysis. J Immunol Res. 2020;2020:8893483. doi: 10.1155/2020/8893483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang S, Li Y, Dai L, et al. Safety and immunogenicity of a recombinant tandem-repeat dimeric RBD protein vaccine against COVID-19 in adults: pooled analysis of two randomized, double-blind, placebo-controlled, phase 1 and 2 trials. Lancet Infect Dis. 2021;21(8):1–13. doi: 10.1016/S1473-3099(21)00127-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forni G, Mantovani A, Forni G, et al. COVID-19 vaccines: where we stand and challenges ahead. Cell Death Diff. 2021;28(2):626–639. doi: 10.1038/s41418-020-00720-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahase E. Covid-19: novavax vaccine efficacy is 86% against UK variant and 60% against South African variant. BMJ. 2021;372:n296. doi: 10.1136/bmj.n296. [DOI] [PubMed] [Google Scholar]
- 33.Callaway E, Mallapaty SJN. Novavax offers first evidence that COVID vaccines protect people against variants. Nature. 2021;590(7844):17. doi: 10.1038/d41586-021-00268-9. [DOI] [PubMed] [Google Scholar]
- 34.Chang-Monteagudo A, Ochoa-Azze R, Climent-Ruiz Y, et al. A single dose of SARS-CoV-2 FINLAY-FR-1A dimeric-RBD recombinant vaccine enhances neutralization response in COVID-19 convalescents, with excellent safety profile. a preliminary report of an open-label phase 1 clinical trial. medRxiv. 2021 doi: 10.1016/j.lana.2021.100079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, Zeng H, Gu J, Li H, Zheng L, Zou Q. Progress and prospects on vaccine development against SARS-CoV-2. Vaccines. 2020;8(2) doi: 10.3390/vaccines8020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586(7830):516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 37.Liang Z, Zhu H, Wang X, et al. Adjuvants for coronavirus vaccines.Front. Immunol. 2020;11:2896. doi: 10.3389/fimmu.2020.589833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bettini E, Locci M. SARS-CoV-2 mRNA vaccines: immunological mechanism and beyond. Vaccines. 2021;9(2):147. doi: 10.3390/vaccines9020147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amanat F, Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020;52(4):583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Azkur AK, Akdis M, Azkur D, Sokolowska M, van de Veen W, Brüggen M-C, et al. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy. 2020;75(7):1564–1581. doi: 10.1111/all.14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng S, Cao F, Xia Y, Gao XD, Dai L, Yan J, et al. Particulate Alum via Pickering Emulsion for an Enhanced COVID‐19 Vaccine Adjuvant. Adv Mater. 2020;32(40) doi: 10.1002/adma.202004210. [DOI] [PubMed] [Google Scholar]
- 42.Adney DR, Wang L, van Doremalen N, et al. Efficacy of an adjuvanted middle east respiratory syndrome coronavirus spike protein vaccine in dromedary camels and alpacas. Viruses. 2019;11(3) doi: 10.3390/v11030212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuo T-Y, Lin M-Y, Coffman RL, et al. Development of CpG-adjuvanted stable pre-fusion SARS-CoV-2 spike antigen as a subunit vaccine against COVID-19. Sci Rep. 2020;10(1):1–10. doi: 10.1038/s41598-020-77077-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi S, Zhu H, Xia X, Liang Z, Ma X, Sun BJV. Vaccine adjuvants: Understanding the structure and mechanism of adjuvanticity. Vaccine. 2019;37(24):3167–3178. doi: 10.1016/j.vaccine.2019.04.055. [DOI] [PubMed] [Google Scholar]
- 45.Shen X, Tang H, McDanal C, Wagh K, Fischer W, Theiler J, et al. SARS-CoV-2 variant B. 1.1. 7 is susceptible to neutralizing antibodies elicited by ancestral Spike vaccines. Cell Host Microbe. 2021.14;29(4):529-539. [DOI] [PMC free article] [PubMed]
- 46.Altmann DM, Boyton RJ, Beale R. Immunity to SARS-CoV-2 variants of concern. Science. 2021;371(6534):1103. doi: 10.1126/science.abg7404. [DOI] [PubMed] [Google Scholar]
- 47.Rees-Spear C, Muir L, Griffith SA, et al. The impact of spike mutations on SARS-CoV-2 neutralization. bioRxiv. 2021 doi: 10.1016/j.celrep.2021.108890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu J, Peng P, Wang K, Fang L, Luo F-y, Jin A-s, et al. Emerging SARS-CoV-2 variants reduce neutralization sensitivity to convalescent sera and monoclonal antibodies. Cell Mol Immunol. 2021;18(4):1061–1063. doi: 10.1038/s41423-021-00648-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang P, Nair MS, Liu L, et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593(7857):130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 50.Cherian S, Potdar V, Jadhav S, et al. Convergent evolution of SARS-CoV-2 spike mutations, L452R, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. bioRxiv. 2021 doi: 10.3390/microorganisms9071542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gómez CE, Perdiguero B, Esteban M. Emerging SARS-CoV-2 variants and impact in global vaccination programs against SARS-CoV-2/COVID-19. Vaccines. 2021;9(3):243. doi: 10.3390/vaccines9030243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar V, Singh J, Hasnain SE, Sundar D. Possible link between higher transmissibility of B.1.617 and B.1.1.7 variants of SARS-CoV-2 and increased structural stability of its spike protein and hACE2 affinity. bioRxiv. 2021 doi: 10.3390/ijms22179131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Planas D, Bruel T, Grzelak L, et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat Med. 2021;27(5):917–924. doi: 10.1038/s41591-021-01318-5. [DOI] [PubMed] [Google Scholar]
- 54.Singh J, Samal J, Kumar V, et al. Structure-function analyses of new SARS-CoV-2 variants B. 1.1. 7, B. 1.351 and B. 1.1. 28.1: clinical, diagnostic. Therap Public Health Implic. 2021;13(3):439. doi: 10.3390/v13030439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang G-L, Wang Z-Y, Duan L-J, et al. Susceptibility of circulating SARS-CoV-2 variants to neutralization. N Engl J Med. 2021;384(24):2354–2356. doi: 10.1056/NEJMc2103022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koyama T, Platt D, Parida L. Variant analysis of SARS-CoV-2 genomes. Bull World Health Organ. 2020;98(7):495–504. doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gupta RK. Will SARS-CoV-2 variants of concern affect the promise of vaccines? Nat Rev Immunol. 2021;21(6):340–341. doi: 10.1038/s41577-021-00556-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Slavov SN, Leister Patané JS, Santos Bezerra Rd, et al. Genomic monitoring unveil the early detection of the SARS-CoV-2 B.1.351 lineage (20H/501Y.V2) in Brazil. J Med Virol. 2021 doi: 10.1002/jmv.27190. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mallm J-P, Bundschuh C, Kim H, et al. Local emergence and decline of a SARS-CoV-2 variant with mutations L452R and N501Y in the spike protein. medRxiv. 2021 [Google Scholar]
- 60.Volz E, Hill V, McCrone JT, et al. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2021;184(1):64–75. doi: 10.1016/j.cell.2020.11.020. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoffmann M, Arora P, Groß R, et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184(9):2384–2393. doi: 10.1016/j.cell.2021.03.036. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moyo-Gwete T, Madzivhandila M, Makhado Z, et al. SARS-CoV-2 501Y.V2 (B.1.351) elicits cross-reactive neutralizing antibodies. bioRxiv. 2021 doi: 10.1056/NEJMc2104192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou D, Dejnirattisai W, Supasa P, et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184(9):2348–2361. doi: 10.1016/j.cell.2021.02.037. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rees-Spear C, Muir L, Griffith SA, et al. The effect of spike mutations on SARS-CoV-2 neutralization. Cell Rep. 2021;34(12) doi: 10.1016/j.celrep.2021.108890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen X, Tang H, Pajon R, et al. Neutralization of SARS-CoV-2 variants B.1.429 and B.1.351. N Engl J Med. 2021;384(24):2352–2354. doi: 10.1056/NEJMc2103740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shinde V, Bhikha S, Hoosain Z, et al. Efficacy of NVX-CoV2373 Covid-19 Vaccine against the B.1.351 Variant. N Engl J Med. 2021;1384(20):1899–1909. doi: 10.1056/NEJMoa2103055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wibmer CK, Ayres F, Hermanus T, et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med. 2021;27(4):622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 68.Hoffmann M, Arora P, Groß R, et al. SARS-CoV-2 variants B.1.351 and B.1.1.248: escape from therapeutic antibodies and antibodies induced by infection and vaccination. bioRxiv. 2021 [Google Scholar]
- 69.Focosi D, Maggi F. Neutralising antibody escape of SARS-CoV-2 spike protein: risk assessment for antibody-based Covid-19 therapeutics and vaccines. Rev Med Virol. 2021 doi: 10.1002/rmv.2231. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fontanet A, Autran B, Lina B, Kieny MP, Karim SSA, Sridhar DJTL. SARS-CoV-2 variants and ending the COVID-19 pandemic. Lancet. 2021;397(10278):952–954. doi: 10.1016/S0140-6736(21)00370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Davis C, Logan N, Tyson G, et al. Reduced neutralization of the Delta (B.1.617.2) SARS-CoV-2 variant of concern following vaccination. Nature. 2021;596:276–280. doi: 10.1371/journal.ppat.1010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lazarevic I, Pravica V, Miljanovic D, Cupic M. Immune evasion of SARS-CoV-2 emerging variants: what have we learnt so far? Viruses. 2021;13(7) doi: 10.3390/v13071192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lustig Y, Zuckerman N, Nemet I, et al. Neutralizing capacity against Delta (B.1.617.2) and other variants of concern following Comirnaty (BNT162b2, BioNTech/Pfizer) vaccination in health care workers. Israel. 2021;26(26) doi: 10.2807/1560-7917.ES.2021.26.26.2100557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kupferschmidt K, Wadman M. Delta variant triggers new phase in the pandemic. Science. 2021;372(6549):1375. [Google Scholar]
- 75.Romero PE, Dávila-Barclay A, Salvatierra G, et al. The emergence of SARS-CoV-2 variant lambda (C.37) in South America. medRxiv. 2021 doi: 10.1128/Spectrum.00789-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tada T, Zhou H, Dcosta BM, Samanovic MI, Mulligan MJ, Landau NR. SARS-CoV-2 lambda variant remains susceptible to neutralization by mRNA vaccine-elicited antibodies and convalescent serum. bioRxiv. 2021 [Google Scholar]
- 77.Acevedo ML, Alonso-Palomares L, Bustamante A, et al. Infectivity and immune escape of the new SARS-CoV-2 variant of interest Lambda. medRxiv. 2021 [Google Scholar]
- 78.Weisblum Y, Schmidt F, Zhang F, DaSilva J, Poston D, Lorenzi JC, et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020;9 doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen RE, Zhang X, Case JB, et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat Med. 2021;27(4):717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xie X, Liu Y, Liu J, Zhang X, Zou J, Fontes-Garfias CR, et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat Med. 2021;27(4):620–621. doi: 10.1038/s41591-021-01270-4. [DOI] [PubMed] [Google Scholar]
- 81.Collier D, Meng B, Ferreira I, Datir R, Temperton NJ, Elmer A, et al. Impact of SARS-CoV-2 B. 1.1. 7 Spike variant on neutralisation potency of sera from individuals vaccinated with Pfizer vaccine BNT162b2. medRxiv. 2021 [Google Scholar]
- 82.Wu K, Werner AP, Moliva JI, et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv. 2021 [Google Scholar]
- 83.Garcia-Beltran WF, Lam EC, St. Denis K, et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184(9):2372–2383. doi: 10.1016/j.cell.2021.03.013. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mlcochova P, Kemp S, Dhar MS, Papa G, Meng B, Mishra S, et al. SARS-CoV-2 B.1.617.2 Delta variant emergence, replication and sensitivity to neutralising antibodies. bioRxiv. 2021 [Google Scholar]